Supplemental Digital Content is available in the text
Keywords: chronic thromboembolic pulmonary hypertension, circRNA–miRNA–mRNA network, circular RNAs, functional enrichment
Abstract
Background:
Chronic thromboembolic pulmonary hypertension (CTEPH) is a rare but debilitating and life-threatening complication of acute pulmonary embolism. Circular RNAs (circRNAs), presenting as covalently closed continuous loops, are RNA molecules with covalently joined 3′- and 5′-ends formed by back-splicing events. circRNAs may be significant biological molecules to understand disease mechanisms and to identify biomarkers for disease diagnosis and therapy. The aim of this study was to investigate the potential roles of circRNAs in CTEPH.
Methods:
Ten human blood samples (5 each from CTEPH and control groups) were included in the Agilent circRNA chip. The differentially expressed circRNAs were evaluated using t test, with significance set at a P value of < .05. A functional enrichment analysis for differentially expressed circRNAs was performed using DAVID online tools, and a Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis for target genes of miRNAs was performed using the R package clusterProfiler. Furthermore, miRNAs that interacted with differentially expressed circRNAs were predicted using the miRanda package. mRNAs that had clear biological functions and were regulated by miRNAs were predicted using miRWalk2.0 and then combined into a circRNA–miRNA–mRNA network.
Results:
In total, 351 differentially expressed circRNAs (122 upregulated and 229 downregulated) between CTEPH and control groups were obtained; among these circRNAs, hsa_circ_0002062 and hsa_circ_0022342 might be important because they can regulate 761 (e.g., hsa-miR-942–5p) and 453 (e.g., hsa-miR-940) miRNAs, respectively. Target genes (e.g., cyclin-dependent kinase 6) of hsa-miR-942–5p were mainly enriched in cancer-related pathways, whereas target genes (e.g., CRK-Like Proto-Oncogene, Adaptor Protein) of hsa-miR-940 were enriched in the ErbB signaling pathway. Therefore, these pathways are potentially important in CTEPH.
Conclusions:
Our findings suggested that hsa_circ_0002062 and hsa_circ_0022342 may be key circRNAs for CTEPH development and that their targeted regulation may be an effective approach for treating CTEPH.
1. Introduction
Chronic thromboembolic pulmonary hypertension (CTEPH), a rare but debilitating and life-threatening complication of acute pulmonary embolism, is caused by persistent obstruction of pulmonary arteries and progressive vascular remodeling.[1] The estimated prevalence of CTEPH is 0.1% to 9.1% at 2 years after acute pulmonary embolism.[2,3] The risk factors for CTEPH include some clinical disorders, such as inflammatory bowel disease, splenectomy, and myeloproliferative disorders.[4–7] Patients with CTEPH exhibit a poor prognosis unless they receive treatment at an early stage.[8] Thus, early diagnosis and treatment of this disease are important.
Circular RNAs (circRNAs), presenting as covalently closed continuous loops, are RNA molecules with covalently joined 3′- and 5′-ends formed by back-splicing events.[9] Recently, circRNAs have increasingly been recognized as an abundant class of regulatory transcripts primarily derived from protein-coding exons.[10,11] CircRNAs are characterized by scrambled exons, and until 20 years ago, they were mostly misinterpreted as resulting from splicing errors.[12] Only recently, circRNAs have been rediscovered from RNA-seq data.[10,11,13] Further, circRNAs functioning as sponges of miRNAs are believed to regulate the expression of miRNA targets, thus contributing to the competing endogenous RNA network.[14] Some studies have demonstrated that CDR1as functions as a sponge for miR-7, whereas circRNA generated from the Sry gene (circSry) functions as a sponge for miR-138.[15,16] In addition, circRNAs play key roles in diseases, especially in cancer, and thus constitute new potential diagnostic and therapeutic targets for diseases.[17,18] Lukiw [19] have suggested that deficits in other circRNA-mediated “miRNA sponging systems” and ambient upregulation of specific inducible miRNAs help in explaining downregulated gene expression in the brains of patients with sporadic Alzheimer disease. Some other studies have indicated that circRNAs are involved in the development of nervous system disorders and atherosclerosis.[20,21] Therefore, circRNAs may be significant biological molecules to understand disease mechanisms and to identify biomarkers for disease diagnosis and therapy.[14]
To investigate the potential roles of circRNAs in CTEPH, we performed a microarray analysis to identify differentially expressed circRNAs in a CTEPH group compared with those in a control group. These circRNAs were then subjected to functional enrichment analyses, including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. Furthermore, miRNAs regulated by differentially expressed circRNAs were predicted, and enrichment analyses for the target genes of these miRNAs were also performed.
2. Methods
2.1. CircRNA expression profile data
Peripheral blood samples from 5 CTEPH patients at Beijing Chao-Yang Hospital, Capital Medical University, and 5 healthy controls who had undergone a routine physical examination at a physical examination center from March 2016 to April 2016 were collected. The criterion for inclusion in the CTEPH group was a previous diagnosed of CTEPH by right heart cardiac catheterization, ventilation-perfusion lung scans, or computed tomography (CT) pulmonary angiography (CTPA). The criterion for exclusion from the CTEPH group was the presence of a disease of the circulatory system, such as malignant tumor, hypertension, diabetes mellitus, coronary heart disease, or cerebrovascular disease. The control group comprised healthy controls with age and sex matched to those in the CTEPH group and normal routine blood test/routine urine test/biochemical test/carcinoembryonic antigen (CEA)/alpha-fetoprotein (AFP)/erythrocyte sedimentation rate (ESR)/chest X-ray. This study was approved by the Ethics Committee of Beijing Chao-Yang Hospital, Capital Medical University. This study was approved by the ethics review board of our hospital, and the requirement to obtain informed written consent was waived. Demographic information on the patients is summarized in Table 1. No significant difference was observed in risk factors for CTEPH between the CTEPH and control groups (P > .1).
Table 1.
The baseline characteristics of the 10 samples.
Total RNAs were extracted from the samples using RNAprep Pure Blood Kit (Tiangen Biotech Co., Ltd., Beijing, China) according to the manufacturer's protocol. First-strand cDNA was synthesized using first-strand enzyme mix with random primers containing the T7 RNA polymerase promoter sequence. RNA in the DNA–RNA hybrid was transcribed to second-strand cDNA, and double-stranded DNA was synthesized. Subsequently, using second-strand cDNA as a template, cRNA was synthesized using T7 enzyme mix. Next, cRNA was purified and subjected to quantification and quality control. Reverse transcription of cRNA was then performed using random primers and CbcScript II reverse transcriptase. The obtained cDNA was purified and quantified. Subsequently, using cDNA, random primers, and DNA polymerase (Klenow fragment), the other chain of cDNA was synthesized and fluorescently labeled with dNTP (Cy3-dCTP). Lastly, the products were purified and quantified. This fluorescently labeled DNA was used subsequently for microarray hybridization.
2.2. Screening of differentially expressed circRNAs
CircRNA gene expression profiles (Agilent circRNA chip) were preprocessed using the Feature Extraction package, and subsequently, the data were normalized using the GeneSpring GX package. The CTEPH and control group samples could be clearly identified in the expression profile matrix file. Subsequently, the differentially expressed circRNAs between the 2 groups were evaluated using t test, with statistical significance set at a P value of <.05.
2.3. miRNA prediction of differentially expressed circRNAs
CircRNAs can regulate mRNA translation indirectly via combining with targeted miRNAs. The miRNA prediction of differentially expressed circRNAs was performed using the miRanda package (http://www.microrna.org/microrna/home.do).[22] The top 10 key target miRNAs based on the number of circRNAs associated with them were selected for analysis. On the basis of these findings, the miRNA–circRNA network was constructed using Cytoscape software.[23]
2.4. Construction of the circRNA–miRNA–mRNA network
mRNAs that had clear biological functions and were regulated by miRNAs were predicted using miRWalk2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/).[24,25] The results were also combined with the former predicted miRNA–circRNA results into a circRNA–miRNA–mRNA network.
2.5. Functional enrichment analysis for differentially expressed circRNAs and target genes of miRNAs
GO is a tool used for gene annotation by collecting defined, structured, controlled vocabulary.[26] KEGG is a database used to categorize associated gene sets into appropriate pathways.[27] DAVID is an integrated data-mining environment to analyze gene lists.[28]
GO and KEGG pathway enrichment analyses were performed using DAVID online tools (Version 6.8, https://david-d.ncifcrf.gov/), with the classification stringency set to “medium.” P value of <.05 and a count of >2 were regarded as the cut-off values.
KEGG pathway enrichment analysis for target genes of miRNAs was performed using the R package clusterProfiler.[29] A P value of <.01 was set as the threshold value.
3. Results
3.1. Screening of differentially expressed circRNAs
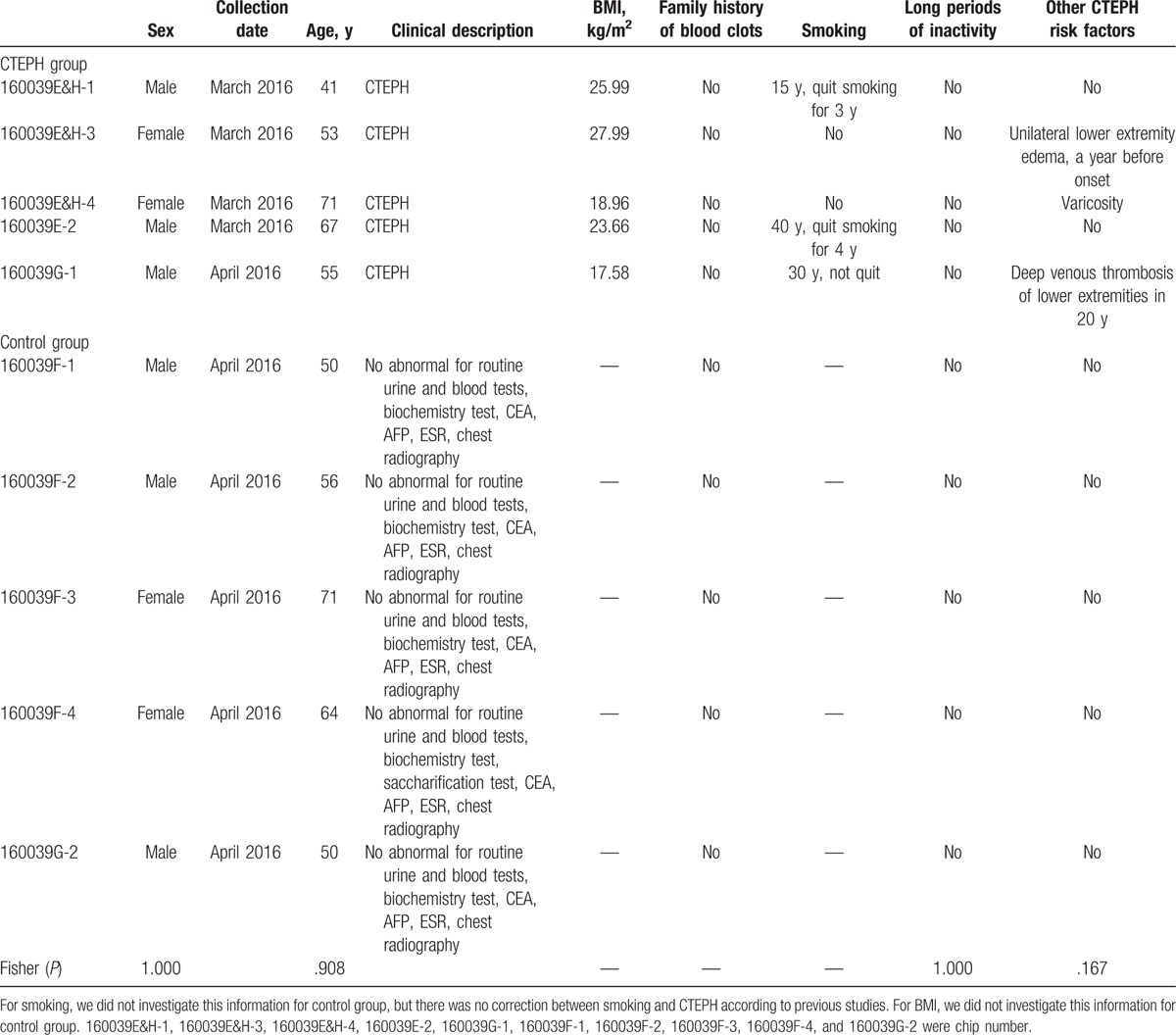
In the CTEPH group, 351 differentially expressed circRNAs (122 upregulated and 229 downregulated) compared with those in the control group were identified. The heat map is shown in Fig. 1, and information on these 351 differentially expressed circRNAs is summarized in Supplemental Table 1.
Figure 1.
(A) A heat map of 351 differentially expressed circRNAs. (B) A heat map of the top 10 upregulated and top 10 downregulated differentially expressed circRNAs. Green represents a low expression level, whereas red represents a high expression level.
3.2. miRNA prediction of differentially expressed circRNAs
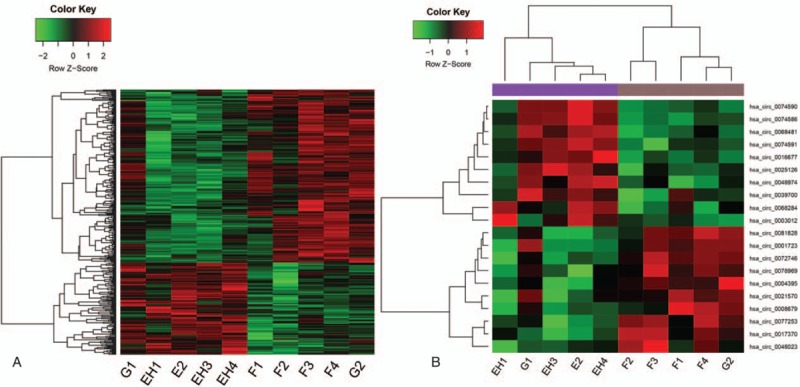
The results for predicting miRNAs associated with differentially expressed circRNAs (limited to circRNAs with >100 predicted associated miRNAs) are summarized in Table 2. The circRNAs hsa_circ_0002062 and hsa_circ_0022342 were associated with most miRNAs (761 and 453, respectively).
Table 2.
The results for miRNA prediction of differentially expressed circRNA with the number of miRNA greater than 100.
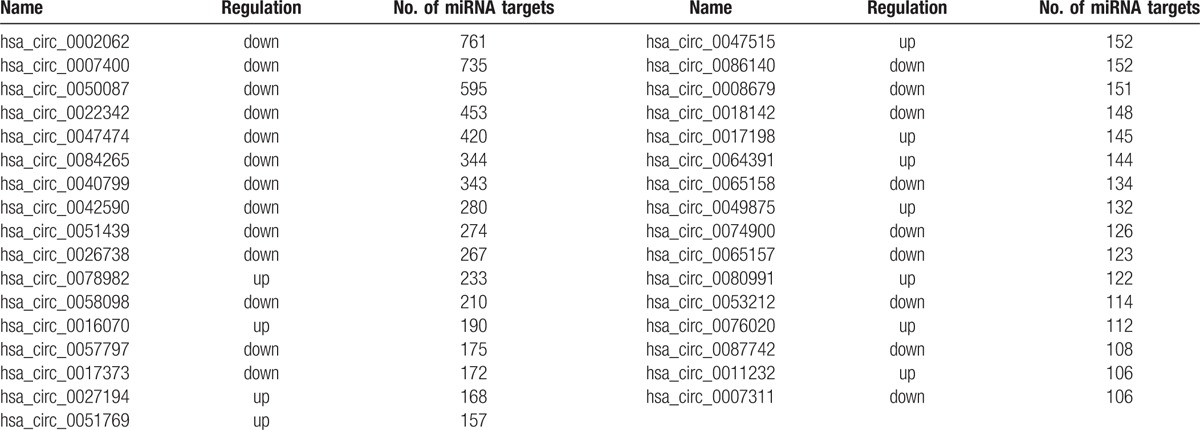
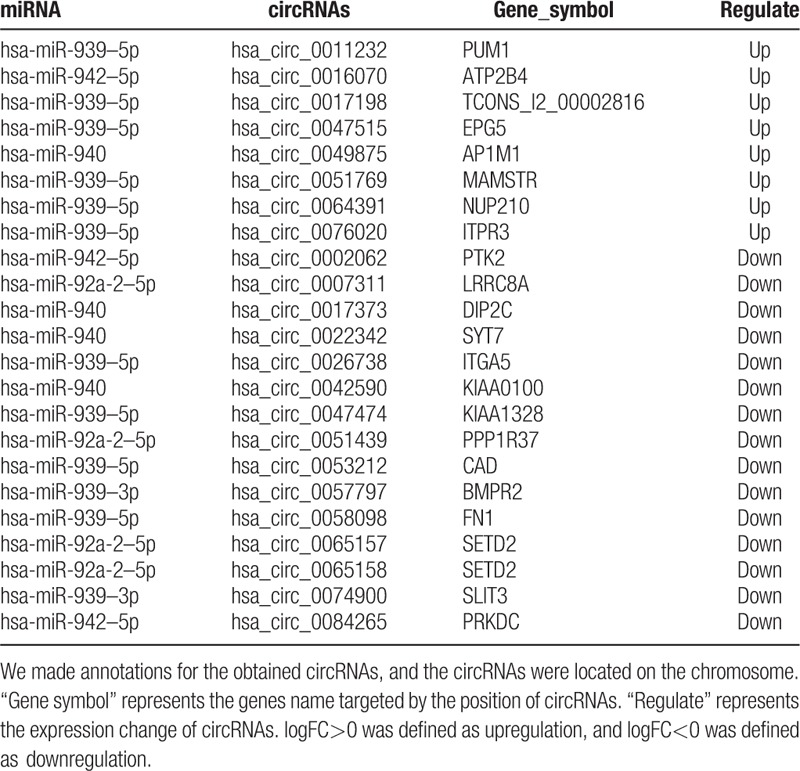
The miRNA–circRNA network is shown in Fig. 2. The 23 circRNAs shown in Fig. 2 are the differentially expressed circRNAs associated with >100 miRNAs provided in Table 2. From their status, we inferred that these 23 circRNAs were the key circRNAs. These 23 key circRNAs and the miRNAs regulated by them are summarized in Table 3. hsa_circ_0011232, hsa_circ_0017198, hsa_circ_0047515, hsa_circ_0051769, hsa_circ_0064391, hsa_circ_0076020, hsa_circ_0026738, hsa_circ_0047474, hsa_circ_0053212, and hsa_circ_0058098 regulated hsa-miR-939–5p; hsa_circ_0016070, hsa_circ_0002062, and hsa_circ_0084265 regulated hsa-miR-942–5p; hsa_circ_0049875, hsa_circ_0017373, hsa_circ_0022342, and hsa_circ_0042590 regulated hsa-miR-940; hsa_circ_0007311, hsa_circ_0051439, hsa_circ_0065157, and hsa_circ_0065158 regulated hsa-miR-92a-2–5p, and hsa_circ_0057797 and hsa_circ_0074900 regulated hsa-miR-939–3p. These circRNAs were related to cell movement and cell migration, as revealed by the enrichment analysis of the circRNA sequence annotation results.
Figure 2.
The miRNA–circRNA network. Green hexagon: upregulated differentially expressed circRNA; orange hexagon: downregulated differentially expressed circRNA; white diamond: miRNA.
Table 3.
The 23 key circRNAs and miRNAs regulated by these circRNAs.
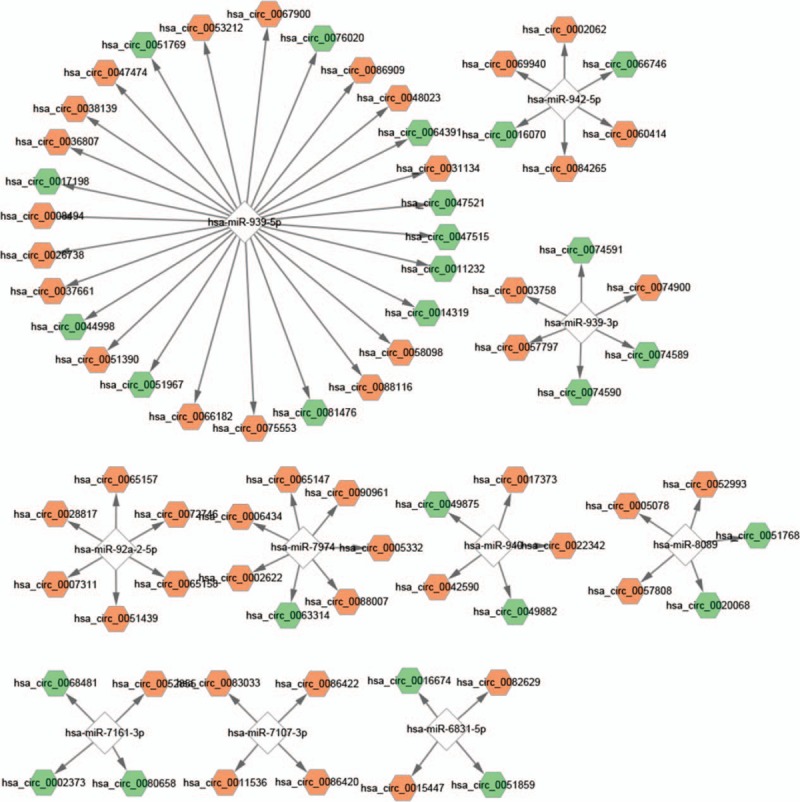
3.3. Construction of the circRNA–miRNA–mRNA network
The target genes of miRNAs were predicted, and the circRNA–miRNA–mRNA network is shown in Fig. 3. It shows that each of the 10 miRNAs (hsa-miR-939–5p, hsa-miR-939–3p, hsa-miR-942–5p, hsa-miR-940, hsa-miR-92a-2–5p, hsa-miR-7107–3p, hsa-miR-7161–3p, hsa-miR-6831–5p, hsa-miR-7974, and hsa-miR-8089) were regulated by circRNAs, which then regulated the target genes of miRNAs.
Figure 3.
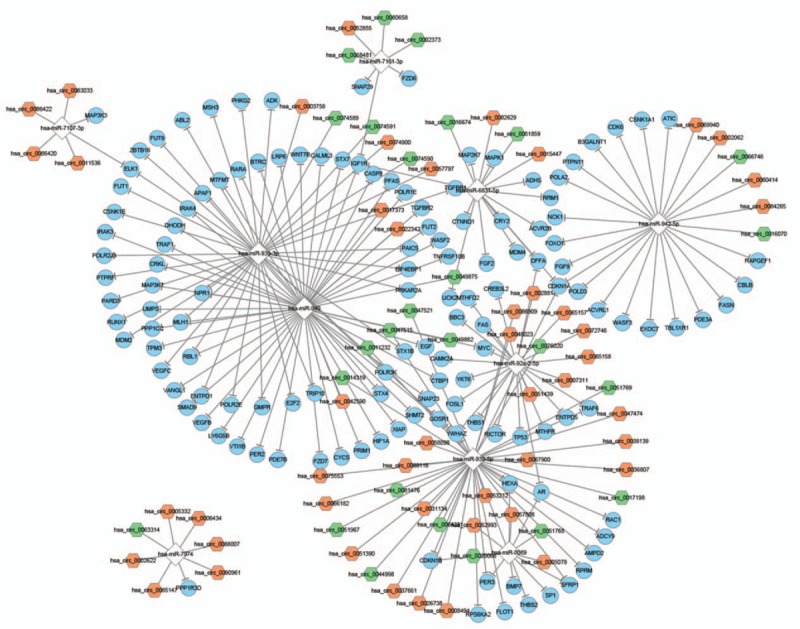
The circRNA–miRNA–mRNA network. Green hexagon: upregulated differentially expressed circRNA; orange hexagon: downregulated differentially expressed circRNA; white diamond: miRNA; blue circle: mRNA regulated by miRNA.
3.4. Functional enrichment analysis for differentially expressed circRNAs and target genes of miRNAs
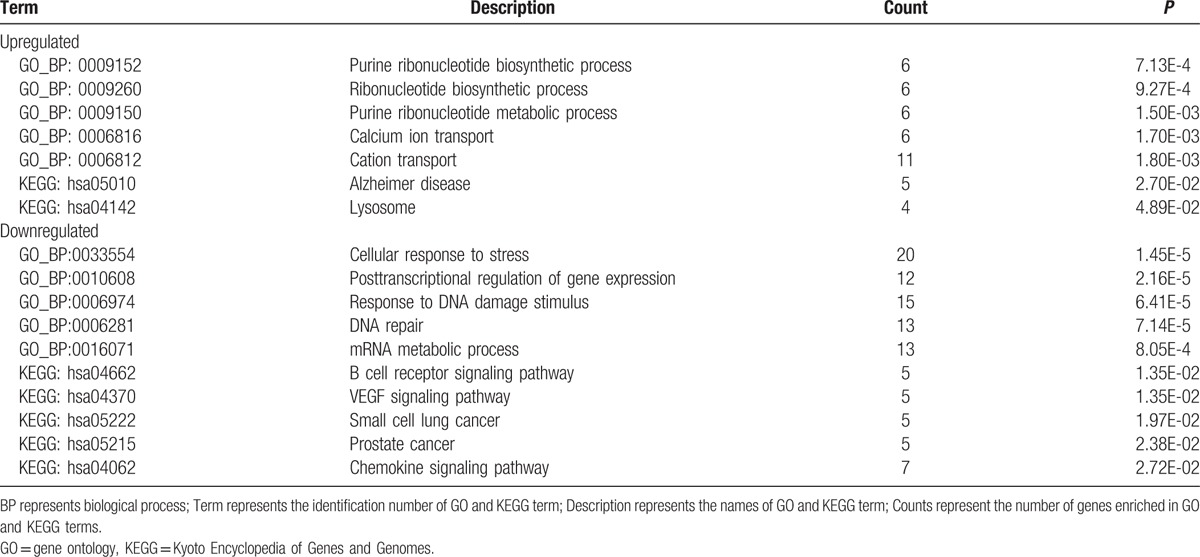
The results of GO and KEGG pathway enrichment analyses for differentially expressed circRNAs are summarized in Table 4. Upregulated circRNAs were mainly enriched in the purine ribonucleotide biosynthetic process and ribonucleotide biosynthetic process. Downregulated circRNAs were mainly enriched in cellular response to stress, post-transcriptional regulation of gene expression, response to DNA damage stimulus, DNA repair, and mRNA metabolic processes.
Table 4.
GO and KEGG pathway enrichment analysis for differentially expressed circRNA.
Furthermore, target genes of six miRNAs (hsa-miR-942–5p, hsa-miR-940, hsa-miR-939–3p, hsa-miR-92a-2–5p, hsa-miR-8089, and hsa-miR-6831–5p) were significantly enriched into function terms (Fig. 4). For example, the target genes [e.g., cyclin-dependent kinase 6 (CDK6)] of hsa-miR-942–5p were mainly enriched in cancer-related pathways, whereas the target genes (e.g., CRK-Like Proto-Oncogene, Adaptor Protein) of hsa-miR-940 were enriched in the ErbB signaling pathway, among others (Supplemental Table 2). Thus, these pathways were considered as potentially important in CTEPH.
Figure 4.
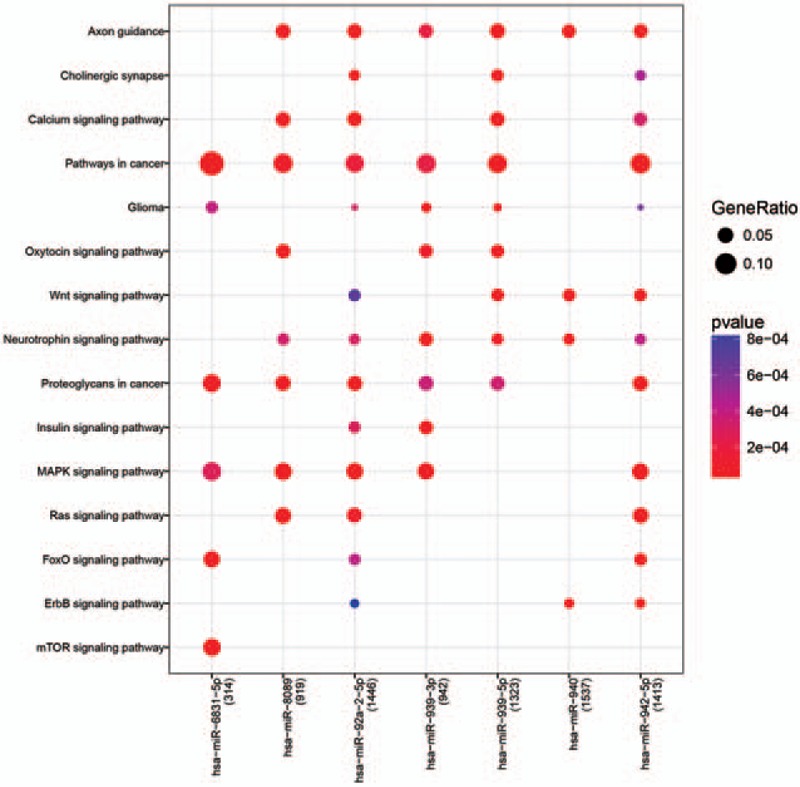
KEGG pathway enrichment analyses for target genes of miRNAs. Node size: gene ratio; node color: P value.
4. Discussion
CircRNAs represent a new special class of endogenous noncoding RNAs. Some studies have shown that circRNAs can function as miRNA sponges or regulate parent gene expression to influence disease initiation and progression.[18,30] In the present study, we analyzed differentially expressed circRNAs in the CTEPH group compared with those in the control group. In total, 351 differentially expressed circRNAs (122 upregulated and 229 downregulated) were identified, which suggested that circRNAs play a role in CTEPH.
Furthermore, in the present study, upregulated circRNAs were found to be mainly enriched in the purine ribonucleotide biosynthetic process and ribonucleotide biosynthetic process. Downregulated circRNAs were mainly enriched in cellular response to stress, post-transcriptional regulation of gene expression, response to DNA damage stimulus, DNA repair, and mRNA metabolic process. Therefore, we inferred that upregulated circRNAs may function in CTEPH mainly by affecting the ribonucleotide biosynthetic process, whereas downregulated circRNAs may function mainly by regulating cellular response to stress, response to DNA damage stimulus, and gene expression. However, additional studies to confirm these findings are warranted.
The circRNA–miRNA–mRNA network (Fig. 3) revealed that each miRNA (hsa-miR-939–5p, hsa-miR-939–3p, hsa-miR-942–5p, hsa-miR-940, hsa-miR-92a-2–5p, hsa-miR-7107–3p, hsa-miR-7161–3p, hsa-miR-6831–5p, hsa-miR-7974, and hsa-miR-8089) in this network is regulated by circRNAs, which in turn regulates the target genes of miRNAs. Furthermore, hsa_circ_0002062 and hsa_circ_0022342 targeted more miRNAs (761 and 453, respectively). To our knowledge, few studies have focused on these miRNAs, but miR-940 has been studied in several diseases, such as pancreatic ductal adenocarcinoma, hepatocellular carcinoma, and prostate cancer.[31–33] Furthermore, 1 study has reported that HIV-1 Vpr can induce miR-942–5p expression, activate NF-κB signaling, and inhibit Kaposi sarcoma associated herpesvirus lytic replication.[34] Rothman et al[35] highlighted the potential of using miRNA signatures as diagnostic and prognostic tools in pulmonary hypertension. Hansen et al[17] have suggested that miR-7 is closely coupled to ciRS-7 and that the fine-tuning of the miR-7/miR-671/ciRS-7 axis probably plays profound roles in diseases such as cancer. However, the functions of these potential miRNAs and circRNAs in CTEPH were unclear. Owing to our limited understanding of miRNAs and circRNAs, more studies on the interactions between them are needed. Improving our understanding of the disease-related mechanisms of circRNAs should help improve the diagnosis and prevention of such diseases.[14]
In the present study, target genes of hsa-miR-942–5p were mainly enriched in cancer-related pathways. Furthermore, CDK6, a target gene of hsa-miR-942–5p, was enriched in cancer-related pathways (Supplemental Table 2). Auger et al[36] have reported that the presence of an organized thrombus in major pulmonary arteries is typically associated with diseases, such as lung cancer. Although no previous studies have reported the direct associations between cancer-related pathways and CTEPH, we inferred that such pathways and axon guidance might be related to CTEPH, based on this previous study by Auger et al.[36] We searched the relevant literature and found that CDK6 was involved in the cell cycle and proliferation in pulmonary hypertension.[37] Combining these findings with the results of our present study showing that hsa_circ_0002062 regulated hsa-miR-942–5p, we suggested that circRNAs function in CTEPH via hsa_circ_0002062–hsa-miR-942–5p–CDK6–pathways involved in cancer.
In addition, target genes of hsa-miR-940 were enriched in the ErbB signaling pathway. CRKL, one target gene of hsa-miR-940, was enriched in the ErbB signaling pathway in the present study (Supplemental Table 2). Grant et al[38] have indicated that modulation of the ErbB signaling pathway can lead to increased apoptosis and loss of clonogenic survival, and cell proliferation has also been shown to be related to pulmonary hypertension.[39,40] Although no previous studies have reported the direct associations between the ErbB signaling pathway and CTEPH, we inferred that the ErbB signaling pathway might be related to CTEPH. From a literature search, we identified several articles reporting that CRKL overexpression can promote cell proliferation in many diseases.[41,42] Combined with the results of our present study showing that hsa_circ_002234 regulated hsa-miR-940, we suggested that circRNAs play potential roles in CTEPH via the hsa_circ_0022342–hsa-miR-940–CRKL–ErbB signaling pathway.
In conclusion, we inferred that hsa_circ_0002062–hsa-miR-942–5p-CDK6 pathways in cancer and the hsa_circ_0022342–hsa-miR-940–CRKL–ErbB signaling pathway may be key mechanisms in CTEPH development. The identification of novel differentially expressed circRNAs is a key step to obtaining a better understanding of CTEPH. However, because of our limited knowledge of circRNAs, more studies are needed for verifying our results. A limitation of the present study is the small sample size. Thus, we plan to collect more samples to support the conclusions of the present study.
Supplementary Material
Supplementary Material
Footnotes
Abbreviations: CircRNAs = Circular RNAs, CTEPH = Chronic thromboembolic pulmonary hypertension, GO = Gene Ontology, KEGG = Kyoto Encyclopedia of Genes and Genomes.
The authors identified 351 differentially expressed circRNAs in CTEPH compared with those in the control group.
Ten miRNAs were regulated by circRNAs, which then regulated target genes in the network.
CircRNAs may have potential roles in the diagnosis and treatment of CTEPH.
Funding/support: This study was supported by the National Natural Science Foundation of China (81300044, 81270117, 81570049, 81200042, 81200041, and 31670928), Beijing Natural Science Foundation (7162069 and 7152062), Beijing Municipal Administration of Hospitals’ Youth Programme (QML20160301), the National Key Research and Development Plan of China (2016YFC0905600), and the open project of Beijing Key Laboratory of Respiratory and Pulmonary Circulation Disorders (2014HXFB03).
The authors declare that they have no competing interest.
Supplemental Digital Content is available for this article.
References
- [1].Hoeper MM, Madani MM, Nakanishi N, et al. Chronic thromboembolic pulmonary hypertension. Lancet Respir Med 2014;2:573–82. [DOI] [PubMed] [Google Scholar]
- [2].Dentali F, Donadini M, Gianni M, et al. Incidence of chronic pulmonary hypertension in patients with previous pulmonary embolism. Thromb Res 2009;124:256–8. [DOI] [PubMed] [Google Scholar]
- [3].Lang IM, Pesavento R, Bonderman D, et al. Risk factors and basic mechanisms of chronic thromboembolic pulmonary hypertension: a current understanding. Eur Respir J 2013;41:462–8. [DOI] [PubMed] [Google Scholar]
- [4].Bonderman D, Turecek PL, Jakowitsch J, et al. High prevalence of elevated clotting factor VIII in chronic thromboembolic pulmonary hypertension. Thromb Haemost 2003;90:372–6. [DOI] [PubMed] [Google Scholar]
- [5].Jaïs X, Ioos V, Jardim C, et al. Splenectomy and chronic thromboembolic pulmonary hypertension. Thorax 2005;60:1031–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bonderman D, Jakowitsch J, Adlbrecht C, et al. Medical conditions increasing the risk of chronic thromboembolic pulmonary hypertension. Thromb Haemost 2005;93:512. [DOI] [PubMed] [Google Scholar]
- [7].Bonderman D, Wilkens H, Wakounig S, et al. Risk factors for chronic thromboembolic pulmonary hypertension. Eur Respir J 2009;33:325–31. [DOI] [PubMed] [Google Scholar]
- [8].Ghofrani H-A, D’Armini AM, Grimminger F, et al. Riociguat for the treatment of chronic thromboembolic pulmonary hypertension. N Engl J Med 2013;369:319–29. [DOI] [PubMed] [Google Scholar]
- [9].Wu HJ, Zhang CY, Zhang S, et al. Microarray expression profile of circular RNAs in heart tissue of mice with myocardial infarction-induced heart failure. Cell Physiol Biochem 2016;39:205–16. [DOI] [PubMed] [Google Scholar]
- [10].Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nigro JM, Cho KR, Fearon ER, et al. Scrambled exons. Cell 1991;64:607–13. [DOI] [PubMed] [Google Scholar]
- [13].Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333–8. [DOI] [PubMed] [Google Scholar]
- [14].Wu H-J, Zhang C-Y, Zhang S, et al. Microarray expression profile of circular RNAs in heart tissue of mice with myocardial infarction-induced heart failure. Cell Physiol Biochem 2016;39:205–16. [DOI] [PubMed] [Google Scholar]
- [15].Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333. [DOI] [PubMed] [Google Scholar]
- [16].Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384–8. [DOI] [PubMed] [Google Scholar]
- [17].Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res 2013;73:5609. [DOI] [PubMed] [Google Scholar]
- [18].Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett 2015;365:141–8. [DOI] [PubMed] [Google Scholar]
- [19].Lukiw WJ. Circular RNA (circRNA) in Alzheimer's disease (AD). Front Genet 2013;4:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Burd CE, Jeck WR, Liu Y, et al. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. Plos Genet 2010;6:e1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chen YT, Rettig WJ, Yenamandra AK, et al. Cerebellar degeneration-related antigen: a highly conserved neuroectodermal marker mapped to chromosomes X in human and mouse. Proc Natl Acad Sci U S A 1990;87:3077–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].John B, Enright AJ, Aravin A, et al. Human microRNA targets. PLoS Biol 2004;2:e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol 2011;696:291–303. [DOI] [PubMed] [Google Scholar]
- [24].Dweep H, Gretz N. miRWalk2. 0: a comprehensive atlas of microRNA-target interactions. Nat Meth 2015;12:697–797. [DOI] [PubMed] [Google Scholar]
- [25].He X, Zhang J. Why do hubs tend to be essential in protein networks? PLoS Genet 2006;2:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Huang DW, Sherman BT, Tan Q, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 2007;8:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yu G, Wang L-G, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 2012;16:284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256–64. [DOI] [PubMed] [Google Scholar]
- [31].Rajendiran S, Parwani AV, Hare RJ, et al. MicroRNA-940 suppresses prostate cancer migration and invasion by regulating MIEN1. Mol Cancer 2014;13:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Song B, Zhang C, Li G, et al. MiR-940 inhibited pancreatic ductal adenocarcinoma growth by targeting MyD88. Cell Physiol Biochem 2015;35:1167–77. [DOI] [PubMed] [Google Scholar]
- [33].Yuan B, Liang Y, Wang D, et al. MiRMiR0 inhibits hepatocellular carcinoma growth and correlates with prognosis of hepatocellular carcinoma patients. Cancer Sci 2015;106:819–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Yan Q, Shen C, Qin J, et al. HIV-1 Vpr inhibits Kaposi's sarcoma-associated herpesvirus lytic replication by inducing microrna miR-942-5p and activating NF-κB signaling. J Virol 2016;90:8739–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rothman AM, Rhodes C, Condliffe R. Investigating the diagnostic potential of circulating miRNA signatures in pulmonary arterial hypertension. C21 Applying’omics Technologies to Reveal Gene Regulators and Biomarkers in Lung Development and Disease. Am Thoracic Soc 2016;193:A4622–22. [Google Scholar]
- [36].Auger WR, Kim NH, Kerr KM, et al. Chronic thromboembolic pulmonary hypertension. Clin Chest Med 2007;28:255–69. [DOI] [PubMed] [Google Scholar]
- [37].White K, Loscalzo J, Chan SY. Holding our breath: the emerging and anticipated roles of microRNA in pulmonary hypertension. Pulm Circ 2012;2:278–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Grant S, Qiao L, Dent P. Roles of ERBB family receptor tyrosine kinases, and downstream signaling pathways, in the control of cell growth and survival. Front Biosci 2002;7:d376–89. [DOI] [PubMed] [Google Scholar]
- [39].Yang X, Long L, Southwood M, et al. Dysfunctional Smad signaling contributes to abnormal smooth muscle cell proliferation in familial pulmonary arterial hypertension. Circ Res 2005;96:1053–63. [DOI] [PubMed] [Google Scholar]
- [40].Perros F, Dorfmüller P, Souza R, et al. Fractalkine-induced smooth muscle cell proliferation in pulmonary hypertension. Eur Respir J 2007;29:937–43. [DOI] [PubMed] [Google Scholar]
- [41].Wang Y, Dong Qz, Fu L, et al. Overexpression of crkl correlates with poor prognosis and cell proliferation in non-small cell lung cancer. Mol Carcinog 2013;52:890–9. [DOI] [PubMed] [Google Scholar]
- [42].Wang J, Chen X, Li P, et al. CRKL promotes cell proliferation in gastric cancer and is negatively regulated by miR-126. Chem Biol Interact 2013;206:230–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


