Abstract
Phenol is a hazardous chemical known to be widely distributed in aquatic environments. Biodegradation is an attractive option for removal of phenol from water sources. Acinetobacter sp. DW-1 isolated from drinking water biofilters can use phenol as a sole carbon and energy source. In this study, we found that Immobilized Acinetobacter sp. DW-1cells were effective in biodegradation of phenol. In addition, we performed proteome and transcriptome analysis of Acinetobacter sp. DW-1 during phenol biodegradation. The results showed that Acinetobacter sp. DW-1 degrades phenol mainly by the ortho pathway because of the induction of phenol hydroxylase, catechol-1,2-dioxygenase. Furthermore, some novel candidate proteins (OsmC-like family protein, MetA-pathway of phenol degradation family protein, fimbrial protein and coenzyme F390 synthetase) and transcriptional regulators (GntR/LuxR/CRP/FNR/TetR/Fis family transcriptional regulator) were successfully identified to be potentially involved in phenol biodegradation. In particular, MetA-pathway of phenol degradation family protein and fimbrial protein showed a strong positive correlation with phenol biodegradation, and Fis family transcriptional regulator is likely to exert its effect as activators of gene expression. This study provides valuable clues for identifying global proteins and genes involved in phenol biodegradation and provides a fundamental platform for further studies to reveal the phenol degradation mechanism of Acinetobacter sp.
Introduction
Phenol is a toxic compound that is widely distributed in nature, especially in aquatic environments. Phenolic compounds are used as raw materials for the production of various products in different industries, including oil refinement, pharmaceuticals, pesticides, resin products, steel mills, coke oven plants, and leather production1, 2. However, because of overuse and uncontrolled emissions, phenol has drawn increasing attention as a threat to human health and is on the priority list of highly hazardous chemicals in China. In the National Chinese Standards for Drinking Water Quality (GB5749-2006), the acceptable concentration of volatile phenol is limited to 2 ppb. Therefore, it is necessary to remove phenol from the aquatic environment. Biodegradation technology is a potentially attractive tool for removal of environmental pollutants because it has low cost, saves energy, is efficient, and does not lead to secondary pollution3.
Acinetobacter sp. DW-1(GenBank accession number KU499947), which was isolated from a biological active carbon (BAC) filter in a drinking water plant in our laboratory4, is able to grow on mineral salt medium (MSM) with phenol as a sole carbon and energy source. Acinetobacter spp. are commonly found in soil and aquatic environments, where they comprise a significant part of the total heterotrophic, aerobic population. Moreover, they are able to grow in low nutrient media containing inorganic sources of nitrogen and a variety of simple compounds as carbon and energy source5. Acinetobacter spp. have been attracting increasing attention in both environmental and biotechnological applications6. Previous studies demonstrated that immobilized cells often exhibited better performance in biodegradation than free-bacteria7, 8. Moreover, immobilized bacteria could strengthen the resistance to environmental stress compared with free bacteria9. Therefore, it is of great importance to investigate the phenol biodegradation capacity of immobilized Acinetobacter sp. DW-1. Meanwhile, antimicrobial susceptibility data for Acinetobacter spp. isolated from drinking water environment is still scarce. Therefore the goal of the current antimicrobial susceptibility experiments was to determine the controllability of this strain when it is applied in water treatment.
To date, the aerobic catabolic pathway for phenol has been extensively investigated in the Pseudomonas 10–12 and Acinetobacter 13, 14 genera which often isolated from polluted or contaminated environmental samples. Therefore, they had a strong innate tolerance to pollutants from different sources, and can metabolize these pollutants. However, how the indigenous bacterium which isolated from low-nutrient and micro-pollution environment responses to phenol, and the reasons for variations in the level of expression of important enzymes and regulatory factors during catabolism are still unclear. More work is necessary to thoroughly characterize the catabolic pathways of phenol and global metabolism to elucidate phenol degradation mechanism. To the best of our knowledge, proteome and transcriptome approaches were powerful to discover molecular markers and elucidation of functional mechanisms15, 16. However, using alone approach could not obtain sufficient information. Different types of data often provide complementary information to the elucidation of mechanisms, so that the detection bias from each of the technologies can be avoided17. They can provide detailed and comprehensive information on the biodegradation process18, 19. However, currently the information on phenol biodegradation by Acinetobacter sp. was mainly evaluated by proteome20–22, there is no reports about using transcriptome approach to investigate phenol biodegradation by Acinetobacter sp. Therefore, it is of interest to determine the catabolic pathway and to elucidate the mechanism of phenol biodegradation by Acinetobacter sp. DW-1 using both proteomic and transcriptomic analyses. Proteomic techniques are effective methods for the detection of detailed differences in protein expression under alternative growth condition23, 24. Among proteomic techniques, two-dimensional electrophoresis (2-DE) is widely used to analyze bacterial proteins; for example, the log to stationary growth phase in Lactobacillus plantarum cells25, differentiation of low-Mr-secreted protein species in mycobacteria26, and response to alkaline stress induced by root canal bacteria in biofilms27 have been analyzed by proteomic methods. Moreover, differential gene expression in response to phenol and catechol has revealed various metabolic activities for the degradation of aromatic compounds in Bacillus subtilis 28. Similarly, transcriptome sequencing is a good tool to comprehensively study the differences in genes expressed under different treatments29–32.
The aim of this study was to better understand the biofilm-forming ability of Acinetobacter sp. DW-1 on carrier materials, the controllability of this strain, and then to find novel candidate proteins or related genes so as to further reveal phenol degradation mechanism, ultimately to evaluate the potential for its practical use in bioremediation applications. These aims were achieved using drug sensitivity test and by comparing the proteome and transcriptome of strains grown on different carbon sources and analyzing the expression of major metabolic enzymes during the phenol degradation process. To the best of our knowledge, this is the first attempt to systematically investigate the biofilm-forming ability of the native bacterium isolated from drinking water biofilter on carrier materials, and to assess the controllability of indigenous bacterium before practical application. This study provides abundant both proteome and transcriptome data for phenol biodegradation by Acinetobacter sp. The candidate genes involved in the biosynthesis and transcriptional regulation of Acinetobacter sp. were obtained in this study. This provides information that increases our understanding of the mechanism of phenol regulation by Acinetobacter sp. The global gene regulation and interplay during phenol degradation process remain largely unaddressed. With the present study we provide novel information about the gene regulation relevant to such a biodegradation process by Acinetobacter sp. The comparative analysis of the proteome and transcriptome data of phenol biodegradation by Acinetobacter sp. DW-1, lays the foundation for elucidating the mechanism of phenol degradation, and thus, enables future improvements in phenol biodegradation.
Materials and Methods
Immobilization of Acinetobacter sp. DW-1 cells
Prepared sterilized polyhedron hollow polypropylene balls (Jingying, Yixing, China) were placed into a 500 mL flask containing 200 mL of MSM medium supplemented with 4 mM phenol as a sole carbon and energy source. This mixture was incubated without motion at 30 °C to allow Acinetobacter sp. DW-1 isolated in our laboratory cells to attach to the polyhedron hollow polypropylene balls. The balls were withdrawn at 24, 48, and 72 h using sterile forceps. The balls were then rinsed with sterile MSM to remove any Acinetobacter sp. DW-1 cells not sufficiently adhered to their surfaces. Next, the rinsed balls were examined by scanning electron microscopy (SEM).
Detection of biofilm by scanning electron microscopy and confocal laser scanning microscopy
Acinetobacter sp. DW-1 cells immobilized on polyhedron hollow polypropylene balls were examined using a S-3000N scanning electron microscope (Hitachi, Tokyo, Japan). In preparations, cells were fixed in 0.1 M phosphate buffer solution (PBS) containing 3% (v/v) glutaraldehyde at 4 °C for 5 h. The samples were then washed with PBS (pH 7.0) six times (20 min each). The samples were then gradually dehydrated with ethanol. Dehydrated cells were filtered through a 0.2 µm polycarbonate filter, dried with a CO2-critical point dryer, coated with gold and subsequently observed by SEM at 20 kV. Confocal laser scanning microscopy (CLSM) (Zeiss, Berlin, Germany) analysis was performed using a LIVE/DEAD® BacLight™ Bacterial Viability and Counting Kit (Invitrogen, Carlsbad, USA) with an appropriate mixture of the SYTO 9 and propidium iodide (PI).
Adennosine tri-phosphate (ATP) analysis
ATP measurements to estimate the active biomass of Acinetobacter sp. DW-1 immobilized on polyhedron hollow polypropylene balls were obtained. For the preparations, the balls were withdrawn from the incubated flask, and then rinsed with PBS three times to remove the unattached cells. Subsequently, the rinsed polyhedron hollow polypropylene balls with PBS into an ultrasonic cleaner for 5 min. The supernatant was used for ATP measurement which was developed by our laboratory as described elsewhere in detail33.
Investigation of phenol biodegradation by immobilized Acinetobacter sp. DW-1 cells
Six glass columns (inner diameter = 40 cm) with a working volume of 450 mL each were filled with sterile polyhedron hollow polypropylene balls. Three columns were used as controls, whereas the other three were used to immobilize Acinetobacter sp. DW-1. Sand-filtered water samples with initial phenol concentrations of 4, 2, and 1 mM were applied to the columns using a circulating water pump (Sensen, Zhoushan, China). The empty bed contact time was 18 min. The entire water treatment process was maintained at room temperature. Effluent water from samples with different initial phenol concentrations was collected for phenol detection after 1 h. The phenol concentration was determined as described elsewhere4.
Transmission electron microscopy
Acinetobacter sp. DW-1 cultures were grown in MSM supplemented with 4 mM phenol as a single carbon and energy source at 30 °C for 30 h; then, several drops of cell suspension were applied to a collodion-coated copper grid of mesh size 200 for 5 min. After a brief wash with water, the grid was immersed in filtered 3% phosphotungstic acid (pH 7.0) for 2 min. The grid was then removed, the excess stain was wicked away with filter paper, and the grid was washed with water and dried at room temperature for 45 min. Specimens were observed under an H7500 transmission electron microscope (TEM; Hitachi) operating at 80 kV.
Antimicrobial susceptibility test
Antibiotic susceptibility tests were performed using the agar disk diffusion method34, following the guidelines of the Clinical and Laboratory Standards Institute. Nineteen antibiotic-containing disks (Oxoid, Basingstoke, UK) were performed for 17 antibiotics that listed in Supplementary Information. Antimicrobial susceptibility tests were performed as previously described35.
Bacterial culture conditions and phenol biodegradation
Proteins prepared from Acinetobacter sp. DW-1 cells were inoculated into R2A fluid nutrient medium36 and incubated until the culture reached exponential phase; then, 200 μL of R2A-preculture was inoculated into an Erlenmayer flask (1000 mL) containing MSM supplemented with acetate (4 mM) as the sole carbon source. The composition of MSM was as follows (mg L−1): MgSO4.7H2O, 0.1; NaCl, 0.2; NH4Cl, 0.5; Na2HPO4.12H2O, 0.5; KH2PO4, 0.5; FeCl3.6H2O, 0.1; and CaSO4.H2O, 0.1. Cultures were incubated at 30 °C and shaken at 120 rpm in an environmental chamber until they reached standardized optical densities at of 0.7–0.8 600 nm (OD600). Subsequently, phenol was added to the cultures to obtain a final concentration of 4 mM. Unsupplemented cultures served as controls. Cells from the control and phenol- supplemented cultures were harvested when the phenol biodegradation rate reached a maximum. However, to determine growth curves, we performed viable cell counts during the phenol biodegradation process by withdrawing 1 mL samples of culture every 2 h under sterile conditions, diluting them up to 106 in Eppendorf tubes, and plating the dilutions in triplicate on Plate Count Agar using a glass L-shaped rod. After 48 h of incubation, colonies were counted to identify dilutions producing colony numbers between 40 and 400 (to facilitate interpretation of results). Phenol degradation was evaluated by collecting samples of MSM solution at every two hours for acetate as well as phenol quantification. The determination of acetate and phenol concentration were performed as described previously37, 38. Meanwhile, the major intermediate products of phenol which including catechol and cis,cis-muconate were also investigated during the biodegradation process, and the detection methods of them were according to previous studies39, 40.
Preparation of crude protein extracts
Cells were harvested by centrifugation at 12,000 g for 10 min at 4 °C, washed twice in MSM, and sonicated with an ultrasonic processor (Sonics VCX130PB, USA) for a total of 20 min at 20 kHz with intervals of 1 min, where the cells were kept on ice. The sonicated solution was then clarified by centrifugation at 12,000 g for 1 h at 4 °C. The protein concentration was determined using a Non-Interfering Protein Concentration Determination Kit (Sangon, Shanghai, China).
Conditions for 2-DE
Protein samples (800 µg) were mixed with rehydration solution (0.2% (v/v) Bio-Lyte ampholyte (pH 3–10), 8 M urea, 4% (w/v) CHAPS, 1% (w/v) DTT, and 0.01% (w/v) bromophenol blue) in total volume of 350 µL. Immobiline TM Dry Strip IPG strips (17 cm, pH 4–7) were passively hydrated at 16 °C for 13 h. IEF and two-dimensional SDS-polyacrylamide gel electrophoresis (PAGE) were performed as described elsewhere in detail41.
Image analysis and statistics
Gels were stained with Coomassie brilliant blue G-250, and triplicate gels were scanned using a densitometric Image Scanner (GE Healthcare). The raw images were analyzed using Image Master TM platinum version 7.0 (GE Healthcare). For each comparison of cells grown on acetate with those grown on acetate plus phenol), the 2-D gels were analyzed in triplicate. The method to screen for phenol-induced proteins was consulted previous literature42.
Identification of proteins by MS
To identify differentially expressed proteins, spots of interest were excised manually from Coomassie brilliant blue stained polyacrylamide gels and subjected to in-gel digestion as previously described43. The excised proteins were analyzed using a 4700 MALDI-TOF/TOF proteomics analyzer (Applied Biosystems). A peptide mass fingerprinting search and a combined search (+MS/MS) were performed using GPS Explorer™ software (Applied Biosystems) in non-redundant NCBI database of proteins using MASCOT searching engine as described previously44.
RNA extraction and sequencing
Acinetobacter sp. strain DW-1 cells in exponential phase growing on acetate or acetate plus phenol were collected and used for total RNA extraction. RNA of each sample was extracted using a MiniBEST Universal RNA Extraction Kit (TaKaRa, Dalian, China) according to the manufacturer’ instructions. The assessing of RNA concentration and quality and RNA sequencing as described previously45.
Transcriptome assembly and analysis
Raw sequences were first quality-filtered, and the differential gene expression analysis was then performed. The differentially expressed genes were subjected to GO (http://www.geneontology.org/) and KEEG (http://www.genome.jp/kegg/) analyses. Differentially expressed genes (DEGs) were obtained based on the Reads Per Kilobase of exon model per Million (RPKM), followed by a multiple hypothesis testing, false discovery rate (FDR) control to correct the p-value. Genes with an FDR value < 0.05 and |log2FC| > 1 were classified as differentially expressed.
Validation of RT-qPCR
After RNA extraction, complementary DNA(cDNA) was synthesized using total RNA with a PrimeScript RT reagent kit (TaKaRa, Dalian, China), and then the cDNA product was amplified by RT-PCR. 16sRNA was used as an internal reference in all reactions. Real-time quantification of mRNA was performed on an Eppendorf Realplex (4) PCR system (Eppendorf, Hamburg, Germany) using SYBR Premix ExTaq II kits (TaKaRa, Dalian, China). The comparative Ct method was employed for quantification of target gene expression that was normalized to 16sRNA expression and relative to the calibrator. Data were expressed as the fold change (T/CK) = 2−ΔΔCt. A list of all primers used in this study is presented in Supplementary Table S6.
Results and Discussion
Detection of Acinetobacter sp. DW-1 biofilm formation and phenol degradation test
According to previous experiment (Supplementary methods 1.2), we found that immobilized cells of Acinetobacter sp. DW-1 showed better activity for the phenol biodegradation than free cells (Supplementary Fig. S1). Therefore, we chose immobilized cells to study in this study. SEM visualization demonstrated that Acinetobacter sp. DW-1 successfully attached to polyhedron hollow polypropylene balls and grew well (Fig. 1A). The surface of the polyhedron hollow polypropylene balls was fully covered with Acinetobacter sp. DW-1cells after 96 h immobilization. Confocal laser scanning microscopy confirmed that the quantity of Acinetobacter sp. DW-1 cells on the polypropylene balls increased over time; however, the number of dead cells (red spots) also increased gradually (Fig. 1B). The results were in agreement with those obtained by measuring the ATP concentration, which demonstrated that the highest biological activity was obtained for Acinetobacter sp. DW-1 cultivated for 72 h, and that the biological activity decreased when the cultures were grown for 96 h (Table 1). Figure 1B shows that the quantity of Acinetobacter sp. DW-1 cells increased as the cultivation increased, but the number of dead cells (red spots) also increased gradually. The results agree with the ATP results (Table 1), where Acinetobacter sp. DW-1 cultivated for 72 h showed the highest biological activity. However, the biological activity decreased when the cultures were grown to 96 h (Table 1). TEM images of Acinetobacter sp. DW-1 (Fig. 2) revealed anchor-like appendages connecting cells, which may contribute to the capability of Acinetobacter sp. DW-1 to adhere to solid surfaces46. These results indicated that the Acinetobacter sp. DW-1 biofilm reached an optimal state after 72 h of cultivation. Therefore, we selected this stage for assessment of phenol biodegradation by the attached biomass.
Figure 1.
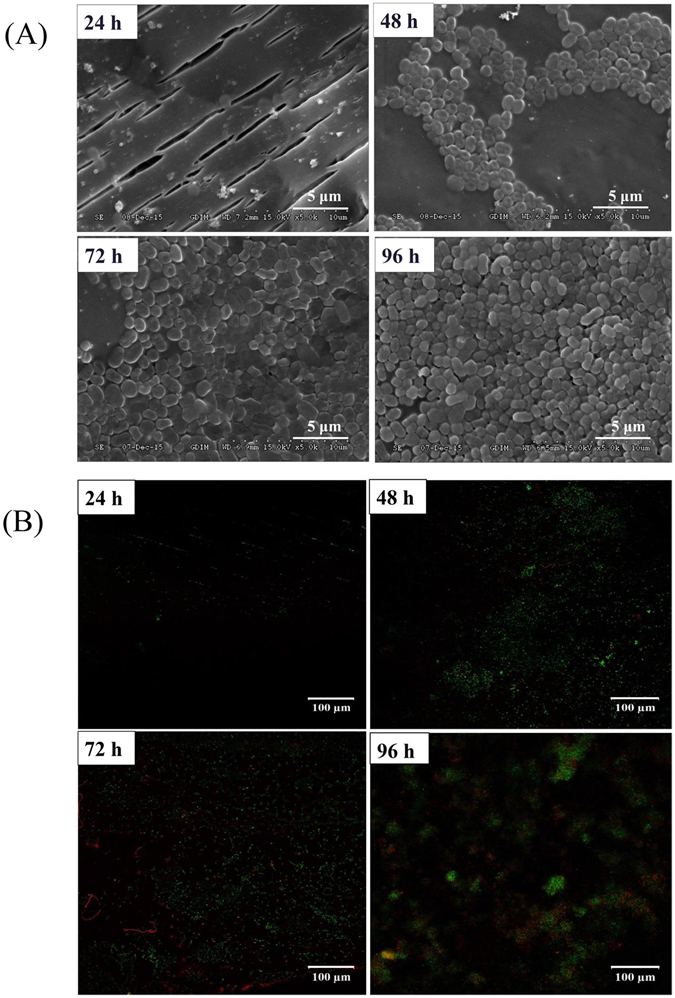
Growth of an Acinetobacter sp. DW-1 biofilm on polyhedron hollow polypropylene balls visualized by scanning electron microscopy (A) and confocal laser scanning microscopy (B).
Table 1.
Adenosine tri-phosphate (ATP) analysis of Acinetobacter sp. DW-1 immobilized on polyhedron hollow polypropylene balls at different times.
| Time (h) | 24 | 48 | 72 | 96 |
|---|---|---|---|---|
| ATP (mol/g) | 8.41 × 10−8 ± 1.23 | 3.36 × 10−7 ± 4.16 | 2.11 × 10−6 ± 5.02 | 6.28 × 10−8 ± 3.46 |
Figure 2.
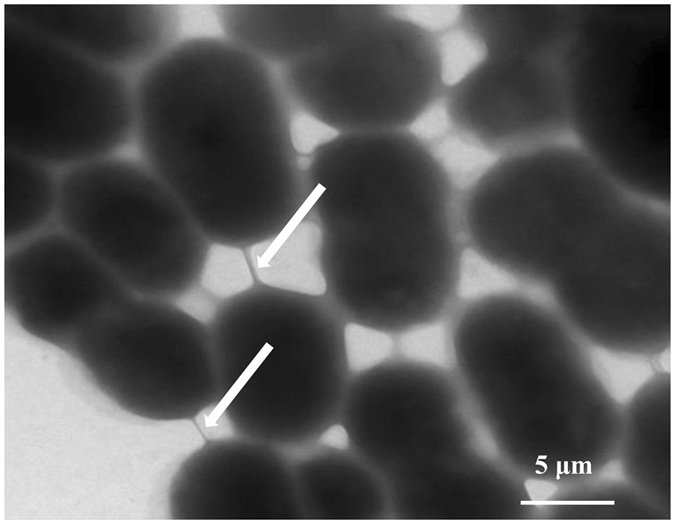
TEM image of Acinetobacter sp. DW-1 grown in MSM supplemented with 4 mM phenol as single carbon and energy source. The arrows indicate anchor-like appendages among cells.
Acinetobacter sp. DW-1 immobilized on polyhedron hollow polypropylene balls exhibited good performance in phenol biodegradation (Fig. 3). At a phenol concentration of 1 mM, Acinetobacter sp. DW-1 degraded approximately 80% of phenol, and at 4 mM, approximately 33%. The results from the control samples indicated that the polyhedron hollow polypropylene balls adsorbed negligible amounts of phenol, demonstrating their suitability as a material for investigation of degradation by Acinetobacter sp. DW-1. Therefore, our results indicate that Acinetobacter sp. DW-1 has a strong ability to form biofilms and degrade phenol.
Figure 3.
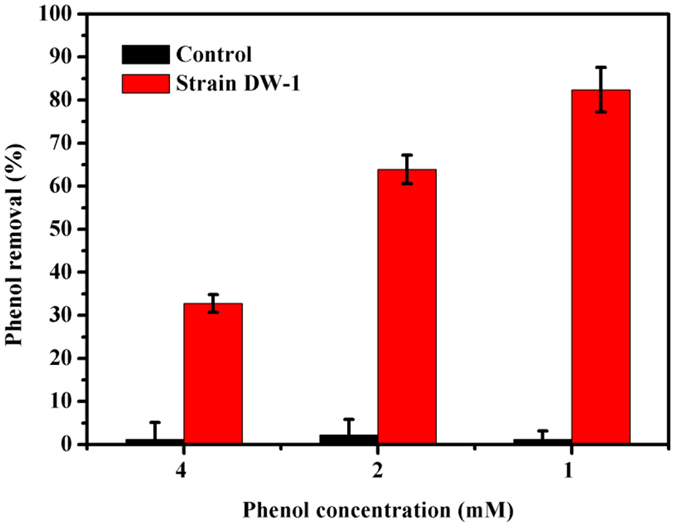
Phenol degradation by Acinetobacter sp. DW-1 immobilized on polyhedron hollow polypropylene balls.
Antimicrobial susceptibility test
Acinetobacter sp. DW-1 was susceptible to the majority of antimicrobials included in the test panel. However, it was resistant to chloramphenicol and aztreonam (Table 2). The resistance profile of Acinetobacter sp. DW-1, which was isolated from an activated carbon filter in a drinking water plant, was different from that of clinically isolated Acinetobacter strains. Previous reports indicate that imipenem is highly active against all Acinetobacter isolates obtained from patients47–49. However, in the present study, Acinetobacter sp. DW-1 was susceptible to imipenem, but resistant to chloramphenicol. These results were consistent with those obtained using Acinetobacter sp. isolated from wastewater50, mineral water51 and drinking water52, all of which showed resistance to chloramphenicol. In general, strain DW-1 was susceptible to antibiotics and, therefore, controllable. Moreover, as an indigenous bacterium from an aquatic environment, strain DW-1 was able to adapt to oligotrophic conditions. In summary, strain DW-1 shows strong potential for application in the removal of phenol from water.
Table 2.
Results of antibiotic susceptibility tests of Acinetobacter sp. DW-1.
| Antimicrobial agents | Breakpoints (mm) | Tolerance (Breakpoints (mm)) | ||
|---|---|---|---|---|
| Susceptible (S) | Intermediate (I) | Resistant (R) | ||
| Kanamycin(K) | ≥18 | 14–17 | ≤13 | S(26.96) |
| Streptomycin(S) | ≥15 | 11–14 | ≤10 | S(16.09) |
| Tetracycline(TE) | ≥19 | 15–18 | ≤14 | S(20.02) |
| Ampicillin(AMP) | ≥17 | 14–16 | ≤13 | S(14.72) |
| Chloramphenicol(C) | ≥26 | 20–25 | ≤19 | R(15.84) |
| Ciprofloxacin(CIP) | ≥21 | 16–20 | ≤15 | S(25.69) |
| Meropenem(MEM) | ≥16 | 14–15 | ≤13 | S(29.55) |
| Cephalothin(KF) | ≥18 | 15–17 | ≤14 | S(21.52) |
| Tobramycin(TOB) | ≥15 | 13–14 | ≤12 | S(21.61) |
| Gentamicin(CN) | ≥15 | 13–14 | ≤12 | S(20.21) |
| Ceftazidime(CAZ) | ≥21 | 18–20 | ≤17 | I(18.86) |
| Cefotaxime(CTX) | ≥23 | 15–22 | ≤14 | I(17.67) |
| Imipenem(IPM) | ≥16 | 14–15 | ≤13 | S(28.67) |
| Cefepime(FEP) | ≥18 | 15–17 | ≤14 | S(19.45) |
| Trimethoprim-Sulfamethoxazoie(SXT) | ≥16 | 11–15 | ≤10 | S(19.07) |
| Levofloxacin(LEV) | ≥17 | 14–16 | ≤13 | S(29.38) |
| Aztreonam (ATM) | ≥21 | 18–20 | ≤17 | R(0) |
Bacterial strains and culture conditions
To evaluate phenol-induced alterations occurring in the proteome, of Acinetobacter sp. DW-1, samples were collected during the exponential growth phase from cultures grown on acetate, or acetate plus phenol, for proteomic analysis. These conditions were selected with the aim of obtaining relevant information about the global response of proteins in cells adapted to consumption of phenol. Acinetobacter sp. DW-1 required 30 h to degrade 4 mM acetate in its growth medium (Fig. 4). However, when the cultures were provided with a mixture of acetate and phenol, the degradation rate of acetate was reduced compared to growth on acetate as the single carbon source (Fig. 5). Therefore, high concentrations of phenol inhibit the growth of cells, as previously reported53. By contrast, phenol consumption rates remained low until acetate levels in the growth media were depleted, after which the rate of phenol degradation increased rapidly (Fig. 5). These results are consistent with previous reports demonstrating a hierarchy of substance preference: benzoate > acetate > phenol54. In addition, the major intermediate products of phenol including catechol and cis,cis-muconate were observed in medium simultaneously (Supplementary Figure 2). However, the concentrations of intermediates were always less than 10 mg/L under the conditions studied, and they quickly disappeared when phenol completely degraded. This data is agreement with previous report55.
Figure 4.
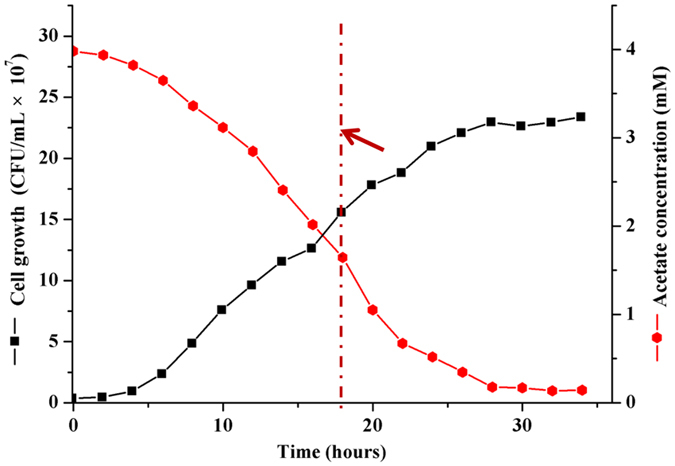
Growth curves of Acinetobacter sp. DW-1. Cells were grown at 30 °C in MSM with 4 mM acetate as the sole carbon and energy source. Growth was followed using the plate counting method. Cells were harvested from cultures for subsequent experiments after 18 h incubation (red arrow).
Figure 5.
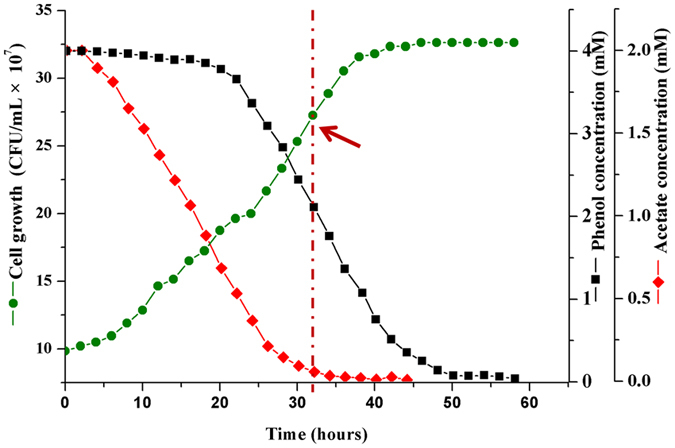
Growth curves of Acinetobacter sp. DW-1. Cells were grown at 30 °C in MSM with 4 mM acetate and 4 mM phenol. Growth was assessed using the plate counting method. Cells were harvested from cultures that after 32 h incubation (red arrow) following phenol supplementation for subsequent experiments.
Identification of proteins by mass spectrometry
To determine which proteins were induced by culture of Acinetobacter sp. DW-1 with phenol, comparative 2-DE was performed in the pH range of 4–7 using crude extract from cells grown on acetate as controls (Fig. 6A). Because phenol is metabolized to acetyl CoA, which then enters the tricarboxylic acid cycle, acetate was a useful carbon source for comparison with the known proteomic effects of phenol. Additionally, phenol is always decomposed into acetate, succinate, pyruvic acid, and acetaldehyde via the ortho- and meta-pathways during phenol biodegradation.
Figure 6.
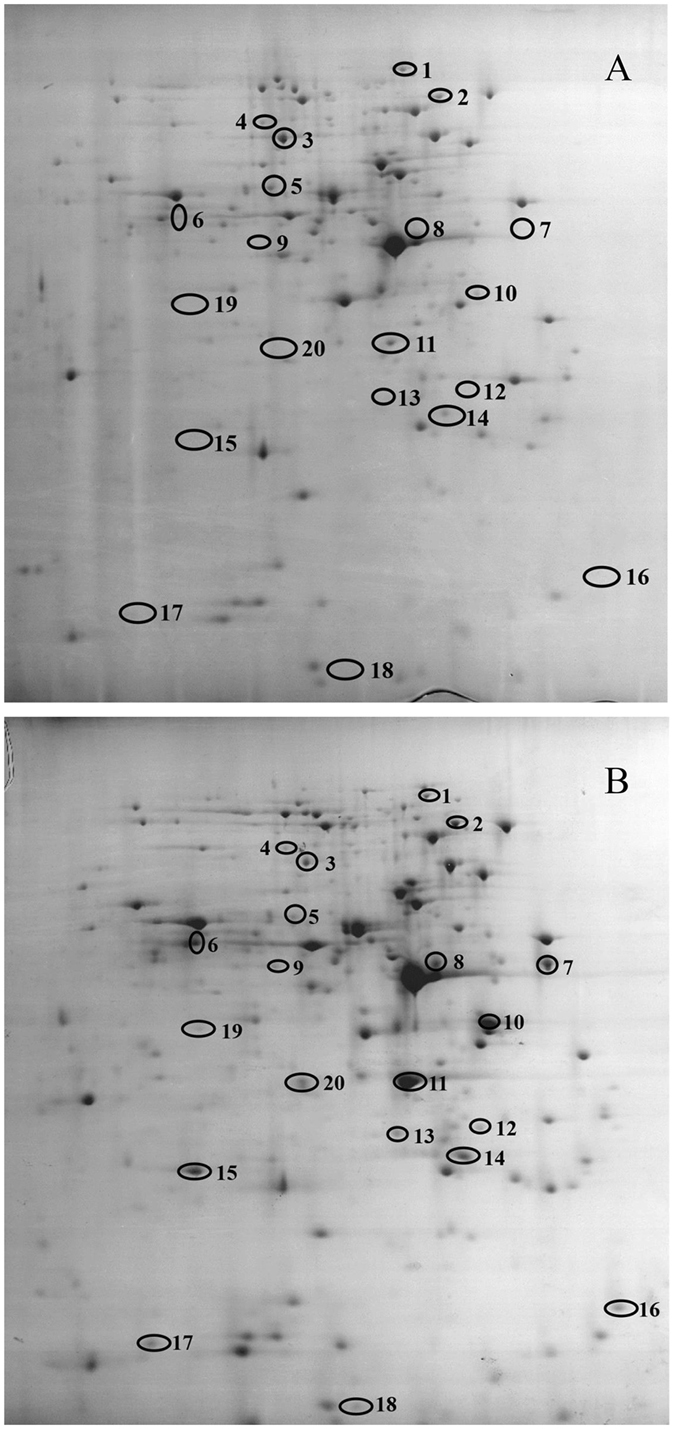
2-D PAGE gels of soluble proteins from Acinetobacter sp. DW-1 cells grown in MSM supplemented with 4 mM acetate (A) or 4 mM acetate and 4 mM phenol (B). pH 4–7 (from right to left).
As shown in Fig. 6, bacterium grown on acetate plus phenol expressed more proteins than bacterium grown on acetate. Over 500 proteins were separated on the pH 4–7 acetate 2-D gels, and over 600 proteins were separated on the acetate plus phenol 2-D gels. In total 20 protein spots with significant differences between acetate and acetate plus phenol 2-D gels were identified by MALDI-TOF/TOF MS (Table 3), with 15 up-regulated protein spots, and 11 spots that were only present in samples grown on acetate plus phenol. This difference was partly attributed to the ability of acetate to easily enter most central metabolic cycles because of its C2 molecule. To grow on a phenol substrate, Acinetobacter sp. DW-1 must activate additional enzymes (Table 3). Among the proteins identified in cells growing on phenol, six protein spots corresponded to enzymes involved in the phenol catabolic pathway. Most of these enzymes were found only in samples grown on phenol-containing media, including phenol hydroxylase, 3-oxoadipate CoA-transferase subunit B, 3-oxoadipate CoA-transferase subunit A, and the muconate and chloromuconate cycloisomerases family protein, 3-oxoadipyl-CoA thiolase. Among these enzymes, previous studies indicate that phenol hydroxylases are the key rate-limiting enzymes catalyzing the initial reaction in phenol degradation, and can be classified into three types: single-, double-, and multi-component56, 57. Multi-component phenol hydroxylases are widely distributed in gram-negative bacteria, such as Pseudomonas sp. OX1 and Acinetobacter radioresistens 58, 59. Two-component phenol hydroxylases are exclusively distributed in gram-positive bacteria, such as Bacillus thermoglucosidasius A760. A single-chain phenol hydroxylases isolated from the eukaryote Trichosporon cutaneum has been reported61. Catechol-1, 2-dioxygenase (C12O) is another important enzyme in the phenol degradation pathway, as the second enzyme of phenol metabolism, and has been used as a marker for this pathway62. Catechol is cleaved by C12O, and the cleavage product, cis, cis-muconate, is transformed via 1-ketoadipate-enol-lactone to succinate and acetyl CoA63. In the present study, the expression of C12O in samples grown on phenol-containing medium was 24.5042-fold higher than that in cells grown on acetate alone. C12O has been discovered in many bacteria grown on aromatic compounds, including phenol, as the sole carbon source. Interestingly, muconate and chloromuconate cycloisomerase family protein was also upregulated when bacteria grew on acetate plus phenol. Muconate cycloisomerase (EC 5.5.1.-) is the third enzyme of phenol metabolism62, and has been purified from extracts of Rhodococcus erythropolis 1CP cells grown on benzoate. Furthermore, 3-oxoadipate CoA-transferase subunit B (Fragment) has been described among benzoate-induced proteins of Pseudomonas. putida KT244012, and among aromatic and choloroaromatic-induced proteins of Pseudomonas sp. Strain B13, which are necessary to reach the Krebs cycle after the convergence of pathways used for the degradation of aromatic compounds64. The function of 3-oxoadipate CoA-transferase, an important enzyme in benzoate catabolism, is to catalyze the conversion of 3-oxadipate to 3-oxoadipyl-CoA, in the catechol branch of the beta-ketoadipate pathwaty. Moreover, it is necessary in the chloroaromatic degradation pathway65.
Table 3.
Identification and characterization of differential expression proteins by MALDI/TOF/TOF MS between acetate or phenol as the sole carbon.
| Identified proteins | Mas scorea | Spot No.b | GI No. | Calculated pI value c | Sequence coverage (%) | Mass (Da)d | Proteins location | Express ion of acetate/acetate plus phenol (fold change) |
|---|---|---|---|---|---|---|---|---|
| Aconitate hydratase B | 169 | 1 | 1310689 | 4.99 | 22 | 95384 | Cytoplasm | ↓ 2.67829 |
| TonB-dependent Receptor Plug domain protein | 136 | 2 | 1310709 | 5.10 | 28 | 36470 | Outer membrane | ↓ 2.01854 |
| Urocanate hydratase OS | 353 | 3 | 470 | 5.63 | 26 | 59646 | Cytoplasm | ↓ 2.73441 |
| 60 kDa chaperonin, groL | 193 | 4 | 903916 | 4.92 | 24 | 57189 | Cytoplasm | ↓ 1.98799 |
| Serine hydroxymethyltransferase, | 173 | 5 | 557600 | 5.44 | 31 | 45195 | Cytoplasm | ↓ 2.10096 |
| 3-oxoadipyl-CoA thiolase | 270 | 6 | 1310697 | 5.89 | 51 | 41279 | Cytoplasm | ↑ 2.18868 |
| Phenol hydroxylase P5 protein | 124 | 7 | 1310911 | 4.53 | 18 | 39500 | Cytoplasm | ↑ Only in acetate + phenol |
| Pirin family protein OS | 147 | 8 | 1310689 | 4.96 | 13 | 35227 | Cytoplasm | ↑ Only in acetate + phenol |
| Muconate and chloromuconate cycloisomerases family protein | 196 | 9 | 1343073 | 5.41 | 27 | 39275 | Cytoplasm | ↑ Only in acetate + phenol |
| Catechol 1,2-dioxygenase, catA | 140 | 10 | 1343073 | 4.70 | 37 | 33526 | Cytoplasm | ↑ 24.5042 |
| MetA-pathway of phenol degradation family protein OS | 81 | 11 | 1358412 | 5.30 | 26 | 32212 | Outer membrane | ↑ 9.57105 |
| 3-oxoadipate CoA-transferase subunit B (Fragment) | 181 | 12 | 1310659 | 4.86 | 42 | 22789 | Cytoplasm | ↑ Only in acetate + phenol |
| 3-oxoadipate CoA-transferase subunit A (Fragment) | 149 | 13 | 1310720 | 7.66 | 46 | 19886 | Cytoplasm | ↑ Only in acetate + phenol |
| Uncharacterized protein | 133 | 14 | 1148157 | 4.85 | 19 | 23098 | Cytoplasm | ↑ 1.81059 |
| Nitroreductase family protein | 118 | 15 | 1310689 | 5.83 | 41 | 21938 | Cytoplasm | ↑ Only in acetate + phenol |
| Fimbrial protein | 205 | 16 | 575584 | 7.77 | 39 | 13897 | Outer membrane | ↑ Only in acetate + phenol |
| OsmC-like family protein | 176 | 17 | 1310689 | 5.71 | 66 | 14449 | Cytoplasm | ↑ Only in acetate + phenol |
| Twitching mobility protein, pilT | 40 | 18 | 1310605 | 6.27 | 23 | 35731 | Cytoplasm | ↑ Only in acetate + phenol |
| Cupin domain protein | 112 | 19 | 1310689 | 5.37 | 25 | 26710 | Cytoplasm | ↑ Only in acetate + phenol |
| Isocitrate lyase | 216 | 20 | 470 | 5.30 | 19 | 59646 | Cytoplasm | ↑ Only in acetate + phenol |
aMOWSE scores of the highest confident matches (P < 0.05).
bThe number refers to the spot numbers as given in Fig. 6.
cpI, the predicted isoelectric point calculated from the protein sequence.
dThe predicted molecular weight calculated from the protein sequence.
In this study, 3-oxoadipyl-CoA thiolase was up-regulated by 2.18868-fold in bacteria grown on acetate plus phenol versus those grown on acetate alone. The function of 3-oxoadipyl-CoA thiolase is to catalyze the transformation of 3-oxoadipyl-CoA via the 3-oxoadipate pathway, forming acetyl-CoA. 3-oxoadipyl-CoA thiolases from various gram-negative have been biochemically characterized including those of Pseudomonas knackmussii B1363 and P. putida PRS200066. To date, 3-oxoadipyl-CoA thiolases have been discovered in only a few gram-positive organisms, including Rhodococcus opacus1CP67 and Rhodococcus jostii RHA168.
Interestingly, the enzyme of the MetA-pathway of phenol degradation family protein was up-regulated by 9.57105 fold in cells grown on phenol plus acetate compared with those grown on acetate alone. The specific function of this protein has not been documented.
Proteins from spot numbers 8, 15, 16, 17, 18, 19 and 20 (Table 3) were only expressed when bacteria were grown on acetate plus phenol, and these proteins included fimbrial protein. Previous studies have demonstrated that fimbrial proteins are generally insoluble and resistant to chemical and proteolytic degradation69, and this protein may be predicted to contribute to enhanced immune responses. Therefore, we can infer that high concentrations of phenol may be toxic to bacterial cells.
Other proteins not known to be involved in phenol catabolism were also upregulated in Acinetobacter sp. DW-1 grown on phenol-containing medium. These proteins included OsmC-like family proteins, which was consistent with the finding that the toluene catabolic plasmid pD2RT of Pseudomonas migulae strain D2RT, which encodes an OsmC-like protein is up-regulated when this bacterium is grown on toluene, Thus these proteins likely play a role in the detoxification of organic peroxides70.
Isocitrate lyase was also highly up-regulated in Acinetobacter sp. DW-1 grown on acetate supplemented with phenol. Isocitrate lyase has been reported to be responsible for the glyoxylate bypass, a unique metabolic pathway71, and it is also up-regulated during hexadecane degradation by Acinetobacter oleivorans DR1. These results suggest that bacterial cells need to synthesize cellular precursors from carbon at a rapid rate during detoxification of organic compounds72.
Nitroreductases, a group of enzymes that catalyze the reduction of nitro groups on aromatic compounds, were also upregulated in Acinetobacter sp. DW-1 grown on phenol-containing medium. These proteins have recently attracted enormous interest because of their central role in mediating nitroaromatic toxicity. In particular, they were mainly investigated in TNT73 and RDX74 degrading strains. However, the detailed mechanism of their involvement in degradation of phenol by bacteria has not been documented. Some studies have reported that nitroreductase family proteins could be induced in response to catechol, 2-methoxyhydroquinone, and chromanon23, 28, although other reports indicate that nitroreductase family proteins are not induced within a short time of phenol stress28. We hypothesize that the nitroreductase upregulation in Acinectobacter sp. DW-1 grown on acetate supplemented with phenol resulted from the decomposition of phenol into catechol, catalyzed by phenol hydroxylase.
Interestingly, the abundance of other proteins including Cupin domain protein OS and Twitching mobility protein were also increased in samples from bacteria grown on acetate plus phenol. To date, no correlation between these proteins and biodegradation of organic matter has been documented.
Five proteins were found to be downregulated in Acinetobacter sp. DW-1 grown on phenol-containing medium. Among these, aconitate hydratase, plays a critical role in the tricarboxylic acid (TCA) cycle by catalyzing the citrate to isocitrate reaction. The TCA cycle responsible for energy supply. The presence of phenol toxic to bacterial cells, leading to inhibition of their growth and metabolism and accounting for the reduction in this enzyme of the TCA cycle.
TonB-dependent receptors have been reported to mediate the transport of siderophore into the periplasm of gram-negative bacteria. A complex of three membrane-spanning proteins TonB, ExbB, and ExbD couples the chemiosmotic protential of the cytoplasmic membrane with siderophore uptake across the outer membrane. The function of 60 kDa chaperonin is to prevent misfolding and promote refolding and proper assembly of unfolded polypeptides generated under stress conditions (http://www.uniprot.org/uniprot/A0A010LHT0). Elucidation of the reason for its downregulation as a result of exposure to phenol will require further study.
Transcriptome sequences assembly and analysis
To reveal the mechanism of phenol biodegradation by Acinetobacter sp. DW-1, we performed comparisons of the transcriptome of Acinetobacter sp. DW-1 grown on acetate and acetate plus phenol. The high-throughput RNA-seq generated a total length of 1208514000 and 1792215250 clean reads from control samples CK-1and CK-2, respectively. A total length of 1459341500 and 1393273750 clean reads was obtained from samples T-1 and T-2, respectively. The Q20 of all of these samples reached 97%, which indicated the high quality of the transcriptome sequencing. Biosample (SAMN05919934, SAMN05919933, SAMN05792302 and SAMN05710740) and Bioproject (PRJNA350871) information has been submitted to NCBI.
Subsequently, 959 unigenes were annotated and divided into 21 cluster of orthologous groups of proteins (COG) (Fig. 7). The largest cluster was R (general function prediction only), followed by C (energy production and conversion). These unigenes are known to be involved in the degradation and metabolism of benzene rings, the TCA cycle, amino acids and lipids, and they are involved in the metabolic processes of Acinetobacter sp. DW-1. The relatively high frequency of S (function unknown) indicates that many novel genes still require further identification in Acinetobacter sp. DW-1. The GO database annotated 903 unigenes (Supplementary Table S1). These unigenes were classified as having biological process, cellular component, and molecular functions. The high proportion of unigenes involved in catalytic activity function, binding, metabolic processes, cellular processes and single-organism processes implied the presence of various types of secondary metabolites and a complex regulation mechanism in phenol biodegradation by Acinetobacter sp. DW-1(Supplementary Table S2).
Figure 7.
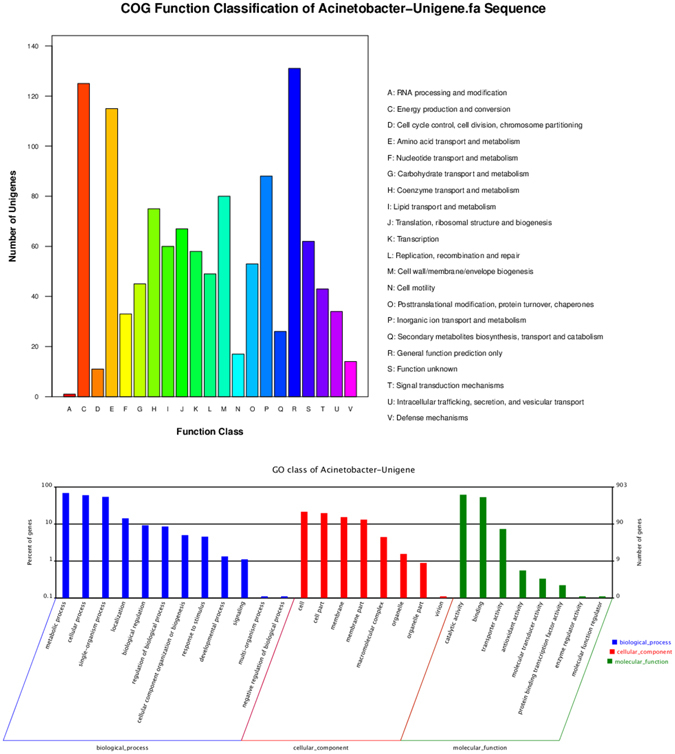
GO and COG function classifications of the unigenes of CK and T samples.
Differentially expressed genes (DEGs) were observed during phenol biodegradation. Among the DEGs, 694 genes were found to be differentially up-regulated genes (DUGs), and 340 genes were identified as differentially down-regulated genes (DDGs). The data, including the gene ID, gene length, FRKM in the control and treatment groups, log2FC (T/CK), FDR, and annotation of all of the DEGs are provided in Supplementary Table S3. GO analysis indicated that DEGs were members of 22 classes. The number of up-regulated genes in each functional category is shown in Supplementary Table S4. Among the 694 up-regulated genes, genes encoding metabolic processes comprised the highest percentage (201 genes, approximately 16% of all up-regulated genes), whereas genes related to catalytic activity comprised approximately 15% of all up-regulated genes (182 genes). Genes encoding proteins involved in cellular processes (179 genes), binding (155 genes), single-organism processes (146 genes), cell (80 genes), cell parts (75 genes), and membrane (39 genes) were also up-regulated. In addition, genes classified in the category of genes of unknown function represented a considerable portion of the induced genes (26 genes). The numbers of down-regulated genes in each functional category are shown in Supplementary Table S4. Among the down-regulated genes, those involved in metabolic processes were the dominant category (63 genes; 15%), followed by genes involved in single-organism processes (56 genes; 10%), catalytic activity (51 genes), binding (48 genes), and cellular processes (45 genes), and membrane (22 genes).
Additionally, 331 DEGS were annotated by the KEGG database. The high proportion of metabolic pathways (19.94%) and the biosynthesis of secondary metabolite pathways (10.88%) among all of the DEGs indicated the production of various types of metabolites during phenol biodegradation process (Supplementary Table S5). It is more interesting that phenol degradation pathway only belongs to metabolic pathways. During phenol biodegradation, genes involved in responses to stimuli showed significantly increased expression, including Unigene0000224, Unigene0000852, Unigene0002231, Unigene0001700, Unigene0000948, Unigene0000663 and Unigene0001896. Thus high concentrations of phenol cause physiologic stress in cells of Acinetobacter sp. DW-1.
The phenol degradation pathway is a part of the benzoate degradation pathway in the KEGG database (ko00362), and crucial genes in this metabolic pathway were found to be up-regulated (Table 4), including Unigene0000836 (phenol hydroxylase), Unigene0001131 (catechol 1, 2-dioxygenase), Unigene0001341 (muconate cycloisomerase), Unigene0000980 (3-carboxy-cis,cis-muconate cycloisomerase) and Unigene0000678(3-oxoadipate CoA-transferase). The MetA-pathway of phenol degradation family protein was also up-regulated by 2.3-fold compared to the expression in the CK groups. In addition, phenol degradation protein involved in meta-pathway of phenol degradation was up-regulated 37.3-fold. It has been reported that cells grown on sufficiently high concentrations of sodium benzoate simultaneously induce catechol 1,2-dioxygenase and catechol 2,3-dioxygenase, indicating that they can degrade sodium benzoate through both the ortho- and the meta- cleavage pathways75. In addition, proteome analysis indicates that Pseudomonas sp. K82 has three main metabolic pathways for degradation of aromatic compounds, and could induce a specific metabolic pathway for each compound. Moreover, the catechol 2, 3-dioxygenase pathway was the major pathway induced by aniline exposure in Pseudomonas sp. K82 and the catechol 1, 2-dioxygenase pathway was the secondary pathway whereas the catechol 1,2-dioxygenase pathway was the major pathway induced by benzoate exposure76. In addition, the results of enzyme assays showed that the efficiency of biodegradation of phenol by meta-pathways was higher than that by ortho-pathways77. Therefore, we infer from the data produced in the present study that Acinetobacter sp. DW-1 utilizes phenol primarily through the orth-pathway, but that it can also degrade phenol through the meta-pathway in some conditions.
Table 4.
Functions of key genes detected by RNA-Seq.
| Gene ID | Description | COG no. | Fold changes (log2 Ratio(T/CK)) | KEGG no. | EC no. | Predicted function |
|---|---|---|---|---|---|---|
| Energy production and conversion | ||||||
| Unigene0002417 | phenol degradation protein | COG4313 | 5.217441542 | N/A | N/A | Protein involved in meta-pathway of phenol degradation |
| Unigene0002240 | bacterial Cytochrome Ubiquinol Oxidase family protein | COG1271 | 4.535288588 | K00425 | [EC:1.10.3.-] | establishment of localization |
| Unigene0001576 | phenol 2-monooxygenase | COG2871 | 6.942107699 | N/A | N/A | oxidoreductase activity |
| Unigene0002249 | ATP F0F1 synthase subunit alpha | COG0056 | 2.197069232 | K02111 | [EC:3.6.3.14] | cation-transporting ATPase activity |
| Secondary metabolites biosynthesis, transport and catabolism | ||||||
| Unigene0001131 | catechol 1,2-dioxygenase | COG3485 | 4.94 | K03381 | [EC:1.13.11.1] | catechol-containing compound metabolic process |
| Unigene0000378 | protocatechuate 3,4-dioxygenase | COG3485 | 5.587102309 | K00448 | [EC:1.13.11.3] | cellular aromatic compound metabolic process |
| Unigene0001606 | coenzyme F390 synthetase | COG1541 | −15.2013869 | K01912 | [EC:6.2.1.30] | aromatic compound catabolic process |
| Unigene0000908 | 3-carboxy-cis,cis-muconate cycloisomerase | COG0015 | 4.01609738 | N/A | N/A | catechol-containing compound catabolic process |
| Unigene0000803 | benzoate 12 dioxygenase alpha subunit | COG4638 | −2.879859554 | K05549 | [EC:1.14.12.10] | cellular aromatic compound metabolic process |
| Unigene0001123 | ligand-gated channel protein | COG4774 | −3.646712089 | K02014 | N/A | iron chelate transport |
| Unigene0000980 | acetyl-CoA acetyltransferase | COG0183 | −1.173807482 | K00632 | [EC:2.3.1.16] | Acetyl-CoA acetyltransferase |
| Unigene0000678 | 3-oxoadipate CoA-transferase | COG1788 | 4.294322399 | K01031 | [EC:2.8.3.6] | Acyl CoA:acetate/3-ketoacid CoA transferase, alpha subunit |
| Unigene0001528 | ABC transporter ATP-binding protein, partial | COG1127 | 1.998799132 | K02065 | N/A | ABC-type transport system involved in resistance to organic solvents |
| Cell wall/membrane/envelope biogenesis; General function prediction only | ||||||
| Unigene0001341 | muconate cycloisomerase | COG4948 | 2.49 | K01856 | [EC:5.5.1.1] | L-alanine-DL-glutamate epimerase and related enzymes of enolase superfamily |
| Unigene0000039 | nitroreductase family protein | COG3560 | 4.047415045 | K07078 | N/A | catalytic activity |
| Unigene0000969 | quercetin 2,3-dioxygenase | COG1741 | 3.101493853 | K06911 | N/A | None predicted |
| Posttranslational modification, protein turnover, chaperones | ||||||
| Unigene0000948 | osmotically inducible protein C | COG1765 | 3.54518103 | K07397 | N/A | response to stress |
| Translation, ribosomal structure and biogenesis; Inorganic ion transport and metabolism | ||||||
| Unigene0002513 | 50 S ribosomal protein L10, partial | COG0244 | 2.634136949 | K02864 | N/A | rRNA binding; structural molecule activity |
| Unigene0001303 | 50 S ribosomal protein L2 | COG0090 | 2.264955891 | K02886 | N/A | RNA binding; structural molecule activity |
| Unigene0000305 | 30 S ribosomal protein S13 | COG0099 | 2.001593462 | K02952 | N/A | RNA binding; structural molecule activity |
| Unigene0000065 | 30 S ribosomal protein S3, partial | COG0092 | 2.392017692 | K02982 | N/A | poly(A) RNA binding; structural molecule activity |
| Unigene0000602 | 50 S ribosomal protein L7/L12 | COG0222 | 2.765846319 | K02935 | N/A | None predicted |
| Unigene0000077 | ATPase, partial | COG2217 | 5.545384277 | K01533 | [EC:3.6.3.4] | transition metal ion binding,copper-transporting ATPase activity |
| Unigene0000176 | E1-E2 ATPase | COG2217 | 3.350705804 | K01533 | [EC:3.6.3.4] | intrinsic component of membrane, copper ion transmembrane transport |
| Coenzyme transport and metabolism; Lipid transport and metabolism | ||||||
| Unigene0001464 | lipoyl synthase | COG0320 | −3.178920684 | K03644 | [EC:2.8.1.8] | cation binding; sulfurtransferase activity; |
| Unigene0001096 | 3-oxoacyl-ACP synthase | COG0332 | 1.099154317 | K00648 | [EC:2.3.1.180] | fatty acid metabolic process |
| Unigene0002287 | fatty acid desaturase family protein | COG3239 | 1.092701954 | K00508 | [EC:1.14.19.3] | none predicted |
| Transcription; Defense mechanism; Signal transduction mechanisms | ||||||
| Unigene0001039 | LysR family transcriptional regulator | COG0583 | 2.992016687 | N/A | N/A | cellular macromolecule biosynthetic process |
| Unigene0000056 | TetR family transcriptional regulator | COG1309 | −0.379311349 | NA | NA | nucleic acid binding |
| Unigene0000663 | Predicted transcriptional regulator,BolA superfamily | COG5007 | 3.489769782 | NA | NA | cellular response to stress |
| Unigene0000886 | AraC family transcriptional regulator | COG2207 | −1.360719141 | NA | NA | DNA binding |
| Unigene0001910 | GntR family transcriptional regulator | COG2186 | −0.97172089 | K05799 | NA | transcriptional repressor for pyruvate dehydrogenase complex |
| Unigene0002400 | CRP/FNR family transcriptional regulator | COG0664 | −0.045371841 | K10914 | NA | nucleic acid binding |
| Unigene0000781 | HTH-type transcriptional regulator | COG0789 | 1.939857402 | N/A | N/A | regulation of transcription, DNA-templated |
| Unigene0000852 | serine hydrolase | COG1680 | 1.512798695 | N/A | N/A | response to toxic substance |
| No related COG | ||||||
| Unigene0000054 | phenol hydroxylase | COG0543 | 15.87929606 | K03380 | [EC:1.14.13.17] | 2-polyprenylphenol hydroxylase and related flavodoxin oxidoreductases |
| Unigene0002446 | metA-pathway of phenol degradation family protein | N/A | 1.177456264 | N/A | N/A | None predicted |
| Unigene0001290 | biofilm synthesis protein | N/A | 4.094059655 | N/A | N/A | transmembrane transporter activity |
| Unigene0001394 | fimbrial protein | N/A | 15.21820373 | N/A | N/A | cell projection |
| Unigene0000307 | membrane protein | N/A | 4.679820328 | N/A | N/A | intrinsic component of membrane |
| Unigene0002168 | Fis family transcriptional regulator | COG1690 | −6.6691460517 | K14415 | NA | gene expression |
| Unigene0000243 | LuxR family transcriptional regulator | NA | −0.652472163 | NA | NA | nucleic acid binding |
N/A: Not available.
Protocatechuate 3,4-dioxygenase, which catalyzes the aromatic ring cleavage of 3,4-dihydroxy benzoate by incorporating both atoms of molecular oxygen to yield β-carboxy-cis,cis-muconate was highly up-regulated. Nitroreductase is typically reported to be up-regulated in the presence of toxic substances. Biofilm synthesis protein and membrane protein were strongly up-regulated. Biofilm synthesis proteins such as Unigene0001290 and Unigene0001946 were up-regulated. Biofilm synthesis protein is predicted to have transmenbrane transporter activity and thus to be responsible for nutrient transport and processing. In addition, many membrane proteins were also found markedly up-regulated. For instance, the membrane proteins which were responsible for protein binding (Unigene0000178), establishment of localization (Unigene0000385), transition metal ion binding (Unigene0000415), nucleic acid binding(Unigene0001807), and iron-sulfur cluster binding (iron-sulfur cluster binding). Furthermore, integral membrane TerC family protein (Unigene0002265), outer membrane autotransporter barrel domain protein (Unigene0000415) and outer membrane OmpA family protein (Unigene0001866) were also up-regulated. Additionally, it is reported that some cell surface proteins such as fimbrial protein were found to be implicated in biofilm formation78. In this study, fimbrial protein was markedly up-regulated in both 2D-gel based proteomic analysis and RNA-seq analysis. In particular, its expression was 38,165-fold higher than that the CK groups in transcriptome analysis. It is reported that fimbrial protein could confer the ability to bind to specific surfaces79, and thus its upregulation makes biofilm easier to form. To the best of our knowledge, the up-regulation of membrane proteins were to defense the toxicity from higher concentration of phenol22. High concentrations of phenol as a toxicant might stimulate immune responses and the defense function of cells, which leads to the formation of biofilm. Additionally, there were many transcriptionl regulators were involved in phenol biodegradation (Table 4), such as LysR/AraC/LuxR/TetR/Fur/GntR/Fis/CRP/FNR/CysB family transcriptional regulators, even some putative transcriptional regulator. LysR family transcriptional regulators were reported to be activators of expression of genes encoding enzymes that are associated with ortho-cleavage through the β-ketoadipate pathway, including the gene of catBCA 80, 81. In this study, LysR family transcriptional regulators was detected up-regulate similarly. Preliminary data for AraC family transcriptional regulators indicated that they were involved in expression control of a catabolic operon, and generally acted as transcription activators in the presence of a chemical effector molecule82. There was few example about TetR family involved in aromatic compouds83. In this study, TetR family transcriptional regulator was down-regulate, the result was in accord with the primary data TetR-type regulator was a repressor for the expression of the genes for aromatic compounds84–86. GntR family members were also assumed as repressors to control the degradation of aromatic compounds in the absence of the pathway substrate81. In our study, GntR family transcriptional regulator was detected down-regulate 1.96 fold, which was consistent with previous reports that it played a repressor. There was little information about the CRP/FNR family transcriptional regulator involved in aromatic compounds. It has been reported that the CRP/FNR regulator activates expression of the hbaR gene which allowed transcription of the 4-hydroxybenzoate degradation genes in Rhodopseudomonas palustris under anaerobic conditions87. However, in our study, the function of the FNR regulator need to be further verified. Fis regulators are global regulators of transcription in Escherichia coli in the adjustment of cellular metabolism to varying growth. But the function of Fis regulators during phenol biodegradation need to be further studied88. Similarly, according to preliminary data, the LuxR-type regulators are generally functioning as transcriptional activators, the accurate function of this regulator in phenol metabolism will attract more attention89. HTH-type transcriptional regulator always bind transcriptional activators with the target DNA, which induce transcription initiation by interacting with the aromatic substrate90. In addition, the genes related basic metabolism were also increased, including 50 S ribosomal protein, 30 S ribosomal protein, ATPase and fatty acid desaturase family protein. However, some stress response genes were up-regulated, which indicated the toxicity of phenol to cells during phenol degradation process.
Validation of RT-qPCR
The RNA-seq data were validated by RT-qPCR selecting 22 DEGs (Table 4) that were crucial for phenol biodegradation. The log2 fold change in gene expression between the two conditions demonstrated significant correlation between RNA-seq and RT-qPCR. In particular, the results for the relative expression of these genes using RNA-seq and RT-qPCR showed over 80% consistency, indicating the reliability of the RNA-seq analysis (Fig. 8).
Figure 8.
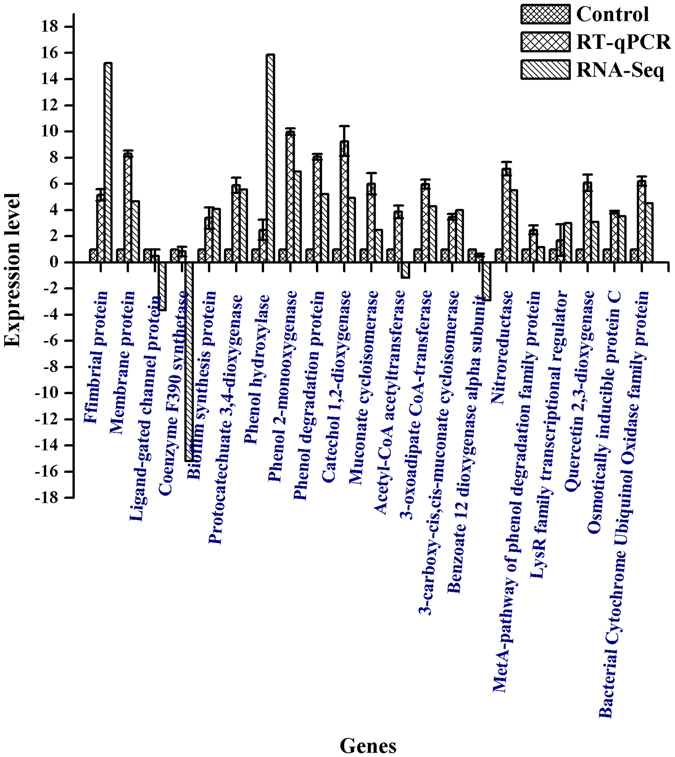
Validation of transcriptome data by quantitative RT-PCR analysis. Log2 fold changes calculated with both RT-qPCR and RNA-Seq analyses
Metabolism pathway analysis
According to the identified catabolic proteins and genes, phenol metabolism related genes were constructed basing on the location of the protein after searching in the KEGG database as shown in Fig. 9. The most important phenol catabolic enzymes were identified: phenol hydroxylase, catechol 1, 2-dioxygenase, muconate cycloisomerases, 3-oxoadipate CoA-transferase, acetyl-CoA acyltransferase, 3-oxoadipyl-CoA thiolase. In addition to these genes, transcriptional regulator indicated obvious expression in Table 4 and Fig. 9, which involved in regulating the expression of key genes in phenol metabolic pathway, and the function of some new transicriptional regulators need to be further confirmed. Meanwhile, membrane related proteins such as biofilm synthesis protein and membrane protein showed increased expression, ribosomal proteins also upregulated differentially. Results of pathway analysis indicated that this strain catabolized phenol via the β-ketoadipate pathway, also known as the “ortho” route. This is consistent with previous studies, indicating that Acinetobacter sp. usually catalyze phenol as sole carbon and energy source by the ortho-cleavage pathway13, 91. However, some genes related aromatic compound metabolic process were found up-regulated, especially the phenol degradation protein which involved in the meta-pathway of phenol degradation. Thus, we could infer that meta-cleavage pathway may exist.
Figure 9.
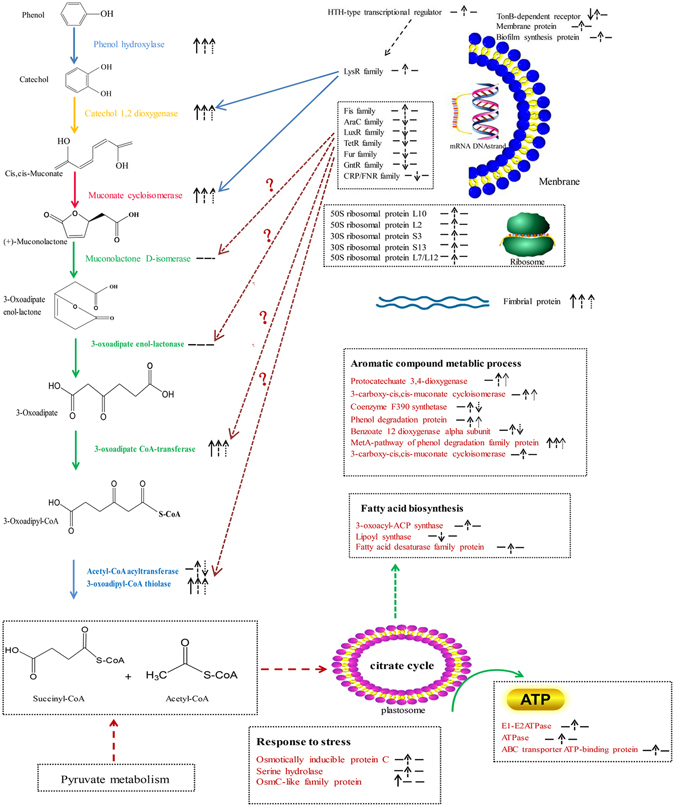
The metabolic pathway of phenol biodegradation. The proteins and genes identified by 2-D electrophoresis and RNA-seq, and the variation of them in the state of regulation are indicated by arrows(up/down), “ ”represents the state of proteins detected by 2-D electrophoresis, “
”represents the state of proteins detected by 2-D electrophoresis, “ ”represents the state of genes detected by RNA-seq. “
”represents the state of genes detected by RNA-seq. “ ”represents the state of proteins or genes validated by RT-qPCR. “−” represents none detected.
”represents the state of proteins or genes validated by RT-qPCR. “−” represents none detected.
In summary, Acinetobacter sp. DW-1 has the potential to be applied in bioremediation. From the proteomic and transcriptomic analysis, we determined that Acinetobacter sp. DW-1 degraded phenol mainly by the ortho pathway because of the induction of phenol hydroxylase and catechol-1, 2-dioxygenase. There is also evidence for induction of the meta-pathway of phenol biodegradation by Acinetobacter sp. DW-1, as the expression of phenol degradation protein involved in meta-pathway of phenol degradation increased. Despite the abundance of catabolic proteins, some novel candidate proteins (OsmC-like family protein, MetA-pathway of phenol degradation family protein, fimbrial protein, coenzyme F390 synthetase, nitroreductase family protein) and low abundance transcriptional regulators (GntR/LuxR/CRP/FNR/TetR/Fis family transcriptional regulator) were successfully identified, especially MetA-pathway of phenol degradation family protein, it is firstly reported dramatically up-regulated in phenol biodegradation, and the specific function of this protein need to be further studied. In addition, fimbrial protein in outer membrane was also dramatically up-regulated and has not been reported in phenol degradation related literatures. FNR and Fis family transcriptional regulators were also not documented before during phenol biodegradation. These findings provide novel insights and improved understanding of the phenol biodegradation mechanisms of Acinetobacter sp. DW-1. However, further study of the detailed roles and factors regulating these proteins in Acinetobacter sp. and of mechanism of phenol biodegradation are urgently required.
Conclusion
This study provides a detailed view of biofilm formation of Acinetobacter sp. DW-1 on a carrier material consisting of polyhedron hollow polypropylene balls. The biofilm activity was optimal when the cells were immobilized for approximately 72 h. The phenol degradation test indicated that the immobilized Acinetobacter sp. DW-1 cells exhibited high phenol degradation ability and thus could potentially be applied to bioremediation in phenol-polluted drinking water. Additionally, it is the first time to simultaneously use both proteomic and transcriptomic approaches to investigate phenol metabolism. The transcriptomics and proteomics analysis data indicated that there was a significant correlation between them. However, transcriptomic analysis obtained less-soluble membrane proteins and low-abundance transcriptional regulators which had not been found in proteomic analysis with its inherent limitation. Whereas, proteomic analysis verified the expression of genes which were also detected by transcriptome sequencing. Omics analysis showed that Acinetobacter sp. DW-1 degrades phenol mainly by the ortho pathway, some novel candidate proteins (OsmC-like family protein, MetA-pathway of phenol degradation family protein, fimbrial protein and coenzyme F390 synthetase) and transcriptional regulators (GntR/LuxR/CRP/FNR/TetR/Fis family transcriptional regulator) were identified to be potentially involved in phenol biodegradation, especially MetA-pathway of phenol degradation family protein and fimbrial protein showed a strong positive correlation with phenol biodegradation, and Fis family transcriptional regulator is likely to exert its effect as activators of gene expression. However, the specific function of these candidate proteins and genes in phenol biodegradation requires further confirmation.
Electronic supplementary material
Acknowledgements
We would like to acknowledge the financial support of the Science and Technology Planning Project of Guangdong Province (2012 A032300018), Guangdong province science and technology plan projects (201313040400010) and (2014A020219005).
Author Contributions
Conceived and designed the experiments: Q.W., J.Z., and Q.G. Performed the experiments: Q.G. and W.G. Analyzed the data: Q.G. and M.S. Contributed reagents/materials/analysis tools: H.W. Contributed to the writing of the manuscript: Q.G. and Q.W.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-04187-6
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Senthilvelan T, Kanagaraj J, Panda RC, Mandal A. Biodegradation of phenol by mixed microbial culture: an eco-friendly approach for the pollution reduction. Clean Technologies and Environmental Policy. 2014;16:113–126. doi: 10.1007/s10098-013-0598-2. [DOI] [Google Scholar]
- 2.Mukherjee S, Kumar S, Misra AK, Fan M. Removal of phenols from water environment by activated carbon, bagasse ash and wood charcoal. Chemical Engineering Journal. 2007;129:133–142. doi: 10.1016/j.cej.2006.10.030. [DOI] [Google Scholar]
- 3.Jiang Y, Yang K, Wang H, Shang Y, Yang X. Characteristics of phenol degradation in saline conditions of a halophilic strain JS3 isolated from industrial activated sludge. Marine pollution bulletin. 2015;99:230–234. doi: 10.1016/j.marpolbul.2015.07.021. [DOI] [PubMed] [Google Scholar]
- 4.Gu Q, et al. Community analysis and recovery of phenol-degrading bacteria from drinking water biofilters. Frontiers in Microbiology. 2016;7:495. doi: 10.3389/fmicb.2016.00495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LaCroix SJ, Cabelli VJ. Membrane filter method for enumeration of Acinetobacter calcoaceticus from environmental waters. Applied and environmental microbiology. 1982;43:90–96. doi: 10.1128/aem.43.1.90-96.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung J, Park W. Acinetobacter species as model microorganisms in environmental microbiology: current state and perspectives. Applied microbiology and biotechnology. 2015;99:2533–2548. doi: 10.1007/s00253-015-6439-y. [DOI] [PubMed] [Google Scholar]
- 7.Chung T-P, Tseng H-Y, Juang R-S. Mass transfer effect and intermediate detection for phenol degradation in immobilized Pseudomonas putida systems. Process Biochemistry. 2003;38:1497–1507. doi: 10.1016/S0032-9592(03)00038-4. [DOI] [Google Scholar]
- 8.Mordocco A, Kuek C, Jenkins R. Continuous degradation of phenol at low concentration using immobilized Pseudomonas putida. Enzyme and Microbial Technology. 1999;25:530–536. doi: 10.1016/S0141-0229(99)00078-2. [DOI] [Google Scholar]
- 9.Wang Y, et al. Biodegradation of phenol by free and immobilized Acinetobacter sp.strain PD12. Journal of Environmental Sciences. 2007;19:222–225. doi: 10.1016/S1001-0742(07)60036-9. [DOI] [PubMed] [Google Scholar]
- 10.Santos PM, Benndorf D, Sá-Correia I. Insights into Pseudomonas putida KT2440 response to phenol-induced stress by quantitative proteomics. Proteomics. 2004;4:2640–2652. doi: 10.1002/pmic.200300793. [DOI] [PubMed] [Google Scholar]
- 11.Kurbatov L, Albrecht D, Herrmann H, Petruschka L. Analysis of the proteome of Pseudomonas putida KT2440 grown on different sources of carbon and energy. Environmental microbiology. 2006;8:466–478. doi: 10.1111/j.1462-2920.2005.00913.x. [DOI] [PubMed] [Google Scholar]
- 12.Kim YH, et al. Analysis of aromatic catabolic pathways in Pseudomonas putida KT 2440 using a combined proteomic approach: 2-DE/MS and cleavable isotope-coded affinity tag analysis. Proteomics. 2006;6:1301–1318. doi: 10.1002/pmic.200500329. [DOI] [PubMed] [Google Scholar]
- 13.Giuffrida MG, et al. Media containing aromatic compounds induce peculiar proteins in Acinetobacter radioresistens, as revealed by proteome analysis. Electrophoresis. 2001;22:1705–1711. doi: 10.1002/1522-2683(200105)22:9<1705::AID-ELPS1705>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 14.Anli G, Lim CJ. Proteome analysis of the adaptation of a phenol-degrading bacterium Acinetobacter sp. EDP3 to the variation of phenol loadings. Chinese Journal of Chemical Engineering. 2007;15:781–787. doi: 10.1016/S1004-9541(08)60002-6. [DOI] [Google Scholar]
- 15.Griffin TJ, et al. Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Molecular & Cellular Proteomics. 2002;1:323–333. doi: 10.1074/mcp.M200001-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Kim SJ, Kweon OG, Cerniglia CE. Proteomic applications to elucidate bacterial aromatic hydrocarbon metabolic pathways. Current Opinion in Microbiology. 2009;12:301–309. doi: 10.1016/j.mib.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 17.Zhang W, Li F, Nie L. Integrating multiple ‘omics’ analysis for microbial biology: application and methodologies. Microbiology. 2010;156:287–301. doi: 10.1099/mic.0.034793-0. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen VD, et al. Transcriptome and proteome analyses in response to 2-methylhydroquinone and 6-brom-2-vinyl-chroman-4-on reveal different degradation systems involved in the catabolism of aromatic compounds in Bacillus subtilis. Proteomics. 2007;7:1391–1408. doi: 10.1002/pmic.200700008. [DOI] [PubMed] [Google Scholar]
- 19.Duy NV, et al. The proteome and transcriptome analysis of Bacillus subtilis in response to salicylic acid. Proteomics. 2007;7:698–710. doi: 10.1002/pmic.200600706. [DOI] [PubMed] [Google Scholar]
- 20.Mazzoli R, et al. Degradation of aromatic compounds by Acinetobacter radioresistens S13: growth characteristics on single substrates and mixtures. Archives of Microbiology. 2007;188:55–68. doi: 10.1007/s00203-007-0223-z. [DOI] [PubMed] [Google Scholar]
- 21.Giuffrida MG, et al. Media containing aromatic compounds induce peculiar proteins in Acinetobacter radioresistens, as revealed by proteome analysis. Electrophoresis. 2001;22:1705–1711. doi: 10.1002/1522-2683(200105)22:9<1705::AID-ELPS1705>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 22.Geng A, Lim CJ. Proteome Analysis of the Adaptation of a Phenol-Degrading Bacterium Acinetobacter sp. EDP3 to the Variation of Phenol Loadings. Chinese Journal of Chemical Engineering. 2007;15:781–787. doi: 10.1016/S1004-9541(08)60002-6. [DOI] [Google Scholar]
- 23.Van Duy N, et al. Transcriptome and proteome analyses in response to 2-methylhydroquinone and 6-brom-2-vinyl-chroman-4-on reveal different degradation systems involved in the catabolism of aromatic compounds in Bacillus subtilis. Proteomics. 2007;7:1391–1408. doi: 10.1002/pmic.200700008. [DOI] [PubMed] [Google Scholar]
- 24.Sprenger RR, et al. Comparative proteomics of human endothelial cell caveolae and rafts using two-dimensional gel electrophoresis and mass spectrometry. Electrophoresis. 2004;25:156–172. doi: 10.1002/elps.200305675. [DOI] [PubMed] [Google Scholar]
- 25.Cohen DP, et al. Proteomic analysis of log to stationary growth phase Lactobacillus plantarum cells and a 2-DE database. Proteomics. 2006;6:6485–6493. doi: 10.1002/pmic.200600361. [DOI] [PubMed] [Google Scholar]
- 26.Lange S, et al. Analysis of protein species differentiation among mycobacterial low-Mr-secreted proteins by narrow pH range Immobiline gel 2-DE-MALDI-MS. Journal of proteomics. 2014;97:235–244. doi: 10.1016/j.jprot.2013.06.036. [DOI] [PubMed] [Google Scholar]
- 27.Chávez de Paz LE, Bergenholtz G, Dahlén G, Svensäter G. Response to alkaline stress by root canal bacteria in biofilms. International endodontic journal. 2007;40:344–355. doi: 10.1111/j.1365-2591.2006.01226.x. [DOI] [PubMed] [Google Scholar]
- 28.Tam LT, et al. Differential gene expression in response to phenol and catechol reveals different metabolic activities for the degradation of aromatic compounds in Bacillus subtilis. Environmental microbiology. 2006;8:1408–1427. doi: 10.1111/j.1462-2920.2006.01034.x. [DOI] [PubMed] [Google Scholar]
- 29.Jin M, Yu X, Chen X, Zeng R. Pseudomonas putida IOFA1 transcriptome profiling reveals a metabolic pathway involved in formaldehyde degradation. Process Biochemistry. 2015;51:220–228. doi: 10.1016/j.procbio.2015.11.036. [DOI] [Google Scholar]
- 30.Hervás AB, Canosa I, Santero E. Transcriptome Analysis of Pseudomonas putida in Response to Nitrogen Availability. Journal of Bacteriology. 2008;190:416–420. doi: 10.1128/JB.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao, X., Qi, Y., Xu, C., Yang, Y. & Wang, J. Transcriptome and metabolome responses of Shewanella oneidensis MR-1 to methyl orange under microaerophilic and aerobic conditions. Applied Microbiology & Biotechnology 1–10 (2017). [DOI] [PubMed]
- 32.Kim, J. & Lorenzo, V. D. Deep Sequencing to Study Microbial Transcriptomic Responses to Hydrocarbon Degradation/Production/Stress[M]. Humana Press (2015).
- 33.Zhang Y, Wu Q, Zhang J, Yang X. Effects of ozone on membrane permeability and ultrastructure in Pseudomonas aeruginosa. Journal of applied microbiology. 2011;111:1006–1015. doi: 10.1111/j.1365-2672.2011.05113.x. [DOI] [PubMed] [Google Scholar]
- 34.Bauer A, Kirby W, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. American journal of clinical pathology. 1966;45:493. [PubMed] [Google Scholar]
- 35.Kovačević J, Mesak LR, Allen KJ. Occurrence and characterization of Listeria spp. in ready-to-eat retail foods from Vancouver, British Columbia. Food microbiology. 2012;30:372–378. doi: 10.1016/j.fm.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 36.Massa S, Caruso M, Trovatelli F, Tosques M. Comparison of plate count agar and R2A medium for enumeration of heterotrophic bacteria in natural mineral water. World Journal of Microbiology and Biotechnology. 1998;14:727–730. doi: 10.1023/A:1008893627877. [DOI] [Google Scholar]
- 37.Ampe F, Lindley ND. Acetate utilization is inhibited by benzoate in Alcaligenes eutrophus: evidence for transcriptional control of the expression of acoE coding for acetyl coenzyme A synthetase. Journal of bacteriology. 1995;177:5826–5833. doi: 10.1128/jb.177.20.5826-5833.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu Q, et al. Community Analysis and Recovery of Phenol-degrading Bacteria from Drinking Water Biofilters. Frontiers in microbiology. 2016;7:495. doi: 10.3389/fmicb.2016.00495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee BL, Ong HY, Shi CY, Ong CN. Simultaneous determination of hydroquinone, catechol and phenol in urine using high-performance liquid chromatography with fluorimetric detection. Journal of Chromatography A. 1993;619:259–266. doi: 10.1016/0378-4347(93)80115-K. [DOI] [PubMed] [Google Scholar]
- 40.Yoshikawa N, Mizuno S, Ohta K, Suzuki M. Microbial production of cis,cis -muconic acid. Applied microbiology and biotechnology. 1988;28:20–25. [Google Scholar]
- 41.Cao, B. & Loh, K. Catabolic pathways and cellular responses of Pseudomonas putida P8 during growth on benzoate with a proteomics approach. Biotechnology & Bioengineering101, 1297 (2008). [DOI] [PubMed]
- 42.Ye Y, et al. Identification of potential virulence factors of Cronobacter sakazakii isolates by comparative proteomic analysis. International journal of food microbiology. 2016;217:182–188. doi: 10.1016/j.ijfoodmicro.2015.08.025. [DOI] [PubMed] [Google Scholar]
- 43.Roma-Rodrigues C, Santos PM, Benndorf D, Rapp E, Sá-Correia I. Response of Pseudomonas putida KT2440 to phenol at the level of membrane proteome. Journal of proteomics. 2010;73:1461–1478. doi: 10.1016/j.jprot.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 44.Maldonado AM, Echevarria-Zomeno S, Jean-Baptiste S, Hernandez M, Jorrin-Novo JV. Evaluation of three different protocols of protein extraction for Arabidopsis thaliana leaf proteome analysis by two-dimensional electrophoresis. Journal of proteomics. 2008;71:461–472. doi: 10.1016/j.jprot.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 45.Xie Z, Lin W, Luo J. Promotion of microalgal growth by co-culturing with Cellvibrio pealriver using xylan as feedstock. Bioresource technology. 2016;200:1050–1054. doi: 10.1016/j.biortech.2015.10.058. [DOI] [PubMed] [Google Scholar]
- 46.Ishii Si, Koki J, Unno H, Hori K. Two morphological types of cell appendages on a strongly adhesive bacterium, Acinetobacter sp. strain Tol 5. Applied and environmental microbiology. 2004;70:5026–5029. doi: 10.1128/AEM.70.8.5026-5029.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wood CA, Reboli AC. Infections caused by imipenem-resistant Acinetobacter calcoaceticus biotype anitratus. Journal of Infectious Diseases. 1993;168:1602–1603. doi: 10.1093/infdis/168.6.1602. [DOI] [PubMed] [Google Scholar]
- 48.Rao RS, et al. Correlation between biofilm production and multiple drug resistance in imipenem resistant clinical isolates of Acinetobacter baumannii. Indian journal of medical microbiology. 2008;26:333. doi: 10.4103/0255-0857.38876. [DOI] [PubMed] [Google Scholar]
- 49.Levin AS, Levy CE, Manrique AEI, Medeiros EA, Costa SF. Severe nosocomial infections with imipenem-resistant Acinetobacter baumannii treated with ampicillin/sulbactam. International journal of antimicrobial agents. 2003;21:58–62. doi: 10.1016/S0924-8579(02)00276-5. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y, Marrs CF, Simon C, Xi C. Wastewater treatment contributes to selective increase of antibiotic resistance among Acinetobacter spp. Science of the Total Environment. 2009;407:3702–3706. doi: 10.1016/j.scitotenv.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 51.Messi P, Guerrieri E, Bondi M. Antibiotic resistance and antibacterial activity in heterotrophic bacteria of mineral water origin. Science of the total environment. 2005;346:213–219. doi: 10.1016/j.scitotenv.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 52.Papandreou S, Pagonopoulou O, Vantarakis A, Papapetropoulou M. Multiantibiotic resistance of gram-negative bacteria isolated from drinking water samples in southwest Greece. Journal of chemotherapy. 2000;12:267–273. doi: 10.1179/joc.2000.12.4.267. [DOI] [PubMed] [Google Scholar]
- 53.Santos PM, et al. Mechanistic insights into the global response to phenol in the phenol-biodegrading strain Pseudomonas sp. M1 revealed by quantitative proteomics. Omics: a journal of integrative biology. 2007;11:233–251. doi: 10.1089/omi.2007.0009. [DOI] [PubMed] [Google Scholar]
- 54.Mazzoli R, et al. Degradation of aromatic compounds by Acinetobacter radioresistens S13: growth characteristics on single substrates and mixtures. Archives of microbiology. 2007;188:55–68. doi: 10.1007/s00203-007-0223-z. [DOI] [PubMed] [Google Scholar]
- 55.Alva VA, Peyton BM. Phenol and catechol biodegradation by the haloalkaliphile Halomonas campisalis: influence of pH and salinity. Environmental Science & Technology. 2003;37:4397–4402. doi: 10.1021/es0341844. [DOI] [PubMed] [Google Scholar]
- 56.Basile LA, Erijman L. Maintenance of phenol hydroxylase genotypes at high diversity in bioreactors exposed to step increases in phenol loading. FEMS microbiology ecology. 2010;73:336–348. doi: 10.1111/j.1574-6941.2010.00898.x. [DOI] [PubMed] [Google Scholar]
- 57.Peters M, et al. Acquisition of a deliberately introduced phenol degradation operon, pheBA, by different indigenous Pseudomonas species. Applied and environmental microbiology. 1997;63:4899–4906. doi: 10.1128/aem.63.12.4899-4906.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pessione E, et al. Phenol hydroxylase from Acinetobacter radioresistens is a multicomponent enzyme. European Journal of Biochemistry. 1999;265:549–555. doi: 10.1046/j.1432-1327.1999.00720.x. [DOI] [PubMed] [Google Scholar]
- 59.Izzo V, et al. PHK from phenol hydroxylase of Pseudomonas sp. OX1. Insight into the role of an accessory protein in bacterial multicomponent monooxygenases. Archives of biochemistry and biophysics. 2011;505:48–59. doi: 10.1016/j.abb.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 60.Kirchner U, Westphal AH, Müller R, van Berkel WJ. Phenol hydroxylase from Bacillus thermoglucosidasius A7, a two-protein component monooxygenase with a dual role for FAD. Journal of Biological Chemistry. 2003;278:47545–47553. doi: 10.1074/jbc.M307397200. [DOI] [PubMed] [Google Scholar]
- 61.Neujahr HY, Gaal A. Phenol hydroxylase from yeast. European Journal of Biochemistry. 1973;35:386–400. doi: 10.1111/j.1432-1033.1973.tb02851.x. [DOI] [PubMed] [Google Scholar]
- 62.Spånning Å, Neujahr HY. The effect of glucose on enzyme activities and phenol utilization in Trichosporon cutaneum grown in continuous culture. Journal of general microbiology. 1990;136:1491–1495. doi: 10.1099/00221287-136-8-1491. [DOI] [Google Scholar]
- 63.Kasak L, Hôrak R, Nurk A, Talvik K, Kivisaar M. Regulation of the catechol 1, 2-dioxygenase-and phenol monooxygenase-encoding pheBA operon in Pseudomonas putida PaW85. Journal of bacteriology. 1993;175:8038–8042. doi: 10.1128/jb.175.24.8038-8042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaschabek SR, Kuhn B, Müller D, Schmidt E, Reineke W. Degradation of aromatics and chloroaromatics by Pseudomonas sp. strain B13: purification and characterization of 3-oxoadipate: succinyl-coenzyme A (CoA) transferase and 3-oxoadipyl-CoA thiolase. Journal of bacteriology. 2002;184:207–215. doi: 10.1128/JB.184.1.207-215.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abbai NS, Govender A, Shaik R, Pillay B. Pyrosequence analysis of unamplified and whole genome amplified DNA from hydrocarbon-contaminated groundwater. Molecular biotechnology. 2012;50:39–48. doi: 10.1007/s12033-011-9412-8. [DOI] [PubMed] [Google Scholar]
- 66.Parales R, Harwood C. Characterization of the genes encoding beta-ketoadipate: succinyl-coenzyme A transferase in Pseudomonas putida. Journal of bacteriology. 1992;174:4657–4666. doi: 10.1128/jb.174.14.4657-4666.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eulberg D, Lakner S, Golovleva LA, Schlömann M. Characterization of a protocatechuate catabolic gene cluster from Rhodococcus opacus 1CP: evidence for a merged enzyme with 4-carboxymuconolactone-decarboxylating and 3-oxoadipate enol-lactone-hydrolyzing activity. Journal of bacteriology. 1998;180:1072–1081. doi: 10.1128/jb.180.5.1072-1081.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Patrauchan MA, et al. Catabolism of benzoate and phthalate in Rhodococcus sp. strain RHA1: redundancies and convergence. Journal of bacteriology. 2005;187:4050–4063. doi: 10.1128/JB.187.12.4050-4063.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klemm, P. Fimbriae Adhesion, Genetics, Biogenesis, and Vaccines. CRC Press 9 (1994).
- 70.Dubbs JM, Mongkolsuk S. Peroxiredoxins in Bacterial Antioxidant Defense in Peroxiredoxin Systems. Sub-cellular biochemistry. 2007;44:143–193. doi: 10.1007/978-1-4020-6051-9_7. [DOI] [PubMed] [Google Scholar]
- 71.Schaible UE, Kaufmann SH. A nutritive view on the host–pathogen interplay. TRENDS in Microbiology. 2005;13:373–380. doi: 10.1016/j.tim.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 72.Jung J, Noh J, Park W. Physiological and metabolic responses for hexadecane degradation in Acinetobacter oleivorans DR1. The Journal of Microbiology. 2011;49:208–215. doi: 10.1007/s12275-011-0395-8. [DOI] [PubMed] [Google Scholar]
- 73.Caballero A, Lázaro JJ, Ramos JL, Esteve-Núñez A. PnrA, a new nitroreductase-family enzyme in the TNT-degrading strain Pseudomonas putida JLR11. Environmental microbiology. 2005;7:1211–1219. doi: 10.1111/j.1462-2920.2005.00801.x. [DOI] [PubMed] [Google Scholar]
- 74.Nejidat A, Kafka L, Tekoah Y, Ronen Z. Effect of organic and inorganic nitrogenous compounds on RDX degradation and cytochrome P-450 expression in Rhodococcus strain YH1. Biodegradation. 2008;19:313–320. doi: 10.1007/s10532-007-9137-3. [DOI] [PubMed] [Google Scholar]
- 75.Loh K-C, Chua S-S. Ortho pathway of benzoate degradation in Pseudomonas putida: induction of meta pathway at high substrate concentrations. Enzyme and Microbial Technology. 2002;30:620–626. doi: 10.1016/S0141-0229(02)00016-9. [DOI] [Google Scholar]
- 76.Kim SI, et al. Proteome analysis of Pseudomonas sp. K82 biodegradation pathways. Proteomics. 2004;4:3610–3621. doi: 10.1002/pmic.200400977. [DOI] [PubMed] [Google Scholar]
- 77.Chen Y, Liu H, Chen H. Characterization of phenol biodegradation by Comamonas testosteroni ZD4-1 and Pseudomonas aeruginosa ZD4-3. Biomedical and environmental sciences: BES. 2003;16:163–172. [PubMed] [Google Scholar]
- 78.Cabral MP, et al. Proteomic and Functional Analyses Reveal a Unique Lifestyle for Acinetobacter baumannii Biofilms and a Key Role for Histidine Metabolism. Journal of Proteome Research. 2011;10:3399–3417. doi: 10.1021/pr101299j. [DOI] [PubMed] [Google Scholar]
- 79.Schembri MA, Klemm P. Heterobinary adhesins based on the Escherichia coli FimH fimbrial protein. Appl Environ Microbiol. 1998;64:1628–1633. doi: 10.1128/aem.64.5.1628-1633.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gerischer U. Specific and global regulation of genes associated with the degradation of aromatic compounds in bacteria. Journal of Molecular Microbiology & Biotechnology. 2002;4:111–121. [PubMed] [Google Scholar]
- 81.Birgit Morawski AS, Nicholas Ornston L. Repression of Acinetobacter vanillate demethylase synthesis by VanR, a member of the GntR family of transcriptional regulators. FEMS microbiology letters. 2000;187(1):65–68. doi: 10.1111/j.1574-6968.2000.tb09138.x. [DOI] [PubMed] [Google Scholar]
- 82.Inouye SANaTN. Molecular cloning of gene xylS of the TOL plasmid: evidence for positive regulation of the xylDEGF operon by xylS. Journal of bacteriology. 1981;148(2):413–418. doi: 10.1128/jb.148.2.413-418.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tropel, D. & van der Meer, J. R. Bacterial transcriptional regulators for degradation pathways of aromatic compounds. Microbiology and molecular biology reviews: MMBR68, 474-500, table of contents, doi:10.1128/MMBR.68.3.474-500.2004 (2004). [DOI] [PMC free article] [PubMed]
- 84.Eaton RW. p-Cymene catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA encoding conversion of p-cymene to p-cumate. Journal of Bacteriology. 1997;179:3171–3180. doi: 10.1128/jb.179.10.3171-3180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ampe FNDL. Acetate utilization is inhibited by benzoate in Alcaligenes eutrophus: evidence for transcriptional control of the expression of acoE coding for acetyl coenzyme A synthetase. Journal of Bacteriology. 1995;177:5826–5833. doi: 10.1128/jb.177.20.5826-5833.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brinkrolf K, Brune I, Tauch A. Transcriptional regulation of catabolic pathways for aromatic compounds in Corynebacterium glutamicum.[J] Genetics & Molecular Research Gmr. 2006;5(4):773–789. [PubMed] [Google Scholar]
- 87.Paul, G. Egland & C. S. H. HbaR, a 4-Hydroxybenzoate Sensor and FNR-CRP Superfamily Member, Regulates Anaerobic 4-Hydroxybenzoate Degradation by Rhodopseudomonas palustris. Journal of bacteriology182(1), 100–106, 100–106 (2000). [DOI] [PMC free article] [PubMed]
- 88.Nasser W, Schneider R, Travers A, Muskhelishvili G. CRP Modulates fis Transcription by Alternate Formation of Activating and Repressing Nucleoprotein Complexes. Journal of Biological Chemistry. 2001;276:17878–17886. doi: 10.1074/jbc.M100632200. [DOI] [PubMed] [Google Scholar]
- 89.Cramer A, Gerstmeir R, Schaffer S, Bott M, Eikmanns BJ. Identification of RamA, a Novel LuxR-Type Transcriptional Regulator of Genes Involved in Acetate Metabolism of Corynebacterium glutamicum. Journal of Bacteriology. 2006;188:2554–2567. doi: 10.1128/JB.188.7.2554-2567.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Prieto EDaMA. Bacterial promoters triggering biodegradation of aromatic pollutants. Current Opinion in Biotechnology. 2000;11(5):467–475. doi: 10.1016/S0958-1669(00)00126-9. [DOI] [PubMed] [Google Scholar]
- 91.Paller G, Hommel RK, Kleber HP. Phenol degradation by Acinetobacter calcoaceticus NCIB 8250. Journal of basic microbiology. 1995;35:325–335. doi: 10.1002/jobm.3620350508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


