Combining high-coverage whole-genome sequencing with functional analyses, Giessler et al. demonstrate that tumor initiation and long-term tumor formation in human colorectal cancer are driven by multiple genomic subclones and that the functional heterogeneity of colorectal cancer tumor clone–initiating cells is not based on genomic architecture.
Abstract
A hierarchically organized cell compartment drives colorectal cancer (CRC) progression. Genetic barcoding allows monitoring of the clonal output of tumorigenic cells without prospective isolation. In this study, we asked whether tumor clone-initiating cells (TcICs) were genetically heterogeneous and whether differences in self-renewal and activation reflected differential kinetics among individual subclones or functional hierarchies within subclones. Monitoring genomic subclone kinetics in three patient tumors and corresponding serial xenografts and spheroids by high-coverage whole-genome sequencing, clustering of genetic aberrations, subclone combinatorics, and mutational signature analysis revealed at least two to four genetic subclones per sample. Long-term growth in serial xenografts and spheroids was driven by multiple genomic subclones with profoundly differing growth dynamics and hence different quantitative contributions over time. Strikingly, genetic barcoding demonstrated stable functional heterogeneity of CRC TcICs during serial xenografting despite near-complete changes in genomic subclone contribution. This demonstrates that functional heterogeneity is, at least frequently, present within genomic subclones and independent of mutational subclone differences.
Introduction
More than 1,700 somatic mutations in protein-coding regions of the genome have been found in colorectal cancer (CRC) patient tumors and metastases, with an average of 80–90 single-nucleotide variants (SNVs), which largely differ among individual patients (Wood et al., 2007). Only ∼32 genes are recurrently mutated in tumors from different CRC patients, but a low incidence rate does not preclude functional relevance of a gene mutation (Cancer Genome Atlas Network, 2012). The acquisition of multiple genetic lesions in protooncogenes and tumor suppressors drives the dominating clones during cancer development (Nowell, 1976; Fearon and Vogelstein, 1990). Importantly, this does not occur in a fully linear order but in a branched, evolutionary process, resulting in multiple coexisting clones (Gerlinger et al., 2012; Landau et al., 2013; Siravegna et al., 2015).
Besides their genetic heterogeneity, cells within individual tumors can differ functionally. CRC progression and metastasis formation are driven by tumorigenic cells that are able to generate tumors in immune-deficient mice and are thought to underlie tumor progression, relapse, and spreading (O’Brien et al., 2007; Ricci-Vitiani et al., 2007; Dieter et al., 2011; Merlos-Suárez et al., 2011). These tumorigenic cells share characteristics with normal intestinal epithelial stem cells, including high self-renewal and multilineage differentiation capacity, and can be enriched as spheroids (Vermeulen et al., 2008). Genetic marking allows measurement of clonal output from individual tumor cells after transplantation of a mixture of tumorigenic and nontumorigenic cells because the exact integration site is unique to each transduced cell. As clonal behavior of single tumor cells in total isolation cannot be predicted, we termed tumorigenic cells that give rise to uniquely marked clones as tumor clone–initiating cells (TcICs). Using TcICs as a surrogate to measure the self-renewal and tumor-forming capacity of cell mixtures without single-cell isolation, it has been shown that, like the normal epithelial regenerative compartment, the tumorigenic CRC cell compartment itself is functionally heterogeneous and contains distinct subfractions, which differ in self-renewal capacity and activation kinetics. Highly self-renewing, long-term, active cells on top of a cellular hierarchy drive long-term disease progression and metastasis formation, whereas tumor-transient amplifying cells display limited self-renewal potential, and delayed contributing self-renewing cells contribute solely to tumor formation in secondary or tertiary mice (Dieter et al., 2011). Whether that functional heterogeneity is based on the presence of genetically distinct subclones is unclear.
To understand whether multiple, distinct genomic subclones with TcIC activity exist within individual tumors and whether genetic subclones determine the functional heterogeneity of CRC TcICs, in this study, we combined ultradeep whole-genome sequencing of primary patient tumors and derived serially transplanted xenografts and parallel spheroid cultures with secondary genetic marking.
Results
Genetic makeup of patient tumors as well as derived spheroids and xenografts
To address whether the TcIC compartment in human CRC is genetically heterogeneous, we established spheroid cultures from three CRC patient tumors (P1-TU, P2-TU, and P3-TU), as previously described (Dieter et al., 2011). Spheroid cultures are the most widely used model for enriching tumorigenic cells from primary, patient-derived cancer specimens without the need for cell-surface marker-based sorting strategies (Singh et al., 2004; Fang et al., 2005; Lee et al., 2006; Hermann et al., 2007; O’Brien et al., 2007; Ricci-Vitiani et al., 2007; Todaro et al., 2007; Vermeulen et al., 2008; Dieter et al., 2011; Merlos-Suárez et al., 2011). Early passage spheroid cells derived from each patient were transplanted into immune-deficient NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice and were serially retransplanted for three generations. In parallel, spheroids were cultivated for the duration of the serial xenotransplantations and were harvested simultaneously with each xenograft generation (Fig. 1 A).
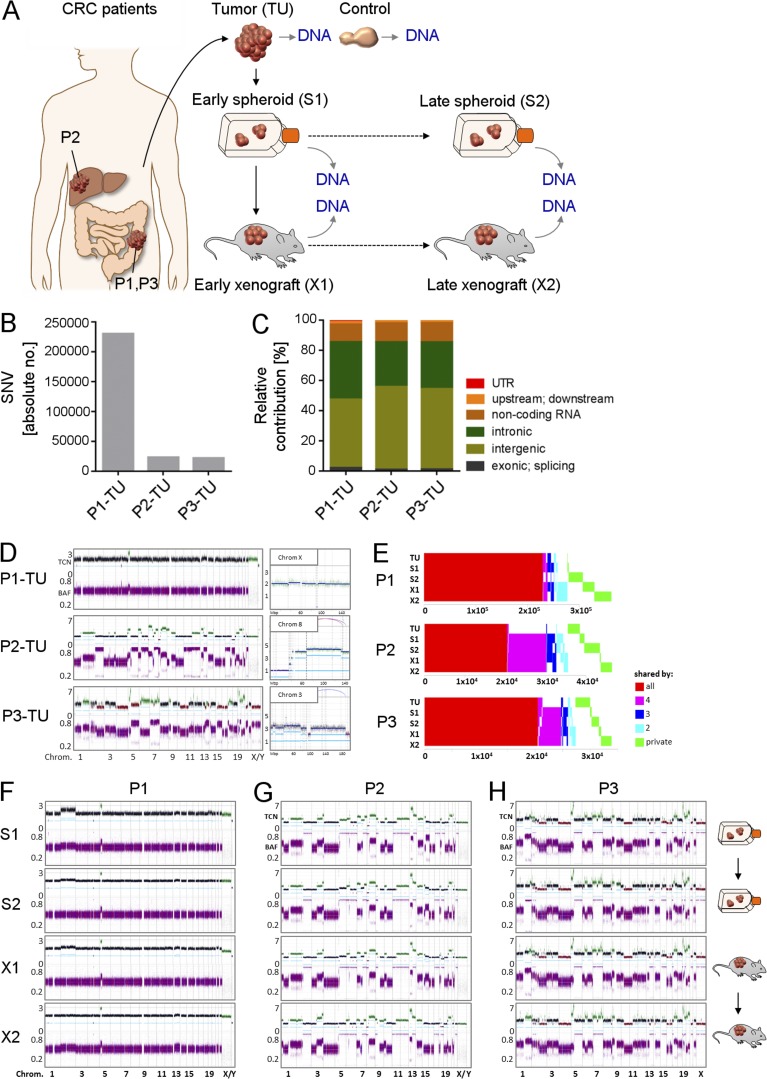
Figure 1.
Genetic makeup of primary CRCs, derived xenografts, and spheroids. (A) High-coverage WGS of primary CRC tissue (TU; n = 3 patients), derived serial xenografts (n = 6; 2 serial mice/patient), and parallel spheroids (n = 6; 2 serial passages/patient). (B) Number of SNVs in three primary tumors. (C) Distribution of SNVs in different genomic regions. (D) CNAs in patient tumors. (Top left) copy numbers with baseline ploidy (black), gains (green), and losses (red). (Bottom left) Raw BAF. (Right) Segments harboring noninteger copy numbers. X axis, genomic location; y axis, copy numbers; light-blue lines, allele-specific copy numbers; dark-blue lines, total copy number; Chrom, chromosome; Mbp, mega base pair. (E) Concordance and discordance of SNVs in serial patient-derived samples. X-axis, number of SNVs. (F–H) Copy number profiles of patient-derived xenografts and spheroid cultures from P1 (F), P2 (G), and P3 (H) as shown in D. (A–H) All experiments were performed independently for three CRC patients.
We first assessed the mutational landscape of primary patient tumors by high-coverage whole-genome sequencing (WGS; 110–126-fold; Fig. 1). We detected 23,121 SNVs, including synonymous and nonsynonymous SNVs in P2-TU and 22,830 in P3-TU. In P1-TU, the absolute number of SNVs was roughly 10-fold higher (232,387), translating into a mutation rate of 100 SNVs per Mbp and reflecting the clinical diagnosis of MLH1-driven Lynch syndrome with an associated microsatellite instability–high (MSI-H) genotype (Fig. 1 B). As expected, in all patients, most SNVs (95–97%) were located in noncoding areas of the genome (Fig. 1 C). Although P1-TU and P2-TU displayed diploid baseline copy numbers, P3-TU contained a triploid genome. In line with the MSI-H phenotype, P1-TU contained only few copy number alterations (CNAs). P2-TU and P3-TU comprised multiple CNAs, leading to their classification as chromosomally instable (CIN) tumors (Fig. 1 D).
Next, we assessed whether the genetic makeup of primary CRCs was maintained in spheroids and xenografts. Subsequently, all samples (early “S1” and late “S2” spheroids; early “X1” and late “X2” xenografts from the first and third xenograft generation, respectively) were sequenced genome wide with high coverage (102–120×). A large fraction of SNV was shared among all samples from an individual patient (P1:63%, P2:44%, and P3:61%). However, 37–56% of SNVs were found only in a subset of each patient’s samples or solely in individual samples (Fig. 1 E). The copy number profiles of spheroids and xenografts indicated the same base ploidies as the respective primary tumors, with a largely stable genome for all samples derived from P1 and highly chromosomally unstable genomes in P2- and P3-derived samples (Fig. 1, F–H). Together, WGS revealed that, although spheroid cultures and xenografts faithfully reflected the general tumor genomic makeup, differences in the individual mutations were present in each sample.
Genomic subclones are highly dynamic in spheroids and xenografts
Throughout this article, the operational term “growth clone” is used to describe a cluster of genomic alterations, i.e., SNV or CNA, with analogous dynamics over time, as defined by their relative contribution within serial sample sets, whereas “genomic subclone” defines cells carrying identical genetic aberrations, i.e., SNVs and CNAs.
In summary, we first used SNVs and CNAs to define “growth clones” with analogous dynamics over time (Fig. 2). For each growth clone, we then calculated the cellular fraction (CF). In a second step, we combined SNV and CNA growth clones, based on their CF, by applying combinatorics and maximum parsimony. This strategy allowed us to set up a model comprising the minimum number of genomic clones, defined as cells carrying identical genetic aberrations, which had to be present in each sample to explain the profound-growth clone kinetics.
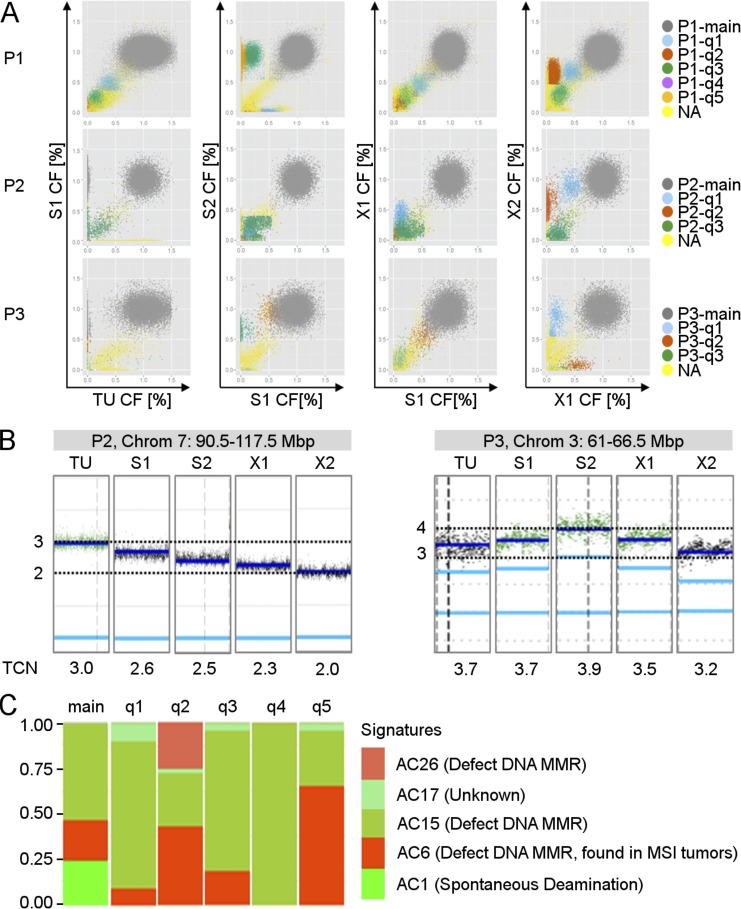
Figure 2.
Patient tumors, spheroids, and xenografts harbor distinct SNV- and CNV-based growth clones. (A) Each panel shows a pairwise comparison of CFs of the SNVs. Each dot represents one SNV. SNVs with similar kinetics over time were assigned to the same growth clone, colored accordingly, and either assigned to the main SNV-based growth clone (main; gray color), which is present in all cells, or to SNV-based growth clones, which are present in only a fraction of cells (q1-qx; colors as indicated). X and y axes, CF. (B) Representative chromosomal segments harboring subclonal copy numbers. Each small vertical panel shows the allele-specific copy number (light-blue line) and TCN (dark-blue line) for the chromosomal region indicated in the gray box above for one sample. Chrom, chromosome; Mbp, mega base pair. (C) Representative mutational profiles for growth clones of P1-X1. Each panel shows the relative signature exposures of one SNV-based growth clone. AC, Alexandrov COSMIC (nomenclature referring to http://cancer.sanger.ac.uk/cosmic/signatures (Alexandrov et al., 2013); Chrom, chromosome. See also Figs. S1 and S3 and Table S1. (A–C) Experiments were performed independently with tumor material from three CRC patients.
To identify SNV-based growth clones, the CF of SNVs was calculated (Materials and methods). Clusters of SNVs with similar CF dynamics over time were detected in all three patients (Fig. 2 A). SNVs contained in the main population represent fully clonal mutations, including alterations in coding regions of commonly mutated genes in CRC, such as APC and CTNNB1 (P1-main), KRAS (P1-main and P2-main), and TP53 (P1-main, P2-main, and P3-main; Table S1). Interestingly, for each patient, we detected three to five distinct subclonal SNV populations containing between 226 and 18,158 individual SNVs distributed throughout the entire genome (Fig. 2 A, Table 1, and Fig. S1). Importantly, for most growth clones, the CFs, i.e., the corrected allele frequencies, were not stable but changed dynamically over time during serial in vitro and in vivo passaging. Distinct growth clones were present at low CFs in S1 and grew out during long-term in vitro culture (e.g., P1-q3, P1-q5, and P3-q3), including SNVs in the coding regions of genes involved in deficient DNA mismatch repair (MMR), e.g., LIG1 (P1-q5). Other growth clones profoundly grew out during xenografting (e.g., P1-q1, P1-q2, P2-q1, P2-q2, and P3-q1; Fig. 2 A), including SNVs located in coding regions of MMR-related genes, e.g., ABL1 (P1-q1), CSDE1, and GLCCI1 (P1-q2), as well as the coding regions of commonly mutated genes in CRC, such as APC (P3-q1; Table S1).
Table 1. CFs of SNV- and CNA-based growth clones.
| Patient | Growth clone | No. SNVs | Chrom | Position | TCN | CF | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| TU | S1 | S2 | X1 | X2 | ||||||
| P1 | Mbp | % | % | % | % | % | ||||
| main | 194.680 | na | na | na | 105 | 100 | 100 | 100 | 100 | |
| P1-q1 | 2.937 | na | na | na | 40 | 45 | 0 | 45 | 65 | |
| P1-q2 | 18.158 | na | na | na | 0 | 5 | 0 | 15 | 60 | |
| P1-q3 | 5.640 | na | na | na | 15 | 20 | 95 | 20 | 25 | |
| P1-q4 | 226 | na | na | na | 35 | 40 | 0 | 30 | 0 | |
| P1-q5 | 15.353 | na | na | na | 0 | 0 | 90 | 0 | 0 | |
| P1-c1 | na | X | 1-155 | 1n | 0 | 10 | 0 | 40 | 10 | |
| P2 | ||||||||||
| main | 14.875 | na | na | na | 95 | 100 | 100 | 100 | 100 | |
| P2-q1 | 796 | na | na | na | 0 | 10 | 10 | 45 | 90 | |
| P2-q2 | 1.174 | na | na | na | 0 | 0 | 0 | 0 | 60 | |
| P2-q3 | 3.106 | na | na | na | 0 | 15 | 20 | 10 | 0 | |
| P2-c1 raw | na | 5 | 63-69 | 3 n | 0 | 70 | 40 | 30 | 10 | |
| P2-c1 raw | na | 5 | 71-180.5 | 3 n | 0 | 70 | 40 | 30 | 10 | |
| P2-c1 raw | na | 7 | 62-70.5 | 3 n | 10 | 60 | 40 | 20 | 0 | |
| P2-c1 raw | na | 7 | 70.5-88.5 | 3 n | 0 | 70 | 40 | 30 | 0 | |
| P2-c1 combined | na | 5 and 7 | na | na | 0 | 70 | 40 | 30 | ≤10 | |
| P2-c2 raw | na | 7 | 90.5-117.5 | 3 n | 100 | 60 | 50 | 30 | 0 | |
| P2-c2 raw | na | 7 | 117.5-159 | 4 n | 100 | 60 | 40 | 30 | 0 | |
| P2-c2 combined | na | 7 | na | na | 100 | 60 | 40-50 | 30 | 0 | |
| P2-c3 raw | na | 18 | 26.5-36.5 | 1 n | 100 | 10 | 10 | 40 | 80 | |
| P2-c3 raw | na | 18 | 38.5-43 | 1 n | 100 | 20 | 20 | 50 | 90 | |
| P2-c3 raw | na | 18 | 48.5-78 | 1 n | 100 | 10 | 10 | 40 | 90 | |
| P2-c3 combined | na | 18 | na | na | 100 | 10 | 10 | 40 | 90 | |
| P2-c4 raw | na | 3 | 95-102.5 | 4 n | 0 | 10 | 0 | 10 | 60 | |
| P2-c4 raw | na | 3 | 105-175.5 | 4 n | 0 | 0 | 0 | 10 | 60 | |
| P2-c4 raw | na | 3 | 175.5-198 | 5 n | 0 | 0 | 0 | 0 | 70 | |
| P2-c4 combined | na | 3 | na | na | 0 | 0 | 0 | 10 | 60 | |
| P2-c5 | na | 8 | 68-131.5 | 5 n | 40 | 0 | 0 | 0 | 0 | |
| P2-c6 | na | 22 | 17.5-20 | 3 n | 50 | 0 | 0 | 0 | 0 | |
| P3 | ||||||||||
| main | 13.625 | na | 13.625 | na | 100 | 100 | 100 | 100 | 100 | |
| P3-q1 | 608 | na | 608 | na | 0 | 10 | 0 | 15 | 85 | |
| P3-q2 | 343 | na | 343 | na | 0 | 55 | 95 | 55 | 10 | |
| P3-q3 | 390 | na | 390 | na | 0 | 0 | 65 | 0 | 0 | |
| P3-c1 raw | na | 3 | 1-7 | 4 n | 30 | 0 | 0 | 0 | 0 | |
| P3-c1 raw | na | 3 | 13-29 | 4 n | 20 | 0 | 0 | 0 | 0 | |
| P3-c1 raw | na | 3 | 29.5-33.5 | 4 n | 20 | 0 | 0 | 0 | 0 | |
| P3-c1 raw | na | 3 | 52.5-61 | 4 n | 30 | 0 | 0 | 0 | 0 | |
| P3-c1 combined | na | 3 | na | na | 20-30 | 0 | 0 | 0 | 0 | |
| P3-c2 | na | 3 | 7-13 | 4 n | 50 | 0 | 0 | 0 | 0 | |
| P3-c3 raw | na | 3 | 40.5-51.4 | 4 n | 30 | 10 | 0 | 10 | 10 | |
| P3-c3 raw | na | 3 | 66.5-75.5 | 4 n | 30 | 0 | 0 | 10 | 10 | |
| P3-c3 combined | na | 3 | na | na | 30 | ≤10 | 0 | 10 | 10 | |
| P3-c4 | na | 3 | 61-66.5 | 4 n | 70 | 70 | 90 | 50 | 20 | |
| P3-c5 | na | 16 | 53.5-56 | 2 n | 100 | 30 | 0 | 10 | 0 | |
| P3-c6 | na | 10 | 75-79 | 3 n | 10 | 0 | 60 | 0 | 0 | |
All noninteger CNA segments included are ≥2.5 Mbp, harbor maximal one noninteger allele, and have a minimum deviation of 30% from both contributing copy numbers in at least one sample (TU, S1, S2, X1, or X2). Breakpoints were rounded to the nearest 0.5 Mbp. Individual “raw” CNA segments were “combined” to CNA-based growth clones. Cellular fractions of raw CNA segments and CNA-based growth clones were rounded to the nearest 10%, cellular fractions of SNV-based growth clones to the neatest 5%. All experiments were performed independently with tumor material from three patients with CRC. c, CNA-based growth clone; na, not applicable; q, SNV-based growth clone.
In addition to SNVs, we used CNAs to identify growth clones. Although the majority of CNAs were clonal (1n, 2n, 3n, etc.), subclonal CNAs with noninteger copy numbers could be identified in all patients (Fig. 1 D, right; and Fig. 2 B). In the next step, CFs were calculated for 25 noninteger CNA segments from the total copy number (TCN) and grouped into 13 CNA-based growth clones, according to concordant clonal dynamics (P1:1, P2:6, and P3:6; Materials and methods). Similar to SNV-based growth clones, the CFs of CNA-based growth clones were not stable in serial passaging but showed highly dynamic kinetics in serial xenografts and spheroids (Fig. 2 B and Table 1).
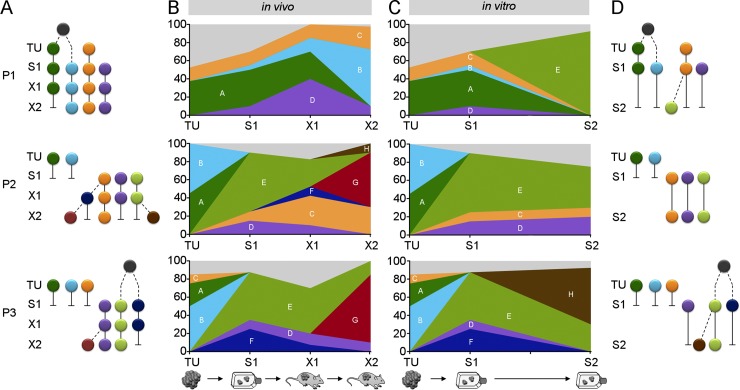
Next, we combined SNV- and CNA-based growth clones, applying combinatorics and maximum parsimony to define the minimum number of genomic subclones that had to be present in each sample to explain the profound growth-clone kinetics (Fig. 3, Table 1, and Fig. S2; Materials and methods). This strategy revealed that all primary tumors contained at least two coexisting genomic subclones. Furthermore, genomic-subclone heterogeneity was detectable in S1 with at least three coexisting genomic subclones. Tumor initiation in X1 was driven by a minimum of three to four coexisting genomic subclones. In addition, multiple genomic subclones were present in X2, demonstrating that long-term tumor formation was driven by coexisting genomic subclones. Interestingly, although the majority of genomic subclones identified in X2 already contributed to X1, their relative contribution changed substantially. Distinct genomic subclones grew out, whereas other genomic subclones eventually decreased or even vanished. Similarly, long-term spheroid cultivation resulted in outgrowth and loss of genomic subclones. Strikingly, within individual patients, completely different genomic subclones grew out and became dominant in vitro or in vivo. In summary, subclone combinatorics revealed that primary patient tumors, spheroids, and xenografts comprised multiple, coexisting genomic subclones. Those subclones were not static but displayed substantial dynamic fluctuation, with different genomic subclones dominating the xenografts and spheroids of the same patient.
Figure 3.
Genomic subclone kinetics in serial in vivo and in vitro passaging. (A) Ancestral trees of identified genomic subclones in patient tumors, early spheroids, and serial xenografts. (B) Relative contribution of genomic subclones from A in each sample. (C) Relative contribution of genomic subclones in patient tumors and serial spheroids. (D) Ancestral trees for genomic subclones shown in (C). Colors, defined genomic subclones; light gray, unidentified genomic subclones; gray circles, common ancestors; dashed lines, phylogeny constructed applying maximum parsimony. See also Fig. S2. (A–D) Experiments were performed independently with tumor material from three CRC patients.
Genomic subclones harbor distinct mutational signatures
Mutational processes leave mutational signatures in the genome, which are characterized by specific nucleotide exchanges in the motif context of given SNVs (Alexandrov et al., 2013). In the main SNV population of all patients, stratified signature analysis identified a clocklike, spontaneous deamination signature, which represents mutations accumulated in a common ancestor before malignant transformation (Alexandrov et al., 2015). Other signatures in the main populations were associated with deficient MMR (P1 and P3), double-strand break (DSB) repair (P2 and P3) or the APOBEC family (P3; Fig. 2 C and Fig. S3). Although most of these signatures were also detectable in corresponding growth clones, the relative contribution of the signatures differed significantly among growth clones. Strikingly, within an individual growth clone, the relative contribution of a given signature remained stable during serial in vivo and in vitro passaging, thereby strongly supporting our strategy of genomic subclone definition. Likewise, we found that individual growth clones harbored private mutations in genes commonly affected in CRC or involved in MMR and DSB repair (Table S1). It is tempting to speculate that such mutations may have driven development of individual genomic subclones; however, that question needs to be addressed in future studies. In summary, mutational signature analysis strongly supported the presence of distinct genomic subclones with different mutational signatures present in serial xenografts and spheroids.
Functional heterogeneity is maintained despite highly variable genomic-subclone contribution
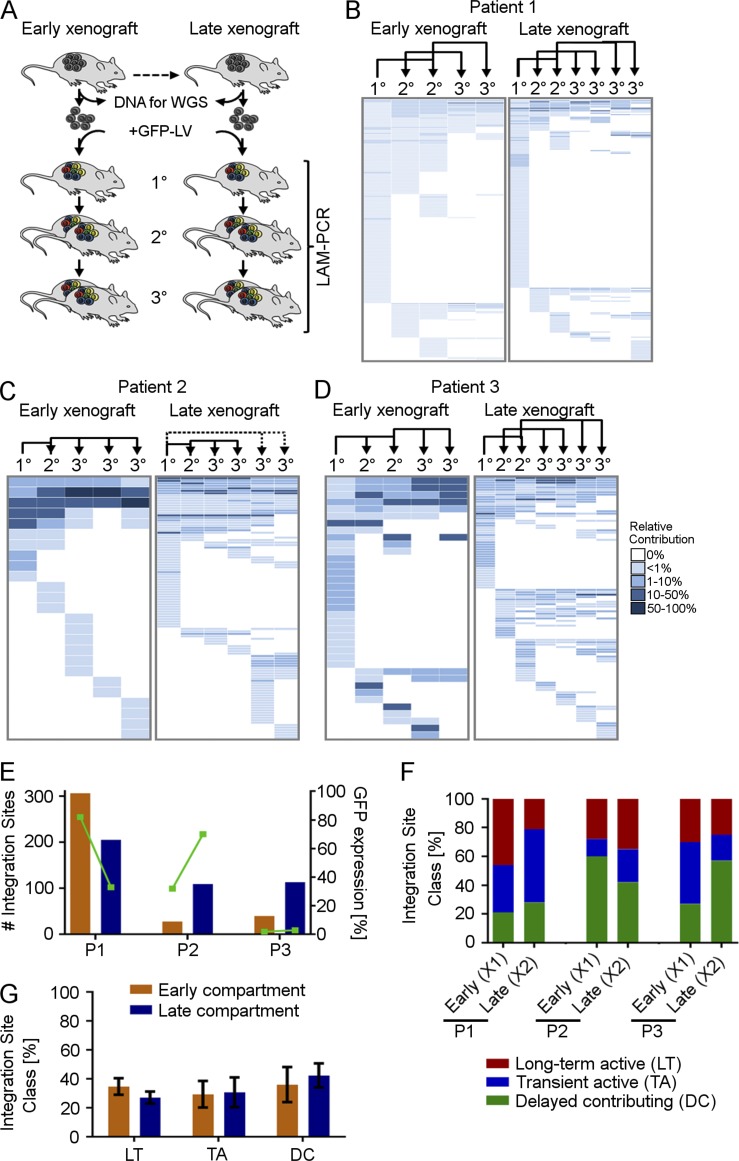
We have shown before that the tumorigenic cell compartment in CRC is hierarchically organized and comprises three distinct classes of cells driving tumor formation in vivo (Dieter et al., 2011): (1) long term active cells with a high self-renewal capability that rebuild tumors after serial transplantation, (2) tumor-transient amplifying cells with no detectable self-renewal potential that only transiently contribute to tumor formation, and (3) delayed contributing, self-renewing tumorigenic cells that do not substantially drive primary tumor growth but are recruited to tumor formation after serial transplantation. To test whether this functional heterogeneity is driven by genetic differences of distinct genomic subclones, we assessed the contribution of these functionally heterogeneous cell fractions during early and late serial xenografting by secondary genetic marking (Fig. 4). Therefore, representative cell aliquots from sequenced X1 and X2 xenografts were transduced with an integrating lentiviral vector and serially transplanted for three successive generations. Because the vector semirandomly integrates into the genome, each transduced cell harbors a unique integration site (IS), which is stably passed onto daughter cells. Consequently, the ISs can be used as individual molecular barcodes for initially transduced clones (Dieter et al., 2011). Because we could not predict the clonal behavior of single cells in complete isolation, we assessed tumorigenic cells giving rise to uniquely marked clones (TcICs) within tumors as surrogates. After tumor formation, genetically marked tumors were harvested and used for highly sensitive linear amplification–mediated PCR (LAM-PCR), followed by amplicon sequencing. IS analysis allowed a determination of the contribution of each individual transduced (TcIC) clone in serial transplants (Fig. 4 A). Interestingly, all samples contained multiple ISs (Fig. 4, B–D). The absolute number of ISs in individual, transduced xenografts from each patient reflected the cellular-transduction rate (Fig. 4 E). The clonal dynamics of marked TcIC clones in serial transplantation demonstrated that, in all three patients, highly proliferative, self-renewing cells; transiently active cells; and delayed contributing cells were present in the early xenografts (X1), demonstrating their hierarchical organization. Strikingly, we detected similar contributions of the different subtypes to the marked serial xenografts derived from late-passage xenografts (X2; Fig. 4 F). Although the relative proportion eventually differed among different patients, the content and functional heterogeneity within the TcICs were maintained in serial xenografts from the same patient, despite profound, near-complete changes in the genomic subclone contribution (Fig. 4 G). These data strongly indicate that functional TcIC hierarchies were present within at least multiple genomic subclones and were not caused by differences in the genetic mutations driving different genomic subclones.
Figure 4.
Functional heterogeneity of TcIC early and late in serial transplantation. (A) Tumor cells of early and late xenograft tumors, used for assessing genetic heterogeneity by WGS, were genetically barcoded and serially transplanted for three mouse generations. The contribution of individually marked cell clones was assessed by LAM-PCR and high-throughput sequencing. Relative contribution of individually marked cell clones to tumor formation in serial transplantation from P1 (B), P2 (C), and P3 (D). Each row displays one unique IS; each column displays one xenograft in serial transplantation. Dotted lines, 2° n/a. Arrows indicate serial transplantation steps. (E) Total number of ISs detected in serial transplants derived from X1 or X2 and corresponding mean GFP expression (green). (F) Relative contribution of functional tumor-initiating cell classes to early and late TcIC compartments in individual xenografts, and (G) on average, from three patients. Error bars represent the SEM. (A–G) All experiments were performed independently with tumor material from three CRC patients.
Discussion
The functional heterogeneity of individual tumors is well established in human CRCs, with a tumorigenic cell fraction with stem-like properties, i.e., self-renewal and differentiation capacity, driving disease progression in serial transplantation models (O’Brien et al., 2007; Ricci-Vitiani et al., 2007). Moreover, others and we have shown that this tumorigenic cell compartment itself is functionally heterogeneous with distinct subclasses differing in their self-renewal and metastasis-forming capacity (Dieter et al., 2011; Kreso et al., 2013). Apart from functional differences, coexisting genetic subclones in individual patient tumors have been described in many cancers (Anderson et al., 2011; Gerlinger et al., 2012; Landau et al., 2013). However, it remained an unsolved question whether functional tumor heterogeneity was based on fixed, differing genetic makeups that conferred functional consequences to subclones, i.e., self-renewal and tumor-initiating capacity.
We demonstrate in this study that distinct genomic subclones are present in CRC primary tumors, xenografts, and spheroids. Assessing the relative contribution of tumor-cell classes differing in their tumor-forming and self-renewal capacity, as described before (Dieter et al., 2011), revealed that the functional heterogeneity was stable even after three rounds of serial transplantation. Self-renewing, long-term active tumorigenic cells were not enriched, demonstrating that serially xenotransplanted tumors were not formed by highly proliferative clones without functional heterogeneity. At the same time, profound changes in genomic subclone contribution resulted in almost complete changes of the subclonal repertoire in these tumor models. These findings strongly indicate that all individual genomic subclones are organized in a functional hierarchy, giving rise to all distinct “private” subtypes of self-renewing tumorigenic cells during serial xenotransplantation. One could speculate that the sequential addition of further relevant mutations to existing genomic subclones could alter the functional heterogeneity of affected clones. Although, in different patients, the neoplastic process is fueled by different sets of driver mutations, the malignant stem-cell compartment in colon cancer consists of hierarchical cell classes with crucial differences in self-renewal and metastatic potential, whose occurrence and kinetics cannot be explained by a stochastic contribution (Dieter et al., 2011). By using concordant dynamics of large numbers of SNVs and CNVs to identify genomic subclones, our study did not find any evidence that sequential accumulation of mutations determines subclone dynamics during the time span of serial transplantation. Of note, the identified subclones preexisted at the time of tumor resection and did not freshly occur during serial in vitro and in vivo passaging because they were defined by large numbers of independent mutations, thereby precluding conclusions regarding the initial transformation process.
Earlier studies using targeted or exome sequencing and SNV genotyping alone or in combination reported stable genetics in serial xenotransplantation and functional heterogeneity in genomically stable, single-cell–derived tumor xenografts (Julien et al., 2012; Kreso et al., 2013; Bossi et al., 2016). Our results indicate that, in those studies, the interclonal dynamics were largely underestimated because of the lower sensitivity for genomic subclone detection. In contrast, more-sensitive WGS revealed highly dynamic genomic subclones in xenografts and spheroid cultures. Tracing clonal dynamics demonstrates that, in competitive environments, multiple, coexisting genomic subclones within individual patient-derived xenografts fuel long-term tumor initiation, suggesting that the long-term tumor-initiating capacity is a basic characteristic of several genomic subclones and not restricted to a specific clone within an individual tumor.
We would like to emphasize that all genomic subclones derive from a common ancestor and are, thereby, of clonal origin by definition. The additional genetic variation generated by accumulation of mutations during further progression enables robust identification of genomic subclones. Sequencing coverage of >100 times and applying a minimalistic model, in this study, allowed us to identify clones with a contribution of at least 5–10% within a given sample. The fact that we detected slightly fewer genomic subclones in primary tumors than in spheroids and xenografts from the same patient can likely be attributed to the lower tumor cell content in patient tumors. Selective engraftment of subclonal tumor-cell populations in xenograft and in vitro models points to initial seeding efficiency as a major bottleneck for studying subclone composition in experimental models (Klco et al., 2014; Eirew et al., 2015; van de Wetering et al., 2015). Notably, in our study, a single-cell suspension was prepared from P1 primary tumor cells, and equal proportions of cells were used for spheroid culture and xenotransplantation and tumor sequencing. In contrast, sequencing of P2 and P3 patient tumor tissue and the generation of serial samples were performed from independently sampled tumor pieces of the same patient’s tumor. Regional differences in subclone composition in the patient tumor (Sottoriva et al., 2015) sampled for tumor tissue sequencing and generation of spheroids most likely contributed to the increased mutational differences among primary tumors and the derived serial samples in our setting.
In agreement with our results, coexistence of multiple genetic subclones and variation of genetic subclones in patient-derived organoid models and xenografts have previously been observed in solid cancer tumors and other entities (Anderson et al., 2011; Schmitz et al., 2011; Klco et al., 2014; van de Wetering et al., 2015), which raises the question of whether specific genetic events pivotally fuel the proliferation and survival of individual subclones. Interestingly, we found only a few mutations in commonly mutated CRC genes or in genes involved in DNA MMR pathways attributable to subclonal SNV populations. Private point mutations present in individual growth clones in relevant genes, including β-catenin or APC, in our study, point toward a functional role for those private mutations in the development of individual genomic subclones. The functional effect of those mutations, however, needs to be addressed in future studies. The majority of the tested potential driver SNVs were fully clonal, which is in line with recent multiregion sequencing studies, indicating that most common mutations in CRCs are shared throughout all sites and are most probably acquired early during cancer progression (Jesinghaus et al., 2015; Kim et al., 2015; Sottoriva et al., 2015). Interestingly, in our study, different genomic subclones dominated in vitro cultures or in vivo xenografts. This may be a consequence of different environmental conditions, including oxygen levels, anchorage-independent growth, and other microenvironmental factors, and opens up an avenue for studying the impact of such factors on subclone dynamics in controlled models (Visvader and Lindeman, 2012; Weiswald et al., 2015).
Two limitations of our study need to be considered. First, our experimental approach required transplantation of human CRC cells under the kidney capsule of highly immune-deficient NSG mice. We and others have previously shown that this strategy allows highly efficient engraftment of primary patient tumor cells (O’Brien et al., 2007; Ricci-Vitiani et al., 2007; Dieter et al., 2011). We did not transplant orthotopically due to the requirement to transplant large numbers of tumor cells to allow for representative and sensitive genetic subclone detection, which is not feasible at orthotopic locations. Orthotopic transplantation of CRC cells may, in theory, provide a more-physiological environment, even though that has not been proven experimentally. Moreover, the role of the immune system cannot be assessed in experimental models that allow in vivo tumor formation by human cancer cells because of the immunosuppression required. Further, the disruption of the original tumor architecture, as well as the transplantation of human tumor cells into a xenogenic environment, may influence the behavior of the assayed tumor-cell population. Second, our study was done with the limited number of three CRC samples. The results observed were very similar among those patients, irrespective of whether primary tumor– or metastasis-derived tissue was analyzed, indicating that our results are at least applicable to a large proportion of CRCs.
Considering the number of cells used in our study in comparison to the overall cell load of a patient’s tumor, many genomic subclones presumably coexist in a patient’s tumor at the time of diagnosis. Consequently, sequential therapy or intermittent drug schedules may cause profound subclone dynamics because of dynamic-adaptation processes (Siravegna et al., 2015). Because functional and genetic heterogeneity within the TcIC compartment is reflected in patient-derived xenografts and TcIC-enriched spheroids, they represent suitable models for identifying subclone selection and the de novo alterations underlying therapy sensitivity and resistance.
In conclusion, in this study, we demonstrated that, in CRC, multiple subclones drive long-term tumor formation. Individual subclones can grow out during serial in vitro and in vivo passaging, and within the same patient, the dominating subclones in the xenografts and spheroids can differ. Strikingly, functional differences and genetic-clone diversity are two independent layers of heterogeneity within individual CRC tumors that need to be addressed therapeutically to overcome treatment resistance.
These findings have important implications for the development and design of future studies targeting tumorigenic CRC cells.
Materials and methods
Isolation and culture of CRC cells
All experiments with human material were performed in accordance with the guidelines of the Declaration of Helsinki and were approved by the ethics committee of the Medical Faculty at the University Heidelberg (323/2004). Informed consent was received from participants before study inclusion. Representative pieces of tumor tissue and matching normal control (P1, whole blood; P2 and P3, normal intestinal mucosa) were collected from patients undergoing surgery at the Department of Surgery, University Hospital (Heidelberg, Germany), and representative pieces were frozen at −80°C for histologic assessment. To obtain single-cell suspensions, tumor tissue was cut into 2–3-mm, small pieces and digested with 0.08 U Dispase (BD) per ml and 50 µM magnesium chloride, supplemented with 1% antibiotic–antimycotic in a rotating incubator for 1.5 h at 37°C. Digested cells were filtered, washed, and cultured in advanced DMEM/F-12 (Gibco/Invitrogen), supplemented with 0.6% glucose, 2 mM PenStrep, and 2 mM l-glutamine (all Invitrogen); 4 µg/ml heparin, 5 mM Hepes, and 4 mg/ml BSA (all Sigma-Aldrich); and 10 ng/ml human fibroblast growth factor and 20 ng/ml epidermal growth factor (R&D Systems) in ultralow attachment flasks (Corning). For each patient, spheroid cultures were established and seeded for serial passaging (105 cells for P1 and P3 and 2 × 105 cells for P2). Spheroid cultures were serially passaged for the entire period of serial xenografting (P1, 190 d, equal to ∼16 passages; P2, 107 d, equal to ∼13 passages; and P3, 142 d, equal to ∼6 passages), and 6–34% (equal to 5 × 105–1.7 × 107 cells) were transferred at each passage step. In total, six spheroid cultures were sequenced by WGS (early and late passage cultures for 3 patients). For P1, the primary tumor sample sequenced was obtained from the single-cell suspension, which was also used for spheroid formation. From the primary tumors of P2 and P3, adjacent tumor pieces were used for spheroid-culture generation or sequencing.
Xenotransplantation
Primary tumor–derived spheroid cells or serial xenograft–derived, singularized tumor cells were suspended in 50 µl culture media, mixed 1:1 with Matrigel (BD), and transplanted under the renal capsule of NSG mice (The Jackson Laboratory), which were housed under pathogen-free conditions, according to the German Animal Protection laws and regulations approved by the ethical committee. For serial whole-genome sequenced xenografts, 105 cells were transplanted into one primary mouse per patient, and after xenograft formation, single-cell suspensions were prepared, and aliquots were serially transplanted or saved for molecular analysis; 13–50% (equal to 1.5 × 106–3.6 × 107) xenograft cells were serially transplanted into one secondary and one tertiary mouse. In total, six xenografts were whole-genome sequenced, i.e., early (first-generation) and late (third-generation) xenografts for three patients. Because mouse DNA lowers the effective sequencing coverage, mouse cells were depleted from xenograft tumors by FACS (BD). To that end, the xenograft tumors were dissected into a xenograft part and a xenograft/kidney-junction zone. Both fractions were dissociated and purified separately. The xenograft/kidney-junction fraction was sorted with a human-specific epithelial cell-adhesion molecule antibody (EBA-1, 1:10; BD). The sorted cells were subsequently combined with the separately digested cell fraction. For the clonal-marking arms, 105 transduced cells from the early or late xenograft were initially transplanted into one primary mouse per patient; 11–50% (equal to 1.3–9 × 106 cells) were serially transplanted for three generations. In total, 12 gene-marked xenografts were analyzed for P1, 11 xenografts were analyzed for P2, and 12 xenografts were analyzed for P3.
Library preparation and WGS
DNA was extracted from primary patient tumors, matching healthy tissues, serial xenografts, and serial spheroids using AllPrep DNA/RNA/miRNA Universal kit (80224; QIAGEN). Identity of samples was ensured by ESSplex (QIAGEN) analyses. Library preparation was performed with the NEBNext DNA Library Prep Master Mix Set (E6040L; Illumina) and the NEBNext Multiplex Oligos (set 1 and set 2; E7335S und E7500S; Illumina). Library concentrations were measured using Qubit 3.0 (Thermo Fisher Scientific), and quality was assured with a Bioanalyzer/TapeStation (Agilent Technologies). Genomic DNA was sequenced on an HiSeq 2000 (Illumina) with 100-bp, paired-end reads (European Genome-Phenome Archive; accession no. EGAS00001001857). To decrease duplicate rates, three libraries were prepared for each individual neoplastic sample and distributed to three to four HiSeq 2000 lanes each, resulting in 10 lanes per sample, sequenced with 100-bp paired-end reads. From control samples, one library was prepared and sequenced on three lanes. Read pairs were mapped to the 1000 Genomes Project phase 2 assembly of the human reference genome (hs37d5) using Burrows-Wheeler Aligner software (version 0.6.2; Li and Durbin, 2009). Duplicates were marked with Picard tools (version 1.90, http://broadinstitute.github.io/picard/). For alignment of xenograft samples, a combined human–mouse reference genome was constructed as a concatenation of the 1000 Genomes Project phase 2 assembly of the human-reference genome (NCBI build 37.1, downloaded from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/reference/phase2_reference_assembly_sequence/hs37d5.fa.gz) and the Ensembl version 73 mouse reference genome (downloaded from ftp://ftp.ensembl.org/pub/release-73/fasta/mus_musculus/dna/Mus_musculus.GRCm38.73.dna.primary_assembly.fa.gz).
SNV identification
Somatic SNVs in tumor, xenograft, and spheroid binary alignment/map files were identified using an in-house–developed pipeline based on SAMtools mpileup and bcftools (version 0.1.19, Li et al., 2009) It involves parameter adjustments with heuristic filtering to allow calling of somatic variants (Jones et al., 2012, 2013). ANNOVAR (Wang et al., 2010) was used to annotate the variants with Gencode version 19. Variant allele frequencies (VAFs) of the SNVs were calculated from the reference and from alternative read counts of the samtools mpileup output. To prevent calling biases in the growth-clone analysis, all high-confidence SNVs that were identified in a subset of samples from one patient were used as a lookup identifier in the other samples, using samtools mpileup, and VAFs were calculated as described for the directly identified variants.
Estimation of CF
The CF of a variant indicates the proportion of tumor cells that carry the variant, which can be calculated from the VAF by correction for tumor-cell content and regional allele-specific copy numbers. The estimation of the CF was performed in a two-step process. First, all VAFs were normalized for tumor-cell content. Tumor-cell content was estimated by allele-specific copy number estimation from sequencing (ACEseq) analyses (described in the Copy number estimation section) as follows: P1-TU, 0.55; P1-S1, 1.0; P1-S2, 1.0; P1-X1, 1.0; P1-X2, 1.0; P2-TU, 0.88; P2-S1, 1.0; P2-S2, 1.0; P2-X1, 1.0; P2-X2, 1.0; P3-TU, 0.55; P3-S1, 1.0; P3-S2, 1.0; P3-X1, 1.0; and P3-X2, 1.0. Second, the VAFs were corrected for regional copy numbers. Only SNVs in regions with clonal copy numbers and fewer than four copies in all five samples per patient were considered for correction. Based on the copy-number state and the tumor-cell content-corrected VAF, it was estimated whether an SNV was most likely found on one, two, or three alleles, i.e., variants in diploid regions with corrected VAFs <0.8 were considered to be present on one allele. In triploid regions, cutoffs were set to 0.5 and 0.8 for one and two alleles, respectively. VAFs of variants in diploid regions found on one allele (i.e., with corrected VAFs <0.8) were multiplied by a factor two. Variants in triploid regions found on one allele were multiplied by three, whereas variants on two alleles in triploid regions required a factor 1.5. Variants in single-copy regions did not need further correction.
Determination of SNV-based growth clones
Scatterplots of the CFs of pairwise combinations from all samples per patient, similar to Fig. 2 A, were used to define SNV-based growth clones. A growth clone was defined as a distinct cluster of SNVs in those scatterplots. For each patient, the main population was defined as the bulk of points distributed around 1.0 in all samples. Clusters found in a single scatterplot were color coded and tracked in all other samples for the respective patient. Integration of information from scatterplots of other pairwise combinations was used to refine the cluster boundaries and to increase cluster separation. SNVs that could not be assigned to any cluster remained in an unassigned state (na). One additional growth clone (P1-q4) was defined during combinatorics. In that case, two genomic subclones (A and B, defined by P1-q1 and P1-q1 + P1-q2) could be tracked to the same founder clone, requiring an additional growth clone to explain the unique SNVs of one of the newly evolved clones (genomic subclone A; see the Genomic subclone combinatorics section). For all SNV-based growth clones, the median CF was calculated and rounded to the nearest 5% precision.
Supervised analysis of mutational signatures
A supervised analysis of mutational signatures determines contributions of known mutational signatures to the mutational catalog, i.e., the counts of the nucleotide exchanges in their motif context, of a given data set. As opposed to an unsupervised analysis of mutational signatures, i.e., a discovery method such as nonnegative matrix factorization as used in (Alexandrov et al., 2013), a supervised analysis requires much less statistical power but will not discover new signatures. Software to carry out such an analysis was written as an R package (YAPSA [Yet Another Package for Signature Analysis]). Because of the low requirements on statistical power, i.e., the number of samples in a cohort and the number of point mutations per sample, the supervised analysis of mutational signatures, as performed by YAPSA, may be run on very small cohorts.
Using YAPSA, a linear combination decomposition of the mutational catalog with known and predefined signatures was computed by nonnegative least squares (NNLS), with its functions implemented in the R package limSolve (Van Den Meersche et al., 2009). To increase specificity, the NNLS algorithm was applied twice: once proposing all signatures supplied by the user to the decomposition, and then, after the first execution, only those signatures with exposures (i.e., contributions in the linear combination) greater than a certain cutoff were kept, and the NNLS was run again with the reduced set of signatures. Because detectability of the different signatures can vary, signature-specific cutoffs were chosen, as determined in a receiver operating characteristic analysis from publicly available data on mutational catalogs of 7,042 cancers (507 from WGS and 6,535 from whole-exome sequencing; Alexandrov et al., 2013) and mutational signatures (http://cancer.sanger.ac.uk/cosmic/signatures). Before running an analysis, YAPSA builds up the mutational catalogs of the samples or cohorts to be analyzed by calling the functions mutationContext and mutationContextMatrix from the R package SomaticSignatures (Gehring et al., 2015).
YAPSA was used to stratify the analysis of mutational signatures. For each patient, the growth clones (as defined in the section Genomic subclones are highly dynamic in spheroids and xenografts) were taken as strata. The stratified analysis was performed as a multistep procedure: (1) to determine which mutational signatures were present in the sample, the supervised analysis of mutational signatures was first run (as described in Genomic subclones harbor distinct mutational signatures section) without any stratification; (2) for every detected SNV in a sample, the stratum it belonged to (i.e., the growth clone, as determined by an analysis of CFs as described in Genomic subclones are highly dynamic in spheroids and xenografts section) was annotated in the initial variant-calling file (vcf-like); (3) for every stratum, a stratum-specific mutational catalog was built (as described in the Supervised analysis of mutational signatures section); and (4) enrichment and depletion patterns for all mutational signatures detected in step 1 were computed from the mutational catalogs of all strata. Step 4, implemented in YAPSA, involved a supervised and constrained analysis of mutational signatures, performed by constrained NNLS, also using limSolve (Van Den Meersche et al., 2009), with the constraint as the sum over the strata of exposures per stratum equaling the exposures computed by the unstratified analysis.
To obtain characteristic enrichment and depletion patterns per signature, cohort-wide averages per stratum over the sample-wise exposures were computed. Differences among strata were tested by Kruskal-Wallis tests and corrected for multiple testing with the Benjamini-Hochberg procedure (P1: AC1, p = 1.331 × 10−4, AC6, p = 1.586 × 10−4, AC15, p = 1.586 × 10−4, AC17, p = 1.586 × 10−4, and AC26, p = 6.680 × 10−3; P2: AC1, 1.416 × 10−3, AC3, 2.611 × 10−3, AC9, 2.626 × 10−3, AC13, 1.416 × 10−3, and AC17, 1.416 × 10−3; and P3: AC1, 3.408 × 10−2, AC3, 3.408 × 10−2, AC8, 5.125 × 10−2, AC9, 5.125 × 10−2, AC13, 4.019 × 10−2, and AC15, 3.112 × 10−1). Absolute and relative exposures per sample and per stratum as well as cohort-wide averages were displayed in integrative figures.
Driver- and signature-related genes
Commonly mutated genes in CRC were defined from The Cancer Genome Atlas (Cancer Genome Atlas Network, 2012), MMR, and DSB homologous recombination or DSB nonhomologous end-joining from the Kyoto Encyclopedia of Genes and Genomes (KEGG; hsa03440, hsa03450, and hsa03430; Kanehisa and Goto, 2000; Kanehisa et al., 2016) and the Gene Ontology (GO; GO0000724, GO0006303, and GO0006298; Ashburner et al., 2000; Gene Ontology Consortium, 2015) databases. Variants were annotated with Combined Annotation-Dependent Depletion scores (version 1.3; Kircher et al., 2014).
Structural variant calling
The CREST algorithm (Wang et al., 2011), with default parameters, was used to call structural variants from the primary alignment data.
Copy number estimation
Copy numbers were estimated using ACEseq. This method uses a coverage ratio of tumor and control over genome windows as well as the B-allele frequency (BAF). In addition to the copy numbers, tumor ploidy and tumor-cell content were estimated in ACEseq. During preprocessing of the data, allele frequencies were obtained for all single-nucleotide polymorphism (SNP) positions recorded in the dbSNP database (Database of Single Nucleotide Polymorphisms [dbSNP]. Bethesda [MD]: National Center for Biotechnology Information, National Library of Medicine. [dbSNP Build ID: 135]. Available from: http://www.ncbi.nlm.nih.gov/SNP/). To improve sensitivity for unbalanced and balanced regions, SNP positions in the control were phased with impute2 (Howie et al., 2009). Additionally, the coverage for 10 kbp windows with sufficient mapping quality and read density was recorded and subsequently corrected for guanine-cytosine content and replication timing to remove coverage changes introduced by those biases.
The genome was segmented using the PSCBS package in R software (Olshen et al., 2011) in consideration of structural-variant breakpoints defined by CREST. Segments were clustered, using k-means clustering, according to their coverage ratio and BAF value, and neighboring segments that fell into the same cluster were joined. Small segments (<9 kbp) were attached to the more-similar neighbor. Finally, tumor-cell content and the ploidy of the samples were estimated by testing how well different tumor-cell content and ploidy combinations explained the data. Segments with balanced BAF were fitted to even-numbered copy-number states, whereas unbalanced segments were allowed to fit to uneven numbers as well. Finally, estimated tumor-cell content and ploidy values were used to compute the total and allele-specific copy number for each segment.
The coverage for the control of P3 was very noisy and prevented a proper estimation of the copy number states. Thus, the 10-kbp control coverage used to obtain the coverage ratio was replaced by an unmatched in-house control. Heterozygous SNPs for BAF estimation were obtained from the original patient control.
For P1-S1, a tetraploid solution allowed a slightly better fit of the segments than a diploid solution did. However, because all other samples for that patient were estimated to be diploid and the SNV mutant-allele frequency distribution does not support tetraploidy, the ploidy for P1-S1 was adjusted accordingly.
Determination of CNA-based growth clones
Genomic segments with indications for subclonal copy number aberrations were used to define CNA-based growth clones. For this, segments ≥2.5 Mbp with noninteger copy numbers (>30% deviation from the nearest integer copy number) in at least one sample were determined systematically. Interruptions <10 kbp were ignored, and segments with two noninteger alleles were excluded. For all remaining segments, genomic breakpoints were extracted. Exact copy numbers were calculated as the mean copy number of all SNPs in a respective region. CFs for the underlying growth clones were estimated for each segment using a minimalistic model with two contributing growth clones differing by a single copy number. Values were rounded to the nearest 10%. Segments with similar CF dynamics over time (deviations ≤10%) were assigned to the same growth clone. In those cases, the CF found in the majority of segments of one growth clone was used for the genomic subclone combinatorics. In cases with equal numbers of segments displaying two different CFs, a CF range was used for genomic subclone combinatorics.
Genomic subclone combinatorics
Genomic subclone combinatorics was used to assign SNV- and CNA-based growth clones to—and thus identify the minimal number of—distinct genomic subclones. The CF was used as the common denominator, and maximum parsimony was applied for the construction of the phylogenetic trees.
Patient 1
In the primary tumor of P1, two growth clones (P1-q1 and P1-q3) were easily identifiable. P1-q1 was assigned to genomic subclone A (Fig. S2 A, dark green) and P1-q3 to genomic subclone C (Fig. S2 A, orange). P1-q2 was first identified in S1. Because P1-q1 and P1-q2 displayed collective outgrowth in X2 to 60–65%, dependence of those growth clones was concluded. In S1, P1-q1 and P1-q2 were, therefore, combined in genomic subclone B with 5% (Fig. S3 A, light blue). In addition, a remaining fraction of P1-q1 was assigned to genomic subclone A. P1-q4 resulted as a logical consequence from the branching of q1 into genomic subclones A and B. SNVs in P1-q1 must represent SNVs present in the founder clone, whereas P1-q2 comprised SNVs present in an individual clone. The vast number of SNVs in P1-q2 suggested that the separation from its ancestor P1-q1 happened a considerable phylogenetic time ago. Thus, we expected an additional SNV growth clone contributing to genomic subclone A, explaining the difference between CFs of P1-q1 and P1-q2. That growth clone was defined by the difference of CFs in S1 (40%) and X1 (30%) and fulfilled the condition of disappearing in X2 because P1-q2 was as prominent as P1-q1, without adding that constraint in the first definition. Genomic subclone C contributed to 20% in S1. The previously undetected growth clone P1-c1 was assigned to genomic subclone D, contributing 10% (Fig. S2 A, purple). In Fig. S2, P1-q3 and P1-q5 were assigned to genomic subclone E with 90–95% CF (Fig. S2 A, light green). In X1 and X2, all growth clones could be explained with previously determined genomic subclones.
Patient 2
All cells in the primary tumor of P2 comprised P2-c2 and P2-c3. In addition, growth clones P2-c5 and P2-c6 were present in TU with a CF of 40 and 50%, respectively. Both were assigned to genomic subclone A (Fig. S2 B, dark green). Genomic subclone B (Fig. S2 B, light blue) was defined by the remaining CFs of P2-c2 and P2-c3. In S1, two SNV-based growth clones were detected (P2-q1 and P2-q3). P2-q1 was present with 10% CF and, therefore, was combined with the CNA-based growth clone P2-c3 (also 10% CF) and was assigned to growth clone C (Fig. S2 B, orange). Growth clone P2-q3 was assigned to the independent genomic subclone D (Fig. S2 B, purple). The independence of P2-q1 and P2-q3 was substantiated by the fact that, in X1 and X2, P2-q1 contributed substantially more than P2-q3, and in S1 and S2, P2-q3 displayed more CF than P2-q1 had. In a model with one genomic subclone harboring both growth clones, the fractions P2-q1 and P2-q3 should be always present with the same CF. In the case of one ancestor and a second, further-developed genomic subclone, ancestral SNVs should be equal or greater in all samples. P2-c1 and P2-c2 were present with 60–70% CF in S1 and, therefore, were assigned to genomic subclone E (Fig. S2 B, light green). An ancestral connection of genomic subclone B and genomic subclone C, as well as genomic subclone B and genomic subclone E with shared CNAs, would have been possible theoretically. Because of the sampling strategy in P2 (adjacent tumor pieces for sequencing and spheroid culture generation), a model with independent genomic subclones was chosen. That decision was supported by the general absence of shared SNV-based growth clones between TU and the remaining samples in this patient and, importantly, did not influence the total number of genomic subclones in those samples. In S2, all growth clones could be explained with previously defined genomic subclones. In X1, P2-c4 was detected with 10%. In X2, CFs of P2-q1 and P2-c3 increased to 90% and P2-c4 to 60%, clearly indicating partial dependence by those growth clones. In X1, P2-c4 was, therefore, assigned to genomic subclone F (Fig. S2 B, dark blue), branching from genomic subclone C, thus, also containing P2-q1 and P2-c3. All remaining genomic subpopulations could be explained with previously defined genomic subclones. In X2, the previously undetected growth clone P2-q2 was detected at CFs of 60% and was, therefore, coassigned with P2-q1, P2-c3, and P2-c4 to genomic subclone G (Fig. S2 B, red), branching from genomic subclone F. Further, growth clone P2-c1 was assigned to genomic subclone H (Fig. S2 B, brown), potentially descending from genomic subclone E.
Patient 3
In the primary tumor of P3, all cells harbored P3-c5. In addition, P3-c1 and P3-c3 were present with 20–30% CF and were assigned to genomic subclone A (Fig. S2 C, dark green). Genomic subclone B harbored P3-c2 and P3-c5 (Fig. S2 C, light blue). P3-c4, which was present with 70% CF, could be assigned to subclones A and B. Genomic subclone C (Fig. S2 C, orange) harbored P3-c5 and P3-c6. In S1, P3-q1 and P3-c3 were assigned to genomic subclone D (Fig. S2 C, purple) based on concordant CFs. Two further genomic subclones were necessary to explain the remaining growth clones (P3-q2, P3-c4, and P3-c5). P3-q2 was assigned to subclone E (Fig. S2 C, light green) and P3-c5 to subclone F (Fig. S2 C, dark blue), both harboring P3-c4, therefore descending from a common ancestor. For the same reasons explained for P2, a model with independent genomic subclones from P3-TU to P3-S1 was chosen. In S2, a previously undetected growth clone, P3-q3, together with P3-c6, was detected at a cellular fraction of 60–65% and was assigned to genomic subclone H (Fig. S2 C, brown), a descendant from genomic subclone E. In X1, all growth clones could be explained with previously identified genomic subclones. In X2, the strong outgrowth of P3-q1 required a genomic subclone purely containing that growth clone (Fig. S2 C, red) and was possibly a descendant from genomic subclone D.
Genetic marking of CRC cells
An enhanced GFP–expressing. vesicular stomatis virus protein G–pseudotyped, third-generation, self-inactivating lentiviral vector, under the control of the human CMV promoter, was produced (Dull et al., 1998) and concentrated through ultracentrifugation. Polyethylenimine (Sigma-Aldrich) was used for the stable genetic marking of CRC cells at multiplicities of infection of 20–30. To that end, dissociated colon cancer cells were transduced with Viromag R/L beads (OZ Biosciences), with transduction efficiencies ranging from 1 to 80%. To assess lentiviral IS, 3′-LAM-PCR was performed and analyzed, as described, using the restriction enzyme MluCI (New England Biolabs, Inc.). The original protocol was adjusted for downstream Illumina sequencing. Raw LAM amplicon sequences were trimmed, aligned to the human genome assembly (GRCh38/hg38), and clustered.
Online supplemental material
Fig. S1 presents the genome-wide distribution of SNVs, and Fig. S2 shows a detailed model of growth clones. Fig. S3 provides mutational signatures for all SNV-based growth clones, and Table S1 (in a separate Excel file) presents mutations in CRC-associated and MMR genes.
Supplementary Material
Acknowledgments
We thank the DKFZ Sample Processing Laboratory for sample preparation, the Genomics and Proteomics Core Facility for sequencing, the Nationales Centrum für Tumorerkrankungen (NCT) tissue bank for histological assessment, the DKFZ Center for Preclinical Research and Central Animal Laboratory, as well as the DKFZ Heidelberg Center for Personalized Oncology (DKFZ-HIPO), and the NCT Precision Oncology Program for technical support (funding through P002). We thank Christian Heyer for his contribution to method development.
This work was supported by the Deutsche Forschungsgemeinschaft (KFO227/BA4806/1-2, SFB873), the Baden-Württemberg Stiftung (P-LS-ASII/33), NCT 3.0 Precision Oncology Program (NCT3.0_2015.4 TransOnco; NCT3.0_2015.54 DysregPT), the EU Framework Program Horizon 2020 (TRANSCAN-2 ERA-NET, TACTIC consortium), and the Deutsche Krebshilfe Prioroty program "Translational Oncology" (Colon-Resist-Net). Data analysis was supported by the Bundesministerium für Bildung und Forschung–funded Heidelberg Center for Human Bioinformatics within the German Network for Bioinformatics Infrastructure (031A537A, 031A537C). K.M. Giessler, T.D. Dubash, and K. Kleinheinz were supported by scholarships from the Deutsches Krebsforschungszentrum, and S.M. Dieter by a fellowship from the Heidelberg School of Oncology. D. Huebschmann is a member of the Hartmut Hoffmann-Berling International Graduate School of Molecular and Cellular Biology and the MD/PhD program at Heidelberg University.
The authors declare no competing financial interests.
Author contributions: K.M. Giessler, C.R. Ball, and H. Glimm designed the project and prepared the manuscript. K.M. Giessler, T.D. Dubash, S.M. Dieter, S. Weber, and C.M. Hoffmann collected the data. K.M. Giessler, C.R. Ball, and H. Glimm analyzed the data. K. Kleinheinz, D. Huebschmann, G.P. Balasubramanian, I. Buchhalter, and N. Paramasivam provided bioinformatical analysis. K.M. Giessler, K. Kleinheinz, and D. Huebschmann performed genomic subclone combinatorics:, and K.M. Giessler, R. Fronza, M. Schmidt, and C.R. Ball provided IS analysis. M. Schneider and A. Ulrich prepared the clinical information and human tissues. W. Weichert performed pathological examinations, and F. Herbst, C. Siegl, S. Fröhling, and C. von Kalle provided important intellectual content. M. Schlesner, R. Eils, and B. Brors supervised the bioinformatical analyses, and C.R. Ball and H. Glimm approved the final manuscript.
Footnotes
Abbreviations used:
- ACEseq
- allele-specific copy number estimation from sequencing
- APOBEC
- apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like
- BAF
- B-allele frequency
- CF
- cellular fraction
- CNA
- copy number alterations
- CRC
- colorectal cancer
- DSB
- double-strand break
- IS
- integration site
- LAM-PCR
- linear amplification–mediated PCR
- MMR
- mismatch repair
- MSI-H
- microsatellite instability high
- NNLS
- nonnegative least squares
- NSG
- NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ
- P1–3
- patients 1–3
- S1
- early spheroid
- S2
- late spheroid
- SNP
- single-nucleotide polymorphism
- SNV
- single-nucleotide variant
- TcIC
- tumor clone-initiating cell
- TCN
- total copy number
- VAF
- variant allele frequency
- WGS
- whole-genome sequencing
- X1
- early xenograft
- X2
- late xenograft
References
- Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.L., et al. ICGC PedBrain . 2013. Signatures of mutational processes in human cancer. Nature. 500:415–421 (published erratum appears in Nature. 2013 502:258). 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov L.B., Jones P.H., Wedge D.C., Sale J.E., Campbell P.J., Nik-Zainal S., and Stratton M.R.. 2015. Clock-like mutational processes in human somatic cells. Nat. Genet. 47:1402–1407. 10.1038/ng.3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K., Lutz C., van Delft F.W., Bateman C.M., Guo Y., Colman S.M., Kempski H., Moorman A.V., Titley I., Swansbury J., et al. . 2011. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 469:356–361. 10.1038/nature09650 [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. The Gene Ontology Consortium . 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25:25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi D., Cicalese A., Dellino G.I., Luzi L., Riva L., D’Alesio C., Diaferia G.R., Carugo A., Cavallaro E., Piccioni R., et al. . 2016. In vivo genetic screens of patient-derived tumors revealed unexpected frailty of the transformed phenotype. Cancer Discov. 6:650–663. 10.1158/2159-8290.CD-15-1200 [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 487:330–337. 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieter S.M., Ball C.R., Hoffmann C.M., Nowrouzi A., Herbst F., Zavidij O., Abel U., Arens A., Weichert W., Brand K., et al. . 2011. Distinct types of tumor-initiating cells form human colon cancer tumors and metastases. Cell Stem Cell. 9:357–365. 10.1016/j.stem.2011.08.010 [DOI] [PubMed] [Google Scholar]
- Dull T., Zufferey R., Kelly M., Mandel R.J., Nguyen M., Trono D., and Naldini L.. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72:8463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirew P., Steif A., Khattra J., Ha G., Yap D., Farahani H., Gelmon K., Chia S., Mar C., Wan A., et al. . 2015. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature. 518:422–426. 10.1038/nature13952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D., Nguyen T.K., Leishear K., Finko R., Kulp A.N., Hotz S., Van Belle P.A., Xu X., Elder D.E., and Herlyn M.. 2005. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 65:9328–9337. 10.1158/0008-5472.CAN-05-1343 [DOI] [PubMed] [Google Scholar]
- Fearon E.R., and Vogelstein B.. 1990. A genetic model for colorectal tumorigenesis. Cell. 61:759–767. 10.1016/0092-8674(90)90186-I [DOI] [PubMed] [Google Scholar]
- Gehring J.S., Fischer B., Lawrence M., and Huber W.. 2015. SomaticSignatures: inferring mutational signatures from single-nucleotide variants. Bioinformatics. 31:3673–3675. 10.1093/bioinformatics/btv408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium 2015. Gene Ontology Consortium: going forward. Nucleic Acids Res. 43:D1049–D1056. 10.1093/nar/gku1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M., Rowan A.J., Horswell S., Larkin J., Endesfelder D., Gronroos E., Martinez P., Matthews N., Stewart A., Tarpey P., et al. . 2012. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366:883–892. 10.1056/NEJMoa1113205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann P.C., Huber S.L., Herrler T., Aicher A., Ellwart J.W., Guba M., Bruns C.J., and Heeschen C.. 2007. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 1:313–323. 10.1016/j.stem.2007.06.002 [DOI] [PubMed] [Google Scholar]
- Howie B.N., Donnelly P., and Marchini J.. 2009. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5:e1000529 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesinghaus M., Wolf T., Pfarr N., Muckenhuber A., Ahadova A., Warth A., Goeppert B., Sers C., Kloor M., Endris V., et al. . 2015. Distinctive spatiotemporal stability of somatic mutations in metastasized microsatellite-stable colorectal cancer. Am. J. Surg. Pathol. 39:1140–1147. 10.1097/PAS.0000000000000423 [DOI] [PubMed] [Google Scholar]
- Jones D.T., Jäger N., Kool M., Zichner T., Hutter B., Sultan M., Cho Y.J., Pugh T.J., Hovestadt V., Stütz A.M., et al. . 2012. Dissecting the genomic complexity underlying medulloblastoma. Nature. 488:100–105. 10.1038/nature11284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.T., Hutter B., Jäger N., Korshunov A., Kool M., Warnatz H.J., Zichner T., Lambert S.R., Ryzhova M., Quang D.A., et al. International Cancer Genome Consortium PedBrain Tumor Project . 2013. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat. Genet. 45:927–932. 10.1038/ng.2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien S., Merino-Trigo A., Lacroix L., Pocard M., Goéré D., Mariani P., Landron S., Bigot L., Nemati F., Dartigues P., et al. . 2012. Characterization of a large panel of patient-derived tumor xenografts representing the clinical heterogeneity of human colorectal cancer. Clin. Cancer Res. 18:5314–5328. 10.1158/1078-0432.CCR-12-0372 [DOI] [PubMed] [Google Scholar]
- Kanehisa M., and Goto S.. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Kawashima M., Furumichi M., and Tanabe M.. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44(D1):D457–D462. 10.1093/nar/gkv1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.M., Jung S.H., An C.H., Lee S.H., Baek I.P., Kim M.S., Park S.W., Rhee J.K., Lee S.H., and Chung Y.J.. 2015. Subclonal genomic architectures of primary and metastatic colorectal cancer based on intratumoral genetic heterogeneity. Clin. Cancer Res. 21:4461–4472. 10.1158/1078-0432.CCR-14-2413 [DOI] [PubMed] [Google Scholar]
- Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., and Shendure J.. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46:310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klco J.M., Spencer D.H., Miller C.A., Griffith M., Lamprecht T.L., O’Laughlin M., Fronick C., Magrini V., Demeter R.T., Fulton R.S., et al. . 2014. Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell. 25:379–392. 10.1016/j.ccr.2014.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreso A., O’Brien C.A., van Galen P., Gan O.I., Notta F., Brown A.M., Ng K., Ma J., Wienholds E., Dunant C., et al. . 2013. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 339:543–548. 10.1126/science.1227670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau D.A., Carter S.L., Stojanov P., McKenna A., Stevenson K., Lawrence M.S., Sougnez C., Stewart C., Sivachenko A., Wang L., et al. . 2013. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 152:714–726. 10.1016/j.cell.2013.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Kotliarova S., Kotliarov Y., Li A., Su Q., Donin N.M., Pastorino S., Purow B.W., Christopher N., Zhang W., et al. . 2006. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 9:391–403. 10.1016/j.ccr.2006.03.030 [DOI] [PubMed] [Google Scholar]
- Li H., and Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.; 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlos-Suárez A., Barriga F.M., Jung P., Iglesias M., Céspedes M.V., Rossell D., Sevillano M., Hernando-Momblona X., da Silva-Diz V., Muñoz P., et al. . 2011. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 8:511–524. 10.1016/j.stem.2011.02.020 [DOI] [PubMed] [Google Scholar]
- Nowell P.C. 1976. The clonal evolution of tumor cell populations. Science. 194:23–28. 10.1126/science.959840 [DOI] [PubMed] [Google Scholar]
- O’Brien C.A., Pollett A., Gallinger S., and Dick J.E.. 2007. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 445:106–110. 10.1038/nature05372 [DOI] [PubMed] [Google Scholar]
- Olshen A.B., Bengtsson H., Neuvial P., Spellman P.T., Olshen R.A., and Seshan V.E.. 2011. Parent-specific copy number in paired tumor-normal studies using circular binary segmentation. Bioinformatics. 27:2038–2046. 10.1093/bioinformatics/btr329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci-Vitiani L., Lombardi D.G., Pilozzi E., Biffoni M., Todaro M., Peschle C., and De Maria R.. 2007. Identification and expansion of human colon-cancer-initiating cells. Nature. 445:111–115. 10.1038/nature05384 [DOI] [PubMed] [Google Scholar]
- Schmitz M., Breithaupt P., Scheidegger N., Cario G., Bonapace L., Meissner B., Mirkowska P., Tchinda J., Niggli F.K., Stanulla M., et al. . 2011. Xenografts of highly resistant leukemia recapitulate the clonal composition of the leukemogenic compartment. Blood. 118:1854–1864. 10.1182/blood-2010-11-320309 [DOI] [PubMed] [Google Scholar]
- Singh S.K., Hawkins C., Clarke I.D., Squire J.A., Bayani J., Hide T., Henkelman R.M., Cusimano M.D., and Dirks P.B.. 2004. Identification of human brain tumour initiating cells. Nature. 432:396–401. 10.1038/nature03128 [DOI] [PubMed] [Google Scholar]
- Siravegna G., Mussolin B., Buscarino M., Corti G., Cassingena A., Crisafulli G., Ponzetti A., Cremolini C., Amatu A., Lauricella C., et al. . 2015. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 21:827 10.1038/nm0715-827b [DOI] [PubMed] [Google Scholar]
- Sottoriva A., Kang H., Ma Z., Graham T.A., Salomon M.P., Zhao J., Marjoram P., Siegmund K., Press M.F., Shibata D., and Curtis C.. 2015. A big bang model of human colorectal tumor growth. Nat. Genet. 47:209–216. 10.1038/ng.3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro M., Alea M.P., Di Stefano A.B., Cammareri P., Vermeulen L., Iovino F., Tripodo C., Russo A., Gulotta G., Medema J.P., and Stassi G.. 2007. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell. 1:389–402. 10.1016/j.stem.2007.08.001 [DOI] [PubMed] [Google Scholar]
- Van Den Meersche K., Soetaert K., and Van Oevelen D.. 2009. xsample(): An R function for sampling linear inverse problems. Journal of Statistical Software. 30:1–15.21666874 [Google Scholar]
- van de Wetering M., Francies H.E., Francis J.M., Bounova G., Iorio F., Pronk A., van Houdt W., van Gorp J., Taylor-Weiner A., Kester L., et al. . 2015. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 161:933–945. 10.1016/j.cell.2015.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen L., Todaro M., de Sousa Mello F., Sprick M.R., Kemper K., Perez Alea M., Richel D.J., Stassi G., and Medema J.P.. 2008. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc. Natl. Acad. Sci. USA. 105:13427–13432 (published erratum appears in Proc. Natl. Acad. Sci. USA. 2009. 106:9534–9535). 10.1073/pnas.0805706105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader J.E., and Lindeman G.J.. 2012. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 10:717–728. 10.1016/j.stem.2012.05.007 [DOI] [PubMed] [Google Scholar]
- Wang J., Mullighan C.G., Easton J., Roberts S., Heatley S.L., Ma J., Rusch M.C., Chen K., Harris C.C., Ding L., et al. . 2011. CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nat. Methods. 8:652–654. 10.1038/nmeth.1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M., and Hakonarson H.. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38:e164 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiswald L.B., Bellet D., and Dangles-Marie V.. 2015. Spherical cancer models in tumor biology. Neoplasia. 17:1–15. 10.1016/j.neo.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood L.D., Parsons D.W., Jones S., Lin J., Sjöblom T., Leary R.J., Shen D., Boca S.M., Barber T., Ptak J., et al. . 2007. The genomic landscapes of human breast and colorectal cancers. Science. 318:1108–1113. 10.1126/science.1145720 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.