Abstract
Synthetic promoter technology offers a framework for designing expression cassettes that could provide precise control of transgene expression. Such artificially designed promoters enable defined transgene regulation, reduce unwanted background expression, and can overcome homology-dependent gene silencing in transgenic plants. In the present study, a synthetic root-specific module was designed using characterized cis-acting elements, fused with minimal promoter (86 bp) from PortUbi882 promoter, and cloned in pCAMBIA1305.1 by replacing CaMV 35S promoter so as to drive GUS expression. Two constructs were made; one had the synthetic module at the 5′ end of the minimal promoter (SynR1), whereas in the other construct, the module was present in both 5′ and 3′ ends (SynR2). Furthermore, the synthetic promoter constructs were transformed in tobacco wherein SynR1 promoter drove constitutive expression, whereas SynR2 conferred root-specific expression though slight leaky expression was present in stem. GUS assay in the roots of transgenic tobacco plants (T1) indicated that SynR2 promoter expressed significantly higher GUS activity than the CaMV 35S promoter. The real-time quantitative PCR (RT-qPCR) analysis of GUS gene further confirmed that SynR2 promoter conferred 2.1-fold higher root-specific expression when compared to CaMV 35S promoter.
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0872-9) contains supplementary material, which is available to authorized users.
Keywords: GUS, Root specific, Synthetic promoter, Tobacco, Transgene expression
Introduction
The use of an appropriate promoter is a key determinant in plant genetic engineering. Though several promoters have been characterized from different sources, there is a necessity for regulatory sequences enabling precise control of gene expression that could be overcome by deploying synthetic promoters. Synthetic promoters can be designed using an array of cis-acting elements from several sources and thereby improve the expression characteristics and also reduce unwanted background expressions (Mehrotra et al. 2011). They help to mitigate several limitations, i.e., they can increase promoter availability, can help to control the expression of multiple transgenes, can help to prevent silencing, and can ensure more refined control of transgene expression in a tissue and environment specific manner.
Synthetic promoters have been used in several studies to either reveal the role of cis regulatory elements or to modulate targeted inducibility, independently, and/or within a specific cis-motif arrangement. Several cis-acting regulatory elements corresponding to light inducibility, developmental stage, tissue specificity, mechanical wounding, sugar sensing, reactive oxygen species, and cold stress have been reported (Comai et al. 1990; Gilmartin and Chua 1990; Ni et al. 1995, 1996; Mitsuhara et al. 1996; Geisler et al. 2006; Mazarei et al. 2008; Zhu et al. 2008). These characterized cis-elements can be effectively employed in conjunction with minimal promoters to engineer targeted transgene expression.
A series of synthetic promoters were developed with inducible cis-elements and their expression was studied transiently (Liu et al. 2011). A synthetic pathogen inducible promoter (SynP-FF) was tested in transgenic canola and could impart resistance against Sclerotinia sclerotiorum (Shokouhifar et al. 2011). Recently, Ranjan and Dey (2012) have tailor-made superior vascular tissue and stress inducible hybrid–synthetic promoters by rearranging Dof-1 motifs based on Caulimovirus. About 188 synthetic plant promoters were categorized and described and their differences when compared to native ones were elaborated recently (Dey et al. 2015). Very recently, Liu and Stewart (2016) presented a detailed review on the recent advances in plant synthetic promoters and transcription factors. However, to our knowledge, there are no reports of synthetic promoters that confer root-specific expression in plants.
In the present study, we designed a synthetic module to confer root-specific gene expression, fused with a minimal promoter from Portubi882 and functionally validated in transgenic tobacco plants. Our results demonstrate that the synthetic promoter confers root-specific transgene expression and could, therefore, be used as a potential root-specific promoter in genetic transformation of dicotyledonous crop plants to impart disease/stress tolerance.
Materials and methods
Designing and synthesis of synthetic root-specific module
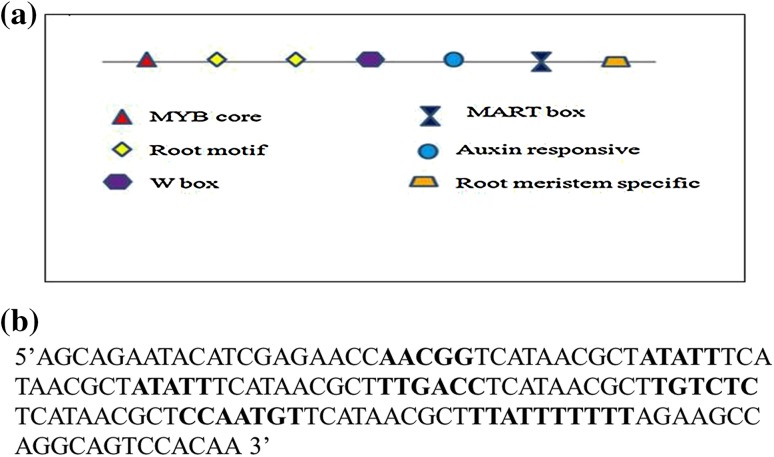
The synthetic root-specific module was designed in such a way to incorporate several cis-acting regulatory elements responsive to tissue specificity, expression enhancement, and stress responsiveness which were identified from literature. The various cis-elements used are listed in Table 1 and the module was named SynR. Since spacing and copy number of the cis-elements are the key players of tunable gene expression, two copies of the root-specific motif (ATATT motif) were incorporated with a spacing of 10 bp between cis-elements. The other cis-acting elements [stress responsive, elicitor responsive, and matrix attachment region (MAR) box] were placed one copy each in the module. The spacer sequence was designed, so that no cis-acting elements were present and the design was checked using Plant cis-acting DNA regulatory elements (PLACE) database (Higo et al. 1999). The synthetic module was custom synthesized (Bioserve, India) and used as a template for cloning. The arrangement of cis-elements in the synthetic module is depicted in Fig. 1a. The pCAMBIA1305.1 vector was used as a backbone for cloning and the CaMV 35S promoter driving the GUS gene and catalase intron was replaced by the synthetic promoters.
Table 1.
Various cis-elements incorporated in the synthetic module (SynR)
| Cis element | Sequence motif | Function | References |
|---|---|---|---|
| ROOTMOTIFTAPOX1 | ATATT | Root specific | Elmayan and Tepfer (1995) |
| MYBCOREATCYCB1 | AACGG | Stress responsive | Planchais et al. (2002) |
| ELRECOREPCRP1 | TTGACC | Elicitor responsive | Rushton et al. (1996) |
| ARFAT | TGTCTC | Auxin responsive | Ulmasov et al. (1999) |
| LEAFYATAG | CCAATGT | Root meristem specific | Kamiya et al. (2003) |
| MARTBOX | TTTATTTTTTT | Enhances expression | Gasser et al. (1989) |
Fig. 1.
Synthetic root module. a Arrangement of cis-elements in the synthetic module. b Nucleotide sequence of the SynR module; the cis-acting elements are in bold letters
Recombinant plasmid constructs
Two synthetic promoters were constructed by fusing the synthetic module either at the 5′ (SynR1) or both at the 5′ and the 3′ end (SynR2) of the minimal promoter (Philip et al. 2013). The primers used for amplifying the minimal promoter and the synthetic module are given in Table 2. The nucleotide sequence of the synthetic root-specific module is given in Fig. 1b.
Table 2.
Primer sequences used in the study
| Primer | Sequence 5′–3′ |
|---|---|
| PMF | GCCGAAGCTTCCAATAAAT |
| PMR | GATCCCATGGGTACATGTCT |
| S1F | GATCGAATTCAGCAGAATAC |
| S1R | GATCAAGCTTCCTGGCTTCT |
| S2F | GATCCCATGGATCGAGAACC |
| S2R | GATCAGATCTTTGTGGACTG |
| GUSR | GATCAATGTCGTGAAAGCCCGCA |
The minimal promoter was amplified with PMF/PMR primers and the synthetic module was amplified with S1F/S1R primers (Table 2). The amplified products were restricted with SacI enzyme for 1 h at 37 °C and separated on 1% agarose gel through electrophoresis. The restricted minimal promoter and synthetic module were eluted and ligated overnight at 4 °C. The ligated mixture was diluted 1:10 ratio and used as a template for PCR to amplify the SynR1 promoter with EcoRI anchored forward primer (S1F) and NcoI anchored reverse primer (PMR) (Table 2).
To prepare the SynR2 promoter, the root-specific synthetic module was fused to the 3′ end of SynR1 promoter. The synthetic module was PCR amplified with S2F/S2R primers (Table 2). In addition, the SynR1 promoter was amplified using S1F/PMR primers (Table 2). The promoters and the synthetic module were restricted with NcoI enzyme at 37 °C for 1 h, and the restricted products were separated in 1% gel through electrophoresis. The restricted products were eluted and ligated overnight at 4 °C. The ligated mixture was diluted 1:10 ratio and used as a template for PCR to amplify the SynR2 promoter with EcoRI anchored forward primer (S1F) and BglII anchored reverse primer (S2R) (Table 2). Table S1 lists the reaction conditions and product sizes for the PCR amplification of the minimal promoter, synthetic module, and promoter-GUS fusion.
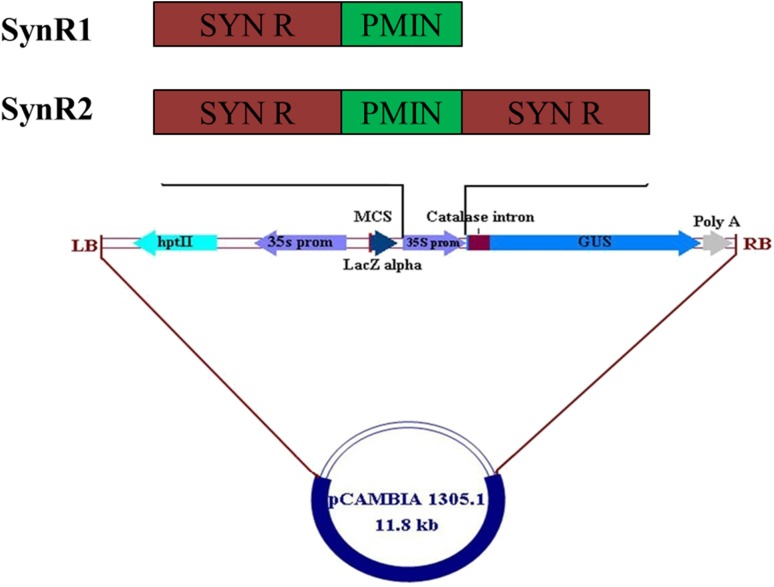
The pCAMBIA1305.1 vector and the amplified products were restricted (EcoRI/NcoI for SynR1 and EcoRI/BglII for SynR2) for 1 h at 37 °C and separated in 1% agarose gel through electrophoresis. The restricted vector and promoters were eluted, ligated overnight at 4 °C and transformed in Escherichia coli DH5α cells. Figure 2 depicts the pictorial representation of the cloning strategy. The recombinant plasmids were isolated and individually mobilized into Agrobacterium LBA4404 for stable transformation.
Fig. 2.
Schematic representations of binary vectors generated with synthetic promoters-GUS fusions (LB left border, MCS multiple cloning site, RB right border)
Plant transformation
Agrobacterium-mediated transformation was performed in tobacco following the procedure described by Horsch et al. (1985). The tobacco leaf discs were individually co-cultivated with Agrobacterium strains harboring each of the recombinant plasmids SynR1, SynR2, and pCAMBIA1305.1 (positive control). Three days after co-cultivation, they were transferred to regeneration medium containing 6-benzylaminopurine (1.0 mg/L) and naphthalene acetic acid (0.1 mg/L). Regenerated shootlets were formed after three rounds of selection with hygromycin antibiotic (30 mg/L), and later, fully rooted plantlets were transferred to pots and maintained in greenhouse conditions for molecular analysis.
PCR confirmation of transgenics
Genomic DNA from putative transgenics was isolated using DNeasy plant mini kit (Qiagen, Germany). PCR was carried out to confirm integration of the promoter and GUS using promoter-specific forward primer (S1F) and GUS-specific reverse primers (GUSR), respectively. The reaction cocktail contained 50 ng of template DNA, 0.25 μM each of the primers, 1.5 mM dNTP (Merck Biosciences, Germany), and one unit of the Taq polymerase enzyme (Merck Biosciences, Germany), and PCR products were separated through electrophoresis.
Expression analysis using RT-qPCR
Total RNA was extracted from the transgenic plants using TRI Reagent (Sigma, USA) and after DNase treatment, cDNA was synthesized using RevertAid first strand cDNA synthesis kit as per manufacturer’s instructions (Thermo Scientific, USA). The GUS primers described in Chakravarthi et al. (2015) were used for RT-qPCR. Initially, the cDNA concentration of each sample was standardized and melt curve analysis was carried out to determine the primer specificity. Each reaction was performed in triplicates on a Step-One plus Real-Time PCR system (Applied Biosystems, Canada) and contained 200 ng cDNA, 12.5 μl of 2X MESA GREEN RT-qPCR master mix plus (Eurogentec, Belgium), 3 pmol each of forward and reverse primers and made up to 25 μl with nuclease free water, and the following were the cycle conditions: 10 min 95 °C; 40 cycles: 15 s 95 °C, 60 s 60 °C. The actin transcript was used as reference gene and the relative GUS expression was calculated by 2−ΔΔCt method (Livak and Schmittgen 2001).
Histochemical and fluorometric GUS assay in transgenic plants
GUS staining was performed by the following Jefferson et al. (1987). Two-month-old transgenic tobacco plants were assayed for GUS expression. The tissues were excised aseptically and washed in 50 mM sodium phosphate buffer prior to incubation in phosphate buffer (with 1% Triton X-100) at 37 °C for 1 h. Later, the explants were transferred to 1 mM X-Gluc staining solution and vacuum infiltrated for 5 min. The tissues were incubated for 16 h at 37 °C. After de-staining in 70% ethanol, the tissues were observed under a stereo light microscope (Zeiss, Germany).
Fluorometric GUS assay was carried out in different transgenic plant parts (leaf, stem, root, and seed) with CaMV 35S transgenics as control (Jefferson et al. 1987). The liberation of 4-methyl umbelliferone (4-MU) was assayed by measuring the fluorescence with excitation at 365 nm and emission at 455 nm in a fluorometer (Promega). The assays were performed in triplicates for six independent transgenic events (per construct) and the whole experiment was repeated twice. GUS activity was calculated as nanomoles of 4-MU hydrolyzed/min/mg of total protein.
Statistical analysis
All the GUS assays were performed in triplicates and subjected to statistical analysis using a one tailed paired Student’s t test. Significance of the treatments was analyzed using one-way analysis of variance (ANOVA). A probability (P) value of ≤0.05 was considered to be statistically significant.
Results
Stable transformation in tobacco
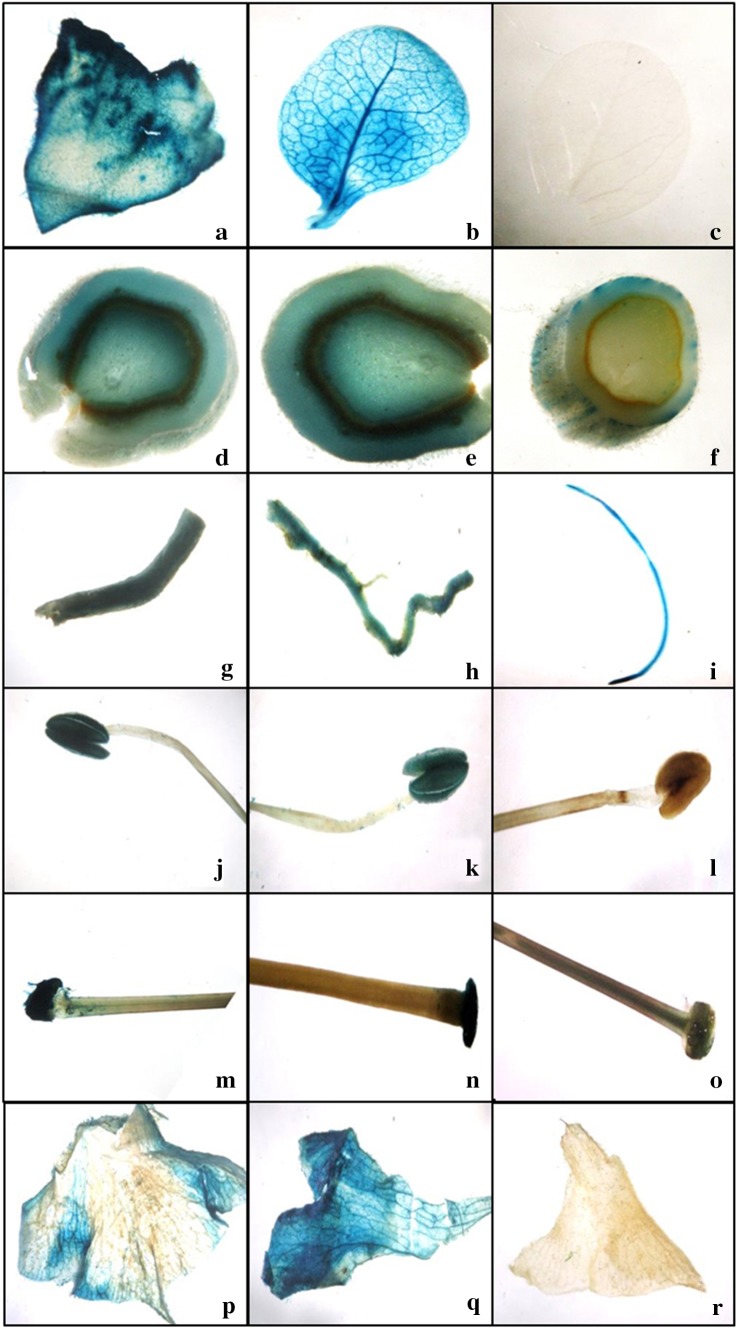
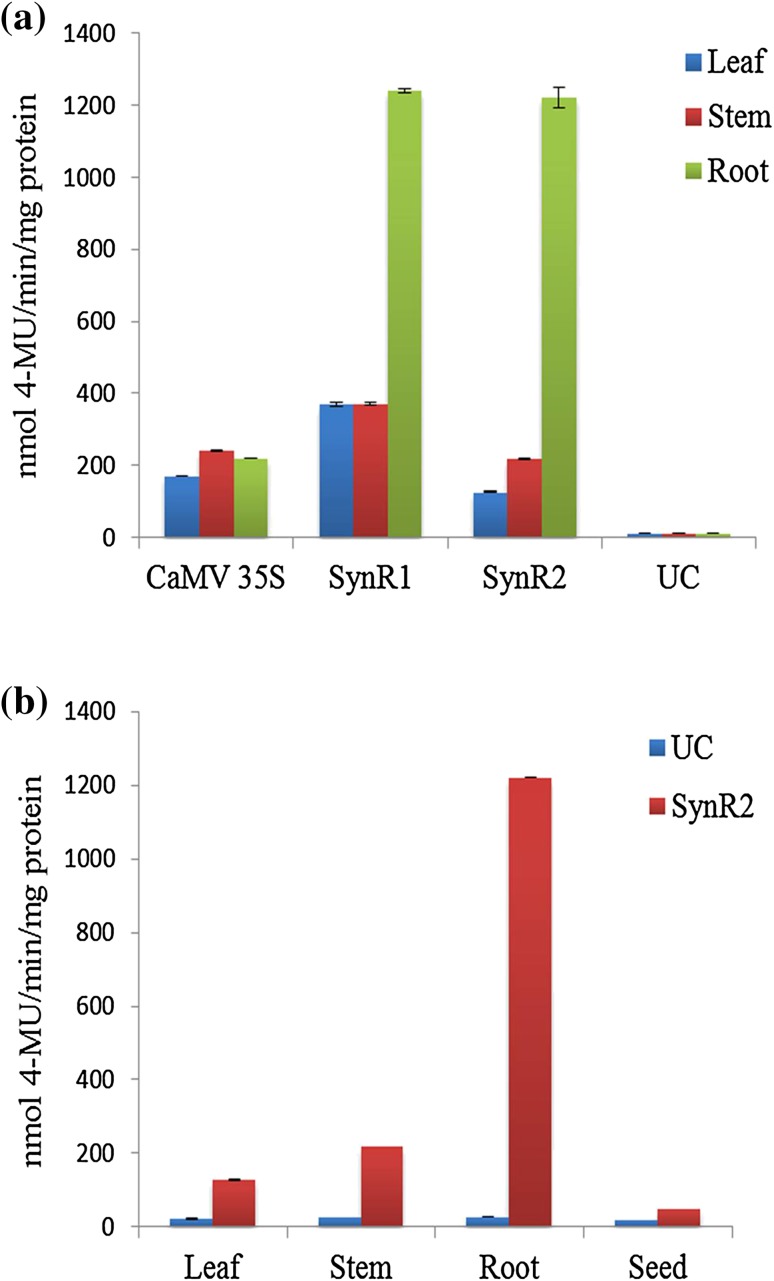
Initially, the recombinant plasmids containing synthetic promoters-GUS gene fusions were introduced into competent Agrobacterium strain LBA4404 and the colonies obtained were screened by polymerase chain reaction (PCR) and the positive ones were used for plant transformation. Transgenic tobacco plants were developed and all the putative transgenics were subjected to PCR with promoter-specific forward and GUS reverse fusion primers. PCR analysis showed that all the putative transgenic plants that survived through stringent hygromycin selection were positive for the transgene integration (Fig S1). The transgenic plants were further subjected to histochemical staining using X-gluc substrate. Figure 3 shows the GUS expression driven by CaMV 35S, SynR1, and SynR2 promoters in different parts of transgenic tobacco. The transgenic events were also analyzed for GUS expression through fluorometric assay and the results showed that in tobacco, SynR2 promoter conferred root specificity, although slight expression was observed in stem (Figs. 3, 4a) and SynR1 promoter drove constitutive expression (Fig. 4a). Transgenic seeds of tobacco driven by SynR2 promoter were also assayed for GUS activity and there was negligible GUS activity in seeds (Fig. 4b).
Fig. 3.
Histochemical localization of GUS activity driven by different promoters in transgenic tobacco plants; CaMV 35S: a mature leaf, d mature stem, g mature root, j anther, m stigma, p petal; SynR1: b mature leaf, e mature stem, h mature root, k anther, n stigma, q petal; SynR2: c mature leaf, f mature stem, i mature root, l anther, o stigma, r petal
Fig. 4.
Fluorometric GUS analysis. a GUS activity in transgenic tobacco plants driven by the synthetic promoters. b GUS activity in seeds; UC: untransformed tobacco plant; each value represents the average of six independent transgenic events; the error bars indicate SD; P ≤ 0.05 was considered statistically significant as determined by Student’s t test
GUS fluorometric assay in T1 transgenic tobacco driven by SynR2 promoter
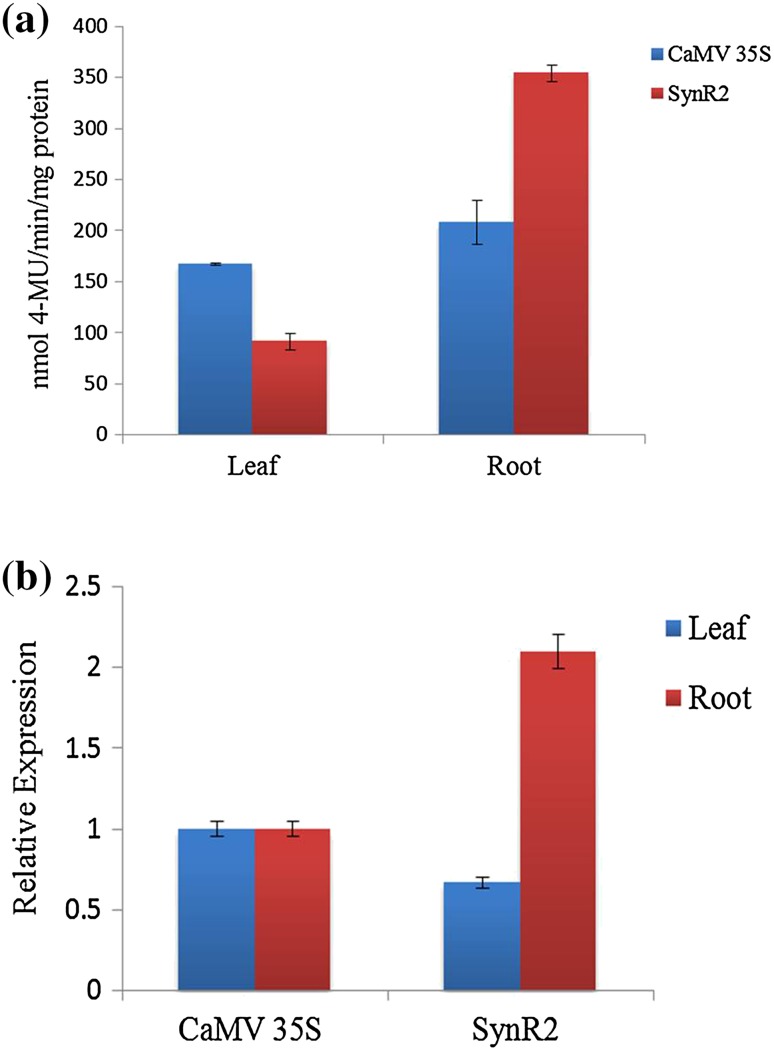
The transgenic seeds of three independent events expressing GUS driven by SynR2 promoter were further germinated in MS media with 25 mg/L hygromycin and the resistant seedlings were planted in transgenic glasshouse for further analysis. Figure 5 shows the GUS expression observed in a transgenic tobacco plantlet driven by SynR2 promoter. The PCR positive transgenic tobacco plants (T1) driven by SynR2 promoter were assayed for GUS activity and the results revealed that in leaf, GUS activity was lower than that of CaMV 35S promoter, whereas in roots, there was an increased GUS activity (1.8 times) in SynR2 promoter driven plants than that of CaMV 35S promoter (Fig. 6a).
Fig. 5.

Histochemical GUS staining of transgenic tobacco plantlet driven by SynR2 promoter
Fig. 6.
Quantification of GUS expression. a Fluorometric GUS analysis in transgenic tobacco (T1) plants driven by SynR2 promoter. b Relative GUS expression driven by SynR2 and CaMV 35S promoters in transgenic tobacco (T1); each value represents the average of three independent transgenic events; the error bars indicate SD
Real-time expression analysis in T1 transgenic tobacco plants driven by SynR2 promoter
RNA was isolated from leaves and roots of 2-month-old CaMV 35S-GUS and SynR2-GUS transgenic plants (three events each), and RT-qPCR was performed. The RT-qPCR results demonstrated that in roots, SynR2 promoter drove 2.1-fold higher GUS expression when compared with CaMV 35S promoter (Fig. 6b). However, in leaves, GUS expression was relatively lower than CaMV 35S promoter (Fig. 6b).
Discussion
Artificial promoters deliver an efficient and flexible approach to regulate transgene expression in a desired spatio-temporal manner and can also greatly reduce the complex expression pattern of natural promoters (Gurr and Rushton 2005; Venter 2007). Studies have demonstrated that individual pathogen responsive cis-acting elements when fused with a minimal promoter can locally direct reporter gene expression in response to pathogens (Cazzonelli and Velten 2008; Mazarei et al. 2008). The CaMV 35S promoter has been a model for cis engineering in plant promoters. Earlier studies conducted by Bhullar et al. (2003, 2007, 2010) have reported that rearrangement of cis-elements in CaMV 35S promoter region creates synthetic CaMV 35S promoters with minimum sequence homology whose transgene activity is equivalent to that of the wild-type CaMV 35S promoter. It was found that defense signaling could be well conserved across species at the promoter element level. An array of cis-acting elements (boxes W1, W2, GCC, JERE, S, Gst1, and D) recognized by specific transcription factors (WRKYs, ERFs, bZIPs, Mybs, Dofs, and bHLHs) can affect local gene expression in plants upon pathogen attack. Hence, synthetic promoters with tetramers of only a single cis element were constructed and the expression was monitored during interactions with several pathogens, including compatible, incompatible, and non-host interactions (Rushton et al. 2002).
The synergistic effect of cis-acting elements was demonstrated by placing eight cis-acting motifs upstream of the TATA box (at the −38 position) of the basal promoter (Sawant et al. 2005). Multimers of the cis-elements were inserted, taking one at a time, such that each of these caused 2–8-fold activations of the basal transcription. The complete module brought enhancement of 110-fold in transcription levels. This study proved that the use of many cis-elements together may provide additional TF-binding sites and contribute towards the stability of pre initiation complex (PIC) at TATA box. A strategy for regulatable gene expression was developed which controlled gene expression through construction of synthetic promoter libraries by making changes in the −35 and −10 consensus sequences of bacterial promoters (Jensen and Hammer 1998; Mijakovic et al. 2005; Hammer et al. 2006). A library of synthetic promoters with varying strengths was constructed through mutagenesis (Alper et al. 2005). Unfortunately, the availability of novel engineered promoters for plant molecular biology seems to be very limited, since the demand for tunable promoters is increasing rapidly. Wang et al. (2015) obtained five novel green tissue-specific synthetic promoters and also developed a feasible method for screening and functional identification of tissue-specific cis-elements with their flanking sequences at the genome level in rice.
The spacing and copy number are the most critical factors in designing a synthetic promoter and determine the strength and spatio-temporal expression of the transgenes. In this study, we designed the synthetic module by keeping the spacing constant between each motif (10 bp). In addition, other beneficial cis-elements like stress responsive element, W box, were incorporated one copy each in the module, so that the promoter would be of great use in developing disease/stress tolerant plants. In addition, the matrix attachment region (MAR) that is known to enhance transgene expression in tobacco (Fukuda and Nishikawa 2003), was included in the module. However, two root motifs (ATATT) were incorporated in the module. It is well known that UTRs enhance transgene expression (Sivamani et al. 2009; Yang et al. 2009). Therefore, in our study, a novel attempt was made to utilize the synthetic module as 5′ UTR (SynR1) as well as both 5′ and 3′ UTR (SynR2) with a view to enhance transgene expression.
Since the effect of spacing between individual cis-acting elements on transgene expression is quite difficult to predict (Wray 1998), it has to be deciphered experimentally. The spacing of cis-acting elements leads to differences in the inducibility of the various promoters for the pathogens tested, the speed of induction, and the basal expression levels. However, in the present investigation, the spacing was kept constant so as to optimize the copy number of the motifs. The root motif was present in two copies in SynR1, whereas four motifs in SynR2. In addition, the minimal promoter contained a root motif.
In the present study, the transgenic tobacco expressing GUS driven by SynR2 promoter showed root-specific expression with a slight leaky expression in stem. However, none of the transgenic tobacco plants transformed with SynR2 promoter showed GUS activity in leaves. The root-specific expression may be attributed to the presence of additional copy of root-specific cis element (ATATT motif) present in the minimal promoter in addition to four motifs already present in SynR2 promoter. The slight expression in stem may be avoided by further careful manipulation of the synthetic root module. The fluorometric analysis was further substantiated through RT-qPCR where a 2.1-fold higher GUS expression was obtained in roots of transgenic tobacco (T1) with SynR2 promoter when compared with CaMV 35S promoter. Though CaMV 35S promoter confers higher expression in dicots, SynR2 promoter was clearly better and the reason may be due to the use of the minimal promoter from a constitutive ubiquitin gene of P. coarctata. Philip et al. (2013) reported that the promoter was, indeed, seven times better than the CaMV 35 promoter.
However, in case of the transgenic tobacco plants transformed with SynR1 promoter, constitutive expression was observed. The failure of the tissue-specific expression may be due to the lesser ATATT motifs (3) when compared to SynR2. Also the minimal promoter which is obtained from a constitutive promoter backbone may be attributed for the constitutive expression. Several factors such as availability of transcription factors, copy number of other cis-elements, orientation, and spacing may also be involved, which can be rectified by careful modification of the promoter module.
Conclusion
To summarize, meticulous selection of cis-elements, function of the motif, spacing, orientation, and copy number are the critical factors that have to be taken care of while designing a synthetic promoter. In addition, generation of huge synthetic promoter libraries and screening them for tissue specificity will aid in constructing tissue-specific synthetic promoters. However, there are several combinatorial mechanisms of transgene regulation and signaling that are largely unclear, which prevents the optimal design of synthetic tissue-specific promoters. Advances in bioinformatics and in depth studies of plant transcription factor networks and unravelling the roles of novel cis- and trans-acting elements could greatly accelerate design strategies for the construction of effective synthetic promoters. A high-throughput promoter designing strategy adhering to all the critical factors mentioned above combined with in silico methods would be a solution to generate synthetic promoters with tunable transgene expression.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are grateful to the Indian Council of Agricultural Research and the Sugarcane Breeding Institute, Coimbatore, for the funding and infrastructure. The first author is also grateful to the São Paulo Research Foundation (FAPESP, Proc. 2015/10855-9) for the postdoctoral research grant.
Abbreviations
- ERF
Ethylene response factor
- DOF
Domain of function
- bHLH
Basic helix loop helix
- GUS
β-Glucuronidase
- X-gluc
5-Bromo-4-chloro-3-indolyl-beta-d-glucuronic acid, cyclohexylammonium salt
Author contributions
NS and MC conceived and designed the research. MC and JAN performed the experiments. MC analyzed the results and drafted the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interests.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0872-9) contains supplementary material, which is available to authorized users.
References
- Alper H, Fischer C, Nevoigt E, Stephanopoulos G. Tuning genetic control through promoter engineering. Proc Natl Acad Sci. 2005;102:12678–12683. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhullar S, Chakravarthy S, Advani S, Datta S, Pental D, Burma PK. Strategies for development of functionally equivalent promoters with minimum sequence homology for transgene expression in plants: cis-elements in a novel DNA context versus domain swapping. Plant Physiol. 2003;132:988–998. doi: 10.1104/pp.103.020602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhullar S, Datta S, Advani S, Chakravarthy S, Gautam T, Pental D, Burma PK. Functional analysis of the cauliflower mosaic virus 35S promoter: re-evaluation of the role of subdomains B5, B4, and B2 in promoter activity. Plant Biotechnol J. 2007;5:696–708. doi: 10.1111/j.1467-7652.2007.00274.x. [DOI] [PubMed] [Google Scholar]
- Bhullar S, Datta S, Burma PK. Delayed trans-inactivation of synthetic domain A 35S promoters by ‘‘Tobacco 271 Locus’’ due to reduced sequence homology. Plant Mol Biol Rep. 2010;29:1–11. doi: 10.1007/s11105-010-0202-4. [DOI] [Google Scholar]
- Cazzonelli CI, Velten EJ. In vivo characterization of plant promoter element interaction using synthetic promoters. Trans Res. 2008;17:437–457. doi: 10.1007/s11248-007-9117-8. [DOI] [PubMed] [Google Scholar]
- Chakravarthi M, Philip A, Subramonian N. Truncated Ubiquitin 5′ regulatory region from Erianthus arundinaceus drives enhanced transgene expression in heterologous systems. Mol Biotechnol. 2015;57:820–835. doi: 10.1007/s12033-015-9875-0. [DOI] [PubMed] [Google Scholar]
- Comai L, Moran P, Maslyar D. Novel and useful properties of a chimeric plant promoter combining CaMV 35S and MAS elements. Plant Mol Biol. 1990;15:373–381. doi: 10.1007/BF00019155. [DOI] [PubMed] [Google Scholar]
- Dey N, Sarkar S, Acharya S, Maiti IB. Synthetic promoters in planta. Planta. 2015;242(5):1077–1094. doi: 10.1007/s00425-015-2377-2. [DOI] [PubMed] [Google Scholar]
- Elmayan T, Tepfer M. Evaluation in tobacco of the organ specificity and strength of the rolD promoter, domain A of the 35S promoter and the 35S2 promoter. Trans Res. 1995;4(6):388–396. doi: 10.1007/BF01973757. [DOI] [PubMed] [Google Scholar]
- Fukuda Y, Nishikawa S. Matrix attachment regions enhance transcription of a downstream transgene and the accessibility of its promoter region to micrococcal nuclease. Plant Mol Biol. 2003;51(5):665–675. doi: 10.1023/A:1022509909838. [DOI] [PubMed] [Google Scholar]
- Gasser SM, Amati BB, Cardenas ME, Hofmann JFX. Studies on scaffold attachment sites and their relation to genome function. Int Rev Cytol. 1989;119:57–96. doi: 10.1016/S0074-7696(08)60649-X. [DOI] [PubMed] [Google Scholar]
- Geisler M, Kleczkowski LA, Karpinski S. A universal algorithm for genome-wide in silico identification of biologically significant gene promoter putative cis-regulatory-elements; identification of new elements for reactive oxygen species and sucrose signaling in Arabidopsis. Plant J. 2006;45(3):384–398. doi: 10.1111/j.1365-313X.2005.02634.x. [DOI] [PubMed] [Google Scholar]
- Gilmartin PM, Chua NH. Localization of a phytochrome-responsive element within the upstream region of pea rbcS-3A. Mol Cell Biol. 1990;10(10):5565–5568. doi: 10.1128/MCB.10.10.5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurr SJ, Rushton PJ. Engineering plants with increased disease resistance: how are we going to express it? Trends Biotechnol. 2005;23:283–290. doi: 10.1016/j.tibtech.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Hammer K, Mijakovic I, Jensen PR. Synthetic promoter libraries—ltuning of gene expression. Trends Biotechnol. 2006;24:53–55. doi: 10.1016/j.tibtech.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucl Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffman NL, Eichholtz D, Rogers SG, Fraley RT. A simple and general method for transferring genes into plants. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PR, Hammer K. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl Environ Microbiol. 1998;64:82–87. doi: 10.1128/aem.64.1.82-87.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya N, Nagasaki H, Morikami A, Sato Y, Matsuoka M. Isolation and characterization of a rice WUSCHEL-type homeobox gene that is specifically expressed in the central cells of a quiescent center in the root apical meristem. Plant J. 2003;35(4):429–441. doi: 10.1046/j.1365-313X.2003.01816.x. [DOI] [PubMed] [Google Scholar]
- Liu W, Stewart CN. Plant synthetic promoters and transcription factors. Curr Opin Biotechnol. 2016;37:36–44. doi: 10.1016/j.copbio.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Liu W, Mazarei M, Rudis MR, Fethe MH, Stewart CN. Rapid in vivo analysis of synthetic promoters for plant pathogen phytosensing. BMC Biotechnol. 2011;11(1):108. doi: 10.1186/1472-6750-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mazarei M, Teplova I, Hajimorad MR, Stewart CN. Pathogen phytosensing: plants to report plant pathogens. Sensors. 2008;8(4):2628–2641. doi: 10.3390/s8042628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra R, Gupta G, Sethi R, Bhalothia P, Kumar N, Mehrotra S. Designer promoter: an artwork of cis engineering. Plant Mol Biol. 2011;75(6):527–536. doi: 10.1007/s11103-011-9755-3. [DOI] [PubMed] [Google Scholar]
- Mijakovic I, Petranovic D, Jensen PR. Tunable promoters in systems biology. Curr Opin Biotechnol. 2005;16:329–335. doi: 10.1016/j.copbio.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Mitsuhara I, Ugaki M, Hirochika H, Ohshima M, Murakami T, Gotoh Y, Katayose Y, Nakamura S, Honkura R, Nishimiya S, Ueno K, Mochizuki A, Tanimoto H, Tsugawa H, Otsuki Y, Ohashi Y. Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol. 1996;37(1):49–59. doi: 10.1093/oxfordjournals.pcp.a028913. [DOI] [PubMed] [Google Scholar]
- Ni M, Cui D, Einstein J, Narasimhulu S, Vergara CE, Gelvin SB. Strength and tissue specificity of chimeric promoter derived from octopine and mannopine synthase gene. Plant J. 1995;7:661–676. doi: 10.1046/j.1365-313X.1995.7040661.x. [DOI] [Google Scholar]
- Ni M, Cui D, Gelvin SB. Sequence-specific interactions of wound-inducible nuclear factors with mannopine synthase 2′ promoter wound-responsive elements. Plant Mol Biol. 1996;30(1):77–96. doi: 10.1007/BF00017804. [DOI] [PubMed] [Google Scholar]
- Philip A, Syamaladevi DP, Chakravarthi M, Gopinath K, Subramonian N. 5′ Regulatory region of ubiquitin 2 gene from Porteresia coarctata makes efficient promoters for transgene expression in monocots and dicots. Plant Cell Rep. 2013;32(8):1199–1210. doi: 10.1007/s00299-013-1416-3. [DOI] [PubMed] [Google Scholar]
- Planchais S, Perennes C, Glab N, Mironov V, Inzé D, Bergounioux C. Characterization of cis-acting element involved in cell cycle phase-independent activation of Arath; CycB1; 1 transcription and identification of putative regulatory proteins. Plant Mol Biol. 2002;50(1):109–125. doi: 10.1023/A:1016018711532. [DOI] [PubMed] [Google Scholar]
- Ranjan R, Dey N. Development of vascular tissue and stress inducible hybrid–synthetic promoters through DOF-1 motifs rearrangement. Cell Biochem Biophy. 2012;63(3):235–245. doi: 10.1007/s12013-012-9359-9. [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 1996;15(5):690–700. [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Reinstadler A, Lipka V, Lippok B, Somssich IE. Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen—and wound-induced signaling. Plant Cell. 2002;14:749–762. doi: 10.1105/tpc.010412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawant S, Kiran K, Mehrotra R, Chaturvedi CP, Ansari SA, Singh P, Lodhi N, Tuli R. A variety of synergistic and antagonistic interactions mediated by cis-acting DNA motifs regulate gene expression in plant cells and modulate stability of the transcription complex formed on a basal promoter. J Exp Bot. 2005;56:2345–2353. doi: 10.1093/jxb/eri227. [DOI] [PubMed] [Google Scholar]
- Shokouhifar F, Zamani MR, Motallebi M. Expression pattern of the synthetic pathogen-inducible promoter (SynP-FF) in the transgenic canola in response to Sclerotinia sclerotiorum. Iranian J Biotechnol. 2011;9(1):1–10. [Google Scholar]
- Sivamani E, Starmer JD, Qu R. Sequence analysis of rice rubi3 promoter gene expression cassettes for improved transgene expression. Plant Sci. 2009;177(6):549–556. doi: 10.1016/j.plantsci.2009.08.006. [DOI] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. Plant J. 1999;19(3):309–319. doi: 10.1046/j.1365-313X.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- Venter M. Synthetic promoters: genetic control through cis engineering. Trends Plant Sci. 2007;12:118–124. doi: 10.1016/j.tplants.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Wang R, Zhu M, Ye R, Liu Z, Zhou F, Chen H, Lin Y. Novel green tissue-specific synthetic promoters and cis-regulatory elements in rice. . Sci Rep. 2015;5:18256. doi: 10.1038/srep18256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. Promoter logic. Science. 1998;279:1871–1872. doi: 10.1126/science.279.5358.1871. [DOI] [PubMed] [Google Scholar]
- Yang L, Wakasa Y, Kawakatsu T, Takaiwa F. The 3′-untranslated region of rice glutelin GluB-1 affects accumulation of heterologous protein in transgenic rice. Biotechnol let. 2009;31(10):1625–1631. doi: 10.1007/s10529-009-0056-8. [DOI] [PubMed] [Google Scholar]
- Zhu J, Jeong JC, Zhu Y, Sokolchik I, Miyazaki S, Zhu JK, Hasegawa PM, Bohnert HJ, Shi H, Yun DJ, Bressan RA. Involvement of Arabidopsis hos15 in histone deacetylation and cold tolerance. Proc Natl Acad Sci. 2008;105:4945–4950. doi: 10.1073/pnas.0801029105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.