Abstract
African green monkeys (AGMs) are a natural host of simian immunodeficiency virus (SIVagm) that do not develop simian AIDS. Adult AGMs naturally have low numbers of CD4+ T cells and a large population of MHC Class II-restricted CD8αα T cells that are generated through CD4 down-regulation in CD4+ T cells. Here we study the functional profiles and SIV infection status in vivo of CD4+ T cells, CD8αα T cells, and CD8αβ T cells in lymph nodes (LN), peripheral blood, and broncheoalveloar lavage (BAL) of AGM and rhesus macaques (where CD4 down regulation is not observed). We show that while the CD8αα T cells in AGM maintain functions associated with CD4+ T cells (including helper T follicular functionality in lymphoid tissues and Th2 responses in BAL), they also accumulate functions normally attributed to canonical CD8+ T cells. These hyperfunctional CD8αα T cells are found to circulate peripherally as well as reside within the lymphoid tissue. Due to their unique combination of CD4 and CD8 T cell effector functions these CD4− CD8αα T cells are likely able to serve as an immunophenotype capable of Th1, Tfh, and CTL functionalities, yet unable to be infected by SIV. These data demonstrate the ambiguity of CD4/CD8 expression in dictating functional capacities of T cells and suggest that accumulation of hyperfunctional CD8αα T cells in AGM may lead to tissue-specific antiviral immune responses in lymphoid follicles which limit SIV replication in this particular anatomical niche.
Introduction
Simian immunodeficiency virus (SIV) infection of non-human primates can results in either pathogenic or non-pathogenic infection. The variability in disease outcomes depends largely on the host species. SIV infection of natural host species, including the African green monkey (AGM) and sooty mangabey (SM), results in a nonprogressive SIV infection with low levels of immune activation (1, 2). In contrast, SIV infection in non-natural species such as the rhesus macaque, and HIV infection of humans, generally results in a pathogenic infection with high levels of immune activation, loss of CD4+ T cells, and progression to AIDS (3). Despite high plasma SIV viral loads in both non-natural and natural host species, progression to AIDS is very rare in the natural host species (4). Thus, contrasting immunological differences between the natural host and non-natural host models may provide us with a better understanding of the mechanisms by which natural hosts have evolved to avoid disease progression.
Previous studies have shown that preservation of key immunological T cell functionality in cells that are resistant to SIV-infection may be critical to the nonprogressive nature of the disease in natural host species (5–7). We have shown that multiple species of natural hosts of SIV have low frequencies of CD4+ T cells and high frequencies of CD4− T cells that have the capability to elicit effector functions normally attributed to CD4+ T cells (6). Recent studies have expanded on these findings demonstrating that some SIV-infected SM have very low numbers of CD4+ T cells, but high numbers of double-negative (DN) T cells that have a diverse T cell receptor repertoire and can exhibit functionality of Th1, Th2, Th17, and Tfh subsets, while maintaining proliferative capacity and resistance to SIV-infection in vivo (5).
The natural host species Chlorocebus (AGM) and Erythrocebus (patas monkeys) that manifest low numbers of CD4+ T cells in adulthood, have large populations of CD8αdim T cells that express CD8αα homodimers and are distinct from the classical CD8 T cells that express the α and β chain of CD8 (6–10). These CD8αdim T cells arise post-thymically upon transcriptional CD4 down-regulation by CD4+ T cells (7). The resulting CD8αα T cells retain many functional characteristics of CD4+ T cells, including MHC class II restriction, and expression of FoxP3, CD40 ligand, IL-17, and/or IL-2 (7). However, because these T cells do not express CD4 mRNA or protein, they are resistant to SIV infection in vivo, with viral infection remaining restricted to CD4+ T cells (7, 11). In addition, recent data from mouse models suggest that down-regulation of CD4 by CD4+ T cells limits the capacity of the resultant T cells from maintaining certain Th2 effector functions, and renders the cells capable of performing canonical CD8 T cell functions (12, 13). The existence of this population of CD8αα T cells, which can perform functions associated with CD4+ T cells, but are not infected with SIV, may underlie in part the non-progressive phenotype of SIV infection in AGMs.
Another, possibly related, manifestation of AGMs and other natural hosts is an apparent lack of both preferential infection of Tfh cells and limited follicular dendritic cell deposition of virus, unlike what is observed in progressive HIV/SIV infections (14–17). It is unclear how or if down-regulation of CD4 in AGM memory CD4+ T cells influences the functionality of Tfh cells or their infectivity in vivo and whether this affects patterns of viral dissemination in lymphoid tissues.
Here we aim to determine whether CD4 down-regulation in AGMs influences the effector functions that are typically associated with Th1, Th2, and Tfh subsets in comparison to rhesus macaque (RM) cell populations that do not down-regulate CD4. Since transcription factors involved in CD4 and CD8 lineage commitment can suppress the functional differentiation of the other cell lineage in murine models, we characterize the relative expression of ThPOK and Runx3 in the AGM T cell subsets. In murine models, a dichotomy between the transcription factors Runx3 and ThPOK dictates T cell phenotype, with ThPOK being important for both the maintenance of CD4 expression and suppression of CD8 expression while Runx3 is important for expression of CD8 and suppression of CD4 expression (18). Further, ThPOK expression in mature CD4+ T cells blocks activation of the cytolytic genes characteristic of CD8 T cells (19). Conversely, Runx3 has been shown to be able to repress ThPOK expression and play an important role in the differentiation and functionality of CD8 single positive cells (20, 21). In these models, the conditional knockout of ThPOK in mature CD4+ T cells induces a downregulation of CD4 and upregulation of CD8, similar to what we observe in AGM (19).
To assess the necessity of CD4 protein and CD8β expression in cells in relation to the cell’s functional capabilities, we characterize expression patterns of cytolytic associated markers in the AGM CD8αα T cell subset. Finally, we aim to decipher whether the functionality of the CD8αα subset may affect the infectivity of LN resident Tfh cells. The results of this study will illuminate how AGMs limit disease progression and inform future HIV therapeutic development.
Materials and Methods
Study Animals & Sample Processing
Twenty-six rhesus macaques (RMs, Macacca mulatta), nineteen African green monkeys (AGMs, Chlorocebus pygerythrus), and five patas monkeys (Erythrocebus patas) were used for this study (Table 1). The RM study cohort was comprised of 14 SIV-infected RMs (8 chronically infected with SIVmac239, 2 chronically infected with SIVmac251, 1 chronically infected with SIVsmE543, and 3 with SIVmac239-associated simian AIDS) and 12 SIV-uninfected RMs. The AGM cohort was comprised of 13 SIV-infected AGM (9 chronically infected with SIVagm90 and 4 naturally infected) and 6 SIV-uninfected AGM. The patas cohort was limited to 5 SIV-uninfected animals. All animals were housed and cared for in accordance with standards outlined in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health in AAALAC-accredited (American Association for Accreditation of Laboratory Animal Care) facilities. The NIAID Division of Intramural Research Animal Care and Use Committees (IACUC) approved all animal procedure protocols (LPD26 and LMM6).
Table 1.
Study Animals
| Animal | Species | Infection Status | Tissues Studied | Disease Statea | Plasma Viremiab | CD4+ T cellsc |
|---|---|---|---|---|---|---|
| RH37360 | RM | SIV− | PBMC | N/A | 0 | 1267 |
| RHDFAi | RM | SIV− | PBMC, BAL | N/A | 0 | 1213 |
| RHF64 | RM | SIV− | PBMC, BAL | N/A | 0 | 1092 |
| RH37033 | RM | SIV− | PBMC, BAL | N/A | 0 | 1461 |
| RH37034 | RM | SIV− | PBMC, BAL | N/A | 0 | 1473 |
| RH37073 | RM | SIV− | PBMC, BAL | N/A | 0 | 512 |
| RHDBV1 | RM | SIV− | LN | N/A | 0 | 693 |
| RHDBXG | RM | SIV− | LN | N/A | 0 | 696 |
| RH485 | RM | SIV− | LN | N/A | 0 | 269 |
| RHDA6A | RM | SIV− | LN | N/A | 0 | 472 |
| RH595 | RM | SIV− | LN | N/A | 0 | 685 |
| RH769 | RM | SIV− | LN | N/A | 0 | 465 |
| RHF98 | RM | SIVmac239 | PBMC | Chronic | 1.90×102 | 923 |
| RHR106 | RM | SIVmac251 | PBMC | Chronic | 6.00×102 | 1137 |
| DE2W | RM | SIVmac239 | PBMC | Chronic | 9.20×105 | 209 |
| RHR444 | RM | SIVmac251 | PBMC, BAL | Chronic | 1.40×105 | 479 |
| RHDCBC | RM | SIVmac239 | PBMC, BAL | Chronic | 2.00×106 | 253 |
| DE1A | RM | SIVmac239 | PBMC, BAL | Chronic | 7.80×104 | 680 |
| RHZG24 | RM | SIVmac239 | PBMC, BAL | Chronic | 2.60×105 | 394 |
| RHZA52 | RM | SIVmac239 | PBMC, BAL | Chronic | 1.40×106 | 406 |
| RHDB4E | RM | SIVmac239 | LN | Chronic | 8.10×105 | 565 |
| RH760 | RM | SIVsmE543 | LN | Chronic | 5.00×103 | 296 |
| RHDB07 | RM | SIVmac239 | LN | Chronic | 5.50×105 | 317 |
| RHDB17 | RM | SIVmac239 | LN | sAIDs (likely Pneumocystis pneumonia) | 9.20×104 | 122 |
| RHCF4J | RM | SIVmac239 | LN | sAIDs (Parainfluenza) | 2.00×105 | 241 |
| RHCF5T | RM | SIVmac239 | LN | sAIDs (Strep bovis) | 8.00×105 | 216 |
| AG33 | AGM | SIV− | PBMC, BAL | N/A | 0 | 373 |
| AG5419 | AGM | SIV− | PBMC, BAL | N/A | 0 | 70 |
| AG5506 | AGM | SIV− | PBMC, BAL | N/A | 0 | 191 |
| AG36 | AGM | SIV− | PBMC, BAL | N/A | 0 | 218 |
| AGY682 | AGM | SIV− | PBMC, BAL | N/A | 0 | 416 |
| AG5339 | AGM | SIV− | LN | N/A | 0 | 500 |
| AG32 | AGM | SIVagm90 | PBMC | Chronic | undetected | 358 |
| AG346 | AGM | SIVagm90 | PBMC, BAL | Chronic | 3.73×103 | 0 |
| AG38 | AGM | SIVagm90 | PBMC, BAL | Chronic | 8.69×104 | 609 |
| AG28 | AGM | SIVagm90 | PBMC, BAL | Chronic | 1.18×105 | 193 |
| AG5431 | AGM | SIVagm90 | PBMC, BAL | Chronic | 5.40×103 | 84 |
| AG5387 | AGM | SIVagm90 | PBMC, BAL | Chronic | 1.39×105 | 224 |
| AG16 | AGM | Naturally Infected | LN | Chronic | 3.19×104 | 404 |
| AG10 | AGM | Naturally Infected | LN | Chronic | 5.39×104 | 233 |
| AG12 | AGM | Naturally Infected | LN | Chronic | 4.77×102 | 132 |
| AG17 | AGM | Naturally Infected | LN | Chronic | 9.70×104 | 483 |
| AG35 | AGM | SIVagm90 | LN | Chronic | 1.97×104 | 533 |
| AG31 | AGM | SIVagm90 | LN | Chronic | undetected | 2001 |
| AG37 | AGM | SIVagm90 | LN | Chronic | 1.62×104 | 409 |
| R294 | Patas | SIV− | PBMC, LN | N/A | 0 | N/D |
| R300 | Patas | SIV− | PBMC, LN | N/A | 0 | N/D |
| R302 | Patas | SIV− | PBMC, LN | N/A | 0 | N/D |
| R304 | Patas | SIV− | PBMC, LN | N/A | 0 | N/D |
| R303 | Patas | SIV− | LN | N/A | 0 | N/D |
N/A = not applicable
copies of viral RNA/ml of plasma
N/D = not determined
Peripheral blood and bronchoalveolar lavage (BAL) were processed and analyzed from fresh samples, while cell suspensions from axillary and inguinal lymph nodes (LNs) were analyzed from frozen samples attained at necropsy. Peripheral blood mononuclear cells (PBMCs) were isolated via standard density gradient centrifugation using lymphocyte separation medium (LSM, MP Biomedicals, Santa Ana, CA). BAL samples were washed twice in complete medium consisting of: Hyclone RPMI 1640 media supplemented with 10% Hyclone Defined Fetal Bovine Serum, 1% Hyclone L-glutamine 200MM, and 1% Hyclone Penicillin/Streptomycin solution (GE Healthcare Bio-Sciences Corp, Marlborough, MA). BAL samples were then filtered and resuspended in complete medium. LNs were processed into single-cell suspensions as described previously (22). To study T cell functionality, we incubated cells from peripheral blood, LN, or BAL overnight at 37°C with medium alone (unstimulated control) or 1 μg/mL of staphylococcus enterotoxin B (SEB) (Sigma, St. Louis, MO) in the presence of 5 μL/mL of CD107a APC monoclonal antibody (H4A3; Biolegend; San Diego, CA), 1 μL/mL BD GolgiStop (BD Biosciences; San Diego, CA), and 1 μg/mL Brefeldin A (Sigma).
Flow cytometry
Immunophenotypic and functional characteristics were analyzed utilizing multicolor flow cytometry. Mononuclear cells were first stained with a live/dead fixable aqua dead cell dye (Life Technologies, Carlsbad, CA). The following Mabs were then used for ex vivo staining of cells: CD3 ALX700 (Clone SP34-2), CD4 BV605 (L200), IFNγ PE-Cy7 (4S.B3), TNFα FITC (MAb11), and Granzyme B PE (GB11) from BD Biosciences; CD8 PB or CD8 BV650 (RPA-T8), CD95 PE-Cy5 (DX2), IL-2 BV785 (MQ1-17H12), Foxp3 ALX488 (206D), IL-5 PE (JES1-39D10), and IL-13 APC (JES10-5A2) from Biolegend; CD28 ECD (28.2) from Beckman Coulter (Brea, CA); and PD-1 PerCP-eFluor710 (eBioJ105), CXCR5 eFluor450 (MU5UBEE), IL-21 PE (eBio3A3-N2), and CD40L APC-eFluor780 (24–31) from Affymetrix/eBioscience (San Diego, CA). Cells were stained for surface markers, washed in PBS, and then permeabilized and washed with the Foxp3/Transcription Factor Staining Buffer Set from Affymetrix/eBioscience before intracellular staining.
All flow samples were run on LSR Fortessa or FACS Aria cytometers (BD Biosciences) and analyzed using the Tree Star FlowJo software version 9.9 (Ashland, OR). A threshold cutoff of 200 cells of the parent population was used for all cell subset analysis. Absolute cell counts were calculated from flow cytometric determination of cell frequencies and complete blood cell absolute lymphocyte counts (Antech, Irvine, CA).
We gated lymphocytes based on size by FSC-A and SSC-A, followed by singlets, live CD3+, memory (CD28+/−CD95+). From this parent population we then gated on CD4+, CD8bright (CD8αβ), CD8dim (CD8αα), and double-negative (DN; cells that expressed neither CD4 nor CD8) cell populations, followed by their corresponding expression of each cytokine or functional marker. When characterizing Tfh cells we used the following gating: lymphocytes/singlets/live CD3+/memory/Tfh (CXCR5+PD1+)/T cell subset.
Real-time PCR analysis of ThPOK and Runx3 expression
Memory CD4+, CD8αβ, and CD8αα T cells were sorted from RM and AGM peripheral blood samples using a FACS Aria II flow cytometer. Total RNA was extracted using TRI reagent (Sigma Aldrich, St. Louis, MO), followed by reverse transcription (Superscript III, Life Technologies). Real-time PCR was performed using Power SYBR Green Master Mix (Applied Biosystems, Foster City, CA) with the following detection primers: ThPOK Forward 5′-GGAGTTCCCAATGGTGAAGA-3′; ThPOK Reverse 5′-TCAGGGACTAGGTGGTTTGC-3′; Runx3 Forward 5′-TTCCTAACTGTTGGCTTTCC-3′; Rux3 Reverse 5′-TAGGTGCTTTCCTGGGTTTA-3′; ACTB forward 5′-ATTGCCGACAGGATGCAGAA-3′, and ACTB reverse 5′-GCTGATCCACATCTGCTGGAA-3′. Amplification was achieved using the following conditions: 95°C holding stage for 10 minutes, followed by 45 cycles at 94°C for 30 seconds, 58°C for 45 seconds, and 72°C for 30 seconds. Relative expression was calculated using the ΔCt method.
Quantitative PCR for SIV DNA
We sorted CD4+ non-Tfh (live, CD3+CD4+CD28+/−CD95+CXCR5−PD1low lymphocytes) and CD4+ Tfh (live, CD3+CD4+CD28+/−CD95+CXCR5+PD1high lymphocytes) cell populations utilizing a BD FACS ARIA II flow cytometer and then lysed the sorted cells with 25 μL of a 1:100 dilution of proteinase K (Roche, Indianapolis, IN) in 10 mM Tris buffer (23). We performed quantitative PCR with 5 μL of cell lysates per reaction using the following reaction conditions: 95°C for 5 minutes and 40 cycles of 95°C for 15 seconds followed by 60°C for 1 minute. We used the TaqMan Gene Expression Master Mix (Life Technologies) with species specific PCR primers and probes used to amplify viral DNA (Table 2). The StepOne Plus PCR machine and software was used to amplify genes of interest (Applied Biosystems, Foster City, CA).
Table 2.
qPCR Primers and Probes
| Virus | Forward Primer | Reverse Primer | Probe |
|---|---|---|---|
| SIVmac | GTCTGCGTCAT(T/C)TGGTGCATTC | CACTAG(C/T)TGTCTCTGCACTAT(A/G)TGTTTTG | CTTC(A/G)TCAGT(C/T)TGTTTCACTTTCTCTTCTGCG |
| SIVsm | GGCAGGAAAATCCCTAGCAG | GCCCTTACTGCCTTCACTCA | AGTCCCTGTTCRGGCGCCAA |
| SIVagm | GTCCAGTCTCAGCATTTACTTG | CGGGCATTGAGGTTTTTCAC | CAGATGTTGAAGCTGACCATTTGGG |
Plasma Viral Load
A real-time RT-PCR assay for quantitation of viral RNA in plasma was performed as previously described (24).
SIV DNAscope & Quantitative Image Analysis
DNAscope in situ hybridization (ISH) was used to detect SIV vDNA positive cells within the LN as previously described (25, 26) and as follows in brief. Inguinal and or axillary LNs were collected at time of necropsy in freshly prepared 4% paraformaldehyde (PFA) for 24 hours, followed by replacement of fixative with 80% ethanol. LN tissues were then paraffin embedded. A sense probe targeting the reverse strand within 943-8792 of SIVagm ver (accession number L40990.1, https://www.ncbi.nlm.nih.gov/nuccore/L40990.1, gag, pol, vif, vpr, tat, rev, and env) was utilized for DNAscope analysis (ACD, Cat. #300031-C1). RNAscope and DNAscope in situ hybridization were performed on subjacent sections of lymph nodes for each sample.
Slides with DNAscope stained tissues were scanned at 40x magnification with an AT2 slide scanner (Aperio Technologies, Vista, CA). Regions of interest (ROIs), including B cell follicles and T cell zone tissue were saved as TIFF images for analysis. We counted hematoxylin-stained nuclei and DAB or Fast-Red-stained vDNA+ cells in B cell follicles and T cell zones with Photoshop CS3 (Adobe Systems, San Jose, CA) and Fovea Pro 4 (Reindeer Graphics, Asheville, NC). The vDNA+ cells were counted as previously described (26). Nuclei were counted with a similar method: duplicating an ROI, thresholding its red color channel, copying and pasting the thresholded image as a new layer on the original image, refining segmentation by morphological opening and closing based on a depth of 1.5 pixels, watershed segmentation, and counting the segmented objects.
Statistical Analyses
Statistical analyses were performed with v6.0 Prism (GraphPad Software, La Jolla, CA). The Wilcoxon matched-pairs signed rank test was used for cell subset comparisons within species and the Mann-Whitney U test was used for cell subset comparisons between species. Pie charts and functionality comparisons were assessed using SPICE v5.3 (National Institute of Allergy and Infectious Diseases). The permutation test within SPICE was run with 10,000 permutations and each species cell subset was treated as an overlay, with each sample grouped. The P-values included with each SPICE analysis reflects the differences between T cell subsets based upon differential combinations of functionalities (i.e. CD40L+CD107+Foxp3−Granzyme B−IFNγ−IL-2−) and not differences of total number of functions (i.e. 1+ function, 2+ functions, etc).
Results
ThPOK and Runx3 are expressed in AGM CD8αα T cells
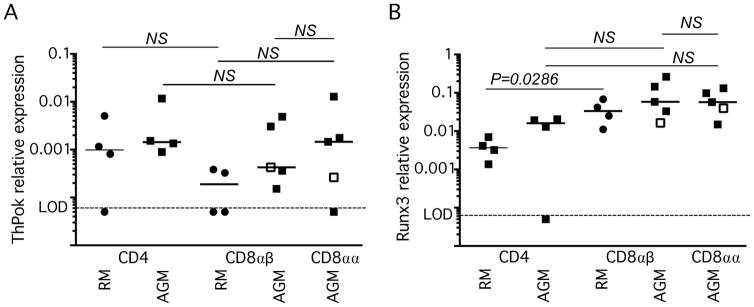
Given the important relationship of ThPOK and Runx3 in the expression and maintenance of CD4 and CD8 in the murine model, we evaluated the relative expression of these transcription factors in the T cell subsets of RMs and AGMs. Using flow cytometric sorting, we isolated memory CD4+, CD8αβ, and CD8αα T cells from the peripheral blood of four SIV-uninfected RM and five AGMs (one SIVagm90 infected, with no CD4+ T cells as previously reported (27)). Surprisingly, unlike observations in mice, all T cell subsets from both AGM and RM expressed similar levels of ThPOK (Fig. 1A). Moreover, in AGM, all T cell subsets, irrespective of CD4/CD8 expression patterns, expressed similar levels of Runx3 (Fig. 1B). RM CD8αβ T cells expressed Runx3 at significantly higher levels than did RM CD4 T cells (P=0.0286, Fig 1B). The lack of expression differences of ThPOK and Runx3 among the AGM T cell subsets may, indeed, contribute to decreased expression of CD4 by CD4+ T cells after their immunological experience.
Fig. 1. ThPOK and Runx3 expression in memory T cell subsets of rhesus macaques (RMs) & African Green Monkeys (AGMs).
(A) ThPOK mRNA and (B) Runx3 mRNA in memory CD4+, CD8αβ, and CD8αα peripheral blood T cells of RMs and AGMs. Expression is relative to β-actin mRNA. All horizontal bars represent the median. Limit of detection (LOD) is <0.0005.
Functionality of PB T cells
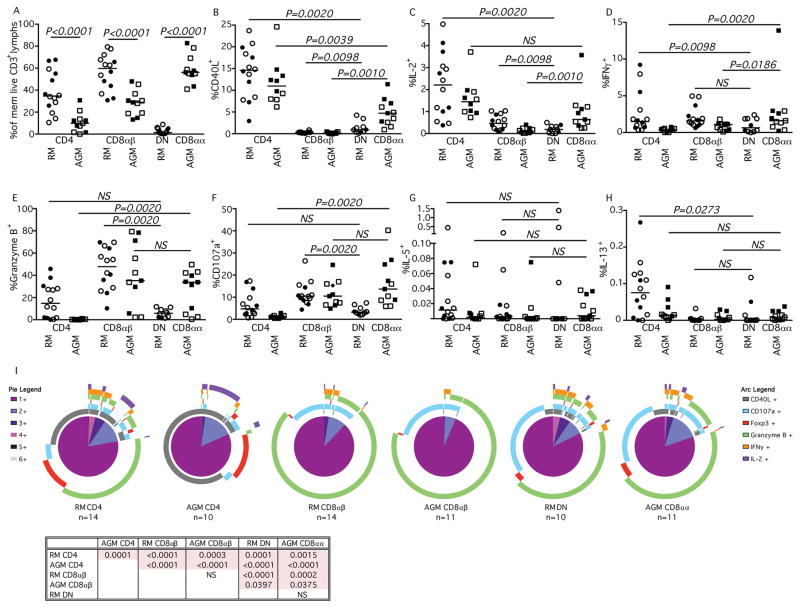
Previously, we reported that AGM have high frequencies of T cells that down regulate CD4 and up regulate the alpha chain of CD8, while maintaining effector functions generally associated with CD4+ T cells (7). Recent studies in mice suggest that Th2 responses require expression of CD4 and that loss of CD4 expression by MHC-II-restricted T cells can be associated with an increased CD8+ T cell associated effector functionality (12, 13). To assess cellular immunophenotype and functionality of T cells from progressive and nonprogressive NHP species, we stimulated peripheral blood mononuclear T cells from rhesus macaques (whose CD4+ T cells maintain expression of CD4), AGM, and patas (whose CD4+ T cells can down regulate CD4 expression) with the super antigen Staphylococcus enterotoxin B (SEB) (Fig. 2 and Fig. S1). From each species, frequencies of memory T cells that expressed CD4, CD8αα, CD8αβ, or were double negative (DN, Fig. 2A) mirrored what we have reported previously. Within these populations, we then measured SEB-induced expression of CD40L (Fig. 2B), IL-2 (Fig. 2C), IFNγ (Fig. 2D), Granzyme B (Fig. 2E), CD107a (Fig. 2F), IL-5 (Fig. 2G), and IL-13 (Fig. 2H) (patas data, Supplemental Fig. 1). Consistent with our previous reports, the CD8αα T cells from AGM, and the CD8αα and DN T cells from patas animals were able to elicit effector functions normally associated with CD4+ T cells (6). While, significantly lower frequencies of CD8αα T cells from AGM expressed CD40L and IL-2 than canonical CD4+ T cells, a significantly higher percentage of them expressed these markers than did the CD8αβ T cells of either RM or AGM. These AGM CD8αα T cells could additionally perform effector functions normally associated with CD8 T cells including expression of IFNγ, Granzyme B, and were able to mobilize CD107a to the cell surface. Indeed, similar frequencies of CD8αα and CD8αβ T cells from AGM expressed Granzyme B and mobilized CD107a after stimulation (Fig. 2E & 2F). Similar results were seen in patas CD8αα T cells that had down-regulated CD4 (Supplemental Fig. 1E & 1F). Thus, these results suggest that in AGM and patas, circulating T cells that down regulate CD4 maintain “CD4+ T cell” functions, while also developing effector functions normally attributed to CD8αβ T cells.
Fig. 2. Peripheral blood T cell functionality in RMs & AGMs.
Characterization of (A) memory T cell lymphocyte subsets and their expression of (B) CD40L, (C) IL-2, (D) IFNγ, (E) Granzyme B, (F) CD107a, (G) IL-5, and (H) IL-13 after SEB stimulation in RM shown in circles and AGM shown in squares. Closed symbols represent SIV-uninfected animals, while open symbols represent SIV-infected animals. (I) SPICE analysis illustrating number of functions in corresponding colored pie sections (legend on the left) and specific functionality as illustrated by the colored arcs (legend on the right). Multifunctional profile differences between cellular subsets (P-values) were assessed by the SPICE software permutation test. All horizontal bars in A–H reflect the median of that T cell subset. All P-values greater than 0.05 are represented as NS (not significant).
Given recent data suggesting that loss of CD4 expression by CD4+ T cells renders T cells incapable of eliciting Th2 types of immune responses (28), we also measured expression of IL-5 and IL-13 post-SEB stimulation. We found that in AGM and patas low frequencies of stimulated T cells that had down regulated CD4 could express IL-13 and IL-5, similar to the frequencies of both CD8αβ T cells and canonical CD4+ T cells (Fig. 2G & 2H). However, it is important to note that there were very low frequencies of T cells that produced either IL-13 and/or IL-5 in peripheral blood.
To categorize CD8αα T cells along the spectrum of canonical CD4 and CD8αβ function, we graphically visualized the peripheral blood flow cytometry data with SPICE and observed the truly intermediary immunophenotype of the AGM CD8αα T cells (Fig. 2I). These cells share functional characteristics of both CD4+ and CD8αβ T cells, but are still significantly functionally distinct. While the CD8αβ T cells of RM and AGM are remarkably similar, their CD4+ T cell subsets are functionally distinct. Surprisingly, the low frequency of CD4/CD8 double negative (DN) T cells in RM and AGM CD8αα T cells share many functional characteristics (P=0.5653, Fig. 2I). The shared functionality of the AGM CD8αα and RM DN T cells demonstrate that seemingly neither the expression of CD4 or the CD8β chain are necessary for the CD4 and CD8 T cell ascribed functions.
Functionality of BAL T cells
Several studies have shown that Th2 type responses are present at higher levels within the lung (reviewed in (29)). Therefore, we extended our analysis to the functionalities of T cell subsets in broncheoalveolar lavage samples (BAL). In AGMs BAL T cells are predominantly comprised of CD8αα cells that have down-regulated CD4 and conventional CD8αβ cells (52.17% and 42.96%, respectively), with few contributing canonical CD4+ T cells (Fig. 3A). We found that stimulated CD8αα T cells in the BAL are capable of retaining CD4+ T cell functionality including CD40L and IL-2 expression (Fig. 3B & 3C). Similar to what we saw in peripheral blood, AGM CD8αα T cells within the BAL also exhibited CD8 T cell functions including the expression of IFNγ (Fig. 3D). Additionally, AGM BAL CD8αα T cells were capable of expressing both IL-5 and IL-13 (Fig. 3E & 3F). Hence, the CD8αα T cells in BAL could certainly perform effector functions associated with CD4+ Th2 cells. SPICE analysis of BAL cellular functionality characterized the CD8αα T cells as again, an intermediate cell type, having functionality of both CD4+ and CD8αβ T cell subsets, with the AGM BAL CD8αα T cell subset being functionally distinct from all other examined T cell subsets, irrespective of species (Fig. 3G).
Fig. 3. BAL T cell functionality in RMs & AGMs.
Characterization of (A) memory T cell lymphocyte subsets and their expression of (B) CD40L, (C) IL-2, (D) IFNγ, E) IL-5, and (F) IL-13 after SEB stimulation in RM shown in circles and AGM shown in squares. Closed symbols represent SIV-uninfected animals, while open symbols represent SIV-infected animals. (G) SPICE analysis illustrating number of functions in corresponding colored pie sections (legend on the left) and specific functionality as illustrated by the colored arcs (legend on the right). Multifunctional profile differences between cellular subsets (P-values) were assessed by the SPICE software permutation test. All horizontal bars in A–F reflect the median of that T cell subset. All P-values greater than 0.05 are represented as NS (not significant).
Functionality of LN T cells
Given the observation of multi-functional CD8αα T cells in both the peripheral blood and BAL, we next sought to assess functionality in the LN – in particular, if the CD8αα T cells could behave as Tfh cells. Tfh cells play an important role in promoting the activation, expansion, and differentiation of B cells, as well as contributing to the formation of the germinal center. The ability of the AGM CD8αα T cells to interact within the immune system with the dual functionality of both CD4+ and CD8αβ T cells could allow them to avoid SIV infection while maintaining protective cytolytic activity. This may be particularly important at immunological sites such as the lymph node that serve as long-term reservoirs for both HIV and SIV infections (30).
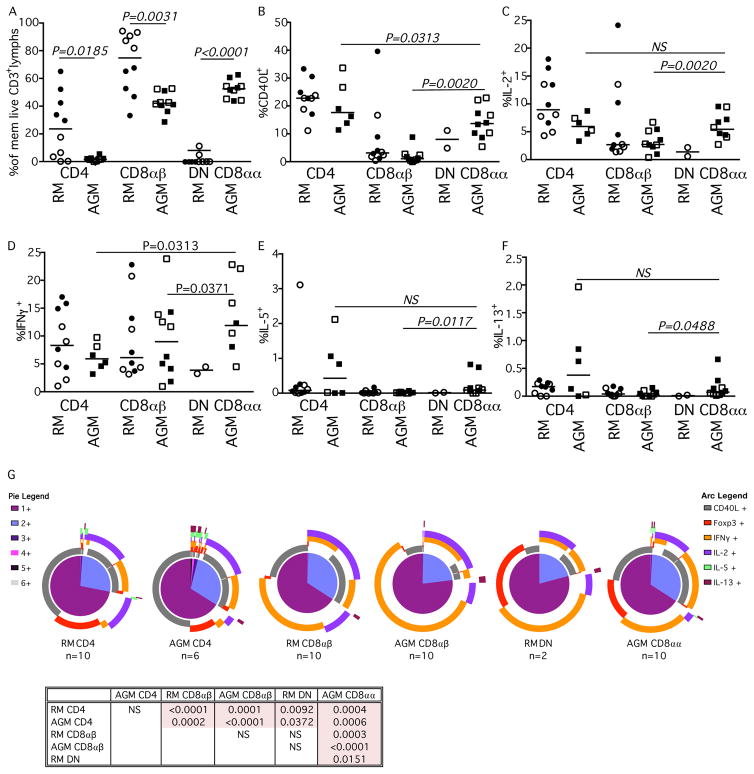
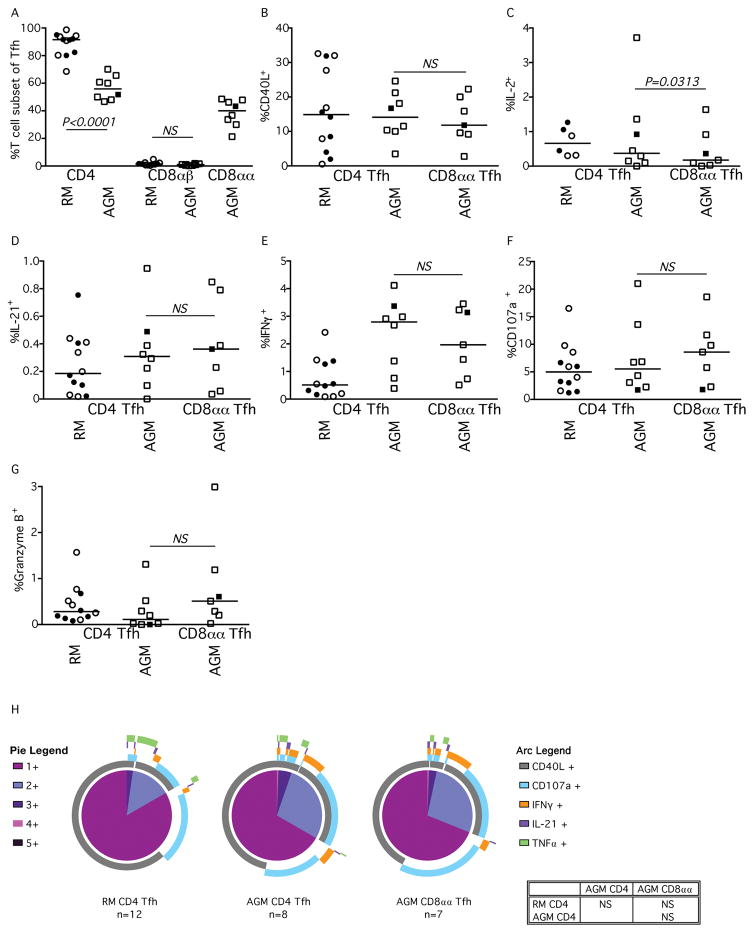
Within the LN of AGM the CD8αα T cells comprised 46.4% to 59.4% of memory T cells (Fig. 4A). The percentage of CD40L and IL-2 expressing CD8αα AGM T cells were between that of CD4+ and the CD8αβ T cell subsets of both AGM and RM (Fig. 4B & 4C). Additionally, the frequencies of AGM CD8αα and AGM CD8αβ cells expressing IFNγ were similar (Fig. 4D). Although frequencies of AGM CD8αα T cells within the LN that could mobilize CD107a to the surface were comparable with the frequencies of CD8αβ T cells (Fig. 4E), fewer CD8αα T cells expressed Granzyme B compared to CD8αβ T cells (Fig. 4F). Analysis of TNFα expression showed no significant differences among AGM T cell subtypes (data not shown). While higher frequencies of AGM CD4+ T cells were phenotypically characterized as Tfh cells (based upon expression of CXCR5 and PD1) compared to CD8αα T cells (Fig. 4G), it was clear that the AGM CD8αα T cells could exhibit Tfh phenotypes which was not observed among CD8αβ T cells of either RM or AGM. Similarly, the highest expression of the Tfh effector cytokine IL-21 was observed in stimulated CD4+ T cells, while IL-21 was practically absent in CD8αβ T cells of either RM or AGM, yet some CD8αα T cells could produce this cytokine (Fig. 4H). Although, these data strongly suggest that AGM CD8αα T cells can behave as Tfh cells, Spice analysis of the overall functionality of the lymphocyte cellular subsets within the LN suggests that the profile of the AGM CD8αα T cells more closely resembles that of the CD8αβ T cell subset of the RM than the CD4+ subset of either species (Fig. 4I).
Fig. 4. Functionality of LN-resident T cell subsets in RMs and AGMs.
Characterization of (A) memory T cell CD4+, CD8αβ, and CD8αα subsets and their expression of (B) CD40L, (C) IL-2, (D) IFNγ, E) CD107a, (F) Granzyme B, (G) Tfh markers CXCR5 and PD-1, (H) IL-21, and TNFα (graphical data not shown) after SEB stimulation in RM (circles) and AGM (squares). Closed symbols represent SIV-uninfected animals, while open symbols represent SIV-infected animals. (I) SPICE analysis illustrating number of functions in corresponding colored pie sections (legend on the left) and specific functionality as illustrated by the colored arcs (legend on the right). Multifunctional profile differences between cellular subsets (P-values) were assessed by the SPICE software permutation test. All horizontal bars in A–H reflect the median of that T cell subset. All P-values greater than 0.05 are represented as NS (not significant).
To better understand the degree to which CD8αα T cells contribute to the overall levels of Tfh cell activity within lymphoid tissues of AGM, we identified Tfh cells (based upon expression of CXCR5 and PD1) and then determined the frequencies of those cells that expressed CD4, CD8αβ, or CD8αα in AGM and RM (Fig. 5A). As expected, in RM the vast majority (median 92%) of all Tfh cells were CD4+ T cells. However, in AGM many (up to 50%) of the Tfh cells were CD8αα T cells (Fig. 5A).
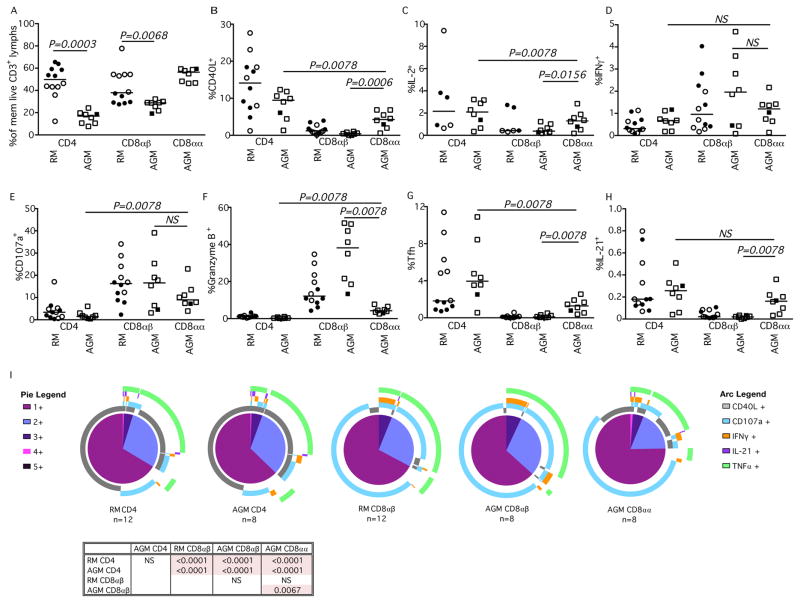
Fig. 5. Functionality of Tfh T-cell subsets of RMs and AGMs.
Percent of (A) CXCR5+PD-1+ memory CD3+ T cell lymphocyte CD4+, CD8αβ, and CD8αα subsets and their expression of (B) CD40L, (C) IL-2, (D) IL-21, E) IFNγ, (F) CD107a, and (G) Granzyme B after SEB stimulation in RM (circles) and AGM (squares). Closed symbols represent SIV-uninfected animals, while open symbols represent SIV-infected animals. (I) SPICE analysis illustrating number of functions in corresponding colored pie sections (legend on the left) and specific functionality as illustrated by the colored arcs (legend on the right). Multifunctional profile differences between Tfh cellular subsets (P-values) were assessed by the SPICE software permutation test. All horizontal bars in A–G reflect the median of that T cell subset. All P-values greater than 0.05 are represented as NS (not significant).
We next sought to examine the functionality of Tfh cells in RM and AGM based upon their differential expression of CD4 or CD8αα (Fig. 5B–H). We examined several important Tfh functions including expression of CD40L (Fig. 5B), IL-2 (Fig. 5C), and IL-21 (Fig. 5D). With the exception of IL-2, the CD8αα Tfh cells were able to elicit similar functional responses when compared to CD4+ Tfh cells of either RM or AGM. We also measured expression of IFNγ (Fig. 5E), mobilization of CD107a (Fig. 5F) and expression of Granzyme B (Fig. 5G) to determine if CD8αα Tfh cells could function cytolytically within the LN. The CD8αα Tfh cells had similar expression of these functions, normally associated with CD8 T cell effector functions, as the CD4+ Tfh cells from both RM and AGM. Indeed, Spice analysis shows that the functionality of the different Tfh cells were comparable (Fig. 5H). Thus the CD8αα Tfh cells did not exhibit the CD4+/CD8+ T cell dual functionality observed among the other memory CD8αα T cells.
Tfh cells are preferentially infected in progressive infection by virtue of the fact that they express CD4, are highly activated, and located in a microenvironment ideally suited for infection. We have previously shown that down-regulation of CD4 influences how T cell subsets are targeted by SIVagm in vivo in peripheral blood (7) and we postulated that CD4 down-regulation might influence infectivity of Tfh cells in AGM lymphoid tissues in vivo. We used DNAscope in situ hybridization to detect and quantify the number of viral DNA+ cells in paraffin-embedded lymph nodes and to determine the anatomic localization of SIVagm-infected cells in distinct LN microenvironments (i.e. B cell follicles or T cell zones). Hemotoxylin counter staining of DNAscope stained tissue sections allowed us to quantitate how many viral DNA+ cells resided within the distinct T cell zone and B cell follicle anatomical sites (Fig. 6A). This analysis revealed that there were comparable numbers of SIV viral DNA+ cells within the lymphoid follicles of AGM lymph nodes compared to the numbers of viral DNA+ cells within the T cell zone (Fig. 6B).
Fig. 6. SIV viral DNA infectivity in the AGM LN.
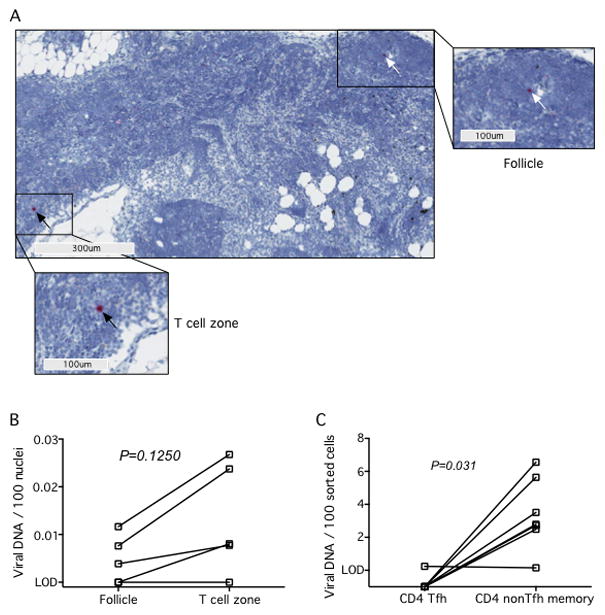
(A) Representative DNAscope image showing SIV vDNA+ cells located within follicles (white arrows) and in the paracortex (black arrows) in a representative AGM at low magnification (main panel, 100μm) and high magnification (300μm). (B) Quantitation of the number of SIV vDNA+ cells located within B cell follicles or T cell zones of AGM by DNAscope ISH staining. The limit of detection (LOD) is less than 1 in 7,000 cells. (B) Memory CD4+ non-Tfh (CXCR5−PD-1low) and memory CD4+ Tfh (CXCR5+PD-1high) were flow cytometrically sorted from LNs of SIV-infected AGM and infection levels of the subsets were determined by quantitative PCR. The LOD is less than 1 in 10,000 cells. Lines connecting data points indicate cells from the same animal (matched pairs). All P values are based on the Wilcoxon matched pairs test.
To further address specific infectivity of the CD4+ Tfh subset we flow cytometrically sorted CD4+ Tfh cells and CD4+ memory T cells that were not Tfh cells (based upon expression of CXCR5, PD1, CD28, and CD95) and assessed infection status by qPCR for SIVagm DNA (Fig. 6C). In doing so, we found very low levels of viral DNA in AGM Tfh cells, with higher levels of viral DNA in lymph node-resident non Tfh memory T cells. Conversely, we have previously shown in RM increased frequencies of SIV-infected CD4+ Tfh cells when compared to non Tfh, memory, CD4+ T cells (15). Collectively, these data demonstrate that SIVagm does not preferentially infect AGM Tfh cells in vivo.
Discussion
Here we have studied the functionality of tissue-resident T cells in AGM based upon differential expression of CD4 and CD8. We find that: i) AGM CD8αα T cells (CD4+ T cells that have down-regulated CD4 expression) express both ThPOK and Runx3 at similar levels as CD4+ and CD8αβ T cells respectively; ii) CD8αα T cells in peripheral blood and tissues maintain effector functions of CD4+ T cells including those of Th2 cells and Tfh cells; iii) CD8αα T cells can also accumulate some effector functions normally associated with cytotoxic CD8αβ T cells; and iv) down-regulation of CD4 by Tfh cells in lymphoid tissue protects these cells from preferential infection by SIVagm. These data show the probable irrelevance of CD4 expression on functionality of T helper cell subsets in primates and how CD8 expression by CD4 T cells allows them to accumulate CD8 T cell functionality.
T helper lymphocyte functionalities are dictated by the differential expression patterns of particular master transcription factors such as T-Bet (Th1), Gata-3 (Th2), FoxP3 (Treg), and RORγt (Th17/Th22) (31). Although these transcription factors mediate the functional responses of their respective cell type, these T cell lineage fates are not fixed (31). While T helper cell functionality might have some degree of plasticity, post-thymic expression of CD4 by these T cells is, in almost every mammalian species, constant. Similar to the control of helper cell functionality, expression patterns of CD4 and CD8 are dictated by differential expression patterns. Here we studied the relative expression the transcription factors ThPOK and Runx3 in sorted AGM and RM T cell subsets and found similar expression levels among all AGM T cell subsets. However, in RM Runx3 was expressed at higher levels in CD8αβ T cells, which is congruent with findings in murines wherein ThPOK and Runx3 serve as master transcriptional factors regulating CD4/CD8 expression and function. Interestingly, the lack of differences in relative ThPOK and Runx3 expression levels among AGM T cell subsets may underlie the CD4 down-regulation and their ability to acquire effector functions normally attributed to cytolytic T cells, as we observe in the peripheral blood, BAL, and LN. ThPOK deleted murine T cells are able to simultaneously maintain MHC-II restriction while maintaining most effector functions of both T helper cells as well as cytotoxic CD8 T cells, including degranulation and expression of Granzyme B (32, 33). While the CD4 down-regulated T cells tended to maintain T helper effector functions in the conditional ThPOK knockout mice, these T cells could not effectively perform Th2 effector functions after parasitic infections and their ability to perform Tfh activity has not thoroughly been studied (28).
Our data shows that the AGM CD8αα T cells in circulation, within the lung, and lymph node are capable of Th1, Th2, Tfh and, to some extent CTL functions. Thus the lack of Th2 activity observed in murine models when CD4 is down-regulated, does not demonstrate an absolute necessity of CD4 for Th2 functionality in AGMs or other primate species. In fact, DN cells of SMs have been shown to have Th1, Th2, Th17, and Tfh functionality despite their lack of CD4 expression (5). Interestingly, Sundaravaradan et al demonstrated that SIV infection augments the ability of DN T cells to upregulate both IL-5 and IL-13 while maintaining IL-4 and IL-10 (5), further demonstrating that CD4 expression is not necessary for Th2 functionality. Furthermore, our analysis of DN and CD8αα T cell subsets in the peripheral blood of patas monkeys demonstrates this functionality may be conserved in CD4− T cell subsets among multiple natural hosts species of SIV (Suppl. Fig. 1).
Increased cytolytic function by AGM CD8αα cells may include an absolute increase in anti-SIV responses, as is observed in controller macaques (34). Indeed, decreased infection of CD4+ Tfh relative to non-Tfh may be a reflection of increased anti-SIV responses resulting in a limitation of SIV infection within lymphoid follicles. Although approximately 50% of all AGM Tfh cells were CD8αα these T cells did not, preferentially, exhibit CTL activity. Thus the lack of preferential infection of AGM Tfh cells does not appear to be mediated by CD8αα T cell clearance. Rather, down regulation of CD4 on these cells in combination with a very low level of viral trapping on the FDC network similar to what we have seen previously in sooty mangabeys is the likely mechanism (15).
These data demonstrate that CD4 expression is not critically important to maintain the effector functions of T helper cells in primates, thus highlighting the importance of CD4 down-regulation for the nonprogressive nature of SIVagm infection of AGM in maintenance of immunological function. We believe these data could lead to novel therapeutic interventions for HIV-infected individuals.
Supplementary Material
Acknowledgments
We thank the veterinary staff at the NIH NIAID Animal Center with special appreciation to Heather Kendall, Joanna Swerczek, and Richard Herbert for their excellent animal care.
Footnotes
Funding for this study was provided in part by the Division of Intramural Research/NIAID/NIH and by federal funds from the National Cancer Institute (NIH Contract HHSN261200800001E). The content of this publication does not necessarily reflect the views or policies of DHHS, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Author Contributions:
Conceived and designed experiments: CLV, JMB. Performed the experiments: CLV, NC, CD, JMB. Data analysis: CLV, NC, AMO, DM. Contributed reagents/materials/analysis tools: JMB, VMH, JDE, SW, CD, DM. Wrote the paper: CLV, JB
Bibliography
- 1.Pandrea I, Apetrei C. Where the wild things are: pathogenesis of SIV infection in African nonhuman primate hosts. Current HIV/AIDS reports. 2010;7:28–36. doi: 10.1007/s11904-009-0034-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sodora DL, Allan JS, Apetrei C, Brenchley JM, Douek DC, Else JG, Estes JD, Hahn BH, Hirsch VM, Kaur A, Kirchhoff F, Muller-Trutwin M, Pandrea I, Schmitz JE, Silvestri G. Toward an AIDS vaccine: lessons from natural simian immunodeficiency virus infections of African nonhuman primate hosts. Nat Med. 2009;15:861–865. doi: 10.1038/nm.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenchley JM, Paiardini M. Immunodeficiency lentiviral infections in natural and non-natural hosts. Blood. 2011;118:847–854. doi: 10.1182/blood-2010-12-325936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.VandeWoude S, Apetrei C. Going wild: lessons from naturally occurring T-lymphotropic lentiviruses. Clinical microbiology reviews. 2006;19:728–762. doi: 10.1128/CMR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sundaravaradan V, Saleem R, Micci L, Gasper MA, Ortiz AM, Else J, Silvestri G, Paiardini M, Aitchison JD, Sodora DL. Multifunctional double-negative T cells in sooty mangabeys mediate T-helper functions irrespective of SIV infection. PLoS Pathog. 2013;9:e1003441. doi: 10.1371/journal.ppat.1003441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vinton C, Klatt NR, Harris LD, Briant JA, Sanders-Beer BE, Herbert R, Woodward R, Silvestri G, Pandrea I, Apetrei C, Hirsch VM, Brenchley JM. CD4-Like Immunological Function by CD4- T Cells in Multiple Natural Hosts of Simian Immunodeficiency Virus. J Virol. 2011;85:8702–8708. doi: 10.1128/JVI.00332-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beaumier CM, Harris LD, Goldstein S, Klatt NR, Whitted S, McGinty J, Apetrei C, Pandrea I, Hirsch VM, Brenchley JM. CD4 downregulation by memory CD4+ T cells in vivo renders African green monkeys resistant to progressive SIVagm infection. Nat Med. 2009;15:879–885. doi: 10.1038/nm.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murayama Y, Amano A, Mukai R, Shibata H, Matsunaga S, Takahashi H, Yoshikawa Y, Hayami M, Noguchi A. CD4 and CD8 expressions in African green monkey helper T lymphocytes: implication for resistance to SIV infection. Int Immunol. 1997;9:843–851. doi: 10.1093/intimm/9.6.843. [DOI] [PubMed] [Google Scholar]
- 9.Murayama Y, Mukai R, Inoue-Murayama M, Yoshikawa Y. An African green monkey lacking peripheral CD4 lymphocytes that retains helper T cell activity and coexists with SIVagm. Clinical and experimental immunology. 1999;117:504–512. doi: 10.1046/j.1365-2249.1999.00999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pandrea IV, Gautam R, Ribeiro RM, Brenchley JM, Butler IF, Pattison M, Rasmussen T, Marx PA, Silvestri G, Lackner AA, Perelson AS, Douek DC, Veazey RS, Apetrei C. Acute Loss of Intestinal CD4+ T Cells Is Not Predictive of Simian Immunodeficiency Virus Virulence. J Immunol. 2007;179:3035–3046. doi: 10.4049/jimmunol.179.5.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitz JE, Ma ZM, Hagan EA, Wilks AB, Furr KL, Linde CH, Zahn RC, Brenchley JM, Miller CJ, Permar SR. Memory CD4(+) T lymphocytes in the gastrointestinal tract are a major source of cell-associated simian immunodeficiency virus in chronic nonpathogenic infection of African green monkeys. Journal of virology. 2012;86:11380–11385. doi: 10.1128/JVI.01556-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carpenter AC, Grainger JR, Xiong Y, Kanno Y, Chu HH, Wang L, Naik S, dos Santos L, Wei L, Jenkins MK, O’Shea JJ, Belkaid Y, Bosselut R. The transcription factors Thpok and LRF are necessary and partly redundant for T helper cell differentiation. Immunity. 2012;37:622–633. doi: 10.1016/j.immuni.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X, Taylor BJ, Sun G, Bosselut R. Analyzing expression of perforin, Runx3, and Thpok genes during positive selection reveals activation of CD8-differentiation programs by MHC II-signaled thymocytes. J Immunol. 2005;175:4465–4474. doi: 10.4049/jimmunol.175.7.4465. [DOI] [PubMed] [Google Scholar]
- 14.Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med. 2013;210:143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brenchley JM, Vinton C, Tabb B, Hao XP, Connick E, Paiardini M, Lifson JD, Silvestri G, Estes JD. Differential infection patterns of CD4+ T cells and lymphoid tissue viral burden distinguish progressive and nonprogressive lentiviral infections. Blood. 2012;120:4172–4181. doi: 10.1182/blood-2012-06-437608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hufert FT, van Lunzen J, Janossy G, Bertram S, Schmitz J, Haller O, Racz P, von Laer D. Germinal centre CD4+ T cells are an important site of HIV replication in vivo. Aids. 1997;11:849–857. doi: 10.1097/00002030-199707000-00003. [DOI] [PubMed] [Google Scholar]
- 17.Smith-Franklin BA, Keele BF, Tew JG, Gartner S, Szakal AK, Estes JD, Thacker TC, Burton GF. Follicular dendritic cells and the persistence of HIV infectivity: the role of antibodies and Fcgamma receptors. J Immunol. 2002;168:2408–2414. doi: 10.4049/jimmunol.168.5.2408. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Wildt KF, Castro E, Xiong Y, Feigenbaum L, Tessarollo L, Bosselut R. The zinc finger transcription factor Zbtb7b represses CD8-lineage gene expression in peripheral CD4+ T cells. Immunity. 2008;29:876–887. doi: 10.1016/j.immuni.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vacchio MS, Wang L, Bouladoux N, Carpenter AC, Xiong Y, Williams LC, Wohlfert E, Song KD, Belkaid Y, Love PE, Bosselut R. A ThPOK-LRF transcriptional node maintains the integrity and effector potential of post-thymic CD4+ T cells. Nature immunology. 2014;15:947–956. doi: 10.1038/ni.2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, Littman DR. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- 21.Djuretic IM, Levanon D, Negreanu V, Groner Y, Rao A, Ansel KM. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nature immunology. 2007;8:145–153. doi: 10.1038/ni1424. [DOI] [PubMed] [Google Scholar]
- 22.Klatt NR, Harris LD, Vinton CL, Sung H, Briant JA, Tabb B, Morcock D, McGinty JW, Lifson JD, Lafont BA, Martin MA, Levine AD, Estes JD, Brenchley JM. Compromised gastrointestinal integrity in pigtail macaques is associated with increased microbial translocation, immune activation, and IL-17 production in the absence of SIV infection. Mucosal Immunol. 2010;3:387–398. doi: 10.1038/mi.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA. HIV Preferentially Infects HIV-Specific CD4+ T-cells. Nature. 2002;417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 24.Goldstein S, Brown CR, Ourmanov I, Pandrea I, Buckler-White A, Erb C, Nandi JS, Foster GJ, Autissier P, Schmitz JE, Hirsch VM. Comparison of simian immunodeficiency virus SIVagmVer replication and CD4+ T-cell dynamics in vervet and sabaeus African green monkeys. J Virol. 2006;80:4868–4877. doi: 10.1128/JVI.80.10.4868-4877.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smedley J, Turkbey B, Bernardo ML, Del Prete GQ, Estes JD, Griffiths GL, Kobayashi H, Choyke PL, Lifson JD, Keele BF. Tracking the luminal exposure and lymphatic drainage pathways of intravaginal and intrarectal inocula used in nonhuman primate models of HIV transmission. PLoS One. 2014;9:e92830. doi: 10.1371/journal.pone.0092830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deleage C, Wietgrefe SW, Del Prete G, Morcock DR, Hao XP, Piatak M, Jr, Bess J, Anderson JL, Perkey KE, Reilly C, McCune JM, Haase AT, Lifson JD, Schacker TW, Estes JD. Defining HIV and SIV Reservoirs in Lymphoid Tissues. Pathogens & immunity. 2016;1:68–106. doi: 10.20411/pai.v1i1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perkins MR, Briant JA, Calantone N, Whitted S, Vinton CL, Klatt NR, Ourmanov I, Ortiz AM, Hirsch VM, Brenchley JM. Homeostatic Cytokines Induce CD4 Downregulation in African Green Monkeys Independently of Antigen Exposure To Generate Simian Immunodeficiency Virus-Resistant CD8alphaalpha T Cells. J Virol. 2014;88:10714–10724. doi: 10.1128/JVI.01331-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, Wildt KF, Zhu J, Zhang X, Feigenbaum L, Tessarollo L, Paul WE, Fowlkes BJ, Bosselut R. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nature immunology. 2008;9:1122–1130. doi: 10.1038/ni.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halim TY, McKenzie AN. New kids on the block: group 2 innate lymphoid cells and type 2 inflammation in the lung. Chest. 2013;144:1681–1686. doi: 10.1378/chest.13-0911. [DOI] [PubMed] [Google Scholar]
- 30.Deleage C, Turkbey B, Estes JD. Imaging lymphoid tissues in nonhuman primates to understand SIV pathogenesis and persistence. Current opinion in virology. 2016;19:77–84. doi: 10.1016/j.coviro.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li P, Spolski R, Liao W, Leonard WJ. Complex interactions of transcription factors in mediating cytokine biology in T cells. Immunological reviews. 2014;261:141–156. doi: 10.1111/imr.12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheroutre H, Husain MM. CD4 CTL: living up to the challenge. Semin Immunol. 2013;25:273–281. doi: 10.1016/j.smim.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mucida D, Husain MM, Muroi S, van Wijk F, Shinnakasu R, Naoe Y, Reis BS, Huang Y, Lambolez F, Docherty M, Attinger A, Shui JW, Kim G, Lena CJ, Sakaguchi S, Miyamoto C, Wang P, Atarashi K, Park Y, Nakayama T, Honda K, Ellmeier W, Kronenberg M, Taniuchi I, Cheroutre H. Transcriptional reprogramming of mature CD4(+) helper T cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nature immunology. 2013;14:281–289. doi: 10.1038/ni.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukazawa Y, Lum R, Okoye AA, Park H, Matsuda K, Bae JY, Hagen SI, Shoemaker R, Deleage C, Lucero C, Morcock D, Swanson T, Legasse AW, Axthelm MK, Hesselgesser J, Geleziunas R, Hirsch VM, Edlefsen PT, Piatak M, Jr, Estes JD, Lifson JD, Picker LJ. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat Med. 2015;21:132–139. doi: 10.1038/nm.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.