An integrated reagent toolkit and streamlined protocols work across diverse plant species to enable sophisticated genome edits.
Abstract
We report a comprehensive toolkit that enables targeted, specific modification of monocot and dicot genomes using a variety of genome engineering approaches. Our reagents, based on transcription activator-like effector nucleases (TALENs) and the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system, are systematized for fast, modular cloning and accommodate diverse regulatory sequences to drive reagent expression. Vectors are optimized to create either single or multiple gene knockouts and large chromosomal deletions. Moreover, integration of geminivirus-based vectors enables precise gene editing through homologous recombination. Regulation of transcription is also possible. A Web-based tool streamlines vector selection and construction. One advantage of our platform is the use of the Csy-type (CRISPR system yersinia) ribonuclease 4 (Csy4) and tRNA processing enzymes to simultaneously express multiple guide RNAs (gRNAs). For example, we demonstrate targeted deletions in up to six genes by expressing 12 gRNAs from a single transcript. Csy4 and tRNA expression systems are almost twice as effective in inducing mutations as gRNAs expressed from individual RNA polymerase III promoters. Mutagenesis can be further enhanced 2.5-fold by incorporating the Trex2 exonuclease. Finally, we demonstrate that Cas9 nickases induce gene targeting at frequencies comparable to native Cas9 when they are delivered on geminivirus replicons. The reagents have been successfully validated in tomato (Solanum lycopersicum), tobacco (Nicotiana tabacum), Medicago truncatula, wheat (Triticum aestivum), and barley (Hordeum vulgare).
INTRODUCTION
Genome engineering is a rapidly emerging discipline that seeks to develop strategies and methods for the efficient, targeted modification of DNA in living cells. Most genome engineering approaches use sequence-specific nucleases (SSNs), such as transcription activator-like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 reagents, to create a targeted DNA double-strand break (DSB) in a genome. DNA repair then achieves the desired DNA sequence modification. DSBs are most frequently repaired by nonhomologous end joining (NHEJ), which can create insertion/deletion (indel) mutations at the break site that knock out gene function. TALEN- or CRISPR/Cas9-mediated gene knockouts have been made in diverse plant species amenable to transformation (Brooks et al., 2014; Fauser et al., 2014; Forner et al., 2015; Gao et al., 2015; Christian et al., 2013; Liang et al., 2014; Shan et al., 2013; Sugano et al., 2014; Xu et al., 2015; Zhang et al., 2014). Traits of commercial value have also been created by targeted knockout of selected genes (Wang et al., 2014; Clasen et al., 2016; Haun et al., 2014; Li et al., 2012, 2016). Alternatively, DSBs can be repaired by homology-dependent repair (HDR), which copies information from a “donor” DNA template. HDR typically occurs at lower frequencies than NHEJ in somatic cells and is more challenging to achieve because it requires introducing into the plant cell both the donor DNA molecule and the SSN expression cassette. As described below, however, novel strategies to deliver these reagents have recently realized some improvements in efficiency (Baltes et al., 2014; Čermák et al., 2015; Fauser et al., 2012; Schiml et al., 2014).
Since the development of TALENs and CRISPR/Cas9 reagents, improvements in the technology have occurred at a rapid pace. For example, indel frequencies have been increased by coupling SSNs with exonucleases (Certo et al., 2012; Kwon et al., 2012), altering guide RNA (gRNA) architecture (Chen et al., 2013) or using specialized promoters (Wang et al., 2015; Yan et al., 2015). Increased specificity has been achieved using paired TALEN or CRISPR/Cas9 nickases (Ran et al., 2013), truncated gRNAs (Fu et al., 2014), or dimeric CRISPR/Cas9 nucleases (Guilinger et al., 2014; Tsai et al., 2014). Other diverse applications have emerged, including targeted regulation of gene expression, targeted epigenetic modification, or site-specific base editing through deamination (La Russa and Qi, 2015; Kungulovski and Jeltsch, 2016; Zong et al., 2017; Shimatani et al., 2017).
CRISPR/Cas9 is particularly useful for multiplexed gene editing because target specificity is determined by short gRNAs rather than proteins that must be engineered for each new target (Baltes and Voytas, 2015). Several systems for simultaneous expression of multiple gRNAs have been developed, and these most often involve the assembly of multiple, individual gRNA expression cassettes, each transcribed from a separate RNA polymerase III (Pol III) promoter (Xing et al., 2014; Ma et al., 2015; Lowder et al., 2015). Multiple gRNAs can also be expressed from a single transcript. Such polycistronic mRNAs are processed posttranscriptionally into individual gRNAs by RNA-cleaving enzymes. These enzymes include the CRISPR-associated RNA endoribonuclease Csy4 from Pseudomonas aeruginosa (Tsai et al., 2014), the tRNA processing enzymes naturally present in the host cells (Xie et al., 2015), and ribozymes (Gao and Zhao, 2014).
One significant remaining challenge in plant genome engineering is achieving high frequency gene editing by HDR. As mentioned above, HDR requires introducing into the plant cell both a donor DNA molecule and a SSN expression cassette. DNA sequence modifications incorporated into the donor template are then copied into the broken chromosome. The in planta gene targeting (GT) approach stably integrates the donor template into the plant genome. This ensures that each cell has at least one copy of the donor, which is released from the chromosome by a SSN, making it available for HDR (Schiml et al., 2014; Fauser et al., 2012). Another approach involves increasing donor template availability by replicating it to high copy number using geminivirus replicons (GVRs) (Baltes et al., 2014). GVRs make it possible to create precise modifications without the need to stably integrate editing reagents into the plant genome (Čermák et al., 2015).
Not all of the methods and approaches described above have been implemented and optimized for use in plants. Although there are several toolsets available for plant genome engineering (Ma et al., 2015; Xing et al., 2014; Lowder et al., 2015), they are limited in that they use a single type of DSB-inducing reagent (e.g., CRISPR/Cas9), a single type of expression system (e.g., Pol III promoters for expression of gRNAs), or they were specialized for a single application (e.g., gene knockouts). Here, we introduce a comprehensive, modular system for plant genome engineering that makes it possible to accomplish gene knockouts, replacements, altered transcriptional regulation, or multiplexed modifications. In addition, the platform can be easily upgraded and adjusted to accommodate novel reagents and approaches as they arise. Our toolkit uses Golden Gate cloning for fast and easy assembly of genome engineering constructs and flexibility in combining different functional modules for different applications. The functional modules include TALE-based reagents (TALENs and TALE activators), CRISPR/Cas9 reagents (nucleases, nickases, catalytically inactive enzymes, and activators), various gRNA expression approaches (single and multi-gRNA expression systems using Pol III or Pol II promoters), donor template plasmids for gene targeting, and a variety of other expression cassettes (Trex2, Csy4, and GFP). Furthermore, a large set of promoters, terminators, codon-optimized genes, and selection markers facilitate engineering dicot or monocot genomes using T-DNA or non-T-DNA vectors for Agrobacterium tumefaciens-mediated, biolistic, or protoplast transformation. We also provide a user-friendly Web portal with protocols, DNA sequences of all reagents, and Web-based tools to assist in construct design. Finally, we validated our reagents in six different plant species and report the use of genome engineering to create modified Medicago truncatula plants.
RESULTS
Direct and Modular Assembly of Genome Engineering Constructs
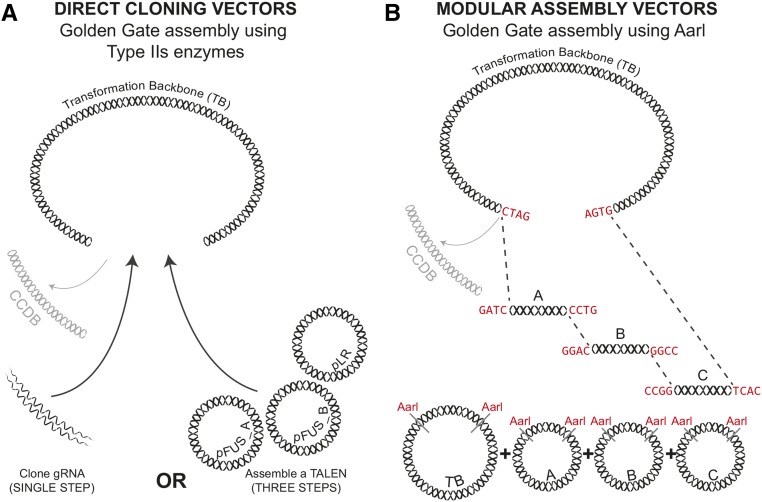
Our plant genome engineering toolkit provides two sets of vectors, designated “direct” and “modular” (Figure 1). Both vector sets can deliver to plant cells either TALENs or CRISPR/Cas9 reagents for creating targeted DNA sequence modifications. The TALEN vectors are fully compatible with our Golden Gate TALEN and TAL Effector Kit 2.0 (Cermak et al., 2011), and they use the highly active Δ152/+63 TALEN architecture (Miller et al., 2011). The CRISPR/Cas9 vectors include either wheat (Triticum aestivum) or Arabidopsis thaliana codon-optimized versions of Cas9 for use in monocots and dicots, respectively. Both versions of Cas9 contain a single C-terminal nuclear localization signal.
Figure 1.
Two Sets of Vectors for Direct Cloning or Modular Assembly of Genome Engineering Reagents.
(A) Direct cloning vectors were designed to speed up the cloning process. Specificity determining elements (gRNAs or TAL repeats) are cloned directly into the transformation backbone (e.g., T-DNA).
(B) The modular assembly vectors enable combination of different functional elements. Specificity determining elements (gRNAs, TAL repeats, and donor templates for gene targeting) are first cloned into intermediate module vectors. Custom-selected modules are then assembled together into the transformation backbone (e.g., T-DNA).
The direct cloning vectors (numbering 31) enable rapid construction of reagents for making targeted gene knockouts. They consist of vector backbones (T-DNA or non-T-DNA), selectable marker expression cassettes, and nucleases (i.e., Cas9 or the TALEN backbone). However, they lack determinants for DNA targeting (i.e., gRNAs or TALE repeats). Single or multiple gRNAs can be added in a single step using a Golden Gate protocol (Engler et al., 2008, 2009) (Figure 1A). Addition of TALENs uses a three-step Golden Gate protocol (see Supplemental Methods for assembly protocols).
The modular vector set uses Golden Gate cloning to create vectors for a diverse range of gene editing applications (Figure 1B; Supplemental Methods, Protocol 5). There are three sets of modular cloning vectors. Module set A contains ready-to-use plasmids (currently numbering 61) with Cas9 or GFP expression cassettes. Module A plasmids are also available that can be used when assembling TALENs with our Golden Gate kit (Cermak et al., 2011). Module B plasmids (currently numbering 22) are used either to add single or multiple gRNAs in a variety of expression formats or to add a second TALEN monomer. Module C plasmids (currently numbering 22) may be used to add donor templates for gene targeting, additional gRNA cassettes, or other expression cassettes, such as GFP, Trex2, or Csy4. Each expression cassette has a similar architecture (Supplemental Figure 1): A common set of restriction enzyme sites are used that allow for easy swapping of regulatory elements (promoters and terminator sequences) or coding sequences. If a reagent from a given set is not desired, there are empty modules to serve as placeholders and still allow for Golden Gate assembly. Finally, a transformation backbone can be selected from a set (currently numbering 33) of T-DNA and non-T-DNA vectors. Standard or GVR vectors are available with or without one of the commonly used antibiotic selectable markers. For most applications, creating a transformation vector using the modular vector set is a two-step cloning process that can be completed in five days; assembly of TALEN-based vectors usually requires an additional cloning step.
To facilitate use of our toolkit, we developed online resources to aid in vector selection and construct design (http://cfans-pmorrell.oit.umn.edu/CRISPR_Multiplex/). Our website provides a complete list of both direct and modular cloning vectors with descriptions of key features, DNA sequence files, and relevant protocols for downloading (see Supplemental Data Set 1 for the list of vectors). Also available online is a “vector selection” tool that uses a dropdown menu to help identify suitable intermediary plasmids needed (e.g., module A, B, or C) or to select a transformation backbone. The vector selection tool then provides a link to the DNA sequence file of the selected plasmid that can be downloaded in GenBank format. For multiplexed gene editing, our “primer design and map construction” tool takes as input a FASTA file with the gRNA target sequences. A list is then prepared of all the primer sequences needed to create a vector that expresses multiple gRNAs using our tRNA, Csy4, or ribozyme expression formats, and a GenBank file is created with the annotated sequence of the specified gRNA array in the selected plasmid backbone.
Targeted Mutagenesis of Genes in M. truncatula Using Direct Vectors
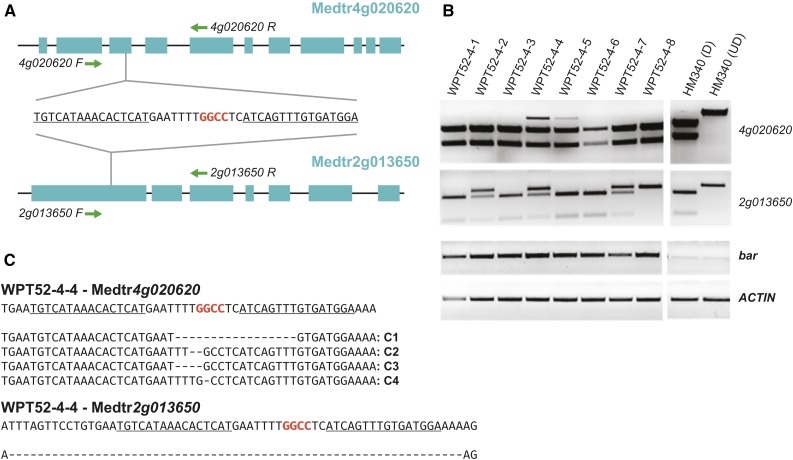
To test the functionality of the direct vectors, we designed a TALEN pair to target two paralogous genes in M. truncatula (Medtr4g020620 and Medtr2g013650) (Figure 2A). The Arabidopsis ortholog is known to play a role in phosphate regulation, and mutants hyperaccumulate phosphate (Park et al., 2014). A T-DNA vector with the TALENs was assembled using our Golden Gate assembly protocol (Supplemental Methods, Protocol 1B) and transformed into M. truncatula. Eleven T0 plants were recovered and screened for targeted mutations at both Medtr4g020620 and Medtr2g013650 using a PCR digestion assay. Two T0 plants (WPT52-4 and WPT52-5) produced PCR amplicons that were resistant to restriction enzyme digestion (at both targets in WPT52-4 and at one target in WPT52-5). Plant WPT52-4 was self-fertilized, and T1 progeny were screened using the same PCR digestion assay. Mutations were confirmed at both loci (Figure 2B). Mutations in Medtr4g020620 were likely somatic, as only two of the tested plants produced digestion-resistant amplicons of varying intensity. However, mutations in Medtr2g013650 segregated as expected for a trait transmitted through the germline, and one out of eight tested plants was a homozygous mutant. This was confirmed by DNA sequencing, which revealed four different Medtr4g020620 deletion alleles in a single plant, and an identical 63-bp deletion in Medtr2g013650 in three of the tested T1 plants (Figure 2C). From the eight T1 plants, we identified a single plant with mutations in both genes. Thus, TALENs work efficiently in our direct vectors and induce heritable mutations.
Figure 2.
TALEN-Mediated Mutagenesis in M. truncatula Using Direct Cloning Vectors.
(A) Maps of two M. truncatula genes targeted for mutagenesis using a single TALEN pair. PCR primers used for screening in (B) are shown as green arrowheads. The sequence of the TALEN target site is shown with TALEN binding sites underlined.
(B) HaeIII PCR digestion screening of eight T1 progeny of plant WPT52-4. The amplified locus is indicated on the right. bar and ACTIN are controls that amplify the transgene and a native gene, respectively. HM340 (D), wild-type product digested with HaeIII; HM340 (UD), undigested wild-type product.
(C) DNA sequences of undigested amplicons from plant WPT52-4-4. The reference sequence of the unmodified locus is shown on the top. TALEN binding sites are underlined. HaeIII restriction site used for the screening is in red.
Expressing Multiple gRNAs by Csy4, tRNA-Processing Enzymes, and Ribozymes
To harness one of the biggest advantages of the CRISPR/Cas9 system, the ability to target multiple sites simultaneously, our modular cloning system provides vectors for multiplexed genome editing. Typically in multiplexed editing, independent Pol III promoters are used to drive expression of each gRNA. One limitation is that Pol III expression requires a specific nucleotide in the first position of the transcript. Furthermore, the size of the final array can be cumbersome due to multiple, repeating promoter sequences. Processing polycistronic transcripts containing multiple gRNAs provides an alternative to the use of independent Pol III promoters. The polycistronic message can be produced from a Pol II promoter that is processed either by the Csy4 protein, tRNA processing enzymes, or ribozymes (Nissim et al., 2014; Aeby et al., 2010; Tang et al., 2016). Pol II promoters provide flexibility in terms of spatial and temporal control of expression in vivo, and they are more likely to produce long transcripts, since Pol II is not hampered by the presence of short internal termination sites, as is the case for Pol III (Arimbasseri et al., 2013). We sought to compare the efficiency of targeted mutagenesis using five different gRNA expression systems, including three different methods for processing polycistronic gRNA transcripts (Figure 3A). All vectors were assembled using our modular cloning system and used two gRNAs to target two sites in the tomato (Solanum lycopersicum) AUXIN RESPONSE FACTOR 8A (ARF8A) gene, a positive regulator of flower development and fruit set (N. Liu et al., 2014).
Figure 3.
Comparison of Different Systems for Expressing Multiple gRNAs.
(A) Structure of six constructs for expressing gRNA9 and gRNA10, which both target the tomato ARF8A gene. CmYLCV, Cestrum yellow leaf curling virus promoter; Csy4, 20-bp Csy4 hairpin; tRNA, 77-bp pre-tRNAGly gene; rb, 15-bp ribozyme cleavage site; ribozyme, 58-bp synthetic ribozyme; AtU6, Arabidopsis U6 promoter; At7SL, Arabidopsis 7SL promoter.
(B) Possible editing outcomes due to expression of both gRNAs. Sequences targeted by gRNA9 and gRNA10 are shown on the top. Cleavage sites are indicated by arrows.
(C) Overall mutation frequencies as determined by deep sequencing. Error bars represent se of three replicates (pools of protoplasts). Statistical significance was determined by Tukey’s test. *P < 0.05 and ***P < 0.001.
We created three vectors that use individual Pol III promoters to express each gRNA: two AtU6 promoters, an AtU6 and At7SL promoter, or AtU6 and At7SL promoters combined with structurally optimized gRNA scaffolds (Chen et al., 2013). The remaining three vectors produce a single transcript with gRNA cassettes separated by 20-bp Csy4 hairpins (Tsai et al., 2014), 77 bp tRNAGly genes (Xie et al., 2015), or 15-bp ribozyme cleavage sites (in addition to a synthetic ribozyme at the 3′ end of the transcript) (Haseloff and Gerlach, 1988; Tang et al., 2016). Thanks to the advantages over Pol III promoters mentioned above, we chose to use a Pol II promoter to drive the expression of the polycistronic gRNAs. Since Cas9 was expressed from a 35S promoter, we chose Cestrum Yellow Leaf Curling Virus promoter (CmYLCV) for gRNA expression to prevent use of duplicate promoters. The CmYLCV promoter drives comparable or higher levels of expression than the 35S or maize (Zea mays) ubiquitin (ZmUbi) promoters in both dicots and monocots (Stavolone et al., 2003), and the intensity of GFP expressed from this promoter was similar to 35S-driven GFP in tomato protoplasts (Supplemental Figure 2). Final vectors were cotransformed into tomato protoplasts along with a YFP expression plasmid. Transformation efficiency approximated 60%, as determined by counting YFP positive cells.
To assess mutation frequencies, the target site was PCR-amplified from DNA prepared from the transformed protoplasts and subjected to Illumina DNA sequencing (Supplemental Table 1). All of the expected types of mutations in the ARF8A locus were revealed in all samples except for the nontransformed control (Figures 3B and 3C; Supplemental Table 2; Supplemental Figures 3 to 7). Interestingly, the overall frequency of mutagenesis was ∼2-fold higher with the Csy4 and tRNA expression systems compared with constructs with two Pol III promoters. Although the overall frequency of mutagenesis in the Csy4 sample was consistently higher than in the tRNA sample across three replicates, the difference was not statistically significant (Figure 3C; Supplemental Table 3). The ribozyme showed lower frequencies of most mutation types, particularly at the 3′-most gRNA target site. We did not observe any enhancement in overall mutation frequencies with the previously reported improved gRNA architecture (Chen et al., 2013). It is important to note that the mutation frequencies were not adjusted to account for the ∼60% transformation efficiency; therefore, they are underestimates.
Deletion between the two CRISPR/Cas9 cleavage sites was the most common mutation in all samples (Supplemental Figure 3). Short indels at either one or both cleavage sites were more than 10-fold less abundant (Supplemental Figures 3 to 6). Inversions of the intervening sequence and deletions combined with insertions were more than 100-fold less frequent (Supplemental Figure 3). Most insertions mapped either to the tomato genome or to the vector, covering almost every region of the vector sequence (Supplemental Figure 7).
Cas9 most frequently creates a blunt-ended DSB three nucleotides upstream of the PAM sequence (Jinek et al., 2012). In our experiments, precise ligation of Cas9-cleaved DNA without the intervening sequence would result in a deletion of 85 bp. Gain or loss of nucleotides due to staggered Cas9 cleavage (Jinek et al., 2012; Li et al., 2015) and/or exonuclease activity could result in shorter or longer deletions. Strikingly, we observed a very high frequency of precise ligation: On average, 83.5% of all reads with long deletions were deletions of 85 bp (Supplemental Figure 8). These results suggest that if a single gRNA was used, most instances of NHEJ repair would not result in a mutation. Therefore, creating deletions with two or more gRNAs should be a more efficient approach to achieve targeted mutagenesis.
We further tested the effectiveness of the polycistronic expression systems using eight gRNAs targeting four genes for deletion in tomato protoplasts. We also monitored the effect of the position of the gRNA in the array. gRNAs 49 to 56 (Supplemental Table 4) were assembled directly into non-T-DNA protoplast expression vectors in two different orders (Figure 4A). In one array, the gRNAs were ordered from 49 to 56, and the order was reversed in the second array. In addition, a vector expressing each gRNA from a separate Pol III promoter was constructed. Each pair of gRNAs was designed to create an ∼3-kb deletion to prevent amplification of the nondeleted loci in subsequent PCR analysis (Figure 4B). The seven vectors were transformed into tomato protoplasts, and the frequencies of deletions induced by the first, last, and one of the internal gRNA pairs in each array were measured by quantitative PCR (Figure 4C). Consistent with the previous experiment, the Csy4 expression system performed best regardless of the position of the gRNA in the array. The tRNA system was the next most effective, followed by the use of individual Pol III promoters; the ribozyme system was the least effective. gRNAs near the end of the Csy4 and tRNA arrays were less efficient in mutagenesis relative to gRNAs in earlier positions for the majority of tested loci, suggesting that position does have an effect on gRNA activity. Nevertheless, regardless of position effects, when Csy4 was used, the frequency of deletions was as high or higher than the use of individual Pol III promoters, confirming the Csy4 is the most efficient system for multiplexed gene editing.
Figure 4.
Evaluation of Eight gRNAs in Various Polycistronic Expression Systems and the Effect of Position in the Array on gRNA Activity.
(A) Structure of seven constructs for expressing gRNAs 49 to 56 targeting four tomato genes for deletion.
(B) Detection of deletions between gRNA sites using qPCR. Each gene is targeted for deletion with two gRNAs designed to create a 3-kb deletion to prevent amplification of the nondeleted, wild-type template. Deletion frequency was quantified by qPCR using primers shown as red and blue arrowheads.
(C) Deletion frequencies in tomato protoplasts as measured by qPCR. Positions of respective gRNAs in the array are specified for each construct. Error bars represent se of two replicates.
Multiplexed Mutagenesis in Tomato, Wheat, and Barley Protoplasts
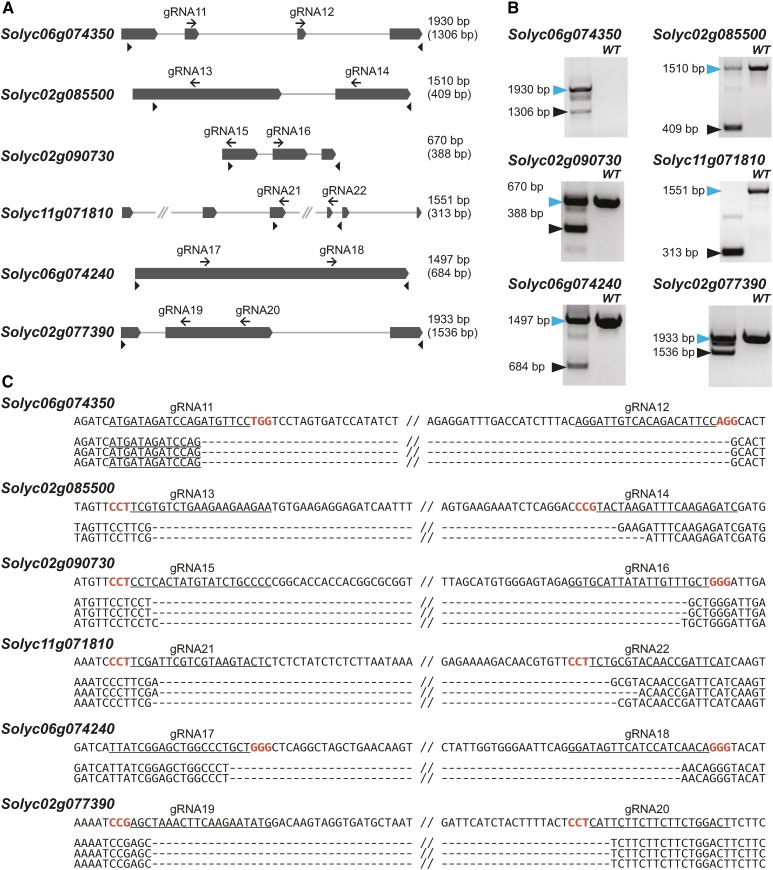
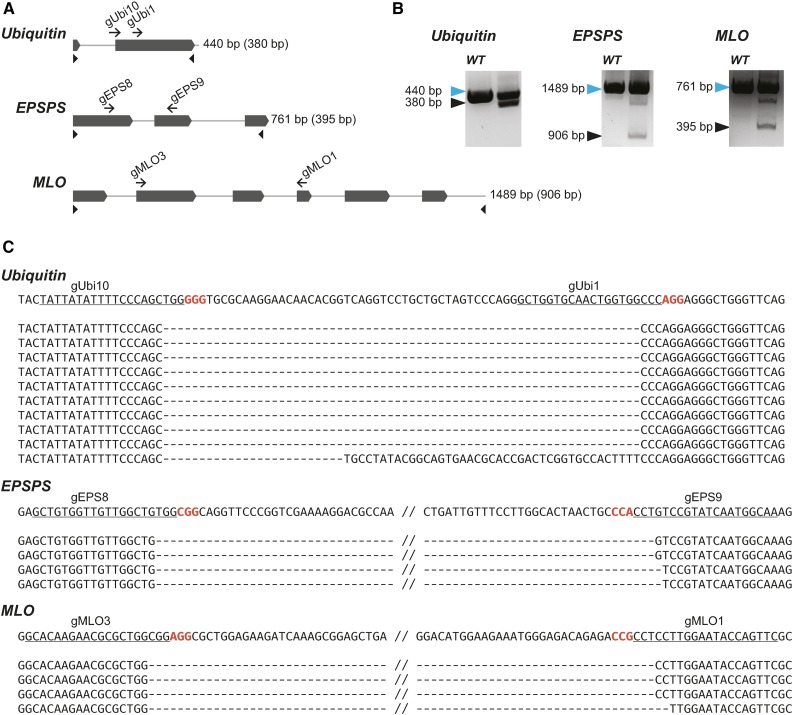
We further explored the effectiveness of the Csy4 system for multiplexing by targeting six genes in tomato (Solyc06g074350, Solyc02g085500, Solyc02g090730, Solyc11g071810, Solyc06g074240, and Solyc02g077390), three genes in wheat (ubiquitin, 5-enolpyruvylshikimate-3-phosphate synthase, and Mildew locus O [TaMLO]), and one gene in barley (Hordeum vulgare; HvMLO). Each gene was targeted for deletion using two gRNAs. To provide expression in cereals, we created vectors with ZmUbi, switchgrass (Panicum virgatum) ubiquitin 1 (PvUbi1), and switchgrass ubiquitin 2 (PvUbi2) promoters (Mann et al., 2011). The sequences of PvUbi1 and PvUbi2 were modified to remove AarI, SapI, and BsaI sites, and the modified promoters were confirmed to drive high levels of GFP expression in wheat scutella (Supplemental Figure 9). Next, we designed Csy4 arrays of 12 gRNAs targeting the six genes in tomato, six gRNAs targeting the three genes in wheat, and two gRNAs targeting the single gene in barley. In tomato, the 35S promoter was used to express Cas9, and the CmYLCV promoter drove expression of the gRNAs. In wheat and barley, ZmUbi was used to drive expression of Cas9, and the gRNA array was under control of PvUbi1. To test whether the PvUbi1 promoter could be substituted with the significantly shorter viral CmYLCV for gRNA expression in monocots, we also designed a vector in which the two gRNAs targeting the barley MLO gene were controlled by the CmYLCV promoter. The final vectors expressing Cas9 and the gRNA array were transformed into tomato, wheat, and barley protoplasts, and PCR was conducted 2 d later to detect targeted deletions. Deletions of the expected size were found in each targeted gene and verified by DNA sequencing (Figures 5 to 7). Both CmYLCV and PvUbi1-driven gRNA arrays induced targeted deletion of the barley MLO gene with similar efficiencies (Figure 7B). Thus, we confirmed that our system is efficient in simultaneously creating multiplexed, targeted gene deletions in both dicots and monocots using up to 12 gRNAs.
Figure 5.
Multiplexed Mutagenesis Using the Csy4 System in Tomato Protoplasts.
(A) Maps of six tomato genes targeted for simultaneous deletion using 12 gRNAs. gRNA sites are shown as black arrows. Arrowheads represent primers used to detect the deletions. Lengths of PCR products from the wild-type locus are shown on the right; lengths of the deletion products are in parentheses.
(B) Deletions of expected length were detected in each of the six genes by PCR. Blue and black arrowheads mark the unmodified and deletion products, respectively. Wild-type DNA from nontransformed cells was used as template.
(C) DNA sequences of representative deletion products. The sequence of the unmodified locus for each of the six genes is shown on the top. gRNA target sites are underlined and PAM sequences are in red.
Figure 7.
Both CmYLCV and PvUbi1 Promoters Are Functional in Barley Protoplasts and Induce Targeted Deletions Using the Csy4 gRNA Expression System.
(A) Map of the barley MLO gene targeted for deletion using two gRNAs. gRNA sites are shown as black arrows. Arrowheads represent primers used to detect the deletions. Length of the PCR product from the wild-type locus is shown on the right; length of the deletion product is in parentheses.
(B) Deletions between the gRNA38 and gRNA39 sites were detected in the barley MLO gene when either the CmYLCV or PvUbi1 promoters were used to express the gRNAs. Blue and black arrowheads mark the unmodified and deletion products, respectively. Two replicates (pools of protoplasts) of the experiment are shown.
(C) DNA sequences of representative deletion products. The sequence of the unmodified locus for each of the three genes is shown on the top. gRNA target sites are underlined and PAM sequences are in red.
Figure 6.
Multiplexed Mutagenesis Using the Csy4 System in Wheat Protoplasts.
(A) Maps of three wheat genes targeted for simultaneous mutagenesis using six gRNAs. gRNA sites are shown as black arrows. Arrowheads represent primers used to detect the deletions. Lengths of PCR products from the wild-type locus are shown on the right; lengths of the deletion products are in parentheses.
(B) Deletions of expected length were detected in each of the six genes by PCR. Blue and black arrowheads mark the unmodified and deletion products, respectively. Wild-type DNA from nontransformed cells was used as template.
(C) DNA sequences of representative deletion products. The sequence of the unmodified locus for each of the three genes is shown on the top. gRNA target sites are underlined and PAM sequences are in red.
Heritable, Multiplexed Mutagenesis in M. truncatula
To demonstrate the efficiency of our Csy4 multiplexing vectors in whole plants, we used six gRNAs to target four M. truncatula genes that encode nodule-specific cysteine-rich (NCR) peptides (Supplemental Figure 10). More than 500 genes make up the NCR family, and their small size, sequence identity, and overall number pose challenges to traditional knockout/knockdown platforms such as transposable elements (Tnt1), RNA interference, and chemical mutagenesis. Three gRNA pairs were designed to each delete one of three loci: NCR53, NCR54 (Medtr4g026818), and NCR55 (Medtr3g014705). The NCR53 locus contains two homologous NCR genes differing by several SNPs (Supplemental Data Set 2) and separated by ∼4 kb. The vector carrying the gRNA array, Cas9 expression cassette, and bar resistance gene was transformed into M. truncatula, and 46 phosphinothricin resistant T0 transformants were recovered and screened by PCR and direct sequencing. Forty-three plants had mutations in NCR53, eight in NCR54, and 12 in NCR55 (Table 1; Supplemental Table 5; Supplemental Data Set 2). The mutations included deletions of the NCR53 and NCR55 genes, resulting from simultaneous cleavage at two sites, as well as short indels in all three loci at a given target site. Three plants had mutations in all three genes, 13 were double mutants, and 27 had mutations in a single gene. Despite its repetitiveness, mutations at the NCR53 locus were exclusively long deletions or inversions between the two cleavage sites in seven out of 46 plants. No mutations were recovered at the gRNA3 site, likely due to two SNPs found at the target site. gRNA1, which targets a site at NCR53, has two mismatches in the seed region to a site at NCR54 and a PAM-disrupting mutation. Similarly, gRNA2 has two mismatches to a site at NCR54, one in each of the 5′ and 3′ ends of the seed region. No off-target mutations were observed from gRNA1 and gRNA2 at the NCR54 locus in the 46 plants, further indicating that two mismatches in the target site are sufficient to prevent off-target activity.
Table 1. Multiplex Targeted Mutagenesis in M. truncatula Using Cas9 and Csy4.
To confirm transmission of the mutations to the next generation, plant #16 (with mutations in NCR53 and NCR54), plant #17 (with mutations in NCR53 and NCR55), plant #27 (with mutations in all three targets), and plant #40 (with mutations in NCR53) were self-fertilized. Seven T1 seedlings from each T0 parent were screened by PCR and DNA sequencing for mutations in the respective genes and for presence of the bar transgene (Supplemental Figures 11A and 11B). Mutations identical to those observed in the T0 parents were found in the NCR53 locus in the progeny of all four tested plants (Supplemental Figure 11A). In two of the four plants (plant #17 and plant #40), all tested progeny only showed the mutated version of the NCR53 locus. Both the double mutant (plant #16) and the triple mutant (plant #27) also transmitted the expected mutations in NCR54 to their progeny, whereas only the triple mutant (plant #27) and not the double mutant (plant #17) transmitted the mutations in NCR55. Although we did not detect the wild-type allele of NCR53 in T0 plant #16 or the wild-type allele of NCR55 in the T0 plant #27, wild-type alleles were recovered in the T1 progeny, suggesting the T0 plants might have been chimeric. Nevertheless, the T-DNA insertion segregated away in several T1 progeny of T0 plants #16, #27 (including a triple mutant 27-6), and #40 (Supplemental Figure 11B), confirming that the mutations in these plants were transmitted through the germline rather than created de novo in the T1. Thus, we have been able to show that mutations in each of the genes are heritable. In addition, we showed that mutations in all three genes can be transmitted simultaneously from a triply mutant parent (plant #27) to its progeny. These results show the feasibility of using the Csy4 system to effectively recover heritable mutations in multiple genes.
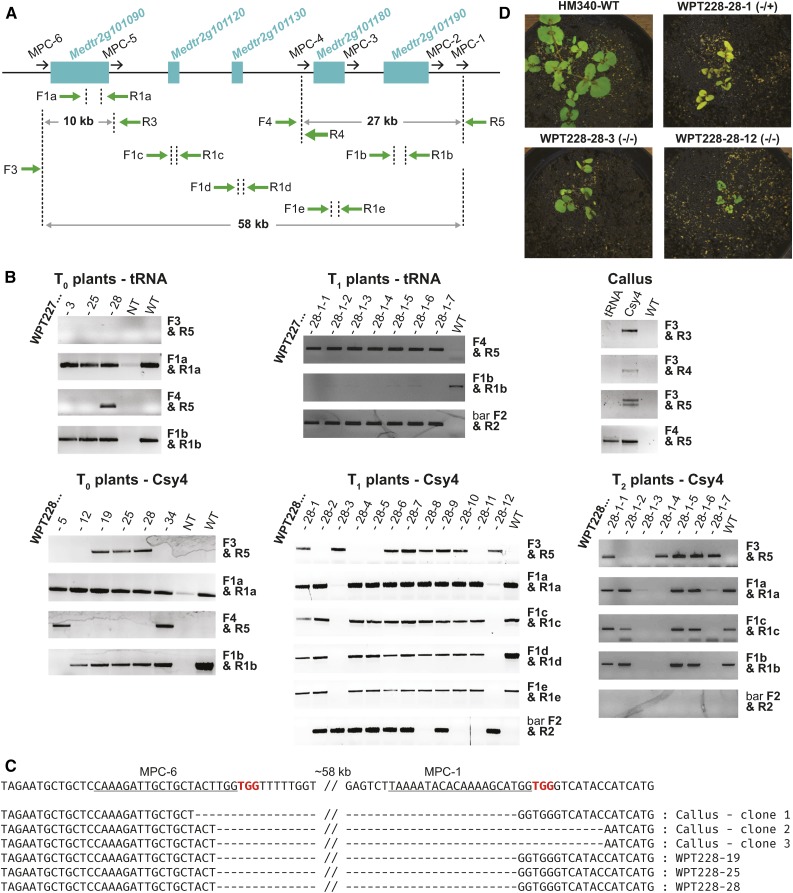
Targeted Deletion of 58 kb in M. truncatula Using the tRNA and Csy4 Multiplexing Systems
To further evaluate the Csy4 and tRNA systems for creating chromosomal deletions in M. truncatula, we designed another set of six gRNAs (MPC1 to MPC6) that target a 58-kb region on chromosome 2 (Figure 8A). The region contains five candidate genes identified by a genome-wide association study to be associated with symbiotic nitrogen fixation (Curtin et al., 2017). The gRNAs were assembled (in the order from 1 to 6) into the direct cloning T-DNA vectors that use either Csy4 or tRNA processing enzymes (see Supplemental Methods, Protocols 3A and 3B), and after transformation, DNA from randomly selected calli was tested by PCR for potential chromosomal deletions. Primers were designed 200 to 300 bp upstream and downstream of various target sites throughout the locus (Figure 8A; Supplemental Table 6). Results from this preliminary assay revealed all four possible deletions (detectable using this set of primers) in the Csy4-transformed calli, whereas only the 27-kb deletion between gRNA sites MPC1 and MPC4 was detected in the calli transformed with the tRNA multiplexing reagents (Figure 8B). Amplicons generated from putative 58-kb deletions were sequence confirmed (Figure 8C). While still in tissue culture, we prescreened 46 T0 plantlets from each of the Csy4 and tRNA transformations for the 27- and 58-kb deletions. The putatively positive plantlets were transferred to soil, grown into mature plants and screened again by PCR. Three Csy4 T0 plants were identified with the 58-kb deletion and two plants with the 27-kb deletion. For the tRNA T0 plants, one plant had the 27-kb deletion (Figure 8B). The wild-type sequence within the 58-kb deleted region could still be amplified with a different set of primers, suggesting that all plants were heterozygous or chimeric. One plant (WPT228-5) appeared to be a T0 homozygous mutant for the 27-kb deletion, since we failed to recover a PCR amplicon from the wild-type allele (Figure 8B).
Figure 8.
Targeted Deletion of 58 kb in M. truncatula.
(A) Map of the region on chromosome 2 targeted for deletion. Cyan rectangles represent individual genes. gRNA sites are shown as black arrows. Primers for detection of deletions are shown below as green arrows.
(B) PCR detection of large deletions in callus tissue, T0, T1, and T2 plants. Primers used to detect deletions of different sizes are indicated on the right of each gel picture. Primer pairs F1a/R1a, F1b/R1b, F1c/R1c, F1d/R1d, and F1e/R1e amplify the nondeleted wild-type sequence. Primers bar F2/R2 amplify the bar transgene. NT, no template control.
(C) DNA sequences of the 58-kb deletion events amplified with primers F3/R5 from callus tissue and T0 plants transformed with the Csy4 vector. The reference sequence of the unmodified locus is shown on the top. gRNA target sites are underlined and PAM sequences are highlighted in red.
(D) Phenotypes of T1 plants heterozygous and homozygous for the 58-kb deletion. Note the dwarf stature of the mutants. Images have been adjusted to minimize soil noise by decreasing the brightness of the white and neutral color channels. All images have been adjusted equally.
To test for heritable transmission of the deletions, T0 plants WPT228-28 (Csy4, heterozygote or chimeric for the 58-kb deletion) and WPT227-28 (tRNA, heterozygote or chimeric for the 27-kb deletion) were self-fertilized, and T1 plants were screened for the segregating chromosomal deletions. Eight out of 12 T1 seedlings produced bands of the expected size for the 58-kb deletion in WPT228-28 progeny, and seven out of seven T1 seedlings showed bands expected for the 27-kb deletion in WPT227-28 progeny, confirming germline transmission in these lines (Figure 8B). In addition, the wild-type allele was not detected in two out of the eight WPT228-28 T1 seedlings, suggesting the two seedlings are homozygous mutants. The bar transgene was not detected in three heterozygous WPT228-28 seedlings, indicating the loss of T-DNA insertion and confirming that the mutations were not created de novo in the T1 (Figure 8B). To determine whether we could recover homozygous mutants in a transgene-free background, T1 plant WPT228-28-1, heterozygous for the 58-kb deletion and lacking the T-DNA insertion, was self-fertilized, and seven T2 seedlings were screened for the presence of the deletion. PCR and sequencing identified two homozygous and three heterozygous nontransgenic plants (Figure 8B).
The lower overall mutation frequencies compared with those observed in the NCR gene editing experiment are likely due to the significantly larger size of deletions being sought. An inverse relationship between Cas9-mediated deletion frequency and size has been observed previously in both mammalian and plant cells (Canver et al., 2014; Xie et al., 2015; Ordon et al., 2017).
Enhancing Targeted Mutagenesis with Trex2
To increase the frequency of NHEJ-induced mutations, sequence-specific nucleases can be coupled with exonucleases to promote end resection and prevent precise ligation. Previously, coexpression of the human 3′ repair exonuclease 2 (Trex2) with a meganuclease resulted in a 25-fold enhancement in gene disruption frequencies in human cells (Certo et al., 2012). In plants, overexpression of exonuclease 1 (Exo1) significantly enhanced the frequency of DSB resection in rice (Oryza sativa) calli (Kwon et al., 2012), and Trex2 increased the mutagenesis frequency when codelivered to plant cells with a meganuclease (Luo et al., 2015). Based on these data and our observation that the majority of Cas9-induced breaks are rejoined precisely, we tested the effect on mutagenesis of expressing Trex2 with Cas9 and a single gRNA. We used gRNAs that target two different sites in the tomato ANT1 gene and one site in the barley MLO gene. Individual gRNAs were cloned under the control of the AtU6 or TaU6 promoters, combined with a dicot or monocot optimized Trex2-P2A-Cas9 fusion, and tested in tomato and barley protoplasts. We saw a modest 1.5- to 2.5-fold increase in indel frequencies in the presence of Trex2 compared with the samples without Trex2 in barley (as measured by the T7EI assay) (Figure 9A). Similar results were obtained in tomato (as measured by both T7EI and DNA sequencing) (Figures 9A and 9B). Sequencing revealed a trend toward longer deletions in the Trex2 samples in both tomato and barley (Figures 9B and 9C). Whereas no deletions ≥10 bp were detected in the samples lacking Trex2, they represented 77% (tomato) and 100% (barley) of all mutations recovered in samples expressing Trex2. Insertions of up to 42 bp were found in samples without Trex2 (Supplemental Figure 12) but not in the samples expressing Trex2, suggesting that deletions were induced preferentially in the presence of the exonuclease.
Figure 9.
Trex2 Enhances Mutagenesis in Tomato and Barley Protoplasts.
(A) T7 endonuclease I assay results for two sites in tomato and one site in barley. Two replicates (pools of protoplasts) and an average fold increase in mutagenesis frequency are shown for the tomato sites. The average fold increase in the two experiments is shown at the top.
(B) DNA sequences of NHEJ-derived mutations found in bacterial clones of PCR products that encompass the gRNA target site in the tomato ANT1 gene. Mutations are shown in 15 of 92 bacterial clones derived from tomato cells transformed with Cas9 and gRNA only and 26 of 88 bacterial clones derived from cells transformed with Cas9, gRNA, and Trex2. Sequences of three clones containing insertions at the gRNA site found in the sample lacking Trex2 are not shown. The reference sequence is shown on the top. The 20-bp gRNA target site is underlined and the PAM sequence is highlighted in red.
(C) DNA sequences of NHEJ-derived mutations found in bacterial clones of PCR products that encompass the gRNA target site in the barley MLO gene. Mutations are shown in 18 bacterial clones derived from barley cells transformed with Cas9 and gRNA only and 19 bacterial clones derived from cells transformed with Cas9, sgRNA, and Trex2. The samples were enriched for mutations by digesting the PCR amplicons with NcoI before cloning. The reference sequence is shown above; the 20-bp gRNA target site is underlined and the PAM is highlighted in red. The sequence of the NcoI recognition site is in italics. The 1-bp insertion is highlighted in green.
Gene Targeting with Cas9 Nickases
Several strategies have been developed to decrease the frequency of off-target mutagenesis by CRISPR/Cas9. Among these are the use of paired Cas9 nickases, which create a DSB by introducing nicks on opposite DNA strands. Paired nickases have on-target cleavage efficiency comparable to wild-type Cas9 in human cells and plants (i.e., Arabidopsis and rice) and reduce off-target activity up to 1500-fold (Ran et al., 2013; Schiml et al., 2014; Mikami et al., 2016). We created Cas9 nickase variants by mutating the catalytic residues in the conserved RuvC (D10A) and HNH (H840A) nuclease domains. Module A vectors were then created that express Cas9 variants with individual or combined mutations (the latter resulting in a catalytically dead Cas9 enzyme).
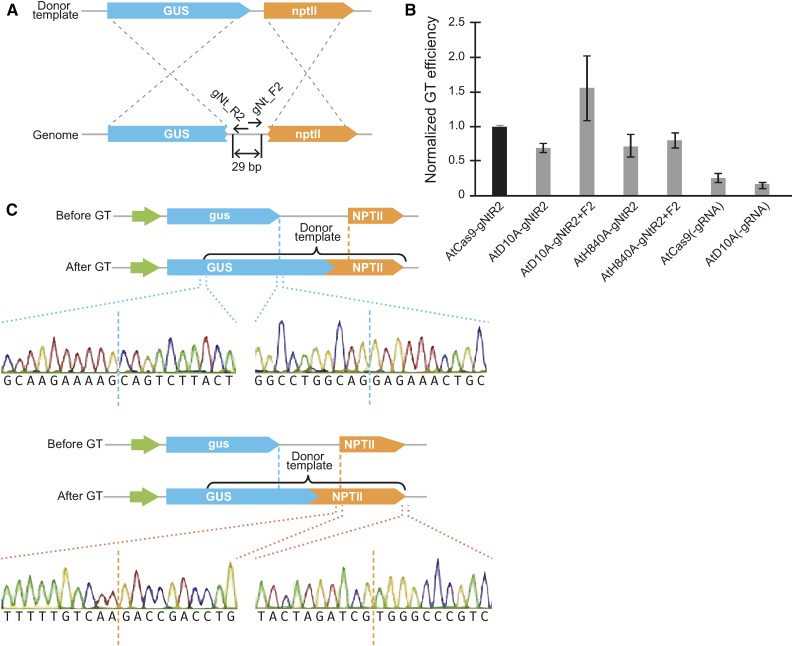
We were interested in exploring whether paired Cas9 nickases can also promote GT in plants using a previously described tobacco (Nicotiana tabacum) transient GT assay. In this assay, a defective gus:nptII transgene is restored by cleavage and homologous recombination (Figure 10A) (Wright et al., 2005; Baltes et al., 2014). We designed a pair of gRNAs in PAM-out orientation that overlapped by 5 bp (−5 bp offset) and should create a 29-bp overhang (Figure 10A). To stimulate GT, the reagents were delivered to the plant tissue on the Bean yellow dwarf virus (BeYDV) replicons (Baltes et al., 2014; Čermák et al., 2015) available in our toolkit as T-DNA transformation backbones. The readout was the number of blue (GUS positive) spots and intensity of staining in tobacco leaves (Supplemental Figure 13A).
Figure 10.
Gene Targeting with Cas9 Nickases and GVRs in Tobacco.
(A) Illustration of the gene targeting approach in tobacco. gRNAs gNt_F2 and gNt_R2 target the defective gus:nptII transgene stably inserted in the tobacco genome. The missing coding sequence is restored through homology directed repair using the donor template, resulting in a functional GUS gene.
(B) Effect of single and double nicks on gene targeting in tobacco. The Cas9 genes and gRNAs were delivered to tobacco leaves on a BeYDV replicon by agroinfiltration. The leaves were stained in an X-Gluc solution 5 d later, and blue (GUS positive) spots were quantified by image analysis. Data were normalized to the Cas9 nuclease sample. Error bars represent the se of 5 independent experiments. AtCas9, Cas9 nuclease; AtD10A, Cas9 D10A nickase; AtH840A, Cas9 H840A nickase; (-gRNA), no gRNA control.
(C) DNA sequences of the left and right recombination junctions from leaves infiltrated with the D10A paired nickase construct. Multiple bacterial clones were sequenced and were identical. Junctions at both ends of each homology arms are shown.
We first compared the GT frequencies induced by a single nick or DSB. The number of GT events induced by single nicks was around 70% of the GT events induced by a DSB (Figure 10B). No significant differences in GT frequencies were observed between the D10A and H840A nickases. Then, we compared the GT efficiencies using paired nickases that induce double nicks resulting in 29-bp 5′ overhangs (AtD10A) and 3′ overhangs (AtH840A). We observed 2-fold increase in GT with 5′ overhangs compared with 3′ overhangs. The number of GT events induced by double nicks creating 5′ overhangs was even higher than the number of GT events induced by the wild-type Cas9 nuclease. This is in agreement with observations in human cells (Ran et al., 2013).
To confirm the nature of the GT events on the molecular level, we extracted DNA from infiltrated tobacco leaves and PCR amplified both junctions of the inserted sequence (Supplemental Figure 13B). All samples except the one that was infiltrated with the vector expressing noncutting gRNAs, yielded products of expected size for both junctions. Six to 10 bacterial clones for each junction from each sample were analyzed by sequencing, which revealed precise integration of the donor template in all clones (Figure 10C). These data suggest that the overall frequency and precision of GT induced by nickases is comparable to the nuclease-based approach, which we have previously shown can be used to efficiently recover plants with heritable modifications created by GT (Baltes et al., 2014; Čermák et al., 2015).
To see whether the nickase approach is applicable for GT in a monocot species, we targeted an in-frame insertion of a T2A-GFP cassette into the third exon of the ubiquitin gene in wheat scutella, using Wheat dwarf virus (WDV) replicons as described by Gil-Humanes et al. (2017). We observed GFP expression consistent with insertion into the ubiquitin locus (Supplemental Figures 14A to 14C), suggesting the feasibility of this approach across different species.
DISCUSSION
Genome engineering promises to advance both basic and applied plant research, and we developed a comprehensive reagent toolkit to make the latest genome engineering technologies available to the plant science community. Both TALENs and CRISPR/Cas9 can be easily combined with a variety of regulatory sequences, transformation vectors, and other DNA modifying enzymes, making it possible to rapidly assemble reagents necessary to make single and multigene knockouts as well as gene replacements.
Using our flexible, modular system, we evaluated four different multi-gRNA expression strategies in tomato protoplasts to achieve multiplexed editing. A polycistronic gRNA transcript, when processed by the bacterial Csy4 ribonuclease, resulted in almost 2 times higher mutation frequencies compared with the commonly used strategy in which each gRNA is expressed from an individual Pol III promoter. Furthermore, the position of the gRNAs in the array has only a modest effect on activity, and the last gRNAs in an array of eight perform comparably to gRNAs expressed independently from Poll III promoters. Although the frequencies observed with Csy4 were comparable to the previously described tRNA processing system (Xie et al., 2015), we favor the Csy4 system because it consistently gave us higher frequencies of deletions in tomato protoplasts, and the multi-gRNA transcripts are shorter and less repetitive (i.e., Csy4 recognition sites are 20 bp versus 77 bp for tRNAs). Although the Csy4 system requires expression of the Csy4 ribonuclease in the plant cell, and no extra factors are required for tRNA processing, the Csy4 gene is relatively short (561 bp = 187 amino acids), and we typically express it as P2A fusion to Cas9 without any observable phenotypic consequences to transgenic plants.
Although we have previously shown that gRNA transcripts processed by ribozymes efficiently induce mutagenesis in rice, tobacco, and Arabidopsis (Tang et al., 2016), of the polycistronic RNA processing mechanisms tested in this study, ribozymes resulted in the lowest mutation frequencies. One difference between the two studies is that we expressed Cas9 and the gRNA array from separate promoters, whereas Tang et al. (2016) used a single transcription unit to express both the Cas9 and the gRNAs. Furthermore, the frequency of deletions created by two gRNAs was not directly measured in the previous study. It is possible that coordinate expression of Cas9 and gRNAs from a single transcription unit, which is not possible in the current version of our cloning system due to its modular structure, might be essential to achieve higher gene editing frequencies using ribozyme processing. Whether this is true remains to be elucidated. Nevertheless, we demonstrate Csy4 and tRNA expression systems efficiently process multi-gRNA transcripts and enable high efficiency multiplexed gene editing.
Analysis of mutations resulting from cleavage of a single gene with two gRNAs revealed that ∼85% of DSBs were precisely ligated. This is consistent with the high frequency of precise repair observed in mouse and human cells (Li et al., 2015; Geisinger et al., 2016). Since DSBs induced by a single gRNA are also likely accurately repaired, we favor the use of two gRNAs to induce loss-of-function mutations. Precise repair can be used to your advantage to make the mutation of choice, and there are no concerns about creating functional gene isoforms through non-frameshift indels or alternative splicing. Furthermore, the targeted deletions can be detected by PCR, making the screening for mutants significantly easier. This is particularly valuable, if not essential, when simultaneously mutagenizing several genes, as screening for loss of restriction enzyme sites, T7 assays, or direct DNA sequencing is time-consuming and costly. Targeted deletions might also be valuable for editing noncoding sequences, including promoters and regulatory sequences, where short indel mutations may not be effective. Moreover, engineering complex traits will likely require an efficient multiplexed editing platform. Here, we show that combinations of different gene deletions can be induced in a single experiment using a single vector, highlighting the potential of this approach to speed up candidate gene evaluation. Using our Csy4-based system, we successfully deleted up to six genes in tomato, wheat, and barley protoplasts and in M. truncatula plants, demonstrating the versatility of this system across a variety of plant species. Finally, the high level of precision in NHEJ repair also opens up an opportunity for targeted and precise gene insertion/replacement, independent of HDR (Geisinger et al., 2016).
As an alternative to knocking out genes with two or more gRNAs, we also show that Trex2, in combination with Cas9, increases the length and frequency (2.5-fold) of NHEJ-mediated deletions in both tomato and barley. A single study that combined Trex2 with Cas9 for gene editing in human cells reported a similar 2.5-fold increase in mutagenesis (Chari et al., 2015). When Trex2 was used in combination with TALENs, a 3- to 4-fold enhancement was observed (Certo et al., 2012; Luo et al., 2015), whereas a 25-fold increase was achieved when a meganuclease was used as the DSB-inducing agent (Certo et al., 2012). The differences in fold enhancement between the three types of nucleases may be caused by different levels of Trex2 activity on different types of DNA ends induced by Cas9 (blunt), TALENs (5′ overhang), or meganucleases (3′ overhang, the preferred substrate of Trex2). Interestingly, Certo et al. (2012) detected a bias toward small deletions when they combined Trex2 with meganucleases and TALENs, but the study of Chari et al. (2015) as well as our data indicate that the deletion length increases when Trex2 is used with the CRISPR/Cas9 system. Nevertheless, deletions seem to be induced preferentially in the presence of Trex2, regardless of the type of nuclease used to create the DSB since the lower frequency of insertions compared with the no Trex2 control is consistent in all studies.
Cas9 nickases are frequently used because they reduce the risk of off-target mutations (Ran et al., 2013; Cho et al., 2014; Shen et al., 2014; Mikami et al., 2016). Single nickases do not induce detectable levels of on-target NHEJ-mediated mutagenesis (Cong et al., 2013; Ran et al., 2013; Fauser et al., 2014), whereas paired nickases, which create two adjacent nicks in the target sequence, induce mutations at efficiencies comparable to native Cas9 (Ran et al., 2013; Schiml et al., 2014; Mikami et al., 2016). Paired nickases also efficiently induce gene targeting in human cells (Mali et al., 2013; Ran et al., 2013), and, here, we demonstrate that paired nickases constructed using our modular system induce gene targeting in tobacco and wheat with efficiencies comparable to the nuclease. Consistent with the data from human cells (Mali et al., 2013; Ran et al., 2013), paired nickases that create 5′ overhangs induced GT more frequently than those creating 3′ overhangs. In a separate study (Fauser et al., 2014), single nickases were found capable of promoting GT in Arabidopsis at frequencies exceeding the nuclease; however, we observed that GT frequencies with single nickases were above background in both tobacco and wheat, but they were lower than the GT frequency achieved with the nuclease and paired nickases. The high frequency of GT observed by Fauser et al. (2014) with a single nick may in part be due the proximity of the donor template, which was integrated into the chromosome adjacent to the nick site. Indeed, significantly lower GT frequencies were observed in human cells when single nickases were used with extrachromosomal donors (Ran et al., 2013), although other factors such as donor architecture and cell type could contribute to the observed differences between studies. Nevertheless, we show that paired nickases, which have been shown to have minimal off-target effects in plants (Mikami et al., 2016), can be used for efficient GT in both monocots and dicots.
A number of different toolkits for plant genome engineering have been published within the past two years (Xing et al., 2014; Xie et al., 2015; Ma et al., 2015; Lowder et al., 2015; Zhang et al., 2016; Liang et al., 2016; Vazquez-Vilar et al., 2016; Kim et al. 2016; Ordon et al., 2017). However, all have limitations: Most do not offer flexibility in the gene editing platform (TALENs or CRISPR/Cas9); some reagents are optimized for a non-target organism (e.g., humans); typically, there is little choice in promoters for expressing targeting reagents; most systems are limited to a single vector type (e.g., T-DNA); applications are limited to NHEJ-mediated gene knockouts; and most reagents are validated in only one or two model species. Although two studies have made their approaches compatible with a wider variety of vectors and reagents, such as the D10A Cas9 nickase and Cas9-based transcriptional activators and repressors (Lowder et al., 2015; Vazquez-Vilar et al., 2016), they remain limited in other aspects. For example, the Lowder et al. (2015) toolkit limits gRNA expression to Pol III promoters, requires an 8-d assembly process, and lacks cross-compatible and adaptable expression vectors; the Vazquez-Vilar et al. (2016) toolkit requires a time-consuming binary cloning approach and lacks substantial evaluation of the multiplexed gene editing vectors. In comparison, our plant genome engineering platform provides the user with an option to choose the desired reagent from a comprehensive set of expression cassettes, including TALENs, TALE-VP64, TALE-VPR, Cas9, D10A Cas9 nickase, H840A Cas9 nickase, dCas9, dCas9-VP64 and dCas9-VPR, GFP, Trex2, and Csy4 with versions codon optimized for use in dicots or monocots. For Cas9-based genome editing, the toolkit enables assembly of vectors with all gRNA expression systems described to date, including two dicot and four monocot Pol III promoters. Furthermore, multiplexed systems based on Csy4, tRNA, and ribozymes are available that can be combined with Pol II promoters. All reagents can be assembled into T-DNA or non-T-DNA vectors with or without one of the three most common plant selectable markers driven by dicot or monocot promoters. Faster direct or flexible modular cloning protocols are available. While previous studies focused on evaluating reagents in the model species Arabidopsis, rice, and tobacco, here we provide evidence that in addition to tobacco, our system is functional and effective in two important dicot and two monocot crop species.
To the best of our knowledge, our toolkit is the first designed to facilitate gene targeting through homologous recombination. We include GVR-based vectors, which were first demonstrated to work in proof of concept experiments in tobacco (Baltes et al., 2014) and more recently to effectively introduce heritable targeted insertions into the tomato genome at frequencies 3- to 9-fold higher than standard T-DNA (Čermák et al., 2015). We have also used WDV replicons to target insertion of a selectable marker into the wheat ubiquitin locus (Gil-Humanes et al., 2017); here, we show GVRs work in combination with paired nickases. Moreover, our modular system is also compatible with the in planta gene targeting approach described earlier (Fauser et al., 2012). An integral element in a gene targeting strategy is the DNA donor template, which carries custom modifications being introduced in the target genome. All of the module C plasmids in our toolkit include a unique restriction site for insertion of the donor template, and we provide protocols for assembly of the donor template plasmids for both the replicon and in planta gene targeting methods (see Supplemental Methods, Protocol 4). All together, our toolkit enables the most efficient plant gene targeting methods currently available.
In addition to the commonly used 35S and maize ubiquitin promoters, the current version of our cloning system includes seven other dicot and three monocot promoters. These include constitutive viral, bacterial (CmYLCV, M24, FMV, and nos), or plant (AtUbi10, OsAct1, PvUbi1, and PvUbi2) promoters, which provide a range of expression strengths in a variety of plant species (Stavolone et al., 2003; Sahoo et al., 2014; Maiti et al., 1997; Norris et al., 1993; McElroy et al., 1990; Mann et al., 2012). We have also included the Arabidopsis Ec1.2 and YAO promoters, which are egg cell and embryo sac/pollen specific, respectively. Although Arabidopsis was not our target species in this study, these two promoters have been shown to enhance mutagenesis in Arabidopsis when used to drive Cas9 expression (Wang et al., 2015; Yan et al., 2015). In our toolkit, these promoters can drive either Cas9 expression or the polycistronic gRNA arrays. Due to the universal structure of the modular vectors, new combinations of promoters and coding sequences can be easily added. The versatility of the toolkit is further increased by its optimization for Golden Gate assembly, one of the most popular DNA assembly methods. We have made the T-DNA backbones derived from the commonly used pCAMBIA vectors free of most relevant type IIs restriction sites, AarI, BsaI, Esp3I, and SapI. AarI, SapI, and BsaI sites were also removed from the CmYLCV, PvUbi1, and PvUbi2 promoters, and the activity of the modified promoters was verified in plants to ensure these modifications do not affect expression levels. Since the Golden Gate modules can be easily modified to contain other genes, the system can be adopted for applications beyond genome engineering, such as synthetic biology.
Finally, the ability to alter transcription levels of endogenous genes is often desirable. Transcriptional activation domains have been fused to both TALEs and Cas9 to enable targeted gene activation (Lowder et al., 2015; Vazquez-Vilar et al., 2016). Although these custom transcription factors have proven effective (W. Liu et al., 2014; Piatek et al., 2015; Lowder et al., 2015; Vazquez-Vilar et al., 2016), they have mostly been tested on reporter genes driven by synthetic or minimal viral promoters. Therefore, the increase in expression levels might not be sufficient to induce phenotypic changes when endogenous genes are targeted (Lowder et al., 2015). To overcome the modest levels of gene activation, Chavez et al. (2015) recently showed that fusions of dCas9 or TALE proteins to a strong transcriptional activator VPR mediate up to 320-fold higher activation of endogenous targets compared with fusions to VP64 alone. Although further optimization of artificial plant transcriptional regulators is needed, at present, we provide modules with fusions of both VP64 and VPR activator domains to TALEs and dCas9 for transcriptional activation, as well as modules with dCas9 only, which has been shown to be equally effective in transcriptional repression as dCas9 fusions to repressor domains (Piatek et al., 2015; Vazquez-Vilar et al., 2016).
In conclusion, we would like to note that due to our toolkit’s modular design, it can be easily updated with new functions, and we intend to incorporate new tools as they become available. Given the ease and speed of reagent construction and the availability of user-friendly protocols and online tools, we hope our platform will accelerate progress in plant functional genomics and crop improvement.
METHODS
Direct and Modular Vector Library Construction
Direct TALEN cloning vectors pDIRECT_37-39J were first constructed by Gibson assembly (Gibson et al., 2009) of PCR products containing parts of pCLEAN-G126 backbone (Thole et al., 2007; GenBank accession no. EU186082), selection marker cassettes, the OsADH 5′ untranslated region amplified from the rice (Oryza sativa cv Nipponbare) genome (Sugio et al., 2008), 35S promoter, δ152/+63 TALE scaffolds including 3xFLAG epitope and SV40 NLS, Esp3I flanked ccdB cassette, BsaI-flanked LacZ cassette, ELD and KKR heterodimeric FokI variants (Doyon et al., 2011), and Arabidopsis thaliana heat shock protein 18.2 (HSP) terminator amplified from the Arabidopsis genome (Nagaya et al., 2010). In the process, Esp3I recognition sites were removed from the nptII gene and the ELD FokI coding sequence, BsaI from the ccdB gene, and the BglII site from the HSP terminator. Unique restriction sites were inserted to flank the promoter and terminator in the dual TALEN expression cassette. TALE sequences from pTAL plasmids (Cermak et al., 2011) were used and 19 bp of sequence immediately upstream of the first Esp3I site in TALEN1 and downstream of the second BsaI site in TALEN2 were recoded to include unique primer binding sites for colony PCR and sequencing of both TALE repeat arrays. pCAMBIA-based vectors pDIRECT_21-23J were constructed accordingly, except they were first made with an AscI and SacI flanked ccdB cassette, later replaced with the AscI/SacI fragment from the pCLEAN-based vectors containing the dual TALEN expression cassette; and unique restriction sites were inserted to flank the selection marker gene. For expression in monocots, the ZmUbi1 promoter was PCR amplified from pAHC25 (Christensen and Quail, 1996) and cloned between AscI and SbfI sites of pDIRECT_21J and pDIRECT_23J to replace the 35S promoter and create pDIRECT_25K and pDIRECT_26K.
To create the first module plasmids, the BsaI LacZ cassette in pTC102 (a derivative of pDIRECT_37J with translation initiation sequence optimized for dicots) was replaced with a second Esp3I ccdB cassette to create pTC163. The first and second TALEN cassettes were PCR amplified from pTC163 (splitting the P2A sequence between the two cassettes and removing the AarI site from it) and a BaeI site containing fragment was amplified from pTC130 (Čermák et al., 2015), using three pairs of primers containing AarI sites with distinct 4-bp overhangs. The first three module plasmids pMOD_A1001, pMOD_B2000, and pMOD_C0000 were constructed by inserting each of these three PCR fragments into BsaXI and PmlI-linearized pTAL3 (Cermak et al., 2011) using Gibson assembly. To generate the first CRISPR/Cas9 modules pMOD_A0101 and pMOD_B2515, PCR fragments containing 35S promoter, AtCas9 gene codon-optimized for Arabidopsis (Fauser et al., 2014) and the HSP terminator, or the AtU6 gRNA cassette were inserted into AarI-linearized pMOD_A1001 and pMOD_B2000 by Gibson assembly. Similarly, an At7SL gRNA cassette was inserted into NaeI and XhoI cut pMOD_C0000 to create pMOD_C2516. All remaining module plasmids were derived from these initial plasmids through replacing the gene cassettes and promoters using the unique restriction sites AscI, SbfI, XhoI, and SacI, or in case of multi-gRNA plasmids, through Golden Gate assembly of PCR products containing the specific elements. TaCas9 was codon optimized for wheat (Triticum aestivum) and Gibson assembled from nine gBlocks (Integrated DNA Technologies). Cas9 nickases and catalytically dead variants were made by Gibson assembly of PCR products containing the specific mutations into the Cas9 nuclease modules. Cas9 and TALE transcriptional activators were constructed by Gibson assembly of fragments into TALEN and dCas9-containing modules. The VPR activators were built first, using the tripartite VPR transcription activation domain described by Chavez et al. (2015). The VP64 activators were derived from the VPR modules by deleting the p65 and RTA domains by restriction cloning. The Csy4 genes, codon-optimized for dicots (tomato [Solanum lycopersicum] + Arabidopsis) and monocots (wheat and rice), were synthesized as gBlocks along with the P2A ribosomal skipping sequence and Gibson assembled into SbfI- and SalI-digested pMOD_A0101 and SbfI- and BstBI-digested pMOD_A1510, respectively. Trex2 was cloned similarly, except it was PCR amplified from pExodus CMV.Trex2 plasmid (Certo et al., 2012) containing the mouse version of the gene that we previously observed had robust activity in plant cells. The CmYLCV (without the AarI site) and FMV promoters were synthesized as gBlocks. The PvUbi1 and PvUbi2 promoters were amplified from pANIC10A (Mann et al., 2012), each as a set of overlapping PCR fragments to enable removal of AarI, BsaI, and SapI sites through Gibson assembly. The Nos promoter and rbcsE9 and 35S terminators were amplified from pFZ19 (Zhang et al., 2010) and pCAMBIA vectors. The remaining promoter fragments were obtained by PCR amplification from the respective genomic DNAs. The tRNAGly sequences used to create the multi-gRNA constructs were amplified from tomato cv MicroTom genomic DNA, while the Csy4 and ribozyme sequences were synthesized as parts of primers for Golden Gate assembly. Finally, the empty modules pMOD_A0000 and pMOD_B0000 were created by ligating annealed oligonucleotides carrying unique restriction sites into the AscI and SacI sites in pMOD_A0101 and AscI and AgeI sites in pMOD_B2103.
The T-DNA transformation backbones were derived from pCAMBIA1300, 2300, and 3300 vectors by Gibson assembly of the CmR/ccdB selection cassette amplified from pMDC32 (Curtis and Grossniklaus, 2003) using primers carrying AarI sites specifying 4-bp overhangs designed to accept inserts from the three modules, into HindIII- and BamHI-digested pCAMBIA backbones. In addition, pTRANS_210d-250d are modified versions of the resulting vectors with AarI, BsaI, Esp3I, and SapI sites removed from the vector backbone by Gibson assembly of backbone fragments overlapping at the mutated sites. pTRANS_250d and pTRANS_260d contain PvUbi1 and PvUbi2 promoters that were directly assembled into these vectors as described above. All non-T-DNA transformation backbones are derived from pTRANS_100. This vector was created de novo by Gibson assembly of two PCR products, one containing the spectinomycin resistance gene and bacterial origin of replication amplified from pTC14 (Cermak et al., 2011) and the other containing the AarI-flanked CmR/ccdB selection cassette from the T-DNA transformation vector pTRANS_210. The selection marker cassettes were then inserted into this vector by restriction cloning from the T-DNA transformation backbones. The GVR transformation vectors were built by Gibson assembly from modified pCAMBIA backbone (without BsaI and Esp3I sites) and geminivirus elements described by Baltes et al. (2014), Čermák et al. (2015), and Gil-Humanes et al. (2017). All CRISPR/Cas9 direct cloning and Cas9 only vectors were assembled by Golden Gate assembly from modules A, B, and C. To create the Cas9 only vectors, the transformation backbones were modified to accept the insert from a single module.
Gene Editing in Medicago truncatula
For targeted mutagenesis of the Medtr4g020620 and Medtr2g013650 genes, a pair of TALENs was assembled into pDIRECT_39J using Protocol 1B (Supplemental Methods), resulting in pSC1 and transformed into an Agrobacterium tumefaciens strain EHA105 for whole-plant transformation of the M. truncatula accession HM340 using an established protocol (Cosson et al., 2006). The resulting plants were screened for putative mutations using a PCR digestion assay with primers listed in Supplemental Table 6 and the HaeIII restriction enzyme. The digestion-resistant products were cloned and several clones were sequenced to identify the mutations. For combinatorial deletion of NCR genes, the six gRNAs were assembled into pDIRECT_23C along with the CmYLCV promoter using Protocol 3A (Supplemental Methods), resulting in pSC2 and transformed into A. tumefaciens EHA105. To detect the targeted gene deletions and short indels at each gRNA site, the three loci were PCR amplified from T0 and T1 plants using primers listed in Supplemental Table 6. The PCR products were first directly sequenced. PCR products from samples containing both the long deletion and nondeletion alleles were gel purified before sequencing and sequenced separately. Samples that showed overlapping sequence traces starting at either gRNA site indicative of indel mutations were cloned and several clones were sequenced to identify the type of mutations. For targeted deletion of the 58-kb region on chromosome 2, the six gRNAs were assembled as Csy4 or tRNA arrays driven by the CmYLCV promoter into pDIRECT_23C using Protocols 3A and 3B (Supplemental Methods), yielding pSC3 and pSC4, respectively, and transformed into M. truncatula accession HM340 as above. Targeted deletions in the resulting T0, T1, and T2 plants were detected using primers listed in Supplemental Table 6, cloned, and sequenced.
Determining the Efficiency of Multi-gRNA Expression Systems in Tomato Protoplasts by Deep Sequencing
To build vector pTC303, expressing the two gRNAs from AtU6 and At7SL promoters, and pTC363 expressing each gRNA from an individual AtU6 promoter, the gRNA spacers were first cloned into pMOD_B2515 (module B with AtU6 promoter, used for both pTC303 and pTC363), pMOD_C2515 (module C with AtU6 promoter, used for pTC363), and pMOD_C2516 (module C with At7SL promoter, used for pTC303) using Protocol 2A (Supplemental Methods). The resulting plasmids were assembled together with pMOD_A0101 into the pTRANS_100 protoplast vector using Protocol 5 (Supplemental Methods). pAH750, expressing the gRNAs with structurally optimized scaffolds from AtU6 and At7SL promoters, was constructed accordingly, except the gRNA spacers were cloned into pMOD_B2515b and pMOD_C2516b, the respective module B and C plasmids that carry the optimized gRNA repeats. The Csy4, tRNA, and ribozyme expression cassettes were created directly in protoplast vectors pDIRECT_10C (Csy4) and pDIRECT_10E (tRNA and ribozyme) using Protocols 3A, 3B, and 3C (Supplemental Methods), resulting in pTC364 (Csy4), pTC365 (tRNA), and pTC366 (ribozyme). Next, protoplasts were isolated from tomato cv MicroTom as described by Zhang et al. (2013), with minor modifications. Leaves from one to two sterile grown (26°C with 16 h light) ∼4-week-old plantlets were sliced into thin strips with a sharp razor, transferred to the enzyme solution (1.0% cellulase R10, 0.25% macerozyme R10, 0.45 M mannitol, 20 mM MES, 1× MS salts, and 0.1% BSA), and incubated 15 h at 25°C and 40 rpm in the dark. The digested product was filtered through a 40-μm cell strainer (Falcon) and pipetted directly on top of 0.55 M sucrose solution. The rest of the protocol followed Zhang et al. (2013), except W5 solution (2 mM MES, 154 mM NaCl, 125 mM CaCl2, and 5 mM KCl) was used as washing buffer. The CRISPR/Cas9 constructs were cotransformed into ∼200,000 cells along with the YFP expression plasmid pZHY162 (Zhang et al., 2013), the protoplasts were resuspended in W5 solution and kept at 25°C in the dark. For all samples 2 d later, the transformation efficiency was determined by counting the YFP-positive cells using image analysis, and genomic DNA was isolated and used as a template for the deep sequencing library preparation, as described by Čermák et al. (2015). Briefly, 40 ng of protoplast genomic DNA was used as template to amplify a 354-bp region of the ARF8A locus encompassing the gRNA9 and gRNA10 target sites with primers TC324F and TC324R (Supplemental Table 6), which have overhangs complementary to Nextera XT indices. PCR products were purified with Agencourt AMPure XP beads (Beckman Coulter) and 5 μL of the purified PCR product was used as template for the second PCR to attach dual indices and Illumina sequencing adapters. PCR products were purified with Agencourt AMPure XP Beads, quantified, and pooled. The pooled libraries were enriched for fragment sizes in the range 250 to 700 bp using BluePippin (Sage Science) and sequenced on an Illumina MiSeq platform (ACGT). An average of 140,000 reads per sample was generated (Supplemental Table 1; http://www.ebi.ac.uk/ena/data/view/PRJEB13819). Sequence data analysis was done as described previously (Čermák et al., 2015), except different sequence variants were manually sorted into specific groups. All data are presented as the mean of three biological replicates (three separate pools of protoplasts) ± se. To test for the significance of the difference between the samples, Tukey test for multiple comparison of means in R (version 3.2.4 RC) was used (Supplemental Table 4).
Testing Multi-gRNA Expression Systems in Tomato Protoplasts by qPCR
The Csy4, tRNA, and ribozyme arrays of eight gRNAs (Supplemental Table 4) were created directly in protoplast vectors pDIRECT_10C (Csy4) and pDIRECT_10E (tRNA and ribozyme) using Protocols 3A, 3B, and 3C (Supplemental Methods), resulting in pTC634 (Csy4, gRNA order 49-56), pTC635 (Csy4, gRNA order 56-49), pTC636 (tRNA, gRNA order 49-56), pTC637 (tRNA, gRNA order 56-49), pTC638 (ribozyme, gRNA order 49-56), and pTC639 (ribozyme, gRNA order 56-49). To construct the vector with eight gRNAs each expressed from an individual AtU6 (Pol III) promoter, four AtU6:gRNA cassettes were first assembled into each pMOD_B2203 and pMOD_C2200 (resulting in pTC643 and pTC644) using Protocol 3, except the first, last, and each reverse primer were redesigned to bind the AtU6 sequence. The AtU6 cassettes were amplified from BanI-digested pTC363 (first cassette), a HindIII and SgrAI fragment from pTC363 (intermediary cassettes), and a BsaHI fragment from pTC363 (last cassette). The 35S terminator was removed from pTC644 by SnaBI/Eco53kI-mediated release of the terminator fragment and religation of the backbone, resulting in pTC645. pTC643 and pTC645 were assembled along with pMOD_A0101 into the non-T-DNA vector pTRANS_100 using Protocol 5, resulting in the final vector pTC646.
The CRISPR/Cas9 constructs were transformed into tomato protoplasts as described above. Purified protoplast DNA was extracted 48 h later and used with Luna Universal qPCR Master Mix (NEB) following the manufacturer’s protocol. All samples were run in technical triplicate of two biological replicates (two separate pools of protoplasts) in a 384-well format on a LightCycler 480 Instrument (Roche). All qPCR primers are listed in Supplemental Table 6. Primers for the genomic locus SGN-U346908 control were previously described (Expósito-Rodríguez et al., 2008). All data are presented as the mean ± se with each sample relative to the levels of total DNA as measured by the genomics locus control.
Gene Editing in Tomato, Wheat, and Barley Protoplasts
To build the tomato construct pTC362 with 12 gRNAs targeting six genes, two arrays of six gRNAs were cloned into pMOD_B2203 (with CmYLCV promoter) and pMOD_C2200 using Protocol 3S1 and the two arrays were assembled together with the Csy4-Cas9 module pMOD_A0501 into the non-T-DNA transformation backbone pTRANS_100 using Protocol 5 (Supplemental Methods). The wheat vector pJG612 was built accordingly using the version of Cas9 optimized for monocots in pMOD_A1510, a single array of six gRNAs driven by the PvUbi1 promoter in pMOD_B2112 and an empty module pMOD_C0000. Similarly, pMOD_A1510, pMOD_B 2112 (PvUbi1), or pMOD_B2103 (CmYLCV), the empty pMOD_C0000 modules, and the transformation backbone pTRANS_100 were used to construct the barley (Hordeum vulgare) vectors expressing two gRNAs from the PvUbi1 (pTC441) or CmYLCV (pTC437) promoters, targeting for deletion the MLO gene. Tomato protoplasts were isolated and transformed as described above. Wheat protoplasts were isolated from ∼10-d-old plantlets of the bread wheat cv Bobwhite 208. Seeds were surface sterilized by rinsing in 70% ethanol for 10 min and soaking for 30 min in a 1% sodium hypochlorite solution and aseptically grown in MS medium at 24°C under a 16-h light cycle. Approximately 20 plantlets were harvested in each experiment, cut into ∼1-mm strips with a razor blade, and digested with an enzyme solution (1.5% cellulase R10, 0.75% macerozyme R10, 0.6 M mannitol, 10 mM MES, 10 mM CaCl2, and 0.1% BSA, pH 5.7). The remaining steps were identical as in the protocol for protoplast isolation from tomato described above. Barley protoplasts were isolated from cv Golden Promise using the same protocol, except leaves from 9- to 14-d-old plantlets (22°C with 16-h light cycle) were used. The CRISPR/Cas9 constructs were cotransformed into ∼200,000 cells along with the GFP expression plasmid pMOD_A3010, and the protoplasts were resuspended in W5 solution and kept at 25°C in the dark. Genomic DNA was isolated 2 d after transformation, and targeted deletions were detected using primers listed in Supplemental Table 6, cloned, and sequenced.
Gene Editing with Trex2
Individual gRNAs were cloned into pMOD_B2515 (AtU6 promoter, for tomato) and pMOD_B2518 (TaU6 promoter, for barley) using Protocol 2A and assembled with 35S:Trex2-P2A-AtCas9 in pMOD_A0901 (for tomato) or ZmUbi:Trex2-P2A-TaCas9 in pMOD_A1910 (for barley) and the empty module pMOD_C0000 into the protoplast transformation backbone pTRANS_100 to create pTC391 (gRNA23, tomato ANT1), pTC393 (gRNA24, tomato ANT1), and pTC436 (gRNA38, barley MLO) using Protocol 5. The control plasmids pTC392 (gRNA23, tomato ANT1), pTC394 (gRNA24, tomato ANT1), and pTC440 (gRNA38, barley MLO) without Trex2 were constructed accordingly, except 35S:AtCas9 in pMOD_A0101 and ZmUbi:TaCas9 in pMOD_A1110 were used instead of the Trex2 containing modules. The constructs were transformed into tomato and barley protoplasts as described above, and genomic DNA was extracted 2 d later. The ANT1 and MLO loci were amplified by PCR (see Supplemental Table 6 for primers), and the gene editing rates were determined using T7 endonuclease I according to the manufacturer’s protocol (NEB). Gel images were analyzed using ImageJ (Schneider et al., 2012). In addition, the ANT1 PCR products were cloned and sequenced to determine both frequency and type of mutations. For barley, the PCR products were first digested with NcoI to enrich for mutations, and only the digestion-resistant product was gel purified, cloned, and sequenced.
Gene Targeting with Cas9 Nickases in Tobacco and Wheat
Individual gRNA spacers were cloned in module B (pMOD_B2515 for tobacco [Nicotiana tabacum] or pMOD_B2518 for wheat) or C (pMOD_C2516 for tobacco or pMOD_C2518 for wheat), downstream of an AtU6 and At7SL or two TaU6 promoters. The GT donors were inserted into the resulting module C plasmids using Protocol 4 (Supplemental Methods). A promoter-less T2A-gfp-nos terminator cassette with 747- and 773-bp-long flanking homology arms, and a 2530-bp-long functional gus:nptII gene fusion were used as donor templates for wheat and tobacco, respectively. Finally, the B and C modules were assembled with selected module A Cas9 variants (nuclease, D10A nickase, H840 nickase, or dead Cas9 expressed from either 35S or ZmUbi promoter into a BeYDV [pTRANS_201] or WDV [pTRANS_203] replicon vectors) and used for Agrobacterium infiltration of tobacco leaves (pJG363, pJG365, pJG367, pJG369, pJG370, pJG380, and pJG382) or biolistic bombardment of wheat scutella (pJG455, pJG456, pJG480, pJG481, pJG484, pJG486, pJG488, and pJG493). Note that plasmids pJG455-pJG493 above used the octopine synthase terminator sequence instead of the more efficient HSP terminator used in the published modules. The TaCas9 coding sequence and the rest of the plasmids were identical with the newer modules included in the current vector set. Four different transgenic lines (Wright et al., 2005) were used to determine gene targeting frequencies in tobacco as previously described (Baltes et al., 2014), with minor modifications. Leaves from 1 to 6 weeks old plants were used for Agrobacterium leaf infiltration. For each leaf, one-half was infiltrated with a control plasmid pLSLZ.D.R (Baltes et al., 2014) that served as a reference to control for transformation efficiency and the other half with one of the CRISPR/Cas9 constructs. Four to six leaves were infiltrated with each construct in each experiment. Five days after infiltration, the leaf tissue was stained in the X-Gluc solution. Whole leaves were scanned and the stained area was quantified by image analysis using ImageJ (Schneider et al., 2012). The GT frequencies were normalized to the pLSLZ.D.R control and displayed relative to the Cas9 nuclease construct. For molecular analysis, genomic DNA was extracted from two infiltrated leaves per sample (three leaf punches were taken from each leaf and pooled) and used as a template for PCR to amplify the left and right recombination junctions using primers listed in Supplemental Table 6. PCR products from both leaf samples were cloned and sequenced. To determine GT frequencies in wheat, immature wheat scutella (0.5–1.5 mm) were isolated from primary tillers harvested 16 d after anthesis and used for biolistic transformation. Scutella isolation and culture conditions were as described by Gil-Humanes et al. (2011). Biolistic bombardment was performed using a PDS-1000 gene gun. Equimolar amounts (1 pmol DNA mg−1 of gold) of plasmid were used for each experiment with 60 μg of gold particles (0.6 μm diameter) per shot. GFP images of transformed tissue were taken 7 d after transformation and GFP positive cells were counted. The GT frequencies were normalized to the Cas9 nuclease construct.
Accession Numbers
The deep sequencing data are available under the European Nucleotide Archive accession number PRJEB13819 (http://www.ebi.ac.uk/ena/data/view/PRJEB13819). All vectors are publicly available from Addgene (plasmid 90997-91225) and the ABRC. Annotated vector sequences are available from Addgene and from http://cfans-pmorrell.oit.umn.edu/CRISPR_Multiplex/.
Supplemental Data
Supplemental Figure 1. Universal architecture of expression cassettes with unique restriction sites shown.
Supplemental Figure 2. GFP expression mediated by different promoter/terminator combinations in tomato protoplasts.
Supplemental Figure 3. Frequencies of mutations induced using different systems for gRNA expression in tomato protoplasts.
Supplemental Figure 4. Short indel sequence variants detected at gRNA site 9.
Supplemental Figure 5. Short indel sequence variants detected at gRNA site 10.
Supplemental Figure 6. Short indel sequence variants detected at both gRNA sites 9 and 10.
Supplemental Figure 7. Insertions from the ARF8A locus mapped to the vector pTC364.
Supplemental Figure 8. Long deletion sequence variants detected between gRNA sites 9 and 10.
Supplemental Figure 9. GFP expression mediated by different promoters in wheat scutella.
Supplemental Figure 10. Multiplexed targeted mutagenesis in Medicago truncatula using Cas9 and Csy4.
Supplemental Figure 11. Analysis of T1 progeny from four Medicago truncatula T0 plants with mutations in NCR53, NCR54, and NCR55.
Supplemental Figure 12. Sequences of insertions detected at the ANT1 site #2 in tomato protoplast samples without Trex2 expression.
Supplemental Figure 13. Detection of gene targeting in tobacco leaves.
Supplemental Figure 14. Gene targeting with Cas9 nickases and geminivirus replicons in wheat.
Supplemental Table 1. Sequencing and read filtering data.
Supplemental Table 2. Frequencies of individual mutation types among all ARF8A sequencing reads.
Supplemental Table 3. Multiple Tukey test results.
Supplemental Table 4. Sequences of the gRNA sites used in the qPCR experiment.
Supplemental Table 5. Genotypes of 46 Medicago truncatula hygromycin resistant T0 plants transformed with a Cas9 Csy4 construct expressing six gRNAs targeting three genes.
Supplemental Table 6. List of primers used in this study.
Supplemental Methods. Protocols for vector assembly.
Supplemental Data Set 1. List of vectors included in the toolset.
Supplemental Data Set 2. Sequences of NCR53, NCR54, and NCR55 loci targeted for deletion in 46 Medicago truncatula T0 plants.
Acknowledgments
We thank members of the Voytas lab for helpful discussions and insights. We particularly thank Lynn Hu for technical assistance, Lázaro Peres (Universidade de São Paulo, Brazil) and Agustin Zsögön (Universidade Federal de Viçosa, Brazil) for input on target genes in tomato, Joseph Guhlin (College of Biological Sciences, University of Minnesota) for input on M. truncatula NCR target genes, Aaron Hummel for providing the plasmids with structurally optimized gRNA scaffolds, Evan Ellison for the GmUbi promoter reagents, and Kit Leffler for help with the figures. This work was funded by grants from the National Science Foundation (IOS-1339209 and IOS-1444511) and the Department of Energy (DE-SC0008769). S.J.C. was supported by the laboratory of Nevin D. Young under National Science Foundation Award IOS-1237993.
AUTHOR CONTRIBUTIONS
T.C. designed the vector system and work plan, and performed the experiments in tomato protoplasts. T.C., J.G.-H., C.G.S., and R.L.G. constructed the vectors. S.J.C. performed the experiments in M. truncatula. J.G.-H. performed the experiments in wheat protoplasts and gene targeting experiments in tobacco and wheat. E.K. performed the experiments in barley protoplasts and carried out the molecular analysis of the gene targeting events in tobacco. R.C. analyzed the deep sequencing data. T.J.Y.K. designed and built the set of online tools. J.J.B. performed the qPCR analysis to assess deletion frequencies. J.W.M. performed the analysis of GUS staining in tobacco leaves. T.C., J.G.H., S.J.C., and D.F.V. wrote the manuscript.
Glossary
- SSN
sequence-specific nuclease
- DSB
double-strand break
- NHEJ
nonhomologous end joining
- HDR
homology-dependent repair
- gRNA
guide RNA
- GT
gene targeting
- GVR
geminivirus replicon
- NCR
nodule-specific cysteine-rich
Footnotes
Articles can be viewed without a subscription.
References
- Aeby E., Ullu E., Yepiskoposyan H., Schimanski B., Roditi I., Mühlemann O., Schneider A. (2010). tRNASec is transcribed by RNA polymerase II in Trypanosoma brucei but not in humans. Nucleic Acids Res. 38: 5833–5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimbasseri A.G., Rijal K., Maraia R.J. (2013). Transcription termination by the eukaryotic RNA polymerase III. Biochim. Biophys. Acta 1829: 318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltes N.J., Gil-Humanes J., Cermak T., Atkins P.A., Voytas D.F. (2014). DNA replicons for plant genome engineering. Plant Cell 26: 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltes N.J., Voytas D.F. (2015). Enabling plant synthetic biology through genome engineering. Trends Biotechnol. 33: 120–131. [DOI] [PubMed] [Google Scholar]
- Brooks C., Nekrasov V., Lippman Z.B., Van Eck J. (2014). Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 166: 1292–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver M.C., Bauer D.E., Dass A., Yien Y.Y., Chung J., Masuda T., Maeda T., Paw B.H., Orkin S.H. (2014). Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J. Biol. Chem. 289: 21312–21324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čermák T., Baltes N.J., Čegan R., Zhang Y., Voytas D.F. (2015). High-frequency, precise modification of the tomato genome. Genome Biol. 16: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T., Doyle E.L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J.A., Somia N.V., Bogdanove A.J., Voytas D.F. (2011). Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certo M.T., et al. (2012). Coupling endonucleases with DNA end-processing enzymes to drive gene disruption. Nat. Methods 9: 973–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clasen B.M., et al. (2016). Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol. J. 14: 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson V., Durand P., d’Erfurth I., Kondorosi A., Ratet P. (2006). Medicago truncatula transformation using leaf explants. Methods Mol. Biol. 343: 115–127. [DOI] [PubMed] [Google Scholar]
- Curtin S.J., Tiffin P., Guhlin J., Trujillo D.I., Burghart L.T., Atkins P., Baltes N.J., Denny R., Voytas D.F., Stupar R.M., Young N.D. (2017). Validating Genome-Wide Association candidates through quantitative variation in nodulation. Plant Physiol. 173: 921–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y., Vo T.D., Mendel M.C., Greenberg S.G., Wang J., Xia D.F., Miller J.C., Urnov F.D., Gregory P.D., Holmes M.C. (2011). Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat. Methods 8: 74–79. [DOI] [PubMed] [Google Scholar]
- Engler C., Gruetzner R., Kandzia R., Marillonnet S. (2009). Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One 4: e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C., Kandzia R., Marillonnet S. (2008). A one pot, one step, precision cloning method with high throughput capability. PLoS One 3: e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expósito-Rodríguez M., Borges A.A., Borges-Pérez A., Pérez J.A. (2008). Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 8: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauser F., Roth N., Pacher M., Ilg G., Sánchez-Fernández R., Biesgen C., Puchta H. (2012). In planta gene targeting. Proc. Natl. Acad. Sci. USA 109: 7535–7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauser F., Schiml S., Puchta H. (2014). Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 79: 348–359. [DOI] [PubMed] [Google Scholar]
- Forner J., Pfeiffer A., Langenecker T., Manavella P.A., Lohmann J.U. (2015). Germline-transmitted genome editing in Arabidopsis thaliana Using TAL-effector-nucleases. PLoS One 10: e0121056 Erratum. PLoS One 10: e0133945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K. (2014). Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Wang G., Ma S., Xie X., Wu X., Zhang X., Wu Y., Zhao P., Xia Q. (2015). CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol. Biol. 87: 99–110. [DOI] [PubMed] [Google Scholar]
- Gao Y., Zhao Y. (2014). Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J. Integr. Plant Biol. 56: 343–349. [DOI] [PubMed] [Google Scholar]
- Geisinger J.M., Turan S., Hernandez S., Spector L.P., Calos M.P. (2016). In vivo blunt-end cloning through CRISPR/Cas9-facilitated non-homologous end-joining. Nucleic Acids Res. 44: e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A. III, Smith H.O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Gil-Humanes J., Ozuna C.V., Marín S., León E., Barro F., Pistón F. (2011). Genetic transformation of wheat: advances in the transformation method and applications for obtaining lines with improved bread-making quality and low toxicity in relation to celiac disease. In Genetic Transformation, Alvarez M., ed (InTech; ), pp. 135–150. [Google Scholar]
- Gil-Humanes J., Wang Y., Liang Z., Shan Q., Ozuna C.V., Sánchez-León S., Baltes N.J., Starker C., Barro F., Gao C., Voytas D.F. (2017). High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 89: 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger J.P., Thompson D.B., Liu D.R. (2014). Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 32: 577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W.L. (1988). Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature 334: 585–591. [DOI] [PubMed] [Google Scholar]
- Haun W., et al. (2014). Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J. 12: 934–940. [DOI] [PubMed] [Google Scholar]
- Chari R., Mali P., Moosburner M., Church G.M. (2015). Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat. Methods 12: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A., et al. (2015). Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12: 326–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Gilbert L.A., Cimini B.A., Schnitzbauer J., Zhang W., Li G.W., Park J., Blackburn E.H., Weissman J.S., Qi L.S., Huang B. (2013). Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155: 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.W., Kim S., Kim Y., Kweon J., Kim H.S., Bae S., Kim J.S. (2014). Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 24: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen A.H., Quail P.H. (1996). Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 5: 213–218. [DOI] [PubMed] [Google Scholar]
- Christian M., Qi Y., Zhang Y., Voytas D.F. (2013). Targeted mutagenesis of Arabidopsis thaliana using engineered TAL effector nucleases. G3 (Bethesda) 3: 1697–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Kim S.T., Ryu J., Choi M.K., Kweon J., Kang B.C., Ahn H.M., Bae S., Kim J., Kim J.S., Kim S.G. (2016). A simple, flexible and high-throughput cloning system for plant genome editing via CRISPR-Cas system. J. Integr. Plant Biol. 58: 705–712. [DOI] [PubMed] [Google Scholar]
- Kungulovski G., Jeltsch A. (2016). Epigenome Editing: State of the Art, Concepts, and Perspectives. Trends Genet. 32: 101–113. [DOI] [PubMed] [Google Scholar]
- Kwon Y.I., Abe K., Osakabe K., Endo M., Nishizawa-Yokoi A., Saika H., Shimada H., Toki S. (2012). Overexpression of OsRecQl4 and/or OsExo1 enhances DSB-induced homologous recombination in rice. Plant Cell Physiol. 53: 2142–2152. [DOI] [PubMed] [Google Scholar]
- La Russa M.F., Qi L.S. (2015). The new state of the art: Cas9 for gene activation and repression. Mol. Cell. Biol. 35: 3800–3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., et al. (2016). Multiplexed, targeted gene editing in Nicotiana benthamiana for glyco-engineering and monoclonal antibody production. Plant Biotechnol. J. 14: 533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Liu B., Spalding M.H., Weeks D.P., Yang B. (2012). High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30: 390–392. [DOI] [PubMed] [Google Scholar]
- Li Y., et al. (2015). A versatile reporter system for CRISPR-mediated chromosomal rearrangements. Genome Biol. 16: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Zhang H., Lou D., Yu D. (2016). Selection of highly efficient sgRNAs for CRISPR/Cas9-based plant genome editing. Sci. Rep. 6: 21451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z., Zhang K., Chen K., Gao C. (2014). Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genomics 41: 63–68. [DOI] [PubMed] [Google Scholar]
- Liu N., Wu S., Van Houten J., Wang Y., Ding B., Fei Z., Clarke T.H., Reed J.W., van der Knaap E. (2014). Down-regulation of AUXIN RESPONSE FACTORS 6 and 8 by microRNA 167 leads to floral development defects and female sterility in tomato. J. Exp. Bot. 65: 2507–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Rudis M.R., Peng Y., Mazarei M., Millwood R.J., Yang J.P., Xu W., Chesnut J.D., Stewart C.N. Jr (2014). Synthetic TAL effectors for targeted enhancement of transgene expression in plants. Plant Biotechnol. J. 12: 436–446. [DOI] [PubMed] [Google Scholar]
- Lowder L.G., Zhang D., Baltes N.J., Paul J.W. III, Tang X., Zheng X., Voytas D.F., Hsieh T.-F., Zhang Y., Qi Y. (2015). A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 169: 971–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Li J., Stoddard T.J., Baltes N.J., Demorest Z.L., Clasen B.M., Coffman A., Retterath A., Mathis L., Voytas D.F., Zhang F. (2015). Non-transgenic plant genome editing using purified sequence-specific nucleases. Mol. Plant 8: 1425–1427. [DOI] [PubMed] [Google Scholar]
- Ma X., et al. (2015). A robustCRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicotplants. Mol. Plant 8: 1474–1484. [DOI] [PubMed] [Google Scholar]
- Maiti I.B., Gowda S., Kiernan J., Ghosh S.K., Shepherd R.J. (1997). Promoter/leader deletion analysis and plant expression vectors with the figwort mosaic virus (FMV) full length transcript (FLt) promoter containing single or double enhancer domains. Transgenic Res. 6: 143–156. [DOI] [PubMed] [Google Scholar]
- Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M. (2013). CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31: 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann D.G., et al. (2011). Switchgrass (Panicum virgatum L.) polyubiquitin gene (PvUbi1 and PvUbi2) promoters for use in plant transformation. BMC Biotechnol. 11: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann D.G.J., Lafayette P.R., Abercrombie L.L., King Z.R., Mazarei M., Halter M.C., Poovaiah C.R., Baxter H., Shen H., Dixon R.A., Parrott W.A., Neal Stewart C. Jr (2012). Gateway-compatible vectors for high-throughput gene functional analysis in switchgrass (Panicum virgatum L.) and other monocot species. Plant Biotechnol. J. 10: 226–236. [DOI] [PubMed] [Google Scholar]
- McElroy D., Zhang W., Cao J., Wu R. (1990). Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 2: 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami M., Toki S., Endo M. (2016). Precision targeted mutagenesis via Cas9 paired nickases in rice. Plant Cell Physiol. 57: 1058–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.C., et al. (2011). A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29: 143–148. [DOI] [PubMed] [Google Scholar]
- Nagaya S., Kawamura K., Shinmyo A., Kato K. (2010). The HSP terminator of Arabidopsis thaliana increases gene expression in plant cells. Plant Cell Physiol. 51: 328–332. [DOI] [PubMed] [Google Scholar]
- Nissim L., Perli S.D., Fridkin A., Perez-Pinera P., Lu T.K. (2014). Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol. Cell 54: 698–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris S.R., Meyer S.E., Callis J. (1993). The intron of Arabidopsis thaliana polyubiquitin genes is conserved in location and is a quantitative determinant of chimeric gene expression. Plant Mol. Biol. 21: 895–906. [DOI] [PubMed] [Google Scholar]
- Ordon J., Gantner J., Kemna J., Schwalgun L., Reschke M., Streubel J., Boch J., Stuttmann J. (2017). Generation of chromosomal deletions in dicotyledonous plants employing a user-friendly genome editing toolkit. Plant J. 89: 155–168. [DOI] [PubMed] [Google Scholar]
- Park B.S., Seo J.S., Chua N.H. (2014). NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 26: 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatek A., Ali Z., Baazim H., Li L., Abulfaraj A., Al-Shareef S., Aouida M., Mahfouz M.M. (2015). RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol. J. 13: 578–589. [DOI] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y., Zhang F. (2013). Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154: 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo D.K., Dey N., Maiti I.B. (2014). pSiM24 is a novel versatile gene expression vector for transient assays as well as stable expression of foreign genes in plants. PLoS One 9: e98988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q., Wang Y., Li J., Zhang Y., Chen K., Liang Z., Zhang K., Liu J., Xi J.J., Qiu J.-L., Gao C. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31: 686–688. [DOI] [PubMed] [Google Scholar]
- Shen B., Zhang W., Zhang J., Zhou J., Wang J., Chen L., Wang L., Hodgkins A., Iyer V., Huang X., Skarnes W.C. (2014). Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods 11: 399–402. [DOI] [PubMed] [Google Scholar]
- Shimatani Z., et al. (2017). Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 35: 441–443. [DOI] [PubMed] [Google Scholar]
- Schiml S., Fauser F., Puchta H. (2014). The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J. 80: 1139–1150. [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavolone L., Kononova M., Pauli S., Ragozzino A., de Haan P., Milligan S., Lawton K., Hohn T. (2003). Cestrum yellow leaf curling virus (CmYLCV) promoter: a new strong constitutive promoter for heterologous gene expression in a wide variety of crops. Plant Mol. Biol. 53: 663–673. [DOI] [PubMed] [Google Scholar]
- Sugano S.S., Shirakawa M., Takagi J., Matsuda Y., Shimada T., Hara-Nishimura I., Kohchi T. (2014). CRISPR/Cas9-mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol. 55: 475–481. [DOI] [PubMed] [Google Scholar]
- Sugio T., Satoh J., Matsuura H., Shinmyo A., Kato K. (2008). The 5′-untranslated region of the Oryza sativa alcohol dehydrogenase gene functions as a translational enhancer in monocotyledonous plant cells. J. Biosci. Bioeng. 105: 300–302. [DOI] [PubMed] [Google Scholar]
- Tang X., Zheng X., Qi Y., Zhang D., Cheng Y., Tang A., Voytas D.F., Zhang Y. (2016). A single transcript CRISPR-Cas9 system for efficient genome editing in plants. Mol. Plant 9: 1088–1091. [DOI] [PubMed] [Google Scholar]
- Thole V., Worland B., Snape J.W., Vain P. (2007). The pCLEAN dual binary vector system for Agrobacterium-mediated plant transformation. Plant Physiol. 145: 1211–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S.Q., Wyvekens N., Khayter C., Foden J.A., Thapar V., Reyon D., Goodwin M.J., Aryee M.J., Joung J.K. (2014). Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 32: 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Vilar M., Bernabé-Orts J.M., Fernandez-Del-Carmen A., Ziarsolo P., Blanca J., Granell A., Orzaez D. (2016). A modular toolbox for gRNA-Cas9 genome engineering in plants based on the GoldenBraid standard. Plant Methods 12: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Cheng X., Shan Q., Zhang Y., Liu J., Gao C., Qiu J.L. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32: 947–951. [DOI] [PubMed] [Google Scholar]
- Wang Z.P., Xing H.L., Dong L., Zhang H.Y., Han C.Y., Wang X.C., Chen Q.J. (2015). Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright D.A., Townsend J.A., Winfrey R.J. Jr., Irwin P.A., Rajagopal J., Lonosky P.M., Hall B.D., Jondle M.D., Voytas D.F. (2005). High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J. 44: 693–705. [DOI] [PubMed] [Google Scholar]
- Xie K., Minkenberg B., Yang Y. (2015). Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA 112: 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H.L., Dong L., Wang Z.P., Zhang H.Y., Han C.Y., Liu B., Wang X.C., Chen Q.J. (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R.F., Li H., Qin R.Y., Li J., Qiu C.H., Yang Y.C., Ma H., Li L., Wei P.C., Yang J.B. (2015). Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci. Rep. 5: 11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Wei S., Wu Y., Hu R., Li H., Yang W., Xie Q. (2015). High efficiency genome editing in Arabidopsis using Yao promoter-driven CRISPR/Cas9 system. Mol. Plant 8: 1820–1823. [DOI] [PubMed] [Google Scholar]
- Zhang F., Maeder M.L., Unger-Wallace E., Hoshaw J.P., Reyon D., Christian M., Li X., Pierick C.J., Dobbs D., Peterson T., Joung J.K., Voytas D.F. (2010). High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc. Natl. Acad. Sci. USA 107: 12028–12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhang J., Wei P., Zhang B., Gou F., Feng Z., Mao Y., Yang L., Zhang H., Xu N., Zhu J.K. (2014). The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12: 797–807. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang F., Li X., Baller J.A., Qi Y., Starker C.G., Bogdanove A.J., Voytas D.F. (2013). Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol. 161: 20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Mao Y., Ha S., Liu W., Botella J.R., Zhu J.K. (2016). A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep. 35: 1519–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Y., Wang Y., Li C., Zhang R., Chen K., Ran Y., Qiu J.L., Wang D., Gao C. (2017). Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 35: 438–440. [DOI] [PubMed] [Google Scholar]