Abstract
Application of the experimental design of genome-wide association studies (GWASs) is now 10 years old (young), and here we review the remarkable range of discoveries it has facilitated in population and complex-trait genetics, the biology of diseases, and translation toward new therapeutics. We predict the likely discoveries in the next 10 years, when GWASs will be based on millions of samples with array data imputed to a large fully sequenced reference panel and on hundreds of thousands of samples with whole-genome sequencing data.
Keywords: genome-wide association study, SNP, obesity, auto-immune disease, schizophrenia, heritability
Main Text
Introduction
Here, we review the remarkable range of discoveries that genome-wide association studies (GWASs) have facilitated in population and complex-trait genetics, the biology of diseases, and translation toward new therapeutics. In the introductory sections, we provide a background for this review, summarize its scope and layout, and revisit the scientific rationale for GWASs. We then review general conclusions that can be drawn from GWAS discoveries across a wide range of traits. We subsequently highlight more specific results of discoveries and methods on the path from GWAS to biology and review progress in three exemplar diseases, namely type 2 diabetes (T2D [MIM: 125853]), auto-immune diseases (MIM: 109100), and schizophrenia (MIM: 181500). We end the review with a number of sections on the limitations of current experimental designs and possible ways to overcome these and a prediction on the future of GWASs for human traits.
Background
Five years ago, a number of us reviewed (and gave our opinion on) the first 5 years of discoveries that came from the experimental design of the GWAS.1 That review sought to set the record straight on the discoveries made by GWASs because at that time, there was still a level of misunderstanding and distrust about the purpose of and discoveries made by GWASs. There is now much more acceptance of the experimental design because the empirical results have been robust and overwhelming, as reviewed here.
Scope and Framework
Data generated from genome-wide SNP surveys have been exploited for addressing many scientific questions other than SNP-trait associations. We do not have the space to give adequate coverage of discoveries in evolutionary and population genetics, nor can we fully cover the many developments in analytic methods, although we will briefly mention some recent developments. The scope of our review is novel discoveries on the genetics and resulting biology of common adult diseases (auto-immune, metabolic, and psychiatric disease in particular) and their risk factors and the wider implications of those discoveries. GWAS discoveries have and are affecting a wide variety of diseases and traits, many of which have been covered in other in-depth reviews. Our focus is on associations between complex traits and SNPs, but we note that there have been many reported associations between traits and copy-number variants (CNVs) and that there are known mechanisms by which CNVs can be associated with disease.2 Results from other genome-wide surveys, including exome and whole-genome sequencing (WGS) studies, are not reviewed here.
GWAS Rationale and Scientific Basis
The GWAS is an experimental design used to detect associations between genetic variants and traits in samples from populations. The primary goal of these studies is to better understand the biology of disease, under the assumption that a better understanding will lead to prevention or better treatment. The path from GWAS to biology is not straightforward because an association between a genetic variant at a genomic locus and a trait is not directly informative with respect to the target gene or the mechanism whereby the variant is associated with phenotypic differences. However, as reviewed herein, new types of data, new molecular technologies, and new analytical methods have provided opportunities to bridge the knowledge gap from sequence to consequence. GWASs have also been successfully implemented for better defining the relative role of genes and the environment in disease risk, assisting in risk prediction (enabling preventative and personalized medicine), and investigating natural selection and population differences (Table 1).
Table 1.
The Role of GWAS SNP Arrays in Human Genetic Discoveries
| Analysis | Purpose | Discoveries |
|---|---|---|
| GWAS | detecting trait-SNP associations | ∼10,000 robust associations with diseases and disorders, quantitative traits, and genomic traits |
| Genome-wide CNV analysis | detecting trait-CNV associations | hundreds of associations with diseases and disorders |
| Genome-wide assessment of LD | quantifying genome architecture | large variation in LD in the genome |
| Estimation of SNP heritabilitya | genetic architecture | large proportion of genetic variation captured by common SNPs |
| Estimation of genetic correlationa | detecting and quantifying pleiotropy | pleiotropy is ubiquitous |
| Polygenic risk scoresa | detecting pleiotropy; validating GWAS discoveries | out-of-sample prediction works as expected; detection of novel trait associations |
| Mendelian randomizationa | testing causal relationships | replication of known causal relationships; empirical evidence of observational associations that are not causal |
| Population differences in allele frequencies | reconstructing human population history; detecting selection | genetic structure can mimic geographical structure; evidence of natural selection |
| Trait GWAS with -omics GWASa | fine-mapping; detecting target genes; function | two-thirds of GWAS-associated loci implicate a gene that is not the nearest gene to the most associated SNP |
These analyses can be performed with GWAS summary statistics.
GWASs to date rely on and exploit linkage disequilibrium (LD), the correlation structure that exists among DNA variants in the current human genome as a result of historical evolutionary forces, particularly finite population size, mutation, recombination rate, and natural selection. The statistical power to detect associations between DNA variants and a trait depends on the experimental sample size, the distribution of effect sizes of (unknown) causal genetic variants that are segregating in the population, the frequency of those variants, and the LD between observed genotyped DNA variants and the unknown causal variants. Therefore, the potential of a GWAS to succeed for a particular trait or disease depends on (1) how many loci affecting the trait segregate in the population, (2) the joint distribution of effect size and allele frequency at those loci (sometimes called genetic architecture), (3) the experimental sample size, (4) the panel of genome-wide variants that are used in the GWAS, and (5) how heterogeneous the trait or disease being studied is. The last relates to both the biology of the trait and the ability to diagnose or measure it with precision.
If the genetic architecture of a particular trait or disease were known, the optimum experiments could be designed to detect specific variants. However, despite many theoretical studies on the likely relationship between allele frequency and trait loci, until the onset of GWASs, there was very little empirical data to validate prediction from theoretical models.
GWASs have been facilitated by the development of relatively inexpensive SNP arrays. Commonly used SNP arrays vary in their content, but they broadly contain 200,000 to more than 2,000,000 SNPs. To date, most genetic variants that have been surveyed through GWASs are common in the population, in that they have a minor allele frequency (MAF) typically larger than 1%. For the purpose of this review, we arbitrarily define common variants to have MAF ≥ 1% and rare variants to have MAF < 1%. The GWAS as an experimental design is more than just an array-based study of common variants. For example, association studies using WGS data are also GWASs. There is a continuum from GWASs based on SNP arrays to those using WGS, and the only difference (apart from cost) is the density of coverage of variation in the genome and the MAF spectrum of the variants.
LD between genetic variants is commonly measured as a squared correlation (r2) because this measure is linear in the sample size required for detecting association between an observed genotyped and an unobserved causal variant. LD r2 can be large only if the allele frequencies at the two loci match,3, 4 and this is the reason why GWASs from common SNP arrays are not powerful enough to detect associations due to rare causal variants (in addition to sample-size considerations; see below). Statistical imputation5, 6, 7 of unobserved variants can recover some of the information lost because of imperfect LD between observed genotypes and unobserved causal variants. Imputation is enabled by the fact that the genotypes of unobserved genetic variants can be predicted by the haplotypes inferred from multiple observed SNPs and the haplotypes observed from a fully sequenced reference panel.
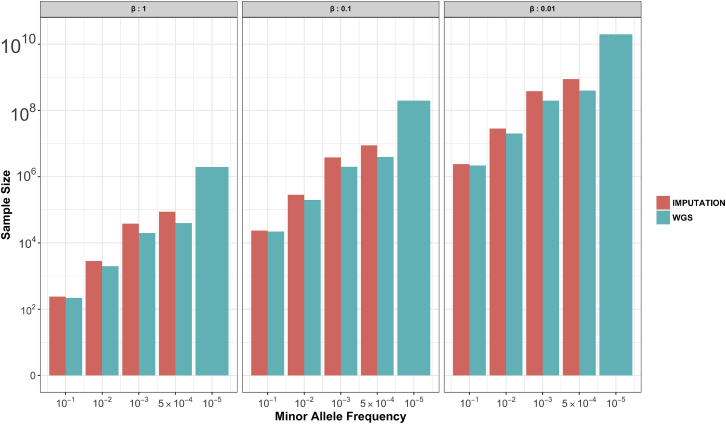
In Figure 1, we summarize power calculations (see Appendix A for theory) of the minimum sample size required for detecting an association as a function of genotype method (SNP array plus imputation or WGS), allele frequency, and effect size. Given that statistical imputation of variants as infrequent as 1/1,000 is still reasonably accurate,8 not much power of detection can be gained from WGS. For ultra-rare variants, for example, those with a frequency of 1/100,000, WGS can identify associations but only when the effect sizes of the polymorphisms (mutations) are very large. For example, for such rare variants with an effect size of 1 phenotypic standard deviation unit (about 7 cm for height or 5 BMI units), a sample size of more than one million is required (i.e., an allelic count of ten). For case-control studies of disease, the effects sizes of β = 0.01, 0.1, and 1 phenotypic standard deviation in Figure 1 correspond approximately to odds ratios of 1.02, 1.2, and 4, respectively, if we assume that both allele frequency and population prevalence are 0.01 or lower.9 Segregation of rare variants with very large effects might be observable in certain families, and then a family-based experimental design would be more efficient at locating and identifying such (near) monogenic traits. In addition, other genome-wide scans, such as WES and WGS studies, allow testing for a burden of rare variants across shared functional units (e.g., genes) in a way that is not accessible to GWASs.
Figure 1.
Minimum Sample Sizes for Detecting Trait-SNP Associations from Imputed and WGS Data
Required sample sizes for detecting association were calculated with (Equation A1), (Equation A2), (Equation A3), (Equation A4), (Equation A5) under the assumption of a type I error rate of 5 × 10−8, 80% power, and Hardy-Weinberg equilibrium. Effect sizes (β) are in phenotypic standard deviation units. For genotyped SNPs imputed to a fully sequenced reference, we have used the average imputation Rimp2 values reported by the Haplotype Reference Consortium8 in their Figure S3. This is a conservative estimate of imputation accuracy because it is based on a less dense genotyping array. For the WGS data, we have assumed no sequencing errors. Note that for some combinations of allele frequency and effect size, the required minimum number of individuals for detecting association exceeds 100 million.
Results in Figure 1 are based on unselected population samples or population-based case-control samples and the detection of association between the trait or disease and the same genetic variant. Power will be increased for highly ascertained cases and enrichment of extreme cases or family-based studies with multiple cases of a rare disease. Furthermore, using WGS data for association analysis of rare variants has the potential to boost power through the combination of alleles of similar impact (e.g., via burden tests across a gene) under the assumption of multiple independent causative variants in a gene region. This strategy is justified from knowledge of monogenic disorders, where it is typical for different variants of the same gene to segregate with disease in different families. However, such tests also have challenges because prior knowledge about function or frequency is required for determining which alleles in a gene should be included in the burden count.
General Results
Complex Traits Are Highly Polygenic
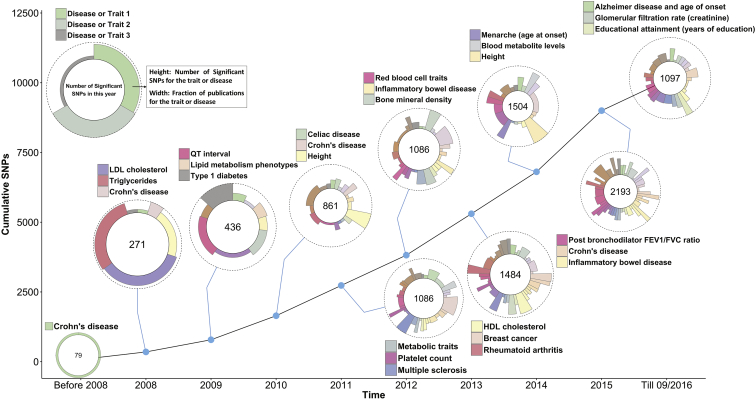
GWAS results have now been reported for hundreds of complex traits across a wide range of domains, including common diseases, quantitative traits that are risk factors for disease, brain imaging phenotypes, genomic measures such as gene expression and DNA methylation, and social and behavioral traits such as subjective well-being and educational attainment. About 10,000 strong associations have been reported between genetic variants and one or more complex traits,10 where “strong” is defined as statistically significant at the genome-wide p value threshold of 5 × 10−8, excluding other genome-wide-significant SNPs in LD (r2 > 0.5) with the strongest association (Figure 2). GWAS associations have proven highly replicable, both within and between populations,11, 12 under the assumption of adequate sample sizes.
Figure 2.
GWAS SNP-Trait Discovery Timeline
Data used for generating the graph were taken from the GWAS Catalogue.10 SNPs and traits were selected according to the following filters. SNPs were selected with a p value < 5 × 10−8. For each trait with two or more selected SNPs, SNPs were removed if they had an LD r2 > 0.5 (calculated from 1000 Genomes phase 3 data) with another selected SNPs and their p value was larger. For each year of discovery, only the top three traits and diseases with the largest number of SNPs are labeled in the circle.
One unambiguous conclusion from GWASs is that for almost any complex trait that has been studied, many loci contribute to standing genetic variation. In other words, for most traits and diseases studied, the mutational target in the genome appears large so that polymorphisms in many genes contribute to genetic variation in the population. This means that, on average, the proportion of variance explained at the individual variants is small. Conversely, as predicted previously,1, 13 this observation implies that larger experimental sample sizes will lead to new discoveries, and that is exactly what has occurred over the last decade. For example, in 2009 the first genomic locus robustly associated with liability to schizophrenia was discovered with a sample of 3,000 cases;14 by 2014, this had increased to 108 with a sample size of 35,000 cases.15 Similarly, when the concept of “missing heritability”16 was introduced, it highlighted that in 2008, only 40 genome-wide-significant SNPs had been identified for height, and together these explained about 5% of heritability.17 In 2014, the number of associated SNPs had increased to ∼700, explaining more than 20% of heritability,18 and from the relationship between sample size and discoveries in the last 10 years, it is reasonable to predict that in the next few years, this will increase to thousands of variants, which will cumulatively explain a substantial proportion (e.g., more than one-third) of heritability.
The term polygenic describes the genetic architecture underpinning variation in a trait between individuals in a population, but what does it mean for each individual? It means that each individual will carry a number of alleles that increase (+) and a number of alleles that decrease (−) the trait or disease risk. There are so many possible combinations of these sets of alleles that each individual is likely to have a unique combination, and in studies designed to detect associated loci, the effect size of each allele is measured across the context of an averaged background, and the effect size of each locus is found to be small.
Pleiotropy Is Pervasive
The number of segregating variants in the human population is large but finite. It is not known what proportion of the segregating variants are associated with complex-trait genetic variation, but the fact that each of the many studied traits is associated with variants at hundreds to thousands of loci in the genome strongly suggests that some of the underlying causal variants are the same. Multiple lines of evidence are consistent with widespread pleiotropy for complex traits. First, Mendelian mutations that cause specific syndromes or diseases are frequently associated with multiple phenotypes in an affected individual. Second, pedigree studies have reported genetic correlations between traits, implying that a number of the same variants affect two or more traits in a consistent direction.19 Third, GWASs have shown that the same genetic variants can be significantly associated with multiple diseases and traits when the phenotypes are measured on different individuals (so that no environmental associations are driving the results).20, 21, 22 In the case of auto-immune diseases, evidence implies that at some loci, the same causal variants are driving the observations of associations across diseases.23, 24, 25 Fourth, analytical methods that estimate genetic correlations from GWAS data have provided evidence for widespread pleiotropy.20, 26
The corollary of pervasive pleiotropy for complex traits is that the paradigm of “one gene, one function, one trait” is the wrong way to view genetic variation in the human genome (and the same applies for studying disease in experimental organisms).27 It also implies that studying traits or disease in isolation with respect to past or present natural selection might lead to the wrong inference. The true nature of the pleiotropy is currently unknown but, in some cases, could imply an impact of the variants on different tissues and/or at different ages.
New Analysis Methodology Underpinning New Discovery
GWAS data have led to new analysis methods that fall into a number of categories depending on their purpose: (1) methods of better modeling population structure and relatedness between individuals in a sample during association analyses,28, 29, 30, 31, 32, 33, 34 (2) methods of detecting novel variants and gene loci on the basis of GWAS summary statistics,35, 36, 37 (3) methods of estimating and partitioning genetic (co)variance,38, 39 and (4) methods of inferring causality.40, 41, 42 In addition, GWAS discoveries and interpretation have benefited substantially from improved algorithms in statistical imputation of unobserved genotypes and statistical imputation of human leukocyte antigen (HLA) genes and amino acid polymorphisms.43, 44, 45, 46
Common Variants Together Tag a Substantial Proportion of Additive Genetic Variance
In addition to enabling the discovery of specific trait-locus associations, GWASs have facilitated estimation of how much of the total additive genetic variation due to segregating variants in the population is tagged by genotyped and imputed SNPs. This quantification of “SNP heritability” is informative with respect to the unknown genetic architecture of the trait. SNP heritability has provided objective guidance to inform decisions about which experimental designs are most efficient at detecting novel trait-locus associations on the basis of empirical data, i.e., increasing sample size of GWASs. Classical estimation of total narrow-sense heritability (estimated from phenotypic records of samples that include family members) captures the total amount of additive genetic variance in the population irrespectively of the joint distribution of allele frequency and effect size47 (we acknowledge a potential for bias by common environmental effects and non-additive genetic variation). In contrast, SNP heritability (estimated from tiny genetic relationships from unrelated individuals) captures only the proportion of additive genetic variance due to LD between the assayed and imputed SNPs and the unknown causal variants. Estimation and partitioning of additive genetic variation for quantitative traits and liability to disease have implied that one-third to two-thirds of genetic variation at causal variants can be tagged by common genotyped and imputed SNPs through LD.1, 48 At present, it is not known how much of the total additive genetic variation is due to causal variants with frequencies less than 1%. Evidence from imputed genotype data for height implies that more additive genetic variation is explained by variants with MAF < 10% than expected under an evolutionary neutral model, consistent with purifying selection of the height-associated loci.49 In the near future, when additive genetic variance will be estimated from WGS data in large samples, the contribution of observed rare and low-frequency variants will be estimated explicitly. Estimates from data available to date provide the first evidence for different genetic architectures between diseases,50 for example, there is more signal from rare variants for amyotrophic lateral sclerosis (motor neuron disease [MIM: 105400]) than for schizophrenia51 and more predicted loci for schizophrenia than for immune disorders52 and hypertension.53
Theoretical and empirical observations suggest a place for non-additive genetic variation, and there have been many largely unsuccessful attempts to detect epistasis with GWAS data. There are a number of likely explanations. First, there is limited evidence that non-additive genetic variation makes up a large fraction of the total genetic variation, so detection requires larger sample sizes than those necessary for main effects. Second, the loss of information due to imperfect LD between genotyped SNPs and causal variants is larger for interactions than for main effects. For example, loss of information for additive effects is proportional to the LD r2, whereas information loss for dominance and additive-additive interaction effects is proportional to r4. The first observation also applies to interactions between genes and environmental factors. One replicable example of epistatic interaction is the ERAP1-HLA interaction for psoriasis (MIM: 177900) and ankylosing spondylitis (MIM: 106300).54
The Utility of GWAS-Derived Genetic Predictors
In 2007, it was shown that one could use GWAS data from human studies to create genetic predictors for disease and other complex traits by estimating the effect size at multiple loci in a discovery sample and using those estimated SNP effects in independent samples13, 55 to generate a polygenic risk score (PRS) per individual. A thorough review of different methods of generating PRSs is outside the scope of this review, but currently the key driving force influencing prediction accuracy is the size of the discovery sample used for estimating the effects of individual variants. PRSs have been applied extensively over the last 5 years, not in a clinical setting for the prediction of a healthy individual’s risk of disease but in applications that facilitate new experimental designs and discoveries. Polygenic predictions are not particularly informative for an individual, but they do explain a sufficient proportion of variation (between 1% and 15% at present for highly polygenic traits without a major gene) to separate groups, for example, samples with the highest and lowest risk. They are also useful for detecting new trait associations by correlating observed phenotypes in a sample or cohort with the genetic prediction of another trait. This design is powerful, because if the discovery sample is fully independent of the new sample, an observed association between a complex trait and a genetic predictor from the discovery sample must be due to genetic factors, given that there are no shared environmental factors. The paradigm of PRSs can also be applied to the prediction of molecular phenotypes such as gene expression, even when they are not observed,56 for mining the human “phenome” for association with predictors derived from diseases and other traits57 or investigating genotype (proxied by PRS) by environment interaction.58
The Public Availability of Data Has Enabled Novel Research and Discoveries
Sharing of genetic data in the gene-mapping community has been a major enabling factor in gene-mapping success. At this point, the vast majority of the available data are from studies of populations of European descent, and it is hoped that data from other ethnic groups will be deposited more extensively in years to come.
The availability of GWAS summary statistics (the effect sizes and their standard errors or p values on millions of SNPs) in the public domain has increased dramatically in the last 5 years, and in 2017 hundreds of such datasets are publicly available.59 There are a number of reasons for this. Previous concerns about potential individual identification from GWAS summary data have proven to be unfounded, either because the sample size from GWAS summary statistics is typically very large or because a simple step such as providing average allele frequencies from a reference sample negates potential identification. The entire genomics field benefits from wide availability of genetic data. When a GWAS is published, full genome-wide summary statistics (at the very least) should be available for uncontrolled download. Funding bodies and journals could play a stronger role in enforcing such a requirement. The availability of summary statistics in the public domain has enabled discoveries of novel associations,37, 60, 61, 62 estimation of SNP-based heritability,63 quantification of pleiotropy across many traits,20, 21, 38 and creation of more accurate prediction scores, as well as follow-up with computational tools, functional assays, and model systems for the identification of candidate genes.
For the near future, the UK Biobank is pushing the barriers further by releasing both genome-wide genotypes and rich phenotypic data on 500,000 people to the international research community.
From GWAS to Biology
By design, associations detected by GWASs do not yield a particular gene target or mechanism. This is in contrast to the detection of Mendelian coding mutations in family studies, where the variant, target gene, and mechanism (change in protein) are identified simultaneously. Moreover, the sheer number of associated variants means that the battery of follow-up functional studies traditionally applied to new discoveries from Mendelian disease is not appropriate or achievable for discoveries of genes associated with complex traits. It should be noted that although the effect sizes of individual genetic variants are small in populations, their effect sizes on molecular phenotypes can be large, and the drug effects of gene targets can also be magnified (e.g., statins). Notably, the last 5 years have witnessed some clever laboratory experiments that have followed up on GWAS association, and these have led to the discovery of the target gene, for example, the targets of the associations between FTO (MIM: 610966) and obesity (MIM: 601665)64 and between the major histocompatibility complex (MHC) and schizophrenia.65 Performing similar or new laboratory experiments for many loci could be possible but would be time consuming and expensive.
Until recently, efforts to understand the biological mechanisms through which these various risk variants act have been thwarted by limitations in the capacity to perform large-scale evaluation of functional impact.66 The advent of sequence-based -omic analyses have been transformative by allowing functional analyses of risk variants to be pursued on the same genome scale (which has fueled their discovery) and allowing mechanistic inferences to be based on the behavior of the full set of risk loci for a given trait.67 The maps of regulatory annotations and connections in disease-relevant tissues, generated by projects such as ENCODE,68 Epigenome RoadMap,69 and GTEx,70 have been crucial to interpretation of the non-coding variants that account for the majority of GWAS-identified risk alleles. Tissue-specific resources could become increasingly important, and for neuro-psychiatric disorders in particular, appropriate human brain resources are essential. New initiatives such as CommonMind and PsychENCODE are providing data and tools for the neuro-psychiatry research community to follow up on GWAS signals. New analytical methods now provide the first steps of functional in silico follow-up by exploiting the availability of resource datasets detailing gene expression, epigenetic marks, 3D chromatin contacts,71 or other genomic annotations, including drug targets. One fertile area of method development is integrating data from GWASs and expression quantitative trait locus (eQTL) studies to identify associations between transcripts and complex traits.56, 61, 62 These methods are useful for prioritizing genes from known GWAS loci for functional follow-up, detecting novel gene-trait associations, and inferring the directions of associations.21, 27, 62 The analytical results that only about one-third of the associated genes are the nearest genes61, 62 are informative for the design of fine-mapping experiments.
One of the ultimate objectives of genetic research is to drive translational advances that enable more effective prevention and/or treatment of disease. Despite the inevitable time lag between basic research discoveries and clinical implementation, a growing number of examples highlight the diverse routes by which human genetics can inform translational medicine.
Three Exemplars of GWAS Success
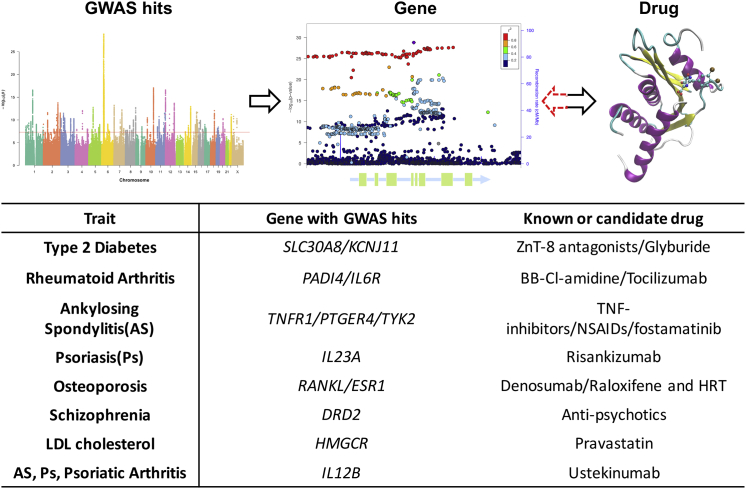
Here, we focus on three examples of adult-onset disease to demonstrate some of the significant advances that have followed as a direct result of GWASs. Figure 3 illustrates examples of an overlap between GWAS signals that are known drug targets. In general, drug targets that are genetically informed have a higher probability of making it to phase III trial or to market, implying potential huge cost savings to the pharmaceutical industry.72
Figure 3.
Examples of Links between GWAS Discoveries and Drugs
Type 2 Diabetes
Variant and Gene Discovery. Scores of genes have been causally implicated in monogenic forms of diabetes (e.g., neonatal diabetes mellitus [MIM: 601410]73), but GWASs have now identified over 100 common variant signals.74, 75, 76 Recent efforts to extend GWASs beyond array-based genotyping and to access a broader range of variants through sequencing (particularly those of lower frequency) have revealed that most genetic variation influencing T2D appears to reside at common variant sites.74, 77 This chimes with the view of T2D as a largely post-reproductive trait and is consistent with a failure to detect compelling empirical evidence that T2D risk alleles have been subject to marked purifying selection.78, 79 In keeping with the age of these common risk alleles (which predates the diaspora of modern humans out of Africa), most common variant associations for T2D are replicated across major ethnic groups.75, 80 However, as increasingly diverse populations are genotyped and sequenced, more ethnic-specific alleles are being identified. Several of these alleles have a relatively large phenotypic impact and have risen to high frequency in specific populations, including variants in PAX4 (MIM: 167413) in East Asians74 and TBC1D4 (MIM: 612465) in Inuit.81 Efforts to identify compelling evidence for gene-gene and gene-environment interactions have been largely unsuccessful.82
From GWAS to Biology. Regulatory information on the key tissues of insulin action (fat, muscle, and liver)82, 83 and equivalent data from pancreatic islet material67, 84 have provided compelling evidence that the variants most strongly associated with T2D (as well as fasting glucose and other related quantitative traits) are preferentially located at active enhancers (particularly stretch enhancers) in pancreatic islets67, 84 and, to a lesser extent, at enhancers active in fat, muscle, and liver.83, 85 Increasing refinement of regulatory annotation has brought more precise localization of these global regulatory effects, for example, emphasizing specific transcription factor genes (such as FOXA2 [MIM: 600288]).86 These patterns of tissue-specific genomic enrichment tie in with studies of the physiological correlates of T2D risk alleles, as observed in physiological data from non-diabetic subjects; these have indicated that, whereas some T2D risk alleles have a primary effect on insulin action, most appear to be associated with reduced insulin secretion.87 These approaches have generated some notable advances, for example, cis-expression mapping has highlighted KLF14 (MIM: 609393) as the mediator of a chromosome 7 T2D signal that is associated with insulin resistance and hyperlipidemia (appropriately, this expression signal is specific to adipose tissue).85 Equivalent data from human islets have characterized the likely effector transcripts at several T2D GWAS loci (such as ZMIZ1 [MIM: 607159], MTNR1B [MIM: 600804], and ADCY5 [MIM: 600293]), where the major impact is to reduce insulin secretion.86, 88 Additional clues to the identification of the causal transcripts at certain GWAS loci have come from examining the credentials of the regional transcripts themselves, assigning candidacy on the basis of known biology (e.g., NOTCH2 [MIM: 600275] and GIPR [MIM: 137241]),89 involvement in related monogenic conditions (WFS1 [MIM: 606201], HNF1 [MIM: 142410], and HNF4A [MIM: 600281]),90, 91 or data from animal models (CDKAL1 [MIM: 611259]).92 Finally, the accumulation of data on coding variants (via exome sequencing and/or exome array genotyping) has highlighted several instances where GWAS signals previously attributed to non-coding variants can be reassigned to causal coding variants (e.g., TM6SF2 [MIM: 606563] 74). For others, such as RREB1 (MIM: 602209), identification of T2D-associated coding variants, statistically independent of the original GWAS signal, flags the likely effector transcripts.74 All in all, it is possible to point to a compelling effector transcript at around one-third of the 100 T2D loci identified by GWASs. These genes represent legitimate targets for detailed empirical validation and mechanistic exploration. They also support efforts, via network-based approaches, to establish the extent to which the biology of T2D predisposition converges onto a restricted set of pathways.
Translation. Examples from T2D research highlight the diverse routes by which human genetics can inform translational medicine: (1) the combination of common-variant GWASs and candidate-gene resequencing has demonstrated that loss-of-function mutations in SLC30A8 (MIM: 611145; encoding a zinc transporter expressed in pancreatic islets) are protective for T2D, leading to efforts by several pharma companies to develop ZnT-8 antagonists;93 (2) the use of genetic variants as instruments that “simulate” variation in environmental and biochemical exposures has clarified the extent to which vitamin D intake, early nutrition, circulating lipid levels, and chronic inflammation play causal roles with respect to the development of T2D94, 95, 96, 97, 98 and has defined the relationship between insulin resistance and the distribution of adipose tissue;99 (3) the identification of genetic variants associated with individual variation in response to commonly used therapeutic agents has refined our understanding of the mechanisms through which those agents operate100,101 and, in some instances, has led to therapeutic optimization on the basis of genetic and/or clinical phenotype;102 and (4) the combination of -omic measurements, longitudinal clinical phenotypes, and GWAS data has highlighted sets of molecules (e.g., branched-chain amino acids) that not only are prospectively associated with T2D progression but could also play a causal role in T2D development and thereby provide valuable clinical tools for stratification and prognostication.103, 104
Auto-immune Diseases
Variant and Gene Discovery. In the last 5 years, GWASs have been undertaken for nearly all major immune-mediated diseases (with sample sizes of tens of thousands of case and control individuals for more common immune-mediated diseases studied either by GWASs or by more targeted chips, such as Illumina’s Immunochip105), resulting in hundreds of associated loci. The development of statistical approaches for cross-disease studies to identify pleiotropic loci has been particularly productive in identifying new genes and in better understanding the pathogenic relatedness of immune-mediated diseases. A recent cross-disease study involving the conditions ankylosing spondylitis (AS [MIM: 106300]), inflammatory bowel disease (IBD [MIM: 266600]), primary sclerosing cholangitis (MIM: 613806), and psoriasis identified without any further genotyping 30 new loci at genome-wide significance.24 Trans-ethnic studies have demonstrated substantial genetic overlap between ethnically remote populations;106, 107, 108 for example, genetic correlations of 0.76 and 0.79 between European and East Asians have been estimated for Crohn disease and ulcerative colitis (UC).106 Trans-ethnic comparisons of associations at shared loci have been quite helpful in pinpointing causal variants; for example, population-specific variation in HLA-DRB1 (MIM: 142857) associations in rheumatoid arthritis (RA [MIM: 180300]) has helped to define the key amino acids underpinning that association.
From GWAS to Biology. GWAS results have made key contributions to deeper biological understanding of immune-mediated diseases in the last 5 years. For example, in a cross-disease study, new loci included genes that for the first time implicate pathogenesis associated with methylation variation (DNMT3A [MIM: 602769] and DNMT3B [MIM: 602900]), bacteria-sensing genes (TLR4 [MIM: 603030]), genes influencing the host microbiome (FUT2 [MIM: 182100]), and NFKB pathway genes (NFKB1 [MIM: 164011], NFKBIA [MIM: 164008], TNFAIP3 [MIM: 191163]).24 Evidence of extensive pleiotropy includes variants that have different directions of association in different diseases or disease-specific variants at the same loci. This has relevance for the likely impact of targeting these loci therapeutically. For example, SNP rs1800693 in the major TNF-receptor gene TNFR1 (MIM: 191190) is associated in different directions with multiple sclerosis (MS [MIM: 126200]) and AS.109 The SNP leads to loss of the transmembrane domain of the receptor, and the risk SNP for MS (protective for AS) leads to increased serum soluble TNF receptor.110 TNF inhibition, including by decoy TNF receptor therapies, is highly effective for AS and many other immune-mediated diseases, but its use can be complicated by de novo development of MS, and in MS itself, it can exacerbate disease. Although this is a retrospective example, it demonstrates the potential of using genetics to predict toxicities. There are several agents in development where the genetics would point to the likelihood of toxicities. Examples include CD40 and its ligand, where SNP rs1883832 in the C allele of CD40 (MIM: 109535) is a risk factor for RA and auto-immune thyroid disorder (AITD) but is protective against MS and IBD, and the PTPN22 (MIM: 600716) variant c.1858C>T (p.Arg620Trp), which increases the risk of type 1 diabetes (MIM: 222100), systemic lupus erythematosus (MIM: 152700), vitiligo (MIM: 606579), AITD, and UC but is protective against Crohn disease.23 At the very least, this suggests that any clinical trials in these conditions should carefully screen for the development of the diseases with the converse genetic associations.
The MHC, as well as the HLA genes encoded within it, is the major locus for the majority of immune-mediated diseases. Although the major highly penetrant HLA types involved in different diseases have long been established, in the last 5 years, the ability to impute the composite amino acids and then test these for disease association has enabled research that has better defined the key components of the HLA proteins involved in disease. In RA, it had been known for roughly 30 years that a sequence of amino acids at positions 70–74 of HLA-DRB1 largely, though not fully, determine the differential association between HLA-DRB1 types and disease.111 Through the use of amino acid imputation and association studies, this “shared epitope” sequence was extended,112 and this information used to provide a molecular explanation for the propensity of peptides with citrullinated component amino acids to induce disease.113 HLA variants have long been known to be major determinants of severe immunologically mediated adverse drug reactions. For example, toxicity to the anti-retroviral abacavir is largely restricted to HLA-B57 carriers. With the use of GWAS and HLA imputation, an HLA-DQA1∗0201-HLA-DRB1∗0701 haplotype has been shown to be strongly associated with the risk of thiopurine-induced pancreatitis, such that homozygotes for this haplotype have a 17% risk of this major side effect.114 It is likely that with the increasing use of genetic profiling in clinical practice, further examples will be identified in coming years.
Translation. GWAS results have already proven highly successful at initiating medication repositioning. For example, GWAS discoveries triggered the repositioning of biological medications targeting components of the IL-23 pathway (including IL-12p40, IL-17, and IL-23p19), and now these are mainstay treatments for psoriasis and psoriatic arthritis (MIM: 607507), are highly effective in AS, and (with the exception of IL-17 blockade) are effective in IBD, as suggested by early studies.115, 116, 117 The annual sales of these medications alone are likely to be greater than the total amount spent on GWASs in the past decade.
Many other GWAS discoveries have stimulated targeted therapy-development programs, a few of which are described here. The discovery of the association between PADI4 (MIM: 605347) and RA provided conclusive evidence that immunological reactions to epitopes that had been citrullinated by PAD enzymes were causatively involved in RA. This led to programs developing PAD inhibitors in RA, and these have shown significant promise.118, 119 Major drug-development programs have been initiated to target the M1 aminopepidase genes ERAP1 (MIM: 606832) and ERAP2 (MIM: 609497) because of their genetic associations with AS, psoriasis, IBD, Behcet disease (MIM: 109650), and the rare condition Birdshot retinopathy (MIM: 605808).120
Bioinformatic follow-up of GWAS results has also been fruitful. For example, Okada et al. screened the overlap between genetic associations and known drug targets to demonstrate that existing RA therapies disproportionately target RA-associated gene products and their interacting protein partners.108 From this, they extrapolated that other agents with high levels of effects on these proteins would be enriched with potential new RA therapies and provided suggestive evidence that CDK4 and CDK6 inhibitors already in use, particularly in oncology, could be effective in RA. These agents have been shown to be effective in the collagen-induced arthritis model of RA and are now in human trials in RA in Japan.
Schizophrenia
Variant and Gene Discovery. Although psychiatric diseases had a slow start in GWAS locus identification, more than 50,000 samples have been genotyped in the last 5 years; the typical linear relationship between sample size and number of loci has been observed, and more than 100 risk loci have been discovered to date. These risk loci are enriched in genes containing de novo mutations in schizophrenia, autism (MIM: 209850), and intellectual disability,15 and several identified loci contain genes relevant to major hypotheses of schizophrenia etiology, including DRD2 (MIM: 126450; the target of anti-psychotic drugs) and genes involved in glutamatergic neurotransmission (GRM3 [MIM: 601115], GRIN2A [MIM: 138253], and GRIA1 [MIM: 138248]), as well as genes that extend previous observations of association with voltage-gated calcium channel subunits (CACNA1C [MIM: 114205], CACNB2 [MIM: 600003], and CACNA1I [MIM: 608230]).15 One of the most striking findings that emerged early in schizophrenia studies—at the stage where there were only a handful of genome-wide-significant loci—was the highly polygenic nature of the common variants contributing to risk.14 This observation has been widely replicated, and estimates are that 71% of 1 Mb genomic regions have at least one variant influencing schizophrenia risk,53 and there is evidence of substantial pleiotropy with other psychiatric disorders.26 However, genetic architecture, described as the mix of rare and common variants, is likely to differ between psychiatric disorders, as is already being observed for the higher rates of rare, de novo penetrant CNVs and single-nucleotide variants found in autism than in schizophrenia or bipolar disorder.121, 122, 123, 124, 125, 126, 127, 128, 129 PRS studies are being utilized extensively to investigate disease heterogeneity and contributions from environmental risk factors.
From GWAS to Biology. Functional follow-up is necessarily more difficult for psychiatric disorders, and to date, bioinformatic analyses have been the key focus providing strategies for prioritization of loci. Schizophrenia risk loci are over-represented in regulatory regions active in the brain15, 130, 131 and are enriched in genes from postsynaptic density, postsynaptic membrane, dendritic spine, axon, and voltage-gated potassium channel pathways, as well as histone H3-K4 methylation132 overlap with pathways identified in rare-variant studies of autism. Prioritization of GWAS results has progressed through integration with eQTL datasets, implicating synaptic genes (SNAP91 [MIM: 607923], TSNARE1, and CLCN4 [MIM 302910]) and genes with roles in neurodevelopment (FURIN [MIM: 136950] and CNTN4 [MIM: 607280]). 3D contacts between risk variants and promoters, explored by chromosome conformation capture (Hi-C) in the subcortical plate and germinal zone of the developing human cortex, supported putative interactions between causal risk variants and promoters in glutamatergic and calcium signaling genes (GRIA1 [MIM: 138248], NLGN1 [MIM: 600568], GRIN2A [MIM: 138253], and CACNA1C [MIM: 114205]), in several genes long implicated in schizophrenia (including DRD2 and DRD6, encoding acetylcholine receptors subunits), and genes SNAP91, TSNARE1, CLCN4, FURIN, and CNTN4.71
Fine-mapping has been accomplished for the strongest, and first identified, association with schizophrenia in the MHC region, a challenge because of its high genic content and high LD. The position of the association signal within the MHC region led to investigation of common structural haplotypes of complement factor 4 genes C4A (MIM: 120810) and C4B (MIM: 120820), combinations of which correlated well with schizophrenia risk and increased C4 expression and showed differential brain expression between case and control individuals.65 Several other complement proteins play a role in synapse elimination, and decreased numbers of synapses have long been suggested as a primary abnormality in schizophrenia. Observations that, in mice, a complement gene that shares features with human C4A and C4B is expressed in neurons and promoted synapse elimination in a developmental brain circuit strongly implicate this gene and its protein.
Translation. No new molecular targets for schizophrenia have been successfully identified since the first antipsychotic drugs were identified several decades ago. The reasons are likely to be manifold, but most drug development for schizophrenia has focused on achieving high-potency drugs for a single target—a methodology successful in many other areas of medicine—which necessitates a choice between the competing hypotheses of schizophrenia pathophysiology. GWAS results have provided unequivocal evidence of polygenicity, and because many of the GWAS loci contain genes that code for proteins among those indicated through multiple prior hypotheses, e.g., dopamine, glutamate, immune modulation, calcium signaling, and nicotinic cholinergic, future drug development could benefit from taking a multi-target approach. A proof-of-concept gene-set enrichment of schizophrenia risk alleles in sets of genes for drug targets identified several potential repurposing opportunities.133 Single-target medications could be appropriate for specific genetic subgroups, although identifying genetic subtypes is not yet part of the clinical trial paradigm.
Discussion
The Present
We have summarized the major kinds of discoveries made from GWASs focusing on adult traits and have reviewed the new biology and emerging translational outcomes for three diseases. Over the last decade, this experimental design has delivered a remarkably diverse set of discoveries in human genetics. For most traits and diseases studied, the mutational target in the genome appears large, in that polymorphisms in many genes contribute genetic variation in the population. Furthermore, the empirical evidence of widespread pleiotropy implies that many segregating variants affect multiple traits. A precise estimate of the proportion of all segregating genetic variants that are “functional” in the context of being associated with one or more traits, conditional on all other causal variants, remains elusive. For the highlighted traits, disorders, and diseases, we have given examples of routes from GWAS to biology and translation. For an experimental design only a decade old, this is an example of rapid translation of genetic findings toward clinical application.
The relationship between sample size and number of risk loci detected varies between traits, but all show a sharp increase at a critical sample size. To date, there has been no trait with evidence of a plateau of the number of risk loci discovered with increasing sample size. For some traits, such as height, schizophrenia, and IBD, discovery samples in the next 5 years are likely to continue to increase, perhaps at a lower rate per additional sample. A diminishing rate of discovery of new loci will provide a more complete picture of genetic architecture and will best satisfy the understanding of contributing biological pathways. According to the knowledge of Mendelian disease, the expectation is that multiple risk variants will be detected within loci that have already been identified. Hence, as sample sizes increase, the new discoveries of associated pathways will be saturated first, followed by genes and lastly variants.
GWASs have been successfully applied to molecular traits such as gene expression, DNA methylation,134 and metabolites.135 Conclusions from these studies are that most molecular phenotypes are just like other complex traits, in that differences between individuals are due to a combination of genetic factors and environmental exposures and that genetic loci can be mapped by GWASs.136 This makes the discovery of causal pathways from genomes to phenomes challenging, in that variation between people in modifiable risk factors might be partially anchored in DNA sequence variation for these “exposures.” Nevertheless, the combination of sequence variation with molecular phenotypes and disease data with novel analytical methods, such as Mendelian randomization,42 has great potential to unravel cause and consequence and to improve phenotypic prediction.137
GWASs to date have been based on SNP arrays designed to tag common variants in the genome. These arrays do not cover all genetic variants in the population, and it would seem natural that future GWASs will be based on WGS. However, the price differential between SNP arrays and WGS is still substantial, and array technology remains more robust than sequencing. Nevertheless, now hundreds of thousands of genomes are being sequenced as part of major initiatives, and the next 5 years will allow direct comparisons of discoveries made from sequencing and array studies. Interestingly, custom arrays without a GWAS “backbone” (such as the Immunochip, Metabochip, and exome-only arrays) have by and large failed to identify rare (MAF < 1%) variants at loci that were initially discovered from a GWAS, one of their aims. The reason for this is not clear. It could be because there are no rare variants of major effect, because the sample size is too small for detecting rare variants and/or estimating their effect size, because the chip coverage of rare variants is inadequate, or because of a combination of these. However, these custom arrays have led to the discovery of new loci and to fine-mapping at existing loci, mostly driven by increasing experimental sample size (see Appendix A on the relationship between sample size, imputation accuracy, and allele frequency on power of detection). A recent study of height using exome SNP arrays and a sample size of ∼700,000 reported 83 height-associated coding variants with a frequency of less than 5% and effect sizes of up to 2 cm.138 These variants each explain, on average, about the same amount of variation as common variants, whose effect sizes are of the order of 1 mm, because it is the combination of frequency and effect size that determines variation (Appendix A).
One limitation of both current array and WGS technology is that the precision of detection of structural variants (indels or inversions > 50 bp) is less than that of SNP detection. New technologies that enable long-range haplotyping are helping to overcome the weakness of short-read technologies, and cheap, genome-wide technologies for structural variants would constitute an important advance.
Fine-mapping of SNP-trait associations is the attempt to identify one or more causal variants that are responsible for the observed GWAS signals. Fine-mapping solely by statistical association is limited by experimental sample size and LD, given that the statistical evidence to separate a causal variant from a variant in LD with it is proportional to n(1 – r2) (see Equation A1 in Appendix A). If causal variants are not in the data (e.g., they have not been genotyped), then the imputation error also limits fine-mapping. With the likely availability of SNP-array-based GWAS data on very large sample sizes and WGS data on large sample sizes, statistical fine-mapping power will improve, and a small number of variants that are in extremely high LD might be identified as a plausible set of variants with a high probability of containing one or more causal variants. The use of additional information, such as prior knowledge of the likely function of specific variants given their location and surrounding DNA motif(s),139, 140 could help to reduce the set of statistical candidates to a smaller number. This is already a fertile area of statistical and bioinformatic research56, 62, 131, 141, 142 bringing together trait or disease GWAS results with those of tissue gene expression. More research on the resolution of fine-mapping is warranted, and this will be fueled by an expected increase in GWAS data on tissue- and cell-specific gene expression.
Most GWASs to date have been conducted on individuals of European descent, but there is a growing number of studies on populations of Asian and African ancestry. Because common variants contribute to the genetic architecture of complex traits, the expectation is that these common variants are evolutionarily old and shared across ethnicities, which is encouraging for generalizing treatments. The clearest demonstration of this, as discussed above, has been for IBD, for which the genetic correlation between Asian and European samples is close to 0.8, even though some individual risk loci differ in frequency or effect size.106 A characteristic of GWAS analyses to date is to strictly exclude individuals outside ethnicity boundaries on the basis of standard deviation units in principal-component dimensions. However, as sample sizes become larger, it is becoming possible to not only utilize but also take advantage of mixed and admixed ethnicity. The differing allele frequencies and LD structure across populations should aid in fine-mapping causal variants. New methods have emerged to deal with these data,75, 143 and we expect that this will be a fertile area of method development and discovery in the coming years.
The Future
Does the GWAS have a future? Extrapolating the discoveries from the last 10 years to the future, if we were to keep with the current experimental strategy of SNP arrays and imputation, then ever-increasing sample sizes would undoubtedly lead to new genetic discoveries, particularly (1) the discovery of more variants and more genes associated with one or more traits, (2) accounting for more genetic variation, (3) more accurate genetic predictors, and (4) a greater ability to evaluate disease heterogeneity and to derive genetically informed diagnoses that might be more aligned to specific treatments. For biological enrichment analyses and the discovery or fine-tuning of pathways involved in quantitative traits and disease, more loci are likely to increase resolution. In fields where diagnostic criteria are not based on biological markers, such as psychiatry, GWASs have turned the field on its head by, for the first time, contributing quantitative data that can be used for completely re-evaluating the relationship of previously distinct disorders.
The future of GWASs will have old and new challenges. With ever larger studies, the new loci identified will typically individually have smaller effect sizes (e.g., less than 0.5 mm for a trait such as height and an odds ratio of 1.01 for common disease) or, for rare variants, will be at very low frequency.138 For disorders with population prevalence of the order of 0.1%, discovery will still be limited by experimental sample size, given that it will take many years to accumulate sample sizes of 100,000 cases or more. One challenge is how such loci can be fine-mapped or studied for mechanism. Upscaling of technology, either through interfacing with sequenced-based -omic data or through upscaling by experimental perturbations (e.g., multiple-locus or genome-wide CRISPR) are likely to be key to overcoming the challenges of small effect size. What is likely to change in the near future is that GWASs by SNP arrays will be gradually replaced by GWASs by WGS, particularly for quantitative traits and very common diseases. Nonetheless, given a finite budget, larger sample sizes that are phenotypically more informative and genotyped on a SNP chip remain a powerful strategy for maximizing discovery. Fifteen years ago, genotyping technology was the limiting step to discovery, but now discovery is limited by phenotypic descriptors that could link with genetic data to allow disease stratification that might be more aligned with treatments. Furthermore, the emphasis in research will need to shift from gene discovery to translation into biological understanding and patient-focused outcomes, such as better diagnostic tests and novel treatments.
In conclusion, the experimental design of GWASs has led to a remarkable range of discoveries in human genetics over the last decade. It has delivered on its original aim of detecting associations between common DNA variants and human disease and disorders. It has led to a better understanding of the genetic architecture of complex traits and therefore of past natural selection on traits associated with fitness. It has led to the discovery of variants, genes, and biological pathways that play a role in specific diseases and disorders. It has led to new discoveries in disease epidemiology and to the discovery or repurposing of candidate therapeutics. As foreshadowed in 2007, it has indeed been a case of drinking from the fire hose.144 For the future, the combination of whole-genome surveys of genetic variation and detailed phenotypic and -omics data on millions of individuals will be a treasure trove for making new fundamental discoveries in human genetics. Some of those discoveries will be wholly unexpected, and others will detect or unravel biological mechanisms. Disease-specific discoveries will continue to spur the development and trials of new therapeutics, the understanding of pathways from sequence to consequence, and for some diseases, prevention or early intervention. In 10 years from now, genomic “personalized” or “precision” medicine is likely to be widespread and will include some applications to common diseases either directly through risk stratification for targeted prevention or intervention strategies or indirectly through new treatments where GWAS results provide the first step in the discovery pipeline. The experimental design of GWASs, which started as a theoretical exercise more than 20 years ago,145 has matured and delivered.
Acknowledgments
This research was supported by the Australian National Health and Medical Research Council (1107258, 1078901, 1078037, 1056929, 1048853, and 1113400), the gs2:Sylvia and Charles Viertel Charitable Foundation, the NIH (GM099568, MH100141-01, MH095034, MH109897, DK098032, DK085545, MH101814, and DKD105535), the Wellcome Trust (090532, 090367, 098381, and 106130), the UK Medical Research Council (L020149, 004422, and J010642), the Innovative Medicines Initiative, and the Accelerating Medicines Partnership (via the Foundation for the NIH). M.I.M. is a Wellcome Trust Senior Investigator. We thank the reviewers for their many helpful comments, which improved the paper. Because of space limitations, we were unable to do full justice to all relevant and important GWAS articles that have been published in the last 10 years. We apologize to many of our colleagues whose work is not cited.
Appendix A: Experimental Power to Detect Association
We re-visit statistical power because the interplay of experimental sample size, causal variant frequency and effect size, and platform (genotypes, imputed genotypes, and whole-genome sequence) remains essential to judging the optimum experimental design for discovery. The power to detect a variant-trait association from LD between an unobserved causal variant and an observed genotype can be quantified in the non-centrality parameter (NCP) of a statistical test to detect association (i.e., the expected value of the test statistic under the alternative hypothesis). It is defined as
| (Equation A1) |
where n is the experimental sample size, q2 is the proportion of phenotypic variance explained by a causal variant in the population, and r2 is the squared LD correlation between the causal variant and a genotyped SNP. This can also be expressed as the proportion of variance explained by the genotyped SNP in the population (R2 = r2q2),
| (Equation A2) |
If the genotypes at the causal locus are in Hardy-Weinberg equilibrium, then
| (Equation A3) |
where β is the effect size of an allele on the phenotype in standard deviation units. This assumes that the analysis for detecting an association is by regression of the phenotype on the genotype count (e.g., zero, one, or two minor alleles). Therefore, when q2 is small,
| (Equation A4) |
The power to detect an association between a trait and an ungenotyped but imputed causal variant is, similarly,
| (Equation A5) |
where Rimp2 is the squared correlation between the actual and imputed genotypes at the locus. These power calculations illustrate the trade-off between sample size, allele frequency, and effect size. In the future, when GWASs are likely to be performed by WGS, the r2 and Rimp2 values in Equations A4 and A5 will be 1 if the causal variant is sequenced without error, and considerable power can be gained to detect association between a trait and a sequenced variant in comparison to having array-based genotyped or imputed data if r2 or Rimp2 is small to modest, but only when the experimental sample size is kept constant. The equation above also demonstrates that the power to detect the association for a rare variant is limited because of its low allele frequency, even if the effect size is larger than that of a common variant. This means that for rare-variant associations, the sample sizes need to be very large or at least comparable to those used for GWASs with common variants, even with WGS data.
Web Resources
GWAS Catalog, https://www.ebi.ac.uk/gwas/
OMIM, http://www.omim.org/
References
- 1.Visscher P.M., Brown M.A., McCarthy M.I., Yang J. Five years of GWAS discovery. Am. J. Hum. Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang F., Gu W., Hurles M.E., Lupski J.R. Copy number variation in human health, disease, and evolution. Annu. Rev. Genomics Hum. Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hudson R.R. Properties of a neutral allele model with intragenic recombination. Theor. Popul. Biol. 1983;23:183–201. doi: 10.1016/0040-5809(83)90013-8. [DOI] [PubMed] [Google Scholar]
- 4.Wray N.R. Allele frequencies and the r2 measure of linkage disequilibrium: impact on design and interpretation of association studies. Twin Res. Hum. Genet. 2005;8:87–94. doi: 10.1375/1832427053738827. [DOI] [PubMed] [Google Scholar]
- 5.Browning S.R., Browning B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 8.McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J., Wray N.R., Visscher P.M. Comparing apples and oranges: equating the power of case-control and quantitative trait association studies. Genet. Epidemiol. 2010;34:254–257. doi: 10.1002/gepi.20456. [DOI] [PubMed] [Google Scholar]
- 10.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torgerson D.G., Ampleford E.J., Chiu G.Y., Gauderman W.J., Gignoux C.R., Graves P.E., Himes B.E., Levin A.M., Mathias R.A., Hancock D.B. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat. Genet. 2011;43:887–892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marigorta U.M., Navarro A. High trans-ethnic replicability of GWAS results implies common causal variants. PLoS Genet. 2013;9:e1003566. doi: 10.1371/journal.pgen.1003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wray N.R., Goddard M.E., Visscher P.M. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007;17:1520–1528. doi: 10.1101/gr.6665407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.International Schizophrenia Consortium Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maher B. Personal genomes: The case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 17.Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood A.R., Esko T., Yang J., Vedantam S., Pers T.H., Gustafsson S., Chu A.Y., Estrada K., Luan J., Kutalik Z. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lynch M., Walsh B. Sinauer Associates; 1998. Genetics and analysis of quantitative traits. [Google Scholar]
- 20.Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.R., ReproGen Consortium. Psychiatric Genomics Consortium. Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3. Duncan L. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pickrell J.K., Berisa T., Liu J.Z., Ségurel L., Tung J.Y., Hinds D.A. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 2016;48:709–717. doi: 10.1038/ng.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sivakumaran S., Agakov F., Theodoratou E., Prendergast J.G., Zgaga L., Manolio T., Rudan I., McKeigue P., Wilson J.F., Campbell H. Abundant pleiotropy in human complex diseases and traits. Am. J. Hum. Genet. 2011;89:607–618. doi: 10.1016/j.ajhg.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parkes M., Cortes A., van Heel D.A., Brown M.A. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat. Rev. Genet. 2013;14:661–673. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 24.Ellinghaus D., Jostins L., Spain S.L., Cortes A., Bethune J., Han B., Park Y.R., Raychaudhuri S., Pouget J.G., Hübenthal M. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat. Genet. 2016;48:510–518. doi: 10.1038/ng.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y.R., Zhao S.D., Li J., Bradfield J.P., Mohebnasab M., Steel L., Kobie J., Abrams D.J., Mentch F.D., Glessner J.T. Genetic sharing and heritability of paediatric age of onset autoimmune diseases. Nat. Commun. 2015;6:8442. doi: 10.1038/ncomms9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cross-Disorder Group of the Psychiatric Genomics Consortium Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat. Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Visscher P.M., Yang J. A plethora of pleiotropy across complex traits. Nat. Genet. 2016;48:707–708. doi: 10.1038/ng.3604. [DOI] [PubMed] [Google Scholar]
- 28.Yu J., Pressoir G., Briggs W.H., Vroh Bi I., Yamasaki M., Doebley J.F., McMullen M.D., Gaut B.S., Nielsen D.M., Holland J.B. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- 29.Kang H.M., Sul J.H., Service S.K., Zaitlen N.A., Kong S.Y., Freimer N.B., Sabatti C., Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lippert C., Listgarten J., Liu Y., Kadie C.M., Davidson R.I., Heckerman D. FaST linear mixed models for genome-wide association studies. Nat. Methods. 2011;8:833–835. doi: 10.1038/nmeth.1681. [DOI] [PubMed] [Google Scholar]
- 31.Zhou X., Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svishcheva G.R., Axenovich T.I., Belonogova N.M., van Duijn C.M., Aulchenko Y.S. Rapid variance components-based method for whole-genome association analysis. Nat. Genet. 2012;44:1166–1170. doi: 10.1038/ng.2410. [DOI] [PubMed] [Google Scholar]
- 33.Yang J., Zaitlen N.A., Goddard M.E., Visscher P.M., Price A.L. Advantages and pitfalls in the application of mixed-model association methods. Nat. Genet. 2014;46:100–106. doi: 10.1038/ng.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loh P.R., Tucker G., Bulik-Sullivan B.K., Vilhjálmsson B.J., Finucane H.K., Salem R.M., Chasman D.I., Ridker P.M., Neale B.M., Berger B. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 2015;47:284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J.Z., McRae A.F., Nyholt D.R., Medland S.E., Wray N.R., Brown K.M., Hayward N.K., Montgomery G.W., Visscher P.M., Martin N.G., Macgregor S., AMFS Investigators A versatile gene-based test for genome-wide association studies. Am. J. Hum. Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li M.X., Gui H.S., Kwan J.S., Sham P.C. GATES: a rapid and powerful gene-based association test using extended Simes procedure. Am. J. Hum. Genet. 2011;88:283–293. doi: 10.1016/j.ajhg.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J., Ferreira T., Morris A.P., Medland S.E., Genetic Investigation of ANthropometric Traits (GIANT) Consortium. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium. Madden P.A., Heath A.C., Martin N.G., Montgomery G.W. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012;44:369–375. doi: 10.1038/ng.2213. S1–S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.R., Anttila V., Xu H., Zang C., Farh K. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J., Benyamin B., McEvoy B.P., Gordon S., Henders A.K., Nyholt D.R., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowden J., Davey Smith G., Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burgess S., Butterworth A., Thompson S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013;37:658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith G.D., Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 43.Jia X., Han B., Onengut-Gumuscu S., Chen W.M., Concannon P.J., Rich S.S., Raychaudhuri S., de Bakker P.I. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS ONE. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang J., Howie B., McCarthy S., Memari Y., Walter K., Min J.L., Danecek P., Malerba G., Trabetti E., Zheng H.F. Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nat. Commun. 2015;6:8111. doi: 10.1038/ncomms9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Browning B.L., Browning S.R. Genotype Imputation with Millions of Reference Samples. Am. J. Hum. Genet. 2016;98:116–126. doi: 10.1016/j.ajhg.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Visscher P.M., Hill W.G., Wray N.R. Heritability in the genomics era--concepts and misconceptions. Nat. Rev. Genet. 2008;9:255–266. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- 48.Yang J., Lee T., Kim J., Cho M.C., Han B.G., Lee J.Y., Lee H.J., Cho S., Kim H. Ubiquitous polygenicity of human complex traits: genome-wide analysis of 49 traits in Koreans. PLoS Genet. 2013;9:e1003355. doi: 10.1371/journal.pgen.1003355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang J., Bakshi A., Zhu Z., Hemani G., Vinkhuyzen A.A., Lee S.H., Robinson M.R., Perry J.R., Nolte I.M., van Vliet-Ostaptchouk J.V. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat. Genet. 2015;47:1114–1120. doi: 10.1038/ng.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moser G., Lee S.H., Hayes B.J., Goddard M.E., Wray N.R., Visscher P.M. Simultaneous discovery, estimation and prediction analysis of complex traits using a bayesian mixture model. PLoS Genet. 2015;11:e1004969. doi: 10.1371/journal.pgen.1004969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Rheenen W., Shatunov A., Dekker A.M., McLaughlin R.L., Diekstra F.P., Pulit S.L., van der Spek R.A., Võsa U., de Jong S., Robinson M.R. Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat. Genet. 2016;48:1043–1048. doi: 10.1038/ng.3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ripke S., O’Dushlaine C., Chambert K., Moran J.L., Kähler A.K., Akterin S., Bergen S.E., Collins A.L., Crowley J.J., Fromer M. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loh P.R., Bhatia G., Gusev A., Finucane H.K., Bulik-Sullivan B.K., Pollack S.J., Schizophrenia Working Group of Psychiatric Genomics Consortium. de Candia T.R., Lee S.H., Wray N.R. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat. Genet. 2015;47:1385–1392. doi: 10.1038/ng.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cortes A., Pulit S.L., Leo P.J., Pointon J.J., Robinson P.C., Weisman M.H., Ward M., Gensler L.S., Zhou X., Garchon H.J. Major histocompatibility complex associations of ankylosing spondylitis are complex and involve further epistasis with ERAP1. Nat. Commun. 2015;6:7146. doi: 10.1038/ncomms8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Evans D.M., Visscher P.M., Wray N.R. Harnessing the information contained within genome-wide association studies to improve individual prediction of complex disease risk. Hum. Mol. Genet. 2009;18:3525–3531. doi: 10.1093/hmg/ddp295. [DOI] [PubMed] [Google Scholar]
- 56.Gamazon E.R., Wheeler H.E., Shah K.P., Mozaffari S.V., Aquino-Michaels K., Carroll R.J., Eyler A.E., Denny J.C., GTEx Consortium. Nicolae D.L. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Evans D.M., Brion M.J., Paternoster L., Kemp J.P., McMahon G., Munafò M., Whitfield J.B., Medland S.E., Montgomery G.W., GIANT Consortium Mining the human phenome using allelic scores that index biological intermediates. PLoS Genet. 2013;9:e1003919. doi: 10.1371/journal.pgen.1003919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tyrrell J., Wood A.R., Ames R.M., Yaghootkar H., Beaumont R.N., Jones S.E., Tuke M.A., Ruth K.S., Freathy R.M., Davey Smith G. Gene-obesogenic environment interactions in the UK Biobank study. Int. J. Epidemiol. 2017;46:559–575. doi: 10.1093/ije/dyw337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng J., Erzurumluoglu A.M., Elsworth B.L., Kemp J.P., Howe L., Haycock P.C., Hemani G., Tansey K., Laurin C., Early Genetics and Lifecourse Epidemiology (EAGLE) Eczema Consortium LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–279. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoffmann T.J., Ehret G.B., Nandakumar P., Ranatunga D., Schaefer C., Kwok P.Y., Iribarren C., Chakravarti A., Risch N. Genome-wide association analyses using electronic health records identify new loci influencing blood pressure variation. Nat. Genet. 2017;49:54–64. doi: 10.1038/ng.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B.W., Jansen R., de Geus E.J., Boomsma D.I., Wright F.A. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu Z., Zhang F., Hu H., Bakshi A., Robinson M.R., Powell J.E., Montgomery G.W., Goddard M.E., Wray N.R., Visscher P.M., Yang J. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 63.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J., Schizophrenia Working Group of the Psychiatric Genomics Consortium. Patterson N., Daly M.J., Price A.L., Neale B.M. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Claussnitzer M., Dankel S.N., Kim K.H., Quon G., Meuleman W., Haugen C., Glunk V., Sousa I.S., Beaudry J.L., Puviindran V. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N. Engl. J. Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sekar A., Bialas A.R., de Rivera H., Davis A., Hammond T.R., Kamitaki N., Tooley K., Presumey J., Baum M., Van Doren V. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Albert F.W., Kruglyak L. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 2015;16:197–212. doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- 67.Pasquali L., Gaulton K.J., Rodríguez-Seguí S.A., Mularoni L., Miguel-Escalada I., Akerman I., Tena J.J., Morán I., Gómez-Marín C., van de Bunt M. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat. Genet. 2014;46:136–143. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roadmap Epigenomics Consortium. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Won H., de la Torre-Ubieta L., Stein J.L., Parikshak N.N., Huang J., Opland C.K., Gandal M.J., Sutton G.J., Hormozdiari F., Lu D. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 2016;538:523–527. doi: 10.1038/nature19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nelson M.R., Tipney H., Painter J.L., Shen J., Nicoletti P., Shen Y., Floratos A., Sham P.C., Li M.J., Wang J. The support of human genetic evidence for approved drug indications. Nat. Genet. 2015;47:856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 73.McDonald T.J., Ellard S. Maturity onset diabetes of the young: identification and diagnosis. Ann. Clin. Biochem. 2013;50:403–415. doi: 10.1177/0004563213483458. [DOI] [PubMed] [Google Scholar]
- 74.Fuchsberger C., Flannick J., Teslovich T.M., Mahajan A., Agarwala V., Gaulton K.J., Ma C., Fontanillas P., Moutsianas L., McCarthy D.J. The genetic architecture of type 2 diabetes. Nature. 2016;536:41–47. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium. Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium. South Asian Type 2 Diabetes (SAT2D) Consortium. Mexican American Type 2 Diabetes (MAT2D) Consortium. Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium. Mahajan A., Go M.J., Zhang W., Below J.E., Gaulton K.J. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segrè A.V., Steinthorsdottir V., Strawbridge R.J., Khan H., Grallert H., Mahajan A. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Steinthorsdottir V., Thorleifsson G., Sulem P., Helgason H., Grarup N., Sigurdsson A., Helgadottir H.T., Johannsdottir H., Magnusson O.T., Gudjonsson S.A. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat. Genet. 2014;46:294–298. doi: 10.1038/ng.2882. [DOI] [PubMed] [Google Scholar]
- 78.Ayub Q., Moutsianas L., Chen Y., Panoutsopoulou K., Colonna V., Pagani L., Prokopenko I., Ritchie G.R., Tyler-Smith C., McCarthy M.I. Revisiting the thrifty gene hypothesis via 65 loci associated with susceptibility to type 2 diabetes. Am. J. Hum. Genet. 2014;94:176–185. doi: 10.1016/j.ajhg.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Field Y., Boyle E.A., Telis N., Gao Z., Gaulton K.J., Golan D., Yengo L., Rocheleau G., Froguel P., McCarthy M.I., Pritchard J.K. Detection of human adaptation during the past 2000 years. Science. 2016;354:760–764. doi: 10.1126/science.aag0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Waters K.M., Stram D.O., Hassanein M.T., Le Marchand L., Wilkens L.R., Maskarinec G., Monroe K.R., Kolonel L.N., Altshuler D., Henderson B.E., Haiman C.A. Consistent association of type 2 diabetes risk variants found in europeans in diverse racial and ethnic groups. PLoS Genet. 2010;6:e1001078. doi: 10.1371/journal.pgen.1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moltke I., Grarup N., Jørgensen M.E., Bjerregaard P., Treebak J.T., Fumagalli M., Korneliussen T.S., Andersen M.A., Nielsen T.S., Krarup N.T. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature. 2014;512:190–193. doi: 10.1038/nature13425. [DOI] [PubMed] [Google Scholar]
- 82.Claussnitzer M., Dankel S.N., Klocke B., Grallert H., Glunk V., Berulava T., Lee H., Oskolkov N., Fadista J., Ehlers K. Leveraging cross-species transcription factor binding site patterns: from diabetes risk loci to disease mechanisms. Cell. 2014;156:343–358. doi: 10.1016/j.cell.2013.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Scott L.J., Erdos M.R., Huyghe J.R., Welch R.P., Beck A.T., Wolford B.N., Chines P.S., Didion J.P., Narisu N., Stringham H.M. The genetic regulatory signature of type 2 diabetes in human skeletal muscle. Nat. Commun. 2016;7:11764. doi: 10.1038/ncomms11764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Parker S.C., Stitzel M.L., Taylor D.L., Orozco J.M., Erdos M.R., Akiyama J.A., van Bueren K.L., Chines P.S., Narisu N., NISC Comparative Sequencing Program Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl. Acad. Sci. USA. 2013;110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Small K.S., Hedman A.K., Grundberg E., Nica A.C., Thorleifsson G., Kong A., Thorsteindottir U., Shin S.Y., Richards H.B., GIANT Consortium Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat. Genet. 2011;43:561–564. doi: 10.1038/ng.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gaulton K.J., Ferreira T., Lee Y., Raimondo A., Mägi R., Reschen M.E., Mahajan A., Locke A., Rayner N.W., Robertson N. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat. Genet. 2015;47:1415–1425. doi: 10.1038/ng.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dimas A.S., Lagou V., Barker A., Knowles J.W., Mägi R., Hivert M.F., Benazzo A., Rybin D., Jackson A.U., Stringham H.M. Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes. 2014;63:2158–2171. doi: 10.2337/db13-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van de Bunt M., Manning Fox J.E., Dai X., Barrett A., Grey C., Li L., Bennett A.J., Johnson P.R., Rajotte R.V., Gaulton K.J. Transcript Expression Data from Human Islets Links Regulatory Signals from Genome-Wide Association Studies for Type 2 Diabetes and Glycemic Traits to Their Downstream Effectors. PLoS Genet. 2015;11:e1005694. doi: 10.1371/journal.pgen.1005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saxena R., Hivert M.F., Langenberg C., Tanaka T., Pankow J.S., Vollenweider P., Lyssenko V., Bouatia-Naji N., Dupuis J., Jackson A.U. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat. Genet. 2010;42:142–148. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kooner J.S., Saleheen D., Sim X., Sehmi J., Zhang W., Frossard P., Been L.F., Chia K.S., Dimas A.S., Hassanali N. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat. Genet. 2011;43:984–989. doi: 10.1038/ng.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sandhu M.S., Weedon M.N., Fawcett K.A., Wasson J., Debenham S.L., Daly A., Lango H., Frayling T.M., Neumann R.J., Sherva R. Common variants in WFS1 confer risk of type 2 diabetes. Nat. Genet. 2007;39:951–953. doi: 10.1038/ng2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wei F.Y., Tomizawa K. Functional loss of Cdkal1, a novel tRNA modification enzyme, causes the development of type 2 diabetes. Endocr. J. 2011;58:819–825. doi: 10.1507/endocrj.ej11-0099. [DOI] [PubMed] [Google Scholar]
- 93.Flannick J., Thorleifsson G., Beer N.L., Jacobs S.B., Grarup N., Burtt N.P., Mahajan A., Fuchsberger C., Atzmon G., Benediktsson R. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat. Genet. 2014;46:357–363. doi: 10.1038/ng.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fall T., Xie W., Poon W., Yaghootkar H., Mägi R., GENESIS Consortium. Knowles J.W., Lyssenko V., Weedon M., Frayling T.M., Ingelsson E. Using Genetic Variants to Assess the Relationship Between Circulating Lipids and Type 2 Diabetes. Diabetes. 2015;64:2676–2684. doi: 10.2337/db14-1710. [DOI] [PubMed] [Google Scholar]
- 95.Horikoshi M., Beaumont R.N., Day F.R., Warrington N.M., Kooijman M.N., Fernandez-Tajes J., Feenstra B., van Zuydam N.R., Gaulton K.J., Grarup N. Genome-wide associations for birth weight and correlations with adult disease. Nature. 2016;538:248–252. doi: 10.1038/nature19806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lotta L.A., Sharp S.J., Burgess S., Perry J.R., Stewart I.D., Willems S.M., Luan J., Ardanaz E., Arriola L., Balkau B. Association Between Low-Density Lipoprotein Cholesterol-Lowering Genetic Variants and Risk of Type 2 Diabetes: A Meta-analysis. JAMA. 2016;316:1383–1391. doi: 10.1001/jama.2016.14568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rafiq S., Melzer D., Weedon M.N., Lango H., Saxena R., Scott L.J., DIAGRAM Consortium. Palmer C.N., Morris A.D., McCarthy Gene variants influencing measures of inflammation or predisposing to autoimmune and inflammatory diseases are not associated with the risk of type 2 diabetes. Diabetologia. 2008;51:2205–2213. doi: 10.1007/s00125-008-1160-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ye Z., Sharp S.J., Burgess S., Scott R.A., Imamura F., InterAct Consortium. Langenberg C., Wareham N.J., Forouhi N.G. Association between circulating 25-hydroxyvitamin D and incident type 2 diabetes: a mendelian randomisation study. Lancet Diabetes Endocrinol. 2015;3:35–42. doi: 10.1016/S2213-8587(14)70184-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lotta L.A., Gulati P., Day F.R., Payne F., Ongen H., van de Bunt M., Gaulton K.J., Eicher J.D., Sharp S.J., Luan J. Integrative genomic analysis implicates limited peripheral adipose storage capacity in the pathogenesis of human insulin resistance. Nat. Genet. 2017;49:17–26. doi: 10.1038/ng.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.GoDARTS and UKPDS Diabetes Pharmacogenetics Study Group. Wellcome Trust Case Control Consortium 2. Zhou K., Bellenguez C., Spencer C.C., Bennett A.J., Coleman R.L., Tavendale R., Hawley S.A., Donnelly L.A. Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat. Genet. 2011;43:117–120. doi: 10.1038/ng.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou K., Yee S.W., Seiser E.L., van Leeuwen N., Tavendale R., Bennett A.J., Groves C.J., Coleman R.L., van der Heijden A.A., Beulens J.W. Variation in the glucose transporter gene SLC2A2 is associated with glycemic response to metformin. Nat. Genet. 2016;48:1055–1059. doi: 10.1038/ng.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gloyn A.L., Pearson E.R., Antcliff J.F., Proks P., Bruining G.J., Slingerland A.S., Howard N., Srinivasan S., Silva J.M., Molnes J. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N. Engl. J. Med. 2004;350:1838–1849. doi: 10.1056/NEJMoa032922. [DOI] [PubMed] [Google Scholar]
- 103.Lynch C.J., Adams S.H. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat. Rev. Endocrinol. 2014;10:723–736. doi: 10.1038/nrendo.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pedersen H.K., Gudmundsdottir V., Nielsen H.B., Hyotylainen T., Nielsen T., Jensen B.A., Forslund K., Hildebrand F., Prifti E., Falony G. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535:376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 105.Cortes A., Brown M.A. Promise and pitfalls of the Immunochip. Arthritis Res. Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu J.Z., van Sommeren S., Huang H., Ng S.C., Alberts R., Takahashi A., Ripke S., Lee J.C., Jostins L., Shah T. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jiang L., Yin J., Ye L., Yang J., Hemani G., Liu A.J., Zou H., He D., Sun L., Zeng X. Novel risk loci for rheumatoid arthritis in Han Chinese and congruence with risk variants in Europeans. Arthritis Rheumatol. 2014;66:1121–1132. doi: 10.1002/art.38353. [DOI] [PubMed] [Google Scholar]
- 108.Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.International Genetics of Ankylosing Spondylitis Consortium (IGAS) Cortes A., Hadler J., Pointon J.P., Robinson P.C., Karaderi T., Leo P., Cremin K., Pryce K., Harris J. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat. Genet. 2013;45:730–738. doi: 10.1038/ng.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gregory A.P., Dendrou C.A., Attfield K.E., Haghikia A., Xifara D.K., Butter F., Poschmann G., Kaur G., Lambert L., Leach O.A. TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis. Nature. 2012;488:508–511. doi: 10.1038/nature11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gregersen P.K., Silver J., Winchester R.J. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 112.Raychaudhuri S., Sandor C., Stahl E.A., Freudenberg J., Lee H.S., Jia X., Alfredsson L., Padyukov L., Klareskog L., Worthington J. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet. 2012;44:291–296. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Scally S.W., Petersen J., Law S.C., Dudek N.L., Nel H.J., Loh K.L., Wijeyewickrema L.C., Eckle S.B., van Heemst J., Pike R.N. A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. J. Exp. Med. 2013;210:2569–2582. doi: 10.1084/jem.20131241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Heap G.A., Weedon M.N., Bewshea C.M., Singh A., Chen M., Satchwell J.B., Vivian J.P., So K., Dubois P.C., Andrews J.M. HLA-DQA1-HLA-DRB1 variants confer susceptibility to pancreatitis induced by thiopurine immunosuppressants. Nat. Genet. 2014;46:1131–1134. doi: 10.1038/ng.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hueber W., Patel D.D., Dryja T., Wright A.M., Koroleva I., Bruin G., Antoni C., Draelos Z., Gold M.H., Psoriasis Study Group Effects of AIN457, a fully human antibody to interleukin-17A, on psoriasis, rheumatoid arthritis, and uveitis. Sci. Transl. Med. 2010;2:52ra72. doi: 10.1126/scitranslmed.3001107. [DOI] [PubMed] [Google Scholar]
- 116.McInnes I.B., Sieper J., Braun J., Emery P., van der Heijde D., Isaacs J.D., Dahmen G., Wollenhaupt J., Schulze-Koops H., Kogan J. Efficacy and safety of secukinumab, a fully human anti-interleukin-17A monoclonal antibody, in patients with moderate-to-severe psoriatic arthritis: a 24-week, randomised, double-blind, placebo-controlled, phase II proof-of-concept trial. Ann. Rheum. Dis. 2014;73:349–356. doi: 10.1136/annrheumdis-2012-202646. [DOI] [PubMed] [Google Scholar]
- 117.Sieper J., Deodhar A., Marzo-Ortega H., Aelion J.A., Blanco R., Jui-Cheng T., Andersson M., Porter B., Richards H.B., MEASURE 2 Study Group Secukinumab efficacy in anti-TNF-naive and anti-TNF-experienced subjects with active ankylosing spondylitis: results from the MEASURE 2 Study. Ann. Rheum. Dis. 2017;76:571–592. doi: 10.1136/annrheumdis-2016-210023. [DOI] [PubMed] [Google Scholar]
- 118.Subramanian V., Knight J.S., Parelkar S., Anguish L., Coonrod S.A., Kaplan M.J., Thompson P.R. Design, synthesis, and biological evaluation of tetrazole analogs of Cl-amidine as protein arginine deiminase inhibitors. J. Med. Chem. 2015;58:1337–1344. doi: 10.1021/jm501636x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kawalkowska J., Quirke A.M., Ghari F., Davis S., Subramanian V., Thompson P.R., Williams R.O., Fischer R., La Thangue N.B., Venables P.J. Abrogation of collagen-induced arthritis by a peptidyl arginine deiminase inhibitor is associated with modulation of T cell-mediated immune responses. Sci. Rep. 2016;6:26430. doi: 10.1038/srep26430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Agrawal N., Brown M.A. Genetic associations and functional characterization of M1 aminopeptidases and immune-mediated diseases. Genes Immun. 2014;15:521–527. doi: 10.1038/gene.2014.46. [DOI] [PubMed] [Google Scholar]
- 121.CNV and Schizophrenia Working Groups of the Psychiatric Genomics Consortium. Psychosis Endophenotypes International Consortium Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet. 2017;49:27–35. doi: 10.1038/ng.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Girard S.L., Gauthier J., Noreau A., Xiong L., Zhou S., Jouan L., Dionne-Laporte A., Spiegelman D., Henrion E., Diallo O. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat. Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- 123.Xu B., Roos J.L., Dexheimer P., Boone B., Plummer B., Levy S., Gogos J.A., Karayiorgou M. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat. Genet. 2011;43:864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fromer M., Pocklington A.J., Kavanagh D.H., Williams H.J., Dwyer S., Gormley P., Georgieva L., Rees E., Palta P., Ruderfer D.M. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.McCarthy S.E., Gillis J., Kramer M., Lihm J., Yoon S., Berstein Y., Mistry M., Pavlidis P., Solomon R., Ghiban E. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol. Psychiatry. 2014;19:652–658. doi: 10.1038/mp.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jiang Y.H., Yuen R.K., Jin X., Wang M., Chen N., Wu X., Ju J., Mei J., Shi Y., He M. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am. J. Hum. Genet. 2013;93:249–263. doi: 10.1016/j.ajhg.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Iossifov I., O’Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Georgieva L., Rees E., Moran J.L., Chambert K.D., Milanova V., Craddock N., Purcell S., Sklar P., McCarroll S., Holmans P. De novo CNVs in bipolar affective disorder and schizophrenia. Hum. Mol. Genet. 2014;23:6677–6683. doi: 10.1093/hmg/ddu379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Roussos P., Mitchell A.C., Voloudakis G., Fullard J.F., Pothula V.M., Tsang J., Stahl E.A., Georgakopoulos A., Ruderfer D.M., Charney A. A role for noncoding variation in schizophrenia. Cell Rep. 2014;9:1417–1429. doi: 10.1016/j.celrep.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gusev A., Lee S.H., Trynka G., Finucane H., Vilhjálmsson B.J., Xu H., Zang C., Ripke S., Bulik-Sullivan B., Stahl E. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am. J. Hum. Genet. 2014;95:535–552. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat. Neurosci. 2015;18:199–209. doi: 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.de Jong S., Vidler L.R., Mokrab Y., Collier D.A., Breen G. Gene-set analysis based on the pharmacological profiles of drugs to identify repurposing opportunities in schizophrenia. J. Psychopharmacol. (Oxford) 2016;30:826–830. doi: 10.1177/0269881116653109. [DOI] [PubMed] [Google Scholar]
- 134.Bell J.T., Pai A.A., Pickrell J.K., Gaffney D.J., Pique-Regi R., Degner J.F., Gilad Y., Pritchard J.K. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kettunen J., Tukiainen T., Sarin A.P., Ortega-Alonso A., Tikkanen E., Lyytikäinen L.P., Kangas A.J., Soininen P., Würtz P., Silander K. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat. Genet. 2012;44:269–276. doi: 10.1038/ng.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shah S., McRae A.F., Marioni R.E., Harris S.E., Gibson J., Henders A.K., Redmond P., Cox S.R., Pattie A., Corley J. Genetic and environmental exposures constrain epigenetic drift over the human life course. Genome Res. 2014;24:1725–1733. doi: 10.1101/gr.176933.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shah S., Bonder M.J., Marioni R.E., Zhu Z., McRae A.F., Zhernakova A., Harris S.E., Liewald D., Henders A.K., Mendelson M.M. Improving Phenotypic Prediction by Combining Genetic and Epigenetic Associations. Am. J. Hum. Genet. 2015;97:75–85. doi: 10.1016/j.ajhg.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Marouli E., Graff M., Medina-Gomez C., Lo K.S., Wood A.R., Kjaer T.R., Fine R.S., Lu Y., Schurmann C., Highland H.M. Rare and low-frequency coding variants alter human adult height. Nature. 2017;542:186–190. doi: 10.1038/nature21039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Farh K.K., Marson A., Zhu J., Kleinewietfeld M., Housley W.J., Beik S., Shoresh N., Whitton H., Ryan R.J., Shishkin A.A. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Spain S.L., Barrett J.C. Strategies for fine-mapping complex traits. Hum. Mol. Genet. 2015;24(R1):R111–R119. doi: 10.1093/hmg/ddv260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.He X., Fuller C.K., Song Y., Meng Q., Zhang B., Yang X., Li H. Sherlock: detecting gene-disease associations by matching patterns of expression QTL and GWAS. Am. J. Hum. Genet. 2013;92:667–680. doi: 10.1016/j.ajhg.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hormozdiari F., Kostem E., Kang E.Y., Pasaniuc B., Eskin E. Identifying causal variants at loci with multiple signals of association. Genetics. 2014;198:497–508. doi: 10.1534/genetics.114.167908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Morris A.P. Transethnic meta-analysis of genomewide association studies. Genet. Epidemiol. 2011;35:809–822. doi: 10.1002/gepi.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hunter D.J., Kraft P. Drinking from the fire hose--statistical issues in genomewide association studies. N. Engl. J. Med. 2007;357:436–439. doi: 10.1056/NEJMp078120. [DOI] [PubMed] [Google Scholar]
- 145.Risch N., Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]