Abstract
DNA-scaffolded oligodeoxyriboside fluorophores (ODFs) were used as the reporters in turn-on sensing of enzymatic bond-cleaving activity. A tetramer ODF of pyrene deoxynucleosides displayed high quenching efficiency when conjugated via ester linkages with a dabcyl quencher, and yielded large signal increases with several enzymes in vitro and in intact human cells.
Fluorophore-labeled substrates and sensor systems are widely used in enzyme-substrate activity assays, enzyme discovery, identification and classification of enzymes, enzyme engineering and drug discovery.1–5 Fluorogenic substrates for many classes of enzymes have been developed, including substrates for proteases, glycosidases, and esterases.
Esterases and lipases belong to the hydrolase super-family and catalyze the hydrolysis of esters. These enzymes are widely useful in organic synthesis,6 in industrial applications, and are important drug targets7–9 and prodrug activators.10,11 Esterases are mainly localized in the endoplasmic reticulum and cytosol of various tissues, and are important in the detoxification and metabolism of many drugs.11,12 Lipases are important in lipid transfer among organisms, and are essential for animals in the utilization of dietary lipids in lipid metabolism.13 Esterases and lipases share a general common catalytic structure in their substrates, but esterases prefer substrates in the soluble state, while lipases have higher activities towards aggregated substrates.14 Esterase-triggered fluorescent sensors have been reported for high-throughput enzyme and substrate screening,15–18 biomolecular imaging,19 and in vitro and in vivo metal ion sensing.20 However, in many existing traditional fluorophore-based esterase sensors, complicated design and synthetic strategies, poor water solubility, and low fluorescence turn-on ratios present barriers towards their practical application in biological systems.
To address some of the limitations of standard fluorophores and reporters, we have developed oligodeoxyfluorosides (ODFs), in which multiple adjacent chromophores replace the bases in DNA.21–23 The DNA phosphodiester backbone acts as a scaffold, holding these fluorophores at close proximity and promoting electronic interactions. In earlier studies, multiple forms of energy and excitation transfer have been observed, including excimer and exciplex formation.24–26 ODFs show promise as discrete fluorescent dyes, with useful characteristics including large Stokes shifts of more than 100 nm. Moreover, since a broad-spectrum set of ODFs that can be excited at one wavelength has been identified,27 these compounds offer the possibility of simultaneous multicolor imaging in biological systems.
Here we describe a new type of excimer-based esterase and lipase sensor design, in which a tetrapyrene ODF is used as the fluorophore. Dabcyl is placed at the 5′-end of the ODF sequence to act as a quencher, conjugated to the reporter fluorophore by an ester linkage. Upon exposure to esterase or lipase activity, the quencher is released, resulting in an efficient fluorescence light-up signal in vitro and in cells. The results suggest the future possibility of multienzyme, multicolor sensing with ODF reporters.
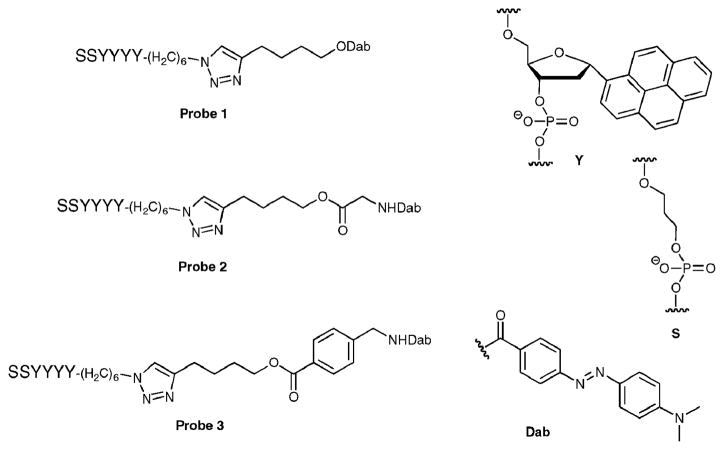
We designed three ODF-based esterase/lipase sensors with the tetrapyrene reporter (Fig. 1). The excitation maximum wavelength of this fluorophore (YYYY, assembled on a DNA synthesizer) is 340 nm and the maximum emission wavelength is 480 nm (Φem = 0.1528), characteristic of the pyrene excimer. Two C3 spacers were placed at the 3′-end to increase aqueous solubility, and an azide group was placed at the 5′-end for conjugation of the substrate-quencher moiety. Dabcyl was selected as the quencher; previous studies have shown that this moiety quenches the emission of excimers and exciplexes efficiently in a sequence dependent manner.26,28 Three different ester substrate designs were used. In probe 1, the ester bond was attached directly to dabcyl, giving the smallest possible substrate structure. In probe 2, a glycine moiety was inserted between the ester bond and dabcyl to mimic an aliphatic ester substrate. In probe 3, a benzoic acid derivative was inserted at the same position to present an aromatic ester substrate. An alkyne group was introduced in this substrate-quencher moiety for conjugation to the 5′-azido-ODF via Huisgen–Sharpless “click” chemistry.
Fig. 1.
Structure of ODF-based fluorescent probes 1, 2 and 3, along with components Y, S, and quencher (Dab).
The syntheses of the alkyne quencher compounds 1, 2 and 3 and of the three probes are shown in ESI† (Schemes S1–S5).
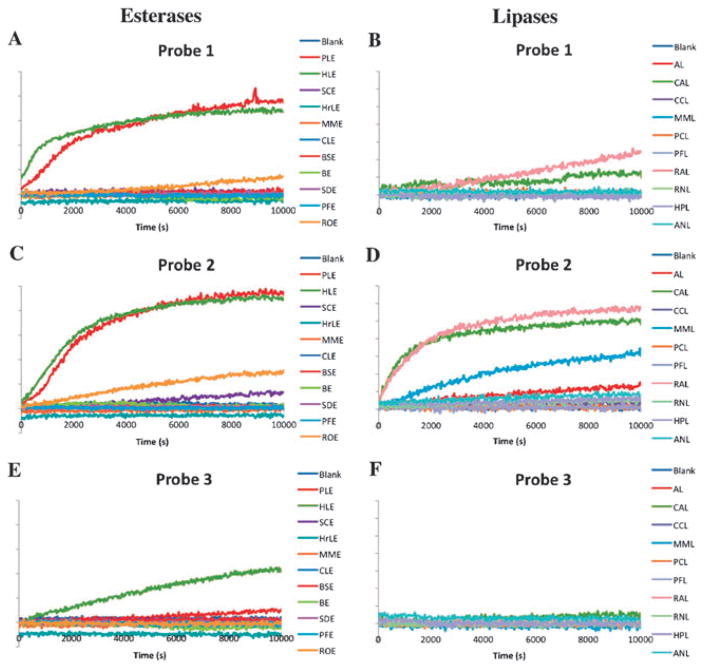
The esterase/lipase screening experiments were performed with a fluorescence microplate reader; each reaction well contained 0.5 μMof probe and 30 μgmL−1 of enzyme in pH 7.4 PBS buffer (25 °C). Screening results showed that 4 of 11 esterases displayed activity for at least one probe, while 7 of 10 lipases tested exhibited observable activity (Fig. 2). All three probes were activated by at least one enzyme. Among all esterases that showed activities, most were not specific to only one probe, although selectivities were observed. The SCE enzyme was selective for probe 2, although the activity was low. ROE displayed a similar level of activity to both probes 1 and 2, but no activity with probe 3. PLE and HLE were active for all three probes, with higher activities for probes 1 and 2 compared to probe 3.
Fig. 2.
Enzymatic screening results. Each reaction well contained 0.5 μM probe and 30 μg mL−1 enzyme in pH 7.4 PBS buffer at 25 °C. Samples were excited at 340 nm, and fluorescence emission was monitored at 480 nm. The blank control experiments contained 0.5 μM of the respective probe in pH 7.4 PBS buffer. (a) Probe 1 treated with esterases; (b) Probe 1 treated with lipases; (c) Probe 2 treated with esterases; (d) Probe 2 treated with lipases; (e) Probe 3 treated with esterases; (f) Probe 3 treated with lipases. Esterase and lipase abbreviations are given in ESI†.
Lipases showed a different spectrum of probe preference; all active lipases showed selectivity towards probe 2. CAL and RAL had low activities towards probe 1, and none of the lipases tested displayed appreciable activity towards probe 3. Overall, these results indicate that lipases preferred the aliphatic ester substrates, and the flexibility of the substrate appears to be important. Esterases are also very sensitive to the size of the substrates.
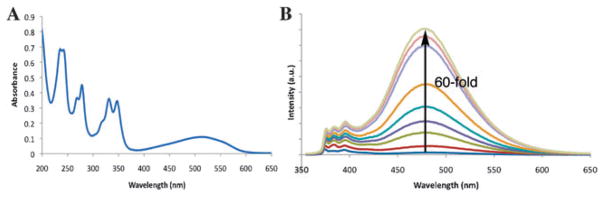
Next, the probe 1–PLE substrate–enzyme pair was chosen for further fluorescence studies (Fig. 3). The increase in the pyrene excimer fluorescence peak at 480 nm could be observed at 1 nM probe concentration using the fluorometer, and green fluorescence could be seen with the naked eye at 0.1 μMprobe. Dabcyl had a highly efficient quenching effect on the oligopyrene excimer, as revealed by the magnitude of fluorescence enhancement. A 60-fold excimer fluorescence enhancement upon treatment with PLE was observed when the concentration of ODF was in the range of 0.1–0.5 μM. The increase in fluorescence of the monomer at 395 nm (the residual pyrene monomer emission band) was only about 10-fold, consistent with studies showing delocalized excimers to be quenched more efficiently than localized monomer emission states.26,28–30 When the probe concentration was higher than 1.0 μM, greater excimer enhancement (110-fold) and weaker monomer enhancement (7-fold) were observed (ESI†, Fig. S3). At the higher concentration it is possible that the multipyrene reporters aggregate, resulting in stronger quenching28 and increased enhancement upon reaction.
Fig. 3.
UV spectrum (a) and fluorescence turn-on (b) of probe 1. The enzymatic fluorescence assay was performed using 0.33 μM probe, 30 μg mL−1 enzyme in a pH 7.4 PBS buffer at room temperature. The time course was measured over 2 h.
The kinetics of the catalytic process are complicated by apparently tight enzyme binding and by this hypothesized ODF aggregation at high concentration. When enzyme concentration was high, an apparent enzyme/ODF complex was observed, showing a blue shift in its fluorescence spectrum (see ESI†, Fig. S4), which may be caused by alteration of pyrene π–π stacking.31 High probe concentrations resulted in a slower initial reaction rate, but the aggregation of the ODF (which can enhance quenching efficiency26,28–30) resulted in a higher excimer turn-on ratio.
Comparison studies were made with fluorescein diacetate (FDA, a commercial esterase reporter) and probe 1. FDA is not fluorescent in the inactivated form, while the fluorescence of our probes is small but non-zero. Thus the background of our probes is higher than that of unreacted FDA. FDA has poor aqueous solubility; thus the enzymatic assay was performed with PLE in 5% DMSO in pH 7.4 PBS buffer. To maintain similar experimental conditions, probe 1 was also dissolved in the same solvent system at the same concentration for this assay. The results showed that probe 1 did not have as high a turn-on ratio as FDA, but the reaction rates were comparable (ESI†, Fig. S5). Notably, FDA is hydrolytically unstable while the ODF probe is not: a stability test in 1% DMSO in PBS buffer (pH 7.4) showed significant decomposition for FDA at room temperature, while probe 1 did not decompose measurably after 24 h (ESI†, Fig. S6).
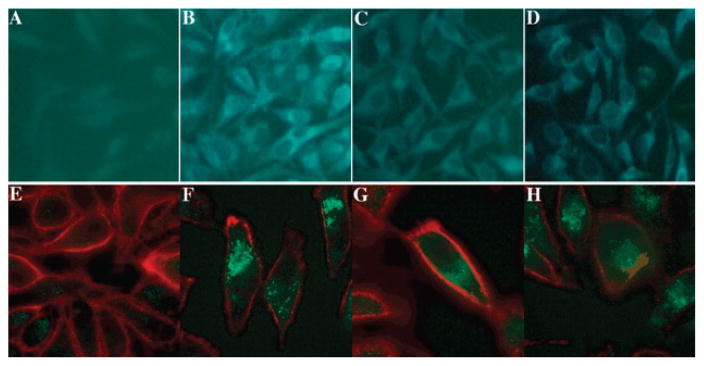
To determine if the probes were active in live cells, HeLa cells were incubated with probes 1 and 2. Results indicate that both probes 1 and 2 could permeate cellular membranes, and are effectively turned on in vivo (Fig. 4 and S7 †). Probe 2 showed a brighter fluorescence turn-on signal than probe 1, consistent with the earlier screening results where probe 2 was found to be a better substrate for esterases and lipases.
Fig. 4.
Images of esterase sensing in HeLa cells. (a–d) True color images of HeLa cells under 20X microscope. (a) Negative control (HeLa cells only); (b) positive control (incubated with fluorophore 5′-SSYYYY-3′, the product of enzymatic cleavage); (c) incubated with probe 1; (d) incubated with probe 2. (e–h) False-color overlay images of HeLa cells under 63X confocal microscope, showing outer cell membrane marker (red; wheat germ agglutinin Alexa Fluor 633 membrane dye) alongside probe signals (cyan). (e) Negative control (HeLa cells only); (f) positive control (incubated with fluorophore 5′-SSYYYY-3′); (g) incubated with probe 1; (h) incubated with probe 2.
Compared to traditional fluorophore-based sensors, our new ODF-based fluorescence sensor design for esterases/lipases has several advantages. We used ODF as the fluorophore of choice, which confers good aqueous solubility due to the negatively charged DNA backbone, enhanced quenching characteristics, and ease of synthesis on an automated DNA synthesizer. Our design provides a modular structure whereby the ODF fluorophore could be easily constructed with multiple sequences27 to give other emission colors in the sensor without the need to modify conjugation chemistries.
Also unlike previous sensors in which the enzymatic activity is dependent on the structure of the fluorophore, the current modular design allows more freedom of substrate selection. Indeed, our results have already shown that different enzymes have different preferences for substrates. By changing only the substrate moiety, we may potentially find specific substrates for specific enzymes of interest. This can greatly facilitate substrate and enzyme screening in drug discovery and biological studies. In addition, quenching efficiency may well be improved by selecting a more efficient quencher,32,33 which can easily be varied without altering the remainder of the structure. The current proof-of-principle suggests that the favorable photophysical properties of ODFs might be harnessed to design sensors in a variety of colors, all excited at a single wavelength, by simply changing the ODF sequences. This presents substantial potential for use in real-time multi-enzyme, multi-substrate assays in complex biological systems.
Supplementary Material
Acknowledgments
This work was supported by the U.S. National Institutes of Health (GM067201). YNT acknowledges an A*STAR NSS scholarship.
Footnotes
Electronic supplementary information (ESI) available: Details of organic and oligonucleotide synthesis, enzymatic screening, cell culture and stability tests. See DOI: 10.1039/b926338a
Notes and references
- 1.Goddard JP, Reymond JL. J Am Chem Soc. 2004;126:11116–11117. doi: 10.1021/ja0478330. [DOI] [PubMed] [Google Scholar]
- 2.Goddard JP, Reymond JL. Trends Biotechnol. 2004;22:363–370. doi: 10.1016/j.tibtech.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Kohl T, Heinze KG, Kuhlemann R, Koltermann A, Schwille P. Proc Natl Acad Sci U S A. 2002;99:12161–12166. doi: 10.1073/pnas.192433499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao W, Xing B, Tsien RY, Rao J. J Am Chem Soc. 2003;125:11146–11147. doi: 10.1021/ja036126o. [DOI] [PubMed] [Google Scholar]
- 5.Fonovic M, Bogyo M. Curr Pharm Des. 2007;13:253–261. doi: 10.2174/138161207779313623. [DOI] [PubMed] [Google Scholar]
- 6.Boland W, Frossl C, Lorenz M. Synthesis. 1991:1049–1072. [Google Scholar]
- 7.Munoz-Torrero D. Curr Med Chem. 2008;15:2433–2455. doi: 10.2174/092986708785909067. [DOI] [PubMed] [Google Scholar]
- 8.Vandevoorde S. Curr Top Med Chem. 2008;8:247–267. doi: 10.2174/156802608783498005. [DOI] [PubMed] [Google Scholar]
- 9.Cummings BS. Biochem Pharmacol. 2007;74:949–959. doi: 10.1016/j.bcp.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 10.Liederer BM, Borchardt RT. J Pharm Sci. 2006;95:1177–1195. doi: 10.1002/jps.20542. [DOI] [PubMed] [Google Scholar]
- 11.Lavis LD. ACS Chem Biol. 2008;3:203–206. doi: 10.1021/cb800065s. [DOI] [PubMed] [Google Scholar]
- 12.Satoh T, Hosokawa M. Annu Rev Pharmacol Toxicol. 1998;38:257–288. doi: 10.1146/annurev.pharmtox.38.1.257. [DOI] [PubMed] [Google Scholar]
- 13.Gilham D, Lehner R. Methods. 2005;36:139–147. doi: 10.1016/j.ymeth.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Fojan P, Jonson PH, Petersen MT, Petersen SB. Biochimie. 2000;82:1033–1041. doi: 10.1016/s0300-9084(00)01188-3. [DOI] [PubMed] [Google Scholar]
- 15.Grognux J, Reymond JL. Mol BioSyst. 2006;2:492–498. doi: 10.1039/b609275f. [DOI] [PubMed] [Google Scholar]
- 16.Sicart R, Collin MP, Reymond JL. Biotechnol J. 2007;2:221–231. doi: 10.1002/biot.200600181. [DOI] [PubMed] [Google Scholar]
- 17.Yang Y, Babiak P, Reymond JL. Org Biomol Chem. 2006;4:1746–1754. doi: 10.1039/b601151a. [DOI] [PubMed] [Google Scholar]
- 18.Wichmann O, Gelb MH, Schultz C. Chem Bio Chem. 2007;8:1555–1569. doi: 10.1002/cbic.200600462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lavis LD, Chao TY, Raines RT. ACS Chem Biol. 2006;1:252–260. doi: 10.1021/cb600132m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodroofe CC, Lippard SJ. J Am Chem Soc. 2003;125:11458–11459. doi: 10.1021/ja0364930. [DOI] [PubMed] [Google Scholar]
- 21.Gao J, Strassler C, Tahmassebi D, Kool ET. J Am Chem Soc. 2002;124:11590–11591. doi: 10.1021/ja027197a. [DOI] [PubMed] [Google Scholar]
- 22.Gao J, Watanabe S, Kool ET. J Am Chem Soc. 2004;126:12748–12749. doi: 10.1021/ja046910o. [DOI] [PubMed] [Google Scholar]
- 23.Cuppoletti A, Cho Y, Park JS, Strassler C, Kool ET. Bioconjugate Chem. 2005;16:528–534. doi: 10.1021/bc0497766. [DOI] [PubMed] [Google Scholar]
- 24.Wilson JN, Gao J, Kool ET. Tetrahedron. 2007;63:3427–3433. doi: 10.1016/j.tet.2006.07.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teo YN, Kool ET. Bioconjugate Chem. 2009;20:2371–2380. doi: 10.1021/bc9003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teo YN, Wilson JN, Kool ET. Chem–Eur J. 2009;15:11551–11558. doi: 10.1002/chem.200901607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teo YN, Wilson JN, Kool ET. J Am Chem Soc. 2009;131:3923–3933. doi: 10.1021/ja805502k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson JN, Teo YN, Kool ET. J Am Chem Soc. 2007;129:15426–15427. doi: 10.1021/ja075968a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conlon P, Yang CJ, Wu Y, Chen Y, Martinez K, Kim Y, Stevens N, Marti AA, Jockusch S, Turro NJ, Tan W. J Am Chem Soc. 2008;130:336–342. doi: 10.1021/ja076411y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bouquin N, Malinovskii VL, Haner R. Chem Commun. 2008:1974–1976. doi: 10.1039/b802193g. [DOI] [PubMed] [Google Scholar]
- 31.Winnik FM. Chem Rev. 1993;93:587–614. [Google Scholar]
- 32.Johansson MK, Cook RM. Chem–Eur J. 2003;9:3466–3471. doi: 10.1002/chem.200304941. [DOI] [PubMed] [Google Scholar]
- 33.Johansson MK. Methods Mol Biol. 2006;335:17–29. doi: 10.1385/1-59745-069-3:17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.