Abstract
Saporin L3 from the leaves of the common soapwort is a catalyst for hydrolytic depurination of adenine from RNA. Saporin L3 is a type 1 ribosome inactivating protein (RIP) composed only of a catalytic domain. Other RIPs have been used in immunotoxin cancer therapy, but off-target effects have limited their development. In the current study, we use transition state theory to understand the chemical mechanism and transition state structure of saporin L3. In favorable cases, transition state structures guide the design of transition state analogues as inhibitors. Kinetic isotope effects (KIEs) were determined for an A14C mutant of saporin L3. To permit KIE measurements, small stem–loop RNAs that contain an AGGG tetraloop structure were enzymatically synthesized with the single adenylate bearing specific isotopic substitutions. KIEs were measured and corrected for forward commitment to obtain intrinsic values. A model of the transition state structure for depurination of stem–loop RNA (5′-GGGAGGGCCC-3′) by saporin L3 was determined by matching KIE values predicted via quantum chemical calculations to a family of intrinsic KIEs. This model indicates saporin L3 displays a late transition state with the N-ribosidic bond to the adenine nearly cleaved, and the attacking water nucleophile weakly bonded to the ribosyl anomeric carbon. The transition state retains partial ribocation character, a feature common to most N-ribosyl transferases. However, the transition state geometry for saporin L3 is distinct from ricin A-chain, the only other RIP whose transition state is known.
Graphical Abstract

Ribosome inactivating proteins (RIPs) are enzymes that irreversibly depurinate adenine from the conserved sarcin–ricin loop of rRNA.1 Some RIPs are highly specific for a single adenine while others are found to remove more than one adenine per ribosome.2,3 Other RIPs remove adenine not only from RNA, but also from DNA and other polynucleotides;4,5 as such they are referred to as polynucleotide adenine glycosylases.6 RIPs have been isolated from various plant tissues including leaves, bulbs, and seeds; the fruiting bodies of mushrooms; and other fungi. Most RIPs have been characterized with respect to their antiviral,7 antifungal,8 insecticidal,9 abortifacient,10 immunomodulatory,11 antitumor,12,13 and neurotoxic14,15 properties.
Many RIPs display extremely high enzymatic efficiency. For example, each ricin A-chain molecule can depurinate thousands of ribosomes per minute, to give a kcat/KM value of 2.6 × 108 M−1 s−1 on rabbit reticulocyte 80S ribosomes, a near diffusion-limited reaction indicative of a highly evolved catalyst.16,17 Accordingly, RIPs offer attractive properties when considered as potential therapeutic agents against cancers.
Several RIPs, including ricin, saporin, gelonin, and pokeweed antiviral protein, have been employed in the preparation of immunotoxins (ITs), carrying the toxin covalently linked to an antibody.18–20 The antibody component specifically targets the cell responsible for diseases and delivers the toxin to the cell. Several clinical trials have reported promising results for the application of ITs against solid tumors,21,22 bladder tumors,23,24 hematological malignancies,25 anti-inflammatory conditions,26–28 anti-spasm centers,29 ophthalmology disorders,30,31 and in pain killing.32 Impressive initial responses have been obtained in cancer therapy, particularly in hematological malignancies.25
Compared with conventional anticancer therapeutics, RIP-containing ITs show advantages of specificity, as well as acting on both dividing and non-dividing cancer cells. Since RIPs (e.g., saporin L3) target multiple adenine depurination sites, there is little possibility for the development of resistance. So far, the use of ITs in cancer therapy has not fully met the expectations from cellular studies. Two main challenges reported in clinical therapy are immunogenicity and vascular leak syndrome (VLS). VLS is caused by imperfect targeting such that excess RIP constructs, or RIP constructs lost to circulation following tumor lysis, enter the capillary bed with the loss of vascular integrity.33 VLS is the dose-limiting toxicity of immunotherapy. As a result, there is growing interest in the design of potent inhibitors of RIP toxin activity, which can function as rescue treatments following IT therapy. Neutralizing the ITs that are in excess or released from tumors upon lysis is expected to reduce the side effects.
Current inhibitor designs for RIPs are targeted to ricin (the A-chain, termed RTA) and include transition state (TS) analogues,33 pterin, and purine scaffolds;34–36 rationally designed peptides;37 and aptamers selected via in vitro screening.38 Pteroic acid is a modest inhibitor of ricin discovered by virtual screening and has an IC50 of 600 μM.36 A series of inhibitors was designed based on structural data for pterin binding in the specificity pocket, with the most potent inhibitor in this study displaying an IC50 of 240 μM.39 Millard’s group reported a small molecule which protected 20% of cells from death from ricin constructs at a drug concentration of 300 nM, by preventing an active site residue from adopting the active conformation.40 The strongest inhibitor reported for ricin A-chain has a Kd value of 26 nM and was a mimic of the ribocation TS of the ricin reaction determined at pH 4.0.41 Unfortunately, these inhibitors bind tightly to ricin only at low pH and do not protect eukaryotic ribosomes against ricin A-chain at physiological pH values. However, some of the TS analogue inhibitors designed for ricin show tight binding to saporin L3 with Kd values as low as 2.3 nM, making them potentially useful as a cancer therapy when administered in conjunction with saporin IT.42
Among the type 1 RIPs, saporin S6 isolated from Saponaria officinalis seeds has been used the most frequently to construct anti-cancer ITs. Although saporin S6 is reported to have high catalytic activity and killing efficiency of target cells after conjugation, its catalytic activity is less than that of saporin L3. In addition, saporin S6 is not inhibited by the transition state analogues designed against ricin A-chain that are effective for saporin L3.29 Herein, we focus on the TS structure of saporin L3 as an essential step in understanding its catalytic mechanism, and to provide information for the future design of inhibitors. Saporin L3 is not well characterized, and we focus our attention on its TS structure, as isozymes of target enzymes are by now well-known to have distinct transition states and are therefore susceptible to specific design of transition state analogues.
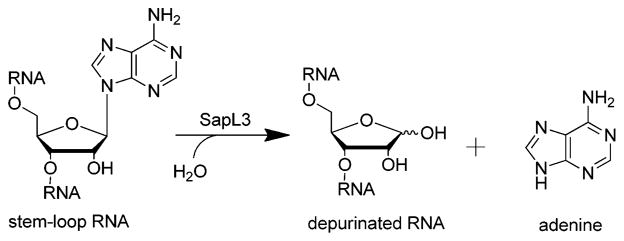
Saporin L3 is a monomeric protein extracted from Saponaria officinalis leaves and is one of the most potent RIPs. Saporin L3 catalyzes the depurination of adenine residues from the conserved GAGA tetraloop of the sarcin-ricin loop (SRL) within eukaryotic rRNA, as well as depurination at other sites including rRNAs (Scheme 1). To permit isotopic labeling at specific positions and adenine release from a single site, we used a stem–loop RNA substrate (5′-GGGAGGGCCC-3′) containing an AGGG tetraloop with a single A as a substrate for saporin L3. We mutated and expressed saporin L3 as the A14C mutant since the native protein contains no Cys residues, and Cys14 provides a surface-accessible site for eventual linking of antibodies or nucleic acid elements for recognition of target cancer cells.
Scheme 1.
Depurination of RNA by Saporin L3
Kinetic isotope effects (KIEs) provide the only experimental approach currently available to study the TS structure (e.g., bond distances, angles, and charges) of enzymatic reactions. Intrinsic KIEs were determined for saporin L3 A14C using labeled stem–loop RNA substrates. The saporin TS structure was analyzed by a quantum computational approach in which computed TS models were constrained by experimental KIEs data. The TS structure of saporin L3 and its electrostatic potential surface, along with crystallographic data, provide an unprecedented tool for understanding catalysis and offer insight into tight-binding inhibitor design of TS analogues.
RESULTS AND DISCUSSION
Synthesis of Saporin Substrate
Substrate specificity studies with saporin L3 using a single adenosine at different positions in a tetraloop indicated the stem–loop 5′-GGGAGGGCCC-3′(A10) is a slow substrate. As slow turnover is a desirable feature in the measurement of KIEs,43 A10 was used as the stem–loop substrate to study the saporin depurination reaction. A10 was synthesized by in vitro transcription with the A position labeled with [1′-3H], [1′-14C], [5′-3H], [5′-14C, 7-15N], [5′-14C, 9-15N], [1′-14C, 9-15N], and [4′-3H].
Kinetic Isotope Effects
A family of KIEs at atomic positions surrounding the N-ribosidic bond of the A10 substrate was obtained for saporin L3 depurination. The Vmax/KM 3H KIE experiments were carried out under competitive conditions with [5′-14C] as the substrate remote label. The 15N and 14C KIEs were measured with [5′-3H] labeled substrate at the remote site and were corrected for the remote [5′-3H] KIE using eq 1. Remote 3H KIEs can report on distortional binding effects and therefore must be corrected for to obtain intrinsic values. The individual isotopic substitutions and measured KIEs are reported in Table 1.
Table 1.
KIEs for RNA Depurination Catalyzed by Saporin L3 A14C
| RNA 10-mer | type of KIE | experimental KIE | intrinsic KIE | computed KIE |
|---|---|---|---|---|
| [1′-14C], [5′-3H] | primary 14C | 1.051 ± 0.008a | 1.052 ± 0.008 | 1.052 |
| [9-15N, 1′-14C], [5′-3H] | combined 14C + 15N | 1.062 ± 0.009a | 1.064 ± 0.009 | 1.073 |
| [1′-3H], [5′-14C] | α-secondary 3H | 1.044 ± 0.004 | 1.045 ± 0.004 | 1.048 |
| [4′-3H], [5′-14C] | γ-secondary 3H | 0.961 ± 0.004 | 0.960 ± 0.004 | 0.943 |
| [5′-3H], [5′-14C] | δ-secondary 3H | 1.040 ± 0.006 | 1.041 ± 0.006 | 1.042 |
| [7-15N, 5′-14C], [5′-3H] | secondary 15N | 0.974 ± 0.007a | 0.973 ± 0.007 | 0.984 |
| [9-15N, 5′-14C], [5′-3H] | primary 15N | 1.022 ± 0.009a | 1.023 ± 0.009 | 1.022 |
KIEs were corrected for the 5′-3H KIE according to the expression: KIEexperimental = KIEobserved(5′-3H KIE).
| (1) |
Forward Commitment Factor
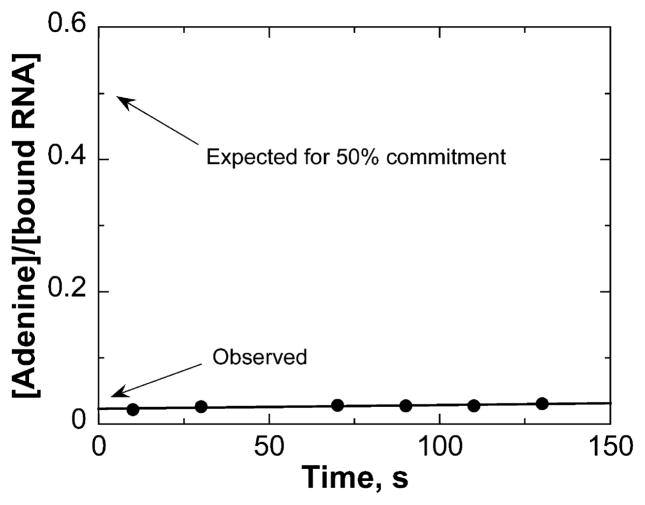
Intrinsic KIEs are useful in defining TS structure; however they can be masked by commitment factors. The forward commitment factor (Cf) reflects the probability of the Michaelis complex decomposing to unreacted substrates versus crossing the barrier to product formation. We determined Cf using the isotope trapping method developed by Rose.44 From a 40 μM preformed complex (Kd = 2 μM), a rapid-mix dilution into excess RNA allowed the monitoring of radiolabeled adenine formation. The data were plotted as a function of time, and Cf was calculated using eqs 2 and 3 (Figure 1). The amount of 3H-RNA in the ES* complex was obtained from the equilibrium constant for A10 binding to saporin L3 A14C. In eq 2, extrapolation to zero time allows determination of the fraction of bound A10 converted to product (adenine). The Cf for the saporin L3 A14C reaction was determined to be 0.024, which indicates only 2.4% of the initial complex is converted to product. The small value for Cf indicates that the Michaelis complex of saporin-RNA converts to product slower than equilibration of bound RNA with unbound RNA. These data are consistent with the relatively slow kcat of 14.6 min−1 for this complex.
Figure 1.
Commitment to catalysis for the saporin A14C-RNA complex at pH 7.2. The complex of saporin and [1′-3H]RNA was either terminated or diluted with a large excess of unlabeled RNA at 3 s. Aliquots were removed at the indicated time points and analyzed for product formation. If there is no significant commitment to catalysis, the labeled RNA in the Michaelis complex will be displaced by the unlabeled RNA. If there is significant commitment to catalysis, the amount of labeled products formed will be greater. The ordinate intercept is indicated for a 50% Cf for comparison.
| (2) |
| (3) |
Intrinsic KIEs and Their Significance
Intrinsic KIEs for the saporin L3 A14C reaction were obtained by correcting experimental KIEs for Cf using eq 4.
| (4) |
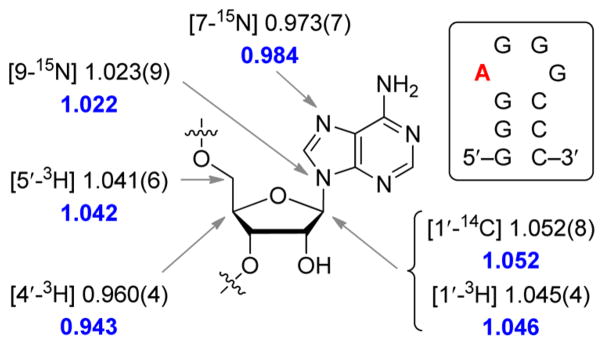
Intrinsic KIEs were determined to be 1.052 ± 0.008 for the primary anomeric carbon 1′-14C, 1.023 ± 0.009 for the primary 9-15N, 1.045 ± 0.004 for the secondary 1′-3H, 0.973 ± 0.007 for the remote 7-15N, 0.960 ± 0.004 for the 4′-3H, and 1.041 ± 0.006 for the remote 5′-3H (Table 1 and Figure 2). A control KIE using combined [9-15N, 1′-14C] additionally validated the individual measurements, a finding also consistent with a single concerted transition state.
Figure 2.
Intrinsic KIEs for saporin L3. The A (red) in the stem–loop RNA 10-mer is the site of depurination for which the KIEs were measured. Intrinsic KIEs are reported for specific positions with uncertainty (standard deviation) in the third decimal place given in parentheses. Calculated KIEs for the TS are shown in blue.
The primary 1′-14C KIE at the reaction center indicates SN1 or SN2 hybridization of that carbon at the TS.45 For a fully associative SN2 TS, the primary 1′-14C KIE can be as large as 12% due to participation at the carbon center from both the leaving group and nucleophile. The primary 1′-14C KIE in an SN1 mechanism is small and can be near unity because of loss of the leaving group bond order and reaction coordinate motion. The intrinsic primary 1′-14C KIE for saporin L3 depurination is 1.052, indicating significant nucleophilic involvement at the TS, but less than a pure SN2 mechanism. The primary 9-15N KIE of 1.023 is similar to that expected for a fully broken C–N bond at the TS. The inverse 7-15N KIE of 0.973 is consistent with protonation of N7 at the TS, resulting from increased bond order vibrational stiffness at the N7 position. Protonation of N7 at the TS prevents negative charge buildup in the purine leaving group upon breaking the C1′–N9 bond.
Although TS information cannot be obtained from the crystal structures, it is helpful to see that crystallographic studies of saporin L3 with TS analogues contain the elements needed to form this TS structure.46 The crystal structure reveals the mechanism of adenine leaving group activation by π-stacking interactions between the adenine ring, Tyr123 and Tyr73. The N7-protonated adenine is stabilized by the backbone carbonyl oxygen of Glu121. The normal α-secondary 1′-3H KIE reflects an increase in the out-of-plane C1′–H1′ bending mode as C1′ rehybridizes from sp3 to sp2. For example, the intrinsic 1′-3H KIE of human PNP of 1.184 is consistent with a fully dissociative SN1 TS with full ribocation character.47 The small α-secondary 1′-3H KIE of 4.5% for saporin L3 A14C clearly indicates less developed ribocationic character at the TS. The extent of bond contribution to C1′ from both leaving group and nucleophile can be estimated via computational analysis of potential TS structures when constrained by the intrinsic KIEs.
Comparison of KIEs for Saporin and Ricin
Despite the 30% sequence identity shared between saporin and ricin, and the similarity of their reactions, substantial differences in TS structures are suggested by intrinsic KIE values (Table 2).
Table 2.
Intrinsic KIEs for Saporin and Ricin
| 10-mer | saporin with RNA | ricin with RNAa | ricin with DNAb |
|---|---|---|---|
| [1′-14C], [5′-3H] | 1.052 ± 0.008 | 0.993 ± 0.004 | 1.015 ± 0.001 |
| [9-15N, 1′-14C], [5′-3H] | 1.064 ± 0.009 | 1.012 ± 0.004 | |
| [1′-3H], [5′-14C] | 1.045 ± 0.004 | 1.163 ± 0.009 | 1.187 ± 0.008 |
| [4′-3H], [5′-14C] | 0.960 ± 0.004 | 0.992 ± 0.004 | |
| [5′-3H], [5′-14C] | 1.041 ± 0.006 | 0.996 ± 0.003 | 1.014 ± 0.004 |
| [7-15N, 5′-14C], [5′-3H] | 0.973 ± 0.007 | 0.981 ± 0.008 | |
| [9-15N, 5′-14C], [5′-3H] | 1.023 ± 0.009 | 1.016 ± 0.005 | 1.023 ± 0.004 |
The intrinsic 1′-14C KIE for the ricin A-chain-catalyzed depurination of RNA and DNA were 0.993 and 1.015, respectively. For RNA depurination by ricin, the TS involves a dissociative ribocation. The intrinsic primary 1′-14C KIE for saporin L3 depurination is 1.052, indicating significant nucleophilic involvement at the TS, but less than a pure SN2 mechanism. Also, comparing the 1′-3H KIEs of saporin and ricin, the smaller α-secondary hydrogen KIEs of saporin L3 indicate a less dissociative TS. Similar 9-15N KIEs for saporin L3 and ricin indicate a fully broken C–N bond at both TSs. The large 5′-3H KIE for saporin reveals a significant distortion in the vibrational modes of 5′-hydrogens between the free substrate and the TS. The inverse 7-15N KIEs reveal N7 protonation at the TS for both enzymes.
Computational Modeling for the Saporin L3 TS
A TS model consistent with the intrinsic KIEs was determined using density functional theory calculations (B3LYP/6-31G*) as implemented in Gaussian 09. The TS model includes unmodified substrates, one water molecule, and active site residue Glu174 involved in water nucleophile activation. Bond distances of rC–N and rC–O from 1.6 to 3.0 Å were fixed at 0.2 Å steps in an initial approach to predict the enzymatic TS structures. Several features of the TS structure were varied, including systematic variation of bond distances along the reaction coordinate, inclusion of the nucleophilic water molecule, and the protonation state of the adenine N7. The best matches between the intrinsic and calculated KIEs were found with TS structures having bond distances of 1.8–2.2 Å to the attacking water nucleophile (rC–O) and 2.6–2.8 Å for the bond distance to the departing adenine nucleophile (rC–N).
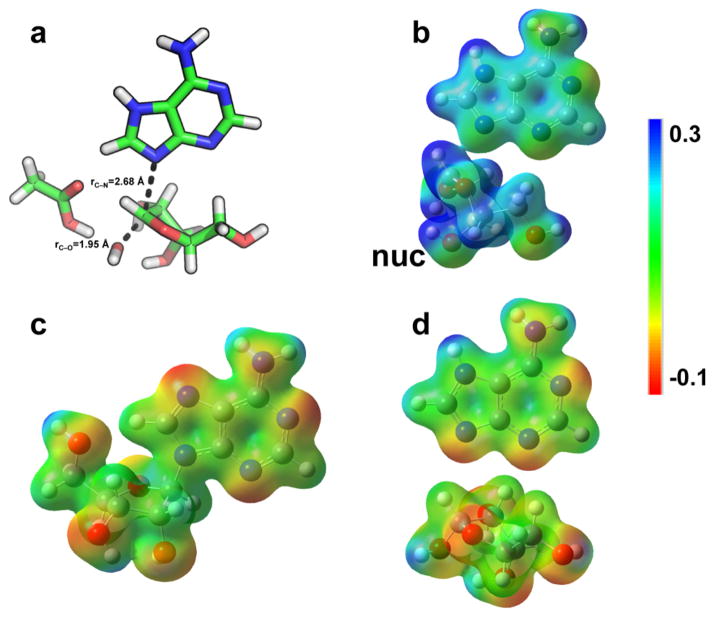
To better simulate the enzyme active site, TS structures that best matched the intrinsic KIEs in the gas-phase calculations were recalculated using a polarizable continuum model (PCM) with two different solvents: acetone and water (with dielectric constants of 20.49 and 78.36). The best match to the intrinsic KIEs was obtained with either gas-phase or water calculations (Table 1 and Figure 1). The single TS structure that best matched the intrinsic KIEs exhibited a near complete loss of the C1′–N9 bond with a 2.68 Å distance and displayed a stronger bond order to the attacking water nucleophile with a C1′–O bond distance of 1.95 Å at the TS (Figure 3).
Figure 3.
Transition structure geometry, molecular electrostatic potential surfaces (MEPs), and NBO charges for the reaction of saporin L3 A14C. Panel a shows the geometry of the transition state. Panels b, c, and d show the MEPs of the TS, GS, and products, respectively. nuc = nucleophilic water at the TS. MEPs were calculated using the CUBE subprogram of Gaussian 03 for the optimized geometry at the B3LYP/6-31G(d,p) level of theory and visualized with Molekel 4.0 at a density of 0.02 electron/Å3.
Molecular Electrostatic Potential Energy Surface Analysis
Using the structure of the TS as a stationary point permits calculation of the wave function for the TS and extraction of the molecular electrostatic potentials (MEPs). The MEPs defines the TS relative charge together with bond distances and angles. The changes in electrostatic potential and natural bond orbitals (NBO) of the ground state, TS, and products are shown in Figure 3. The late TS for saporin L3 causes the electron density at the TS to be more similar to product than to substrate. The N7 protonation of adenine is an important characteristic of the TS, as it assists leaving group dissociation by neutralizing the leaving group, which otherwise would need to overcome the unfavorable barrier of ribocation–adenine anion separation at the TS. Electrons from ribose flow to adenine as the C–N bond breaks, and this increases the pKa of N7, leading to its protonation.
Comparing the TS with a TS Analogue
Some inhibitors designed to match the ricin A-chain TS (obtained at pH 4) are also good inhibitors of saporin L3 at neutral pH values.42 A four-base cyclic, covalently closed RNA construct with a stem–loop structure containing a TS mimic for the ricin A-chain depurination site was the best inhibitor at 2.3 nM for saporin L3.42 Saporin L3, unlike ricin A-chain, can also be inhibited by RNA analogue constructs without the hairpin step-loop structure. TS analogue constructs based on the TS of ricin A-chain also had Kd values between 3 and 9 nM for saporin L3.42 In addition to the powerful inhibition at physiological pH, these inhibitors protected rabbit reticulocyte ribosomes against inactivation by saporin L3.42
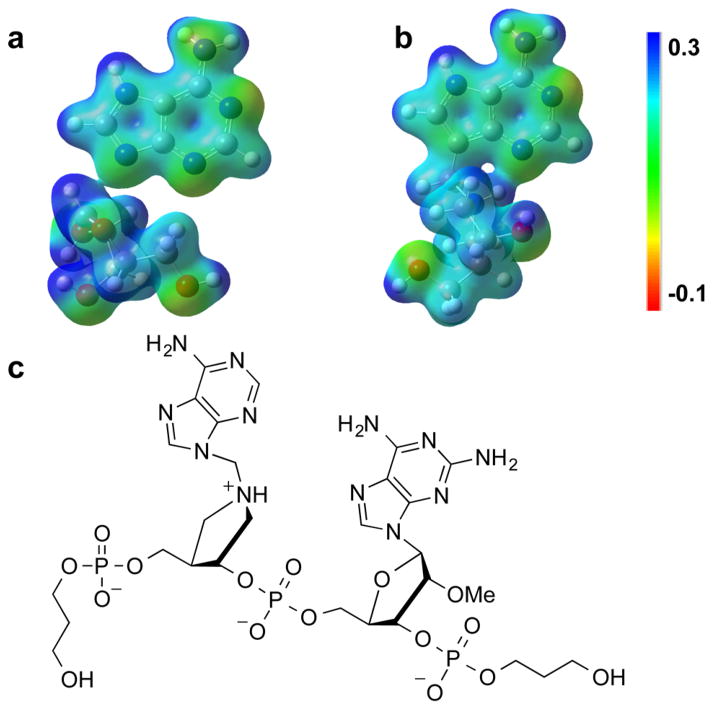
With knowledge of the saporin L3 TS, it is of interest to compare the MEP for the saporin L3 TS to that of a small, linear TS analogue construct that binds tightly to saporin L3, but not to ricin A-chain (Figure 4). The MEP surface for JMG29642 reveals a distinct electrostatic similarity between the TS structure and the inhibitor. The binding affinity of TS analogues is related to how closely their electrostatics matches that of the TS structure.
Figure 4.
Comparison of MEPs for the transition state (a) of saporin A14C, and the transition-state mimic center of JMG296 (b), a 3.3 nM TS analogue. The Kd values are dissociation constants for the inhibitors following slow-onset inhibition. The full chemical structure of JMG296 is shown in the panel c.
In the JMG296 inhibitor design, the N9 of adenine is replaced with carbon to create a stable 9-deazaadenine mimic of the glycosidic bond and the N7 is protonated.50 The increased distance between the ribocation mimic and the 9-deazaadenine by a methylene group provides geometric approximation of a fully dissociative TS. Ribocationic charge at C1′ was achieved by N1′. The MEP of this inhibitor provides a sufficiently good match to the MEP of the TS to provide a binding affinity of 105 for the KM/Kd ratio.42 Despite this powerful binding, this inhibitor lacks the electrostatic potential provided by the 2′-hydroxyribosyl. This omission is a necessary chemical feature to provide chemical stability of the inhibitor. These changes, together with other ribocation scaffolds, might be considered for future inhibitor design to enhance binding affinity to saporin L3.
Conclusion
KIEs were determined for the depurination of stem–loop RNA by saporin L3 A14C. Stem-loop RNA (5′-GGGAGGGCCC-3′) was synthesized with isotopic labels in the A-depurination position and used as substrates for KIE analysis. TS structure was analyzed by a quantum computational approach. Our analysis indicates the leaving group and attacking water nucleophile are positioned 2.68 and 1.95 Å (respectively) from the ribosyl carbon reaction center at the TS. Knowledge of the TS structure and MEP for the saporin L3 reaction reveals distinct transition states among the RIPs. Our model of the saporin L3 TS shows high similarity to a nanomolar TS analogue inhibitor. The TS structure and MEP provide information to enable the design of additional potent inhibitors. Potent inhibitors of saporin L3 may be of utility in the implementation of IT cancer therapy.
METHODS
Materials
Radiolabeled glucose and ribose substrates were purchased from American Radiolabeled Chemicals Inc. Enzymes used in the synthesis of ATP from glucose or ribose were purchased from Sigma or expressed in our laboratory (e.g., ribokinase, phosphoribosyl pyrophosphate (PRPP) synthase, and adenine phosphoribosyltransferase (APRTase)). The T7 RNA synthesis kit and RNA-free reagents used in RNA handling were purchased from Ambion. The DNA template was obtained from Integrated DNA Technologies, Inc. Saporin L3 A14C was purified as previously reported.51
Synthesis of 7-15N and 9-15N Adenines
[7-15N]Adenine was synthesized from 4,6-diamino-5-[15N]nitrosopyrimidine using a method adapted from Sethi et al. (Supporting Information Figure S1).52 [9-15N]Adenine was synthesized from [4-15N]diamino-6-chloropyrimidine using methods adapted from Sethi et al.52 and Jones et al. (Supporting Information Figure S2).53
Synthesis of Radiolabeled ATPs
[1′-3H]ATP, [1′-14C]ATP, [5′-14C]ATP, [5′-3H]ATP, and [4′-3H]ATP were synthesized enzymatically from [1-3H]ribose, [2-14C]glucose, [6-14C]glucose, [6-3H]glucose, and [5-3H]glucose, respectively (Supporting Information Figure S3). [7-15N, 5′-14C]ATP, [9-15N, 1′-14C]ATP, and [9-15N, 5′-14C]ATP were synthesized from [6-14C]glucose, [1-3H]ribose, and [7-15N], [9-15N] adenine, respectively. The reaction mixture for the synthesis of radiolabeled ATPs from ribose contained 100 mM phosphate buffer, at a pH of 7.4, 2 mM adenine, 10 mM MgCl2, 0.1 mM ATP, 20 mM PEP, and 20 μCi of labeled D-ribose in a reaction volume of 1 mL. Ribokinase (5 units), myokinase (2 units), pyruvate kinase (2.5 units), PRPP synthase (5 units), and APRTase (2 units) were added to initiate the reaction. The reaction mixture for the synthesis of radiolabeled ATPs from D-glucose contained 100 mM phosphate buffer, at a pH of 7.4, 50 mM glycylglycine, 50 mM KCl, 20 mM MgCl2, 1 mM glucose, 40 mM PEP, 1 mM DTT, 2 mM NADP+, 2 mM adenine, and 20 μCi of labeled D-glucose in a reaction of 1 mL. The reaction was initiated by adding myokinase (4 units), pyruvate kinase (3 units), phosphoriboisomerase (5 units), glucose-6-phosphate dehydrogenase and phosphogluconic acid dehydrogenase (1 unit each), APRTase and PRPPase (5 units each), and hexokinase (1 unit). Reaction mixtures were incubated for 12 h at 37 °C, and the ATP was purified by reverse phase HPLC on a waters C18 Deltapak column with a linear gradient from 2 to 50% acetonitrile in 8 mM tetrabutylammonium bisulfate and 50 mM potassium monophosphate, at a pH of 6.0 over 50 min at a flow rate of 1 mL/min, and then desalted on the same column eluting with 25% acetonitrile.
Synthesis of Radiolabeled RNA
Isotopically labeled stem loop RNAs were synthesized by incorporating labeled ATPs via T7 RNA polymerase in 20 μL reaction mixtures containing 3 μM duplex 5′-TAATACGACTCACTATAGGGAGGGCCC-3′ and 5′-GGGCCCTCCCTATAGTGAGTCGTATTA-3′, 10 mM GTP, 5 mM CTP, 1 mM radiolabeled ATP, two units of T7 polymerase, and incubated at 37 °C for 4 h. The RNA was purified by electrophoresis with TBE buffer on 15% acrylamide gel. The RNA was eluted from the gel with 0.5 M NH4Ac, 1 mM EDTA, and 0.2% SDS, then precipitated by adding 2.5 volumes of ethanol and centrifuging 30 min at 4 °C. The RNA pellets were washed with 75% ethanol, spun down, and stored in TE (Tris-EDTA) buffer.
Kinetic Isotope Effects Measurement
Kinetic isotope effects were measured in 10 mM sodium phosphate buffer and 1 mM EDTA, at a pH of 7.2 at 37 °C containing 3H and 14C-labeled RNA. One label in the RNA was at the position of interest for KIE measurement and the other at a remote site. For example, measurement of the [1′-3H] KIE was carried out under competitive conditions54 with a [5′-14C, 1′-1H] labeled substrate. The 15N KIEs were measuring using [5′-14C] to represent the isotopic 15N label and the silent 14N label was represented by [5′-3H] at the remote site. The reaction mixture was prepared in a total volume of 310 μL (at least 1 × 106 cpm). The first portion of 60 μL was incubated at 37 °C for 3 h in the absence of saporin L3 as a control. The second portion of 60 μL was incubated with 30 nM saporin L3 at 37 °C for 3 h to give 100% completion. The third portion containing three parts of 60 μL reaction mixtures was incubated at 37 °C for 20 min by addition of 1 nM saporin L3 to give 20–30% completion. The reactions were stopped by heating to 95 °C for 10 min. Addition of 0.6 M NaOH and incubation at 37 °C overnight converted RNA to nucleotides and depurinated RNA into nucleotides and ribose phosphates. The reactions were neutralized with formic acid. Reactions were passed through charcoal columns and eluted with 50 mM ribose in 50 mM potassium phosphate buffer at a pH of 6.0.43 Fractions (2 mL) of the eluate were collected, dried by speedvac, resuspended in 200 μL of H2O, mixed with 10 mL of liquid scintillation counter cocktail (Ultima GoldTM) and counted in a Wallac 1414 LSC, PerkinElmer scintillation counter.
3H and 14C counts for each sample were calculated according to eq 5 and eq 6. Here, the 14C ratio for cpm A to cpm B used a pure 14C labeled RNA standard. The cpm A and cpm B channels were set such that all 3H counts appear in cpm A and only 14C counts appear in cpm B.
| (5) |
| (6) |
The KIE values were determined by the partial and complete reaction based on the eq 7:54
| (7) |
where f is the fraction of the reaction conversion and Rf and Ro are the ratios of heavy to light at partial and 100% reaction conversion.
Measurement of Commitment Factor
Forward commitment for depurination by saporin L3 was measured by an isotope trapping experiment under rapid-mixing pre-steady-state conditions.44,55 Forward commitment is the ratio of k3 and k2 in the reaction scheme below:which is the fraction of the A10 bound to the ES* complex to convert to product (k3 relative to k2) in the first catalytic turnover.
Briefly, 20 μL of 60 μM enzyme was rapidly mixed with 20 μL of 40 μM [1′-3H]RNA (4 × 106 cpm) in 10 mM sodium phosphate buffer and 1 mM EDTA, at a pH of 7.2 for 3 s. The reaction mixture was diluted immediately by a 1000-fold molar excess of unlabeled RNA substrate. Then, 150 μL aliquots were collected every 20 s for 2 min and quenched with 0.6 M NaOH. The mixture was neutralized by formic acid and then passed through the charcoal column and eluted with 50 mM ribose in 50 mM potassium phosphate buffer at a pH of 6.0. Then, 2 mL fractions of the flow through were collected and prepared for the radioactive analysis.
Computational Modeling of Transition-State Structure
All structure optimizations and energy and frequency calculations were performed using density functional theory in B3LYP and using a 6–31g* basis set as implemented in Gaussian 09.56 The crystal structure of the E. coli 23s rRNA GAGA tetraloop (PDB: 483D) was used as the initial geometry for the reactant free solution (ground-state). It was located as a global minimum, and frequency calculations performed on the optimized structure contained no imaginary frequencies.57 A previously published crystal structure of saporin with an RNA dimer at the catalytic site (PDB: 3HIT) was used as the starting point reactant geometry for the transition state calculations.46 The transition state structures were optimized from a fully formed intermediate by varying C1–hydroxyl and C1–N9 bond distances as well as the N7 protonation state. Bond frequencies for ground and transition states were calculated with Gaussian 09 and used as an input to the ISOEFF98 program for KIE computations.58,59 The molecular electrostatic potential surfaces (MEPs) were calculated with the CUBE program from Gaussian and the files were visualized with GaussView 3.0.
Supplementary Material
Acknowledgments
Funding
This work was supported in part by Grants CA072444 and GM41916 from the National Institutes of Health and by funds provided by the Albert Einstein College of Medicine.
The authors thank Q. Du (Einstein) and E. Burgos (Einstein) for providing the APRTase and ribokinase for ATP synthesis and P. C. Tyler and G. B. Evans of the Ferrier Research Institute, Victoria University of Wellington, New Zealand for the synthesis of JMG 296, used in this study.
ABBREVIATIONS
- RIPs
ribosome inactivating proteins
- KIE
kinetic isotope effect
- TS
transition state
Footnotes
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschem-bio.5b01069.
Complete experimental procedures for the synthesis of [9-15N] adenine and [7-15N] adenine and a summary of the approach to synthesis of isotopically labeled stem–loop RNAs (PDF)
The authors declare no competing financial interest.
The nomenclature for saporin isozymes has been changed for those of leaf (L) origin. The saporin L3, described here, was previously known as saporin-L1 and is called that in early literature.42 This protein has been reassigned as saporin L3, the nomenclature used here.
References
- 1.Endo Y, Tsurugi K. RNA N-glycosidase activity of ricin A-chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J Biol Chem. 1987;262:8128–8130. [PubMed] [Google Scholar]
- 2.Barbieri L, Gorini P, Valbonesi P, Castiglioni P, Stirpe F. Unexpected activity of saporins. Nature. 1994;372:624. doi: 10.1038/372624a0. [DOI] [PubMed] [Google Scholar]
- 3.Liu RS, Yang JH, Liu WY. Isolation and enzymatic characterization of lamjapin, the first ribosome-inactivating protein from cryptogamic algal plant (Laminaria japonica A) Eur J Biochem. 2002;269:4746–4752. doi: 10.1046/j.1432-1033.2002.03165.x. [DOI] [PubMed] [Google Scholar]
- 4.Barbieri L, Valbonesi P, Bonora E, Gorini P, Bolognesi A, Stirpe F. Polynucleotide:adenosine glycosidase activity of ribosome-inactivating proteins: effect on DNA, RNA and poly(A) Nucleic Acids Res. 1997;25:518–522. doi: 10.1093/nar/25.3.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbieri L, Brigotti M, Perocco P, Carnicelli D, Ciani M, Mercatali L, Stirpe F. Ribosome-inactivating proteins depurinate poly(ADP-ribosyl)ated poly(ADP-ribose) polymerase and have transforming activity for 3T3 fibroblasts. FEBS Lett. 2003;538:178–182. doi: 10.1016/s0014-5793(03)00176-5. [DOI] [PubMed] [Google Scholar]
- 6.Barbieri L, Valbonesi P, Bondioli M, Alvarez ML, Dal Monte P, Landini MP, Stirpe F. Adenine glycosylase activity in mammalian tissues: an equivalent of ribosome-inactivating proteins. FEBS Lett. 2001;505:196–197. doi: 10.1016/s0014-5793(01)02789-2. [DOI] [PubMed] [Google Scholar]
- 7.Parikh BA, Tumer NE. Antiviral activity of ribosome inactivating proteins in medicine. Mini-Rev Med Chem. 2004;4:523–543. doi: 10.2174/1389557043403800. [DOI] [PubMed] [Google Scholar]
- 8.Ng TB. Antifungal proteins and peptides of leguminous and non-leguminous origins. Peptides. 2004;25:1215–1222. doi: 10.1016/j.peptides.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Carlini CR, Grossi-de-Sa MF. Plant toxic proteins with insecticidal properties. A review on their potentialities as bioinsecticides. Toxicon. 2002;40:1515–1539. doi: 10.1016/s0041-0101(02)00240-4. [DOI] [PubMed] [Google Scholar]
- 10.Ng TB, Chan WY, Yeung HW. Proteins with abortifacient, ribosome inactivating, immunomodulatory, antitumor and anti-AIDS activities from Cucurbitaceae plants. Gen Pharmacol. 1992;23:579–590. doi: 10.1016/0306-3623(92)90131-3. [DOI] [PubMed] [Google Scholar]
- 11.Surendranath K, Karande AA. A neutralizing antibody to the a chain of abrin inhibits abrin toxicity both in vitro and in vivo. Clin Vaccine Immunol. 2008;15:737–743. doi: 10.1128/CVI.00254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zarovni VN, Vago R, Solda T, Monaco L, Fabbrini MS. Saporin as a novel suicide gene in anticancer gene therapy. Cancer Gene Ther. 2007;14:165–173. doi: 10.1038/sj.cgt.7700998. [DOI] [PubMed] [Google Scholar]
- 13.Mohamedali KA, Poblenz AT, Sikes CR, Navone NM, Thorpe PE, Darnay BG, Rosenblum MG. Inhibition of prostate tumor growth and bone remodeling by the vascular targeting agent VEGF121/rGel. Cancer Res. 2006;66:10919–10928. doi: 10.1158/0008-5472.CAN-06-0459. [DOI] [PubMed] [Google Scholar]
- 14.Wiley RG. Substance P receptor-expressing dorsal horn neurons: lessons from the targeted cytotoxin, substance P-saporin. Pain. 2008;136:7–10. doi: 10.1016/j.pain.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Sha O, Kwong WH, Pang Cho EY, Wai Yew DT, Ng TB. Different neuronal toxicity of single-chain ribosome-inactivating proteins on the rat retina. Toxicon. 2008;51:45–53. doi: 10.1016/j.toxicon.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Olsnes S, Fernandez-Puentes C, Carrasco L, Vazquez D. Ribosome inactivation by the toxic lectins abrin and ricin. Kinetics of the enzymic activity of the toxin A-chains. Eur J Biochem. 1975;60:281–288. doi: 10.1111/j.1432-1033.1975.tb21001.x. [DOI] [PubMed] [Google Scholar]
- 17.Sturm MB, Schramm VL. Detecting ricin: sensitive luminescent assay for ricin A-chain ribosome depurination kinetics. Anal Chem. 2009;81:2847–2853. doi: 10.1021/ac8026433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cumber AJ, Forrester JA, Foxwell BM, Ross WC, Thorpe PE. Preparation of antibody-toxin conjugates. Methods Enzymol. 1985;112:207–225. doi: 10.1016/s0076-6879(85)12018-5. [DOI] [PubMed] [Google Scholar]
- 19.Vitetta ES, Uhr JW. Immunotoxins. Annu Rev Immunol. 1985;3:197–212. doi: 10.1146/annurev.iy.03.040185.001213. [DOI] [PubMed] [Google Scholar]
- 20.Thorpe PE, Brown AN, Bremner JA, Jr, Foxwell BM, Stirpe F. An immunotoxin composed of monoclonal anti-Thy 1.1 antibody and a ribosome-inactivating protein from Saponaria officinalis: potent antitumor effects in vitro and in vivo. J Natl Cancer Inst. 1985;75:151–159. [PubMed] [Google Scholar]
- 21.Theuer CP, Pastan I. Immunotoxins and recombinant toxins in the treatment of solid carcinomas. Am J Surg. 1993;166:284–288. doi: 10.1016/s0002-9610(05)80975-4. [DOI] [PubMed] [Google Scholar]
- 22.Oldfield EH, Youle RJ. Immunotoxins for brain tumor therapy. Curr Top Microbiol Immunol. 1998;234:97–114. doi: 10.1007/978-3-642-72153-3_7. [DOI] [PubMed] [Google Scholar]
- 23.Yu L, Gu F, Zhang C, Xie S, Guo Y. Targeted diagnosis and treatment of superficial bladder cancer with monoclonal antibody BDI-1. Chin Med J. 1998;111:404–407. [PubMed] [Google Scholar]
- 24.Zang Z, Xu H, Yu L, Yang D, Xie S, Shi Y, Li Z, Li J, Wang J, Li M, Guo Y, Gu F. Intravesical immunotoxin as adjuvant therapy to prevent the recurrence of bladder cancer. Chin Med J. 2000;113:1002–1006. [PubMed] [Google Scholar]
- 25.Frankel AE, Neville DM, Bugge TA, Kreitman RJ, Leppla SH. Immunotoxin therapy of hematologic malignancies. Semin Oncol. 2003;30:545–557. doi: 10.1016/s0093-7754(03)00241-0. [DOI] [PubMed] [Google Scholar]
- 26.Thepen T, van Vuuren AJ, Kiekens RC, Damen CA, Vooijs WC, van De Winkel JG. Resolution of cutaneous inflammation after local elimination of macrophages. Nat Biotechnol. 2000;18:48–51. doi: 10.1038/71908. [DOI] [PubMed] [Google Scholar]
- 27.van Roon JA, van Vuuren AJ, Wijngaarden S, Jacobs KM, Bijlsma JW, Lafeber FP, Thepen T, van de Winkel JG. Selective elimination of synovial inflammatory macrophages in rheumatoid arthritis by an Fcgamma receptor I-directed immunotoxin. Arthritis Rheum. 2003;48:1229–1238. doi: 10.1002/art.10940. [DOI] [PubMed] [Google Scholar]
- 28.Kanai T, Watanabe M, Okazawa A, Sato T, Yamazaki M, Okamoto S, Ishii H, Totsuka T, Iiyama R, Okamoto R, Ikeda M, Kurimoto M, Takeda K, Akira S, Hibi T. Macrophage-derived IL-18-mediated intestinal inflammation in the murine model of Crohn’s disease. Gastroenterology. 2001;121:875–888. doi: 10.1053/gast.2001.28021. [DOI] [PubMed] [Google Scholar]
- 29.Bolognesi A, Polito L. Immunotoxins and other conjugates: pre-clinical studies. Mini-Rev Med Chem. 2004;4:563–583. doi: 10.2174/1389557043403864. [DOI] [PubMed] [Google Scholar]
- 30.Clark DS, Emery JM, Munsell MF. Inhibition of posterior capsule opacification with an immunotoxin specific for lens epithelial cells: 24 month clinical results. J Cataract Refractive Surg. 1998;24:1614–1620. doi: 10.1016/s0886-3350(98)80352-0. [DOI] [PubMed] [Google Scholar]
- 31.Harrison AR, Skladzien S, Christiansen SP, McLoon LK. Myotoxic effects of the skeletal muscle-specific immunotoxin, ricin-mAb35, on orbicularis oculi muscle after eyelid injections in rabbits. Opthal Plast Reconstr Surg. 2004;20:312–316. doi: 10.1097/01.iop.0000131732.78227.6d. [DOI] [PubMed] [Google Scholar]
- 32.Ralston HJ., 3rd Pain and the primate thalamus. Prog Brain Res. 2005;149:1–10. doi: 10.1016/S0079-6123(05)49001-9. [DOI] [PubMed] [Google Scholar]
- 33.Roday S, Amukele T, Evans GB, Tyler PC, Furneaux RH, Schramm VL. Inhibition of ricin A-chain with pyrrolidine mimics of the oxacarbenium ion transition state. Biochemistry. 2004;43:4923–4933. doi: 10.1021/bi0498499. [DOI] [PubMed] [Google Scholar]
- 34.Bai Y, Monzingo AF, Robertus JD. The X-ray structure of ricin A chain with a novel inhibitor. Arch Biochem Biophys. 2009;483:23–28. doi: 10.1016/j.abb.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 35.Miller DJ, Ravikumar K, Shen H, Suh JK, Kerwin SM, Robertus JD. Structure-based design and characterization of novel platforms for ricin and shiga toxin inhibition. J Med Chem. 2002;45:90–98. doi: 10.1021/jm010186s. [DOI] [PubMed] [Google Scholar]
- 36.Yan X, Hollis T, Svinth M, Day P, Monzingo AF, Milne GW, Robertus JD. Structure-based identification of a ricin inhibitor. J Mol Biol. 1997;266:1043–1049. doi: 10.1006/jmbi.1996.0865. [DOI] [PubMed] [Google Scholar]
- 37.Wang ST, Feng JN, Guo JW, Li Y, Sun YX, Qin WS, Hu MR, Shen BF. Structural-based rational design of an antagonist peptide that inhibits the ribosome-inactivating activity of ricin A chain. Int J Pept Res Ther. 2005;11:211–218. [Google Scholar]
- 38.Hesselberth JR, Miller D, Robertus J, Ellington AD. In vitro selection of RNA molecules that inhibit the activity of ricin A-chain. J Biol Chem. 2000;275:4937–4942. doi: 10.1074/jbc.275.7.4937. [DOI] [PubMed] [Google Scholar]
- 39.Jasheway K, Pruet J, Anslyn EV, Robertus JD. Structure-based design of ricin inhibitors. Toxins. 2011;3:1233–1248. doi: 10.3390/toxins3101233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pang YP, Park JG, Wang S, Vummenthala A, Mishra RK, McLaughlin JE, Di R, Kahn JN, Tumer NE, Janosi L, Davis J, Millard CB. Small-molecule inhibitor leads of ribosome-inactivating proteins developed using the doorstop approach. PLoS One. 2011;6:e17883. doi: 10.1371/journal.pone.0017883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sturm MB, Roday S, Schramm VL. Circular DNA and DNA/RNA hybrid molecules as scaffolds for ricin inhibitor design. J Am Chem Soc. 2007;129:5544–5550. doi: 10.1021/ja068054h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sturm MB, Tyler PC, Evans GB, Schramm VL. Transition state analogues rescue ribosomes from saporin-L1 ribosome inactivating protein. Biochemistry. 2009;48:9941–9948. doi: 10.1021/bi901425h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen XY, Link TM, Schramm VL. Ricin A-chain: kinetics, mechanism, and RNA stem–loop inhibitors. Biochemistry. 1998;37:11605–11613. doi: 10.1021/bi980990p. [DOI] [PubMed] [Google Scholar]
- 44.Arose I. Partition analysis: detecting enzyme reaction cycle intermediates. Methods Enzymol. 1995;249:315–340. doi: 10.1016/0076-6879(95)49040-x. [DOI] [PubMed] [Google Scholar]
- 45.Berti PJ, Tanaka KSE. Transition state analysis using multiple kinetic isotope effects: Mechanisms of enzymatic and non-enzymatic glycoside hydrolysis and transfer. Adv Phys Org Chem. 2002;37:239–314. [Google Scholar]
- 46.Ho MC, Sturm MB, Almo SC, Schramm VL. Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins. Proc Natl Acad Sci U S A. 2009;106:20276–20281. doi: 10.1073/pnas.0911606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lewandowicz A, Schramm VL. Transition state analysis for human and Plasmodium falciparum purine nucleoside phosphorylases. Biochemistry. 2004;43:1458–1468. doi: 10.1021/bi0359123. [DOI] [PubMed] [Google Scholar]
- 48.Chen XY, Berti PJ, Schramm VL. Ricin A-chain: Kinetic isotope effects and transition state structure with stem–loop RNA. J Am Chem Soc. 2000;122:1609–1617. [Google Scholar]
- 49.Chen XY, Berti PJ, Schramm VL. Transition State Analysis for Depurination of DNA by Ricin A-Chain. J Am Chem Soc. 2000;122:6527–6534. [Google Scholar]
- 50.Sauve AA, Cahill SM, Zech SG, Basso LA, Lewandowicz A, Santos DS, Grubmeyer C, Evans GB, Furneaux RH, Tyler PC, McDermott A, Girvin ME, Schramm VL. Ionic states of substrates and transition state analogues at the catalytic sites of N-ribosyltransferases. Biochemistry. 2003;42:5694–5705. doi: 10.1021/bi034003a. [DOI] [PubMed] [Google Scholar]
- 51.Yuan H, Du Q, Sturm MB, Schramm VL. Soapwort Saporin L3 Expression in Yeast, Mutagenesis and RNA Substrate Specificity. Biochemistry. 2015;54:4565–4574. doi: 10.1021/acs.biochem.5b00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sethi SK, Gupta SP, Jenkins EE, Whitehead CW, Townsend LB, McCloskey JA. Mass spectrometry of nucleic acid constituents. Electron ionization spectra of selectively labeled adenines. J Am Chem Soc. 1982;104:3349–3353. [Google Scholar]
- 53.Shallop AJ, Gaffney BL, Jones RA. Use of 13C as an Indirect Tag in 15N Specifically Labeled Nucleosides. Syntheses of [8-13C-1,7,NH2-15N3]Adenosine, - Guanosine, and Their Deoxy Analogues. J Org Chem. 2003;68:8657–8661. doi: 10.1021/jo0345446. [DOI] [PubMed] [Google Scholar]
- 54.CooK PF, Cleland WW. Enzyme Kinetics and Mechanism. Taylor & Francis Group, LLC; Oxfordshire, United Kingdom: 2007. [Google Scholar]
- 55.Markham GD, Parkin DW, Mentch F, Schramm VL. A kinetic isotope effect study and transition state analysis of the S-adenosylmethionine synthetase reaction. J Biol Chem. 1987;262:5609–5615. [PubMed] [Google Scholar]
- 56.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA, Jr, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam NJ, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ. Gaussian 09. Gaussian, Inc; Wallingford, CT: 2009. [Google Scholar]
- 57.Correll CC, Wool IG, Munishkin A. The two faces of the Escherichia coli 23 S rRNA sarcin/ricin domain: the structure at 1.11 Å resolution. J Mol Biol. 1999;292:275–287. doi: 10.1006/jmbi.1999.3072. [DOI] [PubMed] [Google Scholar]
- 58.Anisimov V, Paneth P. ISOEFF98. A program for studies of isotope effects using Hessian modifications. J Math Chem. 1999;26:75–86. [Google Scholar]
- 59.Scott AP, Radom L. Harmonic vibrational frequencies: An evaluation of Hartree-Fock, Møller-Plesset, quadratic configuration interaction, density functional theory, and semiempirical scale factors. J Phys Chem. 1996;100:16502–16513. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.