Abstract
Massively parallel sequencing technologies provide great opportunities for discovering rare susceptibility variants involved in complex disease etiology via large-scale imputation and exome and whole-genome sequence-based association studies. Due to modest effect sizes, large sample sizes of tens to hundreds of thousands of individuals are required for adequately powered studies. Current analytical tools are obsolete when it comes to handling these large datasets. To facilitate the analysis of large-scale sequence-based studies, we developed SEQSpark which implements parallel processing based on Spark to increase the speed and efficiency of performing data quality control, annotation, and association analysis. To demonstrate the versatility and speed of SEQSpark, we analyzed whole-genome sequence data from the UK10K, testing for associations with waist-to-hip ratios. The analysis, which was completed in 1.5 hr, included loading data, annotation, principal component analysis, and single variant and rare variant aggregate association analysis of >9 million variants. For rare variant aggregate analysis, an exome-wide significant association (p < 2.5 × 10−6) was observed with CCDC62 (SKAT-O [p = 6.89 × 10−7], combined multivariate collapsing [p = 1.48 × 10−6], and burden of rare variants [p = 1.48 × 10−6]). SEQSpark was also used to analyze 50,000 simulated exomes and it required 1.75 hr for the analysis of a quantitative trait using several rare variant aggregate association methods. Additionally, the performance of SEQSpark was compared to Variant Association Tools and PLINK/SEQ. SEQSpark was always faster and in some situations computation was reduced to a hundredth of the time. SEQSpark will empower large sequence-based epidemiological studies to quickly elucidate genetic variation involved in the etiology of complex traits.
Keywords: CCDC62, complex traits, computational tools, imputed data, quality control, rare variant aggregate association analysis, Spark, UK10K, waist-to-hip ratio, whole-genome sequence data
Main Text
Massively parallel sequencing technologies are generating an unprecedented amount of sequence data on various kinds of samples including human exomes and genomes. Many rare variant association methods have been developed to elucidate the underlying disease etiology using large-scale population-based sequence datasets.1, 2, 3, 4, 5 Although some findings are promising,6 statistical power analyses performed with simulated data demonstrate that large sample sizes of tens or even hundreds of thousands of individuals are required for adequately powered studies.7, 8
Large-scale genetic epidemiological studies are currently ongoing, including the Trans-Omics for Precision Medicine program (TopMed) (see Web Resources) and UK BioBank9 studies. Additional large-scale genetic epidemiological studies are emerging that will generate whole-genome sequence (WGS) data or impute WGS data into existing genotype array data to better understand the genetic etiology of complex traits.
It is problematic to analyze large datasets of massively parallel sequence data given the limitations of current analytic tools for annotation, data quality control, and association testing.9, 10 Analytic tools such as PLINK/SEQ and Variant Association Tools (VAT)11 are designed to run on a single computer/processor, with poor support for parallel computation. For example, PLINK/SEQ is a single threaded program, and VAT can be multi-threaded on a single server for some tasks. None of the existing tools can leverage the computational resources of a cluster of multiple servers. Backend database systems for current tools are also obsolete, usually relying on a single flat file or a file-based relational database like SQLite, which is not suitable for high input/output applications.
To address these issues, we developed SEQSpark, a new analysis tool for large-scale sequence data quality control, annotation, and rare variant association analysis. SEQSpark is based on Spark, a fast engine for large-scale data processing. The Spark platform was selected because it has a simple parallel computation model and it has nearly unlimited scalability, allowing for expanded computational resources, e.g., number of servers within a cluster to enhance the performance without modification of the software. Spark makes use of the Hadoop file system (HDFS), a distributed file system that allows for data storage in a cluster environment (Figure 1A). An additional advantage of Spark is its ability to store data in memory, allowing for many magnitudes faster analysis speeds compared to software that accesses data from hard drives. Spark can efficiently make use of today’s servers that have tens of gigabytes of memory.
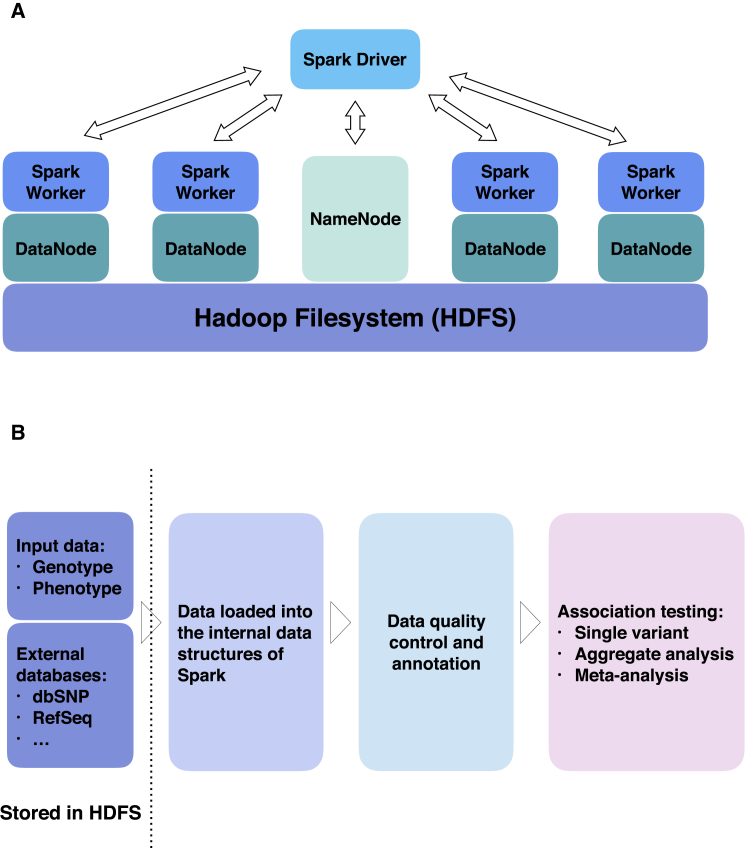
Figure 1.
Spark Architecture and SEQSpark Workflow
(A) Interaction of the Spark components—driver and workers and the Hadoop filesystem (HDFS) components—NameNode and DataNodes. The NameNode is the master node and manages the file system’s meta-data. A file in the HDFS can be split into several blocks and those blocks are stored in a set of slave nodes (DataNodes). The NameNode determines the mapping of the blocks to the DataNodes, while the DataNodes performs the read and write operations within the file system. The Spark driver talks with the HDFS NameNode and obtains the meta-data from NameNode and then distributes the jobs to the Spark workers.
(B) SEQSpark workflow that begins with importing data and databases (used for annotation). The data are loaded into the internal data structures of Spark. Data quality control and annotation can be performed followed by association testing.
SEQSpark splits large datasets into many small blocks that are stored across an entire cluster of servers. The blocks (Figure 1A) can then be accessed and processed simultaneously, so the speed is enhanced by a factor that is proportional to the number of blocks. Variants in different blocks can be analyzed together when necessary. The enhancement in speed is dependent on the hardware resources in a cluster, i.e., the total number of processors, size of memory, and number of disks, but is not limited by the computation power and input/output throughput of a single server. By carefully implementing algorithms in the map-reduce form compatible with the processing engine, we overcome the computational and input/output bottleneck that is experienced by other tools.
SEQSpark uses the memory caching advantage of Spark and outperforms PLINK/SEQ and VAT even in a single-server environment. PLINK/SEQ and VAT both use the file-based relational database, SQLite, which can considerably reduce computational speed, because it lacks optimization for large datasets and high-frequency access to data. SEQSpark also takes advantage of the sparse data structure of genotype data.6
SEQSpark (Figure 1B) performs data quality control based on genotype and variant level metrics, e.g., read depth, quality score. Using a variety of databases, annotation and bioinformatics evaluation is performed at both variant and gene/region levels. Allele frequencies and bioinformatics scores from external databases can be used as weights in subsequent association tests. SEQSpark implements both single variant association tests and rare variant aggregate association methods, e.g., combined multivariate collapsing (CMC),1 burden of rare variants (BRV),2, 12 variable threshold (VT),4 sequence kernel association test (SKAT),5 and SKAT-optimal (SKAT-O).13 All methods are implemented in a regression framework so that important covariates can be included in the analysis and gene × gene and gene × environment interactions can be investigated. Conditional regression can also be performed to tease apart, for example, associations with susceptibility variants from those due to linkage disequilibrium. The speed and versatility of SEQSpark make it ideal for the analysis of small- to large-scale genetic studies of complex traits.
WGS data from the UK10K and 50,000 simulated exomes were analyzed on a small cluster to demonstrate the versatility and speed of SEQSpark. The cluster consists of eight servers each with two AMD Opteron 8-core CPUs, 64 GB memory, and three 4 TB SATA hard drives. For the analysis, the number of blocks was set to 5,120. When comparing the performance of SEQSpark to PLINK/SEQ and VAT, 2,000 simulated exomes were analyzed on a workstation which consists of two Intel Xeon 8-core CPUs with hyper-threading turned on (32 virtual cores in total); eight 8 GB DDR3 memory sticks (64 GB in total); and six 1 TB SATA hard drives. For SEQSpark the number of blocks for the analysis was set to 1,024. We recommended to set the number of blocks for an analysis to a value so that each block contains 32–128 megabytes of data. It is preferable to have more blocks when using a very large cluster with many CPUs so that each block has fewer megabytes of data. No modifications were made to the workstation’s hardware to run SEQSpark and it could even be run on a laptop. Cloud computing can also be used to analyze data using SEQSpark. The speed of SEQSpark can be increased not only by adding additional CPUs but also by increasing the number of hard drives per server, although increasing the number of CPUs will have a greater impact on the speed than the number of hard drives.
Analysis of UK10K data and the 50,000 simulated exomes were not performed using either PLINK/SEQ or VAT due to the extended computational time necessary to perform data analysis. For PLINK/SEQ and VAT, considerable computational time was needed to load these two datasets; e.g., for the WGS UK10K data it took PLINK/SEQ 8.5 hr and VAT 9.3 hr to load chromosome 1, and for the 50,000 simulated exomes PLINK/SEQ took 6.6 hr and VAT 26.3 hr to load the chromosome 1 data. The loading time depends not only on the size of the file but also the number of variant sites and genotypes. Although the simulated exome data contains 3× the number of genotypes of the UK10K, PLINK/SEQ can load this dataset quicker because there are fewer variant sites, where the loading time for VAT is impacted by the number of genotypes. In contrast to the loading times for VAT and PLINK/SEQ, it took SEQSpark 2.3 and 5.0 min to load the chromosome 1 data for the UK10K and 50,000 simulated exomes, respectively. Therefore, to compare the three programs in a reasonable time frame, 2,000 exomes were generated and analyzed.
Analysis of waist-to-hip ratio was performed using WGS data from the Avon Longitudinal Study of Parents and Children (ALSPAC) cohort which was included in the UK10K. This study includes 1,927 individuals of which 1,811 individuals had waist-to-hip ratio (WHR) data available for analysis. The generation of the WGS data as well as the quality control, which was performed before distribution, has been previously described.14 Functional annotation was performed to determine gene boundaries and to classify coding variants, i.e., splice sites, nonsense, missense, and frameshift indels. Ti/Tv ratio was calculated for variants with an MAF < 0.01 and ≥ 0.01. It took 12 s to calculate Ti/Tv ratios and 1.4 min to annotate the rare variants (MAF < 0.01). Table 1 contains the benchmark times to complete each step of the analysis including benchmark times for each association method.
Table 1.
Benchmarks for SEQSpark Analysis of UK10K Hip-to-Waist Ratio Data
| Variants: MAF ≥ 0.01a | Rare Variants: MAF < 0.01b | |
|---|---|---|
| Load datac | 21.75 min | 16.25 min |
| Annotation | N/A | 1.40 min |
| Ti/Tv ratio | 1.92 min | 0.20 min |
| PCAd | 11.65 min | N/A |
| Single variant | 16.03 min | N/A |
| CMC | N/A | 0.22 min |
| BRV | N/A | 0.23 min |
| VT | N/A | 7.90 min |
| SKAT | N/A | 0.18 min |
| SKAT-O | N/A | 0.22 min |
Quality control was performed using data from 1,927 individuals with WGS data. PCA was performed using 1,811 individuals who had data on WHRs and association analysis was performed using 1,798 individuals with WHRs data who were not outliers in the PC analysis.
A total of 9,332,772 variants with an MAF ≥ 0.01 analyzed.
A total of 542,616 rare variants within coding regions were loaded and after annotation, a total of 163,578 missense, splice-site, frameshift, and nonsense variants in 18,011 genes were available for analysis.
The dataset size is 669.4 GB in LZ4 compression format.
Ten PCs were generated using all variants with an MAF ≥ 0.01.
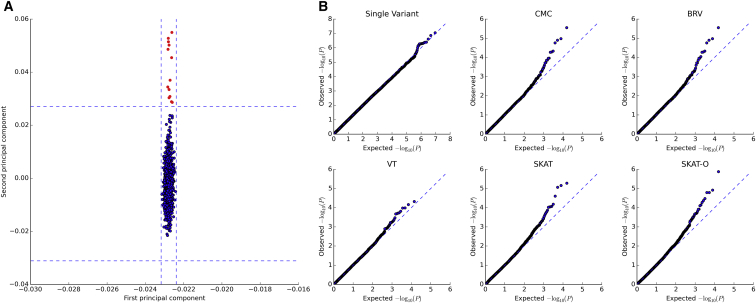
To determine whether there were outliers and to control for population substructure in the analysis, the first ten principal components (PCs) were generated for the 1,811 samples with WHR phenotype data using variants with an MAF ≥ 0.01. Individuals with a first or second PC value that was more than 4 standard deviations (SDs) from the mean were removed before analysis. Although for the first PC, all values were within 4 SDs of the mean, 13 individuals had a second PC value that was >4 SDs from the mean and therefore were removed from further analysis (Figure 2A). Stepwise regression was used to determine which covariates should be adjusted for in the analysis. Covariates age (p = 0.0362), sex (p < 2.0 × 10−16), and body mass index (BMI) (p < 2.0 × 10−16) were significant. Residuals were generated for analysis adjusting for these covariates. To evaluate whether inclusion of PCs aided in controlling for population substructure, analysis was performed without including any PCs, including the first PC, including the first and second PCs, and lastly including the first, second, and third PCs. When analysis was performed without inclusion of PCs, the lambda values were as follows: single variant (λ = 0.9991), CMC (λ = 1.0267), BRV (λ = 1.0302), VT (λ = 1.0096), SKAT (λ = 1.0669), and SKAT-0 (λ = 0.9603). Although for some tests, λ is slightly inflated, inclusion of additional PCs did not reduce the λ values and therefore the analysis was performed without inclusion of any PCs. It can be observed from the quantile-quantile plots that type one error is well controlled (Figure 2B).
Figure 2.
UK10K Waist-to-Hip Ratio Data Scatterplot of the First Two Principal Components and Quantile-Quantile Plots for the Association Analyses
(A) First two PCs for the WGS data from 1,811 UK10K study subjects with WHR data. The PCs were constructed using variants with an MAF ≥ 0.01. For the first PC, μ = −0.0228 and STD = 9.8292 × 10−5 while for second PC, μ = −0.0021 and STD = 0.0073. The dashes outline the 4 STDs for the first and second PCs. 13 individuals which are shown in red fall outside of 4 STDs for the second PC and were removed from additional analysis.
(B) Quantile-quantile plots for each association analysis performed: single variants, CMC, BRV, VT, SKAT, and SKAT-O.
Single variant analysis was performed using the score test assuming an additive model for all variants with an MAF > 0.01. It took 16.3 min to perform the analysis for 9,332,772 variant sites. None of the single variant analyses reached the genome-wide significance level of p < 5.0 × 10−8. This is not surprising given the modest sample size of 1,798 study subjects used for the analysis.
For each gene region, missense, nonsense, splice-site, or frameshift variants with an MAF < 0.01 were selected to perform aggregate rare variant association analysis using CMC, BRV, VT, SKAT, and SKAT-O. For CMC and BRV, a score test was performed. For gene-based aggregate rare variant association analysis of 15,937 genes, each with two or more variant sites and at least three alternative alleles, it took 14 s to perform the fixed effect BRV test and 11 s to perform the random effects SKAT.
For the gene-based rare variant aggregate analysis, an association was observed with CCDC62 (MIM: 613481) that met exome-wide significance (p < 2.5 × 10−6) with SKAT-O (p = 6.89 × 10−7), CMC (p = 1.48 × 10−6), and BRV(p = 1.48 × 10−6) and suggestive evidence of association with VT (p = 2.15 × 10−5) and SKAT (p = 5.50 × 10−6). To validate the results obtained from SEQSpark, the analysis of rare variants in CCDC62 was also performed using VAT and PLINK/SEQ. The results were the same, except for CMC and BRV, because VAT implements the Wald test instead of the score test. When analysis was performed using the Wald test in SEQSpark, identical results were obtained for VAT and SEQSpark (CMC [p = 1.39 × 10−6]; BRV [p = 1.39 × 10−6]). PLINK/SEQ produced results only for SKAT, since WHR is a quantitative trait. The results for SKAT were identical for the three programs. There have been no previous reports of common or rare variants in CCDC62 being associated with WHR or obesity. There are also no functional studies that demonstrate the involvement of CCDC62 in metabolism. It is important that this association is replicated in an independent sample to determine whether rare variants in CCDC62 are truly associated with WHR.
To benchmark and evaluate the performance of PLINK/SEQ, VAT, and SEQSpark, we simulated exome data using non-Finnish European allele frequencies obtained from 33,370 individuals from ExAC. Two samples were generated, one with 50,000 exomes to demonstrate the capabilities of SEQSpark and the other with 2,000 exomes to compare the performance and benchmark SEQSpark, PLINK/SEQ, and VAT. Genotype data were generated assuming Hardy-Weinberg equilibrium (HWE) and missing genotypes were introduced using a probability of 1%. In all a total of 3,681,143 variants in 18,295 genes were generated for the 22 autosomes for the 50,000 exomes and 872,218 variants and 18,295 genes for the sample of 2,000 exomes. To generate realistic sequence data to analyze and perform data quality control, we generated sequence data with variable read depths as well as genotype quality scores. For each genotype, read depth information was generated by drawing from a negative binomial (r,p) where r = 7 is the number of failures until the experiment is stopped and p = 0.2 is the probability of failure, that led to ∼2% of the genotypes having a read depth of <8×. GQ scores were also generated using a two-step sampling procedure: first for 95% of the genotypes the GQ was set to 99; for the remaining genotypes the GQ scores is obtained by drawing from a uniform (a,b) distribution where a = 0 is the starting value and b = 100 is the ending value, leading to ∼1% of the GQ scores being less than 20.
None of the phenotype data were generated to be in association with the genotypes, i.e., data are generated under the null hypothesis of no association. For each sample that exome data were generated, age, sex, a quantitative trait, and a dichotomous trait were assigned. Age was generated using a negative binomial (r,p) distribution where r = 20 and p = 0.3, while the quantitative trait was simulated using a normal (μ,σ2) distribution where the mean μ = 25 and the variance σ2 = 3. Of the samples, 50% were assigned to be male. For the qualitative trait, 50% of the samples were assigned to be case subjects and the other half control subjects. When the quantitative trait was analyzed, three covariates (sex, age, and the binary trait) were included in the analysis, while for the analysis of the qualitative trait, the three covariates that were included in the analysis are sex, age, and the quantitative trait.
First the 50,000 simulated exomes were loaded (44.6 min) and annotated (12.0 min) using SEQSpark on a small cluster. The simulated case-control and quantitative trait data were analyzed in a regression framework including covariates using several rare variant aggregate association tests (CMC, BRV, SKAT, and SKAT-O), obtaining p values analytically. A total of 18,219 genes, with at least two missense, nonsense, or splice site variants sites with an MAF < 0.01 and at least three alternative alleles, were analyzed. The analysis times varied for the same test depending on whether a binary or quantitative trait was analyzed. SKAT-O took more than twice the time to analyze the case-control data than the quantitative trait data. On the other hand, analysis of the case-control data was quicker for fixed-effect tests CMC and BRV and the random effects test SKAT than the analysis of the quantitative trait data. The analysis time for CMC, BRV, SKAT, and SKAT-O combined for the quantitative trait was 48.4 min and for the qualitative trait 72.4 min. Table 2 displays the benchmarks for the analysis of the 50,000 simulated exomes.
Table 2.
Benchmarks for SEQSpark Analysis of 50,000 Simulated Exomes
| Analysis Times for Rare Variantsa | ||
|---|---|---|
| Load datab | 44.63 min | |
| Annotation | 12.00 min | |
| Quantitative | Case-Control | |
| CMC | 14.40 min | 12.72 min |
| BRV | 3.82 min | 2.97 min |
| SKAT | 6.25 min | 5.92 min |
| SKAT-O | 24.12 min | 51.13 min |
A total of 3,681,143 variants were loaded and after annotation, a total of 1,420,128 missense, splice-site, and nonsense variants with a MAF of < 0.01 in 18,219 genes with ≥2 variant sites and ≥3 alternative alleles were available for analysis.
The dataset size is 659.6 GB in LZ4 compression format.
The first step in analysis was to load the data into each of the software packages. This process was most lengthy for VAT, taking slightly more than 1 hr to perform, was faster for PLINK/SEQ taking almost 40 min, and the quickest for SEQSpark run on a cluster taking slightly over 4 min. Performing quality control of the genotype data, i.e., removal of genotypes with a read depth <8× and/or genotype quality score (GQ) < 20, took 48 min using PLINK/SEQ, 37 min for VAT, 7 min for SEQSpark on a single server, and 3 min when SEQSpark was run on a cluster. For calculation of Ti/Tv ratios, it took PLINK/SEQ 57 min, VAT 11 min, and SEQSpark run on a cluster 8 s. The calculations of allele frequencies took more than 1 hr for PLINK/SEQ, 41 min for VAT, and 5 min for SEQSpark on the server and 1 min on the cluster. Table 3 displays benchmark times for loading, annotation, quality control, and data exploration.
Table 3.
Benchmark for Performing Quality Control
| PLINK/SEQa | VATa | SEQSpark Single Servera | SEQSpark Custerb | |
|---|---|---|---|---|
| Load data | 38.75 min | 61.75 min | 5.67 min | 4.35 min |
| Annotation | N/Ac | 3.32 min | 1.42 min | 1.38 min |
| Genotype and variant removald | 48.43 min | 36.67 min | 6.75 min | 2.57 min |
| Calculation of Ti/Tv ratios | 56.54 min | 10.83 min | 1.48 min | 0.13 min |
| Calculation of allele frequencies | 62.5 min | 40.43 min | 5.03 min | 1.30 min |
Exome variant data were generated for a total of 2,000 samples using ExAC non-Finnish European allele frequencies. A total of 872,218 variants were generated with genotype-specific read depths and quality scores.
All software, PLINK/SEQ, VAT, and SEQSpark were run on a single server.
SEQSpark was also run on a cluster and is the only software which has this capability.
PLINK/SEQ does not have a separate annotation step.
Those genotypes with a read depth of <8× and/or GQ score <20 were removed.
Association testing of the 2,000 exomes was performed using several commonly used aggregate rare variant association tests, i.e., CMC, VT, SKAT, SKAT-O, BRV, “Burden Test” for both quantitative and qualitative traits controlling for confounders. For the BRV and CMC, SEQSpark implemented the score test to perform gene-based rare variant association testing. Missense, nonsense, or splice-site with an MAF < 0.01 were analyzed using RefSeq gene boundaries to determine which variants to analyze in aggregate. The p values for the tests were obtained either analytically or empirically using adaptive permutation (Table 4). For adaptive permutation, the number of permutations each program uses are not equivalent; PLINK/SEQ performs far fewer permutations than either VAT or SEQSpark which can reduce the accuracy of empirical p values (Table 4). For example, for case-control data for the Burden Test, PLINK/SEQ ran 912K permutations while SEQSpark performed 38,203K permutations. PLINK/SEQ is more limited in the analyses that it can perform compared to VAT and SEQSpark. For PLINK/SEQ, quantitative trait analysis is limited to SKAT and for qualitative trait fixed effect tests p values can be obtained only using adaptive permutation. When analysis was performed with SKAT for a quantitative trait obtaining analytical p values, it takes 70 min for PLINK/SEQ, 36 min for VAT, and 44 s for SEQSpark run on a server and 12 s when analysis with SEQSpark was performed on a cluster. SKAT-O was also used to perform analysis and PLINK/SEQ failed to produce results, while VAT took 46 min and SEQSpark run on a cluster took 21 s. To perform the Burden Test for case control data implementing adaptive permutation using PLINK/SEQ took more than 100 min, VAT took more than 24 hr, and SEQSpark on a cluster took 39 min. It should be noted that PLINK/SEQ performs a “Burden Test” which like the BRV analyzes the allele counts within a genomic region. In addition to running quicker than PLINK/SEQ, SEQSpark performed many more permutations. The analytic BRV took 23 min to run on VAT and 1 min and 12 s to perform analysis with SEQSpark on a server and cluster, respectively (Table 4).
Table 4.
Benchmarks for Performing Aggregate Rare Variant Association Analysis
| PLINK/SEQa | VATa | SEQSpark Single Servera | SEQSpark Clusterb | |
|---|---|---|---|---|
| Quantitative Trait - Analytical P Values | ||||
| CMC | N/Ac | 19.17 min | 0.73 min | 0.25 min |
| Burden Test | N/Ac | 19.65 min | 0.75 min | 0.23 min |
| SKAT | 70 min | 36.43 min | 0.73 min | 0.20 min |
| SKAT-O | failedd | 45.52 min | 1.27 min | 0.35 min |
| Case-Control - Analytical P Values | ||||
| CMC | N/Ac | 21.90 min | 0.67 min | 0.23 min |
| Burden test | N/Ac | 22.99 min | 1.05 min | 0.20 min |
| SKAT | 70 min | 46.71 min | 0.70 min | 0.23 min |
| SKAT-O | failedd | 49.49 min | 1.18 min | 0.45 min |
| Quantitative Trait – Empirical P Values | ||||
| CMC | N/Ac | 81.89 min 38,793 ke | 13.08 min 30,942 ke | 5.33 min 3,0675 ke |
| Burden test | N/Ac | 81.65 min 38,769 ke | 12.88 min 29,097 ke | 4.98 min 28,902 ke |
| VT | N/Ac | 46.95 min 19,853 ke | 25.23 min 3,0171 ke | 12.62 min 3,0278 ke |
| Case-Control - Empirical P Values | ||||
| CMC | N/Ac | >24 hr | 96.18 min 37,846Ke | 38.90 min 37,522Ke |
| Burden testf | 100.9 min 912Ke | >24 hr | 97.23 min 38,224Ke | 39.25 min 38,203Ke |
| VT | 62 min not reportede | failedd | 81.82 min 2,7043Ke | 34.72 min 26,852Ke |
A total of 2,000 samples were analyzed for both quantitative and binary traits. Using allele frequencies from ExAC non-Finnish European, 760,133 rare variants were generated (MAF < 0.05) in autosomal 18,295 genes. All splice-site, missense, and nonsense variants were analyzed. For all tests, variants with an MAF of < 0.01 in genes with ≥2 variant sites and ≥3 alternative alleles were analyzed (n = 17,322 genes) except for the VT where an MAF < 0.05 was used to select variants for analysis and 17,517 genes with ≥2 variant sites and ≥3 alternative alleles were analyzed. Abbreviation: k, thousand.
All software, PLINK/SEQ, VAT, and SEQSpark were run on a single server.
SEQSpark was also run on a cluster and is the only software that has this capability.
The program cannot perform this test/analysis.
Program unable to complete analysis.
Total number of permutations in the thousands (k) performed to obtain empirical p values.
For PLINK/SEQ there are two versions of the burden test (burden and burden 1), the analysis time and number of permutations shown here are for “burden” for VAT and SEQSpark the BRV test was performed. PLINK/SEQ can only perform the Burden Tests for case-control data obtaining empirical p values.
For the benchmarking of SEQSpark, PLINK/SEQ, and VAT, we used a server with 32 virtual cores, 64 gigabytes of memory, and 6 hard drives. Increasing the number of cores will not impact the speed of PLINK/SEQ since it uses only a single core, and it cannot make use of additional memory. Increasing the number of hard drives will impact the time that is needed to load the data and thus reduce computational time. For VAT, increasing the number of hard drives will also decrease the time which is needed to load the data and increasing the numbers of cores and memory will reduce the time needed to perform association testing. For SEQSpark, increasing the number of cores, memory, and hard drives will have a greater impact on the speed to load data and perform quality control, annotation, and association analysis compared to either PLINK/SEQ or VAT (data not shown). Using the available resources on the server, SEQSpark was faster for each benchmark than PLINK/SEQ or VAT. For example, to calculate allele frequencies SEQSpark was 48 times faster than PLINK/SEQ and 31 times faster than VAT (Table 4). For performing association analysis using SKAT for quantitative trait data, SEQSpark was 350 times faster then PLINK/SEQ and 182 times faster than VAT (Table 4). For both PLINK/SEQ and VAT, analysis is limited to a single server, but for SEQSpark analysis can be performed on a cluster. We selected a very small cluster of multiple servers to perform the benchmarks to demonstrate that even research groups with limited computational resources can use SEQSpark to increase computational speeds. It can be observed that even when a small cluster is used, there is a great reduction in the time required to perform the analysis compared to when SEQSpark is run on a server. For example, when SKAT-O was used to analyze quantitative trait data, analysis using SEQSpark on a cluster was 4.6 times faster than performing the analysis on the server. Naturally if a cluster is used with additional resources, e.g., cores, memory, hard drives, analysis can be performed much faster, making it possible to analyze large datasets of tens to hundreds of thousands of individuals in a short time span.
Through the analysis of the WHR UK10K WGS data and the analysis of the simulated exome data, we demonstrated the versatility of SEQSpark, but the data analysis and benchmarks do not demonstrate all capabilities of SEQSpark. To perform annotation, the program is distributed with many commonly used databases such as dbSNP, ExAC, gnomAD, and dbNSFP which provides functional evaluation using a variety of conservation and bioinformatics tools.15, 16, 17 SEQSpark also allows users to upload additional databases from either the public domain or those they created. Data quality control can be performed using all matrices which are annotated in the user’s variant call format (VCF) file or computed from the data. For all matrices summary statistics including HWE can be calculated on the entire dataset or data subsets and the user can specify which samples, genotype, and variant sites to remove from the analysis. Variants can be pruned to remove or greatly reduce inter-marker linkage disequilibrium to generate a set of variants which can be used for a variety of purposes including PCA. Duplicate samples and related individuals can be identified using the KING algorithm.18 Genomic sex can be used to verify that the correct sex was specified for each sample. PCA can be performed for data quality control to detect samples which are outliers due to problems with sequence data quality or membership in another population.
For association analyses, the user can select a subset of variants to analyze, e.g., based upon regions, frequency, or functional annotations. Often information on covariates is missing for a subset of individuals and instead of removing these samples from the analysis, the missing quantitative covariate can be replaced by the mean value for the sample. Outliers can be winsorized and for traits that violate normality, quantile normalization can be performed. For quantitative traits residuals are generated for analysis.
Statistical association testing is performed within the regression framework. This allows for easy control of potential confounders, testing for interactions and performing conditional analysis. Additionally, when performing association analysis, components from PCA19 can be included in the regression model to control for population substructure and admixture. Two versions of SKAT and SKAT-O can be used to perform the analysis either using “Liu modified method” or the “Davies method” to generate the cumulative distribution function of the null distribution. For single variant and aggregate rare variant association fixed effect tests, i.e., CMC, BRV, either the score or Wald tests can be used in a regression framework. Analysis can be performed by obtaining either analytical p values or empirical p values using adaptive permutation. Except for the CMC, a weighted rare variant association analysis can be performed using weights obtained from allele frequencies from the sample, e.g., controls3 or entire samples20 or allele frequencies obtained from external sources such as ExAC or gnomAD.16 Additionally, functional annotation can be used to weight variants, e.g., c-scores from CADD.21 For missing data, to avoid increased type I error rates for aggregate rare variant association tests, missing genotypes are replaced by dosages obtained from observed allele frequencies.12 In addition to analyzing genotypes, dosages22 from imputed data can also be analyzed either as single variants or in an aggregate rare variant association analysis. Meta-analysis can also be performed to combine results from different studies or populations.23
SEQSpark is ideal to use for the analysis of large-scale genetic epidemiological studies. It has higher computational efficiency for data quality control, annotation, and association analysis than other available software. The computational speed of SEQSpark is greater even when run on the same hardware as other programs. However, unlike other genetic association software programs which are limited to performing analysis on a single server, SEQSpark is scalable and can be run in a multiple-server environment, greatly increasing its computational speed and ability to handle large genetic datasets consisting of tens to hundreds of thousands of individuals. Due to its versatility and speed, SEQSpark will meet the demands of data analysis for emerging large-scale studies of imputed and massively parallel sequence data. The SEQSpark software package and documentation are publicly available online.
Acknowledgments
We would like to thank Katherine Montague for her thoughtful editing and Dr. Andrew DeWan for his suggestions which aided in improving the article. This work was supported by National Human Genome Research Institute grant HG008972.
Published: June 29, 2017
Web Resources
ALSPAC, http://www.bristol.ac.uk/alspac/
dbNSFP v.2.0, https://sites.google.com/site/jpopgen/dbNSFP
ExAC Browser, http://exac.broadinstitute.org/
gnomAD Browser, http://gnomad.broadinstitute.org/
Hadoop, http://hadoop.apache.org/
OMIM, http://www.omim.org/
PLINK/SEQ, https://atgu.mgh.harvard.edu/plinkseq/ /
Spark, http://spark.apache.org/
TopMed, https://www.nhlbi.nih.gov/research/resources/nhlbi-precision-medicine-initiative/topmed
UK10K Consortium, http://www.uk10k.org/
Variant Association Tools (VAT), http://varianttools.sourceforge.net/Association/HomePage
References
- 1.Li B., Leal S.M. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morris A.P., Zeggini E. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet. Epidemiol. 2010;34:188–193. doi: 10.1002/gepi.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Madsen B.E., Browning S.R. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Price A.L., Kryukov G.V., de Bakker P.I.W., Purcell S.M., Staples J., Wei L.-J., Sunyaev S.R. Pooled association tests for rare variants in exon-resequencing studies. Am. J. Hum. Genet. 2010;86:832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu M.C., Lee S., Cai T., Li Y., Boehnke M., Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schick U.M., Auer P.L., Bis J.C., Lin H., Wei P., Pankratz N., Lange L.A., Brody J., Stitziel N.O., Kim D.S., Cohorts for Heart and Aging Research in Genomic Epidemiology. National Heart, Lung, and Blood Institute GO Exome Sequencing Project Association of exome sequences with plasma C-reactive protein levels in >9000 participants. Hum. Mol. Genet. 2015;24:559–571. doi: 10.1093/hmg/ddu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Auer P.L., Reiner A.P., Wang G., Kang H.M., Abecasis G.R., Altshuler D., Bamshad M.J., Nickerson D.A., Tracy R.P., Rich S.S., Leal S.M., NHLBI GO Exome Sequencing Project Guidelines for large-scale sequence-based complex trait association studies: lessons learned from the NHLBI Exome Sequencing Project. Am. J. Hum. Genet. 2016;99:791–801. doi: 10.1016/j.ajhg.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuk O., Schaffner S.F., Samocha K., Do R., Hechter E., Kathiresan S., Daly M.J., Neale B.M., Sunyaev S.R., Lander E.S. Searching for missing heritability: designing rare variant association studies. Proc. Natl. Acad. Sci. USA. 2014;111:E455–E464. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trehearne A. Genetics, lifestyle and environment. UK Biobank is an open access resource following the lives of 500,000 participants to improve the health of future generations. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2016;59:361–367. doi: 10.1007/s00103-015-2297-0. [DOI] [PubMed] [Google Scholar]
- 10.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38 doi: 10.1093/nar/gkq603. e164–e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang G.T., Peng B., Leal S.M. Variant association tools for quality control and analysis of large-scale sequence and genotyping array data. Am. J. Hum. Genet. 2014;94:770–783. doi: 10.1016/j.ajhg.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Auer P.L., Wang G., Leal S.M. Testing for rare variant associations in the presence of missing data. Genet. Epidemiol. 2013;37:529–538. doi: 10.1002/gepi.21736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S., Wu M.C., Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics. 2012;13:762–775. doi: 10.1093/biostatistics/kxs014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walter K., Min J.L., Huang J., Crooks L., Memari Y., McCarthy S., Perry J.R., Xu C., Futema M., Lawson D., UK10K Consortium The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X., Wu C., Li C., Boerwinkle E. dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum. Mutat. 2016;37:235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.-M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 20.Lin D.-Y., Tang Z.-Z. A general framework for detecting disease associations with rare variants in sequencing studies. Am. J. Hum. Genet. 2011;89:354–367. doi: 10.1016/j.ajhg.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y., Willer C., Sanna S., Abecasis G. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu D.J., Peloso G.M., Zhan X., Holmen O.L., Zawistowski M., Feng S., Nikpay M., Auer P.L., Goel A., Zhang H. Meta-analysis of gene-level tests for rare variant association. Nat. Genet. 2014;46:200–204. doi: 10.1038/ng.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]