Abstract
Long non-coding RNAs (lncRNAs) are remarkably powerful, flexible and pervasive cellular regulators. With the help of cheaper RNA-seq, high-throughput screening of lncRNAs has become widely applied and has identified large numbers of specific lncRNAs in various physiological or pathological processes. Vision is known to be a complex and vital perception that comprises 80% of the sensory information we receive. A consensus has been reached that normal visual maintenance and impairment are primarily driven by gene regulation. Recently, it has become understood that lncRNAs are key regulators in most biological processes, including cell proliferation, apoptosis, differentiation, immune responses, oxidative stress and inflammation. Our review is intended to provide insight towards a comprehensive view of the precise modulation of lncRNAs in visual maintenance and impairment. We also highlight the challenges and future directions in conducting lncRNA studies, particularly in patients whose lncRNAs may hold expanded promise for diagnostic, prognostic and therapeutic applications.
Keywords: Long noncoding RNAs, Visual system, Ocular diseases, Visual maintenance, Visual impairment
Introduction
Long non-coding RNAs (lncRNAs) represent a class of DNA transcripts longer than 200 nucleotides with limited protein-coding potential. Research has focused on their functional regulation of gene expression at the epigenetic, transcriptional and post-transcriptional levels. Scientists have hypothesised that lncRNAs work by acting as signals, chaperones, scaffolds or decomposers that could conduct mRNA degeneration by producing endogenous siRNA.
Vision is a complex and vital perception. More than 80% of the sensory information we receive relies on the function of the visual system, which expertly manages large amounts of information rapidly. Moreover, precise spatial orientation and daily activities such as walking are primarily dependent on visual cues.
Various eye diseases lead to visual impairment and vision loss and can have huge detrimental impacts on daily life. Visual impairment not only causes suffering for patients but also increases familial and social burdens, generating adverse social and economic consequences.1 Total annual costs for visually impaired persons were estimated at €12 749 million in Europe. The main cost components of visual impairment in the community were ‘loss of income’ (23–43% of community costs), ‘burden on career’ (24–39%) and ‘paid assistance’ (13–29%).
A consensus has been reached that normal visual maintenance and visual impairment are primarily determined by the precise regulation of gene expression that affects diverse biological processes, including cell proliferation, apoptosis, cell specification, immune responses, oxidative stress and inflammation. Recently, lncRNAs are known to be key regulators in many of the biological processes mentioned above.
Currently, lncRNAs are known to participate in almost every aspect of visual impairment, including ocular malignancy, diabetic retinopathy (DR), ocular neovascularisation, ocular degeneration, proliferative vitreoretinopathy (PVR) and rare eye diseases. In this review, we focused on recent studies associated with lncRNAs in normal eye development and dysfunction and illustrated the role of some specific lncRNAs in the precise modulation of visual maintenance and impairment.
The role of lncRNAs in the precise regulation of visual maintenance
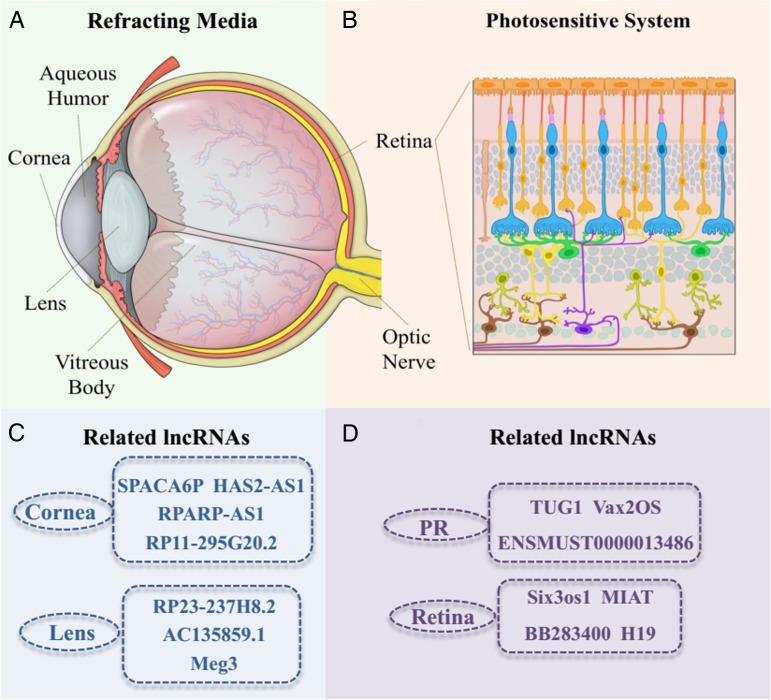
The visual system consists of two parts: (A) the refracting media and (B) the photosensitive system. The refracting media includes the cornea, aqueous humour, lens and vitreous body. When the eyes look at a foreign object, the object reflects light through the cornea and aqueous humour, lens and vitreous body, focusing the image on the retinal fovea. The photosensitive system is mainly composed of retina and the optic nerve connecting to the visual centre (figure 1).
Figure 1.
Landscape of long non-coding RNAs (lncRNAs) expression in visual maintenance. Visual system consists two parts: (A) Refracting media, including cornea, aqueous humour, lens and vitreous body. (B) Photosensitive system mainly composed of retina and optic nerve connecting with visual centre. Overview of the related lncRNAs in visual system was displayed in the lower half of the figure.
Precise modulation of the photosensitive system by lncRNAs
Confirmation of regulation by lncRNAs in the retina
Liao and colleagues conducted the first computational annotation of the functions of lncRNAs based on public microarray expression profiles. They reannotated Affymetrix Mouse Genome Array data to predict the probable functions of 340 lncRNAs referring to gene ontology (GO), biological process annotations and genomic adjacency. The functions of lncRNAs mainly involve organ or tissue development, especially eye and neuron development.2–4 This research leads us to focus on the importance of lncRNAs in normal eye development, the foundation of visual maintenance.
To confirm this finding and to verify it in different mammalian species, RNA sequencing of eye tissue from diverse species was conducted by Mustafi et al. They identified a group of 18 highly conserved large intergenic non-coding RNAs (lincRNAs) in different mammalian species as well as in adult human retinal neurons whose retinas are composed of varied rod/cone photoreceptor contents. In total, 16 of the 18 lincRNAs were conserved in human retinal tissue and other mammalian species, and 14 of these were conserved in the macular region as well. This finding is also verified by the restricted tissue expression profiles of lincRNAs exhibited in mouse retina, further confirming their crucial role in retina development across species. With the help of in situ hybridisation, evidence from research studies indicates that these highly conserved lincRNAs are localised to specific retinal layers, especially to the photoreceptor layer. This finding revealed that lncRNAs could be indispensable for retinal development and visual maintenance.5
Role of specific lncRNAs in the retina
Many lncRNAs have important roles in determining retinal cell fate specification. We listed several of them, hoping to give an example for future study.
Six3os1, an evolutionary conserved lncRNA, was identified to be localised in mice retina by FISH. To determine whether Six3os1 plays an important role in retinal development, Rapicavoli and colleagues conducted overexpression and knockdown analysis of Six3os1. Results revealed that the upregulation of Six3os inhibited normal photoreceptors' cell specification, whereas the knockdown of Six3os prevented the differentiation of bipolar cell and Müller glial. They demonstrated a novel mechanism by which promoter-associated lncRNAs could modulate the activity of their associated protein-coding genes, not by modulating gene expression levels. This trans regulation mode of Six3os with proteins may cast new light on the precise control of neural development.6
MIAT, which is selectively expressed in a subset of progenitors and retinal precursor cells, was identified to play a critical role in regulating mammalian retinal cell specification. Results found that the knockdown of MIAT in the developing retinas inhibited amacrine cell and Müller glia differentiation. Furthermore, the overexpression of MIAT–IRES–GFP phenocopies the effects of the knockdown of MIAT by inducing mislocalisation of this nuclear-retained lncRNA. With the help of the IRES–GFP fusion approach, the specific domains of MIAT that are required for repressing both amacrine and Müller glial differentiation were identified.7
lncRNA BB283400 To explore the molecular mechanism that controls the uniformity of various retina layers, Krol and colleagues8 have portrayed a regulatory network consisting of lncRNA BB283400, a retina-specific lncRNA, RNA helicase Ddx3x and miR-183/96/182; this network controls the timing of miR-183/96/182 accumulation in photoreceptors. In this network, Ddx3x prevents pri-miR-183/96/182 from becoming mature miRNAs in postnatal photoreceptors. This block is relieved by lncRNA BB283400, which antagonises the repressive effect of Ddx3x. Dysregulation of this network damages the regional variation in the thickness of the photoreceptor layer and may secondarily damage the thickness of the adjacent inner nuclear layer.
lncRNA TUG1 was expressed in the developing and adult retinal tissues to promote the production of rod photoreceptors. In the newborn retina, knockdown of TUG1 resulted in malformed or absent outer segments of transfected photoreceptors compared with normal photoreceptors. Additionally, the transfected rod photoreceptors showed a defect in migration into the outer nuclear layer, ectopic expression of cone-specific markers and increased apoptosis. In all, TUG1 is necessary for the proper formation of photoreceptors in the developing rodent retina, though the mechanism remains unclear.9
Vax2os is a retina-specific lncRNA whose expression is restricted to the mouse ventral retina.10 Meola et al demonstrated that spatiotemporal misexpression of Vax2os disturbs the progression of the cell cycle in photoreceptor progenitor cells. In particular, the overexpression of Vax2os in postnatal mouse retina blocks photoreceptor progenitors from progression towards their committed fate and consequently delays their differentiation process. Compared with control retinas, this perturbation is accompanied by an increase in the apoptosis of the photoreceptor cells, without affecting the developed adult retina. Similar results are observed in mouse photoreceptor-derived 661W cells, in which Vax2os overexpression results in similar impairment of the cell cycle progression and cell differentiation. Based on these results, it can be concluded that Vax2os participated in the regulation of cell cycle progression in photoreceptor progenitor cells.11
ENSMUST0000013486 Circadian oscillation is critical for modulating metabolic demands in the retina. Circadian shedding of retinal photoreceptor cell discs with subsequent phagocytosis by the neighbouring retinal pigmented epithelium (RPE) is essential for the removal of toxic metabolites and lifelong survival of these postmitotic neurons. Defects in photoreceptor phagocytosis would cause severe retinal pathology, but the biochemical mechanisms remain poorly defined. RNA-seq revealed oscillating expression of 16 lincRNAs that are critical for circadian gene expression. Pathway analysis implicates circadian rhythm signalling as the top biological process involving these differentially expressed transcripts.12 Among these lincRNAs, ENSMUST0000013486 displayed dramatically differential expression, with a 9.8-fold enrichment at 9.0 hours compared with 1.5 hours.
Precise modulation of refracting media by lncRNAs
lncRNAs play important roles not only in retinal development but also in refracting media, such as in the lens and cornea. To illustrate the transcriptional changes during the lens differentiation process, Hoang et al conducted high-throughput RNA-seq of neonatal mouse lens epithelial cells and lens fibre cells. Eighty-six differentially expressed lncRNAs were identified through comprehensively comparing the transcriptomes of these two cell types, such as RP23–237H8.2, AC135859.1, AL663030.1 and Meg3. These differentially expressed lncRNAs participated in several important signalling pathways, such as lens structural components, organelle loss and denucleation. lncRNAs were also new breakthroughs for studying lens-related visual impairments.13 14
Corneal-specific epithelial cells play an important role in maintaining corneal transparency and preventing vision loss. Joo et al 15 studied the lncRNA profiles of the corneal epithelium and focused on a small group of lncRNAs that exhibit splicing changes attributed to PNN knockdown, which is responsible for the maintenance of epithelial phenotypes.16 This subset exhibited significant expression changes after the perturbation of PNN, including the lncRNAs SPACA6P, HAS2-AS1, RPARP-AS1, RP11-295G20.2 and NUTM2a-AS1. Although incomplete, their findings paved the way for the potential relevance of lncRNAs in the maintenance of cornea transparency (table 1).
Table 1.
The role of long non-coding RNAs (lncRNAs) in the precise regulation of visual maintenance
| Official symbol | Also known as | Genomic location | Cell specific | Function |
|---|---|---|---|---|
| Six3os1 | Rncr1; Six3os; D17Mgi26; E130112H22Rik | 17 E4 | Retinal progenitors | Six3os1 was the first identified lncRNA that coeffect with its coding gene to play an important role in retinal development |
| MIAT | RNCR2; GOMAFU; C22orf35; LINC00066; NCRNA00066; lncRNA-MIAT | 22q12.1 | Progenitors and retinal precursor cells | MIAT plays a critical role in regulating mammalian retinal cell fate specification |
| BB283400 | Rncr4; BF465573 | Chromosome 6 | Retina | BB283400 was a retina-specific lncRNA that controls the uniformity of various retina layers |
| TUG1 | TI-227H; LINC00080; NCRNA00080 | 22q12.2 | Rod photoreceptors | TUG1 is necessary for the proper formation of photoreceptors in the developing rodent retina |
| Vax2OS | Vas2os; Vax2os1; Vax2os2 | 6 C3 | Photoreceptor progenitor cells | Vax2os is involved in the control of cell cycle progression of photoreceptor progenitor cells in the ventral retina |
| ENSMUST0000013486 | – | – | Photoreceptor phagocytosis | ENSMUST0000013486c is critical for circadian gene expression |
| H19 | ASM; BWS; WT2; ASM1; D11S813E; LINC00008; NCRNA00008 | 11p15.5 | – | H19 was reported to be female-biased specifically in the female mouse eye |
The role of lncRNAs in various types of visual impairment
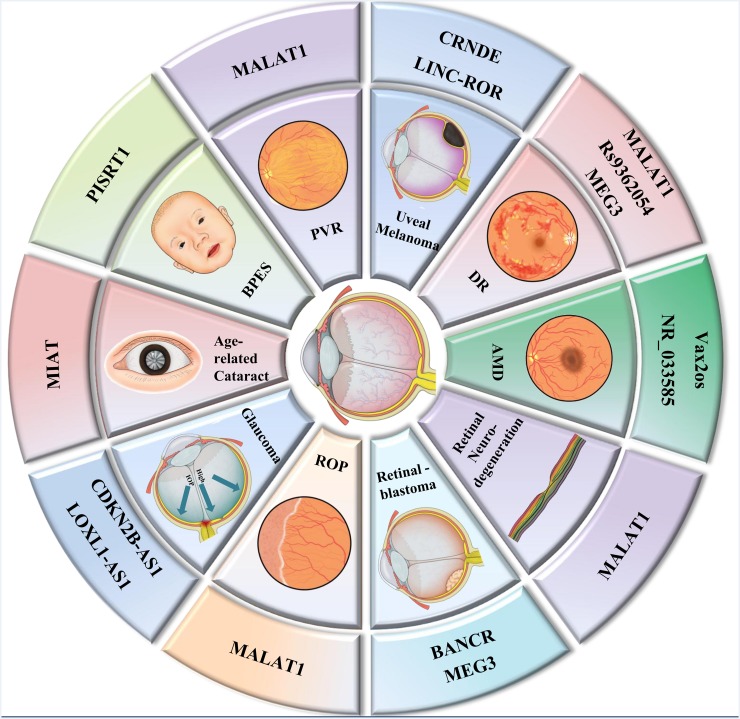
The WHO published new data on the prevalence of global blindness in 2010. According to the updated data, 285 million people are visually impaired, of whom 246 million people suffer from low vision and 39 million are blind. Cataract is the major cause for global blindness (51%), especially in the low/middle-income countries. Glaucoma (8%), age-related macular degeneration (AMD) (5%) and DR (1%), along with cataracts, account for 65% of all blindness in the world (figure 2).
Figure 2.
Long non-coding RNAs (lncRNAs) in various kinds of visual impairment. This figure displayed lncRNAs in most visual impairments mentioned in this article, including (A) blepharophimosis, ptosis and epicanthus inversus syndrome (BPES), (B) age-related cataract, (C) glaucoma, (D) proliferative vitreoretinopathy (PVR), (E) uveal melanoma, (F) retinoblastoma, (G) age-related macular degeneration (AMD), (H) retinal neurodegeneration, (I) retinopathy of prematurity (ROP) and (J) diabetic retinopathy (DR).
Ocular malignancy
Uveal melanoma
Uveal melanoma, the most common eye malignancy in adults with the prevalence of approximately 1 in 10 000,17 leads to severe visual impairment and can even be fatal in more than half of patients. The common treatments for primary uveal melanoma are surgery and radiotherapy, but metastasis is treatment refractory.
To comprehensively understand the genetic characteristics of uveal melanoma, Furney et al conducted SNP arrays and whole-genome sequencing of 12 primary uveal melanomas. SF3B1 mutations that related to differential alternative splicing of lncRNA CRNDE were identified and further confirmed to be associated with good prognosis in an extensive cohort of 105 samples.18 CRNDE has been reported to promote glioma cell proliferation and invasion through mTOR signalling.19 In addition, CRNDE functions primarily via epigenetic mechanisms, particularly through modulating the methylation status of histones.20
Fan et al 21 found that another lncRNA LINC-ROR and its target gene TESC were both significantly upregulated in 3 ocular melanoma cell lines and 20 ocular melanoma specimens compared with normal cells or adjacent normal tissues, respectively. They speculate LINC-ROR acts as an oncogenic lncRNA, activating the TESC promoter through demethylation by repelling the histone G9A methyltransferase. Therefore, suppression of LINC-ROR could serve as a potential treatment to restrain tumour growth and metastasis.
Retinoblastoma
Retinoblastoma is a rare form of retina malignancy but is the most common cancer of the eye in children, which is around 1 in 20 000 in live births,22 and it is almost exclusively found in young children. Therefore, instead of simple operations such as eye salvage, elucidating the underlying genetic mechanisms of retinoblastoma and its progression is of utmost importance.
Recent evidence shows that lncRNA BANCR is critical in the proliferation and metastasis in malignant melanoma and lung cancer via the MAPK pathway.23–25 lncRNA BANCR is overexpressed in both retinoblastoma tissues and cell lines and is associated with tumour size, choroidal invasion and optic nerve invasion. Knocking down lncRNA BANCR expression significantly suppressed the retinoblastoma cell proliferation, migration and invasion in vitro. BANCR has also been found participating in gastric cancer cells through the regulation of nuclear factor-kB1.26 However, little is known about the significance of lncRNA BANCR in retinoblastoma. lncRNA BANCR is found to be overexpressed in retinoblastoma tissues and cell lines in 60 retinoblastoma samples and normal retina samples, and its expression is associated with tumour size and choroidal and optic nerve invasion. Moreover, patients with high levels of lncRNA BANCR expression had poorer survival than those with lower levels. Multivariate analysis showed that increased lncRNA BANCR expression was an independent poor prognostic factor for patients with retinoblastoma. Furthermore, knocking down lncRNA BANCR expression significantly suppressed the retinoblastoma cell proliferation, migration and invasion in vitro. In conclusion, lncRNA BANCR plays a significant role in retinoblastoma aggressiveness and prognosis, and it may act as a promising target for therapeutic strategy and prognostic prediction.27
Unlike BANCR, MEG3 is regarded as a tumour suppressor. Gao and Lu28 found that MEG3 is significantly decreased in retinoblastoma samples and that the reduction is associated with a poor prognosis. Further studies have proven that MEG3 suppresses retinoblastoma progression, nominating MEG3 a molecular therapeutic target.
Diabetic retinopathy
DR is one of the most common peripheral vascular complications in chronic diabetes. The prevalence of DR is 3.5–5.7% among US29 adults and 1.1–1.3% among total population in China.30 Elucidation of genetic susceptibility factors for DR is important to completely understand its pathogenesis and may help to define possible treatments.
Yan et al performed lncRNA microarray analysis in a mouse model of streptozotocin (STZ)-induced diabetes. In addition, MALAT1, a conserved lncRNA, was also significantly upregulated in a RF/6A cell model of hyperglycaemia, in aqueous humour samples and fibrovascular membranes of patients with diabetes.31 GO analysis was targeted to the eye development process, while pathway analysis was mostly enriched in the axon guidance signalling pathway. Taken together, these results support the hypothesis that MALAT1 dysregulation is a potential molecular mechanism of DR pathogenesis.32
The MALAT1 level is also found to be significantly upregulated in the retinas of db/db mice, a type II diabetic model. However, the role of MALAT1 in retinal vasculature remodelling still remains elusive. To illustrate the function of MALAT1, MALAT1 knockdown was carried out, and it was found that the downregulation could obviously ameliorate DR, which is characterised by pericyte loss, capillary degeneration, microvascular leakage and retinal inflammation.
Moreover, MALAT1 knockdown could regulate retinal endothelial cell proliferation, migration and tube formation in vitro33 by functioning as a competing endogenous RNA in the regulation of vascular endothelial growth factor (VEGF) levels by miR-150-5p.34 The upregulation of tumour necrosis factor-α (TNF-α) and ICAM-1 was inhibited by MIAT knockdown, thereby alleviating vascular leakage and inflammation.35 MALAT1 upregulation represents a critical pathogenic mechanism for diabetes-induced microvascular dysfunction. Inhibition of MALAT1 may serve as a potential target for antiangiogenic therapy for diabetes-related microvascular complications.33
lncRNA MEG3 plays an important role in diabetes-related microvascular dysfunction as well. MEG3 is significantly downregulated in the retinas of STZ-induced diabetic mice and in endothelial cells upon high glucose and oxidative stress. MEG3 knockdown aggravates retinal vessel dysfunction in vivo and retinal endothelial cell in vitro.36
To see whether lncRNAs function in patients with DR, scientists conducted a three-stage genome-wide association study (GWAS) to identify DR susceptibility loci in 837 Japanese patients with type II diabetes and in 1149 controls. The most probable loci in their association analysis was a lincRNA rs9362054 in an intron of RP1-90L14.1. The present study raises the possibility that the dysregulation of key lncRNAs plays a role in the susceptibility of patients with DR.37
Ocular neovascularisation
Ocular pathological neovascularisation is a complex process that is the most prevalent cause of vision impairment, such as corneal neovascularisation, retinopathy of prematurity (ROP) and AMD. Here, we provide evidence that lncRNAs play a regulatory role in altering the balance of pro-angiogenic and antiangiogenic factors in the eye.
Choroidal neovascularisation (CNV) represents the basic pathological changes in neovascular AMD, a leading cause of visual impairment in elderly individuals. Xu et al analysed RNA expression profiles and found that the expression of VEGF and two lncRNAs, Vax2os1 and Vax2os2, was significantly upregulated in the aqueous humour of CNV AMD patients. In murine ocular neovascular models, Vax2os1 and Vax2os2 were also found to be significantly upregulated. These findings suggest a potential role of lncRNAs as biomarkers,38 and they may be employed for CNV treatment.
Corneal neovascularisation, which is mostly caused by chronic hypoxia or various inflammatory stimuli, leads to visual impairment or even blindness.39 Huang et al identified 154 differentially expressed lncRNAs between vascularised and normal corneas, including 60 downregulated lncRNAs and 94 upregulated lncRNAs. GO enrichment analysis of lncRNA coexpressed mRNAs indicated that the biological modules were correlated with the extracellular region, DNA binding and the immune response. KEGG pathway analysis indicated that ‘pathways in cancer’ was the most enriched signalling pathway. The lncRNA NR_033585 was significantly upregulated in neovascularised corneas and acts as a pro-angiogenic factor, such as VEGF, whereas the lincRNA chr8:129102060–129109035 reverse strand was found to be markedly downregulated in neovascularised corneas, exhibiting antiangiogenic effects.40
In ROP, endothelial cells express relatively high levels of the conserved lncRNAs MALAT1, TUG1, MEG3, linc00657 and linc00493. MALAT1 was significantly increased by hypoxia and controls a phenotypic switch in endothelial cells. Silencing of MALAT1 induced a promigratory response and promoted basal sprouting and migration, whereas proliferation of endothelial cells was inhibited. When angiogenesis was further stimulated by VEGF, MALAT1 knockdown caused discontinuous sprouts indicative of defective proliferation of stalk cells. In vivo studies confirmed that genetic ablation of MALAT1 inhibited proliferation of endothelial cells and reduced neonatal retina vascularisation. Knocking down MALAT1 shifts the balance from a proliferative to a migratory endothelial cell phenotype in vitro, and its genetic deletion restrains vascular growth in vivo.41
Additional work is required to define the precise role of lncRNAs in the angiogenesis cascade and to investigate whether altering the expression of lncRNAs could become a potential therapeutic strategy for treating patients with ocular neovascular diseases.
Ocular degeneration
Ocular degeneration is a group of visual impairments that is associated with ageing and involves different ocular tissues, such as the lens and retina.
Age-related cataracts
Age-related cataract is among the most common chronic disorders of ageing and is the leading disorder that results in blindness worldwide. To explore lncRNAs in the physiopathological mechanism of cataract, Shen et al determined lncRNA expression profiles in the transparent and age-matched cataractous human lenses42 and identified 38 lncRNAs that were differentially expressed. One of the most highly abundant lncRNAs, MIAT, was specifically upregulated both in the plasma fraction of whole blood and in the aqueous humour of cataract patients. The results suggest that MIAT is a cataract-specific biomarker.
Posterior capsule opacification (PCO) is a common complication of cataract surgery that is associated with abnormal production of inflammatory factors and oxidative stress. MIAT knockdown could repress TNF-α-induced abnormal proliferation and migration of human lens epithelial cells (HLECs). Additionally, MIAT could affect the proliferation, apoptosis and migration of HLECs upon oxidative stress, suggesting a potential role of MIAT in PCO-related pathological processes. Moreover, they found that MIAT acted as a ceRNA by forming a feedback loop with Akt and miR-150-5p to regulate HLEC function.
Retinal neurodegeneration
A previous study reveals that MALAT1 expression levels are significantly upregulated in diabetic retinas. Yao et al investigated the role of MALAT1 in the retinas of optic nerve transection rat and mouse models of retinal neurodegeneration.43 MALAT1 expression is significantly upregulated in the retinas, cultured Müller cells and primary retinal ganglion cells (RGCs) upon stress. MALAT1 knockdown decreases reactive gliosis, Müller cell activation and RGC survival in vivo and in vitro through CREB signalling. Thus, the experimental evidence suggests that MALAT1 dysregulation is crucial in neurodegenerative processes.
Glaucoma
The number of people with glaucoma worldwide will increase to 111.8 million in 2040,44 disproportionally affecting people residing in Asia and Africa. Primary open-angle glaucoma (POAG) is the predominant subtype of glaucoma, with a characteristic acquired loss of optic nerve fibres. Such loss develops in the corresponding loss of the visual field.
Intraocular pressure (IOP) is just one of the risk factors, and the underlying genetic predisposing factors are gaining increased attention. In recent studies, the pathogenesis of glaucoma is found to be related to lncRNAs. Evidence from recent studies has shown that genetic variants at the chromosome 9p21 locus execute predominant roles in the development and progression of POAG, including the CDKN2B-AS1 and CDKN2B genes and SIX1/SIX6.45
CDKN2B-AS1, located on chromosome 9p21.3, is a genetic susceptibility locus for several age-related diseases, such as coronary artery disease, type II diabetes and Alzheimer's disease.2–4 Therefore, a group of scientists explored the association between ten CDKN2B-AS1 SNPs and glaucoma, another age-related ocular disease, among 2947 POAG cases from the GLAUGEN study and NEIGHBOR consortium. Their research suggests that the CDKN2B-AS1 SNPs modulate the vulnerability of the optic nerve undergoing glaucomatous change.
Analysis of 9 of the 10 CDKN2B-AS1 SNPs associated with reduced disease risk showed that POAG patients carrying these minor alleles had a smaller cup-to-disc ratio (VCDR), despite higher IOP. Only one SNP is associated with increased disease risk with a larger VCDR, despite having lower IOP. Carriers of this adverse SNP seem to be predisposed to the development of POAG at lower IOP levels and exhibit stronger associations with normal-pressure glaucoma (NPG, defined by IOP <22 mm Hg) and advanced glaucoma phenotypes. This may be of relevance when establishing target IOP levels in patients carrying these risk alleles.46 47
In addition, scientists have confirmed five variants of lncRNA CDKN2B-AS1, which have been reported as a significant locus associated with glaucoma in the Caucasian population, in 2219 Japanese individuals.48
For African-Americans, Liu et al conducted an investigation of CDNK2B-AS1 and SIX1/SIX6 regions, the known genetic risk factors for POAG, in two populations of African ancestry. In the subgroup analyses, significant associations were raised for rs10965245 in the CDKN2B-AS1 region with high-pressure glaucoma and rs11849906 in the SIX1/SIX6 region with NPG. No significant association was identified with any loci in the Ghanaian samples reported. Meanwhile, POAG genetic susceptibility alleles associated with Caucasians appear to play a greatly reduced role in populations of African ancestry. Thus, the components of POAG susceptibility associated with lncRNAs among different human races remain to be identified.49
Based on the large-scale studies of lncRNAs in patients with POAG above, a meta-analysis of two independent GWAS for POAG was conducted and followed by an NPG subgroup analysis. The data set of this meta-analysis was composed of the GLAUGEN and NEIGHBOR (3146 cases and 3487 controls). Significant associations between two loci and POAG have been identified: the CDKN2BAS region on 9p21 and the SIX1/SIX6 region on chromosome 14q23. In subgroup analysis, two loci were significantly associated with NPG: 9p21 containing the CDKN2BAS gene and a probable regulatory region on 8q22. Both NPG loci were also nominally associated with a second type of glaucoma, exfoliation syndrome (XFS) glaucoma, suggesting that these loci might contribute more generally to optic nerve degeneration in glaucoma.50
To identify the accurate working mechanisms of CDKN2B-AS region, Gao and colleagues used a transgenic mouse model in which 70 kb of murine chromosome 4 containing the gene for lncRNA CDKN2B-AS were deleted, which is syntenic to human chromosome 9p21. This transgenic mouse model was carried out to study whether this deletion leads to a discernible phenotype in ocular structures implicated in glaucoma. The results showed that CDKN2B knockdown had no deleterious consequences for the retina but that these mice are more vulnerable to RGC loss in response to elevated IOP compared with wild-type animals.51 All of this evidence further implies a key regulatory role for CDKN2B-AS in modulating optic nerve degeneration.
Exfoliation syndrome
XFS is a common, age-related, systemic fibrillinopathy. XFS is characterised by the accumulation of protein clumps, which can block the normal drainage of aqueous humour, greatly increasing the risk of exfoliation glaucoma, a major cause of irreversible blindness in the elderly. To identify the genomic basis of XFS, the entire LOXL1 genomic locus was nominated as candidate functional variants in 50 black South African XFS cases and 50 matched controls. The finding was replicated in US Caucasian (91 cases/1031 controls), German (771 cases/1365 controls) and Japanese (1484 cases/1188 controls) populations. The region of peak association lies upstream of LOXL1-AS1, an lncRNA transcribed from the opposite strand of LOXL1. Similar dysregulation of LOXL1-AS1 expression is also significantly altered in HLECs under oxidative stress and in human Schlemm's canal endothelial cells with cyclic mechanical stress. Taken together, these findings support a functional role for the lncRNA LOXL1-AS1 in the cellular stress response and in XFS pathogenesis.52
Proliferative vitreoretinopathy
PVR is a disease that develops as a complication of retinal detachment and vitreoretinal surgery that results in severe vision reduction. Zhou et al attempted to determine the role of lncRNAs in PVR. Microarray analysis revealed that 78 lncRNAs were abnormally expressed in the ERMs of PVR patients, including 48 upregulated and 30 downregulated lncRNA transcripts. They subsequently focus on one lncRNA, MALAT1, whose upregulation contributed to the proliferation and migration of retinal pigment epithelium, formation of ERM, and many other types of PVR pathogenesis. Moreover, MALAT1 upregulation also appeared in the cellular and plasma fraction of peripheral blood in patients with PVR and was obviously reduced after PVR operation. Therefore, MALAT1 is a potential prognostic indicator and a target for the diagnosis and gene therapy for patients with PVR.53
Rare eye diseases
Rare eye diseases, diseases with extremely low incidence account for 0.65–1% of the total population, could be a perfect model in excavating the functions of lncRNAs in visual impairments due to the extreme phenotypes of rare eye diseases.54
MOMO syndrome
Macrosomia, obesity, macrocephaly and ocular abnormalities (MOMO) syndrome is an extremely rare genetic disorder that has been diagnosed in only six cases around the world, and it occurs in 1 in 100 million births. Although no chromosomal or molecular abnormality has been identified so far, Vu's team revealed a disruption of a novel gene named LINC00237 that is expressed in lymphocytes of control individuals, while normal transcripts were absent in lymphocytes of this patient with MOMO. We suspect that LINC00237 could play a role in the pathogenesis of this syndrome, but its function remains unknown.55
BPES
Blepharophimosis, ptosis and epicanthus inversus syndrome (BPES) is a rare autosomal-dominant congenital disorder. In type I BPES, eyelid abnormalities are associated with ovarian failure. Type II BPES shows only the eyelid defects. Mutations in FOXL2, a gene located at 3q23, have been shown to attribute to the syndrome. Molecular studies discovered a novel 63.2 kb deletion involving a non-protein-coding gene (PISRT1). This novel deletion could be involved in FOXL2 regulation and constitutes the smallest deletion described in a female with BPES. In cases of apparently balanced ‘de novo’ translocation, only a 5-6% risk of phenotype alterations have been described56 (table 2).
Table 2.
The role of long non-coding RNAs (lncRNAs) in various kinds of visual impairment
| Official symbol | Also known as | Genomic location | Related disease | Alteration | Function |
|---|---|---|---|---|---|
| CRNDE | CRNDEP; PNAS-108; lincIRX5; LINC00180; NCRNA00180 | 16q12.2 | Uveal Melanoma |
Dys regulation |
CRNDE were associated with good prognosis via epigenetic mechanisms, particularly through histone methylation or demethylation by the PRC2 or CoREST complexes. |
| LINC-ROR | ROR; lincRNA-RoR | 18q21.31 | Uveal melanoma |
Upregulation | LINC-ROR acts as an oncogenic lncRNA, activating the TESC promoter by repelling the histone G9A methyltransferase and promoting the release of histone H3K9. Suppression of ROR could reduce tumour growth and metastasis as reported. |
| BANCR | LINC00586 | Chr 9 | Retinoblastoma | Up regulation |
BANCR is overexpressed and associated with tumour size, choroidal invasion, and optic nerve invasion. Moreover, patients with high levels of lncRNA BANCR expression had poorer survival than those with lower levels of lncRNA BANCR expression. |
| PISRT1 | NCRNA00195 | 3q23 | BPES | Dys regulation |
PISRT1 deletion could be involved in FOXL2 regulation and constitutes the smallest deletion described in a female with BPES. |
| LINC00237 | NCRNA00237 | 20p11.23 | MOMO syndrome |
– | LINC00237 was expressed in lymphocytes of control individuals while normal transcripts were absent in lymphocytes of our MOMO patient. |
| MALAT1 | HCN; NEAT2; PRO2853; mascRNA; LINC00047; NCRNA00047 | 11q13.1 | Diabetic retinopathy |
Up regulation |
MALAT1 downregulation could obviously ameliorate DR by functioning as a competing endogenous RNA in the regulation of VEGF levels through miR-150-5p. |
| Retinopathy of prematurity | Up regulation |
Knocking down of MALAT1 tips the balance from a proliferative to a migratory endothelial cell phenotype in vitro, and its genetic deletion restrains vascular growth in vivo. | |||
| Retinal neurodegeneration |
Up regulation |
MALAT1 knockdown reduces reactive gliosis, Müller cell activation, and RGC survival in vivo and in vitro. MALAT1 regulates Müller and RGC function through CREB signalling. | |||
| Proliferative vitreoretinopathy | Up regulation |
MALAT1 related to RPE proliferation and migration, promotion of ERM formation, and PVR pathogenesis. MALAT1 upregulation appeared in the cellular and plasma fraction of peripheral blood in PVR patients as well and obviously reduced after PVR operation. | |||
| rs9362054 | – | Chr 6 | Diabetic retinopathy |
Dys regulation |
Top signal of DR susceptibility loci. |
| Vax2os | Vas2os; Vax2os1; Vax2os2 | 6 C3 | Ocular neovascularisation |
Up regulation |
Vax2os significantly upregulated in the aqueous humour of patients with CNV AMD and murine ocular neovascular models. |
| NR_033585 | – | – | Corneal neovascularisation |
Up regulation |
The lncRNA NR_033585 was significantly upregulated in vascularised corneas and presented a similar expression pattern as pro-angiogenic factors. |
| Chr8:129102060–129109035 | – | – | Corneal neovascularisation |
Down regulation |
lincRNA chr8:129102060–129109035 reverse strand was found to be markedly downregulated in vascularised corneas. |
| CDKN2B-AS1 | ANRIL; p15AS; PCAT12; CDKN2BAS; CDKN2B-AS; NCRNA00089 | 9p21.3 | Glaucoma | Dys regulation |
CDKN2B-AS1 is the genetic susceptibility locus for glaucoma, influencing the nearby CDKN2A and CDKN2B genes via regulatory mechanisms, which can influence cell proliferation and senescence. |
| LOXL1-AS1 | – | 15q24.1 | Exfoliation glaucoma |
Dys regulation |
LOXL1-AS1 manipulates the activity of promoter in LOXL1 region. |
| MIAT | RNCR2; GOMAFU; C22orf35; LINC00066; NCRNA00066; lncRNA-MIAT | 22q12.1 | Age-related cataract |
Up regulation |
MIAT knockdown could repress TNF-α-induced abnormal proliferation and migration of HLECs, by acting as a ceRNA and formed a feedback loop with Akt and miR-150-5p. |
| MEG3 | GTL2; FP504; prebp1; PRO0518; PRO2160; LINC00023; NCRNA00023 | 14q32 | Diabetic retinopathy |
Down regulation |
MEG3 knockdown aggravates retinal vessel dysfunction in vivo, and also regulates retinal endothelial cell proliferation, migration and tube formation in vitro. |
| Retinoblastoma | Down regulation |
MEG3 is significantly downregulated and that the reduced expression is associated with a poor prognosis among patients with retinoblastoma. |
AMD, age-related macular degeneration; BPES, blepharophimosis, ptosis and epicanthus inversus syndrome; CNV, choroidal neovascularisation; HLEC, human lens epithelial cell; RPE, retinal pigmented epithelium; TNF-α, tumour necrosis factor α; VEGF, vascular endothelial growth factor.
Discussion
lncRNAs staged on postgenome era: scientific demands and technical innovations
With the draft publication of human genome project in 2001, lncRNAs came into view for the first time.57 However, scientists paid little attention to lncRNAs, which have been identified as ‘genome garbage’.58 This phenomenon could be partially ascribed to the eager concern on protein-coding RNAs as well as on sequencing limitations at that time.
As time passed, when unexplainable questions arose from the known sequence, scientists returned to the lncRNAs that have been ignored for a long time.
Researchers rectified lncRNAs as precious genomic members after the launching of the ENCODE project in 2003. lncRNAs not only exceed mRNAs in quantity but also participate in species evolution and various biological processes.59 They perform together with DNAs and proteins, ensuring harmonious operations of life.
The evolution in high-throughput sequencing technology has made it possible for scientists to conduct cheaper but more accurate studies in identifying functional lncRNAs and lucubrating their possible roles.60 From 2003 to 2010, we focused on circulation, immunity and tumours by regarding life as a whole.61 Thus, relatively fewer studies concerning the visual system were published during this period.
Characteristics of visual system endowed it the breakthrough of lncRNA research
As precision medicine has advanced, the visual system shifted to the frontier of lncRNAs research due to its irreplaceable uniqueness.
Thanks to its barrier from the systemic circulation and from the immune system, the visual system is capable of blocking sequencing noise from other tissues or organs.62 So far, the most remarkable challenge in studying lncRNAs in other tissues has been to identify the real functional lncRNAs instead of disturbances. It seems that including more individuals in the data analysis is the only method for reducing data noise, despite the excessively high financial cost of such work. However, the natural characteristics of the visual system make it the best tissue for these types of analyses and this realisation has been a breakthrough in the transcriptomic analysis of lncRNAs. Additionally, ophthalmological operations have a leadership position in the era of minimally invasive surgery. Precise sampling brought about incomparable sequencing reliability and validity.
We believe that the visual system will always be at the forefront of lncRNA research. The first reason is that almost every part of the visual system can be observed directly with the help of simple devices, such as slit lamp and ophthalmoscope. We can obtain intuitionistic images such as anterior segment photographs and fundus photographs to monitor the progress of visual development and impairment. Clinical efficacy evaluations can also be carried out easily.
Second, the visual system has obvious phenotypes such as the integrity and transparency of the cornea, abnormal ocular or eyelid position, the shape of the anterior chamber and iris, and the colour of the lens and image of fundus, in contrast to the potentially ambiguous evaluation criteria of visceral lesions. These characteristics perfectly match the current, predominant challenges in the study of lncRNAs, that is, the difficulty in identifying the possible functions of lncRNAs simply in accordance with their nucleotide sequences.63 Unlike protein-coding sequences, most lncRNAs are poorly conserved among different species and evolve rapidly.64 These distinctive features make the visual system a perfect model for lncRNA functional studies because phenotypic changes are easy to assess and diagnose.
Researchers are consistently troubled by the data noise in lncRNA analyses, even in strict pairwise control studies. However, we can solve this problem by simply obtaining data separately on the two eyes in a single individual because one eye presents the most accurate control for the other. Using one eye to control for the other not only provides a self-control but also eliminates the disturbance of a variety of factors that would be present when comparing tissues from separate individuals, namely individual differences, temporal variations, environmental changes, self-healing tendencies and periodic impairments.
As reported, almost every tissue in the visual system is incapable of self-repair, giving rise to irreversible visual impairments. Although ocular regenerative medicine led by stem cell studies has thrived in recent years, we have only harvested limited effects in cornea and lens. In particular, the retinal regeneration field experienced various challenges and has made little progress. Most bionic prostheses failed in clinical trials as well. Therefore, a conceptual transition from ‘postdisaster reconstruction’ to ‘predisaster prevention’ is almost inevitable. Since lncRNAs were proposed as strong regulators in nearly every aspect of pathophysiology, the modulation of lncRNAs participating in the upstream link or even in the outset of visual development and impairment should be our focus.
Future directions and challenges
Visual system will pioneer prospective studies in lncRNA research
In the first stage of lncRNA research, from approximately 2003 until now, most studies favoured cross-sectional gene expression. Two or more groups were recruited for high-throughput or microarray sequencing to determine the responsible lncRNAs. We expect to see the visual system leading the arrival of a new era in lncRNA research by changing the traditional research model. By establishing the dynamic study of different stages of visual development and in various visual impairment models, we can construct the landscape of lncRNAs in visual development and impairment. This landscape may engender a comprehensive understanding of functional lncRNAs in the specific timeline of biological processes and at different locations in the visual system. We need to conduct prospective studies by regulating lncRNAs to obtain accurate research results. With the help of these data, meaningful clinical significance may be achieved to guide clinical practice from prevention to treatment.
Promising vision rescuer: gene therapy targeting lncRNAs
With the deepening of lncRNA research, scientists look forward to developing successful genetic treatments that target lncRNAs. It is even conceivable to produce antagoNATs that target the responsible lncRNAs as a direct therapeutic approach. The visual system will be best for developing genetic treatments, considering the following reasons. As stated above, gene therapy in the visual system will not cause side effects to the systemic function due to the natural blood–brain barrier. Additionally, significantly increased efficacy could be obtained as a result of the relative separation from the external environment.
Several researches emerged in genetic treatments that target lncRNAs in visual system, including lncRNA-MIAT in microvascular dysfunction,34 MALAT1 in diabetes-related microvascular complications33 and PVR diseases,53 lncRNA-GAS5 in hypertension-induced neovascularisation and capillary leakage.65 These results encouraged us to make more efforts in exploring the potential therapeutic roles of lncRNAs in visual system. Genetic therapy targeting lncRNAs in visual system calls for further clinical trials and applications. We believe that researchers will endeavour in making lncRNAs the promising vision rescuer in the near future.
Challenges to technical approaches in lncRNA studies
Although bioinformatics is the experimental approach favoured by biologists worldwide, there are still several obstacles to be overcome. Above all, high-throughput screening is affordable and currently widely used. However, the veracity of biological correlation analysis remains questionable. Elucidating the subtler cis-regulatory and trans-regulatory roles of lncRNAs may require technological advances in both high-throughput identification of lncRNA binding sites and in high-resolution in vivo imaging at the subcellular level.
Chen et al 66 published their results that brought a new dawn to solve the problems in high-resolution imaging at the subcellular level in vivo. They developed a single-molecule imaging-based MERFISH technology that allowed the simultaneous detection of the expression and localisation 100–1000 RNA molecules in cells at same time.
Rapicavoli's group has demonstrated a novel tool for lucubrating nuclear-retained lncRNAs, which occupies the majority of mammalian lncRNAs. Considering that conventional RNA interference approaches like shRNA-mediated degradation are less effective at downregulating nuclear-retained lncRNAs, IRES-GFP sequence-mediated mislocalisation of nuclear-retained lncRNAs may achieve domain-restrained knockdown effect.7 They have reported that fusion of a nuclear-retained lncRNA to an IRES-GFP sequence can mislocalise lncRNAs to the cytoplasmic ribosome and away from nucleus, which were their normal cellular functional sites. The IRES-GFP method produces phenotypes that largely overlap with those seen with shRNA-mediated knockdown. Therefore, IRES-GFP may serve as a new tool for functional analysis of these molecules.
Although the MERFISH and IRES-GFP technology has been a major step forward in studying RNA molecule function, it is not enough. We need better tools to explore the universe of lncRNAs.
Footnotes
Contributors: WP performed the literature collection. PW wrote the manuscript. ZY and SW contributed to intellectual discussions. All authors gave the final approval of the manuscript submission.
Funding: ZYs laboratory is supported by research grants from the Guangdong Province National Natural Science Foundation (2014A030308016).
Disclaimer: The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Lafuma A, Brézin A, Lopatriello S, Hieke K, Hutchinson J, Mimaud V, Berdeaux G. Evaluation of non-medical costs associated with visual impairment in four European countries: France, Italy, Germany and the UK. Pharmacoeconomics 2006;24:193–205. 10.2165/00019053-200624020-00007 [DOI] [PubMed] [Google Scholar]
- 2. Popov N, Gil J. Epigenetic regulation of the INK4b-ARF-INK4a locus: in sickness and in health. Epigenetics 2010;5:685–90. 10.4161/epi.5.8.12996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li Z, Yu X, Shen J. ANRIL: a pivotal tumor suppressor long non-coding RNA in human cancers. Tumour Biol 2016;37:5657–61. 10.1007/s13277-016-4808-5 [DOI] [PubMed] [Google Scholar]
- 4. Zhou X, Han X, Wittfeldt A, Sun J, Liu C, Wang X, Gan LM, Cao H, Liang Z. Long non-coding RNA ANRIL regulates inflammatory responses as a novel component of NF-κB pathway. RNA Biol 2016;13:98–108. 10.1080/15476286.2015.1122164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mustafi D, Kevany BM, Bai X, Maeda T, Sears JE, Khalil AM, Palczewski K. Evolutionarily conserved long intergenic non-coding RNAs in the eye. Hum Mol Genet 2013;22:2992–3002. 10.1093/hmg/ddt156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rapicavoli NA, Poth EM, Zhu H, Blackshaw S. The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural Dev 2011;6:32 10.1186/1749-8104-6-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rapicavoli NA, Poth EM, Blackshaw S. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev Biol 2010;10:49 10.1186/1471-213X-10-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krol J, Krol I, Alvarez CP, Fiscella M, Hierlemann A, Roska B, Filipowicz W. A network comprising short and long noncoding RNAs and RNA helicase controls mouse retina architecture. Nat Commun 2015;6:7305 10.1038/ncomms8305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol 2005;15:501–12. 10.1016/j.cub.2005.02.027 [DOI] [PubMed] [Google Scholar]
- 10. Rapicavoli NA, Blackshaw S. New meaning in the message: noncoding RNAs and their role in retinal development. Dev Dyn 2009;238:2103–14. 10.1002/dvdy.21844 [DOI] [PubMed] [Google Scholar]
- 11. Meola N, Pizzo M, Alfano G, Surace EM, Banfi S. The long noncoding RNA Vax2os1 controls the cell cycle progression of photoreceptor progenitors in the mouse retina. RNA 2012;18:111–23. 10.1261/rna.029454.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mustafi D, Kevany BM, Genoud C, Bai X, Palczewski K. Photoreceptor phagocytosis is mediated by phosphoinositide signaling. FASEB J 2013;27:4585–95. 10.1096/fj.13-237537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoang TV, Kumar PK, Sutharzan S, Tsonis PA, Liang C, Robinson ML. Comparative transcriptome analysis of epithelial and fiber cells in newborn mouse lenses with RNA sequencing. Mol Vis 2014;20:1491–517. [PMC free article] [PubMed] [Google Scholar]
- 14. Wormstone IM, Wride MA. The ocular lens: a classic model for development, physiology and disease. Philos Trans R Soc Lond, B, Biol Sci 2011;366:1190–2. 10.1098/rstb.2010.0377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Joo JH, Ryu D, Peng Q, Sugrue SP. Role of Pnn in alternative splicing of a specific subset of lncRNAs of the corneal epithelium. Mol Vis 2014;20:1629–42. [PMC free article] [PubMed] [Google Scholar]
- 16. Joo JH, Correia GP, Li JL, Lopez MC, Baker HV, Sugrue SP. Transcriptomic analysis of PNN- and ESRP1-regulated alternative pre-mRNA splicing in human corneal epithelial cells. Invest Ophthalmol Vis Sci 2013;54:697–707. 10.1167/iovs.12-10695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Andreoli MT, Mieler WF, Leiderman YI. Epidemiological trends in uveal melanoma. Br J Ophthalmol 2015;99:1550–3. 10.1136/bjophthalmol-2015-306810 [DOI] [PubMed] [Google Scholar]
- 18. Furney SJ, Pedersen M, Gentien D, Dumont AG, Rapinat A, Desjardins L, Turajlic S, Piperno-Neumann S, de la Grange P, Roman-Roman S, Stern MH, Marais R. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov 2013;3:1122–9. 10.1158/2159-8290.CD-13-0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Y, Wang Y, Li J, Zhang Y, Yin H, Han B. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Lett 2015;367:122–8. 10.1016/j.canlet.2015.03.027 [DOI] [PubMed] [Google Scholar]
- 20. Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea MD, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA 2009;106:11667–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fan J, Xing Y, Wen X, Jia R, Ni H, He J, Ding X, Pan H, Qian G, Ge S, Hoffman AR, Zhang H, Fan X. Long non-coding RNA ROR decoys gene-specific histone methylation to promote tumorigenesis. Genome Biol 2015;16:139 10.1186/s13059-015-0705-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. ElZomor H, Taha H, Aleieldin A, Nour R, Zaghloul MS, Fawzi M, Kamel A, Alfaar AS. High risk retinoblastoma: prevalence and success of treatment in developing countries. Ophthalmic Genet 2015;36:287–9. 10.3109/13816810.2015.1016241 [DOI] [PubMed] [Google Scholar]
- 23. Li R, Zhang L, Jia L, Duan Y, Li Y, Bao L, Sha N. Long non-coding RNA BANCR promotes proliferation in malignant melanoma by regulating MAPK pathway activation. PLoS ONE 2014;9:e100893 10.1371/journal.pone.0100893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jiang W, Zhang D, Xu B, Wu Z, Liu S, Zhang L, Tian Y, Han X, Tian D. Long non-coding RNA BANCR promotes proliferation and migration of lung carcinoma via MAPK pathways. Biomed Pharmacother 2015;69:90–5. 10.1016/j.biopha.2014.11.027 [DOI] [PubMed] [Google Scholar]
- 25. Flockhart RJ, Webster DE, Qu K, Mascarenhas N, Kovalski J, Kretz M, Khavari PA. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res 2012;22:1006–14. 10.1101/gr.140061.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang ZX, Liu ZQ, Jiang B, Lu XY, Ning XF, Yuan CT, Wang AL. BRAF activated non-coding RNA (BANCR) promoting gastric cancer cells proliferation via regulation of NF-κB1. Biochem Biophys Res Commun 2015;465:225–31. 10.1016/j.bbrc.2015.07.158 [DOI] [PubMed] [Google Scholar]
- 27. Su S, Gao J, Wang T, Wang J, Li H, Wang Z. Long non-coding RNA BANCR regulates growth and metastasis and is associated with poor prognosis in retinoblastoma. Tumour Biol 2015;36:7205–11. 10.1007/s13277-015-3413-3 [DOI] [PubMed] [Google Scholar]
- 28. Gao Y, Lu X. Decreased expression of MEG3 contributes to retinoblastoma progression and affects retinoblastoma cell growth by regulating the activity of Wnt/β-catenin pathway. Tumour Biol 2016;37:1461–9. 10.1007/s13277-015-4564-y [DOI] [PubMed] [Google Scholar]
- 29. Zhang X, Saaddine JB, Chou CF, Cotch MF, Cheng YJ, Geiss LS, Gregg EW, Albright AL, Klein BE, Klein R. Prevalence of diabetic retinopathy in the United States, 2005-2008. JAMA 2010;304:649–56. 10.1001/jama.2010.1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu L, Wu X, Liu L, Geng J, Yuan Z, Shan Z, Chen L. Prevalence of diabetic retinopathy in mainland China: a meta-analysis. PLoS ONE 2012;7:e45264 10.1371/journal.pone.0045264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yan B, Tao ZF, Li XM, Zhang H, Yao J, Jiang Q. Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Invest Ophthalmol Vis Sci 2014;55:941–51. 10.1167/iovs.13-13221 [DOI] [PubMed] [Google Scholar]
- 32. Li F, Wen X, Zhang H, Fan X. Novel insights into the role of long noncoding RNA in ocular diseases. Int J Mol Sci 2016;17;478 10.3390/ijms17040478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li YJ, Yan B, Jiang Q. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis 2014;5:e1506 10.1038/cddis.2014.466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li YJ, Tao ZF, Song YC, Chen Q, Jiang Q. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res 2015;116:1143–56. 10.1161/CIRCRESAHA.116.305510 [DOI] [PubMed] [Google Scholar]
- 35. Jaé N, Dimmeler S. Long noncoding RNAs in diabetic retinopathy. Circ Res 2015;116:1104–6. 10.1161/CIRCRESAHA.115.306051 [DOI] [PubMed] [Google Scholar]
- 36. Qiu GZ, Tian W, Fu HT, Li CP, Liu B. Long noncoding RNA-MEG3 is involved in diabetes mellitus-related microvascular dysfunction. Biochem Biophys Res Commun 2016;471:135–41. 10.1016/j.bbrc.2016.01.164 [DOI] [PubMed] [Google Scholar]
- 37. Awata T, Yamashita H, Kurihara S, Morita-Ohkubo T, Miyashita Y, Katayama S, Mori K, Yoneya S, Kohda M, Okazaki Y, Maruyama T, Shimada A, Yasuda K, Nishida N, Tokunaga K, Koike A. A genome-wide association study for diabetic retinopathy in a Japanese population: potential association with a long intergenic non-coding RNA. PLoS ONE 2014;9:e111715 10.1371/journal.pone.0111715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xu XD, Li KR, Li XM, Yao J, Qin J, Yan B. Long non-coding RNAs: new players in ocular neovascularization. Mol Biol Rep 2014;41:4493–505. 10.1007/s11033-014-3320-5 [DOI] [PubMed] [Google Scholar]
- 39. Gupta D, Illingworth C. Treatments for corneal neovascularization: a review. Cornea 2011;30:927–38. 10.1097/ICO.0b013e318201405a [DOI] [PubMed] [Google Scholar]
- 40. Huang J, Li YJ, Liu JY, Zhang YY, Li XM, Wang LN, Yao J, Jiang Q, Yan B. Identification of corneal neovascularization-related long noncoding RNAs through microarray analysis. Cornea 2015;34:580–7. 10.1097/ICO.0000000000000389 [DOI] [PubMed] [Google Scholar]
- 41. Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, Boon RA, Dimmeler S. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res 2014;114:1389–97. 10.1161/CIRCRESAHA.114.303265 [DOI] [PubMed] [Google Scholar]
- 42. Shen Y, Dong LF, Zhou RM, Yao J, Song YC, Yang H, Jiang Q, Yan B. Role of long non-coding RNA MIAT in proliferation, apoptosis and migration of lens epithelial cells: a clinical and in vitro study. J Cell Mol Med 2016;20:537–48. 10.1111/jcmm.12755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yao J, Wang XQ, Li YJ, Shan K, Yang H, Wang YN, Yao MD, Liu C, Li XM, Shen Y, Liu JY, Cheng H, Yuan J, Zhang YY, Jiang Q, Yan B. Long non-coding RNA MALAT1 regulates retinal neurodegeneration through CREB signaling. EMBO Mol Med 2016;8:1113 10.15252/emmm.201606749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tham YC, Li X, Wong TY, Quigley HA, Aung T, Cheng CY. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology 2014;121:2081–90. 10.1016/j.ophtha.2014.05.013 [DOI] [PubMed] [Google Scholar]
- 45. Wan P, Su W, Zhuo Y. The role of long noncoding RNAs in neurodegenerative diseases. Mol Neurobiol 2016. http://dx.doi.org/10.1007/s12035-016-9793-6 [DOI] [PubMed] [Google Scholar]
- 46. Burdon KP, Crawford A, Casson RJ, Hewitt AW, Landers J, Danoy P, Mackey DA, Mitchell P, Healey PR, Craig JE. Glaucoma risk alleles at CDKN2B-AS1 are associated with lower intraocular pressure, normal-tension glaucoma, and advanced glaucoma. Ophthalmology 2012;119:1539–45. 10.1016/j.ophtha.2012.02.004 [DOI] [PubMed] [Google Scholar]
- 47. Pasquale LR, Loomis SJ, Kang JH, Yaspan BL, Abdrabou W, Budenz DL, Chen TC, Delbono E, Friedman DS, Gaasterland D, Gaasterland T, Grosskreutz CL, Lee RK, Lichter PR, Liu Y, McCarty CA, Moroi SE, Olson LM, Realini T, Rhee DJ, Schuman JS, Singh K, Vollrath D, Wollstein G, Zack DJ, Allingham RR, Pericak-Vance MA, Weinreb RN, Zhang K, Hauser MA, Richards JE, Haines JL, Wiggs JL. CDKN2B-AS1 genotype-glaucoma feature correlations in primary open-angle glaucoma patients from the United States. Am J Ophthalmol 2013;155:342–53.e5. 10.1016/j.ajo.2012.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nakano M, Ikeda Y, Tokuda Y, Fuwa M, Omi N, Ueno M, Imai K, Adachi H, Kageyama M, Mori K, Kinoshita S, Tashiro K. Common variants in CDKN2B-AS1 associated with optic-nerve vulnerability of glaucoma identified by genome-wide association studies in Japanese. PLoS ONE 2012;7:e33389 10.1371/journal.pone.0033389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu Y, Hauser MA, Akafo SK, Qin X, Miura S, Gibson JR, Wheeler J, Gaasterland DE, Challa P, Herndon LW, Ritch R, Moroi SE, Pasquale LR, Girkin CA, Budenz DL, Wiggs JL, Richards JE, Ashley-Koch AE, Allingham RR. Investigation of known genetic risk factors for primary open angle glaucoma in two populations of African ancestry. Invest Ophthalmol Vis Sci 2013;54:6248–54. 10.1167/iovs.13-12779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wiggs JL, Yaspan BL, Hauser MA, Kang JH, Allingham RR, Olson LM, Abdrabou W, Fan BJ, Wang DY, Brodeur W, Budenz DL, Caprioli J, Crenshaw A, Crooks K, Delbono E, Doheny KF, Friedman DS, Gaasterland D, Gaasterland T, Laurie C, Lee RK, Lichter PR, Loomis S, Liu Y, Medeiros FA, McCarty C, Mirel D, Moroi SE, Musch DC, Realini A, Rozsa FW, Schuman JS, Scott K, Singh K, Stein JD, Trager EH, Vanveldhuisen P, Vollrath D, Wollstein G, Yoneyama S, Zhang K, Weinreb RN, Ernst J, Kellis M, Masuda T, Zack D, Richards JE, Pericak-Vance M, Pasquale LR, Haines JL. Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet 2012;8:e1002654 10.1371/journal.pgen.1002654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gao S, Jakobs TC. Mice homozygous for a deletion in the glaucoma susceptibility locus INK4 show increased vulnerability of retinal ganglion cells to elevated intraocular pressure. Am J Pathol 2016;186:985–1005. 10.1016/j.ajpath.2015.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hauser MA, Aboobakar IF, Liu Y, Miura S, Whigham BT, Challa P, Wheeler J, Williams A, Santiago-Turla C, Qin X, Rautenbach RM, Ziskind A, Ramsay M, Uebe S, Song L, Safi A, Vithana EN, Mizoguchi T, Nakano S, Kubota T, Hayashi K, Manabe S, Kazama S, Mori Y, Miyata K, Yoshimura N, Reis A, Crawford GE, Pasutto F, Carmichael TR, Williams SE, Ozaki M, Aung T, Khor CC, Stamer WD, Ashley-Koch AE, Allingham RR. Genetic variants and cellular stressors associated with exfoliation syndrome modulate promoter activity of a lncRNA within the LOXL1 locus. Hum Mol Genet 2015;24:6552–63. 10.1093/hmg/ddv347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou RM, Wang XQ, Yao J, Shen Y, Chen SN, Yang H, Jiang Q, Yan B. Identification and characterization of proliferative retinopathy-related long noncoding RNAs. Biochem Biophys Res Commun 2015;465:324–30. 10.1016/j.bbrc.2015.07.120 [DOI] [PubMed] [Google Scholar]
- 54. Lin H, Long E, Chen W, Liu Y. Documenting rare disease data in China. Science 2015;349:1064 10.1126/science.349.6252.1064-b [DOI] [PubMed] [Google Scholar]
- 55. Vu PY, Toutain J, Cappellen D, Delrue MA, Daoud H, El MAA, Barat P, Montaubin O, Bonnet F, Dai ZQ, Philippe C, Tran CT, Rooryck C, Arveiler B, Saura R, Briault S, Lacombe D, Taine L. A homozygous balanced reciprocal translocation suggests LINC00237 as a candidate gene for MOMO (macrosomia, obesity, macrocephaly, and ocular abnormalities) syndrome. Am J Med Genet A 2012;158A:2849–56. 10.1002/ajmg.a.35694 [DOI] [PubMed] [Google Scholar]
- 56. González-González C, García-Hoyos M, Hernaez CR, Arroyo DC, González FC, Lorda SI, Sánchez-Escribano F. Microdeletion found by array-CGH in girl with blepharophimosis syndrome and apparently balanced translocation t(3;15)(q23;q25). Ophthalmic Genet 2012;33:107–10. 10.3109/13816810.2011.634879 [DOI] [PubMed] [Google Scholar]
- 57. Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet 2002;36:233–78. 10.1146/annurev.genet.36.042902.092433 [DOI] [PubMed] [Google Scholar]
- 58. Zhang X, Zhang M, Li D, He W, Peng J, Betzig E, Xu P. Highly photostable, reversibly photoswitchable fluorescent protein with high contrast ratio for live-cell superresolution microscopy. Proc Natl Acad Sci USA 2016;113:10364–9. 10.1073/pnas.1611038113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Anguera MC, Ma W, Clift D, Namekawa S, Kelleher RJ, Lee JT. Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. PLoS Genet 2011;7:e1002248 10.1371/journal.pgen.1002248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, Mitra S, Mohammed A, James AR, Hoberg E, Moustakas A, Gyllensten U, Jones SJ, Gustafsson CM, Sims AH, Westerlund F, Gorab E, Kanduri C. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat Commun 2015;6:7743 10.1038/ncomms8743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hu YW, Zhao JY, Li SF, Huang JL, Qiu YR, Ma X, Wu SG, Chen ZP, Hu YR, Yang JY, Wang YC, Gao JJ, Sha YH, Zheng L, Wang Q. RP5-833A20.1/miR-382-5p/NFIA-dependent signal transduction pathway contributes to the regulation of cholesterol homeostasis and inflammatory reaction. Arterioscler Thromb Vasc Biol 2015;35:87–101. 10.1161/ATVBAHA.114.304296 [DOI] [PubMed] [Google Scholar]
- 62. McMillan K, Bostani M, Cagnon C, Zankl M, Sepahdari AR, McNitt-Gray M. Size-specific, scanner-independent organ dose estimates in contiguous axial and helical head CT examinations. 2014;41:121909 10.1118/1.4901517 [DOI] [PubMed] [Google Scholar]
- 63. Chodroff RA, Goodstadt L, Sirey TM, Oliver PL, Davies KE, Green ED, Molnár Z, Ponting CP. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol 2010;11:R72 10.1186/gb-2010-11-7-r72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kutter C, Watt S, Stefflova K, Wilson MD, Goncalves A, Ponting CP, Odom DT, Marques AC. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet 2012;8:e1002841 10.1371/journal.pgen.1002841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang YN, Shan K, Yao MD, Yao J, Wang JJ, Li X, Liu B, Zhang YY, Ji Y, Jiang Q, Yan B. Long Noncoding RNA-GAS5: A Novel Regulator of Hypertension-Induced Vascular Remodeling. Hypertension 2016;68:736–48. 10.1161/HYPERTENSIONAHA.116.07259 [DOI] [PubMed] [Google Scholar]
- 66. Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 2015;348:aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]