Abstract
Interstitial fibrosis and tubular atrophy (IFTA) is found in approximately 25% of 1-year biopsies posttransplant. It is known that IFTA correlates with decreased graft survival when histological evidence of inflammation is present. Identifying the mechanistic etiology of IFTA is important to understanding why long-term graft survival has not changed as expected despite improved immunosuppression and dramatically reduced rates of clinical acute rejection (AR) (Services UDoHaH. http://www.ustransplant.org/annual_reports/current/509a_ki.htm). Gene expression profiles of 234 graft biopsy samples were obtained with matching clinical and outcome data. Eighty-one IFTA biopsies were divided into subphenotypes by degree of histological inflammation: IFTA with AR, IFTA with inflammation, and IFTA without inflammation. Samples with AR (n = 54) and normally functioning transplants (TX; n = 99) were used in comparisons. A novel analysis using gene coexpression networks revealed that all IFTA phenotypes were strongly enriched for dysregulated gene pathways and these were shared with the biopsy profiles of AR, including IFTA samples without histological evidence of inflammation. Thus, by molecular profiling we demonstrate that most IFTA samples have ongoing immune-mediated injury or chronic rejection that is more sensitively detected by gene expression profiling. These molecular biopsy profiles correlated with future graft loss in IFTA samples without inflammation.
Introduction
Interstitial fibrosis and tubular atrophy (IFTA) describes a common histological abnormality seen in kidney transplant biopsies in which normal cortical structures are replaced by interstitial fibrosis. IFTA, when accompanied by histological evidence of inflammation, correlates with decreased graft survival (1–3). IFTA is evident histologically in 25% or more of 1-year surveillance biopsies despite concomitant stable renal function (4,5). Identifying the etiologic mechanisms of IFTA is important to better understand why 10-year graft survival has not improved significantly despite improved immunosuppression protocols and a dramatic decrease in the incidence of clinical acute rejection (AR) (6–8).
Acute T cell–mediated rejection (TCMR), presenting as either AR or subclinical acute rejection (subAR, histological AR without graft dysfunction only demonstrated by surveillance biopsies), is clearly linked to a higher risk of IFTA (3,9,10). In a study of 797 recipients, early episodes of AR led to more fibrosis and inflammation in 1- and 2-year protocol biopsies than those without an occurrence of AR. AR episodes followed by abnormal histology also resulted in reduced graft survival (9). Likewise, subAR also increases the risks of developing IFTA and graft loss and occurs in as many as 20% of surveillance biopsies done in the first year posttransplant (1,11–15). Given these strong associations of AR and subAR with the future development of IFTA, we questioned whether IFTA biopsies contained unrecognized cellular rejection. In our model, IFTA marks chronically uncontrolled rejection, and its development may associate with a higher risk of graft failure.
We performed gene expression profiling on 234 kidney graft biopsies obtained for both surveillance and cause from over 1000 patients at seven transplant centers with matching clinical and outcome data. Eighty-one samples were given a diagnosis of IFTA, in which there was histological evidence of IFTA without a clear etiology (i.e. BK nephropathy or recurrent glomerulonephritis). These IFTA samples were then classified into subphenotypes based on the degree of inflammation identified on light histology, including IFTA with concomitant acute rejection (IFTA with AR; n = 29), IFTA with inflammation (n = 10), and IFTA without inflammation (n = 42). Samples with biopsy-proven AR (n = 54) and normally functioning transplants (TX; n = 99) were included for comparison. Confirmatory outcome data were obtained by data query to the United Network for Organ Sharing. The gene expression results were validated using a published dataset derived from an independent, external cohort of late biopsies (Gene Expression Omnibus [GEO]; GSE21374) (16,17).
By molecular biopsy profiling we found that differential gene expression in all IFTA phenotypes was strongly enriched for the same dysregulated gene profiles seen in AR biopsies. All IFTA phenotypes (n = 81) demonstrated as much as 81% commonality in differentially expressed genes with AR, and a strong enrichment for AR immune/inflammatory and metabolic/tissue integrity molecular pathways. This finding was true even for IFTA samples without any histological evidence of inflammation (n = 42), a group currently thought to be low risk for graft loss. Thus, molecular profiling indicated that most IFTA samples have ongoing and often subclinical immune-mediated injury that is more sensitively detected with gene expression profiling than by light histology. Furthermore, in IFTA samples without histological evidence of inflammation, we found that the relative expression of AR-affiliated genes correlated with a higher risk of graft loss at 5 or more years.
Methods
Study population
Two hundred thirty-four kidney allograft biopsies were collected as part of an National Institutes of Health–funded Transplant Genomics Collaborative Group from 2005 to 2011 by protocol or “for cause” from 210 patients from seven clinical centers. More than one biopsy from the same patient was included only if there was a change in pathology. The only exclusions were biopsies that did not conform to the study’s inclusion/exclusion criteria (Appendix S1), such as a diagnosis of BK nephritis or recurrent glomerulonephritis (n = 5). Each biopsy was reviewed locally as well as by a blinded central pathologist (LG) with no clinical information provided. When there was a discrepancy between the two reports, the senior investigator (DRS) reviewed the histology slides and reached a conclusion including discussion and agreement with the pathologists as necessary. The phenotypes were defined as follows: AR is biopsy-proven TCMR with a rising serum creatinine; IFTA with inflammation is Banff IFTA+i; IFTA with AR are cases where local and central pathology reviews called both present and TX are controls based on surveillance biopsies done from 1 to 2 years. Institutional review boards approved all research protocols.
Analysis of phenotypic data
ANOVA and chi-squared tests were used to detect differences in continuous and categorical variables between phenotypes and p-values were adjusted with Bonferroni correction for multiple hypothesis testing. Less than 1% of the phenotypic features were missing. Survival curve analysis was performed on death-censored data using JMP software (SAS, Cary, NC) and Wilcoxon’s ranked tests. Hazard ratios for clinical phenotypic characteristics were calculated using a Cox proportional hazards model adjusting for multiple clinical variables: age, sex, race/ethnicity, time posttransplant, C4d, donor age, BMI, and phenotypes (see Results and Appendix S2).
Differential gene expression and pathway mapping
Microarray protocols are in Appendix S1 and array data is available online (NCBI’s Gene Expression Omnibus database; http://www.ncbi.nlm.nih.gov/geo/; Accession number GSE GSE76882). Differentially expressed genes (DEGs) between phenotypes were determined by two-sample t-tests with False Discovery Rates (FDRs) calculated using the method of Storey et al (18) to account for multiple hypothesis testing. Immune pathway mapping and gene set enrichment for biological processes were performed using gene ontology (GO) and Ingenuity Pathway Analysis. To avoid false-positive enrichment based on cell type, kidney gene expression (as found in our biopsy dataset) was used as the background gene set.
Gene Coexpression Network Analysis
By having gene expression profiles for many samples, we can look for pairs of genes that demonstrate a similar expression pattern across samples (two genes in which the transcript levels rise and fall together across the samples). These two genes are called “coexpressed genes.” Gene coexpression is of biological interest since it suggests a relationship among coexpressed genes. A gene coexpression network (GCN) is simply an undirected graph where each node corresponds to a gene, and each gene is linked to other genes by an edge if there exists a statistically significant coexpression. GCNs do not attempt to infer a causal relationship between genes and the edges represent only a correlation in gene expression across samples.
GCNs can separate groups of similar-behaving (and likely to be biologically related) genes from a larger gene set, and do so without the introduction of user bias when groups of genes are identified based on investigator interpretations of external data and immune paradigms. Thus, these groups of genes or GCNs help identify related genes with a specific function within the framework of a larger biological process (e.g. coexpressed immunoglobulin genes within a large set of genes differentially expressed in AR). In this study, we built GCNs from IFTA and AR differentially expressed genes, and thus delineated the biological processes that define these phenotypes. The mathematical model and full explanation for GCN construction is outlined in Appendix S1, Section 4.
Results
Patient characteristics and outcomes
A total of 234 biopsies (114 surveillance, 120 “for cause”) comprise this retrospective study (54 AR, 42 IFTA without inflammation, 10 IFTA with inflammation, 29 IFTA with AR, and 99 TX; Table 1). Twenty-one of the participants had two biopsies analyzed, but the biopsies were taken at different time points and demonstrated a change in pathology. Only the phenotype at the time of most recent biopsy was used to calculate survival analysis. Thirty-three (44%) of all IFTA samples were classified as mild (Banff Grade 1: IFTA without inflammation = 45%; IFTA+AR = 41%; IFTA+i = 50%). Twenty-eight (40%) of IFTA samples were classified as moderate (Grade 2; IFTA without inflammation = 40%; IFTA+AR = 36%; IFTA+i = 50%). The remaining 11 (16%) were classified as severe IFTA (Grade 3). There were no differences in IFTA grades by subgroups (p = 0.67).
Table 1.
Demographics and outcomes of 210 participants grouped by histological phenotypes
| AR | IFTA without inflammation | IFTA with AR | IFTA with inflammation | TX | Group compare1 | |
|---|---|---|---|---|---|---|
| Donors | ||||||
| Age (mean ± SE) | 39 ± 2 | 40 ± 2 | 35 ± 3 | 41 ± 5 | 41 ± 2 | 0.8 |
| Female2 | 22 (47%) | 23 (58%) | 12 (43%) | 5 (50%) | 37 (47%) | 0.61 |
| Black | 2 (4%) | 5 (13%) | 1 (4%) | 0 | 4 (5%) | 0.39 |
| Recipients | ||||||
| Total number of patients | 50 | 40 | 28 | 10 | 82 | N/A |
| Total number of biopsies | 54 | 42 | 29 | 10 | 99 | N/A |
| IFTA Grade (Banff)2 1/2/3 | N/A | 18/16/6 | 9/8/5 | 4/4/0 | N/A | 0.67 |
| Protocol biopsy | 2/50 (4%) | 13/40 (33%) | 1/28 (4%) | 3/10 (30%) | 72/82 (88%) | <0.0001 |
| Age (mean ± SE) | 46 ± 2 | 44 ± 2 | 40 ± 3 | 49 ± 5 | 50 ± 2 | 0.02 |
| Female | 15 (30%) | 19 (48%) | 12 (43%) | 3 (30%) | 28 (34%) | 0.55 |
| African American | 6 (12%) | 4 (10%) | 5 (18%) | 1 (10%) | 6 (7%) | 0.64 |
| Diabetes2 | 5 (10%) | 4 (11%) | 3 (11%) | 2 (20%) | 11 (13%) | 0.91 |
| Deceased donor2 | 30 (60%) | 24 (62%) | 16 (64%) | 7 (78%) | 43 (52%) | 0.57 |
| HLA mm (mean ± SE)3 | 3.8 ± 0.3 | 3.3 ± 0.3 | 3.1 ± 0.4 | 3.7 ± 0.7 | 3.6 ± 0.2 | 0.54 |
| PRA ≥ 202 | 11 (22%) | 8 (22%) | 4 (17%) | 1 (13%) | 11 (13%) | 0.77 |
| Induction therapy4 | 42 (84%) | 34 (85%) | 25 (89%) | 10 (100%) | 62 (76%) | 0.24 |
| C4d positive | 11 (22%) | 2 (5%) | 5 (18%) | 1 (10%) | 1 (1%) | <0.0001 |
| Borderline or suspicious for acute cellular rejection (according to local pathologist) | N.A. | 14 (33%) | N.A. | 6 (60%) | 2 (2%) | <0.0001 |
| Time to biopsy (median days; interquartile range) | 385; 105–1159 | 1105; 377–2875 | 1719; 1178–2977 | 489; 231–1692 | 376; 362–425 | <0.0001 |
| Biopsy >12 months | 27 (54%) | 34 (85%) | 27 (96%) | 6 (60%) | 60 (73%) | <0.0001 |
| Death-censored graft loss | 15 (32%) | 14 (35%) | 11 (38%) | 3 (30%) | 0 | N/A |
| Time to death-censored graft loss | 1450 ± 334 | 2935 ± 346 | 2747 ± 390 | 1708 ± 747 | N.A. | 0.015 |
| Time from biopsy to graft loss | 665 ± 183 | 452 ± 189 | 678 ± 213 | 412 ± 408 | N.A. | 0.78 |
| Death | 12 (24%) | 6 (15%) | 2 (7%) | 1 (10%) | 3 (4%) | N/A |
| Time to death | 1304 ± 273 | 1813 ± 385 | 1417 ± 667 | 1324 ± 944 | 1549 ± 408 | 0.87 |
| Clinical center enrollment | ||||||
| CCF | 9 (18%) | 5 (12.5%) | 2 (7%) | 2 (20%) | 12 (15%) | N/A |
| SGH | 10 (20%) | 7 (17.5%) | 2 (7%) | 2 (20%) | 48 (58%) | N/A |
| SVMC | 20 (40%) | 14 (35%) | 15 (54%) | 4 (40%) | 2 (2%) | N/A |
| MC | 1 (2%) | 5 (12.5%) | 0 | 2 (20%) | 14 (17%) | N/A |
| UCHSC | 6 (12%) | 7 (17.5%) | 5 (18%) | 0 | 3 (4%) | N/A |
| UM | 1 (2%) | 2 (5%) | 4 (14%) | 0 | 0 | N/A |
| NU | 3 (6%) | 0 | 0 | 0 | 3 (4%) | N/A |
AR, acute rejection; CCF, Cleveland Clinic Foundation; HLA, human leukocyte antigen; IFTA, interstitial fibrosis and tubular atrophy; MC, Mayo Clinic, Phoenix; mm, mismatch; N/A, not applicable; PRA, panel reactive antibody; SE, standard error; SGH, Scripps Green Hospital; SVMC, Saint Vincent’s Medical Center, Los Angeles; TX, Treatment group with excellent functioning kidney; UCHSC, University of Colorado Health Sciences Center; UM, University of Michigan; NU, Northwestern University.
Analysis method: analysis of variance for quantitative data (probability > F statistic), Pearson’s chi-squared test for dichotomous data. Significant intergroup comparisons found in Appendix S2.
Missing data: 9 (3%) diabetes, 5 (2%) donor sex, 6 (2%) donor race, 27 (10%) PRA studies, 11 (4%) deceased donor, IFTA grade 4 (5%).
Typed HLA antigens: HLA-A1, HLA-A2, HLA-B1, HLA-B2, HLA-DR1, HLA-DR2.
Induction therapy includes the following: anti-thymocyte globulin (Thymoglobulin), muromonab-CD3 (OKT3), basiliximab (Simulect), daclizumab (Zenapax), and alemtuzumab (Campath).
Median follow-up time was 1613 days posttransplant (≈4.4 years). Only one patient was lost to follow-up. There were no differences in age, sex, % African American, % diabetics, number of HLA mismatches, or % deceased donors across phenotypes. There were a total of 24 deaths, but no significant differences in mortality in the “non-TX” groups according to survival analyses.
Median time to biopsy was 420 days (374 and 1200 days for surveillance and “for cause,” respectively). The times to biopsy were significantly greater for AR (800 ± 164), IFTA without inflammation (1796 ± 178), IFTA with inflammation (1008 ± 356), and IFTA with AR (2121 ± 213) when compared to the TX phenotype (603 ± 127 days) (p < 0.0001). In over half of the subjects with AR, onset was >12 months posttransplant.
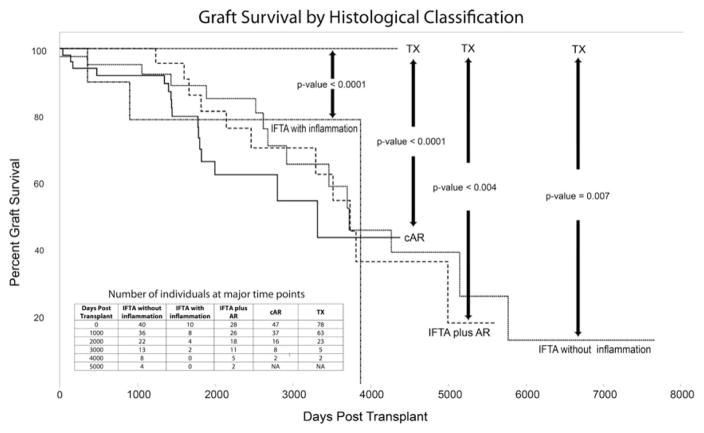
After censoring death, 43/210 (20%) had graft loss with a median time of 1885 days (≈5.2 years; 43–9302 days). Graft survival was significantly lower in subjects with AR, IFTA with AR, IFTA with inflammation, and IFTA without inflammation in comparison to TX (Figure 1). Despite differences in graft loss risk, times from biopsy to graft loss did not significantly differ by phenotype: IFTA with inflammation (412 days), IFTA without inflammation (452 days), AR (665 days), and IFTA with AR (678 days) (p = 0.78).
Figure 1. Graft survival according to histological phenotype.
Interstitial fibrosis and tubular atrophy (IFTA) samples were classified into three subphenotypes according to the degree of inflammation: IFTA plus clinical acute rejection (AR), IFTA with inflammation, and IFTA without inflammation. Biopsies with only AR and normally functioning transplants (TX) were used for survival comparisons. The figure shows graft survival according to these phenotypes in days posttransplant. The insert table shows the number of subjects at key time points by phenotypes.
A Cox proportional hazards model was also used to examine the effect of various clinical variables on survival times. We created a model including the following variables: time from transplant to biopsy, phenotype, age, sex, black race, diabetes, C4d status, and donor age (Appendix S2, Section 1). Of these variables, only days from transplant to biopsy (p < 0.0001), phenotype (p < 0.0001), and recipient age (p = 0.04) were found to be statistically significant. We then adjusted the above survival curves for age and time of biopsy posttransplant using a Stratified Cox model. In the adjusted model, both AR and IFTA phenotypes showed the same results of equally poor long-term graft survival rates (Appendix S2, Section 2).
A majority (n = 84; 71%) of the “for cause” biopsies and a minority (n = 21; 19%) of the protocol biopsies had C4d staining performed. There was no difference in death-censored graft survival between those with positive versus negative C4d staining (p = 0.3). The calculated Cox hazard ratios for C4d positivity versus negativity were not statistically significant (CI: 0.58–4.2) (Appendix S2, Section 1). The majority of the samples with future graft loss were C4d negative (74%). We do not have donor-specific antibody (DSA) data. These biopsies were collected prior to the current practices of measuring serial DSAs.
Gene expression comparison between AR and IFTA samples
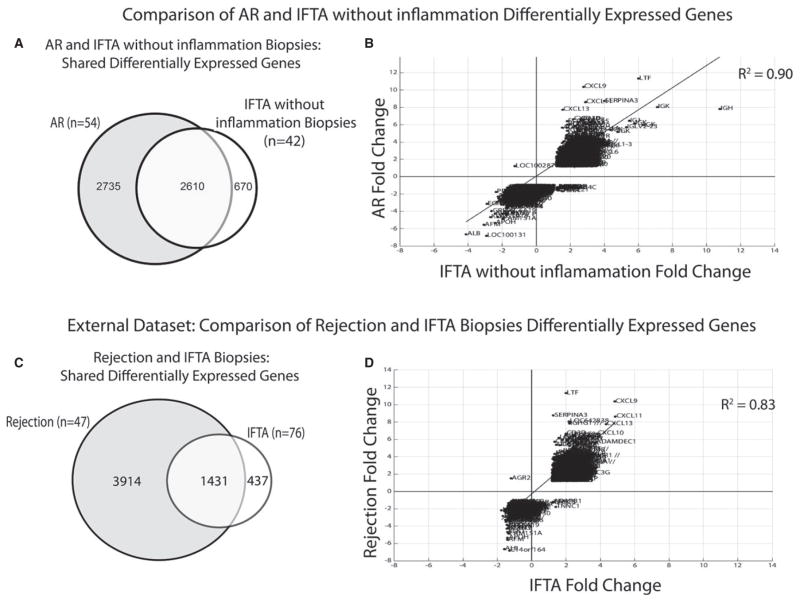
Four gene expression profiles were created by independently comparing each histological phenotype (AR, IFTA with AR, IFTA with inflammation, and IFTA without inflammation) to the controls (TX). A threshold calculated FDR of <0.05 and fold change (FC) of >1.2 was used (full gene lists; Appendix S3). The majority (72–81%) of DEGs in biopsies with IFTA and histological evidence of inflammation were common to AR DEGs (Table 2). Surprisingly, DEGs for IFTA without inflammation were also highly shared with AR (80%; Figure 2A) and differentially expressed in a concordant pattern (Figure 2B). Moreover, 25 of the top 50 IFTA without inflammation DEGs (ranked by absolute FC) were shared with the top 50 for AR. A literature review of the top IFTA without inflammation DEGs showed that these have been associated with AR in prior studies (Table 3) (16,19–43). Finally, there was strong enrichment for AR immune/inflammatory and metabolic molecular pathways using Ingenuity gene set enrichment tools (Table 4).
Table 2.
Shared differentially expressed transcripts between IFTA subphenotypes (IFTA plus AR, IFTA with inflammation, and IFTA without inflammation) and clinical acute rejection (cAR)1
| All samples with IFTA (n = 78) | IFTA without inflammation (n = 40) | IFTA with inflammation (n = 10) | IFTA plus AR (n = 28) | |
|---|---|---|---|---|
| Number of DEGs | 4705 | 3280 | 1513 | 6229 |
| Number (%) shared with cAR differentially expressed transcript list | 3817 (81%) | 2610 (80%) | 1040 (69%) | 4466 (72%) |
AR, acute rejection; cAR, clinical acute rejection; DEGs, differentially expressed genes; IFTA, interstitial fibrosis and tubular atrophy; FC, fold-change; FDR, false discovery rate.
In comparison of AR samples to patients with normal, well-functioning transplants (control; TX), there were 5345 differentially expressed transcripts (FDR*<0.05; FC*>1.2). This table shows the large number and percent of gene transcripts shared between cAR and each IFTA subphenotype.
Figure 2. Differentially expressed genes shared between IFTA and AR.
(A) Venn diagram showing differentially expressed genes (DEGs) shared between interstitial fibrosis and tubular atrophy (IFTA) without inflammation and clinical acute rejection (AR). (B) Plots the differential fold changes in gene expression (DEGs) comparing IFTA without inflammation versus AR. A linear regression line and R2 statistic demonstrates a highly concordant direction of gene expression between phenotypes; (C) and (D) repeat and validate the analysis using an independent, external dataset. Note 1: Differentially expressed genes and fold changes are calculated in relation to normal transplants (TX) defined by stable function and light histology. Note 2: The subphenotypes of IFTA with and without inflammation were not available for the external data set.
Table 3.
IFTA without inflammation differentially expressed genes (DEGs) (threshold FDR<0.05) ranked by absolute fold change are shared with AR1
| IFTA gene | IFTA FC | Function | External literature AR citings | AR DEG rank | Significance |
|---|---|---|---|---|---|
| IGHC | 2.7–10.8 | Ig heavy chain production | Tissue (19,20) | 6 | Indicates immunoglobulin producing allograft infiltrating B cells |
| IGLC (9)1 | 2.6–7.1 | Ig light chain production | Tissue (19,20) | 5 | Indicates immunoglobulin producing allograft infiltrating B cells |
| LTF | 6.1 | Innate anti-microbial, inflammation-related | Tissue (16,20–24) | 1 | Literature review connects with kidney injury and #1 AR gene, which links AR and IFTA to injury |
| IGJ | 5.9 | Ig linker protein | Tissue (20) | 12 | Indicates Immunoglobulin producing allograft infiltrating B cells |
| ALB | −4.1 | Main protein in blood, carrier protein for steroids, fatty acids and hormones. | Tissue (25) | 8 | Previously showed to be decreased in AR (25) and kidney injury, (35) likely reflects metabolic disturbance. |
| SERPINA3 | 4.0 | Protease inhibitor, cleaves PMN cathepsin G and MC chymase | Tissue (16,23,24,26,27) | 3 | Literature review associates with kidney injury and cell turnover. As the #3 AR gene, connects AR and IFTA to injury |
| CXCL6 | 3.5 | Interacts with CXCR1 and 2, chemoattractant for PMNs | Tissue (16) | 248 | Indicates likely neutrophil allograft chemotaxis |
| IL7R | 3.4 | Cell surface marker for memory T cells, important specifically for T cell development and VDJ recombination. | N/A | 44 | Signifies the presence memory T cells and may indicate ongoing activation and/or development. |
| DARC | 3.3 | Promiscuous chemokine-binding, acts as chemokine scavenging and decoy receptor, regulating chemokine bioavailability and likely leukocyte recruitment | N/A | 113 | Indicative of ongoing immune mechanisms |
| CCL5 | 3.2 | A.k.a. RANTES, chemokine secreted late after T cell activation, induced by IFN-γ, secreted by CD8+ T cells. Chemotactic for T cells, eosinophils, and basophils. Activates NK cells. | Tissue (19,21,28–30) | 14 | Strongly associated with AR and indicative of Th1 and NK cell activation |
| CPA3 | 3.2 | Mast cell carboxypeptidase A, degrades chymases and tryptases | N/A | 112 | Indicates the presence of mast cells in IFTA and AR, which may be causative of interstitial fibrosis |
| SLPI | 3.2 | Secreted leukocyte serine protease inhibitor, likely protects epithelial surfaces from endogenous proteolytic enzymes. | N/A | 38 | Likely parallels immune response to limit damage by leukocytes, specifically neutrophils |
| CORO1A | 3.1 | Interacts with actin and involved in a number of cellular processes such as cell locomotion, phagocytosis and NK cell cytotoxicity. Deficiency in CORO1 associated with TSCID. | Tissue (19) | 29 | Indicative of ongoing immune mechanisms |
| ISG20 | 3.1 | Interferon-induced antiviral exoribonuclease with antiviral activity (56) | Tissue (19,21,25,31,32) | 22 | Induced by IFN-γ and strongly associated with AR |
| AFM | −3.1 | Albumin family member, may transport vitamin E. | Blood (33) Tissue (16,34,35) |
26 | Previously showed to be decreased in AR (25) and kidney injury, (35) likely reflects metabolic disturbance. High coexpression with ALB gene. |
| REG1A | 3.0 | Associated with brain and pancreas regeneration | NA | 75 | N/A |
| CD2 | 3.0 | Costimulatory and cell adhesion molecule on NK and T cells | Tissue (19,25,31) | 24 | Strongly associated with AR and indicative of Th1 and NK cell activation |
| CD52 | 2.9 | A.k.a. CAMPATH-1 antigen. Unknown function on mature lymphocytes | N/A | 17 | Indicates presence of mature lymphocytes |
| EGF | −2.9 | Epidermal growth factor | Tissue (36) | 177 | Indicates decreased growth and proliferation factor, which may contribute to kidney injury |
| CXCL11 | 2.9 | CXCR3 ligand, Chemokine, IFN-γ inducible | Urine (37) Tissue (28,29) |
4 | Induced by IFN-γ and strongly associated with AR |
| CXCL1 | 2.9 | Secreted growth factor that signals through CXCR2. Has chemotactic activity for PMNs. | Tissue (16,38) | 99 | Similar to CXCL6 above, indicates likely neutrophil chemotaxis |
| EVI2B | 2.8 | N/A | N/A | 34 | N/A |
| CXCL9 | 2.8 | CXCR3 ligand, Chemokine, IFN-γ inducible | Tissue (28,29,32,39) Urine (37,40–43) |
2 | Induced by IFN-γ and strongly associated with AR, suggests TCMR |
| GZMA | 2.8 | Fond in CTL and NK cell cytolytic granules, responsible for cytotoxicity of these | Tissue (20,21,25,28–30) | 15 | Found in NK and CTLs, strongly associated with AR and indicative of TCMR |
For each gene in the interstitial fibrosis and tubular atrophy (IFTA) ranked gene list, the table gives the function and its association with acute rejection (AR). The third column provides the biological function, and often demonstrates a gene with a role in immune response and inflammation. The fourth column provides literatures references where the genes are linked to AR in prior studies. The fifth column gives the ranking in the AR ranked gene list and thus demonstrates that these genes are also some of the most highly ranked AR genes. FC, fold change; FDR, false discovery rate; N/A, not applicable; PMN, polymorphonuclear leukocytes.
Table 4.
Results of pathway and gene enrichment tool analysis for cAR and IFTA without inflammation differentially expressed transcripts1
| cAR | IFTA without inflammation | ||
|---|---|---|---|
|
|
|
||
| Canonical Pathway Analysis | p-value2 | Canonical Pathway Analysis | p-value2 |
| Communication between innate and adaptive immune cells | 2.00E-13 | Granulocyte adhesion and diapedesis | 3.16E-12 |
| Allograft rejection signaling | 6.31E-12 | Antigen presentation pathway | 3.98E-12 |
| Antigen presentation pathway | 7.94E-11 | Allograft rejection signaling | 1.51E-09 |
| Dendritic cell maturation | 1.23E-10 | Dendritic cell maturation | 1.51E-09 |
| Graft-versus-host disease signaling | 1.38E-10 | Agranulocyte adhesion and diapedesis | 2.19E-09 |
| FXR/RXR activation | 2.29E-09 | B cell development | 6.91E-09 |
| B cell development | 4.90E-09 | Role of NFAT in regulation of the immune response | 1.74E-08 |
| LPS/IL-1 mediated inhibition of RXR function | 1.02E-08 | Communication between innate and adaptive immune cells | 5.01E-08 |
| OX40 signaling pathway | 3.02E-08 | OX40 signaling pathway | 2.57E-07 |
| Crosstalk between dendritic cells and natural killer cells | 3.02E-08 | Complement system | 3.55E-06 |
|
| |||
| Activated upstream regulator analysis2 | Activated upstream regulator analysis2 | ||
|
| |||
| IFN-γ | 7.22E-79 | IFN-γ | 1.42E-63 |
| TNF | 5.00E-66 | TNF | 5.21E-46 |
| IL-4 | 1.80E-54 | IL-10 | 1.60E-39 |
| IL-1B | 1.77E-48 | IL-1B | 3.09E-37 |
| IFN-α | 1.27E-45 | CD40LG | 1.62E-34 |
| IL-10 | 1.60E-43 | TGF-B1 | 1.11E-30 |
| STAT3 | 7.44E-43 | IL-4 | 1.53E-30 |
| IL-6 | 1.54E-41 | IL-2 | 1.11E-29 |
| STAT1 | 1.59E-41 | IL-6 | 1.10E-28 |
|
| |||
| Inhibited upstream regulator analysis2 | Inhibited upstream regulator analysis2 | ||
|
| |||
| MAPK1 | 1.31E-28 | IL-10RA | 8.89E-20 |
| IL-1RN | 2.13E-27 | PTGER4 | 7.25E-18 |
| IL-10RA | 4.79E-27 | IL-1RN | 1.52E-16 |
| PPARA | 5.83E-21 | CD3 | 8.64E-15 |
| PTGER4 | 3.75E-20 | SOCS1 | 1.00E-14 |
| TRIM24 | 2.84E-19 | PRDM1 | 1.66E-13 |
| NKX2-3 | 5.04E-19 | Nr1 h | 2.04E-13 |
| PRDM1 | 2.40E-18 | NKX2-3 | 2.24E-11 |
cAR, clinical acute rejection; IFN, interferon; IFTA, interstitial fibrosis and tubular atrophy; TNF, tumor necrosis factor.
Mapping of AR and IFTA without inflammation differentially expressed genes (DEGs) to canonical functional biological pathways was performed using Ingenuity Pathway Analysis (IPA). Enrichment of these DEGs for immune and biological pathways was performed by using genes significantly expressed in the kidney as the background. Pathways or genes highlighted in gray are shared between AR and IFTA without inflammation. These data emphasize the high level of shared immune/inflammatory-based pathways according to unbiased pathway enrichment tools.
Benjamini-Hochberg correction applied to p-values account for multiple test comparisons.
These findings were then validated using a publically available gene expression dataset that consisted of 105 “for cause” late biopsies taken between 1 and 31 years posttransplant (GEO; GSE21374).(16) Using this external dataset and our thresholds for FDR and FC, we found that 2523 transcripts (1868 genes) were differentially expressed in subjects with IFTA (Appendix S4). Subphenotypes of IFTA with or without inflammation and IFTA with AR were not specifically described. Nonetheless, DEGs in the external dataset were highly shared with our AR and IFTA biopsy profiles (77%; Figure 2C) and differentially expressed in the same concordant patterns (Figure 2D).
Development of “rejection” GCNs
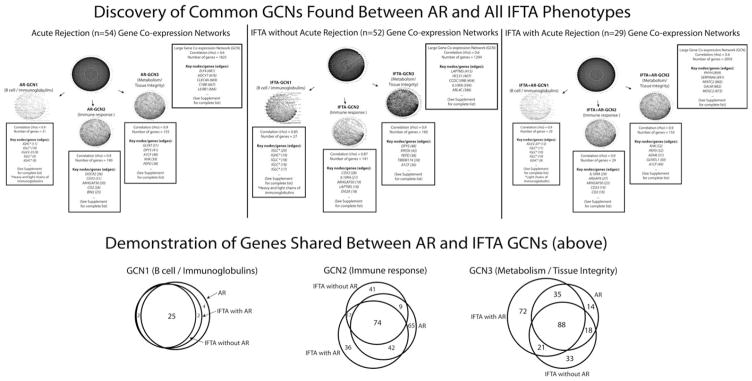
GCNs were created using the DEGs from (1) AR biopsies, (2) IFTA with AR, and (3) IFTA without AR samples. Our intent was to identify groups of genes indicative of discrete acute rejection mechanisms, and then determine and compare the expression of these gene groups in IFTA samples. Using a relatively low coexpression threshold (0.6), a large network of 1825 AR genes was formed (Appendix S5). “Hub” transcripts, or “highly connected” genes with the most connections to other genes in a network, were also determined. Increasing the stringency of the coexpression threshold in order to identify smaller, tighter clusters of coexpressed genes resulted in three major dense networks of AR GCNs (Figure 3; Appendix S5). The same procedure applied to the IFTA samples identified the same three networks as found with the AR samples, reflecting their highly shared molecular mechanisms, and this was confirmed in the external dataset (Appendix S4).
Figure 3. Gene coexpression networks (GCNs).
GCNs were discovered in an unbiased manner using the coexpression of differentially expressed genes for biopsies with clinical acute rejection (AR), interstitial fibrosis and tubular atrophy (IFTA) without AR (i.e. without inflammation), and IFTA with AR (i.e. with inflammation). A number of GCN correlation thresholds (ranging from R2 values of 0.6 to 0.9) were tested to examine both loose and tight networks of coexpressed genes. With an increase in the correlation coefficient threshold, a large GCN network split into three smaller and tighter clusters with common biological functions for each. Genes with the most connections (i.e. edges) to other genes in a network are given for each GCN.
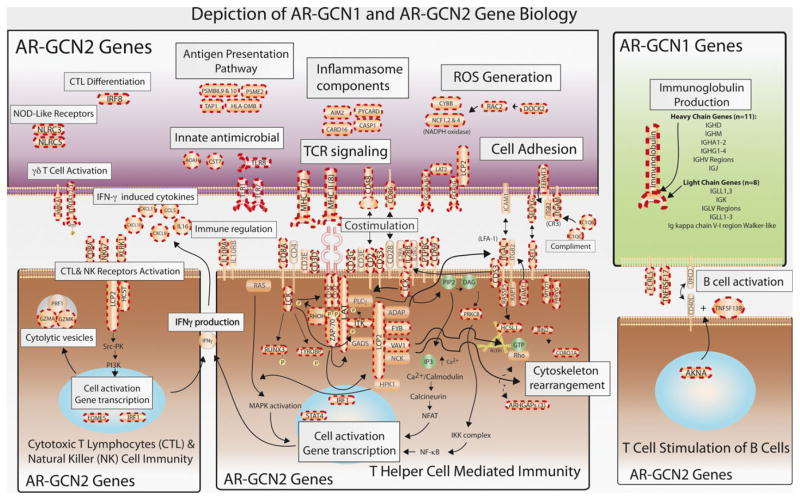
The first network, named AR-GCN1, consisted of only 27 upregulated transcripts, of which 25 were immunoglobulin (93%). The two remaining genes, TNFRSF17 and FCRL5, are B cell receptor–associated transcripts critical for B cell activation. As expected, our biopsies with pathology-defined TCMR contain B cells (44). The second network (AR-GCN2) consisted of 190 genes, all upregulated in AR. One hundred eighty-six of these genes (93%) had known biological functions identifiably related to T cell immune responses and inflammation (Appendix S6). Figure 4 illustrates the function and connection of the AR-GCN1 and AR-GCN2 genes. The illustration includes 107 (56%) of the AR-GCN2 genes. The gene set defining AR-GCN2 was also independently validated using the external GEO data.
Figure 4. Biological functions of clinical acute rejection–gene coexpression network 1 (AR-GCN1) and AR-GCN2 genes.
The figure illustrates the biological functions of 107 (56%) of the AR-GCN2 (immune response/inflammation) genes and all 31 of the ARGCN1 (B cell/immunoglobulin production) genes. The genes in the illustration with dashed red border are present in the GCNs. It is important to note that these genes are essentially the same in IFTA-GCN2.
AR-GCN3 consisted of 186 genes that mapped functionally to cellular metabolism/tissue integrity (Appendix S6). Eighty-nine (48%) of these genes were found to code enzymes important in amino acid turnover, glucose and fatty acid metabolism, and energy production. Twenty-five (13%) coded for proteins involved in cellular detoxification, and 33 (18%) were membrane transporters of various important solutes, organic anions, and drugs. Importantly, all the AR-GCN3 genes are downregulated.
Shared expression of the three key GCNs discovered in AR patients in the IFTA samples
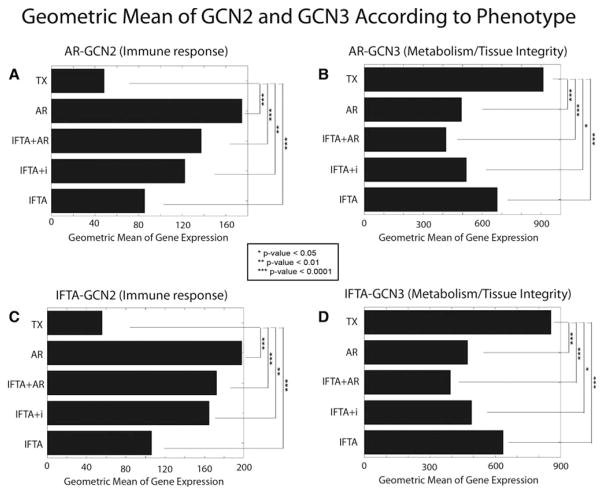
The geometric means of the AR and IFTA GCN genes were next determined for all the IFTA phenotypes. Among the IFTA phenotypes, the geometric mean of GCN2 transcripts (immune response) was highest in samples with IFTA and concomitant histological AR (Figure 5; p = 0.0001 when compared TX). The changes were second highest in IFTA with inflammation samples, and lowest in IFTA without inflammation samples. Of note, the expression in IFTA without inflammation was still significantly higher than TX (p = 0.003), which demonstrates the key point of the increased sensitivity of gene expression profiling to detect an ongoing immune response and inflammation. The geometric means of the metabolism/tissue integrity–related AR-GCN3 genes showed the same hierarchy in the inverse direction compared to TX controls from the lowest in IFTA plus AR, higher in IFTA with inflammation, and highest in IFTA without inflammation (Figure 5). Thus, metabolic and tissue integrity gene dysregulation tracks with degrees of inflammation.
Figure 5. Using the geometric means for each gene coexpression network (GCN) to rank the impact by phenotype.
(A) Geometric means of AR-GCN2 transcripts (immune response/inflammation) correlated with the degree of histological inflammation: clinical acute rejection (AR) > interstitial fibrosis and tubular atrophy (IFTA) with AR > IFTA with inflammation (IFTA+i) > IFTA without inflammation > transplants with stable function and normal histology (TX). Note that the geometric mean of AR-GCN2 in IFTA without inflammation was still significantly higher than TX (p = 0.003). (B) In contrast, the geometric means of AR-GCN3 transcripts (metabolism/tissue integrity) were inversely related to inflammation: TX > IFTA without inflammation > AR > IFTA with inflammation > IFTA plus AR. (C and D) Same analyses using the IFTA-GCNs. *p-value < 0.05, **p-value < 0.01, ***p-value < 0.0001.
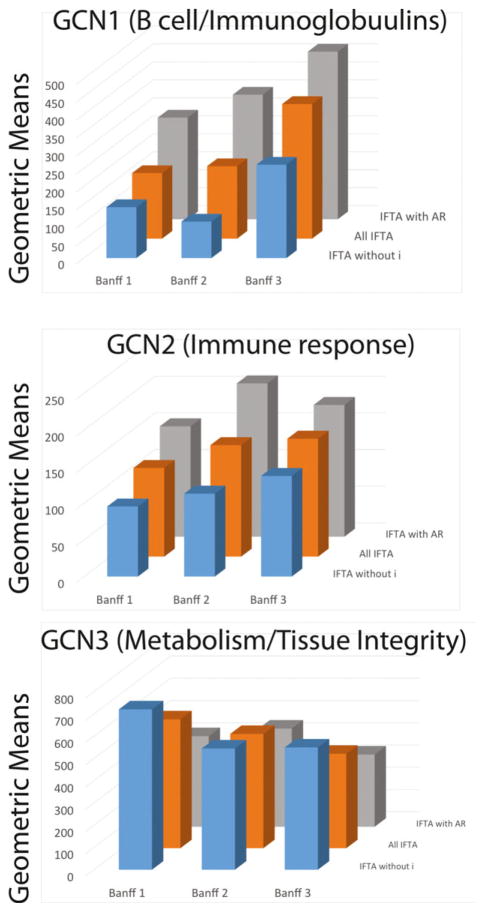
Next, we examined the geometric means according to IFTA grades: Banff 1 (mild), 2 (moderate), and 3 (severe). The geometric means of GCN1 and GCN2 increase in relation to both the degree of inflammation and the severity of IFTA (Figure 6). Likewise, the geometric mean of GCN3 decreases with both the degree of inflammation and the extent of IFTA.
Figure 6. Correlations between biopsy histology, Banff interstitial fibrosis and tubular atrophy (IFTA) grades, and the geometric means of the three IFTA–coexpression networks (GCNs).
The geometric means (y-axis) are plotted as a function of three interstitial fibrosis and tubular atrophy (IFTA) phenotypes: IFTA with AR, all IFTA biopsies, and IFTA without inflammation (IFTA without i) on the z-axis. In parallel, the geometric means are plotted as a function of Banff IFTA severity grades (x-axis).
IFTA-GCNs correlate with graft loss in biopsies with IFTA and no inflammation
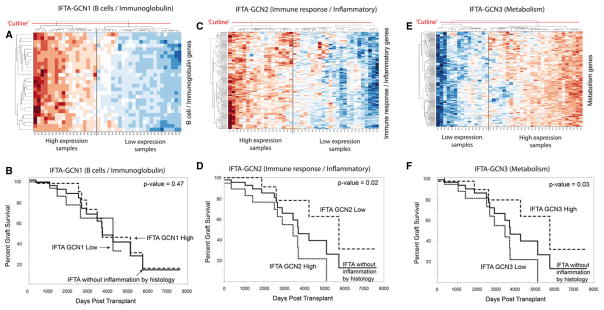
First, we clustered IFTA samples without inflammation into sample clusters based on the relative gene expression of the three IFTA-GCN transcript lists (note: IFTA-GCNs are highly matched to the AR-GCNs). The heat maps in Figure 7 show that the samples clearly separate based on the expression of each GCN. Second, we separated the samples into two clusters for each GCN: IFTA GCN-high and IFTA GCN-low. We then compared graft survival for each sample cluster (Figure 7). Our results show significantly increased rates of graft loss in patients with IFTA without inflammation based on IFTA GCN2 (p = 0.02) and GCN3 (p = 0.03). No correlation to graft loss is seen with GCN1 (p = 0.47). Thus, gene expression profiling detects correlations with graft loss risk for individual patients that are not detected by histology.
Figure 7. Graft survival of subjects with IFTA without inflammation according to expression of our three gene coexpression networks (GCNs).
(A) Interstitial fibrosis and tubular atrophy (IFTA) without inflammation samples clustered into two clusters based on high versus low expression of GCN1 (B cell/immunoglobulin genes). (B) High versus low expression of GCN1 did not demonstrate a difference in graft survival (p = 0.47). (C and D) In contrast, when this analysis is repeated using GCN2 (immune response/inflammatory), graft survival of subjects with IFTA without inflammation correlates with relative expression of GCN2 (p = 0.02). (E and F) Relative expression of GCN3 (metabolism/tissue integrity) also correlates with graft survival (p = 0.03).
A set of 224 differentially expressed genes distinguish two groups of IFTA without inflammation biopsies with higher versus lower risk of graft loss
In the subset of IFTA patients without inflammation (n = 40), we determined differential gene expression between patients with and without graft loss (n = 14 vs. 26; 35% vs. 65%). This analysis revealed 224 differentially expressed transcripts (FDR<0.05) (Appendix S7). One hundred twenty-five (57%) of these genes were common to the top-ranked AR DEGs. Many of these DEGs have been identified in previous studies of acute and chronic transplant injury associated with graft loss (e.g. LTF, SERPINA3, CXCL6, MMP7, AFM, ISG20, and CXCL1; Table 3). Figure 2B and D shows these genes are also among the most “upregulated” genes in AR.
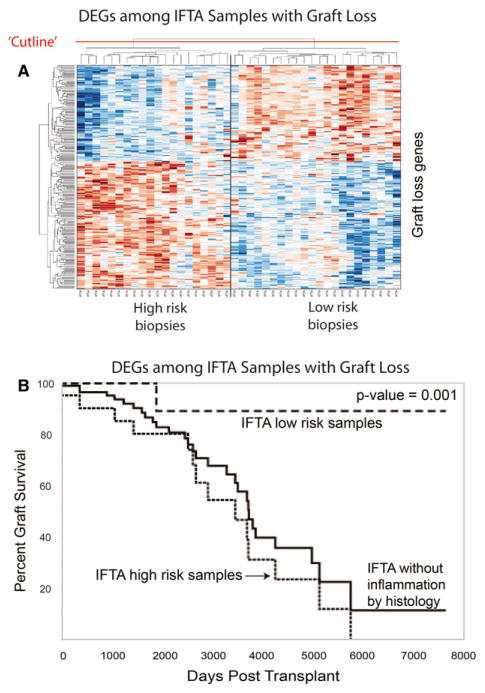
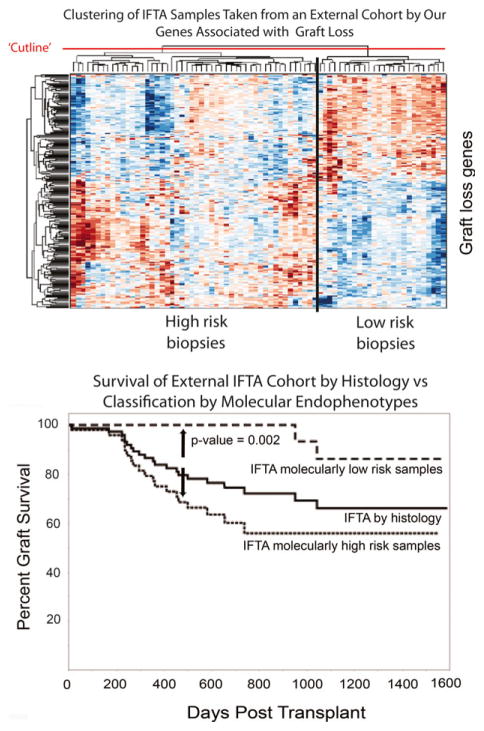
To determine whether these 224 DEGs could delineate IFTA without inflammation patients into groups at high versus low risk of graft loss, we clustered all 40 samples based on the expression of these 224 transcripts. Using complete linkage, hierarchical clustering, two groups were identified with the expected differences in survival curves (Figure 8). Enrichment of these 224 genes for well-known immune rejection and inflammation pathways, and the ability of these genes to cluster our study population into subgroups at high and low risk graft loss, provides both biological and technical plausibility to their discovery. Next, we validated these results in an independent, external cohort of “late” biopsies with IFTA (n = 105) (GEO#: GSE21374). The expression of these 224 genes was also able to separate this external cohort into high and low risk phenotypes (Figure 9).
Figure 8. Graft survival of subjects with IFTA without inflammation correlates with the expression of 224 differentially expressed “high risk” genes.
(A) Interstitial fibrosis and tubular atrophy (IFTA) without inflammation samples clustered into high versus low risk clusters based on expression of 224 differentially expressed transcripts. (B) The high versus low risk sample clusters correlate with graft survival (p = 0.001). DEGs, differentially expressed genes.
Figure 9. Validating the correlation between high risk gene expression and graft survival using an independent external dataset.
Interstitial fibrosis and tubular atrophy (IFTA) biopsies from an external dataset (GEO accession number: GSE21374) (16) were clustered into high and low risk subgroups based on expression of the same 224 transcripts that correlated with graft loss. Again, two subject clusters were identified with marked difference in survival curves (p = 0.002). Note that the subphenotypes of IFTA with and without inflammation were not available for this external dataset.
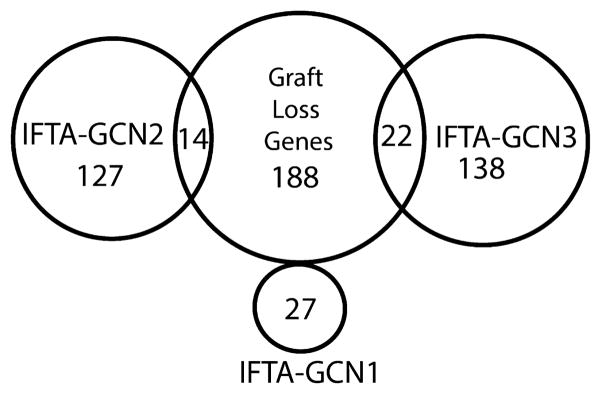
Given that GCNs 2 and 3 correlate with graft survival, we examined the overlap of the GCN-defining genes and the 224 graft loss set (Figure 10). The results reveal 188 nonoverlapping genes that refine the GCN classifiers for graft loss (Appendix S7). Pathway enrichment analysis (GO) demonstrated the highest correlations with immune responses (p = 3.2 × 10 −9), cytokine-mediated signaling (p = 2.8 × 10 −6), interferon gamma (IFN-γ) signaling (p = 1.7 × 10 −5) and antigen presentation via MHC class I (p = 2.2 × 10 −5) There was no overlap with GCN1 (B cell genes).
Figure 10.
Venn diagram demonstrating the overlap of the 224 differentially expressed genes associated with graft loss to the genes comprising the three interstitial fibrosis and tubular atrophy–gene coexpression networks IFTA-GCNs.
Finally, the majority (n = 84; 71%) of the “for cause” biopsies and a minority (n = 21; 19%) of the protocol biopsies had C4d staining performed. Seven hundred fifty-six genes were differentially expressed between C4d-stained positive versus negative (Appendix S3). Seventeen of these 756 genes were shared with the 224 graft loss genes, including two HLA molecules (HLA-F,-G), three proteasome subunits (PSMB8, 9, 10), and TAP1—genes that are all in GCN2 (T cell–mediated immune response) and consistent with activated interferon signaling and antigen presentation. Indeed, pathway enrichment analysis using gene ontology of the 17 overlapping genes showed the highest correlations with type I interferon signaling (p = 1.98 × 10 −11) and antigen processing and presentation (p = 8.8 × 10 −7). None were linked mechanistically to B cell networks.
Discussion
In this multicenter, retrospective analysis, we used gene expression profiles and multiple bioinformatics tools to show that all the biopsies with IFTA (n = 81) demonstrate strong molecular evidence of immune rejection, injury, and decreased metabolism/tissue integrity. This finding was true for biopsies of IFTA without histological inflammation (n = 40). In all cases, IFTA was defined by biopsy histology without identifiable causes present (i.e. BK nephritis or recurrent disease). We used a novel bioinformatic method called Gene Coexpression Network analysis (GCN) to identify the underlying biological networks without introducing any user selection bias. A key point is that the molecular GCNs identified in IFTA were essentially the same as found for biopsies with AR. The relative expression of differentially expressed genes comprising the GCNs correlated with graft loss and the severity of IFTA based on Banff grades. These findings indicate that IFTA biopsies, in which there is no other explanation for pathogenesis, demonstrate evidence of ongoing, cellular immune-mediated injury that is more sensitively detected with gene expression than by light histology.
There were several salient findings in the clinical data. First, patients with a histological diagnosis of AR or IFTA at any time posttransplant demonstrate decreased graft survival compared to those with normal biopsies (TX). Second, our cohorts show 51% of AR and 99% of “IFTA with AR” samples were diagnosed >1 year posttransplant. This finding confirms Scientific Registry for Transplant Recipients and DeKAF data and growing evidence that AR episodes often occur late posttransplant in both adult (9,45,46) and pediatric populations (47).
In this study we describe a network of objectively identified, tightly coexpressed genes with clear biological function related to T cell–driven immunity and inflammation (GCN2; Figure 4). The geometric means of these genes correlated with histologically identified inflammation and Banff IFTA grades: AR > IFTA with AR > IFTA with inflammation >IFTA without inflammation (Figures 5 and 6), indicating the increased expression of cellular immune response genes. A relevant study listed 28 genes that could most successfully predict AR versus non-AR status that included biopsies with both antibody-mediated rejection (ABMR) and TCMR (6). Of these 28 genes, 26 (93%) were found in the top 150 differentially expressed genes in IFTA without inflammation. Nineteen (68%) were found in the GCN2s for both AR and IFTA. Several of these genes, including CXCL9, CXCL11, GZMA, and CCL5, were the most differentially expressed genes in IFTA without inflammation (Table 3). Our results are also consistent with a recent study of 33 kidney biopsies with “IFTA and inflammation” demonstrating an increase in the expression of genes associated with both B and cytotoxic T cells (47). Although we cannot say that the expression of these genes causes IFTA, our study demonstrates that graft loss rates and IFTA grades are associated with higher relative expression of these genes and this is equally true for the subset of patients with IFTA without inflammation. Our hypothesis is that AR and IFTA phenotypes are different stages along the arc of the same alloimmune process.
Since GCN2 was identified objectively based on gene coexpression, the comprising genes, particularly those with a high number of connections to other genes, may provide new mechanistic and biological understanding of acute and chronic rejection (Figures 2 and 4). For examples, dedicator of cytokinesis 2 (DOCK2) is the most connected AR-GCN2 hub gene (Figure 3) and ranked 15 and 10, respectively, in the IFTA-GCN2 hub genes in our data and the external dataset (Appendix S5). DOCK2 is critical to lymphocyte homing and the formation of immunological synapses. Deficiency of DOCK2 attenuates AR in mouse cardiac allografts (48). Another AR-GCN2 hub gene, the IL10RAα, codes for a receptor to the potent anti-inflammatory cytokine, IL-10. It is also identified in the IFTA-GCN2s for our data and the external dataset. IL-10 expression has been associated with acute rejection (21,25,49) and the overexpression of IL-10 improved renal function and survival in rat rejection models (31). IL-10 expression parallels Th1 cytokine expression, suggesting a protective mechanism limiting the immune response (50).
In contrast, we demonstrated an inverse relationship between the metabolism/tissue integrity network (GCN3) to histological inflammation and IFTA grades, results consistent with previously published data (35,51). Similar to GCN2, we revealed that many IFTA samples without histological inflammation had higher rates of graft loss correlating with decreased GCN3 gene expression. The biological function of GCN3 genes may explain the response to immune-mediated tissue injury. For example, PEPD and XPNPEP2 code for enzymes important to regulating collagen metabolism. Decreased expression of these genes may contribute to fibrosis. MME encodes for neutral endopeptidase, a protein that inactivates several peptide hormones including angiotensin II and glucagon. Deficiency in MME leads to fetal membranous glomerulopathy (52). The key point is that therapeutic targeting of the metabolic/functional impacts of rejection on tissue integrity may ultimately turn out to be another effective strategy to preserve graft function and survival.
Our model is that perpetual T cell–driven immune activation and inflammation due to ineffective immunosuppression leads to cell breakdown, release of alloantigens, and the creation of an inflammatory milieu that promotes T cell– mediated B cell activation including production of DSAs. For example, B cell activating factor (TNFSF13B) was found in the GCN2 while its receptor (TNFRSF17) clustered tightly among the GCN1 genes. The AT-Hook Transcription Factor (AKNA) was found in GCN2, and has been shown to upregulate transcription of the receptor–ligand pair CD40 and CD40L, an essential interaction for B cell activation and antibody isotype switching (53,54). Another GCN2 gene, SLAMF8, plays a role in B lineage development and modulation of B cell activation through B cell receptor signaling (55). Finally, the GCN2 gene, RANTES (CCL5), is involved in activation of both T and B cells and immunoglobulin switching in B cells (56).
Consistent with our model, molecular profiling demonstrates that the relative expression of genes related to immunoglobulin production (GCN1) did not independently correlate with graft loss or worse outcomes for either AR or IFTA phenotypes. However, our model recognizes the close connections between humoral and T cell immunity. Although ABMR has been associated with IFTA and increased risk of graft loss (57), the majority of patients with de novo DSA (dnDSA) followed for 5 years or more do not lose their grafts (58,59). Other studies demonstrate that (1) the development of dnDSA correlates with medication nonadherence and AR episodes, (2) dnDSA correlate with transplant glomerulopathy but not IFTA, and (3) biopsies with ABMR frequently show concomitant histological evidence of TCMR (60–63). Our gene expression and functional mapping are consistent with this literature by showing a high correlation between C4d staining and T cell immune networks.
The major limitation in this retrospective, longitudinal study is that the majority of patients had a single biopsy. These biopsies only provide a cross-sectional view of pathology on a large population of transplant patients with known outcomes. This is not a prospective study that follows patients from the time of transplantation, obtains multiple biopsies and gene profiles, and monitors patient events and other variables over time. Thus, although our IFTA samples demonstrated strong evidence for cellular rejection and inflammation at the time of biopsy, there may have been preceding nonimmunological insults that also played a role in the development of IFTA prior to the biopsy. Likewise, we do not have any data on medication nonadherence. However, our model is that chronic rejection leads to tissue injury and IFTA. The corollary is that chronic rejection is the result of inadequate immunosuppression. Thus, whether inadequate immunosuppression was the decision of a physician to reduce dosing or due to patient medication nonadherence is not relevant to our conclusions. Another limitation is that the overall percentage of African Americans in this study was less than the percentage that receive kidney transplants (10% vs. 34%) (64). Finally, this study cannot account for the possibility of ABMR coexisting with TCMR in some biopsies. At the time this study was designed, dnDSA were not routinely measured except when pathologists found positive C4d staining. Moreover, the Banff criteria at that time did not include the current metrics for defining ABMR on biopsies.
This study demonstrates that IFTA biopsies without alternative explanations for pathogenesis (i.e. BK or recurrent disease) reveal differential gene expression evidence of ongoing cellular immune-mediated injury. Specifically, GCNs and the mapping of genes to functional pathways demonstrate significant molecular overlap to profiles of AR biopsies, supporting our model that IFTA is a manifestation of chronic rejection. The connection between AR and IFTA profiles is true even for biopsies of IFTA without inflammation. Expression of GCN2 (immune response) and GCN3 (metabolism/tissue integrity) genes correlates with increased risk of graft loss. Furthermore, a set of 224 genes differentially expressed with graft loss refines the functional pathways found by GCN analysis. The clinical relevance is that a future prospective trial may demonstrate that informing immunosuppressive and monitoring protocols for individual patients based on serial gene expression profiling of biopsies improves long-term clinical outcomes.
Supplementary Material
Expanded methods sections.
Association of clinical or histological factors with graft loss.
Differentially expressed genes according to phenotype.
External data interstitial fibrosis and tubular atrophy validation.
Genes of clinical acute rejection interstitial fibrosis and tubular atrophy and interstitial fibrosis and tubular atrophy plus clinical acute rejection gene coexpression networks.
Appendix S6: Literature review of GCN2 and GCN3.
Appendix S7: Genes associated with graft loss in interstitial fibrosis and tubular atrophy samples.
Acknowledgments
We could not have done this work without the hard work of many nurses, transplant and research coordinators that were instrumental in the collection and local processing of all the samples. We also acknowledge the patients that consented to the work. We would like to acknowledge the contributions of Drs. Roy First and Stan Rose in a critical reading of this manuscript. National Institutes of Health funding was provided by U19 AI063603 (D.R.S., S.M.K., T.G., T.S.M., S.R.H.) and CTSA KL2 TR001112 (B.M.).
Abbreviations
- ABMR
antibody-mediated rejection
- ANOVA
analysis of variance
- AR
clinical acute rejection
- DEG
differentially expressed gene
- dnDSA
de novo donor-specific antibody
- DSA
donor-specific antibody
- FC
fold change
- FDR
false discovery rate
- GCN
gene coexpression network
- GEO
Gene Expression Omnibus
- HLA
human leukocyte antigen
- IFN-γ
interferon-gamma
- IFTA
interstitial fibrosis and tubular atrophy
- N/A
not applicable
- subAR
subclinical acute rejection
- TCMR
T cell–mediated rejection
- TX
Treatment group with excellent functioning kidney
Footnotes
Disclosure
The authors of this manuscript have conflicts of interest to disclose as described by the American Journal of Transplantation. D.R.S. and M.M.A. are the founding scientists of Transplant Genomics Inc. S.M.K. and S.R.H. have stock equity and receive consulting fees from TGI. T.G. is partially supported by salary funding from TGI to The Scripps Research Institute. The other authors have no conflicts of interest to disclose.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Mannon RB, Matas AJ, Grande J, et al. Inflammation in areas of tubular atrophy in kidney allograft biopsies: A potent predictor of allograft failure. Am J Transplant. 2010;10:2066–2073. doi: 10.1111/j.1600-6143.2010.03240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park WD, Griffin MD, Cornell LD, Cosio FG, Stegall MD. Fibrosis with inflammation at one year predicts transplant functional decline. J Am Soc Nephrol. 2010;21:1987–1997. doi: 10.1681/ASN.2010010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cosio FG, Grande JP, Wadei H, Larson TS, Griffin MD, Stegall MD. Predicting subsequent decline in kidney allograft function from early surveillance biopsies. Am J Transplant. 2005;5:2464–2472. doi: 10.1111/j.1600-6143.2005.01050.x. [DOI] [PubMed] [Google Scholar]
- 4.Heilman RL, Smith ML, Kurian SM, et al. Transplanting kidneys from deceased donors with severe acute kidney injury. Am J Transplant. 2015;15:2143–2151. doi: 10.1111/ajt.13260. [DOI] [PubMed] [Google Scholar]
- 5.Loupy A, Vernerey D, Tinel C, et al. Subclinical rejection phenotypes at 1 year post-transplant and outcome of kidney allografts. J Am Soc Nephrol. 2015;26:1721–1731. doi: 10.1681/ASN.2014040399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meier-Kriesche HU, Schold JD, Srinivas TR, Kaplan B. Lack of improvement in renal allograft survival despite a marked decrease in acute rejection rates over the most recent era. Am J Transplant. 2004;4:378–383. doi: 10.1111/j.1600-6143.2004.00332.x. [DOI] [PubMed] [Google Scholar]
- 7.Hart A, Smith JM, Skeans MA, et al. Kidney. Am J Transplant. 2016;16:11–46. doi: 10.1111/ajt.13666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joosten SA, van Kooten C, Sijpkens YW, de Fijter JW, Paul LC. The pathobiology of chronic allograft nephropathy: Immunemediated damage and accelerated aging. Kidney Int. 2004;65:1556–1559. doi: 10.1111/j.1523-1755.2004.05410.x. [DOI] [PubMed] [Google Scholar]
- 9.El Ters M, Grande JP, Keddis MT, et al. Kidney allograft survival after acute rejection, the value of follow-up biopsies. Am J Transplant. 2013;13:2334–2341. doi: 10.1111/ajt.12370. [DOI] [PubMed] [Google Scholar]
- 10.Nickerson PW, Rush DN. Rejection: An integrated response. Am J Transplant. 2013;13:2239–2240. doi: 10.1111/ajt.12365. [DOI] [PubMed] [Google Scholar]
- 11.Nankivell BJ, Chapman JR. The significance of subclinical rejection and the value of protocol biopsies. Am J Transplant. 2006;6:2006–2012. doi: 10.1111/j.1600-6143.2006.01436.x. [DOI] [PubMed] [Google Scholar]
- 12.Legendre C, Thervet E, Skhiri H, et al. Histologic features of chronic allograft nephropathy revealed by protocol biopsies in kidney transplant recipients. Transplantation. 1998;65:1506–1509. doi: 10.1097/00007890-199806150-00020. [DOI] [PubMed] [Google Scholar]
- 13.Seron D, Moreso F. Protocol biopsies in renal transplantation: Prognostic value of structural monitoring. Kidney Int. 2007;72:690–697. doi: 10.1038/sj.ki.5002396. [DOI] [PubMed] [Google Scholar]
- 14.Heilman RL, Devarapalli Y, Chakkera HA, et al. Impact of subclinical inflammation on the development of interstitial fibrosis and tubular atrophy in kidney transplant recipients. Am J Transplant. 2010;10:563–570. doi: 10.1111/j.1600-6143.2009.02966.x. [DOI] [PubMed] [Google Scholar]
- 15.Rush DN, Henry SF, Jeffery JR, Schroeder TJ, Gough J. Histological findings in early routine biopsies of stable renal allograft recipients. Transplantation. 1994;57:208–211. doi: 10.1097/00007890-199401001-00009. [DOI] [PubMed] [Google Scholar]
- 16.Einecke G, Reeve J, Sis B, et al. A molecular classifier for predicting future graft loss in late kidney transplant biopsies. J Clin Invest. 2010;120:1862–1872. doi: 10.1172/JCI41789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halloran PF, Pereira AB, Chang J, et al. Potential impact of microarray diagnosis of T cell-mediated rejection in kidney transplants: The INTERCOM study. Am J Transplant. 2013;13:2352–2363. doi: 10.1111/ajt.12387. [DOI] [PubMed] [Google Scholar]
- 18.Storey JD. A direct approach to false discovery rates. J R Stat Soc Series B Stat Methodol. 2002;64:479–498. [Google Scholar]
- 19.Morgun A, Shulzhenko N, Perez-Diez A, et al. Molecular profiling improves diagnoses of rejection and infection in transplanted organs. Circ Res. 2006;98:e74–e83. doi: 10.1161/01.res.0000228714.15691.8a. [DOI] [PubMed] [Google Scholar]
- 20.Sarwal M, Chua MS, Kambham N, et al. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med. 2003;349:125–138. doi: 10.1056/NEJMoa035588. [DOI] [PubMed] [Google Scholar]
- 21.Saint-Mezard P, Berthier CC, Zhang H, et al. Analysis of independent microarray datasets of renal biopsies identifies a robust transcript signature of acute allograft rejection. Transpl Int. 2009;22:293–302. doi: 10.1111/j.1432-2277.2008.00790.x. [DOI] [PubMed] [Google Scholar]
- 22.Hauser P, Schwarz C, Mitterbauer C, et al. Genome-wide geneexpression patterns of donor kidney biopsies distinguish primary allograft function. Lab Invest. 2004;84:353–361. doi: 10.1038/labinvest.3700037. [DOI] [PubMed] [Google Scholar]
- 23.Naesens M, Li L, Ying L, et al. Expression of complement components differs between kidney allografts from living and deceased donors. J Am Soc Nephrol. 2009;20:1839–1851. doi: 10.1681/ASN.2008111145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Famulski KS, Reeve J, de Freitas DG, Kreepala C, Chang J, Halloran PF. Kidney transplants with progressing chronic diseases express high levels of acute kidney injury transcripts. Am J Transplant. 2013;13:634–644. doi: 10.1111/ajt.12080. [DOI] [PubMed] [Google Scholar]
- 25.Flechner SM, Kurian SM, Head SR, et al. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant. 2004;4:1475–1489. doi: 10.1111/j.1600-6143.2004.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kainz A, Mitterbauer C, Hauser P, et al. Alterations in gene expression in cadaveric vs. live donor kidneys suggest impaired tubular counterbalance of oxidative stress at implantation. Am J Transplant. 2004;4:1595–1604. doi: 10.1111/j.1600-6143.2004.00554.x. [DOI] [PubMed] [Google Scholar]
- 27.Famulski KS, de Freitas DG, Kreepala C, et al. Molecular phenotypes of acute kidney injury in kidney transplants. J Am Soc Nephrol. 2012;23:948–958. doi: 10.1681/ASN.2011090887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reeve J, Einecke G, Mengel M, et al. Diagnosing rejection in renal transplants: A comparison of molecular- and histopathologybased approaches. Am J Transplant. 2009;9:1802–1810. doi: 10.1111/j.1600-6143.2009.02694.x. [DOI] [PubMed] [Google Scholar]
- 29.Famulski KS, Einecke G, Reeve J, et al. Changes in the transcriptome in allograft rejection: IFN-gamma-induced transcripts in mouse kidney allografts. Am J Transplant. 2006;6:1342–1354. doi: 10.1111/j.1600-6143.2006.01337.x. [DOI] [PubMed] [Google Scholar]
- 30.Einecke G, Melk A, Ramassar V, et al. Expression of CTL associated transcripts precedes the development of tubulitis in T-cell mediated kidney graft rejection. Am J Transplant. 2005;5:1827–1836. doi: 10.1111/j.1600-6143.2005.00974.x. [DOI] [PubMed] [Google Scholar]
- 31.Chen R, Sigdel TK, Li L, et al. Differentially expressed RNA from public microarray data identifies serum protein biomarkers for cross-organ transplant rejection and other conditions. PLoS Comput Biol. 2010;6:e1000940. doi: 10.1371/journal.pcbi.1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khatri P, Roedder S, Kimura N, et al. A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation. J Exp Med. 2013;210:2205–2221. doi: 10.1084/jem.20122709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freue GV, Sasaki M, Meredith A, et al. Proteomic signatures in plasma during early acute renal allograft rejection. Mol Cell Proteomics. 2010;9:1954–1967. doi: 10.1074/mcp.M110.000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perco P, Pleban C, Kainz A, Lukas A, Mayer B, Oberbauer R. Gene expression and biomarkers in renal transplant ischemia reperfusion injury. Transpl Int. 2007;20:2–11. doi: 10.1111/j.1432-2277.2006.00376.x. [DOI] [PubMed] [Google Scholar]
- 35.Bunnag S, Einecke G, Reeve J, et al. Molecular correlates of renal function in kidney transplant biopsies. J Am Soc Nephrol. 2009;20:1149–1160. doi: 10.1681/ASN.2008080863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stein-Oakley AN, Tzanidis A, Fuller PJ, Jablonski P, Thomson NM. Expression and distribution of epidermal growth factor in acute and chronic renal allograft rejection. Kidney Int. 1994;46:1207–1215. doi: 10.1038/ki.1994.386. [DOI] [PubMed] [Google Scholar]
- 37.Hu H, Aizenstein BD, Puchalski A, Burmania JA, Hamawy MM, Knechtle SJ. Elevation of CXCR3-binding chemokines in urine indicates acute renal-allograft dysfunction. Am J Transplant. 2004;4:432–437. doi: 10.1111/j.1600-6143.2004.00354.x. [DOI] [PubMed] [Google Scholar]
- 38.Stroo I, Stokman G, Teske GJ, et al. Chemokine expression in renal ischemia/reperfusion injury is most profound during the reparative phase. Int Immunol. 2010;22:433–442. doi: 10.1093/intimm/dxq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akalin E, Hendrix RC, Polavarapu RG, et al. Gene expression analysis in human renal allograft biopsy samples using high-density oligoarray technology. Transplantation. 2001;72:948–953. doi: 10.1097/00007890-200109150-00034. [DOI] [PubMed] [Google Scholar]
- 40.Hu H, Kwun J, Aizenstein BD, Knechtle SJ. Noninvasive detection of acute and chronic injuries in human renal transplant by elevation of multiple cytokines/chemokines in urine. Transplantation. 2009;87:1814–1820. doi: 10.1097/TP.0b013e3181a66b3e. [DOI] [PubMed] [Google Scholar]
- 41.Jackson JA, Kim EJ, Begley B, et al. Urinary chemokines CXCL9 and CXCL10 are noninvasive markers of renal allograft rejection and BK viral infection. Am J Transplant. 2011;11:2228–2234. doi: 10.1111/j.1600-6143.2011.03680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaub S, Nickerson P, Rush D, et al. Urinary CXCL9 and CXCL10 levels correlate with the extent of subclinical tubulitis. Am J Transplant. 2009;9:1347–1353. doi: 10.1111/j.1600-6143.2009.02645.x. [DOI] [PubMed] [Google Scholar]
- 43.Hauser IA, Spiegler S, Kiss E, et al. Prediction of acute renal allograft rejection by urinary monokine induced by IFN-gamma (MIG) J Am Soc Nephrol. 2005;16:1849–1858. doi: 10.1681/ASN.2004100836. [DOI] [PubMed] [Google Scholar]
- 44.Carpio VN, de Noronha IL, Martins HL, et al. Expression patterns of B cells in acute kidney transplant rejection. Exp Clin Transplant. 2014;12:405–414. [PubMed] [Google Scholar]
- 45.Rodrigues CA, Franco MF, Cristelli MP, Pestana JOM, Tedesco-Silva HJ. Clinicopathological characteristics and effect of late acute rejection on renal transplant outcomes. Transplantation. 2014;98:885–892. doi: 10.1097/TP.0000000000000145. [DOI] [PubMed] [Google Scholar]
- 46.Gourishankar S, Leduc R, Connett J, et al. Pathological and clinical characterization of the “troubled transplant”: Data from the DeKAF study. Am J Transplant. 2010;10:324–330. doi: 10.1111/j.1600-6143.2009.02954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eid L, Tuchman S, Moudgil A. Late acute rejection: Incidence, risk factors, and effect on graft survival and function. Pediatr Transplant. 2014;18:155–162. doi: 10.1111/petr.12203. [DOI] [PubMed] [Google Scholar]
- 48.Jiang H, Pan F, Erickson LM, et al. Deletion of DOCK2, a regulator of the actin cytoskeleton in lymphocytes, suppresses cardiac allograft rejection. J Exp Med. 2005;202:1121–1130. doi: 10.1084/jem.20050911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spivey TL, Uccellini L, Ascierto ML, et al. Gene expression profiling in acute allograft rejection: Challenging the immunologic constant of rejection hypothesis. J Transl Med. 2011;9:174. doi: 10.1186/1479-5876-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edemir B, Kurian SM, Eisenacher M, et al. Activation of counter-regulatory mechanisms in a rat renal acute rejection model. BMC Genom. 2008;9:71. doi: 10.1186/1471-2164-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Einecke G, Broderick G, Sis B, Halloran PF. Early loss of renal transcripts in kidney allografts: Relationship to the development of histologic lesions and alloimmune effector mechanisms. Am J Transplant. 2007;7:1121–1130. doi: 10.1111/j.1600-6143.2007.01797.x. [DOI] [PubMed] [Google Scholar]
- 52.Ronco P, Debiec H. Molecular pathomechanisms of membranous nephropathy: From Heymann nephritis to alloimmunization. J Am Soc Nephrol. 2005;16:1205–1213. doi: 10.1681/ASN.2004121080. [DOI] [PubMed] [Google Scholar]
- 53.Allen RC, Armitage RJ, Conley ME, et al. CD40 ligand gene defects responsible for X-linked hyper-IgM syndrome. Science. 1993;259:990–993. doi: 10.1126/science.7679801. [DOI] [PubMed] [Google Scholar]
- 54.van Kooten C, Banchereau J. CD40-CD40 ligand. J Leukoc Biol. 2000;67:2–17. doi: 10.1002/jlb.67.1.2. [DOI] [PubMed] [Google Scholar]
- 55.Kingsbury GA, Feeney LA, Nong Y, et al. Cloning, expression, and function of BLAME, a novel member of the CD2 family. J Immunol. 2001;166:5675–5680. doi: 10.4049/jimmunol.166.9.5675. [DOI] [PubMed] [Google Scholar]
- 56.Kimata H, Yoshida A, Ishioka C, Fujimoto M, Lindley I, Furusho K. RANTES and macrophage inflammatory protein 1 alpha selectively enhance immunoglobulin (IgE) and IgG4 production by human B cells. J Exp Med. 1996;183:2397–2402. doi: 10.1084/jem.183.5.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Halloran PF, Chang J, Famulski K, et al. Disappearance of T cellmediated rejection despite continued antibody-mediated rejection in late kidney transplant recipients. J Am Soc Nephrol. 2015;26:1711–1720. doi: 10.1681/ASN.2014060588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Terasaki PI, Ozawa M. Predicting kidney graft failure by HLA antibodies: A prospective trial. Am J Transplant. 2004;4:438–443. doi: 10.1111/j.1600-6143.2004.00360.x. [DOI] [PubMed] [Google Scholar]
- 59.Wiebe C, Gibson IW, Blydt-Hansen TD, et al. Evolution and clinical pathologic correlations of de novo donor-specific HLA antibody post kidney transplant. Am J Transplant. 2012;12:1157–1167. doi: 10.1111/j.1600-6143.2012.04013.x. [DOI] [PubMed] [Google Scholar]
- 60.Wiebe C, Gibson IW, Blydt-Hansen TD, et al. Rates and determinants of progression to graft failure in kidney allograft recipients with de novo donor-specific antibody. Am J Transplant. 2015;15:2921–2930. doi: 10.1111/ajt.13347. [DOI] [PubMed] [Google Scholar]
- 61.Nickeleit V, Andreoni K. The classification and treatment of antibody-mediated renal allograft injury: Where do we stand? Kidney Int. 2007;71:7–11. doi: 10.1038/sj.ki.5002003. [DOI] [PubMed] [Google Scholar]
- 62.Nickeleit V, Mihatsch MJ. Kidney transplants, antibodies and rejection: Is C4d a magic marker? Nephrol Dial Transplant. 2003;18:2232–2239. doi: 10.1093/ndt/gfg304. [DOI] [PubMed] [Google Scholar]
- 63.Magil AB, Tinckam KJ. Focal peritubular capillary C4d deposition in acute rejection. Nephrol Dial Transplant. 2006;21:1382–1388. doi: 10.1093/ndt/gfk028. [DOI] [PubMed] [Google Scholar]
- 64.Matas AJ, Smith JM, Skeans MA, et al. OPTN/SRTR 2012 Annual Data Report: Kidney. Am J Transplant. 2014;14(suppl 1):11–44. doi: 10.1111/ajt.12579. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded methods sections.
Association of clinical or histological factors with graft loss.
Differentially expressed genes according to phenotype.
External data interstitial fibrosis and tubular atrophy validation.
Genes of clinical acute rejection interstitial fibrosis and tubular atrophy and interstitial fibrosis and tubular atrophy plus clinical acute rejection gene coexpression networks.
Appendix S6: Literature review of GCN2 and GCN3.
Appendix S7: Genes associated with graft loss in interstitial fibrosis and tubular atrophy samples.