CCL21Ser is one of the ligands for chemokine receptor CCR7. Kozai et al. report that CCL21Ser is essential for the accumulation of developing thymocytes in the thymic medulla and the establishment of self-tolerance in T cells, indicating a functional inequality among CCR7 ligands in vivo.
Abstract
The chemokine receptor CCR7 directs T cell relocation into and within lymphoid organs, including the migration of developing thymocytes into the thymic medulla. However, how three functional CCR7 ligands in mouse, CCL19, CCL21Ser, and CCL21Leu, divide their roles in immune organs is unclear. By producing mice specifically deficient in CCL21Ser, we show that CCL21Ser is essential for the accumulation of positively selected thymocytes in the thymic medulla. CCL21Ser-deficient mice were impaired in the medullary deletion of self-reactive thymocytes and developed autoimmune dacryoadenitis. T cell accumulation in the lymph nodes was also defective. These results indicate a nonredundant role of CCL21Ser in the establishment of self-tolerance in T cells in the thymic medulla, and reveal a functional inequality among CCR7 ligands in vivo.
Introduction
C-C chemokine receptor 7 (CCR7) is a chemokine receptor that plays a pivotal role in guiding T cell migration in health and disease (Yoshida et al., 1997; Förster et al., 1999). In the thymus, positive selection–inducing TCR signals elevate CCR7 expression in immature thymocytes that are generated in the thymic cortex (Ngo et al., 1998; Ueno et al., 2004), whereas a fraction of medullary thymic epithelial cells (mTECs) constitutively express the ligands for CCR7 (Ueno et al., 2002; Lkhagvasuren et al., 2013). Consequently, CCR7-expressing positively selected thymocytes are attracted to the medullary region in the thymus (Ueno et al., 2004; Witt et al., 2005; Ehrlich et al., 2009). The thymic medulla provides the microenvironment for developing thymocytes to establish self-tolerance, by negatively selecting self-reactive thymocytes and promoting the generation of regulatory T cells (Kyewski and Klein, 2006; Anderson and Takahama, 2012). In mice deficient in CCR7, thymocyte accumulation in the thymic medulla is defective (Ueno et al., 2004), so that T cells are unable to establish medullary self-tolerance, thereby causing autoimmune diseases (Kurobe et al., 2006; Davalos-Misslitz et al., 2007; Martin et al., 2009).
CCL19 and CCL21 are the functional ligands for CCR7 to attract immune cells (Yoshida et al., 1998). Genes that encode CCL19 and CCL21 are well conserved among vertebrate species in their sequences and in their mutual proximity in the genome (Lu et al., 2012; Nomiyama et al., 2013; Fig. S1 A). In the mouse, three distinct genes, Ccl21a, Ccl21b, and Ccl21c, all of which are localized in chromosome 4, encode CCL21 proteins (Vassileva et al., 1999; Nomiyama et al., 2013; Fig. S1, A and B). Mouse Ccl21a gene is proximal to the Ccl19 gene, similar to Ccl21 genes in other species, including human, whereas Ccl21b and Ccl21c genes are distant from Ccl19 and Ccl21a genes (Fig. S1 B). CCL21 protein encoded by mouse Ccl21a gene differs from Ccl21b- and Ccl21c-derived proteins in 1 aa; Ccl21a gene encodes the CCL21Ser protein, whereas both Ccl21b and Ccl21c genes encode the CCL21Leu protein (Nakano and Gunn, 2001; Chen et al., 2002; Lo et al., 2003). The expression of the CCL21Ser-encoding Ccl21a gene is predominantly detectable in immune organs, including the thymus and the lymph nodes, whereas the expression of CCL21Leu-encoding Ccl21b and/or Ccl21c transcripts (Ccl21b/c) is predominantly detectable in nonimmune organs, including the lung (Lo et al., 2003). CCL21Ser and CCL21Leu proteins are functionally equivalent in attracting CCR7-expressing cells (Chen et al., 2002).
Naturally occurring plt/plt (PLT) mutant mouse carries a genomic deletion that causes the lack of proximally localized Ccl19 and Ccl21a loci (Nakano et al., 1998; Luther et al., 2000; Nakano and Gunn, 2001). The reduction of Ccl21b/c expression in immune organs is detected in PLT mice (Nakano et al., 1998; Nakano and Gunn, 2001), which may be due to the loss of a distant cis-regulatory element that is localized within the deleted sequences in the PLT mutation, and/or the loss of cross-reactively detected Ccl21a transcripts predominantly expressed in immune organs (Nakano et al., 1998). Because of the defective expression of CCR7 ligands in immune organs, PLT mice are essentially similar to CCR7-deficient mice in terms of defective medullary accumulation of positively selected thymocytes and impaired self-tolerance in T cells, reinforcing the involvement of these CCR7 ligands in the cortex-to-medulla migration of developing thymocytes to establish self-tolerance (Kurobe et al., 2006). In contrast, mice specifically deficient in CCL19 are neither defective in thymic medulla formation nor prone to autoimmune disease, suggesting that CCL19 alone is not necessary for the medulla migration of thymocytes and the establishment of self-tolerance in T cells (Link et al., 2007; Britschgi et al., 2010). It is thus possible that the loss of any one or two of among the three CCR7 ligands may be compensated by the remaining ligands, so that like CCL19-deficient mice, mice deficient in CCL21Ser and/or CCL21Leu may be undisturbed in the establishment in the thymic medulla of self-tolerance in T cells. Alternatively, unlike CCL19, CCL21Ser and/or CCL21Leu may play a nonredundant role in the medulla migration of thymocytes. Thus, whether any one of the CCR7 ligands plays a unique role in the thymus and how CCL21Ser and/or CCL21Leu contribute to the medulla migration of thymocytes and the establishment of self-tolerance in T cells have remained unanswered.
We report herein the generation of mice in that the CCL21Ser-encoding Ccl21a sequence is specifically deleted. The mice lacked Ccl21a but not Ccl19 or Ccl21b/c transcripts. In these Ccl21a-deficient mice, positively selected thymocytes failed to accumulate in the thymic medulla, and medullary deletion of self-reactive thymocytes was defective. The Ccl21a-deficient mice developed autoimmune dacryoadenitis. These results reveal a nonredundant role of CCL21Ser-encoding Ccl21a in the establishment of self-tolerance in T cells in the thymic medulla.
Results and discussion
Generation of Ccl21a-deficient mice
To generate mice specifically deficient in the CCL21Ser-encoding Ccl21a gene, a targeting vector that contained the gene encoding the tandem dimeric tomato fluorescence protein (tdTomato) at the translation initiation site of the Ccl21a gene along with neighboring genomic sequences was introduced into TT2 embryonic stem cells for homologous recombination (Fig. S1 B). Southern blot analysis, PCR analysis, and sequence analysis of genomic DNA isolated from the tails of offspring mice indicated successful germline recombination at the Ccl21a locus as designed, without affecting Ccl21b and Ccl21c loci (Fig. S1, C and D). Because the two independent mouse lines obtained in this study have exhibited essentially identical phenotypes, we will show the results of one line of Ccl21a-deficient mice.
Ccl21a-deficient mice lack Ccl21a but not Ccl19 or Ccl21b/c
To measure the expression of CCL21Ser-encoding Ccl21a and CCL21Leu-encoding Ccl21b and/or Ccl21c (Ccl21b/c) transcripts, we designed PCR primers specific for Ccl21a and Ccl21b/c sequences. Evaluation of PCR conditions using Ccl21a-containing and Ccl21b/c-containing plasmids demonstrated that the detection of Ccl21a and Ccl21b/c was similarly specific at ∼103–104-fold signal-to-noise ratio (Fig. S1 E). Quantitative RT-PCR analysis using these PCR conditions showed that Ccl21a gene expression was severely defective in various immune organs of Ccl21a-deficient mice and PLT mice (Fig. S1 F). Sequence analysis of trace amounts of PCR products detectable by Ccl21a-specific RT-PCR of the samples derived from Ccl21a-deficient mice showed that those products entirely represented the amplification of Ccl21b/c but not Ccl21a cDNA (unpublished data), confirming that Ccl21a gene expression is lost in Ccl21a-deficient mice. On the other hand, the expression of Ccl21b/c gene predominantly detected in the lung was not reduced in Ccl21a-deficient mice or PLT mice (Fig. S1 G), indicating that Ccl21b/c gene expression is not lost in Ccl21a-deficient mice. Unlike in PLT mice, the expression of Ccl19 in immune organs was not lost in Ccl21a-deficient mice (Fig. S1 H). These results indicate that the Ccl21a-deficient mice newly generated in this study specifically lack the expression of the CCL21Ser-encoding Ccl21a gene, but not the CCL21Leu-encoding Ccl21b/c or Ccl19 gene.
Ccl21a-deficient mice are defective in the expression of CCL21 protein in mTECs
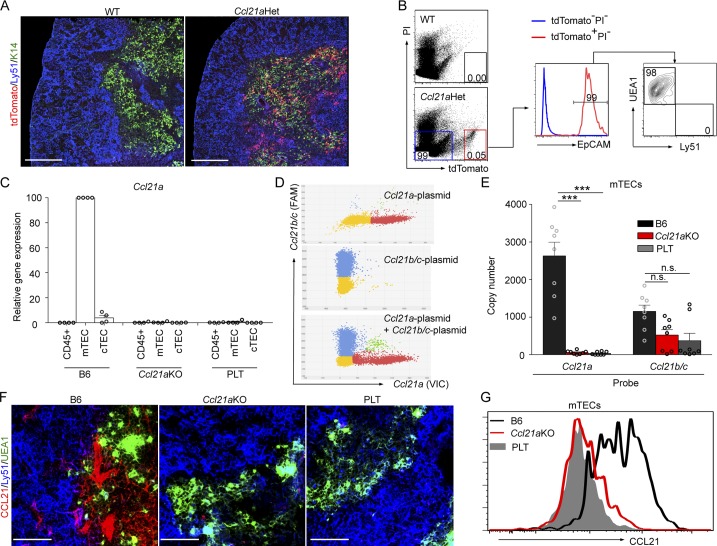
In the thymus, CCL21 protein is predominantly detectable in the medullary region (Ueno et al., 2002, 2004; Misslitz et al., 2004). The detection of tdTomato fluorescence protein in mice that heterozygously carried the Ccl21a-deficient allele indicated the transcription of the Ccl21a gene in the thymic medulla (Fig. 1 A). Flow cytometric analysis demonstrated that the tdTomato fluorescence in the thymus was predominantly detectable in mTECs (Fig. 1 B; and Fig. S2, A and C). Quantitative RT-PCR analysis of cDNAs extracted from isolated cells supported the predominant expression of Ccl21a gene in mTECs rather than cortical thymic epithelial cells (cTECs) or thymocytes (Fig. 1 C). By measuring transcript copy numbers using digital PCR analysis (Fig. 1 D), Ccl21a and Ccl21b/c were readily detected in mTECs (Fig. 1 E), but not cTECs or CD45−EpCAM− non-TECs (Fig. S2 B). The expression of Ccl21a but not Ccl21b/c was severely diminished in mTECs of Ccl21a-deficient mice, in quantitative RT-PCR analysis and in digital PCR analysis (Fig. 1, C–E). Antibody staining of the thymic sections with anti-CCL21 antibody, which detected both CCL21Ser and CCL21Leu proteins, showed that the majority of CCL21 proteins were lost in the medullary region in the thymus of Ccl21a-deficient mice (Fig. 1 F). Flow cytometric analysis supported that CCL21 protein levels expressed in mTECs were severely reduced in Ccl21a-deficient mice (Fig. 1 G). The amount of Ccl21b/c mRNA produced by mTECs in Ccl21a-deficient mice (Fig. 1 E) might be translated inefficiently leading to an amount of CCL21 proteins below the threshold, detectable by the anti-CCL21 antibody used in this study (Fig. 1, F and G). In contrast to PLT mice, Ccl19 mRNA in mTECs nor CCL19 protein in the thymic medulla were detected in Ccl21a-deficient mice (Fig. S2, D and E). These results indicate that mTECs in Ccl21a-deficient mice are severely and specifically defective in the expression of Ccl21a gene and CCL21 proteins. These results also suggest that most CCL21 proteins expressed in the thymic medulla are CCL21Ser, derived from Ccl21a gene, rather than CCL21Leu, derived from Ccl21b or Ccl21c gene.
Figure 1.
CCL21 expression in thymus of Ccl21a-deficient mice. (A) Immunofluorescence analysis of tdTomato (red), Ly51 (blue), and K14 (green) in the thymic sections of WT mice and heterozygous mice carrying one targeted allele (Ccl21aHet). Bars, 300 µm. Representative results from three independent experiments. (B) Flow cytometric analysis of liberase-digested thymic cells from WT and Ccl21aHet mice. Dot plots show tdTomato and propidium iodide (PI) expression in total cells (left), and the histogram shows EpCAM expression in tdTomato−PI− (blue) and tdTomato+PI− (red) cells. The contour plot shows UEA1 reactivity and Ly51 expression in tdTomato+EpCAM+ (red) TECs. Numbers indicate frequency of cells within the areas. Representative results from three independent experiments are shown. (C) Relative mRNA level (means and SEs; n = 4; four independent experiments) of Ccl21a in CD45+ thymocytes, CD45−EpCAM+UEA1+Ly51− mTECs, and CD45−EpCAM+UEA1−Ly51+ cTECs isolated from indicated mice. mRNA levels were normalized to Gapdh mRNA levels and are shown relative to the levels in the thymus of B6 mice. Open circles indicate individual data. (D) Digital PCR analysis of Ccl21a and Ccl21b/c cDNA-containing plasmid DNAs (104 copies) using VIC-labeled Ccl21a and FAM-labeled Ccl21b/c probes. (E) Digital PCR analysis of Ccl21a and Ccl21b/c expression in mTECs isolated from indicated mice. Means and SEs (n = 8) from eight independent experiments are shown. Open circles indicate individual data. ***, P < 0.001; n.s., not significant (unpaired Student’s t test). (F) Immunofluorescence analysis of CCL21 (red), Ly51 (blue), and UEA1-binding molecules (green) in the thymic sections of indicated mice. Bars, 75 µm. Representative results from three independent experiments. (G) Flow cytometric analysis of CCL21 expression in CD45−EpCAM+UEA1+Ly51− mTECs from indicated mice. Representative results from three independent experiments are shown.
It was previously noted that scattered signals of CCL21 proteins are detectable in the thymic cortex, predominantly in the deep cortex (Misslitz et al., 2004; Ueno et al., 2004). Those cortical CCL21 proteins were lost in Ccl21aKO mice (Fig. S2 F), whereas tdTomato+ cells in Ccl21a-tdTomato-knock-in mice were detectable predominantly in the thymic medulla and barely in the cortical region (Fig. 1 A and Fig. S2 F), suggesting that most CCL21 proteins detectable in the thymic cortex are derived from CCL21Ser proteins secreted in mTECs.
Defective thymocyte accumulation in thymic medulla of Ccl21a-deficient mice
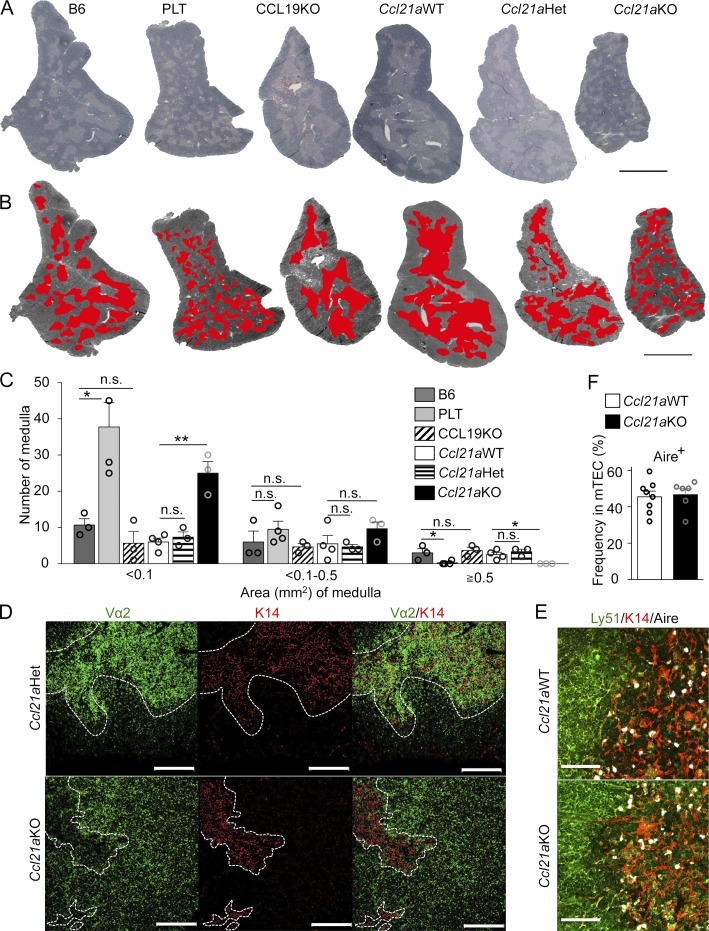
We next examined whether the medullary accumulation of developing thymocytes was defective in Ccl21a-deficient mice, as was observed in CCR7-deficient mice or PLT mice (Ueno et al., 2002; Misslitz et al., 2004; Nitta et al., 2009). To do so, we first analyzed the hematoxylin and eosin–stained thymic sections of Ccl21a-deficient mice (Fig. 2 A). We found that the medullary regions in the thymus of Ccl21a-deficient mice were small and scattered; this was observed in the thymus of PLT mice, but not in the thymus of CCL19-deficient mice or control littermates (Fig. 2 A). Image analysis of the thymic sections demonstrated that the thymic medullas in Ccl21a-deficient mice were significantly (P < 0.05) enriched in small regions and devoid of large ones, like the thymic medullas in PLT mice and unlike those in CCL19-deficient mice (Fig. 2, B and C). Total medullary areas in the largest coronal thymic sections were comparable among the mice analyzed in this study (Fig. S2 G), highlighting that CCL21Ser but not CCL19 affects the size of individual medullary areas in the thymus.
Figure 2.
Thymic medulla in Ccl21a-deficient mice. (A and B) Hematoxylin and eosin staining of thymic sections of 5-wk-old mice. Bar, 2 mm. The medullary regions were highlighted in red. Representative data from three independent experiments are shown (B). (C) Number of the medullary regions of a given size in the largest coronal thymic sections of 5-wk-old indicated mice. The x-axis indicates area (mm2) of individual medulla and the y-axis indicates the number (means and SEs; n = 3) of medullary regions. Open circles indicate individual data. *, P < 0.05; **, P < 0.01; n.s., not significant (unpaired Student’s t test). (D) T cell–depleted bone marrow cells from OT1-TCR-transgenic Rag1-deficient β2m-deficient mice were transferred into lethally irradiated control (Ccl21aHet) mice or Ccl21aKO mice. Thymic sections were analyzed for TCR-Vα2 (green) and K14 (red). Areas surrounded by white line indicate thymic medullary region. Bars, 300 µm. Representative data from at least three independent experiments are shown. (E) Immunofluorescence analysis of Ly51 (green), K14 (red), and Aire (white) in thymic sections of indicated mice. Bars, 75 µm. Representative results from three independent experiments are shown. (F) Flow cytometric analysis of Aire expression in CD45−EpCAM+UEA1+Ly51− mTECs from 5–6-wk-old Ccl21aWT mice (open bar) and Ccl21aKO mice (filled bar). Graph indicates frequency (means and SEs; n = 6–8) of Aire+ cells in mTECs. Open circles indicate individual data.
We then analyzed the distribution of thymocytes in the thymic microenvironments. Immunofluorescence staining for monoclonal OT-I-TCR-Vα2high cells in the thymus of Ccl21a-deficient mice that were reconstituted with bone marrow cells from OT-I-TCR–transgenic Rag1-deficient β2m-deficient mice showed that Vα2high mature thymocytes distributed predominantly in the medullary region in the thymus of control mice but were mainly detectable in the cortical region in the thymus of Ccl21a-deficient mice (Fig. 2 D). In addition, in the polyclonal situation, the density of the numbers of CD4+CD8− and CD4−CD8+ thymocytes in the thymic medulla was reduced in Ccl21a-deficient mice but not CCL19-deficient mice (Fig. S2 H). The defect in the distribution of mature thymocytes in Ccl21a-deficient mice was similar to the defective medullary accumulation previously reported in CCR7-deficient mice and PLT mice (Ueno et al., 2004; Nitta et al., 2009). These results indicate that the medullary accumulation of mature thymocytes is impaired in the thymic microenvironments of Ccl21a-deficient mice.
Nonetheless, the medullary regions were present albeit small (Fig. 2, A–D), and Aire-expressing mTECs were readily detectable in Ccl21a-deficient mice (Fig. 2, E and F), suggesting that the maturation of mTECs to give rise to Aire-expressing mTECs is not defective in Ccl21a-deficient mice.
Thymocyte development and peripheral T cell distribution in Ccl21a-deficient mice
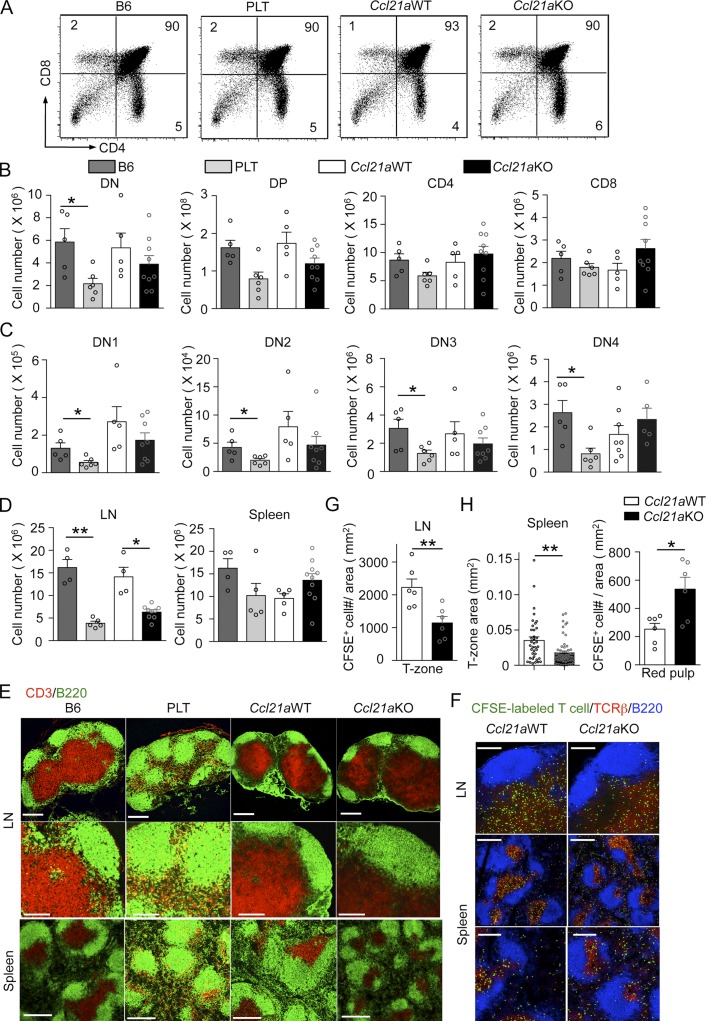
Despite the defective accumulation of developing thymocytes in the thymic medulla, CD4/CD8 profiles of thymocytes and the cellularity of CD4−CD8−, CD4+CD8+, CD4+CD8−TCRβhigh, and CD4−CD8+TCRβhigh thymocytes were comparable in Ccl21a-deficient mice and control mice (Fig. 3, A and B). Interestingly, unlike PLT mice and CCR7-deficient mice, in which the seeding of lymphoid progenitor cells into the thymus is defective (Misslitz et al., 2004; Liu et al., 2005), and like CCL19-deficient mice (Link et al., 2007), the numbers of CD4−CD8− (double negative; DN) thymocytes and their DN1 (CD25−CD44+), DN2 (CD25+CD44+), DN3 (CD25+CD44−), and DN4 (CD25−CD44−) subpopulations were not reduced in Ccl21a-deficient mice compared with those in control mice (Fig. 3, B and C; and Fig. S3). The cellularity of thymic B cells was reduced in Ccl21a-deficient mice (Fig. S2 I). These results indicate that the development of the various thymocyte subpopulations is not disturbed in Ccl21a-deficient mice, and suggest that the absence of Ccl21a-encoding CCL21Ser alone does not severely diminish the seeding of lymphoid progenitor cells into the postnatal thymus.
Figure 3.
T cell development in Ccl21a-deficient mice. (A) Flow cytometric analysis of thymocytes from 5-wk-old female mice. Shown are representative dot plot profiles of CD4 and CD8 expression in PI− viable cells from five independent experiments. Numbers in dot plots indicate frequency of cells within indicated area. (B–D) Cell numbers of indicated thymocyte subpopulations (B and C) and TCRβ+B220− T cells in the LNs and the spleen (D) are plotted (means and SEs; n = 4–10). Thymocyte subpopulations examined are CD4−CD8− (DN), CD4+CD8+ (DP), CD4+CD8−TCRβhigh (CD4), and CD4−CD8+TCRβhigh (CD8) PI− viable cells (B), and CD25−CD44+CD4−CD8− (DN1), CD25+CD44+CD4−CD8− (DN2), CD25+CD44−CD4−CD8− (DN3), and CD25−CD44−CD4−CD8− (DN4) PI− viable cells. Open circles indicate individual data. *, P < 0.05; **, P < 0.01 (unpaired Student’s t test). (E) Immunofluorescence analysis of CD3 (red) and B220 (green) in the LNs and spleens of indicated mice. Bars, 300 µm (top and bottom) and 150 µm (middle). Representative results from three independent experiments are shown. (F-H) T cell migration assay. CFSE-labeled TCRαβ+ B6 T cells were intravenously transferred into indicated mice. (F) The sections of the LN and the spleens were analyzed for CFSE (green), TCRβ (red), and B220 (blue). Bars, 150 µm (top and bottom) and 300 µm (middle). (G and H) T- and B-zones were identified with anti-TCRβ and anti-B220, respectively. Numbers of CFSE-labeled T cells in 1 mm2 of T-zone area in the LNs are shown (G). (left) The size (mm2) of T-zones per mm2 of the spleen sections; (right) number of CFSE-labeled T cells per mm2 of red pulp (non-T- and non-B-zone) in the spleen sections (H). Means and SEs from three different sections in two independent experiments are shown. Open circles indicate individual data. *, P < 0.05. **, P < 0.01 (unpaired Student’s t test).
In the secondary lymphoid organs, the cellularity of T cells was reduced in the lymph nodes but not in the spleen in Ccl21a-deficient mice, as was observed in PLT mice (Fig. 3 D). Immunofluorescence analysis of the distribution of endogenous or transferred T cells showed that T cells in the lymph nodes were less densely accumulated in the T cell zones of the lymph nodes in Ccl21a-deficient mice than in control mice (Fig. 3, E–G; and Fig. S2 J), whereas T cells in the spleen were less densely detected in the white pulp and more densely accumulated in the red pulp in Ccl21a-deficient mice than in control mice (Fig. 3, E and H). These results indicate that in Ccl21a-deficient mice, T cells are defective in the accumulation in the T cell zones.
Autoimmune dacryoadenitis in Ccl21a-deficient mice
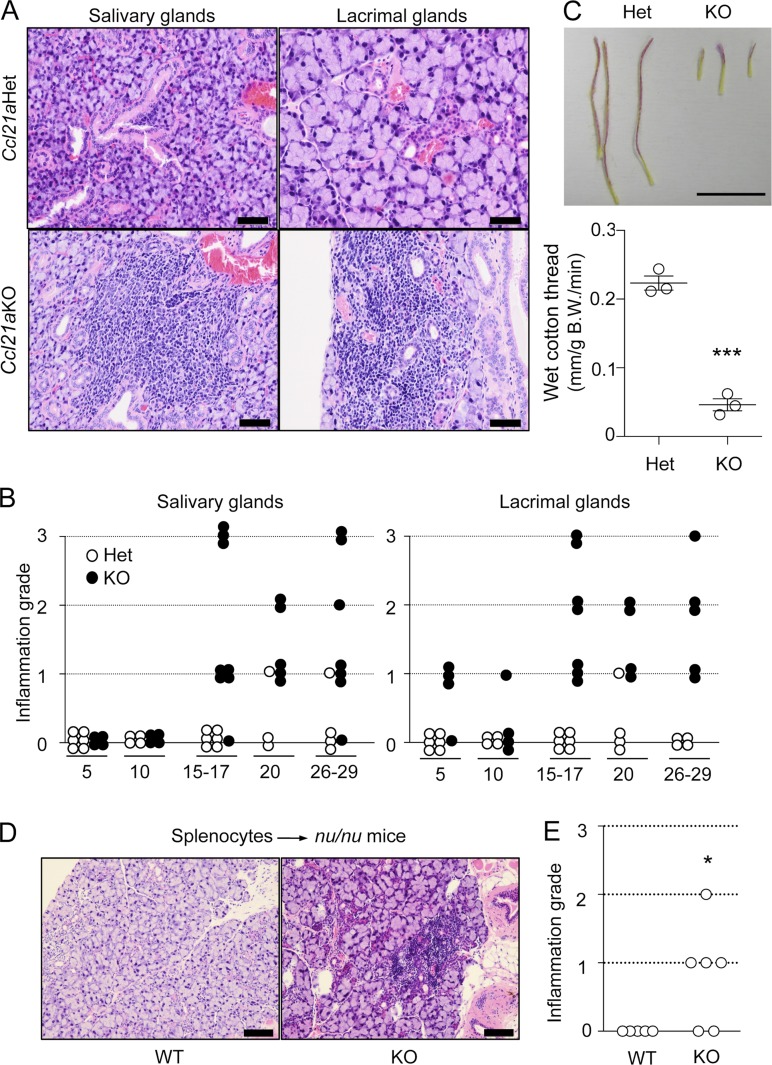
Systemic examination of hematoxylin and eosin–stained sections revealed that lymphocyte infiltration was detectable in the lacrimal glands and the salivary glands in Ccl21a-deficient mice (Fig. 4 A), as was observed in PLT mice and CCR7-deficient mice (Kurobe et al., 2006; Davalos-Misslitz et al., 2007). The inflammation in those exocrine glands became obvious in most individual mice by 20 wk old (Fig. 4 B). The production of tears decreased in Ccl21a-deficient mice, indicating functional damage in the lacrimal glands (Fig. 4 C). Adoptive transfer of spleen cells from Ccl21a-deficient mice reproduced the inflammation in the lacrimal glands in athymic nude mice, indicating the contribution of T cell–mediated autoimmunity to dacryoadenitis in Ccl21a-deficient mice (Fig. 4, D and E). The lacrimal and salivary glands exhibit no statistically significant inflammation in CCL19-deficient mice (n = 7, Mann-Whitney test; and not depicted). These results demonstrate that like PLT mice and CCR7-deficient mice, Ccl21a-deficient mice are prone to autoimmune dacryoadenitis.
Figure 4.
Autoimmune dacryoadenitis in Ccl21a-deficient mice. (A) Hematoxylin and eosin staining of salivary and lacrimal glands from indicated mice (females) at 17 wk of age. Bars, 200 µm. Representative results from at least three independent experiments are shown. (B) Histological grading of inflammatory lesions in Ccl21aHet mice (open circles) and Ccl21aKO mice (filled circles). The x-axis shows ages in weeks, and the y-axis shows inflammation grade (0, none; 1, mild; 2, moderate; and 3, severe) from at least three independent experiments. Symbols indicate data from individual mice. (C) Tear secretion for 3 min was measured by ZONE-QUICK™, which is cotton thread containing phenol red. The color of the cotton thread changes from yellow to red on absorbing tears (top). Bar, 1 cm. Graph indicates the length of cotton thread that absorbed tears per minute normalized to body weight. Symbols indicate data from individual mice. ***, P < 0.001 (unpaired Student’s t test). (D and E) Splenocytes from Ccl21aWT or Ccl21aKO mice were intravenously transferred into B6-nu/nu mice. Lacrimal glands were stained with hematoxylin and eosin (D). Bars, 200 µm. Histological scores of inflammatory lesions are shown (E). Symbols indicate data from individual mice. *, P < 0.05 (unpaired Student’s t test).
Defective negative selection of thymocytes in Ccl21a-deficient mice
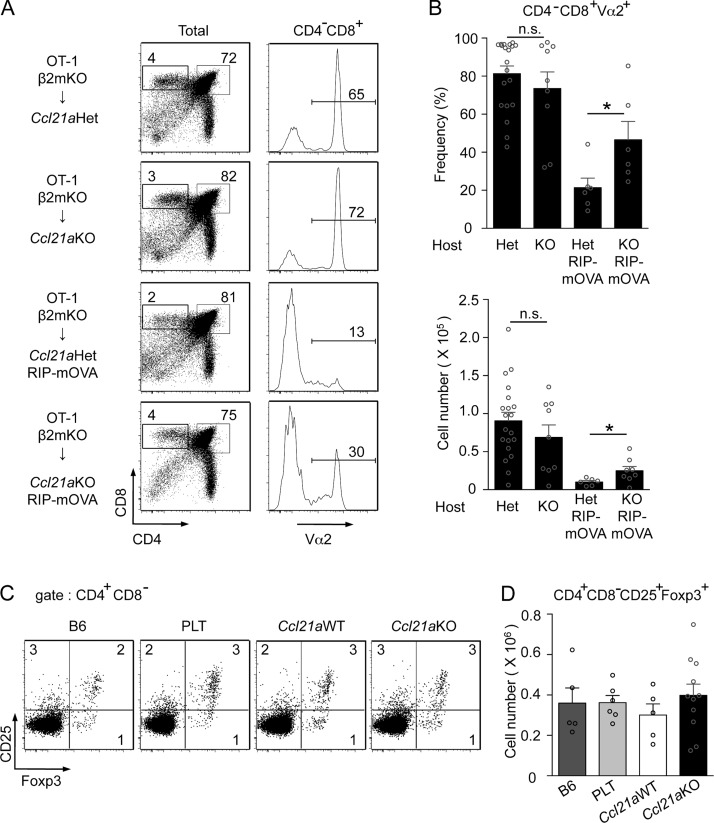
Our results showed that Ccl21a-deficient mice were defective in the medullary accumulation of developing thymocytes and were prone to autoimmune dacryoadenitis. It was therefore reasonable to speculate that the establishment of self-tolerance in T cells in the thymic medulla was defective in Ccl21a-deficient mice. To test this possibility, we finally examined whether Ccl21a-deficient mice were defective in the negative selection of thymocytes and regulatory T cell generation in the thymus. To analyze how mTEC-mediated negative selection of thymocytes was affected in the thymic medulla of Ccl21a-deficient mice, Ccl21a-deficient mice were crossed with RIP-mOVA-transgenic mice, in which membrane-bound chicken ovalbumin antigen was expressed in mTECs, and were lethally irradiated for the reconstitution with bone marrow cells from OT-I-TCR-transgenic β2m-deficient mice (Fig. 5 A). We found that RIP-mOVA–dependent negative selection of CD4−CD8+TCR-Vα2+ mature thymocytes was significantly (P < 0.05), albeit incompletely, disturbed in the thymus of Ccl21a-deficient mice (Fig. 5, A and B). However, the cellularity of CD4+CD8− CD25+Foxp3+ cells was not diminished in the thymus of Ccl21a-deficient mice (Fig. 5, C and D). These results indicate that Ccl21a-deficient mice are defective in the mTEC-dependent negative selection of thymocytes but not in the generation of regulatory T cells in the thymus.
Figure 5.
Negative selection and regulatory T cell development in the thymus of Ccl21a-deficient mice. (A and B) T cell–depleted bone marrow cells from OT-I-TCR-transgenic β2m-deficient mice were transferred into lethally irradiated RIP-mOVA-transgenic Ccl21aHet mice or Ccl21aKO mice. (A) Dot plots show CD4 and CD8 expression profiles in PI− viable thymocytes. Histograms show TCR-Vα2 expression in CD4−CD8+PI− thymocytes. Numbers indicate frequency of cells within indicated area. Representative results from five independent experiments are shown. (B) Shown are frequency (means and SEs; n = 6–20) of TCR-Vα2high cells within CD4−CD8+ cells (top) and absolute numbers of CD4−CD8+TCR-Va2+ thymocytes (bottom). Open circles indicate individual data. *, P < 0.05; n.s., not significant (unpaired Student’s t test). (C) Dot plots of Foxp3 and CD25 expression in CD4+CD8− thymocytes in indicated mice (5-wk-old female). Numbers in dot plots indicate frequency of cells within indicated area. Representative results from five independent experiments are shown. (D) Absolute numbers (means and SEs; n = 5–10) of CD4+CD8−CD25+Foxp3+ thymocytes in indicated mice. Open circles indicate individual data. Data are not significantly different among the groups (unpaired Student’s t test).
Discussion
The present results demonstrate that mice specifically deficient in Ccl21a-encoded CCL21Ser, one of the three CCR7 ligands, are defective in the medullary accumulation of positively selected thymocytes and the establishment of self-tolerance in T cells. It was previously unknown whether any one of the three CCR7 ligands in mice plays a unique role in immune system development, including T cell development in the thymus. This study reveals that the CCR7 ligand, Ccl21a-encoded CCL21Ser, plays a nonredundant and major role in the migration of positively selected thymocytes into the thymic medulla and the establishment of self-tolerance in T cells.
A previous study reported that CCL19 is dispensable for the thymocyte migration into the thymic medulla and for the T cell migration into T cell zones in the lymph nodes (Link et al., 2007). It was therefore possible that CCL21 and CCL19 might play an equivalent and mutually compensatory role in attracting T cells. However, CCL19 and CCL21 proteins have distinct structural properties and CCR7-stimulating capability, despite sharing the same signaling receptor CCR7 (Nagira et al., 1997; Bardi et al., 2001; Kohout et al., 2004). It was shown that CCL19 is essential for the homeostatic maintenance of T cell numbers in the secondary lymphoid organs (Link et al., 2007). Our results highlight the disparity between the in vivo roles of CCL21 and CCL19, by indicating that unlike CCL19, Ccl21a-encoded CCL21Ser plays an essential role in guiding the migration of T cells in the thymus and the secondary lymphoid organs.
Unlike CCL21Ser and CCL19, CCL21Leu and CCL21Leu-encoding Ccl21b and CCL21c genes are present only in mouse and not in many other species, including human (Fig. S1 A). Nonetheless, the function of CCL21Leu in mouse remains unclear. It was reported that CCL21Ser and CCL21Leu exhibit an equivalent ability to recruit T cells into the pancreatic islets in transgenic overexpression mouse models (Chen et al., 2002). However, it was also suggested that CCL21Ser and CCL21Leu might participate differently in the recruitment of T cells into the lung (Lo et al., 2003). Our results further indicate that CCL21Leu does not seem to contribute to the medullary migration of developing thymocytes in the thymus, even though CCL21Leu-encoding Ccl21b/c mRNA transcripts being still detectable in mTECs isolated from either normal or Ccl21a-deficient mice.
Our results also show that Ccl21a-deficient mice develop autoimmune exocrinopathy with dacryoadenitis, and that this exocrinopathy is reproduced in T cell-lacking athymic nude mice that are adoptively transferred with spleen cells from Ccl21a-deficient mice. These results indicate that T cells generated in the thymus without CCL21Ser fail to establish self-tolerance to the exocrine tissues. Interestingly, genome-wide association studies of human populations have revealed that single-nucleotide polymorphisms in the single human Ccl21 gene are associated with autoimmune diseases, including rheumatoid arthritis and dermatomyositis (Raychaudhuri et al., 2008; Orozco et al., 2010; Stahl et al., 2010; Sundqvist et al., 2011; Miller et al., 2013; Chen et al., 2015). However, the association between CCL19 polymorphisms and autoimmune diseases remains unclear. Thus, CCL21 rather than CCL19 appears to be important for the establishment of self-tolerance in both human and mouse.
Finally, our results show that in Ccl21a-deficient mice, T cells are abnormally distributed between the white pulp and the red pulp in the spleen and are defective in the accumulation in the T cell zones in the lymph nodes, which was not previously detected in CCL19-deficient mice (Link et al., 2007). The results also demonstrate the separation between T cell zones and B cell zones in Ccl21a-deficient mice. Further analysis of the lymph nodes and the spleens in Ccl21a-deficient mice versus PLT mice and CCL19-deficient mice, especially for the behavior and distribution of T cells and other immune cells during immune responses, should help clarify the role of CCL21Ser in the secondary lymphoid organs.
In conclusion, we have demonstrated that CCL21Ser in mouse plays a nonredundant role in T cell migration and self-tolerance establishment in T cells, by newly generating mice in that the CCL21Ser-encoding Ccl21a gene is specifically deleted. CCR7 and its ligands are important for various aspects of immune system development and immune response in health and disease. The role of CCL21 and CCR7 interactions has also been noted in cancer metastasis and tumor-induced immune modulation (Müller et al., 2001; Shields et al., 2010; Riedel et al., 2016). The Ccl21a-deficient mice generated in this study will be useful for further studies of the roles of CCL21Ser and other CCR7 ligands in vivo.
Materials and methods
Mice
C57BL/6 (B6) mice and OT-I-TCR transgenic Rag1-deficient mice were obtained from SLC and Taconic, respectively. B6-plt/plt (PLT) mice (Nakano et al., 1998), CCL19-KO mice (Link et al., 2007), RIP-mOVA transgenic mice (Kurts et al., 1996), and β2m-deficient mice (Koller et al., 1990) were described previously. B6-nude (nu/nu) mice were obtained from Riken Bio-Resource Center through the MEXT National Bio-Resource Project, Japan. All mouse experiments were performed with consent from the Animal Experimentation Committee of the University of Tokushima (#13116) and the Institutional Animal Care and Use Committee of Institute of Physical and Chemical Research Kobe Branch (AH13-03).
Generation of Ccl21a-deficient mice
The targeting vector shown in Fig. 1 B was prepared by subcloning Ccl21a-containing mouse genomic BAC fragments (Advanced GenoTechs) and tdTomato-encoding cDNA (Clontech) into a plasmid containing a pgk-neo cassette. The linearized targeting vector was introduced into TT2 ES cells (Yagi et al., 1993). Targeted alleles were screened by genomic PCR analysis and Southern blot analysis. Mice (accession no. CDB1030K) are available to the scientific community (http://www2.clst.riken.jp/arg/mutant%20mice%20list.html). The primers used for genotyping PCR were as follows; WT-F, 5′-CTGGTCTCATCCTCAACTCA-3′; WT-R, 5′-TGTAACCCTAGGATTGTAGG-3′; and tdTomato-R, 5′-GGTCTTGAACTCCACCAGGT-3′.
Southern blotting
Genomic DNA extracted from the liver was digested with KpnI, electrophoresed in 1% agarose, and transferred to nylon membrane (Hybond-N+; GE Healthcare). Probe was labeled with a PCR DIG Probe Synthesis kit (Roche) and hybridization was detected using anti–DIG-AP Fab fragment (Roche), CDP-STAR (Roche), and Light Capture II (Atto).
Quantitative RT-PCR analysis
Total cellular RNA was reverse-transcribed (RT) with oligo-dT primers and SuperScript III reverse transcription (Invitrogen). Quantitative real-time PCR was performed using SYBR Premix Ex Taq (TaKaRa) and 7900HT Sequence Detection System (Applied Biosystems). The primers used were as follows: Ccl21a, 5′-AAGGCAGTGATGGAGGGGGT-3′ and 5′-CTTAGAGTGCTTCCGGGGTG-3′; Ccl21b/c, 5′-AAGGCAGTGATGGAGGGGGA-3′ and 5′-GGCTTAGAGTGCTTCCGGGGTA-3′; Ccl19, 5′-CTGCCTCAGATTATCTGCCAT-3′ and 5′-AGGTAGCGGAAGGCTTTCAC-3′; and Gapdh 5′-CCGGTGCTGAGTATGTCGTG-3′ and 5′-CAGTCTTCTGGGTGGCAGTG-3′. Amplified products were confirmed to be single bands in gel electrophoresis and normalized to the amount of Gapdh amplification products.
Digital PCR analysis
Amplified DNA was detected with VIC-labeled Ccl21a and FAM-labeled Ccl21b/c probes. Amplification primers were 5′-GGCTATAGGAAGCAAGAACCAAGTT-3′ and 5′-CATAGCTCAGGCTTAGAGTGCTT-3′. Detection probes were 5′-CAATCCTGTTCTCACCCCG-3′ for Ccl21a and 5′-CAATCCTGTTCTTACCCCG-3′ for Ccl21b/c. PCR products were analyzed using QuantStudio 3D Digital PCR Chip Loader, GeneAmp PCR System 9700, and QuantStudio 3D Digital PCR System (Applied Biosystems).
Bone marrow chimeras
Bone marrow cells from OT-I-TCR–transgenic β2m-deficient mice were magnetically depleted of T cells by using anti-CD90.2 microbeads (Miltenyi Biotec). Recipient mice were irradiated with 10 Gy x-rays and injected with T cell–depleted bone marrow cells. Mice were analyzed 4–5 wk after the reconstitution.
T cell migration assay
TCRαβ-expressing T cells were enriched in spleens and peripheral lymph nodes of B6 mice by depletion of B220-, NK1.1-, TCRδ-, CD11b-, and CD11c-expressing cells using Microbeads (Miltenyi Biotec) and were >95% TCRβ+. The cells were labeled with 2 µM CFSE, and 2 × 107 cells were intravenously transferred into recipient mice. Spleens and lymph nodes were frozen either 1 h or 15 h after the transfer. T- and B-zone were determined by anti-TCRβ and anti-B220 staining, respectively, of the tissues. T- and B-zone areas (mm2) were computed by using ImageJ software (National Institutes of Health). Non-T- and non-B-zone areas were calculated by subtracting T-zone and B-zone areas from the whole area in each image.
Adoptive cell transfer
Splenocytes (5 × 106) from Ccl21a-deficient mice or control mice were intravenously injected into B6-nude mice. Tissues fixed in 4% phosphate-buffered formaldehyde, pH 7.2, were analyzed 8 wk after the cell transfer.
Flow cytometric analysis and cell sorting
For the analysis of thymic epithelial cells (TECs), minced thymuses were digested with 1 unit/ml Liberase (Roche) in the presence of 0.01% DNase I (Roche). Single-cell suspensions were stained with antibodies specific for CD326 (EpCAM; BioLegend), CD45 (eBioscience), and CD249 (Ly51, eBioscience), and for the reactivity with UEA-1 (Vector Laboratories). For the analysis of Aire and CCL21, surface-stained cells were fixed in 4% (g/vol) paraformaldehyde, permeabilized in 0.1% saponin, and stained with anti-Aire (5H12; eBioscience) antibody or anti-CCL21 (AAM27; Bio-Rad Laboratories) antibody. For the analysis of thymocytes, splenocytes, and lymph node cells, cells were surface-stained with the indicated antibodies. For the intracellular staining of Foxp3, the surface-stained cells were fixed and permeabilized by using a Foxp3 Staining Buffer Set (eBioscience) and stained with anti-Foxp3 antibody (eBioscience). For the isolation of TECs, CD45− cells were enriched in magnetic bead conjugated anti-CD45 antibody (Miltenyi Biotec). Multicolor flow cytometry and cell sorting were performed on FACSAria II (BD).
Immunofluorescence analysis
Paraformaldehyde-fixed frozen tissues embedded in OCT compound (Sakura Finetek) were sliced into 5-µm-thick sections. Lymph nodes were sliced into 10-µm-thick sections. The sections were stained with anti-CCL21 (Bio-Rad Laboratories), anti-CCL19 (R&B System), anti-Aire (eBioscience), anti-Ly51 (eBioscience), anti-K14 (Covance), anti-B220, anti-CD3, anti-CD4, anti-CD8, or anti-TCR-Vα2 (eBioscience) antibodies and UEA-1 (Vector Laboratories). Images were analyzed with a TSC SP8 confocal laser scanning microscope (Leica).
Hematoxylin and eosin staining
Paraformaldehyde-fixed and frozen sections were stained with hematoxylin and eosin (Muto Pure Chemicals Co.) and observed under an Eclipse E1000 microscope (Nikon). The size (mm2) of medullary regions in the thymic sections was measured by using ImageJ software.
Tear secretion
Tear secretion in 3 min was measured by using cotton thread that contained phenol red (Zone-Quick; Showa Yakuhin Kako). The length of cotton thread that absorbed tears per minute was normalized to body weight.
Inflammation grade
Histological grading of inflammatory lesions was performed as described previously (Kohashi et al., 2008). A score of 1 indicates that one to five foci composed of more than 20 mononuclear cells per focus were seen; a score of 2 indicates that more than five such foci were seen, but without significant parenchymal damage; and a score of 3 indicates degeneration of parenchymal tissue.
Statistical analysis
Statistical significance was accessed using the two-tailed unpaired Student’s t test with Welch’s correction for unequal variances.
Online supplemental material
Fig. S1 shows the generation of Ccl21a-knockout mice. Fig. S2 shows additional phenotypes of Ccl21a-knockout mice. Fig. S3 shows the cellularity of thymocytes in CCL19-deficient thymus.
Supplementary Material
Acknowledgments
We thank Drs. Masayuki Miyasaka and Mie Sakata for support in the analysis of lymphocyte migration in secondary lymphoid organs and in the immunofluorescence analysis of thymic sections, respectively; Ms. Hitomi Kyuma for assistance in genotyping animals; and Dr. Kenta Kondo for reading the manuscript.
This study was supported by grants from MEXT-JSPS (24111004 and 16H02630 to Y. Takahama, 25860361 and 15K19130 to I. Ohigashi, and 16K19157 to M. Kozai).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- CCR7
- C-C chemokine receptor 7
- cTEC
- cortical thymic epithelial cell
- mTEC
- medullary thymic epithelial cell
- RIP-mOVA
- membrane-bound chicken ovalbumin under the control of rat insulin promoter
References
- Anderson G., and Takahama Y.. 2012. Thymic epithelial cells: working class heroes for T cell development and repertoire selection. Trends Immunol. 33:256–263. 10.1016/j.it.2012.03.005 [DOI] [PubMed] [Google Scholar]
- Bardi G., Lipp M., Baggiolini M., and Loetscher P.. 2001. The T cell chemokine receptor CCR7 is internalized on stimulation with ELC, but not with SLC. Eur. J. Immunol. 31:3291–3297. [DOI] [PubMed] [Google Scholar]
- Britschgi M.R., Favre S., and Luther S.A.. 2010. CCL21 is sufficient to mediate DC migration, maturation and function in the absence of CCL19. Eur. J. Immunol. 40:1266–1271. 10.1002/eji.200939921 [DOI] [PubMed] [Google Scholar]
- Chen S.C., Vassileva G., Kinsley D., Holzmann S., Manfra D., Wiekowski M.T., Romani N., and Lira S.A.. 2002. Ectopic expression of the murine chemokines CCL21a and CCL21b induces the formation of lymph node-like structures in pancreas, but not skin, of transgenic mice. J. Immunol. 168:1001–1008. 10.4049/jimmunol.168.3.1001 [DOI] [PubMed] [Google Scholar]
- Chen S., Wang Q., Wu C.Y., Wu Q.J., Li Y., Wu Z.Y., Li P., Sun F., Zheng W.J., Deng C.W., et al. 2015. A single-nucleotide polymorphism of CCL21 rs951005 T>C is associated with susceptibility of polymyositis and such patients with interstitial lung disease in a Chinese Han population. Clin. Exp. Rheumatol. 33:639–646. [PubMed] [Google Scholar]
- Davalos-Misslitz A.C., Rieckenberg J., Willenzon S., Worbs T., Kremmer E., Bernhardt G., and Förster R.. 2007. Generalized multi-organ autoimmunity in CCR7-deficient mice. Eur. J. Immunol. 37:613–622. 10.1002/eji.200636656 [DOI] [PubMed] [Google Scholar]
- Ehrlich L.I., Oh D.Y., Weissman I.L., and Lewis R.S.. 2009. Differential contribution of chemotaxis and substrate restriction to segregation of immature and mature thymocytes. Immunity. 31:986–998. 10.1016/j.immuni.2009.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster R., Schubel A., Breitfeld D., Kremmer E., Renner-Müller I., Wolf E., and Lipp M.. 1999. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 99:23–33. 10.1016/S0092-8674(00)80059-8 [DOI] [PubMed] [Google Scholar]
- Kohashi M., Ishimaru N., Arakaki R., and Hayashi Y.. 2008. Effective treatment with oral administration of rebamipide in a mouse model of Sjögren’s syndrome. Arthritis Rheum. 58:389–400. 10.1002/art.23163 [DOI] [PubMed] [Google Scholar]
- Kohout T.A., Nicholas S.L., Perry S.J., Reinhart G., Junger S., and Struthers R.S.. 2004. Differential desensitization, receptor phosphorylation, beta-arrestin recruitment, and ERK1/2 activation by the two endogenous ligands for the CC chemokine receptor 7. J. Biol. Chem. 279:23214–23222. 10.1074/jbc.M402125200 [DOI] [PubMed] [Google Scholar]
- Koller B.H., Marrack P., Kappler J.W., and Smithies O.. 1990. Normal development of mice deficient in β 2M, MHC class I proteins, and CD8+ T cells. Science. 248:1227–1230. 10.1126/science.2112266 [DOI] [PubMed] [Google Scholar]
- Kurobe H., Liu C., Ueno T., Saito F., Ohigashi I., Seach N., Arakaki R., Hayashi Y., Kitagawa T., Lipp M., et al. 2006. CCR7-dependent cortex-to-medulla migration of positively selected thymocytes is essential for establishing central tolerance. Immunity. 24:165–177. 10.1016/j.immuni.2005.12.011 [DOI] [PubMed] [Google Scholar]
- Kurts C., Heath W.R., Carbone F.R., Allison J., Miller J.F., and Kosaka H.. 1996. Constitutive class I-restricted exogenous presentation of self antigens in vivo. J. Exp. Med. 184:923–930. 10.1084/jem.184.3.923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyewski B., and Klein L.. 2006. A central role for central tolerance. Annu. Rev. Immunol. 24:571–606. 10.1146/annurev.immunol.23.021704.115601 [DOI] [PubMed] [Google Scholar]
- Link A., Vogt T.K., Favre S., Britschgi M.R., Acha-Orbea H., Hinz B., Cyster J.G., and Luther S.A.. 2007. Fibroblastic reticular cells in lymph nodes regulate the homeostasis of naive T cells. Nat. Immunol. 8:1255–1265. 10.1038/ni1513 [DOI] [PubMed] [Google Scholar]
- Liu C., Ueno T., Kuse S., Saito F., Nitta T., Piali L., Nakano H., Kakiuchi T., Lipp M., Hollander G.A., and Takahama Y.. 2005. The role of CCL21 in recruitment of T-precursor cells to fetal thymi. Blood. 105:31–39. 10.1182/blood-2004-04-1369 [DOI] [PubMed] [Google Scholar]
- Lkhagvasuren E., Sakata M., Ohigashi I., and Takahama Y.. 2013. Lymphotoxin β receptor regulates the development of CCL21-expressing subset of postnatal medullary thymic epithelial cells. J. Immunol. 190:5110–5117. 10.4049/jimmunol.1203203 [DOI] [PubMed] [Google Scholar]
- Lo J.C., Chin R.K., Lee Y., Kang H.S., Wang Y., Weinstock J.V., Banks T., Ware C.F., Franzoso G., and Fu Y.X.. 2003. Differential regulation of CCL21 in lymphoid/nonlymphoid tissues for effectively attracting T cells to peripheral tissues. J. Clin. Invest. 112:1495–1505. 10.1172/JCI19188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu I.N., Chiang B.L., Lou K.L., Huang P.T., Yao C.C., Wang J.S., Lin L.D., Jeng J.H., and Chang B.E.. 2012. Cloning, expression and characterization of CCL21 and CCL25 chemokines in zebrafish. Dev. Comp. Immunol. 38:203–214. 10.1016/j.dci.2012.07.003 [DOI] [PubMed] [Google Scholar]
- Luther S.A., Tang H.L., Hyman P.L., Farr A.G., and Cyster J.G.. 2000. Coexpression of the chemokines ELC and SLC by T zone stromal cells and deletion of the ELC gene in the plt/plt mouse. Proc. Natl. Acad. Sci. USA. 97:12694–12699. 10.1073/pnas.97.23.12694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A.P., Marinkovic T., Canasto-Chibuque C., Latif R., Unkeless J.C., Davies T.F., Takahama Y., Furtado G.C., and Lira S.A.. 2009. CCR7 deficiency in NOD mice leads to thyroiditis and primary hypothyroidism. J. Immunol. 183:3073–3080. 10.4049/jimmunol.0900275 [DOI] [PubMed] [Google Scholar]
- Miller F.W., Cooper R.G., Vencovský J., Rider L.G., Danko K., Wedderburn L.R., Lundberg I.E., Pachman L.M., Reed A.M., Ytterberg S.R., et al. Myositis Genetics Consortium . 2013. Genome-wide association study of dermatomyositis reveals genetic overlap with other autoimmune disorders. Arthritis Rheum. 65:3239–3247. 10.1002/art.38137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misslitz A., Pabst O., Hintzen G., Ohl L., Kremmer E., Petrie H.T., and Förster R.. 2004. Thymic T cell development and progenitor localization depend on CCR7. J. Exp. Med. 200:481–491. 10.1084/jem.20040383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A., Homey B., Soto H., Ge N., Catron D., Buchanan M.E., McClanahan T., Murphy E., Yuan W., Wagner S.N., et al. 2001. Involvement of chemokine receptors in breast cancer metastasis. Nature. 410:50–56. 10.1038/35065016 [DOI] [PubMed] [Google Scholar]
- Nagira M., Imai T., Hieshima K., Kusuda J., Ridanpää M., Takagi S., Nishimura M., Kakizaki M., Nomiyama H., and Yoshie O.. 1997. Molecular cloning of a novel human CC chemokine secondary lymphoid-tissue chemokine that is a potent chemoattractant for lymphocytes and mapped to chromosome 9p13. J. Biol. Chem. 272:19518–19524. 10.1074/jbc.272.31.19518 [DOI] [PubMed] [Google Scholar]
- Nakano H., and Gunn M.D.. 2001. Gene duplications at the chemokine locus on mouse chromosome 4: multiple strain-specific haplotypes and the deletion of secondary lymphoid-organ chemokine and EBI-1 ligand chemokine genes in the plt mutation. J. Immunol. 166:361–369. 10.4049/jimmunol.166.1.361 [DOI] [PubMed] [Google Scholar]
- Nakano H., Mori S., Yonekawa H., Nariuchi H., Matsuzawa A., and Kakiuchi T.. 1998. A novel mutant gene involved in T-lymphocyte-specific homing into peripheral lymphoid organs on mouse chromosome 4. Blood. 91:2886–2895. [PubMed] [Google Scholar]
- Ngo V.N., Tang H.L., and Cyster J.G.. 1998. Epstein-Barr virus-induced molecule 1 ligand chemokine is expressed by dendritic cells in lymphoid tissues and strongly attracts naive T cells and activated B cells. J. Exp. Med. 188:181–191. 10.1084/jem.188.1.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta T., Nitta S., Lei Y., Lipp M., and Takahama Y.. 2009. CCR7-mediated migration of developing thymocytes to the medulla is essential for negative selection to tissue-restricted antigens. Proc. Natl. Acad. Sci. USA. 106:17129–17133. 10.1073/pnas.0906956106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomiyama H., Osada N., and Yoshie O.. 2013. Systematic classification of vertebrate chemokines based on conserved synteny and evolutionary history. Genes Cells. 18:1–16. 10.1111/gtc.12013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco G., Eyre S., Hinks A., Ke X., Wilson A.G., Bax D.E., Morgan A.W., Emery P., Steer S., Hocking L., et al. Wellcome Trust Case Control consortium YEAR Consortium . 2010. Association of CD40 with rheumatoid arthritis confirmed in a large UK case-control study. Ann. Rheum. Dis. 69:813–816. 10.1136/ard.2009.109579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S., Remmers E.F., Lee A.T., Hackett R., Guiducci C., Burtt N.P., Gianniny L., Korman B.D., Padyukov L., Kurreeman F.A., et al. 2008. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat. Genet. 40:1216–1223. 10.1038/ng.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel A., Shorthouse D., Haas L., Hall B.A., and Shields J.. 2016. Tumor-induced stromal reprogramming drives lymph node transformation. Nat. Immunol. 17:1118–1127. 10.1038/ni.3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields J.D., Kourtis I.C., Tomei A.A., Roberts J.M., and Swartz M.A.. 2010. Induction of lymphoidlike stroma and immune escape by tumors that express the chemokine CCL21. Science. 328:749–752. 10.1126/science.1185837 [DOI] [PubMed] [Google Scholar]
- Stahl E.A., Raychaudhuri S., Remmers E.F., Xie G., Eyre S., Thomson B.P., Li Y., Kurreeman F.A., Zhernakova A., Hinks A., et al. YEAR Consortium . 2010. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 42:508–514. 10.1038/ng.582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundqvist J., Falconer H., Seddighzadeh M., Vodolazkaia A., Fassbender A., Kyama C., Bokor A., Stephansson O., Padyukov L., Gemzell-Danielsson K., and D’Hooghe T.M.. 2011. Endometriosis and autoimmune disease: association of susceptibility to moderate/severe endometriosis with CCL21 and HLA-DRB1. Fertil. Steril. 95:437–440. 10.1016/j.fertnstert.2010.07.1060 [DOI] [PubMed] [Google Scholar]
- Ueno T., Hara K., Willis M.S., Malin M.A., Höpken U.E., Gray D.H., Matsushima K., Lipp M., Springer T.A., Boyd R.L., et al. 2002. Role for CCR7 ligands in the emigration of newly generated T lymphocytes from the neonatal thymus. Immunity. 16:205–218. 10.1016/S1074-7613(02)00267-4 [DOI] [PubMed] [Google Scholar]
- Ueno T., Saito F., Gray D.H., Kuse S., Hieshima K., Nakano H., Kakiuchi T., Lipp M., Boyd R.L., and Takahama Y.. 2004. CCR7 signals are essential for cortex-medulla migration of developing thymocytes. J. Exp. Med. 200:493–505. 10.1084/jem.20040643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassileva G., Soto H., Zlotnik A., Nakano H., Kakiuchi T., Hedrick J.A., and Lira S.A.. 1999. The reduced expression of 6Ckine in the plt mouse results from the deletion of one of two 6Ckine genes. J. Exp. Med. 190:1183–1188. 10.1084/jem.190.8.1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt C.M., Raychaudhuri S., Schaefer B., Chakraborty A.K., and Robey E.A.. 2005. Directed migration of positively selected thymocytes visualized in real time. PLoS Biol. 3:e160 10.1371/journal.pbio.0030160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T., Tokunaga T., Furuta Y., Nada S., Yoshida M., Tsukada T., Saga Y., Takeda N., Ikawa Y., and Aizawa S.. 1993. A novel ES cell line, TT2, with high germline-differentiating potency. Anal. Biochem. 214:70–76. 10.1006/abio.1993.1458 [DOI] [PubMed] [Google Scholar]
- Yoshida R., Imai T., Hieshima K., Kusuda J., Baba M., Kitaura M., Nishimura M., Kakizaki M., Nomiyama H., and Yoshie O.. 1997. Molecular cloning of a novel human CC chemokine EBI1-ligand chemokine that is a specific functional ligand for EBI1, CCR7. J. Biol. Chem. 272:13803–13809. 10.1074/jbc.272.21.13803 [DOI] [PubMed] [Google Scholar]
- Yoshida R., Nagira M., Kitaura M., Imagawa N., Imai T., and Yoshie O.. 1998. Secondary lymphoid-tissue chemokine is a functional ligand for the CC chemokine receptor CCR7. J. Biol. Chem. 273:7118–7122. 10.1074/jbc.273.12.7118 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.