Deep evolutionary analysis of the highly conserved miR390-TAS3-ARF pathway identifies the point of emergence and adaption of each of the components in the pathway.
Abstract
In plants, miR390 directs the production of tasiRNAs from TRANS-ACTING SIRNA3 (TAS3) transcripts to regulate AUXIN RESPONSIVE FACTOR (ARF) genes, critical for auxin signaling; these tasiRNAs are known as tasiARFs. To understand the evolution of this miR390-TAS3-ARF pathway, we characterized homologs of these three genes from thousands of plant species, from bryophytes to angiosperms. We found the lower-stem region of MIR390 genes, critical for accurate DICER-LIKE1 processing, is conserved in sequence in seed plants. We propose a model for the transition of functional tasiRNA sequences in TAS3 genes occurred at the emergence of vascular plants, in which the two miR390 target sites of TAS3 genes showed distinct pairing patterns. Based on the cleavability of miR390 target sites and the distance between target site and tasiARF, we inferred a potential bidirectional processing mechanism exists for some TAS3 genes. We also demonstrated a tight mutual selection between tasiARF and its target genes and that ARGONAUTE7, the partner of miR390, was specified later than other factors in the pathway. All these data illuminate the evolutionary path of the miR390-TAS3-ARF pathway in land plants and demonstrate the significant variation that occurs in this functionally important and archetypal regulatory circuit.
INTRODUCTION
In plants, small RNAs (sRNAs) play crucial regulatory roles in growth and development, resistance to abiotic and biotic stresses, and reproduction (Chen, 2009; Axtell, 2013; Bartel, 2009). Based on features such as their biogenesis and function, sRNAs are classified into two major groups, microRNAs (miRNAs) and small interfering RNAs (siRNAs). miRNAs are generated from precursor mRNAs that fold back to form double-stranded stem-loop structures, while siRNAs are produced from double-stranded RNAs biosynthesized secondarily by RNA-dependent RNA polymerase (RDR) (Axtell, 2013). Trans-acting small interfering RNAs (tasiRNAs) are a special type of small RNAs found only in plants, so far. Precursor genes of tasiRNAs (TAS genes) are sliced in a miRNA-directed event, and the cleaved fragment is made double-stranded by RDR6; the resulting double-stranded RNA is chopped by DICER-LIKE4 (DCL4) into 21-nucleotide siRNAs that map back to the precursors in a head-to-tail arrangement initiating from the miRNA cleavage site (Allen et al., 2005; Yoshikawa et al., 2005).
Among plant TAS genes, the most well studied is TAS3; its transcript bears two target sites of miR390, generating tasiRNAs via the so-called “two-hit” mechanism (Axtell et al., 2006). The conserved, resulting tasiRNA is known as “tasiARF” as it targets AUXIN RESPONSIVE FACTOR (ARF) genes (Allen et al., 2005; Axtell et al., 2006). To date, there are two kinds of TAS3 genes described in plants; one contains a single, centrally located tasiARF, while the other generates two tasiARFs. These are denoted as TAS3-short (TAS3S) and TAS3-long (TAS3L), respectively (Xia et al., 2015b). In TAS3L, only the 3′ miR390 target site is cleavable, and this sets the phase of tasiRNA production, giving rise to the two in-phase tasiARFs (Allen et al., 2005; Axtell et al., 2006). The 5′ target site of TAS3L is usually noncleavable because of the presence of a central mismatch (10th position) in the pairing of miR390 and target site (Axtell et al., 2006). The 5′ target sequence serves as an important binding site of ARGONAUTE7 (AGO7), a specialized protein partner of miR390 (Montgomery et al., 2008a). By contrast, both target sites of TAS3S are cleavable, and both can potentially initiate tasiRNA generation (Howell et al., 2007; Xia et al., 2012, 2015b). The single tasiARF of TAS3S is in near-perfect phasing to both miR390 sites as there is only a 2-nucleotide difference between the phase registers set by the two target sites (Xia et al., 2012, 2015b).
Auxin, a plant hormone, has a role in seemingly most aspects of plant growth and development. The small class of ARF transcription factors can either activate or repress expression of downstream auxin-regulated genes through protein-protein interactions with auxin/indole-3-acetic acid (AUX/IAA) family members (Guilfoyle and Hagen, 2007). Most plant genomes contain ∼10 to 30 ARF genes; for example, there are 23 members in the model plant Arabidopsis thaliana. ARFs are split into three lineages: ARF5/6/7/8 (Clade A), ARF1/2/3/4/9 (Clade B), and ARF10/16/17 (Clade C), which can be traced back to the origin of the land plants (Finet et al., 2012). The TAS3-derived tasiARF specifically targets ARF genes of Clade B (ARF2/3/4). This miR390-TAS3-ARF pathway is of critical function in the regulation of plant growth and development, including leaf morphology, developmental timing and patterning, and lateral root growth (Garcia et al., 2006; Fahlgren et al., 2006; Adenot et al., 2006; Marin et al., 2010; Hunter et al., 2006). It was recently found that ARF3, with the transcription factor INDEHISCENT, comprises an alternative auxin-sensing mechanism (Simonini et al., 2016). Loss-of-function mutants of AGO7, the specialized AGO partner of miR390, show varying degrees of growth and developmental disorders due to the malfunction of the tasiARF pathway (Yifhar et al., 2012; Zhou et al., 2013). For example, maize (Zea mays) ago7 (rgd2) plants develop cylindrical leaves that maintain dorsiventral polarity (Douglas et al., 2010), and Medicago truncatula ago7 (lobed leaflet1) mutant plants displayed lobed leaf margins and extra lateral leaflets (Zhou et al., 2013).
All three components of the pathway, miR390, TAS3, and ARFs, are present in the oldest land plants, liverworts (Krasnikova et al., 2013; Finet et al., 2012), and every component is uniquely represented, i.e., the components include (1) a MIRNA gene, (2) the only noncoding gene (TAS3) conserved throughout land plant genomes, and (3) a protein-coding gene. To date, few evolutionary analyses have focused on conserved noncoding genes (mostly miRNAs), and those are mostly limited to just a few species. Interestingly, in bryophytes, the TAS3 genes are different from those found in flowering plants. Although bryophyte TAS3 genes also have two miR390 target sites, they generate tasiRNAs targeting not only ARF genes but also AP2 genes (described, for example, in the moss Physcomitrella patens) (Axtell et al., 2007). Moreover, the bryophyte ARF-targeting tasiRNA is a different sequence compared with the tasiARF in flowering plants (Allen et al., 2005; Axtell et al., 2007). How and when this transition in TAS3 gene composition occurred in the evolution of land plants is fascinating but unknown. We recently characterized ∼20 TAS3 genes in the gymnosperm Norway spruce (Picea abies), demonstrating diverse features of these genes distinct from those characterized in flowering plants (Xia et al., 2015a). In this study, we aimed to understand the evolutionary history of and critical changes in the miR390-TAS3-ARF pathway for the major lineages of land plants. We used >150 plant genomes and the large data set from the 1000 Plant Transcriptome (1KP) project (Matasci et al., 2014), in combination with additional sequencing data and computational approaches; these resources identified hundreds of MIR390 genes and thousands of TAS3 and ARF genes, across numerous plant species. From these data, we illustrated with high resolution the dynamic nature of the evolutionary route of the miR390-TAS3-ARF pathway, revealing new regulatory features of the three critical components of the pathway.
RESULTS
Gene Identification from Genomic and Transcriptomic Data
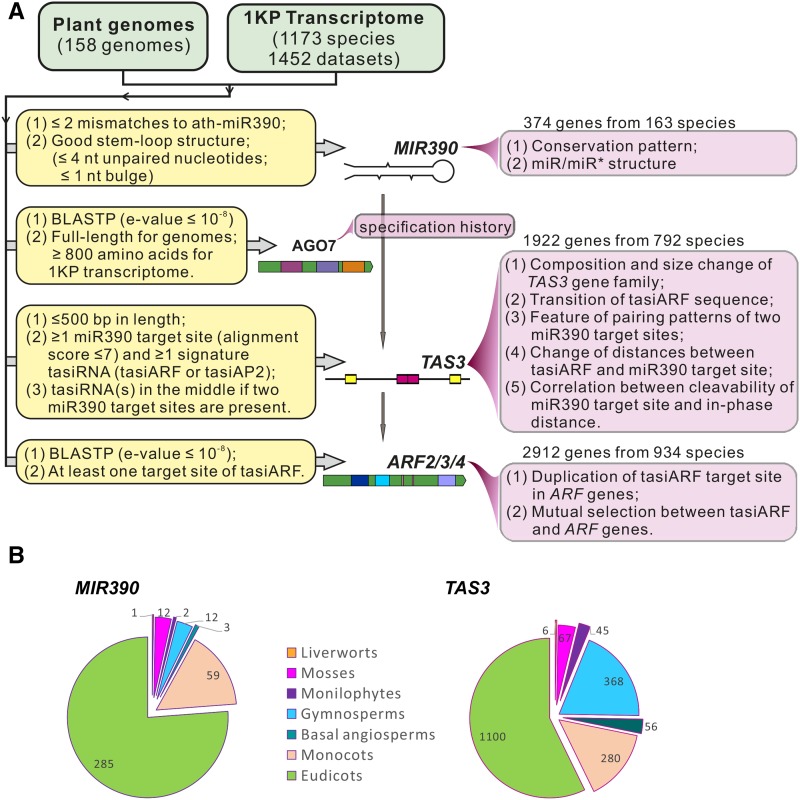
miR390-TAS3-ARF comprise a regulatory pathway highly conserved in plants. To maximize the possibility of characterizing the full diversity of the three main components of this pathway (miR390, TAS3, and ARF genes), we collected 158 sequenced plant genomes, ranging from liverworts to angiosperms, plus the 1KP data (Matasci et al., 2014). The analysis workflow and detailed criteria used for homologous gene identification are depicted in Figure 1A. For example, for the identification of TAS3 genes, only genomic loci from sequenced genomes (≤500 bp) or transcripts (from 1KP data) containing at least one miR390 target site and one tasiRNA targeting an ARF gene were considered valid for our analysis. Using bioinformatics tools in combination with customized scripts (see Methods), we identified 374 MIR390 genes from 163 plant species (Supplemental Data Set 1), 1922 TAS3 genes from 792 species, and 2912 ARF genes (targets of tasiRNAs) from 934 species (Figure 1A; Supplemental Data Sets 1 to 5). These genes were subjected to a series of comparisons and statistical analyses to evaluate the evolutionary changes of the three components of the pathway. Given the uneven composition of species from major plant lineages, we classified all the plant species into one of seven groups (liverworts, mosses, monilophytes or ferns, gymnosperms, basal angiosperms, monocots, and eudicots); each group was considered independently in our subsequent assessments (Figure 1B). Monocots and eudicots accounted for the two largest groups of species and yielded the vast majority of MIR390 genes; many fewer were identified in the liverwort, monilophyte, and basal angiosperm groups (Figure 1B). Similarly, most of the TAS3 genes identified were from angiosperms, although there were many from gymnosperms as well (Figure 1B). We were unable to identify homologs of MIR390 or TAS3 genes in five algal genomes, consistent with the earlier conclusion that the miR390-TAS3-ARF pathway originated in land plants (Krasnikova et al., 2013). In Arabidopsis, the proper execution of the miR390-TAS3-ARF pathway requires that miR390 is loaded into a specific and highly selective AGO partner protein, AGO7 (Montgomery et al., 2008a). Therefore, we also retrieved all the AGO homologs (≥800 amino acids) to trace the history of AGO7 in plants (Figure 1A, described below).
Figure 1.
Homologous Gene Identification from Genomic and Transcriptomic Data.
(A) General workflow of the identification of homologs of MIR390, AGO7, TAS3, and ARF2, ARF3, or ARF4 (ARF2/3/4). Main criteria used in gene identification are listed in yellow boxes, and downstream bioinformatics and statistical analyses conducted for each gene family are itemized in light-brown boxes.
(B) Proportion of MIR390 and TAS3 genes in different lineages of land plants, organized as seven groups of species. For TAS3, two genes found in the only lycophyte (P. drummondii) were grouped in the monilophytes for simplicity.
The Lower Stem Region of MIR390 Is under Selection for Conservation
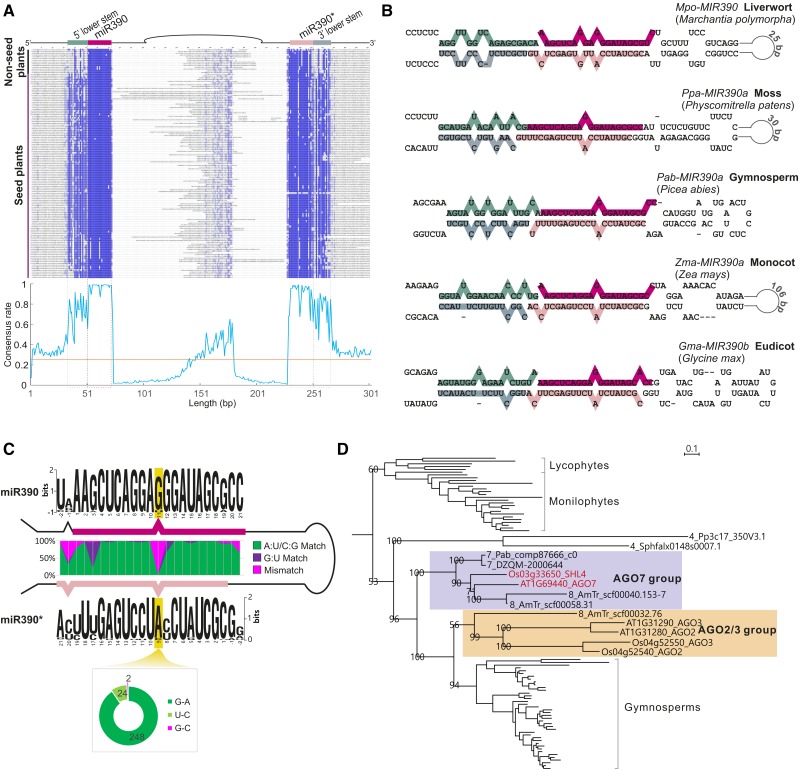
miR390 is one of the most ancient miRNAs and is well conserved in land plants. During the course of evolution, MIRNA genes (i.e., the precursor mRNAs) were relatively labile, typically displaying conservation only in the sequences of the miRNA and miRNA* in the foldback region (Jones-Rhoades et al., 2006; Fahlgren et al., 2010; Ma et al., 2010). Indeed, in our analysis, the sequences of miR390 and miR390* were extremely conserved in land plants, as shown in the sequence alignment in Figure 2A. Interestingly, in addition to the miR390/miR390* region, we identified another two regions of relatively high conservation in the precursors (Figure 2A). These sequences forming the lower stem of the MIR390 stem-loop structure (Figure 2B) displayed a substantially greater consensus, especially within the seed plants, than any other regions of the precursors outside of the miR390/miR390* duplex (Figure 2A).
Figure 2.
Conservation of MIR390 and the Specification of AGO7 in Land Plants.
(A) Nucleotide sequence alignment of MIR390 precursor genes (±50 bp before/after the miR390/miR390* region) with different sequence regions denoted above. The consensus rate of diversity of each position in the alignment is shown in the plot below with the orange line indicating the 25% level, since in a sequence randomized by neutral evolution, each nucleotide (A/U/C/G) would comprise 25% of each position.
(B) Examples of stem-loop structures of MIR390 precursor transcripts. The miRNA and lower-stem regions are indicated according to the colors shown in the top of (A).
(C) Structure conservation of the miR390/miR390* duplex. Sequence logos were generated using WebLogo. Different nucleotide pairings at each position between the miR390 and miR390* are indicated by different colors, with A:U/C:G matches denoted in green, G:U matches in purple, and all mismatches in pink. The yellow shading highlights position 11 of the duplex. The plot at the bottom shows the composition of different parings at the position 11.
(D) Enlarged subtree of the AGO2/3/7 clade from the tree of Supplemental Figure 2. Subgroups AGO7 and AGO2/3 are highlighted, with Arabidopsis and rice AGO7 copies labeled in red. Scale bar of branch length is 0.1. The branch length values are the mean expected rates of substitution per site.
For the effective loading of miR390 to AGO7, the initial adenine “A” of the miR390 sequence and the central region of the miR390/miR390* duplex are critical (Endo et al., 2013). We checked the duplex structures of all identified MIR390 precursors. Indeed, the first nucleotide of miR390 was exclusively an “A,” and the central region of the duplex, especially the mismatch at position 11 (relative to miR390), was extremely conserved as well (Figure 2C). Although other features appeared to be conserved in the duplex, such as the mismatch at position −1 and G:U wobble at position 3, none was as exclusively conserved among all the plant lineages as the mismatch at position 11 (Supplemental Figure 1), suggesting that the central mismatch region is functionally essential in all the land plants. A detailed analysis of position 11 revealed that only two out of the 374 miR390/miR390* duplexes were a G:C pair. The vast majority contained a G-A mismatch, but the U-C mispairing also accounted for an appreciable percentage of the mismatches in this position (24 of 372 mismatches, 6.45%; Figure 2C). In the context of AGO7 loading, these findings indicate that, along with the predominant G-A mismatch, a U-C mismatch is a permissible interaction at position 11.
Specification of AGO7 Occurred Late, Possibly with the Appearance of Seed Plants
AGO7 is an indispensable component for the proper function of the miR390-TAS3-ARF pathway; thus, we also investigated the evolutionary history of AGO7. To complement our recent survey of AGO proteins that mainly focused on flowering plants (Zhang et al., 2015), our analyses here focused on AGO proteins from nonflowering plants. We identified 237 AGO protein sequences with ≥800 amino acids, and these were used for the construction of a phylogenetic tree in combination with AGO proteins from three representative angiosperms: Amborella trichopoda, rice (Oryza sativa), and Arabidopsis. As previously documented (Vaucheret, 2008; Mallory and Vaucheret, 2010; Zhang et al., 2015), AGO proteins clustered into three major clades, AGO1/5/10, AGO2/3/7, and AGO4/6/8/9 (Supplemental Figure 2 and Supplemental File 1). The AGO2/3/7 clade consisted of members all from vascular plants, except two moss AGO proteins (4_Pp3c17_350V3.1 from P. patens and 4_Sphflax0148s0007.1 from Sphagnum fallax) (Figure 2D); we interpreted this as an indication that the ancestor of the AGO2/3/7 clade likely separated from the AGO1/5/10 clade in mosses. Also, AGO7 was apparently not specified until the emergence of gymnosperms, as only gymnosperm and Amborella trichopoda AGOs joined the eudicot AGO7 copies to form a subclade (Figure 2D). These results suggest that the specific partner AGO of miR390, AGO7, emerged much later than the miRNA and the pathway, possibly to enable unique functions of the miR390-TAS3-ARF pathway in seed and flowering plants.
Ancient Origin of TasiRNA-Mediated Regulation of ARFs
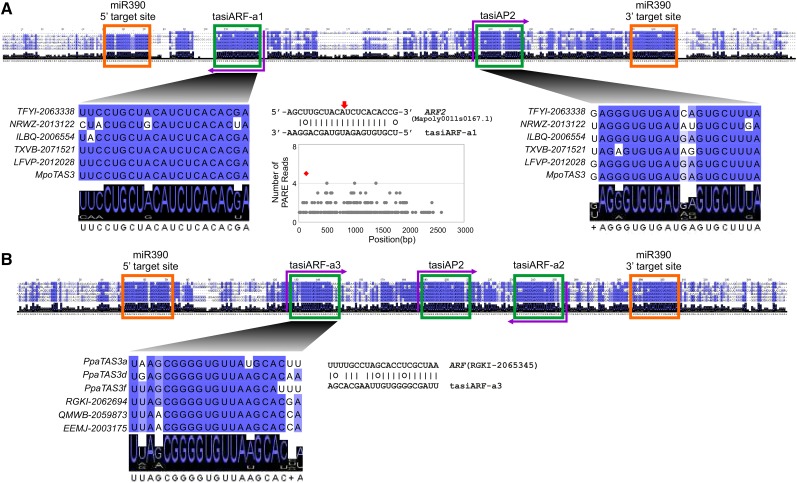
TAS3 genes in bryophytes were first characterized in the moss P. patens, consisting in that genome of a small family of six genes (Axtell et al., 2007; Arif et al., 2012). Many TAS3 genes were subsequently described in mosses (Krasnikova et al., 2013). All known moss TAS3 genes have similar sequence components: two miR390 target sites, a tasiRNA that targets AP2 genes (tasiAP2), and a tasiRNA that targets ARF genes (tasiARF) (Axtell et al., 2007; Krasnikova et al., 2013). A TAS3 gene was also identified in a liverwort Marchantia polymorpha, representing the most ancient extant lineage of land plants (Krasnikova et al., 2013). This TAS3 gene was described as producing only a single tasiRNA, with a sequence similar to that of the moss tasiAP2 (Krasnikova et al., 2013). Here, we found five TAS3 genes from liverwort species in addition to that of M. polymorpha. Sequence alignment of these six liverwort genes revealed the presence of another conserved region, aside from the two miR390 target sites and the previously described tasiAP2, that could also produce a siRNA (Figure 3A). Analyses of public sRNA data from M. polymorpha showed that a highly abundant tasiRNA was produced from the antisense strand of this siRNA site. This tasiRNA (hereafter, “tasiARF-a1”) was predicted to target an ARF2 gene in M. polymorpha (Mapoly0011s0167.1), with the cleavage of the target site confirmed by robust PARE data (category 0; Figure 3A). While the previously described tasiAP2 site is highly conserved, we were unable to validate its target interaction in M. polymorpha, despite combining whole-genome target analysis with sRNA and PARE data. No AP2 homolog was predicted as a target of the tasiRNA even using relaxed prediction criteria (alignment score ≤ 7). Furthermore, when checking M. polymorpha homologs of moss AP2 genes that are validated targets of moss tasiARF-a1, we still found no tasiAP2 target sites (data not shown). Moreover, the tasiAP2 analog was of low abundance and had potential target genes other than AP2 genes (Supplemental Data Set 6), suggesting that the tasiAP2 analog may not be functional or have a function other than targeting AP2 genes in M. polymorpha. Therefore, the miR390-TAS3 machinery likely originated to regulate ARF genes, and not AP2 genes, unlike previous reports.
Figure 3.
TAS3 Originated to Regulate ARF Genes in Land Plants.
(A) Conserved motifs in TAS3 transcripts from liverworts. Purple arrows indicate the encoded strand of tasiRNAs; the left-pointing arrow indicates that the functional tasiRNA is located on the antisense strand, and the right-pointing arrow indicates that the tasiRNA is on the sense strand. tasiARF-a1 encoded in liverwort TAS3 genes targets ARF genes with the tasiRNA:target pairing (cleavage site marked with a red arrow) and validating, experimentally derived PARE data shown in the middle T-plot. The red dot in the plot of PARE data marks the cleavage site directed by tasiARF-a1.
(B) Conserved motifs in a few representative TAS3 genes in mosses. Besides tasiARF-a2, previously reported to target ARF genes, another tasiRNA, denoted as tasiARF-a3, was predicted to target ARF genes. The tasiARF-a2 and tasiARF-a3 sequences are encoded in the antisense and sense strands of TAS3 transcripts, respectively.
For the bryophytes, we identified a large number of TAS3 genes including 67 genes from 36 moss species, in addition to the six liverwort TAS3 copies described above. For the 67 genes, we built a multiple sequence alignment; from this, conserved sequence motifs, including the two miR390 target sites, and both tasiAP2 and tasiARF, were detected as previously reported and as observed in the mosses (Supplemental Figure 3A). In addition, we identified another tasiRNA site that was conserved only in a subset of the moss TAS3 genes. Target predictions indicated that this tasiRNA may target ARF genes as well, and it is conserved in only a few members of the TAS3 family. For instance, three TAS3 genes of P. patens (a/d/f) encode this tasiRNA sequence, but TAS3b/c/e lack it (Figure 3B; Supplemental Figure 3A). In contrast to the previously identified tasiARF (tasiARF-a2, on the 3′ end), which was produced in the antisense strand and in phase with the 3′ miR390 target site, the newly identified tasiARF-a3 was located in the sense strand and in phase with the 5′ miR390 target site (Figure 3B). These three tasiARFs in liverworts (tasiARF-a1) and mosses (tasiARF-a2, -a3) had no sequence similarity, originated from either strand of TAS3 genes, and targeted different regions of ARF genes—all characteristics consistent with independent origins.
To infer the possible evolutionary path of TAS3 in bryophytes, we constructed a phylogenetic tree using their TAS3 genes. The phylogenetic tree (Supplemental Figure 3B) yielded three major classes: class I contained all six liverwort TAS3 genes (tasiAP2 and tasiARF-a1); class II included moss TAS3 genes containing tasiAP2 and tasiARF-a2; and class III comprised moss TAS3s with tasiAP2, tasiARF-a2, and tasiARF-a3. Intriguingly, class II was closer to liverwort TAS3 genes (class I), indicating that class III TAS3 genes likely evolved after the appearance of the class II TAS3 genes, which raises an interesting question of the origin of tasiARF-a3.
TasiARF in Vascular Plants Likely Evolved from the Duplication of the 5′ miR390 Target Site
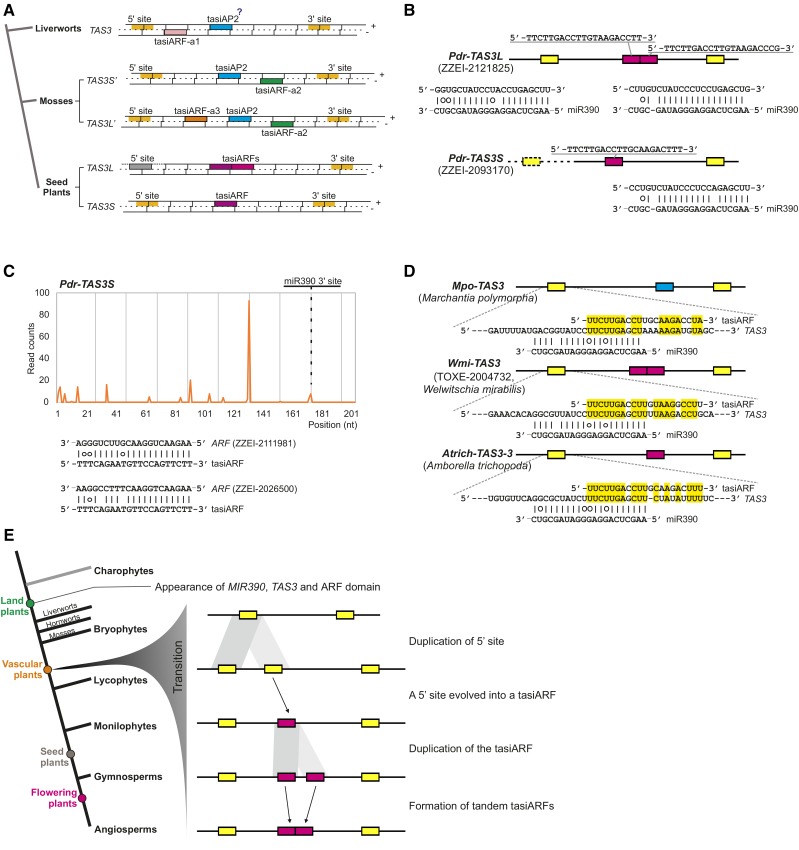
TAS3 genes found in seed plants are different from the bryophyte TAS3s. As summarized in Figure 4A, two types of TAS3 genes, TAS3L with two tandem tasiARFs and TAS3S with one tasiARF, have been previously characterized in gymnosperms and many angiosperms. Despite a similar arrangement of two miR390 target sites, the near-identical tasiARFs in TAS3L and TAS3S are distinct from moss tasiARFs (tasiARF-a1, tasiARF-a2, and tasiARF-a3) in bryophyte TAS3 genes, in terms of sequence, position, and strand (Figure 4A). These differences suggest a significant change occurred during TAS3 evolution in land plants. To better understand when this change happened, we cataloged TAS3 genes with tasiARFs. We found two TAS3 genes from a lycophyte Phylloglossum drummondii, one with two tasiARFs (Pdr-TAS3L) and the other with a single tasiARF (Pdr-TAS3S) (Figure 4B); the cDNA sequence of Pdr-TAS3S was too short to include the 5′ miR390 target site. We generated sRNA sequencing data that confirmed the phased generation of tasiARFs from Pdr-TAS3L (Figure 4C). Both tasiARFs were predicted to target two ARF genes found among the cDNA sequences from the same species (Figure 4C). Therefore, we believe that this transformation of TAS3 genes, and particularly the tasiARF transition, occurred after the divergence of mosses and before or in lycophytes, perhaps with the emergence of vascular plants. Accordingly, we termed the TAS3 genes producing these characteristic tasiARFs (i.e., not tasiARF-a1/a2/a3) “vascular TAS3” genes.
Figure 4.
The Inferred Evolutionary Progression of TAS3 Genes in Land Plants.
(A) A summary of TAS3 gene structures observed in land plants. Colored bars denote different features, as indicated; the gray 5′ miR390 site is not cleaved. The question mark indicates that the function of tasiAP2 (targeting AP2 genes) could not be validated in liverworts.
(B) Two TAS3 gene structures found in the lycophyte species P. drummondii.
(C) TAS3 transcripts produce tasiARFs to regulate ARF genes in P. drummondii. The dotted line and box denote that the identified cDNA sequence of Pdr-TAS3S was too short to include the 5′ miR390 target site.
(D) TasiARF shows sequence similarity to the region partially covering the 5′ miR390 target site of cognate TAS3 genes. Three representative TAS3 genes from different species are displayed here. Identical nucleotides between tasiARF and the region partially covering the 5′ miR390 target site are highlighted in yellow.
(E) An evolutionary model for the divergence of TAS3 genes in land plants. The tasiARF sequence originated from the duplication of the 5′ miR390 target site and the TAS3S genes (with a single tasiARF) might be the ancestor of the TAS3L genes (with two tandem tasiARFs).
We next asked how the transition of TAS3 genes happened. In other words, how was this signature tasiARF sequence generated in vascular plants? We compared the tasiARF sequence to available cDNA or genome sequences. We found the tasiARF sequence shared substantial sequence similarity with a region partially overlapping the 5′ miR390 target site from the cognate TAS3 gene in some species, as exemplified in a few TAS3 genes shown in Figure 4D. In a TAS3 gene of the liverwort M. polymorpha (Mpo-TAS3), the tasiARF sequence had 15 nucleotides of identity with the 5′ miR390 target sequence, with an overlap of 11 nucleotides (Figure 4D). This sequence similarity was even greater in TAS3 genes in a monotypic gymnosperm, Welwitschia mirabilis (Wmi-TAS3), and the basal angiosperm A. trichopoda (Atrich-TAS3) (Figure 4D). This finding of sequence similarity is consistent with a hypothesis that the tasiARF was derived from the 5′ miR390 target site from TAS3.
We previously reported that the genome of the gymnosperm Norway spruce includes a large number of TAS3 genes, among which many have noncanonical sequence features (Xia et al., 2015a). We extended this observation to other plant species, finding TAS3 genes with varied motif structures in our large data set (Supplemental Figure 4). For example, some TAS3 genes had two 5′ or 3′ target sites due to short sequence duplications; others had two or three nonadjacent tasiARFs. We propose a model consistent with these extant TAS3 arrangements for the tasiARF transition from bryophyte TAS3 genes to vascular TAS3 genes (Figure 4E). In the first step, the 5′ miR390 target site of a bryophyte TAS3 gene was duplicated through segmental duplication, as evidenced in a couple of gymnosperm TAS3 genes. Next, the miR390 target site in the middle evolved into a tasiARF and was retained because of its essential function, yielding the short TAS3 gene (TAS3S); after this, two tasiARFs in a single TAS3 gene resulted from the duplication of tasiARF. Finally, the gap between the two tasiARFs was lost, forming a tandem repeat of tasiARFs, yielding the long TAS3 gene (TAS3L) present in vascular plants. This series of steps is consistent with the TAS3 variants present in plant genomes (Supplemental Figure 4).
Distinct Pairing Patterns of Two miR390 Target Sites
TAS3 genes usually comprise a small gene family in plants. For instance, in bryophytes, only one TAS3 gene was identified in M. polymorpha, and six TAS3 copies were found in P. patens. For comparison, there are three TAS3 copies in Arabidopsis, five in rice, and nine in maize—all vascular plants. Comparing across the 157 vascular plants with full-genome sequences that we utilized, we found that this size of the TAS3 gene family is maintained across angiosperms, with most having fewer than ten TAS3 genes and a mean of four genes (Supplemental Figure 5). In gymnosperms, the TAS3 family is substantially larger. The five gymnosperm species surveyed each had at least 28 TAS3 genes, with Pinus taeda encoding as many as 71 TAS3 copies. Notably, almost all of the vascular species had both variants of TAS3 genes (TAS3L and TAS3S) (Supplemental Figure 5), from which we infer that these two types of TAS3 genes likely have nonredundant functions.
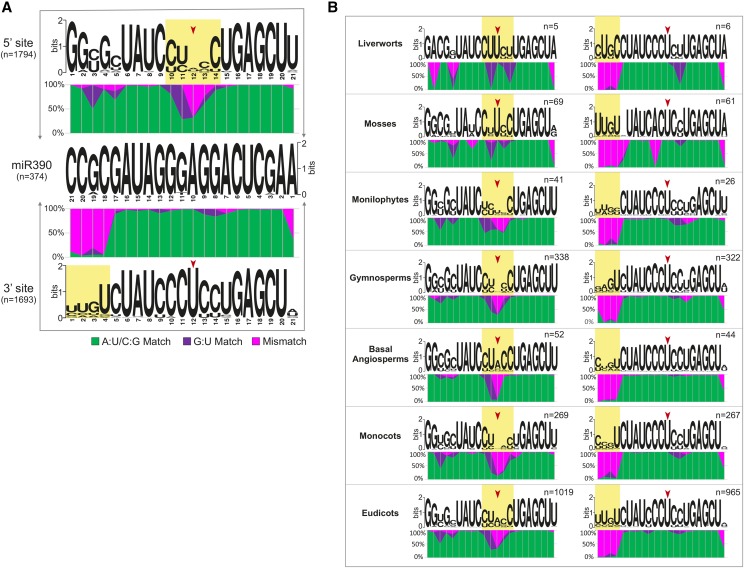
We next evaluated how the pairing patterns for miR390 target sites of TAS3 genes changed in land plants. We identified 3487 target sites of miR390 in 1922 TAS3 copies, including 1794 5′ sites and 1693 3′ sites (Supplemental Data Set 4). These 5′ and 3′ miR390 sites showed different patterns of pairing with miR390, a miRNA with a highly conserved sequence (Figure 5A). In general, the majority of the 5′ sites encoded a central, 10th position mismatch, while the last four nucleotides of the pairing (18th to 21st, relative to the 5′ end of miR390) were always mismatched in the 3′ target site (Figure 5A). More specifically, the middle region (8th to 12th nucleotides) of the 5′ target site was of greater nucleotide diversity, with the 10th position generally unpaired and the 11th position predominantly a G:U pair. By contrast, the 5′ five nucleotides (17th to 21st, relative to the 5′ end of miR390) of the 3′ target sites varied substantially in sequence, with the last four (18th to 21st) always unpaired with miR390. The final nucleotide of the 3′ site (1st relative to miR390) was not well conserved at all and was maintained as a mismatch with miR390, unlike the 5′ site (Figure 5A).
Figure 5.
Pairing Features and Evolutionary Variation of the Two Target Sites of miR390 in TAS3 Genes.
(A) Distinct pairing patterns of the two miR390 target sites in TAS3 genes. Sequence logos were generated using WebLogo. Different nucleotide pairings at each position in the target site (compared with the highly conserved miR390 sequence in the middle) are indicated by different colors, with A:U/C:G matches denoted in green, G:U matches in purple, and all mismatches in pink. The red arrow marks the 10th position, relative to the 5′ end of miR390. The yellow shading indicates regions of substantially imperfect pairing. The upper graph shows the 5′ target site of TAS3, the lower graph shows the 3′ target site; the number of sequences (n) analyzed is indicated for each panel.
(B) Variation in the pairing of the two miR390 target sites in TAS3 genes in different species or lineages of land plants. The images are as described for (A), but the left graph shows the analysis of the 5′ target sites of TAS3, and the right graph shows the analysis of the 3′ target sites.
To assess the history of diversification of the pairing between miR390 and its target sites in TAS3 genes, we grouped all identified miR390 target sites according to the seven lineages of land plants described above, and we generated similar plots to represent miR390-TAS3 pairing. We observed substantial variation in pairing in the 5′ site, especially for the middle region (8th to 12th positions) (Figure 5B). Interestingly, the position most important for AGO-mediated slicing, the 10th position (of miR390), was always matched in bryophytes. In later-diverged species, the mismatch at this position appeared and seemed preferentially retained, as the proportion of mismatches gradually increased over plant evolution. This was particularly noticeable in the basal angiosperms and monocots in which there were almost no matched interactions at this position. For the 11th position of the 5′ site, G:U pairing predominated in all the lineages (Figure 5B). The plant groups showed little variation in the main features of the 3′ site, including the 5′ end mismatch region, the perfect match in the middle, and the high proportion of mismatches for the final nucleotide (except for the monilophytes) (Figure 5B).
Evolutionary Dynamic Distances between TasiARFs and miR390 Target Sites
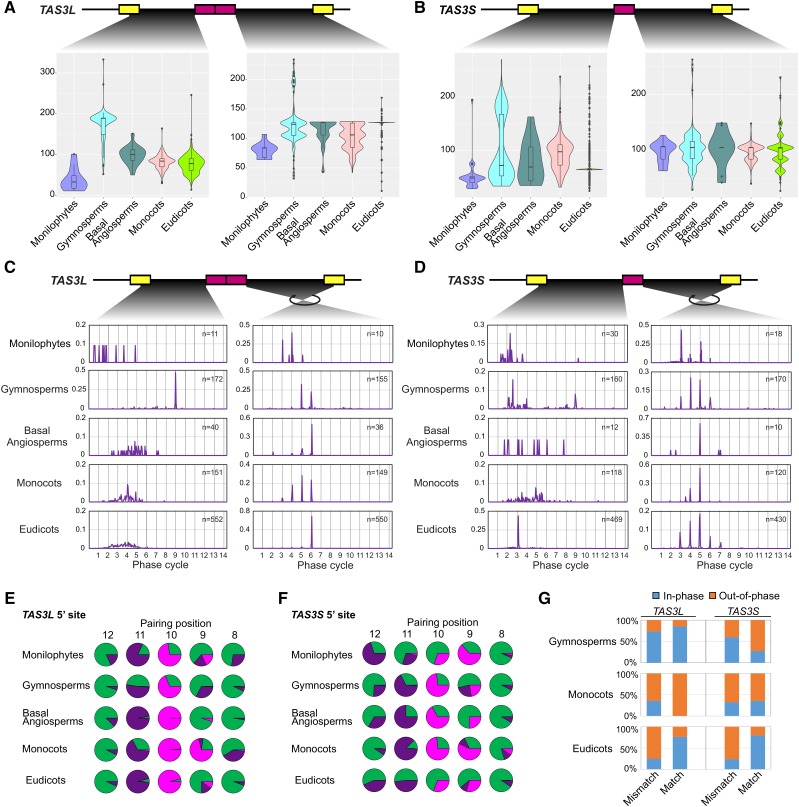
The tasiARF is another functionally essential component of the pathway of our investigation. To correctly generate the tasiARF, this siRNA needs to be in phase with a miR390 target site; in other words, the distance from the cleavage site of a miR390 target site to the end of the tasiARF must be a multiple of 21 nucleotides. Therefore, we calculated the distances and evaluated their evolutionary changes from both 5′ and 3′ miR390 target sites to the tasiARF ends. Given that the tasiARF in vascular plants is distinct from tasiARF-a1, tasiARF-a2, and tasiARF-a3 found in bryophytes, which themselves vary substantially, and given the large number of TAS3 genes identified for vascular plants, we performed distance analyses only for vascular TAS3 genes.
Overall, there was substantial variation in the tasiARF distances (5′ site to tasiARF and tasiARF to 3′ site) in all lineages of vascular plants, with the exception of the eudicots, in which the tasiARF to 3′-site distance of TAS3L and the 5′ site to tasiARF distance of TAS3S were highly consistent in length (Figures 6A and 6B). For TAS3L, both distances were significantly shorter in the monilophytes, but the gymnosperms had a much longer 5′ site to tasiARF region compared with other lineages (Figure 6A). Monilophyte TAS3S also had a shorter 5′ site to tasiARF region, but the tasiARF to 3′-site distance was more or less similar to those of other lineages (Figure 6B).
Figure 6.
The Distances between the Two Target Sites of miR390 and the Central tasiARF Are under Strong Selection.
(A) and (B) Variation of the distances between two miR390 target sites and tasiARF of TAS3L (A) or TAS3S (B) genes in different lineages of vascular plants. In both panels, the lower graphs contain violin plots for each lineage representing the distribution of these distances; internal boxes represent the median as a heavy line surrounded by a box defining the upper and lower quartiles.
(C) and (D) Distribution of the distances between two miR390 target sites and tasiARF of TAS3L (C) or TAS3S (D) genes in different lineages of vascular plants. The y axis is the percentage of TAS3 genes with distances occurring within a given position (the x axis). The 21-nucleotide phased positions (phase “cycles”) are marked as gray gridlines.
(E) and (F) Variation in pairing of the 8th to 12th nucleotide positions (relative to the 5′ end of miR390) of the 5′ target site of TAS3L (E) and TAS3S (F). The type of miR390-TAS3 pairing observed at different nucleotide positions, relative to the 5′ end of miR390, are shown with A:U/C:G matches denoted in green, G:U matches in purple, and all mismatches in pink.
(G) Ratio of the 5′ miR390 target sites in phase or out of phase to the tasiARF in terms of different nucleotide pairing at the 10th position (match, U; mismatches, A, C, and G as the 10th nucleotide of miR390 is “A”).
Next, we assessed the distance from the tasiARFs to a miR390 cleavage site in terms of the phase cycles of phased siRNAs, to determine which site was the trigger. The tasiARFs of TAS3L were mostly out of phase with the 5′ target site, with the exception of those from gymnosperms (see below for a more granular analysis of the relationships between cleavability of the 5′ miR390 site and phasing of the tasiARF in gymnosperms). In gymnosperms, the tasiARFs were consistently positioned at the 9th cycles according to the cleavage site of the 5′ site (Figure 6C, left). By contrast, the TAS3L tasiARFs were consistently in phase with the 3′ site; in other words, the distances of the 3′ site to tasiARF were almost uniformly a multiple of 21 nucleotides, despite considerable length variation in some groups (Figure 6C). For TAS3S, its tasiARF was largely not in phase with the 5′ site, except in the eudicots, which had a consistent 5′ site to tasiARF distance of approximately three cycles, or 65 nucleotides. As with TAS3L, although variation in the length was observed for the 3′ site to tasiARF region of TAS3S, the distance was almost uniformly phased as well, i.e., a multiple of 21 nucleotides (Figure 6D). These results indicated that the 3′ site is the main trigger site of tasiARF generation in both TAS3L and TAS3S, but the gymnosperm TAS3L and eudicot TAS3S likely also generate tasiARFs triggered by the 5′ miR390 target site.
Cleavability of the 5′ Site and Its in-Phase TasiARF Are Selected Coordinately in Eudicots
The noncleavability of the 5′ miR390 target site is functionally important for its role as a binding site of the miR390-RISC complex; this noncleavability results from the presence of a mismatch at the 10th position of the target site pairing (Montgomery et al., 2008b; Axtell et al., 2006). As mentioned before, our analysis of the middle region of the miR390:target pairing of the 5′ site (Figure 5B) demonstrated that, consistent with previous studies, the 10th position mismatch is indeed conserved in the majority of the TAS3 genes in vascular plants. However, we also observed that a not insignificant fraction of interactions of the 10th position of miR390 with TAS3 are perfectly paired, especially in monilophytes, gymnosperms, and eudicots (Figure 5B). Given the finding that the tasiARF in gymnosperm TAS3L and eudicot TAS3S copies are mostly in phase with the 5′ site as well, it is conceivable that the portion of loci with a matched 10th position is contributed by the 5′ sites capable of setting the phase of the tasiARF. To check this possibility, we separated the 5′ sites of TAS3L from those of TAS3S and focused our analyses on the middle region (8th to 12th positions, relative to the miRNA), as shown in Figures 6E and 6F. Although the general pattern was similar for TAS3L and TAS3S, i.e., a predominant 10th position mismatch in most lineages and preferential 11th position G:U pairing, we found a few dissimilarities between TAS3L and TAS3S in the pairing at these positions. Most noticeable was the level of perfect matches at the 10th position for TAS3S compared with the majority of mismatches in TAS3L at the same position (Figure 6E). We then asked whether those 5′ sites in phase to tasiARF were more likely to display a 10th position perfect match. We divided these sequences into two groups, the matched group (with a “U” matching the 10th position “A” of miR390) and the mismatched group (“A,” “C,” or “G”). The matched group had a much higher proportion of in-phase sites in eudicots (Figure 6G), suggesting that the in-phase and cleavable 5′ site was coordinately selected during TAS3 evolution in eudicots.
Strong Mutual Selection between TasiARF and Its Target Site in ARF Genes
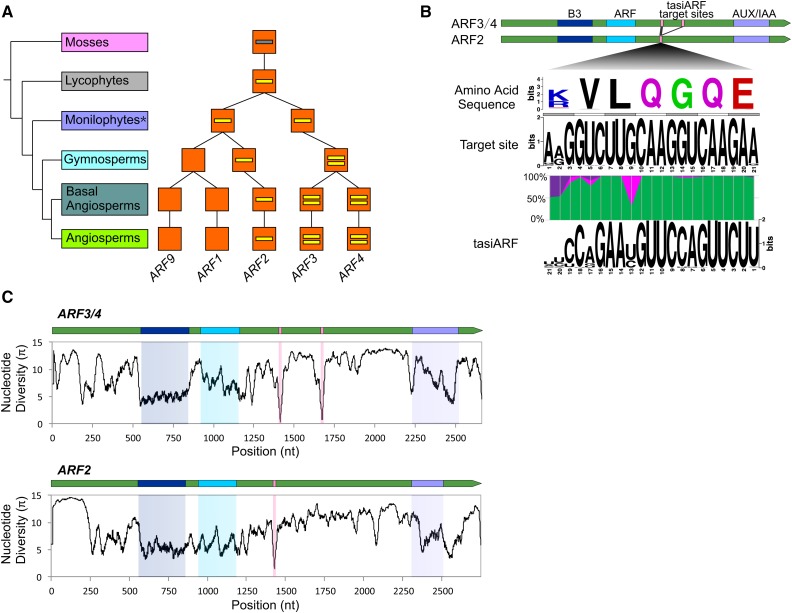
The miR390-TAS3-ARF pathway exerts its function via the silencing of a subgroup of ARF genes, ARF2, ARF3, and ARF4 in Arabidopsis (Allen et al., 2005). In Arabidopsis, the ARF genes are classified into three clades: Clade A (ARF5, ARF6, ARF7, and ARF 8), Clade B (ARF1, ARF2, ARF3, ARF4, and ARF9), and Clade C (ARF10, ARF16, and ARF17) (Finet et al., 2012). The vascular tasiARFs target ARF2, ARF3, and ARF4 of Clade B, the ancestor of which likely emerged in liverworts (Finet et al., 2012). Typically, ARF2 has a single target site for the tasiARF, while ARF3 and ARF4 have two target sites (Allen et al., 2005; Axtell et al., 2006). As described above, the ARF genes in Clade B of bryophytes were regulated by tasiARF-a1 to -a3; thereafter in evolution, this group was targeted by the tasiARF that emerged in vascular plants. However, we found that some Clade B genes from mosses (for example, from P. patens) include sequences analogous to the target site of the vascular tasiARF, suggesting that this target site predates the emergence of the tasiARF of vascular TAS3 genes (Supplemental Figure 6). Combining these data with the ARF evolution history illustrated in Finet et al. (2012), we summarized the likely path of diversification of tasiARF target sites during the evolution of the Clade B ARF genes (Figure 7A). The interaction pattern of tasiARF with ARF genes was likely formed in lycophytes, with only one target site. In monilophytes, genes in Clade B were targeted at a single site in most species, but a few species displayed dual target sites. Thereafter, in evolutionary terms, this dual targeting was maintained in the subclade and likely eventually gave rise to the ARF3/ARF4 genes, while the single targeting was selectively retained in the ARF2 subclade, but lost in the ARF1/ARF9 group (Figure 7A).
Figure 7.
Evolutionary Diversification of tasiARF Target Sites in ARF Genes.
(A) Evolution of the number of tasiARF target sites in plant ARF genes. The evolutionary route of ARF genes was adapted from Finet et al. (2012). The number of short yellow lines in orange boxes denote the number of tasiARF target sites. The gray line means that there are potential tasiARF target sites in ARF genes in mosses. In monilophytes (marked with an asterisk), some ARF3/ARF4 homologous genes have already evolved two tasiARF target sites.
(B) Sequence features of the target site of tasiARF in ARF genes and their encoded proteins. Gene structures of tasiARF-targeted ARF2/ARF3/ARF4 are displayed on the top, including the encoded protein motifs, with the tasiARF target site indicated as pink bars. The target site encodes a short peptide with a consensus sequence of K/RVLQGQE, as indicated with the encoding sequence. Pairing between tasiARF and its target site is color-coded with A:U/C:G matches denoted in green, G:U matches in purple, and all mismatches in pink.
(C) Distribution of nucleotide diversity along tasiARF-targeted ARF2/ARF3/ARF4 genes, with the encoded functional domains and tasiARF target site marked in colors according to those in (B).
The target sites of the vascular tasiARF were located in the middle region between two functional domains (ARF and AUX/IAA) of ARF2/ARF3/ARF4 genes (Figure 7B). We recently reported that the target sites of the miR482/miR2118 family display significant sequence variation at positions matching the 3rd nucleotide of codons, implying a strong selection from the functionally important P-loop motif of NB-LRR proteins that shapes miRNA-target pairing (Zhang et al., 2016). By contrast, the tasiARF sequence is of much lower sequence divergence, and it did not show a pattern like the miR482/miR2118 family, indicating the selection on tasiARF pairing is distinct from that in the miR482/mrR2118 case. Similarly, the tasiARF target sites, unlike the miR390 target sites in TAS3 genes which are relatively diverse, are less divergent in sequence and consistently encode the amino acid sequence K/RVLQGQE (Figure 7B). We also assessed nucleotide diversity of the ARF2, ARF3, and ARF4 genes and found that the three functional domains were, as expected, of relatively low nucleotide diversity. However, the tasiARF target sites (one in ARF2 and two in ARF3/ARF4) showed substantially lower nucleotide diversity, even compared with the encoded, conserved functional domain, indicating strong conservative selection (Figure 7C). Given that tasiARF sequences in TAS3 genes are also highly conserved in vascular plants, we hypothesize that there is strong mutual selection between tasiARF in TAS3 genes and its target sites in ARF genes.
DISCUSSION
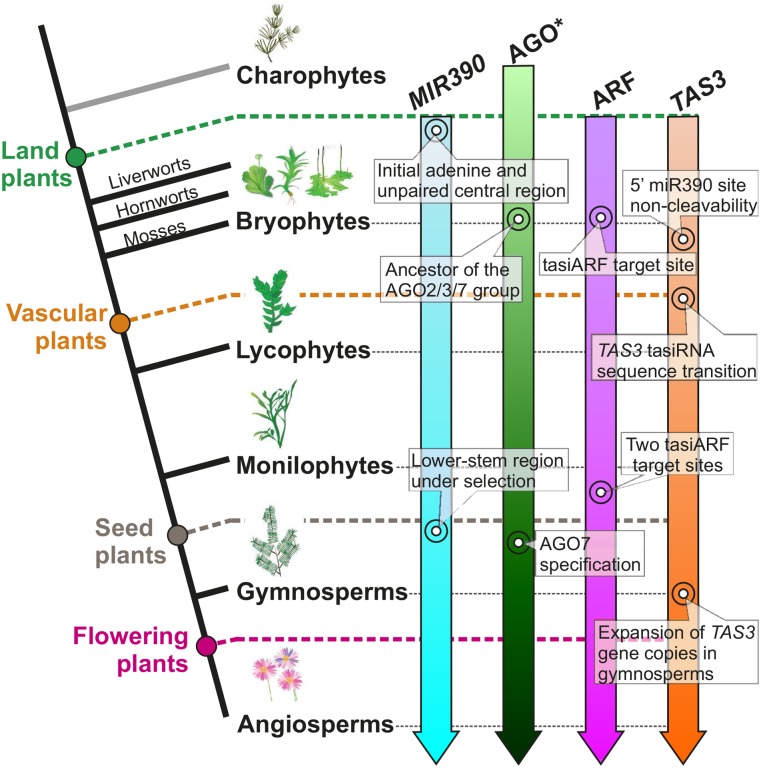
The miR390-TAS3-ARF pathway is a highly conserved, functionally important, and archetypal regulatory circuit in land plants. Taking advantage of vast amounts of publicly available genome and transcriptome data, we demonstrated the dynamic evolutionary nature of the pathway, and we uncovered new features of the three key components of the pathway (Figure 8).
Figure 8.
Evolutionary Emergence of the miR390-TAS3-ARF Pathway in Land Plants.
A simplified phylogenetic tree of plants is drawn on the left. The evolution of each component of the miR390-TAS3-ARF pathway is denoted by arrows of different color on the right. Major evolutionary events are labeled at the corresponding points (referring to the phylogenetic tree). “AGO*” denotes that AGO proteins emerged much earlier than the appearance of plants.
Conservation of MIR390 and Specification of Its Partner AGO7
In plants, miRNA/miRNA* duplexes are released by two sequential cuts of their hairpin precursors by DCL1; these cuts occur directionally, either base-to-loop or loop-to-base (Bologna et al., 2009, 2013). For base-to-loop processing, the first cut is defined by the distance (∼15 nucleotides) from the miRNA/miRNA* duplex to a large loop at the base (Werner et al., 2010; Song et al., 2010; Mateos et al., 2010). miR390 is one such base-to-loop-processed miRNA, with the first cut by DCL1 occurring at a position ∼15 nucleotides from a basal unpaired region (>4 nucleotides) (Bologna et al., 2013). We unexpectedly found conservation in this lower-stem region in seed plants, indicating that selection can maintain bases in the hairpin outside of the miRNA/miRNA*. The conservation in the MIR390 lower stem likely maintains a consistent distance of ∼15 nucleotides to ensure the accuracy of the first cut by DCL1 of the precursor. The conservation of this paired region occurs for many other miRNAs in plants (Chorostecki et al., 2017).
For the proper function of miR390, it must be loaded to its specific partner AGO7. The specificity of this interaction is presumably influenced by features of miR390, including the 5′ nucleotide “A” and the mismatched central region of the miR390/miR390* duplex. We found that these two features are extremely conserved for MIR390 genes in all land plants. However, the phylogenic analysis of AGO7 suggested a more recent origin, coincident with the appearance of seed plants and much later than miR390. Thus, the ancestor of AGO7 already had the capacity to recognize unique features of miR390 prior to the specification of AGO7. Since seed plants are complex organisms in which auxin plays broad regulatory roles, the specificity of AGO7 likely helps accommodate these roles via the miR390-TAS3-ARF pathway. Alternatively, as the phased siRNA regulatory network expanded apparently at least when gymnosperms emerged to include many protein-coding genes (Xia et al., 2015a), perhaps the emergence of AGO7 specified for miR390 separated or compartmentalized the miR390 pathway relative to the many other phasiRNA-generating targets. This compartmentalization may have allowed the two-hit biogenesis of miR390-TAS3 to persist in an evolutionary sense, even as other PHAS loci initiated by 22-nucleotide, one-hit miRNA triggers became more prevalent.
Evolutionary Route and Diversification of TAS3 Genes
The presence of miR390 and TAS3 was tracked back to liverworts (Lin et al., 2016), while the ARF domain encoded by ARF genes likely first appeared in land plants (Finet et al., 2012). We demonstrate that TAS3 in liverworts produces tasiRNAs to target ARF genes, suggesting this was the earliest function of TAS3, a key function maintained throughout land plants. We also observed in liverworts the conservation of another TAS3-derived tasiRNA that, in mosses, targets AP2 genes (referred to as tasiAP2), but we were unable to confirm this function in liverworts. It is possible that this tasiRNA in liverwort TAS3 genes emerged before the appearance of tasiAP2 target sites in AP2 genes or that this tasiRNA has an unidentifiable function or target.
Although the role of TAS3 in regulating ARF genes is conserved across land plants, the bryophyte TAS3 genes are structurally different from those in vascular plants (Axtell et al., 2006). In other words, tasiARFs are different in sequence between bryophytes and vascular plants. In our model for tasiARF evolution, the tasiARF was derived from the duplication of the 5′ target site of miR390, and the short TAS3 variant (TAS3S) is the ancestor of the long TAS3 (TAS3L). We identified the vascular TAS3 in a lycophyte, indicating that the transition of tasiRNA sequences is likely associated with the development of vascular tissue in plants, as lycophytes were among the first vascular plants on earth. Measured across the vascular plants, there are nearly always two types of TAS3 genes (TAS3S and TAS3L) present in each plant genome and totaling approximately four members in most species. The deep conservation of these structures suggests they are not functionally redundant. Future work could address why, perhaps by selective deletion of these two types using CRISPR/Cas9. Another striking observation was that the TAS3 copy number is significantly expanded in conifers, reminiscent of the expansion of NB-LRR-targeting miRNAs (Xia et al., 2015a). Despite evidence of whole-genome duplications in spruce (Li et al., 2015), the >10-fold higher copy number in conifers relative to angiosperms is extraordinary.
One of the major differences between the two main mechanisms of tasiRNA/phasiRNA biogenesis (“one-hit” and “two-hit” models) is the direction of tasiRNA production. In the two-hit model, tasiRNAs are produced in a 3′ to 5′ direction, in contrast to the predominant 5′ to 3′ Dicer processing (i.e., the one-hit model). miR390-TAS3 is the quintessential two-hit locus, yet its 3′ to 5′ processing is distinctive and rare. Evolutionary analyses of miR390-TAS3 pairing revealed two distinct patterns of pairing of the two target sites: (1) a conserved mismatched region mainly caused by the 10th position of the 5′ site (previously known; see below), and (2) an open, unpaired region in the 3′ end of the 3′ target site (from our study). The wide conservation in vascular plants of these features implies functional relevance. Studies in Arabidopsis have shown that the noncleavability of the 5′ site, caused by the central mismatch (10th position), is essential, mediating miR390 binding via AGO7 (Rajeswaran and Pooggin, 2012). Changing the 10th mismatch into a perfect match compromises tasiRNA biogenesis (Axtell et al., 2006; Montgomery et al., 2008a). However, a substantial portion of TAS3 genes, especially the TAS3S subset, have a cleavable 5′ site (A:U pair at the 10th position), and many of these sites, particularly in eudicots, trigger tasiARF production. This indicates that the noncleavability of the 5′ site is helpful but not necessary for tasiARF production. Another notable feature of the 5′ site pairing is the predominant G:U pairing at the 11th position; this preferential wobble pairing might be helpful for maintaining the noncleavability of the 5′ site, which is believed to be mainly caused by the 10th position mismatch. By contrast, the pairing of the 3′ site has a consistently matched middle region, but an open, unpaired 3′ end region. The paired middle region could ensure the cleavage of the 3′ site and make it the typical trigger site for secondary tasiRNAs. The 3′ end open region may direct the 3′ to 5′ production of TAS3 tasiRNAs. Perhaps after cleavage, the 3′ end open region makes the cleaved mRNA end more accessible to RDR6 to facilitate downstream tasiRNA production.
Besides the cleavability of the target site, the distance between the miR390 target site and tasiARF also appears to be a determinant for phasiRNA biogenesis. TasiARF production requires distances in multiples of 21 nucleotides from the cleavage site (“in register”). We showed that the distance of the 3′ side of TAS3 is more consistently a multiple of 21 nucleotides despite considerable length variation; the 3′ site also displayed fewer 10th position mismatches (i.e., better cleavability). However, we noted several exceptions. The TAS3L in gymnosperms and the TAS3S in eudicots had a highly consistent distance on the 5′ side, in approximate phase with tasiARF, and the cleavability of the 5′ site was coordinately selected with the in-phase distance to the tasiARF in eudicots, suggesting that the 5′ site in those TAS3 genes is likely to serve as a trigger site of tasiARF production as well. Therefore, our results suggest that some TAS3 loci in vascular plants are likely bidirectionally processed, consistent with the observation of the original bidirectional processing of functional tasiRNAs in bryophytes (Axtell et al., 2006). For instance, the two target sites of TAS3 in P. patens are both cleavable, and tasiARF-a2 is in phase with the 3′ site, while tasiARF-a3 is in phase with the 5′ site. This bidirectional processing thus yields additional questions about this “two-hit” mechanism. How is the activity of the two sites coordinated? Does cleavage occur simultaneously at both sites or one site at a time?
Selection between TasiARF and Target Sites in ARF Genes
miRNAs, tasiRNAs, and other type of sRNAs together with their target genes each represent a pair of partners, functioning via their interactions, based on sequence complementarity. Few studies have deeply investigated this sRNA:target partnership over evolutionary time. In the case of the widely conserved miR482/2118 family, we previously described selection from target protein-coding genes to miRNAs; in that case, the essential function of the P-loop encoded in NB-LRR genes, targeted by miR482/2118, is most important, as miRNA variation matches a degenerate nucleotide change at the third position of each codon in the target gene (Zhang et al., 2016). In this study, we detected a distinct pattern of selection between tasiARF and target site in ARF genes in vascular plants. Both were depleted of variation, indicating a strong mutual selection. The tasiARF target site sequences in ARF genes showed no periodical variation (at the third position), indicating that the target site sequence is not under strong selection at the amino acid level, in accordance with the location of the tasiARF target site between two encoded domains of ARF proteins, the ARF domain and AUX/IAA domain. The target site in the middle region is of less functional importance at the protein level. This is in contrast with the location of miR482/2118 target site in a functionally critical domain (Zhang et al., 2016). However, the sequence variation (nucleotide diversity) of the tasiARF target sites in ARF genes is dramatically lower than that of other gene regions, even the conserved functional protein domains, suggesting that the tasiARF target site is under a selective force stronger than that experienced by the encoded protein domains. Combined with the fact that tasiARF sequences in vascular TAS3 genes demonstrate substantially less sequence variation, we believe that there is a robust selective connection between tasiARF and its target site in ARF genes, which permits little sequence variation in either component over evolutionary time.
METHODS
Genome Sequences and 1KP Data
Genome sequences of 158 species were retrieved from either the Phytozome or NCBI. The assembled transcriptome data of the 1000 Plant Transcriptome Project (1KP) was kindly shared by the Wang lab at the University of Alberta, Canada (Matasci et al., 2014).
NGS Data and Analyses
RNA of Phylloglossum drummondii was extracted using PureLink Plant RNA Reagent. A sRNA library was constructed using the Illumina TruSeq sRNA kit and sequenced on the Illumina HiSeq platform at the University of Delaware.
sRNA and PARE data of Marchantia polymorpha were retrieved from NCBI Short Read Archive under accession numbers SRR2179617 and SRR2179371, respectively (Lin et al., 2016). sRNA reads were mapped to reference genome or transcripts by Bowtie (Langmead et al., 2009), and PARE data were analyzed using Cleaveland 2.0 (Addo-Quaye et al., 2009).
Homologous Gene Identification
For the identification of MIR390 genes, mature sequences of miR390 were retrieved from miRBase and used to search for homologous sequences using FASTA36 allowing two mismatches. After that, ±500-bp sequence was excerpted for each homologous sequence from reference sequences and used for the evaluation of secondary structure. Only those genomic loci or transcripts with a stem loop structure (≤4-nucleotide mismatches and ≤1-nucleotide bulge) and with the mature miRNA in the 5′ arm were regarded as bona fide MIR390 genes.
For the identification of TAS3 genes, ≤500-bp genomic loci (for genomes) or EST sequences (for transcriptome data) with evidence of at least one miR390 target site and at least one signature tasiRNA (tasiARF for vascular plants, tasiAP2 or tasiARF-a2 for bryophytes) were considered as TAS3 candidates. Their identities as TAS3 genes were further assessed by manual sequence comparisons. The tool MEME (Bailey et al., 2009) was also used to profile the signature sequence motifs of TAS3 genes.
To identify tasiARF-targeted ARF genes, first, Arabidopsis thaliana and subsequently rice (Oryza sativa) ARF proteins were used as bait sequences to identify ARF homologous genes, using either TBLASTN for annotated genomes or 1KP transcriptome data or genBlast (She et al., 2011) for unannotated genomes. Second, TargetFinder (https://github.com/carringtonlab/TargetFinder) was used to identify tasiARF-targeted ARF genes. Third, ARF3/ARF4 and ARF2 genes were distinguished by the number of target sites as ARF3/ARF4 genes have two tasiARF target sites and ARF2 genes have only one target site. AGO proteins were identified using BLASTP for selected annotated genomes or TBLASTN for 1KP data using Arabidopsis and rice AGO proteins as bait sequences. Only full-length AGO protein sequences from sequenced genomes and AGO sequences with ≥800 amino acids from the 1KP data were chosen for subsequent phylogenetic tree construction.
Multiple Sequence Alignment and Tree Construction
Sequences of ARGONAUTE proteins (≥800 amino acids), annotated from transcripts and genomes, were aligned using MUSCLE v3.8.31 with default parameters (Edgar, 2004). The regions poorly aligned were trimmed using trimAl v1.4 (Capella-Gutiérrez et al., 2009), and the trimmed alignments (Supplemental File 1) were used for construction of a maximum likelihood tree using RAxML v8.1.1 under the GTRCAT model (Stamatakis, 2014). For a tree of bryophyte TAS3 genes (Supplemental Figure 3B), the nucleotide sequences of those genes were aligned and edited similarly (Supplemental File 2), and the maximum likelihood tree was made using RAxML under the PROTGAMMAAUTO model. For each tree, 1000 replicates were conducted to generate bootstrap values. The trees were viewed using Dendroscope v3.5.7 (Huson and Scornavacca, 2012).
Jalview was used for the viewing of alignment results (Waterhouse et al., 2009). The R package was used to make violin plots and conduct statistical analyses. Sequence logos of sRNA and target sites were generated using Weblogo (Crooks et al., 2004). To calculate the nucleotide diversity (π) of ARF genes, the amino acid sequences of ARFs were generated by translation of the genes, aligned using MUSCLE, and then the protein sequence alignment was used to generate the alignment of nucleotide sequences using PAL2NAL (Suyama et al., 2006). Poorly aligned regions, those with <30% nucleotide coverage, were removed, and finally the nucleotide diversity (π) at a single nucleotide level was calculated using a 20-nucleotide sliding window.
Accession Numbers
Sequence data from this article can be found in the NCBI Gene Expression Omnibus under accession number GSE90706 for the sRNA data from P. drummondii. The accession numbers for the analyzed AGO and TAS3 sequences can be found in Supplemental Files 1 and 2, respectively, and those for the ARF genes are in Supplemental Data Set 5. The entire precursor sequences for miR390 are given in Supplemental Data Set 1.
Supplemental Data
Supplemental Figure 1. Variation of miR390 and miR390* pairing in the miR390/miR390* duplex in different groups of plants.
Supplemental Figure 2. Phylogenetic tree of AGO proteins from plants prior to angiosperms (except including Amborella trichopoda, Oryza sativa, and Arabidopsis thaliana).
Supplemental Figure 3. Evolution of bryophyte TAS3 genes.
Supplemental Figure 4. TAS3 genes with abnormal structures from vascular plants.
Supplemental Figure 5. Numbers of TAS3 genes found in species with sequenced genomes.
Supplemental Figure 6. Potential target sites of vascular tasiARF matched to ARF proteins of Physcomitrella patens.
Supplemental Data Set 1. MIR390 genes identified.
Supplemental Data Set 2. TAS3 genes identified in bryophytes.
Supplemental Data Set 3. TAS3 genes identified in vascular plants.
Supplemental Data Set 4. TasiARF-targeted ARF genes in vascular plants.
Supplemental Data Set 5. Sequences of tasiARF-targeted ARF genes.
Supplemental Data Set 6. Predicted target genes of Marchantia tasiAP2.
Supplemental File 1. Text file of the AGO protein alignment used for the phylogenetic analysis in Supplemental Figure 2.
Supplemental File 2. Text file of the TAS3 nucleotide alignment used for the phylogenetic analysis in Supplemental Figure 3B.
Acknowledgments
We thank members of the Meyers lab for helpful discussions and input. This study was supported by U.S. National Science Foundation, Division of Integrative Organismal Systems Award 1257869 and the Chinese Thousand Young Talents Program. We thank Dennis Stevenson and Ryan Lister for assistance in obtaining P. drummondii material. We also thank Gane Ka-Shu Wang for assistance with access to the 1KP data.
AUTHOR CONTRIBUTIONS
R.X. and B.C.M. conceived the study. R.X. and J.X. collected and generated the data and did the data analyses. R.X. and B.C.M. wrote the article. All authors read and approved the final manuscript.
Glossary
- sRNA
small RNA
- miRNA
microRNA
- siRNA
small interfering RNA
- tasiRNA
trans-acting small interfering RNA
References
- Addo-Quaye C., Miller W., Axtell M.J. (2009). CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25: 130–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adenot X., Elmayan T., Lauressergues D., Boutet S., Bouché N., Gasciolli V., Vaucheret H. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16: 927–932. [DOI] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221. [DOI] [PubMed] [Google Scholar]
- Arif M.A., Fattash I., Ma Z., Cho S.H., Beike A.K., Reski R., Axtell M.J., Frank W. (2012). DICER-LIKE3 activity in Physcomitrella patens DICER-LIKE4 mutants causes severe developmental dysfunction and sterility. Mol. Plant 5: 1281–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J. (2013). Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 64: 137–159. [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Jan C., Rajagopalan R., Bartel D.P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577. [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Snyder J.A., Bartel D.P. (2007). Common functions for diverse small RNAs of land plants. Plant Cell 19: 1750–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37: W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna N.G., Mateos J.L., Bresso E.G., Palatnik J.F. (2009). A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J. 28: 3646–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna N.G., Schapire A.L., Zhai J., Chorostecki U., Boisbouvier J., Meyers B.C., Palatnik J.F. (2013). Multiple RNA recognition patterns during microRNA biogenesis in plants. Genome Res. 23: 1675–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2009). Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 25: 21–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorostecki U., Moro B., Rojas A.M.L., Debernardi J.M., Schapire A.L., Notredame C., Palatnik J.F. (2017). Evolutionary footprints reveal insights into plant microRNA biogenesis. Plant Cell 29: 1248–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. (2004). WebLogo: a sequence logo generator. Genome Res. 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas R.N., Wiley D., Sarkar A., Springer N., Timmermans M.C.P., Scanlon M.J. (2010). ragged seedling2 encodes an ARGONAUTE7-Like protein required for mediolateral expansion, but not dorsiventrality, of maize leaves. Plant Cell 22: 1441–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y., Iwakawa H.O., Tomari Y. (2013). Arabidopsis ARGONAUTE7 selects miR390 through multiple checkpoints during RISC assembly. EMBO Rep. 14: 652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N., Jogdeo S., Kasschau K.D., Sullivan C.M., Chapman E.J., Laubinger S., Smith L.M., Dasenko M., Givan S.A., Weigel D., Carrington J.C. (2010). MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell 22: 1074–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N., Montgomery T.A., Howell M.D., Allen E., Dvorak S.K., Alexander A.L., Carrington J.C. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16: 939–944. [DOI] [PubMed] [Google Scholar]
- Finet C., Berne-Dedieu A., Scutt C.P., Marlétaz F. (2012). Evolution of the ARF gene family in land plants: old domains, new tricks. Mol. Biol. Evol. 30: 45–56. [DOI] [PubMed] [Google Scholar]
- Garcia D., Collier S.A., Byrne M.E., Martienssen R.A. (2006). Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 16: 933–938. [DOI] [PubMed] [Google Scholar]
- Guilfoyle T.J., Hagen G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10: 453–460. [DOI] [PubMed] [Google Scholar]
- Howell M.D., Fahlgren N., Chapman E.J., Cumbie J.S., Sullivan C.M., Givan S.A., Kasschau K.D., Carrington J.C. (2007). Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19: 926–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C., Willmann M.R., Wu G., Yoshikawa M., de la Luz Gutiérrez-Nava M., Poethig S.R. (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133: 2973–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D.H., Scornavacca C. (2012). Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61: 1061–1067. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P., Bartel B. (2006). MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 57: 19–53. [DOI] [PubMed] [Google Scholar]
- Krasnikova M.S., Goryunov D.V., Troitsky A.V., Solovyev A.G., Ozerova L.V., Morozov S.Y. (2013). Peculiar evolutionary history of miR390-guided TAS3-like genes in land plants. Sci. World J. 2013: 924153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Baniaga A.E., Sessa E.B., Scascitelli M., Graham S.W., Rieseberg L.H., Barker M.S. (2015). Early genome duplications in conifers and other seed plants. Sci. Adv. 1: e1501084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P.C., et al. (2016). Identification of miRNAs and their targets in the liverwort Marchantia polymorpha by integrating RNA-Seq and degradome analyses. Plant Cell Physiol. 57: 339–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Coruh C., Axtell M.J. (2010). Arabidopsis lyrata small RNAs: transient MIRNA and small interfering RNA loci within the Arabidopsis genus. Plant Cell 22: 1090–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A., Vaucheret H. (2010). Form, function, and regulation of ARGONAUTE proteins. Plant Cell 22: 3879–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E., Jouannet V., Herz A., Lokerse A.S., Weijers D., Vaucheret H., Nussaume L., Crespi M.D., Maizel A. (2010). miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matasci N., et al. (2014). Data access for the 1,000 Plants (1KP) project. Gigascience 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos J.L., Bologna N.G., Chorostecki U., Palatnik J.F. (2010). Identification of microRNA processing determinants by random mutagenesis of Arabidopsis MIR172a precursor. Curr. Biol. 20: 49–54. [DOI] [PubMed] [Google Scholar]
- Montgomery T.A., Howell M.D., Cuperus J.T., Li D., Hansen J.E., Alexander A.L., Chapman E.J., Fahlgren N., Allen E., Carrington J.C. (2008a). Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141. [DOI] [PubMed] [Google Scholar]
- Montgomery T.A., Yoo S.J., Fahlgren N., Gilbert S.D., Howell M.D., Sullivan C.M., Alexander A., Nguyen G., Allen E., Ahn J.H., Carrington J.C. (2008b). AGO1-miR173 complex initiates phased siRNA formation in plants. Proc. Natl. Acad. Sci. USA 105: 20055–20062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeswaran R., Pooggin M.M. (2012). RDR6-mediated synthesis of complementary RNA is terminated by miRNA stably bound to template RNA. Nucleic Acids Res. 40: 594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She R., Chu J.S.C., Uyar B., Wang J., Wang K., Chen N. (2011). genBlastG: using BLAST searches to build homologous gene models. Bioinformatics 27: 2141–2143. [DOI] [PubMed] [Google Scholar]
- Simonini S., Deb J., Moubayidin L., Stephenson P., Valluru M., Freire-rios A., Sorefan K., Weijers D., Østergaard L. (2016). A noncanonical auxin-sensing mechanism is required for organ morphogenesis in Arabidopsis. Genes Dev. 30: 2286–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Axtell M.J., Fedoroff N.V. (2010). RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr. Biol. 20: 37–41. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M., Torrents D., Bork P. (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34: W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. (2008). Plant ARGONAUTES. Trends Plant Sci. 13: 350–358. [DOI] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. (2009). Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S., Wollmann H., Schneeberger K., Weigel D. (2010). Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr. Biol. 20: 42–48. [DOI] [PubMed] [Google Scholar]
- Xia R., Xu J., Arikit S., Meyers B.C. (2015a). Extensive families of miRNAs and PHAS loci in norway spruce demonstrate the origins of complex phasiRNA networks in seed plants. Mol. Biol. Evol. 32: 2905–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R., Ye S., Liu Z., Meyers B.C., Liu Z. (2015b). Novel and recently evolved microRNA clusters regulate expansive F-BOX gene networks through phased small interfering RNAs in wild diploid strawberry. Plant Physiol. 169: 594–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R., Zhu H., An Y.Q., Beers E.P., Liu Z. (2012). Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol. 13: R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yifhar T., Pekker I., Peled D., Friedlander G., Pistunov A., Sabban M., Wachsman G., Alvarez J.P., Amsellem Z., Eshed Y. (2012). Failure of the tomato trans-acting short interfering RNA program to regulate AUXIN RESPONSE FACTOR3 and ARF4 underlies the wiry leaf syndrome. Plant Cell 24: 3575–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M.Y., Poethig R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19: 2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Xia R., Meyers B.C., Walbot V. (2015). Evolution, functions, and mysteries of plant ARGONAUTE proteins. Curr. Opin. Plant Biol. 27: 84–90. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Xia R., Kuang H., Meyers B.C. (2016). The diversification of plant NBS-LRR defense genes directs the evolution of microRNAs that target them. Mol. Biol. Evol. 33: 2692–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., et al. (2013). The trans-acting short interfering RNA3 pathway and no apical meristem antagonistically regulate leaf margin development and lateral organ separation, as revealed by analysis of an argonaute7/lobed leaflet1 mutant in Medicago truncatula. Plant Cell 25: 4845–4862. [DOI] [PMC free article] [PubMed] [Google Scholar]