CD177 presents antigens in allo- and autoimmune diseases on the neutrophil surface. Eulenberg-Gustavus et al. show that epigenetic silencing causes CD177negative neutrophils, whereas a novel pattern of monoallelic CD177 expression results in a variable percentage of CD177positive neutrophils in bimodal individuals.
Abstract
CD177 presents antigens in allo- and autoimmune diseases on the neutrophil surface. Individuals can be either CD177-deficient or harbor distinct CD177neg and CD177pos neutrophil subsets. We studied mechanisms controlling subset-restricted CD177 expression in bimodal individuals. CD177pos, but not CD177neg neutrophils, produced CD177 protein and mRNA. Haplotype analysis indicated a unique monoallelic CD177 expression pattern, where the offspring stably transcribed either the maternal or paternal allele. Hematopoietic stem cells expressed both CD177 alleles and silenced one copy during neutrophil differentiation. ChIP and reporter assays in HeLa cells with monoallelic CD177 expression showed that methylation reduced reporter activity, whereas demethylation caused biallelic CD177 expression. HeLa cell transfection with c-Jun and c-Fos increased CD177 mRNA. Importantly, CD177pos human neutrophils, but not CD177neg neutrophils, showed a euchromatic CD177 promoter, unmethylated CpGs, and c-Jun and c-Fos binding. We describe epigenetic mechanisms explaining the two distinct CD177 neutrophil subsets and a novel monoallelic CD177 expression pattern that does not follow classical random monoallelic expression or imprinting.
Introduction
CD177 is a neutrophil-specific receptor with clinical significance in allo- and autoimmune diseases (Stroncek, 2007). CD177 harbors the human neutrophil antigen 2 (HNA-2; formerly neutrophil antigen B1 or NB1). Alloantibodies to HNA-2, either actively generated or acquired from blood products, can cause severe neutropenia in newborns (Lalezari et al., 1971), graft failure in bone marrow recipients (Stroncek et al., 1993), drug-induced neutropenia (Stroncek et al., 1994), and transfusion-related lung injury (TRALI; Bux et al., 1996; Sachs et al., 2006). In addition, CD177 presents proteinase 3 (PR3), a major autoantigen in antineutrophil cytoplasmic antibodies (ANCA)-associated small vessel vasculitis on the neutrophil surface (Bauer et al., 2007; von Vietinghoff et al., 2007). PR3-ANCA bind to membrane-exposed PR3 (mPR3) and activate the neutrophil—a central process for subsequent vascular injury (Kettritz, 2012). Above and beyond its role in immune conditions, CD177 facilitates neutrophil endothelial transmigration involving heterophilic PECAM-1 interactions and catalytic membrane PR3 activity (Sachs et al., 2007; Bayat et al., 2010; Kuckleburg et al., 2012).
CD177 is a glycosyl-phosphatidyl inositol–linked glycoprotein of the urokinase plasminogen activator receptor/CD59/Ly6 snake toxin superfamily (Kissel et al., 2001). CD177 protein is stored in secondary granules and is presented on the neutrophil plasma membrane (von Vietinghoff et al., 2008; Rørvig et al., 2013). CD177 shows a peculiar distribution pattern in that ∼95% of all healthy individuals produce CD177pos neutrophils, whereas 5% are CD177 deficient (Goldschmeding et al., 1992; Matsuo et al., 2000; Li et al., 2015; Wu et al., 2016). However, even the CD177-producing individuals show a stable bimodal CD177 pattern over time, with distinct membrane CD177neg and CD177pos (mCD177) neutrophil subsets. The size of the latter ranges from a few, to >90% (Goldschmeding et al., 1992). Despite the strong clinical implications, the mechanisms controlling subset-restricted CD177 gene expression in bimodal individuals are incompletely understood.
Most diploid cells express both parental alleles of each gene, whereas some genes are monoallelically expressed. To date, two basic monoallelic expression (MAE) mechanisms in autosomes are distinguished. Imprinting describes a parent-of-origin MAE with exclusive silencing of either the maternal or the paternal allele (Barlow and Bartolomei, 2014). In contrast, classical random MAE occurs stochastically, so that clonal cells derived from a given cell type express either the maternal or the paternal, or even both parental alleles (Gimelbrant et al., 2007; Deng et al., 2014).
We CD177-phenotyped a large cohort of healthy individuals and studied genetic and epigenetic mechanisms that control CD177 expression using sorted neutrophil CD177 subsets, parent-offspring trios, neutrophil-differentiated CD34+ hematopoietic stem cells (HSCs), neonatal cord blood neutrophils, and a HeLa cell model. We demonstrate that disparate CpG and histone methylation, and AP-1 (c-Jun/c-Fos) transcription factor (TF) binding explain the two distinct CD177 neutrophil subsets. We observed MAE of CD177 with a novel pattern that follows neither classical random MAE nor imprinting.
Results
Neutrophil mCD177 phenotypes in normal controls
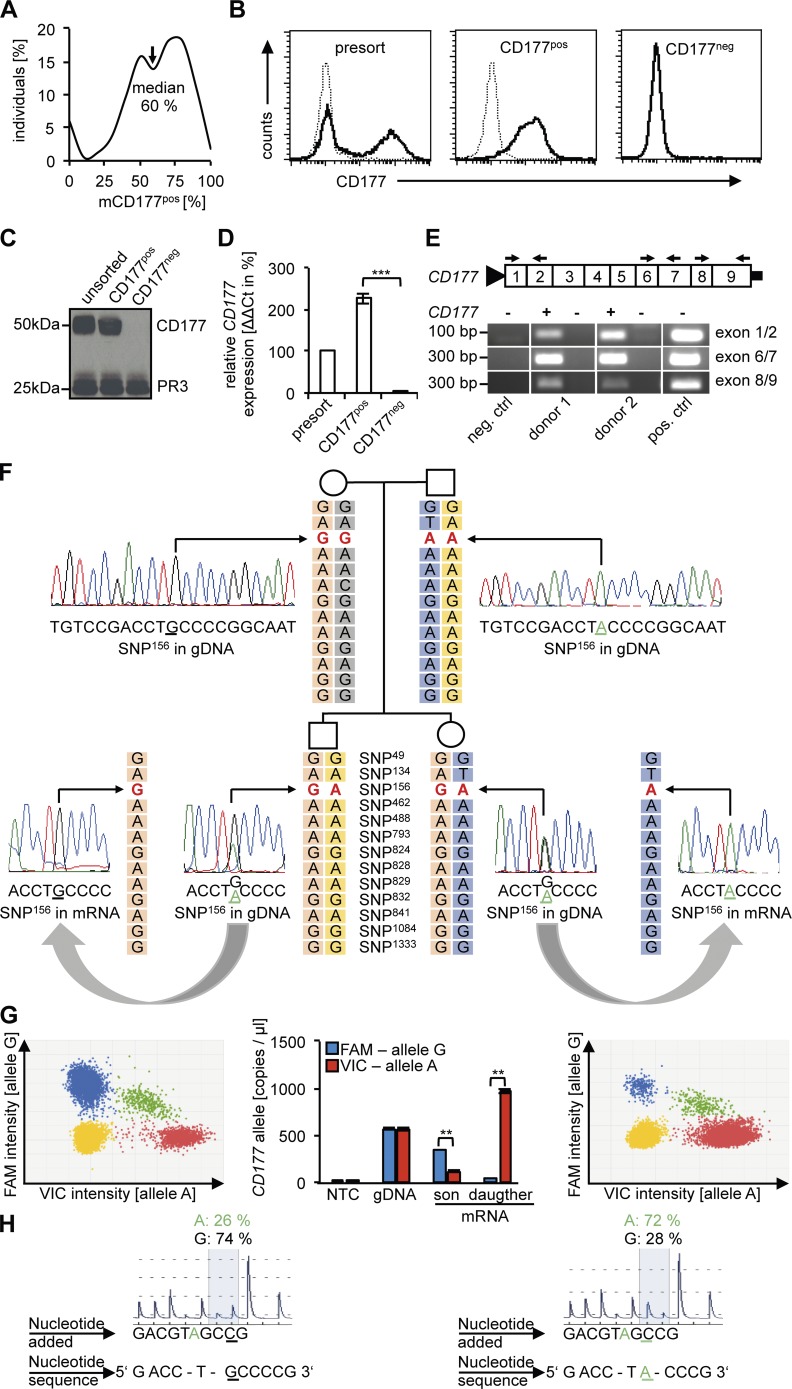
We first characterized the CD177 neutrophil phenotype in 165 normal individuals (66 men and 99 women). We assessed mCD177 on viable blood neutrophils by flow cytometry and found that 6% (10/165) of the individuals were completely CD177 deficient. In contrast, 94% (155/165) showed a bimodal pattern with varying percentages of mCD177pos neutrophils. The median size of the mCD177pos subset was 60% (Fig. 1 A). Our subsequent studies focused on the molecular basis for the CD177 bimodality that involves the vast majority of healthy individuals and has major disease implications for allo- and autoantigen presentation.
Figure 1.
CD177pos neutrophils from bimodal individuals contain CD177 protein and express only one CD177 allele either of paternal or maternal origin, whereas CD177neg neutrophils produce neither CD177 protein nor mRNA. (A) The percentage of CD177pos neutrophils by flow cytometry in 165 normal controls is shown as percentage of individuals per decile. (B) A typical flow cytometry experiment before (presort) and after magnetic cell sorting yielding CD177pos and CD177neg neutrophil subsets is given. Samples obtained as shown in B were subjected (C) to immunoblotting for CD177 and proteinase 3 (PR3) as a control protein (n = 4 different donors), and (D) to CD177 gene expression analysis by qRT-PCR. CD177 mRNA of presorted neutrophils was set at 100% (n = 3 different donors). (E) RT-PCR with different CD177 exon-spanning primers (black arrows) was performed in CD177pos and CD177neg neutrophils from two neutrophil donors to exclude truncated CD177 mRNAs (n = 2 different donors). (F) An example for monoallelic CD177 gene expression in a family with two offspring is depicted. Haplotype analysis showed an informative heterozygous GA SNP (in red, rs45571738) in the genomic DNA (gDNA) of both offspring whereas maternal gDNA showed homozygous GG and paternal gDNA AA. At mRNA, the son monoallelically expressed the maternal G and the daughter the paternal A allele as demonstrated by Sanger sequencing. The parental haplotype that is inherited by the offspring is boxed in light orange (maternal) and blue (paternal). (G) Digital PCR for the monoallelically expressed G allele for SNP156 in the son (left) and A allele in the daughter (right). Dot plots are depicted with each dot representing one of the 20,000 analyzed wells on the digital PCR chip. Blue dots represent the G (FAM-labeled) and red dots the A (VIC-labeled) allele, whereas yellow and green dots do not allow unequivocal allele identification. Quantitative assessment (middle) shows 75% G allele expression for the son and 96% for the daughter. gDNA was assessed as a control showing even distribution of both alleles (n = 2). (H) Pyrosequencing reveals MAE of the G allele (74%) for the son and the A allele (72%) for the daughter, respectively. Note that quantitative assays for assessment of MAE use a threshold of ∼70% for the monallelic call (Reinius and Sandberg, 2015). Data in D were displayed as mean± SEM and were analyzed using one-way ANOVA. ***, P < 0.001. Data in G were given as Poisson plus ratio with confidence intervals and were analyzed using Student’s t test. **, P < 0.01.
CD177 protein and mRNA expression is restricted to the CD177pos neutrophil subset
To gain insight into the molecular basis of the two distinct mCD177 phenotypes, we analyzed CD177 protein and mRNA expression in bimodal mCD177 individuals. For these studies, we isolated highly pure CD177neg and CD177pos blood neutrophils by magnetic cell sorting (Fig. 1 B). Immunoblotting showed CD177 protein (Fig. 1 C) and qRT-PCR detected full-length mRNA (Fig. 1 D) in CD177pos, but not in CD177neg neutrophils. To precisely check CD177 transcription in CD177neg neutrophils, we used three different primer pairs to exclude truncated CD177 mRNAs by qRT-PCR (Fig. 1 E). Thus, CD177 full-length mRNA is actively transcribed in CD177pos neutrophils, whereas both mRNA and protein are absent in the CD177neg subset.
CD177 is monoallelically expressed in neutrophils and neutrophil-differentiated CD34+ HSCs
The human CD177 gene resides on chromosome 19q13.31, 8.4-kb downstream to a CD177 pseudogene (Bettinotti et al., 2002; Caruccio et al., 2006; Li et al., 2015). We next investigated the CD177 gene locus to determine whether or not structural aberrations, such as frame shifts, deletions, or alternative splice sites explained the lack of CD177 mRNA in CD177neg neutrophils. We sequenced the CD177 gene in the two separated CD177 subsets (mCD177neg and mCD177pos neutrophils) and detected no DNA mutations or major substitutions in the nine exons, exon–intron transitions, 5′ UTR (−1,000 bp), and in 3′ UTR (+200 bp). In addition, a genome-wide single nucleotide polymorphisms (SNPs) analysis using an Affimetrix SNP array 6.0, revealed an almost identical SNP and copy number variation (CNV) concordance in both neutrophil subsets (five biological replicates, 99.9% concordance by Fisher’s exact test; Fig. S1).
Because no CD177 DNA sequence alterations or CNVs were found to explain the distinct CD177 mRNA and protein patterns, we performed a systematic haplotype analysis for the CD177 locus with 17 trios from 12 healthy families. We first sequenced the CD177 exons from parents and offspring. Long-range PCR was used to amplify the complete CD177 gene, thereby excluding the highly homologous pseudogene (exons 4–9) in reverse orientation on the antisense strand. Sequencing and SNP analysis of the PCR-amplicon (13 kb) detected all 9 CD177 exons confirming that the product was not contaminated by the CD177 pseudogene. Within the CD177 exons, we found 22 heterozygous SNPs in 15/17 offspring (Table S1) ranging from 1 to 6 in a given child. The remaining two offspring had noninformative homozygous SNPs. Only heterozygous SNPs analyzed in DNA and corresponding mRNA from neutrophils enabled us to distinguish the two parental CD177 alleles and to determine whether or not both alleles were actively transcribed. To our surprise, we found MAE in all 15 offspring with heterozygous SNPs. The parental origin of the actively transcribed allele was identified in 12/15 children, whereas three offspring had no informative SNP. 3/5 daughters expressed the maternal and 2/5 the paternal CD177 allele, whereas 5/7 sons expressed the maternal and 2/7 the paternal allele. These data indicate that the parental choice occurred independently of the offspring’s gender. Remarkably, siblings from two of the 12 families inherited the expressed CD177 allele from a different parent (Fig. 1 F and S2 A). One of these two families allowed sibling testing that confirmed the full-sibling status (Fig. S2 A). We complemented Sanger sequencing in the family shown in Fig. 1 F, with more quantitative approaches, namely digital PCR with informative SNP-specific probes and pyrosequencing, respectively. Results from both assays were >70% cut-off used for a monoallelic call (Reinius and Sandberg, 2015) and strengthen the observation of monoallelic CD177 expression (Fig. 1, G and H). The haplotypes of all remaining trios are shown in Fig. S1 B. We then studied MAE in a given individual over a 7-d period at days 0, 3, and 7, respectively. We observed that the allelic choice was stable and did not change over the observation period (Table 1). Together, these data indicate monoallelic CD177 expression in CD177pos neutrophils. The expressed CD177 transcript in a given individual was derived randomly either from the maternal or paternal allele with variation even within one family and the allelic choice remained stable over time.
Table 1. Monoallelic CD177 expression and the allelic choice over a 7-d period in five different individuals.
| Individual | Allele in gDNA | SNP position | Expressed CD177 allele | ||
|---|---|---|---|---|---|
| day 0 | day 3 | day 7 | |||
| 1 | G/C | SNP49 | G | G | G |
| 2 | C/A | SNP793 | A | A | A |
| C/G | SNP824 | G | G | G | |
| C/A | SNP828 | A | A | A | |
| T/A | SNP829 | A | A | A | |
| A/G | SNP832 | G | G | G | |
| G/A | SNP841 | A | A | A | |
| 3 | G/C | SNP49 | G | G | G |
| 4 | A/T | SNP134 | T | T | T |
| G/A | SNP156 | A | A | A | |
| C/A | SNP793 | A | A | A | |
| G/A | SNP1333 | A | A | A | |
| 5 | C/G | SNP824 | G | G | G |
| C/A | SNP828 | A | A | A | |
| T/A | SNP829 | A | A | A | |
| A/G | SNP832 | G | G | G | |
| G/A | SNP841 | A | A | A | |
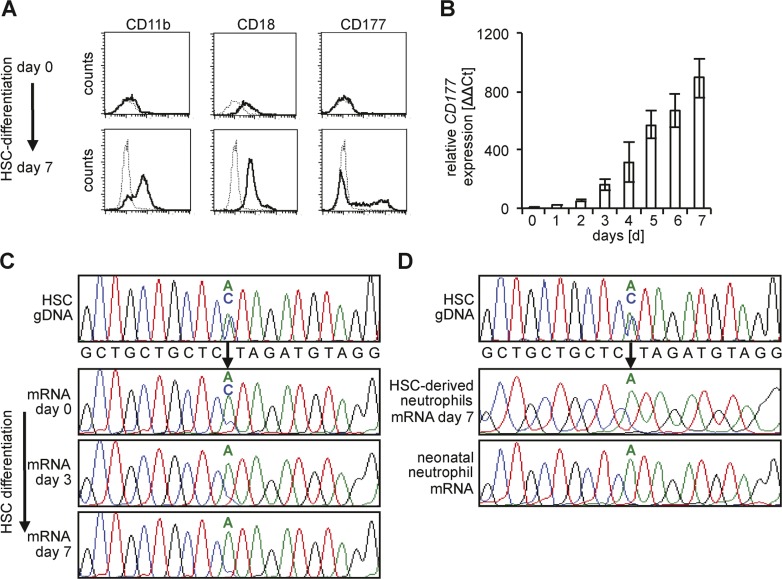
To elucidate the mechanism of CD177 MAE, we studied CD34+ hematopoietic stem cell (HSC)–derived neutrophils and neonatal neutrophils of the identical donor. We questioned whether or not CD177 MAE was already determined in the pluripotent HSCs. Thus, we selected umbilical cord blood donors with heterozygous DNA SNPs that allowed us to determine whether or not both alleles were expressed in the CD177 mRNA transcripts. G-CSF induced neutrophil differentiation and the HSC-derived neutrophils expectantly expressed the maturation markers CD11b and CD18 on the surface, showed bimodal CD177 populations (Fig. 2 A), and progressively transcribed CD177 mRNA over the 7-d observation period (Fig. 2 B). Analyzing heterozygous SNPs in CD177 DNA and mRNA by sequencing, we found that both CD177 alleles were transcribed in undifferentiated CD34+ HSCs. However, biallelic CD177 expression changed to MAE with neutrophil differentiation at day 7 (Fig. 2 C). When we compared neutrophil-differentiated HSCs with the mature neonatal neutrophils obtained from the same cord blood, we validated MAE and found that the identical CD177 allele was monoallelically transcribed in in vitro G-CSF differentiated and in in vivo natively differentiated neutrophils (Fig. 2 D). Our findings indicate that one of two CD177 alleles is silenced during neutrophil differentiation resulting in MAE in mature neutrophils. These observations support the notion that epigenetic mechanisms control the CD177 allele-silencing process.
Figure 2.
Monoallelic CD177 mRNA expression in neutrophil-differentiated CD34+ HSC and neonatal neutrophils. CD34+ HSC were differentiated over 7 d into neutrophils and (A) acquired neutrophil surface markers CD11b and CD18, together with a bimodal CD177 phenotype. A representative flow cytometry analysis from four independent differentiation experiments is shown. Solid lines represent staining with the specific antibodies and dotted lines represent the isotype control. (B) CD177 gene expression increased progressively in CD34+ HSCs during 7 d of neutrophil differentiation (qRT-PCR; n = 4 different CD34+ HSC donors). (C) Sanger sequencing of genomic DNA (gDNA) and mRNA from CD34+ HSC showed a heterozygous AC SNP793 (rs10425835) in gDNA and biallelic CD177 mRNA expression in undifferentiated cells at day 0. The C allele was silenced during neutrophil differentiation up to day 7 (n = 4 different CD34+ HSC donors). (D) Analysis of neutrophil-differentiated CD34+ HSCs and neonatal neutrophils from the same cord blood indicate an informative heterozygous AC SNP793 in the gDNA from CD34+ HSCs.
Epigenetic modifications control CD177 gene expression in a HeLa cell model
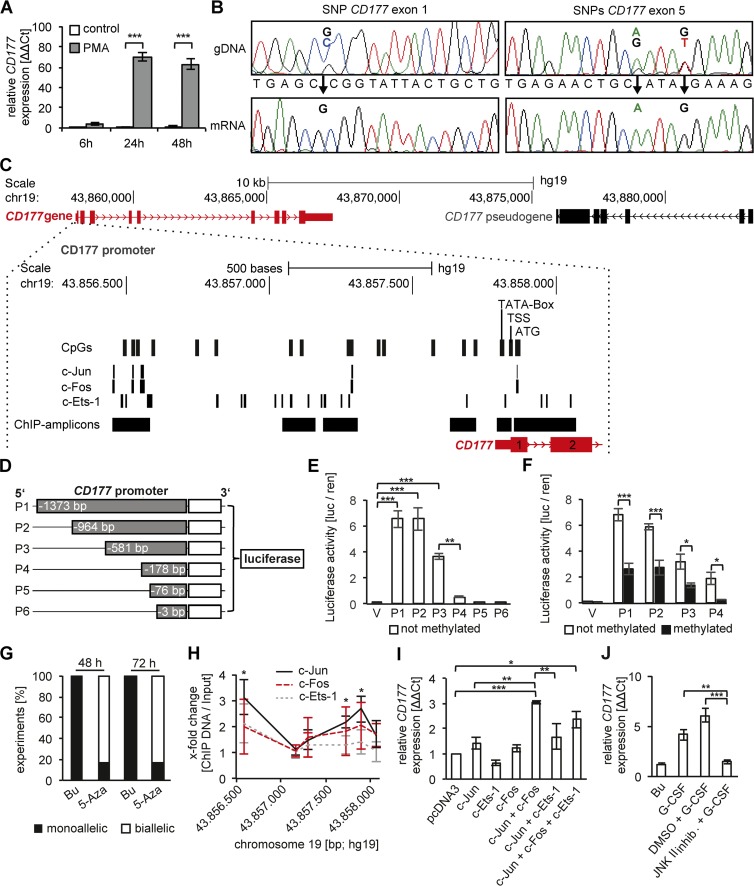
Neutrophils are short-lived and hard to transfect cells that are not suitable for genetic manipulation. Looking for alternative cell models, we successfully induced CD177 transcription in HeLa cells by phorbol myristate acetate (PMA) treatment (Fig. 3 A). A SNP analysis by CD177 exon and mRNA sequencing showed that PMA-stimulated HeLa cells expressed CD177 in a monoallelic fashion similar to CD177pos neutrophils (Fig. 3 B).
Figure 3.
CD177 gene expression, promoter analysis, DNA methylation, and transcription factor binding in a HeLa cell model. (A) CD177 gene expression was induced in PMA-treated HeLa cells as shown by qRT-PCR (n = 3 independent experiments). (B) CD177 haplotype analysis was performed by Sanger sequencing the genomic DNA (gDNA) and mRNA from HeLa cells (n = 6 independent experiments). Three heterozygous SNPs in the gDNA, one in exon 1 (SNP49 GC) and two in exon 5 (SNP652 AG and SNP656 GT), were found and established monoallelic CD177 gene expression. (C) A schematic overview of the CD177 gene (red), its promoter, and the pseudogene (black) is depicted. The CD177 promoter region is enlarged.16 CpG dinucleotides, the c-Jun, c-Fos, and c-Ets-1 TF binding sites (PROMO database), the TATA box, the transcription start site (TSS), the translation start (ATG), and PCR amplicons of the ChIPed DNA are indicated. (D) The CD177 promoter constructs P1 to P6 were cloned into the CpG-free luciferase vector. (E) Luciferase reporter assays were performed with the unmethylated CD177 promoter constructs P1 to P6 (n = 4 independent experiments). CD177 promoter activity was calculated from the firefly and Renilla luciferase signal. (F) Luciferase reporter assays were performed with P1 to P4 promoter constructs that were either left unmethylated (open bars) or were methylated (black bars) by incubation with methyltransferase (n = 7 independent experiments). (G) PMA-stimulated HeLa cells were treated with buffer (Bu) or 10 µM 5′-Aza-2-deoxycytidine (5-Aza) for 48 and 72 h, respectively (n = 6 independent experiments). Allele expression was analyzed using heterozygous CD177 SNPs (SNP49 rs45441892, SNP652 rs199668750, SNP656 rs200662237) in gDNA and the corresponding mRNA. Black indicates the percentage of experiments where monoallelic CD177 gene expression was detected and white indicates the percentage of experiments where biallelic CD177 allele expression was detected. (H) ChIP assays were performed using antibodies to c-Jun, c-Fos, and c-Ets-1 in PMA-stimulated HeLa cells. (I) Nonactivated HeLa cells were transfected with empty pcDNA3 vector, c-Jun, c-Fos, c-Ets-1, and combinations thereof (n = 3 independent experiments). CD177 gene expression was determined by qRT-PCR. (J) CD177 gene expression by qRT-PCR was assessed in human blood neutrophils stimulated with buffer control (Bu) or 100 ng/ml G-CSF for 90 min, respectively. When indicated, G-CSF–stimulated cells were pretreated with buffer control (DMSO) or 20 µM JNK II inhibitor (CAS 129–56-6) for 30 min, respectively (n = 3 independent experiments). Data in A, E, F, I, and J were displayed as mean ± SEM and were analyzed using one-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data in H were displayed ± SEM and were analyzed using Student’s t test. *, P < 0.05.
To characterize the CD177 locus, we dissected the molecular properties of the regulatory promoter region. We performed 5′ RACE and determined the transcription start site (TSS) of the CD177 gene at 29 bp upstream of the ATG translation start site. In the CD177 promoter, we detected a GC-rich element and 16 CpG dinucleotides. We next used the PROMO database (Messeguer et al., 2002) and rVISTA (Loots and Ovcharenko, 2004) to predict TF binding sites in the promoter region of interest. We found several conserved binding sites for the AP-1 family members c-Jun and c-Fos that were shown to be involved in PMA-activated gene transcription (Guerrini et al., 1996). In addition, c-Ets-1 binding sites were also predicted and of potential interest as a result of reported interactions with the AP-1 TF family (Kumari et al., 2015; Fig. 3 C).
First, we studied the effect of CpG-methylation on CD177 allele expression. We cloned the 1,373-bp full-length CD177 promoter with 16 CpGs, as well as shorter promoter regions into a CpG-dinucleotide lacking vector (Klug and Rehli, 2006). The constructs were used in HeLa luciferase reporter assays to study transcriptional activity (Fig. 3 D). The highest CD177 reporter activity was associated with the 1,373 and 964 bp constructs and a significant loss of reporter activity was observed by shortening the construct to 178 bp (Fig. 3 E). To address the effect of CpG-methylation on CD177 reporter activity, we treated constructs 1–4, each in the CpG-free vector, with methyltransferase. Methylation significantly reduced reporter activity (Fig. 3 F). These findings indicate that CpG-methylation in CD177 regulatory promoter sequence silences CD177 allele expression. In a different approach, we treated HeLa cells with the demethylation agent 5-Aza-2-deoxycytidin. By CD177 mRNA sequencing, we found that demethylation converted monoallelic into biallelic CD177 expression demonstrating that CpG-methylation caused indeed silencing of one CD177 allele (Fig. 3 G).
CpG-methylation affects TF binding to gene promoters. We performed ChIP for c-Jun, c-Fos, and c-Ets-1 on chromatin of PMA-stimulated HeLa cells. We found binding of all three TFs to CD177 promoter regions with c-Jun binding reaching statistical significance (Fig. 3 H). To test whether or not c-Jun, c-Fos, and c-Ets-1 control CD177 transcription, we transfected HeLa cells with the TFs and quantified CD177 mRNA by qRT-PCR. We observed that the combination of c-Jun and c-Fos caused strongly induced CD177 mRNA. In contrast, c-Ets-1 transfection alone did not affect CD177 transcription. Moreover, the combination of c-Ets-1 with the AP-1 TFs showed no additive effect (Fig. 3 I). Finally, pharmacologic c-Jun N-terminal kinase inhibition significantly reduced G-CSF–induced CD177 expression in human neutrophils suggesting that c-Jun controls CD177 expression also in neutrophils (Fig. 3 J).
CpG, histone methylation, and TF binding are differentially regulated in the two CD177 neutrophil subsets
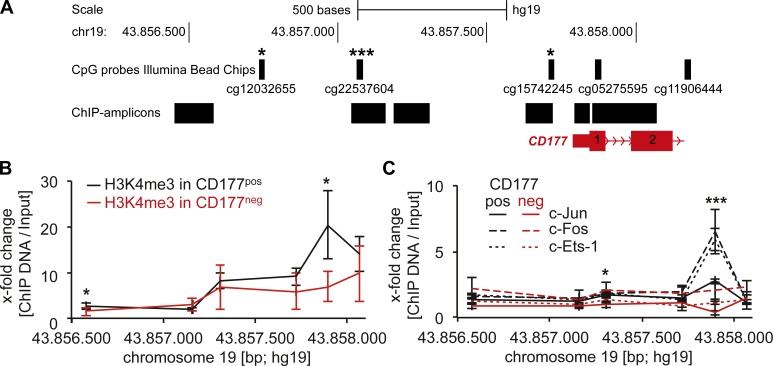
Our analysis established the importance of CpG methylation, and AP-1 TF binding in the CD177 promoter for CD177 gene expression in HeLa cells. We reasoned that if these epigenetic marks provided the basis for the bimodal CD177 expression pattern in neutrophils, they should be differentially regulated in the two separated CD177 neutrophil subsets. Thus, we addressed genome-wide DNA methylation of the two CD177 neutrophil subsets unbiased by Illumina Infinium Human Methylation 450 BeadChips. The probes on this chip covered three of the 16 CpG-methylation sites of the CD177 promoter and two further downstream in CD177 gene body. We found significantly increased CpG methylation in the CD177 promoter of CD177neg neutrophils (paired Student’s t test; Fig. 4 A and Table S2). H3K4me3 ChIP assay showed euchromatin in the CD177 promoter of CD177pos neutrophils, whereas nonenriched H3K4me3 indicated heterochromatin conformation in CD177neg cells (Fig. 4 B). ChIP analysis revealed significantly stronger binding of c-Jun, c-Fos and c-Ets-1 in the CD177 promoter of CD177pos neutrophils, whereas no signal was obtained in the CD177neg cells (Fig. 4 C). In summary, our data generated in HeLa cells and neutrophils showed that CpG and histone methylation regulated AP-1 TF-binding in the regulatory CD177 promoter region thereby controlling CD177 gene expression.
Figure 4.
Differential CpG and histone methylation, and AP-1 TF binding in the CD177 promoter of CD177neg and CD177pos neutrophil subsets. (A) Whole-genome DNA methylation analysis in CD177neg versus CD177pos neutrophils separated from CD177 bimodal individuals by magnetic cell sorting was performed using Illumina methylation chips. The localization of the 5 CpG probes (cg, CpG cluster number) within the CD177 promoter and in the CD177 gene that were present on the chip are indicated. Three of the five CpGs were significantly more methylated in the CD177neg neutrophils compared with the CD177pos subset (n = 6 different neutrophil donors). (B) ChIP assays were performed with the histone H3K4me3 mark for open chromatin in the CD177 promoter region of CD177pos (black lines) and CD177neg (red lines) neutrophils separated from a CD177 bimodal donor (n = 3 different neutrophil donors). (C) ChIP analysis for c-Jun, c-Fos, and c-Ets-1 binding was performed in CD177pos (black lines) and CD177neg (red lines) neutrophils separated from CD177 bimodal donors using specific antibodies to c-Jun, c-Fos, and c-Ets-1, respectively (n = 4 different neutrophil donors). Data in A, B, and C were analyzed using Student’s t test. *, P < 0.05; ***, P < 0.001; ±SEM is shown.
Discussion
Our study revealed several important new findings. First, our haplotype studies in family trios shows monoallelic CD177 gene expression with a novel pattern that differs from classical imprinting and random MAE. Using CD34+ HSCs, we observed that one CD177 allele was silenced during neutrophil differentiation. Second, we show that, in contrast to CD177pos neutrophils, the CD177neg subset transcribed neither full-length nor truncated CD177 mRNAs. Third, studies in HeLa cells supported the notion that CD177 allele silencing involved DNA methylation in the CD177 promoter region thereby regulating c-Jun and c-Fos binding and CD177 gene transcription. Finally, and importantly, these mechanisms were differentially executed in the two CD177 neutrophil subsets thereby explaining the bimodal CD177 phenotype.
Almost half a century ago, clinicians were made aware of HNA-2 (named NB1 at that time), because alloantibodies to this neutrophil antigen caused neonatal neutropenia (Lalezari et al., 1971). Later, researchers showed that the CD177 glycoprotein not only harbored the HNA-2 alloantigen, but also that CD177 presented the PR3 autoantigen on the neutrophil surface (Bauer et al., 2007; von Vietinghoff et al., 2007). Alloantibodies to HNA-2 cause neutropenia (Lalezari et al., 1971; Stroncek et al., 1993, 1994) and TRALI (Sachs et al., 2006) and autoantibodies to PR3 are associated with systemic small-vessel vasculitis (van der Woude et al., 1985; Lüdemann et al., 1990). Because these conditions are associated with substantial mortality, understanding CD177 expression is clinically highly relevant. Complete CD177 deficiency is of particular interest for HNA-2 alloantibody generation because nonexpressers become immunized with pregnancies and transfusions, and the resulting alloantibodies cause disease in HNA-2 expressing individuals. Investigators showed that the genetic basis for CD177 deficiency were SNP variants leading to truncated mRNAs with premature stop codons (Kissel et al., 2002). Additionally, either the homozygous nonsense SNP (829T) that also resulted in a premature stop codon or the heterozygous SNP (829AT) where the 829A allele harbored an additional base deletion resulted in a premature stop codon (Li et al., 2015). Very recently, DNA deep sequencing confirmed this homozygous nonsense SNP (in this study named g7497T) and found that the T allele was derived from gene conversion of the neighboring CD177 pseudogene with the CD177 gene (Wu et al., 2016). 6% of the individuals in our healthy Caucasian cohort were CD177 deficient, whereas 94% produced a variable percentage of CD177pos neutrophils with a median of 60%. This bimodal CD177 expression pattern is of particular interest in patients with ANCA vasculitis, where PR3 is a major autoantigen. Membrane-presented PR3 (mPR3) is recognized by PR3-ANCA that then activates the neutrophil, leading to vascular inflammation (Kettritz, 2012). We found previously that CD177 acts as a PR3-presenting receptor supporting high mPR3 levels (von Vietinghoff et al., 2007). The CD177–PR3 complex recruits additional transmembrane molecules into a larger signaling complex (Jerke et al., 2011). The bimodal CD177 expression pattern therefore results in CD177neg/mPR3low and CD177pos/mPR3high neutrophil subsets. Of clinical importance, a larger percentage of CD177pos/mPR3high neutrophils is found in ANCA vasculitis patients and is associated with worse clinical outcome (Witko-Sarsat et al., 1999; Rarok et al., 2002; Schreiber et al., 2005; Hu et al., 2009; Abdgawad et al., 2010). The molecular basis for the two distinct CD177 subsets, and therefore for the two mPR3 phenotypes is incompletely understood. We confirm data from Wolff et al. (2003), who showed that sorted CD177neg neutrophils from bimodal individuals lacked full-length CD177 mRNA by PCR. We extend these data showing that multiple qRT-PCRs with exon-spanning primers excluded truncated mRNAs. In contrast, Wu et al. (2016) detected full-length mRNA in CD177neg sorted cells of bimodal individuals by deep sequencing. The differences could be related to different sorting algorithms and that very small amounts of contaminating material from CD177pos cells were analyzed by highly sensitive deep sequencing. Moreover, the g7497 SNP in the CD177neg sorted neutrophils in the Wu study was not homozygous for the T allele therefore not explaining the lack of CD177 protein. For individuals with bimodal CD177 neutrophils in our study, we observed that those who were homozygous for g7497A (SNP829) and therefore harbored two alleles without potential stop codons showed monoallelic CD177 expression (Fig. S1). Another interesting phenomenon that was not focus of our study is the occurrence of two CD177pos peaks. Our observations are in agreement with the literature in that ∼5% of individuals show this CD177 pattern (Moritz et al., 2010; Meyerson et al., 2013; Wu et al., 2016). The molecular basis of this phenomenon is unclear and needs to be elucidated in future studies.
MAE can follow a parent-of-origin pattern, where either the maternal or the paternal allele is imprinted (silenced), and all cells of a specific tissue, or even of the entire organism, actively express exclusively the other parental allele (Reik and Walter, 2001; Peters, 2014). In contrast, silencing of one allele can occur in a random fashion first recognized for X chromosomes, where female cells inactivate either the maternal or paternal X (Lyon, 1986). Recently, it became clear that random MAE also occurs in ∼5–10% of the autosomal loci (Gimelbrant et al., 2007). Our haplotype analysis revealed monoallelic CD177 gene expression in blood neutrophils, where all neutrophils of a given individual had uniformly silenced either their maternal or paternal CD177 allele. This issue was best illustrated in two siblings from one family who both showed MAE, but had silenced the opposite parental allele. These findings suggest a novel MAE pattern not described before. It was not truly imprinting because we observed either maternal or paternal silenced alleles. The pattern is also not truly random MAE because this pattern implies that different clonal populations derived from a particular cell type express stochastically the maternal or the paternal, and some even both alleles (Deng et al., 2014). Based on Sanger sequencing, together with two quantitative assays that used thresholds from the literature of 70% for a monoallelic call (Reinius and Sandberg, 2015), we did not find a mixed expression of maternal and paternal alleles within the entire neutrophil population of a given individual. However, if the quantitative assays were interpreted as strongly biased CD177 transcriptional activity based on the inherited allele, two CD177pos neutrophil populations with markedly different CD177 mRNA and protein would result. In this case, one would possibly expect a more skewed peak of the CD177pos neutrophils in the flow cytometry experiments. Neutrophils comprise the largest leukocyte subset with the shortest lifespan and an estimated daily production rate of 1011 cells (Tak et al., 2013). Despite the rapid neutrophil turnover, we found that the allelic choice did not change over time when measured in a given individual. We observed that undifferentiated CD34+ HSC, the neutrophil progenitors, transcribed both CD177 alleles and that one allele was silenced during neutrophil differentiation. Thus, the decision as to which of the two parental alleles is silenced has to be made 1011 times per day lifelong. The fact that the choice of the activated allele remains constant during neutrophil differentiation and maturation suggests a strongly regulated silencing mechanism. CD177 is the first neutrophil gene shown to follow such a stringent and stable MAE. Interestingly, CD177 is an antigen receptor and the initial autosomal random MAE examples came from the antigen receptors of B and T cells (Pernis et al., 1965; Rajewsky, 1996). It was subsequently suggested that surface receptors are prone to autosomal MAE (Chess et al., 1994; Pereira et al., 2003; Gimelbrant et al., 2007). Biological effects could be mediated either through dosage differences or by expressing only one out of two functionally nonequivalent parental alleles. Conceptually, these effects would increase phenotypic variation and cellular diversity, but also harbor a risk when only a dysfunctional allele is expressed (Reinius and Sandberg, 2015). It is conceivable that MAE of CD177 has clinical relevance. In ANCA autoimmune vasculitis, heterozygous CD177 SNPs or mutations and the choice of which allele is expressed could affect PR3 binding and thereby shape antigenicity and binding of PR3-ANCA. In fact, genes with MAE are particularly prone to mutations (Savova et al., 2016). Heterozygous CD177 mutations and the allelic choice could also lead to incomplete penetrance of disease phenotypes. Similar mechanisms could be at work for the presentation of CD177 alloantigen epitopes.
After having excluded DNA sequence abnormalities and CNV, epigenetic modifications became candidates for allelic CD177 gene silencing (Reik and Walter, 2001; Eckersley-Maslin and Spector, 2014; Reinius and Sandberg, 2015; Gendrel et al., 2016). Neutrophils rapidly undergo apoptosis in vivo and in culture, preventing epigenetic studies in these short-lived primary cells (Fig. S2 B). To identify these silencing mechanisms, we took advantage from the observation that activated HeLa cells expressed CD177 in a monoallelic manner. We characterized the promoter region upstream of the CD177 TSS that was important for gene expression and contained 16 CpGs. Our data indicate that in vitro DNA methylation at CpG dinucleotides in the promoter controlled CD177 gene expression by c-Jun and c-Fos. Interestingly, pharmacologic HeLa cell demethylation, certainly not exclusively acting on the CD177 promoter, activated biallelic CD177 expression providing additional evidence for a role of DNA methylation in gene and single allele silencing. These data support the contention that DNA methylation at CpGs in the CD177 promoter silenced two alleles in CD177neg and one allele in the CD177pos neutrophil subset, respectively. Most importantly, these epigenetic control mechanisms were differentially regulated in the two CD177 neutrophil subsets. The promoter of the CD177neg subset showed significantly increased CpG and histone methylation and abrogated AP-1 TF binding. In contrast, the CD177pos neutrophil subset actively transcribed CD177 in a monoallelic fashion.
In summary, we have identified DNA, histone methylation and AP-1 TF binding that control CD177 gene expression and explain the two distinct CD177 neutrophil subsets in bimodal individuals that are of clinical significance. In addition, we describe a novel MAE pattern for CD177 that differs from classical imprinting and random MAE in its stable choice of either parental allele.
Materials and methods
Neutrophil isolation
Blood neutrophils from healthy human donors were obtained after due approval by IRB (Institutional Charité University Ethics Committee, EA1/277/11 and EA3/010/06) and after written, informed consent, and isolated as described previously (von Vietinghoff et al., 2007).
HeLa cell culture, stimulation, and transfection
HeLa cells were cultured in DMEM supplemented with 10% FCS, 100 U/ml penicillin, and 100 µg/ml streptomycin and treated with 100 ng/ml PMA and 10 µM 5′Aza-2-deoxycytidine (Sigma-Aldrich) as indicated. HeLa cells were transiently transfected with 67 ng c-Jun, c-Fos, and c-Ets-1 plasmids, or empty pcDNA3 vector control, respectively. All cells received a total of 2 µg plasmid using Fugene HD (Roche) according to the manufacturer’s instructions.
Flow cytometry and magnetic neutrophil sorting
CD177 membrane expression was assessed by flow cytometry of either isolated neutrophils or in 100 µl whole blood after hypotonic red blood cell lysis using a FITC-labeled anti-CD177 antibody (clone MEM166; Biozol) as described before (von Vietinghoff et al., 2007). CD177 neutrophil subsets were purified using anti-CD177 abs (clone MEM166; Exbio), anti-CD66b (Dako), anti-CD9 abs (Beckman Coulter), rat anti–mouse IgG microbeads (Miltenyi Biotech), and LD columns (Miltenyi Biotech) according to the manufacturer's instructions. Highly pure neutrophils were prepared by density gradient purification, followed by depletion of CD9-positive eosinophils. CD177pos neutrophils were thereafter positively selected for CD177. CD177neg neutrophils were positively selected from this CD177-depleted preparation for CD66b. Mature neutrophil-differentiated CD34+ HSC were purified by FACS using a directly labeled FITC anti-CD16 antibody (BioLegend) and an anti-CD177 antibody (Santa Cruz Biotechnology). When indicated neutrophils were stimulated with 100 µg/ml human granulocyte colony-stimulating factor (G-CSF; PeproTech) or in combination with 20 µM c-Jun N-terminal kinase inhibitor (JNK Inhibitor II, CAS 129–56-6; Merck). 10 µM 5′Aza-2-deoxycytidine was performed as described for HeLa cells. Apoptosis was measured with Annexin V staining (BD) according to the manufacturer’s manual and using flow cytometry.
Sibling testing
DNA testing was performed by an accredited laboratory for forensic genetics (Institut für Medizinische Molekulare Diagnostik und Forensische Genetik, Berlin, Germany) using the Power Plex ESX-17 system (Promega).
Differentiation of CD34+ stem cells from umbilical cord blood into neutrophils
CD34+ stem cells were isolated from cord blood, expanded, and differentiated as described previously (von Vietinghoff et al., 2007). Neonatal neutrophils were isolated from cord blood by density gradient centrifugation as described for blood neutrophils.
Isolation of DNA, mRNA, cDNA generation, and sequencing
Total RNA was extracted using QIAzol and the RNeasy Purification kit (QIAGEN). RNA was treated by deoxyribonuclease I (QIAGEN). cDNA synthesis was performed using hexanucleotide primers and Superscript III RT following the manufacturers protocol (Thermo Fisher Scientific). Genomic DNA was isolated using 5 M sodium perchlorate solution, DNA extraction by chloroform and DNA precipitation by isopropanol. For methylation array analysis genomic DNA was isolated using QIAGEN genomic DNA Isolation kit (QIAGEN). CD177 long templates were amplified by PCR using 250 ng genomic DNA, specific CD177-43.857.788-F and CD177-43.870.898-R primers and Elongase (Thermo Fisher Scientific) in a total volume of 25 µl. PCR products were purified with S.N.A.P. Purification kit (Life Technologies GmbH). CD177 short products were amplified from 20 ng of CD177 long templates or 50–100 ng cDNA with specific primers (Table S3) using Advantage-GC 2 PCR kit (Takara Bio Inc.). CD177 PCR products were sequenced according to Sanger method using the Big Dye Terminator Cycle Sequencing kit v1.1 and analyzed on a 3130xl Genetic Analyzer (both from Applied Biosystems). Quantitative allele analysis by pyrosequencing was performed by the bioanalytical services laboratory (Varionostic GmbH) using four different SNP assays with template-specific PCR and sequencing primers (Table S3) and the Q24 System (QIAGEN). All SNP analyses were generated by PyroMark Q24 software.
Quantitative RT-PCR
For quantitative RT-PCR (qRT-PCR) TaqMan technology (Applied Biosystems) was used with oligonucleotides and probes for human CD177 and human 18S (Table S3), and TaqMan Fast Universal PCR Master Mix (Applied Biosystems). Each sample was measured in triplicate and expression levels were normalized to 18S expression. Quantification was performed using an Applied Biosystems 7500 Sequence detector and data were analyzed using SDS 7500 software and ΔΔCt comparative analysis as described by Applied Biosystems.
Digital PCR
Quantification of allele-specific mRNA expression was performed using the QuantStudio 3D Digital PCR System (Thermo Fisher Scientific). SNP49 (rs45441892) was detected by the human TaqMan SNP Genotyping Assay C_27850890_10 and SNP156 (rs45571738) by the human TaqMan SNP Genotyping Assay C_27837236_10, respectively. Each reaction mix for one QuantStudio 3D Digital PCR chip v2 consisted of QuantStudio 3D Digital PCR Master Mix v2 (Thermo Fisher Scientific), specific human TaqMan SNP Genotyping Assay (primer/probe mix), template cDNA or gDNA with final concentration at 3.45 ng/µl and PCR-grade water in a final volume of 14.5 µl. PCR conditions were as follows: stage 1 enzyme activation, 96°C for 10 min; stage 2 annealing/extension and new denaturation at 54°C (SNP49) or 58°C (SNP156) for 2 min and 98°C for 30 s (39 cycles); stage 3 annealing/extension 60°C for 2 min and storage at 10°C. Each sample was analyzed using a separate 20,000 times compartmented QuantStudio 3D Digital PCR chip v2 and the AnalysisSuite Software (Thermo Fisher Scientific). The expressed allele amounts were presented as copy numbers per microliter PCR mixture.
DNA and cDNA sequencing
DNA and cDNA sequencing was performed from the CD177 PCR products treated with shrimp alkaline phosphatase (SAP; Affymetrix) and exonuclease I (NEB). After enzyme inactivation, the PCR products were subjected to DNA sequencing using BigDye Terminator v1.1 Cycle Sequencing kit from Applied Biosystems. Purification of the samples was done with ZR DNA Sequencing Clean-up kit (ZYMO Research). DNA sequencing was performed with a 16-capillary sequencer 3130xl Genetic Analyzer (Applied Biosystems).
5′ RACE PCR
5′ RACE PCR was performed from neutrophil total RNA using QIAzol (QIAGEN), the GeneRacer kit, specific primers (Table S3), and reverse transcribed using Superscript III RT (Life Technologies GmbH). PCR fragments were cloned into a pDrive vector (QIAGEN) and sequenced as described in the previous section.
DNA methylation analysis
Genomic DNA was purified by using QIAGEN genomic DNA Isolation kit (QIAGEN). Genome-wide methylation analysis was performed by LIFE & BRAIN GmbH using an Infinium Human Methylation 450 BeadChip (Illumina).
SDS-PAGE, Coomassie staining, and immunoblot analysis
Cell lysis, SDS-PAGE, and immunoblots were performed as previously described (Jerke et al., 2015). Immunoblots were developed with a monoclonal rabbit anti-PR3 antibody (clone 4A5; Wieslab), an anti-CD177 mAb (clone MEM166, Exbio), or a polyclonal anti-CD177 (R&D Systems) as indicated and visualized by enhanced chemiluminescence (Thermo Fisher Scientific).
Reporter assays
104 cells per well were seeded into 96-well plates 24 h before transfection. HeLa cells were transiently transfected with the indicated CD177 promoter plasmids, as well as with empty CpG-free methylation vector in equal molarities (55.14 amol). The CD177 promoter luciferase constructs were transfected with Fugene HD according to the manufacturer’s recommendations (Roche). For controlling the transfection efficiency, we transfected each well with 8 ng pRL-TK (Promega). Cell lysates were analyzed using the Dual-Luciferase Reporter Assay System (Promega) and the Infinite M200 PRO Multimode Microplate Reader from Tecan. To normalize reporter data, a quotient of the luciferase and the Renilla signal was calculated for each well.
Chromatin immunoprecipitation
3 × 107 neutrophils or 3 × 107 PMA-stimulated HeLa cells were treated with 0.4% formaldehyde to cross-link proteins and DNA. Chromatin was sonicated with Bioruptor Plus sonication device (Diagenode). 4 × 107 magnetic Dynabeads M-280 sheep anti–rabbit IgG (Thermo Fisher Scientific) were overnight preincubated with 5 µg anti-H3K4me3, 10 µg anti–c-Jun, 10 µg anti-c-Fos, 10 µg anti–c-Ets-1 or 5 µg/10 µg anti-IgG control ChIP-tested antibodies (Abcam). After washing of Dynabead-antibody samples, 50 µg chromatin was added to each sample and incubated overnight. 1 and 10% of the chromatin samples served as input. Antibody-Dynabeads bound DNA was eluted in 1% SDS buffer and incubated with RNase A (Thermo Fisher Scientific) and Proteinase K (NEB). DNA was isolated by phenol/chloroform extraction and analyzed by quantitative RT-PCR using Fast SYBR Green Master Mix and specific and control primers (Table S3) on an ABI Prism 7500 Fast System (Applied Biosystems). The relative signals of the specific PCR product was expressed as percent of input chromatin using raw Ct values and was normalized for the control IgG.
Statistical analysis
Results are given as mean ± SEM. Comparisons between multiple groups were done using ANOVA and appropriate post-hoc tests or differences between experimental groups were analyzed by Student’s t test. Differences were considered significant if P < 0.05. The dPCR results were given as poisson plus ratio with confidence intervals according to the AnalysisSuite Software (Thermo Fisher Scientific).
Online supplemental material
Fig. S1 shows the genome-wide single nucleotide polymorphisms (SNPs) analysis of sorted CD177neg and CD177pos neutrophil subsets and the haplotype analysis of the CD177 gene. Fig. S2 presents haplotype analyses and sibling testing of an informative family with respect to the allelic choice of the CD177 MAE as well as 5-Aza-2-deoxycytidin (5-Aza) treatment of primary human neutrophils. Table S1 lists SNPs identified in the CD177 exons by sequencing. Table S2 shows probes in the CD177 promoter and gene body that are covered by the Methylation450 Bead Chip. Table S3 lists all oligonucleotides for PCR assays.
Supplementary Material
Acknowledgments
We thank Prof. Dr. Michael Rehli (Department of Internal Medicine III, University Hospital Regensburg, F.-J.-Strauss Allee 11, D-93053 Regensburg) for the CpG free luciferase reporter vector.
This work was supported by DFG grant KE 576/8-1 and an ECRC grant.
The authors declare no competing financial interests.
Author contributions: R. Kettritz, S. Bähring, P.G. Maass, and C. Eulenberg-Gustavus designed experiments and analyzed data. C. Eulenberg-Gustavus performed experiments and P.G. Maass and S. Bähring assisted with experiments. R. Kettritz and C. Eulenberg-Gustavus wrote the manuscript and R. Kettritz funded the study. F.C. Luft, P.G. Maass, S. Bähring, R. Kettritz, and C. Eulenberg-Gustavus revised the manuscript.
Footnotes
Abbreviations used:
- ANCA
- anti-neutrophil cytoplasmic autoantibodies
- HSC
- hematopoietic stem cell
- MAE
- monoallelic expression
- SNP
- single nucleotide polymorphism
- TF
- transcription factor
References
- Abdgawad M., Gunnarsson L., Bengtsson A.A., Geborek P., Nilsson L., Segelmark M., and Hellmark T.. 2010. Elevated neutrophil membrane expression of proteinase 3 is dependent upon CD177 expression. Clin. Exp. Immunol. 161:89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow D.P., and Bartolomei M.S.. 2014. Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 6:5–10. 10.1101/cshperspect.a018382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S., Abdgawad M., Gunnarsson L., Segelmark M., Tapper H., and Hellmark T.. 2007. Proteinase 3 and CD177 are expressed on the plasma membrane of the same subset of neutrophils. J. Leukoc. Biol. 81:458–464. 10.1189/jlb.0806514 [DOI] [PubMed] [Google Scholar]
- Bayat B., Werth S., Sachs U.J.H., Newman D.K., Newman P.J., and Santoso S.. 2010. Neutrophil transmigration mediated by the neutrophil-specific antigen CD177 is influenced by the endothelial S536N dimorphism of platelet endothelial cell adhesion molecule-1. J. Immunol. 184:3889–3896. 10.4049/jimmunol.0903136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettinotti M.P., Olsen A., and Stroncek D.. 2002. The use of bioinformatics to identify the genomic structure of the gene that encodes neutrophil antigen NB1, CD177. Clin. Immunol. 102:138–144. 10.1006/clim.2001.5154 [DOI] [PubMed] [Google Scholar]
- Bux J., Becker F., Seeger W., Kilpatrick D., Chapman J., and Waters A.. 1996. Transfusion-related acute lung injury due to HLA-A2-specific antibodies in recipient and NB1-specific antibodies in donor blood. Br. J. Haematol. 93:707–713. 10.1046/j.1365-2141.1996.d01-1703.x [DOI] [PubMed] [Google Scholar]
- Caruccio L., Bettinotti M., Director-Myska A.E., Arthur D.C., and Stroncek D.. 2006. The gene overexpressed in polycythemia rubra vera, PRV-1, and the gene encoding a neutrophil alloantigen, NB1, are alleles of a single gene, CD177, in chromosome band 19q13.31. Transfusion. 46:441–447. 10.1111/j.1537-2995.2006.00741.x [DOI] [PubMed] [Google Scholar]
- Chess A., Simon I., Cedar H., and Axel R.. 1994. Allelic inactivation regulates olfactory receptor gene expression. Cell. 78:823–834. 10.1016/S0092-8674(94)90562-2 [DOI] [PubMed] [Google Scholar]
- Deng Q., Ramsköld D., Reinius B., and Sandberg R.. 2014. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 343:193–196. 10.1126/science.1245316 [DOI] [PubMed] [Google Scholar]
- Eckersley-Maslin M.A., and Spector D.L.. 2014. Random monoallelic expression: regulating gene expression one allele at a time. Trends Genet. 30:237–244. 10.1016/j.tig.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A.V., Marion-Poll L., Katoh K., and Heard E.. 2016. Random monoallelic expression of genes on autosomes: Parallels with X-chromosome inactivation. Semin. Cell Dev. Biol. 56:100–110. 10.1016/j.semcdb.2016.04.007 [DOI] [PubMed] [Google Scholar]
- Gimelbrant A., Hutchinson J.N., Thompson B.R., and Chess A.. 2007. Widespread monoallelic expression on human autosomes. Science. 318:1136–1140. 10.1126/science.1148910 [DOI] [PubMed] [Google Scholar]
- Goldschmeding R., van Dalen C.M., Faber N., Calafat J., Huizinga T.W., van der Schoot C.E., Clement L.T., and von dem Borne A.E.. 1992. Further characterization of the NB 1 antigen as a variably expressed 56-62 kD GPI-linked glycoprotein of plasma membranes and specific granules of neutrophils. Br. J. Haematol. 81:336–345. 10.1111/j.1365-2141.1992.tb08237.x [DOI] [PubMed] [Google Scholar]
- Guerrini L., Casalino L., Corti A., and Blasi F.. 1996. NF-kappa B-mediated regulation of urokinase gene expression by PMA and TNF-alpha in human A549 cells. FEBS Lett. 393:69–73. 10.1016/0014-5793(96)00854-X [DOI] [PubMed] [Google Scholar]
- Hu N., Westra J., Huitema M.G., Bijl M., Brouwer E., Stegeman C.A., Heeringa P., Limburg P.C., and Kallenberg C.G.. 2009. Coexpression of CD177 and membrane proteinase 3 on neutrophils in antineutrophil cytoplasmic autoantibody-associated systemic vasculitis: anti-proteinase 3-mediated neutrophil activation is independent of the role of CD177-expressing neutrophils. Arthritis Rheum. 60:1548–1557. 10.1002/art.24442 [DOI] [PubMed] [Google Scholar]
- Jerke U., Rolle S., Dittmar G., Bayat B., Santoso S., Sporbert A., Luft F., and Kettritz R.. 2011. Complement receptor Mac-1 is an adaptor for NB1 (CD177)-mediated PR3-ANCA neutrophil activation. J. Biol. Chem. 286:7070–7081. 10.1074/jbc.M110.171256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerke U., Hernandez D.P., Beaudette P., Korkmaz B., Dittmar G., and Kettritz R.. 2015. Neutrophil serine proteases exert proteolytic activity on endothelial cells. Kidney Int. 88:764–775. 10.1038/ki.2015.159 [DOI] [PubMed] [Google Scholar]
- Kettritz R. 2012. How anti-neutrophil cytoplasmic autoantibodies activate neutrophils. Clin. Exp. Immunol. 169:220–228. 10.1111/j.1365-2249.2012.04615.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissel K., Santoso S., Hofmann C., Stroncek D., and Bux J.. 2001. Molecular basis of the neutrophil glycoprotein NB1 (CD177) involved in the pathogenesis of immune neutropenias and transfusion reactions. Eur. J. Immunol. 31:1301–1309. [DOI] [PubMed] [Google Scholar]
- Kissel K., Scheffler S., Kerowgan M., and Bux J.. 2002. Molecular basis of NB1 (HNA-2a, CD177) deficiency. Blood. 99:4231–4233. 10.1182/blood.V99.11.4231 [DOI] [PubMed] [Google Scholar]
- Klug M., and Rehli M.. 2006. Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector. Epigenetics. 1:127–130. 10.4161/epi.1.3.3327 [DOI] [PubMed] [Google Scholar]
- Kuckleburg C.J., Tilkens S.B., Santoso S., and Newman P.J.. 2012. Proteinase 3 contributes to transendothelial migration of NB1-positive neutrophils. J. Immunol. 188:2419–2426. 10.4049/jimmunol.1102540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S., Saradhi M., Rana M., Chatterjee S., Aumercier M., Mukhopadhyay G., and Tyagi R.K.. 2015. Pregnane and Xenobiotic Receptor gene expression in liver cells is modulated by Ets-1 in synchrony with transcription factors Pax5, LEF-1 and c-Jun. Exp. Cell Res. 330:398–411. 10.1016/j.yexcr.2014.09.020 [DOI] [PubMed] [Google Scholar]
- Lalezari P., Murphy G.B., and Allen F.H. Jr. 1971. NB1, a new neutrophil-specific antigen involved in the pathogenesis of neonatal neutropenia. J. Clin. Invest. 50:1108–1115. 10.1172/JCI106582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Mair D.C., Schuller R.M., Li L., and Wu J.. 2015. Genetic mechanism of human neutrophil antigen 2 deficiency and expression variations. PLoS Genet. 11:e1005255 10.1371/journal.pgen.1005255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loots G.G., and Ovcharenko I.. 2004. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 32(Web Server):W217-21 10.1093/nar/gkh383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüdemann J., Utecht B., and Gross W.L.. 1990. Anti-neutrophil cytoplasm antibodies in Wegener’s granulomatosis recognize an elastinolytic enzyme. J. Exp. Med. 171:357–362. 10.1084/jem.171.1.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M.F. 1986. X chromosomes and dosage compensation. Nature. 320:313 10.1038/320313b0 [DOI] [PubMed] [Google Scholar]
- Matsuo K., Lin A., Procter J.L., Clement L., and Stroncek D.. 2000. Variations in the expression of granulocyte antigen NB1. Transfusion. 40:654–662. 10.1046/j.1537-2995.2000.40060654.x [DOI] [PubMed] [Google Scholar]
- Messeguer X., Escudero R., Farré D., Núñez O., Martínez J., and Albà M.M.. 2002. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 18:333–334. 10.1093/bioinformatics/18.2.333 [DOI] [PubMed] [Google Scholar]
- Meyerson H.J., Osei E., Schweitzer K., Blidaru G., Edinger A., and Balog A.. 2013. CD177 expression on neutrophils: in search of a clonal assay for myeloid neoplasia by flow cytometry. Am. J. Clin. Pathol. 140:658–669. 10.1309/AJCPDFBEBQZW1OI7 [DOI] [PubMed] [Google Scholar]
- Moritz E., Chiba A.K., Kimura E.Y., Albuquerque D., Guirão F.P., Yamamoto M., Costa F.F., and Bordin J.O.. 2010. Molecular studies reveal that A134T, G156A and G1333A SNPs in the CD177 gene are associated with atypical expression of human neutrophil antigen-2. Vox Sang. 98:160–166. 10.1111/j.1423-0410.2009.01233.x [DOI] [PubMed] [Google Scholar]
- Pereira J.P., Girard R., Chaby R., Cumano A., and Vieira P.. 2003. Monoallelic expression of the murine gene encoding Toll-like receptor 4. Nat. Immunol. 4:464–470. 10.1038/ni917 [DOI] [PubMed] [Google Scholar]
- Pernis B., Chiappino G., Kelus A.S., and Gell P.G.. 1965. Cellular localization of immunoglobulins with different allotypic specificities in rabbit lymphoid tissues. J. Exp. Med. 122:853–876. 10.1084/jem.122.5.853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J. 2014. The role of genomic imprinting in biology and disease: an expanding view. Nat. Rev. Genet. 15:517–530. 10.1038/nrg3766 [DOI] [PubMed] [Google Scholar]
- Rajewsky K. 1996. Clonal selection and learning in the antibody system. Nature. 381:751–758. 10.1038/381751a0 [DOI] [PubMed] [Google Scholar]
- Rarok A.A., Stegeman C.A., Limburg P.C., and Kallenberg C.G.M.. 2002. Neutrophil membrane expression of proteinase 3 (PR3) is related to relapse in PR3-ANCA-associated vasculitis. J. Am. Soc. Nephrol. 13:2232–2238. 10.1097/01.ASN.0000028642.26222.00 [DOI] [PubMed] [Google Scholar]
- Reik W., and Walter J.. 2001. Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2:21–32. 10.1038/35047554 [DOI] [PubMed] [Google Scholar]
- Reinius B., and Sandberg R.. 2015. Random monoallelic expression of autosomal genes: stochastic transcription and allele-level regulation. Nat. Rev. Genet. 16:653–664. 10.1038/nrg3888 [DOI] [PubMed] [Google Scholar]
- Rørvig S., Østergaard O., Heegaard N.H., and Borregaard N.. 2013. Proteome profiling of human neutrophil granule subsets, secretory vesicles, and cell membrane: correlation with transcriptome profiling of neutrophil precursors. J. Leukoc. Biol. 94:711–721. 10.1189/jlb.1212619 [DOI] [PubMed] [Google Scholar]
- Sachs U.J., Hattar K., Weissmann N., Bohle R.M., Weiss T., Sibelius U., and Bux J.. 2006. Antibody-induced neutrophil activation as a trigger for transfusion-related acute lung injury in an ex vivo rat lung model. Blood. 107:1217–1219. 10.1182/blood-2005-04-1744 [DOI] [PubMed] [Google Scholar]
- Sachs U.J., Andrei-Selmer C.L., Maniar A., Weiss T., Paddock C., Orlova V.V., Choi E.Y., Newman P.J., Preissner K.T., Chavakis T., and Santoso S.. 2007. The neutrophil-specific antigen CD177 is a counter-receptor for platelet endothelial cell adhesion molecule-1 (CD31). J. Biol. Chem. 282:23603–23612. 10.1074/jbc.M701120200 [DOI] [PubMed] [Google Scholar]
- Savova V., Chun S., Sohail M., McCole R.B., Witwicki R., Gai L., Lenz T.L., Wu C.T., Sunyaev S.R., and Gimelbrant A.A.. 2016. Genes with monoallelic expression contribute disproportionately to genetic diversity in humans. Nat. Genet. 48:231–237. 10.1038/ng.3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber A., Otto B., Ju X., Zenke M., Goebel U., Luft F.C., and Kettritz R.. 2005. Membrane proteinase 3 expression in patients with Wegener’s granulomatosis and in human hematopoietic stem cell-derived neutrophils. J. Am. Soc. Nephrol. 16:2216–2224. 10.1681/ASN.2004070609 [DOI] [PubMed] [Google Scholar]
- Stroncek D.F. 2007. Neutrophil-specific antigen HNA-2a, NB1 glycoprotein, and CD177. Curr. Opin. Hematol. 14:688–693. 10.1097/MOH.0b013e3282efed9e [DOI] [PubMed] [Google Scholar]
- Stroncek D.F., Shapiro R.S., Filipovich A.H., Plachta L.B., and Clay M.E.. 1993. Prolonged neutropenia resulting from antibodies to neutrophil-specific antigen NB1 following marrow transplantation. Transfusion. 33:158–163. 10.1046/j.1537-2995.1993.33293158050.x [DOI] [PubMed] [Google Scholar]
- Stroncek D.F., Herr G.P., Maguire R.B., Eiber G., and Clement L.T.. 1994. Characterization of the neutrophil molecules identified by quinine-dependent antibodies from two patients. Transfusion. 34:980–985. 10.1046/j.1537-2995.1994.341195065037.x [DOI] [PubMed] [Google Scholar]
- Tak T., Tesselaar K., Pillay J., Borghans J.A., and Koenderman L.. 2013. What’s your age again? Determination of human neutrophil half-lives revisited. J. Leukoc. Biol. 94:595–601. 10.1189/jlb.1112571 [DOI] [PubMed] [Google Scholar]
- van der Woude F.J., Rasmussen N., Lobatto S., Wiik A., Permin H., van Es L.A., van der Giessen M., van der Hem G.K., and The T.H.. 1985. Autoantibodies against neutrophils and monocytes: tool for diagnosis and marker of disease activity in Wegener’s granulomatosis. Lancet. 1:425–429. 10.1016/S0140-6736(85)91147-X [DOI] [PubMed] [Google Scholar]
- von Vietinghoff S., Tunnemann G., Eulenberg C., Wellner M., Cristina Cardoso M., Luft F.C., and Kettritz R.. 2007. NB1 mediates surface expression of the ANCA antigen proteinase 3 on human neutrophils. Blood. 109:4487–4493. 10.1182/blood-2006-10-055327 [DOI] [PubMed] [Google Scholar]
- von Vietinghoff S., Eulenberg C., Wellner M., Luft F.C., and Kettritz R.. 2008. Neutrophil surface presentation of the anti-neutrophil cytoplasmic antibody-antigen proteinase 3 depends on N-terminal processing. Clin. Exp. Immunol. 152:508–516. 10.1111/j.1365-2249.2008.03663.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witko-Sarsat V., Lesavre P., Lopez S., Bessou G., Hieblot C., Prum B., Noël L.H., Guillevin L., Ravaud P., Sermet-Gaudelus I., et al. . 1999. A large subset of neutrophils expressing membrane proteinase 3 is a risk factor for vasculitis and rheumatoid arthritis. J. Am. Soc. Nephrol. 10:1224–1233. [DOI] [PubMed] [Google Scholar]
- Wolff J., Brendel C., Fink L., Bohle R.M., Kissel K., and Bux J.. 2003. Lack of NB1 GP (CD177/HNA-2a) gene transcription in NB1 GP-neutrophils from NB1 GP-expressing individuals and association of low expression with NB1 gene polymorphisms. Blood. 102:731–733. 10.1182/blood-2002-09-2831 [DOI] [PubMed] [Google Scholar]
- Wu Z., Liang R., Ohnesorg T., Cho V., Lam W., Abhayaratna W.P., Gatenby P.A., Perera C., Zhang Y., Whittle B., et al. . 2016. Heterogeneity of Human Neutrophil CD177 Expression Results from CD177P1 Pseudogene Conversion. PLoS Genet. 12:e1006067 10.1371/journal.pgen.1006067 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.