miR159 functions upstream of miR156 to modulate vegetative phase change in Arabidopsis.
Abstract
Temporally regulated microRNAs have been identified as master regulators of developmental timing in both animals and plants. In plants, vegetative development is regulated by a temporal decrease in miR156 level, but how this decreased expression is initiated and then maintained during shoot development remains elusive. Here, we show that miR159 is required for the correct timing of vegetative development in Arabidopsis thaliana. Loss of miR159 increases miR156 level throughout shoot development and delays vegetative development, whereas overexpression of miR159 slightly accelerated vegetative development. The repression of miR156 by miR159 is predominantly mediated by MYB33, an R2R3 MYB domain transcription factor targeted by miR159. Loss of MYB33 led to subtle precocious vegetative phase change phenotypes in spite of the significant downregulation of miR156. MYB33 simultaneously promotes the transcription of MIR156A and MIR156C, as well as their target, SPL9, by directly binding to the promoters of these three genes. Rather than acting as major players in vegetative phase change in Arabidopsis, our results suggest that miR159 and MYB33 function as modifiers of vegetative phase change; i.e., miR159 facilitates vegetative phase change by repressing MYB33 expression, thus preventing MYB33 from hyperactivating miR156 expression throughout shoot development to ensure correct timing of the juvenile-to-adult transition in Arabidopsis.
INTRODUCTION
The founding microRNAs (miRNAs), lin-4 (Lee et al., 1993) and let-7 (Reinhart et al., 2000), were initially identified as key regulators of the juvenile-to-adult transition in Caenorhabditis elegans. More recent work in plants has also shown that two miRNAs, miR156 and miR172, regulate developmental transitions as a part of a regulatory circuit (Wu and Poethig, 2006; Wu et al., 2009). In plants, shoot development can be divided into a juvenile vegetative phase, an adult vegetative phase, and a reproductive phase (Poethig, 1990; Kerstetter and Poethig, 1998). The transition from the juvenile vegetative phase to the adult vegetative phase is referred to as vegetative phase change. In Arabidopsis thaliana, vegetative phase change is characterized by changes in the production of trichomes on the abaxial side of the leaf blade, an increase in the leaf length/width ratio, an increase in the degree of serration of the leaf margin, and a decrease in cell size (Telfer et al., 1997; Tsukaya et al., 2000; Usami et al., 2009). Genetic and molecular analysis demonstrates that miR156 regulates vegetative phase change by repressing plant-specific SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factors (Wu et al., 2009). The expression of miR156 declines gradually as plants progress through a juvenile phase of development to an adult phase of development, whereas the expression of its targets increases during this process (Wu and Poethig, 2006; Wang et al., 2009; Wu et al., 2009). In addition to this temporally regulated decrease in miR156 expression, miR156 expression is also regulated by exogenous cues, such as temperature (Lee et al., 2010; Xin et al., 2010; Yu et al., 2012), phosphate starvation (Hsieh et al., 2009), CO2 concentration (May et al., 2013), and sugar (Yang et al., 2013; Yu et al., 2013). The B3 domain transcription factor FUSCA3 (Wang and Perry, 2013), two MADS box transcription factors, AGL15 and AGL18 (Serivichyaswat et al., 2015), the Polycomb Group Repressive Complex 1 (PRC1) component AtBMI1 (Picó et al., 2015), the PRC2 component FIE (Xu et al., 2016a), and the SWI2/SNF2 chromatin remodeling ATPase BRAHMA (Xu et al., 2016b) regulate the levels of miR156 by directly binding to the promoter regions of genes encoding this miRNA, but how miR156 expression is regulated throughout the plant life cycle, and in response to these cues, is still poorly understood.
miR159 is an evolutionarily conserved miRNA that targets R2R3 MYB domain transcription factors (Park et al., 2002; Rhoades et al., 2002; Axtell and Bartel, 2005). In Arabidopsis, these targets include MYB33 and MYB65, which act in the endosperm and in anthers to promote programmed cell death (Alonso-Peral et al., 2010). Most of the miR159 in Arabidopsis is produced by two genes, MIR159A and MIR159B (Allen et al., 2007). These genes are expressed in mature embryos and in most postembryonic organs and strongly repress MYB33 and MYB65. The miR159-MYB33/MYB65 pathway does not play a major functional role in rosette development in Arabidopsis (Li et al., 2016). However, plants doubly mutant for mir159a and mir159b have a pleiotropic vegetative and reproductive phenotype that is completely attributable to the overexpression of these two transcription factors (Allen et al., 2007).
We became interested in the role of miR159 in vegetative phase change because of studies suggesting that it is involved in the response to the hormone gibberellic acid (GA), a known regulator of vegetative phase change in Arabidopsis and maize (Zea mays; Evans and Poethig, 1995; Telfer et al., 1997; Achard et al., 2004). Here, we were unable to demonstrate a role for miR159 in the response to GA, but found that plants deficient for miR159 have a prolonged juvenile phase due to elevated levels of miR156. We show that the elevated level of miR156 in this double mutant is attributable to the overexpression of MIR156A and MIR156C, and that both genes are direct transcriptional targets of MYB33. Loss of function of MYB33 caused only subtle vegetative phase change phenotypes despite the fact that miR156 was significantly decreased. This result indicates that MYB33 itself plays a minor role in vegetative phase change; instead, it functions as a modifier of vegetative phase change. Our results demonstrate that the repression of MYB33 by miR159 during vegetative development is important to prevent hyperactivation of miR156 and to ensure vegetative phase change occurs at the appropriate time.
RESULTS
miR159 Promotes Vegetative Phase Change Independently of GA
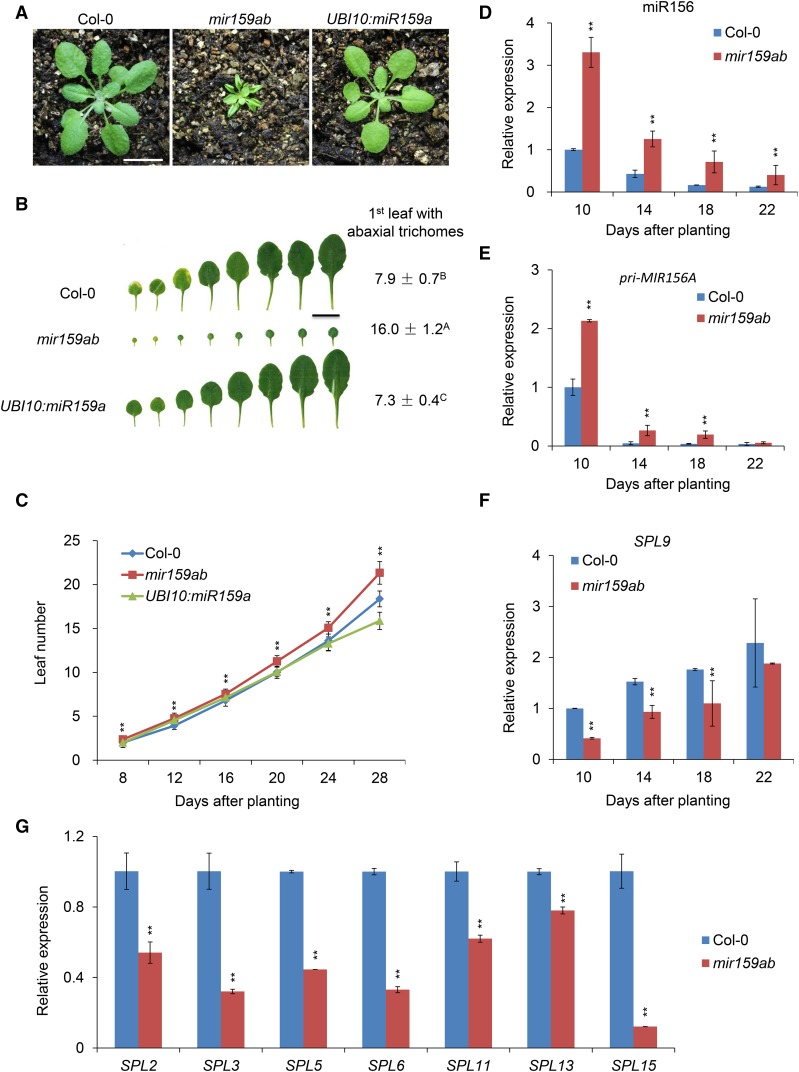
To determine if miR159 has a role in vegetative phase change in Arabidopsis, we characterized phenotypes of plants mutant for both MIR159A and MIR159B (mir159ab), which lack miR159 (Supplemental Figure 1A), as well as of transgenic plants overexpressing miR159 (UBI10:miR159a) under the control of the constitutive UBI10 promoter from Arabidopsis. The leaves of mir159ab mutants were much smaller and rounder than those of the wild type, and they produced abaxial trichomes on leaf 16, compared with leaf 7.9 in wild-type plants (Figures 1A and 1B). mir159ab also had a significantly faster rate of leaf initiation than wild-type plants (Figure 1C). Plants transformed with UBI10:miR159a were morphologically similar to the wild type but produced abaxial trichomes significantly earlier than normal (7.3 ± 0.4 versus 7.9 ± 0.7) (Figures 1A and 1B). Although the difference in abaxial trichome production between UBI10:miR159a and the wild type was small, it was statistically significant and reproducible in our growth conditions. The rate of leaf initiation in UBI10:miR159a was indistinguishable from the wild type (Figure 1C). The observation that UBI10:miR159a has a relatively small effect on shoot development is consistent with previous studies showing that the amount of miR159 present in wild-type rosettes is sufficient to nearly completely repress the expression of its targets (Millar and Gubler, 2005; Reyes and Chua, 2007; Alonso-Peral et al., 2010). Thus, an additional increase in the level of miR159 is not expected to have a major effect on rosette development. On the other hand, the effect of mir159ab on abaxial trichome production and leaf shape demonstrates that miR159 is required for the correct timing of vegetative phase change.
Figure 1.
miR159 Regulates Vegetative Phase Change in Arabidopsis by Affecting miR156 and SPLs.
(A) Twenty-one-day-old Col-0, mir159ab, and UBI10:miR159a plants grown in short days. Bar = 1 cm.
(B) Leaf shape and abaxial trichome phenotypes of fully expanded rosette leaves of Col-0, mir159ab, and UBI10:miR159a in short days. Numbers indicate the first leaf with abaxial trichomes. Different capital letters indicate significant difference between genotypes using one-way ANOVA at P < 0.01 (n = 30 plants, ±sd; Supplemental File 1). Bar = 1 cm.
(C) Leaf initiation rate of Col-0, mir159ab, and UBI10:miR159a in short days. Leaf numbers were scored at 8, 12, 16, 20, 24, and 28 d after planting. Asterisks indicate significant difference from Col-0 using Student’s t test (P < 0.01, n = 30 plants, ±sd).
(D) to (F) The expression of miR156 (D), Pri-MIR156A (E), and SPL9 (F) at different developmental stages in Col-0 and mir159ab plants assessed using qRT-PCR. About 2-mm shoot apices were collected at 10, 14, 18, and 22 d after planting in short days. Asterisks indicate significant difference from Col-0 using Student’s t test (P < 0.01).
(G) The expression of different SPL genes in 10-d-old Col-0 and mir159ab plants assessed using qRT-PCR. About 2-mm shoot apices were collected at 10 d after planting in short days. Asterisks indicate significant difference from Col-0 using Student’s t test (P < 0.01).
All qRT-PCR data ([D] to [G]) represent the mean of three biological replicates (experiments were repeated at different times, the same for the following figures); values were normalized to the 10-d-old wild type (±sd).
It has been reported that miR159 mediates the response of plants to GA (Achard et al., 2004), although this function was not confirmed in a subsequent study (Alonso-Peral et al., 2010). To determine if miR159 is required for the effect of GA on vegetative phase change (Telfer et al., 1997), we examined the effect of exogenous GA on the phenotype of the wild type, mir159ab, and plants transformed with a miR159-insensitive MYB33 (mMYB33) genomic construct. GA accelerated the production of abaxial trichomes in wild-type plants and completely corrected the abaxial trichome phenotype of mir159ab and mMYB33, but did not correct the morphological defects of these lines (Supplemental Figures 1B and 1C). GA also had no effect on miR159 expression (Supplemental Figure 1A). These results indicate that GA and miR159 regulate vegetative phase change by independent mechanisms.
miR159 Constitutively Represses the Expression of miR156
Next, we asked if the delayed vegetative phase change phenotype of mir159ab is attributable to changes in the expression of genes in the miR156-SPL pathway. RNA gel blots and qRT-PCR indicated that miR156 was significantly elevated in mir159ab (Supplemental Figure 1A; Figure 1D). The abundance of the primary transcript of MIR156A (pri-MIR156A) was also elevated in mir159ab at different developmental stages (Figure 1E), suggesting that the effect of this genotype on miR156 levels is attributable to an increase in the transcription of genes encoding miR156, not to a change in the processing of their primary transcripts. However, mir159ab had no effect on the temporal expression patterns of miR156, pri-MIR156A, or its target, SPL9 (Figures 1D to 1F). This result is consistent with the observation that the abundance of miR159 and its target, MYB33, does not change temporally after germination (Alonso-Peral et al., 2012) (Supplemental Figures 2A and 2B). Thus, miR159 represses miR156, but is not responsible for the temporal expression pattern of miR156 and its target during shoot development (Figures 1D to 1F). Elevated levels of miR156 were also correlated with a significant reduction in the levels of most of its targets examined, such as SPL2, SPL3, SPL5, SPL6, SPL9, SPL11, SPL13, and SPL15 (Figure 1G).
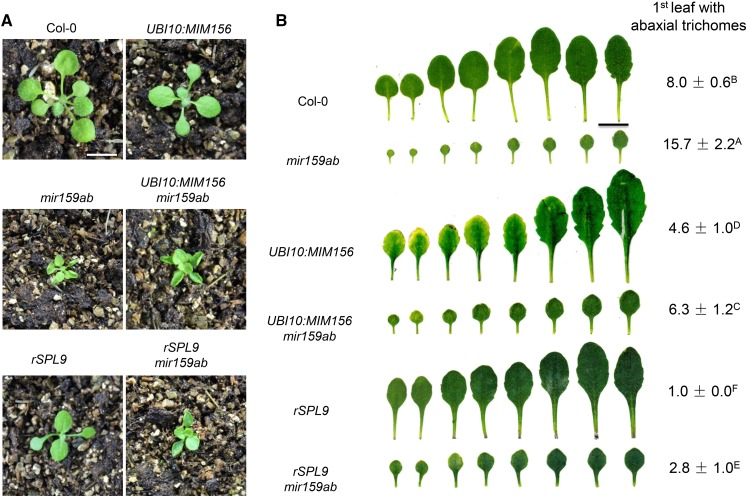
To determine if the effect of mir159ab on vegetative phase change is attributable to the elevated level of miR156, we crossed a UBI10:MIM156 transgene into this double mutant. UBI10:MIM156 acts a sponge for miR156 and provides a convenient way of reducing the activity of this miRNA (Franco-Zorrilla et al., 2007). UBI10:MIM156 had little or no effect on the leaf morphology of mir159ab (Figures 2A and 2B). However, UBI10:MIM156 accelerated abaxial trichome production in a wild-type Columbia (Col) background and was epistatic to mir159ab; the UBI10:MIM156 mir159ab plants produced abaxial trichomes on leaf 6.3, which was significantly earlier than in both mir159ab and Col (Figure 2B). As a further test of the hypothesis that the miR156-SPL pathway acts downstream of miR159, we examined the phenotype of rSPL9 mir159ab plants. rSPL9 is a miR156-resistant version of SPL9 expressed under the regulation of its endogenous promoter (Wu et al., 2009). Consistent with the effect of UBI10:MIM156 on the phenotype of mir159ab, we found that rSPL9 was completely epistatic to miR159ab with respect to abaxial trichome production, and slightly modified the shape, but not the size of mir159ab leaves (Figure 2B). Thus, the miR156-SPL pathway is required for effect of mir159ab on abaxial trichome production, but not for its effect on leaf morphology.
Figure 2.
miR156 and SPL Genes Act Downstream of miR159.
(A) Eighteen-day-old Col-0, UBI10:MIM156, mir159ab, UBI10:MIM156 mir159ab, pSPL9:rSPL9 (rSPL9), and pSPL9:rSPL9 mir159ab (rSPL9 mir159ab) plants grown in short days. Bar = 1 cm.
(B) Leaf shape and abaxial trichome phenotypes of fully expanded rosette leaves of Col-0, UBI10:MIM156, mir159ab, UBI10:MIM156 mir159ab, pSPL9:rSPL9, and pSPL9:rSPL9 mir159ab in short days. UBI10:MIM156 and pSPL9:rSPL9 significantly rescued the mir159ab phenotype. Numbers indicate the first leaf with abaxial trichomes. Different capital letters indicate significant difference between genotypes using one-way ANOVA at P < 0.01(n = 30 plants, ±sd; Supplemental File 1). Bar = 1 cm.
The miR156-SPL pathway acts, in part, by promoting the expression of miR172, which in turn represses the expression of several AP2-like genes, including TOE1 and TOE2. If the elevated levels of miR156 in mir159ab are functionally significant, the expression of these genes should be affected in this mutant. qRT-PCR analysis revealed that miR172 and pri-MIR172B were reduced (Supplemental Figures 3A and 3B), whereas the TOE2 transcript was significantly elevated, in mir159ab (Supplemental Figure 3C). Additionally, we found that a transgene overexpressing miR172, as well as mutations in toe1 and toe2, suppressed the effect of mir159ab on abaxial trichome production (Supplemental Figures 3D and 3E). These results provide further support for the conclusion that the miR156-SPL pathway acts downstream of miR159.
MYB33 Directly Promotes the Transcription of MIR156A and MIR156C
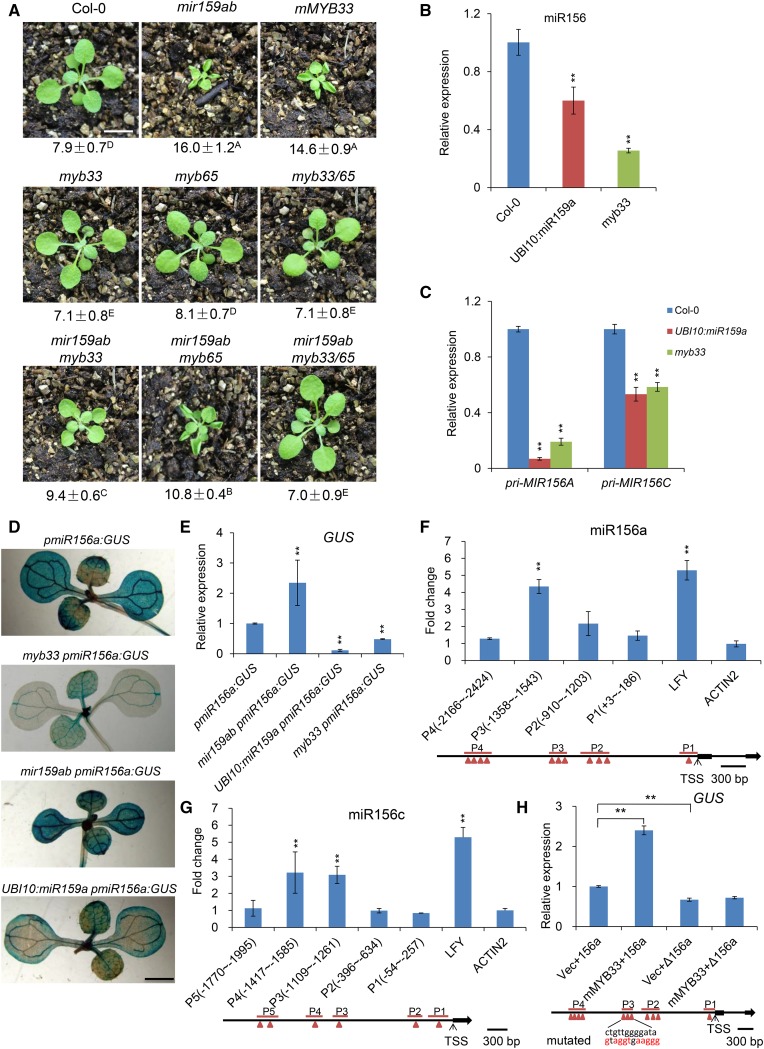
To identify which miR159-targeted MYB genes are responsible for the delayed vegetative phase change of mir159ab, we generated mir159ab myb33, mir159ab myb65, and mir159ab myb33 myb65 mutants. myb33 rescued both the leaf shape and the trichome defects of mir159ab to a greater extent than did myb65 (Figure 3A), and myb33 myb65 fully rescued both of these phenotypes (Figure 3A; Supplemental Figure 4). This result confirms a previous study indicating that MYB33 and MYB65 are the major targets of miR159, and are largely responsible for the phenotype of mir159ab (Allen et al., 2007). It also demonstrates that MYB33 is more important than MYB65 for the effect of mir159ab on vegetative phase change.
Figure 3.
MYB33 Is the Primary Gene Responsible for the Phenotype of mir159ab by Directly Binding to the Promoter of miR156a and miR156c to Regulate miR156 Transcription.
(A) Eighteen-day-old Col-0, mir159ab, mMYB33, myb33, myb65, myb33 myb65 (myb33/65), mir159ab myb33, mir159ab myb65, and mir159ab myb33 myb65 plants grown in short days. Numbers indicate the first leaf with abaxial trichomes. Different capital letters indicate significant difference between genotypes using one-way ANOVA at P < 0.01(n = 30 plants, ±sd; Supplemental File 1). Bar = 1 cm.
(B) The expression level of mature miR156 in Col-0, UBI10:miR159a, and myb33 plants in short days. Ten-day-old whole seedlings were sampled and analyzed. Asterisks indicate significant difference from Col-0 using Student’s t test (P < 0.01). All qRT-PCR data represent the mean of three biological replicates; values were normalized to the wild type (±sd).
(C) The expression level of Pri-MIR156A and Pri-MIR156C in Col-0, UBI10:miR159a, and myb33 plants in short days. Ten-day-old whole seedlings were sampled and analyzed. Asterisks indicate significant difference from Col-0 using Student’s t test (P < 0.01). All qRT-PCR data represent the mean of three biological replicates; values were normalized to the wild type (±sd).
(D) GUS staining analysis of 10-d-old pmiR156a:GUS, mir159ab pmiR156a:GUS, myb33 pmiR156a:GUS, and UBI10:miR159a pmiR156a:GUS plants in short days. Bar = 0.2 cm.
(E) Quantitative analysis of GUS expression in 10-d-old pmiR156a:GUS, mir159ab pmiR156a:GUS, myb33 pmiR156a:GUS, and UBI10:miR159a pmiR156a:GUS plants in short days using qRT-PCR method. All results were normalized to that of pmiR156a:GUS in the wild-type background. Asterisks indicate significant difference from pmiR156a:GUS using Student’s t test (P < 0.01). All qRT-PCR data represent the mean of three biological replicates ±sd.
(F) ChIP-qPCR analysis of MYB33 binding sites in the promoter region of miR156a. Chromatin from 10-d-old pMYB33:3×FLAG-mMYB33-3′UTR and pMYB33:HA-mMYB33-3′UTR seedlings grown in short days was immunoprecipitated with a polyclonal antibody to FLAG. Primers surrounding the putative MYB33 binding sites were designed upstream of the TSS of miR156a. The immunoprecipitated values were first normalized to the input values then divided by the pMYB33:HA-mMYB33-3′UTR to get a fold enrichment. The numbers represent the fold change relative to pMYB33:HA-mMYB33-3′UTR samples. Values are the mean of three biological replicates. P1 to P4 denote different PCR-amplified regions in the promoter region of miR156a. LFY was used as a positive control, whereas ACTIN2 was a negative control. Asterisks indicate significant difference from the value of ACTIN2 using Student’s t test (P < 0.01). Red triangles indicate the predicted MYB binding sites.
(G) ChIP-qPCR analysis of MYB33 binding sites in the promoter region of miR156c. Chromatin from 10-d-old pMYB33:3×FLAG-mMYB33-3′UTR and pMYB33:HA-mMYB33-3′UTR seedlings grown in short days was immunoprecipitated with a polyclonal antibody to FLAG. Primers surrounding the putative MYB33 binding sites in the miR156c promoter region were designed upstream of the TSS of miR156c. Values are the mean of three biological replicates. P1 to P5 denote different PCR-amplified regions in the promoter region of miR156c. LFY was used as a positive control, whereas ACTIN2 was a negative control. Asterisks indicate significant difference from the value of ACTIN2 using Student’s t test (P < 0.01). Red triangles indicate the predicted MYB binding sites.
(H) Activation of MIR156A transcription by direct binding of MYB33 to the cis-regulatory sequence in the promoter of MIR156A. GUS expression was quantitated using qRT-PCR in leaves of N. benthamiana infiltrated with Agrobacterium with different combinations of constructs. pmiR156a:GUS (156a), MIR156A transcriptional reporter construct; pΔmiR156a:GUS (Δ156a), MIR156A transcriptional reporter construct with a mutated cis-regulatory sequence to which MYB33 binds; UBI10:mMYB33, miR159-insensitive overexpression vector; pSY06, the control vector. Vec+156a (pSY06 + pmiR156a:GUS), mMYB33+156a (UBI10:mMYB33 + pmiR156a:GUS), Vec+Δ156a (pSY06 + pΔmiR156a:GUS), mMYB33+Δ156a (UBI10:mMYB33 + pΔmiR156a:GUS). Red letters indicate the mutated bases. Values were normalized to that of Vec+156a and are the mean of two biological replicates; asterisks indicate significant difference from Vec+156a using Student’s t test (P < 0.01).
MYB33 is strongly repressed by miR159 in the rosette (Millar and Gubler, 2005; Alonso-Peral et al., 2010), and the miR159-MYB33/MYB65 pathway was reported to play no role in rosette development (Li et al., 2016). However, our observation that both UBI10:miR159a (Figure 1B) and myb33 (Figure 3A) accelerate abaxial trichome production, albeit weakly, indicates that MYB33 is still active and functional in this tissue. This conclusion was further supported by the phenotype of myb33 and UBI10:miR159a plants under different light intensities in short days. When grown at a light intensity of 110 μmol/m2/s, wild-type plants produced abaxial trichomes on leaf 10.6, whereas myb33 and UBI10:miR159a produced abaxial trichomes on leaves 8.8 and 9.5, respectively (Supplemental Figure 5). The difference in abaxial trichome production between myb33, UBI10:miR159a, and the wild type was less obvious at a light intensity of 145 μmol/m2/s, where wild-type plants produced abaxial trichomes on leaf 7.1, whereas myb33 and UBI10:miR159a produced abaxial trichomes on leaves 6.6 and 6.7, respectively (Supplemental Figure 5). This result suggests that the function of the miR159-MYB33 pathway in vegetative phase change may be dependent on light intensity, which is consistent with the effect of different light intensities on the seed fertility of myb33/myb65 mutants (Millar and Gubler, 2005). It also indicates that MYB33 delays the vegetative phase change in Arabidopsis.
To determine if the abaxial trichome phenotype of UBI10:miR159a and myb33 is attributable to a decrease in miR156 expression, we examined the levels of miR156, pri-MIR156A, and pri-MIR156C in these genotypes. MIR156A and MIR156C are the major sources of mature miR156 in Arabidopsis (Yang et al., 2013). The levels of miR156, pri-MIR156A, and pri-MIR156C were reduced significantly in 10-d-old seedlings of both UBI10:miR159a and myb33 (Figures 3B and 3C). We confirmed this result by examining the effect of mir159ab, myb33, and UBI10:miR159a on the expression of pmiR156a:GUS, a reporter line previously used to show the transcription of MIR156A in Arabidopsis (Yang et al., 2013). GUS activity was elevated in mir159ab and was reduced in myb33 and UBI10:miR159a (Figures 3D and 3E), particularly in cotyledons. Thus, MYB33 directly or indirectly regulates the transcription of MIR156A and MIR156C in young seedlings.
To investigate if MYB33 is bound to MIR156A and MIR156C in vivo, we generated lines transformed with a construct for expression of a miR159-insensitive MYB33 protein tagged with either 3×FLAG (pMYB33:3×FLAG-mMYB33) or the HA epitope (pMYB33:HA-mMYB33). Both pMYB33:3×FLAG-mMYB33 and pMYB33:HA-mMYB33 transgenic plants exhibited similar phenotypes to the mMBY33 plant (Supplemental Figure 6), indicating that the 3×FLAG-mMYB33 and HA-mMYB33 fusion proteins are biologically functional in plants. Chromatin from the pMYB33:3×FLAG-mMYB33 and pMYB33:HA-mMYB33 lines was immunoprecipitated with an antibody to FLAG, and DNA was then amplified using primers surrounding putative MYB protein binding sites in the promoter regions of MIR156A and MIR156C (Supplemental Table 1). qPCR showed that MYB33 is bound to a site 1300 bp upstream of the transcription start site (TSS) of MIR156A (Figure 3F) and to sites 1200 and 1400 bp upstream of the TSS of MIR156C (Figure 3G), suggesting that MYB33 directly regulates the transcription of these genes. To test the significance of direct binding of MYB33 to the cis-regulatory element in the promoters of MIR156-encoding loci, we constructed a mutated pΔmiR156a:GUS vector in which the ctgttggggata sequence (P3) in the MIR156A promoter was mutated to gtaggtgaaggg (Figure 3H). We also generated a construct overexpressing the miR159-insensitive MYB33 cDNA (mMYB33) under the control of the UBI10 promoter from Arabidopsis. We then infiltrated Nicotiana benthamiana leaves with Agrobacterium tumefaciens harboring different combinations of the control vector (Vec), pmiR156a:GUS (156a), mMYB33 overexpression vector, and pΔmiR156a:GUS (Δ156a). qRT-PCR analysis of GUS mRNA in the leaves transformed with different constructs indicated that GUS mRNA was significantly elevated, by ∼2.4-fold, in leaves transformed with mMYB33 and pmiR156a:GUS, whereas the elevation of GUS mRNA was abolished in the leaves transformed with mMYB33 and pΔmiR156a:GUS in which the cis-regulatory sequence to which MYB33 was bound was mutated (Figure 3H). This result implies that MYB33 promotes the transcription of the MIR156A gene by directly binding to its promoter.
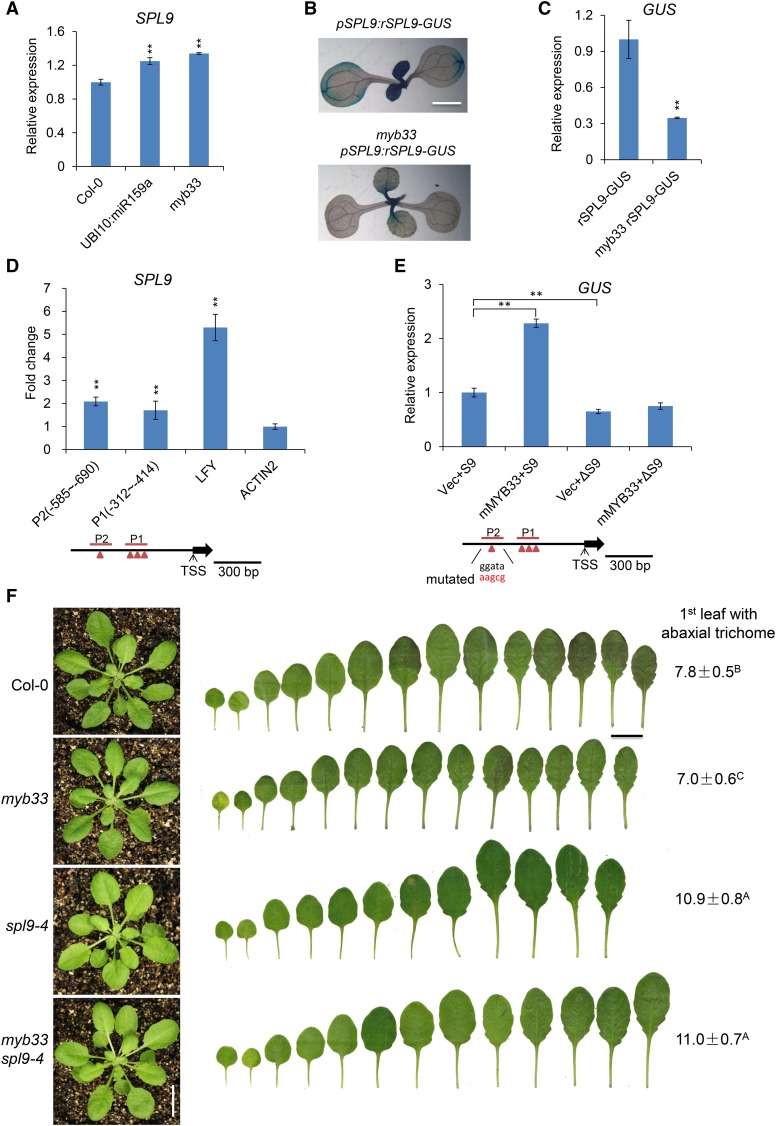
MYB33 Promotes the Transcription of SPL9
Although miR156 was reduced significantly in both myb33 and in plants expressing UBI10:miR159a (Figures 3B and 3C), these plants had a relatively subtle vegetative phase change phenotype (Figures 1B and 3A). This discrepancy prompted us to examine the effect of myb33 and UBI10:miR159a on the expression of SPL9, a direct target of miR156 that plays a major role in vegetative phase change (Wu et al., 2009), as well as some other SPL genes. qRT-PCR analysis of myb33 and UBI10:miR159a revealed a small but statistically significant increase in SPL9 transcripts in these lines (Figure 4A), and SPL2, SPL3, SPL13, and SPL15 were elevated ∼2-fold in myb33 (Supplemental Figure 7). However, we obtained the opposite result when we examined the effect of myb33 on the expression of a miR156-resistant SPL9 reporter (pSPL9:rSPL9-GUS) (Yang et al., 2011). GUS expression was lower in myb33 pSPL9:rSPL9-GUS plants than in wild-type plants containing pSPL9:rSPL9-GUS (Figures 4B and 4C). Chromatin immunoprecipitation (ChIP) analysis of the pMYB33:3×FLAG-mMYB33 line showed that MYB33 is bound to a site 600 bp upstream of the TSS of SPL9 (Figure 4D), suggesting that MYB33 directly regulates SPL9 transcription. Because SPL-GUS activity was reduced ∼3-fold in myb33 pSPL9:rSPL9-GUS (Figure 4C), whereas the endogenous SPL9 transcript was only slightly elevated in myb33 and UBI10:miR159a (Figure 4A), we think that it is more likely that MYB33 promotes, rather than represses, the transcription of SPL9. To test our hypothesis, we generated transcriptional reporter constructs for SPL9 (pSPL9:GUS) with normal or mutated versions of the cis-regulatory sequence to which MYB33 was bound (P2) (pΔSPL9:GUS). We then infiltrated N. benthamiana leaves with Agrobacterium harboring different combinations of the control vector (Vec), pSPL9:GUS (S9), mMYB33 overexpression vector, and pΔSPL9:GUS (ΔS9). qRT-PCR analysis of GUS mRNA in the leaves transformed with different combinations of constructs indicated that GUS mRNA was significantly elevated, by ∼2.2-fold, in leaves transformed with mMYB33 and pSPL9:GUS (S9), but not in leaves transformed with mMYB33 and pΔSPL9:GUS (ΔS9) (Figure 4E). Therefore, MYB33 promotes SPL9 transcription by directly binding to its promoter.
Figure 4.
MYB33 Directly Regulates SPL9 Expression.
(A) The expression level of SPL9 in 10-d-old Col-0, UBI10:miR159a, and myb33 seedlings grown in short days. Values are the means of three biological replicates. Asterisks indicate significant difference from Col-0 using Student’s t test (P < 0.01).
(B) GUS staining analyses of 10-d-old pSPL9:rSPL9-GUS and myb33 pSPL9:rSPL9-GUS plants grown in short days.
(C) Quantitative qRT-PCR analysis of GUS expression in 10-d-old pSPL9:rSPL9-GUS plants in Col-0 and myb33 backgrounds grown in short days. Asterisks indicate significant difference from rSPL9-GUS using Student’s t test (P < 0.01).
(D) ChIP-qPCR analysis of MYB33 binding sites in the promoter of SPL9. Chromatin from 10-d-old pMYB33:3×FLAG-mMYB33-3′UTR and pMYB33:HA-mMYB33-3′UTR seedlings grown in short days was immunoprecipitated with a polyclonal antibody to FLAG. Primers surrounding the putative MYB33 binding sites in the SPL9 promoter region were designed upstream of the TSS of SPL9. Values are the means of three biological replicates, each having three technical replicates. P1 and P2 denote different PCR-amplified regions in the promoter region of SPL9. LFY was used as a positive control, whereas ACTIN2 was a negative control. Asterisks indicate significant difference from the value of ACTIN2 using Student’s t test (P < 0.01). Red triangles indicated the predicted MYB binding sites.
(E) Activation of SPL9 transcription by direct binding of MYB33 to the cis-regulatory sequence in the SPL9 promoter. GUS expression was quantified using qRT-PCR in leaves of N. benthamiana infiltrated with Agrobacterium with different combinations of constructs. pSPL9:GUS (S9), SPL9 transcriptional reporter construct; pΔSPL9:GUS (ΔS9), SPL9 transcriptional reporter construct with a mutated version of the cis-regulatory sequence to which MYB33 binds; UBI10:mMYB33, miR159-insensitive overexpression vector; pSY06, the control vector. Vec+S9 (pSY06 + pSPL9:GUS), mMYB33+S9 (UBI10:mMYB33 + pSPL9:GUS), Vec+ΔS9 (pSY06 + pΔSPL9:GUS), mMYB33+ΔS9 (UBI10:mMYB33 + pΔSPL9:GUS). Red letters indicate the mutated bases. Values were normalized to that of Vec+S9 and are the means of two biological replicates; asterisks indicate significant difference from Vec+S9 using Student’s t test (P < 0.01).
(F) spl9-4 is epistatic to myb33. Leaf shape and abaxial trichome phenotypes of Col-0, myb33, spl9-4, and myb33 spl9-4 in short days. Numbers indicate the first leaf with abaxial trichomes. Different capital letters indicate significant difference between genotypes using one-way ANOVA at P < 0.01(n = 20 plants, ±sd; Supplemental File 1). Bar = 1 cm.
The difference between the effect of myb33 on the SPL9 transcript and its effect on the miR156-insensitive pSPL9:rSPL9-GUS reporter can be explained by the effect of myb33 on miR156. The reduced level of miR156 in myb33 should increase the abundance of the endogenous SPL9 transcript, thus compensating for the effect of myb33 on SPL9 transcription. Note that the reduced level of miR156 in myb33 should not affect the expression of pSPL9:rSPL9-GUS because this transgene is insensitive to miR156. We conclude that the decrease in miR156 levels in myb33 does not produce a major increase in SPL9 expression because myb33 also reduces SPL9 transcription, and as a result, myb33 has a relatively small effect on vegetative phase change (Figure 3A). To test this hypothesis, we asked if SPL9 is required for the early abaxial trichome phenotype of myb33 (Figure 4F). myb33 spl9-4 double mutants produced abaxial trichomes on leaf 11.0 ± 0.7, not significantly different than spl9-4 (10.9 ± 0.8) (Figure 4F). Thus, spl9-4 is epistatic to myb33. This result is consistent with the evidence that SPL9 transcript is elevated in myb33 and suggests that MYB33 regulates vegetative phase change primarily through its effect on miR156 expression.
DISCUSSION
miR159 is an evolutionarily conserved miRNA that is expressed at high levels in many organs and tissues in Arabidopsis (Axtell and Bartel, 2005; Taylor et al., 2014). Previous studies have suggested that miR159 functions both as a switch, completely repressing the expression of its targets in some tissues but not in others, and as a tuning mechanism, modulating the expression of its targets in response to endogenous and exogenous signals (Alonso-Peral et al., 2012). For example, during seed and flower development, miR159 restricts the expression of MYB33/MYB65 to the aleurone and anthers, respectively, where they promote programmed cell death (Millar and Gubler, 2005; Allen et al., 2007; Alonso-Peral et al., 2010). It is also thought to completely repress the expression of its targets during the vegetative phase of shoot development (Alonso-Peral et al., 2010). However, during seed germination, miR159 regulates the sensitivity of seedlings to ABA (Reyes and Chua, 2007) by modulating, but not completely repressing, the expression of MYB33 (Millar and Gubler, 2005; Alonso-Peral et al., 2012).
Here, we show that miR159 also plays a role in vegetative phase change. Mutants deficient for miR159 have a prolonged juvenile phase that is attributable to the elevated expression of miR156 throughout shoot development. This defect is a consequence of the overexpression of MIR156A and MIR156C, whose transcription is promoted by MYB33, a direct target of miR159. However, the extent to which miR159 and MYB33 contribute to the temporal regulation of MIR156A and MIR156C is unclear. Neither of these genes is temporally expressed during shoot development, and the myb33 mutant displays a very weak defect in the timing of vegetative phase change, particularly in comparison to the mir159ab mutant, in which MYB33 is overexpressed. These observations suggest that MYB33 is actually deleterious to vegetative phase change and that miR159 promotes vegetative phase change by preventing MYB33 from activating MIR156A and MIR156C transcription during shoot development; i.e., miR159 has a permissive, rather than a directive, role in vegetative phase change. Therefore, miR159 and MYB33 could be considered as modifiers but not major players in vegetative phase change. Presumably, MYB33 could not have evolved to promote MIR156A and MIR156C transcription if this activity was phenotypic neutral. Consequently, this hypothesis implies that MYB33 regulation of MIR156A and MIR156C might be important at some other stages of the plant life cycle, perhaps in anthers where MYB33 is highly expressed and contributes to pollen development.
An alternative possibility is that MYB33 promotes the transcription of MIR156A and MIR156C throughout shoot development, but does not contribute to their downregulation during vegetative phase change. In this regard, it is important to emphasize that although MIR156A and MIR156C expression is repressed during the adult phase, these genes are expressed in adult shoots and still contribute to shoot development during this phase, as evident from the phenotype of the mir156a mir156c mutant (Yang et al., 2013). In this scenario, miR159 does not completely repress MYB33 activity during vegetative development but sets a threshold for its expression. Small changes in the expression of miR159 could therefore have significant effects on the timing of vegetative phase change, as illustrated in Figure 5. This hypothesis is supported by the observation that loss of MYB33 expression slightly accelerates abaxial trichome production, demonstrating that MYB33 is active during shoot development. It is also noteworthy that MYB33 not only promotes the transcription of MIR156A and MIR156C, but the transcription of SPL9, a direct target of miR156. These functions are counterintuitive because they have opposite effect on SPL9 expression. On the other hand, this regulatory network enables MYB33 to regulate SPL9 expression by a variety of mechanisms and implies that MYB33 is a regulator of SPL9, thus facilitating vegetative phase change.
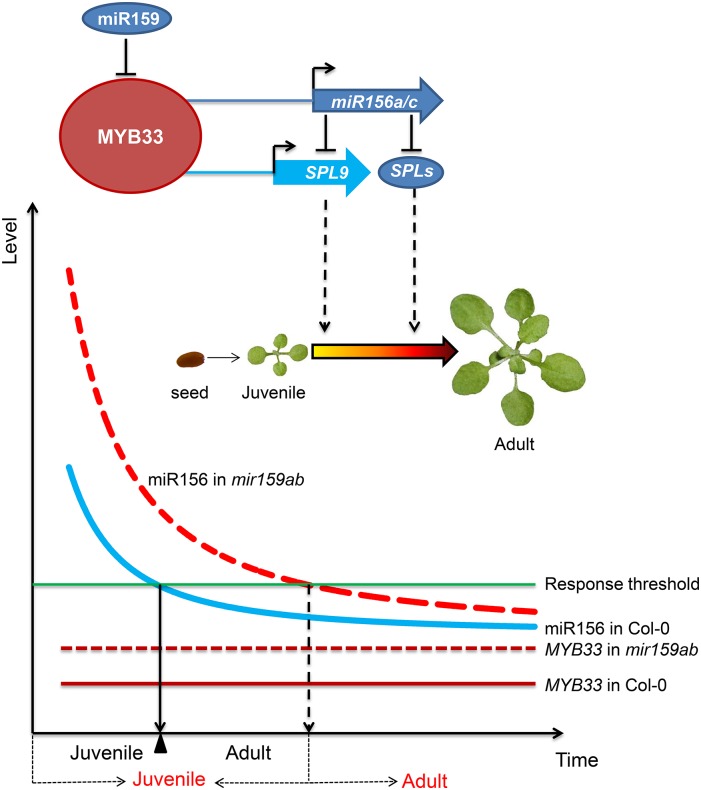
Figure 5.
A Model for the Repression of miR156 by miR159 to Control the Correct Timing of Juvenile-to-Adult Transition in Arabidopsis.
MYB33, a target of miR159, serves as an activator of miR156a and miR156c as well as SPL9 to regulate vegetative phase change in Arabidopsis. In the myb33 mutant, reduction in miR156 causes elevated levels of SPL9; in the meantime, the direct transcriptional activation of SPL9 by MYB33 is also compromised. The reduced level of miR156 increases the abundance of the endogenous SPL9 transcript, thus compensating for the effect of myb33 on SPL9 transcription. In the wild type, miR159 does not completely repress MYB33 activity during vegetative development but sets a threshold for its expression. MYB33 (solid red line) also creates a threshold level of miR156 (solid blue curve) for plants to ensure that vegetative phase change is initiated at the right time. In the mir159ab double mutant, due to the absence of miR159 regulation, MYB33 (dotted red line) is derepressed, and it is highly and ectopically expressed in incorrect cells and tissues to activate the expression of miR156 (dotted red curve) to delay vegetative phase change. x axis: time from juvenile to adult. Black letters under the x axis indicate time from juvenile to adult in the wild type, and red letters indicate time from juvenile to adult in miR159ab mutant. y axis, miR156 and MYB33 expression level.
These and other results (Alonso-Peral et al., 2012) reveal that the function of miR159 is more complex than originally thought. Rather than acting solely to turn MYB33 expression off or on in particular tissues or at particular times in development, miR159 modulates the expression of MYB33 (Alonso-Peral et al., 2012) and serves as a thresholding factor to restrict the expression of this potentially deleterious regulator to a low but functional level in tissues in which its function is required.
METHODS
Plant Materials and Growth Conditions
All genetic stocks used in this study were in a Col background. 35S:miR156a, pmiR156a:GUS, pEG302:rSPL9, pSPL9:rSPL9-GUS, and toe1/2 (toe1-2 toe2-1) were seed stocks as described previously (Wu et al., 2009; Yang et al., 2013); pMYB33:mMYB33 (mMYB33), pMYB33:MYB33-GUS (MYB33-GUS), pMYB33:mMYB33-GUS (mMYB33-GUS), and mir159ab were a kind gift from Anthony A. Millar (Canberra, Australia). myb33 (SALK_065473), myb65 (SALK_063552), and ga1-6 were ordered from the Arabidopsis Biological Resource Center, and ga1-6 was backcrossed to Col-0 six times. MIM156, miR159a, and miR172b coding sequences were PCR amplified. MIM156 was cloned into the BamHI and SmaI sites, and miR159a and miR172b were cloned into the PstI and SmaI sites in the pSY06 expression vector under the control of the Arabidopsis thaliana UBI10 promoter to generate UBI10:MIM156, UBI10:miR159a, and UBI10:miR172b constructs. To generate pMYB33:3×FLAG-mMYB33-3′UTR and pMYB33:HA-mMYB33-3′UTR constructs, the genomic coding sequence plus the 3′ sequence of mMYB33 was amplified with PCR using pMYB33:mMYB33 plasmid as the template and was fused to 3×FLAG and HA first and then was put under the control of the MYB33 native promoter, and the sequences were finally cloned into the BamHI site in the pCAMBIA3300 vector. Seeds were grown in a mixture of soil and vermiculite (1:1) and left at 4°C for 2 d before transfer to the growth chamber. Plants were grown under short-day conditions (10 h light/14 h dark, 130 μmol/m2/s) at 22°C. All of the experiments done in this study were repeated two to three times under the same conditions.
Plant age was measured from the time when seeds were transferred to the growth chamber. Abaxial trichomes were scored 2 to 3 weeks after planting with a stereomicroscope. For leaf shape analysis, fully expanded leaves were removed, attached to cardboard with double-sided tape, flattened with transparent tape, and then scanned in a digital scanner.
GA Treatment
For phenotypic characterization, 6-d-old plants, including Col-0, mir159ab, mMYB33, and myb33 plants, were sprayed with 100 μM GA in 2% ethanol and 2% ethanol (mock) once. The abaxial trichomes and the length and width of leaves were scored when leaves were fully expanded. For gene expression analysis, 14-d-old wild-type (Col-0), ga1-6, and mir159ab plants were sprayed with 100 μM GA or mock treated. The GA-treated plants and the mock-treated plants were harvested 8 h after treatment to analyze the expression of miRNAs.
Small RNA Gel Blot and qRT-PCR Analyses
Small RNA gel blotting was conducted following the protocol described previously (Wu and Poethig, 2006). Tyr-tRNA was used as the loading control. About 2-mm shoot apices of 10-, 14-, 18-, and 22-d-old Col-0, mir159ab, and UBI10:miR159a plants were harvested. Total RNA was isolated using TRIzol reagent and then treated with DNaseI to remove genomic DNA. The first-strand cDNA was synthesized using a TAKARA first-strand cDNA synthesis kit. For the synthesis of the first cDNA strand of miRNA, the reverse transcription primers were designed as previously reported (Varkonyi-Gasic et al., 2007). Reverse transcription was done as follows: one cycle at 16°C for 30 min, 60 cycles at 30°C for 30 s, 42°C for 30 s, and 50°C for 1 s, followed by incubation at 85°C for 5 min to inactivate the reverse transcriptase. Real-time PCR was performed using diluted cDNA on Step One Plus (ABI) real-time PCR machine. Gene-specific primers (Supplemental Table 2) were designed using Beacon Designer 7 software. The Arabidopsis GAPC and eIF4A1 genes (Supplemental Table 2) were selected as the internal controls, and SNOR101 was selected as the internal control for miRNA quantitation. qRT-PCR was performed in 20 μL Eva Green PCR mixtures including 2 μL 10×ExTaq PCR buffer, 1 μL 20×Eva green dye, 1 μL primer mix, 1 μL template, 0.5 units HS ExTaq, and double distilled water.
GUS Staining
GUS activity was visualized by staining the whole plants at the vegetative stage from different transgenic GUS reporter lines. Plants were treated in 90% cold acetone for 10 min; after removal of acetone, plants were washed with staining buffer without X-Gluc [0.0216 M NaH2PO4, 0.029 Na2HPO4, 2 mM K3Fe(CN)6, 2 mM K4Fe(CN)6, and 1 mL/L Triton X-100] and then put under vacuum for 10 min in the staining buffer. After incubation with staining buffer at 37°C for 2 h, stained plants were cleared in 75% (v/v) ethanol and photographed using a Leica microscope. The expression of the GUS gene from different reporter lines was measured using qRT-PCR.
ChIP-qPCR Analysis
The promoter sequence of MIR156A was analyzed using PLACE and Athmap, and the predicted MYB-protein binding sites were identified as shown in Supplemental Table 1. ChIP-qPCR primers were designed surrounding these putative MYB binding sites. More than 3 g of 10-d-old pMYB33:3×FLAG-mMYB33-3′UTR and pMYB33:HA-mMYB33-3′UTR transgenic seedlings were harvested and then carefully ground to fine powder in liquid nitrogen. ChIP was performed as described previously (Gendrel et al., 2005) using an anti-FLAG polyclonal antibody (Sigma-Aldrich; F7425) for the chromatin isolated from pMYB33:3×FLAG-mMYB33-3′UTR and pMYB33:HA-mMYB33-3′UTR transgenic plants. ChIP signals from pMYB33:3×FLAG-mMYB33-3′UTR and pMYB33:HA-mMYB33-3′UTR were first normalized to their corresponding input, and then the normalized value of pMYB33:3×FLAG-mMYB33-3′UTR was divided by that of pMYB33:HA-mMYB33-3′UTR to get a fold enrichment. The LFY gene was used as a positive control for MYB33 binding, whereas ACTIN2 was used as a negative control.
Experiments to Test the Activation of MIR156A and SPL9 by MYB33
To generate the MIR156A reporter construct with a mutated cis-regulatory sequence to which MYB33 is bound (pΔmiR156a:GUS), the putative MYB33 binding sequence (ctgttggggata) at the P3 site in MIR156A promoter was mutated to gtaggtgaaggg using overlapping PCR, and the mutated MIR156A promoter was then cloned into the pmiR156a:GUS vector as reported previously (Yang et al., 2013). To generate the SPL9 transcriptional reporter construct (pSPL9:GUS), a 2.4-kb genomic fragment upstream and a 0.8-kb genomic fragment downstream of the SPL9 open reading frame were cloned into the NcoI/EcoRI and PmlI/BstEII sites in the pCAMBIA3301 vector, respectively. The mutated version of the SPL9 transcriptional reporter construct (pΔSPL9:GUS) was generated by replacing the predicted MYB33 binding site (ggata) at the P2 site in SPL9 promoter with aagcg using overlapping PCR. The construct overexpressing MYB33 (UBI10:mMYB33) was generated by putting the miR159-insensitive MYB33 cDNA under the control of the Arabidopsis Ubi10 promoter in the pSY06 vector. All vectors, including the control vector (pSY06), UBI10:mMYB33, p156a:GUS, pΔ156a:GUS, pSPL9:GUS, and pΔSPL9:GUS, were transformed into Agrobacterium tumefaciens strain GV3101. The transformed Agrobacterium was injected into the leaves of Nicotiana benthamiana. The transformed N. benthamiana was first placed in the dark for 12 h and then grown in the greenhouse (22°C, 12 h light/12 h dark) for 48 h. Six leaves from each group were sampled, and their GUS expression was analyzed using qRT-PCR. The N. benthamiana NbEF1α gene was used as an internal control.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MIR159A, AT1G73687; MIR159B, AT1G18075; MYB33, AT5G06100; MYB65, AT3G11440; MIR156A, AT2G25095; MIR156C, AT4G31877; SPL2, AT5G43270; SPL3, AT2G33810; SPL5, AT3G15270; SPL6, AT1G69170; SPL9, AT2G42200; SPL11, AT1G27360; SPL13, AT5G50570; SPL15, AT3G57920; MIR172B, AT5G04275; TOE1, AT2G28550; TOE2, AT5G60120; LFY, AT5G61850; ACTIN2, AT3G18780; GAPC, AT3G04120; and eIF4A1, AT3G13920.
Supplemental Data
Supplemental Figure 1. GA and miR159 act in parallel pathways to regulate vegetative phase change.
Supplemental Figure 2. Expression levels of MYB33 and miR159 in Col-0 and mir159ab plants at different development stages in short days.
Supplemental Figure 3. miR172 and its targets, TOE1 and TOE2, act downstream of miR159.
Supplemental Figure 4. MYB33 and MYB65 act redundantly to be responsible for the mir159ab mutant phenotype.
Supplemental Figure 5. Effect of different light intensity on vegetative phase change phenotypes of myb33 and UBI10:miR159a.
Supplemental Figure 6. The phenotype of 30-d-old Col-0, mMYB33, 3×FLAG-mMYB33, and HA-mMYB33 plants.
Supplemental Figure 7. Expression analysis of SPL genes in Col-0 and myb33 plants.
Supplemental Table 1. ChIP-PCR-amplified regions and predicted MYB binding sites at the miR156a, miR156c, and SPL9 loci.
Supplemental Table 2. Primers used in this study.
Supplemental File 1. Tables of statistical analysis results.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31470063 and 31271313), the Zhejiang Natural Science Foundation (LR13C060001), a start-up fund from Zhejiang Agriculture and Forestry University (2012FR025,2013FR083), and the 1000 Youth Talents Program in China (2034020065) to G.W. and C.G. Work in the Poethig lab was supported by a grant from the National Institutes of Health (NIH GM51893). We thank Anthony A. Millar (Canberra, Australia) for providing mMYB33, MYB33-GUS, mMYB33-GUS, and mir159ab seeds and constructs. We thank members in the Wu lab for useful comments on the manuscript.
AUTHOR CONTRIBUTIONS
C.G. and G.W. designed the experiments. C.G., Y.X., M.S., Y.L., X.W., Z.Z., H.W., and G.W. performed the experiments. C.G., R.S.P., and G.W. analyzed the data. G.W. and R.S.P. wrote the manuscript.
Glossary
- miRNA
microRNA
- GA
gibberellic acid
- TSS
transcription start site
- ChIP
chromatin immunoprecipitation
References
- Achard P., Herr A., Baulcombe D.C., Harberd N.P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131: 3357–3365. [DOI] [PubMed] [Google Scholar]
- Allen R.S., Li J., Stahle M.I., Dubroué A., Gubler F., Millar A.A. (2007). Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc. Natl. Acad. Sci. USA 104: 16371–16376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Peral M.M., Sun C., Millar A.A. (2012). MicroRNA159 can act as a switch or tuning microRNA independently of its abundance in Arabidopsis. PLoS One 7: e34751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Peral M.M., Li J., Li Y., Allen R.S., Schnippenkoetter W., Ohms S., White R.G., Millar A.A. (2010). The microRNA159-regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis. Plant Physiol. 154: 757–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J., Bartel D.P. (2005). Antiquity of microRNAs and their targets in land plants. Plant Cell 17: 1658–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.M., Poethig R.S. (1995). Gibberellins promote vegetative phase change and reproductive maturity in maize. Plant Physiol. 108: 475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., García J.A., Paz-Ares J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037. [DOI] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Martienssen R., Colot V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2: 213–218. [DOI] [PubMed] [Google Scholar]
- Hsieh L.C., Lin S.I., Shih A.C.C., Chen J.W., Lin W.Y., Tseng C.Y., Li W.H., Chiou T.J. (2009). Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 151: 2120–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter R.A., Poethig R.S. (1998). The specification of leaf identity during shoot development. Annu. Rev. Cell Dev. Biol. 14: 373–398. [DOI] [PubMed] [Google Scholar]
- Lee H., Yoo S.J., Lee J.H., Kim W., Yoo S.K., Fitzgerald H., Carrington J.C., Ahn J.H. (2010). Genetic framework for flowering-time regulation by ambient temperature-responsive miRNAs in Arabidopsis. Nucleic Acids Res. 38: 3081–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854. [DOI] [PubMed] [Google Scholar]
- Li Y., Alonso-Peral M., Wong G., Wang M.-B., Millar A.A. (2016). Ubiquitous miR159 repression of MYB33/65 in Arabidopsis rosettes is robust and is not perturbed by a wide range of stresses. BMC Plant Biol. 16: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May P., Liao W., Wu Y., Shuai B., McCombie W.R., Zhang M.Q., Liu Q.A. (2013). The effects of carbon dioxide and temperature on microRNA expression in Arabidopsis development. Nat. Commun. 4: 2145. [DOI] [PubMed] [Google Scholar]
- Millar A.A., Gubler F. (2005). The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17: 705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W., Li J., Song R., Messing J., Chen X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12: 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picó S., Ortiz-Marchena M.I., Merini W., Calonje M. (2015). Deciphering the role of Polycomb Repressive Complex 1 (PRC1) variants in regulating the acquisition of flowering competence in Arabidopsis. Plant Physiol. 168: 1286–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig R.S. (1990). Phase change and the regulation of shoot morphogenesis in plants. Science 250: 923–930. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. [DOI] [PubMed] [Google Scholar]
- Reyes J.L., Chua N.H. (2007). ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 49: 592–606. [DOI] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D.P. (2002). Prediction of plant microRNA targets. Cell 110: 513–520. [DOI] [PubMed] [Google Scholar]
- Serivichyaswat P., Ryu H.S., Kim W., Kim S., Chung K.S., Kim J.J., Ahn J.H. (2015). Expression of the floral repressor miRNA156 is positively regulated by the AGAMOUS-like proteins AGL15 and AGL18. Mol. Cells 38: 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R.S., Tarver J.E., Hiscock S.J., Donoghue P.C. (2014). Evolutionary history of plant microRNAs. Trends Plant Sci. 19: 175–182. [DOI] [PubMed] [Google Scholar]
- Telfer A., Bollman K.M., Poethig R.S. (1997). Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124: 645–654. [DOI] [PubMed] [Google Scholar]
- Tsukaya H., Shoda K., Kim G.T., Uchimiya H. (2000). Heteroblasty in Arabidopsis thaliana (L.) Heynh. Planta 210: 536–542. [DOI] [PubMed] [Google Scholar]
- Usami T., Horiguchi G., Yano S., Tsukaya H. (2009). The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development 136: 955–964. [DOI] [PubMed] [Google Scholar]
- Varkonyi-Gasic E., Wu R., Wood M., Walton E.F., Hellens R.P. (2007). Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Perry S.E. (2013). Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol. 161: 1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.W., Czech B., Weigel D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749. [DOI] [PubMed] [Google Scholar]
- Wu G., Poethig R.S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Park M.Y., Conway S.R., Wang J.W., Weigel D., Poethig R.S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M., Wang Y., Yao Y., Xie C., Peng H., Ni Z., Sun Q. (2010). Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol. 10: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Hu T., Smith M.R., Poethig R.S. (2016a). Epigenetic regulation of vegetative phase change in Arabidopsis. Plant Cell 28: 28–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Guo C., Zhou B., Li C., Wang H., Zheng B., Ding H., Zhu Z., Peragine A., Cui Y., Poethig S., Wu G. (2016b). Regulation of vegetative phase change by SWI2/SNF2 chromatin remodeling ATPase BRAHMA. Plant Physiol. 172: 2416–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Xu M., Koo Y., He J., Poethig R.S. (2013). Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. eLife 2: e00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Conway S.R., Poethig R.S. (2011). Vegetative phase change is mediated by a leaf-derived signal that represses the transcription of miR156. Development 138: 245–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Cao L., Zhou C.M., Zhang T.Q., Lian H., Sun Y., Wu J., Huang J., Wang G., Wang J.W. (2013). Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. eLife 2: e00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Wang H., Lu Y., de Ruiter M., Cariaso M., Prins M., van Tunen A., He Y. (2012). Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. J. Exp. Bot. 63: 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]