Abstract
Biomarkers are measurable indicators of a biological state. As our understanding of diseases meliorates, it is generally accepted that early diagnosis renders the best chance to cure a disease. In the context of proteomics, the discovery phase of identifying bonafide biomarkers and the ensuing validation phase involving large cohort of patient samples are impeded by the complexity of bodily fluid samples. High abundant proteins found in blood plasma make it difficult for the detection of low abundant proteins that may be potential biomarkers. Extracellular vesicles (EVs) have reignited interest in the field of biomarker discovery. EVs contain a tissue-type signature wherein a rich cargo of proteins and RNA are selectively packaged. In addition, as EVs are membranous structures, the luminal contents are protected from degradation by extracellular proteases and are highly stable in storage conditions. Interestingly, an appealing feature of EV-based biomarker analysis is the significant reduction in the sample complexity compared to whole bodily fluids. With these prescribed attributes, which are the rate-limiting factors of traditional biomarker analysis, there is immense potential for the use of EVs for biomarker detection in clinical settings. This review will discuss the current issues with biomarker analysis and the potential use of EVs as reservoirs of disease biomarkers.
Keywords: exosomes, extracellular vesicles, microvesicles, biomarkers, bodily fluids
Introduction
Biomarkers are tissues and/or bodily fluid-based measurable indicators of a biological condition [1][2]. Biomarkers include DNA, RNA, proteins and metabolites that can reflect an individual’s state of health or disease [3, 4]. The National Institutes of Health Biomarkers Definitions Working Group in 1998 defined a biomarker as “a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention” [5]. Based on their utility in practice, biomarkers can provide insights on diagnosis, prognosis, regression or response to treatment of a disease [6, 7]. Currently, invasive and non-invasive procedures are often employed to identify biomarkers [8]. In the context of cancer, tumour tissues are obtained by invasive biopsies for biomarker analysis. In cases where tumours or diseased tissue (e.g., neurodegenerative diseases) are inaccessible, non-invasive methodologies are relied upon [9]. Non-invasive methods of analysis include the detection of the biomarker(s) in bodily fluids. One of the attractive features of using bodily fluids is that it also allows for the early diagnosis of the disease [8]. A variety of bodily fluid samples including serum, plasma, urine, cerebrospinal fluid, saliva, ascites and amniotic fluid have been commonly analysed for both identification and validation of potential biomarkers [3].
Current state of biomarker analysis
The idea of monitoring serum proteins from blood samples is well established in terms of monitoring tumour growth: for instance, the detection of prostate specific antigen (PSA) for prostate cancer, carbohydrate antigen 125 (CA125) for ovarian cancer and carcinoembryonic antigen (CEA) for multiple cancers [9]. Unfortunately, this approach is not always plausible due to the paucity of tumor specific biomarkers. Compared to other bodily fluids, plasma or serum samples are considered ideal for mining biomarkers for clinical practice as blood contains a sampling of every tissue in the body and can be considered as a universal source of biomarkers [10, 11]. However, proteomic strategies have proven to be difficult in plasma mainly due to the vast dynamic range of proteins contained within plasma [10–12]. With 99% of plasma protein mass made up with 22 abundant proteins, it remains difficult to detect other low abundant proteins [10]. Abundant plasma proteins such as albumin, fibrinogen, immunoglobulins, transferrin, lipoproteins and plasminogen alone account for >90% of the total protein content in blood. Several methods based on immunoaffinity and columns have been developed to deplete the high abundant plasma proteins including albumin and immunoglobulins [13–16]. Even though the methods can successfully deplete some of the abundant proteins, plasma still remains a complex fluid encompassing proteins from many normal tissues and cell types. Additionally, the immunodepletion methods suffer from nonspecific binding, increases the variability between samples and thus sacrifices biomarker screening efficiency [10, 11, 17].
To circumvent the complexity of analysing bodily fluids, the discovery phase is usually carried out in less complex samples such as panel of cell lines. The ensuing validation of potential biomarkers is performed by targeted analysis in bodily fluid samples or in tissue sections [8]. To quantitatively measure the potential biomarker of interest in control and disease samples, immunoassays are employed, ELISA being the gold standard [10]. These current approaches are constrained with the limit of signal detection for clinical biomarkers and the lack of well characterized highly specific monoclonal antibodies [18]. With all the issues described in identifying biomarkers in the discovery phase by proteomic strategies, extracellular vesicles can potentially overcome these challenges and have reignited interest in biomarkers. This review will focus on the utility of extracellular vesicle fractions from bodily fluids for biomarker analysis.
Extracellular vesicles
Extracellular vesicles (EVs) are secreted by various cells into the microenvironment. Based on the biogenesis, EVs can be classified into three categories; apoptotic blebs, shedding microvesicles (SMVs) or ectosomes and exosomes [19, 20]. SMVs (100–1000 nm in diameter) occur through the budding of the plasma membrane [21–24]. Under pro-apoptotic stimuli, a cell undergoes apoptosis and subsequently releases apoptotic blebs [25–27]. Exosomes are membranous nanovesicles with a diameter range of 40–150 nm and float in sucrose gradients at a density range from 1.13 to 1.19 g/mL [19, 28]. Even though multiple attributes of exosomes are poorly understood, exosomes remain the most studied among the EVs [29]. The nomenclature of EVs is yet to be standardized and thus allowed for confusion in vesicle naming [20]. In addition, no vesicle type could be isolated to homogeneity due to the unavailability of molecular markers that differentiate the classes of EVs [30]. For this reason, most of the EV-based studies have isolated a mixed population of vesicles irrespective of the sample source (bodily fluids or cell culture media). However, in the context of biomarkers, the inability of purifying vesicle types to homogeneity may not be a concern as far as patient samples can be differentiated from healthy volunteers.
Exosomes emerge from the endocytic pathway. Invagination of the plasma membrane results in early endosomes that matures into late endosomes or multivesicular bodies (MVBs). Inward budding of the limiting late endosomal membrane forms the intraluminal vesicles (ILVs) [31, 32]. The ILV’s are released into the extracellular space by the fusion of MVBs with the plasma membrane and are called as exosomes thereafter [19, 31, 33]. Exosomes have been reported to be secreted by a wide range of cells including immune, neuronal and cancer cells [34–38]. Similar to cells, exosomes have a lipid bilayer membrane consisting of a range of lipids and proteins [39] as well as luminal proteins and RNA [19]. The molecular content of exosomes and the biological function that they perform are influenced by the cell type of origin [40]. For example, exosomes released by dendritic cells contain co-stimulatory proteins necessary for T-cell activation while most exosomes derived from tumour cells do not [41]. Similarly, platelet-derived exosomes promote tumour progression and metastasis of lung cancer cells [42] while ovarian cancer exosomes stimulate angiogenesis [43]. Most importantly, exosomes are shown to contain oncoproteins which can be transferred to target cells upon fusion/uptake [44]. Exosomes are shown to regulate many biological functions including, immune response regulation [45, 46], antigen presentation [47, 48], the transfer of RNA and proteins [49, 50], transfer of infectious cargo [35, 51, 52], non-classical secretion of proteins [53] and intercellular communication [54–57]. Also, exosomes have been involved in the progression of disease by the transfer of oncogenes in cancer [56] and pathogenic proteins between neurons in neurodegeneration [58–60]. The release of exosomes into the extracellular space affords an opportunity to examine exosomes in body fluids such as blood, urine and malignant ascites [61]. Accessing these bioactive vesicles in a non-invasive manner may lead to potential biomarkers for diagnostic and prognostic purposes.
For the purpose of this review, all the vesicles will be referred to as EVs from here on.
Advantages of EVs in biomarker analysis
As EVs circulating in blood are likely to have been derived from various tissues throughout the body, the isolation of cell specific EVs can provide us with information on a specific pathological condition [19, 62]. Bodily fluids contain EVs and creates unparalleled opportunities to exploit these vesicles as reservoirs of disease biomarkers [62, 63]. Disease derived EVs can relay information which can be used to determine the state or progress of disease as well as optimal modes of treatment [64]. Importantly, exosome based biomarker analysis have many significant advantages compared to conventional strategies which are discussed further (Table 1).
Table 1.
Advantages of EVs as potential source of biomarkers
|
EVs contain disease causing proteins
As EVs may contain disease causing proteins including mutant proteins/RNA, assaying for mutant or disease causing proteins/RNA as disease biomarkers may provide the required specificity for a biomarker test [8]. Wang et al. used MS-based selected reaction monitoring (SRM) and were able to distinguish between wild type and mutant forms of the KRAS protein (G12D) in cell lines, tissues samples and bodily fluids [65]. At least in this context, the study showed that the SRM technique could easily be used on complex biological samples with high sensitivity (~10 fmol). Mathivanan et al. identified 57 mutated proteins from the secretome of 18 cell lines representing different stages and underlying mutation status of colorectal cancer using MS [66]. The possibility of an altered extracellular localization of a mutated protein allows biomedical researchers to exploit such mutant proteins as cancer biomarkers. As wild type proteins can also be expressed in multiple tissues, using them as candidate biomarkers of a disease often lacks the specificity. Currently, CEA is the most widely used biomarker associated with colorectal cancer screening. However, the lack of sensitivity and specificity of the test renders it unsuitable for clinical screening. Elevated serum levels of CEA are not only detected in colorectal cancer patients but also in lung, cervix [67], breast [68], gastric [69] and pancreatic [70] cancer patients. The use of mutant proteins that are also drivers of the disease may provide the much needed specificity that seems to lack from wild type proteins like CEA. Similarly, mRNA of the fusion gene TMPRSS2:ERG was detected in EVs isolated from the urine of prostate cancer patients [71]. In addition, oncogenic receptor EGFRvIII is shown to be released by EVs [41, 44, 72]. Based on these observations, MS techniques including SRM can be utilized for the identification of disease causing proteins in EVs.
EVs are reservoirs of disease biomarkers
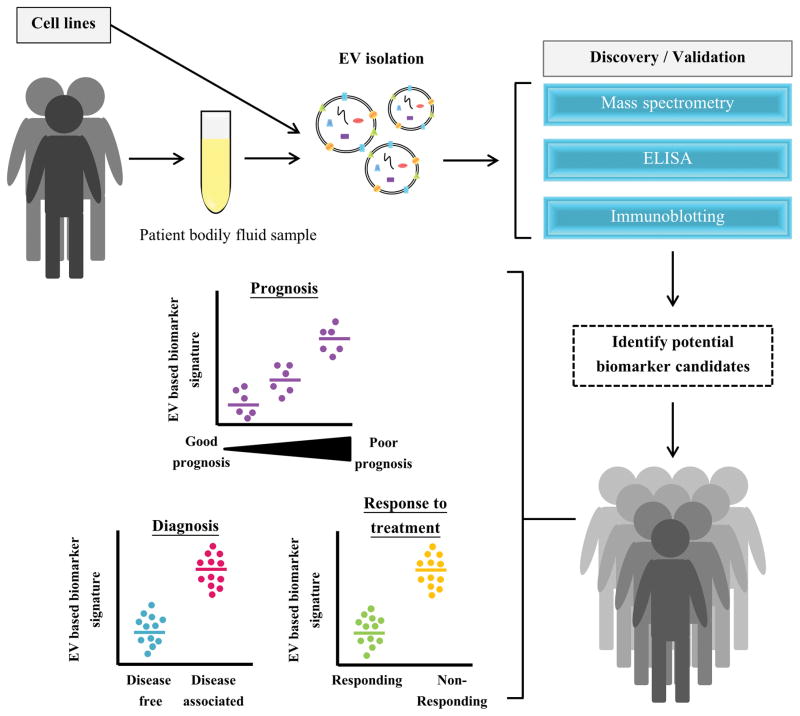
EV-based biomarker analyses are carried out with the ultimate aim to identify biomarkers for early diagnosis and prognosis of disease conditions [73, 74] (Fig. 1). Recently, the use of EVs as indicators for response to treatment has also gained significant interest [75, 76]. Ideally, EV-based protein signatures could predict the outcome of a treatment thereby allowing for strategizing treatment options as well as reducing significant costs associated with the treatment that is bound to fail.
Figure 1. An outline how EVs can be exploited as a potential source of biomarkers in the clinic.
EVs can be isolated from cell lines and/or patient samples for identifying potential biomarkers. Following this, shortlisted candidate biomarkers can be validated in large patient cohorts. With the use of EV based biomarkers, there is immense potential for disease diagnosis, prognosis and prediction of response to treatment.
Currently, the diagnosis and surveillance of prostate cancer utilizes PSA [77, 78]. However, PSA lacks specificity for prostate cancer and hence has the risk of over diagnosis and overtreatment [77, 78]. Nilsson and colleagues identified two distinct potential mRNA biomarkers, PCA-3 and TMPRSS2, in prostate cancer EVs which can be utilised in prostate cancer diagnosis [64, 71]. Similarly, Duijvesz and colleagues observed the proteomic profile of EVs from four prostate cancer epithelial cell lines PNT2C2, RWPE-1, PC346C and VCaP [77]. Following tryptic digestion and LC-MS/MS analysis, 1494 non-redundant peptides were identified. The authors have validated PDCD6IP, FASN, XPO1 and ENO1 by Western blotting and immunohistochemistry. While PDCD6IP and ENO1 are most often identified in EVs (ExoCarta [79] and Vesiclepedia [80]) and may lack the specificity, the other two proteins could be potential biomarkers. However, independent validation of the candidate markers is still needed on large patient cohorts.
It has been previously reported that EGFRvIII containing EVs could be detected in the serum of glioblastoma patients [41]. Shao and colleagues utilised a rapid and highly sensitive technique for the isolation and protein typing of EVs from glioblastoma patient blood samples [75]. EVs were labelled with target specific magnetic nanoparticles and introduced into a microfluidic chip and detected via miniaturized nuclear magnetic resonance system (μNMR). This system was able to differentiate glioblastoma EVs from non-tumour host derived EVs and predict treatment response. Reflective of the protein profiles of the parental cells, glioblastoma derived EVs exhibited elevated levels of EGFR, EGFRvIII, PDPN and IDH1 R132H compared to control EVs [75]. Hence, the detection of tumour derived EV signature via a blood test has the potential to provide diagnostic, prognostic and survival value [41, 56, 75, 81].
Peinado and colleagues isolated EVs from blood plasma of patients with different clinical stages of melanoma [56]. While the quantity of EVs was not affected significantly, the protein content within EVs increased in advanced melanoma patients. Additionally, patients with low protein content within EVs exhibited an increased survival advantage over patients with protein rich EVs. Following tryptic digestion and MS analysis, melanoma cell-derived EV protein signature (human and mice) was identified. Compared to the control EVs, TYRP2, VLA-4 and HSP70 were highly abundant in advanced melanoma cell-derived EVs. Interestingly, an isoform of HSP70 was identified in advanced melanoma cell-derived EVs which has been reported to be involved in cellular transformation [56, 82].
EpCAM-based immunoaffinity pull down of EVs derived from colorectal cancer patient plasma revealed a statistically significant amount of EpCAM-positive EVs as compared to healthy controls [83]. These findings were consistent with Taylor and colleagues in the context of ovarian cancer where the levels of EpCAM-positive EVs correlated with clinical stages of patients [84]. In addition, circulating EVs derived from cancer patients were correlated with disease prognosis and survival [83]. Consistent with previous studies, there were no observed differences in disease free survival between patients with high and low levels of circulating EVs. However, patients with high levels of circulating EVs were shown to have a shorter overall survival compared to patients will low EV levels. After 36 months of follow up, the overall survival of patients with high and low levels of EVs was 55 and 89%, respectively [83].
The possibility of exploiting urinary EVs as source of disease biomarkers beyond the urinary tract was investigated by Li and colleagues [85]. The proteome of urinary EVs from both healthy and non-small cell lung cancer (NSCLC) patients were examined. Urinary EVs isolated by ultracentrifugation were separated by 1-D SDS-PAGE, subjected to tryptic digestion followed by nano-HPLC-chip-MS/MS. Among the 18 proteins identified, leucine-rich α-2-glycoprotein (LRG1) was chosen to be investigated further and Western blotting confirmed the presence of LRG1 in association with EVs in both subjects with approximately a 6-fold increase in NSCLC patient urinary EVs. Additionally, immunohistochemistry was performed on NSCLC tissue sections showing high LRG1 expression levels compared to adjacent non tumour lung tissue. A combination of these findings implies the use of urinary EVs as biomarkers and a non-invasive tool for the clinic [85, 86].
EVs isolated from bodily fluids are less complex than whole bodily fluids
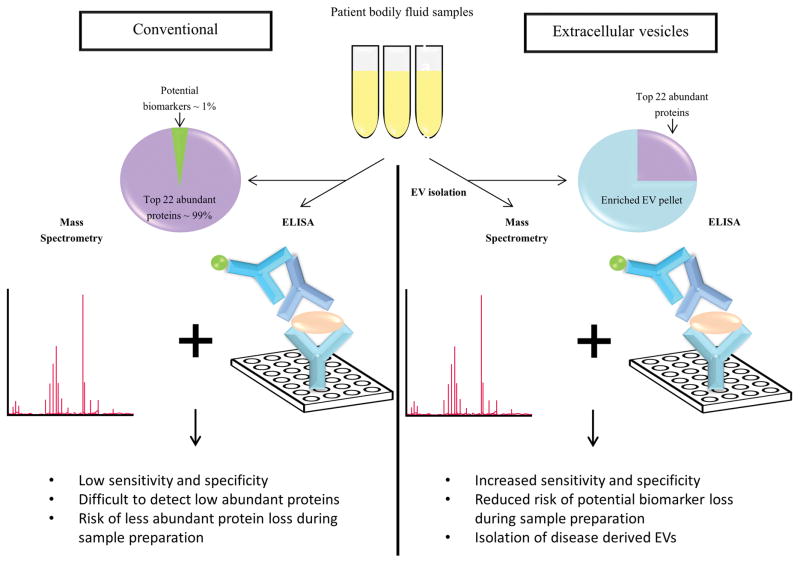
The complexity of bodily fluids is a rate limiting step in the identification and validation of biomarkers in conventional analysis. When the EV fraction of any bodily fluid is targeted, the complexity of the sample is significantly reduced (Fig. 2). For instance, Kalra et al. isolated EVs from plasma and established the depletion of high abundant plasma proteins [63]. Antibody-based high abundant protein depletion strategies may create variability between patient samples by non-specifically depleting low abundant proteins. Whilst different EV isolation methods may yield varying results [87, 88], it can be speculated that the same centrifugation based method within a set of patient samples seldom create any variability.
Figure 2. Conventional versus EV-based methods for the discovery and validation of biomarkers.
Conventional method of biomarker discovery and validation via mass spectrometry and/or ELISA are challenged by high abundant proteins in bodily fluid samples thereby hindering the detection of biomarkers. In contrast, utilizing EVs decreases the complexity of the sample by depleting the high abundant proteins.
Membranous EVs are highly stable
Biorepositories store patient samples for decades. The stability of EVs and their luminal cargo in stored patient samples is critical for biomarker analysis. Sokolova and colleagues assessed the stability of EVs isolated from three different cell lines (HEK293T, ECFC and MSC) in PBS at 37°C, 4°C and −20°C [89]. Nanoparticle tracking analysis revealed a decrease in EV size in both 37°C and 4°C with a slower rate of degradation at 4°C. Notably, multiple freeze thawing of EVs showed no change in EV size indicating that freezing EVs is the best suited method to store EVs [89].
To assess the stability of EVs in plasma, Kalra and colleagues isolated LIM1863 colorectal cell-derived EVs and spiked them in plasma [63]. EV samples were stored with and without protease inhibitors at 37°C, 4°C, −20°C and −80°C and samples were obtained at various time points up to 3 months. Western blot analysis revealed that all recovered EV samples were positive for the EV marker TSG101 indicating EVs are stable at least for 3 months. Consistent with Sokolova and colleagues [89], the results showed that EVs stored at −80°C are highly stable compared to the other storage conditions [63]. These studies confirm that EVs provide a stable environment for luminal proteins and RNA. Even though the stability of luminal protein such as TSG101 is analysed, none of the studies have analysed the stability of EV membrane proteins thus warranting further research.
Isolation of EVs in clinical settings
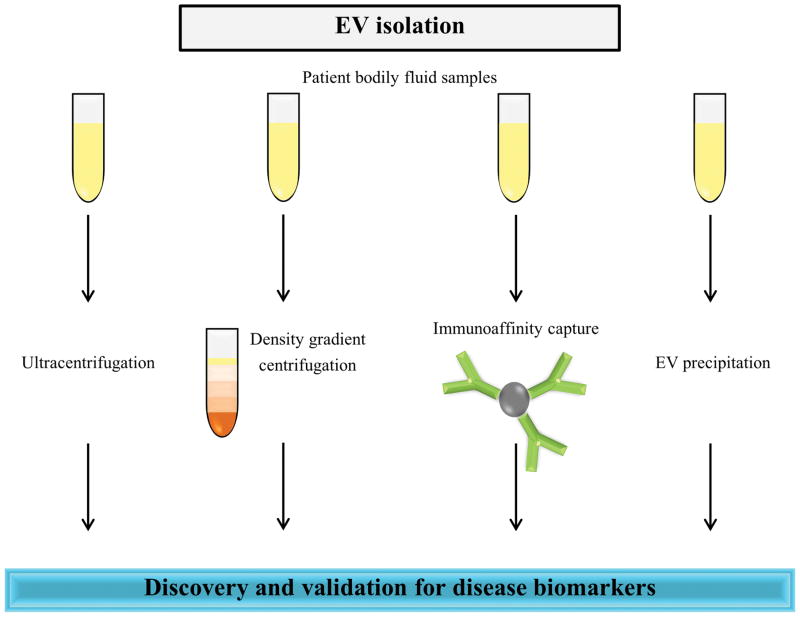
Although there has been growing interest in the potential use of EVs for disease biomarkers, a general consensus of EV isolation protocol is yet to be achieved [20, 90]. Body fluid-based EV isolation traditionally involves any of the three commonly used methods including differential centrifugation coupled with ultracentrifugation, immunoaffinity pull-down and density gradient separation [19, 20, 63]. In addition to these procedures, several commercial kits have also been developed by vendors and are routinely utilised (Fig. 3). Multiple studies have compared EV isolation methodologies to assess the respective procedures in the context of purity of the isolated population of vesicles and the robustness of the technique. Tauro et al. compared commonly used methods to isolate EVs from cell conditioned media and highlighted that immunoaffinity pull-down method isolated EVs containing multiple EV markers [88]. In case of plasma, Kalra et al. compared the commonly used methods to isolate EVs and concluded that density gradient separation method yields EVs devoid of protein contaminants [63]. The study employed transmission electron microscopy, Western blotting, atomic force microscopy and MS analysis to confirm that density gradient centrifugation can purify EVs devoid of high abundant proteins thereby reducing the complexity. Similarly, Alvarez and colleagues subjected urine samples to 6 different EV isolation methods and identified a modified ExoQuick-TC protocol as the better method in achieving the highest EV yield and purity compared to standard ExoQuick-TC, ultracentrifugation with or without 30% sucrose cushion or filtration and nanomembrane ultrafiltration methods [91]. Similar to other body fluid analysis, urine EV purity is also affected by high abundant soluble urinary proteins such as THP [91].
Figure 3. Possible EV isolation techniques for biomarker analysis.
Potential EV isolation strategies such as ultracentrifugation alone or coupled with density gradient centrifugation, immunoaffinity capture and EV precipitation can be employed for the detection of EV based biomarkers.
While ultracentrifugation is highly utilised for isolation of EVs in research laboratories, it is laborious and time consuming [63, 92]. In addition, patient blood samples are usually limited in volume and may be inadequate to be utilised for ultracentrifugation. This impediment may be overcome by using EV precipitation reagents, such as ExoQuick, which can be easily utilised without ultracentrifugation [91]. Importantly, the EV precipitation methods demand low volume of patient samples. In addition, they are simple, fast and only require a common centrifuge making them ideal methods to be utilised in clinical settings. Even though precipitation methods can isolate EVs that can potentially discriminate between normal and disease samples in the validation phase, it is generally accepted that the purity of isolated vesicles is poor. We emphasise caution in the use of precipitation techniques for discovery phase studies for identification of biomarkers. In such cases, ultracentrifugation or density gradient centrifugation-based methods may be considered as it yields sufficient protein quantities suitable for MS. In spite of possessing several advantages over the conventional biomarker analysis, the field of EVs also face few challenges. A robust method of isolating EVs from complex bodily fluids such as plasma is still lacking. In addition, more analysis is needed to understand the impact of the various methods of sample collection, processing and storage on EV stability.
Conclusions
In the recent past, biomarker analyses have been impeded by the complexity of bodily fluids including plasma, serum, urine and cerebrospinal fluid. Potential disease biomarkers are often in low abundance and are readily masked by high abundant proteins in the bodily fluids. EVs contain a rich cargo of proteins, RNA, lipids and metabolites which are specifically sorted and often reflect the biological state of the cell type of origin. In addition, EVs contain a lipid bilayer and are highly stable in stored conditions. All these attributes make them ideal choice for biomarker analysis. Isolation of the EV fraction in bodily fluids readily depletes most of the high abundant proteins thereby significantly reducing the complexity of the sample. Hence, EVs-based biomarker analyses have immense potential to be translated into clinical settings. However, as the field is still in infancy, the isolation protocols are still being optimized and more robust methods are needed to harness the true potential in the clinical scenario.
Statement of clinical relevance.
Recent extracellular vesicles-based studies have reignited interest in the field of biomarker discovery and validation. Extracellular vesicles, including exosomes, contain a rich cargo of proteins and RNA that are reflective of the host cell type and are selectively packaged. As extracellular vesicles are membranous structures, the luminal contents are protected from degradation by extracellular proteases and are highly stable in storage conditions. Interestingly, an appealing feature of extracellular vesicle-based biomarker analysis is the significant reduction in the sample complexity compared to whole bodily fluids. With these prescribed attributes, which are the rate-limiting factors of traditional biomarker analysis, there is immense potential for the use of extracellular vesicles for biomarker detection in the clinical settings.
Acknowledgments
SM is supported by the Australian NH&MRC fellowship (1016599), Australian Research Council Discovery project grant (DP130100535), Ramaciotti Establishment Grant and Award U54-DA036134 supported by the NIH Common Fund through the Office of Strategic Coordination/Office of the NIH Director. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- MVB
Multivesicular body
- EV
Extracellular vesicle
- ILV
Intraluminal vesicle
- PSA
Prostate specific antigen
- CEA
Carcinoembryonic antigen
- CA125
Carbohydrate antigen 125
- CSF
Cerebrospinal fluid
- SMVs
Shedding microvesicles
- NSCLC
Non-small cell lung cancer
- SRM
Selected reaction monitoring
Footnotes
Conflict of interest statement
The authors have declared no conflict of interest
References
- 1.Hulka BS, Wilcosky TC, Griffith JD. Biological markers in epidemiology. Oxford University Press; Nueva York: 1990. [Google Scholar]
- 2.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 3.Magni F, Van Der Burgt YE, Chinello C, Mainini V, et al. Biomarkers discovery by peptide and protein profiling in biological fluids based on functionalized magnetic beads purification and mass spectrometry. Blood Transfus. 2010;8:s92. doi: 10.2450/2010.015S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn SM, Simpson RJ. Body fluid proteomics: Prospects for biomarker discovery. Proteomics Clin Appl. 2007;1:1004–1015. doi: 10.1002/prca.200700217. [DOI] [PubMed] [Google Scholar]
- 5.Strimbu K, Tavel JA. What are biomarkers? Curr Opin HIV AIDS. 2010;5:463–466. doi: 10.1097/COH.0b013e32833ed177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayeux R. Biomarkers: potential uses and limitations. NeuroRx. 2004;1:182–188. doi: 10.1602/neurorx.1.2.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perera FP, Weinstein IB. Molecular epidemiology: recent advances and future directions. Carcinogenesis. 2000;21:517–524. doi: 10.1093/carcin/21.3.517. [DOI] [PubMed] [Google Scholar]
- 8.Mathivanan S. Quest for Cancer Biomarkers: Assaying Mutant Proteins and RNA that Provides the Much Needed Specificity. J Proteomics Bioinform. 2012;5:xiii–xvii. [Google Scholar]
- 9.Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 10.Lescuyer P, Hochstrasser D, Rabilloud T. How shall we use the proteomics toolbox for biomarker discovery? J Proteome Res. 2007;6:3371–3376. doi: 10.1021/pr0702060. [DOI] [PubMed] [Google Scholar]
- 11.Good DM, Thongboonkerd V, Novak J, Bascands JL, et al. Body fluid proteomics for biomarker discovery: lessons from the past hold the key to success in the future. J Proteome Res. 2007;6:4549–4555. doi: 10.1021/pr070529w. [DOI] [PubMed] [Google Scholar]
- 12.Hori SS, Gambhir SS. Mathematical model identifies blood biomarker–based early cancer detection strategies and limitations. Sci Transl Med. 2011;3:109ra116. doi: 10.1126/scitranslmed.3003110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magagnotti C, Fermo I, Carletti RM, Ferrari M, Bachi A. Comparison of different depletion strategies for improving resolution of the human urine proteome. Clin Chem Lab Med. 2010;48:531–535. doi: 10.1515/CCLM.2010.109. [DOI] [PubMed] [Google Scholar]
- 14.Bandow JE. Comparison of protein enrichment strategies for proteome analysis of plasma. Proteomics. 2010;10:1416–1425. doi: 10.1002/pmic.200900431. [DOI] [PubMed] [Google Scholar]
- 15.Ahmed N, Barker G, Oliva K, Garfin D, et al. An approach to remove albumin for the proteomic analysis of low abundance biomarkers in human serum. Proteomics. 2003;3:1980–1987. doi: 10.1002/pmic.200300465. [DOI] [PubMed] [Google Scholar]
- 16.Bjorhall K, Miliotis T, Davidsson P. Comparison of different depletion strategies for improved resolution in proteomic analysis of human serum samples. Proteomics. 2005;5:307–317. doi: 10.1002/pmic.200400900. [DOI] [PubMed] [Google Scholar]
- 17.Yocum AK, Yu K, Oe T, Blair IA. Effect of immunoaffinity depletion of human serum during proteomic investigations. J Proteome Res. 2005;4:1722–1731. doi: 10.1021/pr0501721. [DOI] [PubMed] [Google Scholar]
- 18.Brooks JD. Translational genomics: the challenge of developing cancer biomarkers. Genome Res. 2012;22:183–187. doi: 10.1101/gr.124347.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Simpson RJ, Mathivanan S. Extracellular Microvesicles: The Need for Internationally Recognised Nomenclature and Stringent Purification Criteria. J Proteomics Bioinform. 2012;5:ii–ii. [Google Scholar]
- 21.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Cocucci E, Racchetti G, Podini P, Meldolesi J. Enlargeosome traffic: exocytosis triggered by various signals is followed by endocytosis, membrane shedding or both. Traffic. 2007;8:742–757. doi: 10.1111/j.1600-0854.2007.00566.x. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi T, Okamoto H, Yamada JI, Setaka M, Kwan T. Vesiculation of platelet plasma membranes. Dilauroylglycerophosphocholine-induced shedding of a platelet plasma membrane fraction enriched in acetylcholinesterase activity. Biochim Biophys Acta. 1984;778:210–218. doi: 10.1016/0005-2736(84)90464-4. [DOI] [PubMed] [Google Scholar]
- 24.Dolo V, Li R, Dillinger M, Flati S, et al. Enrichment and localization of ganglioside G(D3) and caveolin-1 in shed tumor cell membrane vesicles. Biochim Biophys Acta. 2000;1486:265–274. doi: 10.1016/s1388-1981(00)00063-9. [DOI] [PubMed] [Google Scholar]
- 25.Hristov M, Erl W, Linder S, Weber PC. Apoptotic bodies from endothelial cells enhance the number and initiate the differentiation of human endothelial progenitor cells in vitro. Blood. 2004;104:2761–2766. doi: 10.1182/blood-2003-10-3614. [DOI] [PubMed] [Google Scholar]
- 26.Boulanger CM, Amabile N, Tedgui A. Circulating microparticles a potential prognostic marker for atherosclerotic vascular disease. Hypertension. 2006;48:180–186. doi: 10.1161/01.HYP.0000231507.00962.b5. [DOI] [PubMed] [Google Scholar]
- 27.Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2:ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 28.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 29.Gangoda L, Boukouris S, Liem M, Kalra H, Mathivanan S. Extracellular vesicles including exosomes are mediators of signal transduction: Are they protective or pathogenic? Proteomics. 2015;15:260–271. doi: 10.1002/pmic.201400234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lotvall J, Hill AF, Hochberg F, Buzas EI, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J Cell Biol. 1985;101:942–948. doi: 10.1083/jcb.101.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trowbridge I, Collawn J, Hopkins C. Signal-dependent membrane protein trafficking in the endocytic pathway. Annu Rev Cell Biol. 1993;9:129–161. doi: 10.1146/annurev.cb.09.110193.001021. [DOI] [PubMed] [Google Scholar]
- 33.Johnstone RM, Adam M, Hammond J, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262:9412–9420. [PubMed] [Google Scholar]
- 34.Kleijmeer MJ, Stoorvogel W, Griffith JM, Yoshie O, Geuze HJ. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J Biol Chem. 1998;273:20121–20127. doi: 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]
- 35.Fevrier B, Vilette D, Archer F, Loew D, et al. Cells release prions in association with exosomes. Proc Natl Acad Sci U S A. 2004;101:9683–9688. doi: 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mallegol J, Van Niel G, Heyman M. Phenotypic and functional characterization of intestinal epithelial exosomes. Blood Cells Mol Dis. 2005;35:11–16. doi: 10.1016/j.bcmd.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 37.Wolfers J, Lozier A, Raposo G, Regnault A, et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat Med. 2001;7:297–303. doi: 10.1038/85438. [DOI] [PubMed] [Google Scholar]
- 38.Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6:267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 39.Simpson RJ, Kalra H, Mathivanan S. ExoCarta as a resource for exosomal research. J Extracell Vesicles. 2012;1:18374. doi: 10.3402/jev.v1i0.18374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathivanan S, Lim JW, Tauro BJ, Ji H, et al. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol Cell Proteomics. 2010;9:197–208. doi: 10.1074/mcp.M900152-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skog J, Wurdinger T, van Rijn S, Meijer DH, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janowska-Wieczorek A, Wysoczynski M, Kijowski J, Marquez-Curtis L, et al. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. Int J Cancer. 2005;113:752–760. doi: 10.1002/ijc.20657. [DOI] [PubMed] [Google Scholar]
- 43.Millimaggi D, Mari M, D’Ascenzo S, Carosa E, et al. Tumor vesicle-associated CD147 modulates the angiogenic capability of endothelial cells. Neoplasia. 2007;9:349–357. doi: 10.1593/neo.07133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 45.Zitvogel L, Regnault A, Lozier A, Wolfers J, et al. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell derived exosomes. Nat Med. 1998;4:594–600. doi: 10.1038/nm0598-594. [DOI] [PubMed] [Google Scholar]
- 46.Théry C, Boussac M, Véron P, Ricciardi-Castagnoli P, et al. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 47.Karlsson M, Lundin S, Dahlgren U, Kahu H, et al. “Tolerosomes” are produced by intestinal epithelial cells. Eur J Immunol. 2001;31:2892–2900. doi: 10.1002/1521-4141(2001010)31:10<2892::aid-immu2892>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 48.Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hunter MP, Ismail N, Zhang X, Aguda BD, et al. Detection of microRNA expression in human peripheral blood microvesicles. PloS One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valadi H, Ekström K, Bossios A, Sjöstrand M, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 51.Robertson C, Booth SA, Beniac DR, Coulthart MB, et al. Cellular prion protein is released on exosomes from activated platelets. Blood. 2006;107:3907–3911. doi: 10.1182/blood-2005-02-0802. [DOI] [PubMed] [Google Scholar]
- 52.Nguyen DG, Booth A, Gould SJ, Hildreth JE. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J Biol Chem. 2003;278:52347–52354. doi: 10.1074/jbc.M309009200. [DOI] [PubMed] [Google Scholar]
- 53.Amzallag N, Passer BJ, Allanic D, Segura E, et al. TSAP6 facilitates the secretion of translationally controlled tumor protein/histamine-releasing factor via a nonclassical pathway. J Biol Chem. 2004;279:46104–46112. doi: 10.1074/jbc.M404850200. [DOI] [PubMed] [Google Scholar]
- 54.Clayton A, Turkes A, Dewitt S, Steadman R, et al. Adhesion and signaling by B cell-derived exosomes: the role of integrins. FASEB J. 2004;18:977–979. doi: 10.1096/fj.03-1094fje. [DOI] [PubMed] [Google Scholar]
- 55.Denzer K, van Eijk M, Kleijmeer MJ, Jakobson E, et al. Follicular dendritic cells carry MHC class II-expressing microvesicles at their surface. J Immunol. 2000;165:1259–1265. doi: 10.4049/jimmunol.165.3.1259. [DOI] [PubMed] [Google Scholar]
- 56.Peinado H, Alečković M, Lavotshkin S, Matei I, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cossetti C, Iraci N, Mercer TR, Leonardi T, et al. Extracellular Vesicles from Neural Stem Cells Transfer IFN-gamma via Ifngr1 to Activate Stat1 Signaling in Target Cells. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alvarez-Erviti L, Seow Y, Schapira AH, Gardiner C, et al. Lysosomal dysfunction increases exosome-mediated alpha-synuclein release and transmission. Neurobiol Dis. 2011;42:360–367. doi: 10.1016/j.nbd.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vella LJ, Sharples RA, Nisbet RM, Cappai R, Hill AF. The role of exosomes in the processing of proteins associated with neurodegenerative diseases. Eur Biophys J. 2008;37:323–332. doi: 10.1007/s00249-007-0246-z. [DOI] [PubMed] [Google Scholar]
- 60.Bellingham SA, Guo BB, Coleman BM, Hill AF. Exosomes: vehicles for the transfer of toxic proteins associated with neurodegenerative diseases? Front Physiol. 2012;3:124. doi: 10.3389/fphys.2012.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6:267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 62.Properzi F, Logozzi M, Fais S. Exosomes: the future of biomarkers in medicine. Biomark Med. 2013;7:769–778. doi: 10.2217/bmm.13.63. [DOI] [PubMed] [Google Scholar]
- 63.Kalra H, Adda CG, Liem M, Ang CS, et al. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics. 2013;13:3354–3364. doi: 10.1002/pmic.201300282. [DOI] [PubMed] [Google Scholar]
- 64.Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med. 2013;91:431–437. doi: 10.1007/s00109-013-1020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Q, Chaerkady R, Wu J, Hwang HJ, et al. Mutant proteins as cancer-specific biomarkers. Proc Natl Acad Sci U S A. 2011;108:2444–2449. doi: 10.1073/pnas.1019203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mathivanan S, Ji H, Tauro BJ, Chen YS, Simpson RJ. Identifying mutated proteins secreted by colon cancer cell lines using mass spectrometry. J Proteomics. 2012;76:141–149. doi: 10.1016/j.jprot.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 67.Goldenberg DM, Sharkey RM, Primus FJ. Carcinoembryonic antigen in histopathology: immunoperoxidase staining of conventional tissue sections. J Natl Cancer Inst. 1976;57:11–22. doi: 10.1093/jnci/57.1.11. [DOI] [PubMed] [Google Scholar]
- 68.Smith TJ, Davidson NE, Schapira DV, Grunfeld E, et al. American Society of Clinical Oncology 1998 update of recommended breast cancer surveillance guidelines. J Clin Oncol. 1999;17:1080–1082. doi: 10.1200/JCO.1999.17.3.1080. [DOI] [PubMed] [Google Scholar]
- 69.Marrelli D, Roviello F, De Stefano A, Farnetani M, et al. Prognostic significance of CEA, CA 19-9 and CA 72-4 preoperative serum levels in gastric carcinoma. Oncology. 1999;57:55–62. doi: 10.1159/000012001. [DOI] [PubMed] [Google Scholar]
- 70.Nazli O, Bozdag AD, Tansug T, Kir R, Kaymak E. The diagnostic importance of CEA and CA 19-9 for the early diagnosis of pancreatic carcinoma. Hepatogastroenterology. 2000;47:1750–1752. [PubMed] [Google Scholar]
- 71.Nilsson J, Skog J, Nordstrand A, Baranov V, et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer. 2009;100:1603–1607. doi: 10.1038/sj.bjc.6605058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Graner MW, Alzate O, Dechkovskaia AM, Keene JD, et al. Proteomic and immunologic analyses of brain tumor exosomes. FASEB J. 2008 doi: 10.1096/fj.08-122184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lv LL, Cao YH, Pan MM, Liu H, et al. CD2AP mRNA in urinary exosome as biomarker of kidney disease. Clinica chimica acta; international journal of clinical chemistry. 2014;428:26–31. doi: 10.1016/j.cca.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 74.Kawikova I, Askenase PW. Diagnostic and therapeutic potentials of exosomes in CNS diseases. Brain Res. 2014 doi: 10.1016/j.brainres.2014.09.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shao H, Chung J, Balaj L, Charest A, et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med. 2012;18:1835–1840. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hong CS, Muller L, Whiteside TL, Boyiadzis M. Plasma exosomes as markers of therapeutic response in patients with acute myeloid leukemia. Front Immunol. 2014;5:160. doi: 10.3389/fimmu.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Duijvesz D, Burnum-Johnson KE, Gritsenko MA, Hoogland AM, et al. Proteomic profiling of exosomes leads to the identification of novel biomarkers for prostate cancer. PloS One. 2013;8:e82589. doi: 10.1371/journal.pone.0082589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Duijvesz D, Luider T, Bangma CH, Jenster G. Exosomes as biomarker treasure chests for prostate cancer. Eur Urol. 2011;59:823–831. doi: 10.1016/j.eururo.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 79.Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40:D1241–1244. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kalra H, Simpson RJ, Ji H, Aikawa E, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10:e1001450. doi: 10.1371/journal.pbio.1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Iero M, Valenti R, Huber V, Filipazzi P, et al. Tumour-released exosomes and their implications in cancer immunity. Cell Death Differ. 2008;15:80–88. doi: 10.1038/sj.cdd.4402237. [DOI] [PubMed] [Google Scholar]
- 82.Grammatikakis N, Vultur A, Ramana CV, Siganou A, et al. The role of Hsp90N, a new member of the Hsp90 family, in signal transduction and neoplastic transformation. J Biol Chem. 2002;277:8312–8320. doi: 10.1074/jbc.M109200200. [DOI] [PubMed] [Google Scholar]
- 83.Silva J, Garcia V, Rodriguez M, Compte M, et al. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosomes Cancer. 2012;51:409–418. doi: 10.1002/gcc.21926. [DOI] [PubMed] [Google Scholar]
- 84.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 85.Li Y, Zhang Y, Qiu F, Qiu Z. Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis. 2011;32:1976–1983. doi: 10.1002/elps.201000598. [DOI] [PubMed] [Google Scholar]
- 86.Conde-Vancells J, Rodriguez-Suarez E, Gonzalez E, Berisa A, et al. Candidate biomarkers in exosome-like vesicles purified from rat and mouse urine samples. Proteomics Clin Appl. 2010;4:416–425. doi: 10.1002/prca.200900103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kalra H, Adda CG, Liem M, Ang CS, et al. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics. 2013;13:3354–3364. doi: 10.1002/pmic.201300282. [DOI] [PubMed] [Google Scholar]
- 88.Tauro BJ, Greening DW, Mathias RA, Ji H, et al. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56:293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 89.Sokolova V, Ludwig AK, Hornung S, Rotan O, et al. Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids Surf B Biointerfaces. 2011;87:146–150. doi: 10.1016/j.colsurfb.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 90.Musante L, Tataruch DE, Holthofer H. Use and isolation of urinary exosomes as biomarkers for diabetic nephropathy. Front Endocrinol. 2014;5:149. doi: 10.3389/fendo.2014.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Alvarez ML, Khosroheidari M, Ravi RK, DiStefano JK. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012;82:1024–1032. doi: 10.1038/ki.2012.256. [DOI] [PubMed] [Google Scholar]
- 92.Dear JW, Street JM, Bailey MA. Urinary exosomes: a reservoir for biomarker discovery and potential mediators of intrarenal signalling. Proteomics. 2013;13:1572–1580. doi: 10.1002/pmic.201200285. [DOI] [PubMed] [Google Scholar]