Class III WRKY transcription factors play positive roles in brassinosteroid-regulated growth and a negative role in drought response by cooperating with BES1.
Abstract
Plant steroid hormones, brassinosteroids (BRs), play important roles in growth and development. BR signaling controls the activities of BRASSINOSTERIOD INSENSITIVE1-EMS-SUPPRESSOR1/BRASSINAZOLE-RESISTANT1 (BES1/BZR1) family transcription factors. Besides the role in promoting growth, BRs are also implicated in plant responses to drought stress. However, the molecular mechanisms by which BRs regulate drought response have just begun to be revealed. The functions of WRKY transcription factors in BR-regulated plant growth have not been established, although their roles in stress responses are well documented. Here, we found that three Arabidopsis thaliana group III WRKY transcription factors, WRKY46, WRKY54, and WRKY70, are involved in both BR-regulated plant growth and drought response as the wrky46 wrky54 wrky70 triple mutant has defects in BR-regulated growth and is more tolerant to drought stress. RNA-sequencing analysis revealed global roles of WRKY46, WRKY54, and WRKY70 in promoting BR-mediated gene expression and inhibiting drought responsive genes. WRKY54 directly interacts with BES1 to cooperatively regulate the expression of target genes. In addition, WRKY54 is phosphorylated and destabilized by GSK3-like kinase BR-INSENSITIVE2, a negative regulator in the BR pathway. Our results therefore establish WRKY46/54/70 as important signaling components that are positively involved in BR-regulated growth and negatively involved in drought responses.
INTRODUCTION
Plant steroid hormones, brassinosteroids (BRs), modulate multiple plant growth and developmental processes, including cell elongation and division, vascular differentiation, senescence, photomorphogenesis, and response to biotic and abiotic stresses (Li et al., 1996; Szekeres et al., 1996; Li and Chory, 1997). Over the past decades, extensive genetic and molecular studies, particularly in Arabidopsis thaliana, have revealed the BR signaling pathway. BRs are perceived by the plasma membrane-localized receptor kinase BRASSINOSTERIOD INSENSITIVE1 (BRI1) and coreceptor BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1); the BR signal is transduced through various intermediates including the negative acting GSK3-like kinase BR-INSENSITIVE2 (BIN2) to downstream BES1/BZR1 family transcription factors (TFs), which regulate the expression of thousands of genes for BR response (Clouse et al., 1996; Li and Chory, 1997; Li et al., 2001, 2002; He et al., 2002; Li and Nam, 2002; Nam and Li, 2002; Wang et al., 2002; Yin et al., 2002; Zhao et al., 2002; Clouse, 2011; Guo et al., 2013).
BRs interact extensively with gibberellic acid (GA) in the regulation of plant growth (Bai et al., 2012; Gallego-Bartolomé et al., 2012; Li et al., 2012; Tong et al., 2014; Unterholzner et al., 2015; Shahnejat-Bushehri et al., 2016). In addition to the critical role in the plant growth and development, BRs are also involved in a wide range of stress responses, such as cold stress, drought, oxidative stress, high salt, high temperature, heavy metal, and pathogen attack (Krishna, 2003; Hao et al., 2013; Rajewska et al., 2016). Earlier studies suggested positive roles of BRs in drought tolerance in wheat (Triticum aestivum), Arabidopsis, and Brassica napus (Sairam, 1994; Kagale et al., 2007). For example, overexpression of Arabidopsis BR biosynthetic gene AtDWARF4 in B. napus resulted in enhanced tolerance to drought (Sahni et al., 2016). However, genetic studies also indicated a negative role of BRs or BR signaling in drought responses. Loss-of-function BR mutants showed increased tolerance to drought (Beste et al., 2011; Northey et al., 2016; Nolan et al., 2017; Ye et al., 2017), and RNA interference-mediated knockdown of BRI1 in Brachypodium distachyon led to enhanced drought tolerance and elevated expression of drought-regulated genes (Feng et al., 2015). Recent studies have started to reveal mechanisms of BR-abiotic stress signaling. BIN2 phosphorylates and positively regulates SnRK2.2 and 2.3 as well as ABSCISIC ACID INSENSITIVE5 (ABI5) involved in drought/abscisic acid signaling (Cai et al., 2014; Hu and Yu, 2014). Abscisic acid induces the expression of OsREM4.1, a membrane-anchored protein that inhibits BR signaling by inhibiting BRI1-BAK1 complex formation (Clouse, 2016; Gui et al., 2016). More recently, it was found that RD26, a NAC transcription factor, mediates crosstalk between BR and drought pathways through reciprocal inhibition between RD26 and BES1 transcriptional activities (Ye et al., 2017). Under drought or starvation conditions, BES1 is targeted to selective autophagy through the actions of SINAT E3 ubiquitin ligase and ubiquitin receptor protein DSK2, thereby balancing plant growth and stress responses (Nolan et al., 2017; Yang et al., 2017).
The WRKY family TFs are only found in higher plants and are composed of over 70 members in Arabidopsis (Ulker and Somssich, 2004). This family of TFs contains a well conserved WRKY domain, which binds to the W-box [(T)TGACC/T] in the target gene promoters (Eulgem and Somssich, 2007), and a zinc finger motif at its C terminus, either CX4-5CX22-23HXH (CCHH, X denotes any amino acid, 4-5/22-23 indicate the number of amino acids) or CX7CX23HXC (CCHC) (Eulgem et al., 2000). The WRKY family is categorized into three groups according to the number of WRKY domains and the structure of zinc finger (Rushton et al., 2010). WRKY46, WRKY54, and WRKY70 belong to the group III with one WRKY domain and CCHC zinc finger motif (Eulgem et al., 2000). Many studies have indicated that WRKY TFs play crucial roles in plant innate immunity as well as abiotic responses (Eulgem et al., 2000; Li et al., 2006, 2013; Eulgem and Somssich, 2007; Murray et al., 2007; Ulker et al., 2007; Higashi et al., 2008; Ren et al., 2010; Rushton et al., 2010; Chen et al., 2012; Hu et al., 2012; Chujo et al., 2014). It is known that WRKY TFs can control multiple plant responses via transcriptional reprogramming (Rushton et al., 2010). For instance, WRKY46 participated in basal defense against bacteria Pseudomonas syringae since gain-of-function WRK46 plants were more resistant to the bacteria (Hu et al., 2012). In addition, WRKY46 was found to have dual roles in regulating plant responses to drought and salt stress as the overexpression of WRKY46 resulted in hypersensitivity to drought and salt stress with a higher rate of water loss (Ding et al., 2014b). Microarray analysis showed that WRKY46 regulates a number of genes in cellular osmoprotection and redox homeostasis under dehydration stress (Ding et al., 2014b). Similarly, a wrky54 wrky70 double mutant showed increased tolerance to osmotic stress, which was accompanied by enhanced stomatal closure and improved water retention, suggesting that WRKY54 and WRKY70 cooperate as negative regulators of osmotic stress in Arabidopsis (Li et al., 2013). Although the role of WRKY family TFs in stress responses is well established, their role in hormone-regulated plant growth remains to be investigated.
In this study, we found that Arabidopsis WRKY46, WRKY54, and WRKY70 were induced by BRs and play positive roles in BR-regulated plant growth. Moreover, we showed that WRKY46, WRKY54, and WRKY70 negatively regulate drought tolerance, consistent with their previously described role in stress response. RNA-sequencing (RNA-seq) analysis indicated that WRKY46, WRKY54, and WRKY70 negatively regulate dehydration-responsive gene expression while promoting BR-regulated gene expression. Furthermore, we demonstrated that WRKY54 interacts with BES1 to control the expression of BR-regulated and dehydration-responsive genes. Our results thus revealed the dual roles of WRKY46/54/70 in plant growth and drought responses by cooperating with BR-regulated transcription factor BES1.
RESULTS
WRKY46, WRKY54, and WRKY70 Are Positive Regulators in the BR Pathway
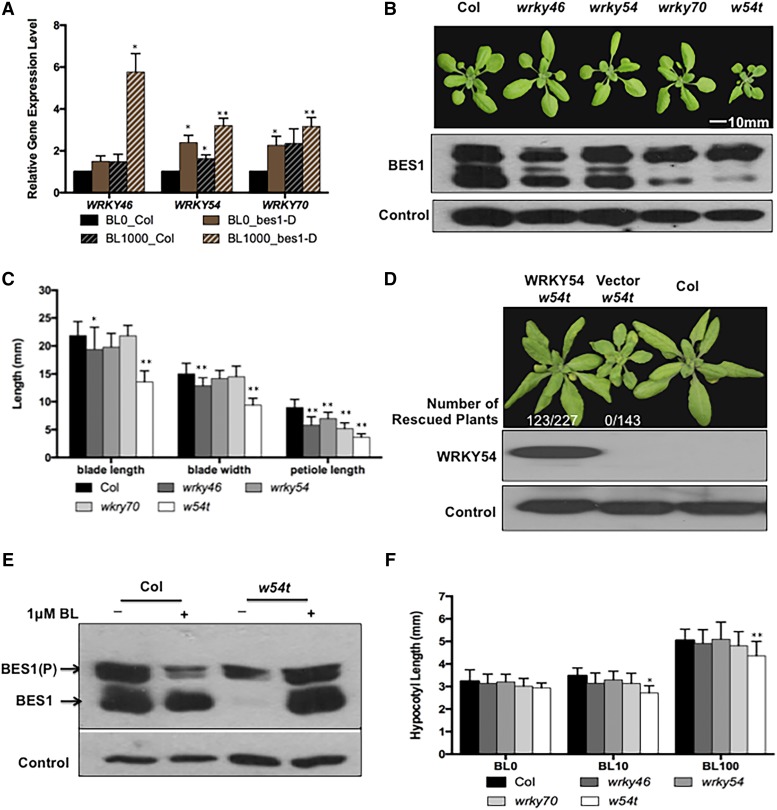
Our previously published microarray data showed that the expression levels of WRKY46, WRKY54, and WRKY70 were induced by BRs in wild-type seedlings and also increased in bes1-D mutants treated with or without brassinolide (BL), the most active BR (Noguchi et al., 2000; Li et al., 2010). To confirm this result, WRKY46, WRKY54, and WRKY70 mRNA levels were determined in 4-week-old wild-type and bes1-D mutants with or without BL treatment by RT-qPCR. Consistent with previous microarray data, WRKY46/54/70 transcript levels were increased by 1.5- to 6-fold in adult wild-type and bes1-D plants after BL treatment (Figure 1A). These results indicate that BRs promote the expression of WRKY46/54/70.
Figure 1.
WRKY46, WRKY54, and WRKY70 Function Redundantly and Play Positive Roles in the BR Pathway.
(A) WRKY46, WRKY54, and WRKY70 mRNA levels were determined in the wild type and bes1-D treated with 1 μM BL or mock control for 2.5 h. The averages and sd were derived from three biological replicates.
(B) Top: The growth phenotype of 3-week-old wild type, wrky46, wrky54, wrky70, and wrky46 wrky54 wrky70 triple mutant (abbreviated as w54t in all figures). Bottom: BES1 protein levels were determined by immunoblot and a loading control was shown at the bottom.
(C) The measurement of blade lengths, blade widths, and petiole lengths of the sixth leaves. Error bars indicate sd, n = 13 (*P < 0.05, **P < 0.01; Student’s t test).
(D) Transgenic complementation of w54t mutant with PWRKY54:WRKY54-FLAG fusion gene and empty vector as the control. Top: Four-week-old wild-type transgenic plants with vector (w54t) or WRKY54 (w54t) are shown. Bottom: WRKY54 protein accumulation was detected in the transgenic plants by immunoblot with anti-FLAG antibody and HERK1 loading control was shown at the bottom.
(E) BES1 protein accumulation was determined in 4-week-old w54t leaves soaked in 0.5× liquid MS medium with 1 μM BL or DMSO for 30 min.
(F) Hypocotyl lengths of 5-d-old seedlings grown on 0.5× MS medium with 0, 10, and 100 nM BL. Mean was calculated and the sd was also presented. Error bars indicate sd (*P < 0.05, **P < 0.01; Student’s t test).
To determine the biological functions of WRKY46/54/70 in the BR pathway, we obtained T-DNA insertion lines for these genes (Supplemental Figure 1A). Single knockout mutants for wrky46, wrky54, or wrky70 did not show any obvious growth phenotype compared with the wild type (Figure 1B). Since WRKY46, WRKY54, and WRKY70 have high similarities in protein sequences (Supplemental Figure 1B) and might function redundantly, we generated wrky46 wrky54, wrky46 wrky70, and wrky54 wrky70 double mutants to determine their role in plant growth. The double mutants showed a slightly reduced-growth phenotype compared with wild-type or the single mutants (Supplemental Figure 2A). We then generated wrky46 wrky54 wrky70 triple mutants (w54t), which displayed a stronger reduction in growth with shorter blade lengths, blade widths, and petiole lengths (Figures 1B and 1C). Moreover, w54t has a dwarf phenotype at the flowering stage (Supplemental Figure 2B).
Genetic complementation experiments were performed to confirm that the w54t mutant phenotype is caused by loss of function of these genes. Expression of WRKY54 in w54t mutant rescued the mutant phenotype, as 123 out of 227 transgenic plant showed a clear wild-type-like phenotype, whereas none of the 143 w54t plant lines transformed with control vector showed a rescued phenotype (Figure 1D; Supplemental Figure 2C).
To further determine if other Class III members (WRKY30, WRKY41, and WRKY53) contribute to plant growth, we constructed a sextuple mutant wrky46 wrky54 wrky70 wrky30 wrky41 wrky53 (wrkyS) and found that the sextuple mutants have a slightly stronger growth phenotype than w54t triple mutants (Supplemental Figures 3A to 3C), suggesting that WRKY30, WRKY41, and WRKY53 play some role in vegetative growth. Taken together, these genetic results indicate that WRKY46/54/70, together with other group III WRKY TFs, function redundantly and play a positive role in plant growth.
We then monitored BES1 protein levels, a well-established marker for the BR pathway (Yin et al., 2002, 2005). BES1 levels, particularly the dephosphorylated form, decreased significantly in 4-week-old w54t plants compared with the wild type, whereas the single mutants had only slightly reduced BES1 levels (Figure 1B, middle and lower panels). The reduction of BES1 protein might be due to reduced BR biosynthesis or signaling.
To elucidate the mechanism underlying the altered BES1 protein levels, the expression of BR biosynthesis genes, DWF4, DET2, and CPD, was determined in the w54t mutants (Kim et al., 2005). The mRNA levels of DWF4, DET2, and CPD decreased 1- to 5-fold in the triple mutant compared with the wild type (Supplemental Figure 4A). The reduction of BR biosynthesis genes in w54t prompted us to determine the endogenous levels of BRs in wild-type and w54t plants (Xin et al., 2013). The amount of BL was below detectable levels in adult leaves, but the level of castasterone (CS), a precursor of BL, was reduced slightly, by 10%, in w54t compared with the wild type (Supplemental Figure 4B). The sextuple mutant wrkyS showed a 20% decrease in CS levels accompanied by a stronger reduction in growth (Supplemental Figure 4B). The levels of 6-deoxoCS, the precursor of CS, which is ∼50 to 100 times more abundant than CS, does not seem to significantly change in the mutants (Supplemental Figures 4C and 4D).
We also examined the BES1 protein phosphorylation status and level in w54t mutant in response to BL. Application of exogenous BL restored the BES1 protein level in w54t to the wild type level after 0.5 h BL treatment (Figure 1E). However, when grown in the presence of different concentrations of BL, the w54t mutants showed decreased sensitivity to BL compared with the wild type with shorter hypocotyls, although BL could restore the BES1 protein in w54t to the wild-type level (Figures 1E and 1F; Supplemental Figures 5A and 5B).
We determined mutant responses to other plant hormones and found that the w54t as well as single mutants have normal response to auxin and ethylene in hypocotyl elongation assays (Supplemental Figure 5B) (Smalle et al., 1997). It appears that w54t mutants also have reduced hypocotyl elongation in response to GA, consistent with recent findings that BRs can function upstream of GA to regulate cell elongation (Supplemental Figure 5B) (Tong et al., 2014; Unterholzner et al., 2015). The fact that BES1 levels could be restored by BL treatment yet the w54t mutant still displayed decreased BL responses suggests that WRKY46/54/70 might play a pivotal role in BR signaling.
WRKY46/54/70 Are Required for the Regulation of BR/BES1 Target Genes
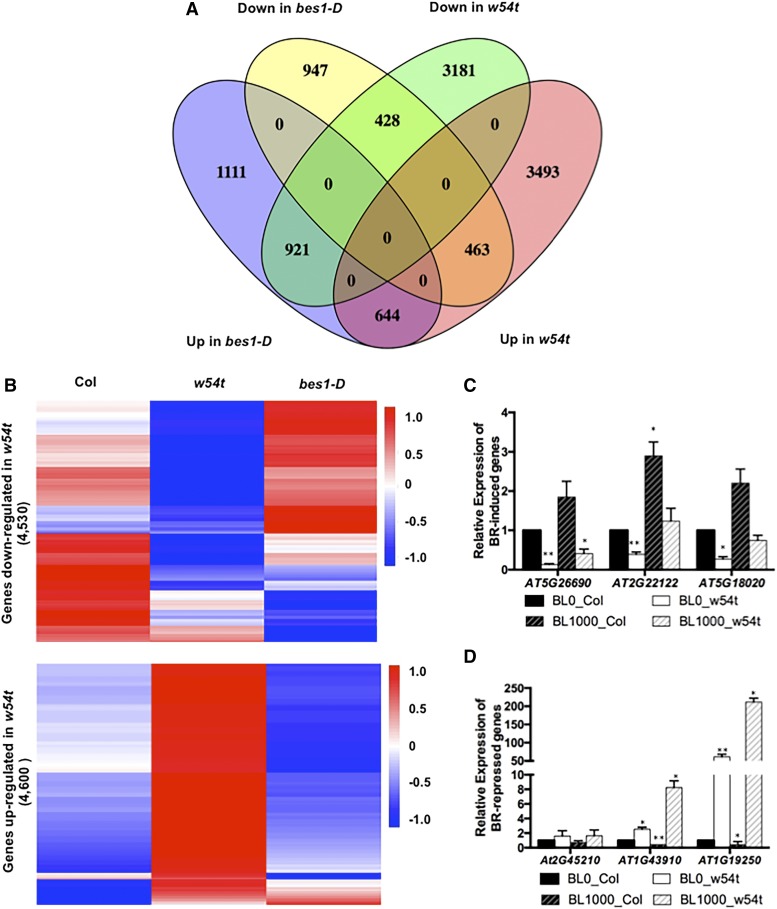
BES1/BZR1 interact with other transcription factors, such as MYB30, PHYTOCHROME INTERACTING FACTOR4, and HOMEO-DOMAIN-LEUCINE ZIPPER PROTEIN OF ARABIDOPSIS THALIANA1 to control BR-regulated gene expression (Li et al., 2009; Li et al., 2012; Oh et al., 2012; Zhang et al., 2014). We hypothesized that WRKY46/54/70 might also function as cofactors of BES1 to regulate BR target genes. To test this idea, we first performed RNA-seq analysis with 4-week-old adult plants of w54t and analyzed the overlap of the genes differentially expressed in the triple mutant with those affected in bes1-D, a gain-of-function mutant in BES1, to determine if WRKY46/54/70 regulate the expression of BR/BES1 target genes. A significant portion of genes up- or downregulated in the w54t mutant are down- or upregulated, respectively, in bes1-D (Figures 2A and 2B; Supplemental Data Sets 1 and 2). The results suggest that WRKY46/54/70 positively participate in BES1-regulated gene expression. Similar results were observed in the wrkyS mutant (Supplemental Figures 6A and 6B).
Figure 2.
WRKY46, WRKY54, and WRKY70 Regulate the Expression of BR Target Genes.
(A) Venn diagram showing overlaps among genes up- or downregulated in w54t with those differentially expressed in bes1-D.
(B) Clustering analysis of genes differentially expressed in w54t under control conditions within the wild type, w54t, and bes1-D. Values indicate normalized expression levels.
(C) The expression of genes downregulated in w54t was examined using 4-week-old plants treated with or without 1 μM BL. The averages and sd are derived from three biological replicates.
(D) The expression of genes upregulated in w54t was examined as described in (C).
To confirm the effect of WRKY46/54/70 on the transcriptional regulation on BR targets, we used RT-qPCR to examine the expression of several genes differentially expressed in w54t that are also regulated by BRs, as reported in our previous global gene expression analysis (Supplemental Table 1) (Yu et al., 2011; Wang et al., 2014). All three of the BR-induced genes tested have compromised induction by BL in w54t (Figure 2C). Similarly, three of the BR-repressed genes that were examined are upregulated in w54t (Figure 2D). The results indicate that WRKY46/54/70 are required for the expression of BR-regulated gene expression, confirming that WRKY46/54/70 function positively in BR signaling.
BES1 Cooperates with WRKY54 to Regulate the Transcription of BR Target Genes
Given the strong effect of w54t mutants on BR regulated gene expression, we tested if WRKY46/54/70 interact with BES1 to cooperatively modulate BR-regulated gene expression. We first chose WRKY54 as a representative TF of the WRKY46/54/70 family to investigate the interaction between WRKYs and BES1. Yeast two-hybrid assays demonstrated an interaction between BES1 and WRKY54 (Supplemental Figure 7A), which was confirmed by glutathione S-transferase (GST) pull-down using maltose binding protein (MBP)-tagged BES1 protein and GST-tagged WRKY54 (Supplemental Figure 7B).
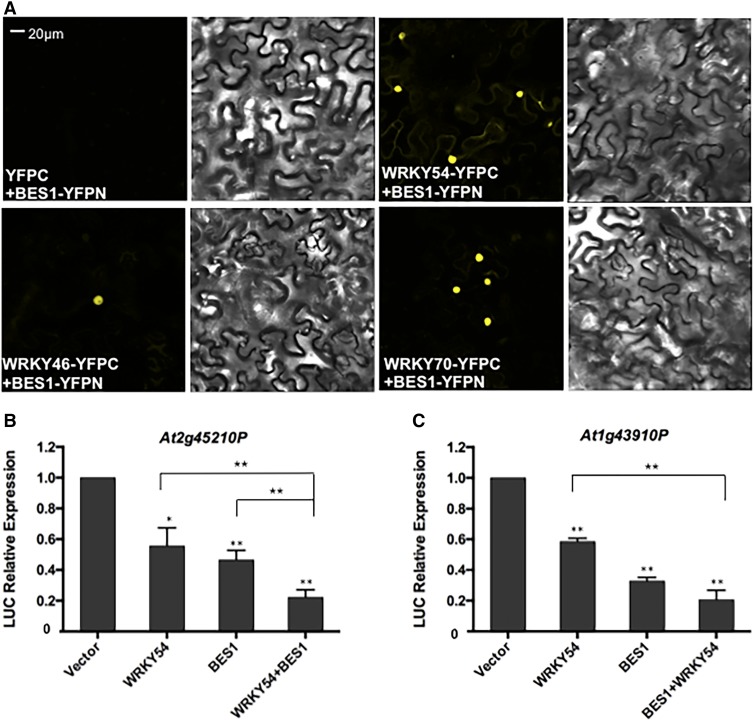
We next tested the interaction between BES1 and WRKY54 in vivo by biomolecular fluorescence (BiFC) assay with BES1 fused to the N terminus of YFP (YFPN) and WRKY54 fused to the C terminus of YFP (YFPC). When coexpressed in Nicotiana benthamiana, BES1-YFPN and WRKY54-YFPC resulted in reconstituted YFP signal (Figure 3A). However, no fluorescence signal was observed in negative controls where WRKY54-YFPC was coexpressed with YFPN or BES1-YFPN was expressed with YFPC (Figure 3A; Supplemental Figure 7F). These results confirm that WRKY54 interacts with BES1 in vivo. Similar results were obtained for WRKY46 and WRKY70 in BiFC assays, indicating that these TFs also interact with BES1 (Figure 3A).
Figure 3.
WRKY46, WRKY54, and WRKY70 Directly Interact with BES1 Both in Vivo and in Vitro.
(A) WRKY46/54/70 interact with BES1 by BiFC assay in vivo. Cotransformation of WRKY46/54/70-YFPC and BES1-YFPN led to the reconstitution of YFP signal, whereas no signal was detected when BES1-YFPN and YFPC or WRKY46/54/70-YFPC and YFPN were coexpressed (Supplemental Figure 7F). The experiments were performed twice with similar results.
(B) and (C) Transient expression of LUC driven by the BR-regulated gene promoters of At2g45210 (B) and At1g43910 (C). The mean and sd were derived from three biological repeats.
To test our hypothesis that WRKY54 and BES1 cooperate in the regulation of BR target genes, two BR-repressed genes (At2g45210 and At1g43910) were used to generate promoter-luciferase (LUC) reporter constructs for transient gene expression analysis in N. benthamiana. BES1 or WRKY54 alone repressed reporter activity to ∼50% of the control level and the reporter activity was further reduced to ∼20% when both BES1 and WRKY54 were coexpressed (Figures 3B and 3C), supporting a role of BES1-WRKY54 interaction in repressing target gene expression. Taken together, our results indicate that BES1 and WRKY54 interact with each other and cooperate to regulate the expression of BR target genes.
WRKY46, WRKY54, and WRKY70 Play Negative Roles in the Drought Response and Repress Dehydration-Inducible Gene Expression
WRKY54 and WRKY70 were previously identified as negative regulators of osmotic stress tolerance in Arabidopsis (Li et al., 2013). To test whether WRKY46/54/70 regulate drought tolerance, the wild type, wrky46, wrky54, and wrky70 single mutants, wrky46 wrky54, wrky46 wrky70, and wrky54 wrky70 double mutants, and w54t triple mutants were subjected to drought survival assays. After drought and rewatering, w54t mutants exhibited significantly higher survival rates than the wild type, the single mutants, or the double mutants (Figures 4A and 4B; Supplemental Figure 8). The results indicate that WRKY46/54/70 negatively regulate drought stress responses.
Figure 4.
WRKY46, WRKY54, and WRKY 70 Play Negative Roles in Drought Response.
(A) Phenotypes of wild-type, wrky46, wrky54, wrky70, and w54t plants before drought (top), after drought (middle), and 2 d after rewatering (bottom).
(B) The survival rate after recovery was determined. The mean and sd were from three biological repeats.
(C) Venn diagram showing comparisons among genes differentially expressed in w54t and genes up- or downregulated by dehydration in the wild type.
(D) Clustering of dehydration downregulated genes in the wild type and w54t mutants under control conditions (W) or dehydration (D).
(E) Clustering of dehydration upregulated genes in the wild type and w54t mutants under control conditions (W) or dehydration (D). Values indicate normalized expression levels.
To reveal the mechanism of WRKY46/54/70 function in the drought response, we performed global gene expression studies using 4-week-old wild-type and w54t plants under control and dehydration conditions by RNA-seq. After a 4-h dehydration, 310 genes were induced and 244 genes were repressed in wild-type plants (Figure 4C; Supplemental Data Set 3). Consistent with the strong phenotype of w54t mutants in growth and drought response, 4600 genes were upregulated and 4530 genes were downregulated in w54t mutants (Figure 2A; Supplemental Data Set 2). Many of the genes differentially expressed in w54t mutants are involved in responses to various stresses and cellular processes (Supplemental Figure 9). Among these, 156 dehydration-repressed genes were constitutively downregulated and 164 dehydration-induced genes were constitutively upregulated in w54t mutants without dehydration treatments, which were further decreased or increased by dehydration, respectively (Figures 4D and 4E). These results were consistent with our observation that w54t was more tolerant to drought stress. We compared the genes differentially regulated in w54t under dehydration conditions (143 genes downregulated in w54t upon dehydration and 235 genes upregulated in w54t upon dehydration) with those differentially expressed in bes1-D and found that there was significant overlap between these two data sets (Supplemental Figure 8D). Moreover, 55.2% of genes downregulated in w54t under dehydration condition were upregulated in bes1-D, but only 3% of the genes were downregulated in bes1-D. Similarly, 25.1% of genes upregulated in w54t under dehydration condition were downregulated in bes1-D, whereas ∼14% were upregulated in bes1-D. The results suggest that WRKY46/54/70 indeed play important roles in BR-regulated drought tolerance.
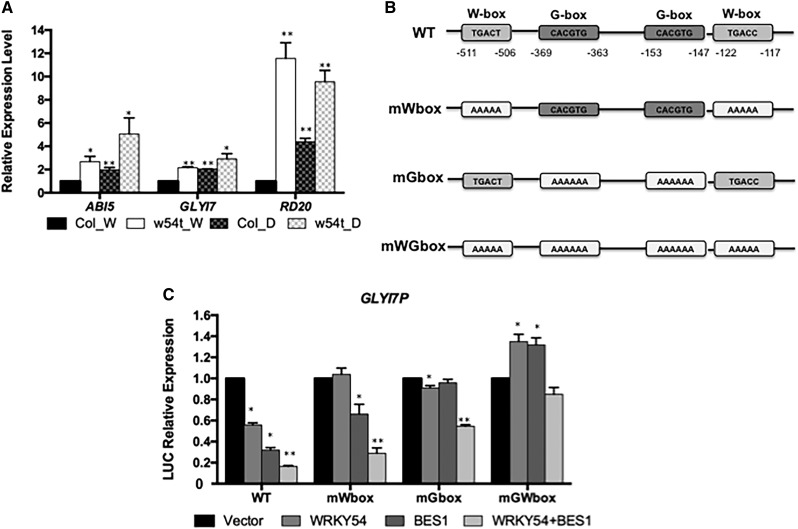
To confirm our RNA-seq data, three dehydration-induced genes, ABI5, GLYOXYLASE I 7 (GLYI7), and RESPONSIVE TO DESICCATION 20 (RD20), were chosen for qPCR validation (Fujita et al., 2005; Yuan et al., 2014; Pinedo et al., 2015). The expression of these genes increased significantly in w54t mutants with or without dehydration (Figure 5A). Next, we investigated if BES1 and WRKY54 cooperate in the regulation of dehydration-induced genes using LUC reporter assays with the GLY17 promoter (Figures 5B and 5C). BES1 or WRKY54 individually resulted in ∼2-fold reduction in reporter activity, and coexpression of BES1 and WRKY54 led to a further reduction, showing a 4-fold reduction in reporter activity (Figure 5C). The W-box [(T)TGACC/T] and G-box (CACGTG) were previously shown to be conserved binding motifs for WRKY TFs and BES1, respectively (Eulgem and Somssich, 2007; Yu et al., 2011). To test if the repression effect of WRKY54 and BES1 on the dehydration-inducible gene is through binding to the W-box and G-box, GLYI7 promoter containing mutated W-box, G-box, or both were fused with LUC and coexpressed with BES1 or WRKY54 alone or together (Figure 5B). The results showed that W-box mutation disrupted WRKY54-mediated repression of the GLYI7 promoter (Figure 5C). Similarly, mutation of the G-box abrogated the effect of BES1 on GLY17 promoter activity. The simultaneous mutation of the W-box and G-box motifs completely reversed the repressive effect of both BES1 and WRKY54 on GLYI7 expression (Figure 5C). Taken together, these results indicate that WRKY46/54/70 negatively modulate drought tolerance and likely cooperate with BES1 to repress drought-inducible genes by binding to the W-box and G-box, respectively.
Figure 5.
WRKY54 and BES1 Cooperate to Negatively Regulate Dehydration-Induced Genes.
(A) The expression of dehydration-inducible genes, ABI5, GLYI7, and RD20, was determined in the wild type and w54t by RT-qPCR under control conditions (W) or dehydration (D). Error bars indicate sd.
(B) Schematic diagram of the promoter region of GLYI7. Wild-type W-box and G-box are indicated. mWbox, the mutation of W-box; mGbox, the mutation of G-box; mGWbox, the mutation of both G-box and W-box.
(C) Transient expression of GLYI7P-fLUC and mutated W-box or G-box or both of GLYI7P-fLUC was determined in the presence of WRKY54 and/or BES1 in protoplasts. Error bars indicate sd (*P < 0.05, **P < 0.01; Student’s t test).
WRKY54 Is Phosphorylated and Destabilized by BIN2 Kinase
BIN2, a glycogen synthase kinase-3 like kinase, functions as a negative regulator in the BR pathway. Substrates of BIN2 share a consensus motif S/TXXXS/T, where S/T denotes serine or threonine and X can be any amino acid (Zhao et al., 2002). WRKY54 protein has 29 putative BIN2 phosphorylation sites, suggesting that it might be a substrate of BIN2 (Supplemental Figure 7C). Yeast two-hybrid assays indicated that BIN2 and WRKY54 indeed interacted with each other (Supplemental Figure 7D). GST pull-down assays showed that GST-WRKY54, but not GST alone, pulled down a significant amount of MBP-BIN2 (Supplemental Figure 7E). BiFC assays further indicated the direct interaction between WRKY54/WRKY46/WRKY70 and BIN2 (Figure 6A). These results suggest that WRKY54 and its homologs directly interact with BIN2.
Figure 6.
BIN2 Kinase Phosphorylates and Destabilizes WRKY54 Protein.
(A) WRKY46/54/70 interact with BIN2 by BiFC assay in vivo. Cotransformation of WRKY46/54/70-YFPC and BIN2-YFPN led to the reconstitution of YFP signal, whereas no signal was detected when BIN2-YFPN and YFPC or WRKY46/54/70-YFPC and YFPN were coexpressed (Supplemental Figure 7F). The experiments were performed twice with similar results.
(B) In vitro kinase assays show BIN2 phosphorylates WRKY54/46/70 (top). The loading controls of MBP, MBP-WRKY54/46/70, and GST-BIN2 by CBB staining are shown in bottom panel.
(C) The phosphorylation of WRKY54 by BIN2 was inhibited with the increasing concentrations of bikinin (top). The loading controls are shown on the bottom.
(D) The WRKY54 protein level was detected in indicated BR mutants and wild type with WRKY54 antibody. The w54t mutant was used as a negative control.
(E) WRKY54 protein accumulated upon BL treatment. Two-week-old wild-type seedlings were treated with or without 1 μM BL for indicated time and used to prepare protein to detect WRKY54 (top), BES1 (middle), and a control protein (bottom).
To test if WRKY46/54/70 are substrates of BIN2, we then performed in vitro kinase assays with 32P-labeled ATP. MBP-tagged WRKY54 could be phosphorylated by GST-BIN2 kinase and bikinin, an inhibitor of BIN2 kinase, inhibited the phosphorylation of WRKY54 and BIN2 autophosphorylation (Figures 6B and 6C) (De Rybel et al., 2009). These results indicate that WRKY54 is a substrate of BIN2. Similar results were obtained for WRKY46 and WRKY70, indicating that these TFs are also phosphorylated by BIN2 kinase (Figure 6B; Supplemental Figure 7G).
Several previous reports indicated that BIN2 phosphorylation can lead to protein destabilization in vivo (Youn and Kim, 2015). In order to determine the biological function of BIN2 phosphorylation on WRKY54 in vivo, the stability of WRKY54 in BIN2 gain-of-function (bin2-1) and loss-of-function (bin2-3 bil1 bil2) mutants was examined by immunoblotting with a WRKY54 antibody we developed (Supplemental Figures 10A and 10B). As shown in Figure 6D, WRKY54 protein increased by more than 3-fold in bin2-3 bil1 bil2 triple mutants and decreased by half in bin2-1 compared with the wild type (Figure 6D). These results suggest that WRKY54 stability is negatively correlated with BIN2 abundance in vivo. To confirm these results, we examined WRKY54 accumulation in wild-type plants after treatment with 1 μM BL, which inhibits BIN2 kinase activity. WRKY54 protein accumulated to ∼2.2-fold after 4 h of BL treatment (Figure 6E). These results illustrate that WRKY54 is involved in the BR pathway and can be regulated by BRs at both transcriptional and posttranscriptional levels.
The roles of WRKY and BES1 in the crosstalk of plant growth and stress response prompted us to examine the protein level of WRKY54 and BES1 in response to drought stress. Water was withheld from 4-week-old wild-type plants, and control or drought-treated samples were collected 8 to 10 d after withholding water. As shown in Figure 7A, the protein levels of WRKY54 and BES1 started to decrease 9 d after drought treatment and WRKY54 protein was almost undetectable at the 10-d time point (Figure 7A). These results suggest that WRKY54 plays a vital role in the coordination of plant growth and drought stress response.
Figure 7.
A Working Model of WRKY46/54/70 Function in Plant Growth and Stress Response.
(A) WRKY54 and BES1 protein decreased with increasing drought treatment time. The 8d, 9d, and 10d indicate days of drought treatment or controls.
(B) A working model of WRKY46/54/70 in BR-regulated growth and drought stress response. WRKY46/54/70 are regulated by BR signaling through BIN2 and BES1, and cooperate with BES1 to promote plant growth and inhibit drought responses. WRKY46/54/70 also slightly promote BR biosynthesis. Under normal growth conditions (left), WRKY46/54/70 and BES1 positively coregulate growth-related genes and negatively control the expression of drought-responsive genes to promote growth. BRs regulate WRKY46/54/70 both transcriptionally and posttranscriptionally through BES1 and BIN2, respectively. Under drought stress conditions (right), WRKY46/54/70 and BES1 protein are destabilized, which leads to repression of growth-related genes and alleviation of WRKY46/54/70’s inhibitory effect on drought-related genes, leading to reduced growth and increased drought tolerance.
DISCUSSION
WRKY transcription factors, found exclusively in the plant kingdom, integrate various signaling pathways to modulate numerous processes including stress responses, nutrient deprivation, senescence, seed and trichome development, and embryogenesis (Hinderhofer and Zentgraf, 2001; Johnson et al., 2002; Miao et al., 2004; Ulker et al., 2007; Zhou et al., 2011; Besseau et al., 2012; Ding et al., 2014a). Many Arabidopsis WRKY genes are regulated by bacterial pathogen or salicylic acid treatment (Dong et al., 2003), and genetic studies have indicated that WRKY transcription factors can regulate plant defense either positively or negatively (Pandey and Somssich, 2009). WRKY TFs also regulate abiotic stress responses including drought, salinity, radiation, and cold (Banerjee and Roychoudhury, 2015). However, the role of WRKYs in BR regulation of plant growth has remained unclear.
Here, we found that Group III WRKY transcription factors play an important role in BR-regulated plant growth as w54t triple mutants displayed a dwarf phenotype and compromised BR responses. Our results suggest that WRKY46/54/70 play positive roles in plant growth mainly by regulating BR signaling with a smaller effect on BR biosynthesis. The role of WRKY46/54/70 in BR signaling is supported by its reduced response in hypocotyl elongation (Figure 1) and significant overlap between genes differentially expressed in w54t and in bes1-D mutants (Figure 2). Moreover, WRKY54 affects BR-regulated genes by interacting and cooperating with BES1 (Figure 3). Our results therefore establish that WRKY46/54/70 promote BR signaling and are required for optimal plant growth.
The regulation of WRKY54 by BIN2 kinase, a negative regulator in the BR pathway, provides further support for its involvement in BR signaling. BIN2, a GSK3-like kinase, plays diverse roles in cellular processes including BR signaling by phosphorylating an array of substrates, leading to functional consequences such as altered protein stability (Youn and Kim, 2015). Here, we identified WRKY54 as a substrate of BIN2 kinase and BIN2 phosphorylation led to destabilization of the WRKY54 protein (Figure 6). It is possible that BIN2 phosphorylation of WRKY54 functions to release the inhibitory effect of WRKY54 on the transcription of drought-responsive genes during drought stress (Zhang et al., 2009).
Our global gene expression studies revealed the molecular basis for the function of WRKY46/54/70 in drought responses. WRKY54 and WRKY70 were reported to act as negative regulators in osmotic stress tolerance in Arabidopsis (Li et al., 2013). WRKY46 is induced by drought stress and was found to regulate osmotic stress responses (Ding et al., 2014b). Consistent with these reports, we found that wrky46 wrky54 wrky70 triple mutants were more tolerant to drought stress compared with the wild type, suggesting that they negatively regulate the drought response (Figure 4). Consistent with the mutant phenotype, we found that ∼53% dehydration-induced genes are upregulated and 64% of dehydration-repressed genes are downregulated in w54t mutants (Figure 4). Our results therefore establish WRKY46/54/70 as important negative regulators for drought tolerance that at least partially mediate BR repression of drought responses.
Interestingly, WRKY54 cooperates with BES1 in the regulation of both BR- and dehydration-regulated genes (Figures 3 and 5). Previous studies revealed that WRKY responds to various environmental signals or plant developmental processes through physical interaction with a wide range of proteins related to signaling, transcription, and chromatin remodeling (Chi et al., 2013). Likewise, BES1/BZR1 interact with multiple cofactors to control BR-regulated plant growth and development (Guo et al., 2013). This study established that BES1-WRKY54 interactions play important roles in BR-regulated plant growth and drought responses (Figure 7).
In summary, we demonstrated that WRKY46, WRKY54, and WRKY70 are involved in BR-regulated plant growth by regulating BR signaling through cooperation with BES1. In addition, WRKY46, WRKY54, and WRKY70 negatively regulate drought tolerance by inhibiting dehydration-inducible gene expression. Future identification of WRKY54 interacting partners and target genes can further our understanding of the mechanisms by which WRKY regulates BR-regulated growth and drought responses.
METHODS
Plant Materials, Growth Conditions, and Hormone Responses
Arabidopsis thaliana ecotype Columbia (Col-0) was used as the wild type. T-DNA insertion lines, wrky46 (SALK_134310), wrky54 (CS873142), and wrky70 (SALK_025198), were obtained from the Arabidopsis Biological Resource Center. Seeds were sterilized by 70% (v/v) ethanol and 0.1% (v/v) Triton X-100. All of the plants were grown on 0.5× Murashige and Skoog (MS) plates with 1% sucrose under long-day conditions (16-h fluorescent light/8-h dark) at 22°C. BL (10 and 100 nM) was added to the 0.5× MS agar plates. The average hypocotyl lengths were measured using 15 samples and repeated three times. Fourteen-day-old seedlings were transferred to soil and grown under the same condition in growth chambers.
Drought Stress Treatment
For drought treatment, soil was weighed in each pot before transferring the seedlings to make sure each pot has the same amount of the soil and same volume of water in the flat. Seedlings were grown on 0.5× MS medium for 2 weeks and then transferred into the weighed soil. Plants were watered once per week after transferring into soil and then water was withheld for 2 weeks. The survival rates are scored based upon plants that had survived 2 d after rewatering from three biological replicates. Each biological repeat had four or five pots for each genotype. All of the pots were randomly distributed in the flat and were rotated frequently during drought stress to minimize the effect from growth environment (Shi et al., 2015). Similar results were obtained for at least three repeats at different times. Three biological replicates were performed each time with three technical replicates (one pot/technical replicate).
Plasmid Construction and Protein-Protein Interaction Assays
The DNA primer sequences used for this study are listed in Supplemental Table 2. For the yeast two-hybrid assays, WRKY54 was cloned into both GAL4 bait and prey vectors. BES1 and BIN2 were cloned into GAL4 bait and prey vectors, respectively (Clontech). The constructs were transformed into yeast strain Y187 and the lacZ reporter assay was conducted using X-gal according to the manufacturer’s protocol (Clontech). For GST pull-down assay, WRKY54 fused to GST was cloned into both pET42a and purified with glutathione agarose beads (Sigma-Aldrich). BIN2 and BES1 were fused with MBP and purified with amylose resin (NEB). GST pull-down assays were performed as described (Yin et al., 2002). For BiFC assays, the N terminus (amino acids 1–174) or C terminus (amino acids 175–239) of YFP vectors was as described (Yu et al., 2008). The full-length coding regions of WRKY54 and BES1 were cloned into YFPC and YFPN and then transformed into Agrobacterium tumefaciens strains GV3101. BiFC assay were performed as described (Wang et al., 2014).
In Vitro Kinase Assay
For the in vitro kinase assay, MBP and MBP-WRKY54 were incubated with GST-BIN2 kinase in 20 μL of kinase buffer (20 mM Tris, pH 7.5, 100 mM NaCl, 12 mM MgCl2, and 10 μCi [γ-32P]ATP (Yin et al., 2002). After incubation at 37°C for 1 h, 20 μL 2× SDS buffer were added to stop the reaction and then the samples were boiled for 5 min. Proteins were resolved by SDS-PAGE gel and phosphorylation signal was detected by Typhoon/Image Quant TL.
Gene Expression Analysis and Luciferase Assay
For the gene expression analysis, 1000 nM BL was sprayed on 4-week-old bes1-D, wrky46 wrky54 wrky70, and wild-type plants. DMSO was used as the control. Plant tissues were collected after 2.5 h of treatment and total RNA was extracted using TRIzol reagent (Thermo Fisher) and the RNeasy Mini Kit (Qiagen). SYBR Green PCR Master Mix (Applied Biosystems) was used in qPCR analysis and qPCR samples were run on Mx4000 multiplex qPCR system (Stratagene) with three technical replicates. UBQ5 was used as the internal control. Similar results were obtained from three biological replicates.
For the transient expression of BR-regulated genes, At2g45210 and At1g43910 promoters were fused with the LUC reporter gene. WRKY54 and BES1 coding regions were cloned into pZP211 vector and transformed into Agrobacterium. Equal amounts of Agrobacterium cells transformed with BES1 or WRKY54 or BES1 and WRKY54 were injected into tobacco (Nicotiana benthamiana) leaves. The luciferase activities were measured with the luciferase assay system from Promega and Berthold Centro LB960 luminometer. The luciferase data were normalized to the total protein content.
For the transient expression of dehydration-inducible genes, GLYI7 promoter driving firefly LUC and CaMV35S driven REN was constructed in the same plasmid and transformed into Arabidopsis protoplasts with WRKY54 or BES1 alone or together. Protoplasts were prepared based on the protocol from Yoo et al. (2007). After 16 h incubation, protoplasts were collected and the dual-luciferase assay system from Promega was used to measure the activity of firefly LUC and renilla luciferase (REN) sequentially using a Berthold Centro LB960 luminometer. The ratio of LUC/REN was calculated and the relative ratio was used as the final measurement.
Determination of Endogenous BR Levels
The quantification of endogenous BRs was performed based on the method reported previously with some simplifications in sample pretreatment (Xin et al., 2013). The harvested plant materials were first ground to a fine powder with a MM-400 mixer milling (Retsch). One hundred milligrams of the powder was extracted with 1 mL of 90% aqueous methanol (methanol) in ultrasonic bath for 1 h. Simultaneously, D3-BL, D3-CS, and D3-6-deoxo-CS were added to the extract as internal standards for measurement of BRs. After the MCX cartridge (3 mL, 60 mg; Waters) was activated and equilibrated with 2 mL of methanol, water, and 40% methanol in sequence, the crude extracts redissolved in 40% methanol were loaded onto the cartridge. Then the MCX cartridge was washed with 2 mL of 10% methanol, followed by 40% methanol in sequence. At last, BRs were eluted with methanol. After being dried with an N2 stream, the eluent was redissolved with anhydrous acetonitrile to be derivatized with DMAPBA prior to UPLC-MS/MS analysis. BR analysis was performed on a quadrupole linear ion trap hybrid MS (QTRAP 5500; AB SCIEX) equipped with an electrospray ionization source coupled with a UPLC (Waters). The UPLC inlet method, ESI source parameters, MRM transitions, and the related compound-dependent parameters were set as described in a previous report (Xin et al., 2013). As for 6-deoxo-CS or D3-6-deoxo-CS, the MRM transition 580.4>176.1 or 583.4>176.1 was used for quantification and 580.4>190.1 or 583.4>190.1 for qualification. The collision energies were set as 60 and 50 V for the transitions, respectively.
Phylogenetic Analysis
The phylogenetic tree of the six WRKY genes was generated using Clustal Omega (Sievers et al., 2011). The alignment can be found in Supplemental File 1.
qPCR Measurement
PCR was performed in a 20-μL reaction containing SYBR Green PCR Master Mix (Applied Biosystems), cDNA, and primers (listed in Supplemental Table 2) and measured with Stratagene Mx4000 qPCR machine.
WRKY54 Antibody Generation and Purification
Serum was generated from rabbit after multiple injections of MBP-WRKY54 (full-length) protein as the antigen. WRKY54 antibody was then purified from the serum with CNBr-activated sepharose. The beads were incubated with 2 mg MBP-WRKY54 protein in 5 mL coupling buffer (0.125 M phosphate, pH 8.3) overnight at 4°C. Then beads were transferred to a 2.5-cm column and equilibrated with 10 mL PBS. Rabbit serum (10 mL) was diluted with 3 volumes PBS and applied to the column. The beads in the column were washed with 30 mL PBS buffer. The bound antibody was eluted with 225 μL glycine⋅Cl (pH 2.0) directly into the tube with 25 μL neutralizing buffer (1 M Tris, pH 8.0).
Dehydration RNA-Seq and Data Analysis
Three biological replicates of 4-week-old wrky46 wrky54 wrky70 and wild-type plants were grown in soil under long-day conditions (16 h light/8 h dark). The whole rosette leaves were cut and placed in empty Petri dish (150 × 15 mm) as dehydration treatment or in Petri dish with moistened Kimwipes as mock control. Each Petri dish consisted of leaves from three or four plants and was considered as one biological replicate. The Petri dish was sealed with Parafilm and left for 4 h. Tissue was then collected and processed for RNA extraction using Trizol and RNeasy Mini Kit (Qiagen) with on-column DNase digestion and cleaned up with column, following the manufacturer’s instructions.
Library preparation and RNA-seq were performed by BGI Americas using an Illumina HiSeq 2000 with 50-bp single-end reads and ∼30 million reads per sample. Raw RNA-seq reads were subjected to quality checking and trimming. The trimmed reads of each sample were aligned to the public available reference genome of Arabidopsis (TAIR10) using GSNAP. The alignment coordinates of uniquely aligned reads to the reference genome were used for lookup and read count tallies were computed for each annotated gene. Finally, RNA-seq reads were used to identify differentially expressed genes with R package DESeq2 for comparison between wrky46 wrky54 wrky70 and the wild type that were subjected for control or dehydration treatment. Normalization was conducted by DESeq2, which automatically corrects for biases introduced by differences in the total numbers of uniquely mapped reads in each sample. Normalized read counts were used to calculate fold changes and statistical significance (Data2Bio). Clustering was performed using the ‘aheatmap’ function of the NMF package in R and log2 reads per million mapped reads values were used for clustering analysis. Gene Ontology analysis was performed using BiNGO software.
Accession Numbers
RNA-seq data from this article can be found in the Gene Expression Omnibus (GEO: GSE93420). The accession numbers for the studied genes are as follows: WRKY30, At5g24110; WRKY41, At4g11070; WRKY46, At2g46460; WRKY53, At4g23810; WRKY54, At2g40750; and WRKY70, At3g56400.
Supplemental Data
Supplemental Figure 1. WRKY T-DNA insertion mutants.
Supplemental Figure 2. The wrky mutants displayed a dwarf phenotype.
Supplemental Figure 3. Group III WRKY (WRKY30/41/46/53/54/70) proteins function redundantly and play positive roles in plant growth.
Supplemental Figure 4. The wrky mutants have slightly reduced endogenous BR levels.
Supplemental Figure 5. Different hormonal responses of wild-type, wrky46, wrky54, wrky70, and w54t plants.
Supplemental Figure 6. Clustering analysis of genes differentially expressed in wrkyS mutants.
Supplemental Figure 7. WRKY46/54/70 interact with BES1/BIN2 and are phosphorylated by BIN2.
Supplemental Figure 8. The drought phenotype of wrky single, double, and triple mutants.
Supplemental Figure 9. Gene Ontology analysis of 9130 genes differentially expressed in w54t mutants.
Supplemental Figure 10. WRKY54 antibody test in wrky single, triple, and sextuple mutants.
Supplemental Table 1. Annotation of the genes used in Figures 2C and 2D.
Supplemental Table 2. Primers and mutant promoter sequences used in this study.
Supplemental Data Set 1. Genes up- or downregulated in bes1-D.
Supplemental Data Set 2. Genes up- or downregulated in wrky46 wrky54 wrky70 triple mutant.
Supplemental Data Set 3. Genes up- or downregulated by dehydration treatment in wild-type plants.
Supplemental File 1. Text file of alignment used for phylogenetic tree in Supplemental Figure 1B.
Acknowledgments
We thank Data2Bio (Ames, IA) for performing RNA-seq analysis and Tadao Asami (University of Tokyo) for providing BRZ. The work is supported by grants from the National Science Foundation (IOS-1257631), the National Institutes of Health (1R01GM120316-01A1), and by the Plant Sciences Institute at Iowa State University. P.X. and J. Chu are supported by National Natural Science Foundation of China Grant 31470433. J. Chen was partially supported by fellowship from China Scholar Council.
AUTHOR CONTRIBUTIONS
J. Chen performed most of the experiments unless indicated as follows. J. Chen and T.M.N. conducted RNA-seq experiments and analyzed RNA-seq data with M.Z. and Z.L. H.Y. was involved in generating the mutants. T.M.N. performed the confocal microscopy in BiFC assays. H.T., C.C., P.X., and J. Chu conducted the BR measurements and analyzed the data. J. Chen and Y.Y. wrote the article with input from other coauthors.
Glossary
- BR
brassinosteroid
- TFs
transcription factors
- GA
gibberellic acid
- BL
brassinolide
- CS
castasterone
- BiFC
biomolecular fluorescence
- MS
Murashige and Skoog
References
- Bai M.Y., Shang J.X., Oh E., Fan M., Bai Y., Zentella R., Sun T.P., Wang Z.Y. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14: 810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Roychoudhury A. (2015). WRKY proteins: signaling and regulation of expression during abiotic stress responses. Sci. World J. 2015: 807560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besseau S., Li J., Palva E.T. (2012). WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 63: 2667–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beste L., Nahar N., Dalman K., Fujioka S., Jonsson L., Dutta P.C., Sitbon F. (2011). Synthesis of hydroxylated sterols in transgenic Arabidopsis plants alters growth and steroid metabolism. Plant Physiol. 157: 426–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z., Liu J., Wang H., Yang C., Chen Y., Li Y., Pan S., Dong R., Tang G., Barajas-Lopez Jde.D., Fujii H., Wang X. (2014). GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 9651–9656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Song Y., Li S., Zhang L., Zou C., Yu D. (2012). The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 1819: 120–128. [DOI] [PubMed] [Google Scholar]
- Chi Y., Yang Y., Zhou Y., Zhou J., Fan B., Yu J.Q., Chen Z. (2013). Protein-protein interactions in the regulation of WRKY transcription factors. Mol. Plant 6: 287–300. [DOI] [PubMed] [Google Scholar]
- Chujo T., Miyamoto K., Ogawa S., Masuda Y., Shimizu T., Kishi-Kaboshi M., Takahashi A., Nishizawa Y., Minami E., Nojiri H., Yamane H., Okada K. (2014). Overexpression of phosphomimic mutated OsWRKY53 leads to enhanced blast resistance in rice. PLoS One 9: e98737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S.D. (2011). Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23: 1219–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S.D. (2016). Brassinosteroid/abscisic acid antagonism in balancing growth and stress. Dev. Cell 38: 118–120. [DOI] [PubMed] [Google Scholar]
- Clouse S.D., Langford M., McMorris T.C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111: 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B., et al. (2009). Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem. Biol. 16: 594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z.J., Yan J.Y., Li G.X., Wu Z.C., Zhang S.Q., Zheng S.J. (2014a). WRKY41 controls Arabidopsis seed dormancy via direct regulation of ABI3 transcript levels not downstream of ABA. Plant J. 79: 810–823. [DOI] [PubMed] [Google Scholar]
- Ding Z.J., Yan J.Y., Xu X.Y., Yu D.Q., Li G.X., Zhang S.Q., Zheng S.J. (2014b). Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis. Plant J. 79: 13–27. [DOI] [PubMed] [Google Scholar]
- Dong J., Chen C., Chen Z. (2003). Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 51: 21–37. [DOI] [PubMed] [Google Scholar]
- Eulgem T., Somssich I.E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10: 366–371. [DOI] [PubMed] [Google Scholar]
- Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5: 199–206. [DOI] [PubMed] [Google Scholar]
- Feng Y., Yin Y., Fei S. (2015). Down-regulation of BdBRI1, a putative brassinosteroid receptor gene produces a dwarf phenotype with enhanced drought tolerance in Brachypodium distachyon. Plant Sci. 234: 163–173. [DOI] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Satoh R., Maruyama K., Parvez M.M., Seki M., Hiratsu K., Ohme-Takagi M., Shinozaki K., Yamaguchi-Shinozaki K. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17: 3470–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Minguet E.G., Grau-Enguix F., Abbas M., Locascio A., Thomas S.G., Alabadí D., Blázquez M.A. (2012). Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 13446–13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui J., Zheng S., Liu C., Shen J., Li J., Li L. (2016). OsREM4.1 interacts with OsSERK1 to coordinate the interlinking between abscisic acid and brassinosteroid signaling in rice. Dev. Cell 38: 201–213. [DOI] [PubMed] [Google Scholar]
- Guo H., Li L., Aluru M., Aluru S., Yin Y. (2013). Mechanisms and networks for brassinosteroid regulated gene expression. Curr. Opin. Plant Biol. 16: 545–553. [DOI] [PubMed] [Google Scholar]
- Hao J., Yin Y., Fei S.Z. (2013). Brassinosteroid signaling network: implications on yield and stress tolerance. Plant Cell Rep. 32: 1017–1030. [DOI] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Yang Y., Li J., Wang Z.Y. (2002). The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 10185–10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi K., Ishiga Y., Inagaki Y., Toyoda K., Shiraishi T., Ichinose Y. (2008). Modulation of defense signal transduction by flagellin-induced WRKY41 transcription factor in Arabidopsis thaliana. Mol. Genet. Genomics 279: 303–312. [DOI] [PubMed] [Google Scholar]
- Hinderhofer K., Zentgraf U. (2001). Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta 213: 469–473. [DOI] [PubMed] [Google Scholar]
- Hu Y., Dong Q., Yu D. (2012). Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 185-186: 288–297. [DOI] [PubMed] [Google Scholar]
- Hu Y., Yu D. (2014). BRASSINOSTEROID INSENSITIVE2 interacts with ABSCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. Plant Cell 26: 4394–4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C.S., Kolevski B., Smyth D.R. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14: 1359–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S., Divi U.K., Krochko J.E., Keller W.A., Krishna P. (2007). Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 225: 353–364. [DOI] [PubMed] [Google Scholar]
- Kim T.W., Hwang J.Y., Kim Y.S., Joo S.H., Chang S.C., Lee J.S., Takatsuto S., Kim S.K. (2005). Arabidopsis CYP85A2, a cytochrome P450, mediates the Baeyer-Villiger oxidation of castasterone to brassinolide in brassinosteroid biosynthesis. Plant Cell 17: 2397–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna P. (2003). Brassinosteroid-mediated stress responses. J. Plant Growth Regul. 22: 289–297. [DOI] [PubMed] [Google Scholar]
- Li J., Chory J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938. [DOI] [PubMed] [Google Scholar]
- Li J., Nam K.H. (2002). Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295: 1299–1301. [DOI] [PubMed] [Google Scholar]
- Li J., Nam K.H., Vafeados D., Chory J. (2001). BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 127: 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Brader G., Kariola T., Palva E.T. (2006). WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 46: 477–491. [DOI] [PubMed] [Google Scholar]
- Li J., Wen J., Lease K.A., Doke J.T., Tax F.E., Walker J.C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222. [DOI] [PubMed] [Google Scholar]
- Li J., Besseau S., Törönen P., Sipari N., Kollist H., Holm L., Palva E.T. (2013). Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 200: 457–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Nagpal P., Vitart V., McMorris T.C., Chory J. (1996). A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272: 398–401. [DOI] [PubMed] [Google Scholar]
- Li L., Ye H., Guo H., Yin Y. (2010). Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proc. Natl. Acad. Sci. USA 107: 3918–3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Yu X., Thompson A., Guo M., Yoshida S., Asami T., Chory J., Yin Y. (2009). Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 58: 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.F., Wang C., Jiang L., Li S., Sun S.S., He J.X. (2012). An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci. Signal. 5: ra72. [DOI] [PubMed] [Google Scholar]
- Miao Y., Laun T., Zimmermann P., Zentgraf U. (2004). Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 55: 853–867. [DOI] [PubMed] [Google Scholar]
- Murray S.L., Ingle R.A., Petersen L.N., Denby K.J. (2007). Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol. Plant Microbe Interact. 20: 1431–1438. [DOI] [PubMed] [Google Scholar]
- Nam K.H., Li J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212. [DOI] [PubMed] [Google Scholar]
- Noguchi T., Fujioka S., Choe S., Takatsuto S., Tax F.E., Yoshida S., Feldmann K.A. (2000). Biosynthetic pathways of brassinolide in Arabidopsis. Plant Physiol. 124: 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan T.M., Brennan B., Yang M., Chen J., Zhang M., Li Z., Wang X., Bassham D.C., Walley J., Yin Y. (2017). Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev. Cell 41: 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northey J.G., Liang S., Jamshed M., Deb S., Foo E., Reid J.B., McCourt P., Samuel M.A. (2016). Farnesylation mediates brassinosteroid biosynthesis to regulate abscisic acid responses. Nat Plants 2: 16114. [DOI] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Wang Z.Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S.P., Somssich I.E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150: 1648–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinedo I., Ledger T., Greve M., Poupin M.J. (2015). Burkholderia phytofirmans PsJN induces long-term metabolic and transcriptional changes involved in Arabidopsis thaliana salt tolerance. Front. Plant Sci. 6: 466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewska I., Talarek M., Bajguz A. (2016). Brassinosteroids and response of plants to heavy metals action. Front. Plant Sci. 7: 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Chen Z., Liu Y., Zhang H., Zhang M., Liu Q., Hong X., Zhu J.K., Gong Z. (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63: 417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton P.J., Somssich I.E., Ringler P., Shen Q.J. (2010). WRKY transcription factors. Trends Plant Sci. 15: 247–258. [DOI] [PubMed] [Google Scholar]
- Sahni S., Prasad B.D., Liu Q., Grbic V., Sharpe A., Singh S.P., Krishna P. (2016). Overexpression of the brassinosteroid biosynthetic gene DWF4 in Brassica napus simultaneously increases seed yield and stress tolerance. Sci. Rep. 6: 28298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sairam R.K. (1994). Effects of homobrassinolide application on plant metabolism and grain yield under irrigated and moisture-stress conditions of two wheat varieties. Plant Growth Regul. 14: 173–181. [Google Scholar]
- Shahnejat-Bushehri S., Tarkowska D., Sakuraba Y., Balazadeh S. (2016). Arabidopsis NAC transcription factor JUB1 regulates GA/BR metabolism and signalling. Nat. Plants 2: 16013. [DOI] [PubMed] [Google Scholar]
- Shi H., Chen Y., Qian Y., Chan Z. (2015). Low Temperature-Induced 30 (LTI30) positively regulates drought stress resistance in Arabidopsis: effect on abscisic acid sensitivity and hydrogen peroxide accumulation. Front. Plant Sci. 6: 893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J.D., Higgins D.G. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J., Haegman M., Kurepa J., Van Montagu M., Straeten D.V. (1997). Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proc. Natl. Acad. Sci. USA 94: 2756–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M., Németh K., Koncz-Kálmán Z., Mathur J., Kauschmann A., Altmann T., Rédei G.P., Nagy F., Schell J., Koncz C. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171–182. [DOI] [PubMed] [Google Scholar]
- Tong H., Xiao Y., Liu D., Gao S., Liu L., Yin Y., Jin Y., Qian Q., Chu C. (2014). Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 26: 4376–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulker B., Somssich I.E. (2004). WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 7: 491–498. [DOI] [PubMed] [Google Scholar]
- Ulker B., Shahid Mukhtar M., Somssich I.E. (2007). The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 226: 125–137. [DOI] [PubMed] [Google Scholar]
- Unterholzner S.J., Rozhon W., Papacek M., Ciomas J., Lange T., Kugler K.G., Mayer K.F., Sieberer T., Poppenberger B. (2015). Brassinosteroids are master regulators of gibberellin biosynthesis in Arabidopsis. Plant Cell 27: 2261–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chen J., Xie Z., Liu S., Nolan T., Ye H., Zhang M., Guo H., Schnable P.S., Li Z., Yin Y. (2014). Histone lysine methyltransferase SDG8 is involved in brassinosteroid-regulated gene expression in Arabidopsis thaliana. Mol. Plant 7: 1303–1315. [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Chory J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513. [DOI] [PubMed] [Google Scholar]
- Xin P., Yan J., Fan J., Chu J., Yan C. (2013). An improved simplified high-sensitivity quantification method for determining brassinosteroids in different tissues of rice and Arabidopsis. Plant Physiol. 162: 2056–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Li C., Cai Z., Hu Y., Nolan T., Yu F., Yin Y., Xie Q., Tang G., Wang X. (2017). SINAT E3 ligases control the light-mediated stability of the brassinosteroid-activated transcription factor BES1 in Arabidopsis. Dev. Cell 41: 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H., et al. (2017). RD26 mediates crosstalk between drought and brassinosteroid signalling pathways. Nat. Commun. 8: 14573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J. (2005). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259. [DOI] [PubMed] [Google Scholar]
- Yin Y., Wang Z.Y., Mora-Garcia S., Li J., Yoshida S., Asami T., Chory J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Youn J.H., Kim T.W. (2015). Functional insights of plant GSK3-like kinases: multi-taskers in diverse cellular signal transduction pathways. Mol. Plant 8: 552–565. [DOI] [PubMed] [Google Scholar]
- Yu X., Li L., Li L., Guo M., Chory J., Yin Y. (2008). Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 7618–7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Li L., Zola J., Aluru M., Ye H., Foudree A., Guo H., Anderson S., Aluru S., Liu P., Rodermel S., Yin Y. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65: 634–646. [DOI] [PubMed] [Google Scholar]
- Yuan X., Li Y., Liu S., Xia F., Li X., Qi B. (2014). Accumulation of eicosapolyenoic acids enhances sensitivity to abscisic acid and mitigates the effects of drought in transgenic Arabidopsis thaliana. J. Exp. Bot. 65: 1637–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Ye H., Guo H., Johnson A., Zhang M., Lin H., Yin Y. (2014). Transcription factor HAT1 is phosphorylated by BIN2 kinase and mediates brassinosteroid repressed gene expression in Arabidopsis. Plant J. 77: 59–70. [DOI] [PubMed] [Google Scholar]
- Zhang S., Cai Z., Wang X. (2009). The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc. Natl. Acad. Sci. USA 106: 4543–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Peng P., Schmitz R.J., Decker A.D., Tax F.E., Li J. (2002). Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol. 130: 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Jiang Y., Yu D. (2011). WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells 31: 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]