Cytokinin signaling, in combination with epigenetic regulation, de novo activates WUSCHEL expression during axillary meristem initiation to enable shoot branching.
Abstract
The homeodomain transcription factor WUSCHEL (WUS) defines the shoot stem cell niche, but the mechanisms underlying the establishment of WUS expression remain unclear. Here, we show that cytokinin signaling precedes WUS expression in leaf axils and activates WUS expression de novo in the leaf axil to promote axillary meristem initiation. Furthermore, type-B Arabidopsis response regulator proteins, which are transcriptional activators in the cytokinin signaling pathway, directly bind to the WUS promoter and activate its expression. Finally, we show that cytokinin activation of WUS in the leaf axil correlates with increased histone acetylation and methylation markers associated with transcriptional activation, supporting the fact that WUS expression requires a permissive epigenetic environment to restrict it to highly defined meristematic tissues. Taken together, these findings explain how cytokinin regulates axillary meristem initiation and establish a mechanistic framework for the postembryonic establishment of the shoot stem cell niche.
INTRODUCTION
Plant meristems are responsible for organogenesis, which can continue throughout the life of the plant. Shoot meristems harbor stem cells located in the central zone (CZ), and these cells have a lower cell division rate. Some daughter cells of the CZ cells replenish themselves in the CZ, and other cells that are closer to the peripheral zone form the organ primordia. The organizing center (OC) contains a small group of cells underneath the CZ and maintains the stem character of stem cells in the CZ. A key factor regulating shoot stem cell specification is the WUSCHEL (WUS) homeobox transcription factor (Laux et al., 1996; Mayer et al., 1998). WUS is expressed in the OC, which comprises the stem cell niche of shoot meristems, including the shoot apical meristems (SAMs), axillary meristems (AMs), floral meristems (FMs), and adventitious shoot meristems.
Although we do not know how WUS expression is established in each type of shoot meristem, extensive studies have shown both how WUS promotes shoot meristems and how WUS expression is maintained in them. WUS function requires interaction with transcriptional regulators of the HAIRY MERISTEM family (Zhou et al., 2015). WUS migrates from the OC to the CZ to activate the expression of the negative regulator CLAVATA3 (CLV3), which encodes a secreted peptide (Fletcher et al., 1999; Yadav et al., 2011; Daum et al., 2014). The CLV3 peptide activates CLV1, a transmembrane receptor kinase expressed in the OC, and this kinase inhibits WUS expression via a yet unknown signaling cascade (Clark et al., 1997; Ogawa et al., 2008). Thus, the WUS-CLV feedback loop forms a self-correcting mechanism that maintains a stem cell pool of constant size (Brand et al., 2000; Schoof et al., 2000).
In addition, phytohormones also help maintain shoot stem cell homeostasis. A positive-feedback loop between WUS function and cytokinin provides positional cues for shoot meristem patterning (Leibfried et al., 2005; Gordon et al., 2009; Chickarmane et al., 2012), although the underlying molecular mechanism has not been fully resolved. A number of additional WUS targets have been identified (Leibfried et al., 2005; Busch et al., 2010; Yadav et al., 2013), supporting the idea that WUS functions as a central regulator of stem cells. In contrast to the indeterminate SAM and AM, the determinate FM only transiently maintains WUS expression. The MADS transcription factor AGAMOUS (AG) is activated by WUS in the FM, and AG in turn terminates WUS expression and thus determines floral bud growth (Lenhard et al., 2001; Lohmann et al., 2001; Liu et al., 2011). However, there is a major gap in our understanding of the mechanisms that establish the initial expression of WUS.
In addition to the SAM formed during embryogenesis, AMs form in the axils of leaves and develop into buds to enable branching (McConnell and Barton, 1998; Wang et al., 2016). Iterative branching in perennial plants can lead to thousands of terminal branches and thus determines aerial plant architecture (Wang and Li, 2008). The AM has the same developmental potential as the SAM to maintain itself and to initiate new organs. Genetic studies in Arabidopsis thaliana and other species have shown that AM initiation is regulated by several transcription factors, such as LATERAL SUPPRESSOR (LAS), REGULATOR OF AXILLARY MERISTEMS (RAX), and REVOLUTA (REV) (Talbert et al., 1995; Greb et al., 2003; Müller et al., 2006). Genetic and molecular studies revealed direct and indirect interactions among these genes in a regulatory network (Raman et al., 2008; Tian et al., 2014). WUS is expressed in the AM as in the SAM, but how WUS expression is established during AM initiation remains enigmatic. The FM shares many similarities with the AM and has been suggested to be a specialized AM (Long and Barton, 2000). How WUS expression is established in the FM is also unknown. Our recent work showed that initiation of both the AM and the FM requires a cytokinin signaling pulse (Han et al., 2014; Wang et al., 2014b).
In this study, we report that cytokinin promotes WUS expression to establish stem cell niches de novo during AM initiation. We then show that type-B Arabidopsis response regulator proteins (ARRs), which mediate the transcriptional response to cytokinin (Argyros et al., 2008), bind to the WUS promoter to activate its expression. We also show that WUS activation requires permissive chromatin modifications. In summary, we have provided a model for a direct molecular link explaining how shoot stem cell niches are established.
RESULTS
WUS Expression Is Activated de Novo during AM Initiation
To determine when WUS expression is established during AM initiation, we monitored the dynamics of leaf axil WUS expression in plants grown under short-day conditions for 28 d. Using the ProWUS:DsRed-N7 reporter line for WUS expression (Gordon et al., 2007), we found that WUS expression is activated de novo prior to AM initiation. We did not detect DsRed expression in young leaf axils. We observed that DsRed expression in cells of the subepidermal layer started in the axil protrusions of the thirteenth oldest leaf primordium (P13) (Figure 1A) and increased in P14 and older leaf axils (Figure 1B), a time when AMs are morphologically detectable (Long and Barton, 2000; Greb et al., 2003). At this stage, WUS is expressed in a small group of cells below the second cell layer of the AM (Figures 1A and 1B), resembling WUS expression in the SAM (Mayer et al., 1998). Thus, similar to the de novo activation of WUS expression during FM initiation (Mayer et al., 1998), de novo activation of WUS expression also occurs during AM initiation.
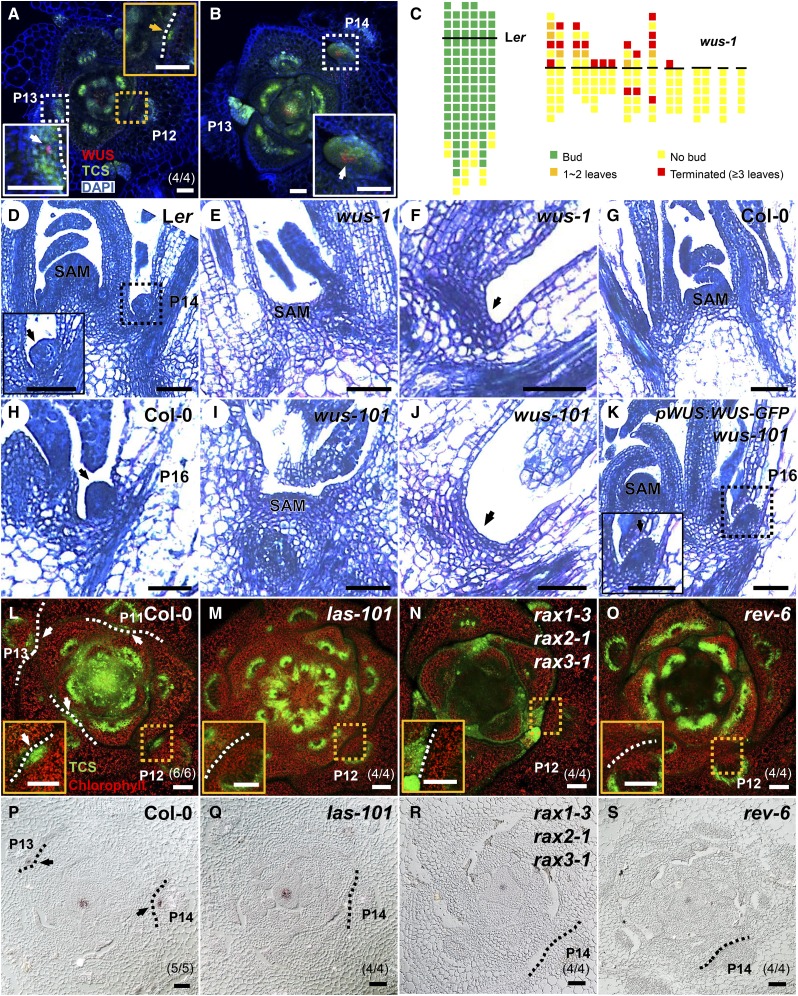
Figure 1.
Cytokinin Signaling Precedes and Overlaps with WUS Expression prior to AM Initiation.
(A) and (B) Serial transverse sections through a wild-type vegetative shoot apex showing expression of ProTCS:GFP-ER (green) and ProWUS:DsRed-N7 (red) in leaf axils. The orange arrow indicates GFP in the leaf axil, and white arrows indicate overlapping leaf axil GFP and DsRed signals. Sections are ordered from apical (A) to basal (B) parts from the same plant.
(C) Schematic diagram of axillary buds of Ler and wus-1 plants. The thick black horizontal line represents the border between the youngest rosette leaf and the oldest cauline leaf. For Ler, each column represents a single plant, and each square within a column represents an individual leaf axil. For wus-1, each column represents a single main branch, and branches from a single plant are grouped together. The bottom row represents the oldest rosette leaf axils, with progressively younger leaves above. Green, presence of an axillary bud; yellow, absence of an axillary bud; orange, one or two leaves in place of an axillary bud; red, a terminated axillary bud (with three or more leaves) in any particular leaf axil.
(D) to (K) Longitudinal sections of vegetative shoot apices. Images show a protruding AM in the leaf axil of Ler (D), Col-0 (H), and ProWUS:WUS-GFP wus-101 (K), but the lack of an AM in the leaf axil of wus-1 (F) and wus-101 (J). Note the normal SAM in Ler (D), Col-0 (G), and ProWUS:WUS-GFP wus-101 (K) in contrast to flat shoot apices in wus-1 (E) and wus-101 (I). Black arrows indicate leaf axils.
(L) to (O) Expression of ProTCS:GFP-ER (green) in leaf axils. Images show transverse sections through shoot apices of Col-0 (L), las-101 (M), rax1-3 rax2-1 rax3-1 (N), and rev-6 mutants (O). White arrows indicate GFP in leaf axils.
(P) to (S) In situ hybridization of WUS in the shoot apex. Images show transverse sections from Col-0 (P), las-101 (Q), rax1-3 rax2-1 rax3-1 (R), and rev-6 mutants (S). Black arrows indicate WUS signal in leaf axils.
Dotted boxes indicate the locations of the regions magnified in the insets. Dotted lines indicate the outlines of leaf axils. Pn indicates leaf primordium number, and (m/n) indicates m in n of biological repeats showing the displayed features. Bars = 50 μm.
It has long been known that WUS is required for SAM and FM function (Laux et al., 1996), but it remains unknown if WUS is required for AM integrity. Therefore, we analyzed bud development in the wus-1 and wus-101 mutants. The wus mutants have a bushy phenotype due to repetitive initiation of defective shoot meristems (Laux et al., 1996). We carefully analyzed these defective shoot meristems and found that they all originated from leaf axils and were defective AMs. Because of their defective SAM, wus plants have reduced apical dominance and enhanced (terminated) axillary bud outgrowth. On the other hand, both wus-1 and wus-101 plants have a dramatic axillary bud formation defect (Figures 1C to 1J). We followed anatomical changes at the leaf axils of wild-type and wus plants. In P14 to P16 stage wild-type leaf axils, cells with a denser staining content form morphologically distinguishable bumps, which mark the first morphological change associated with AM development (Figures 1D and 1H). In both wus-1 and wus-101 plants, dense staining cells and AM structure are lost in comparable stage leaf axils at a high frequency (Figures 1F and 1J). Consistently, there is a dramatic reduction of axillary buds in wus-1 (Figure 1C), with 77% (87 out of 113) of leaf axils not supporting the formation of axillary buds (Figure 1F; Supplemental Figure 1B), 6% (7 out of 113) of leaf axils forming one or two leaf-like structures (Supplemental Figure 1C), and 17% (19 out of 113) of leaf axils forming terminated buds (with three or more leaves). The combination of reduced apical dominance and outgrowth of occasionally formed but terminated buds led to the observed bushy phenotype. Notably, the AM initiation defect of wus-101 can be rescued by ProWUS:WUS-GFP (Figure 1K) (Daum et al., 2014). Nevertheless, the wus-1 and wus-101 plants also have severe SAM defects, making it difficult to exclude an indirect effect of the SAM on AM initiation.
The expression of the meristematic gene SHOOT MERISTEMLESS (STM) marks leaf axil AM progenitor cells (Grbić and Bleecker, 2000; Long and Barton, 2000; Greb et al., 2003; Shi et al., 2016). Examination of accumulation of the STM transcript in the wus-1 mutant by RNA in situ hybridization revealed a leaf axil expression pattern that is similar to the pattern in Ler wild-type plants (Supplemental Figure 2), suggesting that the maintenance of meristematic cells does not rely on WUS.
Furthermore, WUS overexpression induced ectopic AM initiation. We used an inducible WUS overexpression line, pga6-1 (Zuo et al., 2002), in which β-estradiol induces constitutive WUS overexpression, and a hormone-free leaf culture system (Wang et al., 2014b; Shi et al., 2016), in which we can quantify and live-image AM initiation. In isolated pga6-1 leaves in culture, we found that β-estradiol induction can lead to the formation of additional AMs in the leaf axil (Supplemental Figures 3A and 3B). Ectopic WUS expression also leads to ectopic AM initiation in cotyledon axils, a phenotype indicating that AM initiation was enhanced (Wang et al., 2014a). Whereas wild-type Arabidopsis cotyledons lack axillary buds, we found that axillary buds could form in over 50% (n > 10) of cotyledon axils after WUS induction (Supplemental Figure 3C). To confirm that the ectopic AM initiation phenotype was due to WUS overexpression, but not potential second-site mutations, we generated an independent dexamethasone-inducible WUS overexpression line, ProUBQ10:WUS-GR. In ProUBQ10:WUS-GR leaf axils, we similarly observed ectopic AM formation after WUS induction (Supplemental Figure 3D). Additionally, a constitutive WUS overexpression line sef, which was isolated as an activation tagging mutant (Xu et al., 2005), showed multiple buds or branches per leaf axil (Supplemental Figures 3E and 3F). Taken together, these findings indicated that WUS expression promoted AM initiation (Figure 1C).
Leaf Axil Cytokinin Signaling Precedes de Novo WUS Activation
Our recent work shows that a cytokinin signaling pulse occurs prior to AM initiation (Han et al., 2014; Wang et al., 2014b). We also used a cytokinin analog and a cytokinin antagonist to test whether AM initiation required cytokinin signaling. Treatment with the cytokinin analog 6-benzylaminopurine (BAP) caused production of multiple axillary buds (Supplemental Figures 4A to 4C). Treatment of detached leaf axils with the phenylquinazoline compound S-4893, a noncompetitive cytokinin antagonist that targets cytokinin receptors (Arata et al., 2010), severely compromised axillary bud formation (Supplemental Figure 4D).
To determine the dynamics of local cytokinin signaling and WUS expression in a developmental context, we examined the timing and location of expression of ProTCS:GFP-ER (Müller and Sheen, 2008), a cytokinin signaling reporter, in combination with ProWUS:DsRed-N7. Image analysis indicated that the leaf axil cytokinin signaling pulse emerged earlier than, and overlapped with, WUS expression during AM initiation (Figures 1A and 1B). Note that TCS signals in the leaf axil were substantially stronger than that in the SAM (Supplemental Figure 5) (Wang et al., 2014b), implying that the AM and SAM require different levels of cytokinin signaling. Taken together, these results indicated that de novo WUS activation during AM initiation is associated with a prior cytokinin signaling pulse in the same cells.
We also analyzed whether perturbed leaf axil cytokinin signaling was associated with defective WUS activation. AM initiation is compromised in the las, rax, and rev mutants (Talbert et al., 1995; Greb et al., 2003; Müller et al., 2006). Expression of the meristematic cell marker STM is maintained in las and rev mutants (Greb et al., 2003; Shi et al., 2016), suggesting that at least partial AM progenitor cell specification occurs in these mutants. We found that the leaf axils of these mutants lack the cytokinin signaling pulse (Figures 1L to 1O; Supplemental Figure 6A). In addition, we could not detect WUS expression in the leaf axils of these mutants (Figures 1P to 1S; Supplemental Figure 6B). Thus, lack of leaf axil cytokinin signaling associated with leaf axil WUS activation. Because cytokinin treatment can rescue AM initiation defects in rax mutants (Wang et al., 2014b), we speculate that cytokinin signaling activates WUS expression to enable AM initiation.
Cytokinin Activates WUS Expression
To test whether cytokinin signaling can activate WUS expression, we treated shoot tissues with the cytokinin analog BAP at a concentration of 0.89 μM, which is within physiological levels (Corbesier et al., 2003). To enrich leaf axil tissues, we removed leaves and used the remaining shoot tissue for gene expression analysis by RT-PCR with a limited number of cycles (see Methods). We found that a 4-h BAP treatment rapidly activated WUS expression, even in the presence of the protein synthesis inhibitor cycloheximide (CHX) (Figure 2A), suggesting that activation of WUS does not require de novo protein synthesis. Similarly, BAP activation of WUS expression was also found in the inflorescence (Supplemental Figure 7) (Gordon et al., 2009; Chickarmane et al., 2012), even when we used a physiological concentration of BAP (see Methods for details).
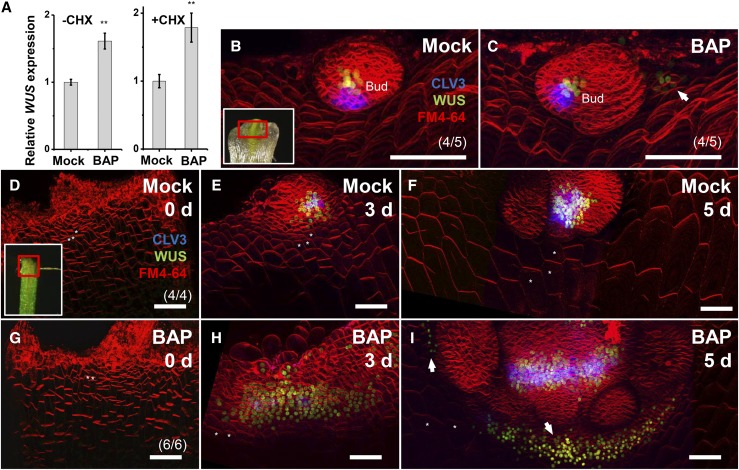
Figure 2.
Induction of WUS Expression by Cytokinin Treatment.
(A) A 4-h 0.89 μM BAP treatment induced WUS expression in leaf-removed shoot apex tissues in both the absence and the presence of CHX. Error bars indicate the sd of three biological replicates, run in triplicate. **P < 0.01 (Student’s t test).
(B) to (I) Time-lapse images of WUS expression in leaf axils. BAP treatment caused rapid induction of ectopic ProWUS:DsRed-N7 in mature leaf axils (C) but delayed induction in immature leaf axils ([G] to [I]).
(B) and (C) A 24-h 0.89 μM BAP treatment (C), but not mock treatment (B), induced ectopic expression of ProWUS:DsRed-N7 (green) in the leaf axil of an isolated P15 stage leaf (arrow).
(D) to (I) BAP treatment alters the WUS expression level, but not its timing. Time-lapse images showing expression of ProWUS:DsRed-N7 (green) and ProCLV3:GFP-ER (blue) in isolated P9 leaf axil centers after mock treatment ([D] to [F]) or 0.89 μM BAP treatment ([G] to [I]). BAP treatment caused the leaf axil center WUS expression domain to enlarge and activated ectopic WUS expression centers (arrows), but did not activate precocious WUS expression.
The regions bordered by the red boxes in the insets in (B) and (D) roughly correspond to the imaged region. Asterisks in (D) to (F) and (G) to (I) label the same cells in corresponding time points. The cell membrane was labeled using FM4-64 (red). Bars = 50 μm.
By live-imaging the expression of the ProWUS:DsRed-N7 reporter and a functional ProWUS:WUS-GFP reporter (Daum et al., 2014), we found that BAP activated ectopic WUS expression centers de novo and substantially enlarged the endogenous WUS expressing domain. We employed a hormone-free leaf culture system to live-image AM initiation (Wang et al., 2014b; Shi et al., 2016). In isolated P15+ stage leaves, a 24-h BAP treatment induced multiple de novo centers of WUS expression in the leaf axil, so that the leaf axils formed multiple meristems, in contrast to the single meristem formed in untreated leaves (Figures 2B and 2C; Supplemental Figures 4A to 4C). In the leaf axil center where WUS normally is expressed, we observed a substantial enlargement of the WUS expression domain (Figures 2D to 2F; Supplemental Figures 8A to 8C, 9A to 9C, and 10). The expression of WUS was maintained in the center of the leaf axil until a visible axillary bud formed (Figures 2D to 2F). These results showed that local cytokinin signaling induced WUS expression in leaf axils to promote AM initiation. Consistent with this, mutants defective in cytokinin synthesis, perception, or signaling show defects in AM initiation (Wang et al., 2014b; Müller et al., 2015).
In contrast to mature leaves (P15+), ectopic cytokinin treatment of immature leaf axils or other tissues did not lead to precocious WUS activation. In isolated P8 to P10 stage immature leaves, the expression of ProWUS:DsRed-N7 was detectable in the center of the leaf axil at ∼52 h in culture without the addition of BAP (Figures 2D to 2F; Supplemental Figures 8A to 8C and 9A to 9C). The expression of ProCLV3:GFP-ER, a WUS target, was detected 18 h later in the cells on top of the WUS-expressing cells (Supplemental Figures 9A to 9C). BAP treatment did not induce precocious WUS (and CLV3) expression (Supplemental Figure 9G). Nevertheless, BAP treatment induced additional de novo centers of WUS and CLV3 expression accompanying the emergence of the central expression domain (Figure 2I). Also, the region of ProWUS:DsRed-N7 and ProCLV3:GFP-ER expression in the central domain was substantially enlarged after BAP treatment (Figures 2G to 2I; Supplemental Figures 8D to 8F, 9D to 9F, and 10). In particular, BAP induced ectopic WUS expression in the epidermal cell layer (compared with Supplemental Figures 9B and 9E). The ectopic epidermal expression of WUS in the center of the leaf axil diminished when CLV3 expression appeared (Supplemental Figure 9F). This may be explained by CLV3 inhibition of WUS expression (Gaillochet and Lohmann, 2015). The BAP treatment did not induce WUS expression in differentiated cells, such as leaf blade cells. Thus, our imaging results also suggested that the precise activation of WUS by cytokinins depended on the developmental stage and cell location (i.e., cell type).
WUS Expression Requires a Permissive Epigenetic Environment
Recent studies shows that the histone modification marker histone H3 lysine 27 trimethylation (H3K27me3), which is associated with transcriptional repression, is highly enriched at the WUS locus in mature leaves, which have no WUS expression (Li et al., 2011; Liu et al., 2011). In addition, we found that histone H3 lysine 4 trimethylation (H3K4me3), a histone modification marker associated with transcriptional activation, was enriched at the WUS locus in inflorescences containing WUS-expressing cells but not in mature leaves lacking WUS-expressing cells (Supplemental Figure 11).
Stage-specific sensitivity of leaf axil cells to BAP treatment suggested that epigenetic modifications may change during leaf maturation. To test this hypothesis, we isolated the basal 2- to 3-mm leaf axil tissues from immature and mature leaves and analyzed histone modifications. To accommodate the limited sample amount, we used the ultralow input micrococcal nuclease-based native chromatin immunoprecipitation (ULI-NChIP) protocol (Brind’Amour et al., 2015). We found that the WUS genomic region showed higher levels of H3K27me3 and lower levels H3 acetylation in early stage (P8 to P10) leaf axils than in late stage (P15 to P17) axils (Figures 3A to 3D). Histone acetylation increases the accessibility of DNA inside chromatin and promotes gene expression (Charron et al., 2009). Thus, the observed temporal histone modification changes correlate with activation of WUS expression in mature leaves, and stringent epigenetic modifications may prevent cytokinins from activating WUS expression in immature leaf axils and differentiated cells.
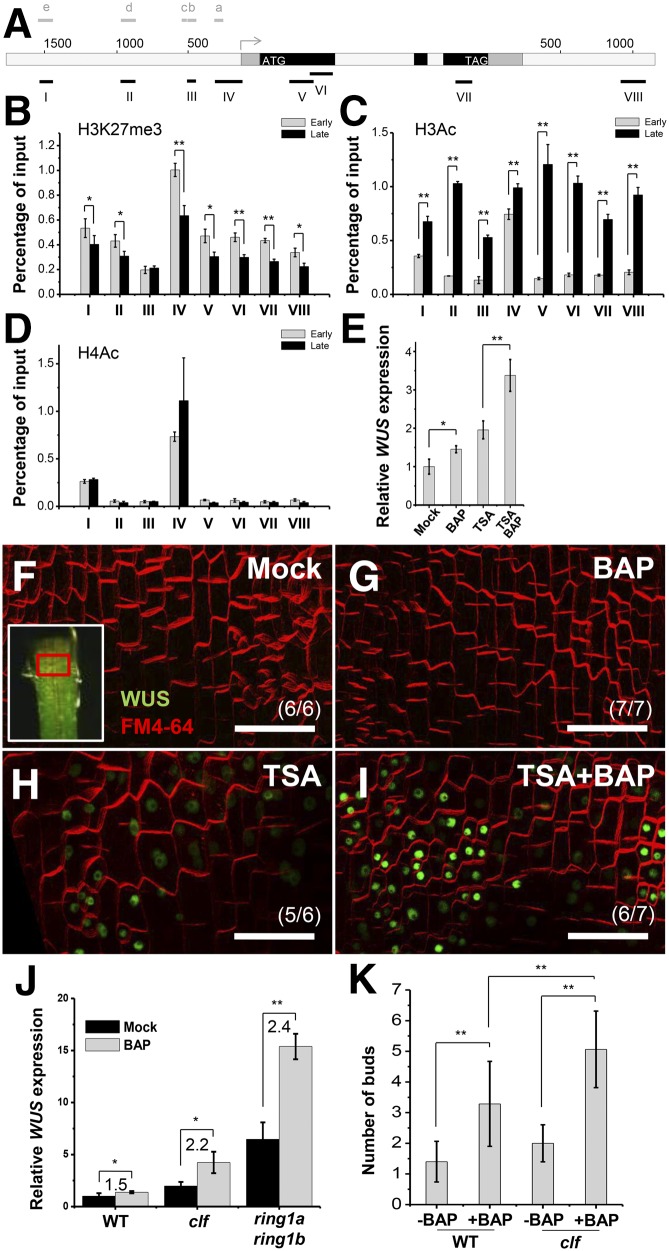
Figure 3.
The Epigenetic Environment Affects Cytokinin Induction of WUS Expression.
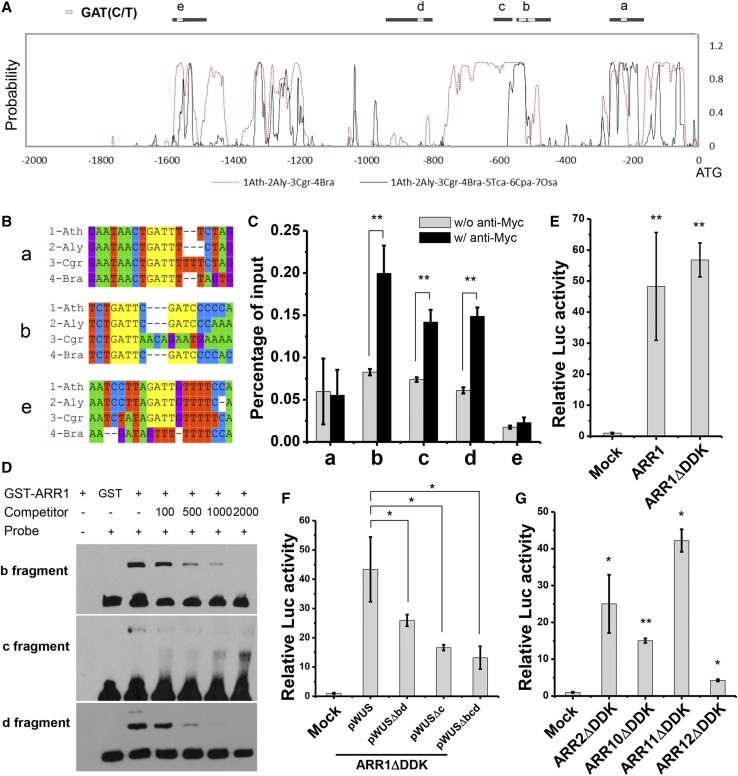
(A) A diagram of the WUS genomic region with an arrow representing the transcription start site. Black, coding sequences; dark gray, untranslated regions; light gray, intron/intergenic regions; ATG and TAG, start and stop codons. Black bold lines with Roman numerals indicate fragments amplified by ChIP-qPCR. Gray bold lines with a to e indicate fragments showing in Figure 5A.
(B) to (D) The comparison of histone modifications in early and late stage of leaf axils. Images show ULI-NChIP with H3K27me3 (B), and histone H3 (C) and H4 (D) acetylation of WUS genomic regions using early stage (P8 to P10) and late stage (P15 to P17) leaf axil tissues. Error bars represent sd, which was calculated from three technical replicates. Two biological replicates gave similar results.
(E) Expression of WUS after a 4-h 0.89 μM BAP and/or 1 μM TSA treatment in shoot apex tissues. Leaves were removed before RNA isolation. Error bars indicate the sd of three biological replicates, run in triplicate.
(F) to (I) Expression of ProWUS:DsRed-N7 (green) in P9 leaf petiole cells. Images show WUS expression 15 h after mock (F), 0.89 μM BAP (G), 1 μM TSA (H), or BAP and TSA (I) treatment. The regions bordered by the red box in the insets in (F) roughly correspond to the imaged region. Note that differentiated petiole cells were imaged. Bars = 50 μm.
(J) A 0.89 μM BAP treatment induced WUS expression in Col-0 wild-type, clf-29, and ring1a ring1b seedlings. Error bars indicate the sd of three biological replicates, run in triplicate.
(K) The number of buds in isolated leaf axils of Col-0 or clf-29 mutants after a 2-week mock or BAP treatment. Error bars indicate the sd (n ≥ 10). *P < 0.05 and **P < 0.01 (Student’s t test).
We next ectopically enhanced histone acetylation by applying the histone deacetylation inhibitor trichostatin A (TSA). TSA treatment, but not BAP treatment, increased histone H3 and H4 acetylation of the WUS genomic region (Supplemental Figure 12), which allows transcription. Whereas TSA treatment alone mildly increased the WUS expression level, which may be due to endogenous cytokinins, a 4-h cotreatment with BAP and TSA caused a 3.5-fold increase in WUS expression (Figure 3E). At the cellular level, we found that TSA enabled rapid de novo activation of WUS expression by BAP in differentiated leaf petiole cells (Figures 3F to 3I), explaining the dramatic increase of WUS levels by cotreatment with BAP and TSA.
We also used mutants that are defective in epigenetic repression of gene expression. The polycomb repressive complex (PRC) establishes the H3K27me3 mark and a repressive chromatin configuration (Schuettengruber et al., 2007). CURLY LEAF (CLF) is a PRC2 core component catalyzing H3K27me3, and RING1a and RING1b are PRC1 core components that bind to H3K27me3, inhibiting transcription (Goodrich et al., 1997; Argyros et al., 2008; Xu and Shen, 2008). We detected precocious WUS expression in young leaf axils of the clf-29 mutant (Supplemental Figures 13A to 13B). In the ring1a ring1b mutant, we detected WUS expression in widely observed ectopic meristems (Supplemental Figure 13C). Furthermore, we found that the induction of WUS expression by BAP was significantly enhanced in these mutants (Figure 3J). Consistent with this, BAP induced more buds in isolated leaf axils of clf-29 plants than in wild-type plants (Figure 3K). These results indicated that the PRC-mediated H3K27me3 repressed WUS in earlier leaf axil and differentiated tissues. Taken together, these results supported the idea that the induction of WUS required a permissive chromatin configuration.
ARR1 Activates WUS Expression
ARR1 is a typical type-B ARR mediating the transcriptional response to cytokinin (Argyros et al., 2008). We previously showed that the arr1-4 loss-of-function mutant is defective in AM initiation (Wang et al., 2014b). Using a ProARR1:GFP-N7 reporter line, we observed ARR1 expression in the leaf axil, prior to and during AM initiation (Figure 4A). Furthermore, cell type-specific transcriptome data (Tian et al., 2014) show that many cytokinin signaling components, including other type-B ARRs, are expressed in the leaf axil (Supplemental Figure 14).
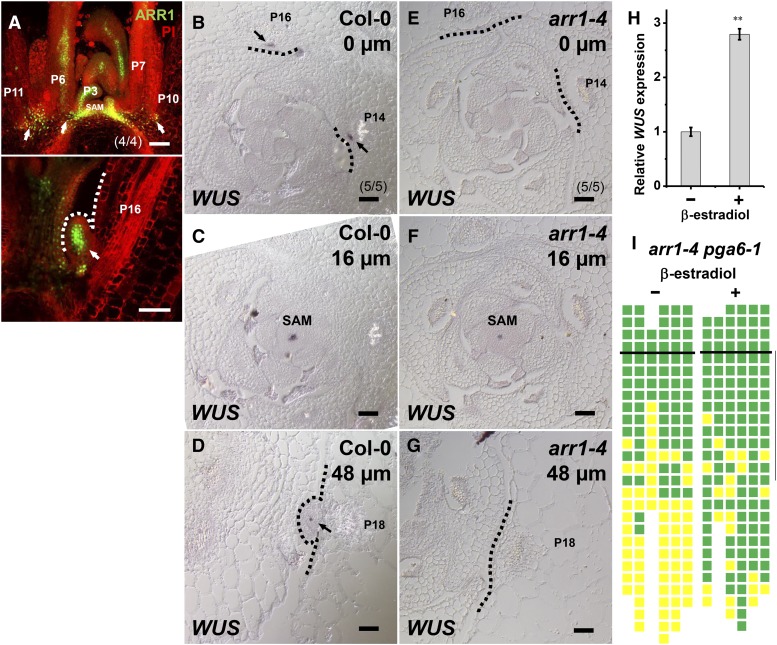
Figure 4.
Activation of WUS Expression by ARR1 Is Required for AM Initiation.
(A) Expression of ProARR1:GFP-N7 in the leaf axil. Images show longitudinal sections through a vegetative shoot apex demonstrating expression of ProARR1:GFP-N7 (green) in the leaf axil prior to (upper panel) and during (lower panel) AM initiation. Arrows indicate GFP in leaf axils. The dotted line indicates the outline of the bulged meristem. PI, propidium iodide.
(B) to (G) In situ hybridization of WUS. Images show serial transverse sections of vegetative shoot apexes in Col-0 ([B] to [D]) and the arr1-4 mutant ([E] to [G]). Arrows indicate WUS signal in leaf axils, and dotted lines indicate the outlines of leaf axils. Sections are ordered from apical ([B] and [E]) to basal ([D] and [G]). The approximate distance from the summit of the SAM to section is given in the upper right-hand corner of each image. Note that (D) and (G) are from regions more distant from the center of the same plants shown in (B) and (C), and (E) and (F). Bars in (A) to (G) = 50 μm.
(H) Expression of WUS after 8-h mock treatment or induction of ARR1ΔDDK in leaf-removed shoot tissues. Error bars indicate the sd of three biological replicates, run in triplicate. **P < 0.01 (Student’s t test).
(I) Schematic diagram of axillary buds of arr1-4 pga6-1 mutants with or without β-estradiol induction to activate WUS overexpression. See the legend to Figure 1C for a description of symbols. Green indicates the presence of an axillary bud, and yellow indicates the absence of an axillary bud. Plants were grown under short-day conditions for 15 d without treatment; leaf axil regions were treated with 10 μM β-estradiol every other day for another 15 d and then shifted to long-day conditions without treatment until axillary buds were counted. The vertical line indicates leaves initiated during β-estradiol treatment.
To test whether ARR1 was required for WUS activation in the leaf axil, we analyzed WUS expression in the arr1-4 mutant. We could not detect WUS expression in the leaf axil in arr1-4 plants (Figures 4B to 4G). We next tested whether ARR1 can activate WUS expression during AM initiation using an inducible cytokinin-independent ARR1ΔDDK-MYC line, in which the N-terminal region encompassing the DDK domain was deleted (Guan et al., 2014). Phosphorylation of the Asp residue in the receiver domain activates the ability of the protein to promote the transcription of target genes. Because the DDK domain functions as a negative regulatory motif, its removal causes constitutive activation of transcription in the absence of cytokinin (Sakai et al., 2001). We found that activation of ARR1ΔDDK-MYC resulted in rapid induction of WUS expression both in shoot apex tissues and in the inflorescence within 8 h (Figure 4H; Supplemental Figure 15). Consistent with enhanced WUS expression, ectopic ARR1ΔDDK-MYC promoted AM initiation and bud outgrowth, resulting in a bushy phenotype. However, we did not observe ectopic meristem in leaves or on the stem, supporting the idea that WUS expression activation requires a permissive epigenetic environment.
We tested whether AM initiation defects in arr1-4 can be rescued by restoring WUS expression using the inducible WUS overexpression line pga6-1 (Zuo et al., 2002). In untreated arr1-4 pga6-1 plants, very few of the first ∼10 rosette leaves, which were formed during the first 2 weeks of vegetative development, develop axillary buds (Figure 4I). Starting at 15 d after germination, we treated shoot apexes of arr1-4 pga6-1 plants with β-estradiol to induce WUS expression. We found that induction of WUS expression rescued the axillary bud formation defects (Figure 4I; Supplemental Figure 16). In addition, a substantial portion of the rosette leaves formed prior to treatment supported the formation of axillary buds after WUS induction (Figure 4I), suggesting that mature leaf axil cells of arr1-4 plants are competent to respond to WUS activity. On the other hand, we did not observe precocious axillary bud formation after β-estradiol induction of WUS expression, suggesting that WUS expression is not sufficient for AM initiation. Taken together, these results indicated that ARR1 regulated AM initiation through activating WUS expression in the leaf axil. Because ARR10, 11, and 12 function redundantly with ARR1 in promoting AM initiation (Wang et al., 2014b), these related type-B ARRs may also promote WUS expression.
Type-B ARRs Bind to the WUS Promoter to Activate Its Expression
Because BAP induction of WUS did not require de novo protein synthesis (Figure 2A), we speculated that ARR1 and related type-B ARRs could bind directly to the WUS promoter region. Based on sequence conservation and the existence of the putative ARR1 binding site GAT(T/C) (Figures 5A and 5B; Supplemental Figure 17) (Sakai et al., 2000), we selected five regions of the WUS locus (a–e) for analysis.
Figure 5.
Direct Interaction of ARR1 with the WUS Promoter Region.
(A) and (B) Phylogenetic footprinting analysis of the WUS genomic region. Black bold lines with a to e indicate fragments amplified by ChIP-qPCR, and white boxes represent the GAT(T/C) in (A), with adjacent sequences shown in (B). Green, adenine; red, thymine; blue, cytosine; purple, guanine; yellow, conserved GAT(T/C) domain highlight.
(C) ChIP of ARR1ΔDDK-MYC protein with WUS chromatin regions. Shoot tissues (with leaves removed) were used.
(D) EMSA of ARRM-GST with the WUS genomic regions. ARRM indicates the DNA binding domain of ARR1.
(E) to (G) Ratio of firefly luciferase (Luc) to Renilla luciferase (Ren) activity in Arabidopsis protoplasts cotransformed with different reporter and effector construct combinations. Error bars in (C) to (G) indicate the sd of three biological replicates run in triplicate. *P < 0.05 and **P < 0.01 (Student’s t test).
Chromatin immunoprecipitation (ChIP) assays using either shoot tissues with leaves removed or inflorescence tissues showed that ARR1ΔDDK-MYC strongly associated with regions b, c, and d of the WUS promoter region (Figure 5C). To test whether ARR1 can directly bind to these regions, we performed an independent electrophoretic mobility shift assay (EMSA) and confirmed that the DNA binding domain of ARR1 bound to regions b, c, and d (Figure 5D; Supplemental Figure 18). Region c showed a lower affinity for the ARR1 protein than did regions b and d.
We also examined type-B ARR activation of WUS expression using a transient transfection assay, chosen because transiently expressed reporter constructs would lack epigenetic modifications that might interfere with ARR binding. Consistent with our ChIP and EMSA results, a transient transfection assay in protoplasts demonstrated that ARR1 activated the WUS promoter (Figure 5E; Supplemental Figures 19A and 19C). Regions b, c, and d are redundantly required for ARR1 activation (Figure 5F; Supplemental Figures 19B and 19D). Several other related type-B ARRs (Argyros et al., 2008) are expressed in the leaf axil (Supplemental Figure 14), and their mutations can enhance AM initiation defects in arr1 mutants (Wang et al., 2014b). We then tested whether ARR2, 10, 11, and 12 could also directly activate WUS expression in protoplasts. Our results indicated that all of these type-B ARRs activated WUS expression, although to different extents (Figure 5G; Supplemental Figure 19E). This is consistent with their redundant roles in promoting AM initiation. Taken together, these results indicated that ARR1 and other related type-B ARRs can bind to the promoter of WUS to activate its expression.
DISCUSSION
WUS Expression Is Activated de Novo by Cytokinin Signaling during AM Initiation
Tremendous interest has focused on understanding how stem cells are specified in both animals and plants. Stem cell niches provide a microenvironment for stem cell fate determination and the establishment of stem cell niches is central to stem cell biology. In Arabidopsis, the expression of WUS defines shoot stem cell niches (Mayer et al., 1998), as shown by overexpression analyses (Zuo et al., 2002; Xu et al., 2005). Thus, understanding how WUS expression is regulated both spatially and temporally provides key information on stem cell niche specification. Several types of shoot meristems can form in plants. The SAM is established during embryonic development and AMs initiate post embryonically from the axils of leaves. Cells in AMs and the SAM have similar potential for indeterminate growth. After the floral transition, determinate FMs form and initiate a limited number of floral organs. Adventitious shoot meristems may also form, especially in tissue culture conditions. To understand how each type of shoot meristem is established, it is important to understand the mechanism of activation of WUS expression during shoot meristem initiation.
In this study, we focused on the de novo activation of WUS expression during AM initiation. We previously showed that cytokinin promotes AM initiation (Han et al., 2014; Wang et al., 2014b). During AM initiation, the meristematic gene STM is highly expressed in the leaf axil (Grbić and Bleecker, 2000; Long and Barton, 2000; Greb et al., 2003; Shi et al., 2016). The leaf axil cytokinin signaling pulse may result from increased STM expression because STM can promote cytokinin biosynthesis (Jasinski et al., 2005; Yanai et al., 2005). In this study, we show that leaf axil cytokinin signaling directly increases the expression of WUS, which defines stem cell niches and completes AM initiation (Figure 6). The expression of cytokinin signaling pathway genes in the leaf axil (Figure 4A; Supplemental Figure 14), together with the STM-promoted cytokinin biosynthesis in the leaf axil, leads to the observed leaf axil-specific cytokinin signaling pulse and, more importantly, to de novo activation of WUS expression in the leaf axil (Figure 1). This activation is mediated by direct binding of ARR1 and related type-B ARRs, to the WUS promoter region (Figure 5). The same regulatory principle likely functions during FM initiation (Supplemental Figures 7 and 15). In fact, floral bud development is also compromised in mutants defective in cytokinin synthesis, perception, or signaling, with phenotypes including defective floral bud initiation, fewer flowers, and early termination of inflorescences (Higuchi et al., 2004; Nishimura et al., 2004; Argyros et al., 2008; Tokunaga et al., 2012). The same regulatory circuits may also contribute to the de novo activation of WUS expression during embryonic SAM and adventitious shoot meristem formation. Recent studies have proposed that SAM maintenance requires cytokinin signaling (Gordon et al., 2009; Chickarmane et al., 2012). Therefore, the same regulatory principle may function during the maintenance of WUS expression. However, as shown by the ProTCS:GFP reporter, the SAM has much weaker cytokinin signaling than the leaf axil (Supplemental Figure 5), resulting from a lower cytokinin concentration and/or a weaker cytokinin response. This suggests that type-B ARR-based transcriptional activation may be insufficient to maintain WUS expression. Whether other regulatory mechanisms exist for maintenance of WUS expression in established shoot meristems remains to be answered.
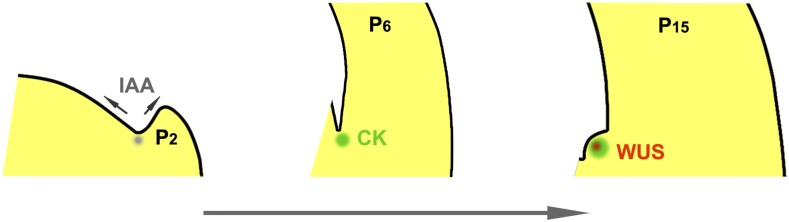
Figure 6.
A Developmental Framework of AM Initiation.
In the leaf axil region, an auxin minimum (gray) at the early stage and a subsequent cytokinin signaling pulse (green) are required for AM initiation (Wang et al., 2014b). Later, cytokinin signaling de novo induces WUS expression (red) to activate the stem cell niche and complete AM initiation. IAA, indole acetic acid; CK, cytokinin.
WUS Is Required for AM Initiation and Integrity
In wus mutants, axillary buds are either absent or replaced by leaf-like structures that obviously lack indeterminate growth (Figures 1C to 1K; Supplemental Figure 1). A substantial portion (77%) of leaf axils are empty in wus-1, and most leaf axils lack discernible shoot meristem structure in wus mutants (Figures 1D to 1K), indicating that WUS is required for AM initiation. Although the wus-1 and wus-101 alleles we used are strong alleles with severe SAM defects, leaf patterning was mostly unaffected (Laux et al., 1996). Nevertheless, it is possible that the general effect of WUS on SAM function has an indirect role on AM initiation in the leaf axil. Recent studies also showed that formation of functional tiller buds in rice (Oryza sativa) requires the rice ortholog of WUS (Lu et al., 2015; Tanaka et al., 2015). In Arabidopsis wus mutants, leaf-like structures or even-terminal branches can still form in a portion of leaf axils (23% in wus-1; Figure 1C). Thus, additional regulators redundantly promote stem cell activities. Nevertheless, yet unknown stem cell activators are insufficient to maintain stem cell homeostasis and indeterminacy (Figure 1C).
It is plausible that WUS is required for stem cell initiation to promote AM initiation. Alternatively, defective stem cell homeostasis alone could explain the lack of AM in wus mutants. Recent work on embryonic shoot stem cell initiation indicated that WUS is dispensable for stem cell initiation and that several members of the WUSCHEL-RELATED HOMEOBOX gene family redundantly function in this process in embryos (Zhang et al., 2017). It remains to be tested whether WUS is required for stem cell initiation in the AM, in addition to its role in stem cell homeostasis.
STM is also necessary for AM initiation (Shi et al., 2016), and the expression of STM is maintained in the leaf axil in wus-1 (Supplemental Figure 2). In contrast to the de novo activation of WUS, STM expression is maintained in the leaf axil (Grbić and Bleecker, 2000; Long and Barton, 2000; Greb et al., 2003; Shi et al., 2016). In fact, the expression patterns of WUS and STM are very different between AM initiation and embryogenesis. During embryogenesis, the onset of WUS expression at the 16-cell stage is much earlier than STM initiation at the late globular stage, when the embryo consists of ∼100 cells (Lenhard and Laux, 1999). The continuous STM expression during AM initiation is consistent with the STM functions in shielding meristematic cells from differentiation, allowing later stem cell initiation (Shi et al., 2016). Nevertheless, STM may also provide rudimentary stem cell initiation activity in the absence of WUS (Brand et al., 2002).
Type-B ARRs Bind to the WUS Genomic Region
In addition to conducting genetic analysis, we provided multiple lines of evidence to support direct binding of ARR1 and related type-B ARRs, to the WUS promoter. ChIP, EMSA, and protoplast transactivation assays all demonstrated ARR binding to three discrete regions (b, c, and d) of the WUS promoter (Figure 5). Whereas regions b and d contain the canonical ARR1 binding core motif GAT(T/C) (Sakai et al., 2000), region c lacks this motif. EMSA showed weaker binding of ARR1 to region c compared with regions b and d (Figure 5D), implying an alternative DNA binding mechanism. Promoter deletion analysis indicated that these three regions are redundantly required for ARR activation of WUS expression (Figure 5F). The wus-6 hypomorphic allele provides additional support for the importance of type-B ARR binding sites. In wus-6, a 7-kb T-DNA insertion separates the type-B ARR binding sites from the WUS open reading frame. The T-DNA insertion also caused a 95-bp deletion that partially overlaps with region b. The expression of WUS is substantially reduced in wus-6 (Hamada et al., 2000), suggesting these evolutionarily conserved type-B ARR binding regions and/or additional upstream regions are important for WUS expression.
Notably, a WUS promoter lacking all three regions still showed activity, although the activity was much reduced (Figure 5F), indicating the existence of additional regulatory regions. One such candidate is a 57-bp region between regions c and d. A previous study showed that this region, when present in tandem and fused to a minimal CaMV 35S promoter, could drive WUS expression in the FM (Bäurle and Laux, 2005). Promoter deletion analysis also showed that flanking regions of the 57-bp core sequence, which covers regions c and d, were required for optimal promoter activity (Bäurle and Laux, 2005). This 57-bp region likely is sufficient for the maintenance of WUS expression, as this assay was done in wild-type plants with functional FMs and endogenous WUS expression, but is insufficient for de novo activation of WUS expression. It again highlights that de novo activation of WUS expression prior to meristem initiation and maintenance of WUS expression in established meristems could use different molecular mechanisms.
Epigenetic Regulation Restricts WUS Expression
In addition to type-B ARR regulation, our work suggests that spatiotemporal epigenetic regulation refines WUS expression. Thus, hormones and epigenetic factors act in concert to govern formation of the lateral shoot stem cell niche. Although leaf axils have highly defined cytokinin signaling (Figure 1), additional cytokinin signaling centers exist in plants (Müller and Sheen, 2008). Type-B ARRs also have broad expression (Figure 4A; Supplemental Figure 14) (Mason et al., 2004). However, most cytokinin signaling centers do not activate WUS expression. Instead, in addition to cytokinin signaling, existing meristematic tissues are likely required to restrict spatiotemporal WUS expression. Our data indicated that increasing the chromatin accessibility by TSA treatment or in PRC mutants led to precocious and ectopic WUS expression and ectopic meristem formation following cytokinin treatment (Figures 3E to 3K; Supplemental Figure 13). Following TSA treatment of wild-type plants or PRC mutants, the onset of WUS expression in the leaf axil occurred earlier, and ectopic WUS appeared in differentiated cells (Figures 3F to 3I; Supplemental Figure 13) (Bratzel et al., 2010), resulting in ectopic meristems (Figure 3K; Supplemental Figure 13C). Epigenetic regulation is involved in the termination of WUS expression in the FM (Liu et al., 2011), and related mechanisms may suppress WUS expression in immature leave axil cells and in differentiated tissues. During leaf maturation, leaf axil cells divide (Wang et al., 2014b; Shi et al., 2016), and epigenetic modifications change (Figures 3A to 3D). This observation suggests a cell division-dependent induction, as was recently found in FM termination (Sun et al., 2014). Cell division may dilute inhibitory cis-acting marks and/or trans-acting factors so that the chromatin environment would be permissive for WUS expression. BAP does not affect prohibiting factors but TSA removes such factors (Figures 3E to 3I; Supplemental Figure 12). TSA treatment leads to different histone acetylation profiles of the WUS promoter than do endogenous regulators, indicating different site specificity. Nevertheless, TSA was efficient in conditioning BAP activation of WUS. The enrichment of H3K27me3, a marker of transcriptional repression, at the WUS locus in mature leaves and the enrichment of H3K4me3, a transcriptional activation marker, in inflorescences may be causal in the regulation of WUS expression (Supplemental Figure 11). Alternatively, these markers may simply reflect transcription status. The H3K4me3 binding protein REPRESSOR OF WUSCHEL1 may contribute to the epigenetic regulation of WUS in the leaf axil (Han et al., 2008; Zhang et al., 2015).
The activity of the AM determines plant architecture and crop yield (McSteen and Leyser, 2005; Wang and Li, 2008; Yang and Jiao, 2016). The finding that cytokinin activates WUS expression provides insight into how shoot stem cell niches are established and may ultimately facilitate the manipulation of plant architecture to enhance crop yield.
METHODS
Plant Materials and Treatment Conditions
Arabidopsis thaliana ecotypes Col-0, Ler, and Ws were used as wild-type controls. Arabidopsis plants were grown under short-day conditions (8 h light and 16 h dark at 22°C) for 30 d and then under long-day conditions (16 h light and 8 h dark at 22°C) to induce flowering before axillary buds were counted. The transgenic lines ProTCS:GFP, ARR1ΔDDK-MYC, ProWUS:WUS-GFP wus-101 (GK870H12), and ProUBQ10:WUS-GR are in the Col-0 background (Müller and Sheen, 2008; Daum et al., 2014; Guan et al., 2014), and ProCLV3:GFP-ER ProWUS:DsRed-N7 is in the Ler background (Reddy et al., 2004). The wus-101, arr1-4, clf-29, and ring1a ring1b mutants are in the Col-0 background (Goodrich et al., 1997; Argyros et al., 2008; Xu and Shen, 2008), the wus-1 mutant is in the Ler background (Laux et al., 1996), and the pga6-1 and sef mutants are in the Ws background (Zuo et al., 2002; Xu et al., 2005). The las, rax, and rev mutants have been previously described (Talbert et al., 1995; Greb et al., 2003; Müller et al., 2006). For in vitro leaf culture, P8 to P11 leaves were taken from plants grown on Murashige and Skoog medium in short-day conditions for 15 to 17 d. Detached leaves were cultured on Murashige and Skoog medium supplemented with 0.1 mg/L inositol acid and 0.5 mg/L folic acid under short-day conditions (Steeves et al., 1957; Wang et al., 2014b).
For chemical treatments, 0.89 μM BAP (Sigma-Aldrich), 30 μM S-4893 (3-[(6-chloro-4-phenylquinazolin-2-yl) amino] propan-1-ol; Vitas-M Laboratory) (Arata et al., 2010), 10 μM CHX (Sigma-Aldrich), and/or 1 μM TSA (Sigma-Aldrich) were used to treat 3-week-old short-day grown plants. For detached leaf culture, the solution was added to the leaf axil region. For seedlings and inflorescences, tissues were soaked in the solution. For inducible WUS expression, pga6-1 and arr1-4 pga6-1 plants were grown under short-day conditions for 15 d, treated with 10 μM β-estradiol every other day for 15 d, and then shifted to long-day conditions without β-estradiol treatment until axillary buds were counted (Zuo et al., 2002; Wang et al., 2014b). We used 6 to 16 plants for phenotypic analysis (Figures 1C, 3K, and 4I; Supplemental Figures 3 and 7).
In Situ Hybridization and Microscopy
In situ hybridization was performed as previously described (Wang et al., 2014b). The digoxigenin-labeled WUS probe contained nucleotides 382 to 1075 bp downstream of the start codon. Shoots were fixed and sectioned following previously described methods (Wang et al., 2014b). Images of sections and plants were taken by a Nikon SMZ1000 stereoscopic microscope or an Olympus BX60 microscope equipped with a Nikon DS-Ri1 camera. Scanning electron microscopy was performed using a Hitachi S-3000N variable pressure scanning electron microscope after standard tissue preparation (Wang et al., 2014b). For confocal microscopy, sample preparation was performed as previously described (Wang et al., 2014b). Images were taken with a Nikon A1 confocal microscope. Excitation and detection window setups for GFP, DsRed, 4′,6-diamidino-2-phenylindole, DsRed, GFP, FM4-64 (to label the cell membrane), and autofluorescence were previously described (Qi et al., 2014; Wang et al., 2014b). Ten to twenty replicates (in three batches) were analyzed.
RT-PCR and RT-qPCR
Total RNA was extracted from shoot tissues (with leaves removed), mature leaves, or inflorescences with the AxyPrep Multisource RNA MiniPrep kit (Corning). Arabidopsis shoot tissue is mainly composed of leaves, making leaf axil tissues low in abundance. As WUS expression is restricted to leaf axils, where axillary buds form, we removed leaves to enrich leaf axil tissues with WUS expression, so that its expression could be reliably detected. First-strand cDNA synthesis was performed using the TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen). Quantitative RT-PCR was performed on a Bio-Rad CFX96 real-time detection system using the KAPA SYBR FAST qPCR kit (KAPA Biosystems) (Tian et al., 2014). ACTIN2 was used as the reference gene to normalize the relative expression for quantitative RT-PCR analysis. RNAs from three batches of independently prepared plant materials (biological replicates with using different plants), each run in triplicate (technical replication), were analyzed. Mean and sd of biological replicates were used to present the data (Figures 2–5).
Phylogenetic Footprinting
The sequences of ∼2000 bp upstream of the WUS start codon were aligned, and the degree of sequence divergence was quantified across for seven seed plant species (Arabidopsis thaliana, Arabidopsis lyrata, Capsella grandiflora, Brassica rapa, Theobroma cacao, Carica papaya, and Oryza sativa). All sequences were obtained from Phytozome 11.0 (https://phytozome.jgi.doe.gov/pz/portal.html). Sequence alignment (Supplemental File 1) was performed with ClustalX version 2.1 (Larkin et al., 2007) with gap opening = 10, gap extension = 0.2, delay divergent sequence = 30%, and turning off of negative matrix and Gonnet series for protein weight matrix. The phylogenetic tree for WUS (Supplemental File 1) was calculated with MEGA version 6.06 based on protein sequences (Tamura et al., 2013) using the neighbor-joining method (Saitou and Nei, 1987). Confidence intervals on phylogenies were inferred by the bootstrap method (Felsenstein, 1985). The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and are in units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated. BigFoot, a Bayesian alignment and phylogenetic footprinting software, was then used to align genomic sequences and score the degree of conservation with default settings (Satija et al., 2009).
ChIP
ChIP experiments were performed according to published protocols (Han et al., 2014). Inflorescence or shoot tissues (with leaves removed) of ARR1ΔDDK-MYC plants were induced with 10 μM β-estradiol for 8 h. Samples of more than 800 mg of tissue were harvested and fixed with 1% (v/v) 4°C formaldehyde at 4°C (Han et al., 2014). Immunoprecipitations were performed with or without anti-MYC (ab9132; Abcam). The precipitated DNA was isolated, purified, and used as a template for quantitative RT-qPCR. For detection of histone modifications, seedlings, inflorescence, and leaves from Col-0 plants, and anti-H3K4m3 (ab8580; Abcam), anti-H3K27me3 (07-449; Millipore), anti-acetyl-Histone H3 (06-599; Millipore), and anti-acetyl-Histone H4 (06-598; Millipore) were used.
ULI-NChIP was performed according to published protocols with modifications (Brind’Amour et al., 2015). The basal 2 to 3 mm of leaf axil tissues from early stage (P8 to P10) or late stage (P15 to P17) leaves were was isolated from 4- to 5-week-old wild-type Col-0 plants. Tissues from 30 to 40 leaf axils were used for each replicate. Tissues were fully ground with in 30 μL Galbraith buffer (45 mM MgCl2, 30 mM sodium citrate, and 20 mM MES, pH 7.0) in a 1.5-mL tube. The pestle was washed with additional 20 μL Galbraith buffer into the same tube. Nuclei were spun down at 1000g for 10 min at 4°C. The supernatant was discarded, and the sediment was resuspended with 50 μL nuclear isolation buffer (NUC-101; Sigma-Aldrich). The subsequent chromatin preparation was based on micrococcal nuclease fragmentation at 37°C for 7 min. Chromatin was precleared with 10 μL of 1:1 protein A:protein G Dynabeads (Life Technologies) and then immunoprecipitated with 1 μg of antibody in antibody-bead complexes at 4°C overnight. Protein-DNA-bead complexes were washed twice with 400 μL low salt wash buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, and 0.1% SDS) and twice with high-salt wash buffer (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 1% Triton X-100, and 0.1% SDS). Protein-DNA complexes were eluted in 30 μL ChIP elution buffer (100 mM NaHCO3 and 1% SDS). DNA was purified using phenol/chloroform and ethanol precipitation (Brind’Amour et al., 2015). The concentration of purified DNA was measured using the Qubit dsDNA HS Assay kit (1674653; Thermal Fisher) before the DNA was used as the template for PCR analysis.
EMSA
The DNA binding domain of ARR1 (ARRM; amino acids 236–299) fused with the GST tag was produced in a prokaryotic expression system as previously described (Tian et al., 2014). Biotin-labeled probes were amplified using 5′ biotin-labeled primers synthesized by Sangon Biotech, and corresponding competitor probes were amplified using primers of the same sequences without labeling. Binding reactions and competition experiments were performed as described (Tian et al., 2014).
Transient Expression in Protoplasts
Full-length coding sequences of ARR1, 460 bp downstream of the start codon of ARR1 (ARR1ΔDDK), 433 bp downstream of the start codon of ARR2 (ARR2ΔDDK), 400 bp downstream of the start codon of ARR10 (ARR10ΔDDK), 382 bp downstream of the start codon of ARR11 (ARR11ΔDDK), and 400 bp downstream of the start codon of ARR12 (ARR12ΔDDK) were amplified from Arabidopsis cDNA by PCR and inserted between the KpnI and BstBI sites of the ProUC19-p35S-FLAG-RBS vector (Feng et al., 2012). To generate ProWUS:Luc, 1708 bp upstream of the start codon of WUS was amplified and inserted between the EcoRI and SacI sites of the ProFRK1:Luc vector (Feng et al., 2012). WUS promoter deletions (ProWUSΔ:Luc) were obtained by inverse PCR using the ProWUS:Luc as the template. The conserved GAT(C/T) motifs were removed in ProWUSΔbd (Supplemental Figure 19B), and −617 to −599 bp were removed in ProWUSΔc.
Arabidopsis protoplasts were isolated from leaves of plants grown under short-day conditions for 5 to 6 weeks. The ProUC19-Pro35S-ARRs-FLAG-RBS vector was cotransformed with ProWUS:Luc (firefly luciferase) and Pro35S:Ren (Renilla luciferase) into protoplasts and incubated at room temperature overnight under weak light. The relative Luc activity (as relative Luc/Ren ratio) was detected with the dual-luciferase report assay system (Promega) and using a Promega GLOMAX 96 microplate luminometer. The efficiency of transient expression was quantified by western immunoblotting using anti-FLAG (A8592; Sigma-Aldrich).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database and/or the Arabidopsis Genome Initiative database under the following accession numbers: ACTIN2 (At3g18780), ARR1 (At3g16857), ARR2 (At4g16110), ARR10 (At4g31920), ARR11 (AT1G67710), ARR12 (At2g25180), CLF (At2g23380), CLV3 (At2g27250), LAS (AT1G55580), WUS/PGA6 (At2g17950), RAX1 (AT5G23000), RAX2 (AT2G36890), RAX3 (AT3G49690), REV (AT5G60690), RING1A (AT5G44280), RING1B (AT1G03770), STM (At1g62360), and UBQ10 (AT4G05320).
Supplemental Data
Supplemental Figure 1. Defective Axillary Bud Formation in the wus-1 Mutant.
Supplemental Figure 2. In Situ Hybridization of STM in the wus-1 Mutant.
Supplemental Figure 3. Bud Formation after WUS Overexpression.
Supplemental Figure 4. Regulation of Axillary Bud Initiation by Cytokinin.
Supplemental Figure 5. The Cytokinin Signaling Pulse Is Much Stronger in the Leaf Axil Than in the SAM.
Supplemental Figure 6. Lack of the Leaf Axil Cytokinin Signaling Pulse and WUS Expression in the rax1-3 Mutant.
Supplemental Figure 7. BAP Induction of WUS Expression in the Inflorescence.
Supplemental Figure 8. Expression of WUS-GFP in Response to BAP Treatment in Immature Leaf Axils.
Supplemental Figure 9. Expression of WUS and CLV3 in Response to BAP Treatment in the Center Region of Immature Leaves.
Supplemental Figure 10. BAP Treatment Enlarges the Expression Domain of WUS and CLV3.
Supplemental Figure 11. H3K4me3 Is Associated with the WUS Chromatin Region.
Supplemental Figure 12. Histone Acetylation of the WUS Genomic Region Increases Following TSA Treatment.
Supplemental Figure 13. In Situ Hybridization of WUS in clf-29 and ring1a ring1b Mutants.
Supplemental Figure 14. Expression of Cytokinin Signaling and Biosynthesis Genes in the Leaf Axil.
Supplemental Figure 15. ARR1 Induction of WUS in the Inflorescence.
Supplemental Figure 16. Buds Formed by WUS Induction Are Identical to Normal Buds.
Supplemental Figure 17. Coding Sequence-Based Phylogenetic Tree of WUS from Seven Seed Plant Species.
Supplemental Figure 18. ARR1 No Longer Binds the Mutated Region b in an EMSA.
Supplemental Figure 19. Type-B ARRs Induce WUS Expression.
Supplemental Table 1. List of Primers.
Supplemental File 1. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 17.
Acknowledgments
We thank Ying Wang for critical reading of the manuscript, Kang Chong, Klaus Theres, Lin Xu, Yunyuan Xu, and Jianru Zuo for seeds, Jian-Min Zhou for the help with the transient expression assay, and Xian Sheng Zhang for exchanging unpublished results. This work was funded by National Natural Science Foundation of China (NSFC) Grant 31430010, by National Basic Research Program of China (973 Program) Grant 2014CB943500, by the National Program for Support of Top-Notch Young Professionals, and by the State Key Laboratory of Plant Genomics. C.T. is a member of the Youth Innovation Promotion Association of the Chinese Academy of Sciences (2017139).
AUTHOR CONTRIBUTIONS
Y.J. conceived the project. J.W., C.T., C.Z., B.S., and X.C. performed experiments and analyzed data. T.-Q.Z., Z.Z., and J.-W.W. contributed reagents. J.W. and Y.J. wrote the manuscript.
Glossary
- CZ
central zone
- OC
organizing center
- SAM
shoot apical meristem
- AM
axillary meristem
- FM
floral meristem
- BAP
6-benzylaminopurine
- CHX
cycloheximide
- ULI-NChIP
ultralow input micrococcal nuclease-based native chromatin immunoprecipitation
- TSA
trichostatin A
- ChIP
chromatin immunoprecipitation
- EMSA
electrophoretic mobility shift assay
Footnotes
Articles can be viewed without a subscription.
References
- Arata Y., Nagasawa-Iida A., Uneme H., Nakajima H., Kakimoto T., Sato R. (2010). The phenylquinazoline compound S-4893 is a non-competitive cytokinin antagonist that targets Arabidopsis cytokinin receptor CRE1 and promotes root growth in Arabidopsis and rice. Plant Cell Physiol. 51: 2047–2059. [DOI] [PubMed] [Google Scholar]
- Argyros R.D., Mathews D.E., Chiang Y.H., Palmer C.M., Thibault D.M., Etheridge N., Argyros D.A., Mason M.G., Kieber J.J., Schaller G.E. (2008). Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäurle I., Laux T. (2005). Regulation of WUSCHEL transcription in the stem cell niche of the Arabidopsis shoot meristem. Plant Cell 17: 2271–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U., Grünewald M., Hobe M., Simon R. (2002). Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 129: 565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U., Fletcher J.C., Hobe M., Meyerowitz E.M., Simon R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619. [DOI] [PubMed] [Google Scholar]
- Bratzel F., López-Torrejón G., Koch M., Del Pozo J.C., Calonje M. (2010). Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr. Biol. 20: 1853–1859. [DOI] [PubMed] [Google Scholar]
- Brind’Amour J., Liu S., Hudson M., Chen C., Karimi M.M., Lorincz M.C. (2015). An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat. Commun. 6: 6033. [DOI] [PubMed] [Google Scholar]
- Busch W., et al. (2010). Transcriptional control of a plant stem cell niche. Dev. Cell 18: 849–861. [DOI] [PubMed] [Google Scholar]
- Charron J.B., He H., Elling A.A., Deng X.W. (2009). Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell 21: 3732–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chickarmane V.S., Gordon S.P., Tarr P.T., Heisler M.G., Meyerowitz E.M. (2012). Cytokinin signaling as a positional cue for patterning the apical-basal axis of the growing Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 109: 4002–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.E., Williams R.W., Meyerowitz E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585. [DOI] [PubMed] [Google Scholar]
- Corbesier L., Prinsen E., Jacqmard A., Lejeune P., Van Onckelen H., Périlleux C., Bernier G. (2003). Cytokinin levels in leaves, leaf exudate and shoot apical meristem of Arabidopsis thaliana during floral transition. J. Exp. Bot. 54: 2511–2517. [DOI] [PubMed] [Google Scholar]
- Daum G., Medzihradszky A., Suzaki T., Lohmann J.U. (2014). A mechanistic framework for noncell autonomous stem cell induction in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 14619–14624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: An approach using the Bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Feng F., Yang F., Rong W., Wu X., Zhang J., Chen S., He C., Zhou J.M. (2012). A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature 485: 114–118. [DOI] [PubMed] [Google Scholar]
- Fletcher J.C., Brand U., Running M.P., Simon R., Meyerowitz E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914. [DOI] [PubMed] [Google Scholar]
- Gaillochet C., Lohmann J.U. (2015). The never-ending story: from pluripotency to plant developmental plasticity. Development 142: 2237–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J., Puangsomlee P., Martin M., Long D., Meyerowitz E.M., Coupland G. (1997). A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51. [DOI] [PubMed] [Google Scholar]
- Gordon S.P., Chickarmane V.S., Ohno C., Meyerowitz E.M. (2009). Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 106: 16529–16534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S.P., Heisler M.G., Reddy G.V., Ohno C., Das P., Meyerowitz E.M. (2007). Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 134: 3539–3548. [DOI] [PubMed] [Google Scholar]
- Grbić V., Bleecker A.B. (2000). Axillary meristem development in Arabidopsis thaliana. Plant J. 21: 215–223. [DOI] [PubMed] [Google Scholar]
- Greb T., Clarenz O., Schafer E., Müller D., Herrero R., Schmitz G., Theres K. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17: 1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan C., Wang X., Feng J., Hong S., Liang Y., Ren B., Zuo J. (2014). Cytokinin antagonizes abscisic acid-mediated inhibition of cotyledon greening by promoting the degradation of abscisic acid insensitive5 protein in Arabidopsis. Plant Physiol. 164: 1515–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S., Onouchi H., Tanaka H., Kudo M., Liu Y.G., Shibata D., MacHida C., Machida Y. (2000). Mutations in the WUSCHEL gene of Arabidopsis thaliana result in the development of shoots without juvenile leaves. Plant J. 24: 91–101. [DOI] [PubMed] [Google Scholar]
- Han P., Li Q., Zhu Y.X. (2008). Mutation of Arabidopsis BARD1 causes meristem defects by failing to confine WUSCHEL expression to the organizing center. Plant Cell 20: 1482–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Zhang C., Yang H., Jiao Y. (2014). Cytokinin pathway mediates APETALA1 function in the establishment of determinate floral meristems in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 6840–6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M., et al. (2004). In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl. Acad. Sci. USA 101: 8821–8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., Phillips A., Hedden P., Tsiantis M. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15: 1560–1565. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Laux T., Mayer K.F., Berger J., Jürgens G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96. [DOI] [PubMed] [Google Scholar]
- Leibfried A., To J.P., Busch W., Stehling S., Kehle A., Demar M., Kieber J.J., Lohmann J.U. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175. [DOI] [PubMed] [Google Scholar]
- Lenhard M., Laux T. (1999). Shoot meristem formation and maintenance. Curr. Opin. Plant Biol. 2: 44–50. [DOI] [PubMed] [Google Scholar]
- Lenhard M., Bohnert A., Jürgens G., Laux T. (2001). Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105: 805–814. [DOI] [PubMed] [Google Scholar]
- Li W., Liu H., Cheng Z.J., Su Y.H., Han H.N., Zhang Y., Zhang X.S. (2011). DNA methylation and histone modifications regulate de novo shoot regeneration in Arabidopsis by modulating WUSCHEL expression and auxin signaling. PLoS Genet. 7: e1002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Kim Y.J., Müller R., Yumul R.E., Liu C., Pan Y., Cao X., Goodrich J., Chen X. (2011). AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 23: 3654–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann J.U., Hong R.L., Hobe M., Busch M.A., Parcy F., Simon R., Weigel D. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105: 793–803. [DOI] [PubMed] [Google Scholar]
- Long J., Barton M.K. (2000). Initiation of axillary and floral meristems in Arabidopsis. Dev. Biol. 218: 341–353. [DOI] [PubMed] [Google Scholar]
- Lu Z., Shao G., Xiong J., Jiao Y., Wang J., Liu G., Meng X., Liang Y., Xiong G., Wang Y., Li J. (2015). MONOCULM 3, an ortholog of WUSCHEL in rice, is required for tiller bud formation. J. Genet. Genomics 42: 71–78. [DOI] [PubMed] [Google Scholar]
- Müller B., Sheen J. (2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller D., Schmitz G., Theres K. (2006). Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18: 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller D., Waldie T., Miyawaki K., To J.P., Melnyk C.W., Kieber J.J., Kakimoto T., Leyser O. (2015). Cytokinin is required for escape but not release from auxin mediated apical dominance. Plant J. 82: 874–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason M.G., Li J., Mathews D.E., Kieber J.J., Schaller G.E. (2004). Type-B response regulators display overlapping expression patterns in Arabidopsis. Plant Physiol. 135: 927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K.F., Schoof H., Haecker A., Lenhard M., Jürgens G., Laux T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815. [DOI] [PubMed] [Google Scholar]
- McConnell J.R., Barton M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125: 2935–2942. [DOI] [PubMed] [Google Scholar]
- McSteen P., Leyser O. (2005). Shoot branching. Annu. Rev. Plant Biol. 56: 353–374. [DOI] [PubMed] [Google Scholar]
- Nishimura C., Ohashi Y., Sato S., Kato T., Tabata S., Ueguchi C. (2004). Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16: 1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Shinohara H., Sakagami Y., Matsubayashi Y. (2008). Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319: 294. [DOI] [PubMed] [Google Scholar]
- Qi J., Wang Y., Yu T., Cunha A., Wu B., Vernoux T., Meyerowitz E., Jiao Y. (2014). Auxin depletion from leaf primordia contributes to organ patterning. Proc. Natl. Acad. Sci. USA 111: 18769–18774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S., Greb T., Peaucelle A., Blein T., Laufs P., Theres K. (2008). Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 55: 65–76. [DOI] [PubMed] [Google Scholar]
- Reddy G.V., Heisler M.G., Ehrhardt D.W., Meyerowitz E.M. (2004). Real-time lineage analysis reveals oriented cell divisions associated with morphogenesis at the shoot apex of Arabidopsis thaliana. Development 131: 4225–4237. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Sakai H., Aoyama T., Oka A. (2000). Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. 24: 703–711. [DOI] [PubMed] [Google Scholar]
- Sakai H., Honma T., Aoyama T., Sato S., Kato T., Tabata S., Oka A. (2001). ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521. [DOI] [PubMed] [Google Scholar]
- Satija R., Novák A., Miklós I., Lyngsø R., Hein J. (2009). BigFoot: Bayesian alignment and phylogenetic footprinting with MCMC. BMC Evol. Biol. 9: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K.F., Jürgens G., Laux T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., Cavalli G. (2007). Genome regulation by polycomb and trithorax proteins. Cell 128: 735–745. [DOI] [PubMed] [Google Scholar]
- Shi B., et al. (2016). Two-step regulation of a meristematic cell population acting in shoot branching in Arabidopsis. PLoS Genet. 12: e1006168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves T.A., Gabriel H.P., Steeves M.W. (1957). Growth in sterile culture of excised leaves of flowering plants. Science 126: 350–351. [DOI] [PubMed] [Google Scholar]
- Sun B., Looi L.S., Guo S., He Z., Gan E.S., Huang J., Xu Y., Wee W.Y., Ito T. (2014). Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science 343: 1248559. [DOI] [PubMed] [Google Scholar]
- Talbert P.B., Adler H.T., Parks D.W., Comai L. (1995). The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121: 2723–2735. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka W., Ohmori Y., Ushijima T., Matsusaka H., Matsushita T., Kumamaru T., Kawano S., Hirano H.Y. (2015). Axillary meristem formation in rice requires the WUSCHEL ortholog TILLERS ABSENT1. Plant Cell 27: 1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C., et al. (2014). An organ boundary-enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol. Syst. Biol. 10: 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga H., Kojima M., Kuroha T., Ishida T., Sugimoto K., Kiba T., Sakakibara H. (2012). Arabidopsis lonely guy (LOG) multiple mutants reveal a central role of the LOG-dependent pathway in cytokinin activation. Plant J. 69: 355–365. [DOI] [PubMed] [Google Scholar]
- Wang Q., Hasson A., Rossmann S., Theres K. (2016). Divide et impera: boundaries shape the plant body and initiate new meristems. New Phytol. 209: 485–498. [DOI] [PubMed] [Google Scholar]
- Wang Q., Kohlen W., Rossmann S., Vernoux T., Theres K. (2014a). Auxin depletion from the leaf axil conditions competence for axillary meristem formation in Arabidopsis and tomato. Plant Cell 26: 2068–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li J. (2008). Molecular basis of plant architecture. Annu. Rev. Plant Biol. 59: 253–279. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang J., Shi B., Yu T., Qi J., Meyerowitz E.M., Jiao Y. (2014b). The stem cell niche in leaf axils is established by auxin and cytokinin in Arabidopsis. Plant Cell 26: 2055–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Shen W.H. (2008). Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr. Biol. 18: 1966–1971. [DOI] [PubMed] [Google Scholar]
- Xu Y.Y., Wang X.M., Li J., Li J.H., Wu J.S., Walker J.C., Xu Z.H., Chong K. (2005). Activation of the WUS gene induces ectopic initiation of floral meristems on mature stem surface in Arabidopsis thaliana. Plant Mol. Biol. 57: 773–784. [DOI] [PubMed] [Google Scholar]
- Yadav R.K., Perales M., Gruel J., Girke T., Jönsson H., Reddy G.V. (2011). WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 25: 2025–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R.K., Perales M., Gruel J., Ohno C., Heisler M., Girke T., Jönsson H., Reddy G.V. (2013). Plant stem cell maintenance involves direct transcriptional repression of differentiation program. Mol. Syst. Biol. 9: 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai O., Shani E., Dolezal K., Tarkowski P., Sablowski R., Sandberg G., Samach A., Ori N. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15: 1566–1571. [DOI] [PubMed] [Google Scholar]
- Yang M., Jiao Y. (2016). Regulation of axillary meristem initiation by transcription factors and plant hormones. Front. Plant Sci. 7: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Jiao Y., Liu Z., Zhu Y.X. (2015). ROW1 maintains quiescent centre identity by confining WOX5 expression to specific cells. Nat. Commun. 6: 6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Tucker E., Hermann M., Laux T. (2017). A molecular framework for the embryonic initiation of shoot meristem stem cells. Dev. Cell 40: 264–277.e4. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Liu X., Engstrom E.M., Nimchuk Z.L., Pruneda-Paz J.L., Tarr P.T., Yan A., Kay S.A., Meyerowitz E.M. (2015). Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 517: 377–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E., Pauling L. (1965). Evolutionary divergence and convergence in proteins. In Evolving Genes and Proteins, V. Vogel and H.J. Bryson, eds (Academic Press), pp. 97–166. [Google Scholar]
- Zuo J., Niu Q.W., Frugis G., Chua N.H. (2002). The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J. 30: 349–359. [DOI] [PubMed] [Google Scholar]