Chromosome ends are sequestered from aberrant DNA repair activities within the DNA binding channel of the Ku heterodimer.
Abstract
Telomeres form specialized chromatin that protects natural chromosome termini from being recognized as DNA double-strand breaks. Plants possess unusual blunt-ended telomeres that are unable to form t-loops or complex with single-strand DNA binding proteins, raising the question of the mechanism behind their protection. We have previously suggested that blunt-ended telomeres in Arabidopsis thaliana are protected by Ku, a DNA repair factor with a high affinity for DNA ends. In nonhomologous end joining, Ku loads onto broken DNA via a channel consisting of positively charged amino acids. Here, we demonstrate that while association of Ku with plant telomeres also depends on this channel, Ku’s requirements for DNA binding differ between DNA repair and telomere protection. We show that a Ku complex proficient in DNA loading but impaired in translocation along DNA is able to protect blunt-ended telomeres but is deficient in DNA repair. This suggests that Ku physically sequesters blunt-ended telomeres within its DNA binding channel, shielding them from other DNA repair machineries.
INTRODUCTION
Natural chromosome termini in linear eukaryotic genomes formally resemble DNA double-strand breaks (DSBs). Therefore, chromosome ends are protected from DNA damage sensors by telomeres, specialized nucleoprotein structures composed of telomeric DNA and associated telomere binding proteins. Impaired telomere function triggers a strong DNA damage response that can lead to cell senescence and genome instability via end-to-end chromosome fusions. The majority of eukaryotic telomeres end with single-stranded DNA protrusions called G-overhangs, which are either sequestered in complexes with specialized telomere binding proteins or within DNA secondary structures known as t-loops (Fulcher et al., 2014; Arnoult and Karlseder, 2015; Martínez and Blasco, 2015). Thus, G-overhangs are thought to be necessary for telomere protection. However, a substantial portion of telomeres in angiosperm plants are either blunt-ended or possess G-overhangs that are only a few nucleotides long, too short for stable t-loop formation, or single-stranded protein binding (Kazda et al., 2012). It has been proposed that these blunt-ended telomeres are produced by leading strand replication and, in contrast to other eukaryotes, do not undergo postreplicative resection to form G-overhangs (Kazda et al., 2012; Derboven et al., 2014). It remains unclear how these blunt-ended telomeres are protected from being recognized as DSBs.
Integrity of these blunt-ended telomeres depends on Ku, a key component of the classical nonhomologous end joining (C-NHEJ) DSB repair pathway. Ku is a DNA binding complex with a high affinity for DNA ends. It consists of Ku70 and Ku80 subunits that form a ring-like structure with a preformed channel that slides onto free DNA ends (Walker et al., 2001; Fell and Schild-Poulter, 2015). Ku has multiple functions in C-NHEJ. It acts as a DNA damage sensor that can localize to a DSB within 5 s of its induction (Mari et al., 2006). Ku binding to DNA stabilizes DSBs by preventing excessive nucleolytic resection. In a subsequent step, Ku translocates inwards along DNA to free the broken ends for processing and ligation. Concomitantly, Ku promotes these reactions by recruiting both DNA processing factors required for generating ligatable ends as well as the ligase complex that concludes repair (Fell and Schild-Poulter, 2015).
Inactivation of Ku in Arabidopsis thaliana leads to telomerase-dependent telomere elongation, resection of blunt-ended telomeres by exonuclease 1, and increased telomeric recombination (Bundock et al., 2002; Riha et al., 2002; Gallego et al., 2003; Riha and Shippen, 2003; Zellinger et al., 2007; Kazda et al., 2012). Ku is also important for proper telomere function in many other eukaryotes, including mice, humans, yeast, fungi, and even Drosophila melanogaster, whose telomeres are not formed by arrays of canonical telomeric repeats, but rather by retrotransposons (Samper et al., 2000; d’Adda di Fagagna et al., 2001; Gravel et al., 1998; Maringele and Lydall, 2002; Celli et al., 2006; Wang et al., 2009; Chico et al., 2011; de Sena-Tomás et al., 2015; Melnikova et al., 2005). One of the most common consequences of Ku depletion is nucleolytic resection of chromosome ends and increased telomeric recombination, which, in some organisms, leads to a massive loss of telomeric DNA and ensuing cell death (Wang et al., 2009; de Sena-Tomás et al., 2015). Thus, Ku appears to be an evolutionarily conserved telomere protector. Considering Ku’s importance in C-NHEJ, a DNA repair mechanism that can produce deleterious end-to-end chromosome fusions, the function of Ku at telomeres is puzzling and the mechanism by which it protects telomeres remains unresolved.
Studies in budding yeast and human cells revealed that Ku can interact with telomeric chromatin either by direct binding to telomeric DNA or via interaction with telomere associated proteins such as Sir4 or TRF2 (Hsu et al., 1999; Hass and Zappulla, 2015). In Saccharomyces cerevisiae, mutations that abolish Ku’s DNA binding cause deficiencies in both C-NHEJ and telomere protection (Lopez et al., 2011; Pfingsten et al., 2012). Thus, at least in budding yeast, Ku may utilize a similar mode of action at DSBs and telomeres, as its ability to load onto DNA appears to be essential for both processes. Whether this applies to other organisms remains to be established.
The dependence of Arabidopsis blunt-ended telomeres on Ku provides an important clue to its function at telomeres. It suggests that Ku may bind to the very end of telomeric DNA, sterically hindering access of DNA repair enzymes to the chromosome termini, effectively blocking resection and G-overhang formation. It further implies that, in contrast to C-NHEJ, Ku does not translocate inward onto telomeric DNA and remains, at least temporarily, tethered to telomere termini. In this study, we investigated how Ku protects telomeres in Arabidopsis by analyzing Ku complexes with modified DNA binding properties. We determined that, as in budding yeast, both DNA repair and telomere protection rely on the ability of Ku to bind to DNA. In addition, we discovered that a Ku complex proficient in DNA loading, but impaired in docking or sliding, is still fully proficient in protecting blunt ended telomeres, but deficient in DNA repair. This observation supports the hypothesis that Ku sequesters chromosome termini within its DNA loading channel and protects them from nuclease processing. Furthermore, our data indicate that the role of Ku in telomere length regulation is partially uncoupled from its function in chromosome end protection.
RESULTS
Ku Loading onto DNA Is Required for DNA Repair and Telomere Protection
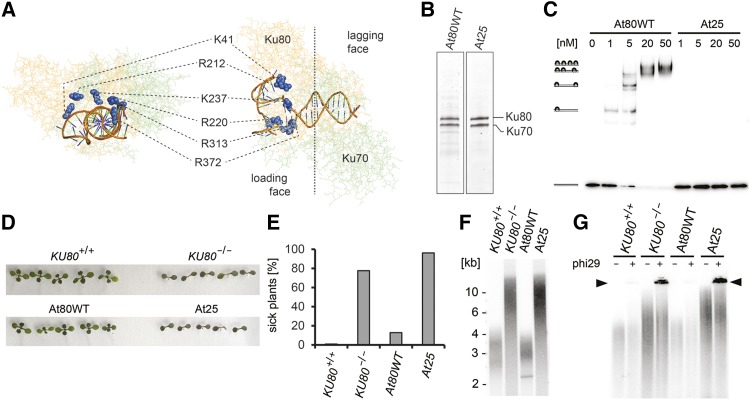
To determine whether Ku binding to DNA is required for protection of blunt-ended telomeres, we generated Arabidopsis lines in which the endogenous Ku was replaced by a DNA binding deficient variant. The crystal structure of human Ku showed that the preformed channel consists of positively charged residues that can accommodate 14 bp of duplex DNA (Walker et al., 2001). Ku loads onto DNA with the Ku80 side of the dimer first (loading face); as a result, Ku70 is positioned proximally to the DNA end (lagging face). We reasoned that modification of the charge on the entry to the channel may abolish Ku binding to DNA. Based on the crystal structure of the human Ku-DNA complex (Walker et al., 2001), we modeled the structure of Arabidopsis Ku to identify conserved charged residues within the channel (Figure 1A). Six residues in the Ku80 subunit located in the leading part of the channel, K41, R212, R220, K237, R313, and R372, were converted to negatively charged glutamines (Figure 1A; Supplemental Table 1). We were able to reconstitute the mutant Ku complex, named At25, after coexpression of the Ku80 and the Ku70 subunits in insect cells (At25), demonstrating that these six substitutions do not affect dimerization (Figure 1B; Supplemental Figure 1). Electrophoretic mobility shift assay (EMSA) showed that At25 is unable to bind to a linear 98-bp DNA fragment, while up to four wild-type Ku molecules (AtWT) associated with this substrate (Figure 1C).
Figure 1.
Ku DNA Binding Is Essential for DNA Repair and Telomere Maintenance.
(A) Predicted structure of the Arabidopsis Ku-DNA complex. Amino acid residues located at the entrance to the central channel are highlighted in blue.
(B) Reconstituted Ku complexes separated by SDS-PAGE and visualized by Coomassie blue staining.
(C) EMSA of a radioactively labeled 98-bp DNA probe (0.3 nM) with increasing concentrations of recombinant Ku complexes. Positions of Ku-DNA complexes with different stoichiometries are indicated on the left side of the gel.
(D) Bleomycin growth assay. Eleven-day-old seedlings grown on media supplemented with 150 ng/mL bleomycin. Seedlings were analyzed for formation of true leaves.
(E) Quantification of the bleomycin growth assay showing the frequency of seedlings with no or rudimentary true leaves. At least 100 seedlings were scored for each category.
(F) Terminal restriction fragment analysis with Tru1I-digested genomic DNA.
(G) Formation of telomeric circles assessed by t-circle amplification assay. Reactions without phi29 polymerase were run in parallel as a control. The signal from t-circles is indicated by the arrowhead. Two independent complementation lines were analyzed; data presented here were obtained with the lines 40-1 and 19-1 (Supplemental Figure 2).
To assess the functionality of the At25 Ku complex in plants, we complemented Arabidopsis lines carrying a disruption in the KU80 gene with ectopically expressed At25 or a wild-type variant of Ku80 (Supplemental Figure 2). Arabidopsis Ku mutants are sensitive to bleomycin, a radiomimetic drug that induces DSBs (West et al., 2002). A seedling growth assay on agar plates with bleomycin showed growth retardation of Arabidopsis At25 mutants comparable to Ku-null plants (Figures 1D and 1E), indicating a deficiency in DNA repair. Inactivation of Ku in Arabidopsis leads to a 3-fold increase in telomere length and elevated telomeric recombination that results in the excision of extrachromosomal telomeric-circles (t-circles) (Riha et al., 2002; Gallego et al., 2003; Zellinger et al., 2007). While complementation of the KU80-null mutation with the wild-type KU80 prevented telomere elongation and t-circle formation, no suppression of these telomeric phenotypes was observed in plants complemented with the At25 construct (Figures 1F and 1G). These data demonstrate that Ku binding to DNA is required for DNA damage repair as well as telomere maintenance.
Partially Impaired DNA Binding Renders the Ku Complex Dysfunctional in DNA Repair
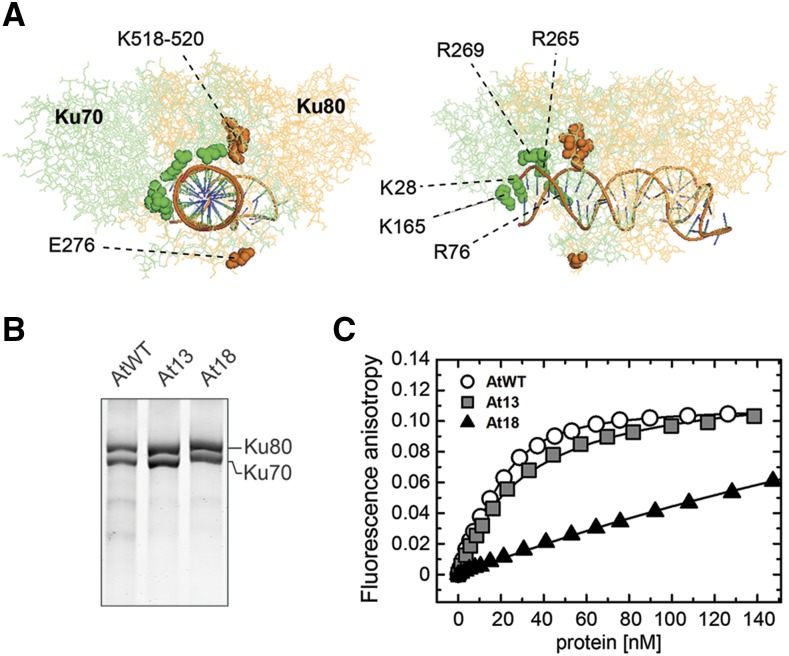
We next asked whether DNA repair and telomere protection have the same requirements for Ku-DNA interaction. To accomplish this, we created a Ku complex with a lower affinity for DNA. We reasoned that modification of the charge in the lagging part of the channel may impair, but not fully abolish, DNA binding. Based on phylogenetic comparisons and structural modeling, we identified four conserved charged residues in Arabidopsis Ku80 (E276, K518, L519, and K520) and five residues in Ku70 (K28, R76, K165, R265, and R269) located in the lagging part of the channel (Figure 2A; Supplemental Table 1 and Supplemental Figure 3). We generated two Ku complexes with swapped charges at these conserved residues: At13 with four substitutions in Ku80 (E276R, K518E, L519E, and K520E) and At18 with five substitutions in Ku70 (K28E, R76E, K165E, R265E, and R269E; Supplemental Table 1; Figure 2B). DNA binding was quantified by fluorescence anisotropy using a 25-bp duplex DNA probe sufficient for association of a single Ku heterodimer. While DNA binding of At13 was only slightly affected (Kd of At13 = 31 ± 1 nM compared with AtWT = 23 ± 1 nM), At18 showed a substantially reduced association with DNA (estimated Kd of At18 > 400 nM) (Figure 2C).
Figure 2.
Ku Complexes with Mutations in the Lagging Part of the Channel.
(A) Predicted structure of the Arabidopsis Ku-DNA complex with amino acid residues mutated in the At13 (orange) and the At18 (green) complexes indicated.
(B) Reconstituted Ku complexes separated by SDS-PAGE and visualized by Coomassie blue staining.
(C) Ku binding to duplex DNA measured by fluorescence anisotropy. The measurements were performed by adding Ku to a fluorescein-labeled DNA duplex (5 nM). Average fluorescence anisotropy values of three replicates were plotted to form binding isotherms.
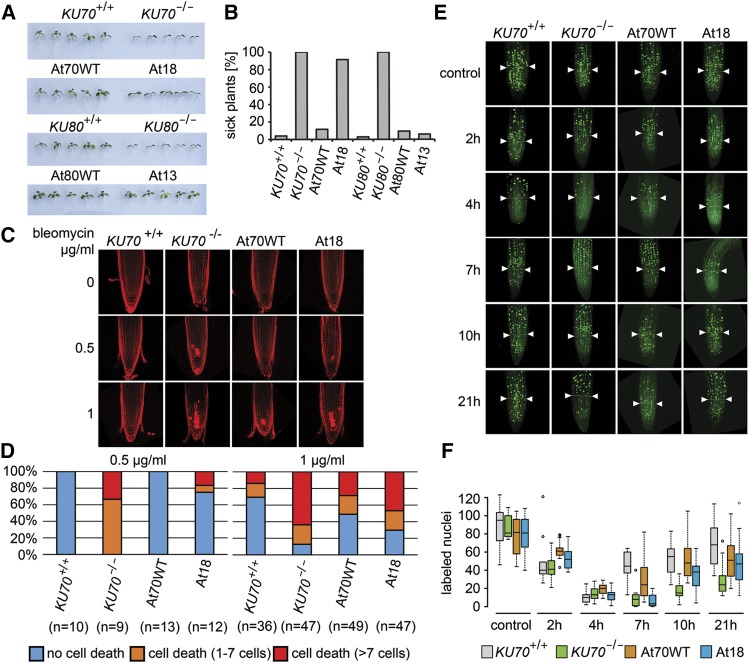
We generated Arabidopsis lines in which the endogenous KU70 or KU80 genes were inactivated and complemented with their respective mutated variants At18 or At13:myc, or wild-type controls (KU70, KU80:myc) (Supplemental Figure 4). Complemented lines exhibiting wild-type levels of Ku were selected for further analyses (Supplemental Figure 4). Using a seedling growth assay, At18 seedlings showed retarded growth on media containing bleomycin indicating their sensitivity to genotoxic treatment. At18 seedlings cultivated on bleomycin appeared slightly larger than Ku70-null mutants, but, in contrast to wild-type controls, they formed no or only rudimentary true leaves (Figures 3A and 3B; Supplemental Figure 5). The growth of At13 lines was comparable to wild-type controls, suggesting their proficiency in DNA repair. Plant growth retardation in response to genotoxic stress is caused by preferential cell death of stem cells in meristematic tissues, and Ku deficiency exacerbates this sensitivity (Fulcher and Sablowski, 2009). Therefore, we quantified cell death in the root apical meristem of At18 plants 24 h after treatment with high doses of bleomycin (Figures 3C and 3D). At18 mutants exhibited a frequency of cell death higher than the wild type but less than Ku70-null plants, corroborating data from the seedling growth assay.
Figure 3.
DSB Repair in Plants Carrying At13 or At18 Ku Complexes.
(A) Eleven-day-old seedlings of complemented lines, wild-type plants, and Ku70 and Ku80 null mutants grown on media supplemented with 150 ng/mL bleomycin.
(B) Quantification of the bleomycin growth assay showing the frequency of seedlings with no or rudimentary true leaves. At least 100 seedlings from two independent complementation lines were scored for each category.
(C) Bleomycin-induced cell death in the root stem cell niche determined by propidium iodide, which stains the interior of dead cells. Representative confocal images of propidium iodide-stained root tips from seedlings incubated for 24 h in the presence of bleomycin.
(D) Quantification of the cell death assay. Root tips were scored for the number of dead cells and divided into three categories (no cell death, 1–7 dead cells, >7 dead cells). Plots show the frequency of each category in the analyzed genotypes. The number of roots scored for each genotype is indicated in parentheses.
(E) Analysis of DNA damage checkpoint by EdU labeling. Representative examples of EdU-stained root tips for each genotype at given time points. Boundaries defining meristematic cells counted in this experiment are indicated by arrowheads.
(F) Quantification of replicating cells per root meristem during the time course after bleomycin treatment. Box plots indicate median, 25th and 75th percentile from 5 to 19 sampled roots.
DNA damage induces cell cycle arrest, which resumes following DNA repair. To examine the effect of At18 mutations on DNA damage checkpoint integrity and repair efficiency, we performed a time-course analysis of replicating cells in the root meristem after DSB induction. Seedlings were incubated for 2 h with bleomycin, sampled at given time points, and incubated for 60 min with EdU to label cells undergoing DNA replication. Analysis of meristematic cells in wild-type roots showed a massive decline in S-phase cells 4 h after bleomycin treatment, followed by a detectable resumption in the cell cycle 7 h after treatment. In contrast, both Ku70-null and At18 lines still showed severe cell cycle inhibition at this time point and gradually resumed DNA replication only at later time points; this resumption appeared to be slower in Ku70-null than in At18 mutants (Figures 3E and 3F). These data show that At18 and Ku70-null mutants have an intact DNA damage checkpoint, but are delayed in repair.
The Ku Complex Partly Impaired in DNA Binding Is Proficient in Telomere Protection
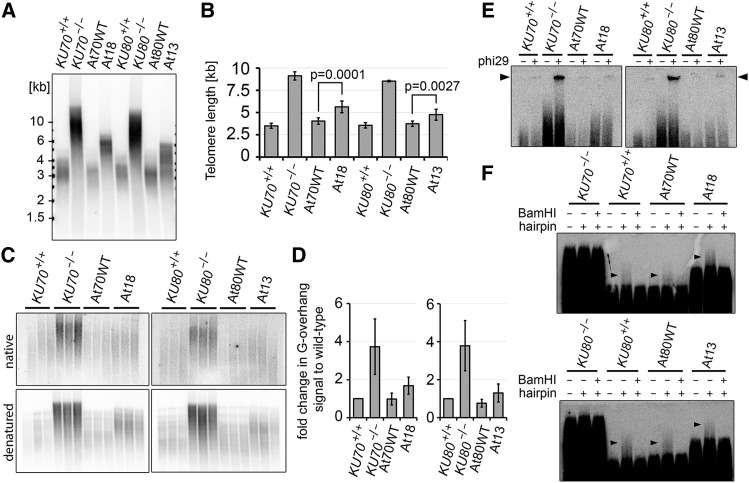
We next analyzed the effect of At18 and At13 complexes on telomere maintenance. Complementation of ku70 and ku80 null mutations with their respective wild-type constructs fully inhibited the telomere elongation observed in Ku-null plants (Figures 4A and 4B). Telomeres in lines complemented with the At18 and At13:myc constructs were substantially shorter than telomeres in Ku-null mutants, but remained on average 1.3× longer than telomeres in lines complemented with the wild-type constructs. Ku deficiency in Arabidopsis also leads to the extension of telomeric G-overhangs through nuclease resection and telomerase elongation (Kazda et al., 2012). In-gel hybridization analysis revealed that both At18 and At13 Ku complexes prevented excessive G-overhang elongation, although a slight increase in the G-overhang signal was detected in At18 and At13 mutants in comparison with control lines (Figures 4C and 4D). These data indicate that At18 and At13 Ku complexes are largely proficient in inhibiting the extension of G-overhangs and duplex telomeres.
Figure 4.
Telomere Maintenance in At13 and At18 Mutants.
(A) Terminal restriction fragment analysis with Tru1I-digested genomic DNA from At18 and At13 plants and corresponding controls.
(B) Quantification of telomere length from TRF blots by TeloTool software (Göhring et al., 2014). Error bars represent sd from at least three independent samples.
(C) G-overhang analysis by the in gel hybridization technique. Restriction fragments generated by HindIII were separated by electrophoresis, and gels were first hybridized under nondenaturing conditions (top panels) than denatured and hybridized again (bottom panels).
(D) Quantification of G-overhang signal from native gels. Signals were normalized to the wild type; error bars represent sd from three independent samples.
(E) The presence of t-circles in At18 and At13 lines detected by t-circle amplification assay. Reactions without phi29 polymerase were run in parallel as a control. The signal from t-circles is indicated by the arrowhead.
(F) Integrity of blunt-ended telomeres in At18 and At13 lines determined by the hairpin ligation assay. TRFs from blunt-ended telomeres that were ligated with a hairpin migrate slower than bulk telomeres (arrowheads). The signal from blunt-ended telomeres is sensitive to cleavage of the hairpin with BamHI.
Ku deficiency in Arabidopsis leads to telomere deprotection, manifested by nucleolytic resection of blunt-ended telomeres, and increased excision of t-circles via homologous recombination (Zellinger et al., 2007; Kazda et al., 2012). While we readily detected signals from t-circles in Ku-null plants, only a weak signal, comparable to wild-type controls, was detected in plants complemented with the At13 and At18 constructs (Figure 4E; Supplemental Figure 6). Next, we examined At13 and At18 plants for the presence of blunt-ended telomeres by ligating blunt-ended hairpins to genomic DNA. Ligation of such hairpins to blunt-ended telomeres covalently links complementary telomeric strands. Restriction fragments containing such telomeres can be separated from nonligated telomeres by alkaline electrophoresis. Slow migrating terminal restriction fragments sensitive to cleavage of the hairpin were detected in both At13 and At18 mutants, demonstrating the presence of blunt ended telomeres (Figure 4F). No such signals were present in Ku-null plants. Taken together, the normal level t-circles and the integrity of blunt-ended telomeres demonstrate that both mutant complexes are fully proficient in telomere protection.
DNA Binding Is Required for Ku to Associate with Telomeres
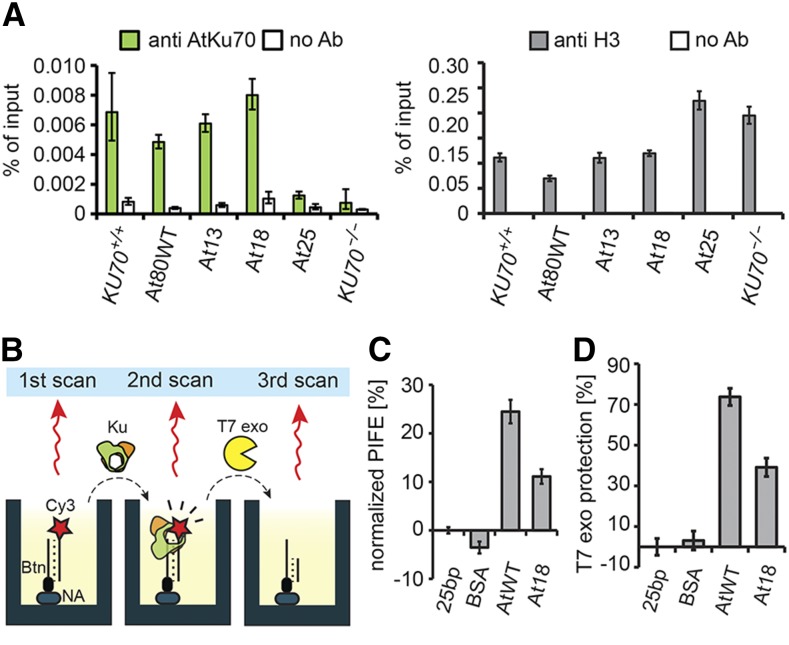
A role of Ku in protecting blunt-ended telomeres predicts its localization to telomeric chromatin. In support of this prediction, chromatin immunoprecipitation (ChIP) with a Ku70 antibody showed an enrichment of telomeric DNA in the immunoprecipitated fraction (Figure 5A). The inability of the At25 Ku complex to complement telomeric phenotypes indicates that telomere protection in Arabidopsis relies on the ability of Ku to load directly onto telomeric DNA. Indeed, no enrichment of telomeric DNA in the ChIP fraction was detected in At25 plants (Figure 5A). However, despite its severely reduced ability to bind DNA, the At18 complex associated to telomeric chromatin as well as wild-type or At13 Ku complexes. While this result is consistent with the observed proficiency of At18 in telomere protection, it also suggests that even a relatively weak Ku-DNA interaction is sufficient for association with telomeric chromatin.
Figure 5.
Association of Ku Complexes with Telomeres and Their Ability to Protect DNA Ends.
(A) Association of Ku with telomeres assessed by ChIP with Ku70 and histone H3 antibodies. Control reaction with no antibody was run in parallel. Error bars represent sd from three technical replications. Increased amount of telomeric DNA immunoprecipitated by histone H3 antibody in At25 and KU70-null plants reflects longer telomeres in these mutants.
(B) Diagram depicting individual steps of the nuclease protection assay coupled to PIFE.
(C) Association of Ku with Cy3-labeled 25-bp DNA determined by PIFE.
(D) Nuclease protection measured as the intensity of Cy3 signal after T7exo treatment relative to DNA probe without added protein. Data in (C) and (D) are derived from consecutive measurements of the same samples. Error bars indicate sd from at least three replicas.
The At18 Ku Complex Can Protect DNA Ends from Nuclease Resection in Vitro
One explanation for the correlation between telomere protection and the ability of Ku to bind DNA is that Ku may physically sequester blunt-ended telomeres, shielding them from nucleolytic degradation. To test whether Ku can inhibit resection, we developed a nuclease protection assay utilizing protein-induced fluorescence enhancement (PIFE), a phenomenon whereby an increase in fluorescence is observed when a protein binds to a nucleic acid in the vicinity of a fluorophore. Thus, PIFE can be used to measure association of a protein with DNA (Figure 5B) (Stennett et al., 2015; Valuchova et al., 2016). We linked a linear 25-bp dsDNA at one end to a microtiter well (and have hence named this assay mwPIFE) (Valuchova et al., 2016), while the opposite end was labeled with Cy3 and available for Ku binding. The addition of reconstituted Ku to the well yielded a roughly 25% PIFE signal confirming efficient binding of Ku to the Cy3-labeled oligo. The At18 Ku complex yielded only a 10% PIFE signal (Figure 5C), which is consistent with the reduced binding of At18 to DNA as determined by fluorescence anisotropy (Figure 2C). These preformed Ku-DNA complexes were then treated with the 5′ to 3′ T7 exonuclease and nuclease protection was measured as the fraction of fluorescence remaining after Cy3 removal by the nuclease. This experimental setup enabled us to measure DNA end protection as a function of Ku binding. Both wild-type Ku and At18 complexes showed a substantial protection of 5′ DNA termini from nucleolytic resection (Figure 5D); the reduction in end protection of At18 relative to the wild type was proportional to its reduced ability to bind DNA as determined by mwPIFE (Figures 5C and 5D). This in vitro data demonstrate that the wild type, as well as At18 Ku, can protect blunt-ended DNA termini from removal of the terminal 5′ nucleotide by an exonuclease.
The At18 Ku Complex Is Impaired in Translocation onto DNA
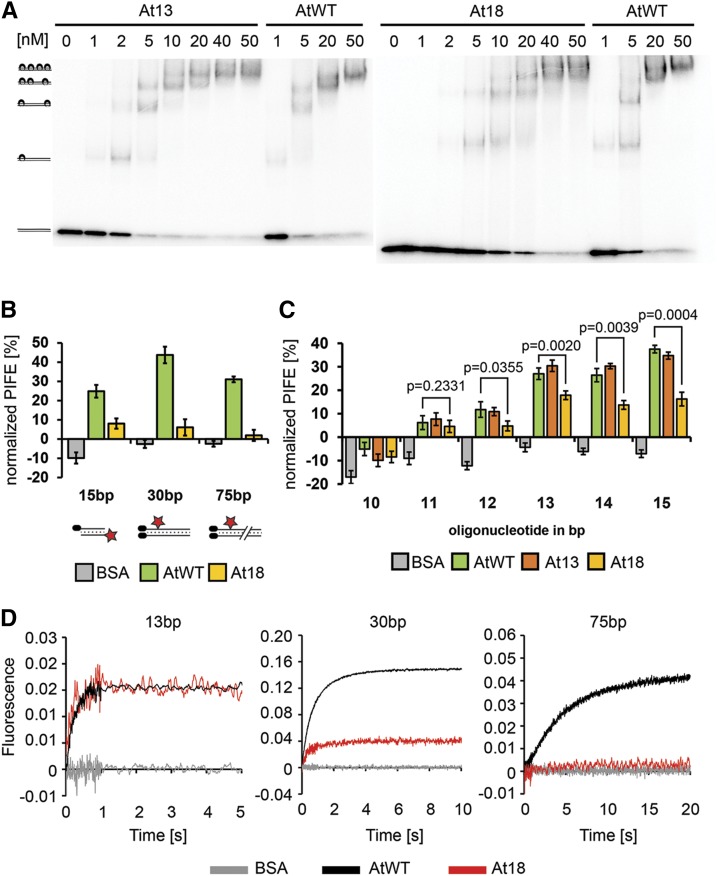
The weak DNA binding activity of the At18 complex is in stark contrast with its ability to bind and protect Arabidopsis telomeres. One possible explanation for this conundrum is that only a partial loading of Ku onto telomeric DNA is sufficient for telomere protection. This is consistent with the position of amino acid substitutions in the lagging part of the DNA loading channel in At18. To characterize the DNA binding properties of At18 in detail, we performed EMSA with a 98-bp DNA probe. This DNA fragment should be able to accommodate up to 4 Ku complexes, resulting in five bands in an EMSA (Figure 6A). Since Ku loads on DNA exclusively through free ends, the two most retarded products, consisting of a DNA probe and three or four Ku molecules, reflect the ability of the Ku complex to load and translocate along DNA. Wild-type and At13 Ku complexes exhibited similar EMSA binding patterns and readily formed four Ku/oligo products at higher concentrations. In contrast, At18 formed higher order complexes less efficiently (Figure 6A).
Figure 6.
At18 Complex Is Impaired in Docking or Sliding on DNA.
(A) EMSA of radioactively labeled 98-bp DNA probe (0.3 nM) with increasing concentrations of recombinant Ku complexes. Positions of Ku-DNA complexes with different stoichiometries are indicated on the left side of the gel.
(B) Steady state interaction of Ku at different positions along DNA determined by mwPIFE. PIFE was calculated as the relative difference in fluorescence upon protein binding. BSA was used as control. Standard deviations from three independent measurements are indicated. Cy3-labeled 15-bp, 30-bp, and 75-bp DNA probes used in the experiment are depicted below the chart.
(C) Analysis of Ku-DNA interaction by mwPIFE with 5′-end labeled DNA probes ranging from 10 to 15 bp. Standard deviations from three independent measurements are indicated. P values show significance of difference between wild-type and At18 complexes determined by two-tailed t test.
(D) Kinetic analysis of Ku-DNA binding by stop-flow using the 13-, 30-, and 75-bp DNA probes. Each trace represents the average of at least five individual measurements. Kinetic curves were scaled according to the data obtained with the 13-bp probe to normalize for the signal amplitude of the At18 variant.
The Ku/DNA association is presumably a multistep process that includes an initial interaction and entry of DNA into the Ku channel, stabilization of this interaction, possibly through conformational changes in the Ku complex, and sliding of Ku along DNA. The reduced ability of At18 complexes to form the three to four Ku/oligo products in an EMSA may either be due to a reduced ability of the mutant complexes to translocate along DNA or due to impaired loading of Ku onto DNA ends. These two parameters are likely coupled, as the translocation of Ku along DNA will stabilize the Ku-DNA association, thereby improving loading. To separately assess the different Ku-DNA binding steps, we designed a set of protein-DNA interaction assays using mwPIFE. Different stages of the Ku-DNA association were monitored by placing a Cy3 fluorophore at increasing distances from the free DNA end: either terminally located at the 5′ end of a 15-bp DNA probe or placed internally 15 nucleotides from the end of a 30-bp DNA probe and 60 bp from the end of a 75-bp probe. The presence of wild-type or At13 Ku complexes usually yielded a 20 to 50% PIFE signal, depending on the probe, while the addition of BSA did not result in any fluorescence enhancement (Figure 6B). The At18 complex yielded significantly less PIFE signals than wild-type Ku with all DNA probes. The decrease was most remarkable with the 75-bp probe, where we observed a 16-fold reduction, whereas only a 3-fold reduction was detected with the end-labeled 15-nucleotide probe. These data indicate that At18 is impaired in the later steps of DNA binding and/or translocation. To further assess the effect of At18 mutations specifically on the initial Ku-DNA interaction, we tested the association of Ku with the shortest possible DNA substrate that does not permit translocation. The minimal DNA length requirement for human Ku binding was reported to be 14 bp (Yoo et al., 1999). Therefore, we assayed PIFE on double-stranded oligonucleotides ranging from 10 to 15 bp (Figure 6C). A PIFE signal was observed with 11- to 15-bp oligonucleotides and increased with the length of the DNA substrate. While there was a negligible difference between At18 and wild-type Ku in binding to 11-bp DNA, this difference became more significant with longer oligonucleotides. This supports the hypothesis that the initial Ku-DNA interaction step is less compromised in the At18 mutant than later steps.
Next, we performed kinetic analysis of the Ku-DNA interaction using a stopped flow system, where Ku was rapidly mixed with DNA and PIFE was measured in millisecond intervals for up to 20 s. We used the same set of DNA substrates as in the steady state studies, with Cy3 placed at varying distances from the free end (Figure 6D), while the opposite end was biotinylated and blocked with neutravidin. As expected, detection of a PIFE signal was fastest at the terminally labeled probe, with signal saturation within 1 s, whereas the fluorescence probe 60 bp from the end did not reach saturation even after 20 s (Figure 6D). Kinetic curves were measured for different protein/DNA molar ratios, and these data were used to calculate the kinetic phases of the Ku-DNA association. Binding to the 13 bp terminally labeled DNA showed a single fast kinetic phase that likely reflects the initial interaction of Ku with DNA (Table 1). The binding curve with the 30-bp probe containing Cy3 15 bp from the free terminus exhibited two kinetic phases, where the first fast phase represents the initial binding interaction and the second slower phase may reflect docking or sliding of Ku onto DNA. The 75-bp DNA substrate with Cy3 placed 60 bp from the free end cannot record initial binding events but clearly showed a slow kinetic step that likely corresponds to sliding of Ku along DNA.
Table 1. Kinetic Model of Ku-DNA Association.
| Binding |
Docking/Sliding |
|||
|---|---|---|---|---|
| Ku_oligo | kon (nM−1 s−1) | koff (s−1) | Kd (nM) | kobs (s−1) |
| AtWT_13bp | 0.017 ± 0.010 | 2.2 ± 0.8 | 131 | Not observed |
| At18_13bp | 0.011 ± 0.006 | 1.6 ± 0.4 | 151 | Not observed |
| AtWT_30bp | 0.009 ± 0.002 | 1.1 ± 0.2 | 121 | 0.63 ± 0.26 |
| At18_30bp | 0.009 ± 0.006 | 1.3 ± 0.2 | 143 | Not observed |
| AtWT_75bp | – | – | – | 0.28 ± 0.04 |
| At18_75bp | – | – | – | Not observed |
Kinetics measurements with three different DNA substrates identified the occurrence of at least two kinetic phases during Ku-DNA association: a fast initial binding followed by a docking/sliding phase. Individual values of the rate of Ku-DNA association (k1) and dissociation (k-1) and equilibrium dissociation constant (Kd
= k-1/k1) evaluated from the concentration dependence of the observed rate of the initial binding phase are shown. The rate of docking/sliding phase represents the value of the second exponential phase observed in the kinetic data (kobs, the combination of individual rate constants of unknown mechanism of sliding). The docking/sliding phase observed with the 30-bp probe may represent different processes than the sliding phase detected with the 75-bp probe. The kinetic model of Ku binding to DNA is represented by the following equation: 
In agreement with the steady state measurements, the PIFE signal of the At18 complex observed in the stopped flow system was weaker than wild-type Ku. The kinetics of the Ku-DNA interaction were also altered (Figure 6D). While At18 showed the initial interaction phase with both 13- and 30-bp substrates, the fluorescence trace with the 30-bp probe exhibited only the fast phase while the slow docking or sliding phase was missing (Table 1). No meaningful signal was detected with the 75-bp probe (Figure 6D). These data support the view that the At18 mutations in the lagging portion of the channel permit Ku to load onto DNA but impair later steps that involve docking and sliding.
DISCUSSION
Ku is involved in two seemingly contradictory processes: telomere protection and DSB repair via NHEJ. Nevertheless, in some respects, the telomeric function of Ku parallels its role in DSB repair. For example, Ku was reported to block Exo1 resection and homologous recombination mediated repair of single-ended DSBs induced during replication in Mre11-deficient yeast (Balestrini et al., 2013). This is analogous to the situation at telomeres, where Ku prevents C-strand resection, recombination, and induction of DNA damage response (Gravel et al., 1998; Maringele and Lydall, 2002). Thus, telomere protection by Ku may represent the initial steps of C-NHEJ, where tethering of Ku to a DNA end creates a physical barrier inhibiting end processing and alternative DNA repair pathways. Blunt-ended chromosome termini produced by leading strand replication represent a perfect substrate for Ku, making this a plausible mechanism for protecting blunt-ended telomeres in plants.
In order to dissect the mechanism of Ku’s action at telomeres, we created Ku complexes with modified DNA binding properties and examined their proficiency in DNA repair and telomere maintenance. Our protein-engineering strategy was based on the modification of different parts of the DNA binding channel. Modification of the charge at the entry to the channel abolished DNA binding, but mutations in the lagging part of the channel still allowed Ku to load onto DNA. While mutations in At13 are distal to DNA and have negligible effect, At18 substitutions are more proximal to DNA in the structural model and strongly impaired DNA binding. Kinetic experiments revealed three steps in the Ku-DNA interaction. The first, fast step likely represents the initial contact of Ku with DNA. We propose that the second, slower phase reflects the docking of Ku onto DNA, accompanied by a conformational change in the complex. Indeed, visualization of the human Ku complex by electron microscopy and DNA cross-linking experiments indicates that the C-terminal SAP domain of Ku70 undergoes a major rearrangement upon DNA binding (Rivera-Calzada et al., 2007; Lehman et al., 2008). The sliding of Ku along DNA represents the slowest kinetic phase. The lack of both docking and sliding phases in At18 points to an impaired transition to a more stable Ku-DNA complex and inefficient sliding. Our data show that protection of Arabidopsis telomeres requires Ku to bind DNA through the central channel but is less sensitive to defects in Ku docking and sliding, and provides strong support to the hypothesis that loading of Ku onto the chromosome end and its sequestration within the DNA loading channel is sufficient for telomere protection. Consistent with this hypothesis, the At18 complex efficiently associates with telomeric chromatin and protects blunt-ended DNA substrates in vitro despite being impaired in DNA binding.
The sensitivity of plants carrying At18 to bleomycin-induced DSBs is in accordance with the observed biochemical behavior of the complex. Inefficient sliding of Ku away from a DSB may lead to occlusion of DNA termini and aberrant assembly of downstream NHEJ factors. DNA-PKcs, the core NHEJ component in vertebrates, binds extreme termini of the DSB and promotes internalization of Ku (Yoo and Dynan, 1999; Turchi et al., 2000). While DNA-PKcs is absent in Arabidopsis, displacement of Ku from the DSB is also expected to occur during the assembly of the evolutionary conserved ligase IV-XRCC4 complex (Kysela et al., 2003; Hammel et al., 2011). Furthermore, DSB occlusion by Ku may hinder access of factors involved in alternative DNA repair pathways and hence block its efficient repair (Wang et al., 2006; Fattah et al., 2010; Balestrini et al., 2013).
It has been estimated that ∼10% of Arabidopsis telomeres are blunt-ended, while 40% undergo limited 5′ to 3′ resection resulting in one- to three-nucleotide long G-overhangs (Kazda et al., 2012). Such a residual nucleolytic processing could be caused by the limited ability of Ku to translocate away from chromosome termini upon loading onto telomeres. Such an entrapment of Ku at telomere ends may not only inhibit resection and recombination, but also effectively block NHEJ (Supplemental Figure 7). Ku loading or sliding along DNA can be inhibited by DNA-bound proteins (Roberts and Ramsden, 2007). Thus, the translocation of Ku onto telomeric DNA may be restricted by telomere-associated proteins, such as TRF2, Taz1, or Rap1. Notably, inactivation of these proteins leads to massive chromosome end-to-end fusions via C-NHEJ (Ferreira and Cooper, 2001; Smogorzewska et al., 2002; Pardo and Marcand, 2005) and in vitro experiments with reconstituted telomeres indicate that TRF2/RAP1 modulates NHEJ activity at the stage of Ku loading and DNA-PKcs activation (Bombarde et al., 2010). An alternative, though not mutually exclusive, model of telomere protection may involve binding of multiple Ku molecules to a single chromosome terminus (Riha et al., 2006). While ligation is stimulated at DSBs occupied by one to two Ku molecules, it is inhibited by loading of multiple Ku complexes (Kysela et al., 2003). Ku is locally concentrated at telomeric chromatin through association with telomere binding proteins (Hsu et al., 1999; Martin et al., 1999), which may facilitate the loading of multiple Ku molecules leading to the occlusion of chromosome ends. The affinity of Ku for DNA termini can be further modified by redox potential (Andrews et al., 2006) or posttranslational modifications (Lee et al., 2016), offering additional means for regulating Ku activities depending on the local chromatin environment. The activity of Ku at telomeres can be further modulated through direct association with telomere binding proteins; a recent study in human suggests that binding of TRF2 to the lagging face of the Ku complex blocks NHEJ by preventing synapsis of broken ends (Ribes-Zamora et al., 2013).
Besides mediating protection of blunt-ended telomeres, Ku is also a negative regulator of telomerase in Arabidopsis (Bundock et al., 2002; Riha et al., 2002; Gallego et al., 2003; Riha and Shippen, 2003). It has been proposed that the dramatic elongation of Ku-depleted telomeres is caused by an increase in telomerase’s access to unprotected chromosome termini (Kazda et al., 2012). However, the extension of telomeres and G-overhangs we observed in plants expressing At13 and At18 Ku complexes suggests a more specific mechanism of telomerase regulation that is, at least partially, uncoupled from telomere protection. Arabidopsis possesses two divergent telomerase RNA genes. While TER1 is the canonical telomerase subunit, TER2 RNA acts as a telomerase inhibitor by sequestering telomerase reverse transcriptase in a nonproductive ribonucleoprotein complex (Cifuentes-Rojas et al., 2011, 2012), and Ku has been reported to associate with TER2. Hence, in an analogy to budding yeast, Ku may facilitate recruitment of the nonproductive TER2 telomerase, thereby inhibiting telomere extension by the functional TER1 complex.
Ku is a multifaceted protein complex that represents the major end binding activity to duplex nucleic acids within cells. As such, it has acquired a number of functions throughout evolution, ranging from DNA repair and transcription to regulation of virus pathogenicity (Masson et al., 2007; Fell and Schild-Poulter, 2015). The most perplexing is its counterintuitive involvement in telomere biology that seemingly contradicts its canonical role in C-NHEJ. In this study, we show that the distinct cellular functions of Ku, i.e., telomere protection and NHEJ, may be achieved by modulating its DNA binding properties. A more precise knowledge of Ku-nucleic acid binding requirements in different processes may open interesting avenues for pharmacological interventions in areas such as tumor and antiviral therapies, aging, or genome editing.
METHODS
Plant Material
The Arabidopsis thaliana ecotype Col-0 ku70-2 (SALK_123114) and ku80-7 (SALK_112921) lines were obtained from the Nottingham Arabidopsis Stock Centre. Plants were grown in phytotrons under LED illumination (white 77%/red 20%/infrared 3%; 150 μmol m−2 s−1) with 16/8-h light/dark regime. Genotyping was performed by PCR with primers indicated in Supplemental Table 1.
Generation of Complementation Vectors and Transgenic Plants
The AtKu70 promoter as well as the first exon and first intron were amplified from genomic DNA of Arabidopsis Col-0 using primers AtKu705’F/AtKu705’R (Supplemental Table 1) and cloned into pGEM-T (Promega). AtKu70 cDNA was amplified using GEM_cF WT/GEM_cR or GEM_cF#2/GEM_cR primers for Ku70WT or At18, respectively. The AtKU70 3′ untranslated region was amplified from gDNA of Arabidopsis Col-0 using GEM_uF/GEM_uR primers. pGEM-T containing the AtKU70 promoter region was digested with XbaI and respective cDNA fragments and 3′ untranslated region was cloned using In-Fusion HD cloning kit (Clontech). XbaI restriction site was removed by site-directed mutagenesis with primers GEM_noXba_F/GEM_noXba_R. AtKu70 was amplified using AtKu705’F/GEM_uR from pGEM-T vector and subsequently cloned into the SmaI site, introduced using site-directed mutagenesis, into the plant binary vector pCBK05 (Riha et al., 2002) using pCBKSma_F/pCBKSma_R. AtKu80WT and AtKu13 cDNAs were amplified using primers Ku80cSap_F/Ku80cBbvCI_r. Plant binary vector pCBA02 containing the AtKU80 gene with 12x Myc-tag at the C terminus was digested with SapI/BbvCI restriction enzymes and AtKu80 cDNA fragments were cloned using In-Fusion HD cloning kit (Clontech).
Plant Transformation
Binary vectors were electroporated into Agrobacterium tumefaciens GV3101 and Arabidopsis plants heterozygous for ku70-2 or ku80-7 were transformed by the floral dip method. Transformed plants were selected on soil sprayed with 40 μg/mL BASTA. Protein levels in transgenic plants were examined by immunoblotting using rabbit anti-AtKu70 (custom made against the entire Ku70 protein) and rabbit anti-c-Myc A-14 (Santa Cruz) and detected on LiCor Odyssey using goat IRDye 800 CW α- rabbit.
Protein Expression and Purification
The expression vector pFastBac Dual (Invitrogen) was used to coexpress Arabidopsis Ku70 and His-tagged Ku80 subunits in insect cells using a baculovirus expression system. The pFastBac Dual-based plasmids were transposed to EMBacY bacmid in Escherichia coli and subsequently transfected to Sf9 cells for baculovirus generation. Tni cells were infected with virus and harvested on DPA3. Cells were resuspended in lysis buffer (50 mM Tris-Cl, 250 mM KCl, 10% v/v glycerol, and 1 mM DTT, pH 8.0) supplemented with protease inhibitors (Roche) and frozen in liquid nitrogen. Thawed cells were spun and Ku was bound to His Mag Sepharose Ni (GE Healthcare). Beads were washed in lysis buffer containing 50 mM imidazole, and Ku was eluted in lysis buffer with 250 mM imidazole. Proteins were filtered using Nanosep centrifugal columns (Pall) and stored at 4°C.
EMSA
DNA binding substrate was a nontelomeric PCR fragment amplified from Arabidopsis cDNA by qF1/qR1 primers (Fermentas). Blunt-ended products served as substrate for kinase end labeling with 32P-ATP. Increasing amounts of Ku protein were incubated with DNA (0.3 nM) in a final volume of 20 μL binding buffer (35 mM Tris, 5% glycerol, 150 mM KCl, 1 mM EDTA, and 0.1 mM DTT). Binding was performed at room temperature for 30 min. Samples were loaded onto native polyacrylamide gel (5% polyacrylamide 29:1 [Sigma-Aldrich], 0.5× TBE, 0.33% APS, and 0.06% Temed) that had undergone pre-electrophoresis at 160 V for 10 min at 4°C in 0.5× TBE. Electrophoresis continued with samples in the same conditions for 3 h. Gels were dried and exposed to phospho-screen (GE Healthcare or Amersham). The phospho‐screens were scanned with Molecular Imager FX or Pharo Plus FX (Bio-Rad).
Fluorescence Anisotropy
To measure Ku binding to DNA that was conjugated with fluorescein using fluorescence anisotropy, the excitation wavelength was set to 494 nm and emission wavelength to 521 nm with the width of slits at 9 nm. The integration time was 3 s. The titration of 1 μM Ku solution into 5 nM DNA solution (25 mM Tris and 150 mM KCl, pH 8.0) was performed in a 10 × 4-mm quartz-glass cuvette with a chamber for magnetic bar stirrer at 25°C. A cuvette was equilibrated 10 min prior each titration experiment. All fluorescence measurements were performed on a FluoroMax-4 spectrofluorometer (Horiba Jobin Yvon) equipped with automatically adjustable polarizers for excitation and emission controlled by Origin-based FluorEssence software (version 2.1.6). Fluorescence anisotropy was measured three times and averaged for each point. Equilibrium dissociation constant values were calculated using a one-site binding model by SigmaPlot 12 (Systat Software).
mwPIFE Assay and Nuclease Protection Assay
mwPIFE was performed as previously described (Valuchova et al., 2016). Fluorescence (F1) of the DNA probe was measured in 100 μL EMSA buffer using FLUOstar Omega (BMG Labtech; scan width 6 mm with 10 × 10 scan points and 10 flashes per point, excitation 540-10 BP filter, emission 580-10 BP filter, gain 2800). The buffer was replaced with 100 μL of EMSA buffer with Ku and plates were incubated for 30 min in the dark prior to the second fluorescence measurement (F2). PIFE in each well was determined with the formula: PIFE = (F2 − F1)/F1 * 100 [%], and this value was further normalized by subtracting PIFE in a control well without a protein. All measurements were performed in triplicate. The first steps of the T7 nuclease protection assay represent a standard mwPIFE reaction in EMSA buffer supplemented with 10 mM magnesium acetate. Following the PIFE measurement, the Ku-DNA mixture (15 and 2.5 pmol, respectively, in 100 μL per well) was treated with 10 units of T7 Exonuclease (New England Biolabs) for 1 h at 37°C. Control reactions without T7 exonuclease were run in parallel. After washing wells three times with 200 μL of the EMSA buffer, the fluorescence (F3) was measured in 100 μL of EMSA buffer. For all replicas, we subtracted F3 from F1 and calculated the average decrease in fluorescence, ΔF. The T7 exonuclease-induced decrease in fluorescence was calculated by subtracting ΔF values from T7 exonuclease-treated wells from nontreated wells. The T7 exonuclease-induced decrease in fluorescence in wells with free DNA probe represented no protection. Relative protection of the tested proteins was calculated with the formula (decrease_no protein – decrease_protein)*100/deacrease_no protein [%].
Stopped-Flow PIFE
Stop-flow PIFE was measured in a three syringe SFM-300 device equipped with a microcuvette µFC 08 cell (8 µL) and combined with a manual monochromator spectrometer MOS-200 equipped with a Xe arc lamp (BioLogic). The instrument was operated by a BioKine 32 v 4.63. We applied excitation light at 547 nm and detected emission using a 585/65 ET band-pass filter (AHF Analysentechnik) in 1-ms intervals for 1-s and 10-ms intervals for 20 s. The measurements were performed in the following buffer (150 nM KCl, 35 mM Tris-Cl pH 8.15, 1 mM EDTA, 0.1 mM DTT, 22.4 mM imidazole, and 5.90% glycerol). Protein solution (250 nM; the first syringe) was premixed with buffer (the second syringe) in a ratio from 0:10 to 10:0, and the premix was then mixed with 25 nM DNA oligonucleotide and 250 nM NeutrAvidin (Thermo Scientific; the third syringe) in a 1:1 ratio. An average trace was composed of at least five traces and was used for analysis. The fluorescence traces were fitted to single or double exponentials using KinTek Global Kinetic Explorer version 5.2 providing the observed rate constants and amplitudes of individual kinetic phases.
ChIP
Immunoprecipitation was done according to Kaufmann et al. (2010). Tissue of 0.7 to 0.9 g of ∼10-d-old seedlings was used. Crude nuclear pellet was resuspended in 2 mL of Sonic buffer and transferred to two 1-mL milliTUBES with AFA fiber (Covaris). Chromatin was sonicated three times for 2 min using S220 Focused-ultrasonicator Covaris (peak power 140, cycles/burst 200, duty factor 5.0). Supernatant was diluted 10 times using modified IP buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 10 µM ZnSO4, and 0.1% Triton X-100). After preclearing with 20 μL of IP buffer equilibrated Protein G Mag Sepharose slurry (GE Healthcare), 100 μL was saved as input and the rest was divided into three equal parts for overnight immunoprecipitation at 4°C. No-antibody, 5 μL of anti-histone H3 antibody (1 mg/mL; ab1791; Abcam), and 10 μL of rabbit anti-AtKu70 (custom made against the whole Ku70 protein) were added to respective parts. Twenty microliters of IP buffer equilibrated Protein G Mag Sepharose slurry was added to each part and incubated at 4°C for 4 h. Protein-DNA complexes were eluted and incubated with 1 μL of RNase A (10 mg/mL) at 37°C for 1 h and with 2 μL of Prot. K (20 mg/mL) at 55°C for 1 h. After overnight de-cross-linking at 65°C, phenol-chloroform extraction, isolated DNA was dissolved in 50 μL of 5 mM Tris-Cl, pH 8.0. Quantification of telomeric DNA was performed by qPCR as described (Vaquero-Sedas and Vega-Palas, 2014). DNA purified from ChIP (1 µL) was used in 20 μL of qPCR reaction containing 1× LightCycler 480 High Resolution Melting Master mix, 3 mM MgCl2, and 0.25 µM each TelA and TelB primers (Supplemental Table 1). Reactions were performed in technical triplicate and percent input method was used for calculation.
DNA Repair and Checkpoint Assays
The seedling growth assay was performed by germinating sterilized seeds on 0.5× Murashige and Skoog (MS) plates supplemented with 150 ng/mL bleomycin (Calbiochem). Seedlings were scored 10 d after germination. The cell death assay was performed as described (Fulcher and Sablowski, 2009). For the DNA damage checkpoint assay, 3-d-old seedlings were incubated for 2 h in 0.5× MS media supplemented with bleomycin (2 µg/mL), followed by a time course recovery in 0.5× MS media followed by a 1-h treatment with 5 µM EdU (Click-iT EdU Alexa Fluor 488 imaging kit; Molecular Probes). Seedlings were treated in fixative solution (ethanol:AcOH, 3:1), subjected to Click-iT chemistry and 4′,6-diamidino-2-phenylindole counterstaining, and mounted on microscopic slides prepared in 50% glycerol. Root tips were imaged with a Zeiss LSM 700 confocal microscope, and the number of nuclei in S-phase was counted in the lower half of the root tip in the region from the tip to the 15th ± 2 cells distal from the quiescent center.
Telomere Structure Analysis
Terminal restriction fragment analysis, t-circle amplification, G-overhang analysis, and the assay for detecting blunt-ended telomeres were performed as previously described (Zellinger et al., 2007; Kazda et al., 2012). Telomere length was determined by TeloTool (Göhring et al., 2014).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: KU70 (At1g16970) and KU80 (At1g48050).
Supplemental Data
Supplemental Figure 1. Separation of reconstituted Ku complexes by native gel electrophoresis.
Supplemental Figure 2. Complementation of Arabidopsis ku80 mutants with the At25 Ku complex.
Supplemental Figure 3. Structure-guided mutagenesis of the lagging face of the Ku complex.
Supplemental Figure 4. Complementation of Arabidopsis with the At13 and At18 Ku complexes.
Supplemental Figure 5. Growth phenotype of mutant and complemented lines.
Supplemental Figure 6. T-circle amplification assay performed with an independent set of samples.
Supplemental Figure 7. Model of telomere protection by Ku.
Supplemental Table 1. Mutant Ku complexes created in this study.
Supplemental Table 2. Primers and oligonucleotides used in this study.
Acknowledgments
We thank Bojan Zagrovic and Anton Polyansky for their expertise in protein modeling and Matt Watson for valuable comments on the manuscript. Expression of recombinant Ku was carried out at the VBCF Protein Technologies Facility. This work was supported by the Austrian Science Fund (Y418-B03), by an EMBO Installation Grant (1304130933), and by the Ministry of Education, Youth, and Sports of the Czech Republic under Project CEITEC 2020 (LQ1601), RECETOX (LO1214) and RECETOX (LM2015051). C.H. and E.J. were supported by the Czech Science Foundation (GAP205/12/0550).
AUTHOR CONTRIBUTIONS
S.V., J.F., and K.R. designed experiments. S.V., J.F., E.J., C.H., P.S.-B., and Z.P. performed experiments and data analysis. S.V. and K.R. wrote the manuscript.
Glossary
- DSB
double-strand break
- C-NHEJ
classical nonhomologous end joining
- EMSA
electrophoretic mobility shift assay
- ChIP
chromatin immunoprecipitation
- PIFE
protein-induced fluorescence enhancement
Footnotes
Articles can be viewed without a subscription.
References
- Andrews B.J., Lehman J.A., Turchi J.J. (2006). Kinetic analysis of the Ku-DNA binding activity reveals a redox-dependent alteration in protein structure that stimulates dissociation of the Ku-DNA complex. J. Biol. Chem. 281: 13596–13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult N., Karlseder J. (2015). Complex interactions between the DNA-damage response and mammalian telomeres. Nat. Struct. Mol. Biol. 22: 859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrini A., Ristic D., Dionne I., Liu X.Z., Wyman C., Wellinger R.J., Petrini J.H. (2013). The Ku heterodimer and the metabolism of single-ended DNA double-strand breaks. Cell Reports 3: 2033–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarde O., Boby C., Gomez D., Frit P., Giraud-Panis M.J., Gilson E., Salles B., Calsou P. (2010). TRF2/RAP1 and DNA-PK mediate a double protection against joining at telomeric ends. EMBO J. 29: 1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock P., van Attikum H., Hooykaas P. (2002). Increased telomere length and hypersensitivity to DNA damaging agents in an Arabidopsis KU70 mutant. Nucleic Acids Res. 30: 3395–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli G.B., Denchi E.L., de Lange T. (2006). Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat. Cell Biol. 8: 885–890. [DOI] [PubMed] [Google Scholar]
- Chico L., Ciudad T., Hsu M., Lue N.F., Larriba G. (2011). The Candida albicans Ku70 modulates telomere length and structure by regulating both telomerase and recombination. PLoS One 6: e23732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes-Rojas C., Kannan K., Tseng L., Shippen D.E. (2011). Two RNA subunits and POT1a are components of Arabidopsis telomerase. Proc. Natl. Acad. Sci. USA 108: 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cifuentes-Rojas C., Nelson A.D., Boltz K.A., Kannan K., She X., Shippen D.E. (2012). An alternative telomerase RNA in Arabidopsis modulates enzyme activity in response to DNA damage. Genes Dev. 26: 2512–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Adda di Fagagna F., Hande M.P., Tong W.M., Roth D., Lansdorp P.M., Wang Z.Q., Jackson S.P. (2001). Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol. 11: 1192–1196. [DOI] [PubMed] [Google Scholar]
- Derboven E., Ekker H., Kusenda B., Bulankova P., Riha K. (2014). Role of STN1 and DNA polymerase α in telomere stability and genome-wide replication in Arabidopsis. PLoS Genet. 10: e1004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sena-Tomás C., Yu E.Y., Calzada A., Holloman W.K., Lue N.F., Pérez-Martín J. (2015). Fungal Ku prevents permanent cell cycle arrest by suppressing DNA damage signaling at telomeres. Nucleic Acids Res. 43: 2138–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattah F., Lee E.H., Weisensel N., Wang Y., Lichter N., Hendrickson E.A. (2010). Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet. 6: e1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell V.L., Schild-Poulter C. (2015). The Ku heterodimer: function in DNA repair and beyond. Mutat. Res. Rev. Mutat. Res. 763: 15–29. [DOI] [PubMed] [Google Scholar]
- Ferreira M.G., Cooper J.P. (2001). The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol. Cell 7: 55–63. [DOI] [PubMed] [Google Scholar]
- Fulcher N., Sablowski R. (2009). Hypersensitivity to DNA damage in plant stem cell niches. Proc. Natl. Acad. Sci. USA 106: 20984–20988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher N., Derboven E., Valuchova S., Riha K. (2014). If the cap fits, wear it: an overview of telomeric structures over evolution. Cell. Mol. Life Sci. 71: 847–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M.E., Jalut N., White C.I. (2003). Telomerase dependence of telomere lengthening in Ku80 mutant Arabidopsis. Plant Cell 15: 782–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göhring J., Fulcher N., Jacak J., Riha K. (2014). TeloTool: a new tool for telomere length measurement from terminal restriction fragment analysis with improved probe intensity correction. Nucleic Acids Res. 42: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S., Larrivée M., Labrecque P., Wellinger R.J. (1998). Yeast Ku as a regulator of chromosomal DNA end structure. Science 280: 741–744. [DOI] [PubMed] [Google Scholar]
- Hammel M., et al. (2011). XRCC4 protein interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double strand break repair. J. Biol. Chem. 286: 32638–32650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass E.P., Zappulla D.C. (2015). The Ku subunit of telomerase binds Sir4 to recruit telomerase to lengthen telomeres in S. cerevisiae. eLife 4: 10.7554/eLife.07750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H.L., Gilley D., Blackburn E.H., Chen D.J. (1999). Ku is associated with the telomere in mammals. Proc. Natl. Acad. Sci. USA 96: 12454–12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J.M., Østerås M., Farinelli L., Krajewski P., Angenent G.C. (2010). Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat. Protoc. 5: 457–472. [DOI] [PubMed] [Google Scholar]
- Kazda A., Zellinger B., Rössler M., Derboven E., Kusenda B., Riha K. (2012). Chromosome end protection by blunt-ended telomeres. Genes Dev. 26: 1703–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kysela B., Doherty A.J., Chovanec M., Stiff T., Ameer-Beg S.M., Vojnovic B., Girard P.M., Jeggo P.A. (2003). Ku stimulation of DNA ligase IV-dependent ligation requires inward movement along the DNA molecule. J. Biol. Chem. 278: 22466–22474. [DOI] [PubMed] [Google Scholar]
- Lee K.J., Saha J., Sun J., Fattah K.R., Wang S.C., Jakob B., Chi L., Wang S.Y., Taucher-Scholz G., Davis A.J., Chen D.J. (2016). Phosphorylation of Ku dictates DNA double-strand break (DSB) repair pathway choice in S phase. Nucleic Acids Res. 44: 1732–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman J.A., Hoelz D.J., Turchi J.J. (2008). DNA-dependent conformational changes in the Ku heterodimer. Biochemistry 47: 4359–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez C.R., Ribes-Zamora A., Indiviglio S.M., Williams C.L., Haricharan S., Bertuch A.A. (2011). Ku must load directly onto the chromosome end in order to mediate its telomeric functions. PLoS Genet. 7: e1002233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari P.O., Florea B.I., Persengiev S.P., Verkaik N.S., Brüggenwirth H.T., Modesti M., Giglia-Mari G., Bezstarosti K., Demmers J.A., Luider T.M., Houtsmuller A.B., van Gent D.C. (2006). Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl. Acad. Sci. USA 103: 18597–18602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele L., Lydall D. (2002). EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 16: 1919–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S.G., Laroche T., Suka N., Grunstein M., Gasser S.M. (1999). Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 97: 621–633. [DOI] [PubMed] [Google Scholar]
- Martínez P., Blasco M.A. (2015). Replicating through telomeres: a means to an end. Trends Biochem. Sci. 40: 504–515. [DOI] [PubMed] [Google Scholar]
- Masson C., Bury-Moné S., Guiot E., Saez-Cirion A., Schoëvaërt-Brossault D., Brachet-Ducos C., Delelis O., Subra F., Jeanson-Leh L., Mouscadet J.F. (2007). Ku80 participates in the targeting of retroviral transgenes to the chromatin of CHO cells. J. Virol. 81: 7924–7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikova L., Biessmann H., Georgiev P. (2005). The Ku protein complex is involved in length regulation of Drosophila telomeres. Genetics 170: 221–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo B., Marcand S. (2005). Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J. 24: 3117–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfingsten J.S., Goodrich K.J., Taabazuing C., Ouenzar F., Chartrand P., Cech T.R. (2012). Mutually exclusive binding of telomerase RNA and DNA by Ku alters telomerase recruitment model. Cell 148: 922–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribes-Zamora A., Indiviglio S.M., Mihalek I., Williams C.L., Bertuch A.A. (2013). TRF2 interaction with Ku heterotetramerization interface gives insight into c-NHEJ prevention at human telomeres. Cell Reports 5: 194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha K., Shippen D.E. (2003). Ku is required for telomeric C-rich strand maintenance but not for end-to-end chromosome fusions in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 611–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha K., Heacock M.L., Shippen D.E. (2006). The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology. Annu. Rev. Genet. 40: 237–277. [DOI] [PubMed] [Google Scholar]
- Riha K., Watson J.M., Parkey J., Shippen D.E. (2002). Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 21: 2819–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Calzada A., Spagnolo L., Pearl L.H., Llorca O. (2007). Structural model of full-length human Ku70-Ku80 heterodimer and its recognition of DNA and DNA-PKcs. EMBO Rep. 8: 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S.A., Ramsden D.A. (2007). Loading of the nonhomologous end joining factor, Ku, on protein-occluded DNA ends. J. Biol. Chem. 282: 10605–10613. [DOI] [PubMed] [Google Scholar]
- Samper E., Goytisolo F.A., Slijepcevic P., van Buul P.P., Blasco M.A. (2000). Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 1: 244–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A., Karlseder J., Holtgreve-Grez H., Jauch A., de Lange T. (2002). DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr. Biol. 12: 1635–1644. [DOI] [PubMed] [Google Scholar]
- Stennett E.M., Ciuba M.A., Lin S., Levitus M. (2015). Demystifying PIFE: the photophysics behind the protein-induced fluorescence enhancement phenomenon in Cy3. J. Phys. Chem. Lett. 6: 1819–1823. [DOI] [PubMed] [Google Scholar]
- Turchi J.J., Henkels K.M., Zhou Y. (2000). Cisplatin-DNA adducts inhibit translocation of the Ku subunits of DNA-PK. Nucleic Acids Res. 28: 4634–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valuchova S., Fulnecek J., Petrov A.P., Tripsianes K., Riha K. (2016). A rapid method for detecting protein-nucleic acid interactions by protein induced fluorescence enhancement. Sci. Rep. 6: 39653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero-Sedas M.I., Vega-Palas M.A. (2014). Determination of Arabidopsis thaliana telomere length by PCR. Sci. Rep. 4: 5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J.R., Corpina R.A., Goldberg J. (2001). Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412: 607–614. [DOI] [PubMed] [Google Scholar]
- Wang M., Wu W., Wu W., Rosidi B., Zhang L., Wang H., Iliakis G. (2006). PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 34: 6170–6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ghosh G., Hendrickson E.A. (2009). Ku86 represses lethal telomere deletion events in human somatic cells. Proc. Natl. Acad. Sci. USA 106: 12430–12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West C.E., Waterworth W.M., Story G.W., Sunderland P.A., Jiang Q., Bray C.M. (2002). Disruption of the Arabidopsis AtKu80 gene demonstrates an essential role for AtKu80 protein in efficient repair of DNA double-strand breaks in vivo. Plant J. 31: 517–528. [DOI] [PubMed] [Google Scholar]
- Yoo S., Dynan W.S. (1999). Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKcs induces inward translocation of Ku protein. Nucleic Acids Res. 27: 4679–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S., Kimzey A., Dynan W.S. (1999). Photocross-linking of an oriented DNA repair complex. Ku bound at a single DNA end. J. Biol. Chem. 274: 20034–20039. [DOI] [PubMed] [Google Scholar]
- Zellinger B., Akimcheva S., Puizina J., Schirato M., Riha K. (2007). Ku suppresses formation of telomeric circles and alternative telomere lengthening in Arabidopsis. Mol. Cell 27: 163–169. [DOI] [PubMed] [Google Scholar]