Abstract
Purpose
Proteomic analysis of blood proteins in DBS is gaining attention as a possible replacement for measurements in plasma/serum collected by venipuncture. We aimed to develop and provisionally validate a nanoflow LC-PRM-MS method for clinical use.
Experimental design
We used Skyline to develop a nanoflow LC-PRM-MS method to quantify glycated hemoglobin-β, apolipoprotein A-I, and apolipoprotein B in DBS. Precision, linearity, interferences, and stability and were determined and the method was used to analyze samples from 36 human volunteers. The method was compared with clinically validated measurements in paired blood collected via venipuncture.
Results
The method was relatively precise for these proteins (10–11% CV) and linear across the normal concentration ranges of these proteins. Interference from high total serum protein concentration (>8 g/dL) was noted for apolipoprotein A-I and apolipoprotein B. Proteins in DBS were stable for 14 d at temperatures below 25°C and trypsinized samples were stable for 48 h at 7°C. There was moderate correlation with clinical methods (r = 0.783–0.858) and significant bias in individual samples.
Conclusions and clinical relevance
Although the method had adequate precision and linearity for a biomarker, the accuracy compared with clinically validated assays raises concerns regarding the use of DBS compared with venipuncture for clinical use.
Keywords: LC-MS/MS, biomarkers, diabetes, multiplexing, apolipoproteins, dried blood spots, quantification, method evaluation, provisional validation
1. Introduction
Nearly 29 million Americans have diabetes and those with the disease are almost twice as likely to die from a heart attack or stroke compared to the general population [1]. Insulin resistance that accompanies type-2 diabetes can contribute to dyslipidemias that result in cardiovascular disease (CVD) [2]. Traditionally, plasma concentrations of cholesterol in high-density and low-density lipoprotein particles have been the mainstay for evaluating CVD risk [3]. However, recent studies suggest that plasma concentrations of apolipoprotein B (apoB) and apolipoprotein A-I (apoA-I) may be superior [4, 5]. Elevated plasma glucose concentrations lead to higher concentrations of hemoglobin with a β chain that has been glycated at the N-terminal valine (HbA1c), which is used as a surrogate for 3–4 month glucose control and has been accepted as a diagnostic biomarker for diabetes [6]. Analysis of these biomarkers requires a venous blood draw and multiple testing platforms, which can be uncomfortable for some patients and may be too expensive for large epidemiological studies or clinical trials.
Capillary blood collection via DBS is an established method of sample collection for the quantification of small molecules in whole blood [7–9] using immunoassays and liquid chromatography-tandem mass spectrometry [10]. Guthrie developed the first clinical application of DBS nearly 50 years ago to quantify phenylalanine levels in infants to diagnose phenylketonuria [11]. Since this time, clinical methodologies to detect other inborn errors in metabolism and other diseases have been developed utilizing DBS [9]. Several methodologies have been developed for analyzing proteins in DBS, including ceruloplasmin (Wilson’s disease), hemoglobinopathies, thyroid-stimulating hormone, and insulin-like growth factor-I [12–17]. However, multiplex MS-based assays to quantify numerous proteins in DBS have only been recently described. Chambers, et al., demonstrated proof-of-principal assays utilizing highly multiplexed LC-MS/MS analysis of DBS to identify 100s of whole blood proteins with the ability to quantify up to 97 of these proteins [18–20].
As an alternative to venipuncture, sample collection via lancet and spotting requires minimal training. Importantly, upon drying, most analytes remain stable over a broad temperature range when properly packaged in sealed containers with desiccant [7]. Additionally, because the samples are dry, they minimize exposure to potential biohazards and reduce processing and logistics costs [9]. As a result, DBS are an interesting alternative to venous blood draws for large-scale epidemiological studies, pharmaceutical trials, and for clinicians caring for patients in remote locations.
LC-MS/MS is well-suited for the analysis of proteins in complex mixtures [21], including serum and plasma [22–25]. Targeted MS methods can overcome important limitations of some immunoassays, including antibody cross-reactivity and interfering endogenous antibodies [26]. Isotope-labeled internal standards in targeted MS methods normalize ion suppression and fluctuations in instrument performance, allowing for the specific and robust quantification of plasma and serum proteins, as well as the capability to multiplex the analysis of numerous plasma proteins in a single assay [27, 28]. Indeed, targeted, multiplexed LC-MS/MS methods can be used to quantify CVD-associated proteins in human plasma or serum with good correlation to clinical immunoassays [29–32]. It has not been determined if the LC-MS/MS quantification of proteins in DBS from capillary blood also correlates well with clinical assays.
We present a method for the quantification of apoA-I, apoB, and HbA1c in DBS made from EDTA anticoagulated-whole blood (EDTA-WB) and capillary blood using nanoflow LC-PRM-MS, which has recently been shown to be equivalent to LC-MRM-MS for quantification of proteins using bottom-up proteomic methods [33]. The method was evaluated for clinical use according to recently published guidelines from our group [34] and was compared with established clinical assays using paired specimens collected by venipuncture to evaluate the analytical performance characteristics of DBS analysis by LC-PRM-MS. Precision and bias targets were set using biological variability, which corresponds to model 2 of the Stockholm Conference on Quality Specifications in Laboratory Medicine [35–37].
2. Materials and Methods
2.1 Chemicals and human samples
All chemicals were obtained from Fisher Scientific unless otherwise noted. Human specimens were used in accordance with policies established by the Human Subjects Division at the University of Washington. Stable isotope-labeled internal standard peptides (SIS, IS_pep) contained heavy lysine or arginine at the C-terminus and were synthesized in-house or obtained from New England Peptide. Quantification of SIS using amino acid analysis was performed by the University of California, Davis Molecular Structure facility. Isotopically labeled 15N-apoA-I protein (IS_prot) was produced recombinantly via a bacterial expression system as described [38].
2.2 Clinical assays
Automated clinical analyzers were used to quantify plasma concentrations in calibrators and samples of apoA-I and apoB [Siemens BNII nephelometer], HbA1c [Bio-Rad DC-10 HPLC], total cholesterol, high density lipoprotein cholesterol (HDL-C), non-HDL-C (calculated as total cholesterol minus HDL-C), triglycerides, and low density lipoprotein cholesterol (LDL-C, direct and calculated via the Friedewald equation) [Beckman AU 680] (Supplementary Table 1). The Variant II A1c assay is certified by the NGSP and traceable to the DCCT reference and the Siemens BN-II assays for apoA-I and apoB are traceable to WHO reference materials SP1-01 and SP3-07, respectively.
2.3 Dried blood spot and matched plasma sample preparation, extraction, digestion, and analysis
Details for the preparation of DBS and matched plasma and their subsequent extraction and digestion are provided in Supplementary Information. Briefly, EDTA-WB or capillary-WB was spotted onto Whatman 903 filter paper and allowed to dry for 4 h at room temperature. One punch (3.2 mm) from each spot was placed in a microcentrifuge tube and proteins were extracted and denatured from the filter paper in 100 mM ammonium bicarbonate/trifluoroethanol (AMBIC/TFE). Alternatively, 10 µL of plasma was added to a microcentrifuge tube and subsequently diluted in AMBIC/TFE. The samples were then reduced with dithiothreitol, and acetylated with iodoacetamide. The TFE was diluted to ~5% and the proteins were proteolyzed using trypsin for 20 hours with agitation. Following digestion, the samples were desalted using solid phase extraction and dried down in a Speedvac. Prior to analysis, the samples were reconstituted in a small volume of water/acetonitrile/trifluoroacetic acid. Details of the chromatographic conditions (Easy nLC 1000, Thermo) and mass spectrometer settings (Q Exactive Plus, Thermo) are listed in Supplementary Tables 2 and 3.
2.4 Preparation of dried blood spots for assay development and provisional validation
Details regarding the preparation of standards and the evaluation procedures are provided in the Supplementary Information. The provisional validation procedures are briefly summarized in Table 1 [34] and used EDTA-WB samples from a healthy volunteer (Standard H) and a pool of leftover samples from patients with HbA1c >10% and LDL-C >150 mg/dL (Standard D). Linearity and lower limits of quantification were assessed using mixtures of Standard H and Standard D and mixtures of the Standards with chicken EDTA-WB (Pel-Freez Biologicals) that were spotted onto filter paper. Concentrations of proteins in Standards H and D were determined using the traceable clinical assays described above and a single point calibrator (SPC) was prepared using a one-to-one mixture of Standard H and Standard D.
Table 1.
Provisional validation criteria for the nanoflowLC-PRM-MS method to quantify apoA-1, apoB, and HbA1c in dried blood spots.
| Experiment | Description | Criteria | Figures of Merit |
|---|---|---|---|
| Precision | Healthy and diseased pools analyzed 5 times over the course of 5 days. | Coefficients of Variation within day (CVIntra), between day (CVInter) and total (CVTot) as the sum of squares. | Both CVIntra and CVInter< 20% |
| Linearity | Healthy and diseased samples admixed: Healthy, 3:1, 1:1, 1:3 and Diseased | CV and bias of admix samples and correlation of PAR to extrapolated values obtained from reference methods | Bias, CV <20% |
| Lower limit of quantification | Healthy and diseased pools diluted 1:1, 1:2, 1:4, 1:6 1:8 and 1:10 in chicken (Gallus gallus) EDTA-whole blood | CV of triplicate diluted samples compared to concentration of apoA-1, apoB and HbA1c incorporating dilution factors for samples. | Recovery 100% ± 20%, CV < 25% |
| Sample Stability | Stability of Healthy DBS across range of temperatures and sample stability in auto-sampler after 48 hour period | CV of triplicate samples and mean comparison to spot storage conditions and after 48 hours in auto-sampler | p > 0.05, CV <20% |
2.5 Peptide selection
Peptides used for absolute quantification of apoA-I (P02647), apoB (P04114) and HbA1c (glycated hemoglobin-β) (P68871) were based on FASTA sequences obtained from UniProtKB/Swis-Prot database [39]. The sequences were imported into Skyline [40] and tryptically digested in silico to generate precursor ion inclusion lists that also contained instrument control parameters for Xcalibur to detect peptides using PRM-MS. The N-terminal peptide of hemoglobin-β (VHLTPEEK) was used to quantify HbA1c, which was calculated as the ratio of the peak area of the glycated peptide to the sum of the peak areas for the glycated and unglycated peptides. Transition ion ratios [Supplemental Table 4] were monitored during method comparison to identify sample-specific interferences [41].
2.6 Data analysis
Raw mass spectrometry data (Thermo) were exported to Skyline-daily (version 3.1.1.8663) for identification of transitions and peak area integration. Data were exported in .csv file format and analyzed in Excel 2010 [Ver. 14.0.7145.5000 (64-bit)]. Comparative statistical analyses using T-test (two-tailed, unequal variance) or single factor ANOVA were performed in Microsoft Excel with the Analysis ToolPak Add-In. Analysis of linearity was performed in GraphPad Prism (Ver. 6.0g). Method comparison of human samples was performed using R statistical software package (Ver. 3.3.2).
3. Results
3.1 Assay development
There were 17 and 240 tryptic peptides between 7 and 25 residues in length for apoA-I and apoB, respectively, excluding signal sequences from the FASTA files. One apoA-I peptide was not detected in DBS. Many peptides from apoB were excluded from consideration: 58 peptides contained non-ideal amino acids; 53 were not detected in DBS; 86 had interfering chromatographic peaks and/or very low signal intensity; 7 peptides were unstable in the autosampler after 48 h (>50% reduction in peak area between injections); and 20 had poor mass accuracy (2 < ppm < 4). A small set of DBS made from EDTA-WB was used to identify the peptides in apoA-I and apoB that had the highest correlation with plasma protein concentrations measured by immunoassay. From these experiments, 2 apoA-I peptides and 5 apoB peptides were selected and stable isotope-labeled internal standard peptides were synthesized (Supplementary Table 5). Only two of the apoB peptides were used in the final quantitative assay, while the other peptides were reserved as a failsafe in the event that sample-specific interferences were observed with the primary quantifying peptides, though none were detected based on monitoring of transition ion ratios (Supplementary Table 4). Five other 15N-apoA-I peptides were used to monitor retention time drifts and adjust the scheduled PRM method (Supplementary Table 5).
Instrument parameters predicted by Skyline were utilized for all peptides and empirical adjustment of these parameters did not improve signal intensity (data not shown) [42, 43]. Time-course digestion experiments indicated that peptides used in the quantification of these three proteins achieved steady state by 20 hours and previous work by our group indicated that internal standard peptides should be added before digestion [43] (Supplementary Figure 1).
3.2 Single point calibrator
The total imprecision of the quantification of HbA1c was determined using a 5×5 analysis to be 22–24% CV (Table 2). Imprecision was significantly improved to 11% CV by incorporation of a SPC in the assay, which reduced within-day variability (9–10% to 8% CV) and markedly reduced between-day variability (20–22% to 8% CV) (Table 2). The total imprecision for the assay of apoA-I and apoB that used stable isotope labeled internal standard peptides (IS_pep) added prior to digestion improved from 12–13% CV to 10–11% CV when including a SPC in the assay (Table 2). A similar reduction in variability due to the inclusion of an SPC was observed for the assay of apoA-I and apoB that used 15N-apoA-I internal standard protein (IS_prot) (Supplementary Table 6). However, the imprecision of the DBS assay did exceed the target that was developed for clinical assays based on biological variation (%CV) of 3.3, 3.5 and 0.9 for apoA-I, apoB and HbA1c, respectively [36, 37].
Table 2.
Imprecision with and without calibrator
| Imprecision Without Calibratora | Imprecision With Calibratorb | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Healthy | Diseased | Healthy | Diseased | |||||||||
| CVc | Intra | Inter | Total | Intra | Inter | Total | Intra | Inter | Total | Intra | Inter | Total |
| HbA1c | 9% | 20% | 22% | 10% | 22% | 24% | 8% | 8% | 11% | 8% | 8% | 11% |
| ApoA-I | 7% | 11% | 13% | 6% | 11% | 12% | 7% | 7% | 10% | 6% | 8% | 10% |
| ApoB | 6% | 10% | 12% | 7% | 10% | 12% | 6% | 9% | 11% | 7% | 9% | 11% |
Imprecision of the peak area ratio: endogenous peptide peak area divided by IS_pep peak area. Glycated VHLTPEEK divided by sum of glycated and non-glycated VHLTPEEK.
Imprecision of the normalized peak area ratio: peak area ratio of each sample divided by the peak area ratio of the single point calibrator then multiplied by the concentration of the SPC.
Coefficients of variation (CV) were calculated as the mean intra-assay variability determined from five replicates on the same day (over five days), the mean inter-assay variability determined from one replicate measured each day for five days (over five replicates each day), and the total variability (CVintra2 + CVinter2)1/2.
3.3 Other performance characteristics
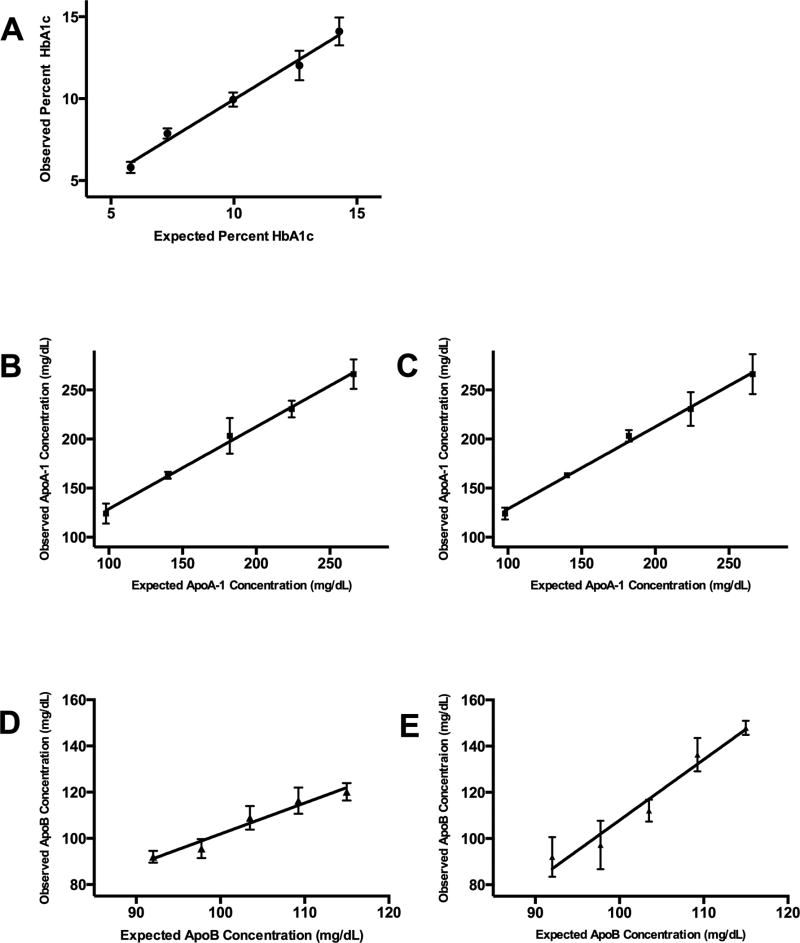
The assay of each protein was linear across normal ranges for HbA1c, 5.8 to 14.1% [6–9% CV, r2=0.9899]; for apoA-I, 98–266 mg/dL [6–9% CV, r2=0.9953]; and for apoB, 92–115 mg/dL [3–5% CV, r2=0.9607] (Figure 2; Supplementary Table 7). The lower limit of quantification for apoA-I and apoB was 14 mg/dL (6% CV) and 8.3 mg/dL (12% CV), respectively (Supplementary Tables 8 and 9). Significant interference from high total protein (>8 g/dL) was observed for apoA-I and apoB, with percent recoveries of 120% and 122%, respectively. Interference was less significant for HbA1c (115%) (Supplementary Table 10). Importantly, interference from elevated total protein was reduced in assays that used IS_prot. Interference due to triglycerides was not possible to assess for apoB due the presence of apoB in the triglyceride-rich spiking solution. No interference was observed due to high triglycerides for either apoA-I or HbA1c (97% and 101%, respectively) and similar data were seen when using IS_prot. When DBS were incubated in sealed plastic bags containing desiccant for up to 14 d at different temperatures (−80°C to 37°C), the proteins appeared to be stable up to 25°C, but not at 37°C (Supplementary Tables 11 and 12). Interestingly, there was no instability of apoA-I or apoB apparent when using IS_prot, which suggests that matrix effects, and not protein instability, might lead to the observed temperature-dependent bias in the method that uses IS_pep. Peptides were stable in the autosampler (7°C) for 48 h (Supplementary Table 13).
Figure 2. Linearity studies.
The results of linear regression of the observed concentrations of each protein in different mixtures of EDTA-WB spotted onto DBS paper vs. expected concentration are shown for each protein: (A) HbA1c, (B) apoA-I normalized to internal standard peptides (IS_pep), (C) apoA-I normalized to IS_prot, (D) apoB normalized to (IS_pep) and (E) apoB normalized to 15N-apoA-I peptides (IS_prot).
3.4 Correlation with clinical methods
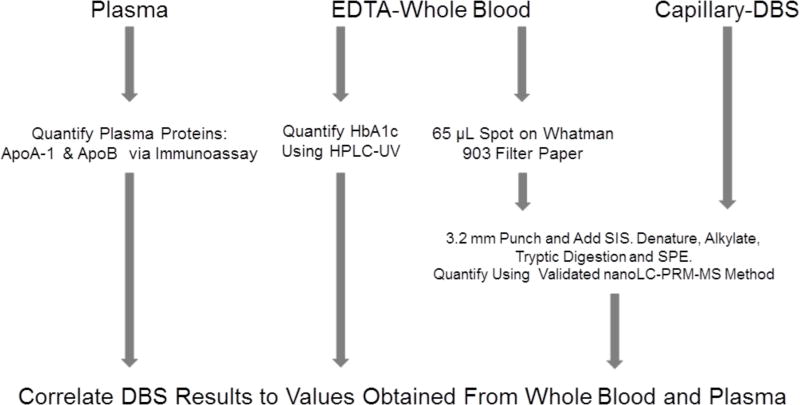
With the aim of developing a clinically useful method, we compared the LC-PRM-MS assay of HbA1c, apoA-I, and apoB in DBS to clinically validated methods. DBS made with EDTA-WB (65 µL per spot), DBS made with capillary blood, EDTA plasma samples, and EDTA-WB samples were obtained from 36 human volunteers (Figure 1) (n=29 for matched plasma experiments). The plasma concentrations of apoA-I and apoB were measured using nephelometry (~5% CV) and HbAlc in WB was quantified via HPLC-UV (~2% CV).
Figure 1. Experimental workflow.
In 36 ETDA whole blood samples, plasma concentrations of apoA-I and apoB were measured by nephelometry and HbA1c concentrations were measured by HPLC-UV. The same 36 samples were used to make EDTA-WB DBS. Paired capillary DBS samples were collected at the same time.
The correlation of HbA1c concentration measured in capillary DBS with HbA1c concentration determined in EDTA-WB [r=0.858, mean bias of −0.4% HbA1c, limits of agreement (LoA) of −1.8 – 1.0% HbA1c, mean percent bias (MPB) of 11%] was similar to the correlation observed for EDTA-WB DBS and EDTA-WB [r=0.845, mean bias of 0.1% HbA1c, LoA of −1.3 – 1.4% HbA1c, MPB of 9%] (Figure 3, Supplementary Tables 14 and 15, Supplementary Figure 2). Bias of the DBS assay in quantifying HbA1c was higher than the target that was developed for clinical assays based on biological variation (Bias of 1.5%) [36, 37].
Figure 3. Method comparison.
The results of linear regression of protein concentrations measured using nanoflow LC-MS/MS and IS_pep vs. clinical measures for each of the proteins are shown: (A) percent HbA1c in capillary DBS, (B) percent HbA1c in EDTA-WB DBS, (C) capillary DBS apoA-I concentration (mg/dL), (D) EDTA-WB DBS apoA-I concentration (mg/dL), (E) matched plasma apoA-I concentration (mg/dL), (F) capillary DBS apoB concentration (mg/dL) and (G) EDTA-WB DBS apoB concentration (mg/dL), (H) matched plasma apoB concentration (mg/dL). Dashed lines represent the 95% confidence interval for least squares regression.
ApoA-I concentrations measured in capillary DBS were better correlated with nephelometric measurements [r=0.822, mean bias of 11.6 mg/dL, LoA of −34.9 – 58.2 mg/dL, MPB of 12%] than were EDTA-WB DBS [r=0.783, mean bias of 24.6 mg/dL, LoA of −28.6 – 77.7 mg/dL, MPB of 12%]. ApoA-I concentrations measured in matched plasma were better correlated with nephelometric measurements than DBS [r=0.818, mean bias of 11.8 mg/dL, LoA of −46.7 – 23.1 mg/dL, MPB 12%] (Figure 3, Supplementary Tables 14 and 15, Supplementary Figure 2). Similar correlations were observed when using IS_prot, with the exception of matched plasma, which performed considerably better than IS_pep [r=0.924, mean bias −6.5 mg/dL, LoA of −25.5–12.6 mg/dL, MPB of 6%) (Figure 3, Supplementary Table 15, Supplementary Figure 2). Regardless, the bias of the DBS assay was higher than the target bias of 3.7%. The correlation of apoA-I concentration between EDTA-WB and capillary DBS was lower [IS_pep=0.784, IS_prot=0.754] than when comparing either to immunoassay (Supplementary Table 16). The correlation between capillary DBS measurements and HDL-C was higher than the correlation between EDTA-WB DBS and HDL-C [0.825 and 0.726, respectively].
Similar data were observed for apoB, with capillary DBS measurements correlating with nephelometry [r=0.830, mean bias of 1.1 mg/dL, LoA of −26.3 – 28.5 mg/dL, MPB of 12%] better than EDTA-WB DBS [r=0.809, mean bias of 9.0 mg/dL, LoA of −33.1 – 51.2 mg/dL, MPB of 16%] (Figure 3, Supplementary Table 14, and Supplementary Figure 2). Similar results were observed using IS_prot (Supplementary Table 15). ApoB concentrations measured in matched plasma were much better correlated with nephelometric measurements than DBS [r=0.0.974, mean bias of 0.1 mg/dL, LoA of −9.1 – 9.3 mg/dL, MPB of 4.5%] (Figure 3, Supplementary Table 14, Supplementary Figure 2). While the bias of this assay was within the target bias of 6% when quantifying apoB in plasma, this bias was exceeded when assaying DBS. The correlation of apoB concentrations measured in EDTA-WB DBS with those measured in capillary DBS was higher for IS_pep than for IS_prot [0.882 vs. 0.821] (Supplementary Table 16). The correlation of apoB concentrations in DBS (capillary or EDTA-WB) with LDL-C or non-HDL-C was 0.799–0.829 (Supplementary Table 14).
To rule out sample-specific interferences in the correlation samples, the ratio of the two most abundant transition ions was calculated for all quantification and internal standard peptides analyzed in theses samples and the average absolute difference of the ratios was less than 5% (Supplementary Table 4).
4. Discussion
The application of dried blood spots in the analysis of protein biomarkers is a promising technology due to convenient sample collection, simplified logistics and mitigation of exposure to blood-borne pathogens. Furthermore, mass spectrometry-based analysis of proteins has several advantages over immunoassays [26]. The objective of this study was to evaluate if the quantification of HbA1c, apoA-I, and apoB in DBS obtained from venous or capillary blood using a provisionally validated multiplex nanoflow LC-PRM-MS assay demonstrated promise as a suitable substitute for traditional clinical methods using venipuncture.
4.1 Provisional Validation
Discoveries of novel biomarkers are regularly reported in the literature, however it is rare that these biomarkers make the transition from discovery experiments to utilization in the clinical laboratory. Often, the results of these studies cannot be reproduced in the laboratories seeking to use these protein targets to diagnose and monitor human diseases and this has been attributed, in part, to inadequately evaluated assays [44]. The fundamental hallmarks of any robust and reliable assay are precision, linearity, specificity and stability, and ideally could be determined with only a small number of samples prior to committing the necessary resources for a complete method validation. Provisional validation of the method presented here followed recently outlined recommendations [34].
4.2 Single Point Calibrator
Provisional validation of this method demonstrated that the assay was linear for normal concentration ranges of these proteins. The ability of the assay to accurately determine concentrations in apoA-I- or ApoB-deficient individuals was not assessed in this study. Interference experiments indicated that high total protein resulted in a significant over-recovery of apoA-I and apoB. Interestingly, stability experiments indicated that degradation of the DBS at 37°C may introduce matrix effects that affected digestion and were corrected when normalizing to IS_prot instead of IS_pep. The inter-day and total precision of this assay was within acceptable limits when normalizing with either IS_pep (added prior to digestion) or IS_prot, with notable exceptions for HbA1c, which was resolved by including an SPC with each digestion batch. Indeed, the inter-day and total precision were improved for all proteins with the inclusion of an SPC. However, the bias and imprecision of the DBS assay were higher for these proteins than previously identified targets set based on biological variability [36, 37].
4.3 Internal Standard Protein
Incorporation of isotopically labeled internal standard peptides or proteins in quantitative proteomics experiments normalizes for ion suppression and fluctuations in instrument performance. Furthermore, inclusion of an isotopically labeled protein during digestion further normalizes for variability inherent to enzymatic hydrolysis of peptides [27, 28]. Previous LC-MS/MS methods developed by our group employed an internal standard protein (15N-apoA-I) for absolute and relative quantification of proteins in HDL demonstrated similar precision to stable isotope labeled peptides [32]. The results from this work indicated significant differences in variability and slope from least squares regression in mixing experiments of endogenous apoB peptides normalized to 15N-apoA-I peptides, likely due to matrix effects. Therefore, the type of internal standard utilized in an assay (i.e. stable isotope labeled peptide or protein) should be determined on a case-by-case basis and assessed during method evaluation.
4.4 Comparison of method with clinical assays
In addition to precision and linearity, accuracy is of paramount importance in any assay used for clinical care [45]. To assess the accuracy of nanoflow LC-PRM-MS in DBS, we used our method to measure the concentration of apoA-I, apoB, and HbA1c in samples from 36 healthy volunteers and compared the results to those obtained using immunoassays and HPLC-UV, respectively. We observed linear correlations between the clinical and DBS results, but there was significant scatter around the line of regression, indicating sample-specific disagreement between these methodologies. Overall, bias was more noticeable for capillary DBS when quantifying HbA1c, but higher for EDTA-WB DBS when quantifying apoA-I and apoB. Similar analysis using matched plasma using LC-PRM-MS demonstrated better agreement with nephelometric immunoassay compared to DBS, which suggests that it is not the liquid chromatographic or the tandem mass spectrometric method that causes the disagreements, but instead the blood spots themselves. To rule out interference, monitoring transition ion ratios of the quantifying peptides indicated no sample-specific interferences, however the possibility of physiological or biological variation in the composition of the proteins (e.g., intermediate glycation products) was not ruled out as a potential source of disagreement between the analytical methods
4.4.1 Capillary vs. whole blood
According to CLSI guidelines, proper technique for the collection of capillary blood on DBS paper is crucial to minimize contamination from interstitial and cellular fluids that are released when the dermis is punctured by the lancet [46]. In addition, due to the action of tissues on capillary blood, it is possible that the concentration of a protein in serum or plasma could differ from that in blood drawn from capillaries. However, we expected that EDTA-WB DBS would agree with venipuncture more than capillary DBS, which we did not see for all three proteins [46, 47]. While we took considerable care in the collection of capillary DBS in this study (i.e. trained personnel collected samples), the broad LoA between this assay and clinical methods is concerning.
4.4.2 Hematocrit effect
The inherent inaccurate quantification of analytes in DBS has been attributed to the so-called hematocrit effect [46, 47]. The hematocrit, which is defined as the volume fraction of red blood cells in whole blood, can affect blood viscosity and spreading of blood on the filter paper, potentially resulting in localized variations in the concentration of analytes on the DBS [47]. Using multiple linear regression, we were unable to use the hemoglobin peak area to improve correlation between the blood spot measurements and venipuncture measurements (data not shown).
4.5 Conclusions
Our provisional validation experiments demonstrate considerable promise for the quantification of apoA-I, apoB, and HbA1c in DBS, however when used in a small population of human samples, the method suffered from sample-specific biases that must be resolved if it is to be a substitute for established clinical methodologies. The field is currently investigating different DBS technologies that may mitigate sampling issues and facilitate workflows that will allow DBS to be a suitable replacement for venipuncture.
Supplementary Material
Statement of Clinical Relevance.
Blood collection using dried blood spots (DBS) is an attractive alternative to venipuncture as it is less invasive, requires minimal training, reduces exposure to blood-borne pathogens, and simplifies shipping and storage. DBS are important in newborn screening and are increasingly being used in other clinical settings and pharmaceutical studies as well. They are now receiving attention for their possible use in the quantification of blood proteins for diagnosis and disease monitoring. We have developed and evaluated a mass spectrometric method for the quantification of hemoglobin A1c, apolipoprotein A-I, and apolipoprotein B in DBS. The method is linear and precise, but suffers from poor accuracy when compared with conventional clinically validated assays, which has important implications for the future use of DBS in clinical care.
Acknowledgments
This study was supported by NIH grants T32HL007028, P30DK035816, and R01HL111375.
Conflicts of Interest
The University of Washington receives grant support from Waters, Inc., a manufacturer of mass spectrometers.
Abbreviations
- apoA-I
apolipoprotein A-I
- apoB
apolipoprotein B
- CVD
cardiovascular disease
- DBS
dried blood spots
- EDTA-WB
EDTA anticoagulated whole blood
- HbA1c
hemoglobin A1c
- HDL-C
high density lipoprotein cholesterol
- LDL-C
low density lipoprotein cholesterol
- WB
whole blood
References
- 1.Centers for Disease Control and Prevention, National Diabetes Statistics Report: Estimates of Diabetes and Its Burden in the United States, 2014. US Department of Health and Human Services. 2014:1–12. [Google Scholar]
- 2.Collaboration, T. E. R. F. Diabetes mellitus, fasting blood glucose concentration, and risk of vascular disease: a collaborative meta-analysis of 102 prospective studies. The Lancet. 2010;375:2215–2222. doi: 10.1016/S0140-6736(10)60484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boden WE. High-density lipoprotein cholesterol as an independent risk factor in cardiovascular disease: assessing the data from framingham to the veterans affairs high-density lipoprotein intervention trial. The American Journal of Cardiology. 2000;86:19–22. doi: 10.1016/s0002-9149(00)01464-8. [DOI] [PubMed] [Google Scholar]
- 4.Walldius G, Jungner I. The apoB/apoA-I ratio: a strong, new risk factor for cardiovascular disease and a target for lipid-lowering therapy – a review of the evidence. Journal of Internal Medicine. 2006;259:493–519. doi: 10.1111/j.1365-2796.2006.01643.x. [DOI] [PubMed] [Google Scholar]
- 5.Walldius G, Jungner I, Aastveit AH, Holme I, et al. The apoB/apoA-I ratio is better than the cholesterol ratios to estimate the balance between plasma proatherogenic and antiatherogenic lipoproteins and to predict coronary risk. Clinical chemistry and laboratory medicine : CCLM/FESCC. 2004;42:1355–1363. doi: 10.1515/CCLM.2004.254. [DOI] [PubMed] [Google Scholar]
- 6.Rahbar S. The discovery of glycated hemoglobin: a major event in the study of nonenzymatic chemistry in biological systems. Annals of the New York Academy of Sciences. 2005;1043:9–19. doi: 10.1196/annals.1333.002. [DOI] [PubMed] [Google Scholar]
- 7.Kapur S, Kapur S, Zava D. Cardiometabolic risk factors assessed by a finger stick dried blood spot method. Journal of diabetes science and technology. 2008;2:236–241. doi: 10.1177/193229680800200210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W, Tse FLS. Dried blood spot sampling in combination with LC-MS/MS for quantitative analysis of small molecules. Biomedical chromatography : BMC. 2010;24:49–65. doi: 10.1002/bmc.1367. [DOI] [PubMed] [Google Scholar]
- 9.Tanna S, Lawson G. Analytical methods used in conjunction with dried blood spots. Analytical Methods. 2011;3:1709–1709. [Google Scholar]
- 10.Keevil BG. The analysis of dried blood spot samples using liquid chromatography tandem mass spectrometry. Clinical biochemistry. 2011;44:110–118. doi: 10.1016/j.clinbiochem.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Spooner N. A glowing future for dried blood spot sampling. Bioanalysis. 2010;2:1343–1344. doi: 10.4155/bio.10.111. [DOI] [PubMed] [Google Scholar]
- 12.Boemer F, Ketelslegers O, Minon J-M, Bours V, Schoos R. Newborn Screening for Sickle Cell Disease Using Tandem Mass Spectrometry. Clinical Chemistry. 2008;54:2036–2041. doi: 10.1373/clinchem.2008.106369. [DOI] [PubMed] [Google Scholar]
- 13.Cox H, Rampton J, Eichner D. Quantification of insulin-like growth factor-1 in dried blood spots for detection of growth hormone abuse in sport. Analytical and Bioanalytical Chemistry. 2013;405:1949–1958. doi: 10.1007/s00216-012-6626-y. [DOI] [PubMed] [Google Scholar]
- 14.deWilde A, Sadilkova K, Sadilek M, Vasta V, Hahn SH. Tryptic Peptide Analysis of Ceruloplasmin in Dried Blood Spots Using Liquid Chromatography–Tandem Mass Spectrometry: Application to Newborn Screening. Clinical Chemistry. 2008;54:1961–1968. doi: 10.1373/clinchem.2008.111989. [DOI] [PubMed] [Google Scholar]
- 15.Hofman LF, Foley TP, Henry JJ, Naylor EW. Original Paper: Assays for thyroid-stimulating hormone using dried blood spotted filter paper specimens to screen for hypothyroidism in older children and adults. Journal of Medical Screening. 2003;10:5–10. doi: 10.1258/096914103321610734. [DOI] [PubMed] [Google Scholar]
- 16.Ignjatovic V, Pitt J, Monagle P, Craig JM. The utility of dried blood spots for proteomic studies: Looking forward to looking back. PROTEOMICS – Clinical Applications. 2014;8:896–900. doi: 10.1002/prca.201400042. [DOI] [PubMed] [Google Scholar]
- 17.Pai JK, Cahill LE, Hu FB, Rexrode KM, et al. Hemoglobin A1c Is Associated With Increased Risk of Incident Coronary Heart Disease Among Apparently Healthy, Nondiabetic Men and Women. Journal of the American Heart Association. 2013;2 doi: 10.1161/JAHA.112.000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chambers A, Percy A, Hardie D, Borchers C. Comparison of Proteins in Whole Blood and Dried Blood Spot Samples by LC/MS/MS. Journal of The American Society for Mass Spectrometry. 2013;24:1338–1345. doi: 10.1007/s13361-013-0678-x. [DOI] [PubMed] [Google Scholar]
- 19.Chambers AG, Percy AJ, Yang J, Borchers CH. LC/MRM-MS Enables Precise and Simultaneous Quantification of 97 Proteins in Dried Blood Spots. Molecular & Cellular Proteomics. 2015 doi: 10.1074/mcp.O115.049957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chambers AG, Percy AJ, Yang J, Camenzind AG, Borchers CH. Multiplexed quantitation of endogenous proteins in dried blood spots by multiple reaction monitoring-mass spectrometry. Molecular cellular proteomics MCP. 2013;12:781–791. doi: 10.1074/mcp.M112.022442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aebersold R, Goodlett DR. Mass spectrometry in proteomics. Chemical reviews. 2001;101:269–295. doi: 10.1021/cr990076h. [DOI] [PubMed] [Google Scholar]
- 22.Anderson NL. The Human Plasma Proteome: History, Character, and Diagnostic Prospects. Molecular & Cellular Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 23.Anderson NL. The clinical plasma proteome: a survey of clinical assays for proteins in plasma and serum. Clinical chemistry. 2010;56:177–185. doi: 10.1373/clinchem.2009.126706. [DOI] [PubMed] [Google Scholar]
- 24.Anderson NL, Polanski M, Pieper R, Gatlin T, et al. The human plasma proteome: a nonredundant list developed by combination of four separate sources. Molecular & cellular proteomics : MCP. 2004;3:311–326. doi: 10.1074/mcp.M300127-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Muthusamy B, Hanumanthu G, Suresh S, Rekha B, et al. Plasma Proteome Database as a resource for proteomics research. Proteomics. 2005;5:3531–3536. doi: 10.1002/pmic.200401335. [DOI] [PubMed] [Google Scholar]
- 26.Hoofnagle AN, Wener MH. The fundamental flaws of immunoassays and potential solutions using tandem mass spectrometry. Journal of immunological methods. 2009;347:3–11. doi: 10.1016/j.jim.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoofnagle AN. Quantitative Clinical Proteomics by Liquid Chromatography–Tandem Mass Spectrometry: Assessing the Platform. Clinical Chemistry. 2010;56:161–164. doi: 10.1373/clinchem.2009.134049. [DOI] [PubMed] [Google Scholar]
- 28.Villanueva J, Carrascal M, Abian J. Isotope dilution mass spectrometry for absolute quantification in proteomics: Concepts and strategies. Journal of Proteomics. 2014;96:184–199. doi: 10.1016/j.jprot.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 29.Kay RG, Gregory B, Grace PB, Pleasance S. The application of ultra-performance liquid chromatography/tandem mass spectrometry to the detection and quantitation of apolipoproteins in human serum. Rapid Communications in Mass Spectrometry. 2007;21:2585–2593. doi: 10.1002/rcm.3130. [DOI] [PubMed] [Google Scholar]
- 30.Krastins B, Prakash A, Sarracino DA, Nedelkov D, et al. Rapid development of sensitive, high-throughput, quantitative and highly selective mass spectrometric targeted immunoassays for clinically important proteins in human plasma and serum. Clinical Biochemistry. 2013;46:399–410. doi: 10.1016/j.clinbiochem.2012.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agger SA, Marney LC, Hoofnagle AN. Simultaneous quantification of apolipoprotein A-I and apolipoprotein B by liquid-chromatography-multiple- reaction-monitoring mass spectrometry. Clinical chemistry. 2010;56:1804–1813. doi: 10.1373/clinchem.2010.152264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoofnagle AN, Becker JO, Oda MN, Cavigiolio G, et al. Multiple-reaction monitoring-mass spectrometric assays can accurately measure the relative protein abundance in complex mixtures. Clinical chemistry. 2012;58:777–781. doi: 10.1373/clinchem.2011.173856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ronsein GE, Pamir N, von Haller PD, Kim DS, et al. Parallel reaction monitoring (PRM) and selected reaction monitoring (SRM) exhibit comparable linearity, dynamic range and precision for targeted quantitative HDL proteomics. Journal of Proteomics. 2015;113:388–399. doi: 10.1016/j.jprot.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grant RP, Hoofnagle AN. From Lost in Translation to Paradise Found: Enabling Protein Biomarker Method Transfer by Mass Spectrometry. Clinical Chemistry. 2014;60:941–944. doi: 10.1373/clinchem.2014.224840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandberg S, Fraser Callum G, Horvath Andrea R, Jansen R, et al. Clinical Chemistry and Laboratory Medicine (CCLM) 2015:833. doi: 10.1515/cclm-2015-0067. [DOI] [PubMed] [Google Scholar]
- 36.Ricós C, Alvarez V, Cava F, García-Lario JV, et al. Current databases on biological variation: pros, cons and progress. Scandinavian Journal of Clinical and Laboratory Investigation. 1999;59:491–500. doi: 10.1080/00365519950185229. [DOI] [PubMed] [Google Scholar]
- 37.Perich C, Minchinela J, Ricós C, Fernández-Calle P, et al. Clinical Chemistry and Laboratory Medicine (CCLM) 2015:299. doi: 10.1515/cclm-2014-0739. [DOI] [PubMed] [Google Scholar]
- 38.Ryan RO, Forte TM, Oda MN. Optimized bacterial expression of human apolipoprotein A-I. Protein Expression and Purification. 2003;27:98–103. doi: 10.1016/s1046-5928(02)00568-5. [DOI] [PubMed] [Google Scholar]
- 39.Consortium TU. UniProt: a hub for protein information. Nucleic Acids Research. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacLean B, Tomazela DM, Shulman N, Chambers M, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lynch KL. CLSI C62-A: A New Standard for Clinical Mass Spectrometry. Clinical Chemistry. 2016;62:24–29. doi: 10.1373/clinchem.2015.238626. [DOI] [PubMed] [Google Scholar]
- 42.MacLean B, Tomazela DM, Abbatiello SE, Zhang S, et al. Effect of Collision Energy Optimization on the Measurement of Peptides by Selected Reaction Monitoring (SRM) Mass Spectrometry. Analytical Chemistry. 2010;82:10116–10124. doi: 10.1021/ac102179j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henderson CM, Lutsey PL, Misialek JR, Laha TJ, et al. Measurement by a Novel LC-MS/MS Methodology Reveals Similar Serum Concentrations of Vitamin D–Binding Protein in Blacks and Whites. Clinical Chemistry. 2015 doi: 10.1373/clinchem.2015.244541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature. 2012;483:531–533. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- 45.Carr SA, Abbatiello SE, Ackermann BL, Borchers C, et al. Targeted Peptide Measurements in Biology and Medicine: Best Practices for Mass Spectrometry-based Assay Development Using a Fit-for-Purpose Approach. Molecular & Cellular Proteomics. 2014;13:907–917. doi: 10.1074/mcp.M113.036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner M, Tonoli D, Varesio E, Hopfgartner G. The use of mass spectrometry to analyze dried blood spots. Mass Spectrometry Reviews. 2015:361–438. doi: 10.1002/mas.21441. [DOI] [PubMed] [Google Scholar]
- 47.De Kesel PMM, Sadones N, Capiau S, Lambert WE, Stove CP. Hemato-critical issues in quantitative analysis of dried blood spots: challenges and solutions. Bioanalysis. 2013;5:2023–2041. doi: 10.4155/bio.13.156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.