Stress affects the production and fate of alternative mRNA isoforms to coordinate and fine-tune the adaptation of plants to the environment.
Abstract
Stresses from various environmental challenges continually confront plants, and their responses are important for growth and survival. One molecular response to such challenges involves the alternative polyadenylation of mRNA. In plants, it is unclear how stress affects the production and fate of alternative mRNA isoforms. Using a genome-scale approach, we show that in Arabidopsis thaliana, hypoxia leads to increases in the number of mRNA isoforms with polyadenylated 3′ ends that map to 5′-untranslated regions (UTRs), introns, and protein-coding regions. RNAs with 3′ ends within protein-coding regions and introns were less stable than mRNAs that end at 3′-UTR poly(A) sites. Additionally, these RNA isoforms were underrepresented in polysomes isolated from control and hypoxic plants. By contrast, mRNA isoforms with 3′ ends that lie within annotated 5′-UTRs were overrepresented in polysomes and were as stable as canonical mRNA isoforms. These results indicate that the generation of noncanonical mRNA isoforms is an important feature of the abiotic stress response. The finding that several noncanonical mRNA isoforms are relatively unstable suggests that the production of non-stop and intronic mRNA isoforms may represent a form of negative regulation in plants, providing a conceptual link with mechanisms that generate these isoforms (such as alternative polyadenylation) and RNA surveillance.
INTRODUCTION
A major challenge of modern agriculture is to maintain crop production under adverse environmental conditions. Understanding the different mechanisms by which plants adapt and respond to environmental stresses may lead to novel strategies for crop improvement. Flooding, a compound abiotic stress that often leads to cellular hypoxia and energy depletion, is responsible for significant crop damage and yield losses in most crops (Bailey-Serres and Voesenek, 2008; Bailey-Serres et al., 2012b; Bailey-Serres and Colmer, 2014; Voesenek et al., 2016). The response of plants to hypoxic stress is highly complex and involves changes in mRNA and protein levels and subsequent biochemical and physiological adaptations.
Posttranscriptional regulation plays an important role in the control of gene expression. One mode of control that is accomplished through RNA processing is alternative polyadenylation (APA) (Di Giammartino et al., 2011; Lutz and Moreira, 2011; Shi, 2012; Akman and Erson-Bensan, 2014; de Klerk and ’t Hoen, 2015; Mayr, 2016). Genome-wide studies in several eukaryotes have found that 10 to 50% of genes encode transcripts with multiple poly(A) sites (Tian et al., 2005; Nagalakshmi et al., 2008; Wu et al., 2011; Sherstnev et al., 2012). Poly(A) sites can occur at any position during the transcription of a gene and effectively determine protein length or influence mRNA stability, translation, and/or transport (Lutz and Moreira, 2011; Wu et al., 2011). APA may occur through differential selection of polyadenylation signals in the 3′-untranslated region (UTR) of pre-mRNAs. This results in the production of different mRNA isoforms, which can quantitatively affect protein synthesis, while leaving the encoded protein unchanged (Sun et al., 2012). APA may also involve events that occur in upstream regions (5′-UTRs, introns, or protein-coding regions; these isoforms are collectively called noncanonical in this report). Polyadenylation within the protein-coding region of the pre-mRNA will generate transcripts that lack a translation termination codon (non-stop mRNAs). In yeast (Saccharomyces cerevisiae) and mammals, non-stop RNAs are unstable and rapidly degraded through the non-stop decay mechanism (Frischmeyer et al., 2002; Vasudevan et al., 2002; Akimitsu et al., 2007). Polyadenylation within introns can result in alternative protein product synthesis or affect the stability of the mRNA through the nonsense-mediated mRNA decay (NMD) pathway (Hogg and Goff, 2010; Hwang and Maquat, 2011). Surprisingly, APA within 5′-UTRs also occurs, precluding transcription of the open reading frame.
In the past few years, studies have highlighted the important roles of APA in the control of global physiological events such as cellular proliferation, differentiation, transformation, and organ development (Lutz and Moreira, 2011). This is true for APA events that lead to the production of noncanonical mRNA isoforms, affecting the fate of the transcript and the nature of the products of translation. For example, in yeast, the induction of respiration-dependent growth leads to increases in the levels of several mRNA isoforms derived from polyadenylation within protein-coding regions (Sparks and Dieckmann, 1998). In genome-wide studies in yeast, environmental stresses were found to upregulate the levels of non-stop transcript isoforms generated by polyadenylation within protein-coding regions (Yoon and Brem, 2010). In Arabidopsis thaliana, intronic-APA regulates the balance of non/functional mRNA isoforms encoded by the FCA gene, a central flowering time regulator (Simpson et al., 2003; Duc et al., 2013) and by the gene that encodes RPP7, a receptor involved in defense responses (Tsuchiya and Eulgem, 2013). Using genome-scale approaches, numerous noncanonical mRNA isoforms with polyadenylated 3′ ends that lie within intronic, 5′-UTR, and protein-coding regions have been described in plants (Shen et al., 2011; Wu et al., 2011, 2014). In Arabidopsis, the set of genes that encode mRNAs with polyadenylation sites occurring within protein-coding regions and introns is enriched in those that are associated with stress responses and those that encode putative receptor proteins (Wu et al., 2011). In addition, there is a modest evolutionary conservation of APA events that lead to noncanonical mRNA isoforms in Arabidopsis and Medicago truncatula (Wu et al., 2014). These reports indicate that noncanonical polyadenylated mRNA isoforms are a standard component of the transcriptional output in plants.
We hypothesized that abiotic stress may lead to changes in polyadenylation site usage, leading to mRNA isoforms with differing stabilities or protein-coding capacities. Accordingly, we conducted a study to assess differential poly(A) site usage in Arabidopsis seedlings exposed to hypoxic conditions and to determine the relative stabilities and ribosome associations of mRNA isoforms derived from APA. We found that hypoxia increases the levels of mRNA isoforms with 3′ ends that lie in coding regions (non-stop mRNAs) and within introns. Interestingly, non-stop mRNAs and mRNA isoforms with 3′ ends that lie within introns were less stable than mRNAs with 3′ ends that lie within 3′-UTRs. These mRNA isoforms were less likely to be associated with ribosomes, especially in plants subjected to hypoxic conditions. A number of mRNA isoforms were identified that are generated by APA that have 3′ ends within annotated 5′-UTRs. Remarkably, these transcripts were present in polysomes in seedlings subject to normal conditions, but less under hypoxia. These findings indicate that the production of noncanonical mRNA isoforms is an important component of the response of plants to hypoxia. Moreover, they demonstrate that noncanonical mRNA isoforms have different properties and may be considered as three distinct classes of RNAs. Together, they indicate that production of noncanonical mRNA isoforms is a means by which transcriptional output is directed into decay pathways and is thus a global mechanism for negative gene regulation in plants.
RESULTS
General Experimental Approach for Studying Poly(A) Site Choice
An overview of the experimental approach used to evaluate polyadenylated RNA 3′ ends in Arabidopsis (Col-0) seedlings is summarized in Figure 1. Briefly, total or polysomal RNA was isolated from plants grown under control conditions, after hypoxic stress, or following treatment with the transcriptional inhibitor cordycepin. For each experimental treatment, samples were prepared in triplicate (biological replicates, pool of ∼30 whole plants per condition). Isolated RNA was used to prepare libraries of poly(A) tags (PATs), consisting of short cDNAs of 3′-terminal mRNA sequence primed with a fixed 18-nucleotide oligo(dT) tract. PAT libraries were sequenced on the Illumina platform, yielding 600,000 to 25 million individual sequence reads per library (Supplemental Data Sets 1A and 1B). Pooled libraries were demultiplexed using adaptor barcode sequences and aligned to the Arabidopsis genome.
Figure 1.
General Experimental Approach.
The general experimental approach performed in this study is shown, and three sections are highlighted: poly(A) tag library preparation (left), sequencing data workflow (middle), and downstream analyses (right), including gene expression analysis, distribution of PATs, relative poly(A) site usage, and Gene Ontology enrichment. In the right panel, the relative poly(A) usage was calculated as the ratio of the fractional contribution of individual clusters to total poly(A) usage for a gene after treatment divided by the fractional contribution in control conditions; an example for relative poly(A) usage in the coding region (labeled as C in the equation) is shown.
Aligned reads were analyzed in several ways. First, they were used to assess overall gene expression by counting the total numbers of reads that map to each annotated Arabidopsis gene. The results were compared with similar published RNA-seq data to evaluate the consistency of the treatments. Second, poly(A) site analysis was conducted using a dedicated computational pipeline (Bell et al., 2016). For this, mapped reads were trimmed to a single nucleotide coordinate corresponding to the mRNA-poly(A) junction. These reduced tags were used to define poly(A) sites (PASs) and poly(A) site clusters (PACs), which are sets of individual sites that lie within 24 nucleotides of one another (Supplemental Data Sets 1C and 1D). The collection of experimentally determined PASs and PACs was used to determine the genomic distributions of PATs (Supplemental Data Set 1E) and to quantify relative poly(A) site usage in different biological replicates and experiments (Figure 1, middle and right panels, respectively).
Differential Poly(A) Site Choice in Plants Subjected to Hypoxia
To study hypoxia-induced processes, poly(A) site choice was studied in seedlings exposed to 2 h of hypoxic conditions, as well as in seedlings maintained under nonstress conditions (see Methods). Seedling growth and treatment conditions were identical to those used in prior experiments to monitor dynamics in total mRNA abundance, association with polysomes, and ribosome footprints (Juntawong et al., 2014a). PATs prepared from these plants were used to estimate fold change in transcript abundance in response to hypoxia; the results were compared with previously obtained total cellular poly(A)+ RNA data (Juntawong et al., 2014a) (Figure 2A). There was a good correlation between the data generated by RNA-seq and PAT-seq with samples produced at different times (correlation coefficient r1 = 0.707 and r2 = 0.912, using the whole transcriptome and only genes whose expression change by 2-fold or more, respectively). There were 777 gene transcripts that were significantly upregulated based on PAT counts (fold change > 2, false discovery rate [FDR]-corrected P value < 0.05) under hypoxic conditions; in this group, genes associated with responses to abiotic stresses were significantly overrepresented (Supplemental Data Sets 2A and 2B). Conversely, there were 1166 significantly downregulated gene transcripts based on PAT counts (fold change < −2, FDR-corrected P value < 0.05) in hypoxic conditions; genes associated with membrane functionality were significantly overrepresented in this set (Supplemental Data Sets 2A and 2B). These results corroborate previous reports (Mustroph et al., 2009b, 2014) and establish the utility of PATs for genome-wide studies.
Figure 2.
Genome-Wide Distribution of Poly(A) Site Usage in Response to Hypoxia.
(A) Gene expression comparison under normal and hypoxic conditions. Scatterplot comparing gene expression results from previous published studies using RNA-seq (Juntawong et al., 2014a) and from the assessment of gene expression using PAT-seq (this study). Scatterplot of the log2-transformed expression ratios for genes present in both experiments. The correlation coefficients were calculated using the whole transcriptome (r1) or only values with greater than 2-fold change (r2) from both experiments.
(B) Box plots showing change in usage of different classes of poly(A) sites. For this, the relative contribution that each PAC makes to total poly(A) usage was determined on a gene-by-gene basis, with the ratios of usage in hypoxic and control plants calculated and log2-transformed. “Gene” represents the complete collection of PACs. Vertically oriented numbers near the top indicate the total number of PACs in each class.
(C) Percentage of upregulated and downregulated mRNA isoforms. Values > 2× or < 0.5× of PAC/gene normalized fraction with respect to the control are indicative of more or less relative poly(A) cluster usage, respectively. Statistically significant values were determined using the hypergeometric probability (P value < 0.0001, comparison between 2-fold change usage in a specific isoforms with the overall of the same isoform) and are denoted with asterisks (detailed statistic archived in Supplemental Data Set 4).
(D) Ontology term enrichment according to the BAR Arabidopsis website (http://bar.utoronto.ca/). Only ontology terms significantly enriched were included (P value < 0.01 and 1.5-fold). The purple scale shows the bootstrap values.
(E) Box plots showing changes in overall expression of genes possessing different classes of poly(A) sites, as described in the text. For each gene, the log2-transformed fold change in expression in hypoxic plants was extracted from the data set described (Juntawong et al., 2014a); the bulk values for each class were then presented as shown. “Multiple locations” denotes genes with noncanonical poly(A) sites that lie within more than one genomic location. FC, fold change; r, correlation coefficient; T, total RNA; NS, normal conditions; HS, hypoxic conditions. PACs defined by fewer than 10 PATs in the study were excluded from the analysis.
To determine the effect of hypoxia on poly(A) site choice, individual PACs were evaluated for relative usage under control and hypoxic conditions. Thus, the number of tags that mapped to a given PAC was divided by the total number of tags that map to the associated gene, yielding a value for fractional usage of the PAC. The ratio of this value in hypoxic and control samples was used to evaluate changes in poly(A) site choice in response to hypoxia.
As stated above, poly(A) sites may lie at multiple locations within genic regions (5′-UTR, exon, intron, and 3′-UTR). As polyadenylation within functionally distinct regions may reflect different outcomes, the changes in relative usage of sites in each class in response to hypoxic stress were determined (Figure 2B). For reference, the changes in usage of all sites in the study were displayed as well (“Gene” in Figure 2B). For the collection of all sites, the median ratio of apparent usage in hypoxic plants compared with control plants was close to 0 (Figure 2B). A similar result was seen for sites that map to 3′-UTRs; however, the 25 to 75 percentile range for 3′-UTR-situated sites was much narrower than the same range for all sites, suggesting that most of the variability in poly(A) site choice induced by hypoxia was due to changes in sites that fall in other genic locations. This was indeed the case, as the apparent usages of sites that map to protein coding regions (“CDS” in Figure 2B), 5′-UTRs, and introns were greater in hypoxic plants (Figure 2B).
The increase in levels of mRNA isoforms with 3′ ends in coding sequences (CDSs), 5′-UTRs, and introns is also illustrated by consideration of the fraction of each mRNA isoform whose apparent usage changes by more than 2-fold after exposure to hypoxic conditions (Figure 2C). As seen in Figure 2B, 6% of all mRNAs with 3′ ends in CDSs were upregulated by at least 2-fold, as were more than 10% of all isoforms with 3′ ends in 5′-UTRs and introns, respectively. In contrast, roughly 0.3% of isoforms with 3′ ends in 3′-UTRs were upregulated. For all four classes, the numbers of downregulated isoforms were <4%.
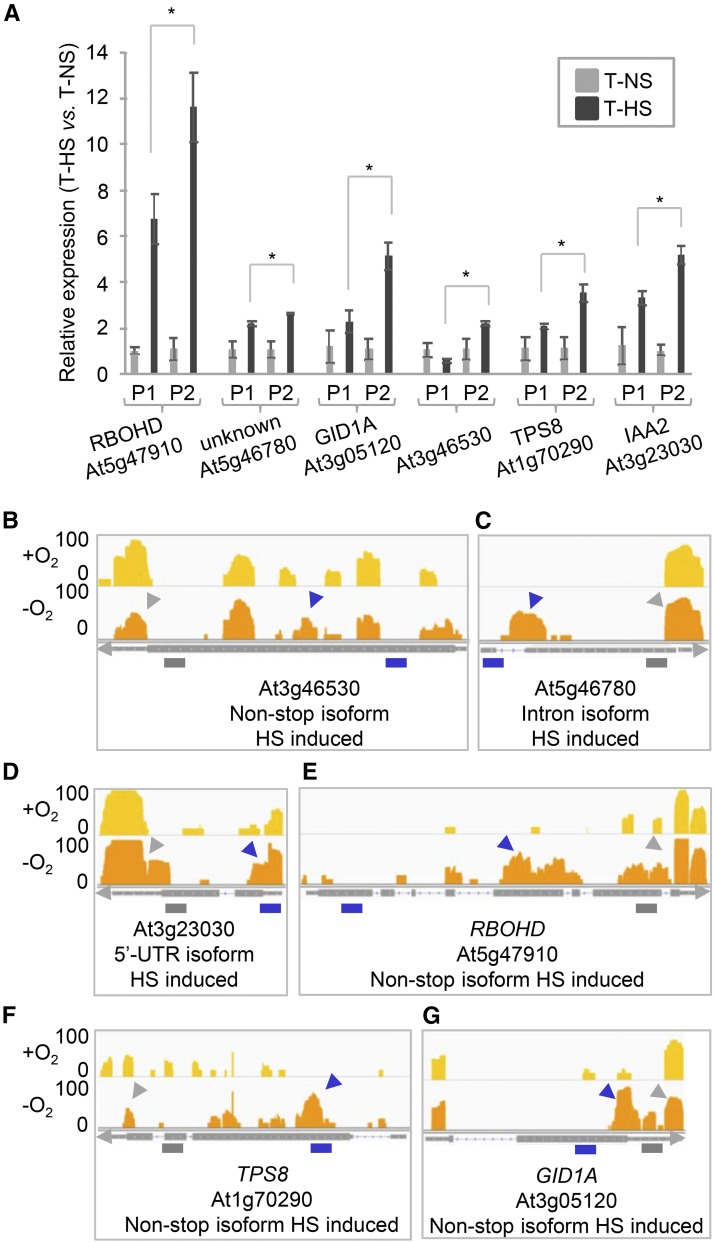
Several individual examples were confirmed using a quantitative RT/PCR approach. For this, one primer set was designed that would query the combined expression of canonical and noncanonical isoforms for a given gene, while another set was designed that would query only canonical isoforms (the locations of amplification products are shown with purple and gray bars in Figures 3B to 3G). As shown in Figure 3A, in all cases there was a discrepancy in the relative changes in mRNA levels determined by the two sets of primers that matched the results seen in the high-throughput sequencing data (Figures 3B to 3G).
Figure 3.
Examples of Differential Poly(A) Site Choice in Hypoxic Plants.
(A) Quantitative RT-PCR analysis of poly(A) site abundance in 3′-UTR (P1) and noncanonical (P2) regions in plants grown in normoxic and hypoxic conditions. Error bars represent sd between three biological replicates (pool of ∼30 whole plants per condition), and asterisks are indicative of statistically significant differences using t test (P < 0.01, three biological replicates).
(B) and (E) to (G) PAT read coverage on selected genes. At3g46530, RBOHD (RESPIRATORY BURST OXIDASE HOMOLOG D; At5g47910), TPS8 (TREHALOSE-6-PHOSPHATASE SYNTHASE S8; At1g70290), and GID1A (GA INSENSITIVE DWARF1A; At3g05120) are examples of a non-stop isoforms induced under hypoxia condition (HS).
(C) At5g46780, an example of an intronic isoform induced under HS.
(D) At3g23030, an example of a 5′-UTR isoform induced under HS. The tracks for each gene in normoxic (+O2) and hypoxic (−O2) conditions are shown in the upper and lower panels, respectively. The purple arrow indicates the region of interest in each gene, and the gray arrow shows the 3′-UTR. The minimum and maximum read value of the scale used for each gene is indicated at the y axis (log2-transformed value). Purple bars represent primers that query the combined expression of canonical and noncanonical isoforms for a given gene (P2), while gray bars illustrate primers designed only for canonical isoforms (P1). T, total RNA; NS, normoxia; HS, hypoxia. Primers are listed in Supplemental Table 1.
To further explore the consequences of APA on gene expression, the overall expression of genes subject to different modes of APA was assessed. For this, genes were classified according to their association with different modes of APA-possessing poly(A) sites that lie within 5′-UTRs, CDSs, or introns. All of these genes also possess poly(A) sites that lie within annotated 3′-UTRs, such that alternative poly(A) site choice involving the generation of canonical and noncanonical mRNAs is possible. Two additional classes of genes were assembled: those with multiple poly(A) sites that lie within more than one noncanonical location (e.g., a site in CDS and a site within an intron) and those with sites that fall only in 3′-UTRs (e.g., genes that do not encode noncanonical mRNA isoforms). For these classes of genes, the log2-fold changes in gene expression reported by Juntawong et al. (2014a) were retrieved and plotted, as shown in Figure 2E. The aggregate overall expression of genes in the different classes was somewhat lower in hypoxic plants, reflecting the genome-wide transcriptional responses noted by others (Mustroph et al., 2009b, 2014; Juntawong et al., 2014a). However, the downregulation was much more pronounced for genes with poly(A) sites that fall within the CDS. Interestingly, genes with sites that fall within 5′-UTRs and introns exhibited very little downregulation compared with the other classes of genes.
To further explore hypoxia-associated poly(A) site choice, those individual sites whose differential usage was the most prominent (FDR-adjusted P value < 0.05, determined using the DEX-seq package) were identified and evaluated. Over 569 sites satisfied this criterion; these sites were associated with 363 genes (Supplemental Data Set 3A). Genes associated with defense and stress responses were significantly overrepresented in this set of 363 genes (Figure 2D, bootstrap > 1.5, P value < 0.01; Supplemental Data Set 3B); in particular, genes encoding putative receptors tended to be affected in hypoxic plants. For these genes, there was a consistent increase in the levels of mRNA isoforms derived from polyadenylation within the CDS; examples are shown in Supplemental Figure 1. Analysis of this set of genes using MapMan indicated an interesting effect of APA on the expression of genes associated with nitrogen metabolism and transport processes. Specifically, the two Arabidopsis genes that encode nitrate reductase (NIA1 and NIA2) showed an unusual hypoxia-induced usage of poly(A) sites that lie within the third and first intron of each gene, respectively (Supplemental Figure 2).
Differential Stabilities of mRNA Isoforms
As stated above, some classes of noncanonical mRNA isoforms are expected to be subject to mRNA surveillance mechanisms. To shed light into this, the relative stabilities of the different classes of mRNA isoforms were determined. For this, the approach taken by Gutierrez et al. (2002) was used. Two-week-old Arabidopsis seedlings were treated with 200 µM cordycepin for 120 min to block nuclear transcription. RNA was isolated and used for PAT-seq library preparation and analyzed as described in the preceding sections.
To confirm that the cordycepin treatment was effective, an analysis of overall steady state mRNA levels was conducted, and the results were compared with those of Gutierrez et al. (2002) and Kim and Chen (2011). For this, relative expression levels were determined using the numbers of PATs that map to individual annotated genes. The rationale for this analysis is that, while most mRNAs will decay within 120 min, the decay of the most unstable mRNAs will be more rapid and will thus stand out when relative expression levels are calculated. This analysis is summarized in Supplemental Data Set 2A. A total of 2378 unstable transcripts could be identified (transcripts that decline in abundance by 2-fold or more within 120 min after the addition of cordycepin). In agreement with the previous studies, the set of genes that encode relatively unstable mRNAs tended to be enriched for transcription factors and responses to hormone stimulus, abiotic and biotic stress, and light stimulus and radiation (Supplemental Data Set 2C). Moreover, there was good agreement between the list of genes encoding unstable mRNAs in this study and those published previously (Supplemental Data Set 2D); 83.2% of the unstable transcripts identified by Gutierrez et al. (2002) and 79.1% of the unstable transcripts identified as unstable in the study by Kim and Chen (2011) were also flagged as unstable here. These results indicate that the cordycepin treatment was effective, and by coupling this inhibitor with the use of PAT libraries to discern the frequency of poly(A) site usage, it should be possible to monitor the relative stabilities of different mRNA isoforms.
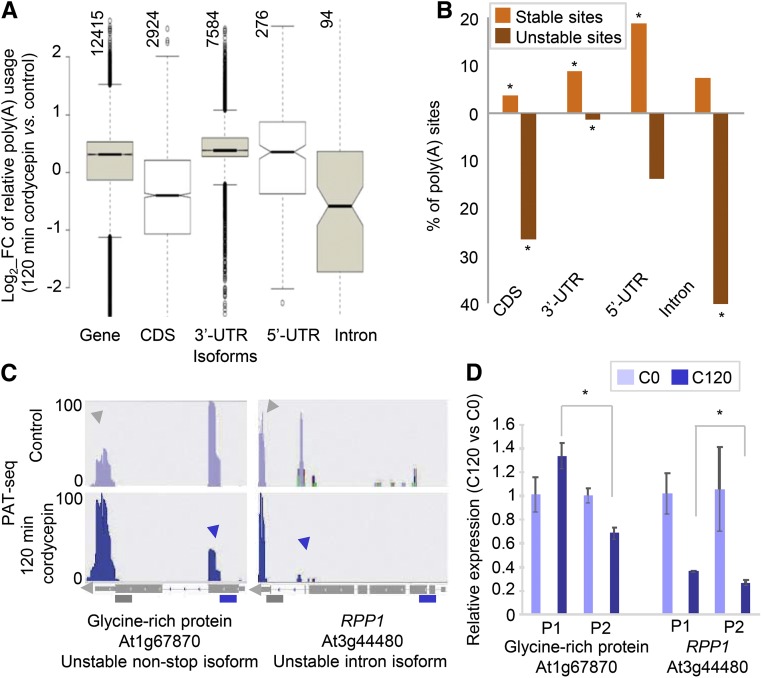
Therefore, the relative stabilities of individual mRNA isoforms were assessed. As was done for the study in Figure 2, the relative usage of each poly(A) site was calculated for the t = 0 (C0) and t = 120 (C120) samples. The ratios of usage in the C0 and C120 samples were determined and log2 transformed. In this analysis, values less than zero indicate lower stabilities (and, hence, a lower contribution of the respective site to the total steady state mRNA level), while values greater than zero suggest a greater stability of the associated mRNA isoform. The values for each class of RNA isoform were then displayed in a box plot (Figure 4A). For all sites taken together, the median value was 0.31. A similar result was obtained for all sites that fell within 3′-UTRs (0.38). Sites that fell within annotated 5′-UTRs had a slightly lower median value (0.36), albeit one that was not significantly different from the median obtained with 3′-UTR sites. In contrast, mRNA isoforms that end within protein-coding regions had a significantly lower value (−0.4). Intronic sites had still lower values (−0.58).
Figure 4.
Analysis of the Stability of Different mRNA Isoforms.
(A) Box plots showing changes in usage of different classes of poly(A) sites after 120 min of treatment with cordycepin. The relative contributions that each PAC makes to total poly(A) usage was determined as in Figure 2B. Vertically oriented numbers indicate the total number of PACs in each class.
(B) Percentage of unstable (down, <0.5×) and stable (up, >2×) mRNA isoforms. Asterisks are indicative of statistically significant differences for each isoform in each group evaluated (less or more stable), determined using the hypergeometric probability (P value < 0.0001).
(C) PAT read coverage on selected genes that illustrate differential isoform stabilities. At1g67870 provides an example of an unstable non-stop isoform and RPP1 (RECOGNITION OF PERONOSPORA PARASITICA1; At3g44480) provides an example of an unstable intronic isoform. The tracks for each gene in control and after 120 min of cordycepin treatment are shown in the upper and lower panels, respectively. The purple and gray arrows indicate the region of interest in each gene and the 3′-UTR, respectively. The read value of the scale used for each gene is indicated on the y axis (log2-transformed value).
(D) Quantitative RT-PCR analysis of poly(A) site abundance in 3′-UTR (P1) and protein-coding (P2) regions in plants grown in control and after 120 min of cordycepin conditions. Error bars represent sd between three biological replicates (pool of ∼30 whole plants per condition), and asterisks are indicative of statistically significant differences using t test (P < 0.01, three biological replicates). Purple and gray bars show the primers (as in Figure 3). C0, control conditions; C120, mRNA after 120 min of cordycepin treatment. PACs defined by fewer than 10 PATs were excluded from the analysis.
The lesser stability of mRNA isoforms with 3′ ends in CDSs and introns is also illustrated when considering the fraction of each mRNA isoform that falls into “unstable” and “stable” categories. If these categories are defined as isoforms whose fractional contribution to all PATs associated with a given gene changes by more than 2-fold, then more than 25% of all non-stop mRNA isoforms (polyadenylated within the CDS and thus lacking a stop codon) fall into the unstable category, while 40% of all isoforms with 3′ ends within introns are unstable (Figure 4B). In contrast, <14 and 2% of mRNA isoforms with 3′ ends in 5′-UTRs or 3′-UTRs, respectively, were unstable, while more than 18 and 8% for each category could be classified as stable, respectively. Some representative examples of unstable mRNA isoforms are shown in Figure 4C; these examples illustrate the general outcome of the assay, showing the disproportionate decrease in some isoforms compared with others. These results further illustrate that mRNAs with 3′ ends within CDSs and introns are, in aggregate, less stable than other mRNA isoforms under standard growth conditions. Analysis by RT-qPCR confirmed these examples (Figure 4D). Altogether, these results indicate that mRNA isoforms derived from polyadenylation within protein-coding regions and introns are less stable than other isoforms.
Differential Translation of mRNA Isoforms upon Hypoxia
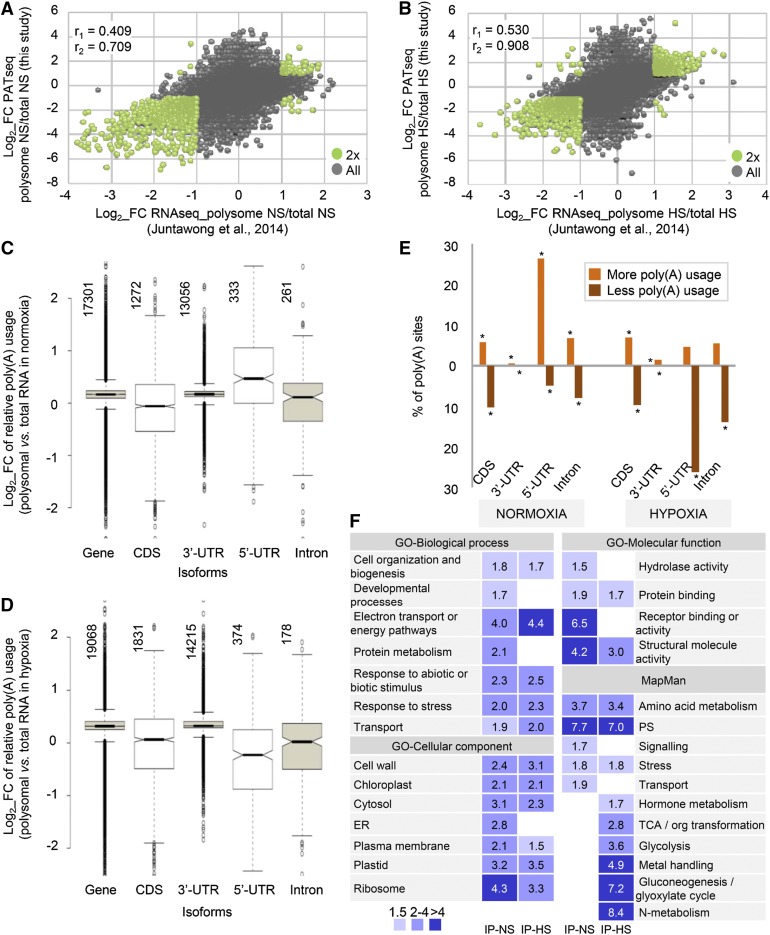
The preceding results indicate that hypoxic conditions lead to increases in the steady state levels of truncated mRNA isoforms derived from proximal poly(A) site choice. These isoforms should not encode full-length polypeptides, raising the question as to whether these isoforms are actively translated. To study this, the poly(A) site profiles were determined for mRNAs associated with polysomes isolated by immunopurification via an epitope-tagged protein of the 60S ribosomal subunit. For this, RNA was purified from polysomes of control and hypoxic plants and PAT libraries were constructed. As before, an analysis geared toward the determination of overall “expression” level was conducted using PATs as the proxy for mRNA transcript. The relative levels of PATs in the total RNA samples were then compared with those from polysomal RNA, and the relative enrichment or depletion of the products of particular genes in the polysomal samples was determined (Supplemental Data Sets 2A, 2E, and 2F). These results were compared with those of previous studies; as seen in Figures 5A and 5B (for normoxia and hypoxia, respectively), there was good agreement with earlier results (Juntawong et al., 2014a).
Figure 5.
Polysome Association of Different mRNA Isoforms in Control and Hypoxic Plants.
(A) and (B) Gene-based determination of polysomal mRNA association under normal and hypoxic conditions. Scatterplots comparing polysomal distributions from (Juntawong et al., 2014a) using RNA-seq and from the assessment of gene expression using PAT-seq (this study). The log2-transformed values for distribution ratios for genes present in both studies were plotted. (A) shows the correlation of polysomal associations in control plants, and (B) shows the correlation in hypoxic plants. The correlation coefficients r1 and r2 were calculated as in Figure 2A.
(C) and (D) Box plots showing changes in poly(A) site usage in total and polysomal RNA in control (C) and hypoxic (D) plants. The relative contributions that each PAC makes to total poly(A) usage was determined on a gene-by-gene basis, as explained before. Vertically oriented numbers indicate the total number of PACs in each class.
(E) Percentage of over- and underrepresented mRNA isoforms in polysomes in control (left panel) and hypoxic (right panel) plants. Values > 2× and < 0.5× of PAC/gene normalized fraction with respect to the control are indicative of more or less relative usage, respectively. Asterisks indicate statistically significant differences for each isoform in each group evaluated (less or more usage), using the hypergeometric probability (P value < 0.0001; Supplemental Data Set 4).
(F) Ontology term enrichment according to the BAR Arabidopsis website (http://bar.utoronto.ca/). Only ontology terms significantly enriched were included (P value < 0.01 and 1.5-fold). The purple scale shows the bootstrap values.
In these plots ([C] and [D]), “Gene” represents the complete collection of PACs. PACs defined by fewer than 10 PATs were excluded from the analysis. FC, fold change; r, correlation coefficient; T, total RNA; IP, polysomal RNA; NS, normal conditions; HS, hypoxic conditions.
Subsequently, the relative abundances of individual mRNA isoforms in RNA isolated from polysomes was calculated and compared with those seen in total RNA to determine if isoforms or classes were more or less prevalent in purified polysomes than in total RNA. In control plants (Figure 5C), the median value for polysome enrichment was close to 0 (“Gene” in Figure 5C). A similar result was seen for mRNA isoforms with 3′ ends that map to 3′-UTRs and introns. The median polysome enrichment for isoforms with 3′ ends that map to protein-coding regions was lower, whereas the enrichment for mRNAs with 3′ ends in 5′-UTRs was higher (Figure 5C). Almost 10% of the individual CDS-situated sites were significantly underrepresented, while ∼5% were overrepresented (Figure 5E, left side). Similar fractions of intron-derived isoforms under- and overrepresented in polysomes were apparent, while there were many more overrepresented 5′-UTR-derived isoforms than underrepresented isoforms (around 25 and 5%, respectively). In contrast, more than 95% of isoforms with 3′ ends in annotated 3′-UTRs were neither over- nor underrepresented in polysomes.
In hypoxic plants, the median value for polysome enrichment was slightly greater than 0 when the entire set of mRNA isoforms was examined (“Gene” in Figure 5D). A similar result was seen for mRNA isoforms with 3′ ends that map to 3′-UTRs. The median polysome enrichments for isoforms with 3′ ends in protein coding regions and introns were lower than the ratios for isoforms with 3′ ends that lie within 3′-UTRs (Figure 5D). Almost 15% of the CDS isoforms were underrepresented in polysomes, while <6% were overrepresented (Figure 5E, right side).
Remarkably, for mRNAs with 3′ ends in 5′-UTRs, the median polysome enrichment in hypoxic plants was much lower than was seen for these isoforms in control plants (compared with “5’-UTR” in Figures 5C and 5D). For these isoforms, almost 30% were underrepresented in polysomes in hypoxic plants (Figure 5E, right side).
To further explore polysome-associated APA under normal and hypoxic conditions, those individual sites whose usages in polysomes and total RNA were significantly different (FDR-adjusted P value < 0.05, determined using the DEX-seq package) were identified and evaluated. A total of 278 sites from 189 genes satisfied these criteria in normal plants, as did 1075 sites involving 684 genes in hypoxic plants (Supplemental Data Sets 3C and 3D). Genes associated with photosynthesis, ribosomes, and stress responses were significantly overrepresented in both sets of genes (Figure 5F, bootstrap > 1.5, P value < 0.01; Supplemental Data Set 3E). Genes associated with intermediary metabolism, including that of hormones, were overrepresented in the list of genes affected by hypoxia-induced changes in polysome association. These results indicate that the production of noncanonical mRNAs seems to have a disparate influence, through both the shunting of transcriptional output into the production of noncanonical mRNAs as well as the diminished translation of these mRNA isoforms, on the functioning of genes that are involved in many aspects of intermediary metabolism and growth.
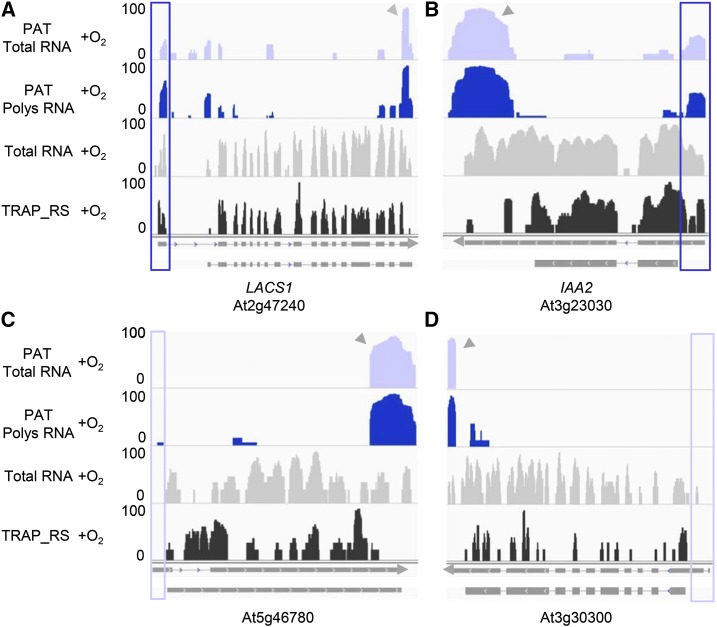
The association of 5′-UTR isoforms with polysomes is unexpected, as these isoforms do not seem to be particularly enriched for so-called upstream open reading frames; only ∼25% of these isoforms have open reading frames of 25 codons or longer that are defined by AUG translation initiation codons and an additional 10% have open reading frames defined by CUG initiation codons. Inspection of ribosome footprinting data (Juntawong et al., 2014a) corroborates this association (Figure 6). For example, a distinct ribosome footprint (in the TRAP_RS) tracks in Figure 6 can be discerned in the At2g47240 and At3g23030 genes, which encode polysome-associated mRNA isoforms derived from polyadenylation within 5′-UTRs (Figures 6A and 6B, respectively). Such footprints are not seen with other genes (such as At5g46780 and At3g30300) that do not encode 5′-UTR isoforms (Figures 6C and 6D). Thus, the association that can be inferred from the results shown in Figure 5 is authentic. The shifts in this association in hypoxic plants suggests the presence of an additional, hitherto unknown, regulatory response involving 5′-UTR mRNA isoforms, perhaps representing an additional contribution that APA makes to stress-related gene regulation.
Figure 6.
Comparisons of PAT-Seq Coverage with Ribosomal Footprint Profiles.
Reads from PAT total RNA, PAT polysomal RNA, RNA-seq total RNA, and translating ribosome affinity purification and footprinting (TRAP_RS) are shown for normoxia.
(A) and (B) LACS1 (LONG-CHAIN ACYL-COA SYNTHASE1; At2g47240) and IAA2 (INDOLE-3-ACETIC ACID INDUCIBLE2; At3g23030) provide examples of genes that encode mRNA with 5′-UTR isoforms associated with polysomes.
(C) and (D) At5g46780 and At3g30300 provide examples of genes that do not encode 5′-UTR isoforms. The purple panels indicate the region of interest in each gene and the gray arrows show the 3′-UTRs. The RNA-seq total RNA and translating ribosome affinity purification and footprinting (TRAP_RS) reads data are from Juntawong et al. (2014a). The read value of the scale used for each gene is indicated on the y axis (log2-transformed value).
DISCUSSION
Distinctions between Different Classes of Noncanonical mRNA Isoforms
In plants, as in other eukaryotes, alternative polyadenylation is a widespread and pervasive process, potentially affecting a large majority of genes for a given species (Shen et al., 2011; Wu et al., 2011, 2014; Sherstnev et al., 2012). In plants, a majority of genes that possess multiple poly(A) sites have at least one site situated upstream of the 3′-UTR, proximal to the associated promoter (Shen et al., 2011; Wu et al., 2011, 2014; Fu et al., 2016). The results presented in this study indicate that the products of proximal poly(A) site choice (collectively, noncanonical mRNA isoforms) may be differentiated according to the location of the poly(A) site within the transcriptional unit.
mRNAs with 3′ Ends That Lie within Protein-Coding Regions
One class of noncanonical mRNA isoform consists of mRNAs with 3′ ends that lie within protein-coding regions. The mRNA isoforms derived from polyadenylation within CDSs will, with few exceptions, lack translation termination codons. In animals and yeast, such RNAs (termed non-stop RNAs) are less stable than canonical mRNAs (Frischmeyer et al., 2002; van Hoof et al., 2002). The decay of non-stop RNAs is mediated by ribosome-associated factors that act to recognize stalled ribosomes and recruit the exosome so as to facilitate degradation of the associated RNA (Klauer and van Hoof, 2012). The results presented in this study confirm that mRNA isoforms derived from polyadenylation within protein-coding regions are generally less stable than other isoforms (Figure 4) and are thus consistent with the presence of a quality control process in plants analogous to the non-stop decay pathway.
In hypoxic conditions, the relative levels of putative non-stop mRNAs in total RNA increase (Figure 2). In addition, in both normoxic and hypoxic conditions, these mRNA isoforms are somewhat underrepresented in polysome-associated RNA populations (Figure 5). This latter consideration, along with the lower stability of these mRNA isoforms, indicates that one aspect of the response of plants to hypoxia is a general redirection, via the process of APA, of transcriptional output into nonproductive outcomes or pathways. In addition to the dynamics of this class of noncanonical mRNA, this redirection has an effect on the overall levels of canonical, functional mRNA isoforms; this is revealed by the decreased levels of functional mRNA isoforms encoded by genes that are affected by APA within protein-coding regions (Figure 2E).
mRNAs with 3′ Ends That Lie within Annotated Introns
Several instances of alternative polyadenylation within introns have been described in plants (Giranton et al., 1995; Bassett et al., 2002; Tang et al., 2002; Simpson et al., 2003; Duc et al., 2013; Tsuchiya and Eulgem, 2013). The distal/proximal choice of poly(A) sites in pre-mRNAs encoded by the gene that encodes lysine-ketoglutarate reductase (LKR) and saccharopine dehydrogenase in cotton (Gossypium hirsutum) determine whether LKR or a bifunctional LKR/saccharopine dehydrogenase polypeptide is produced (Tang et al., 2002). Proximal poly(A) site choice in pre-mRNAs encoded by the Arabidopsis FCA gene leads to the production of a truncated mRNA (with a 3′ end lying within a large intron) and seems to play a negative regulatory role in the expression of this gene (Simpson et al., 2003). A similar scenario and negative regulatory role are operative for the Arabidopsis FPA gene (Duc et al., 2013). Some genes encoding receptors involved in ethylene signaling (Bassett et al., 2002), self-incompatibility (Giranton et al., 1995), and defense signaling (Tsuchiya and Eulgem, 2013) are affected by proximal poly(A) site choice, with proximal polyadenylation seeming to yield truncated mRNAs that may possibly encode shorter polypeptides with some regulatory function. In this study, among genes affected by hypoxia-induced APA within introns are the two Arabidopsis genes that encode nitrate reductase (NIA1 and NIA2); as is the case with other instances listed in the preceding examples, hypoxia induces the production of NIA1/2-derived transcripts that may encode truncated polypeptides.
In all of these cases, the contributions that intronic polyadenylation make to gene regulation have been suggested to involve the production of truncated polypeptides, which may have biochemical activities or may serve as a nonproductive outlet for the transcriptional output of the gene. The results presented here indicate that mRNA isoforms derived from polyadenylation within introns are collectively less stable than canonical mRNA isoforms (Figure 4). This suggests that, as proposed for APA within CDSs, intronic APA may represent a mode of negative regulation that redirects transcriptional output toward mRNAs with lower stability. However, in contrast to what is seen with putative non-stop mRNAs, intronic APA is not associated with a marked decrease in the levels of gene expression (Figure 2E). This suggests that intronic APA is not merely a mechanism for shunting transcriptional output into RNA quality control pathways at the expense of the production of functional mRNAs. Rather, intronic APA may provide a mechanism for generating novel, truncated polypeptides of particular function, and the general lower stability of this class of mRNA isoforms may reflect other aspects of the structures of these isoforms.
The reasons for the reduced stability of mRNA isoforms derived from intronic APA are not entirely clear. As opposed to RNA isoforms derived from included (retained) introns (a common mode of alternative splicing in plants), isoforms with 3′ ends that lie within introns should not have premature translation termination codons in the usual sense (this sense being that they would be followed by an unusually long 3′-UTR with one or more downstream splice junctions). While introns in Arabidopsis that possess poly(A) sites are longer than the typical intron [the average length of introns with poly(A) sites being 270 nucleotides; Wu et al., 2011], they are not so long that polyadenylation within them would lead to unusually long 3′-UTRs (a determinant of NMD; Hogg and Goff, 2010). Specifically, the average 3′-UTR length in the latest version of the TAIR10 Arabidopsis database is 237 nucleotides, whereas the triggering of NMD is usually associated with 3′-UTR lengths of greater than 350 nucleotides (Kalyna et al., 2012). Transcripts derived from intronic polyadenylation are not non-stop RNAs, as they invariably possess one or more in-frame stop codon in the intron-derived part of the 3′-UTR. Other, as yet unidentified, mechanisms that reflect interplay between splicing, polyadenylation, and mRNA turnover may be in force in determining the reduced stability of these mRNA isoforms.
mRNAs with 3′ Ends That Lie within Annotated 5′-UTRs
Perhaps the most enigmatic of the classes of noncanonical mRNA isoforms described in this study are those derived from polyadenylation within 5′-UTRs. The levels of these isoforms increase dramatically in response to hypoxia (Figure 2). However, these isoforms differ little in their stability from isoforms produced by canonical poly(A) site choice (Figure 4). Another surprising feature of mRNAs with 5′-UTR-situated poly(A) sites is the association of some of these RNAs with polysomes in nonstressed conditions (Figure 5). As stated above, these isoforms do not seem to be particularly enriched for annotated upstream open reading frames, and most lack open reading frames of more than 25 codons; there thus seems to be little context by which translation might initiate on these mRNAs. Of all classes of noncanonical mRNAs, the translation of this class is most affected by hypoxia; thus, in normoxic plants, 5′-UTR-derived isoforms are somewhat enriched in polysomes, while in hypoxic plants, they are substantially underrepresented in the set of polysome-associated mRNAs (Figure 5). These characteristics distinguish this class of noncanonical mRNA from other noncanonical mRNA isoforms and raise the possibility that they constitute an as yet unrecognized class of RNA.
While the exact means by which these novel mRNA isoforms are generated are not yet known, one set of results presented here argues against one possible origin. This model holds that these isoforms may be formed by polyadenylation of prematurely terminated Pol II transcripts. The surveillance of premature termination products is linked with a different mode of polyadenylation followed by nuclear exosome-mediated degradation and yields relatively unstable products (Preker et al., 2008; Fox and Mosley, 2016). The observation that Arabidopsis mRNA isoforms derived from polyadenylation within 5′-UTRs are not significantly less stable than canonical mRNA isoforms is not consistent with the idea that the truncated mRNAs are produced by coupled termination and nuclear surveillance.
Other considerations may also be informative as far as the origins and functions of these isoforms. The nature of these isoforms—their enrichment in polysomes in control plants, their relative stabilities compared with other noncanonical isoforms as well as with canonical mRNAs, and the dramatic changes in abundance and polysome association in hypoxic plants—does not easily lend itself to precedent-based interpretations. On one hand, translation of these mRNAs may be fortuitous because of the presumed presence of a 5′-cap as well as a poly(A) tail. However, there is at least a cursory similarity to the identification of ribosome-associated long noncoding RNAs seen in zebra fish. Specifically, in zebra fish, ribosome footprints on long noncoding RNAs resembles the distribution of ribosomes seen along 5′-leader sequences of canonical mRNAs (Chew et al., 2013). Arabidopsis mRNA isoforms with 3′ ends that lie within 5′-UTRs are analogous to mRNA leaders, albeit without significant protein-coding regions. Accordingly, it is possible that these short mRNA isoforms are in fact a subclass of long noncoding RNAs. This possibility is exciting, as it raises the prospect that these mRNA isoforms may play regulatory roles similar to those ascribed to long noncoding RNAs.
Alternative Polyadenylation and the Responses of Plants to Hypoxia
In plants subjected to hypoxic conditions, the abundances of mRNA isoforms derived from polyadenylation at the various proximal sites (5′-UTRs, CDSs, and introns) all increase (Figure 2). The levels of non-stop RNA isoforms increase modestly (albeit significantly), while isoforms with 3′ ends within 5′-UTRs and introns increase substantially. The mechanisms that underlie these increases have not been studied, but other work and considerations that flow from this study support some mechanisms above others. In a general sense, these increases in abundance may be due to increased usage of the respective poly(A) sites, such that more of the transcriptional output is directed toward proximal polyadenylation. Several of the activities of individual polyadenylation complex subunits are affected by phosphorylation, methylation, calmodulin, disulfide remodeling, and ubiquitin (Jacob and Rose, 1984; Thuresson et al., 1994; Colgan et al., 1996; Mizrahi and Moore, 2000; Mouland et al., 2002; Delaney et al., 2006; Ryan, 2007; Addepalli and Hunt, 2008; Ryan and Bauer, 2008; Addepalli et al., 2010; Martin et al., 2010). In addition, alterations in the activities of individual poly(A) complex subunits are often accompanied by large-scale shifts in poly(A) site choice (Jenal et al., 2012; Martin et al., 2012; Thomas et al., 2012; Duc et al., 2013; Li et al., 2015). Hypoxia is associated with transient activation of MPKs and spikes in cytosolic calcium as well as dynamics in reactive oxygen species, along with a reduction in ATP availability and a shift to anaerobic metabolism that can alter cytosolic pH (Chang et al., 2012). All of these signaling events have the potential to alter various subunits of the polyadenylation complex and thus trigger shifts in poly(A) site choice.
Alternatively, the increases in the levels of noncanonical mRNA isoforms in hypoxia may reflect, not increased usage of the associated poly(A) sites, but an increased stability of the various isoforms. For example, the reduction of global protein synthesis by ∼50%, accompanied by the sequestration of mRNAs into cytosolic granules containing Oligouridylate Binding Protein 1C (Sorenson and Bailey-Serres, 2014), may limit the degradation of noncanonical transcripts. The association of RNA quality control processes (in particular, nonsense-mediated decay and non-stop decay) with ribosomes (Simms et al. 2017) and the decreased association of noncanonical mRNA isoforms with polysomes in hypoxic plants (Figure 5) may also contribute to an increase in the levels of noncanonical transcripts. Of course, this assumes that the decay of various noncanonical mRNA isoforms is mediated by these established pathways. For isoforms with 3′ ends that fall within protein coding regions, this seems likely. However, for the other classes of noncanonical mRNAs, this is not as clear. As discussed above, intron-derived isoforms do not have properties consistent with NMD substrates, and 5′-UTR-derived isoforms likewise lack features that are associated with RNA quality control. These caveats notwithstanding, it is probable that alterations in RNA turnover rates contribute to some of the alterations in the levels of noncanonical transcripts in hypoxic plants.
It has been proposed that the production of different mRNAs due to alternative RNA splicing contributes to a fine-tuning of the expression of the associated genes, by shunting transcriptional output into NMD pathways (Green et al., 2003; Lewis et al., 2003; Lareau et al., 2007; Palusa and Reddy, 2010; Kalyna et al., 2012; Drechsel et al., 2013). The results presented here suggest a similar role for APA in plants, especially for events that lead to polyadenylation within protein-coding regions. In plants, genes that encode NBS-LRR (nucleotide binding site-leucine-rich repeat) receptors are subject to APA (Tan et al., 2007), and genes in this set are often affected by polyadenylation within protein coding regions (Thomas et al., 2012; Wu et al., 2014). Many such genes are among those affected by APA in response to hypoxia (Figure 2D). While these proteins are typically associated with innate immunity and defense responses (Marone et al., 2013), there are reports of the involvement of particular NBS-containing receptors with responses to abiotic stress (Huang et al., 2010; Zbierzak et al., 2013; Sarazin et al., 2015). More generally, there is considerable overlap in the molecular responses of plants to biotic and abiotic stress. For example, submergence enhances innate immunity mediated by the transcription factor WKRY22 in Arabidopsis (Hsu et al., 2013). Genes that encode receptors require precise control, and misregulation of these can have detrimental consequences; this is especially true in instances where such genes are overexpressed (Kim et al., 2010; Kato et al., 2014). The results presented here suggest that negative regulation via APA, manifest as indicated in Figure 2E, contributes to the fine-tuning of the expression of genes associated with stress and thus contributes to the global responses of plants to hypoxia.
An additional functional category identified in the meta-analysis as being associated with hypoxia-related APA relates to nitrogen metabolism (MapMan analysis summarized in Figure 2D). This category is flagged largely because of APA involving the two nitrate reductases (NIA1 and NIA2; Supplemental Figure 2). In both instances, poly(A) sites located within introns are utilized in hypoxic plants but not in controls, leading to the production of truncated mRNAs capable of encoding polypeptides consisting of the molybdenum cofactor binding domain, a part of the enzyme capable of generating nitrite in vitro when provided with an external electron transport capacity (Fischer et al., 2005). Nitric oxide enhances the turnover of the key ethylene-responsive transcription factors (ERF-VIIs) that mediate low-oxygen sensing and upregulate the core low oxygen response in hypoxic cells (Gibbs et al., 2011, 2014). Moreover, the nia1nia2 mutant is defective in the ERF-VII and hypoxia mediated positional cue that regulates apical hook opening (Abbas et al., 2015). Nitrate reductase genes are often upregulated under conditions of low oxygen (Mustroph et al., 2010; Juntawong et al., 2014b), and nitrate supplementation alters the physiological responses of plants to low oxygen (Allègre et al., 2004). The induction of the ability to generate truncated nitrate reductase-related polypeptides via APA (Supplemental Figure 2) is yet another aspect of the response of nitrate reductase genes in low oxygen. The significance of this facet of nitrate reductase gene expression is not clear at the moment. Truncated polypeptides may interact with and in some way modify the activities of the full-length enzymes. The truncated polypeptides may retain the nitrite-generating activity in vivo that is seen in vitro (Fischer et al., 2005). This may provide plants with a means by which some nitrate reductase activity can be linked with alternative electron transport capacities under conditions of low oxygen (Bailey-Serres and Voesenek, 2008). Alternatively, these truncated polypeptides may contribute (along with the plant cysteine oxidases) to the rapid oxidation of the N-terminal cysteine of the ERF-VIIs upon re-aeration, which is required to initiate their turnover by the N-end rule pathway (Weits et al., 2014; Voesenek and Bailey-Serres, 2015). The tight control of ERF-VII localization, abundance, and function limits their activation of the transcription of genes encoding enzymes necessary for anaerobic metabolism, which is energetically expensive when oxidative phosphorylation is feasible but a necessity for the maintenance of cell viability during a transient limitation in oxygen (Bailey-Serres and Voesenek, 2008; Bailey-Serres et al., 2012a).
The results presented here reveal a broad-ranging reach of alternative polyadenylation in the response of Arabidopsis to hypoxia. APA generates three distinct classes of noncanonical mRNA, distinguishable by their inherent stabilities, associations with polysomes, and responsiveness to hypoxia. Many of the products of APA are relatively unstable, and their levels and associations with polysomes change substantially under conditions of low oxygen. An important contribution that APA makes to gene regulation in hypoxic plants involves the shunting of transcriptional outputs to nonproductive outcomes; thus, as is seen with alternative splicing, APA constitutes a mode of negative regulation. In the context of the response of plants to hypoxia, this mode of regulation especially affects genes that encode stress-related receptors. Additionally, APA affects the expression of nitrate reductase genes in interesting ways, suggesting that APA plays a role in mediating the links between low oxygen and nitrate signaling or metabolism in plants.
METHODS
Plant Material and Treatment
Arabidopsis thaliana ecotype Col-0 wild-type and transgenic plants expressing NH2-His6-FLAG epitope-tagged ribosomal protein L18 via a near-constitutive promoter (35Spro:FLAG-RPL18) in Col-0 (Zanetti et al., 2005; Mustroph et al., 2009b; Sorenson and Bailey-Serres, 2014) were used. For mRNA polyadenylation site analyses, 35Spro:FLAG-RPL18 seeds were surface sterilized, incubated at 4°C for 2 d, and grown on vertically positioned plates containing solid Murashige and Skoog medium (0.43% Murashige and Skoog salts, pH 5.75, and 29 mM [1% w/v] sucrose) in a growth chamber under long-day conditions (16-h light [∼120 µE s–1 m–2]/8-h dark photoperiod, the bulbs used were Sylvania Octron Eco Fluorescent) at 23°C (Sorenson and Bailey-Serres, 2014). Hypoxic stress was imposed after the end of the light period after 7 d growth by gassing in chambers with humidified 99.99% (v/v) argon for 2 h at 5 to 7 µmol photons m−2 s−1 light; control samples were maintained under the same conditions in chambers open to air as described (Sorenson and Bailey-Serres 2014).
For stability studies, 2-weeks-old Col-0 wild-type plants grown in Murashige and Skoog medium were transferred to a flask with incubation buffer (15 mM sucrose, 1 mM KCl, 1 mM PIPES, and 1 mM sodium citrate, pH 6.5). After a 30-min incubation (time 0), 3′-deoxyadenosine (cordycepin) was added to a final concentration of 200 µM. Tissue samples were harvested after 120 min and quickly frozen in liquid nitrogen (Johnson et al., 2000). Each sample (biological replicate) was the result of pooling ∼30 plants per condition.
In all cases, three biological samples and three independent experiments were performed.
Immunopurification of Polysomes
The immunopurification of ribonucleoprotein complexes from transgenic Arabidopsis line 35Spro:FLAG-RPL18 exposed to hypoxia and control conditions was accomplished as described (Mustroph et al., 2009a).
Poly(A) Tag Library Preparation and Sequencing
Total or polysomal RNA was isolated from Arabidopsis plants after treatment (control, hypoxia, and cordycepin) using the Trizol reagent and RNeasy columns (Qiagen). Quantity and quality measurements were taken using a NanoDrop spectrophotometer. Arabidopsis poly(A) tags (PATs) were generated with 1 µg of total RNA or 0.5 µg of polysomal RNA using Method B1 as described (Ma et al., 2014). The quality of the PAT library was checked with a Bioanalyzer using Agilent High Sensitivity DNA chips (Agilent Technologies), and concentrations were measured with a Qubit fluorometer and a Qubit dsDNA HS assay kit (Life Technologies). The resulting poly(A) tags were sequenced on an Illumina high-throughput sequencing DNA platform. In all cases, three independent biological replicates (pool of ∼30 whole plants) were used.
Poly(A) Tag and Gene Expression Analysis
The sequencing data from PAT-seq were processed using the pipeline as detailed previously (Bell et al., 2016). Briefly, using the CLC Genomics Workbench suite of tools (Qiagen Bioinformatics), the initial sequences obtained were demultiplexed and trimmed to remove the oligo(dT) tracts and sequencing adapters. The processed tags were then mapped to the Arabidopsis reference genome (TAIR10; Berardini et al., 2015) using the current version of CLC Genomics Workbench [individual mapping to evaluate the quality and determine gene expression and pooled sequences to carry out the poly(A) analysis]. The BEDTools suite was used to count PATs that mapped to individual annotated genes and to derive lists of individual PASs and PACs. Only sites and clusters defined by at least 10 individual PATs were retained for subsequent analyses. PAT, PAS, and PAC information was used to carry out three different analyses: (1) to determine PATs, PAS, and PAC distributions on annotated gene regions; e.g., 3′-UTR, 5′-UTR, intron, AMB, and protein-coding regions (AMB from AMBiguous are regions that have different annotations in different transcripts encoded by the same gene); (2) to assess statistically significant differences in poly(A) site usage in a gene-by-gene analysis using the DEX-seq package (Anders et al. 2012); and (3) to determine gene expression using the empirical analysis of DGE (digital gene expression) algorithm in CLC Genomics Workbench. For DGE, genes with a total filter cutoff of 2-fold change and FDR-corrected P values < 0.05 were selected as statistically significant.
A number of ancillary studies were conducted to assess the quality and reproducibility of the libraries prepared and analyzed in this study. These studies are described in the Supplemental Methods and shown in Supplemental Figures 3 to 7.
Quantitative RT-PCR
cDNA was produced with SMARTScribe reverse transcriptase (Clontech), using as starting material 1 to 2 µg of total RNA from three biological replicates. The fragments of interest were amplified by RT-qPCR using specific primers (P1, close to the 3′-UTR; P2, close to the APA-upstream region; and P3, located in APA-5′-UTR upstream region) and SsoAdvanced SYBR Green Supermix (Bio-Rad) as fluorescence dye. ACTIN2 was used as a housekeeping gene to normalize the expression of the genes investigated. The primers used are listed in Supplemental Table 1.
Statistical Analysis
Statistical treatment of the gene expression comparisons used the “exact test” to calculate the P value, assuming that the count data follows a negative binomial distribution (Robinson and Smyth 2008); thus, genes with FDR-corrected P values < 0.05 were selected as statistically significant. The usages of different classes of poly(A) sites were tested for significant differences using a hypergeometric probability test (Supplemental Data Set 4). The statistical method used by DEX-seq was as described (Anders et al. 2012), and an adjusted P value < 0.05 was considered to indicate statistical significance.
Functional Analysis
To search for ontology term enrichment in the different APA-associated gene classes, the classification SuperViewer Tool from Bio-Analytic Resource for plant biology (BAR Arabidopsis website, http://bar.utoronto.ca/ntools/cgi-bin/ntools_classification_superviewer.cgi) was used. Significance threshold of P value < 0.01 and bootstrap > 1.5 were chosen as statistically significant for Gene Ontology and MapMan classification. The ontology term enrichment in differentially expressed genes was also analyzed using DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/home.jsp). Significance threshold of P value < 0.01 and fold enrichment > 1.5 were chosen as statistically significant.
Accession Numbers
The raw data for this study may be found at NCBI SRA (https://submit.ncbi.nlm.nih.gov/subs/sra/) under SRA accession number SRP089899. PASs and PACs that are defined by the poly(A) tags are reported in Supplemental Data Sets 1D and 1E, respectively. The poly(A) metric output for the individual biological replicates comparison using PATapp are reported in Supplemental Data Sets 5A to 5H, and the poly(A) metric output for the comparison with previously published data is included in Supplemental Data Sets 5I and 5J. PACs from 3′ end using RNA-seq that overlap with PACs identify using PAT-seq are reported in Supplemental Data Set 6.
Supplemental Data
Supplemental Figure 1. Examples of hypoxia-induced APA involving genes that encode defense-associated receptors.
Supplemental Figure 2. APA involving the two Arabidopsis genes that encode nitrate reductase.
Supplemental Figure 3. Box plots of the pairwise poly(A) metric comparison between replicates of each sample data set.
Supplemental Figure 4. Box plots of the pairwise comparison of wild-type data sets.
Supplemental Figure 5. Confirmation of poly(A) sites using 3′-RACE.
Supplemental Figure 6. Comparisons of PAT-seq and RNA-seq coverage for poly(A) sites.
Supplemental Figure 7. S1 nuclease protection assay to map the 5′ end of the At1g03610 transcript.
Supplemental Table 1. Summary of oligonucleotides used in this study (5′–3′).
Supplemental Methods. Assessment of libraries and poly(A) site determinations.
Supplemental Data Set 1. Summaries of read mapping outcomes for the different libraries.
Supplemental Data Set 2. Gene expression analysis under different treatments in Arabidopsis thaliana.
Supplemental Data Set 3. DEX-seq analysis output.
Supplemental Data Set 4. Significance of poly(A) cluster usage in different isoforms.
Supplemental Data Set 5. Raw outputs of the analysis presented in Supplemental Figures 3 and 4.
Supplemental Data Set 6. PACs confirmed by RNA-seq.
Acknowledgments
We thank Carol Von Lanken for technical assistance. This work was supported by the National Science Foundation (MCB-1243849 to A.G.H. and MCB-1021969 to J.B.-S.).
AUTHOR CONTRIBUTIONS
All four authors contributed to the conceptualization and design of this study. L.d.L. performed the mRNA stability study and carried out the high-throughput sequencing, analyses, and validation experiments. R.S. performed the hypoxia treatment, polysome purification, and RNA extraction from hypoxic plants and polysomes. A.G.H. helped with the computational and statistical of sequencing data. J.B.-S also contributed to the analysis and presentation of sequencing data. L.d.L. and A.G.H. wrote the manuscript with critical input from R.S. and J.B.-S.
Glossary
- APA
alternative polyadenylation
- UTR
untranslated region
- NMD
nonsense-mediated mRNA decay
- PAT
poly(A) tag
- PAS
poly(A) site
- PAC
poly(A) site cluster
- FDR
false discovery rate
- CDS
coding sequence
- LKR
lysine-ketoglutarate reductase
Footnotes
Articles can be viewed without a subscription.
References
- Abbas M., Berckhan S., Rooney D.J., Gibbs D.J., Vicente Conde J., Sousa Correia C., Bassel G.W., Marín-de la Rosa N., León J., Alabadí D., Blázquez M.A., Holdsworth M.J. (2015). Oxygen sensing coordinates photomorphogenesis to facilitate seedling survival. Curr. Biol. 25: 1483–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addepalli B., Hunt A.G. (2008). Redox and heavy metal effects on the biochemical activities of an Arabidopsis polyadenylation factor subunit. Arch. Biochem. Biophys. 473: 88–95. [DOI] [PubMed] [Google Scholar]
- Addepalli B., Limbach P.A., Hunt A.G. (2010). A disulfide linkage in a CCCH zinc finger motif of an Arabidopsis CPSF30 ortholog. FEBS Lett. 584: 4408–4412. [DOI] [PubMed] [Google Scholar]
- Akimitsu N., Tanaka J., Pelletier J. (2007). Translation of nonSTOP mRNA is repressed post-initiation in mammalian cells. EMBO J. 26: 2327–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akman H.B., Erson-Bensan A.E. (2014). Alternative polyadenylation and its impact on cellular processes. MicroRNA 3: 2–9. [DOI] [PubMed] [Google Scholar]
- Allègre A., Silvestre J., Morard P., Kallerhoff J., Pinelli E. (2004). Nitrate reductase regulation in tomato roots by exogenous nitrate: a possible role in tolerance to long-term root anoxia. J. Exp. Bot. 55: 2625–2634. [DOI] [PubMed] [Google Scholar]
- Anders S., Reyes A., Huber W. (2012). Detecting differential usage of exons from RNA-seq data. Genome Res. 22: 2008–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J., Colmer T.D. (2014). Plant tolerance of flooding stress--recent advances. Plant Cell Environ. 37: 2211–2215. [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J., Fukao T., Gibbs D.J., Holdsworth M.J., Lee S.C., Licausi F., Perata P., Voesenek L.A., van Dongen J.T. (2012a). Making sense of low oxygen sensing. Trends Plant Sci. 17: 129–138. [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J., Lee S.C., Brinton E. (2012b). Waterproofing crops: effective flooding survival strategies. Plant Physiol. 160: 1698–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J., Voesenek L.A. (2008). Flooding stress: acclimations and genetic diversity. Annu. Rev. Plant Biol. 59: 313–339. [DOI] [PubMed] [Google Scholar]
- Bassett C.L., Artlip T.S., Callahan A.M. (2002). Characterization of the peach homologue of the ethylene receptor, PpETR1, reveals some unusual features regarding transcript processing. Planta 215: 679–688. [DOI] [PubMed] [Google Scholar]
- Bell S.A., Shen C., Brown A., Hunt A.G. (2016). Experimental Genome-Wide Determination of RNA Polyadenylation in Chlamydomonas reinhardtii. PLoS One 11: e0146107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini T.Z., Reiser L., Li D., Mezheritsky Y., Muller R., Strait E., Huala E. (2015). The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis 53: 474–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang R., Jang C.J., Branco-Price C., Nghiem P., Bailey-Serres J. (2012). Transient MPK6 activation in response to oxygen deprivation and reoxygenation is mediated by mitochondria and aids seedling survival in Arabidopsis. Plant Mol. Biol. 78: 109–122. [DOI] [PubMed] [Google Scholar]
- Chew G.L., Pauli A., Rinn J.L., Regev A., Schier A.F., Valen E. (2013). Ribosome profiling reveals resemblance between long non-coding RNAs and 5′ leaders of coding RNAs. Development 140: 2828–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan D.F., Murthy K.G., Prives C., Manley J.L. (1996). Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature 384: 282–285. [DOI] [PubMed] [Google Scholar]
- de Klerk E., ’t Hoen P.A. (2015). Alternative mRNA transcription, processing, and translation: insights from RNA sequencing. Trends Genet. 31: 128–139. [DOI] [PubMed] [Google Scholar]
- Delaney K.J., Xu R., Zhang J., Li Q.Q., Yun K.Y., Falcone D.L., Hunt A.G. (2006). Calmodulin interacts with and regulates the RNA-binding activity of an Arabidopsis polyadenylation factor subunit. Plant Physiol. 140: 1507–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino D.C., Nishida K., Manley J.L. (2011). Mechanisms and consequences of alternative polyadenylation. Mol. Cell 43: 853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel G., Kahles A., Kesarwani A.K., Stauffer E., Behr J., Drewe P., Rätsch G., Wachter A. (2013). Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell 25: 3726–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duc C., Sherstnev A., Cole C., Barton G.J., Simpson G.G. (2013). Transcription termination and chimeric RNA formation controlled by Arabidopsis thaliana FPA. PLoS Genet. 9: e1003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K., Barbier G.G., Hecht H.J., Mendel R.R., Campbell W.H., Schwarz G. (2005). Structural basis of eukaryotic nitrate reduction: crystal structures of the nitrate reductase active site. Plant Cell 17: 1167–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox M.J., Mosley A.L. (2016). Rrp6: Integrated roles in nuclear RNA metabolism and transcription termination. Wiley Interdiscip. Rev. RNA 7: 91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer P.A., van Hoof A., O’Donnell K., Guerrerio A.L., Parker R., Dietz H.C. (2002). An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295: 2258–2261. [DOI] [PubMed] [Google Scholar]
- Fu H., Yang D., Su W., Ma L., Shen Y., Ji G., Ye X., Wu X., Li Q.Q. (2016). Genome-wide dynamics of alternative polyadenylation in rice. Genome Res. 26: 1753–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs D.J., Lee S.C., Isa N.M., Gramuglia S., Fukao T., Bassel G.W., Correia C.S., Corbineau F., Theodoulou F.L., Bailey-Serres J., Holdsworth M.J. (2011). Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479: 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs D.J., et al. (2014). Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell 53: 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giranton J.L., Ariza M.J., Dumas C., Cock J.M., Gaude T. (1995). The S locus receptor kinase gene encodes a soluble glycoprotein corresponding to the SKR extracellular domain in Brassica oleracea. Plant J. 8: 827–834. [DOI] [PubMed] [Google Scholar]
- Green R.E., Lewis B.P., Hillman R.T., Blanchette M., Lareau L.F., Garnett A.T., Rio D.C., Brenner S.E. (2003). Widespread predicted nonsense-mediated mRNA decay of alternatively-spliced transcripts of human normal and disease genes. Bioinformatics 19 (suppl. 1): i118–i121. [DOI] [PubMed] [Google Scholar]
- Gutierrez R.A., Ewing R.M., Cherry J.M., Green P.J. (2002). Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc. Natl. Acad. Sci. USA 99: 11513–11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg J.R., Goff S.P. (2010). Upf1 senses 3'UTR length to potentiate mRNA decay. Cell 143: 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F.C., Chou M.Y., Chou S.J., Li Y.R., Peng H.P., Shih M.C. (2013). Submergence confers immunity mediated by the WRKY22 transcription factor in Arabidopsis. Plant Cell 25: 2699–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Li J., Bao F., Zhang X., Yang S. (2010). A gain-of-function mutation in the Arabidopsis disease resistance gene RPP4 confers sensitivity to low temperature. Plant Physiol. 154: 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J., Maquat L.E. (2011). Nonsense-mediated mRNA decay (NMD) in animal embryogenesis: to die or not to die, that is the question. Curr. Opin. Genet. Dev. 21: 422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob S.T., Rose K.M. (1984). Phosphorylation and immunology of poly(A) polymerase. Adv. Enzyme Regul. 22: 485–497. [DOI] [PubMed] [Google Scholar]
- Jenal M., Elkon R., Loayza-Puch F., van Haaften G., Kühn U., Menzies F.M., Oude Vrielink J.A., Bos A.J., Drost J., Rooijers K., Rubinsztein D.C., Agami R. (2012). The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 149: 538–553. [DOI] [PubMed] [Google Scholar]
- Johnson M.A., Perez-Amador M.A., Lidder P., Green P.J. (2000). Mutants of Arabidopsis defective in a sequence-specific mRNA degradation pathway. Proc. Natl. Acad. Sci. USA 97: 13991–13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P., Girke T., Bazin J., Bailey-Serres J. (2014a). Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: E203–E212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P., Sirikhachornkit A., Pimjan R., Sonthirod C., Sangsrakru D., Yoocha T., Tangphatsornruang S., Srinives P. (2014b). Elucidation of the molecular responses to waterlogging in Jatropha roots by transcriptome profiling. Front. Plant Sci. 5: 658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M., et al. (2012). Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 40: 2454–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Saito T., Ito H., Komeda Y., Kato A. (2014). Overexpression of the TIR-X gene results in a dwarf phenotype and activation of defense-related gene expression in Arabidopsis thaliana. J. Plant Physiol. 171: 382–388. [DOI] [PubMed] [Google Scholar]
- Kim E.D., Chen Z.J. (2011). Unstable transcripts in Arabidopsis allotetraploids are associated with nonadditive gene expression in response to abiotic and biotic stresses. PLoS One 6: e24251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Gao F., Bhattacharjee S., Adiasor J.A., Nam J.C., Gassmann W. (2010). The Arabidopsis resistance-like gene SNC1 is activated by mutations in SRFR1 and contributes to resistance to the bacterial effector AvrRps4. PLoS Pathog. 6: e1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauer A.A., van Hoof A. (2012). Degradation of mRNAs that lack a stop codon: a decade of nonstop progress. Wiley Interdiscip. Rev. RNA 3: 649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau L.F., Brooks A.N., Soergel D.A., Meng Q., Brenner S.E. (2007). The coupling of alternative splicing and nonsense-mediated mRNA decay. Adv. Exp. Med. Biol. 623: 190–211. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Green R.E., Brenner S.E. (2003). Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. USA 100: 189–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., You B., Hoque M., Zheng D., Luo W., Ji Z., Park J.Y., Gunderson S.I., Kalsotra A., Manley J.L., Tian B. (2015). Systematic profiling of poly(A)+ transcripts modulated by core 3′ end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS Genet. 11: e1005166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz C.S., Moreira A. (2011). Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. Wiley Interdiscip. Rev. RNA 2: 22–31. [DOI] [PubMed] [Google Scholar]
- Ma L., Guo C., Li Q.Q. (2014). Role of alternative polyadenylation in epigenetic silencing and antisilencing. Proc. Natl. Acad. Sci. USA 111: 9–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marone D., Russo M.A., Laidò G., De Leonardis A.M., Mastrangelo A.M. (2013). Plant nucleotide binding site-leucine-rich repeat (NBS-LRR) genes: active guardians in host defense responses. Int. J. Mol. Sci. 14: 7302–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Gruber A.R., Keller W., Zavolan M. (2012). Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Reports 1: 753–763. [DOI] [PubMed] [Google Scholar]
- Martin G., Ostareck-Lederer A., Chari A., Neuenkirchen N., Dettwiler S., Blank D., Rüegsegger U., Fischer U., Keller W. (2010). Arginine methylation in subunits of mammalian pre-mRNA cleavage factor I. RNA 16: 1646–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C. (2016). Evolution and biological roles of alternative 3'UTRs. Trends Cell Biol. 26: 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrahi N., Moore C. (2000). Posttranslational phosphorylation and ubiquitination of the Saccharomyces cerevisiae Poly(A) polymerase at the S/G(2) stage of the cell cycle. Mol. Cell. Biol. 20: 2794–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouland A.J., Coady M., Yao X.J., Cohen E.A. (2002). Hypophosphorylation of poly(A) polymerase and increased polyadenylation activity are associated with human immunodeficiency virus type 1 Vpr expression. Virology 292: 321–330. [DOI] [PubMed] [Google Scholar]
- Mustroph A., Barding G.A. Jr., Kaiser K.A., Larive C.K., Bailey-Serres J. (2014). Characterization of distinct root and shoot responses to low-oxygen stress in Arabidopsis with a focus on primary C- and N-metabolism. Plant Cell Environ. 37: 2366–2380. [DOI] [PubMed] [Google Scholar]
- Mustroph A., Juntawong P., Bailey-Serres J. (2009a). Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods. Methods Mol. Biol. 553: 109–126. [DOI] [PubMed] [Google Scholar]
- Mustroph A., Lee S.C., Oosumi T., Zanetti M.E., Yang H., Ma K., Yaghoubi-Masihi A., Fukao T., Bailey-Serres J. (2010). Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol. 152: 1484–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A., Zanetti M.E., Jang C.J., Holtan H.E., Repetti P.P., Galbraith D.W., Girke T., Bailey-Serres J. (2009b). Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 18843–18848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., Gerstein M., Snyder M. (2008). The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palusa S.G., Reddy A.S. (2010). Extensive coupling of alternative splicing of pre-mRNAs of serine/arginine (SR) genes with nonsense-mediated decay. New Phytol. 185: 83–89. [DOI] [PubMed] [Google Scholar]
- Preker P., Nielsen J., Kammler S., Lykke-Andersen S., Christensen M.S., Mapendano C.K., Schierup M.H., Jensen T.H. (2008). RNA exosome depletion reveals transcription upstream of active human promoters. Science 322: 1851–1854. [DOI] [PubMed] [Google Scholar]
- Robinson M.D., Smyth G.K. (2008). Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9: 321–332. [DOI] [PubMed] [Google Scholar]
- Ryan K. (2007). Pre-mRNA 3′ cleavage is reversibly inhibited in vitro by cleavage factor dephosphorylation. RNA Biol. 4: 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K., Bauer D.L. (2008). Finishing touches: post-translational modification of protein factors involved in mammalian pre-mRNA 3′ end formation. Int. J. Biochem. Cell Biol. 40: 2384–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarazin V., Duclercq J., Mendou B., Aubanelle L., Nicolas V., Aono M., Pilard S., Guerineau F., Sangwan-Norreel B., Sangwan R.S. (2015). Arabidopsis BNT1, an atypical TIR-NBS-LRR gene, acting as a regulator of the hormonal response to stress. Plant Sci. 239: 216–229. [DOI] [PubMed] [Google Scholar]
- Shen Y., Venu R.C., Nobuta K., Wu X., Notibala V., Demirci C., Meyers B.C., Wang G.L., Ji G., Li Q.Q. (2011). Transcriptome dynamics through alternative polyadenylation in developmental and environmental responses in plants revealed by deep sequencing. Genome Res. 21: 1478–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherstnev A., Duc C., Cole C., Zacharaki V., Hornyik C., Ozsolak F., Milos P.M., Barton G.J., Simpson G.G. (2012). Direct sequencing of Arabidopsis thaliana RNA reveals patterns of cleavage and polyadenylation. Nat. Struct. Mol. Biol. 19: 845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. (2012). Alternative polyadenylation: new insights from global analyses. RNA 18: 2105–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms C.L., Thomas E.N., Zaher H.S. (2017). Ribosome-based quality control of mRNA and nascent peptides. Wiley Interdiscip. Rev. RNA 8: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson G.G., Dijkwel P.P., Quesada V., Henderson I., Dean C. (2003). FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113: 777–787. [DOI] [PubMed] [Google Scholar]
- Sorenson R., Bailey-Serres J. (2014). Selective mRNA sequestration by OLIGOURIDYLATE-BINDING PROTEIN 1 contributes to translational control during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 2373–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks K.A., Dieckmann C.L. (1998). Regulation of poly(A) site choice of several yeast mRNAs. Nucleic Acids Res. 26: 4676–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Fu Y., Li Y., Xu A. (2012). Genome-wide alternative polyadenylation in animals: insights from high-throughput technologies. J. Mol. Cell Biol. 4: 352–361. [DOI] [PubMed] [Google Scholar]
- Tan X., Meyers B.C., Kozik A., West M.A., Morgante M., St Clair D.A., Bent A.F., Michelmore R.W. (2007). Global expression analysis of nucleotide binding site-leucine rich repeat-encoding and related genes in Arabidopsis. BMC Plant Biol. 7: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G., Zhu X., Gakiere B., Levanony H., Kahana A., Galili G. (2002). The bifunctional LKR/SDH locus of plants also encodes a highly active monofunctional lysine-ketoglutarate reductase using a polyadenylation signal located within an intron. Plant Physiol. 130: 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P.E., Wu X., Liu M., Gaffney B., Ji G., Li Q.Q., Hunt A.G. (2012). Genome-wide control of polyadenylation site choice by CPSF30 in Arabidopsis. Plant Cell 24: 4376–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuresson A.C., Aström J., Aström A., Grönvik K.O., Virtanen A. (1994). Multiple forms of poly(A) polymerases in human cells. Proc. Natl. Acad. Sci. USA 91: 979–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B., Hu J., Zhang H., Lutz C.S. (2005). A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 33: 201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T., Eulgem T. (2013). An alternative polyadenylation mechanism coopted to the Arabidopsis RPP7 gene through intronic retrotransposon domestication. Proc. Natl. Acad. Sci. USA 110: E3535–E3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A., Frischmeyer P.A., Dietz H.C., Parker R. (2002). Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295: 2262–2264. [DOI] [PubMed] [Google Scholar]
- Vasudevan S., Peltz S.W., Wilusz C.J. (2002). Non-stop decay--a new mRNA surveillance pathway. BioEssays 24: 785–788. [DOI] [PubMed] [Google Scholar]
- Voesenek L.A., Bailey-Serres J. (2015). Flood adaptive traits and processes: an overview. New Phytol. 206: 57–73. [DOI] [PubMed] [Google Scholar]
- Voesenek L.A., Sasidharan R., Visser E.J., Bailey-Serres J. (2016). Flooding stress signaling through perturbations in oxygen, ethylene, nitric oxide and light. New Phytol. 209: 39–43. [DOI] [PubMed] [Google Scholar]
- Weits D.A., Giuntoli B., Kosmacz M., Parlanti S., Hubberten H.M., Riegler H., Hoefgen R., Perata P., van Dongen J.T., Licausi F. (2014). Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat. Commun. 5: 3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Gaffney B., Hunt A.G., Li Q.Q. (2014). Genome-wide determination of poly(A) sites in Medicago truncatula: evolutionary conservation of alternative poly(A) site choice. BMC Genomics 15: 615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Liu M., Downie B., Liang C., Ji G., Li Q.Q., Hunt A.G. (2011). Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc. Natl. Acad. Sci. USA 108: 12533–12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon O.K., Brem R.B. (2010). Noncanonical transcript forms in yeast and their regulation during environmental stress. RNA 16: 1256–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti M.E., Chang I.F., Gong F., Galbraith D.W., Bailey-Serres J. (2005). Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol. 138: 624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbierzak A.M., Porfirova S., Griebel T., Melzer M., Parker J.E., Dörmann P. (2013). A TIR-NBS protein encoded by Arabidopsis Chilling Sensitive 1 (CHS1) limits chloroplast damage and cell death at low temperature. Plant J. 75: 539–552. [DOI] [PubMed] [Google Scholar]