Abstract
Environmental and metabolic processes shape the profile of glycoprotein glycans expressed by cells, whether in culture, developing tissues, or mature organisms. Quantitative characterization of glycomic changes associated with these conditions has been achieved historically by reductive coupling of oligosaccharides to various fluorophores following release from glycoprotein and subsequent HPLC or capillary electrophoretic separation. Such labeling-based approaches provide a robust means of quantifying glycan amount based on fluorescence yield. Mass spectrometry, on the other hand, has generally been limited to relative quantification in which the contribution of the signal intensity for an individual glycan is expressed as a percent of the signal intensity summed over the total profile. Relative quantification has been valuable for highlighting changes in glycan expression between samples; sensitivity is high, and structural information can be derived by fragmentation. We have investigated whether MS-based glycomics is amenable to absolute quantification by referencing signal intensities to well-characterized oligosaccharide standards. We report the qualification of a set of N-linked oligosaccharide standards by NMR, HPLC, and MS. We also demonstrate the dynamic range, sensitivity, and recovery from complex biological matrices for these standards in their permethylated form. Our results indicate that absolute quantification for MS-based glycomic analysis is reproducible and robust utilizing currently available glycan standards.
Keywords: quantification, N-glycan, permethylation, standard, mass spectrometry
Graphical Abstract

INTRODUCTION
The complete characterization of glycans isolated from biological materials requires multiple complementary analytical approaches. The techniques available currently each possess their own characteristic benefits and limitations. The most structurally informative technique, NMR, requires amounts of material that are frequently not available from biological samples. Other more sensitive methods, such as HPLC (with amperometric or fluorescence detection), gel electrophoresis, capillary electrophoresis (CE), and mass spectrometry (MS), are frequently coupled with orthogonal techniques, such as methylation/linkage analysis, exoglycosidase digestion, or affinity chromatography to characterize glycan structures released from samples at the scale usually encountered in glycomics experiments.1–3 Among these approaches, HPLC or CE coupled to fluorescence detection of tagged glycans possesses the significant advantage that glycan quantification can be achieved by referencing unknown peak heights to a known amount of a tagged standard.4–11 Because fluorescence yield is independent of the glycan structure, a single standard compound is sufficient to quantify all detectable glycans in a sample profile. For tagged glycans, structural characteristics can be further assigned based on exoglycosidase sequencing and on chromatographic retention relative to known standards.12
Because of its sensitivity and speed, as well as the rich structural data obtained by tandem approaches (MS/MS and MSn), mass spectrometry is becoming one of the most broadly employed technologies for glycomic analysis of biological materials.13–27 However, quantification of glycans by MS remains largely underdeveloped.28 Significant questions remain unanswered regarding whether and under what conditions mass spectral signals can be used to quantify absolute amounts of glycan structures. Currently, the predominant practice is for individual components of glycan profiles generated by MS to be quantified relative to each other. This calculation describes each glycan as a percent of the total signal summed across the whole spectrum. Although this normalized parameter has been broadly useful for comparing major changes in profiles across samples or biological conditions, its value for characterizing minor glycans is frequently subject to the undue influence of the major components; teasing apart the impact of variability in numerator and denominator quantification can be difficult for low abundance (but biologically significant) glycans. Generating methods for absolute quantification by MS would allow profile changes to be assessed glycan-by-glycan, independent of variations in the whole profile. A set of well-characterized oligosaccharide standards with demonstrated robustness for quantification of glycans in biological materials is essential to realize this potential.
Underivatized glycans are hydrophilic; their chemistries are dominated by the spatial distribution of hydroxyl, acetamido, and carboxyl moieties. Therefore, the response factors for native glycans in mass spectrometers, which are driven largely by ionization efficiency, vary in proportion to the content of these polar determinants. Permethylation neutralizes these differences, enhancing desolvation and ionization efficiency. Although never rigorously tested, published data has suggested that molar responses for permethylated glycans may be relatively uniform.24,29,30 If true, quantification of MS signals derived from permethylated glycans could be achieved by reference to a single standard or a small set of standards. The aim of this study was to assess the validity of the proposition that permethylation makes it possible to quantify the profile of N-linked glycans in biological materials by reference to a permethylated standard or set of standards. To that end, we have investigated linearity of quantification and recovery from a biological matrix using permethylated glycan standards. We also compare quantification across analytic platforms (1H-NMR, nanospray-iontrap/MS, MALDI-TOF/MS, HPLC with fluorescence detection) to qualify glycan standards for use as quantification standards.
EXPERIMENTAL SECTION
Materials and Reagents
PNGaseF (N-glycanase) was obtained from Prozyme (San Leandro, CA). N-Glycan standards were obtained from the Consortium for Functional Glycomics (CFG) and from The Scripps Research Institute (James Paulson). Sodium hydroxide (50%) was purchased from Fisher Scientific. Sep-Pak C18 disposable extraction columns were obtained from Waters Corporation (Milford, MA, USA). Malto-series oligosaccharides, trypsin, bovine pancreatic ribonuclease B (RNaseB), and all other chemical reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA). Malto-series oligosaccharides were also obtained from Wako Chemicals (Richmond, VA) and Cosmo Bio USA (Carlsbad, CA).
Analytical Platforms
Analytic platforms representative of the major instrumentations currently in use for glycomic analysis were used to analyze glycan standards. Mass spectra were obtained by MALDI-TOF using a Bruker Microflex LRP operating in reflectron mode and by nanospray ionization using a Thermo Fisher LTQ/Orbitrap. Tandem mass spectrometry (MSn) was also performed by NSI-LTQ/Orbitrap. HPLC analysis of fluorescent-labeled glycan standards was performed as previously described using normal phase separations on a TSK-amide-80 column with a 2695 Alliance separations module and 2475 fluorescence detector (Waters, Milford, MA).12,31 1H-NMR spectra were acquired on a Varian Inova 600 MHz spectrometer.
Permethylation
All glycan standards and samples were permethylated prior to MS analysis according to the method of Anumula and Taylor.32 Malto-series oligosaccharide standards were permethylated with 12C-, 13C-, or deuterated methyl iodide (12C-MeI, 13C-MeI, or 12CD3I, respectively).
Glycan Mass Spectrometry
Permethylated glycan samples and standards were analyzed on MALDI-TOF/MS and NSI-MS platforms. For MALDI analysis, permethylated glycans were analyzed in positive ion mode using α-dihyroxybenzoic acid (DHBA, 20 mg/mL solution in 50% methanol:water) as matrix.24 For nanosprayion trap analysis, infusion and ionization conditions were optimized across three isotopic variants (12C-MeI, 13C-MeI, and 12CD3I) of permethylated, equimolar mixtures of malto-series oligosaccharides (maltotetraose, maltopentaose, maltohexaose, and maltoheptaose) by comparing the signal intensities achieved in three different infusion buffers: 1 mM NaOH in 50% methanol, in 50% 2-propanol, and in a mixture of 50% 2-propanol and 50% methanol (1:1, v/v). Maximum signal intensities were achieved by reconstitution and infusion of glycans in 50% methanol containing 1 mM NaOH using a nanospray source at a syringe flow rate of 0.40 μL/min and capillary temperature set to 210 °C.24 The instrument was tuned with permethylated oligosaccharide standards in positive ion mode. For fragmentation by collision-induced dissociation (CID) in MS/MS, normalized collision energy of 30% was applied. Most permethylated glycan components, including malto-series oligosaccharides, were identified as singly, doubly, and triply charged, sodiated species [M + Na] in positive mode. Peaks for all charge states were deconvoluted by charge state and summed for quantification using the Xtract functionality of the Xcalibur software package version 2.0 (Thermo Fisher Scientific) as previously described.33
Analysis of Glycan Standards
Two N-linked glycan standards (standard 107 and 108) were received from the Consortium for Functional Glycomics (CFG) and an additional N-linked glycan standard (standard 121) was received as a gift from Jim Paulson at The Scripps Research Institute (Table 1). Standards 107, 108, and 121 were resuspended in water and dispensed nominally into 2.5 nmol aliquots based on the previous characterization of the supplier. Following drying by vacuum centrifugation, the N-glycan standard aliquots were stored frozen at −80 °C. Aliquots of the N-linked standards were subjected to reductive labeling with 2-aminobenzamide (2-AB) and subsequently analyzed by HPLC coupled to fluorescence detection as previously described.12,31 The structures of the labeled N-linked standards were validated by exoglycosidase sequencing with HPLC and fluorescence detection and by NMR.12,34 For NMR analysis, N-linked glycan standards were deuterium exchanged by dissolving in D2O and lyophilization. Dried samples were resuspended in 80 μL of D2O, and 5 μL of 1% acetone in D2O was added as internal reference. The samples were placed in 3 mm Shigemi NMR tubes, and 1-D Proton, TOCSY, and NOESY NMR spectra, run with water presaturation, and gradient-enhanced COSY and HSQC spectra were acquired on a Varian Inova-600 MHz spectrometer at 25 °C. Chemical shifts were measured relative to internal acetone (δH = 2.218 ppm, δC = 33.00 ppm). Standard Varian acquisition parameters were used (Table S-1). Malto-series oligosaccharide standards were desiccated under vacuum, quantitatively weighed, and dissolved in water to produce stock solutions at 1 mM. The stock solutions were distributed into 10 μmol aliquots and redried by vacuum centrifugation before being stored at −80 °C. Mixtures of malto-series oligosaccharide standards containing various combinations of maltotri-, tetra-, penta-, hexa-, and heptaoli-gosaccharide (dp3–7, respectively) were permethylated and analyzed by NSI-LTQ/Orbitrap MS and MALDI-TOF MS. The malto-series mixtures were also analyzed by fluorescence detection as 2-AB derivatives following HPLC separation.
Table 1.
N-Glycan and Malto-Series Oligosaccharide Standards for Glycan Quantification
| Standard Identifier | Trivial Name | Structurea | Amount Obtained | Native Mass | Permethylated Mass (m+Na)+ |
|---|---|---|---|---|---|
| 107 | NGA2B or A2(2,2)B |

|
9 mg | 1519.56 | 1906.96b |
| 108 | NGA3B(l–4) or A3(2,4,2)B |

|
100 mg | 1722.64 | 2152.09 |
| 121 | A2G2S(6)2 |

|
100 mg | 2222.78 | 2792.38 |
| dp3 | maltotriose |
|
not limited | 504.17 | 681.33 |
| dp4 | maltotetraose |
|
not limited | 666.22 | 885.43 |
| dp5 | maltopentaose |
|
not limited | 828.27 | 1089.53 |
| dp6 | maltohexaose |
|
not limited | 990.33 | 1293.63 |
| dp7 | maltoheptaose |
|
not limited | 1152.38 | 1497.73 |
Monosaccharide symbols in all figures follow the SNFG system.37
Mass of glycan as singly charged sodium adduct permethylated with 12C-MeI.
Quantification of Glycans Released from the Well-Characterized Glycoprotein RNaseB
N-Linked glycans were released from 1 mg of RNaseB glycoprotein (80% purity) by two approaches: (1) glycopeptides were enriched following tryptic digestion of RNaseB by Sep-Pak C18 cartridge chromatography, and the glycans were then released by PNGaseF treatment (18 h at 37 °C), or (2) N-glycans were released by PNGaseF treatment (18 h at 37 °C) of denatured but undigested RNaseB (24, 35). For both approaches, released N-glycans were cleaned up by passage over Sep-Pak C18 and permethylated prior to MS analysis. Permethylated RNaseB glycans were analyzed by NSI-LTQ/Orbitrap MS and quantified relative to a known amount of maltotetraose (10 pmol of dp4) that was permethylated with heavy methyliodine (13C-MeI) and spiked into the sample prior to infusion.
Recovery of Glycan Standards from Biological Matrices
To assess the effect of biological matrices on detection and quantification, permethylated standards were spiked into two different biological samples. First, a mixture of N-glycan standards permethylated with 12C-MeI was added to N-glycans prepared from glycoproteins harvested from Drosophila melanogaster embryos.24 The mixture of N-glycan standards and the Drosophila glycans were each analyzed separately and as a mixture. The N-glycan standards were prepared as an equimolar mixture at a known concentration and were added to the Drosophila glycans to yield standard peak heights comparable in magnitude to the major Drosophila structures. Matrix interference was quantified by calculating the recovery of added standard. Second, a similar experiment was performed using mouse brain N-glycans. Glycoproteins were extracted from the frontal cortex of a single wildtype mouse by the same method used to prepare Drosophila embryo glycoproteins.24 Using this procedure, 51 mg of protein-enriched solids were harvested from 286 mg wet weight of brain, and 3 mg aliquots of the solids were used for N-glycan preparation and analysis. For quantification of mouse brain N-glycans, N-glycan standards and malto-series glycans were permethylated with 13C-MeI and mixed together at known concentrations before being spiked into mouse brain glycans that had been permethylated with 12C-MeI prior to analysis. Matrix interference was quantified by calculating standard recovery, and endogenous mouse brain glycans were quantified relative to each of the spiked standards.
RESULTS AND DISCUSSION
Availability of Glycan Standards
Ideally, a set of well-characterized N-linked glycan standards, obtainable in reasonable amounts and representative of the full range of biologically relevant glycan complexities, would be available for use as quantification standards. Additionally, candidate standards would be most useful if they possessed a free, underivatized reducing end. This preference reflects the broad practice in glycomic analysis of using PNGaseF to release N-linked glycans from glycoproteins, an enzyme that removes the glycan en bloc, generating a glycan with an intact reducing end.35 Although a growing number of commercial and academic sources offer synthetic or purified N-linked glycans in small quantities (less than 10 μg), full qualification of these glycans would significantly deplete the available material. Therefore, we chose to pursue standards that we could acquire in larger quantities. One useful source was the Consortium for Functional Glycomics (CFG), which maintains a reagent bank of oligosaccharides for use by investigators (http://www.functionalglycomics.org). Most of these oligosaccharides carry linker arms on their reducing end for immobilization on surfaces.36 Some, however, are available with free reducing ends (Table 1). Monosaccharide symbols used throughout this work follow the Symbol Nomenclature for Glycans (SNFG) system.37 CFG standards 107 and 108 were made available to us in 100 μg amounts. Our interaction with the CFG also facilitated the acquisition of 9 mg of standard 121, a sialylated complex glycan (courtesy of James Paulson, The Scripps Research Institute). We also acquired a larger amount (>100 mg each) of defined maltose oligosaccharide polymers (Sigma, Wako, and Cosmo Bio) comprising a linear series of size standards from 3 to 7 hexoses in length. We refer to the malto-series oligosaccharides by their degree of polymerization (dp number, dp3–7).
Qualification of Glycan Standards
The N-linked glycan standards 107, 108, and 121 (107–121) were assessed for purity and quantity by several approaches. Each standard was subjected to 1-D and 2-D proton NMR for verification of linkage and anomericity, mass spectrometry (MALDI-TOF and NSI-LTQ/Orbitrap MS) for confirmation of expected mass, and HPLC with reductive labeling for purity by mass. 1-D and 2-D proton NMR of standards 107–121 yielded carbon glycosylation shifts and NOE contacts in agreement with the structures presented in Table 1 (Tables S-1–5 and Figures S-1–3). Sequential exoglycosidase sequencing coupled to HPLC separation of 2-AB-labeled standards 107–121 were also consistent with the proposed structures (Figures S-4–6). Each of the N-linked glycan standards exhibited a predominant species (ranging between 74 and 83% of the total signal intensity by NSI-MS, 68–93% by MALDI-TOF, and 62–93% by HPLC) and an additional minor species corresponding to the absence of one or more nonreducing terminal residues (Figures S-4–8 and Table S-6). The heterogeneity in the standards was comparable across all platforms (MALDI- and NSI-MS, HPLC, and NMR), indicating that it likely reflects the chemical nature of the standard and not an artifact of the analytic approach (Figure 1A). Disassembly of the permethylated N-glycan standards 107, 108, and 121 by NSI-LTQ/Orbitrap MS/MS and MSn yielded fragmentation patterns consistent with the expected structures (Figure S-9).
Figure 1.

Heterogeneity and mass purity of N-glycan standards. Aliquots of N-glycan standards 107, 108, and 121 were analyzed as 2-AB derivatives by HPLC or as permethylated glycans by MALDI-TOF MS and NSI-LTQ/Orbitrap MS. All three analytic platforms revealed the presence of a major component, corresponding to the expected structure, and minor but structurally related components. (A) Plot of the percent of the total signal contributed by the expected structure across all three platforms (mean ± SD, n = 6). The low magnitude of the error indicates that the detected heterogeneity of the standards reflects true structural diversity in the material and was not generated by the analytic technique. (B) Mass purity for the N-glycan standards calculated by referencing 2-AB fluorescence of N-glycan peaks to known amounts of 2-AB-labeled malto-series standards following HPLC separation or by referencing N-glycan signals to an acetone internal standard by NMR. Mass purity values obtained by fluorescence-coupled HPLC and by NMR were averaged for each of the N-glycan samples (mean ± SD, n = 10).
The quantification approach described here assumes high-efficiency permethylation to avoid underpermethylated glycans, which complicate interpretation of MS data. In our hands, careful preparation of permethylation reagents and attention to sample size (no more than 25 μg of a highly glycosylated protein per 550 μL of permethylation reaction) greatly enhances permethylation efficiency.38 Using optimized methods, permethylation can proceed to near completion, even for glycans derived from a biological matrix. For example, following permethylation of rat plasma N-linked glycans, only 1.4% of the endogenous glycan identical to the N-glycan standard 121 was detected as underpermethylated, lacking only a single methyl group (Figure S-10).
To assess the purity of standards 107–121, we quantitatively labeled known amounts of malto-series oligosaccharides (dp3–7) with 2-AB and analyzed them by HPLC coupled to fluorescence detection. Molar response factors were generated for each malto-series standard from the resulting data (Figure S-11). The 5 malto-series standards gave nearly identical results, from which a mean molar response factor was calculated (Table S-6). Fluorescence signals for HPLC-resolved 107–121 were converted to stock concentrations by referencing the detected signal for all glycan peaks to the average malto-series molar response factor and the mass purity of each N-glycan standard was subsequently calculated relative to the mass of the original stock aliquot. Stock concentrations for 107–121 were also independently calculated from NMR data by referencing standard signals to the known amount of acetone added as internal reference. The two methods, HPLC coupled to fluorescence detection and NMR referenced to internal standard, gave mass purity and stock concentrations in good agreement (Figure 1B and Table S-6). For all subsequent experiments in which glycan amounts were calculated by reference to N-glycan standards, signal intensities associated with the major components of the standards, which in all cases corresponded to the expected structures, were utilized. The signal intensities associated with the major components were assigned to represent an adjusted amount of the expected glycans based on the relative abundances of all of the glycans detected in the standard. The consistency of the observed molar response factors for each of the malto-series oligosaccharides and the reproducibility of the stock concentrations measured for standards 107–121 provided a basis for investigating the usefulness of these compounds as mass spectrometry quantification standards.
Linearity of MS and MS/MS Responses
All three N-linked glycan standards gave well-behaved MS responses upon dilution. Linear standard curves were obtained for each structure across 4 orders of magnitude (from 0.5 nM to 6.5 μM at infusion) with regression coefficients (r2) of 0.9958, 0.9856, and 0.9987 for standards 107, 108, and 121, respectively. Combining the data for all three standards, a linear response curve was also observed with an overall regression coefficient of 0.9561 (Figure 2). Linearity of response upon dilution was also observed for the major fragment ions associated with each standard (Figure 3). MS/MS analysis produced consistent ratios between the two major fragment ions for each standard at dilutions spanning 4 orders of magnitude (5.6 ± 0.2 for standard 107, 4.5 ± 0.2 for standard 108, and 4.3 ± 0.1 for standard 121; mean ± SD for n ≥ 5 measurements for each N-glycan standard). The broadly linear responses detected for the N-linked glycan standards indicate that MS-based quantification by standards possesses a useful dynamic range. The conservation of response linearity at MS2 suggests the possibility, although not directly tested here, that quantification may also be achieved utilizing diagnostic fragments that can distinguish between isobaric species.
Figure 2.
Linearity of MS responses for N-glycan standards. Known amounts of N-glycan standards 107, 108, and 121 were permethylated and analyzed as a dilution series ranging from 0.5 nM to 6.5 μM at infusion. Regression analysis of all data points from the three standards yields a high coefficient of correlation (r2 > 0.95) and linearity over 4 orders of magnitude. Black square, standard 107; red diamond, standard 108; gray triangle, standard 121.
Figure 3.

Linearity of MS/MS responses for major N-glycan standard fragments. For each of the N-glycan standards 107, 108, and 121, MS/MS spectra were obtained by NSI-LTQ/Orbitrap mass spectrometry. Signal intensities for the two most abundant fragment ions for each standard were quantified at multiple dilutions of the parent compound. (A) Fragmentation of standard 107 (parent m/z = 946.98, doubly charged) yielded major products at m/z = 835 (black diamonds, loss of 1 HexNAc) and at m/z = 706 (gray circles, loss of 2 HexNAc residues). (B) Fragmentation of standard 108 (parent m/z = 1087.54, triply charged) yielded major products at m/z = 958 (black diamonds, loss of 1 HexNAc) and at m/z = 828 (gray circles, loss of 2 HexNAc residues). (C) Fragmentation of standard 121 (parent m/z = 1407.68, doubly charged) yielded major products at m/z = 1220 (black diamonds, loss of 1 NeuAc, doubly charged) and at m/z = 821 (gray circles, loss of 1 NeuAc, triply charged). For all three N-glycan standards, regression analysis of MS/MS fragment signal intensities as a function of standard concentration at infusion were linear with r2 > 0.996.
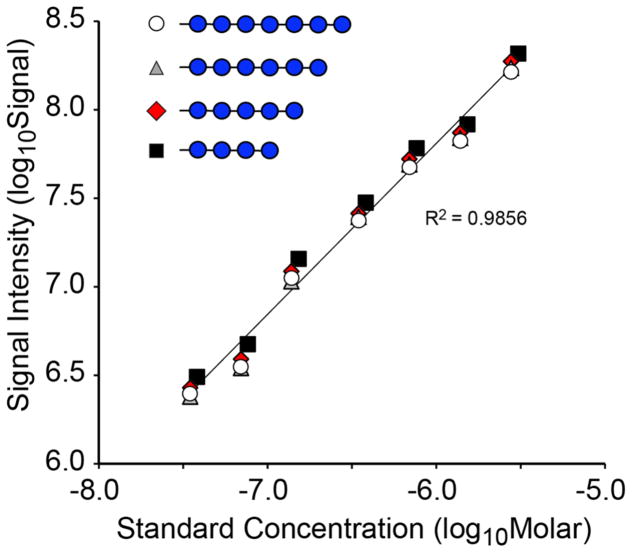
Malto-series oligosaccharides are available in reasonably large amounts at high purity, cover a broad mass range, and can be selected or modified so that they are not isobaric with glycans of interest in the test material. Therefore, malto-series oligosaccharides composed of between 3 and 7 hexose units (dp3–7) were tested for suitability as MS quantification standards. Although the dp3 maltotriose was easily detectable and gave well-behaved MS responses, its small size presented problems for commercially available signal processing software. Specifically, the Xtract functionality of the Xcalibur software package (Thermo-Fisher) routinely underestimated the amount of dp3 oligosaccharide present compared to manual interpretation of the spectra. This underestimation arises from the software’s dependence on using average isotopic distributions characteristic of amino acids for deconvoluting charge states; no currently available commercial deconvolution software has the capacity to use average isotopic distributions of permethylated monosaccharides, which differ from average amino acids with respect to the relative abundance of carbon, nitrogen, hydrogen, and oxygen for charge state deconvolution. The error imposed by matching glycan isotope envelopes with amino acid isotope distributions becomes less pronounced as the glycan structure increases in size. Accordingly, the malto-series oligosaccharides dp4–7 gave well-behaved and consistent MS responses upon dilution. Linear standard curves were obtained for each dp standard across the tested range (from 30 nM to 3 μM at infusion) with regression coefficients of 0.9877, 0.9877, 0.9880, and 0.9869 for dp4–7, respectively. Combining the data for all four malto-series standards, a linear response curve was also observed with an overall regression coefficient of 0.9856 (Figure 4).
Figure 4.
Linearity of MS responses for malto-series standards. Known amounts of malto-series oligosaccharide standards dp4–7 were permethylated and analyzed as a dilution series ranging from 30 nM to 3 μM at infusion. Regression analysis of all data points from the four malto-series standards yields a high coefficient of correlation (r2 > 0.98) and linearity over the assayed range. Black square, dp4; red diamond, dp5; gray triangle, dp6; white circle, dp7.
For the impact of glycan mass on signal response to be investigated directly, the slopes of the standard curves for the three N-glycan standards (107, 108, and 121) and for the 4 malto-series standards (dp4–7) were plotted as a function of their molecular weights (Figure 5). A nominal trend toward increased response was observed as the mass of the standard increases. Although the maximum difference was only 8% across the tested mass range, the difference between the molar response for the largest glycan (standard 121) was statistically significant (p < 0.005) in comparison with all of the other standards except for the standard closest to its own mass (standard 108). Likewise, the difference between the molar response for standard 108 was statistically significant in comparison with all of the other standards except standard 121, albeit at a lower confidence level (p < 0.05). The average slope ± standard deviation was 0.98 ± 0.03 log10(MS signal)·log10(pM)−1 from 885–2831 Da, a mass range that encompasses most N-linked glycans released from biologically relevant glycoprotein sources. The minimal size of the incremental increase in signal response associated with increased glycan mass is consistent with the hypothesis that permethylation diminishes the impact of glycan functional groups (hydroxyl, carboxylate, and acetamido) and molecular shape (linear, branched, bisected, terminally sialylated) on ionization efficiency and further supports the usefulness of glycan standards as quantification tools.
Figure 5.
Molar responses for permethylated N-glycan and malto-series standards are independent of glycan mass. The slopes of the standard curves for N-glycan standards 107, 108, and 121 and malto-series standards dp4–7 were plotted as a function of the mass of the standard. Minimal deviation across the indicated mass range was detected, indicating that permethylation effectively equalizes ionization efficiency across a broad range of glycan structures.
Cross-Platform Performance and Alternative Derivatization of Malto-Series Oligosaccharide Standards
The malto-series oligosaccharides dp3–7 were analyzed by NSI-LTQ/Orbitrap MS, MALDI-TOF MS, and HPLC to assess their performance across commonly used analytic platforms. The five glycans were mixed together at an equimolar ratio except for dp4, which was added at 10% molar excess to distinguish the dp4 peak from others. Once the NSI-MS data were deconvoluted for charge state using Xtract, the relative signal characteristics for the three analytic platforms were well-conserved across the platforms (Figure 6). Molar response factors were calculated as detector signal response per compound concentration (pM) for nine HPLC runs and seven NSI-LTQ/Orbitrap MS runs for each dp (Figure 7). For dp4–7, the molar response factors were 12.6 ± 0.2 and 68.1 ± 2.3 (signal/pM, mean ± SD) by HPLC and NSI-MS, respectively (Table S-6). As discussed above, Xtracted MS signals for dp3 under-represented its abundance, and therefore, the molar response factor for dp3 diverged appreciably from those of the other malto-series glycans. The relatively low and comparable variability of the molar response factors detected by MS and by HPLC support the usefulness of the dp4–7 malto-series oligosaccharides as quantification standards.
Figure 6.
Comparability of signal responses for malto-series standards across three analytic platforms. Malto-series oligosaccharide standards dp3–7 were analyzed as permethylated derivatives by NSI-LTQ/Orbitrap MS ((A) full MS; (B) charge states deconvoluted by Xtract), by MALDI-TOF MS (C), or as 2-AB derivatives by HPLC (D). The malto-series standard mixture was prepared with 10% more dp4 than the other malto-oligosaccharides to readily confirm elution positions and standard identities.
Figure 7.
Molar response factors for malto-series standards measured by HPLC fluorescence detection and mass spectrometry. Molar response factors, calculated as detector signal per pM concentration of standard, were measured for the malto-series oligosaccharide standards dp3–7 at a range of concentrations (35 nM to 3 μM). The mean ± SEM of the molar response factors across the analyzed concentrations for each standard is presented for HPLC coupled to fluorescence detection (black squares; n = 9 for each standard) and for NSI-MS (red diamonds; n = 7 for each standard). For the malto-series standards dp4–7, the relationship between molar response and standard mass is identical for HPLC and MS.
The lack of HexNAc residues in the dp4-7 standards minimizes the likelihood that these compounds would have the same mass/charge ratio as that of N-linked glycans released from glycoproteins isolated from biological sources. However, many biological samples possess varying amounts of a laddered polyhexose contaminant, probably from glycogen or from a cellulosic source, which would be isobaric with the malto-series standards and would negatively impact quantification. The mass of the dp4–7 standards can be shifted away from the endogenous sample hexose ladder by differential permethylation with reagents carrying stable isotopes. To assess the potential impact of alternative permethylation reagents on dp4–7 detection, the malto-series glycans were permethylated with 12C-, 13C-, or deuterated methyl iodide (12C-MeI, 13C-MeI, or CD3I). The signal detected for dp4-7 following permethylation with 12C-MeI or with CD3I was compared to the signal detected for dp4–7 following permethylation with 13C-MeI (Figure 8). Signal intensities did not vary as a result of alternative derivatization, allowing flexibility in positioning the standard peaks relative to peaks of interest in biological samples.
Figure 8.
Permethylation of malto-series standards with methyl iodide containing stable isotopes of carbon or hydrogen yields equivalent MS responses. The indicated malto-series standards were permethylated with 12C-, 13C-, or deuterated methyl iodide (12C-MeI, 13C-MeI, or CD3I, respectively) and analyzed by NSI-LTQ/Orbitrap MS. Instrument responses for standards permethylated with 12C-MeI or CD3I were normalized to the responses obtained for standards permethylated with 13C-MeI, which were set to 100. Normalized responses were averaged for all replicates of the standards permethylated with 12C-MeI or CD3I and are plotted as mean ± SD (n = 3 for each standard). The minor variation around the mean indicates that standards can be permethylated with reagents that induce mass shifts most suitable for the biological sample being studied.
Quantification of RNaseB N-Glycans
RNaseB possesses one high-mannose-type N-linked glycan per molecule of protein, providing a simple test of glycan quantification.39–41 On the basis of the mass of the non-glycosylated protein backbone (RNaseA) and on the mass of the dominant N-linked glycan shown previously to be attached to RNaseB (M5N2), the expected contribution of glycan mass to the total mass of the protein is 8.7%. N-Glycans were prepared from RNaseB by PNGaseF digestion of denatured intact protein or by PNGaseF digestion of tryptic glycopeptides. N-Glycans released from RNaseB by both methods were permethylated and spiked with 13C-labeled maltotetraose (dp4) as an exogenous quantification standard prior to analysis by NSI-MS. Both glycan preparation methods yielded comparable results; quantification by exogenous standard detected 1.37 μg of N-glycan from 16.0 μg of RNaseB, corresponding to 8.6% of the RNaseB mass attributable to glycan, which is 98% of the expected amount.
Recovery and Resolution of Exogenous Standard from Biological Matrices
To assess whether a biological matrix might suppress detection of exogenous standards, the permethylated N-glycan standards 107, 108, and 121 were mixed and spiked into a preparation of permethylated N-linked glycans released from Drosophila melanogaster embryo glycoproteins. This matrix was chosen as a first test of recovery because the three N-glycan standard structures (all complex type) have not previously been detected in Drosophila preparations, an organism that is characterized by a predominance of high-mannose-type glycans.24 The standard mix and the Drosophila glycans were analyzed separately and then in combination (Figure 9). The mean recovery for standard N-glycan signals was 101.6 ± 3.2% relative to the standard mix analyzed alone. For more stringently and comprehensively testing for the recovery of spiked standards, the malto-series standards (dp4–7) and the N-glycan standards (107, 108, and 121) were mixed and permethylated with 13C-MeI before being spiked into N-glycans released from mouse brain glycoproteins that had been permethylated with 12C-MeI. This biological matrix contains high-mannose and complex glycans, providing a source richer in potentially interfering components than the Drosophila material. The mean recovery of all standards together was 90.1 ± 9.3%. The malto-series glycans were recovered at 98.7 ± 6.0%, and the N-glycan standards were recovered at 85.0 ± 5.7%. Thus, signal suppression is not a significant concern when using exogenous glycan standards to quantify N-glycans prepared from biological sources. However, it should be noted that N-glycan standards were recovered at lower efficiency than that of the malto-series glycans, suggesting that the latter may provide more robust quantification.
Figure 9.
Recovery of MS signal responses for exogenous standards mixed into a biological matrix. The N-glycan standards 107, 108, and 121 were permethylated with 13C-MeI and analyzed as a standard mix alone (A). N-Linked glycans prepared from Drosophila melanogaster embryos were permethylated with 12C-MeI and analyzed alone (B) or after being supplemented with the 13C-MeI permethylated N-glycan standard mix (C). Recovery of signal intensities for standards and sample peaks indicates the lack of suppression or matrix interference for quantification by exogenous standard.
To demonstrate directly that an exogenous N-glycan standard can be resolved from the same glycan structure endogenous to a biological matrix, we added N-glycan standard 121 permethylated with 13C-MeI to an aliquot of rat plasma N-glycans permethylated with 12C-MeI. The differentially permethylated, but otherwise structurally identical glycans, were well-resolved as distinct isotopic envelopes, and the Xtract functionality of the Xcalibur software package (Thermo Fisher Scientific) efficiently captured the signal intensity associated with the exogenous and endogenous glycan structures (Figure S-12).
Quantification of Mouse Brain Glycans by N-Glycan and Malto-Series Standards
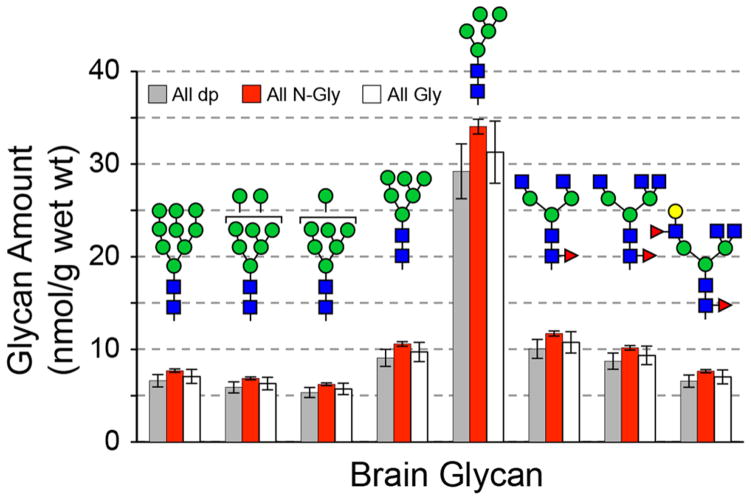
On the basis of the MS signal intensities detected for 44 N-linked glycans harvested from mouse brain, the eight most abundant glycans, defined as those present at greater than 5% of the total glycan profile, were selected for quantification by reference to exogenous standard. Individually, these eight glycans account for between 5 and 26% of the total MS signal intensity associated with the 44 detected glycans, and summed together, they account for 64% of the total glycan MS signal intensity. Malto-series standards (dp4–7) and each of the N-glycan standards (107, 108, and 121) were used as exogenous standards to quantify the eight most abundant glycans. The resulting values for the amount of N-linked glycan in brain (nmol glycan/gram of tissue wet weight) were compiled to determine if the malto-series or N-glycan standards produced substantially different results (Figure 10). The average percent deviation from the mean for measured glycan amount was 9.0 ± 4.5% for amounts determined by reference to the malto-series and N-glycan standards. Therefore, regardless of which type of standard or which individual standard is used for quantification, the resulting abundance measurements vary by less than 10%.
Figure 10.
Quantification of mouse brain N-glycans by reference to exogenous standards. N-Linked glycans were released from mouse brain glycoproteins and permethylated with 12C-MeI. N-Glycan and malto-series exogenous standards were permethylated with 13C-MeI, combined, and spiked into preparations of mouse brain-derived glycans. The eight most abundant mouse brain glycans were quantified by reference to each of the exogenous standards (107, 108, 121, and dp4–7). The calculated amount of each of the brain glycans (mean ± SD) is presented for quantification by reference to malto-series standards (All dp, gray bars; n = 4), N-glycan standards (All N-Gly, red bars; n = 3), or all glycan standards (All Gly, white bars; n = 7). Quantification of brain glycans yielded similar results regardless of which standard set was used.
On the basis of quantification relative to exogenous standards, the data presented here indicate that the N-glycan content of the whole mouse brain is 138 ± 11 nmol oligosaccharide/g of wet weight. Despite the extensive use of the mouse as an experimental system in glycobiology, it is surprisingly difficult to determine from the literature the absolute amount of N-linked glycan present in mouse brain tissue. All published mass spectrometry-based studies express N-linked glycan abundances in mouse brain as normalized distributions of glycans (quantified as % of the total profile) for comparison between wild-type and mutant/experimental samples, further emphasizing the importance of developing a method for quantifying absolute glycan amount.42–45 Nonetheless, assuming that approximately 10% of brain wet weight is protein and that approximately 10–20% of this protein is secreted or membrane bound glycoprotein, and using the average molecular weight of the eight most abundant N-glycans detected in this analysis (1632 Da), the results presented here indicate that 2.2–4.5% of the glycoprotein mass in the mouse brain is attributable to N-linked glycan.20,46–48 Considering a typical membrane protein of 100 kDa, this estimate indicates that 2.2–4.5 kDa of its mass is attributable to glycan, or that, on average, a 100 kDa protein possesses 1.3–2.8 N-linked glycans. This range is reasonable based on the most comprehensive and recent glycoproteomic analysis of rodent brain, which demonstrated that, on average, rodent brain glycoproteins carry 1.7 N-linked glycans per molecule (276 consensus sites utilized across 161 identified proteins, range of 1–27 sites per protein).20 Therefore, quantification by exogenous standard, whether using malto-series glycans or actual N-glycan structures, delivers glycan abundances that are within reason for complex biological samples. An important caveat associated with these calculations is that our MS-based quantification of N-glycans, as well as all other current methods that utilize HPLC or CE coupled to fluorescence detection, are not optimized to detect large polymeric glycans, such as polysialic acid, N-linked keratan sulfate, and polylactosamine repeats. Therefore, the amount of N-glycan in the adult mouse that we report here, as nmol/g of wet weight, may under-report actual brain glycoprotein N-linked glycosylation by the extent to which high molecular weight glycans are invisible to current analytic techniques.
CONCLUSIONS
The current use of mass spectrometry for glycomic analysis has been largely limited to the characterization of fold-differences between control and experimental samples. Although such studies have been invaluable for appreciating the dynamic nature of glycoprotein glycan structural diversity, they have been unable to discern changes in total glycan amount or in the absolute abundance of individual glycans, especially for minor components in complex mixtures. Two related impediments have limited the adoption of quantification by mass spectrometry: the variability of ionization efficiencies across glycan structures and the lack of availability of standard compounds that reflect the diversity of glycan structures found in biological samples.
With regard to the first limitation, permethylation effectively neutralizes any major impact of glycan structural features on ionization. In this study, the slopes of the standard curves were similar for the seven glycans we analyzed. Taken together, this restricted set of glycans possesses characteristics of broader classes of glycans, including linear, branched, bisected, biantennary, triantennary, sialylated, and nonsialylated glycans. The relatively minor changes in standard curve slope or calculated molar responses that were detected fall within a range of variation (<15%) that is acceptable in light of the benefits to be gained through quantification. However, our well-characterized set of N-glycan standards lack multi-antennary, fully extended, and variably sialylated structural topologies as well as diverse combinations of N-acetyl-lactosamine repeats with and without Lewis-type fucosylation. As N-glycan standards of increasing structural diversity become available to the community, it will be important to re-evaluate the constancy of molar responses across increasingly diverse permethylated N-glycans in comparison to those of simpler permethylated linear glycans.
With regard to the second limitation, the past few years have seen increasing effort toward the production of glycan standards for use by the research community. As these resources become available, it will be possible to investigate the value of using a broader range of well-characterized glycans as quantification standards. Synthetic glycans will be especially important for establishing quantification protocols for glycans bearing structural moieties, such as sulfation, that impact ionization because they remain charged after permethylation49,50 However, for neutral glycans or for charged glycans that become neutral upon permethylation (sialylated, phosphorylated, uronylated), well-characterized materials are currently available that can be useful as quantification standards, including N-glycan structures and simpler glycan polymers.
The method described here makes use of malto-series oligosaccharides from a commercial source as quantification standards. These standards, as well as the N-glycan standards procured from noncommercial sources, performed well, exhibiting broad linearity, a lower limit of quantification in a useful range for glycan abundances in biological materials, and acceptable recovery from a complex biological matrix. Although most of the work reported here was performed using nanospray ionization and linear ion or orbital trap mass analyzers, comparable quantification of standards was achieved with MALDI-TOF, NMR, and HPLC, indicating the cross-platform usefulness of the approach. Expanding glycan quantification to mass spectrometry provides another highly sensitive technology platform, along with HPLC and CE coupled to fluorescence detection, for comprehensively exploring glycomic differences in biological samples.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (U54 GM62116 and P41 GM103490 from NIGMS and P01 HL107151 from NHLBI). The authors gratefully acknowledge the assistance in acquiring glycan standards provided by Nahid Razi and by the Steering Committee of the Consortium for Functional Glycomics and by James Paulson of The Scripps Research Institute.
ABBREVIATIONS
- NMR
nuclear magnetic resonance spectroscopy
- HPLC
high-pressure liquid chromatography
- CE
capillary electrophoresis
- MS
mass spectrometry
- MS/MS or MS2
tandem mass spectrometry
- MSn
multidimensional mass spectrometry
- MALDI-TOF
matrix-assisted laser desorption ionization-time-of-flight
- MeI
methyl iodide
- dp
degree of polymerization
- NSI
nanospray ionization
- CFG
Consortium for Functional Glycomics
- 2-AB
2-aminobenzamide
- Hex
hexose
- HexNAc
N-acetylhexosamine
- NeuAc
N-acetylneuraminic acid or sialic acid
Footnotes
Notes
The authors declare no competing financial interest.
Tables and figures presenting: The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.6b00132.
NMR, HPLC, and mass spectrometric characterization for N-glycan standards 107, 108, and 121 as well as for malto-series glycan standards dp3–7(PDF)
References
- 1.Cummings RD, Pierce JM. The challenge and promise of glycomics. Chem Biol. 2014;21(1):1–15. doi: 10.1016/j.chembiol.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geyer H, Geyer R. Strategies for analysis of glycoprotein glycosylation. Biochim Biophys Acta, Proteins Proteomics. 2006;1764(12):1853–69. doi: 10.1016/j.bbapap.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Ito H, Kaji H, Togayachi A, Azadi P, Ishihara M, Geyer R, Galuska C, Geyer H, Kakehi K, Kinoshita M, Karlsson NG, Jin C, Kato K, Yagi H, Kondo S, Kawasaki N, Hashii N, Kolarich D, Stavenhagen K, Packer NH, Thaysen-Andersen M, Nakano M, Taniguchi N, Kurimoto A, Wada Y, Tajiri M, Yang P, Cao W, Li H, Rudd PM, Narimatsu H. Comparison of analytical methods for profiling N- and O-linked glycans from cultured cell lines: HUPO Human Disease Glycomics/Proteome Initiative multi-institutional study. Glycoconjugate J. 2016;33:405. doi: 10.1007/s10719-015-9625-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bigge JC, Patel TP, Bruce JA, Goulding PN, Charles SM, Parekh RB. Nonselective and efficient fluorescent labeling of glycans using 2-amino benzamide and anthranilic acid. Anal Biochem. 1995;230(2):229–38. doi: 10.1006/abio.1995.1468. [DOI] [PubMed] [Google Scholar]
- 5.Guile GR, Rudd PM, Wing DR, Prime SB, Dwek RA. A rapid high-resolution high-performance liquid chromatographic method for separating glycan mixtures and analyzing oligosaccharide profiles. Anal Biochem. 1996;240(2):210–26. doi: 10.1006/abio.1996.0351. [DOI] [PubMed] [Google Scholar]
- 6.Huffman JE, Pucic-Bakovic M, Klaric L, Hennig R, Selman MH, Vuckovic F, Novokmet M, Kristic J, Borowiak M, Muth T, Polasek O, Razdorov G, Gornik O, Plomp R, Theodoratou E, Wright AF, Rudan I, Hayward C, Campbell H, Deelder AM, Reichl U, Aulchenko YS, Rapp E, Wuhrer M, Lauc G. Comparative performance of four methods for high-throughput glycosylation analysis of immunoglobulin G in genetic and epidemiological research. Mol Cell Proteomics. 2014;13(6):1598–610. doi: 10.1074/mcp.M113.037465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knezevic A, Bones J, Kracun SK, Gornik O, Rudd PM, Lauc G. High throughput plasma N-glycome profiling using multiplexed labelling and UPLC with fluorescence detection. Analyst. 2011;136(22):4670–3. doi: 10.1039/c1an15684e. [DOI] [PubMed] [Google Scholar]
- 8.Nagels B, Santens F, Weterings K, Van Damme EJ, Callewaert N. Improved sample preparation for CE-LIF analysis of plant N-glycans. Electrophoresis. 2011;32(24):3482–90. doi: 10.1002/elps.201100268. [DOI] [PubMed] [Google Scholar]
- 9.Novotny MV, Sudor J. High-performance capillary electrophoresis of glycoconjugates. Electrophoresis. 1993;14(5–6):373–89. doi: 10.1002/elps.1150140163. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi N, Nakagawa H, Fujikawa K, Kawamura Y, Tomiya N. Three-dimensional elution mapping of pyridylaminated N-linked neutral and sialyl oligosaccharides. Anal Biochem. 1995;226(1):139–46. doi: 10.1006/abio.1995.1201. [DOI] [PubMed] [Google Scholar]
- 11.Wada Y, Azadi P, Costello CE, Dell A, Dwek RA, Geyer H, Geyer R, Kakehi K, Karlsson NG, Kato K, Kawasaki N, Khoo KH, Kim S, Kondo A, Lattova E, Mechref Y, Miyoshi E, Nakamura K, Narimatsu H, Novotny MV, Packer NH, Perreault H, Peter-Katalinic J, Pohlentz G, Reinhold VN, Rudd PM, Suzuki A, Taniguchi N. Comparison of the methods for profiling glycoprotein glycans–HUPO Human Disease Glycomics/Proteome Initiative multi-institutional study. Glycobiology. 2007;17(4):411–22. doi: 10.1093/glycob/cwl086. [DOI] [PubMed] [Google Scholar]
- 12.Royle L, Radcliffe CM, Dwek RA, Rudd PM. Detailed structural analysis of N-glycans released from glycoproteins in SDS-PAGE gel bands using HPLC combined with exoglycosidase array digestions. Methods Mol Biol. 2006;347:125–43. doi: 10.1385/1-59745-167-3:125. [DOI] [PubMed] [Google Scholar]
- 13.Ashline D, Singh S, Hanneman A, Reinhold V. Congruent strategies for carbohydrate sequencing. 1. Mining structural details by MSn. Anal Chem. 2005;77(19):6250–62. doi: 10.1021/ac050724z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dell A, Morris HR. Glycoprotein structure determination by mass spectrometry. Science. 2001;291(5512):2351–6. doi: 10.1126/science.1058890. [DOI] [PubMed] [Google Scholar]
- 15.Hamouda H, Kaup M, Ullah M, Berger M, Sandig V, Tauber R, Blanchard V. Rapid analysis of cell surface N-glycosylation from living cells using mass spectrometry. J Proteome Res. 2014;13(12):6144–51. doi: 10.1021/pr5003005. [DOI] [PubMed] [Google Scholar]
- 16.Hu Y, Mechref Y. Comparing MALDI-MS, RP-LC-MALDI-MS and RP-LC-ESI-MS glycomic profiles of permethylated N-glycans derived from model glycoproteins and human blood serum. Electrophoresis. 2012;33(12):1768–77. doi: 10.1002/elps.201100703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jang-Lee J, North SJ, Sutton-Smith M, Goldberg D, Panico M, Morris H, Haslam S, Dell A. Glycomic profiling of cells and tissues by mass spectrometry: fingerprinting and sequencing methodologies. Methods Enzymol. 2006;415:59–86. doi: 10.1016/S0076-6879(06)15005-3. [DOI] [PubMed] [Google Scholar]
- 18.Lapadula AJ, Hatcher PJ, Hanneman AJ, Ashline DJ, Zhang H, Reinhold VN. Congruent strategies for carbohydrate sequencing. 3. OSCAR: an algorithm for assigning oligosaccharide topology from MSn data. Anal Chem. 2005;77(19):6271–9. doi: 10.1021/ac050726j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leymarie N, Griffin PJ, Jonscher K, Kolarich D, Orlando R, McComb M, Zaia J, Aguilan J, Alley WR, Altmann F, Ball LE, Basumallick L, Bazemore-Walker CR, Behnken H, Blank MA, Brown KJ, Bunz SC, Cairo CW, Cipollo JF, Daneshfar R, Desaire H, Drake RR, Go EP, Goldman R, Gruber C, Halim A, Hathout Y, Hensbergen PJ, Horn DM, Hurum D, Jabs W, Larson G, Ly M, Mann BF, Marx K, Mechref Y, Meyer B, Moginger U, Neusubeta C, Nilsson J, Novotny MV, Nyalwidhe JO, Packer NH, Pompach P, Reiz B, Resemann A, Rohrer JS, Ruthenbeck A, Sanda M, Schulz JM, Schweiger-Hufnagel U, Sihlbom C, Song E, Staples GO, Suckau D, Tang H, Thaysen-Andersen M, Viner RI, An Y, Valmu L, Wada Y, Watson M, Windwarder M, Whittal R, Wuhrer M, Zhu Y, Zou C. Interlaboratory study on differential analysis of protein glycosylation by mass spectrometry: the ABRF glycoprotein research multi-institutional study 2012. Mol Cell Proteomics. 2013;12(10):2935–51. doi: 10.1074/mcp.M113.030643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker BL, Thaysen-Andersen M, Solis N, Scott NE, Larsen MR, Graham ME, Packer NH, Cordwell SJ. Site-specific glycan-peptide analysis for determination of N-glycoproteome heterogeneity. J Proteome Res. 2013;12(12):5791–800. doi: 10.1021/pr400783j. [DOI] [PubMed] [Google Scholar]
- 21.Walther T, Karamanska R, Chan RW, Chan MC, Jia N, Air G, Hopton C, Wong MP, Dell A, Malik Peiris JS, Haslam SM, Nicholls JM. Glycomic analysis of human respiratory tract tissues and correlation with influenza virus infection. PLoS Pathog. 2013;9(3):e1003223. doi: 10.1371/journal.ppat.1003223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamada K, Hyodo S, Kinoshita M, Hayakawa T, Kakehi K. Hyphenated technique for releasing and MALDI MS analysis of O-glycans in mucin-type glycoprotein samples. Anal Chem. 2010;82(17):7436–43. doi: 10.1021/ac101581n. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Singh S, Reinhold VN. Congruent strategies for carbohydrate sequencing. 2. FragLib: an MSn spectral library. Anal Chem. 2005;77(19):6263–70. doi: 10.1021/ac050725r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aoki K, Perlman M, Lim JM, Cantu R, Wells L, Tiemeyer M. Dynamic developmental elaboration of N-linked glycan complexity in the Drosophila melanogaster embryo. J Biol Chem. 2007;282(12):9127–42. doi: 10.1074/jbc.M606711200. [DOI] [PubMed] [Google Scholar]
- 25.Aoki K, Porterfield M, Lee SS, Dong B, Nguyen K, McGlamry KH, Tiemeyer M. The diversity of O-linked glycans expressed during Drosophila melanogaster development reflects stage-and tissue-specific requirements for cell signaling. J Biol Chem. 2008;283(44):30385–400. doi: 10.1074/jbc.M804925200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atwood JA, 3rd, Cheng L, Alvarez-Manilla G, Warren NL, York WS, Orlando R. Quantitation by isobaric labeling: applications to glycomics. J Proteome Res. 2008;7(1):367–74. doi: 10.1021/pr070476i. [DOI] [PubMed] [Google Scholar]
- 27.Zhang P, Zhang Y, Xue X, Wang C, Wang Z, Huang L. Relative quantitation of glycans using stable isotopic labels 1-(d0/d5) phenyl-3-methyl-5-pyrazolone by mass spectrometry. Anal Biochem. 2011;418(1):1–9. doi: 10.1016/j.ab.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 28.Moh ES, Thaysen-Andersen M, Packer NH. Relative versus absolute quantitation in disease glycomics. Proteomics: Clin Appl. 2015;9(3–4):368–82. doi: 10.1002/prca.201400184. [DOI] [PubMed] [Google Scholar]
- 29.Mechref Y, Kang P, Novotny MV. Solid-phase permethylation for glycomic analysis. Methods Mol Biol. 2009;534:53–64. doi: 10.1007/978-1-59745-022-5_4. [DOI] [PubMed] [Google Scholar]
- 30.Yu SY, Wu SW, Hsiao HH, Khoo KH. Enabling techniques and strategic workflow for sulfoglycomics based on mass spectrometry mapping and sequencing of permethylated sulfated glycans. Glycobiology. 2009;19(10):1136–49. doi: 10.1093/glycob/cwp113. [DOI] [PubMed] [Google Scholar]
- 31.Olajos M, Hajos P, Bonn GK, Guttman A. Sample preparation for the analysis of complex carbohydrates by multicapillary gel electrophoresis with light-emitting diode induced fluorescence detection. Anal Chem. 2008;80(11):4241–6. doi: 10.1021/ac8002598. [DOI] [PubMed] [Google Scholar]
- 32.Anumula KR, Taylor PB. A comprehensive procedure for preparation of partially methylated alditol acetates from glycoprotein carbohydrates. Anal Biochem. 1992;203(1):101–8. doi: 10.1016/0003-2697(92)90048-c. [DOI] [PubMed] [Google Scholar]
- 33.Porterfield M, Zhao P, Han H, Cunningham J, Aoki K, Von Hoff DD, Demeure MJ, Pierce JM, Tiemeyer M, Wells L. Discrimination between adenocarcinoma and normal pancreatic ductal fluid by proteomic and glycomic analysis. J Proteome Res. 2014;13(2):395–407. doi: 10.1021/pr400422g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu J, van Halbeek H. 1H and 13C resonance assignments of a 21-amino acid glycopeptide prepared from human serum transferrin. Carbohydr Res. 1996;296:1–21. doi: 10.1016/s0008-6215(96)00240-6. [DOI] [PubMed] [Google Scholar]
- 35.Tarentino AL, Plummer TH., Jr Enzymatic deglycosylation of asparagine-linked glycans: purification properties, and specificity of oligosaccharide-cleaving enzymes from Flavobacterium meningosepticum. Methods Enzymol. 1994;230:44–57. doi: 10.1016/0076-6879(94)30006-2. [DOI] [PubMed] [Google Scholar]
- 36.Bohorov O, Andersson-Sand H, Hoffmann J, Blixt O. Arraying glycomics: a novel bi-functional spacer for one-step microscale derivatization of free reducing glycans. Glycobiology. 2006;16(12):21C–27C. doi: 10.1093/glycob/cwl044. [DOI] [PubMed] [Google Scholar]
- 37.Varki A, Cummings RD, Aebi M, Packer NH, Seeberger PH, Esko JD, Stanley P, Hart G, Darvill A, Kinoshita T, Prestegard JJ, Schnaar RL, Freeze HH, Marth JD, Bertozzi CR, Etzler ME, Frank M, Vliegenthart JF, Lütteke T, Perez S, Bolton E, Rudd P, Paulson J, Kanehisa M, Toukach P, Aoki-Kinoshita KF, Dell A, Narimatsu H, York W, Taniguchi N, Kornfeld S. Symbol Nomenclature for Graphical Representations of Glycans. Glycobiology. 2015;25(12):1323–1324. doi: 10.1093/glycob/cwv091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nix DB, Kumagai T, Katoh T, Tiemeyer M, Aoki K. Improved in-gel reductive beta-elimination for comprehensive O-linked and sulfo-glycomics by mass spectrometry. J Visualized Exp. 2014;93:e51840. doi: 10.3791/51840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang CJ, Yamashita K, Kobata A. Structural study of the carbohydrate moiety of bovine pancreatic ribonuclease B. J Biochem. 1980;88(1):51–8. [PubMed] [Google Scholar]
- 40.Prien JM, Ashline DJ, Lapadula AJ, Zhang H, Reinhold VN. The high mannose glycans from bovine ribonuclease B isomer characterization by ion trap MS. J Am Soc Mass Spectrom. 2009;20(4):539–56. doi: 10.1016/j.jasms.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tarentino A, Plummer TH, Jr, Maley F. Studies on the oligosaccharide sequence of ribonuclease B. J Biol Chem. 1970;245(16):4150–7. [PubMed] [Google Scholar]
- 42.Nakakita S, Natsuka S, Okamoto J, Ikenaka K, Hase S. Alteration of brain type N-glycans in neurological mutant mouse brain. J Biochem. 2005;138(3):277–83. doi: 10.1093/jb/mvi125. [DOI] [PubMed] [Google Scholar]
- 43.North SJ, Chalabi S, Sutton-Smith M, Dell A. Mouse and human glycomes. In: Cummings RD, Pierce JM, editors. Handbook of Glycomics. Academic Press; Amsterdam: 2009. pp. 263–328. [Google Scholar]
- 44.North SJ, Jang-Lee J, Harrison R, Canis K, Ismail MN, Trollope A, Antonopoulos A, Pang PC, Grassi P, Al-Chalabi S, Etienne AT, Dell A, Haslam SM. Mass spectrometric analysis of mutant mice. Methods Enzymol. 2010;478:27–77. doi: 10.1016/S0076-6879(10)78002-2. [DOI] [PubMed] [Google Scholar]
- 45.Shimizu H, Ochiai K, Ikenaka K, Mikoshiba K, Hase S. Structures of N-linked sugar chains expressed mainly in mouse brain. J Biochem. 1993;114(3):334–8. doi: 10.1093/oxfordjournals.jbchem.a124177. [DOI] [PubMed] [Google Scholar]
- 46.Margolis RK, Preti C, Lai D, Margolis RU. Developmental changes in brain glycoproteins. Brain Res. 1976;112(2):363–9. doi: 10.1016/0006-8993(76)90290-0. [DOI] [PubMed] [Google Scholar]
- 47.Schnaar RL, Gerardy-Schahn R, Hildebrandt H. Sialic acids in the brain: gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol Rev. 2014;94(2):461–518. doi: 10.1152/physrev.00033.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharma K, Schmitt S, Bergner CG, Tyanova S, Kannaiyan N, Manrique-Hoyos N, Kongi K, Cantuti L, Hanisch UK, Philips MA, Rossner MJ, Mann M, Simons M. Cell type- and brain region-resolved mouse brain proteome. Nat Neurosci. 2015;18(12):1819–31. doi: 10.1038/nn.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumagai T, Katoh T, Nix DB, Tiemeyer M, Aoki K. In-gel beta-elimination and aqueous-organic partition for improved O-and sulfoglycomics. Anal Chem. 2013;85(18):8692–9. doi: 10.1021/ac4015935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kurz S, Aoki K, Jin C, Karlsson NG, Tiemeyer M, Wilson IB, Paschinger K. Targeted release and fractionation reveal glucuronylated and sulphated N- and O-glycans in larvae of dipteran insects. J Proteomics. 2015;126:172–88. doi: 10.1016/j.jprot.2015.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.