Synteny-based network analyses provide new insight into the evolution of the MADS-box gene family.
Abstract
Conserved genomic context provides critical information for comparative evolutionary analysis. With the increase in numbers of sequenced plant genomes, synteny analysis can provide new insights into gene family evolution. Here, we exploit a network analysis approach to organize and interpret massive pairwise syntenic relationships. Specifically, we analyzed synteny networks of the MADS-box transcription factor gene family using 51 completed plant genomes. In combination with phylogenetic profiling, several novel evolutionary patterns were inferred and visualized from synteny network clusters. We found lineage-specific clusters that derive from transposition events for the regulators of floral development (APETALA3 and PI) and flowering time (FLC) in the Brassicales and for the regulators of root development (AGL17) in Poales. We also identified two large gene clusters that jointly encompass many key phenotypic regulatory Type II MADS-box gene clades (SEP1, SQUA, TM8, SEP3, FLC, AGL6, and TM3). Gene clustering and gene trees support the idea that these genes are derived from an ancient tandem gene duplication that likely predates the radiation of the seed plants and then expanded by subsequent polyploidy events. We also identified angiosperm-wide conservation of synteny of several other less studied clades. Combined, these findings provide new hypotheses for the genomic origins, biological conservation, and divergence of MADS-box gene family members.
INTRODUCTION
Conserved gene order can be retained for hundreds of millions of years and provides critical information about conserved genomic context and the evolution of genomes and genes. For example, the well-known “Hox gene cluster,” which regulates the animal body plan, is largely collinear across the animal kingdom (Lewis, 1978; Krumlauf, 1994; Ferrier and Holland, 2001). The term synteny was originally defined as a set of genes from two species located on the same chromosome, but not necessarily in the same order (Dewey, 2011; Passarge et al., 1999). However, the current widespread usage of the term synteny, which we adopt, now implies conserved collinearity and genomic context. Synteny data are widely used to establish the occurrence of ancient polyploidy events, to identify chromosomal rearrangements, to examine the expansion and contraction of gene families, and to establish gene orthology (Sampedro et al., 2005; Tang et al., 2008a; Dewey, 2011; Jiao and Paterson, 2014). Synteny likely reflects important relationships between the genomic context of genes both in terms of function and regulation and thus is often used as a “proxy for the conservation or constraint of gene function” (Dewey, 2011; Lv et al., 2011). Syntenic relationships across a wide range of species thus provide crucial information to address fundamental questions on the evolution of gene families that regulate important developmental pathways. For example, the origin of morphological novelty has been linked to the duplication of key regulatory transcription factors in the case of the Hox genes in animals, but also the MADS-box genes in plants (Alvarez-Buylla et al., 2000b; Airoldi and Davies, 2012; Soshnikova et al., 2013). However, gene clusters are frequently dispersed or “broken up” in certain lineages, like the Hox cluster in the genomes of octopus (Lemons and McGinnis, 2006; Duboule, 2007; Albertin et al., 2015) and brachiopods (Schiemann et al., 2017), and this dispersion contributes to divergent gene expression and morphological novelties.
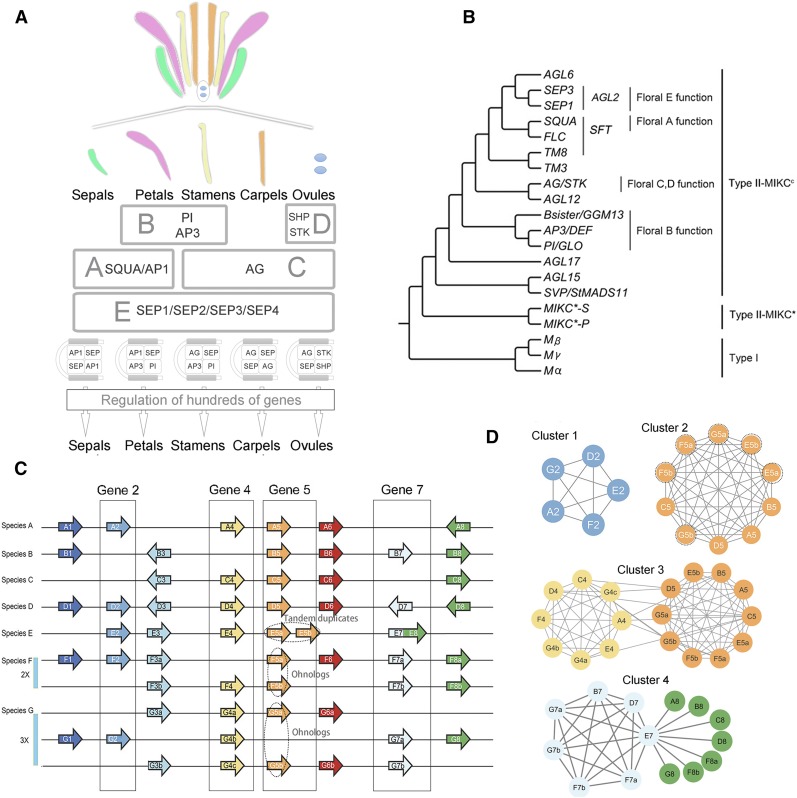
In plants, the MADS-box genes are critical transcription factors that regulate the developmental pattern of the floral organs, the reproductive organs, and other traits (Theissen, 2001; Becker and Theissen, 2003; Smaczniak et al., 2012). For instance, floral organ identity is controlled largely by MADS-box genes, as explained by the ABC(DE) model (Figure 1A) (Coen and Meyerowitz, 1991; Ditta et al., 2004) with, for example, the floral A-, B-, and E-function genes being required for petal identity (Figures 1A and 1B). Synteny data of the MADS-box genes have been used to infer the ancestral genetic composition of the B- and C-function (Causier et al., 2010), and the A- and E-function genes (Ruelens et al., 2013; Sun et al., 2014). However, these studies analyzed only a small number of species (fewer than 10) and the results were displayed as parallel coordinate plots (as in Figure 1C). A systematic comparison of the syntenic relationships for all the MADS-box genes across many plant species has not been done in a single study. That is because this gene family has undergone extensive duplications that have given rise to complicated relationships of orthology, paralogy, and functional homology (Jaramillo and Kramer, 2007). Hence, a systematic investigation in which all the possible syntenic relationships between the family members are sorted and visualized is challenging. With the increase of genomes that are simultaneously analyzed, it becomes increasingly more difficult to organize and display such syntenic relationships. This is due to the ubiquity of ancient and recent polyploidy events, as well as smaller scale events that derive from tandem and transposition duplications (Lynch and Conery, 2000; Bowers et al., 2003; Tang et al., 2008a; Schranz et al., 2012).
Figure 1.
Summary of the MADS-Box Genes and Principles of Phylogenomic Synteny Network Analysis.
(A) Diagram illustrating the ABCDE floral development model. The A- and E-function genes are essential for sepal identity; the A-, B-, and E-function genes for petal identity; the B-, C-, and E-function genes for stamen identity; the C- and E-function genes for carpel identity; and the D-, C-, and E-function genes for ovule identity.
(B) The consensus phylogenetic tree showing the relationships for the different functional gene clades of the MADS-box gene family. The combined clade containing the SQUA-, FLC-, and TM8-like genes is referred to as SFT in this study.
(C) Hypothetical example of a parallel coordinate plot for synteny comparisons across seven species (A–G) for which species F has undergone a whole-genome duplication (WGD = 2x) and species G a whole-genome triplication (WGT = 3x). Examples of tandem duplicates and ohnologs/syntelogs of Gene 5 are indicated by the dotted ovals. Genes 2, 4, 5, and 7 are each boxed as examples of network view of synteny relationships.
(D) Synteny network of Gene 2 (Cluster 1, less conserved), Gene 5 (Cluster 2, highly conserved and includes tandem duplicates and ohnologs), Gene 4 and Gene 5 [Cluster 3, where genes are members of larger gene family and thus are interconnected; in this case, we suppose Gene 4 and Gene 5 belong to a same gene family, which share similar domain(s), and synteny is checked for this whole gene family], and Gene 7 (Cluster 4, where gene E7 represents an actual gene fusion of protein domain from a neighboring gene or is an artifact due to gene misannotation, only synteny of Gene 7 homologs is being checked synteny). Nodes represent syntenic genes, and edges (lines) represent a syntenic connection between two nodes. Edge length in this example is noninformative.
Here, we present a novel approach to cluster synteny networks and then analyze gene ancestry. Instead of presenting syntenic blocks as either parallel coordinate plots (Figure 1C) or pairwise dot plots, we abstracted genome syntenic blocks (derived from intra- and interspecies comparisons) into vertices (nodes or points) and edges (lines between points). Syntelogs (syntenic homologous genes) of a target gene or gene family of interest can be highlighted in one graph without showing the flanking genes (Figure 1D). For example, the syntenic relationships of “Gene 2” across five species (Species A, D, E, F, and G) in Figure 1C can be represented as a cluster of five nodes, with edges representing their syntenic relationships (Figure 1D, Cluster 1). If one gene has undergone an additional duplication event, such as tandem and/or polyploid duplication (for example “Gene 5” that is a tandem-duplicated in Species E [E5a and E5b] and ohnologs [syntelogs derived from polyploidy events] retained in Species F and G in Figure 1C), these duplicated syntelogs are included as nodes rather than adding additional linear panels to a parallel coordinate plot (Figure 1D, Cluster 2).
Potential ancient tandem duplications can also be readily represented and detected by synteny network analyses. Cluster 3 illustrates an example where both “Gene 4” and “Gene 5” genes are found in one cluster (Figure 1D, Cluster 3). Such a result can occur when “Gene 4” and “Gene 5” belong to a same gene family and contain the same protein domain(s). Unlike the tandem duplication of the example of “Gene E5a” and “Gene E5b” (Figure 1D, Cluster 2), they may be derived from an ancient tandem duplication and thus evolved a certain degree of differences at the gene sequence level (and thus may even belong to different clades/subgroups of one gene family). “Gene 4” and “Gene 5” can be calculated as syntenic to each other by synteny detection programs when one of the loci was lost. For example, “Gene A4” is found to be syntenic to “Gene B5” because the best option “Gene B4” may has been lost in Species B. As a result, we obtain a twin cluster layout with more “intra-links” than “inter-links” (Figure 1D, Cluster 3). It is worth mentioning that sometimes one specific node connects (radiates) to other unconnected nodes. For example, node “Gene E7” in Cluster 4 (Figure 1D, Cluster 4) radiates to seven other nodes of syntelogs of “Gene 8,” which belong to another gene family different from the one of “Gene 7.. This is because “Gene E7” contains both domains of “Gene 7” and “Gene 8,” either because of a potential genome misannotation or a real protein domain fusion.
With this background for visualizing synteny networks, we can proceed with their construction and use for understanding evolutionary patterns. We refer readers to a recently published outline of our generalized approach to construct synteny networks (Zhao and Schranz, 2017). The construction of synteny networks uses three main steps: (1) pairwise whole-genome comparisons, (2) detection of syntenic blocks and data fusion, and (3) network clustering. The first two steps provide a database of syntenic relationships between homologous genes for the genomes analyzed using standard programs, such as BLAST (Altschul et al., 1990) for genome comparisons and MCScan (Tang et al., 2008b) for synteny detection. The final step, the network clustering, can make use of a wide range of clustering algorithms and methods (reviewed in Lancichinetti and Fortunato, 2009; Fortunato, 2010) and are at the heart of our synteny network analysis. The resulting clusters can differ from each other according to the methods applied. Here, we use CFinder to cluster our pairwise synteny data, which allows the detection of overlapping communities in network data by using the k-clique percolation method (Palla et al., 2005, 2007). K-clique corresponds to a fully connected subselection of k nodes (e.g., a k-clique of k = 3 is equivalent to a triangle). Two k-cliques are considered adjacent and thus form a k-clique community if they share k-1 nodes (Derényi et al., 2005; Palla et al., 2005).
To illustrate this approach, we analyzed the well-characterized MADS-box gene family. The relationships between the major clades of the plant MADS-box genes have already largely been inferred in various phylogenetic and evolutionary studies (Becker and Theissen, 2003; Martinez-Castilla and Alvarez-Buylla, 2003; Nam et al., 2003, 2004, 2005; Gramzow et al., 2012, 2014; Smaczniak et al., 2012; Kim et al., 2013; Ruelens et al., 2013; Sun et al., 2014; Yu et al., 2016b) (Figure 1B). However, these studies cannot fully resolve some of the deepest nodes of the MADS-box gene tree. The genome of the model plant Arabidopsis thaliana contains a total of 107 MADS-box genes, which derive from multiple gene duplication events (Martinez-Castilla and Alvarez-Buylla, 2003; Parenicová et al., 2003). The MADS-box genes can be divided into two major clades, termed Type I and Type II. The Type II lineage is further divided into the MIKCC- and MIKC*-types (Henschel et al., 2002). The function and evolution of MADS-box genes have been extensively studied, especially the MIKCC types (reviewed in Smaczniak et al., 2012). For convenience, we hereafter refer to the hypothesized common ancestral genes of the SQUA-, FLC-, and TM8-like genes as SFT genes.
Here, we present and discuss the synteny network of all the detected MADS-box genes in 51 plant genomes. This network includes intra- and interspecies syntenic blocks that derive from both shared but also independent polyploidy events in these 51 species. In combination with phylogenetic analysis and phylogenetic profiling (Pellegrini et al., 1999), we could elucidate several previously undetected evolutionary patterns of gene transposition, gene duplication, and shared deep ancestry for different MADS-box gene clades. Our approach sheds new light on the evolutionary trajectory of the MADS-box genes and thereby of the traits they control in different plant lineages. Our approach can be easily applied to other gene families and genomes following the step-by-step workflow given on GitHub (https://github.com/zhaotao1987/SynNet-Pipeline).
RESULTS
Overview of the Synteny Network Pipeline
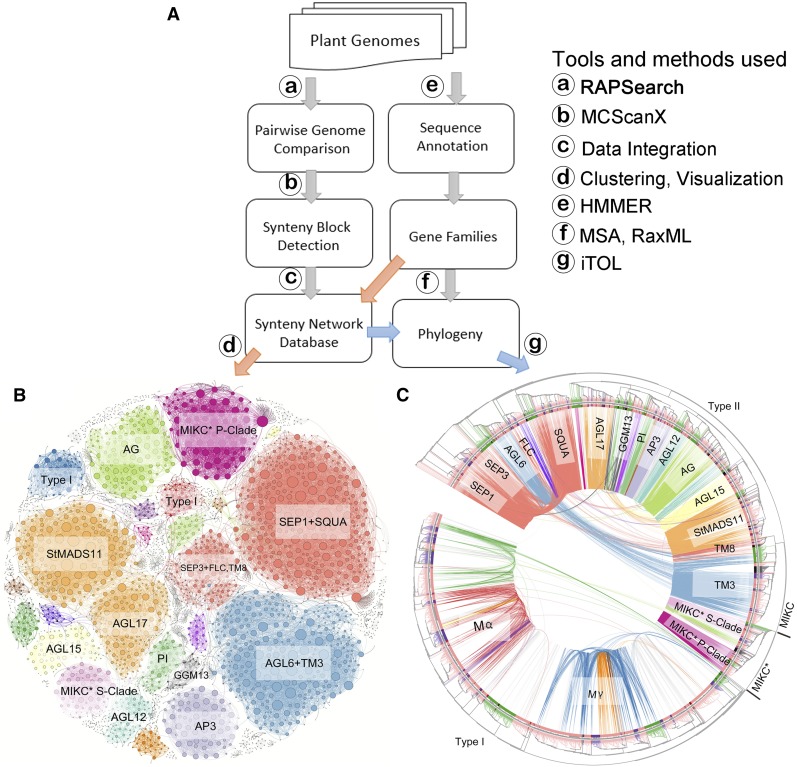
In this study, we analyzed 51 plant genomes covering green algae, mosses, gymnosperms, and angiosperms (Supplemental Table 1 and Supplemental Figure 1). We analyzed all protein models from these genomes for all possible intra- and interspecies whole-genome comparisons (Figure 2A). We then built a database that contains all the links between syntenic gene pairs present in syntenic genomic blocks identified by the tool MCScanX (Tang et al., 2008b; Wang et al., 2012). This database contains in total 921,074 nodes (i.e., genes that were connected by synteny with another gene) and 8,045,487 edges (i.e., pairwise syntenic connections); the data can be downloaded from GitHub (https://github.com/zhaotao1987/SynNet-Pipeline).
Figure 2.
Workflow to Create the Phylogenomic Synteny Network and Example Outputs for the Global MADS-Box Gene Family.
(A) Workflow to create the phylogenomic synteny network. Annotated whole-genome sequences enter the pipeline and are used in two parallel modules. The left panel represents the analysis pipeline for pairwise genome comparisons and synteny calculations (synteny block detection), which creates the global syntenic network database. The right panel depicts the pipeline for a phylogenetic analysis including gene family identification and gene tree construction.
(B) Synteny network of the MADS-box gene family using all the detected syntenic relations in the synteny network database. Communities were rendered based on the clique percolation method at k = 3. The size of each node corresponds to the number of edges it has (node degree). Communities were labeled by the subfamilies/subfamily involved.
(C) Maximum-likelihood gene tree for the MADS-box gene family and syntenic relationships between the genes. The subclades are indicated for the Type I, Type II, and MIKC- and MIKC*-Type II MADS-box genes on the tree. Terminal branch colors represent genes belonging to rosids (light pink), asterids (purple), and monocots (green). Genes belonging to angiosperms in highly informative phylogenetic positions such as A. trichopoda, V. vinifera, B. vulgaris, and N. nucifera are in red and genes of non-angiosperms belonging to Chlamydomonas reinhardtii, Physcomitrella patens, Selaginella moellendorffii, and Picea abies are in black. Each connecting line located inside the inverted circular gene tree indicates a syntenic relationship between two MADS-box genes. The connecting lines are colored according to the discovered communities in (B).
We used this database to investigate the syntenic relationships between the MADS-box genes. To this end, we used HMMER (Finn et al., 2011) to screen the predicted protein sequences of the 51 genomes to identify all the MADS-box genes in these genomes (Supplemental Data Set 1, sheet 1). The resulting list with candidate MADS-box genes was subsequently used to extract the synteny subnetwork for these MADS-box genes from the entire network database. This subnetwork contained 3458 nodes (MADS-box genes) that were linked by 25,500 syntenic edges (Supplemental Data Set 1, sheet 2). We visualized this subnetwork using Gephi (Bastian et al., 2009) and color-coded the clusters using the k-clique percolation clustering method with k = 3 (Figure 2B). This network and its identified clusters give a first impression on how the MADS-box genes are positionally related to each other across all angiosperms lineages (Figure 2B). The network did not contain synteny information that linked to the non-angiosperm species, which is likely due to the extreme phylogenetic distance and the limited sampling of non-angiosperms species. The node size shown indicates the number of connections for each node (Figure 2B). To reveal syntenic relationships between distant gene clades, we then displayed pairwise syntenic relationships between the MADS-box genes in a gene tree that we constructed for the entire gene family (Figure 2C). The colors of the connecting lines indicate again the network communities defined at k = 3 from Figure 2B. Interestingly, we found genes from distal gene clades (shown in Figure 1B) that are syntenically connected, such as SEP1-like (floral E genes) with SQUA-like (floral A genes) genes, AGL6-like with TM3 (SOC1-like) genes, and StMADS11 (SVP-like) with AGL17-like genes (Figures 2B and 2C).
Using CFinder, we detected all cliques of size k = 3 to k = 24 for the MADS-box gene synteny network and the number of k-clique-communities under each k-clique (Supplemental Figure 2A). Each of the community (cluster) sizes under a certain k is shown (Supplemental Figure 2A), which quantifies the strength of the syntenic connections across species. For example, the AP3-like genes of monocot species (green nodes) are only part of a community at relatively low k values (k < 8) (Supplemental Figure 2B). This could be due to several factors, including the larger genome sizes of monocots (making it more difficult to detect synteny), the limited number of monocot genomes included and/or the lack of phylogenetic sampling across the monocots (i.e., there are many Poales genomes included [7/11] but few other monocot lineages [4/11]).
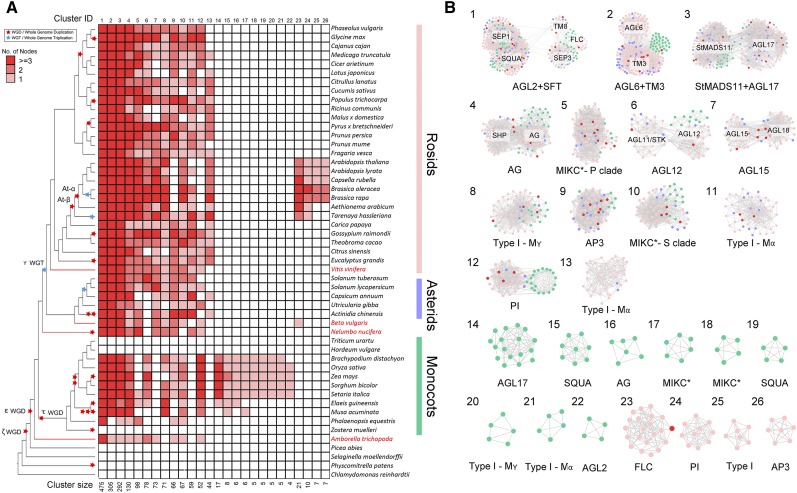
A clique size of k = 3 to 6 was identified to best approximate the true number of communities (Derényi et al., 2005; Palla et al., 2005; Porter et al., 2009; Xie et al., 2013). We obtained 95 clusters using k = 3 (Supplemental Data Set 2), and we used these clusters for phylogenetic profiling (Supplemental Figure 3). Each column depicts a syntenic occurrence for a certain MADS-box gene cluster in each plant species. Thereby the presence/absence of syntenic gene clusters across the 51 analyzed taxa are represented by their respective phylogenetic profiles to determine and infer evolutionary patterns (Supplemental Figure 3). We highlighted 26 relevant (i.e., either broad conservation or lineage specific) clusters in the phylogenetic profile (Figure 3A). For two monocot species, wheat (Triticum urartu) and barley (Hordeum vulgare), we did not find any syntenic regions for any of their MADS-box genes with other plant genomes. This is likely due to the fragmented early-version genome assemblies (partially due to their large genome sizes and transposon expansions) in these two grasses. Using the organic layout function in Cytoscape (Shannon et al., 2003), we further depicted an undirected and unweighted (e.g., edge length of no meaning) network with related gene clade names (Figure 3B). From this, we can then infer the number of syntelogs and relationships among syntelogs generated via polyploidy and tandem duplication events. Below, we highlight three novel insights into the evolution of the MADS-box gene family based on our synteny network cluster analysis: (I) lineage-specific transpositions, (II) ancient tandem gene arrangements, and (III) deep conservation of specific clades across angiosperms.
Figure 3.
Phylogenetic Profiling and Detailed Network Representations for a Set of Selected Important MADS-Box Synteny Network Clusters.
(A) Phylogenetic profiling of 26 example clusters from all the 95 clusters at k = 3 (Supplemental Figure 3). Red-colored cells depict the presence of at least one syntelog in the different species. The phylogenetic profiling approach identified large clusters containing genes from various subfamilies, such as “AGL2+SQUA” (Cluster 1) and “AGL6+TM3” (Cluster 2), or lineage-specific clusters, such as of PI (Cluster 24) and AP3 (Cluster 26) in the Brassicaceae, and “AGL17” (Cluster 14) and “SQUA” (Cluster 15) in monocots. Species names are shown on the right side. The basal rosid V. vinifera, the basal eudicots B. vulgaris and N. nucifera, and the basal angiosperm A. trichopoda are highlighted in red. Red and blue stars on the tree indicate known WGD and whole-genome triplication (WGT) events, respectively, on the phylogenetic tree on the left side. The cluster ID and size are indicated at the top and bottom, respectively. The red color scale of the cell indicates the number of nodes (genes grouping in that cluster) found in a single species.
(B) Clusters from (A), which can be divided into large-conserved clusters (Clusters 1–13) and lineage-specific clusters (monocots [Cluster 14–22] and Brassicales [Cluster 23–26]). The node colors represent rosids (light pink), asterids (blue), and monocots (green); the nodes belonging to A. trichopoda, V. vinifera, B. vulgaris, and N. nucifera are shown in red.
Section I: Lineage-Specific Synteny Relationships
Important angiosperm families (such as Poaceae, Asteraceae, Fabaceae, Brassicaceae, and Solanaceae) are readily identified by unique traits and floral characteristics. These major plant families are also characterized by having independent ancient polyploidy events at their origins (Soltis et al., 2009; Schranz et al., 2012; Tank et al., 2015). Morphological changes could thus be tied to these ancient polyploidy events or specific gene transposition events that place key regulatory factors into new genomic contexts (Soltis et al., 2009; Freeling et al., 2012). Our synteny network approach can identify such lineage-specific transposition events for genes by clustering and phylogenetic profiling.
I.1 B-Function (AP3 and PI) Genes in the Brassicaceae and Cleomaceae Families
The AP3 and PI genes are important for petal and stamen specification (Jack et al., 1992, 1994; Goto and Meyerowitz, 1994; Zhang et al., 2013; Trobner et al., 1992; Sommer et al., 1990). In this study, we found that most AP3 genes reside in a single cluster comprising homologs of both eudicot and monocot species, the basal angiosperm Amborella trichopoda, and the basal eudicot Nelumbo nucifera (Figure 3, Cluster 9). However, the cluster lacks AP3 homologs from the Brassicaceae family (Figure 3, Cluster 9). Instead, the AP3 genes from the Brassicaceae form a separate cluster (Figure 3, Cluster 26) (except for Aethionema arabicum, where the A. arabicum AP3 gene was annotated on a scaffold lacking other genes; gene ID AA1026G00001, highlighted in Supplemental Data Set 1, sheet 1).
A very similar picture emerges for the PI genes: The PI homologs from the analyzed six Brassicaceae species group together with a PI gene from Tarenaya hassleriana (a closely related Cleomaceae species), while the PI homologs from most other species group with a second PI gene from T. hassleriana in another cluster (Figure 3, Cluster 24). To verify this pattern, we investigated the synteny relationships of the PI genes from grapevine (Vitis vinifera; Vv18s0001g01760) and Arabidopsis (AT5G20240) using the Genomicus parallel coordinate plot (Louis et al., 2013). Synteny was not detected with any Brassicaceae species when using the grape homolog of PI (Vv18s0001g01760) (Supplemental Figure 4A), while a unique synteny pattern is shared between the Arabidopsis gene AT5G20240 and the Brassicaceae PI genes (Supplemental Figure 4B).
These two divergent synteny patterns suggest that in both cases (PI and AP3), a gene transposition, a genomic rearrangement event, or extreme genome fractionation led to the unique genomic context seen for both genes in the Brassicaceae. Since one Cleomaceae PI gene belongs to the Brassicaceae PI cluster (Figure 3, Cluster 24) but the Brassicaceae AP3 cluster does not contain a Cleomaceae AP3 gene (Figure 3, Cluster 26), it is clear that PI transposed first and, only later and independently, did AP3 transpose.
I.2 FLC-Like Genes Cluster in Brassicaceae
In Arabidopsis, the FLC gene and its closely related MAF genes are floral repressors and major regulators of flowering time (Michaels and Amasino, 1999; Sheldon et al., 2000). We found a cluster comprising 21 syntelogs of FLC and the MAF genes across the six examined Brassicaceae species and one Cleomaceae species (Tarenaya) (Figure 3, Cluster 23).
This synteny cluster also contains one FLC-like gene from sugar beet (Beta vulgaris). This sugar beet FLC homolog also shares synteny with a cluster comprising StMADS11 (SVP-like) genes, which are found in an array of eudicot species (Figure 3B, Cluster 3; Supplemental Data Set 3). This sugar beet FLC gene thus connects the FLC/MAF genes of the Brassicales lineage with the StMADS11 genes of other eudicots. This highlights that likely a gene transposition or massive genome fractionation process has acted on the ancestral FLC gene in the Brassicales lineage after the split of the early branching papaya (Carica papaya), potentially near the time of the At-β whole-genome duplication (WGD; Edger et al., 2015).
I.3 AGL17-Like Genes Cluster in Monocots
Also, the AGL17-like genes from six monocots specie (Brachypodium distachyon, Oryza sativa, Zea mays, Sorghum bicolor, Setaria italica, and Elaeis guineensis) form a distinct synteny cluster (Figure 3, Cluster 14, size 17). This may be to a specific transposition event and/or due to the ancient τ WGD shared by all monocot species (Jiao et al., 2014).
Section II: Inference of Ancient Tandem Gene Arrangements
Besides the distinctive lineage-specific clusters described above, larger clusters that comprise interconnected subclusters (with a force-directed or organic layout) can also be obtained when using the appropriate clustering methods (such as the k-clique percolation method that allows for community overlapping). As shown in Cluster 3 (Figure 1D), such clusters indicate long-conserved close genomic proximity of the genes involved (representing respective subclusters) and thus helpful for establishing the trajectory of gene evolution.
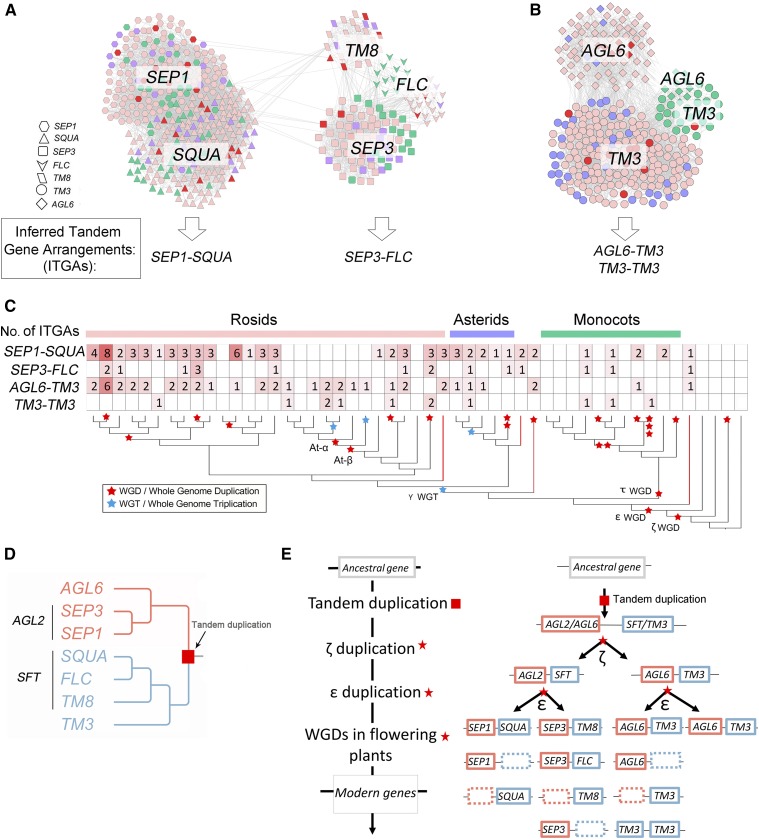
II.1 Angiosperm-Wide Conserved SEP1-SQUA and SEP3-SFT Tandems
The largest cluster (475 nodes) we identified comprises both the AGL2 (SEP)-like and the SFT-like genes (Figures 3B, Cluster 1, and 4A). This cluster can be divided into two subgroups: On the left are the SEP1- and SQUA-like genes, while on the right are the SEP3-, FLC-, and TM8-like genes (Figure 4A). The SEP1- and SQUA-like genes are highly interconnected between and within genomes (Figure 4A) with syntenic orthologs being present for both genes in a wide range of angiosperm species including A. trichopoda, monocots, and eudicots. As exemplified by Cluster 3 in the Introduction (Figure 1D), SEP1- and SQUA-like genes are predominantly found in a tandem gene arrangement in most angiosperm species (Figures 4A and 4C; Supplemental Data Set 3), suggesting that this duplication occurred prior to or at the origin of the angiosperms. For example, there is one SEP1-SQUA tandem gene arrangement in the A. trichopoda and three such tandem gene arrangements in the basal eudicot V. vinifera (Figures 4A and 4C; Supplemental Data Set 3), as a result of the gamma hexaploidization (referred to as γ triplication) in eudicots.
Figure 4.
Evolutionary History of a Major MIKCc-Type MADS-Box Gene Superclade Is Derived from Inferred Tandem Gene Arrangements.
(A) and (B) Close-up of the networks for Cluster 1 (AGL2+SFT) and Cluster 2 (AGL6+TM3) from Figure 3, respectively. Node shapes represent different subfamilies. Node colors represent rosids (light pink), asterids (blue), and monocots (green), and the nodes belonging to A. trichopoda, V. vinifera, B. vulgaris, and N. nucifera are in red. The arrows below the clusters point to the inferred tandem gene arrangements (ITGAs) for the respective three clusters.
(C) Summary of the occurrence of four ITGAs detected in the species analyzed. The two top rows give the number of SEP1-SQUA and SEP3-FLC tandems detected in Cluster 1 (A). While rows three and four give the number of AGL6-TM3 and TM3-TM3 tandems detected in Cluster 2 (B). The species tree is a simplified version (without species names) of the tree used in Figure 3A and Supplemental Figure 1.
(D) Mapping of the tandem duplication event onto the inferred gene tree according to the three ITGAs in (C).
(E) Proposed evolutionary scenario for the origin of the MADS-box gene family clade in (D). One MIKCc-type MADS-box gene initially got tandem duplicated, then this tandem arrangement underwent multiple rounds of polyploidy events and eventually evolved to the superclade depicted in (D).
On the right side of the network in Figure 4A, most eudicot and monocot SEP3-like genes group as a distinct cluster, which is relatively loosely connected to the nodes that represent the FLC-like genes and TM8-like genes (Figure 4A). Similar to the discovery of a SEP1-SQUA tandem, we also identified a SEP3-FLC tandem gene arrangement in 12 eudicots species (Figure 4C; Supplemental Data Set 3). This tandem arrangement was also found twice in monocots, namely, O. sativa and S. bicolor (Figure 4C; Supplemental Data Set 3). However, the SEP3-FLC tandem gene arrangement is found less often than the SEP1-SQUA tandem gene arrangement. Besides this, we found that in A. trichopoda, the SEP3 and TM8 homologs are also arranged in tandem (SEP3-TM8) (Figure 4C; Supplemental Data Set 3). None of the FLC homologs from Brassicaceae and Cleomaceae species are present in the angiosperm-FLC cluster in Figure 4A. As described in Section I, the Brassicales FLC syntelogs form an independent cluster (Figure 3B, Cluster 23).
II.2 Angiosperm-Wide Conserved AGL6-TM3 Tandem
The second largest cluster identified in this study (k = 3, community size: 305) contains the AGL6- and TM3 (SOC1)-like genes (Figures 3B, Cluster 2, and 4B). Like the SEP1-SQUA and SEP3-SFT tandems in Figure 4A, we found that the AGL6-TM3 tandem gene arrangement is widespread across angiosperms (Figures 4B and 4C; Supplemental Data Set 3). For example, there is one AGL6-TM3 tandem gene pair in A. trichopoda and two such tandems in N. nucifera likely due to the most recent WGD this species experienced (Ming et al., 2013; Wang et al., 2013) (Figure 4C; Supplemental Data Set 3). In V. vinifera, we also found two AGL6-TM3 tandems (Figure 4C; Supplemental Data Set 3) that likely originated from the γ triplication after which one tandem lost its AGL6 locus. Like V. vinifera, Theobroma cacao has not undergone any additional WGD after the γ triplication and in this genome two AGL6-TM3 tandems also remain (Figure 4C; Supplemental Data Set 3).
Besides the prevalent AGL6-TM3 tandem gene arrangement found in Figure 4B, we also found the tandem type of TM3-TM3 in 10 species (seven eudicot species and three monocot species) (Figure 4C; Supplemental Data Set 3). Hence, the network has overall more TM3-like genes than AGL6-like genes (Figure 4B).
Section III: Synteny Relations across Angiosperms for Overlooked MADS-Box Gene Clades
In addition to the many functionally characterized MADS genes, a large portion of the gene family members are poorly or not functionally characterized, such as MIKC*-type and Type I genes of the MADS-box gene family. However, the synteny network can provide evidence of synteny conservation for these genes over evolutionary time and thus suggest important conserved gene functions.
III.1 MIKC*-Type Genes
The MIKC*-type genes form a monophyletic clade within the MADS-box genes (Alvarez-Buylla et al., 2000a; Henschel et al., 2002) (Figure 1B), with several of them being reported to play a major role in pollen development (Verelst et al., 2007a, 2007b; Adamczyk and Fernandez, 2009). Using our synteny network analysis, we found two networks that are highly connected and contain (1) the angiosperm AGL30-, AGL65-, and AGL94-like genes (MIKC*-P clade) (Figure 3, Cluster 5) and (2) the AGL66-, AGL67-, and AGL104-like genes (MIKC*-S clade) (Figure 3, Cluster 10), respectively.
Both clusters encompass eudicots and monocot species, as well as A. trichopoda. However, the MIKC*-S cluster appears to have expanded in monocots, while homologs of N. nucifera are absent in this cluster (Figure 3, Cluster 10). This means that both two MIKC* clades are broadly conserved across angiosperms.
Interestingly, MIKC* protein complexes play an essential role in late pollen development in Arabidopsis and the formation of this protein complex requires MIKC* proteins from both clades. For example, the AGL30 and/or AGL65 proteins from the P clade form heterodimers with AGL104 or AGL66, which both group with the S clade (Verelst et al., 2007a, 2007b). This suggests that these two clades (gene clusters) have been functionally retained during angiosperm evolution.
III.2 Type I MADS-Box Genes
Type I MADS-box genes show a higher rate of gene birth and death, often due to gene duplication-transposition, than Type II genes (Nam et al., 2004; Freeling et al., 2008; Wang et al., 2016b). Also, the function of the different Type I genes is generally poorly characterized. However, several Type I genes have been reported to play a role in female gametogenesis, embryogenesis, and seed development (Portereiko et al., 2006; Bemer et al., 2010; Masiero et al., 2011).
With our approach, we found two distinct clusters that contain Type I MADS-box genes (Figure 3, Clusters 8 and 11). For example, the PHERES1 (PHE1/AGL37) genes, which are regulated by genomic imprinting (Köhler et al., 2003), are in the same synteny network as PHERES2 (PHE2/AGL38), AGL35-, and AGL36-like genes, which all belong to the Mγ clade of the Type I MADS-box genes (Figure 3, Cluster 8). Likewise, we found one cluster that contains genes from the Mα clade (Figure 3, Cluster 11).
III.3 StMADS11 (SVP-Like) Genes
In Arabidopsis, the StMADS11 gene clade is composed of two genes called SVP (AGL22) and AGL24. These two genes regulate the transition to flowering in Arabidopsis (Hartmann et al., 2000; Michaels et al., 2003).
We found that the SVP- and AGL24-like genes are contained in one cluster for many of the angiosperms analyzed, which indicates that synteny has been retained for SVP/AFL24 since the last common ancestor of angiosperms (Figure 3B, Cluster 3). It is worth noting that the AGL17-like genes from A. trichopoda, N. nucifera, and most eudicot species form a cluster that is moderately connected to the cluster of StMADS11-like genes (Figure 3B, Cluster 3).
DISCUSSION
Our phylogenomic synteny network analysis provides a novel approach to identify and visualize the relationships of genes of a targeted gene family across a broad range of species (Zhao and Schranz, 2017), which can be used to address fundamental questions on the origin of novel gene functions leading to morphological changes and adaptations. We have provided several new insights into the evolution of the MADS-box gene family from our synteny-based network analyses. These insights, in turn, generate new testable hypotheses on how the genomic context of a gene may (or may not) effect changes in its expression pattern, coexpression with other genes, epigenetic regulation, and ultimately the evolution of plant phenotypes. Some possible hypotheses are discussed below, but first we make a few comments regarding our methodology.
Factors Affecting Synteny Network Analysis
We have presented a methodological roadmap to construct synteny networks and an analysis pipeline, which can now be applied to any gene family across any set of genomes. The power of network analysis is the ability to organize large data sets and provide extrapolation and visualization beyond pairwise contrasts. As more plant species genomes are completed, particularly from underrepresented lineages (such as non-angiosperm species), more robust network inferences can be made. However, our network approach depends on the quality of genome assemblies and their gene annotations. Genome collinearity is de facto more disrupted and difficult to detect in highly fragmented assemblies. Advances in genome sequencing and assembly mean that chromosome level assemblies will be standard in the near future. With these advances, our network approach for synteny comparison will greatly benefit and improve.
The clustering methods used are pivotal for the interpretation of complex synteny networks, as it determines the size and structure of identified clusters. For example, when instead of our k-clique percolation method (at k = 3), other methods are used like k-core decomposition (Alvarez-Hamelin et al., 2006), MCL (Enright et al., 2002), infomap (Rosvall and Bergstrom, 2008), or CNM (Clauset et al., 2004), we would likely have obtained slightly different cluster topologies. Depending on the goals and objectives of a study, the appropriate clustering method should be established.
Lineage-Specific Genomic Context of MADS-Box Genes: Potential Significant Biological Implications
In the model plant Arabidopsis, the B-class AP3 and PI proteins form heterodimers and bind to the CArG-box cis-regulatory elements in promoters (Riechmann et al., 1996; Yang et al., 2003). Heterodimerization and/or homodimerization have contributed to the evolution of the highly diverse flower morphologies in angiosperms (Lee and Irish, 2011; Melzer et al., 2014; Bartlett et al., 2016). Brassicaceae species have rather uniform, or canalized, flowers (typical cross arrangement of the four petals). However, in its closest sister family Cleomaceae, which diverged from each other ∼38 million years ago (Schranz and Mitchell-Olds, 2006; Couvreur et al., 2010), more diverse floral morphologies are observed (Patchell et al., 2011). In this study, we found unique synteny patterns for the T. hassleriana B genes, which is consistent with previous findings (Cheng et al., 2013). One T. hassleriana PI gene resides in the cluster shared with most other eudicots and monocot species, while the other T. hassleriana PI gene sits in a cluster mostly composed of Brassicaceae species (Figure 3, Cluster 24). In Brassicaceae, we find the PI genes only in the new derived syntenic position. Furthermore, only in Brassicaceae we also find a unique syntenic position for the AP3 genes (Figure 3, Cluster 26). SEP- and SQUA-like genes are also involved in petal formation according to the ABC(DE) flowering model (Figure 1A). Moreover, Brassicaceae (and Cleomaceae) species are absent from these AGL2-SFT type of tandems in comparison to other lineages (Figure 4C; Supplemental Data Set 3). It is unclear why the PI, AP3, and SEP3 genes are transposed in the Brassicaceae in comparison to other angiosperms. Potentially higher level inter- and intrachromosomal chromatin interactions between loci, or new cis-regulatory elements, are required for crucifer B-specific gene expression patterns. It will be important to test such hypotheses and if potentially the derived genomic contexts of these genes have contributed to the canalization of the crucifer floral form.
FLC-like genes in the Brassicaceae and Cleomaceae are also in a derived genomic context compared with other angiosperms (Figure 3, Cluster 23). The vernalization process (prolonged cold exposure) is essential for many plants to initiate flowering. In Arabidopsis and other crucifers, this process is mediated by cold-induced epigenetic repression of FLC genes, namely, histone methylation (Bastow et al., 2004), chromatin structure modification with chromatin remodeling protein complexes (Kim and Sung, 2013), and the expression of long noncoding RNAs (Csorba et al., 2014). Genes flanking FLC are epigenetically coordinately regulated (Finnegan et al., 2004). Potentially the evolution of cold-specific epigenetic regulation was facilitated by the new genomic context of FLC-like genes in the Brassicales. It will be important to establish the patterns of epigenetic regulation of FLC-like genes outside of the Brassicales and which aspects are ancestral and which are derived.
A gene transposition event, likely after the split of monocot and eudicot species, has given rise to the specific synteny of the monocot AGL17-like genes found in this study (Figure 3, Cluster 14). In rice, the AGL17/ANR1-like genes are preferentially expressed in root and responsive to various hormone treatments (Puig et al., 2013) and nutrient supply (Yu et al., 2014). Moreover, in rice, the AGL17 clade genes are specific targets of the miR444 miRNA family, and this miRNA family is specific to monocots (Sunkar et al., 2005; Wu et al., 2009; Li et al., 2010). miR444 regulates nutrition signaling and root architecture in a monocot-specific way (Yan et al., 2014), and together with its AGL17 targets, they also play direct control in the rice antiviral pathway (Wang et al., 2016a). The synteny disruption of monocot AGL17-like genes, compared with eudicot species observed in this study, may be correlated with the origin of the miRNA-dependent regulation. Understanding this could be important for understanding the evolution of root architecture and responses to nutrient supplies, such as nitrogen.
Ancient Tandems of MADS-Box Genes
The ancient SEP1-SQUA tandem gene arrangement, as revealed by our angiosperm-wide synteny network analysis (Figure 4A), is in agreement with other studies where the SEP1-SQUA tandem gene arrangement was found in eudicots (Ruelens et al., 2013). Another study also noted that most AP1-like genes (a subclade of the SQUA-like genes) and SEP1-like genes were tightly linked as genomic neighbors since the split of the basal eudicots (Sun et al., 2014). Another example is the ancient tandem arrangement of SEP3-TM8. TM8 was first identified from Solanum lycopersicum (Pnueli et al., 1991), and this clade of genes has been reported to have undergone independent gene loss in different lineages based on phylogenic analyses (Becker and Theissen, 2003; Gramzow and Theißen, 2013). According to the consensus phylogeny based on studies by others (Figure 1B), the TM8-like genes are closely related to TM3-like genes and they both appear to share a common origin with the AGL6-, AGL2-, SQUA-, and FLC-like genes.
Our synteny analysis reveals a broadly conserved, and thus potentially ancient, tandem gene duplication that involves the last common ancestor of all SEP3- and TM8-like genes. Our results are generally consistent with the published results of Ruelens et al. (2013), but extend their model by the inclusion of TM3- and TM8-like genes. Considering that TM8-like genes were already present in the last common ancestor of extant seed plants (Gramzow et al., 2014), it is likely that the SEP3-TM8 tandem is more ancestral than the SEP3-FLC tandem (e.g., as defined by functions). Hence, the FLC-like genes could be derived from a TM8 homolog in an ancestral plant species. According to the network structure and gene copy number of the SEP3-, FLC-, and TM8-like gene clusters, we find that after the split of A. trichopoda from other angiosperms the SEP3- and TM8-like genes generally do not appear as a tandem gene pair within one species and TM8-like homologs tend to be lost from the tandem. This means that the SEP3-TM8/FLC tandem gene pair is more variable than the SEP1-SQUA tandem gene pair. In this study, both the SEP1-SQUA and SEP3-TM8 tandem gene pair were found in A. trichopoda (Figure 4C; Supplemental Data Set 3). Hence, the duplication that led to these two tandems may be the ε WGD event, derived from one ancestral tandem gene pair of AGL2-SFT (Figures 4D and 4E) in a common ancestor of the angiosperms (Jiao et al., 2011; Li et al., 2015).
It is generally thought that the AGL6-like and AGL2 (SEP)-like genes are closely related subfamilies (Figure 1B). It has been hypothesized that the combined ancestral gene of the AGL6- and AGL2-like genes was duplicated in a common ancestor of the seed plants (Spermatophytes) (Zahn et al., 2005; Kim et al., 2013), probably as a result of the ζ WGD (Jiao et al., 2011; Li et al., 2015). By interpreting our synteny networks, we found strong evidence of SEP1-SQUA, SEP3-SFT, and AGL6-TM3 tandems (Figures 4A to 4C) and evidence of monocot TM3-like genes connected to SEP3-, SQUA-, and TM8-like genes (Figure 4A). This enabled us to deduce the deep genealogy and to propose an evolutionary diagram that depicts how one ancestral locus that predates the last common ancestor of all seed plants has given rise to a large MADS-box gene clade with many subfamilies in angiosperms, which includes the AGL2-, AGL6-, SQUA-, TM3-, TM8-, and FLC-like gene clades (Figures 4C to 4E). It can be inferred that in the last common ancestor of seed plants a gene tandem was already present that corresponds with the current AGL2/AGL6-SFT/TM3 tandem gene arrangement (Figures 4C to 4E). The ζ WGD that occurred shortly before the radiation of the extant seed plants (Jiao et al., 2011) is likely causal to the duplication of this original tandem gene pair, after which the AGL2- and AGL6-like genes diverged, as well as the SFT- and TM3-like genes (Figure 4E). As described above, a subsequent more recent WGD (the ε event), which occurred prior to the diversification of the extant angiosperms (Jiao et al., 2011; Li et al., 2015), allowed then the emergence of the SEP1- and SEP3-like genes from the ancestral AGL2 locus, as well as the SQUA-, TM8-, and FLC-like genes from the ancestral SFT gene. During that same period, only one copy of the AGL6-TM3 tandem was retained from the ε WGD (Figure 4E). Altogether, this model hypothesized how one single MIKCc-type MADS-box gene gives birth to a whole superclade of genes composed of AGL2 (SEP)-like, AGL6-like, SFT-like (i.e., SQUA-, FLC-, and TM8-like), and TM3 (SOC1-like) genes/subfamilies due to a tandem duplication and subsequent WGDs.
Plant regulatory genes, such as MADS-box transcription factors, are generally not thought to be organized in coexpressed gene clusters like animal Hox or Para-Hox genes that do show coordinated gene expression (Lewis, 1978; Krumlauf, 1994; Ferrier and Holland, 2001). This could be due to the analysis techniques of plants employed to date, namely, phylogenetic analyses and pairwise synteny analyses, where ancient WGDs can dramatically complicate analyses. More recently, it has become apparent that many plant biosynthetic genes are organized into physical clusters that are coregulated and coexpressed (Boutanaev et al., 2015; Nützmann et al., 2016; Yu et al., 2016a). Often, these biosynthetic clusters are lineage-specific and are not just due to tandem duplication of a single ancestral gene.
With our approach, we have found several examples of highly conserved MADS-box collinearity and of lineage-specific transpositions. MADS protein-protein interactions or gene coexpression data are not obviously consistent with the parallel coregulation model like for animal Hox genes or plant biosynthetic gene clusters. However, potentially high-level chromatin-interacting domains within and between clusters that dictate their relative positions within the nucleus need to be tested for potential coregulatory interactions. Although we describe several interesting patterns of evolution of the MADS-box genes, this is just an example of one gene family across 51 plant species. Thus, we are providing just a proof of concept and a view on the tip of a new genomic iceberg. Our approach is suited for analyzing the positional context of all genes across all completed genomes to examine patterns of genomic conservation and divergence.
METHODS
Plant Genomes Analyzed
In total, 51 plant genomes were included in our analysis (Supplemental Figure 1 and Supplemental Table 1 for detailed information), including 30 rosids, 5 asterids, Beta vulgaris (non-rosid non-asterid), 11 monocots, the early diverging angiosperm (Amborella trichopoda), and a single genome for gymnosperms (Picea abies), club moss (Selaginella moellendorffii), moss (Physcomitrella patens), and green alga (Chlamydomonas reinhardtii). For each genome, all annotated protein sequences (primary transcript only) in a FASTA file and a BED/GFF file indicating gene positions are needed.
Pairwise Whole-Genome Comparisons
Reciprocal all-against-all comparisons between pairwise genomes as well as intraspecies comparisons are needed for synteny block detections. Thus, for 51 species in this study, we need P (51, 2) + 51 = 2, 601 times whole-genome protein comparisons. RAPSearch2 (BLAST-like program, but much more efficient) was used for this task (Zhao et al., 2012).
Syntenic Block Calculation
MCScanX (Tang et al., 2008b; Wang et al., 2012) was used to compute genomic collinearity between all pairwise genome combinations using default parameters (minimum match size for a collinear block = 5 genes, max gaps allowed = 25 genes). The output files from all the intra- and interspecies comparisons were integrated into a single file named “Total_Synteny_Blocks,” including the headers “Block_Index,” “Locus_1,” “Locus_2,” and “Block_Score,” which served as the database file.
Synteny Network for the MADS-Box Gene Family
Candidate MADS-box genes were initially identified using HMMER3.0 with default settings (domain signature PF00319) (Finn et al., 2011) for each of the 51 genomes (Supplemental Data Set 1, sheet 1). Then this gene list containing all candidate MADS-box genes was queried against the “Total_Synteny_Blocks” file. Rows containing at least one MADS-box gene were retrieved into a new file termed “Syntenic_Blocks_MADS-box genes” (Supplemental Data Set 1, sheet 2). This file was then the final synteny network for the MADS-box genes, and the network was imported and visualized in Cytoscape version 3.3.0 (Shannon et al., 2003) and Gephi 0.9.1 (Bastian et al., 2009).
Sequences were labeled based on the Arabidopsis thaliana MADS-box genes plus three representative MADS-box genes that are not represented in Arabidopsis (TM8-gene [GenBank accession number NP_001234105] from Solanum lycopersicum, OsMADS32 gene [GenBank accession number XP_015642650] from rice [Oryza sativa], and TM6 [GenBank accession number AAS46017] from Petunia hybrida) (Lee et al., 2003; Blanc and Wolfe, 2004; Daminato et al., 2014), using BLASTP (Altschul et al., 1990).
Network Clustering
Clique percolation as implemented in CFinder (Derényi et al., 2005; Palla et al., 2005; Fortunato, 2010) was used to locate all possible k-clique communities for the MADS-box gene synteny network to identify communities (clusters of gene nodes). Increasing k values make the communities smaller and more disintegrated but also at the same time more connected.
Phylogenetic Profiling of Clustered Communities
Communities (synteny clusters) derived from a certain k value were extracted, and the node (i.e., gene) composition of each community was then mapped to the phylogenetic tree with 51 species (Smith et al., 2011). Presence (red) or absence (white) of homologs in a cluster was depicted for the different species in the phylogenetic tree, thus creating a phylogenetic profile of a synteny cluster (Supplemental Figure 3). Each column in the illustration represents one community (one synteny cluster), which is labeled at top of the x axis based on its MADS-box name/annotation. Through such clustering and phylogenetic profiling steps, representative communities for the Type II (MIKCC- and MIKC*-type) and Type I MADS-box clades were found and then further analyzed.
Phylogenetic Distance and Tree Construction
Amino acid sequences for the candidate MADS-box genes, both the genes represented in the synteny networks and the genes missing from the networks, were aligned using HmmerAlign (Kristensen et al., 2011). The alignment was then transferred into codon alignment using Pal2nal (Suyama et al., 2006). A phylogenetic tree was computed using RAxML (Stamatakis, 2014) with the GTRCAT (bootstrap = 100). The phylogenetic tree was annotated and depicted using iTOL v3 (Letunic and Bork, 2016).
A script performing the above “Pairwise Whole-Genome Comparisons” and “Syntenic Block Calculation” steps and additional information about the method used in this work can be found at GitHub (https://github.com/zhaotao1987/SynNet-Pipeline).
Accession Numbers
Plant genomes used in this analysis are listed in Supplemental Table 1. All genes analyzed are listed in Supplemental Data Set 1.
Supplemental Data
Supplemental Figure 1. Species used in this study.
Supplemental Figure 2. k-clique percolation of the synteny network for MADS-box genes.
Supplemental Figure 3. Phylogenetic profiling for all the communities for k-clique = 3.
Supplemental Figure 4. Parallel coordinate synteny plots of PI derived from Genomicus.
Supplemental Table 1. Plant genomes used in this analysis.
Supplemental Data Set 1. Candidate MADS-box genes (sheet1) and synteny network for MADS-box genes.
Supplemental Data Set 2. Node list and edge list of the communities at k = 3.
Supplemental Data Set 3. Detailed information for the inferred tandem gene arrangements.
Acknowledgments
T.Z. was supported by the China Scholarship Council, and H.A.v.d.B. and M.E.S. by a Netherlands Scientific Organization (NWO) Vernieuwingsimpuls Vidi grant (numbers 864.10.004 and 864.10.001, respectively). S.d.B. received a NWO Experimental Plant Science Graduate School “Master Talent” fellowship. We thank the three anonymous reviewers for their helpful comments and suggestions for improving the manuscript.
AUTHOR CONTRIBUTIONS
M.E.S. and H.A.v.d.B. designed the research. T.Z. performed the analysis. R.H. and S.d.B. analyzed data. T.Z. and M.E.S. wrote the article. G.C.A. gave suggestions for the draft article. All authors discussed the results, commented on the article, and approved the final version for submission.
Glossary
- WGD
whole-genome duplication
Footnotes
Articles can be viewed without a subscription.
References
- Adamczyk B.J., Fernandez D.E. (2009). MIKC* MADS domain heterodimers are required for pollen maturation and tube growth in Arabidopsis. Plant Physiol. 149: 1713–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airoldi C.A., Davies B. (2012). Gene duplication and the evolution of plant MADS-box transcription factors. J. Genet. Genomics 39: 157–165. [DOI] [PubMed] [Google Scholar]
- Albertin C.B., Simakov O., Mitros T., Wang Z.Y., Pungor J.R., Edsinger-Gonzales E., Brenner S., Ragsdale C.W., Rokhsar D.S. (2015). The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature 524: 220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla E.R., Liljegren S.J., Pelaz S., Gold S.E., Burgeff C., Ditta G.S., Vergara-Silva F., Yanofsky M.F. (2000a). MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant J. 24: 457–466. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla E.R., Pelaz S., Liljegren S.J., Gold S.E., Burgeff C., Ditta G.S., Ribas de Pouplana L., Martínez-Castilla L., Yanofsky M.F. (2000b). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 97: 5328–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Hamelin J.I., Dall’Asta L., Barrat A., Vespignani A. (2006). Large scale networks fingerprinting and visualization using the k-core decomposition. Adv. Neural Inf. Process. Syst. 18: 41. [Google Scholar]
- Bartlett M., Thompson B., Brabazon H., Del Gizzi R., Zhang T., Whipple C. (2016). Evolutionary dynamics of floral homeotic transcription factor protein-protein interactions. Mol. Biol. Evol. 33: 1486–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian M., Heymann S., Jacomy M. (2009). Gephi: an open source software for exploring and manipulating networks. Proc. Int. AAAI Conf. Weblogs Soc. Media 8: 361–362. [Google Scholar]
- Bastow R., Mylne J.S., Lister C., Lippman Z., Martienssen R.A., Dean C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167. [DOI] [PubMed] [Google Scholar]
- Becker A., Theissen G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29: 464–489. [DOI] [PubMed] [Google Scholar]
- Bemer M., Heijmans K., Airoldi C., Davies B., Angenent G.C. (2010). An atlas of type I MADS box gene expression during female gametophyte and seed development in Arabidopsis. Plant Physiol. 154: 287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Wolfe K.H. (2004). Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutanaev A.M., Moses T., Zi J., Nelson D.R., Mugford S.T., Peters R.J., Osbourn A. (2015). Investigation of terpene diversification across multiple sequenced plant genomes. Proc. Natl. Acad. Sci. USA 112: E81–E88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers J.E., Chapman B.A., Rong J., Paterson A.H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438. [DOI] [PubMed] [Google Scholar]
- Causier B., Castillo R., Xue Y., Schwarz-Sommer Z., Davies B. (2010). Tracing the evolution of the floral homeotic B- and C-function genes through genome synteny. Mol. Biol. Evol. 27: 2651–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S., et al. (2013). The Tarenaya hassleriana genome provides insight into reproductive trait and genome evolution of crucifers. Plant Cell 25: 2813–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauset A., Newman M.E., Moore C. (2004). Finding community structure in very large networks. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 70: 066111. [DOI] [PubMed] [Google Scholar]
- Coen E.S., Meyerowitz E.M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37. [DOI] [PubMed] [Google Scholar]
- Couvreur T.L., Franzke A., Al-Shehbaz I.A., Bakker F.T., Koch M.A., Mummenhoff K. (2010). Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae). Mol. Biol. Evol. 27: 55–71. [DOI] [PubMed] [Google Scholar]
- Csorba T., Questa J.I., Sun Q., Dean C. (2014). Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. USA 111: 16160–16165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daminato M., Masiero S., Resentini F., Lovisetto A., Casadoro G. (2014). Characterization of TM8, a MADS-box gene expressed in tomato flowers. BMC Plant Biol. 14: 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derényi I., Palla G., Vicsek T. (2005). Clique percolation in random networks. Phys. Rev. Lett. 94: 160202. [DOI] [PubMed] [Google Scholar]
- Dewey C.N. (2011). Positional orthology: putting genomic evolutionary relationships into context. Brief. Bioinform. 12: 401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Pinyopich A., Robles P., Pelaz S., Yanofsky M.F. (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14: 1935–1940. [DOI] [PubMed] [Google Scholar]
- Duboule D. (2007). The rise and fall of Hox gene clusters. Development 134: 2549–2560. [DOI] [PubMed] [Google Scholar]
- Edger P.P., et al. (2015). The butterfly plant arms-race escalated by gene and genome duplications. Proc. Natl. Acad. Sci. USA 112: 8362–8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A.J., Van Dongen S., Ouzounis C.A. (2002). An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30: 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrier D.E., Holland P.W. (2001). Ancient origin of the Hox gene cluster. Nat. Rev. Genet. 2: 33–38. [DOI] [PubMed] [Google Scholar]
- Finn R.D., Clements J., Eddy S.R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39: W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan E.J., Sheldon C.C., Jardinaud F., Peacock W.J., Dennis E.S. (2004). A cluster of Arabidopsis genes with a coordinate response to an environmental stimulus. Curr. Biol. 14: 911–916. [DOI] [PubMed] [Google Scholar]
- Fortunato S. (2010). Community detection in graphs. Phys. Rep. 486: 75–174. [Google Scholar]
- Freeling M., Lyons E., Pedersen B., Alam M., Ming R., Lisch D. (2008). Many or most genes in Arabidopsis transposed after the origin of the order Brassicales. Genome Res. 18: 1924–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M., Woodhouse M.R., Subramaniam S., Turco G., Lisch D., Schnable J.C. (2012). Fractionation mutagenesis and similar consequences of mechanisms removing dispensable or less-expressed DNA in plants. Curr. Opin. Plant Biol. 15: 131–139. [DOI] [PubMed] [Google Scholar]
- Goto K., Meyerowitz E.M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8: 1548–1560. [DOI] [PubMed] [Google Scholar]
- Gramzow L., Theißen G. (2013). Phylogenomics of MADS-box genes in plants: two opposing life styles in one gene family. Biology (Basel) 2: 1150–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramzow L., Weilandt L., Theißen G. (2014). MADS goes genomic in conifers: towards determining the ancestral set of MADS-box genes in seed plants. Ann. Bot. (Lond.) 114: 1407–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramzow L., Barker E., Schulz C., Ambrose B., Ashton N., Theißen G., Litt A. (2012). Selaginella genome analysis: entering the “Homoplasy Heaven” of the MADS world. Front. Plant Sci. 3: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U., Höhmann S., Nettesheim K., Wisman E., Saedler H., Huijser P. (2000). Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J. 21: 351–360. [DOI] [PubMed] [Google Scholar]
- Henschel K., Kofuji R., Hasebe M., Saedler H., Münster T., Theissen G. (2002). Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Mol. Biol. Evol. 19: 801–814. [DOI] [PubMed] [Google Scholar]
- Jack T., Brockman L.L., Meyerowitz E.M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68: 683–697. [DOI] [PubMed] [Google Scholar]
- Jack T., Fox G.L., Meyerowitz E.M. (1994). Arabidopsis homeotic gene APETALA3 ectopic expression: transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76: 703–716. [DOI] [PubMed] [Google Scholar]
- Jaramillo M.A., Kramer E.M. (2007). Molecular evolution of the petal and stamen identity genes, APETALA3 and PISTILLATA, after petal loss in the Piperales. Mol. Phylogenet. Evol. 44: 598–609. [DOI] [PubMed] [Google Scholar]
- Jiao Y., Paterson A.H. (2014). Polyploidy-associated genome modifications during land plant evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369: 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Li J., Tang H., Paterson A.H. (2014). Integrated syntenic and phylogenomic analyses reveal an ancient genome duplication in monocots. Plant Cell 26: 2792–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100. [DOI] [PubMed] [Google Scholar]
- Kim D.-H., Sung S. (2013). Coordination of the vernalization response through a VIN3 and FLC gene family regulatory network in Arabidopsis. Plant Cell 25: 454–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Soltis P.S., Soltis D.E. (2013). AGL6-like MADS-box genes are sister to AGL2-like MADS-box genes. J. Plant Biol. 56: 315–325. [Google Scholar]
- Köhler C., Hennig L., Spillane C., Pien S., Gruissem W., Grossniklaus U. (2003). The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 17: 1540–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen D.M., Wolf Y.I., Mushegian A.R., Koonin E.V. (2011). Computational methods for Gene Orthology inference. Brief. Bioinform. 12: 379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumlauf R. (1994). Hox genes in vertebrate development. Cell 78: 191–201. [DOI] [PubMed] [Google Scholar]
- Lancichinetti A., Fortunato S. (2009). Community detection algorithms: a comparative analysis. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 80: 056117. [DOI] [PubMed] [Google Scholar]
- Lee H.L., Irish V.F. (2011). Gene duplication and loss in a MADS box gene transcription factor circuit. Mol. Biol. Evol. 28: 3367–3380. [DOI] [PubMed] [Google Scholar]
- Lee S., Kim J., Son J.S., Nam J., Jeong D.H., Lee K., Jang S., Yoo J., Lee J., Lee D.Y., Kang H.G., An G. (2003). Systematic reverse genetic screening of T-DNA tagged genes in rice for functional genomic analyses: MADS-box genes as a test case. Plant Cell Physiol. 44: 1403–1411. [DOI] [PubMed] [Google Scholar]
- Lemons D., McGinnis W. (2006). Genomic evolution of Hox gene clusters. Science 313: 1918–1922. [DOI] [PubMed] [Google Scholar]
- Letunic I., Bork P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44: W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E.B. (1978). A gene complex controlling segmentation in Drosophila. Nature 276: 565–570. [DOI] [PubMed] [Google Scholar]
- Li Y.F., Zheng Y., Addo-Quaye C., Zhang L., Saini A., Jagadeeswaran G., Axtell M.J., Zhang W., Sunkar R. (2010). Transcriptome-wide identification of microRNA targets in rice. Plant J. 62: 742–759. [DOI] [PubMed] [Google Scholar]
- Li Z., Baniaga A.E., Sessa E.B., Scascitelli M., Graham S.W., Rieseberg L.H., Barker M.S. (2015). Early genome duplications in conifers and other seed plants. Sci. Adv. 1: e1501084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis A., Muffato M., Crollius H.R. (2013). Genomicus: five genome browsers for comparative genomics in eukaryota. Nucleic Acids Res. 41: D700–D705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J., Havlak P., Putnam N.H. (2011). Constraints on genes shape long-term conservation of macro-synteny in metazoan genomes. BMC Bioinformatics 12 (suppl. 9): S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Conery J.S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Martinez-Castilla L.P., Alvarez-Buylla E.R. (2003). Adaptive evolution in the Arabidopsis MADS-box gene family inferred from its complete resolved phylogeny. Proc. Natl. Acad. Sci. USA 100: 13407–13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiero S., Colombo L., Grini P.E., Schnittger A., Kater M.M. (2011). The emerging importance of type I MADS box transcription factors for plant reproduction. Plant Cell 23: 865–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer R., Härter A., Rümpler F., Kim S., Soltis P.S., Soltis D.E., Theißen G. (2014). DEF- and GLO-like proteins may have lost most of their interaction partners during angiosperm evolution. Ann. Bot. (Lond.) 114: 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., Ditta G., Gustafson-Brown C., Pelaz S., Yanofsky M., Amasino R.M. (2003). AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J. 33: 867–874. [DOI] [PubMed] [Google Scholar]
- Ming R., et al. (2013). Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biol. 14: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J., dePamphilis C.W., Ma H., Nei M. (2003). Antiquity and evolution of the MADS-box gene family controlling flower development in plants. Mol. Biol. Evol. 20: 1435–1447. [DOI] [PubMed] [Google Scholar]
- Nam J., Kaufmann K., Theissen G., Nei M. (2005). A simple method for predicting the functional differentiation of duplicate genes and its application to MIKC-type MADS-box genes. Nucleic Acids Res. 33: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J., Kim J., Lee S., An G., Ma H., Nei M. (2004). Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc. Natl. Acad. Sci. USA 101: 1910–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nützmann H.W., Huang A., Osbourn A. (2016). Plant metabolic clusters - from genetics to genomics. New Phytol. 211: 771–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palla G., Barabási A.L., Vicsek T. (2007). Quantifying social group evolution. Nature 446: 664–667. [DOI] [PubMed] [Google Scholar]
- Palla G., Derényi I., Farkas I., Vicsek T. (2005). Uncovering the overlapping community structure of complex networks in nature and society. Nature 435: 814–818. [DOI] [PubMed] [Google Scholar]
- Parenicová L., de Folter S., Kieffer M., Horner D.S., Favalli C., Busscher J., Cook H.E., Ingram R.M., Kater M.M., Davies B., Angenent G.C., Colombo L. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15: 1538–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passarge E., Horsthemke B., Farber R.A. (1999). Incorrect use of the term synteny. Nat. Genet. 23: 387. [DOI] [PubMed] [Google Scholar]
- Patchell M.J., Bolton M.C., Mankowski P., Hall J.C. (2011). Comparative floral development in Cleomaceae reveals two distinct pathways leading to monosymmetry. Int. J. Plant Sci. 172: 352–365. [Google Scholar]
- Pellegrini M., Marcotte E.M., Thompson M.J., Eisenberg D., Yeates T.O. (1999). Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl. Acad. Sci. USA 96: 4285–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L., Abu-Abeid M., Zamir D., Nacken W., Schwarz-Sommer Z., Lifschitz E. (1991). The MADS box gene family in tomato: temporal expression during floral development, conserved secondary structures and homology with homeotic genes from Antirrhinum and Arabidopsis. Plant J. 1: 255–266. [PubMed] [Google Scholar]
- Porter M.A., Onnela J.-P., Mucha P.J. (2009). Communities in networks. Not. Am. Math. Soc. 56: 1082–1097. [Google Scholar]
- Portereiko M.F., Lloyd A., Steffen J.G., Punwani J.A., Otsuga D., Drews G.N. (2006). AGL80 is required for central cell and endosperm development in Arabidopsis. Plant Cell 18: 1862–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig J., Meynard D., Khong G.N., Pauluzzi G., Guiderdoni E., Gantet P. (2013). Analysis of the expression of the AGL17-like clade of MADS-box transcription factors in rice. Gene Expr. Patterns 13: 160–170. [DOI] [PubMed] [Google Scholar]
- Riechmann J.L., Krizek B.A., Meyerowitz E.M. (1996). Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA 93: 4793–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosvall M., Bergstrom C.T. (2008). Maps of random walks on complex networks reveal community structure. Proc. Natl. Acad. Sci. 105: 1118–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruelens P., de Maagd R.A., Proost S., Theißen G., Geuten K., Kaufmann K. (2013). FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nat. Commun. 4: 2280. [DOI] [PubMed] [Google Scholar]
- Sampedro J., Lee Y., Carey R.E., dePamphilis C., Cosgrove D.J. (2005). Use of genomic history to improve phylogeny and understanding of births and deaths in a gene family. Plant J. 44: 409–419. [DOI] [PubMed] [Google Scholar]
- Schiemann S.M., Martín-Durán J.M., Børve A., Vellutini B.C., Passamaneck Y.J., Hejnol A. (2017). Clustered brachiopod Hox genes are not expressed collinearly and are associated with lophotrochozoan novelties. Proc. Natl. Acad. Sci. USA 114: E1913–E1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz M.E., Mitchell-Olds T. (2006). Independent ancient polyploidy events in the sister families Brassicaceae and Cleomaceae. Plant Cell 18: 1152–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz M.E., Mohammadin S., Edger P.P. (2012). Ancient whole genome duplications, novelty and diversification: the WGD Radiation Lag-Time Model. Curr. Opin. Plant Biol. 15: 147–153. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C.C., Rouse D.T., Finnegan E.J., Peacock W.J., Dennis E.S. (2000). The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. USA 97: 3753–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C., Immink R.G.H., Angenent G.C., Kaufmann K. (2012). Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139: 3081–3098. [DOI] [PubMed] [Google Scholar]
- Smith S.A., Beaulieu J.M., Stamatakis A., Donoghue M.J. (2011). Understanding angiosperm diversification using small and large phylogenetic trees. Am. J. Bot. 98: 404–414. [DOI] [PubMed] [Google Scholar]
- Soltis D.E., Albert V.A., Leebens-Mack J., Bell C.D., Paterson A.H., Zheng C., Sankoff D., Depamphilis C.W., Wall P.K., Soltis P.S. (2009). Polyploidy and angiosperm diversification. Am. J. Bot. 96: 336–348. [DOI] [PubMed] [Google Scholar]
- Sommer H., Beltran J.P., Huijser P., Pape H., Lonnig W.E., Saedler H., Schwarz-Sommer Z. (1990). Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: the protein shows homology to transcription factors. EMBO J. 9: 605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnikova N., Dewaele R., Janvier P., Krumlauf R., Duboule D. (2013). Duplications of hox gene clusters and the emergence of vertebrates. Dev. Biol. 378: 194–199. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Huang W., Li Z., Song C., Liu D., Liu Y., Hayward A., Liu Y., Huang H., Wang Y. (2014). Functional and evolutionary analysis of the AP1/SEP/AGL6 superclade of MADS-box genes in the basal eudicot Epimedium sagittatum. Ann. Bot. (Lond.) 113: 653–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Girke T., Jain P.K., Zhu J.-K. (2005). Cloning and characterization of microRNAs from rice. Plant Cell 17: 1397–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M., Torrents D., Bork P. (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34: W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Bowers J.E., Wang X., Ming R., Alam M., Paterson A.H. (2008a). Synteny and collinearity in plant genomes. Science 320: 486–488. [DOI] [PubMed] [Google Scholar]
- Tang H., Wang X., Bowers J.E., Ming R., Alam M., Paterson A.H. (2008b). Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 18: 1944–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tank D.C., Eastman J.M., Pennell M.W., Soltis P.S., Soltis D.E., Hinchliff C.E., Brown J.W., Sessa E.B., Harmon L.J. (2015). Nested radiations and the pulse of angiosperm diversification: increased diversification rates often follow whole genome duplications. New Phytol. 207: 454–467. [DOI] [PubMed] [Google Scholar]
- Theissen G. (2001). Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant Biol. 4: 75–85. [DOI] [PubMed] [Google Scholar]
- Trobner W., Ramirez L., Motte P., Hue I., Huijser P., Lonnig W.E., Saedler H., Sommer H., Schwarz-Sommer Z. (1992). GLOBOSA: a homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 11: 4693–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verelst W., Saedler H., Münster T. (2007a). MIKC* MADS-protein complexes bind motifs enriched in the proximal region of late pollen-specific Arabidopsis promoters. Plant Physiol. 143: 447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verelst W., Twell D., de Folter S., Immink R., Saedler H., Münster T. (2007b). MADS-complexes regulate transcriptome dynamics during pollen maturation. Genome Biol. 8: R249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Jiao X., Kong X., Humaira S., Wu Y., Chen X., Fang R., Yan Y. (2016a). A signaling cascade from miR444 to RDR1 in rice antiviral RNA silencing pathway. Plant Physiol. 170: 2365–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ficklin S.P., Wang X., Feltus F.A., Paterson A.H. (2016b). Large-scale gene relocations following an ancient genome triplication associated with the diversification of core eudicots. PLoS One 11: e0155637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tang H., Debarry J.D., Tan X., Li J., Wang X., Lee T.H., Jin H., Marler B., Guo H., Kissinger J.C., Paterson A.H. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40: e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., et al. (2013). The sacred lotus genome provides insights into the evolution of flowering plants. Plant J. 76: 557–567. [DOI] [PubMed] [Google Scholar]
- Wu L., Zhang Q., Zhou H., Ni F., Wu X., Qi Y. (2009). Rice microRNA effector complexes and targets. Plant Cell 21: 3421–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Kelley S., Szymanski B.K. (2013). Overlapping community detection in networks: The state-of-the-art and comparative study. ACM Comput. Surv. 45: 43. [Google Scholar]
- Yan Y., Wang H., Hamera S., Chen X., Fang R. (2014). miR444a has multiple functions in the rice nitrate-signaling pathway. Plant J. 78: 44–55. [DOI] [PubMed] [Google Scholar]
- Yang Y., Fanning L., Jack T. (2003). The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. Plant J. 33: 47–59. [DOI] [PubMed] [Google Scholar]
- Yu C., Su S., Xu Y., Zhao Y., Yan A., Huang L., Ali I., Gan Y. (2014). The effects of fluctuations in the nutrient supply on the expression of five members of the AGL17 clade of MADS-box genes in rice. PLoS One 9: e105597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N., Nützmann H.W., MacDonald J.T., Moore B., Field B., Berriri S., Trick M., Rosser S.J., Kumar S.V., Freemont P.S., Osbourn A. (2016a). Delineation of metabolic gene clusters in plant genomes by chromatin signatures. Nucleic Acids Res. 44: 2255–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Duan X., Zhang R., Fu X., Ye L., Kong H., Xu G., Shan H. (2016b). Prevalent exon-intron structural changes in the APETALA1/FRUITFULL, SEPALLATA, AGAMOUS-LIKE6, and FLOWERING LOCUS C MADS-box gene subfamilies provide new insights into their evolution. Front. Plant Sci. 7: 598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn L.M., Kong H., Leebens-Mack J.H., Kim S., Soltis P.S., Landherr L.L., Soltis D.E., Depamphilis C.W., Ma H. (2005). The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169: 2209–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., et al. (2013). Disruption of the petal identity gene APETALA3-3 is highly correlated with loss of petals within the buttercup family (Ranunculaceae). Proc. Natl. Acad. Sci. USA 110: 5074–5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Schranz M.E. (2017). Network approaches for plant phylogenomic synteny analysis. Curr. Opin. Plant Biol. 36: 129–134. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Tang H., Ye Y. (2012). RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics 28: 125–126. [DOI] [PMC free article] [PubMed] [Google Scholar]