Abstract
We provide a protocol for precision nuclear run-on sequencing (PRO-seq) and its variant, PRO-cap, which map the location of active RNA polymerases (PRO-seq) or transcription start sites (TSSs) (PRO-cap) genome-wide at high resolution. The density of RNA polymerases at a particular genomic locus directly reflects the level of nascent transcription at that region. Nuclei are isolated from cells and, under nuclear run-on conditions, transcriptionally engaged RNA polymerases incorporate one or, at most, a few biotin-labeled nucleotide triphosphates (biotin-NTPs) into the 3′ end of nascent RNA. The biotin-labeled nascent RNA is used to prepare sequencing libraries, which are sequenced from the 3′ end to provide high-resolution positional information for the RNA polymerases. PRO-seq provides much higher sensitivity than ChIP-seq, and it generates a much larger fraction of usable sequence reads than ChIP-seq or NET-seq (native elongating transcript sequencing). Similarly to NET-seq, PRO-seq maps the RNA polymerase at up to base-pair resolution with strand specificity, but unlike NET-seq it does not require immunoprecipitation. With the protocol provided here, PRO-seq (or PRO-cap) libraries for high-throughput sequencing can be generated in 4–5 working days. The method has been applied to human, mouse, Drosophila melanogaster and Caenorhabditis elegans cells and, with slight modifications, to yeast.
INTRODUCTION
The ability to measure the density of RNA polymerase across the genome provides a comprehensive and quantitative snapshot of transcription(Fuda et al., 2009). Collecting a series of these snapshots in response to regulatory switches reveals the identity of genes that respond immediately or secondarily to specific signals, and provides critical insights to the mechanisms of their regulation(Adelman and Lis, 2012). Quantifying RNA polymerase density along the genes is also critical for deciphering the regulatory steps involved in transcription.
In addition to protein coding genes, many other regions in the genome (such as upstream divergent regions, regions downstream of mRNA poly A sites, and enhancers) are transcribed to various extents. Enhancers produce short unstable RNAs (eRNAs) that do not encode proteins(Core et al., 2014) but delineate major hubs of transcription regulation(Heinz et al., 2015). Differential regulation of enhancer mediated transcription is implicated in various diseases(Vahedi et al., 2015), and understanding this regulation is important for deciphering the transcriptional changes in response to developmental, nutritional, and environmental cues. However, sequencing of total RNA by RNA-seq is inefficient in detecting these unstable RNAs.
A number of methods have been described that enrich and sequence nascent RNAs associated with RNA polymerase. These methods are either based on immunoprecipitation of RNA polymerase(Churchman and Weissman, 2011; Larson et al., 2014; Nojima et al., 2015) or are dependent on purification of insoluble chromatin(Weber et al., 2014). Therefore, these methods are highly dependent on antibody specificity or the purity of the chromatin fraction respectively. We have developed nuclear run-on based methods to map active RNA polymerases and their start sites genome-wide at up to base pair resolution(Core et al., 2008; Kwak et al., 2013). In these methods, the endogenous activity of RNA polymerase is used to selectively label nascent RNAs. The ability to affinity-purify nuclear run-on RNA multiple times during the course of library preparation provides an approximate million-fold enrichment of the nascent RNA over other forms of RNA and thereby effectively eliminates background(Core et al., 2008). Furthermore, because the RNA is sequenced, the direction of transcription can be unambiguously identified.
Development of PRO-seq
PRO-seq is based on Global Run-On sequencing (GRO-seq), a genome-wide adaptation of nuclear run-on assays that have been used classically to measure transcription of target genes. In GRO-seq, bromouridine-labeled nascent RNAs are affinity-purified and analyzed by high-throughput sequencing to map RNA polymerase positions. Extremely high sensitivity and specificity is achieved through multiple distinct affinity-purification steps(Core et al., 2008; Jonkers et al., 2014). GRO-seq uses bromouridine as the substrate for the nuclear run-on reaction, enabling RNA polymerase to add multiple nucleotides to the nascent RNA. Therefore, the resolution of GRO-seq is tens of bases.
However, to understand the molecular mechanisms of transcriptional elongation and promoter-proximal pausing, RNA polymerase mapping at base-pair resolution is required. Such resolution enables mechanistic modeling of how DNA sequences, nucleosomes, or other DNA-binding factors affect RNA polymerase elongation and gene expression(Adelman and Lis, 2012; Kwak and Lis, 2013). To achieve base-pair resolution, we used a modified nuclear run-on that limits the number of labeled nucleotides added to the nascent RNA(Core et al., 2008; Hah et al., 2011; Kwak et al., 2013; Larschan et al., 2011; Min et al., 2011). In PRO-seq, biotin-labeled nucleotide triphosphates are provided as the substrates for the nuclear run-on reaction. The incorporation of a biotin-NTP by an RNA polymerase inhibits further incorporation of biotin-NTPs into the nascent RNA. Sequencing from the 3′ end of the nascent transcript therefore identifies the last incorporated NTP and reveals the precise location of the active site of the RNA polymerase engaged with its nascent RNA.
Identification of the precise position of transcription start sites (TSSs) is also important to understand how DNA elements, general transcription factors and transcription activators recruit RNA polymerase to genes and enhancers. RNA polymerase initiates transcription at TSS and quickly transcribes a short region before pausing at a promoter-proximal site. However, because the nascent transcripts are sequenced from the 3′ end in PRO-seq, the positional information of where RNA polymerase began transcription is mostly lost. We therefore developed PRO-cap by modifying the sequencing strategy of PRO-seq to sequence the capped nascent RNA from 5′ end, enabling transcription start sites to be identified at the RNA synthesis level(Kwak et al., 2013).
Overview of the procedure
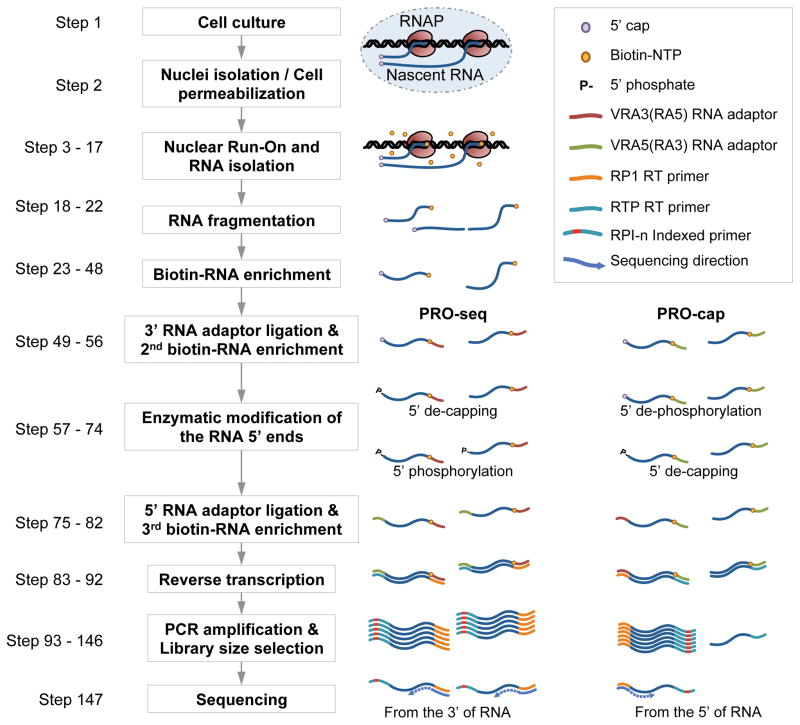
A general overview of the PRO-seq and PRO-cap experimental procedures is shown in Figure A.1. Nuclei from cells are rapidly isolated and native nucleotides are washed away to halt transcription. However, RNA polymerases remain engaged on the DNA and retain their enzymatic activity. Incubation of isolated nuclei with biotin-labeled nucleotide triphosphates allows the RNA polymerases to actively elongate and label the nascent RNA. For PRO-seq only, the labeled nascent RNA is hydrolyzed with NaOH to generate RNA fragments suitable for sequencing (~100 bp in length). The RNA containing a biotin nucleotide is then enriched by affinity purification using streptavidin-coated magnetic beads. The biotin-streptavidin interaction is very stable (Kd ~ 10−14 mol/L), which allows stringent washing of the magnetic beads to minimize contamination with unlabeled RNA. A 3′ sequencing adapter is then ligated to the hydroxyl (OH) group at the 3′end of nascent RNA followed by another affinity purification to further enrich nascent RNA and remove unligated adapter sequences. Preparation for 5′ sequencing adaptor ligation differs for PRO-Seq and PRO-cap. For PRO-seq, the 5′ cap is removed from unhydrolyzed short nascent RNA using either Tobacco acid pyrophosphatase (TAP) or RNA 5′ Pyrophosphohydrolase (RppH). The 5′ OH generated by base hydrolysis is then converted to 5′ phosphate by treatment with T4 polynucleotide kinase (PNK) For PRO-cap, uncapped RNA with a 5′-monophosphate is degraded using 5′-Phosphate-dependent exonuclease. 5′ triphosphates and monophosphates are removed from any remaining contaminating uncapped RNA with Alkaline phosphatase. Only then is the 5′ cap of nascent RNA removed with TAP or RppH treatment. After these chemical modifications, a 5′ sequencing adapter is ligated to the nascent RNA and a third round of affinity purification performed to enrich for nascent RNA with sequencing adapters on both ends. Nascent RNA is then reverse transcribed and test-PCR-amplified to determine an appropriate number of PCR cycles for final amplification; this latter step is critical to avoid over amplification. During the final amplification, barcodes can be added so that multiple libraries can be pooled for sequencing. Finally, the PCR amplified libraries are size-selected for a range of 140–350 bp and sent for high-throughput sequencing. Sequencing depth of 25–50 million for mammalian cells, 10–20 million for organisms with smaller genome size such as Drosophila, and 5–10 million for yeast cells provide useful information. Relatively short read length such as 40–50 bp is sufficient. The sequencing result is a text-based list of short nucleotide sequences and its sequencing quality parameters are provided in a ‘fastq’ format(Cock et al., 2010). The sequences may contain varying lengths of adapter sequences, which are removed, and the adaptor-removed sequences are then aligned to the appropriate genome. Finally, the aligned sequences are used to generate coverage files that can be used to visualize and analyze the data.
Figure A.1. Flowchart of the PRO-seq and PRO-cap protocol.
Stages of the procedure are outlined along with schematics indicating the statuses of the RNA polymerase and the nascent RNA. PRO-seq and PRO-cap share common steps, but are different at the adaptor ligation, reverse transcription, and 5′ modification stages. In PRO-seq, the 3′ RNA adaptor is modified to have the reverse complement sequence of a standard 5′ RNA adaptor. Similarly, the 5′ RNA adaptor and the reverse transcription primer are modified for PRO-seq. This allows 3′ sequencing of the RNA on a standard Illumina platform. For PRO-cap, standard small RNA adaptors are used, and results in a 5′ sequencing as usual. In the 5′ modification step, PRO-seq RNAs are de-capped and re-phosphorylated to ensure all RNA is accessible for 5′ RNA adaptor ligation. PRO-cap RNAs are de-phosphorylated first to convert all forms of 5′ end except for the 5′ capped ends to a 5′ hydroxyl end, in order to restrict the ligation of 5′ adaptor only to the cap containing nascent RNAs after cap removal . Then the 5′ caps are converted to 5′ monophosphates using the tobacco acid pyrophosphatase, and can be ligated to a 5′ RNA adaptor.
Advantages and limitations of PRO-seq
The key advantages of PRO-seq are:
It provides base-pair resolution and strand-specific information of global RNA polymerase occupancy.
Background RNA contamination is hugely reduced due to almost a million fold purification of biotinylated nascent RNAs.
It is highly sensitive in detecting rare and common nascent RNAs with a large dynamic range (>105).
It can identify short unstable nascent RNAs transcribed from enhancer regions.
PRO-seq also has a number of limitations that should be considered when deciding which genome-wide RNA polymerase mapping strategy to use.
In principle, PRO-seq results are ensemble profiles of potentially heterogeneous populations of cells - and this is generally true for all multi-cell, high-throughput sequencing analyses. Unlike mature RNA molecules that are present in multiple copies per cell, RNA polymerase at a specific genomic position can only yield at most two copies of nascent RNA. So while it may be possible to adapt PRO-seq to measure nascent transcript levels for abundantly expressed genes in single cells, genome-wide mapping of RNA polymerase in single cells using PRO-seq would remain a challenge.
PRO-seq detects only the active RNA polymerase and so RNA polymerases in the pre-initiation complex will not be detected. There is also a possibility that other forms of stalled RNA polymerases, such as back-tracked polymerases, may not be detected, although nuclear run-on conditions may allow some of these polymerases to realign the active site through thermal motion. Generally, the signals seen by ChIP-seq of RNA Polymerase II (Pol II) and our genome-wide run-on methods quantitatively agree(Core et al., 2012), so the bulk of Pol II is detectable by GRO- and PRO-seq methods.
Compared to GRO-seq that adds a longer extension to the 3′ end of nascent RNA(Core et al., 2008), PRO-seq only adds one or a few nucleotides in order to provide higher resolution mapping. However, there is a possibility that RNA polymerases positioned very close to the TSS may not be detected because the nascent RNA may not be long enough to be uniquely mapped to the genome. In this case, GRO-seq may provide more accurate quantification of promoter proximal RNA polymerases. Likewise, RNA polymerases positioned in a repetitive sequence region of the genome is difficult to unambiguously map to a particular repeat.
PRO-seq does not distinguish nascent transcription derived from different RNA polymerases (Pol I vs Pol II vs Pol III) unless carried out in the presence of inhibitors of specific RNA polymerases. Also, unlike NET-Seq, nascent RNA associated with specific RNA polymerase modifications (e.g. phosphorylations of the C-terminal domain) cannot be selectively detected using PRO-seq(Nojima et al., 2015).
Applications of PRO-seq and PRO-cap
The most common use of PRO-seq is for the analysis of genome-wide transcription levels with directional information and with higher resolution and sensitivity than an RNA polymerase ChIP-seq assay. PRO-seq provides an independent layer of gene expression analysis distinct from mRNA-seq, revealing the transcriptional stages of regulation before the influence of mRNA processing or stability control. The increased resolution and the directional information become useful in distinguishing upstream divergent (also called upstream antisense) transcription(Core et al., 2008; Seila et al., 2008).
PRO-cap(Kwak et al., 2013) can capture TSS at the nascent RNA synthesis level in contrast to other TSS analyses that use mature RNA(Andersson et al., 2014; Carninci et al., 1996; Forrest et al., 2014). This becomes an advantage in detecting enhancer transcripts(Core et al., 2014; Hah et al., 2013; Wang et al., 2011), upstream antisense transcription(Core et al., 2008; 2014), or other types of unstable transcripts, and avoiding post-transcriptional background capping events(Fejes-Toth et al., 2009).
Alternatives to PRO-seq
RNA polymerase can be mapped genome-wide by a variety of different strategies.
ChIP-seq: In this approach, RNA polymerase proteins are cross-linked to the DNA and then RNA polymerase II (Pol II) is purified by immuno-precipitation. Pol II associated DNA is identified and quantified by high-throughput sequencing thereby providing an estimate of the amount of Pol II at different sites on the genome. The resolution of ChIP is usually limited by the size of the fragmented DNA in the chromatin at the immunoprecipitation step. A variant of this method called ChIP-exo overcomes the resolution limitation by additionally treating the DNA fragments from the Pol II ChIP with a DNA exonuclease(Rhee and Pugh, 2012). The exonuclease digests DNA from the 3′ end of both strands, stopping near the cross-linked polymerase complex. An additional limitation of ChIP-seq is that Pol II-unbound genomic regions that interact with a Pol II-bound region through three-dimensional looping can be falsely identified with this method due to the use of cross-linking. Finally, ChIP-seq will map all forms of Pol II, including transcriptionally inactive Pol IIs and Pol IIs in antisense orientation: therefore, the direction of the transcription is not directly disclosed.
Permanganate footprinting: this method can be used to identify the single stranded DNA created by the transcription bubble formed on DNA by the RNA polymerase. The non-template strand of the DNA is exposed to single-strand specific breakage at T residues through a series of chemical treatments. A method called Permanganate-ChIP-seq couples permanganate footprinting to Pol II ChIP and thereby maps the cleaved ends of the DNA from the single-stranded region of transcription bubbles(Li et al., 2013). This directly maps the transcription active site with high resolution. Permanganate mapping depends on the presence of thymine residues in the non-templated single-stranded DNA of the bubble that are not masked by protein binding. Although the protocol enriches for Pol II in a single chromatin immunoprecipitation, other regions that expose single stranded thymine such as in other DNA-RNA hybrids or intra-strand DNA hairpins could contribute to background.
Native Elongating Transcript sequencing (NET-seq): a number of chromatin bound nascent RNA based methods, including NET-seq and its variants, have been developed for mapping RNA polymerase(Churchman and Weissman, 2011; Mayer et al., 2015; Nojima et al., 2015; Weber et al., 2014). In the original NET-seq protocol(Churchman and Weissman, 2011), the RNA polymerase complex is immunoprecipitated and the co-purified native RNA is sequenced. The 3′ end of the nascent RNA provides base-pair resolution mapping of RNA polymerase. This method is ideally suited to examine the occupancy of differently modified RNA polymerases. In practice, the efficiency of NET-seq relies on the degree of enrichment provided by the single immuno-precipitation step. Because the method detects the 3′ ends of all RNAs that are associated with Pol II, it also captures 3′ ends of intermediates of co-transcriptional splicing and micro-RNA production that can complicate mapping of transcriptionally-engaged Pol II(Nojima et al., 2015).
Experimental Design
Cells
In our lab, we have successfully generated PRO-Seq libraries for cells from plant (unpublished, G.T.B.), yeast (unpublished, G.T.B.), Drosophila(Kwak et al., 2013), and mammals(Core et al., 2014; Mahat et al., 2016). In general, the higher the number of cells the better the PRO-seq read coverage of the genome. However, a minimum of 5–10 million nuclei or permeabilized cells is required for a single PRO-seq library regardless of cell type. In principle, the application of PRO-seq in yeast, including S. pombe and S. cerevisiae, is very similar to that of other organisms; however, some alterations are required in yeast cell permeabilization(García-Martínez et al., 2004), run-on reaction, and post run-on RNA extraction procedures(Collart and Oliviero, 2001); required modifications for yeast are indicated in the appropriate steps of the Procedure.
Sample preparation
Isolation of nuclei for nuclear run-on is a critical step in the procedure not only to preserve the enzymatic activity of the RNA polymerase, but also to capture the precise position of the RNA polymerase on genes. Starting with 10–20 million cells per library is recommended considering the efficiency of nuclei isolation process (~50%). The whole process should take place in the cold room on ice as far as is possible. Isolated nuclei can be resuspended in the glycerol-containing storage buffer, and quickly frozen in liquid nitrogen for long term storage at −80°C. We have used permeabilized cells in PRO-seq as an alternative to isolating nuclei, making handling easier and reducing loss of sample; cell permeabilization has a much higher efficiency (~90%) than nuclei isolation. Cell permeabilization conditions may differ between cell types and may need to be optimized; we provide a general method for permeabilization in the Procedure as well as a version optimized for yeast cells.
Spike-in for library normalization
Disproportionate loss of RNA and/or cDNA can occur during multiple stages of the PRO-seq library preparation, which spans 4–5 days and involves several handling steps. Even with the use of identical starting material, uneven loss of libraries could affect the genome-wide RNA polymerase density between libraries. To control for handling effects on library yield, a small fraction (1–5%) of cells with a distinct genome can be added during library preparation; adding an identical number of spike-in cells to different libraries enables normalization between different conditions. We have used Schizosaccharomyces pombe to normalize Saccharomyces cerevisiae and vice versa, and Drosophila cells to normalize mammalian cells and vice versa. When using the cell-permeabilization approach, spike-in cells should be added and permeabilized together with the experimental cells. For the nuclei isolation approach, spike-in cells should be added to the experimental cells prior to nuclei isolation and dounced together.
Nuclear run-on
In PRO-seq, biotin-NTPs are used as the nuclear run-on substrates. The Km of each of the NTPs as substrates for RNA polymerase lie in the range of 1–20 μM(Job et al., 1984). Therefore, final substrate concentration greater than 1–20 μM range (~25 μM) is, in general, sufficient for each biotin-NTP substrate. Depending on the purpose of the experiment, biotin-NTP substrates can be supplied in different combinations: individual biotin run-on, 4 biotin run-on, 2 biotin run-on or 1 biotin run-on.
Individual-biotin run-on: To obtain the most precise mapping of the RNA polymerase, four separate PRO-seq libraries are made, each supplied with only one type of biotin-NTP in the nuclear run-on reaction. This ensures that the RNA polymerase adds only one, or at most a few (when the polymerase is positioned at multiple stretches of same nucleotide) biotin-NTPs to the nascent RNA. In this case, 4 times more sample is required.
4-Biotin run-on: we found that all 4 biotin-NTPs can be supplied in a single reaction and the Pol II only incorporates one or at most a few bases, giving an equivalent resolution to single biotin run-on. The reason for this is unclear, but we speculate that steric hindrance in the active site of RNA polymerase prevents incorporation of multiple biotinylated nucleotides.
2-Biotin run-on: When the amount of sample or the cost is limiting, unlabeled NTPs can be used in combination with biotin NTPs. Use of biotinylated purine nucleotides (biotin-ATP, biotin-GTP) is more costly than that of the pyrimidine nucleotides (biotin-CTP, biotin-UTP). A combination of biotin-CTP, biotin-UTP, ATP, and GTP can be supplied to the nuclear run-on reaction, providing reasonable resolution and cost.
1-Biotin run-on: If a longer run-on RNA is preferred, combinations of biotin-CTP with unlabeled CTP, UTP, ATP, and GTP can be used effectively in a biotin-NTP form of GRO-seq. This approach can be useful for increasing sequencing coverage of RNA polymerases that reside near the TSSs. While most transcriptionally-engaged RNA polymerases near the 5′ ends reside between 30–60 nucleotides from the TSS(Kwak et al., 2013), RNA polymerases closer to the TSS may fail to map uniquely. Additionally, the longer run-on extensions of nascent RNAs may be desired for distinguishing allele-specific nascent transcription.
PCR amplification of PRO-seq library
When the number of cells and/or nuclei are limited for PRO-seq library preparation, a higher number of PCR amplification cycles will be required to generate sufficient library for reliable quantification and accurate loading into the sequencer. However, a higher number of PCR cycles can result in amplification bias of some sequences. To avoid PCR-induced biases, molecular barcodes(Fu et al., 2014) can be introduced as part of the 3′ RNA adapter, which is ligated to the nascent RNAs. Duplicate reads generated by PCR over-amplification can be identified by identical barcodes and computationally filtered at the stage of mapping the sequenced reads to the genome.
MATERIALS
REAGENTS
CRITICAL: Extreme care should be taken to avoid nuclease contamination. Use nuclease-free reagents and change gloves routinely.
-
Appropriate cell line(s) e.g. K562, GM12878, MCF7, Hela, embryonic stem cells, mouse embryonic fibroblasts, mouse 3T3 cells, Drosophila S2, yeast.
CAUTION: Before use, cells should be checked for contamination.
Chemical stocks
-
Diethyl pyrocarbonate (DEPC; Sigma-Aldrich, cat. no. D5758)
CAUTION: DEPC is toxic and harmful. Proper eyeshield, faceshield, full-face respirator, and gloves are required while handling DEPC.
Sodium chloride, NaCl (Sigma-Aldrich, cat. no. S9888)
Potassium chloride, KCl (Avantor, cat. no. 6858-04)
Magnesium chloride, MgCl2 (Avantor, cat. no. 5958-04)
Sucrose (Sigma-Aldrich, cat. no. S0389)
Calcium chloride, CaCl2 (Sigma-Aldrich, cat. no. C1016)
Magnesium acetate, MgAc2 (Sigma-Aldrich, cat. no. M5661)
Ammonium acetate, NH4Ac (Sigma-Aldrich, cat. no. A1542)
Sodium acetate NaOAc (Sigma-Aldrich, cat. no. S2889)
EDTA (Sigma-Aldrich, cat. no. E9884)
EGTA (Sigma-Aldrich, cat. no. E3889)
Protease inhibitor cocktail, EDTA-free (Roche, cat. no. 11873580001)
Sodium hydroxide, NaOH (Avantor, cat. no. 7708-10)
Triton X-100, (Calbiochem, cat. no. 9410)
Nonidet P40 (NP40) Substitute, (Sigma-Adlrich, cat. no. 11332473001)
Sarkosyl (Sigma-Aldrich, cat. no. L5125)
Tween-20 (Sigma-Aldrich, cat. no. P9416)
Phosphate buffer saline, PBS pH 7.4 (Gibco, cat. no. 10010031).
TRIS (Avantor, cat. no. 4109-02)
Hydrochloric acid, HCl (Avantor, cat. no. 4613-05)
DTT (Sigma-Aldrich, cat. no. D0632)
Betaine (Sigma-Aldrich, cat. no. B0300)
Glycerol (Sigma-Aldrich, cat. no. G5516)
Biotin Nuclear Run-On and enrichment
Biotin-11-ATP (PerkinElmer, cat. no. NEL544001EA)
Biotin-11-CTP (PerkinElmer, cat. no. NEL542001EA)
Biotin-11-GTP (PerkinElmer, cat. no. NEL545001EA)
Biotin-11-UTP (PerkinElmer, cat. no. NEL543001EA)
ATP, 10mM (Roche, cat. no. 11 277 057 001)
GTP, 10mM (Roche, cat. no. 11 277 057 001)
UTP, 10mM (Roche, cat. no. 11 277 057 001)
P-30 column, RNase free (BIORAD, cat. no. 732-6250)
Streptavidin M280 beads (Invitrogen, cat. no. 112.06D)
Reagents for nucleic acid extraction
-
Trizol (Ambion, cat. no. 115596018)
CAUTION: Trizol is harmful and contact with skin, eye or inhalation should be avoided. Use it inside a fume hood.
-
Trizol LS (Ambion, cat. no. 10296028)
CAUTION: Trizol is harmful and contact with skin, eye or inhalation should be avoided. Use it inside a fume hood.
Chloroform (Calbiochem, cat. no. 3150)
GlycoBlue (Ambion, cat. no. AM9515)
Ethanol, 100% (PHARMCO-AAPER, cat. no. 111000200)
Ethanol, 75%(vol/vol)
-
Phenol:Chloroform, Tris buffered (Thermo Scientific, cat. no. 17909)
CAUTION: Phenol:Chloroform is harmful and contact with skin, eye or inhalation should be avoided. Use it inside a fume hood.
-
Phenol, (Ambion, cat. no. 9700).
CAUTION: Phenol is harmful and contact with skin, eye or inhalation should be avoided. Use it inside a fume hood.
Enzymes and recombinant protein reagents
RNase inhibitor, 40 units/μl (Ambion, cat. no. AM2696)
T4 RNA ligase I, 10 units/μl (NEB, cat. no. M0204). Supplied with 10× T4 RNA ligase buffer, 10 mM ATP, and PEG, 50%(wt/vol).
5′-phosphate-dependent exonuclease, 1 unit/μl (Epicenter, cat. no. TER51020) (required for PRO-cap only). Supplied with 10× reaction buffer A.
Alkaline phosphatase, 10 units/μl (NEB, cat. no. M0290) (required for PRO-cap only). Supplied with 10× Alkaline phosphatase buffer. Alternatively, Antarctic phosphatase, 5 units/μl (NEB, cat. no. M0289) can be used.
Tobacco acid pyrophosphatase, 10 units/μl (TAP) (Epicenter, cat. no. T19500). Supplied with 10× TAP buffer. Alternatively, RNA 5′ Pyrophosphohydrolase, 5 units/μl (RppH) (NEB, cat. no. M0356S) can be used with ThermoPol Reaction buffer (NEB, cat. no. B9004S).
T4 polynucleotide kinase, 10 units/μl (PNK) (NEB, cat. no. M0201) (required for PRO-seq only). Supplied with10× PNK buffer.
Superscript III reverse transcriptase (Invitrogen, cat. no. 56575). Supplied with 5× first strand buffer, and 0.1M DTT.
dNTP mix, 12.5 mM each (Roche, cat. no. 03 622 614 001)
Phusion polymerase, 2 units/μl (NEB, cat. no. M0530). Supplied with 5× High-Fidelity buffer.
RNA and DNA oligos. (Custom synthesis from IDT DNA, RNase-free HPLC purified) See Table 1 and Reagent Setup for details. Further information about barcoding and sequencing indexes can be found at http://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/samplepreps_truseq/truseqsampleprep/truseq-library-prep-pooling-guide-15042173-01.pdf
Table 1.
Oligonucleotides required for PRO-seq and PRO-cap.
| Oligo name | Sequence (5′ to 3′) | Purpose | Comments | |
|---|---|---|---|---|
| Oligos for PRO-seq | VRA3 | GAUCGUCGGACUGUAGAACUCUGAAC-/Inverted dT/ | RNA adaptor for ligation to the 3′ end of nascent RNA at step 49 | The 5′ end is phosphorylated and the 3′ end is protected by an inverted dT |
| VRA5 | CCUUGGCACCCGAGAAUUCCA | RNA adaptor for ligation to the 5′ end of nascent RNA at step 75 | The 5′ end is not phosphorylated | |
| RP1 | AATGATACGGCGACCACCGAGATCTACACGTTCAGAGTTCTACAGTCCGA | DNA oligo for reverse transcription of adaptor-ligated nascent RNA at step 84 | ||
| Oligos for PRO-cap | RA3 | UGGAAUUCUCGGGUGCCAAGG-/Inverted dT/ | RNA adaptor for ligation to the 3′ end of nascent RNA at step 49 | The 5′ end is phosphorylated and the 3′ end is protected by an inverted dT |
| RA5 | GUUCAGAGUUCUACAGUCCGACGAUC | RNA adaptor for ligation to the 5′ end of nascent RNA at step 75 | The 5′ end is not phosphorylated | |
| RTP | GCCTTGGCACCCGAGAATTCCA | DNA oligo for reverse transcription of adaptor-ligated nascent RNA at step 84 | ||
| PCR primers for library amplification | RP1 | AATGATACGGCGACCACCGAGATCTACACGTTCAGAGTTCTACAGTCCGA | DNA oligo for PCR amplification of cDNA in both PRO-seq and PRO-cap at steps 94 & 102 | Same as the RT oligo for PRO-seq |
| RPI-n | CAAGCAGAAGACGGCATACGAGAT NNNNNN GTGACTGGAGTT CCTTGGCACCCGAGAATTCCA | DNA oligo with barcodes for PCR amplification of cDNA in both PRO-seq and PRO-cap at steps 94 & 104 | The six Ns represent the barcodes for Illumina TRU-seq multiplexing. For example, the barcode in RPI-1 is CGTGAT |
Electrophoresis
DNA grade agarose (BioRad, cat. no. 161-3102EDU)
Tris/Acetic acid/EDTA (TAE), 50× (BioRad, cat. no. 161-0773). Alternatively, a 50x TAE buffer can be made in house (see Reagent Setup section)
Glacial acetic acid (Sigma-Aldrich, cat. no. 537020) for in house preparation of 50x TAE.
Gel loading dye, Orange G 6× (NEB, cat. no. B7022S)
100 bp DNA ladder (Life Technologies, cat. no. 15628-019)
10 bp DNA ladder (Life Technologies, cat. no. 10821-015). Alternatively, 25 bp DNA ladders (ThermoFisher, cat. no. 10597011) can be used.
SYBRGold nucleic acid gel stain, 6× (Life Technologies, cat. no. S-11494)
Acrylamide (Protogel), 30%(wt/vol) (National Diagnostics, cat. no. EC-980). 30% acrylamide/methylene bisacrylamide solution from other sources is also compatible.
TEMED (BioRad, cat. no. 161-0800)
Ammonium Persulfate (APS), 10%(wt/vol) (BioRad, cat. no. 161-0700). Dissolve in H2O.
Tris/Boric acid/EDTA (TBE), 10× (BioRad, cat. no. 161-0770). Alternatively, a 10× TBE buffer can be made in house (see Reagent Setup section).
Boric acid (Sigma-Aldrich, cat. no. B6768) for in house preparation of 10x TBE.
Software for data analysis
Adaptor removal software, such as ‘cutadapt’(Marcel, 2011) (http://code.google.com/p/cutadapt/)
Mapping or alignment software, such as ‘bwa’(Li and Durbin, 2009) (https://sourceforge.net/projects/bio-bwa/files/) or ‘bowtie’(Langmead et al., 2009) (https://sourceforge.net/projects/bowtie-bio/files/)
Tools for generation of coverage information, such as SAMtools(Li et al., 2009) (https://sourceforge.net/projects/samtools/files/) and BEDTools(Quinlan and Hall, 2010) (https://sourceforge.net/projects/bedtools/files/)
EQUIPMENT
2 heat blocks, one set at 37°C and the other at 65°C, each filled with water equilibrated at the appropriate temperature.
Dounce homogenizer (Wheaton scientific, cat. no. 357546)
Magnetic separator for Streptavidin magnetic beads (Invitrogen, cat. no. K1585-01)
Rotating stand (Thermo Barnstead Labquake rotator, cat. no. 415110)
Refrigerated centrifuge (Eppendorf, cat. no. 5417R)
Microcentrifuge (Eppendorf, cat. no. 5415D)
Speed vac dryer (Thermo Scientific, cat. no. 20-548-134)
Dark Reader transilluminator (Clare Chemical Research, cat. no. DR89X)
REAGENT SETUP
CRITICAL: All reagents, solutions, and buffers should be made with DEPC-treated water
DEPC-H2O
Add 0.1%(vol/vol) DEPC to H2O. Mix overnight then autoclave and filter-sterilize the solution with a 0.22 μm filter. DEPC-H2O can be prepared in advance and stored at room temperature (25°C) for up to a year.
CAUTION: DEPC is toxic and harmful. Proper eyeshield, faceshield, full-face respirator, and gloves are required while handling DEPC.
5M NaCl
Dissolve 14.61 g NaCl in 50 ml H2O with 0.1%(vol/vol) DEPC. Mix overnight and then autoclave and filter.
CRITICAL: 5M NaCl can be prepared in advance and stored at room temperature (25°C) for up to a year.
4M KCl
Dissolve 3.73 g KCl in 50 ml H2O with 0.1%(vol/vol) DEPC, mix overnight then autoclave and filter-sterilize.
CRITICAL: 4M KCl can be prepared in advance and stored at room temperature for up to a year.
1M MgCl2
Dissolve 4.76 g MgCl2 in 50 ml H2O with 0.1%(vol/vol) DEPC, mix overnight then autoclave and filter-sterilize.
CRITICAL: 1M MgCl2 can be prepared in advance and stored at room temperature for up to a year.
1M Sucrose
Dissolve 171.15 g Sucrose in 500 ml H2O with 0.1%(vol/vol) DEPC, mix overnight then autoclave and filter-sterilize.
CRITICAL: 1M Sucrose can be prepared in advance and stored at room temperature for up to a year.
1M CaCl2
Dissolve 5.55 g CaCl2 in 50 ml H2O with 0.1%(vol/vol) DEPC, mix overnight then autoclave and filter-sterilize.
CRITICAL: 1M CaCl2 can be prepared in advance and stored at room temperature for up to a year.
1M MgAc2
Dissolve 7.12 g MgAc2 in 50 ml H2O with 0.1%(vol/vol) DEPC, mix overnight then autoclave and filter-sterilize.
CRITICAL: 1M MgAc2 can be prepared in advance and stored at room temperature for up to a year.
1M NH4Ac
Dissolve 3.85 g NH4Ac in 50 ml H2O with 0.1%(vol/vol) DEPC, mix overnight then autoclave and filter-sterilize.
CRITICAL: 1M NH4Ac can be prepared in advance and stored at room temperature for up to a year.
1M NaOAc, pH 5.3
Dissolve 4.1 g NaOAc in 50 ml H2O and pH to 5.3, add 0.1%(vol/vol) DEPC, mix overnight then autoclave and filter-sterilize.
CRITICAL: 1M NaOAc can be prepared in advance and stored at room temperature for up to a year.
0.5M EDTA
Dissolve 29.22 g EDTA in 100 ml DEPC treated H2O, then autoclave and filter-sterilize.
CRITICAL: 0.5M EDTA can be prepared in advance and stored at room temperature for up to a year.
0.1M EGTA
Dissolve 19.02 g EGTA in 50 ml DEPC treated H2O, then autoclave and filter-sterilize.
CRITICAL: 0.1M EGTA can be prepared in advance and stored at room temperature for up to a year.
1N NaOH
Dissolve 2 g NaOH in 50 ml DEPC treated H2O, filter-sterilize.
CRITICAL: 1N NaOH can be prepared in advance in 50 ul aliquots, stored at −80°C for up to a year. Use freshly thawed aliquot each time.
CAUTION: NaOH is corrosive and contact with skin, eye or inhalation should be avoided.
10% Triton X-100
Dissolve 5 ml of Triton X-100 in 45 ml DEPC H2O and filter-sterilize.
CRITICAL: 10% Triton X-100 can be prepared in advance and stored at room temperature for up to a year.
10% NP40
Dissolve 5 ml of NP40 in 45 ml DEPC H2O and filter-sterilize.
CRITICAL: 10% NP40 can be prepared in advance and stored at room temperature for up to a year.
2% Sarkosyl
Dissolve 1 g of Sarkosyl in 50 ml DEPC H2O and filter-sterilize.
CRITICAL: 2% Sarkosyl can be prepared in advance and stored at room temperature for up to a year. CAUTION: Sarkosyl is an irritant and contact with skin, eye or inhalation should be avoided.
1% Tween-20
issolve 1 ml of Tween-20 in 49 ml DEPC H2O and filter-sterilize.
CRITICAL: 1% Tween-20 can be prepared in advance and stored at room temperature for up to a year.
1M Tris-HCl, pH 6.8
Dissolve 6.06 g TRIS base in 50 ml DEPC H2O, pH to 6.8 with HCl then autoclave and filter-sterilize.
CRITICAL: The buffer can be prepared in advance and stored at room temperature for up to a year.
1M Tris-HCl, pH 7.4
Dissolve 6.06 g TRIS base in 50 ml DEPC H2O, pH to 7.4 with HCl then autoclave and filter-sterilize.
CRITICAL: The buffer can be prepared in advance and stored at room temperature for up to a year.
1M Tris-HCl, pH 8.0
Dissolve 6.06 g TRIS base in 50 ml DEPC H2O, pH to 8.0 with HCl then autoclave and filter-sterilize.
CRITICAL: The buffer can be prepared in advance and stored at room temperature for up to a year.
1M DTT
Dissolve 1.54 g DTT in 10 ml DEPC H2O and filter-sterilize.
CRITICAL: 1M DTT can be prepared in advance and stored at −20°C for up to a year.
1mM Biotin-11-CTP
Mix 10 μl of 10 mM stock in 90 μl DEPC H2O to get 1 mM dilution.
CRITICAL: The buffer can be prepared in advance and stored at 4°C for up to a year.
1mM Biotin-11-UTP
Mix 10 μl of 10 mM stock in 90 μl DEPC H2O to get 1 mM dilution.
CRITICAL: The buffer can be prepared in advance and stored at 4°C for up to a year.
50x TAE
Dissolve 121 g TRIS base, 28.55 g Glacial Acetic acid, and 50 ml 0.5M EDTA in DEPC H2O to final volume of 500 ml then autoclave and filter-sterilize.
CRITICAL: The buffer can be prepared in advance and stored at 4°C for up to a month.
10x TBE
Dissolve 54 g TRIS base, 27.5 g Boric acid, and 20 ml 0.5M EDTA in DEPC H2O to final volume of 500 ml then autoclave and filter-sterilize.
CRITICAL: The buffer can be prepared in advance and stored at 4°C for up to a month.
Douncing buffer (for nuclei isolation)
10 mM Tris-HCl pH 7.4, 300mM sucrose, 3 mM CaCl2, 2 mM MgCl2, 0.1% Triton X-100, 0.5 mM DTT, 1 tablet of protease inhibitors cocktail per 50ml, 4 u/ml RNase inhibitor.
CRITICAL: The buffer without DTT, protease inhibitors, and RNase inhibitor can be prepared and stored at 4°C for up to a month. Add fresh DTT, protease inhibitors, and RNase inhibitor immediately before use.
Permeabilization buffer (for non-yeast cells)
10mM Tris-HCl pH 7.4, 300mM sucrose, 10mM KCl, 5mM MgCl2, 1mM EGTA, 0.05% Tween-20, 0.1% Nonidet P40 substitute, 0.5 mM DTT, 1 tablet of protease inhibitors cocktail per 50ml, 4 u/ml RNase inhibitor.
CRITICAL: The buffer without DTT, protease inhibitors, and RNase inhibitor can be prepared and stored at 4°C for up to a month. Add fresh DTT, protease inhibitors, and RNase inhibitor immediately before use.
Permeabilization buffer (for yeast cells)
0.5% Sarkosyl, 0.5 mM DTT, 1 tablet of protease inhibitors cocktail per 50ml, 4 u/ml RNase inhibitor.
CRITICAL: The buffer without DTT, protease inhibitors, and RNase inhibitor can be prepared and stored at 4°C for up to a month. Add fresh DTT, protease inhibitors, and RNase inhibitor immediately before use.
Storage buffer
10 mM Tris-HCl pH 8.0, 25%(vol/vol) glycerol, 5 mM MgCl2, 0.1 mM EDTA, 5 mM DTT.
CRITICAL: The buffer without DTT can be prepared and stored at 4°C for up to a month. Add fresh DTT immediately before use.
2x Nuclear run-on master mix (for non-yeast cells)
10 mM Tris-HCl pH 8.0, 5 mM MgCl2, 300 mM KCl, and 1 mM DTT.
CRITICAL: The buffer without DTT can be prepared and stored at 4°C for up to a month. Add fresh DTT immediately before use.
2x NRO master mix (for yeast cells)
40 mM Tris-HCl, pH 7.7, 400 mM KCl, 64 mM MgCl2, 1 mM DTT.)
CRITICAL: The buffer without DTT can be prepared and stored at 4°C for up to a month. Add fresh DTT immediately before use.
AES buffer (for yeast cells only)
50 mM NaOAc pH 5.3, 10 mM EDTA, 1% SDS.
High-salt wash buffer
50 mM Tris-HCl pH 7.4, 2 M NaCl, 0.5%(vol/vol) Triton X-100 in DEPC H2O.
CRITICAL: The buffer can be prepared in advance and stored at 4°C for up to a month.
Binding buffer
10 mM Tris-HCl pH 7.4, 300 mM NaCl, 0.1%(vol/vol) Triton X-100 in DEPC H2O.
CRITICAL: The buffer can be prepared in advance and stored at 4°C for up to a month.
Low-salt wash buffer
5 mM Tris-HCl pH 7.4, 0.1%(vol/vol) Triton X-100 in DEPC H2O.
CRITICAL: The buffer can be prepared and stored at 4°C for up to a month.
Pre-washed streptavidin-coated magnetic beads
Take 90 μl of Streptavidin M280 beads per library. Place the beads on the magnetic separator for 1 min and discard the supernatant. Pre-wash by resuspending in 0.1 N NaOH + 50 mM NaCl in DEPC H2O for 1 min, place on the magnetic separator for 1 min, remove supernatant. Wash beads twice with 100 mM NaCl in DEPC H2O. After removing the wash buffer, resuspend the beads in 150 μl of the Binding Buffer and make 3 aliquots of 50 μl each. Scale up accordingly when processing multiple samples.
CRITICAL: The washed beads can be prepared in advance and stored at 4°C for up to a week.
2.2% Agarose gel
3.3 grams DNA grade agarose in 150 ml 1x TAE. Mix by swirling and heat using a lab microwave until the mix bubbles and looks clear.
Gel elution buffer
10 mM Tris-HCl pH 8.0, 0.5 mM NH4Ac, 10 mM MgAc2, 1 mM EDTA, 0.1% SDS.
CRITICAL: The buffer can be prepared in advance and stored at room temperature for up to a month.
DNA and RNA oligos
Oligos for PRO-seq and PRO-cap should be dissolved in DEPC H2O at a concentration of 100 mM. PCR primers should be dissolved in DEPC H2O at a concentration of 25 mM.
CRITICAL: The DNA and RNA oligos can be stored at −80°C for up to 10 years.
Ammonium persulfate
Dissolve 1 g of APS in 10 ml of DEPC-H2O and filter-sterilize the solution with a 0.22-μm filter.
CRITICAL: 10% (wt/vol) APS can be prepared in advance and stored at −20 °C for up to a year.
PROCEDURE
Cell culture | TIMING 24 h
-
1
Seed cells at a concentration that will enable them to reach ~80% confluency in 24 hours. For a PRO-seq experiment using adherent fibroblasts, 4–6 150mm cell culture dishes yield sufficient cells (~107 cells, see Experimental Design for further details). For yeast cells, plate them to ensure they are in the exponential phase of growth (OD600 = 0.5) at the time of harvest.
CAUTION: Check cell lines for mycoplasma contamination before setting up the experiment.
Sample preparation | TIMING: 1 h
CRITICAL: Samples should be prepared in cold room (4°C) to avoid unsolicited run-on.
-
2
Prepare samples by isolating nuclei (Option A) or by cell permeabilization (use Option C for yeast cells and Option B for other cell types).
All centrifugation steps for sample preparation are performed in a cold centrifuge (4°C) at 1000 g (unless stated otherwise) for 5 min.
A. Nuclei isolation
Harvest adherent cells by scraping and centrifuging, and non-adherent cells by centrifuging.
Resuspend the cell pellet in 10 ml ice-cold PBS and centrifuge.
-
Resuspend the cell pellet in ice-cold douncing buffer (1x106 cells/ml).
CRITICAL STEP: If using spike-in cells, add them at this point to the cells resuspended in douncing buffer.
Incubate for 5 min on ice and dounce 25 times using a dounce homogenizer.
Transfer the dounced nuclei to either a 15 or 50 ml conical tube and centrifuge the nuclei.
Wash twice by resuspending the pellet in 5 ml douncing buffer and centrifuging.
-
Resuspend the pellet in storage buffer (5–10x106 nuclei per 100 ml of storage buffer), flash freeze in liquid nitrogen, and store at −80°C.
PAUSE POINT: The nuclei in storage buffer can be stored at −80°C for up to 5 years.
B. Cell permeabilization (non-yeast cells)
Harvest adherent cells by scraping and centrifuging, and non-adherent cells by centrifuging.
Resuspend the cell pellet in 10 ml ice-cold PBS and centrifuge.
-
Resuspend the cell pellet in ice-cold permeabilization buffer (1x106 cells/ml).
CRITICAL STEP: Spike-in cells, if used, should be added to the cells resuspended in permeabilization buffer.
Incubate for 5 min on ice and centrifuge the permeabilized cells.
Wash twice by resuspending in 5 ml permeabilization buffer and centrifuging.
-
Resuspend the cell pellet in storage buffer (5–10x106 permeabilized cells per 100 μl of storage buffer), flash freeze in liquid nitrogen, and store in −80°C.
PAUSE POINT: The permeabilized cells in storage buffer can be stored at −80°C for up to 5 years.
C. Cell permeabilization (optimized for yeast)
Harvest exponentially growing yeast cells by centrifugation at 400 g.
Resuspend the cell pellet in 10 ml ice-cold PBS and centrifuge.
-
Resuspend the cell pellet in ice-cold yeast permeabilization buffer (1x106 cells/ml).
CRITICAL STEP: Spike-in cells, if used, should be added to the cells resuspended in yeast permeabilization buffer.
Incubate for 20 min on ice and centrifuge the cells at 400 g.
-
Resuspend the cell pellet in storage buffer (25–50x106 permeabilized cells per 100 μl of storage buffer), flash freeze in liquid nitrogen, and store in −80°C.
PAUSE POINT: The permeabilized cells in storage buffer can be stored at −80°C for up to 5 years.
Nuclear run-on | TIMING: 1.5 h
-
3
Prepare a 2x nuclear run-on (NRO) master mix; for non-yeast cells, prepare the master mix according to the first table, for yeast cells, use the second table.
Reagents Volume per reaction (μl) Final concentration – 1x (in 200-μl reaction) (mM) Tris-Cl pH 8.0 (1 M) 1 5 MgCl2 (1 M) 0.5 2.5 DTT (0.1 M) 1 0.5 KCl (4 M) 7.5 150 DEPC-H2O 18 Reagents Volume per reaction (μl) Final concentration – 1x (in 200-μl reaction) (mM) Tris-Cl pH 7.7 (1 M) 4 20 MgCl2 (1 M) 6.4 32 DTT (0.1 M) 1 0.5 KCl (4 M) 10 200 DEPC-H2O 6.6 -
4
Depending on the type of run-on experiment (see Experimental Design section), prepare a 2x reaction mix according to Option A (single biotin run-on), Option B (4 biotin run-on), Option C (2 biotin run-on) or Option D (1 biotin run-on). If processing multiple libraries at once, scale up accordingly (Supplementary Table 1).
A. Individual-biotin run-on 2x reaction mix
Transfer a 28 μl aliquot of NRO master mix to each of 4 separate microcentrifuge tubes.
Add 5 μl of biotin-11-ATP (1 mM) to one of the tubes containing NRO master mix. Label this mix “A”
Repeat step ii for the remaining 3 biotin-11-NTPs (1 mM each) and the 3 remaining tubes containing NRO master mix and label them “C”, “G”, “U” accordingly.
Add 15 μl DEPC H2O to all 4 tubes.
Add 2 μl of RNase inhibitor and 50 μl of 2% Sarkosyl to all 4 tubes. From step 5, each tube will be processed as a separate sample.
B. 4-Biotin run-on 2x reaction mix
Transfer a 28 μl aliquot of NRO master mix to a microcentrifuge tube.
Add 5 μl each of all 4 biotin-11-NTPs (1 mM each) to the NRO master mix aliquot.
Add 2 μl of RNase inhibitor and 50 μl of 2% Sarkosyl.
C. 2-Biotin run-on 2x reaction mix
Transfer a 28 μl aliquot of NRO master mix to a microcentrifuge tube.
Add 5 μl each of biotin-11-CTP (1 mM) and biotin-11-UTP (1 mM) to the NRO master mix aliquot.
Add 2.5 μl each of ATP (10 mM) and GTP (10 mM) to the mix.
Add 5 μl DEPC H2O.
Add 2 μl of RNase inhibitor and 50 μl of 2% Sarkosyl.
D. 1-Biotin run-on 2x reaction mix
Transfer a 28 μl aliquot of NRO master mix to a microcentrifuge tube.
Add 5 μl of biotin-11-CTP (1 mM) to the NRO master mix aliquot.
Add 1 μl of CTP (0.05 mM) to the mix.
Add 2.5 μl each of ATP (10 mM), GTP (10 mM), and UTP (10 mM) to the mix.
Add 6.5 μl DEPC H2O.
Add 2 μl of RNase inhibitor and pipette up and down several times.
Add 50 μl of 2% Sarkosyl and pipette up and down 15 times.
-
5
Preheat 100 μl of the appropriate 2x reaction mix prepared in step 4 to 37°C for mammalian cells or 30°C for yeast and insect cells.
-
6
Using a cut-off P200 pipette tip, add 100 μl nuclei or permeabilized cells (in storage buffer from step 2) to 100 μl of preheated 2x reaction mix, gently but thoroughly pipette the reaction 15 times, and place in a heat block at the appropriate temperature.
CRITICAL STEP: Sarkosyl in the 2x reaction mix causes the run-on reaction to become very viscous (except for yeast). When adding the nuclei or permeabilized cells to the reaction mix and when mixing by pipetting up and down, use a wide bore pipette tip or cut the last centimeter off a normal one with ethanol wiped clean scissors or razor blade.
-
7
Incubate for 3 min (5 min for yeast cells), with gentle tapping at the incubation midpoint.
RNA extraction | TIMING: 1 h
-
8
Extract RNA using Option A for non-yeast nuclei or permeabilized cells or Option B for yeast.
A. RNA extraction from non-yeast nuclei or permeabilized cells
Add 500 μl Trizol LS and mix well by vortexing to stop the reaction.
Incubate the homogenized sample for 5 min at room temperature (25°C) to allow the complete dissociation of nucleoprotein complexes and add 130 μl Chloroform.
Vortex sample vigorously for 15 s and incubate at room temperature for 2 to 3 min.
Centrifuge at 14,000 g for 5 min at 4°C, transfer the aqueous phase to a new tube, and add 1 μl GlycoBlue.
B. RNA extraction from yeast cells or nuclei
Pellet cells or nuclei after the run-on reaction at 400 g for 5 mins at 4 °C and quickly resuspend in 500 μl phenol.
Add an equal volume of AES buffer and incubate it at 65°C for 5 min with periodic vortexing. Let the mixture rest on ice for 5 min, and then add 200 μl of chloroform.
Vortex sample vigorously for 15 s and incubate at room temperature for 2 to 3 min.
Centrifuge at 14,000 g for 5 min at 4°C, transfer the aqueous phase to a new tube, and add 1 μl GlycoBlue and NaOAc to 200 mM final concentration.
-
9
Add 2.5x volume of 100% room temperature ethanol & vortex for 10 s.
-
10
Incubate samples at room temperature for 10 min.
-
11
Centrifuge at 14,000 g for 20 min at 4°C. The RNA precipitate forms a gel-like pellet on the side and bottom of the tube.
-
12
Remove supernatant completely.
-
13
Add 750 μl of 75% ethanol.
PAUSE POINT: The RNA pellet in 75% ethanol can be stored up to a week at −80°C.
-
14
Mix by vortexing and centrifuge at 14,000 g for 5 min at 4°C.
-
15
Remove all supernatant.
-
16
Air-dry the RNA pellet for 5–10 min.
CRITICAL STEP: It is important not to let the RNA pellet dry completely as this will greatly decrease its solubility.
-
17
For PRO-seq, re-dissolve the RNA pellet in 20 μl DEPC H2O and proceed to step 18. For PRO-cap, re-dissolve the RNA pellet in 50 μl DEPC H2O and proceed to step 23.
RNA fragmentation by base hydrolysis (PRO-seq only) | Timing: 0.5 h
-
18
Heat denature the RNA at 65°C on a heat block for 40 s and then place the tubes on ice.
-
19
Add 5 μl of ice cold 1 N NaOH and incubate the mixture on ice for 10 min.
-
20
Add 25 μl of 1 M Tris-HCl pH 6.8.
-
21
Perform buffer exchange once by running the 50-μl base-hydrolyzed RNA sample through a P-30 column according to the manufacturer’s instructions.
-
22
Add 1ml RNase inhibitor.
Biotin RNA enrichment | Timing: 3 h
-
23
Mix ~50 ml of the RNA sample from step 22 for PRO-seq or from step 17 for PRO-cap with 50 ml of pre-washed Streptavidin beads.
-
24
Incubate at room temperature on a rotator set at 8 rpm for 20 min.
-
25
Place on magnet for 1 min and remove the liquid.
-
26
Resuspend the beads in 500 μl ice cold High salt wash buffer for a 1 min wash using rotator.
-
27
Place on magnet for 1 min and remove the buffer.
-
28
Repeat steps 26–27 once more.
-
29
Wash two times with 500 μl Binding buffer for a minute and use the magnet to facilitate removal of buffer.
-
30
Wash once with 500 μl Low salt wash buffer.
-
31
Resuspend the beads in 300 μl Trizol and vortex thoroughly.
-
32
Incubate for 3 min at room temperature.
-
33
Add 60 μl chloroform.
CRITICAL STEP: Inaccurate pipetting of chloroform leads to incomplete phase separation of the Trizol when transferring a small volume.
-
34
Vortex thoroughly for more than 20 s and incubate the tubes for 3 min at room temperature.
-
35
Centrifuge at 14,000 g for 5 min, 4°C.
-
36
Transfer ~180 μl of the aqueous layer to a new tube.
-
37
Remove and discard the organic phase, leaving the beads and the unpipetted aqueous phase.
-
38
Extract RNA from the beads once more by repeating steps 31–35.
-
39
Collect ~180 μl of the aqueous layer and combine with the sample from step 36.
-
40
Add 360 μl chloroform to the pooled aqueous layer and vortex.
-
41
Centrifuge at 14,000 g for 5 min, 4°C.
-
42
Transfer ~350 μl of the aqueous layer to a clean tube.
-
43
To the collected aqueous layer, add 1 μl of GlycoBlue, 900 μl of 100% ethanol and vortex.
-
44
Incubate samples at room temperature for 10 min and centrifuge at 14,000 g for 20 min, 4°C.
TROUBLESHOOTING:
-
45
Add 750 ml of 75% ethanol.
PAUSE POINT: The RNA pellet in 75% ethanol can be stored up to a week at −80°C.
-
46
Mix by vortexing and centrifuge at 14,000 g for 5 min, 4°C.
-
47
Remove all residual liquid.
-
48
Air-dry the RNA pellet for 5−10 min.
CRITICAL STEP: Do not re-dissolve in H2O without the RNA adaptor. The RNA pellet is dissolved in small volume of RNA adaptor-containing solution to minimize the adapter ligation reaction volume.
3′ RNA adaptor ligation | Timing: 4.5 h
-
49
Dilute 0.5 μl 100 μM 3′ RNA adaptor in 3.5 μl DEPC H2O. For PRO-seq, use VRA3 RNA adaptor. For PRO-cap, use RA3 RNA adaptor. For processing multiple samples, scale up accordingly.
-
50
Redissolve the RNA pellet from step 48 in 4 μl of the 3′ RNA adaptor dilution.
-
51
Heat denature at 65°C in a heat block for 20 s, then place on ice.
-
52
Make the RNA ligation mix tabulated below. When processing multiple samples, scale up accordingly (Supplementary Table 2).
Reagents Volume per reaction (ml) Final concentration T4 RNA ligase buffer (10x) 1 1x ATP (10 mM) 1 1 mM 50% PEG 2 10 % RNase inhibitor 1 4 units per ml T4 RNA ligase I 1 1 units per ml -
53
Add 6 μl of the mix to the 4 μl of RNA (10 μl final).
CRITICAL STEP: GlycoBlue may form a precipitate in the presence of high PEG concentration, but is not reported to affect the ligation efficiency.
-
54
Incubate at 20°C for 4 hr then place at 4°C until ready to proceed to the next step.
PAUSE POINT: The ligation reaction can be left at 4°C overnight.
Second biotin RNA enrichment | Timing: 3 h
-
55
Bring the volume of the adaptor ligated RNA from step 54 to 50 μl by adding 40 μl DEPC H2O.
-
56
Perform a second biotin enrichment by repeating steps 23–48 with the 50 μl ligated RNA sample.
PAUSE POINT: The RNA pellet in 75% ethanol can be stored up to a week at −80°C.
Enzymatic modification of the RNA 5′ ends | Timing: 3.5–4 h
-
57
Re-dissolve the RNA pellet from step 56 in 5 μl DEPC H2O.
-
58
Heat denature briefly at 65°C in a heat block for 20 secs, then place on ice.
-
59
If performing PRO-cap, degrade the uncapped RNA containing 5′-monophosphate and remove the 5′ triphosphate and monophosphate from uncapped RNA as described in Box 1 (Supplementary Table 3 and 4) before proceeding to step 60. For PRO-seq, continue directly to step 60.
-
60
Prepare 5′ cap repair enzyme mix: depending on the availability of Tobacco acid pyrophosphatase (TAP) or RNA 5′ Pyrophosphohydrolase (RppH). When using TAP, prepare the enzyme mix in the first table. When using RppH, prepare the enzyme mix in the second table. When processing multiple samples, scale up accordingly (Supplementary Table 5).
Reagents Volume per reaction (ml) Final concentration DEPC H2O 3 TAP buffer (10x) 1 1x RNase inhibitor 0.5 2 units per ml TAP 0.5 0.5 units per ml Reagents Volume per reaction (ml) Final concentration DEPC H2O 2.5 ThermoPol Reaction buffer (10x) 1 1x RNase inhibitor 0.5 2 units per ml RppH 1 0.5 units per ml -
61
Add 5μl of the appropriate enzyme mix to the 5μl of RNA from step 58 for PRO-seq or 5μl of RNA from step 24 of Box 1 for PRO-cap.
-
62
Incubate at 37°C for 1 hr.
-
63
For PRO-seq, proceed to step 64 for hydroxyl repair. For PRO-cap, Add 90μl of DEPC H2O to the 10μl of RNA from step 62 and proceed directly to step 67
Box 1. Degradation of RNA containing 5′-monophosphate and removal of 5′-triphosphate and -monophosphate from the RNA for PRO-cap | Timing: 4 h.
-
Prepare the 5′-phosphate-dependent exonuclease enzyme mix. When processing multiple samples, scale up accordingly (Supplementary Table 3)
Reagents Volume per reaction (ml) Final concentration DEPC-H2O 2.5 Buffer A (10x) 1 1x RNase inhibitor 0.5 2 units/ml 5′-phosphate-dependent exonuclease 1 0.1 units/ml Add 5 μl of the mix to the RNA from step 58 of the main Procedure (10μl final)
Incubate at 30°C for 1 hr.
Add 300 μl of Trizol and vortex for 5 s.
Add 60 μl chloroform, vortex for 15 s, and incubate for 2 min at room temperature.
Centrifuge at 14,000 g for 5 min at 4°C.
Transfer ~180 μl aqueous layer to a clean microfuge tube.
Add 180 μl chloroform to the aqueous layer from step 7 and vortex for 5 s.
Centrifuge at 14,000 g for 5 min at 4°C and collect ~180 μl aqueous layer.
Add 0.5 μl GlycoBlue and 450 μl of 100% ethanol to the aqueous layer from step 9, and pellet the RNA by centrifuging at 14,000 g for 20 min at 4°C.
-
Wash the RNA pellet in 75% ethanol by repeating steps 45–48 of the main Procedure.
PAUSE POINT: The RNA pellet in 75% ethanol can be stored up to a week at −80°C.
Re-dissolve the RNA pellet in 5 μl DEPC H2O, and heat denature briefly in 65°C heat block for 20 s, then place on ice.
-
Prepare the alkaline phosphatase enzyme mix. When processing multiple samples, scale up accordingly (Supplementary Table 4).
Reagents Volume per reaction (ml) Final concentration DEPC-H2O 3 Alkaline phosphatase buffer (10x) 1 1x RNase inhibitor 0.5 2 units/ml Alkaline phosphatase 0.5 0.5 units/ml Add 5 μl of the mix to the RNA from step 12 (10μl final)
Incubate at 37°C for 1 hr.
Add 300 μl of Trizol and vortex for 5 s.
Add 60 μl chloroform, vortex for 15 s, and incubate for 2 min at room temperature.
Centrifuge at 14,000 g for 5 min at 4°C.
Transfer ~180 μl aqueous layer to a clean microfuge tube.
Add 180 μl chloroform to the aqueous layer from step 19 and vortex for 5 s.
Centrifuge at 14,000 g for 5 min at 4°C and collect ~180 μl aqueous layer.
Add 0.5 μl GlycoBlue and 450 μl of 100% ethanol to the aqueous layer from step 21, and pellet the RNA by centrifuging at 14,000 g for 20 min at 4°C.
-
Wash the RNA pellet in 75% ethanol by repeating steps 45–48 of the main Procedure.
PAUSE POINT: The RNA pellet in 75% ethanol can be stored up to a week at −80°C.
Re-dissolve the RNA pellet in 5 μl DEPC H2O, and heat denature briefly in 65°C heat block for 20 s, then place on ice. Proceed from step 60 of the main Procedure.
(PRO-seq only) Hydroxyl repair | TIMING: 2 h
-
64
Prepare polynucleotide kinase (PNK) mix as tabulated below. When processing multiple samples, scale up accordingly (Supplementary Table 6).
Reagents Volume per reaction (ml) Final concentration DEPC H2O 65 PNK buffer (10x) 10 1x ATP (10 mM) 10 1 mM RNase inhibitor 2.5 1 units per ml PNK 2.5 0.25 units per ml -
65
Add 90μl of the mix to the 10μl of RNA from step 62.
-
66
Incubate at 37°C for 1 hr.
-
67
Add 300 μl of Trizol and vortex for 5 s.
-
68
Add 60 μl chloroform, vortex for 15 s, and incubate for 2 min at room temperature.
-
69
Centrifuge at 14,000 g for 5 min at 4°C.
-
70
Transfer ~280 μl aqueous layer to a clean microfuge tube.
-
71
Add 280 μl chloroform to the aqueous layer from step 70 and vortex for 5 s.
-
72
Centrifuge at 14,000 g for 5 min at 4°C and transfer ~280 μl aqueous layer to a clean microfuge tube.
-
73
Add 0.5 μl GlycoBlue and 700 μl of 100% ethanol to the aqueous layer from step 72, and pellet the RNA by centrifuging at 14,000 g for 20 min at 4°C.
-
74
Wash the RNA pellet in 75% ethanol by repeating steps 45–48.
PAUSE POINT: The RNA pellet in 75% ethanol can be stored up to a week at −80°C.
CRITICAL STEP: Do not re-dissolve in H2O without the RNA adaptor. The RNA pellet is dissolved in a small volume of RNA adaptor-containing solution to minimize the adapter ligation reaction volume.
5′ RNA adaptor ligation | Timing: 4.5 h
-
75
Dilute 0.5 μl 100 μM 5′ RNA adaptor in 3.5 μl DEPC H2O. For PRO-seq, use VRA5 RNA adaptor. For PRO-cap, use RA5 RNA adaptor. For processing multiple samples, scale up accordingly.
-
76
Redissolve the RNA pellet from step 74 in 4 μl of the 5′ RNA adaptor dilution.
-
77
Heat denature at 65°C in a heat block for 20 s, then place on ice.
-
78
Make the RNA ligation mix as described in step 52. When processing multiple samples, scale up accordingly (Supplementary Table 2).
-
79
Add 6 μl of the RNA ligation mix to the 4 μl of RNA (10 μl final).
CRITICAL STEP: GlycoBlue may form a precipitate in the presence of high PEG concentration, but is not reported to affect the ligation efficiency.
-
80
Incubate at 20°C for 4 hr then place at 4°C until ready to proceed to the next step.
PAUSE POINT: The ligation reaction can be left at 4°C overnight.
Third biotin RNA enrichment | Timing: 3 hr
-
81
Bring the volume of the adaptor ligated RNA to 50 μl by adding 40 μl DEPC H2O.
-
82
Perform a third biotin enrichment by repeating steps 23–48 with the 50 μl ligated RNA sample.
PAUSE POINT: The RNA pellet in 75% ethanol can be stored up to a week at −80°C.
Reverse transcription | Timing: 2 h
-
83
Re-dissolve the RNA pellet in 10 μl DEPC H2O.
-
84
Make reverse transcription (RT) primer mix as shown in the table below.
Component Amount per reaction (μl) Final conc. in 20-μl volume (μM) PRO-seq PRO-cap RP1 reverse transcription primer (100 μM) 0.5 -- 2.5 RTP reverse transcription primer (100 μM) -- 0.5 2.5 12.5 mM dNTP mix 1 1 625 DEPC-H2O 1 1 -
85
Add 2.5 μl of the RT primer mix to the 10 μl of re-dissolved RNA.
-
86
Heat to 70°C for 2 min, chill it on ice for 2 min and briefly spin at 500–1,000g at 25°C for 5 s.
-
87
Prepare the RT buffer mix as tabulated below. When processing multiple samples, scale up accordingly (Supplementary Table 7).
Reagents Volume per reaction (ml) Final concentration First strand buffer (5x) 4 DTT (0.1 M) 1 5 mM RNase inhibitor 1 2 units per ml -
88
Add 6 μl of the RT buffer mix to the 12.5 ul of RNA-primer mix from step 86.
-
89
Incubate for 5 min, 37°C.
-
90
Add 1.5 μl Superscript III reverse transcription enzyme and mix (total 20 μl).
-
91
Incubate for 15 min at 45°C, then 40 min at 50°C, 10 min at 55°C, and 15 min at 70°C.
-
92
Add 6 μl of DEPC H2O to the RT reaction (total 26 μl).
PAUSE POINT: The reverse transcribed cDNA can be stored for a month at −20°C.
Test PCR amplification | Timing: 2 h
-
93
Prepare a series of 4-fold dilutions of the RT sample in H2O as tabulated below. Test PCR amplification (a total of 21 amplification cycles) of these dilutions will be used to determine the appropriate number of PCR cycles to use in full-scale amplification at step 105. The use of 2 μl RT sample in dilution 1 leaves 24 μl RT sample for full-scale amplification at step 105.
Dilution Amount of cDNA Amount of H2O (μl) Equivalent full-scale PCR cycles (step 105) 1 2 μl RT sample 6 17 2 2 μl dilution 1 6 15 3 2 μl dilution 2 6 13 4 2 μl dilution 3 6 11 CRITICAL STEP: As only 6 μl out of the total 8 μl of Dilution 1 is used for PCR, this is equivalent to using 1.5 μl (6/8 x 2) of the original 26 μl RT sample, which is 16 fold less than the remaining 24 μl RT sample. Thus, to account for the 16-fold higher amount of starting material in the full-scale amplification, the number of PCR cycles needs to be reduced by 4 compared to test PCR of Dilution 1 i.e. 17 cycles instead of 21. Each remaining dilution in the 4-fold dilution series will need to be corrected by a further 2 cycles compared to the previous dilution i.e. 15, 13, and 11 cycles for dilutions 2 to 4 respectively.
-
94
Prepare a test PCR mix. One test PCR amplification will be performed for each dilution prepared in step 93 (Supplementary Table 8).
Reagents Volume per reaction (ml) Final concentration DEPC H2O 5 HF buffer (5x) 4 1x Betaine (5 M) 4 1 M dNTP mix (12.5 mM each) 0.4 250 μM each RP1 primer (25 μM) 0.2 250 nM RPI-1 primer (25 μM) 0.2 250 nM Phusion DNA polymerase 0.2 0.02 units per ml -
95
Add 14 μl of the PCR mix to 6 μl of each diluted test sample (Dilution 1, 2, 3, and 4).
-
96
Use the following thermal cycling to perform test PCR amplification (a total of 21 amplification cycles).
Cycle number Denature (95°C) Anneal Extend (72°C) 1 2 min 2 – 6 30 s 56°C for 30 s 30 s 7 – 22 30 s 65°C for 30 s 30 s 23 10 min
Gel analysis of test PCR | Timing: 2 h
-
97
Add 2.2 μl 10x Orange G dye to the 20 μl PCR reactions and load 20 μl of the samples onto a 2.2% agarose gel in 1xTAE.
-
98
Load 8 μl of 100 bp DNA ladder on a separate lane.
-
99
Run the gel at 100 V for 15 min, then run it at 130 V for up to 45 min.
CRITICAL STEP: Orange G dye runs at 50 bp. Stop the electrophoresis before the dye runs out of the gel.
-
100
Add 15 μl of SYBRGold to 150 ml 1x TAE. Place the gel in this solution and stain for 30 min on a shaker.
-
101
Image the gel with 485 nm illumination, or with UV light. Examine the gel and determine the dilution (and therefore the equivalent full-scale PCR amplification cycle) with desired amplification characteristics (sufficient amount of product, not over amplified, and having 50–75% of unused primers). For example, if the lane in the agarose gel with dilution 3 has the desired amplification characteristics, then the optimal number of PCR cycles (optimized cycle, OC) for full-scale amplification is 13 (see step 93). See Figure A.2A and A.2B for an example gel image.
TROUBLESHOOTING:
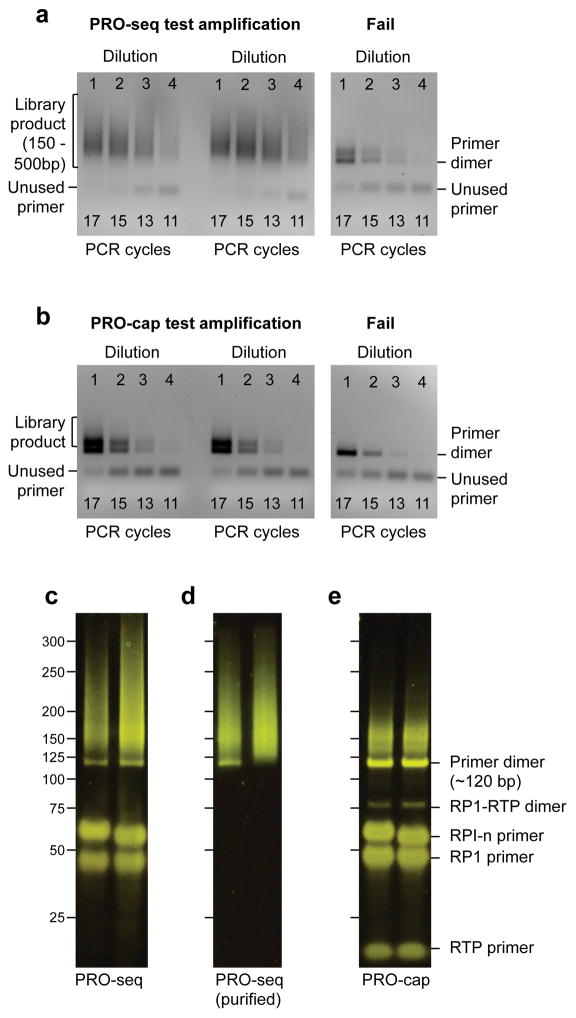
Figure A.2. Gel images of library products.
(A) PRO-seq libraries at the test amplification stage (step 101). 3 different samples are loaded on the agarose gel, each at 4 dilutions. From the left of each series, the 4 dilutions are equivalent to 17, 15, 13, and 11 cycles of full amplification. The gel image in left panel shows two successful PRO-seq libraries. Optimal amplification cycles are determined by comparing both the intensities of library products and the unused primers. Optimal amplification cycles are when 50–75% of the primers remain unused (13 cycles for this example). Gel image of a failed library preparation is shown on the right panel; only primer dimers are detected.
(B) PRO-cap libraries at the test amplification stage (step 101). Library products are smaller in size and amount, because most PRO-cap RNA molecules are from paused RNA polymerase. 15 cycles were optimal for these samples.
(C) Two PRO-seq libraries after full amplification analyzed by 8% PAGE (step 118). Labels on the left indicate DNA sizes. Note the presence of unused primers around 50 bp and primer dimer bands ~120 bp. The library product is the smear above the primer dimer band.
(D) Two PRO-seq libraries after the size selection analyzed by 8% PAGE (step 119). PRO-seq library on the left still has some residual primer dimer.
(E) Two PRO-cap libraries after the full amplification analyzed by 8% PAGE (step 118). Note that there are additional primer and primer dimer bands compared to PRO-seq.
Full-scale PCR amplification | Timing: 2.5 h
-
102
Prepare a full-scale amplification PCR mix according to the table below. For processing multiple samples, scale up accordingly (Supplementary Table 9).
Reagents Volume per reaction (ml) Final concentration DEPC H2O 3 HF buffer (5x) 10 1x Betaine (5 M) 10 1 M dNTP mix (12.5 mM each) 1 250 μM each RP1 primer (25 μM) 0.5 250 nM Phusion DNA polymerase 1 0.04 units per ml -
103
Add 25.5 μl of the PCR mix to the 24 μl of the remaining reverse transcription product (from step 93).
-
104
Add 0.5 μl of a different RPI-n (25 μM) primer to the different libraries so that each library is differentially barcoded.
CRITICAL STEP Different barcodes are only needed if the libraries are to be pooled for sequencing.
-
105
Use the following thermal cycling for full-scale PCR amplification, where OC stands for Optimized Cycle as determined in step 101.
Cycle number Denature (95°C) Anneal Extend (72°C) 1 2 min 2 – 6 30 s 56°C for 30 s 30 s 7 - (OC + 1) 30 s 65°C for 30 s 30 s (OC + 1) - (OC + 2) 10 min PAUSE POINT: The PCR product can be stored up to a month at −20°C.
-
106
Add 231 μl H2O, 18 μl 5 M NaCl, 1 μl GlycoBlue to the 50 μl full amplification PCR product.
-
107
Add 750 μl 100% ethanol and vortex thoroughly.
-
108
Centrifuge at 14,000 g for 20 min, 4°C.
-
109
Remove liquid and the wash the pellet once in 75% ethanol by repeating steps 45–48.
PAUSE POINT: The DNA pellet in 75% ethanol can be stored up to several months at −80°C.
-
110
Re-dissolve the DNA pellet in 18 μl H2O.
Library size selection by PAGE | Timing: 5 h to 1 day
CRITICAL: we describe PAGE purification for size selection of the library. However, a Pippin Prep (Sage Science) can also be used in place of steps 111–130; follow the manufacturer’s instruction for selecting a size range of 140–350 bp.
-
111
Add 2 μl of 10× Orange G loading dye to 18 μl DNA from step 110.
-
112
Prepare a medium size (10 cm running length) Native PAGE gel as below.
Reagents Volume Final concentration DEPC-H2O 31.67 ml Acrylamide (30%) 13.3 ml 8% TBE (5x) 5 ml 0.5x APS (10%) 500 ml 0.1% TEMED 50 ml -
113
Pre-run the gel for 15 min at a constant current of 30 mA.
-
114
Load samples. Also load 2 μl 10 bp DNA ladder and 8 μl of 100 bp DNA ladder.
-
115
Run gel at 15 mA for 30 min until the DNA has entered the gel, and then run at 30 mA for 1.5 hr. Stop electrophoresis 10 minutes after the Orange G dye has run off the gel.
-
116
During the electrophoresis, puncture the bottom of a sterile, nuclease-free, 0.5 ml centrifuge tube using a 21-gauge needle (heated in a bunsen flame) to create a hole or several holes in the bottom of the tube. Place the 0.5 ml microtube into a sterile, round-bottom, nuclease-free, 2 ml microtube.
-
117
After the electrophoresis, pry apart the gel cassette and stain the gel with SYBR Gold (10 μl SybrGold per 100 ml 1× TBE buffer) in a clean container for 5–10 min.
-
118
Visualize the gel on a Dark Reader transilluminator.
-
119
Using a clean scalpel or razor, cut the gel from 140 bp (20 bp just above the 120 bp adapter dimer) up to 350 bp (see Figure A.2C, A.2D, and A.2E).
-
120
Split the gel fragment vertically and place the pieces into the 0.5 ml microtube.
-
121
Centrifuge the stacked tubes at 10,000 g for 2 min at room temperature to shred the gel through the holes into the 2 ml tube (there is no liquid at this point).
-
122
If some gel remains in the top tube, add 100 μl of Gel Elution Buffer and spin at 10,000g again for another 2 min.
-
123
Add 600 μl of Gel Elution Buffer and incubate for 2 hr in a rotating incubator, 37°C.
PAUSE POINT: The elution can continue overnight.
-
124
Spin down gel pieces for 1 min at max speed in a benchtop centrifuge.
-
125
Transfer all liquid possible to a new tube.
-
126
Add 400 μl of Gel Elution Buffer to the remaining gel pieces.
-
127
Incubate for 1 hr in a rotating incubator, 37°C.
-
128
After 1 h incubation, spin at the maximum speed in a benchtop centrifuge for 1 min; take the supernatant and pool with the first elution from step 125.
-
129
Rinse gel pieces with 250 μl H2O, spin and add the rinsed liquid to the pool.
-
130
Transfer the pooled eluate which may contain small pieces of gel debris, to the top of a Spin-X filter.
-
131
Centrifuge the filter for 1–2 min at 6,000–7,000 g, room temperature. Collect the filtrate. If the volume exceeds the filter capacity, use multiple filters or split into batches and repeat filtering a couple of times and pool the filtrates.
-
132
Lyophilize (on medium setting) the sample using a Speed Vac dryer reduce the volume to ~400 μl (takes 45 min–2 hr). If the volume decreases below 400 μl, bring the volume up to 400 μl by adding DEPC-H2O.
-
133
Add an equal volume of buffered phenol:chloroform, and vortex thoroughly.
-
134
Centrifuge at 14,000 g for 5 min, 4°C.
-
135
Collect the aqueous layer in a clean tube.
-
136
Add an equal volume of chloroform to the aqueous layer, and vortex thoroughly.
-
137
Centrifuge at 14,000 g for 5 min, 4°C.
-
138
Collect the aqueous layer in a clean tube.
-
139
Add 1 μl GlycoBlue to the aqueous layer.
-
140
Add 2.5× volume of room temperature 100% ethanol.
-
141
Vortex thoroughly and incubate at room temperature for 10 min.
-
142
Centrifuge at 14,000 g for 20 min, 4°C.
-
143
Remove liquid, and wash the DNA pellet once in 75% ethanol by repeating steps 45–48.
-
144
Re-dissolve the pellet in 12 μl H2O.
-
145
Use 2 μl of the library DNA for quantification using Qubit or Bioanalyzer. The expected concentration of the library is between 1–20 ng/μl.
-
146
If required, dilute the samples to 5 ng/μl. Send ~10 ng to the sequencing facility for sequencing. If the libraries are barcoded, pool the barcoded libraries that are to be sequenced simultaneously.
High-throughput sequencing | Timing: 24 h
-
147
Sequence pooled PRO-seq or PRO-cap libraries using an Illumina TRU-seq compatible sequencing platform. Sequencing depth of ~20 million and ~50 million reads provides good coverage in insect cells and mammalian cells respectively.
Data analysis | TIMING: variable
CRITICAL. In PRO-seq, the 3′ end of the nascent RNA corresponds to the genomic position of the RNA polymerase active site. The modified RNA adaptors were designed to enable sequencing of the reverse complement of nascent RNAs. Therefore, the 3′ end of the reverse complement of the sequencing reads reflects the RNA polymerase active site position. In PRO-cap, conventional RNA adaptors are used, and the 5′ ends of each sequence read reflects the transcription start sites in the same direction. Below, we outline the three major stages of a simple processing pipeline.
-
148
Pre-process the raw sequence data. Filter out low quality reads and trimming potential adaptor sequences (TGGAATTCTCGGGTGCCAAGG) from the sequence reads. Tools such as ‘cutadapt’ are publicly available for this purpose. Depending on the quality of the library, sequences containing only the adaptor sequences (adaptor dimers) may take up to 5% of total reads.
-
149
Map or align the sequence reads to the genomic sequence. Since most of the nascent RNA reads are captured before RNA processing and splicing, they do not contain large gaps in alignment. Therefore, many alignment programs based on the Burrows-Wheeler transformation algorithm such as ‘bwa’ or ‘bowtie’ work well. Reads with multiple alignments are usually discarded, unless they are used for studying specific target regions that are repeated more than once. Sometimes, reads aligning to the ribosomal DNA sequence can be pre-filtered since they can account for 30–40% of all the transcriptional activity. On average, about 55–70% of the raw sequence reads are aligned uniquely to the genome. The alignment results are commonly stored in ‘sam’ or ‘bam’ formats.
-
150
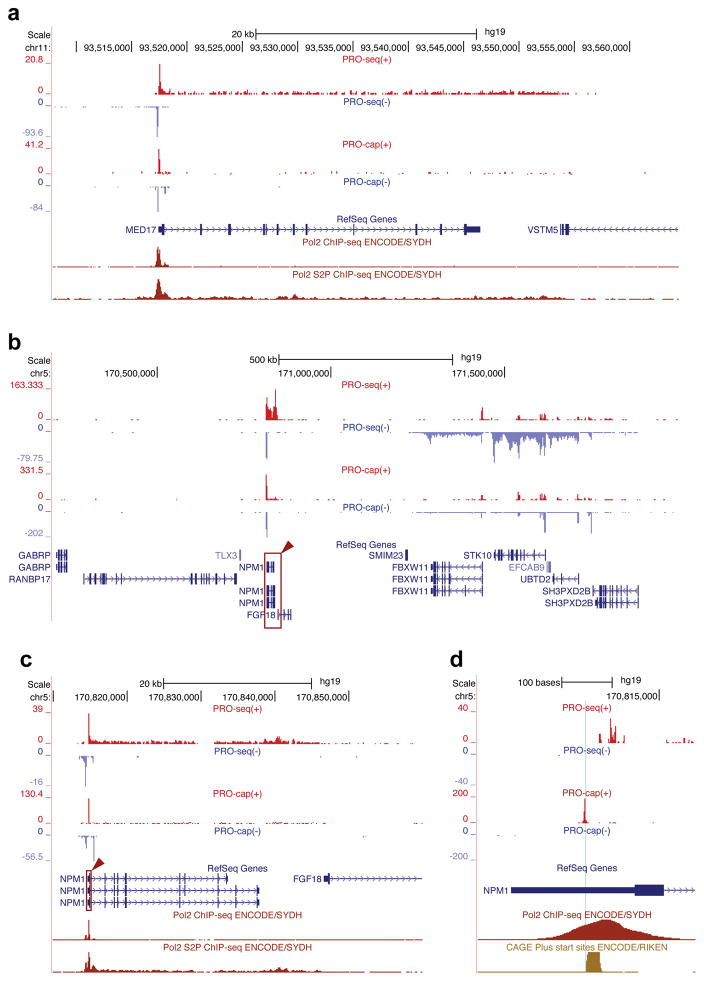
Generate the coverage of the aligned sequence reads. A common way to do this using publicly available tools is as follows: first, sort the ‘bam’ file using ‘samtools sort’; then process the sorted ‘bam’ file using ‘bedtools genomecov’ with ‘-ibam’ (use bam file input), ‘-strand’ (strand specific coverage), and ‘-5′ (5′ position coverage’) options. For the PRO-seq data, swap the plus and minus strand data for the correct orientation. These data can be visualized in genome browsers (Figure A.3), and used in further downstream analyses.
Figure A.3. Genome browser examples of PRO-seq and PRO-cap results.
(A) Sample of PRO-seq and PRO-cap data viewed on the University of California at Santa Cruz (UCSC) genome browser. A region on chromosome 11 encompassing the MED17 gene showing PRO-seq (top), PRO-cap (mid), and Pol II ChIP-seq and Serine-2 phosphorylated Pol II (Pol II S2P) ChIP-seq (bottom) reads aligned to the human genome (hg19). PRO-seq and PRO-cap reads on the plus strand (left to right), red; PRO-seq and PRO-cap reads on the minus strand (right to left), light blue; RefSeq gene annotations, dark blue; and ChIP-seq reads, dark red. The Pol II and Pol II S2P ChIP-seq tracks are from the ENCODE public data on GM12878 cells using 8WG16 and ab5095 antibodies respectively. Y-axis represents raw read counts displayed on the default setting of the UCSC genome browser.
(B) Sample of PRO-seq and PRO-cap data view on the UCSC genome browser. A region on chromosome 5 encompassing the NPM1 gene showing PRO-seq (top) and PRO-cap (bottom) reads aligned to the human genome (hg19). Red boxes with arrowheads mark the region magnified in subsequent panels.
(C) PRO-seq (top), PRO-cap (mid), and Pol II ChIP-seq and Pol II S2P ChIP-seq (bottom) data across the gene body of NPM1 gene.
(D) An up-close view around the annotated TSS of NPM1 gene. ENCODE RIKEN CAGE data (bottom) is shown to illustrate the position of mRNA cap site relative to the PRO-seq, PRO-cap, and Pol II ChIP-seq data around the annotated TSS of NPM1 gene.
TROUBLESHOOTING
Troubleshooting advice is provided in Table 2.
Table 2.
Troubleshooting
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 44 | Small or no pellet | Incomplete precipitation. |
|
| Incomplete separation of the organic phase |
|
||
| Blue jelly ball-like pellet | Incomplete separation of the organic phase. |
|
|
| 101 | No library product or the library product amplifies at later cycles. | Insufficient amount or quality of the nuclei sample. |
|
| RNA degradation. |
|
||
| Incomplete RNA extraction during biotin RNA enrichment. |
|
||
| Inefficient RNA modifying enzymes. |
|
TIMING
Step 1, cell culture: 24 h
Step 2, sample preparation (nuclei isolation/cell permeabalization): 1 h
Steps 3–7, nuclear Run-On: 1.5 h
Steps 8–17, RNA extraction: 1 h
Steps 18–22, RNA fragmentation by base hydrolysis: 0.5 h
Steps 23–48, biotin RNA enrichment: 3 h
Steps 49–54, 3′ RNA adaptor ligation: 0.5 h hands-on, 4.5 h to overnight total (suggested end of the first day)
Steps 55 and 56, second biotin RNA enrichment: 3 h
Steps 57–63, enzymatic modification of the RNA 5′ ends: 3.5–4 h
Steps 64–74, hydroxyl repair (PRO-seq only): 2 h
Steps 75–80, 5′ RNA adaptor ligation: 4.5 h (0.5 h hands-on and 4 h incubation; can be stored overnight (suggested end of the second day)
Steps 81 and 82, third biotin RNA enrichment: 3 h
Steps 83–92, reverse transcription: 2 h
Steps 93–96, test PCR amplification: 2 h
Steps 97–101, test PCR gel analysis: 2 h (Suggested end of the third day)
Steps 102–110, full-scale PCR amplification: 2.5 h
Steps 111–146, library size selection by PAGE: 5 h – 1 day
Step 147, high-throughput sequencing, 24 h
Steps 148–150, data analysis: variable
Box 1, degradation of RNA containing 5′-monophosphate and removal of 5′-triphosphate and monophosphate from the RNA for PRO-cap: 4 h
ANTICIPATED RESULTS
The final product of the PRO-seq or PRO-cap method is genome-wide maps of, respectively, RNA polymerase active sites or transcription start sites (Figure A.3). In general, PRO-seq profile of a gene exhibits several features such as: a) higher read density at the promoter-proximal pause site compared to the gene body representing paused Pol IIs, b) uniform read density across exons and introns, c) reads beyond the polyadenylation site, and d) divergent PRO-seq reads at the promoter of genes in mammals indicating divergent transcription. The enhancer regions, which are present in intergenic and intragenic regions, are also characterized by divergent PRO-seq reads. In PRO-cap, read density is very high at TSS and very low, almost at background level, in gene bodies. An indication of whether library preparation has been successful can be obtained at the Test amplification stage (steps 93–101) through estimates of yield and quality (Figure A.2). Spiky PRO-seq read coverage along the gene body indicates lower complexity of the library, which may arise from use of fewer nuclei or permeabilized cells or by the use of higher number of PCR amplification cycles. Libraries that require fewer amplification cycles (less than 13 cycles) provide high quality results, and those requiring between 14–18 cycles provide meaningful results but with some potential for amplification biases. These amplification biases manifest as low library complexity in general and high repetition of certain sequence reads.
Supplementary Material
References
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kvam C, Kitamura A, Ohsumi T, Okazaki Y, Itoh M, Kamiya M, Shibata K, Sasaki N, Izawa M, et al. High-Efficiency Full-Length cDNA Cloning by Biotinylated CAP Trapper. Genomics. 1996;37:327–336. doi: 10.1006/geno.1996.0567. [DOI] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock PJA, Fields CJ, Goto N, Heuer ML, Rice PM. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. 2010 doi: 10.1093/nar/gkp1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA, Oliviero S. Preparation of Yeast RNA. Hoboken, NJ, USA: John Wiley & Sons, Inc; 2001. [DOI] [PubMed] [Google Scholar]
- Core LJ, Martins AL, Danko CG, Waters CT, Siepel A, Lis JT. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet. 2014;46:1311–1320. doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Gilchrist DA, Fargo DC, Kwak H, Adelman K, Lis JT. Defining the status of RNA polymerase at promoters. Cell Rep. 2012;2:1025–1035. doi: 10.1016/j.celrep.2012.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejes-Toth K, Sotirova V, Sachidanandam R, Assaf G, Hannon GJ, Kapranov P, Foissac S, Willingham AT, Duttagupta R, Dumais E, et al. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest ARR, Kawaji H, Rehli M, Kenneth Baillie J, de Hoon MJL, Haberle V, Lassmann T, Kulakovskiy IV, Lizio M, Itoh M, et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu GK, Xu W, Wilhelmy J, Mindrinos MN, Davis RW, Xiao W, Fodor SPA. Molecular indexing enables quantitative targeted RNA sequencing and reveals poor efficiencies in standard library preparations. Proceedings of the National Academy of Sciences. 2014;111:1891–1896. doi: 10.1073/pnas.1323732111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Martínez J, Aranda A, Pérez-Ortín JE. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Molecular Cell. 2004;15:303–313. doi: 10.1016/j.molcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Research. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job D, Job D, Durand R, Durand R, Job C, Job C, Teissere M, Teissere M. Complex RNA chain elongation kinetics by wheat germ RNA polymerase II. Nucleic Acids Research. 1984;12:3303–3319. doi: 10.1093/nar/12.7.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Kwak H, Lis JT. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. Elife. 2014;3:e02407. doi: 10.7554/eLife.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H, Lis JT. Control of Transcriptional Elongation. Annu Rev Genet. 2013;47:483–508. doi: 10.1146/annurev-genet-110711-155440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H, Fuda NJ, Core LJ, Lis JT. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009 doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E, Bishop EP, Kharchenko PV, Core LJ, Lis JT, Park PJ, Kuroda MI. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature. 2011;471:115–118. doi: 10.1038/nature09757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MH, Mooney RA, Peters JM, Windgassen T, Nayak D, Gross CA, Block SM, Greenleaf WJ, Landick R, Weissman JS. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014;344:1042–1047. doi: 10.1126/science.1251871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics. 1000;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu Y, Rhee HS, Ghosh SKB, Bai L, Pugh BF, Gilmour DS. Kinetic Competition between Elongation Rate and Binding of NELF Controls Promoter-Proximal Pausing. Molecular Cell. 2013;50:711–722. doi: 10.1016/j.molcel.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahat DB, Salamanca HH, Duarte FM, Danko CG, Lis JT. Mammalian Heat Shock Response and Mechanisms Underlying Its Genome-wide Transcriptional Regulation. Molecular Cell. 2016;62:63–78. doi: 10.1016/j.molcel.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal. 2011;17:10–12. [Google Scholar]
- Mayer A, di Iulio J, Maleri S, Eser U, Vierstra J, Reynolds A, Sandstrom R, Stamatoyannopoulos JA, Churchman LS. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell. 2015;161:541–554. doi: 10.1016/j.cell.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes & Development. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T, Gomes T, Grosso ARF, Kimura H, Dye MJ, Dhir S, Carmo-Fonseca M, Proudfoot NJ. Mammalian NET-Seq Reveals Genome-wide Nascent Transcription Coupled to RNA Processing. Cell. 2015;161:526–540. doi: 10.1016/j.cell.2015.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahedi G, Kanno Y, Furumoto Y, Jiang K, Parker SCJ, Erdos MR, Davis SR, Roychoudhuri R, Restifo NP, Gadina M, et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature. 2015;520:558–562. doi: 10.1038/nature14154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CM, Ramachandran S, Henikoff S. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Molecular Cell. 2014;53:819–830. doi: 10.1016/j.molcel.2014.02.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.