CONSTANS forms a trimeric complex with At-NF-YB2/NF-YC3 to efficiently bind the CORE element: the CCT provides trimerization and DNA-binding, akin to NF-YA but divergent in sequence selectivity.
Abstract
Nuclear Factor Y (NF-Y) is a heterotrimeric transcription factor that binds CCAAT elements. The NF-Y trimer is composed of a Histone Fold Domain (HFD) dimer (NF-YB/NF-YC) and NF-YA, which confers DNA sequence specificity. NF-YA shares a conserved domain with the CONSTANS, CONSTANS-LIKE, TOC1 (CCT) proteins. We show that CONSTANS (CO/B-BOX PROTEIN1 BBX1), a master flowering regulator, forms a trimer with Arabidopsis thaliana NF-YB2/NF-YC3 to efficiently bind the CORE element of the FLOWERING LOCUS T promoter. We term this complex NF-CO. Using saturation mutagenesis, electrophoretic mobility shift assays, and RNA-sequencing profiling of co, nf-yb, and nf-yc mutants, we identify CCACA elements as the core NF-CO binding site. CO physically interacts with the same HFD surface required for NF-YA association, as determined by mutations in NF-YB2 and NF-YC9, and tested in vitro and in vivo. The co-7 mutation in the CCT domain, corresponding to an NF-YA arginine directly involved in CCAAT recognition, abolishes NF-CO binding to DNA. In summary, a unifying molecular mechanism of CO function relates it to the NF-YA paradigm, as part of a trimeric complex imparting sequence specificity to HFD/DNA interactions. It is likely that members of the large CCT family participate in similar complexes with At-NF-YB and At-NF-YC, broadening HFD combinatorial possibilities in terms of trimerization, DNA binding specificities, and transcriptional regulation.
INTRODUCTION
In all eukaryotes, the precise regulation of transcription of any given gene is ultimately determined by the combinatorial binding of sequence-specific transcription factors (TFs) to their target sequences within promoters, enhancers, and other genomic regulatory regions. These DNA-protein complexes serve as platforms for recruitment of coactivators, most of which contain enzymatic activities that impact local chromatin organization. Specifically, they either modify DNA directly or the tails of DNA-bound, nucleosomal core histones. Histones have a central, globular Histone Fold Domain (HFD) that is required for heterodimerization, tetramerization, and non-sequence-specific binding to DNA as an octameric structure (Luger et al., 1997). HFDs are not specific to core histones but are shared by other proteins, such as the Nuclear Factor-Y (NF-Y) TF. Canonically, NF-Y is a trimeric complex composed of an NF-YB/NF-YC dimer, homologous to the H2B/H2A type histone heterodimer (Romier et al., 2003), and NF-YA, the subunit conferring sequence specificity (Huber et al., 2012; Nardini et al., 2013). The targeted DNA sequence is the CCAAT box, originally discovered in human promoters, and later found in all eukaryotes. Mechanistically, different experiments in the mammalian system suggested that the CCAAT box and NF-Y serve a “pioneering” role in gene activation, namely, that this TF is able to penetrate “hostile” chromatin territories and set the stage for binding of other TFs required for full gene activation (Fleming et al., 2013; Sherwood et al., 2014; Oldfield et al., 2014). This hypothesis is further supported by recent experiments with mouse zygotes, where NF-Y appears to be the major TF opening chromatin as early as the two- and four-cell stages (Lu et al., 2016).
The genes coding for the three NF-Y subunits have been identified in essentially all eukaryotes and are among the most evolutionarily conserved proteins described to date. Notably, conserved domains include the HFDs, required for heterodimerization and non-sequence-specific DNA binding, and a stretch of 56 amino acids of NF-YA, required for HFD association and specific CCAAT binding. In mammals, invertebrates, and fungi, there are one or two genes coding for each subunit. Instead, plants have dramatically expanded the number of NF-Y genes: Typically, there are 8 to 14 gene family members for each subunit, conferring an enormous combinatorial capacity on the trimer; some are expressed in a tissue-restricted manner, and many are relatively ubiquitous (Gusmaroli et al., 2001, 2002; Stephenson et al., 2007; Siefers et al., 2009; Cao et al., 2011b; Hilioti et al., 2014; Liang et al., 2012, 2014; Quach et al., 2015; Rípodas et al., 2015; Zhang et al., 2015; Qu et al., 2015; Feng et al., 2015; Ren et al., 2016; Malviya et al., 2016; Li et al., 2016; Yang et al., 2016; Zhang et al., 2016). Typical features of other plant TFs, such as the presence of duplicate members with similar functions and neofunctionalization of specific genes, were determined by genetic experiments, mostly performed in Arabidopsis thaliana (reviewed in Laloum et al., 2013; Petroni et al., 2012). A growing body of evidence indicates that specific At-NF-Y subunits are involved in disparate physiological events in plant development, growth, and reproduction, as well as in adaptation to physiological and adverse environmental conditions.
One aspect of plant development in which specific NF-Y genes were shown to be important is the regulation of photoperiod-dependent flowering: At least two At-NF-YB (At-NF-YB2 and At-NF-YB3) and three At-NF-YC subunits (At-NF-YC3, At-NF-YC4, and At-NF-YC9) are involved in floral timing (Wenkel et al., 2006; Kumimoto et al., 2008, 2010; Cao et al., 2014). At-NF-YB and At-NF-YC subunits can physically interact with CONSTANS (CO), which is also an essential regulator of photoperiod-dependent flowering (Putterill et al., 1995; reviewed in Song et al., 2015). CO contains a conserved CCT (CONSTANS, CONSTANS-LIKE, TOC1) domain, which is shared by >30 proteins in Arabidopsis and similar numbers in other angiosperms. Interestingly, the CCT domain is homologous to the HFD interaction and CCAAT binding domain of NF-YA; CO (as well as CO-like and TOC1 proteins) has been shown to bind several NF-Y HFD subunits (Wenkel et al., 2006; Ben-Naim et al., 2006; Kumimoto et al., 2010; Cao et al., 2011a; Li et al., 2011; Chen et al., 2014; Hou et al., 2014) in a CCT domain-dependent manner (Wenkel et al., 2006). The sequence identity/similarity between CO and NF-YA is particularly evident in the subdomain required in NF-YA for CCAAT recognition (Wenkel et al., 2006; Petroni et al., 2012). Furthermore, CO and the HFDs mentioned above participate in the same genetic pathway controlling flowering (Cao et al., 2014), as both CO and NF-Y bind and regulate the expression of FLOWERING LOCUS T (FT; aka, the principal florigen), through a CCAAT box in its enhancer and CO-responsive elements (COREs) in its promoter, respectively (Adrian et al., 2010; Song et al., 2012; Cao et al., 2014; Bu et al., 2014).
COREs have been identified through analysis of evolutionary conservation of the FT promoter in different plant species and through mutagenesis and functional analysis of the promoter in vivo (Adrian et al., 2010; Tiwari et al., 2010; Cao et al., 2014). Despite the wealth of genetic knowledge, the molecular mechanisms of the activity of CO and CO-like proteins are not completely understood (Blackman and Michaels, 2010). In some experiments, CO, and the related TIMING OF CAB EXPRESSION1 (TOC1), were shown to be stand-alone sequence-specific TFs capable of directly binding COREs (Tiwari et al., 2010; Gendron et al., 2012; Abelenda et al., 2016). In others, CO-like proteins compete with NF-YA for association with specific HFDs, thus influencing NF-Y transcriptional activity (Li et al., 2011). Finally, it is also conceivable that CO might form a quaternary complex with NF-Y (through CO/HFD interactions): In this model, CO would essentially act as a coactivator (Cao et al., 2014).
The lack of a general consensus as to the molecular mechanism of CO function, and the observation of the fundamental similarity between NF-YA and CCT conserved domains (Wenkel et al., 2006) drove the experiments reported here. We reasoned that CO, and by inference all CCT proteins, are “NF-YA-likes,” associating with HFD heterodimers, binding to DNA with robust and specific affinity only in the trimeric configuration. We set out biochemical and genetic experiments to test this hypothesis.
RESULTS
The CO/At-NF-YB2/NF-YC3 Trimer Binds to DNA in Vitro
CO was previously shown to interact directly with several NF-YB/NF-YCs (summarized in Supplemental Figure 1). To test whether it forms a DNA binding trimer, we expressed in Escherichia coli and independently purified the CCT domain of CO, previously shown to be sufficient for HFD interactions (Wenkel et al., 2006); in parallel, the HFD dimer At-NF-YB2/NF-YC3 was also produced by coexpressing the subunits (Supplemental Figure 2). We used this specific heterodimer because of the genetic evidence implicating the two genes in the regulation of the timing of flowering, of a known in vivo interaction, and of biochemical data suggesting CO interactions (Wenkel et al., 2006; Kumimoto et al., 2008, 2010). We used the purified proteins in electrophoretic mobility shift assays (EMSAs) with a Cy5-labeled 31-mer oligonucleotide containing the functionally important FT CORE2 (Adrian et al., 2010; Tiwari et al., 2010; Cao et al., 2014). The results demonstrated that a CO/HFD dimer complex, but not CO alone, efficiently bound FT CORE2 (Figure 1A). Note that at high CO concentrations, a very faint, faster-migrating DNA complex could be observed in the absence of HFDs upon long exposures (Supplemental Figure 3). Incubation of CO and HFD subunits with a labeled, functional CCAAT from the FT enhancer (Cao et al., 2014) yielded residual binding only at high CO concentrations. Instead, addition of At-NF-YA2 or At-NF-YA6 generated the NF-Y complex on CCAAT, as expected, but not on CORE2 (Figure 1A). The specificity of the CO/HFD complex was then assayed by competition analysis with different unlabeled oligos containing wild-type or mutant CORE2, CCAAT, or an unrelated sequence. Unlabeled, wild-type CORE2 competitors interfered with binding in a concentration- and position-specific manner; CORE2 oligos with mutations known to reduce FT expression in vivo (Tiwari et al., 2010; Cao et al., 2014), FT CCAAT oligos, or an unrelated sequence did not reduce binding of the labeled probe (Figure 1B). We conclude that CO forms a complex with NF-Y HFD subunits, which binds to the CORE sequence with high affinity and specificity. By analogy to the NF-Y acronym, we refer to the CORE binding trimer as NF-CO.
Figure 1.
CO Binds DNA as a Trimer with At-NF-YB2/NF-YC3 and Recognizes the CORE Element.
(A) CO forms a trimer with At-NF-YB2/NF-YC3 HFD binding to the FT CORE2 element. EMSAs were performed using fluorescently labeled FT CORE2 (lanes 1–14) or FT CCAAT (lanes 15–28) 31-mer oligonucleotide DNA probes (20 nM) by addition of the indicated proteins. CO-CCT (CO) was incubated at increasing concentrations (90, 180, 270, and 360 nM) with the CORE2 probe in the absence (lanes 2–5) or presence (lanes 9–12) of the At-NF-YB2/NF-YC3 HFD dimer (At-NF-YB2/YC3, 60 nM). As controls, At-NF-YA2 or -YA6 (YA2, YA6) was incubated with the CORE2 probe at the highest concentration of the dose curve (360 nM), with or without (−) the HFD dimer (60 nM) (YA2: lanes 13, 6; YA6: lanes 14, 7, respectively). Lane 1: CORE2 probe alone, without protein additions. DNA binding of CO or At-NF-YAs, as indicated (lanes 16–27), was assayed on the FT CCAAT probe in the presence of At-NF-YB2/NF-YC3 (60 nM), with the same protein concentration dose curve (90, 180, 270, and 360 nM). As controls, the FT CCAAT probe was incubated with the HFD dimer alone (60 nM, lane 15) or with At-NF-YA2 protein (360 nM, lane 28). NF-CO and NF-Y/DNA complexes are indicated by closed or open arrowheads. fp, free probe.
(B) EMSA competition analysis of the At-NF-YB2/NF-YC3/CO complex specificity on the labeled CORE2 probe. Top panel: Sequences of the 31-mer CORE2 probe and unlabeled competitor derived from the FT promoter (−172/−141 from ATG). Oligos 1 to 6: CORE2 30-mers and 25-mer, the wild type or mutant was used as unlabeled competitors. The 31-mer derived from the FT enhancer CCAAT sequence and the FT mutant competitor (Cao et al., 2014) are listed below, together with the Hsp70 CCAAT competitor. Sequence identity with the probe is indicated by dots, and 5′ or 3′ sequence extensions, or mutated nucleotides are indicated in capital letters. The previously described TTGTGGTT CORE element (Tiwari et al., 2010) and the CCAAT pentamer are highlighted in bold letters. Bottom panel: EMSA competition analysis was performed by incubation of the CORE2 probe with the trimer composed of indicated subunits (At-NF-YB3/NF-YC3, 60 nM; CO, 180 nM -At-NFY-B2/YC3/CO-: lanes 2–27) in the presence of TE buffer alone (lanes 2 and 27) or with the addition of increasing concentrations of the indicated unlabeled competitors (5× or 25× molar excess; lanes 3–26). Lanes 1 and 28: CORE2 probe alone, without protein addition. The NF-CO/DNA complex is indicated by an arrowhead.
NF-CO Binds the Core Pentamer CCACA, with Preferred Flanking Sequences
To pinpoint precisely the DNA binding requirements of NF-CO, we initially challenged the complex with 10 unlabeled 30-mers containing 3-bp scanning mutations (mI oligos; Figure 2A). Four oligos (mI-3 to 6) showed loss of competition, indicating reduced or absent NF-CO interactions (Figure 2A; Supplemental Figure 4), whereas mutations in the flanking areas had a negligible effect on complex formation. We then dissected this 12-bp central region with six oligos harboring 2-bp mutations (mII oligos). Again, four oligonucleotides (mII-3 to 6) competed poorly, trimming down the targeted element to 8 bp (Figure 2A). Finally, we used 1-bp mutations, changing each of the eight bases to all other three nucleotides (mIII series; Figure 2; Supplemental Figure 4). This led to the definition of a central TGTGG pentanucleotide, or CCACA on the reverse strand, with preferred flanking sequences, as the optimal in vitro binding site of NF-CO (Figure 2C).
Figure 2.
Determination of NF-CO Sequence Specificity in Vitro.
(A) CORE2 competitors and mutagenesis strategy. The unlabeled wild-type CORE2 and 30-mer oligo sequences with mutated nucleotides are shown, as in Figure 1, for the three sets of CORE2 mutant oligos (mI, mII, and mIII). In the bar graphs, mI and mII mutant oligo competitor efficiency (competition) is expressed as ratio of the dose-response curve slope of the mutant versus the wild-type oligo (see Methods). Competition of the wild-type oligo is set as 1. Indicated values represent the mean of three (mI oligos; top panel) or two (mII oligos; bottom panel) series of competition assay experiments (see also Supplemental Figure 4). Error bars indicate ± sd for mI oligos or value ranges for mII oligos. Sequences in red boxes highlight mutations with reduced competition (<0.67 of wild-type oligo efficiency).
(B) and (C) CORE2 mIII mutant oligo EMSA competition results. Competition efficiencies are shown as mean value of three independent series of experiments for each of the mIII single nucleotide mutant oligo, as indicated for (A). For each mutated position, the wild-type oligonucleotide competition value, set as 1, is also shown. Values are also displayed in the bar graph in (C) (average of three independent sets of experiments ± sd). For each nucleotide position, dark and light shaded bars denote mutant and wild-type (asterisk) competitors, respectively (see also Supplemental Figure 4). In (C), the sequence matrix obtained with the mIII competitions (information content) is shown on the right, for the sense (+) and reverse (−) strands of the FT promoter.
In Vivo RNA-Seq Analysis Identifies the CCACA Pentamer in Promoters of Genes Regulated by NF-CO Subunits
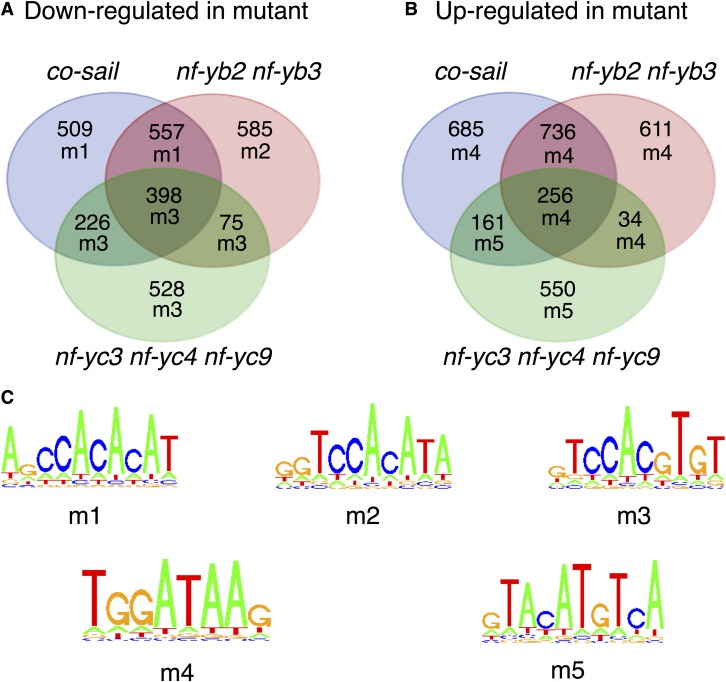
To identify genes regulated by NF-CO subunits in vivo, we performed RNA-seq analysis on previously described, late-flowering co-sail, nf-yb2 nf-yb3, and nf-yc3 nf-yc4 nf-yc9 mutants. The requirement for the double and triple HFD subunit mutants is due to negligible phenotypic effects on flowering timing of single At-NF-YB and single or double At-NF-YC mutants (Kumimoto et al., 2008, 2010). The complete list of genes whose expression was affected in the mutants is in Supplemental Data Set 1. Of the 1690 genes significantly downregulated (false discovery rate < 0.05 in both Limma and DESeq2 analyses; see Methods) in co-sail, 955 were shared in nf-yb2 nf-yb3 (Phypergeometric = <10e-127), 624 in nf-yc3 nf-yc4 nf-yc9 (Phypergeometric = <10e-127), and 398 were common in all three mutants (Phypergeometric = <10e-127) (Figure 3). The overlap among the HFD subunit mutants was somewhat lower, but still striking: 473 of the 1615 downregulated genes in nf-yb2 nf-yb3 are shared with nf-yc3 nf-yc4 nf-yc9 (Phypergeometric = <10e-127). Upregulated gene sets also showed robust overlaps (Figure 3). We did find genes previously known to be dependent upon CO and HFD activity, including FT (Kumimoto et al., 2008, 2010). We validated 16 genes of the cohort downregulated in both nf-yb2 nf-yb3 and co-sail by RT-qPCR analysis, and all showed the expected changes, thus confirming the robustness of the RNA-seq data (Supplemental Figure 5). We then retrieved the promoter sequences (−1 kb to TSS/ATG) of affected genes and analyzed them with Weeder, an algorithm for de novo DNA motif discovery (Pavesi et al., 2004; Zambelli et al., 2014). For genes upregulated in mutants, a matrix resembling a GATA box (Reyes et al., 2004), and a second motif, unrelated to CORE or CCAAT, emerged, suggesting indirect effects on other TFs. On the other hand, three similar, but not identical, matrices emerged in downregulated genes for each data set (Figures 3; Supplemental Figure 6). CCACACA was found in the co-sail and co-sail by nf-yb2 nf-yb3 intersection, which is similar to the “morning element” (Harmer and Kay, 2005; Michael et al., 2008) that is important for circadian clock regulation (Liu et al., 2016). The CCACATA sequence, differing by 1 bp, was found in the nf-yb2 nf-yb3 cohort. Note that in the NF-CO mutagenesis experiments of Figure 2, a C or a T at this position are essentially equivalent. Finally, the CCACGTG motif, resembling a G-box, and previously described in TOC1 and PRR chromatin immunoprecipitation (ChIP)-seq experiments and in promoters of genes upregulated after TOC1 overexpression (Gendron et al., 2012; Liu et al., 2016), was recovered from the nf-yc3 nf-yc4 nf-yc9 cohort and was the most enriched element in intersections involving this cohort. Collectively, these elements all contain or closely resemble the CCACA core motif identified by in vitro EMSAs as optimal for NF-CO. In summary, in vivo RNA-seq analysis was consistent with in vitro biochemical data, both identifying CCACA as the NF-CO matrix.
Figure 3.
Identification of CO and HFD Matrices by RNA-Seq Analysis.
RNA-seq analysis of differentially expressed genes in the co-sail, nf-yb2 nf-yb3, and nf-yc3 nf-yc4 nf-yc9 lines compared with wild-type Arabidopsis. The motifs enriched in the associated promoters are shown for each intersection of coregulated genes.
(A) and (B) Venn diagram showing numbers of differentially expressed genes identified in comparisons between tested lines and wild-type plants, and overlaps between differentially expressed gene sets. For each gene set, an alphanumeric code signifies the most highly enriched motif identified.
(C) Sequence logos describing the motifs identified from analyses of promoters (from −1000 to TSS/ATG) of DE gene sets (Supplemental Figure 6).
Analysis of NF-CO-Regulated Genes
Next, we further analyzed the NF-CO-regulated genes for circadian expression with the Phaser tool, which uses a database derived from microarray analyses of circadian and diurnal gene expression patterns (Mockler et al., 2007). For a long-day light regime, genes upregulated in co-sail and nf-yb2 nf-yb3 were highly significantly enriched for predawn expression (hours 21 to 22 of the long-day cycle). Alternatively, downregulation of expression in these mutants was correlated with morning expressed genes (hours 2–5), as well as distinct peaks for genes normally expressed in the 10th and 15 to 16th hours (Supplemental Figure 7 and Supplemental Data Set 2A). Indeed, a number of genes believed to be involved in circadian regulation (including GIGANTEA, CIRCADIAN CLOCK ASSOCIATED1, LATE ELONGATED HYPOCOTYL, ELONGATED HYPOCOTYL5, CONSTANS-LIKE2, and several pseudo-response regulator [PRR] genes), as well as diurnal markers such as early light-inducible protein genes ELIP1 and ELIP2, showed significant differential expression, often with a relatively large fold change of expression levels between mutants and the wild type (Supplemental Figure 8A). For all mutants tested, and for all intersections between their downregulated genes, multiple Gene Ontology terms related to plastid locations and functions were highly significantly enriched (Supplemental Data Set 2B). For upregulated genes, terms related to the plasma membrane and cell wall, as well as response to carbohydrate stimulus, were consistently overrepresented. No particular enrichment of motifs corresponding to the previously identified CCACA matrices was noted in promoters of plastid genes. Taken together, these observations are consistent with fundamental alterations to the regulation of circadian processes in co-sail and the nf-yb mutants tested, at least under the light conditions employed in our experiments.
We also specifically looked at the expression of paralogs of At-NF-Y subunits and CCT genes. Among the At-NF-Y paralogs, expression of At-NF-YA4/2/5/6/7/9 was detected. CO mRNA itself is expressed at low levels in the wild type, unlike many of the CO-like (COL) and PRR genes, which are more abundantly expressed than At-NF-YA genes (Supplemental Figure 8B and Supplemental Data Set 2C). Few significant changes in expression were observed, with NF-YA4 showing downregulation in co-sail and the nf-yb mutants, At-NF-YB8 and At-NF-YB10 upregulated in nf-yb2 nf-yb3 and nf-yc3 nf-yc4 nf-yc9 mutants, while At-NF-YB7 was downregulated in all mutants tested (Supplemental Figure 8B and Supplemental Data Set 2C). Among the downregulated genes in co-sail, we find many members of the B-box protein (BBX) family (Khanna et al., 2009), mostly those that do not contain a CCT domain. These include BBX19 (At4g38960), whose protein product interacts with CO and whose reduced expression accelerates flowering (Wang et al., 2014), and BBX30 (At4g15248) and BBX31 (At3g21890), which also interact with the CO protein and whose overexpression delays flowering (Graeff et al., 2016), as well as BBX32, which interacts with COL3 to regulate FT (Tripathi et al., 2017).
At-NF-YB2-NF-YB3 Are Essential for CO Recruitment onto the FT Promoter
To verify the importance of HFD subunits for CO association to DNA in vivo, we performed ChIP analysis on Col-0 and nf-yb2 nf-yb3 plants transgenic for CO-YFP/HA under the control of the CaMV 35S promoter (p35S:CO-YFP/HA). We used an HA antibody to assess the overexpression of the transgenes in the two genetic backgrounds (Supplemental Figure 9) and then for ChIP analysis. The immunoprecipitated DNAs were checked by qPCR with three amplicons within the FT locus: at −5.3 kb, a region of the distal promoter which contains the CCAAT box, but no known CORE elements (negative control); at −0.3 kb, a region of the proximal promoter where CORE2 is located and CO is known to bind (Song et al., 2012; Cao et al., 2014); and at +2.0 kb, corresponding to Exon 4 (negative control). Figure 4 shows that CO binding was detected only in the core promoter in wild-type plants, as expected, but not in the nf-yb2 nf-yb3 mutants. Note that the −0.3-kb region contains CORE2, CORE1, and two additional essential CCACA motifs, termed P1/P2 (Adrian et al., 2010), all within 100 bp. With the current precision of the ChIP procedure, it is not possible to discriminate the exact binding site(s) bound by CO, but previous evidence suggests that CO interacts with several CCACA-containing sites in the proximal promoter (Adrian et al., 2010; Cao et al., 2014). Regardless of the specific CCACA(s) bound, these data show that At-NF-YB2 and At-NF-YB3 are required for CO binding at the FT proximal promoter.
Figure 4.
CO Binds the FT Promoter in an At-NF-YB2-NF-YB3-Dependent Manner.
ChIP was performed on Col-0 (parental) and nf-yb2 nf-yb3 plants transgenic for p35S:CO-YFP/HA. Enrichment of the selected segments -CCAAT/-5.3kb, core promoter, exon 4/+2.0kb- were evaluated by qPCR with appropriate amplicons. Error bars indicate se with five biological replicates. In each replicate, three technical replicates were performed. Statistical significance was obtained using Bio-Rad CFX Manager Version 3.0; in each case, the comparison is between the nonimmune control (NIC) and the immunoprecipitation (IP). ***P < 0.001.
Similar HFD Structural Elements Provide Association with NF-YA and CO, and Are Important for the Timing of Flowering
In canonical NF-Y complexes, NF-YA cannot bind to single NF-YB or NF-YC subunits (Kim et al., 1996; Sinha et al., 1996). This result was later rationalized by knowledge of the quaternary NF-Y/CCAAT 3D structure, showing instead that NF-YA binds to a composite surface formed by the α2 helix of NF-YB and the α1/αC helices of NF-YC (Nardini et al., 2013; Huber et al., 2012). In keeping with these data, in a systematic study of subunit interactions using yeast two-hybrid (Y2H) assays, direct interactions between At-NF-YAs and the single HFD subunits were generally not observed (Hackenberg et al., 2012). The original Y2H screenings with CO identified interactions with At-NF-YB or At-NF-YC subunits (Wenkel et al., 2006; Ben-Naim et al., 2006), a result further confirmed by other Y2H studies (Supplemental Figure 1). Because the A1 trimerization domain of NF-YA does not superimpose perfectly with the corresponding area in the CCT domain (Petroni et al., 2012), we wondered whether the CO interaction regions of the HFD heterodimers were equivalent to the ones contacted by At-NF-YA. To evaluate this, we designed specific mutations in At-NF-YC9 and At-NF-YB2 known to affect NF-Y trimer formation and assayed them in vivo and in vitro.
For in vivo testing of At-NF-YC9, we focused on amino acids Phe-151 and Val-153 in the αC helix, mutating these residues to Arg and Lys. These same mutations were originally described for mammalian NF-Y where they eliminated formation of the heterotrimer and DNA binding, but had no impact on formation of the HFD heterodimer (Kim et al., 1996). Note that within the HFD, At-NF-YC9 is identical to At-NF-YC3 and nearly identical to At-NF-YC4 (Figure 5A), and all three proteins have known functional overlap in several processes (Kumimoto et al., 2010; Myers et al., 2016); hence, any data obtained with mutants of the At-NF-YC9 HFD are likely valid for all three proteins. We transformed the nf-yc3 nf-yc4 nf-yc9 triple mutant with At-NF-YC9 under the control of its own promoter, either in the wild-type or F151R/V153K mutant configuration. As shown in Figure 5B, compared with the wild type, triple mutant plants had significantly delayed flowering, as previously reported (Kumimoto et al., 2010). This delay was almost completely reverted by wild-type At-NF-YC9, but not by the At-NF-YC9-F151R/V153K mutant, despite generally robust expression patterns of the mutant transgene (Figure 5C). To check whether the double mutation impaired formation of subunits, we performed Y2H with wild-type and mutant At-NF-YC9. Figure 5D shows that the wild-type At-NF-YC9 interacts with all tested partners (At-NF-YB2, At-NF-YA1, At-NF-YA2, and CO), whereas the mutant only interacted with the HFD partner and was unable to contact At-NF-YAs or CO. As a further control, we used a mutant in a conserved position in the α2 helix (Ile-89) previously shown to affect HFD heterodimerization in mammalian NF-Y subunits (Kim et al., 1996); indeed, the I89D mutant lost At-NF-YB2 interactions in Y2H assays, which agreed with predictions and supported the specificity of the Y2H data.
Figure 5.
Properties of At-NF-YC9 Trimerization Mutants.
(A) Alignment of At-NF-YCs. Multiple sequence alignment of NF-YC protein core domains. Multiple sequence alignment was computed using ClustalW in Geneious version 7.0. Amino acid residue positions of the HFD are indicated for the At-NF-YC9 and human proteins. Amino acids making physical contact with NF-YA are annotated by an asterisk (Nardini et al., 2013). Arrows mark the position of mutated residues, with the closed arrow indicating the conserved phenylalanine required for interaction with NF-YA in mammals, which was mutated in NF-YC9F151R V153K and in the recombinant At-NF-YC9F151R HFD protein. At, Arabidopsis thaliana; Hs, Homo sapiens.
(B) In vivo analysis of timing of flowering. T1 generation flowering time analysis of pNF-YC9:NF-YC9F151R V153K in the nf-yc triple (nf-yc3 nf-yc4 nf-yc9) mutant background. Asterisks represent significant differences derived from one-way ANOVA (P < 0.05) followed by Dunnett’s multiple comparison post hoc tests against the nf-yc triple mutant.
(C) Expression of At-NF-YC9 transgenes in transgenic plants. Protein expression in the plant lines used for the flowering time analysis was analyzed by immunoblot with antibodies directed to a translationally fused HA-epitope (top panel). Protein loading and transfer was assessed by Ponceau staining (bottom panel).
(D) Y2H assays of At-NF-YC9. Full-length NF-YC9 and mutant variants tested using Y2H against empty vector (EV) control, NF-YB2, NF-YA1, NF-YA2, and CO. Note that NF-YC9 has slight autoactivation.
(E) EMSAs on CORE2 and CCAAT of wild-type and mutant At-NF-YC9. Trimerization and DNA binding of the At-NF-YC9F151R mutant (YC9F151R; lanes 7–11) or the wild type (YC9; lanes 2–6) containing HFD dimer (60 nM) was assessed the with the CORE2 probe (lanes 1–14), by addition of the CO subunit at increasing concentrations (90, 180, 270, and 360 nM; lanes 3–6 and 8–11). At-NF-YA2 (YA2) trimerization with the wild-type or mutant dimers (lanes 17–20 and 22–25, respectively) was assessed with the CCAAT probe (lanes 15–28). As negative controls, CO or At-NF-YA2 was added alone to the reaction with the respective probes (lanes 12 and 26). At-NF-YC9 trimer specificity was also assessed by addition of the CO or At-NF-YA2 containing trimers to the CCAAT or CORE2 probe, respectively (lanes 28 and 14). DNA binding of At-NF-YC3 (YC3) containing trimers was also used as internal control (lanes 13 and 27). In lanes 2, 7, 16, and 21, wild-type or mutant HFD dimers were incubated alone with the probe. NF-CO and NF-Y/DNA complexes are indicated by labeled arrowheads. Lanes 1 and 15: CORE2 and CCAAT probes without protein additions. fp, free probe.
Next, we switched to in vitro EMSAs with recombinant proteins. In this case, we generated the single F151R mutation in At-NF-YC9, since mutation of the (nonconserved) Ile residue corresponding to Val-153 (Figure 5A) was previously shown not to impair NF-YA interaction (Romier et al., 2003). This result was later rationalized by the central role played by the perfectly conserved phenylalanine in the nucleation of the hydrophobic core driving the correct positioning of the NF-YC αC helix. The acidic NF-YC αC provides crucial A1 contacts and is further stabilized by NF-YA interactions with main chain atoms of the same Phe residue (Nardini et al., 2013). Coexpression and purification of both wild-type At-NF-YC9 and the F151R mutant with At-NF-YB2 was equally efficient, as expected, indicating similar heterodimerization capacities (Supplemental Figure 2). The purified HFDs were then incubated with either CO or At-NF-YA2 and tested for interaction with the CORE2 and CCAAT probes. As expected, wild-type At-NF-YC9 was able to form efficient DNA binding complexes with either CO or At-NF-YA2 on their respective DNA targets, while the At-NF-YC9F151R mutation led to very inefficient binding to either probe (Figure 5E). Altogether, these data indicate that an At-NF-YC mutation that interfered with heterotrimer formation and CCAAT binding for the canonical NF-Y complex had essentially the same effects on the NF-CO complex at its CORE site.
On the At-NF-YB side, we employed a similar strategy, by targeting the conserved Glu-65 in the α2 helix of At-NF-YB2, corresponding to Glu-90 of mammalian NF-YB. In mammalian NF-Y, this acidic residue provides contacts with two conserved arginines (Arg-249 and Arg-253) of NF-YA helix A1 (Nardini et al., 2013). Similarly to the NF-YC mutagenesis described above, detailed biochemical analyses have previously shown that the mammalian NF-YB E90R mutation does not alter HFD dimerization, but impairs trimerization and DNA binding (Sinha et al., 1996). As expected, both At-NF-YB2 and the At-NF-YB2E65R mutant efficiently heterodimerized with At-NF-YC3 (Supplemental Figure 2), but the E65R mutant lost functional NF-Y binding to CCAAT in EMSAs (Figure 6). Replacing At-NF-YA2 with CO and testing binding to the CORE2 probe gave a similar result: NF-CO binding was reduced, albeit not completely eliminated (Figure 6). Note that this mutant, unlike wild-type At-NF-YB2, could not rescue a late flowering nf-yb2 nf-yb3 mutant (Siriwardana et al., 2016), paralleling the At-NF-YC mutation shown above. In summary, the same conserved residues of At-NF-YC9 αC and At-NF-YB2 α2 are important for trimerization with At-NF-YA and CO, for binding of the NF-Y and NF-CO trimers to their respective DNA sites, and for function in vivo.
Figure 6.
Properties of At-NF-YB2 Trimerization Mutant.
Trimerization and DNA binding of the E65R mutant (YB2E65R; lanes 7–11) or wild-type (lanes 2–6) At-NF-YB2 containing HFD dimer (60 nM) was assessed with the CORE2 probe (lanes 1–14), by addition of the CO subunit at increasing concentrations (90, 180, 270, and 360 nM; lanes 3–6 and 8–11). At-NF-YA2 (YA2) trimerization with the wild-type or mutant dimers (lanes 17–20 and 22–25, respectively) was assessed with the CCAAT probe (lanes 15–28). As negative controls, CO or At-NF-YA2 was added alone to the reaction with the respective probes (lanes 12 and 26). Trimer specificity was assessed by addition of the CO or At-NF-YA2 containing trimers to the CCAAT or CORE2 probe, respectively (lanes 13, 14, 27, and 28 as indicated). In lanes 2, 7, 16, and 21, wild-type or mutant HFD dimers were incubated alone with the probe. NF-CO and NF-Y/DNA complexes are indicated by labeled arrowheads. Lanes 1 and 15: CORE2 and CCAAT probes without protein additions. fp, free probe.
Mutation of a Single Amino Acid in CO That Is Highly Conserved in Both CCT and NF-YA Families Eliminates NF-CO DNA Binding
Having established that the docking sites on the HFD dimers are similar, we switched to analysis of the properties of the CCT. Several single-residue alterations, mutations, or natural variations in the CCT domain of CO and CO-like proteins were previously reported; importantly, these alterations of CCT family members were often pinpointed in genetic screenings as having functional consequences in flowering timing (Distelfeld et al., 2009a, 2009b). In particular, the Arabidopsis co-7 allele is one of the later flowering co alleles (Robson et al., 2001). This mutant allele results in an arginine-to-glutamine change at position 340 in CO, corresponding to mammalian NF-YA Arg-283, a residue known to be important for DNA binding (Xing et al., 1993; Mantovani et al., 1994) and specifically for CCAAT recognition (Nardini et al., 2013; Huber et al., 2012). We produced and purified the co-7 CCT (Supplemental Figure 2) and assayed it with the At-NF-YB2/NF-YC3 dimer for binding to CORE2 in EMSAs. Once again mimicking the canonical NF-Y/CCAAT interaction, NF-CO bound CORE2 with wild-type CO, but not with the R340Q (co-7) mutant protein, even at high concentrations (Figure 7A). We conclude that Arg-340 of the CO CCT domain, corresponding to the perfectly conserved arginine in mammalian, yeast, and plant NF-YA proteins, is equally important in DNA binding. This strongly suggests that the DNA binding subdomains of CO and NF-YA are structurally and mechanistically analogous and likely explains the molecular mechanism of the late flowering phenotypes of co-7 mutant plants.
Figure 7.
The CO CCT Drives Sequence Specificity of NF-CO.
(A) CO mutation R340Q of co-7 abolishes NF-CO DNA binding. Wild-type or R340Q CO was incubated at increasing concentrations (90, 180, 270, and 360 nM) with the CORE2 probe in the absence (−) (lanes 2–9) or presence (lanes 11–18) of the At-NF-YB2/NF-YC3 HFD dimer (60 nM). In lane 10, the At-NF-YB2/NF-YC3 HFD dimer was incubated alone with the probe. Lane 1: probe alone, without protein additions. fp, free probe.
(B) Schematic representation of selected NF-YA interactions with the C2A3 bp of CCAAT. Highlight of NF-YA A2 helix within the NF-Y/DNA 3D structure (PDB: 4AWL), with interactions of Arg-281, Arg-283 (corresponding to CO Arg-340), and Gly-287 (CO Gly-343) amino acid residues (indicated in single letter code: Arg-281, Arg-283, and Gly-287 respectively) with the G2 and A3 nucleotides. NF-YA protein (cyan) and the sugar-phosphate DNA strand backbones are represented as colored strings, with the CCAAT (I) and complementary (J) strands in red and green, respectively. Orientation of DNA strands is indicated. Selected NF-YA residues and nitrogen bases of the C2A3:G2T3 nucleotides are labeled and displayed in ball and stick model in color matching the main chain color code. Gly-287 main chain and Arg-283 side chain contacts with G2 atoms, and Arg-281 side chain with A3, are indicated by gray lines (Nardini et al., 2013). The NF-YB/NF-YC subunits within the 4AWL structure were omitted for clarity. The image was obtained with Protein Workshop (Moreland et al., 2005).
(C) Amino acid sequence alignment of the C-terminal portion of CO CCT domain with mammalian NF-YA homology region is shown, with the proposed sequence-specific interactions, based on the NF-Y/DNA complex crystal structure (PDB: 4AWL). DNA sequence of the NF-CO and NF-Y respective element is shown at the top and bottom of the alignment, with the indicated orientation of the DNA strands and base-pair positions in the bound elements numbered (see text). Side chain interactions of NF-YA with the CCAAT bases are indicated by full lines (bottom). On top of the alignment, dashed lines represent potential CO residues interactions with the CORE matrix. Conserved and nonconserved residues are highlighted in green and blue, respectively. R340Q in co-7 is indicated on top of the alignment. The closed circle represents hydrophobic base stacking interactions of phenylalanine residue side chains with the CA:GT nucleotides. Bold nucleotides highlight the divergence in sequence specificity of the two complexes.
DISCUSSION
Here, we demonstrated that the master flowering regulator CO interacts physically with NF-Y histone-like subunit dimers to form a novel DNA binding sequence-specific trimeric complex, NF-CO (Figure 8). Mutations within the HFDs and CCT domain indicate that the overall modalities of trimerization and DNA binding are similar to the canonical NF-Y. In vitro experiments have defined a DNA sequence matrix that is specifically recognized by NF-CO and is independently recovered through analyses of promoters of genes downregulated in vivo in a co mutant. Indeed, highly significant overlaps of genes dysregulated in co-sail, nf-yb, and nf-yc mutants, as well as ChIP analysis showing lack of CO binding to the FT promoter in the nf-yb2 nf-yb3 mutant, corroborate conclusions from in vitro experiments. Additionally, the recovery of core motifs related to, but distinct from, the NF-CO binding consensus in subsets of the genes differentially expressed in NF-Y HFD subunit mutants is consistent with the possibility that other CCT proteins also form sequence specific DNA binding complexes with NF-Y components.
Figure 8.
Scheme of NF-Y versus NF-CO Specificity.
Association of CO or NF-YA with NF-YB/NF-YC dimers provides robust and specific recognition of the respective DNA element by the trimeric NF-CO and NF-Y complexes.
NF-CO and NF-Y in Flowering
NF-Y subunits have been implicated in a plethora of physiological plant processes. Specifically, there is well established evidence that different NF-Y HFD subunits are involved in the regulation of timing of flowering in Arabidopsis (Wenkel et al., 2006; Chen et al., 2007; Cai et al., 2007; Kumimoto et al., 2008, 2010; Hackenberg et al., 2012; Hou et al., 2014; Cao et al., 2014), rice (Oryza sativa; Dai et al., 2012; Wei et al., 2010; Yan et al., 2011; Feng et al., 2014; Chen et al., 2014; Kim et al., 2016; Hwang et al., 2016; Goretti et al., 2017), wheat (Triticum aestivum; Li et al., 2011), and tomato (Solanum lycopersicum; Ben-Naim et al., 2006). Wenkel et al. originally observed homology between the CCT domain and the conserved domain of NF-YA, suggesting that CO and NF-YA might both bind DNA with NF-YB/NF-YC HFD dimers (Wenkel et al., 2006). However, the CCT domain N-terminal portion is not perfectly superimposable with the NF-YA A1 helix involved in the interactions with the HFD dimer, yet it does share its highly basic nature. Our data (Figures 5 and 6) suggest that the CCT does contact the same acidic surface patch of the HFD dimer recognized by NF-YA A1. A variant in this basic CCT region of both Heading date 1 (Hd1; the rice CO homolog) and OsPRR37 (another rice CCT protein involved in flowering) also impairs HFD interactions (Goretti et al., 2017), further solidifying the idea that multiple CCTs can form NF-CO complexes.
The notion that CO is an NF-YA equivalent, contacting similar HFD surfaces, implies that it could compete for HFD occupancy, and vice versa, that NF-YA could compete with CO. This is consistent with the finding that overexpressing some NF-YAs can cause late flowering (Wenkel et al., 2006; Li et al., 2011; Leyva-González et al., 2012). In turn, this implies that the interpretation of the phenotypes of plants in which NF-YAs or CO, or other CCT proteins, were ubiquitously overexpressed is likely complex, since a change in the levels of either subunit could alter the stoichiometry and function of NF-Y or NF-CO complexes. While interference with HFD dimers remains distinctly possible even at physiological protein concentrations, as a general mechanism of CCT/NF-YA interplay, we note that the FT gene has two canonical sites of regulation for the two trimers: At least one NF-CO site in the core promoter and a canonical CCAAT box in its enhancer, both functionally essential for photoperiod-dependent flowering (Adrian et al., 2010; Tiwari et al., 2010; Cao et al., 2014). Thus, FT appears to be regulated by both NF-Y and NF-CO, sharing common HFD subunits.
CCAAT versus CORE
The sequence targeted by NF-Y [RRCCAAT(C/G)(A/G)] has long been known, thanks to numerous biochemical and genomic studies performed in mammals (Dolfini et al., 2009; Dolfini and Mantovani, 2013). By examining the NF-CO matrices (Figures 2C and 3), one can notice a clear similarity to the NF-Y CCAAT matrix, RRCCAAT(C/G)(A/G), with a deviation of two nucleotides (underlined). Similarity is present at the 5′ end, where the five nucleotides RRCCA are identical, and at the 3′ ends. Given the stunning evolutionary conservation between animal and plant NF-YA proteins, specifically in the sequence-specific DNA binding subdomain, we take for granted that plant NF-Y also binds to sequences centered on the CCAAT pentanucleotide. This was suggested by a previous study showing that the pentanucleotide, but not the mammalian matrix with the flanking nucleotides, was enriched in plant promoters (Siefers et al., 2009). The fundamental differences of the NF-Y and NF-CO matrices are indeed within the central pentanucleotide (CCAAT versus CCACA, respectively). At position 4, a C is crucial for NF-CO, and never an A, as required by NF-Y; the A dominant for NF-CO at position 5 is detrimental in vitro and essentially never found in sites in vivo for NF-Y, at least in mammals. Thus, these two residues are clearly discriminative and divergent for the two complexes, and the respective sequence specificity is expected to drive binding to distinct target sites. In vivo, this was shown on FT (Figure 4; Cao et al., 2014), and in more general terms, it is documented here by the RNA-seq analysis, since mutants of the NF-CO subunits have clearly altered the expression of genes enriched for CORE elements in their promoters.
Schmid et al. (2003) previously showed that, in the apical meristem, co and ft mutants cause almost identical sets of genes to be dysregulated in LD conditions, while Wigge et al. (2005) concluded that the major contribution of CO to floral transition is mediated by its activation of FT. However, this later study, employing leaf tissue, identified over 400 genes that showed CO-dependent dysregulation on transition from short to long day. Interestingly, reanalysis of this expression data (ArrayExpress experiment E-TABM-21) using Limma yielded a significant overlap between these genes and those showing dysregulation in the co-sail mutant in continuous light conditions (Supplemental Data Set 3).
The At-NF-YBs and At-NF-YCs studied in this experiment seem to have a shared impact on CCACGTG (Figure 3), possibly the PRR site (Liu et al., 2016), and CCACA(C/T), possibly the CO and CO-like site. For the time being, it is hard to work out a coherent model of direct and indirect targets because of the overlap between motifs, the combinatorial complexity of the complexes, and the fact that we do not know if a specific combination of HFDs in a CO complex can subtly alter the preferred binding site. We see a strong overrepresentation of plastid genes within the group of genes misregulated in all three mutants analyzed and the best motifs strongly resemble elements known to be involved in diurnal regulation. Genes that are up- or downregulated in co-sail, as well as in HFD mutants after 7 d of continuous light, are strongly enriched for distinct expression times under physiological conditions. Taken together with the similarities in the gene sets observed here and previously (Wigge et al., 2005), these observations suggest that NF-CO has physiological roles that go well beyond induction of flowering and might tie in with fine-tuning of plastid gene expression timing, potentially via PRRs and/or CO-like-containing complexes. Furthermore, the extensive dysregulation of B-box genes might be consistent with a more widespread control on FT- and NF-CO-regulated genes, given the significant level of mutual regulation (Shim et al., 2017).
We were surprised by the overall lack of CCAAT boxes in profiles of the HFD subunit mutants. Note that the presence of the CCAAT pentanucleotide in a partial set of Arabidopsis promoters was detailed before (Siefers et al., 2009). We can offer several nonmutually exclusive explanations. (1) To observe the emergence of CCAAT-dependent promoters, one should analyze NF-YA mutants, as these are the subunits imparting sequence specificity to the trimer. (2) CORE outnumbers CCAAT because CCT encoding genes outnumber NF-YAs by a factor of 4 to 1. (3) CCT genes are overall more abundantly expressed compared with At-NF-YAs; thus, NF-CO complexes could simply be more abundant than NF-Ys, at least in the tissue and under the experimental conditions employed here.
Lessons about NF-CO from the NF-Y/CCAAT Structure
Knowledge of the molecular details of the quaternary 3D NF-Y/CCAAT structures (Nardini et al., 2013; Huber et al., 2012) helps rationalize the NF-CO/DNA interactions. The HFDs are clearly crucial for stable complex formation, making >25 non-sequence-specific contacts with DNA, spanning at least 25 and most likely 30 nucleotides. Indeed, the stabilization of NF-CO DNA binding by HFDs is quite dramatic (Figures 1 and 5 to 7; Supplemental Figure 3). DNA binding of recombinant CO and TOC1 was previously reported, however with very high protein concentrations (Tiwari et al., 2010; Gendron et al., 2012); indeed, we do see this effect in our assays. Intriguingly, this low affinity binding might be consistent with very high overexpression of CO being able to partially rescue late flowering in the nf-yb2 nf-yb3 mutant (Tiwari et al., 2010). However, we provide biochemical evidence that the trimer is a much more efficient DNA binding entity in vitro, and a physiological one in vivo, as the matrices of the HFD mutants resemble those of co. The HFD importance could be linked to their histone-like nature, as they might play the known “pioneer” role of NF-Y in penetration of chromatin territories devoid of positive histone marks (Fleming et al., 2013; Oldfield et al., 2014; Sherwood et al., 2014; Lu et al., 2016).
Structurally, there are two features differentiating CO from NF-YA: (1) the shorter CO A1-A2 linker between the HFD association and DNA binding subdomains, which might severely constrain the flexibility of the complex; (2) the absence in CO of two glycines within the crucial (R)GxGGRF loop of NF-YA (amino acids 283–289), which is (R)VNGRF in CO (amino acids 340–345). Overall, 14 NF-YA amino acids are involved in DNA binding; seven make non-sequence-specific contacts, of which only NF-YA Ser-273 and Arg-288 are conserved (Nardini et al., 2013). Interestingly, of the seven sequence-selective residues, five are conserved in CO (Figure 7C), implying selective pressure to maintain similar, but not identical specificities. Notably, of the shared base pairs in CCAATC and CCACAC (underlined), C2, A3, and C6 are selectively bound by three NF-YA arginines conserved in CO (and in all other CCTs): Arg-283 (CO Arg-340), Arg-281 (CO Arg-338), Arg-274 (CO Arg-331), respectively, with C2 also being contacted by Gly-287 (CO Gly-343). The lack of DNA binding in the CO R340Q mutant protein (co-7) is thus consistent with this arginine providing the same specificity as in NF-YA (Figures 7A to 7C). We are thus tempted to conclude that the two nonconserved residues, corresponding to NF-YA Gly-286 and His-277, dictate the divergence in sequence specificity of NF-CO with respect to NF-YA. Gly-286 is an asparagine in CO, or arginine/lysine in other CCT proteins, which are all bulkier residues in an area where the small glycines of NF-YA allow main chain insertion in the minor groove space (Figure 7B). The CCAAT A4, a C in CORE, is contacted by NF-YA His-277: This is Tyr-334 in CO, a tyrosine, asparagine, or arginine in CO-likes, and a leucine in all TOC1/PRRs (Petroni et al., 2012). We hypothesize that these changes might command yet different selectivity, focused on this specific nucleotide of the pentamer.
Interpretation of Genetic Experiments
The inclusion of NF-CO among sequence-specific complexes containing HFD subunits changes the interpretation of genetic experiments performed on HFD subunits. Since the discovery of the expansion of NF-YB and NF-YC genes in plants, numerous HFD mutants or overexpressors, particularly for NF-YB (also termed Hap3, DTH8, and Ghd8 in rice), were identified and characterized in different species. The obvious molecular interpretation relied on the notion that NF-Y would be crippled or changed in its trimeric assembly, and activity of targeted CCAAT promoters (largely unknown at the moment) altered. The new data indicate that NF-CO would similarly be altered in these HFD mutants or overexpressors. The second important consequence stemming from our data concerns the phenotype of CO mutants and natural variants. Significant evidence already exists that mutations in the CCT domain are functionally important, primarily in the A2 helix and (R)VNGRF motif.
Based on our data, all these variants are now predicted to be loss-of-function (or hypomorphic) DNA binding mutants. For example, VRN2 from wheat (ZCCT1 and 2) blocks flowering in long days until after vernalization (Yan et al., 2004), potentially regulating FT (VRN3 in wheat). Furthermore, in vitro and in planta coimmunoprecipitations have shown that ZCCT proteins interact with selected HFDs and compete with NF-YA for binding (Li et al., 2011). The functionally disruptive, natural variants of VRN2 target either the same arginine as the Arabidopsis co-7 allele (ZCCT1) or the arginine equivalent to CO Arg344 (ZCCT2). In both cases, loss of the arginine eliminates the active roles of ZCCT proteins as repressors of flowering time (Distelfeld et al., 2009a, 2009b). Likewise, the barley PRR7 mutant allele ppd-H1 involves the glycine equivalent to Gly-343 in CO, with a substitution to (a bulky) tryptophan residue (Turner et al., 2005), and the co-9 allele of Arabidopsis (a valine substitution of the perfectly conserved Ala-335; Wenkel et al., 2006) is homologous to the toc1-1 allele of TIMING OF CAB1. Finally, one of the PRR37 natural rice variants, contributing to adaptation of cultivation at different latitudes, harbors a missense L710P that corresponds to the above-mentioned NF-YA crucial residue His-277. A second natural variant is a frameshift mutation at Gln-705 (Koo et al., 2013), which would lose the C-terminal residues of the CCT: Both of these mutations are predicted to be loss of function. Perhaps most intriguing, a third identified variant is on Tyr-704, which becomes a histidine (equivalent to His-271). A histidine at this position is never observed in CCTs but is absolutely conserved in all NF-YAs and required for CCAAT binding (Xing et al., 1993; Mantovani et al., 1994). Might this alter DNA specificity of this variant in a CCAAT-directed way? However, alteration of DNA binding is not the only consequence of mutations in CCT proteins. A rice Hd1 variant with genetic adaptation to flowering in long-day conditions (Mediterranean cultivar) shows deletion of a lysine in the subunit interaction (A1 helix) portion. This protein is unable to associate with OsNF-YB/NF-YC dimers and to bind a conserved CORE element in the Hd3a (FT) promoter (Goretti et al., 2017).
In summary, NF-CO represents a DNA binding complex that includes CO, that may or may not require NF-Y function in genomic contexts, and there is every reason to believe the paradigm is generalizable to other CCT proteins. Our findings indicate a broad change of perspectives in CCT associations. Even more than with NF-Ys, the potential combinatorial diversity of NF-CO complexes is enormous. Searches at The Arabidopsis Information Resource suggest that there are 40 CCT proteins (17 CO-Like with BBX domains) (Khanna et al., 2009). Our data thus represent a considerable broadening of our understanding of combinatorial possibilities of NF-Y and NF-CO complexes in plants. Considering the diverse (and still mostly unknown) roles for CCT proteins, the potential for fine tuning of motif binding depending on specific HFD pairings and trimerization is considerable, and sorting through this complexity represents an important challenge. The biochemical assays shown here open the possibility to molecularly characterize all NF-Y/NF-CO trimeric complexes generated by combinatorial associations. They also set the stage for structural studies to understand the fascinating details of CO/CORE recognition.
METHODS
Protein Production and Purification
The cDNA encoding the CCT domain of CO (amino acids 290–352), with the addition of a 5′ ATG, was obtained by PCR amplification; the cDNA encoding CO CCT amino acids 290 to 352 with the R340Q mutation (Robson et al., 2001), At-NF-YA2 (amino acids 134–207), and At-NF-YA6 (amino acids 170–237) was obtained by gene synthesis (Eurofins Genomics) and cloned into pmcnEA/tH (Diebold et al., 2011) by restriction-end ligation to obtain C-terminal 6His-tag fusions (Siriwardana et al., 2016). At-NF-YB2 mutant cDNA, encoding for amino acids 24 to 116 with residue Glu-65 mutated to Arg was obtained by gene synthesis and subcloned in pET15b to obtain the N-terminal 6His-tag fusion (Siriwardana et al., 2016). At-NF-YC9 cDNA, encoding amino acids 62 to 158 with a 5′ ATG, a 3′ stop codon, and mutant At-NF-YC9 with residue Phe-151 mutated to Arg (NF-YC9F151R) were obtained by gene synthesis and cloned in pmcnYC (Diebold et al., 2011). All constructs were verified by sequencing. 6His-NF-YB2 or 6His-NF-YB2E65R/NF-YC3 soluble HFD dimers were produced by coexpression in Escherichia coli BL21(DE3) and purified by ion metal affinity chromatography as described (Calvenzani et al., 2012). CO-6His and co7-6His were expressed in BL21(DE3)Rosetta by IPTG induction (0.4 mM IPTG for 4 h at 25°C) and purified by ion metal affinity chromatography (HisSelect; Sigma-Aldrich) in buffer A (10 mM Tris-HCl, pH 8.0, 400 mM NaCl, 2 mM MgCl2, and 5 mM imidazole). NF-YA2-6His and NF-YA6-6His were produced in BL21(DE3). Purified proteins were eluted in buffer A containing 100 mM imidazole and dialyzed against buffer B (10 mM Tris-HCl, pH 8.0, 400 mM NaCl, 2 mM DTT, and 10% glycerol).
EMSAs
EMSAs were performed essentially as previously described (Calvenzani et al., 2012; Cao et al., 2014). Heterotrimer formation and DNA binding of wild-type or mutant CO (or NF-YAs) was assessed in the presence of wild-type or mutant NF-YB2/NF-YC3 dimers using Cy5-labeled FT CORE2 (Cy5-AAGAAAAAGATTGTGGTTATGATTTCACCGA) or CCAAT probes (Cao et al., 2014) (Eurofins Genomics). DNA binding reactions (20 nM probe, 12 mM Tris-HCl, pH 8, 50 mM KCl, 62.5 mM NaCl, 0.5 mM EDTA, 5 mM MgCl2, 2.5 mM DTT, 0.2 mg/mL BSA, 5% glycerol, and 6.25 ng/μL poly dA-dT) were added with wild-type or mutant NF-YB2/NF-YC3 HFD dimers (60 nM), in the presence of increasing amounts of the indicated CO or NF-YA proteins. Proteins were premixed in buffer B containing 0.1 mg/mL BSA, then added to DNA binding mixes. After 30 min incubation at 30°C, binding reactions were loaded on 6% polyacrylamide gels and separated by electrophoresis in 0.25× TBE at 4°C. For competition assays, after 10 min incubation at 30°C, binding reactions (containing 60 nM At-NF-YB2/NF-YC3 and 120 nM CO) were supplemented with increasing amounts of indicated unlabeled oligonucleotide competitors or TE buffer and incubated for an additional 45 min at 30°C, then loaded on 6% polyacrylamide, or 2.3% agarose gels in 0.25× TBE for electrophoresis. Fluorescence gel images were obtained and analyzed with a ChemiDoc MP system, and bound DNA complexes were quantified with ImageLab software (Bio-Rad).
Quantification of competition efficiency by mutant CORE2 oligos was performed as follows: Percentage of bound probe was quantified in each lane and plotted versus the competitor concentration (expressed as ratio of the unlabeled versus total oligo concentration). For each oligo, the competitor efficiency represents the slope of the regression line through the competition data points versus the slope of the wild-type oligo competition performed in the same experiment.
RNA-Seq and Bioinformatics Analysis
Seedlings were grown for 7 d on B5 media in continuous white light with standard, 32-W linear florescent tubes (GE product number 26668) producing a light intensity of ∼150 μE. Total RNA was isolated using the E.Z.N.A. Plant RNA Kit from Omega Biotek. To ensure low levels of contaminating rRNA, two rounds of poly(A) mRNA purification were performed using the µMACS mRNA Isolation Kit (Miltenyi Biotech). Indexed RNA-seq libraries were prepared from 100 ng of poly(A) RNA starting material using the NEXTflex Illumina qRNA-Seq Library Prep Kit (Bioo Scientific; catalog no. 5130). Sequencing of 150-bp paired-end reads was performed on an Illumina HiSeq 2500 in rapid output mode at the Texas A&M Agrilife Research Facility (College Station, TX). Sample demultiplexing was performed using CASAVA software v1.8.2, and bcl2fastq was performed using conversion software v1.8.4. Reads were mapped on the reference Arabidopsis thaliana transcriptome (TAIR, version 10) using the bowtie2 program (Langmead and Salzberg, 2012). Estimation of gene expression levels was performed using RSEM (Li and Dewey, 2011). Differential expression analysis was performed applying the latest versions of DESeq2 (Love et al., 2014) and Limma (Ritchie et al., 2015) to RSEM estimated reads counts. Only genes showing a false discovery rate lower than 0.05 according to both tools were considered differentially expressed.
De novo motif discovery was performed with Weeder 2.0 using the default parameters (Pavesi et al., 2004; Zambelli et al., 2014). PScan (Zambelli et al., 2009) was used to generate P values for the enrichment of motif PSSMs generated by Weeder, scanning the same 1-kb intervals upstream of TAIR v10 translation start sites. Analyses of phased gene expression were performed using the Phaser tool associated with the DIURNAL database (Mockler et al., 2007), and heat maps were prepared using the heatmap.2 function from the gplots package for R. Gene Ontology enrichments were estimated using DAVID (Dennis et al., 2003).
ChIP
ChIP experiments were performed according to previous publications (Haring et al., 2007; Cao et al., 2014; Yamaguchi et al., 2014; Pchelintsev et al., 2016) with minor modifications. Briefly, we initially harvested 1.5 to 2 g of 10-d-old, long-day-grown, transgenic (p35S:CO-YFP/HA) seedlings at 14 h after lights on. The p35S:CO-YFP/HA in nf-yb2 nf-yb3 line was generated by crossing nf-yb2 nf-yb3 to a stable, single insertion p35S:CO-YFP/HA line in Col-0 and selecting F3 individuals of the appropriate genotype. Both lines showed accumulation of transgenic, epitope-tagged CO (Supplemental Figure 9). These whole seedlings were then ground to a fine powder in liquid nitrogen. The powder was immediately transferred into 23.5 mL of nuclear isolation buffer (10 mM HEPES, pH 7.6, 400 mM sucrose, 5 mM KCl, 5 mM MgCl2, 5 mM EDTA, 5 mM 2-mercaptoethanol, 1% Triton X-100, 0.4 mM PMSF, 1× protease inhibitor cocktail, and 50 µM MG132) and incubated for 10 min at 4°C. To initiate cross-linking of chromatin complexes, fresh, methanol-free formaldehyde (1.56 mL; catalog no. 28906; Pierce) was added to the above solution and incubated at room temperature (∼22°C) for 10 min. Next, this solution was incubated with 2 M glycine (2 mL) for 5 min to stop the cross-linking reaction. The lysate was then filtered through two layers of Miracloth (catalog no. 475855-1R; Calbiochem) and nuclei were pelleted at 2800g (4000 rpm with 15-cm-diameter rotor) for 10 min at 4°C. Chromatin shearing was then performed using a Bioruptor UCD300 (low power, 12 cycle of 24 s on, 24 s off; Diagenode). Immunoprecipitations were performed using µMACS anti-HA and anti-GFP microbeads, in combination, to improve immunoprecipitation efficiency. For nonimmune controls, the exact same procedure was followed, minus the addition of Miltenyi beads. The immunoprecipitation procedure follows the ChIP protocol described by Miltenyi Biotec. qPCR was performed on a Bio-Rad CFX Connect real-time system with Maxima SYBR Green/ROX qPCR Master Mix (catalog no. K0221; Thermo Fisher Scientific). The qPCR profile was 10 min at 95°C, 45 cycles of 10 s at 95°C, 30 s at 60°C, and 30 s at 72°C, followed by the default dissociation step to generate a melting curve. Primers are listed in Supplemental Table 1. ChIP efficiency was calculated as percentage of input. Statistical analysis and comparisons between samples was performed in the Bio-Rad CFX Manager Software through use of the 2(−ΔΔCT) method.
Immunoblot Analysis
Soluble and nuclear protein fractions were isolated from 10-d-old, long-day-grown seedlings by grinding in sucrose buffer (20 mM Tris, pH 8.0, 330 mM sucrose, 1 mM EDTA, pH 8.0, 5 mM DTT, 1× Protease inhibitor cocktail, and 50 μM MG132), followed by two rounds of low speed centrifugation (1000g for 5 min each) to discard large plant debris. The cleared solution was separated into soluble and nucleus-containing fractions by high-speed centrifugation (20,000g for 30 min). A standard 8% SDS-PAGE gel was loaded with 30 μg total protein for each soluble fraction resuspended, lysed nuclei from the equivalent of 50 mg starting material (∼3.3× concentrated relative to the soluble fraction in cell equivalents). Proteins were transferred to standard PVDF membranes, and the presence of CO-YFP/HA was probed with high affinity anti-HA primary antibodies (catalog no. 11 867 423 001; Roche) and goat, anti-rat, HRP-conjugated secondary antibodies (catalog no. SC-2032; Santa Cruz Biotechnology). A Bio-Rad ChemiDoc XRS imaging system was used for visualizing the protein blot after incubations with ECL plus reagent (catalog no. RPN2132; GE Healthcare).
Cloning
The mutations were made by PCR using appropriate mutagenic primer sequences. Each construct was amplified from Pfu Ultra II (Invitrogen; catalog no.600670) and cloned into the Gateway vector pENTR/D-TOPO (Invitrogen; catalog no.45-0218). All resulting clones were sequenced and, with the exception of the introduced mutations, were identical to the sequences at TAIR (http://www.arabidopsis.org; Huala et al., 2001). Inserts were then cloned into the Y2H expression vectors pDEST 22 or pDEST 32 (Invitrogen). The pNF-YC9:NF-YC9 construct was previously described (Kumimoto et al., 2010). The entry clone pNF-YC9:NF-YC9F151R V153K was cloned into the plant expression vector pEarlyGate301 (Earley et al., 2006).
Plant Transformation, Cultivation, and Flowering Time Experiments
Arabidopsis ecotype Col-0 was the wild type for all experiments. The triple mutant was previously described (Kumimoto et al., 2010). Agrobacterium tumefaciens-mediated floral dipping was used to transform the triple mutant with pNF-YC9:NF-YC9 and pNF-YC9:NF-YC9F151R V153K (Clough and Bent, 1998). All experiments were performed on plants grown in a custom-built walk-in chamber under standard long-day conditions (16 h light/8 h dark, 22°C). Plant growth conditions were as described (Myers et al., 2016). Leaf number at flowering was measured as the total number of rosette and cauline leaves on the primary axis at flowering.
Y2H Analysis
The activation domain or DNA binding domain constructs were introduced into the yeast strain MaV203 (Invitrogen). Y2H assays were performed according to the instructions in the ProQuest manual (Invitrogen). For the X-Gal assay, nitrocellulose membranes were frozen in liquid nitrogen and placed on a filter paper saturated with Z-buffer containing X-Gal (5-bromo-4-chloro-3-indoxyl-β-d-galactopyranoside; Gold Biotechnology; catalog no. Z4281L).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AT5G12840, AT3G05690, AT3G14020, AT5G47640, AT4G14540, AT1G54830, AT5G63470, AT1G08970, AT5G15840, and AT1G65480.
Supplemental Data
Supplemental Figure 1. List of available data on CCT/NF-Y subunits interactions.
Supplemental Figure 2. Coomassie-stained gels of purified proteins samples.
Supplemental Figure 3. CO efficiently binds DNA as a trimer with At-NF-YB2/NF-YC3.
Supplemental Figure 4. EMSA competition analysis of NF-CO sequence specificity.
Supplemental Figure 5. Validation of RNA-seq data.
Supplemental Figure 6. Statistical enrichment of promoter motifs in DE gene sets and subsets.
Supplemental Figure 7. Enrichment of circadian expression phases among genes differentially expressed in co-sail, nf-yb2 nf-yb3, and nf-yc3 nf-yc4 nf-yc9.
Supplemental Figure 8. Fold expression changes of genes in co-sail, nf-yb2 nf-yb3, and nf-yc3 nf-yc4 nf-yc9 lines.
Supplemental Figure 9. Immunoblot analysis of CO-overexpressing lines.
Supplemental Table 1. Oligonucleotides used to amplify regions of the FT gene.
Supplemental Data Set 1. List of differentially expressed genes in RNA-seq analysis.
Supplemental Data Set 2A. Enrichment of peak expression phase (long-day growth) of genes differentially expressed in co-sail, nf-yb2 nf-yb3, and nf-yc3 nf-yc4 nf-yc9 lines.
Supplemental Data Set 2B. List of GO terms showing significant enrichment in sets of genes showing differential expression in co-sail, nf-yb2 nf-yb3, and nf-yc3 nf-yc4 nf-yc9 lines.
Supplemental Data Set 2C. Expression levels by RNA-seq (TPM) for members of the CCT, B-box, NF-YA, NF-YB, and NF-YC gene families.
Supplemental Data Set 3. Intersection of differentially expressed genes in NF-CO subunits mutants with Wigge et al. (2005) expression data.
Acknowledgments
This work was supported by an institutional grant from the Università degli Studi di Milano to N.G. and by National Science Foundation Grant IOS-1149822 to B.F.H.
AUTHOR CONTRIBUTIONS
N.G. performed the biochemical experiments. R.W.K. and C.S. performed the RNA-seq and validation experiments. S.S. performed the ChIP experiments. M.C. and D.S.H. analyzed the RNA-seq experiments. N.G., B.F.H., and R.M. planned the experiments. B.F.H. and R.M. wrote the manuscript.
Glossary
- TF
transcription factors
- EMSA
electrophoretic mobility shift assays
- ChIP
chromatin immunoprecipitation
- Y2H
yeast two-hybrid
Footnotes
Articles can be viewed without a subscription.
References
- Abelenda J.A., Cruz-Oró E., Franco-Zorrilla J.M., Prat S. (2016). Potato StCONSTANS-like1 suppresses storage organ formation by directly activating the FT-like StSP5G repressor. Curr. Biol. 26: 872–881. [DOI] [PubMed] [Google Scholar]
- Adrian J., Farrona S., Reimer J.J., Albani M.C., Coupland G., Turck F. (2010). cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22: 1425–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Naim O., Eshed R., Parnis A., Teper-Bamnolker P., Shalit A., Coupland G., Samach A., Lifschitz E. (2006). The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 46: 462–476. [DOI] [PubMed] [Google Scholar]
- Blackman B.K., Michaels S.D. (2010). Does CONSTANS act as a transcription factor or as a co-activator? The answer may be--yes. New Phytol. 187: 1–3. [DOI] [PubMed] [Google Scholar]
- Bu Z., Yu Y., Li Z., Liu Y., Jiang W., Huang Y., Dong A.W. (2014). Regulation of arabidopsis flowering by the histone mark readers MRG1/2 via interaction with CONSTANS to modulate FT expression. PLoS Genet. 10: e1004617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Ballif J., Endo S., Davis E., Liang M., Chen D., DeWald D., Kreps J., Zhu T., Wu Y. (2007). A putative CCAAT-binding transcription factor is a regulator of flowering timing in Arabidopsis. Plant Physiol. 145: 98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvenzani V., Testoni B., Gusmaroli G., Lorenzo M., Gnesutta N., Petroni K., Mantovani R., Tonelli C. (2012). Interactions and CCAAT-binding of Arabidopsis thaliana NF-Y subunits. PLoS One 7: e42902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Siriwardana C.L., Kumimoto R.W., Holt B.F. III (2011a). Construction of high quality Gateway™ entry libraries and their application to yeast two-hybrid for the monocot model plant Brachypodium distachyon. BMC Biotechnol. 11: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Kumimoto R.W., Siriwardana C.L., Risinger J.R., Holt B.F. III (2011b). Identification and characterization of NF-Y transcription factor families in the monocot model plant Brachypodium distachyon. PLoS One 6: e21805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Kumimoto R.W., Gnesutta N., Calogero A.M., Mantovani R., Holt B.F. III (2014). A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell 26: 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N.Z., Zhang X.Q., Wei P.C., Chen Q.J., Ren F., Chen J., Wang X.C. (2007). AtHAP3b plays a crucial role in the regulation of flowering time in Arabidopsis during osmotic stress. J. Biochem. Mol. Biol. 40: 1083–1089. [DOI] [PubMed] [Google Scholar]
- Chen J., Li X., Cheng C., Wang Y., Qin M., Zhu H., Zeng R., Fu X., Liu Z., Zhang G. (2014). Characterization of epistatic interaction of QTLs LH8 and EH3 controlling heading date in rice. Sci. Rep. 4: 4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Dai X., Ding Y., Tan L., Fu Y., Liu F., Zhu Z., Sun X., Sun X., Gu P., Cai H., Sun C. (2012). LHD1, an allele of DTH8/Ghd8, controls late heading date in common wild rice (Oryza rufipogon). J. Integr. Plant Biol. 54: 790–799. [DOI] [PubMed] [Google Scholar]
- Dennis G. Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. (2003). DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4: 3. [PubMed] [Google Scholar]
- Diebold M.L., Fribourg S., Koch M., Metzger T., Romier C. (2011). Deciphering correct strategies for multiprotein complex assembly by co-expression: application to complexes as large as the histone octamer. J. Struct. Biol. 175: 178–188. [DOI] [PubMed] [Google Scholar]
- Distelfeld A., Tranquilli G., Li C., Yan L., Dubcovsky J. (2009a). Genetic and molecular characterization of the VRN2 loci in tetraploid wheat. Plant Physiol. 149: 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A., Li C., Dubcovsky J. (2009b). Regulation of flowering in temperate cereals. Curr. Opin. Plant Biol. 12: 178–184. [DOI] [PubMed] [Google Scholar]
- Dolfini D., Zambelli F., Pavesi G., Mantovani R. (2009). A perspective of promoter architecture from the CCAAT box. Cell Cycle 8: 4127–4137. [DOI] [PubMed] [Google Scholar]
- Dolfini D., Mantovani R. (2013). Targeting the Y/CCAAT box in cancer: YB-1 (YBX1) or NF-Y? Cell Death Differ. 20: 676–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Feng Z., Zhang L., Yang C., Wu T., Lv J., Chen Y., Liu X., Liu S., Jiang L., Wan J. (2014). EF8 is involved in photoperiodic flowering pathway and chlorophyll biogenesis in rice. Plant Cell Rep. 33: 2003–2014. [DOI] [PubMed] [Google Scholar]
- Feng Z.J., He G.H., Zheng W.J., Lu P.P., Chen M., Gong Y.M., Ma Y.Z., Xu Z.S. (2015). Foxtail millet NF-Y families: genome-wide survey and evolution analyses identified two functional genes important in abiotic stresses. Front. Plant Sci. 6: 1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming J.D., Pavesi G., Benatti P., Imbriano C., Mantovani R., Struhl K. (2013). NF-Y coassociates with FOS at promoters, enhancers, repetitive elements, and inactive chromatin regions, and is stereo-positioned with growth-controlling transcription factors. Genome Res. 23: 1195–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron J.M., Pruneda-Paz J.L., Doherty C.J., Gross A.M., Kang S.E., Kay S.A. (2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 109: 3167–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goretti D., Martignago D., Landini M., Brambilla V., Gómez-Ariza J., Gnesutta N., Galbiati F., Collani S., Takagi H., Terauchi R., Mantovani R., Fornara F. (2017). Transcriptional and post-transcriptional mechanisms Limit Heading Date 1 (Hd1) function to adapt rice to high latitudes. PLoS Genet. 13: e1006530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeff M., Straub D., Eguen T., Dolde U., Rodrigues V., Brandt R., Wenkel S. (2016). MicroProtein-mediated recruitment of CONSTANS into a TOPLESS trimeric complex represses flowering in Arabidopsis. PLoS Genet. 12: e1005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusmaroli G., Tonelli C., Mantovani R. (2001). Regulation of the CCAAT-Binding NF-Y subunits in Arabidopsis thaliana. Gene 264: 173–185. [DOI] [PubMed] [Google Scholar]
- Gusmaroli G., Tonelli C., Mantovani R. (2002). Regulation of novel members of the Arabidopsis thaliana CCAAT-binding nuclear factor Y subunits. Gene 283: 41–48. [DOI] [PubMed] [Google Scholar]
- Hackenberg D., Keetman U., Grimm B. (2012). Homologous NF-YC2 subunit from Arabidopsis and tobacco is activated by photooxidative stress and induces flowering. Int. J. Mol. Sci. 13: 3458–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring M., Offermann S., Danker T., Horst I., Peterhansel C., Stam M. (2007). Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer S.L., Kay S.A. (2005). Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell 17: 1926–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilioti Z., Ganopoulos I., Bossis I., Tsaftaris A. (2014). LEC1-LIKE paralog transcription factor: how to survive extinction and fit in NF-Y protein complex. Gene 543: 220–233. [DOI] [PubMed] [Google Scholar]
- Hou X., Zhou J., Liu C., Liu L., Shen L., Yu H. (2014). Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat. Commun. 5: 4601. [DOI] [PubMed] [Google Scholar]
- Huala E., et al. (2001). The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 29: 102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber E.M., Scharf D.H., Hortschansky P., Groll M., Brakhage A.A. (2012). DNA minor groove sensing and widening by the CCAAT-binding complex. Structure 20: 1757–1768. [DOI] [PubMed] [Google Scholar]
- Hwang Y.H., Kim S.K., Lee K.C., Chung Y.S., Lee J.H., Kim J.K. (2016). Functional conservation of rice OsNF-YB/YC and Arabidopsis AtNF-YB/YC proteins in the regulation of flowering time. Plant Cell Rep. 35: 857–865. [DOI] [PubMed] [Google Scholar]
- Khanna R., Kronmiller B., Maszle D.R., Coupland G., Holm M., Mizuno T., Wu S.H. (2009). The Arabidopsis B-box zinc finger family. Plant Cell 21: 3416–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.K., Park H.Y., Jang Y.H., Lee K.C., Chung Y.S., Lee J.H., Kim J.K. (2016). OsNF-YC2 and OsNF-YC4 proteins inhibit flowering under long-day conditions in rice. Planta 243: 563–576. [DOI] [PubMed] [Google Scholar]
- Kim I.S., Sinha S., de Crombrugghe B., Maity S.N. (1996). Determination of functional domains in the C subunit of the CCAAT-binding factor (CBF) necessary for formation of a CBF-DNA complex: CBF-B interacts simultaneously with both the CBF-A and CBF-C subunits to form a heterotrimeric CBF molecule. Mol. Cell. Biol. 16: 4003–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo B.H., Yoo S.C., Park J.W., Kwon C.T., Lee B.D., An G., Zhang Z., Li J., Li Z., Paek N.C. (2013). Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol. Plant 6: 1877–1888. [DOI] [PubMed] [Google Scholar]
- Kumimoto R.W., Adam L., Hymus G.J., Repetti P.P., Reuber T.L., Marion C.M., Hempel F.D., Ratcliffe O.J. (2008). The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 228: 709–723. [DOI] [PubMed] [Google Scholar]
- Kumimoto R.W., Zhang Y., Siefers N., Holt B.F. III (2010). NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J. 63: 379–391. [DOI] [PubMed] [Google Scholar]
- Laloum T., De Mita S., Gamas P., Baudin M., Niebel A. (2013). CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 18: 157–166. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva-González M.A., Ibarra-Laclette E., Cruz-Ramírez A., Herrera-Estrella L. (2012). Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS One 7: e48138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Distelfeld A., Comis A., Dubcovsky J. (2011). Wheat flowering repressor VRN2 and promoter CO2 compete for interactions with NUCLEAR FACTOR-Y complexes. Plant J. 67: 763–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Li K., Ju Z., Cao D., Fu D., Zhu H., Zhu B., Luo Y. (2016). Genome-wide analysis of tomato NF-Y factors and their role in fruit ripening. BMC Genomics 17: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M., Hole D., Wu J., Blake T., Wu Y. (2012). Expression and functional analysis of NUCLEAR FACTOR-Y, subunit B genes in barley. Planta 235: 779–791. [DOI] [PubMed] [Google Scholar]
- Liang M., Yin X., Lin Z., Zheng Q., Liu G., Zhao G. (2014). Identification and characterization of NF-Y transcription factor families in canola (Brassica napus L.). Planta 239: 107–126. [DOI] [PubMed] [Google Scholar]
- Liu T.L., Newton L., Liu M.J., Shiu S.H., Farré E.M. (2016). A G-box-like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in Arabidopsis. Plant Physiol. 170: 528–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F., Liu Y., Inoue A., Suzuki T., Zhao K., Zhang Y. (2016). Establishing chromatin regulatory landscape during mouse preimplantation development. Cell 165: 1375–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J. (1997). Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260. [DOI] [PubMed] [Google Scholar]
- Malviya N., Jaiswal P., Yadav D. (2016). Genome- wide characterization of Nuclear Factor Y (NF-Y) gene family of sorghum [Sorghum bicolor (L.) Moench]: a bioinformatics approach. Physiol. Mol. Biol. Plants 22: 33–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R., Li X.Y., Pessara U., Hooft van Huisjduijnen R., Benoist C., Mathis D. (1994). Dominant negative analogs of NF-YA. J. Biol. Chem. 269: 20340–20346. [PubMed] [Google Scholar]
- Michael T.P., et al. (2008). Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 4: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler T.C., Michael T.P., Priest H.D., Shen R., Sullivan C.M., Givan S.A., McEntee C., Kay S.A., Chory J. (2007). The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb. Symp. Quant. Biol. 72: 353–363. [DOI] [PubMed] [Google Scholar]
- Moreland J.L., Gramada A., Buzko O.V., Zhang Q., Bourne P.E. (2005). The Molecular Biology Toolkit (MBT): a modular platform for developing molecular visualization applications. BMC Bioinformatics 6: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers Z.A., Kumimoto R.W., Siriwardana C.L., Gayler K.K., Risinger J.R., Pezzetta D., Holt Iii B.F. (2016). NUCLEAR FACTOR Y, Subunit C (NF-YC) transcription factors are positive regulators of photomorphogenesis in Arabidopsis thaliana. PLoS Genet. 12: e1006333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardini M., Gnesutta N., Donati G., Gatta R., Forni C., Fossati A., Vonrhein C., Moras D., Romier C., Bolognesi M., Mantovani R. (2013). Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell 152: 132–143. [DOI] [PubMed] [Google Scholar]
- Oldfield A.J., Yang P., Conway A.E., Cinghu S., Freudenberg J.M., Yellaboina S., Jothi R. (2014). Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Mol. Cell 55: 708–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi G., Mereghetti P., Mauri G., Pesole G. (2004). Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 32: W199–W203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pchelintsev N.A., Adams P.D., Nelson D.M. (2016). Critical parameters for efficient sonication and improved chromatin immunoprecipitation of high molecular weight proteins. PLoS One 11: e0148023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroni K., Kumimoto R.W., Gnesutta N., Calvenzani V., Fornari M., Tonelli C., Holt B.F. III, Mantovani R. (2012). The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 24: 4777–4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J., Robson F., Lee K., Simon R., Coupland G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857. [DOI] [PubMed] [Google Scholar]
- Qu B., He X., Wang J., Zhao Y., Teng W., Shao A., Zhao X., Ma W., Wang J., Li B., Li Z., Tong Y. (2015). A wheat CCAAT box-binding transcription factor increases the grain yield of wheat with less fertilizer input. Plant Physiol. 167: 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach T.N., Nguyen H.T., Valliyodan B., Joshi T., Xu D., Nguyen H.T. (2015). Genome-wide expression analysis of soybean NF-Y genes reveals potential function in development and drought response. Mol. Genet. Genomics 290: 1095–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C., Zhang Z., Wang Y., Li S., Liang Z. (2016). Genome-wide identification and characterization of the NF-Y gene family in grape (Vitis vinifera L.). BMC Genomics 17: 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes J.C., Muro-Pastor M.I., Florencio F.J. (2004). The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 134: 1718–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rípodas C., Castaingts M., Clúa J., Blanco F., Zanetti M.E. (2015). Annotation, phylogeny and expression analysis of the nuclear factor Y gene families in common bean (Phaseolus vulgaris). Front. Plant Sci. 5: 761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson F., Costa M.M., Hepworth S.R., Vizir I., Piñeiro M., Reeves P.H., Putterill J., Coupland G. (2001). Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 28: 619–631. [DOI] [PubMed] [Google Scholar]
- Romier C., Cocchiarella F., Mantovani R., Moras D. (2003). The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 278: 1336–1345. [DOI] [PubMed] [Google Scholar]
- Schmid M., Uhlenhaut N.H., Godard F., Demar M., Bressan R., Weigel D., Lohmann J.U. (2003). Dissection of floral induction pathways using global expression analysis. Development 130: 6001–6012. [DOI] [PubMed] [Google Scholar]
- Sherwood R.I., Hashimoto T., O’Donnell C.W., Lewis S., Barkal A.A., van Hoff J.P., Karun V., Jaakkola T., Gifford D.K. (2014). Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat. Biotechnol. 32: 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J.S., Kubota A., Imaizumi T. (2017). Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol. 173: 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefers N., Dang K.K., Kumimoto R.W., Bynum W.E. IV, Tayrose G., Holt B.F. III (2009). Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 149: 625–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S., Kim I.S., Sohn K.Y., de Crombrugghe B., Maity S.N. (1996). Three classes of mutations in the A subunit of the CCAAT-binding factor CBF delineate functional domains involved in the three-step assembly of the CBF-DNA complex. Mol. Cell. Biol. 16: 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siriwardana C.L., Gnesutta N., Kumimoto R.W., Jones D.S., Myers Z.A., Mantovani R., Holt B.F. III (2016). NUCLEAR FACTOR Y, Subunit A (NF-YA) proteins positively regulate flowering and act through FLOWERING LOCUS T. PLoS Genet. 12: e1006496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Smith R.W., To B.J., Millar A.J., Imaizumi T. (2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Shim J.S., Kinmonth-Schultz H.A., Imaizumi T. (2015). Photoperiodic flowering: time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 66: 441–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson T.J., McIntyre C.L., Collet C., Xue G.P. (2007). Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aestivum. Plant Mol. Biol. 65: 77–92. [DOI] [PubMed] [Google Scholar]
- Tiwari S.B., et al. (2010). The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 187: 57–66. [DOI] [PubMed] [Google Scholar]
- Tripathi P., Carvallo M., Hamilton E.E., Preuss S., Kay S.A. (2017). Arabidopsis B-BOX32 interacts with CONSTANS-LIKE3 to regulate flowering. Proc. Natl. Acad. Sci. USA 114: 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A., Beales J., Faure S., Dunford R.P., Laurie D.A. (2005). The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310: 1031–1034. [DOI] [PubMed] [Google Scholar]
- Wang C.Q., Guthrie C., Sarmast M.K., Dehesh K. (2014). BBX19 interacts with CONSTANS to repress FLOWERING LOCUS T transcription, defining a flowering time checkpoint in Arabidopsis. Plant Cell 26: 3589–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Xu J., Guo H., Jiang L., Chen S., Yu C., Zhou Z., Hu P., Zhai H., Wan J. (2010). DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 153: 1747–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkel S., Turck F., Singer K., Gissot L., Le Gourrierec J., Samach A., Coupland G. (2006). CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18: 2971–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P.A., Kim M.C., Jaeger K.E., Busch W., Schmid M., Lohmann J.U., Weigel D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059. [DOI] [PubMed] [Google Scholar]
- Xing Y., Fikes J.D., Guarente L. (1993). Mutations in yeast HAP2/HAP3 define a hybrid CCAAT box binding domain. EMBO J. 12: 4647–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N., Winter C.M., Wu M.F., Kwon C.S., William D.A., Wagner D. (2014). Protocols: Chromatin immunoprecipitation from Arabidopsis tissues. Arabidopsis Book 12: e0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Loukoianov A., Blechl A., Tranquilli G., Ramakrishna W., SanMiguel P., Bennetzen J.L., Echenique V., Dubcovsky J. (2004). The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303: 1640–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W.H., Wang P., Chen H.X., Zhou H.J., Li Q.P., Wang C.R., Ding Z.H., Zhang Y.S., Yu S.B., Xing Y.Z., Zhang Q.F. (2011). A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 4: 319–330. [DOI] [PubMed] [Google Scholar]
- Yang J., Wan X.L., Guo C., Zhang J.W., Bao M.Z. (2016). Identification and expression analysis of nuclear factor Y families in Prunus mume under different abiotic stresses. Biol. Plant. 60: 419–426. [Google Scholar]
- Zambelli F., Pesole G., Pavesi G. (2014). Using Weeder, Pscan, and PscanChIP for the discovery of enriched transcription factor binding site motifs in nucleotide sequences. Curr. Protoc. Bioinformatics 47: 1–31. [DOI] [PubMed] [Google Scholar]
- Zambelli F., Pesole G., Pavesi G. (2009). Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 37: W247–W252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Han M., Lv Q., Bao F., He Y. (2015). Identification and expression profile analysis of NUCLEAR FACTOR-Y families in Physcomitrella patens. Front. Plant Sci. 6: 642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Li X., Zhang C., Zou H., Wu Z. (2016). Isolation, structural analysis, and expression characteristics of the maize nuclear factor Y gene families. Biochem. Biophys. Res. Commun. 478: 752–758. [DOI] [PubMed] [Google Scholar]