Abstract
In this review, we discuss the genetic architecture of obesity and the metabolic syndrome, highlighting recent advances in identifying genetic variants and loci responsible for a portion of the variation in adiposity traits, serum HDL and triglycerides, blood pressure, and glycemic traits; in other words, the components comprising the metabolic syndrome. We focus particularly on recent progress from large-scale genome-wide association studies. Detailing their successes and how lessons learned can pave the way for future discovery. Results from recent genome-wide association studies coalesce with earlier work suggesting numerous interconnections between obesity and the metabolic syndrome, developed through several potentially pleiotropic effects. We detail recent work by way of a case study on the cadherin 13 gene and its relation with adiponectin in the HyperGEN and the Framingham Heart Studies, and its association with obesity and the metabolic syndrome. We provide also a gene network analysis on recent variants related to obesity and metabolic syndrome discovered through genome-wide association studies.
1. Introduction
Obesity is arguably the most important and, at the same time, the most complex, major public health problem in the United States and elsewhere. Obesity prevalence has increased rapidly over the last several decades [1] and is associated with increased mortality and morbidity from a variety of conditions and disorders [2–4]. The prevalence of obesity and overweight in many countries as diverse as Egypt, Mexico, and South Africa is similar to that of the United States, but rates of increase are higher [5, 6], with some of the most rapid increases in obesity and its co-morbidities seen in Asian populations [7, 8].
Obesity is a central risk factor for a host of metabolic disturbances, primarily in lipid and glucose metabolism. There is ample epidemiologic evidence that adiposity is correlated with adverse lipid profiles and biomarkers of glucose metabolism [9, 10]. In fact, derangements of these variables can cluster together as obesity, dyslipidemia, insulin resistance, and hypertension in what is referred to as the metabolic syndrome (MetS) [11]. Hypertension, hyperlipidemia, impaired glucose tolerance, and obesity are established traditional cardiovascular disease (CVD) risk factors. When these risk factors cluster in one individual, CVD risk increases dramatically. This clustering of risk factors is, in fact, not a rare event but a common cause of CVD in modern society [12, 13]. While this combined phenotype has been described since the late 1980s as the metabolic syndrome, the precise definition has shifted slightly over time, and there have been a number of attempts to develop standardized criteria for its diagnosis. The most widely-used definitions from the World Health Organization (WHO), proposed in 1998 [14], the National Cholesterol Education Program Adult Treatment Panel III (NCEP ATPIII), proposed in 2001 [15], the International Diabetes Federation (IDF) [16], revised in 2005, and the AHA/NHLBI [17], proposed in 2005, are summarized in Table 1. It is clear from inspection of Table 1 that only minor differences exist between the specific components that make up the diagnostic schemes. However, a couple of important differences do exist. First, while the WHO and the IDF require the presence of one essential factor (impaired glucose for the WHO and central adiposity for the IDF), the NCEP ATPIII and AHA/NHLBI equally weight the five criteria listed. Secondly, the definitions differ in their handling of adiposity. While the WHO allows for excess adiposity to be determined either by waist-to-hip ratio (WHR) or body mass index (BMI) as an ancillary criterion, the NCEP ATPIII and AHA/NHLBI use a measure of central adiposity via waist circumference (WC) as one of five components, and the IDF requires excess central adiposity measured via WC as a required criterion. Recently the IDF and the AHA/NHLBI attempted to resolve differences between definitions on the metabolic syndrome, as there continued to be disagreement and confusion on the part of clinicians (see [18]). The resulting definition does not require the presence of abdominal obesity for diagnosis, instead it is one of five criteria, and utilizes population- and country-specific WC cut points. The remaining four criteria are identical to the AHA/NHLBI 2005 definition.
Table 1.
Widely-used definitions of the Metabolic Syndrome
| World Health Organization (adapted from reference [14]) |
| One of the following: |
| Diabetes mellitus |
| Impaired glucose tolerance |
| Impaired fasting glucose |
| Plus any two of the following |
| Waist-to-hip ratio >0.9 (men) or >0.85 (women); BMI>30; or both |
| Triacylglycerols ≥ 1.7 mmol/L; HDL cholesterol <0.9 mmol/L (men) or <1.0 mmol/L (women); or both |
| Blood pressure ≥ 140/90 mmHg |
| Microalbuminuria (urinary albumin excretion rate ≥ 20 μg/min or albumin/creatinine ratio ≥ 30 mg/g) |
| National Cholesterol Education Program Adult Treatment Panel III (adapted from reference [15]) |
| Any three of the following: |
| Fasting glucose ≥ 6.1 mmol/L |
| Waist circumference >102 cm (men) or >88 cm (women) |
| Triacylglycerols ≥ 1.7 mmol/L |
| HDL cholesterol <1.036 mmol/L (men) or <1.295 mmol/L (women) |
| Blood pressure ≥ 130/85 mmHg |
| International Diabetes Federation (adapted from reference [16]) |
| Must have: |
| Waist circumference as defined by sex- and ethnic-specific values |
| Plus any two of the following: |
| Triacylglycerols ≥ 1.7 mmol/L or treatment |
| HDL <1.03 mmol/L (men) or <1.29 mmol/L (women) or treatment |
| Blood pressure ≥ 130/85 mmHg or treatment |
| Fasting plasma glucose ≥ 5.6 mmol/L or diagnosed type 2 diabetes |
| AHA/NHLBI (adapted from reference [17]) |
| Any three of the following: |
| Waist circumference ≥ 102 cm (men) or ≥ 88 cm (women) |
| Triglycerides ≥ 1.7 mmol/L or drug treatment for elevated triglycerides |
| HDL cholesterol <1.03 mmol/L (men) or <1.3 mmol/L (women) or drug treatment for reduced HDL-C |
| Blood pressure ≥ 130 mmHg systolic or ≥ 85 mmHg diastolic or drug treatment |
| Fasting glucose ≥ 5.6 mmol/L or treatment for elevated glucose |
Although estimates of prevalence in different populations are highly dependent on the definition of the metabolic syndrome, the reality is that the current prevalence estimates and future projections are alarming. In fact, age-adjusted estimates from the National Health and Nutrition Examination Survey (NHANES) revealed that approximately 34% of adult Americans aged 20 years or older met the criteria for metabolic syndrome [19]. The prevalence of metabolic syndrome increased with age for males and females 40–59 years of age, about three times as likely as the youngest age group (20–39) to meet the criteria for metabolic syndrome (males: OR=2.70, 95% confidence interval (CI) 1.96–3.73; females: OR=3.20, 95% CI 2.32–4.43). The prevalence of metabolic syndrome also varied by race and ethnicity but the pattern was different in males and females. For example, 25% of African American men met the criteria for metabolic syndrome whereas 37% of non-Hispanic white males met these same criteria. In contrast, although the prevalence did vary by race in females, all differences were non-significant.
A number of studies have reported a positive relationship between the metabolic syndrome and CVD morbidity [20–22]. For example, in the NHANES III [23], the metabolic syndrome was associated with a higher risk of nonfatal myocardial infarction (OR, 2.01; 95% CI, 1.53–2.64) and stroke (OR, 2.16; 95% CI, 1.48–3.16). In other non-US populations, Lakka et al [24] reported a higher risk of coronary mortality associated with the metabolic syndrome (hazard ratio of 4.16; 95% CI, 1.60–10.8) in Finnish adults. Consequently, a major effort should be placed on the detection, prevention, and therapy of the metabolic syndrome. Therapeutic tools are available to successfully deal with a number of the individual components. Specifically, effective drugs are available to lower blood pressure, improve insulin sensitivity, and treat dyslipidemia. In contrast, therapeutic success has not been shared by the other major component of the metabolic syndrome, obesity.
Clearly there is much to be learned about the genetic underpinnings of the components of the metabolic syndrome and there is great hope that discoveries will lead to the development of effective candidates for screening and therapies. With this in mind, the purpose of this review is to describe the current state of the science on the genetic epidemiology of the metabolic syndrome and its components. To accomplish this goal, we first describe the genetic architecture of obesity, a primary component of and a risk factor for the other components of the metabolic syndrome. Next we describe recent progress in genome-wide association studies for obesity and the other metabolic syndrome components. We follow with a discussion of pleiotropic effects and conclude with a case study of some recent work specifically exploring the genetic epidemiology of the metabolic syndrome in the Hypertension Genetic Epidemiology Network (HyperGEN) study (Section 9), as well as provide a gene network analysis of recent GWAS findings (Section 10).
2. The Genetic Architecture of Obesity
Because obesity is a strong risk factor for numerous other metabolic derangements, diabetes, cardiovascular disease, fatty liver disease, various cancers, as well as a host of other morbidities, there is strong motivation to understand its genetic architecture.
Both genetic and environmental factors have been linked to obesity [25]. Heritability estimates for adiposity phenotypes range from 30 to 70% in family and twin studies [26], and multiple quantitative trait loci and candidate genes have been identified [25]. In the current environment termed “obesogenic” and characterized by plentiful inexpensive energy-dense foods and sedentary occupational environments, different constitutions for obesity are evident (monogenic, strong predisposition, moderate predisposition, and resistant) [27]. Despite strong evidence for an underlying genetic component, genes for obesity-related traits using candidate gene and linkage approaches have been difficult to identify and replicate.
3. New Developments in the genetics of Obesity (findings from GWAS)
Genome-wide association studies (GWAS) aimed at discovery of variants associated with adiposity traits, have recently produced many findings, implicating numerous novel genes, owing to the cooperation of large cohort and family studies in meta-analyses of tens of thousands of subjects. Findings are summarized in Table 2.
Table 2.
Adiposity trait loci discovered via genome-wide association studies as of 3/31/10; p<5.0×10−8
| Reported gene | Associated SNPs | Region | Trait | Risk allele | Risk allele frequency† | Reference(s) |
|---|---|---|---|---|---|---|
|
| ||||||
| NEGR1 | rs2815752 | 1p31.1 | BMI | A | 0.637 | [35] |
| rs2568958 | BMI | A | 0.637 | [34] | ||
|
| ||||||
| LYPLAL1 | rs2605100 | 1q41 | WHR | G | 0.686 | [37] |
|
| ||||||
| TMEM18 | rs6548238 | 2p25 | BMI | C | 0.850 | [35] |
| rs7561317 | BMI | G | 0.854 | [34] | ||
|
| ||||||
| ETV5/SFRS10/DGKG | rs7647305 | 3q27 | BMI | C | 0.796 | [34] |
|
| ||||||
| GNPDA2 | rs10938397 | 4p13 | BMI | G | 0.446 | [35] |
|
| ||||||
| TFAP2B | rs987237 | 6p12.3 | WC | G | 0.164 | [37] |
|
| ||||||
| MSRA | rs545854‡ | 8p23.1 | WC | G | 0.183 | [37] |
|
| ||||||
| BDNF/LGR4/LIN7C | rs4923461 | 11p14 | BMI | A | 0.770 | [34] |
|
| ||||||
| MTCH2 | rs10838738 | 11p11.2 | BMI | G | 0.363 | [35] |
|
| ||||||
| FAIM2/BCDIN3D | rs7138803 | 12q13 | BMI | A | 0.345 | [34] |
|
| ||||||
| NRXN3 | rs10146997 | 14q31.1 | WC | G | 0.248 | [32] |
|
| ||||||
| SH2B1/ATP2A1 | rs7498665 | 16p11 | BMI | G | 0.381 | [35], [34] |
|
| ||||||
| MAF | rs1424233 | 16q22-q23 | BMI | A | 0.558 | [33] |
|
| ||||||
| FTO | rs9939609 | 16q12.2 | BMI | A | 0.460 | [35], [28] |
| rs9930506 | BMI | G | 0.482 | [29] | ||
| rs1421085 | BMI | C | 0.460 | [33] | ||
| rs8050136 | BMI | A | 0.460 | [34] | ||
| rs1121980 | BMI | A | 0.482 | [30] | ||
| rs9941349 | ExtrOb | T | 0.473 | [36] | ||
|
| ||||||
| MC4R | rs17782313 | 18q21 | BMI | C | 0.265 | [35], [33], [30] |
| rs12970134 | BMI | A | 0.279 | [34], [31] | ||
|
| ||||||
| KCTD15 | rs29941 | 19q13.11 | BMI | C | 0.677 | [34] |
| rs11084753 | BMI | G | 0.690 | [35] | ||
HapMap release 27, CEU population;
originally rs7826222; WC, waist circumference; BMI, body mass index; WHR, waist-to-hip ratio; ExtrOb, extreme obesity (as defined by bariatric surgery patients)
In two contemporaneous genome-wide association studies in 2007, Frayling et al and Scuteri et al identified the fat mass and obesity associated (FTO) gene as highly significantly related to body mass index (BMI) and waist circumference (WC) [28, 29]. In 2008, three studies identified the melanocortin 4 receptor (MC4R), known before this time to be important in monogenic forms of obesity, as playing a role in common forms of obesity [30, 31]. 2009 was quite a banner year for genetics of common complex obesity, with an additional six genome-wide association studies published, reporting 14 new loci for adiposity traits [32–37]. While many of the biological mechanisms behind these variants remain unknown, as do the actual functional variants, many of the discovered candidates locate near genes that are highly expressed in the brain and hypothalamus, suggesting a role for neuronal control in body weight regulation. For example, our study uncovered a new gene influencing waist circumference, the neurexin 3 (NRXN3) gene, which has been previously implicated in studies of addiction and reward behavior [32]. However, a sobering statistic is that collectively these variants explain only a small proportion of the population variation in adiposity. Given that the heritability of BMI ranges from 30–70%, we should conclude that many more loci remain to be discovered, many which are below the level of detection given current technology. Also of interest and of note is that the vast majority of GWAS conducted thus far have been in populations of European-descent. Similar studies in other populations are just now getting underway and researchers are hopeful that differences in linkage disequilibrium will bring to light new findings.
GWAS have revealed many genomic loci, often surprising, that were not previously identified through traditional linkage and candidate gene studies (see [25] for review of linkage and candidate gene findings). Major insights from these studies include: 1) most complex traits have many associated loci, rather than few; 2) variants have small effects of 1–5%; 3) associated variants usually have allele frequencies greater than 10%; and 4) large samples are needed to detect these associations [38, 39]. Thus, even for the common disease, common variant hypothesis which postulates that common disease-causing alleles found in all populations will be responsible for small effects on a given disease, the research community learned that it needed sample sizes larger than those previously used for linkage and candidate gene studies. For this reason, the past few years has seen the establishment of consortia of studies to maximize sample size, previously unheard of, for the purpose of detecting common genetic variants of small to moderate effect sizes.
4. The Genetic Architecture of the Metabolic Syndrome
Like obesity, the development of metabolic risk factors is likely to involve both genetic and environmental components. Serum lipid levels, including HDL and triglycerides, two components of the metabolic syndrome definitions, are highly heritable, with estimates consistently over 50% [40]. Studies of related individuals have found evidence of significant familial aggregation for the individual components of the metabolic syndrome, including blood lipids [41], blood pressure [42–44], and blood glucose/insulin levels [45, 46]. Further studies have investigated the co-occurrence of risk factors and found evidence that pleiotropy may underlie the clustering [47, 48]. Results from a number of genome-wide linkage analyses provide further evidence of common genetic factors on multiple individual components of the metabolic syndrome [49–51], as well as evidence for clustering [47, 48, 52–57].
5. New Developments in the genetics of the Metabolic Syndrome (findings from GWAS)
Relatively few genome-wide association studies have been published using presence of the metabolic syndrome as the dependent variable. We published a study using data from the Framingham Heart Study investigating the association of variants present on the 500K SNP chip with the metabolic syndrome [58]. We found significant associations in the PTHBI, PAPPA, and FBN3 genes, and in intergenic regions of chromosomes 12 and 14. Results however have yet to be replicated. Follow-up work of ours, using the Hypertension Genetic Epidemiology Network (HyperGEN) data, with replication in the Family Heart Study, is currently being finalized (data as yet unpublished). We have identified several genome-wide significant loci for loadings on latent factors describing obesity/insulin traits, lipid/insulin traits, blood pressure traits, and central obesity traits. Details of these analyses are presented below in the case study.
Far more genome-wide studies have been published investigating the individual components of the metabolic syndrome. Table 3 tallies genome-wide significant and replicated results for blood pressure traits. While loci have been identified for both systolic and diastolic blood pressure, as well as for hypertension affectation status, only two studies have reported genome-significant results. Conversely, genome-wide association studies have been quite successful in identifying and replicating variants for HDL-cholesterol and triglycerides (Table 4) and diabetes traits (Table 5). Like GWAS for obesity, larger and larger consortia are convening, increasing the ability to discover additional variants. Also similar to obesity GWAS results, several of the loci identified to harbor variants associated with common dyslipidemia are also implicated in Mendelian disorders featuring a similar phenotype (e.g. ABCA1). In contrast to results from obesity GWAS, a much greater percentage of interindividual variability is explained, 9.3% and 7.4% for HDL and triglycerides, respectively [59], and ~10% for fasting glucose [60].
Table 3.
Blood pressure/hypertension loci discovered via genome-wide association studies as of 3/31/10; p<5.0×10−8
| Reported gene | Associated SNPs | Region | Trait | Risk allele | Risk allele frequency† | Reference(s) |
|---|---|---|---|---|---|---|
|
| ||||||
| MTHFR/NPPA/CLCN6/NPPB/AGTRAP | rs17367504 | 1p36.22 | SBP | A | 0.832* | [174]* |
| CHB: 0.900 | ||||||
|
| ||||||
| ULK4 | rs9815354 | 3p22.1 | DBP | A | 0.228 | [173] |
|
| ||||||
| FGF5/PRDM8 | rs16998073 | 4q21.21 | DBP | T | 0.192 | [174]* |
| CHB: 0.159 | ||||||
|
| ||||||
| CACNB2 | rs11014166 | 10p12.33 | DBP | A | 0.633 | [173] |
|
| ||||||
| TMEM26/RTKN2/RHOBTB1/ARID5B | rs1530440 | 10q21.2 | DBP | C | 0.839 | [174]* |
| CHB: 0.932 | ||||||
|
| ||||||
| CYP17A1/AS3MT/CNNM2/NT5C2 | rs1004467 | 10q24.32 | SBP | A | 0.916 | [173] |
| rs11191548 | SBP | T | 0.925 | [174]* | ||
| CHB: 0.756 | ||||||
|
| ||||||
| PLAKHA7 | rs381815 | 11p15.1 | SBP | T | 0.297 | [173] |
|
| ||||||
| ATP2B1 | rs2681472 | 12q21.33 | Hyp/DBP | A | 0.885 | [173] |
| rs2681492 | SBP | T | 0.881 | [173] | ||
|
| ||||||
| SH2B3/ATXN2 | rs3184504 | 12q24.12 | SBP/DBP | T | 0.445 | [173] |
| rs653178 | DBP | C | 0.438 | [174]* | ||
| CHB: 0.000 | ||||||
|
| ||||||
| TBX3/TBX5 | rs2384550 | 12q24.21 | DBP | G | 0.633 | [173] |
|
| ||||||
| CYP1A1/CYP1A2/CSK/LMAN1L/CPLX3/ARID3B/ULK3 | rs1378942 | 15q24.1 | DBP | C | 0.321 | [174]* |
| CHB: 0.833 | ||||||
| rs6495122 | DBP | A | 0.379 | [173] | ||
|
| ||||||
| PLCD3/ACBD4/HEXIM1/HEXIM2 | rs12946454 | 17q21.31 | SBP | T | 0.245 | [174]* |
| CHB: 0.202 | ||||||
|
| ||||||
| ZNF652/PHB | rs16948048 | 17q21.32 | DBP | G | 0.416 | [174]* |
| CHB: 0.144 | ||||||
HapMap release 27, CEU population unless otherwise noted;
Replication sample included individuals of Indian Asian ancestry, we include CHB allele frequencies for reference; SBP, systolic blood pressure; DBP, diastolic blood pressure; Hyp, hypertension
Table 4.
HDL-cholesterol and triglyceride loci discovered via genome-wide association studies as of 3/31/10; p<5.0×10−8
| Reported gene | Associated SNPs | Region | Trait | Risk allele | Risk allele frequency† | Reference(s) |
|---|---|---|---|---|---|---|
|
| ||||||
| DOCK7/ANGPTL3/ATG4C | rs10889353 | 1p31.3 | TG | A | 0.664 | [129] |
| rs12130333 | TG | C | 0.777 | [130] | ||
| rs1748195 | TG | C | 0.659 | [131] | ||
|
| ||||||
| GALNT2 | rs2144300 | 1q42.13 | HDL | C | 0.432 | [131] |
| rs4846914 | HDL | G | 0.429 | [62] | ||
| HDL/TG | [130] | |||||
|
| ||||||
| GCKR | rs1260326 | 2p23.3 | TG | T | 0.420 | [62], [128] |
| rs780094 | TG | T | 0.394 | [129], [130], [131], [132] | ||
|
| ||||||
| APOB | rs6754295 | 2p24.1 | HDL/TG | A | 0.788 | [129] |
| rs7557067 | TG | A | 0.798 | [62] | ||
| rs673548 | TG | G | 0.795 | [129], [128] | ||
|
| ||||||
| MLXIPL/BCL7B/TBL2 | rs17145738 | 7q11.23 | TG | C | 0.878 | [131], [130] |
| rs2240466 | TG | G | 0.879 | [129] | ||
| rs3812316 | TG | C | 0.847 | [175] | ||
| rs714052 | TG | A | 0.881 | [62] | ||
|
| ||||||
| LPL | rs10096633 | 8p21.3 | TG | G | 0.858 | [129] |
| rs10503669 | HDL/TG | C | 0.885 | [131] | ||
| rs12678919 | HDL/TG | A | 0.885 | [62] | ||
| rs17482753 | HDL | G | 0.893 | [214] | ||
| TG | T** | 0.107 | [132] | |||
| rs2083637 | HDL | A | 0.726 | [129] | ||
| rs326 | TG | A | 0.708 | [175] | ||
| rs328 | HDL/TG | C | 0.875 | [130] | ||
| rs17411031 | HDL | G** | 0.274 | [132] | ||
|
| ||||||
| XKR6/AMAC1L2 | rs7819412 | 8p23.1 | TG | A | 0.513 | [62] |
|
| ||||||
| TRIB1 | rs17321515 | 8q24.13 | TG | A | 0.571 | [131], [130] |
| rs2954029 | TG | A | 0.617 | [62] | ||
|
| ||||||
| TTC39B | rs471364 | 9p22.3 | HDL | C | 0.106 | [62] |
|
| ||||||
| ABCA1 | rs1883025 | 9q31.1 | HDL | T | 0.199 | [62] |
| rs3890182 | HDL | A | 0.080 | [130] | ||
| rs3905000 | HDL | A | 0.084 | [129] | ||
| rs4149268 | HDL | T | 0.279 | [131] | ||
|
| ||||||
| NR1H3 | rs7120118 | 11p11.2 | HDL | A | 0.699 | [128] |
|
| ||||||
| MADD/FOLH1 | rs7395662 | 11p11.2 | HDL | G | 0.642 | [129] |
|
| ||||||
| FADS1/FADS2/FADS3 | rs174547 | 11q12.2 | HDL/TG | C | 0.344 | [62] |
|
| ||||||
| APOA1/APOC3/APOA4/APOA5/DSCAML1 | rs12272004 | 11q23.3 | TG | A | 0.036 | [129] |
| rs12286037 | TG | T | 0.040 | [131] | ||
| rs28927680 | TG | G | 0.067 | [130] | ||
| rs1558861 | TG | C | 0.111 | [175] | ||
| rs6589566 | TG | G** | 0.071 | [132] | ||
| rs964184 | HDL/TG | G | 0.121 | [62] | ||
|
| ||||||
| MMAB/MVK | rs2338104 | 12q24.11 | HDL | C | 0.482 | [62], [131] |
|
| ||||||
| LIPC | rs10468017 | 15q22.1 | HDL | C | 0.690 | [62] |
| rs1532085 | HDL | G | 0.588 | [128], [129] | ||
| rs1800588 | HDL | C | 0.742 | [130] | ||
| rs4775041 | HDL | G | 0.318 | [131] | ||
| TG | C | 0.682 | [131] | |||
|
| ||||||
| CETP | rs1532624 | 16q13 | HDL | C | 0.540 | [129] |
| rs173539 | HDL | C | 0.633 | [62] | ||
| rs1800775 | HDL | C | 0.487 | [130], [215] | ||
| rs1864163 | HDL | A | 0.259 | [131] | ||
| rs3764261 | HDL | C | 0.655 | [131], [128] | ||
| JPT: 0.800 | [176]* | |||||
| rs9989419 | HDL | A | 0.398 | [214], [131], [132] | ||
| rs7205804 | HDL | G | 0.532 | [175] | ||
|
| ||||||
| LCAT/CTCF/PRMT8 | rs2271293 | 16q22.1 | HDL | G | 0.907 | [62], [129] |
| rs255049 | HDL | A | 0.835 | [128] | ||
|
| ||||||
| LIPG | rs2156552 | 18q21.1 | HDL | A | 0.190 | [131] |
| rs4939883 | HDL | T | 0.190 | [129], [62] | ||
| rs7240405 | HDL | A | 0.186 | [214] | ||
|
| ||||||
| NCAN/CILP2/PBX4 | rs16996148 | 19p13.11 | TG | G | 0.905 | [131], [130] |
| rs17216525 | TG | C | 0.917 | [62] | ||
|
| ||||||
| ANGPTL4 | rs2967605 | 19p13.2 | HDL | T | 0.173 | [62] |
|
| ||||||
| TOMM40-APOE | rs439401 | 19q13.32 | TG | G | 0.625 | [129] |
|
| ||||||
| HNF4A | rs1800961 | 20q13.12 | HDL | T | 0.036 | [62] |
|
| ||||||
| PLTP | rs7679 | 20q13.12 | HDL/TG | C | 0.168 | [62] |
HapMap release 27, CEU population unless otherwise noted;
Includes individuals of non-European descent;
minor allele and MAF reported, risk allele was unclear from paper; HDL, high density lipoprotein cholesterol; TG, triglycerides
Table 5.
Diabetes-related loci discovered via genome-wide association studies as of 3/31/10; p<5.0×10−8
| Reported gene | Associated SNPs | Region | Trait | Risk allele | Risk allele frequency† | Reference(s) |
|---|---|---|---|---|---|---|
|
| ||||||
| NOTCH2/ADAAM30 | rs10923931 | 1p12 | T2DM | T | 0.093 | [61] |
|
| ||||||
| PROX1 | rs340874 | 1q32.3 | FPG | C | 0.562 | [60] |
|
| ||||||
| THADA | rs7578597 | 2p21 | T2DM | T | 0.876 | [61] |
|
| ||||||
| GCKR | rs780094 | 2p23.3 | FPG/FI/HOMA-IR | C | 0.606 | [60] |
|
| ||||||
| G6PC2 | rs560887 | 2q24.3 | FPG | C | 0.674 | [216], [200] |
| FPG/HOMA-B | [60] | |||||
|
| ||||||
| LOC64673/IRS1 | rs2943641 | 2q36.3 | T2DM | C | 0.633 | [217] |
|
| ||||||
| ADAMTS9 | rs4607103 | 3p14.1 | T2DM | C | 0.810 | [61] |
|
| ||||||
| ADCY5 | rs11708067 | 3q21.1 | FPG/HOMA-B | A | 0.774 | [60] |
|
| ||||||
| SLC2A2 | rs11920090 | 3q26.2 | FPG | T | 0.853 | [60] |
|
| ||||||
| IGF2BP2 | rs4402960 | 3q27.2 | T2DM | T | 0.296 | [201], [126], [127] |
| rs6769511 | T2DM | C | JPT: 0.318 | [194]* | ||
|
| ||||||
| WFS1/PPP2R2C | rs4689388 | 4p16.1 | T2DM | T | 0.668 | [217] |
|
| ||||||
| CDKAL1 | rs10946398 | 6p22.3 | T2DM | C | 0.336 | [127] |
| rs4712523 | T2DM | G | 0.336 | [217] | ||
| JPT: 0.511 | [190]* | |||||
| rs4712524 | T2DM | G | JPT: 0.511 | [194]* | ||
| rs6931514 | T2DM | G | 0.279 | [61] | ||
| rs7754840 | T2DM | C | 0.336 | [126], [201] | ||
| rs7756992 | T2DM | G | 0.279 | [191] | ||
|
| ||||||
| GCK | rs4607517 | 7p13 | FPG | A | 0.195 | [200] |
| FPG/HOMA-B | [60] | |||||
|
| ||||||
| JAZF1 | rs864745 | 7p15.1 | T2DM | T | 0.487 | [61] |
|
| ||||||
| DGKB/TMEM195 | rs2191349 | 7p21.2 | FPG/HOMA-B | T | 0.467 | [60] |
|
| ||||||
| ZMAT4 | rs2722425 | 8p11.21 | FPG | A** | 0.093 | [195] |
|
| ||||||
| SLC30A8 | rs11558471 | 8q24.11 | FPG | A | 0.748 | [60] |
| rs13266634 | T2DM | C | 0.761 | [201], [126], [127] | ||
| JPT: 0.556 | [190]* | |||||
|
| ||||||
| CDKN2A/CDKN2B | rs10811661 | 9p21.3 | T2DM | T | 0.801 | [201], [126], [127] |
| rs2383208 | T2DM | A | JPT: 0.568 | [190]* | ||
|
| ||||||
| PTPRD | rs17584499 | 9p24.1 | T2DM | T | CHB: 0.089 | [196]* |
|
| ||||||
| GLIS3 | rs7034200 | 9p24.2 | FPG/HOMA-B | A | 0.525 | [60] |
|
| ||||||
| CDC123/CAMK1D | rs12779790 | 10p13 | T2DM | G | 0.229 | [61] |
|
| ||||||
| HHEX | rs1111875 | 10q23.33 | T2DM | C | 0.584 | [201], [126], [127] |
| JPT: 0.409 | [190]* | |||||
|
| ||||||
| ADRA2A | rs10885122 | 10q25.2 | FPG | G | 0.900 | [60] |
|
| ||||||
| TCF7L2 | rs4506565 | 10q25.2 | T2DM | T | 0.296 | [197] |
| FPG | [60] | |||||
| rs7901695 | T2DM | C | 0.281 | [127] | ||
| rs7903146 | T2DM | T | 0.279 | [191], [198], [192], [201], [126], [61], [217] | ||
| JPT: 0.023 | [190]* | |||||
|
| ||||||
| CRY2 | rs11605924 | 11p11.2 | FPG | A | 0.542 | [60] |
|
| ||||||
| MADD | rs7944584 | 11p11.2 | FPG | A | 0.712 | [60] |
|
| ||||||
| KCNJ11 | rs5215 | 11p15.1 | T2DM | C | 0.398 | [127] |
| rs5219 | T2DM | T | 0.500 | [126], [201] | ||
|
| ||||||
| KCNQ1 | rs2237895 | 11p15.4 | T2DM | C | CHB: NR | [196]* |
| rs2237892 | T2DM | C | JPT: 0.611 | [193]*, [190]* | ||
| rs2283228 | T2DM | A | JPT: 0.593 | [194]* | ||
|
| ||||||
| FADS1 | rs174550 | 11q12.2 | FPG/HOMA-B | T | 0.633 | [60] |
|
| ||||||
| MTNR1B | rs10830963 | 11q14.3 | FPG/HOMA-B | G | 0.300 | [60], [200] |
| rs1387153 | FPG | T | 0.272 | [188] | ||
| rs2166706 | FPG | G | 0.389 | [189]* | ||
| CHB: 0.489 | ||||||
|
| ||||||
| TSPAN8/LGR5 | rs7961581 | 12q21.1 | T2DM | C | 0.252 | [61] |
|
| ||||||
| IGF1 | rs35767 | 12q23.2 | FI/HOMA-IR | G | 0.885 | [60] |
|
| ||||||
| C2CD4B | rs11071657 | 15q22.2 | FPG | A | 0.592 | [60] |
|
| ||||||
| FTO | rs8050136 | 16q12.2 | T2DM | A | 0.460 | [126], [127] |
|
| ||||||
| SRR | rs391300 | 17p13.3 | T2DM | G | CHB: 0.740 | [196]* |
HapMap release 27, CEU population unless otherwise noted;
Includes individuals of non-European descent;
minor allele and MAF reported, risk allele was not reported in paper; T2DM, type 2 diabetes mellitus; FPG, fasting plasma glucose; FI, fasting insulin; HOMA-IR, homeostatic model assessment of insulin resistance; HOMA-B, homeostatic model assessment of beta-cell function; NR, not reported
As with obesity, the advent of the genome-wide association study has revolutionized our ability to find genetic variants associated with the various components of the metabolic syndrome. Results indicate many novel loci harboring these genetic variants, loci likely to have remained undiscovered as part of candidate gene studies due to many of them not being recognized players in known pathways. Notably, in contrast to adiposity GWAS, findings are in some cases generalizable to other ethnic populations, particularly Asian populations, and in other cases have revealed new variants (e.g. KCNQ1). Future work in additional populations is forthcoming, and it will be interesting to see if further generalization is achieved and new variants uncovered.
6. Lessons learned from GWAS will help pave the way for future discovery
Regardless of phenotype, investigators have learned many lessons from genome-wide association studies of common complex traits such as obesity and others that comprise the metabolic syndrome. First, effect sizes for common variants confer small increments in risk and explain relatively little of the inter-individual variation in the trait. Combined with correcting for multiple testing, large consortia are essential to discovery efforts, and with further increases in sample size, we should expect to identify more variants with small(er) effect sizes, as well as rarer variants with possibly larger effect sizes. In fact, several studies have shown that the number of detected variants increases with increasing sample size [61, 62]. Both the discovery of rare variants and identification of functional variants will be facilitated with the 1,000 Genomes Project catalogue (http://www.1000genomes.org), as well as with targeted or whole genome sequencing. Second, the majority of identified variants fall outside of coding regions, underscoring the need for future research to identify functional variants. Third, most of the identified genes or loci were not previously thought to be associated with the biology of the trait. While initially a bit perplexing, this has begun to provide new insights into the biological pathways of disease etiology. Lastly, the exciting discovery that both common complex disease and monogenic disease variants often reside within the same genes suggests that with fine mapping or targeted sequencing we may discover rarer variants of larger effect in these loci. In the future, more precise measurement of phenotypes should reduce the heterogeneity that may be limiting our ability to detect informative loci; future studies should be designed with this in mind and potentially target endophenotypes. Similarly, investigation of gene-environment interaction, which requires harmonized measures of environmental factors, is critical to assessing the contextual milieu in which genetic effects may or may not give rise to specific phenotypes.
7. Obesity and Metabolic Syndrome interconnections
As defined by the NCEP ATPIII, MetS is a combination of three or more risk factors beyond clinical thresholds (larger WC, elevated TG, lower HDL-C, elevated glucose, and elevated BP). In contrast, obesity represents an excess of body fat, as measured by BMI, skin-fold thickness, bioelectrical impedance, or other measures. In clinical practice a simple and successful measure has been the assessment of WC, because an excess of abdominal fat is strongly associated with metabolic risk factors. Obesity per se is a combination of an existing genetic profile that predisposes to obesity and a chronic imbalance between energy intake, energy utilization for basic metabolic processes, and energy expenditure from physical activity [63]. Obesity is viewed as an important cause of insulin resistance in children and associated with dyslipidemia, earlier puberty and menarche in girls, type 2 diabetes, increased incidence of obesity and MetS in adults and long-term vascular complications [64–71].
Reaven [72] hypothesized that insulin dysregulation was the underlying disorder in the syndrome. Haffner et al [73] suggested that fasting insulin, serving as surrogate of insulin resistance, predicted the development of dyslipidemia, hypertension and T2DM. Other studies nevertheless showed that insulin and measures of obesity, BMI and WHR, had joint contributions to MetS [74]. Many later studies have shown that fasting insulin and fasting glucose are correlated with obesity measures as well as with measures of lipids in the blood [18, 75–90]. For example, Han et al [91] reported that BMI and insulin had similar areas under the receiver operating characteristic curves (0.74 and 0.76, respectively). In addition, 32% of the obese subjects (BMI≥30 kg/m2) with high WC from the San Antonio Heart Study developed MetS, compared with 10% of participants with both low BMI and low WC. Ascaso et al [92] showed that subjects with high WC had a prevalence of insulin resistance of 54.6% as measured by HOMA-IR ≥ 3.8, compared to 31.7% on those with normal WC. During that time and later, several studies showed the fundamental importance of obesity in the foundation of MetS by the way of multivariate analysis of several traits related with obesity and central obesity, insulin and glucose, lipid profiles and blood pressure [93–95]. Arnlöv et al [96] in a follow-up study of more than 30 years of middle-aged men with MetS reached the conclusion that these subjects with MetS had increased risk for cardiovascular events and total death regardless of BMI status. Therefore how strong is obesity a predisposing condition for MetS? And what might be the mechanism of action? Multiple hypotheses have been proposed. The insulin hypothesis states that obesity typically results in insulin and leptin resistance and a shift from expansion of subcutaneous fat to deposition of abdominal and ectopic fat [97], which leads to metabolic dysregulation, elevated fatty acids, and increased pro-inflammatory adipokines. Although, it is important to note that not all obese individuals are insulin resistant [98]. An alternative hypothesis proposes that the metabolic syndrome starts with an obesity-associated metabolic dysfunction, where chronic macronutrient and/or lipid overload (associated with adiposity) induce cellular stress that initiates and propagates an inflammatory cycle [99, 100]. Studies have reported that a postprandial hyperinsulinemia exists in obesity, in cases when fasting insulin levels are normal [101–103]. For more information on diet and metabolic syndrome see a detailed review in this issue by Djoussé at al [104].
McGarry (2001) in his Banting Lecture posited that an abnormal accumulation of fat in muscle and other tissues plays an important role in the etiology of insulin resistance and possibly also in the demise of the β-cell in type 2 diabetes [105]. This thinking coalesces with the MetS obesity hypothesis. Despite these two main hypotheses on the origin of MetS, obesity or insulin resistance or a combination of the two, Carr and Brunzell [106] underline familial combined hyperlipidemia (FCHL) as a subgroup of subjects that have MetS and a special increment of apolipoprotein B. Hopkins et al, and Hunt et al [107, 108] have shown that in patients with FCHL, abdominal adiposity and insulin resistance do not fully explain the elevated levels of apo B. In contrast, evidence exist that there is a very small group of subjects that are obese yet metabolically healthy [109]. Because obesity intersects with many metabolic pathways, the possible confounders are numerous, making it difficult to discriminate among them based on their importance. Furthermore, because MetS represents an atherogenic combination of central obesity, dyslipidemia, glucose intolerance and/or insulin resistance and high BP disorders, it is quite complex to explain. This complexity provides a great challenge for basic and clinical research. Together with challenges it opens opportunities for new targets of therapy for the metabolic syndrome [110].
The development of obesity as a chronic disease has a strong genetic component, although recent increases in prevalence do not reflect changes in population genetics. Therefore obesity is arguably the result of the obesogenic environment and gene-by-environment interactions, reflecting the selectivity to deposit fat efficiently that is maladaptive in the conditions of today’s energy-dense foods and sedentary lifestyles [111].
Adipose tissue may be the origin of one or more interconnections between obesity and the metabolic syndrome, as adipose tissue serves as storage and also as a place where lipids are mobilized [112, 113]. Free fatty acid (FFA) production and reduced FFA oxidation may obstruct insulin activity at the tissue level, especially where intramuscular fat is present. Moreover, the adipose tissue produces several adipokines that serve as a communication link with several important tissues including the liver (adiponectin, IL6), brain (adiponectin, IL6, leptin), vasculature (adiponectin, MCP-1, IL6, PAI1, SAA), muscle (adiponectin, IL6, MCP1), β-cells (IL6), and the reproductive tract (leptin) [114]. Abdominally obese individuals show higher levels of CRP, IL-6, TNF-α and reduced levels of adiponectin. Plasminogen activator inhibitor-1 (PAI-1) is an important regulator of the endogenous fibrinolytic and modulates thrombosis progression. PAI-1 and IL6 show strong correlations with measures of obesity [114–117]. We have reported that PAI-1 and IL6 clustered in two MetS latent factors related to obesity and lipids due to this correlation [116]. An accumulation of visceral fat accompanied with insulin resistance, rising blood pressure, and a prothrombotic and inflammatory profile suggests that adipose tissue is causally involved in the pathophysiology of MetS.
8. Evidence for Pleiotropic Genetic Effects on Adiposity Related Phenotypes and Metabolic Syndrome Related Traits
The heritability of the NCEP ATPIII-defined metabolic syndrome has been estimated to be about 30% [118], and principal component analysis combining these risk factors support the idea that the clustering or correlation among them is heritable and has a genetic basis [119]. Moreover, Kullo et al [120] investigated the degree to which pleiotropy contributes to the correlation between lipid traits related to the metabolic syndrome in the Genetic Epidemiology Network of Atherosclerosis (GENOA) cohort of hypertensive sibships and reported that pleiotropy contributes to the additive genetic variation in three correlated lipid traits – high density lipoprotein cholesterol, triglycerides, and low density lipoprotein particle size [121].
Genes that influence (have penetrance on) multiple phenotypes can be said to have pleiotropic effects, even if the effect may not be directly on the outcome variables. Genes that influence multiple outcomes may be highly desirable targets for intervention, and may identify points of connection between different pathways. Indeed, there are examples of individual genes found to have pleiotropic effects on the metabolic syndrome suite of phenotypes. NR3C1, which encodes the glucocorticoid receptor, has been associated with obesity, hypertension, and insulin resistance, and ADIPOQ has been associated with diabetes, hypertension, and dyslipidemia [122–125]. Findings from the recent genome-wide association studies reviewed here (Tables 2–5) have identified many new trait genes for obesity and central adiposity, blood pressure, lipids, and glycemic traits, some of which may have pleiotropic effects across phenotypic domains. In fact, it has been demonstrated that the first major well-replicated adiposity gene found by GWAS, the FTO gene, not only influences BMI, but may also predispose to type 2 diabetes [28, 29, 126, 127]. Other examples include loci containing the FADS1, GCKR, and MADD genes, all of which have been associated with type 2 diabetes [60] and fasting lipid concentrations [62, 128–132]. Lastly, pleiotropic relationships between the metabolic syndrome and coronary heart disease have also been revealed [133–136]. Elucidating these relationships is an important step in defining the genetic architecture of these correlated traits.
9. A case study: CDH13 gene in association with obesity and metabolic syndrome in the HyperGEN and Framingham studies
In this section we provide novel experimental information from the HyperGEN and Framingham studies on the CDH13 (cadherin 13) gene and MetS. We assess the association of CDH13 variants with obesity and MetS by means of latent factors resulting from factor analysis of a group of risk factors for MetS (Table 6). The HyperGEN study is part of the NHLBI Family Blood Pressure Program (FBPP, see Williams et al [137] and Province et al [138] for detailed descriptions). Briefly, sibships were selected so that at least two individuals per sibship were hypertensive prior to age 60. Parents and offspring of some of the hypertensive sibs, as well as a random sample of unrelated African Americans and whites were also recruited, totaling 4,781 participants. Those with genotype data and non-missing measures on 11 important metabolic and adiposity traits were utilized in this analysis [n=1935 (910 African American, 1025 white)]. Traits utilized include BMI (kg/m2), WC measured at the level of the umbilicus (cm); waist-to-hip ratio (WHR); fasting (≥12 hours) glucose (mg/dL); fasting (≥12 hours) insulin (μU/mL), LDL cholesterol (mg/dL), HDL cholesterol (mg/dL), triglycerides (mg/dL), sitting systolic blood pressure (SBP, mm Hg), diastolic blood pressure (DBP, mm Hg), and percent body fat (PBF) derived from the bioelectric impedance measurements based on the Lukaski formula [139]. All traits were adjusted for age within gender strata, and transformations were implemented when needed, followed by standardization of each trait to a mean 0 and variance 1. The transformed traits were included in multivariate factor analyses, and 4 factors were produced representing obesity and insulin (OBS-INS), lipids and insulin (LIP-INS), blood pressure (BP) and central obesity (CENT-OBS) factors. A detailed account on preparing phenotypes is provided in Kraja et al [93]. Previously we demonstrated that obesity and hypertension were the most important factors contributing to the MetS in the HyperGEN Study. Whites tended to have a higher prevalence of deleterious triglyceride and HDL levels than African Americans. The prevalence of MetS was 34% in African Americans and 39% in whites. Participants with complete phenotypic data had a mean age of 46 (SD 13) years in African Americans, and a mean of 51 (SD 14) years in whites. Overall, when compared to whites, African Americans tended to have a higher BMI, fasting glucose levels, fasting insulin levels, SBP, DBP, WC, and PBF; similar LDL levels and WHR; and lower TG and HDL levels. Similar trends were reported also in the FBPP study with a sample of 13,592 participants including African Americans, whites and Asians [140]. The Framingham Heart Study (FHS) is a generational cohort study which aims to identify the common factors that contribute to CVD by following its longitudinal development in participants who at enrollment had not yet developed overt symptoms of CVD or suffered a heart attack or stroke [141, 142]. We have shown in another publication [58] the trends of MetS prevalence in FHS which were comparable with the MetS prevalence in the general US population. We applied similar adjustments in the FHS data to the ones described above for the HyperGEN data.
Table 6.
Loadings of factors produced for each study using VARIMAX rotation in each factor analysis. In the case of variables standardized to a mean 0 and variance 1, loadings represent correlation coefficients of contributing variables into the latent factor.
| Study | Factor | Label* | BMI | WC | WHR | INS | GLUC | LDL | HDL | TG | SBP | DBP | PBF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HyperGEN (African Americans) | 1 | OBS-INS | 0.95 | 0.86 | 0.32 | 0.42 | −0.20 | 0.13 | 0.71 | ||||
|
| |||||||||||||
| 2 | BP | 0.99 | 0.76 | ||||||||||
|
| |||||||||||||
| 3 | LIP-IND | 0.22 | 0.24 | 0.31 | 0.52 | −0.43 | 0.27 | −0.57 | 0.64 | 0.19 | |||
|
| |||||||||||||
| 4 | CENT-OBS | 0.40 | 0.90 | 0.15 | −0.12 | 0.11 | |||||||
|
| |||||||||||||
| HyperGEN (whites) | 1 | OBS-INS | 0.94 | 0.86 | 0.40 | 0.46 | −0.26 | −0.11 | 0.11 | 0.14 | 0.78 | ||
|
| |||||||||||||
| 2 | LIP-INS | 0.23 | 0.21 | 0.27 | 0.51 | −0.27 | −0.68 | 0.64 | 0.17 | ||||
|
| |||||||||||||
| 3 | BP | 0.13 | −0.28 | 0.88 | 0.76 | ||||||||
|
| |||||||||||||
| 4 | CENT-OBS | 0.47 | 0.69 | −0.10 | 0.16 | ||||||||
|
| |||||||||||||
| Framingham Offspring (whites) | 1 | OBS-INS | 0.88 | 0.95 | 0.77 | 0.53 | −0.44 | −0.19 | 0.16 | 0.12 | ** | ||
|
| |||||||||||||
| 2 | LIP-INS | 0.12 | 0.11 | 0.16 | 0.43 | −0.34 | −0.83 | 0.83 | |||||
|
| |||||||||||||
| 3 | BP | 0.11 | −0.15 | 0.05 | 0.13 | 0.89 | 0.89 | ||||||
|
| |||||||||||||
| 4 | −0.15 | 0.17 | 0.97 | 0.16 | |||||||||
|
| |||||||||||||
| Framingham 3rd generation (whites) | 1 | OBS-INS | 0.92 | 0.92 | ** | 0.59 | −0.51 | −0.21 | 0.18 | 0.17 | 0.11 | ** | |
|
| |||||||||||||
| 2 | LIP-INS | 0.15 | 0.14 | −0.15 | 0.17 | 0.90 | 0.90 | ||||||
|
| |||||||||||||
| 3 | BP | 0.10 | 0.39 | −0.20 | −0.84 | 0.82 | |||||||
|
| |||||||||||||
| 4 | 0.10 | 0.10 | 0.18 | 0.97 | 0.17 | ||||||||
Notes: Loadings ≥ 0.40 are considered significant and are represented by shaded boxes; Loading coefficients <0.1 are not reported. BMI, body mass index; WC, waist circumference; WHR, waist-to-hip ratio; GLUC, glucose; LDL, Low density lipoprotein cholesterol; HDL, high density lipoprotein cholesterol; TG, triglycerides; SBP, systolic blood pressure; DBP, diastolic blood pressure; PBF, percent body fat
OBS-INS, obesity insulin factor; LIP-INS, lipids insulin factor; BP, blood pressure factor; CENT-OBS, central obesity factor;
Variable not present in data.
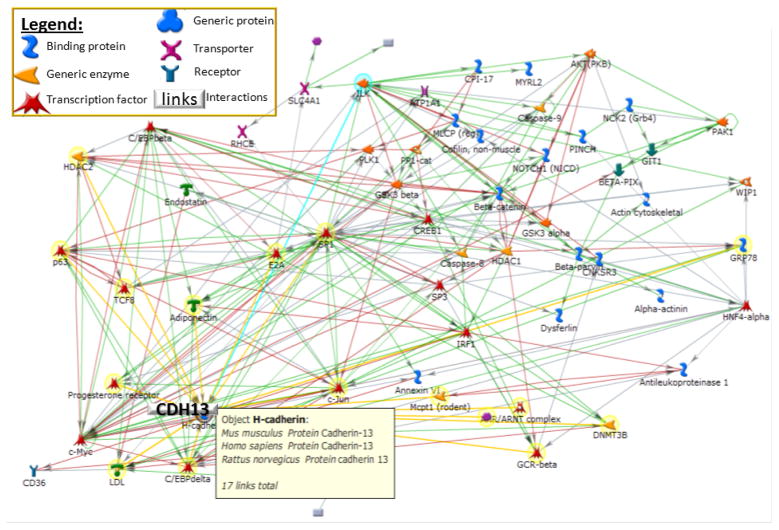
In previous work, using the HyperGEN data to investigate echocardiographic and metabolic syndrome features, we reported a LOD score of 2.8 on chromosome 16 in whites for a multivariate factor related to cardiovascular disease, namely “Left Ventricular wall thickness” [143]. This multivariate factor explained variation in left ventricular mass index (LVMI), diastolic posterior wall thickness (PWT), diastolic relative wall thickness (RWT), and left ventricular mid-wall shortening (MWS) [144]. This result was particularly interesting because the QTL for “LV wall thickness” on chromosome 16q24.2-q24.3 reached its local maximum LOD score at microsatellite marker D16S402, which is positioned within the 5th intron of the cadherin 13 gene (CDH13), implicated in heart and vascular remodeling [145, 146]. The CDH13 gene is a putative mediator of cell-cell interaction in the heart and may act as a negative regulator of neural cell growth. The gene locus is hypermethylated or deleted in breast, ovarian and lung cancers. It has been considered an LDL receptor [147] and has also been associated with BP [148]. Almost a decade ago this gene was identified as a receptor of adiponectin [149, 150]. Adiponectin is an adipokine produced in adipose tissue, and is believed to sensitize the body to insulin. Adiponectin receptors AdipoR1 and AdipoR2 are considered to play a special role in T2DM and CVD [151–158]. A number of interactions of adiponectin have already been confirmed via the curated literature as shown in Figure 1. However, it is unclear if these interactions, including CDH13 as a new receptor of adiponectin, contribute substantially to obesity and metabolic syndrome phenotypes.
Figure 1.
A network of genes that includes 17 interactions with Cadherin 13 gene (CDH13), comprising Adiponectin. Highlighted in yellow are direct interactions with CDH13. (The network is built using the curated literature of GeneGO Inc.)
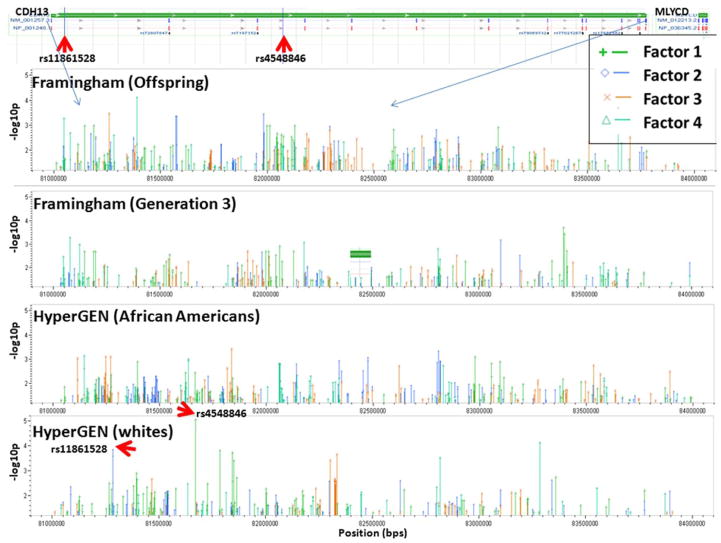
We studied the association of SNPs located within the CDH13 gene and the surrounding area using genotyped (not imputed) data, by selecting a 2 MB region on chromosome 16 (82MB–84MB) based on reference assembly build 36.3 of NCBI (accessed on April 12, 2010). Genotyping platforms used were Affymetrix 5.0 chip 500K Array Set for HyperGEN whites, Affymetrix 6.0 chip 1M Array Set for HyperGEN African Americans; and for the Framingham Heart Study genotyping GeneChip® Human Mapping 500K Array Set (Nsp and Sty), and the 50K Human Gene Focused Panel of Affymetrix platform. As shown in Table 6, latent factor scores that represented contributions of risk factors for obesity and insulin measures, dyslipidemia and insulin measures, blood pressure, and central obesity were used in mixed model analysis (SAS version 9.2), for testing the hypothesis that CDH13 is associated, under an additive genetic model, with obesity and/or other metabolic syndrome risk variables. Pedigree ID was included as a random effect in the model to correct for familial relationships. Figure 2 shows the results of these analyses with a baseline above a negative log10 p-value of 1.3 (p < 0.05). Two SNPs from the HyperGEN whites showed significant associations, rs4548846 with the OBS-INS latent factor (p-value 9.14×10−6, r2=0.019, β= −0.997, se=0.2236, MAF=1.03%), and rs11861528 with the LIP-INS latent factor (p-value 0.00014, r2=0.014, β=0.221, se=0.0579, MAF=9.1%). There were also many SNPs with p-values <0.05, although they were not significant after correcting for multiple comparisons. Rs4548846 had a p-value of 0.03 in the HyperGEN African Americans (MAF 9.46%); rs11861528 was genotyped only in whites. Thus, although CDH13 has a demonstrated importance on obesity, metabolic syndrome, and diabetes, our significant associations between CDH13 variants and latent factors of obesity and metabolic syndrome were relatively weak with the exception of the two SNPs reported. Similarly weak significant results were also found when testing the association of several variants in the AdipoR1 and AdipoR2 genes in association with T2DM in a French case control study (n=1,498), as well as a Greek CVD case control study (n=68) [153, 157]. However, AdipoR2 variants were found to contribute to variation in hepatic fat accumulation (n=302 Finnish; replication: n=619 Swedish, and n=3,050 Finnish individuals) [154]. It is possible that the role of adiponectin and its receptors including CDH13 play a more indirect role, possibly through anti-inflammatory, anti-atherogenic, and anti-diabetic properties. Adiponectin has been considered a key molecule in MetS and viewed as a possible therapeutic target. Our results indicate that CDH13 variants have only small effects on the phenotypes studied. More recent studies place CDH13 in the role of a signaling receptor participating in recognition of the environment and regulation of cell motility, proliferation, and phenotype, controlling and guiding tissue architecture [159–162]. Because this gene is highly expressed in the heart (see www.genecards.org), it will be of interest to see if CDH13 variants associate more significantly with echocardiographic variables in the HyperGEN study, especially for LV Wall thickness, which will be focus of future work.
Figure 2.
Association tests results expressed as negative log10 p-value (> 1.3) of a number of SNPs selected from a region on chromosome 16, starting at 82MBs–84MBs, which comprises CDH13. The association tests were implemented with latent factors result of factor analysis on a number of risk variables for obesity and metabolic syndrome (see text).
10. Meta-analysis of genetic findings in obesity, metabolic syndrome and pathways
Meta-analyses of genetic studies have become the norm in order to increase the power to discover variants with small effects on obesity and the metabolic syndrome. Prior to this time, the most authoritative summary on discovery of obesity genes was the human obesity gene map, a yearly review of cumulative findings on obesity genetic research, which published its 12th and final issue in 2006 [25]. The final issue reported 11 genes as leading to single-gene mutations obesity cases, 50 loci related to Mendelian segregation, 244 genes which when mutated or expressed as transgenes in animal models affected weight or adiposity, and 22 gene polymorphisms replicated in at least 5 studies. In addition, it reported 408 animal QTLs and 253 human QTLs humans, 52 of which were replicated by two or more studies.
An important review on the genetics of MetS findings is the publication by Terán-García and Bouchard in 2007 [118], where they summarized 14 MetS related genes and 38 QTLs. In 2008, Joy et al [163] provided an alternative review of genetic findings on candidate genes that may contribute to components of MetS, namely adipokines, lipoproteins, inflammation, adipose, glucose, and energy metabolism. Other publications have summarized important work in animal models [164, 165].
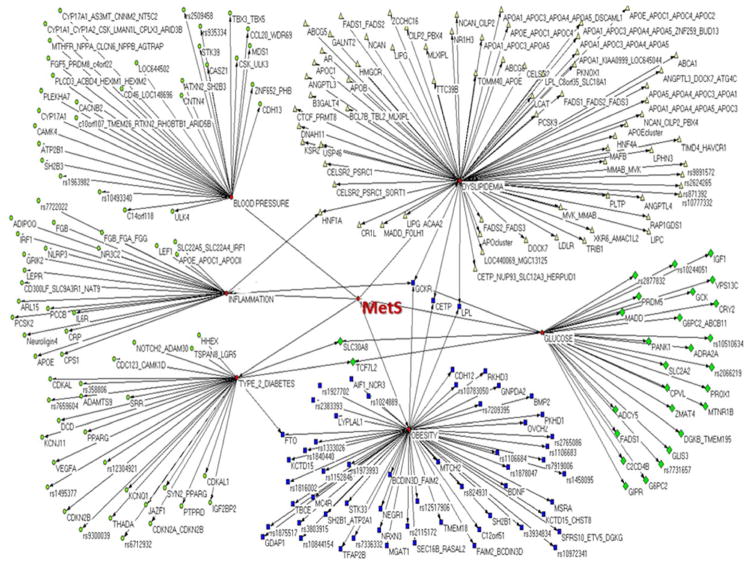
In recent years several studies have combined evidence from different candidate gene work and genome-wide association studies of obesity and MetS via meta-analysis. Results have been more productive than expected in terms of discovery; however, as we noted earlier, findings indicate that obesity and MetS are quite complex and a substantial number of gene variants appear to be involved in disease development. Hinney et al [166, 167], supporting a polygenic view of obesity in humans, provided a comprehensive overview of two gene variants (MC4R and FTO) with small but replicable effects on obesity. Not only do the results from the recent studies cited herein suggest obesity and MetS arise from a combination of more common variants of relatively small effect, but also that several causative alleles may be quite rare and beneath the detectable limit of current genome-wide studies. Genome-wide and candidate gene studies have recently reported several genetic variants associated with adiposity and obesity [28, 30, 32, 34, 35, 37, 168–171], blood pressure [172–174], dyslipidemia [62, 128, 129, 132, 175–180], inflammation and prothrombosis [155, 181–186], and insulin resistance, diabetes, and glucose intolerance [60, 61, 126, 187–204]. Using these data and the database and tools created by Hindorff et al [205–207] (see www.genome.gov/gwastudies), we conducted an analysis of recent genetic findings on obesity and metabolic syndrome. The database contains data from studies that attempted to assay at least 100,000 SNPs in the initial stage, and results were limited to those with p-values <1.0×10-5. Based on the phenotypes studied in the reported GWAS, we grouped the results into the following domains: obesity, dyslipidemia, type 2 diabetes, glucose, blood pressure, or inflammation. Of these, adiposity and obesity included a total of 58 unique intragenic SNPs or genes; dyslipidemia, 74; glucose studies, 29; type 2 diabetes, 29; blood pressure, 30; and inflammation, 24. Our results are summarized in Figure 3, which shows gene names, and in cases when the SNP was intragenic, their rs numbers in association with a specific domain. We observe that the CETP, GCKR, LPL, FTO, HNF1A, SLC30A8, and TCF7L2 genes have connections between at least two of the domains, with GCKR reported to connect to 4 domains. Kraja et al [208] recently reported for the STAMPEED consortium a meta-analysis of 7 studies for the bivariate analysis on components of MetS and MetS itself (a total of 11 traits, n=22,161 participants). This analysis, which also took into account pleiotropic effects of the most significant variants, identified a number of associations at or near the genes LPL, CETP, APOA5 (and its cluster, namely ZNF259 and BUD13), and GCKR. In addition they reported a number of variants for MetS components in LIPC, TRIB1, LOC100128354/MTNR1B, ABCB11, and in LOC100129150.
Figure 3.
A network of genes or intragenic SNPs reported from the most recent GWAS in relation to obesity and metabolic syndrome. Results represented here include those that attempted to assay at least 100,000 SNPs in the initial stage, and are limited to those with p-values <1.0×10−5. (Source: http://www.genome.gov/gwastudies/, accessed on April 05, 2010).
In parallel with work on metabolic syndrome in humans, several animal models are used to study potential causative gene polymorphisms. For example Su et al [165] identified 15 QTLs for obesity in mouse models, and using comparative genomics narrowed their findings to candidate genes (referred by their locations in human genome mapping), to Apcs on chromosome 1 (1q21-q23), Ppargc1a on chromosome 4 (4p15.1), Ucp1 on chromosome 4 (4q28-q31), Angptl6 on chromosome 19 (19p13.2), and Lpin1 on chromosome 2 (2p25.1). Lawson and Cheverud [164] report 31 adiposity QTLs in their recent work on obesity genes utilizing a different mouse model. This article is part of a series of review papers on MetS, and has an in-depth analysis of MetS components and several candidate gene findings for obesity and metabolic syndrome in murine models and their comparative action in humans. These findings underscore that complex traits such as obesity and metabolic syndrome have an intricate genetic background and must be viewed in their complexity.
11. Future challenges
There are several advances that parallel each other in understanding the genetic basis of common complex disease. A drastic increase of DNA sequence information by recent introduction of instruments capable of producing millions of bases of DNA sequence in a single run is rapidly changing the landscape of genetics [209]. Massive genotyping and sequencing efforts require enormous computing resources to analyze, access, and archive these data. Earlier programs that were designed to work with single computers and one sequential run of a program are capable of perform these analyses. Instead, researchers are utilizing parallel computing by distributing the work to large numbers of computing servers (cloud computing) [210]. Studies are taking advantage of the wealth of phenotype data collected and reanalyzing them in association with genotype and sequence data. However, advancements in genotyping have lead to challenges in the phenotype realm. For example, while BMI as a measure correlates very well with obesity, it is an imperfect measure, and it is becoming increasingly clear that future studies will need to limit heterogeneity by collecting data which more precisely measures adiposity or endophenotypes related to adiposity. Not only do we need “deep sequencing,” but also “deep phenotyping” [211]. While several multivariate methods have been applied to analyze obesity and the other components of the metabolic syndrome, more comprehensive statistical methodologies are needed especially to find useful subsets in these massively large accumulations of data. As stated earlier, findings from GWAS explain only a relatively small percentage of interindividual variation in obesity and metabolic syndrome traits. We believe that information from other methods including gene expression, copy number variation, micro RNA regulation [212], methylation [213], special treatment of rare variants and study of other regulatory processes, as well as information from proteomics, and from comprehensive databases on modules, pathways and networks of genes and proteins will contribute to further understanding of the true functioning of genes that increase susceptibility to obesity and the metabolic syndrome.
Acknowledgments
This work was supported by the NIH Family Blood Pressure Program grant U01HL54473. We acknowledge the Framingham Heart Study, the NHLBI and Boston University for providing access to the FHS data through dbGAP of NCBI.
Footnotes
Conflicts of interest
None to declare
References
- 1.Ogden CL, et al. Prevalence of overweight and obesity in the United States, 1999–2004. JAMA. 2006;295(13):1549–55. doi: 10.1001/jama.295.13.1549. [DOI] [PubMed] [Google Scholar]
- 2.Calle EE, et al. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N Engl J Med. 2003;348(17):1625–38. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- 3.Gregg EW, et al. Secular trends in cardiovascular disease risk factors according to body mass index in US adults. Jama. 2005;293(15):1868–74. doi: 10.1001/jama.293.15.1868. [DOI] [PubMed] [Google Scholar]
- 4.Kopelman PG. Obesity as a medical problem. Nature. 2000;404(6778):635–43. doi: 10.1038/35007508. [DOI] [PubMed] [Google Scholar]
- 5.Mendez MA, Monteiro CA, Popkin BM. Overweight exceeds underweight among women in most developing countries. Am J Clin Nutr. 2005;81(3):714–21. doi: 10.1093/ajcn/81.3.714. [DOI] [PubMed] [Google Scholar]
- 6.Popkin BM. The shift in stages of the nutrition transition in the developing world differs from past experiences! Public Health Nutr. 2002;5(1A):205–14. doi: 10.1079/PHN2001295. [DOI] [PubMed] [Google Scholar]
- 7.Adair LS. Dramatic rise in overweight and obesity in adult filipino women and risk of hypertension. Obes Res. 2004;12(8):1335–41. doi: 10.1038/oby.2004.168. [DOI] [PubMed] [Google Scholar]
- 8.Wang H, et al. Trends in the distribution of body mass index among Chinese adults, aged 20–45 years (1989–2000) Int J Obes (Lond) 2007;31(2):272–8. doi: 10.1038/sj.ijo.0803416. [DOI] [PubMed] [Google Scholar]
- 9.Brown CD, et al. Body mass index and the prevalence of hypertension and dyslipidemia. Obes Res. 2000;8(9):605–19. doi: 10.1038/oby.2000.79. [DOI] [PubMed] [Google Scholar]
- 10.Ferrannini E, Camastra S. Relationship between impaired glucose tolerance, non-insulin-dependent diabetes mellitus and obesity. Eur J Clin Invest. 1998;28(Suppl 2):3–6. doi: 10.1046/j.1365-2362.1998.0280s2003.x. discussion 6–7. [DOI] [PubMed] [Google Scholar]
- 11.Ford ES, Giles WH, Dietz WH. Prevalence of the metabolic syndrome among US adults: findings from the third National Health and Nutrition Examination Survey. Jama. 2002;287(3):356–9. doi: 10.1001/jama.287.3.356. [DOI] [PubMed] [Google Scholar]
- 12.Martins D, et al. The relative risk of cardiovascular death among racial and ethnic minorities with metabolic syndrome: data from the NHANES-II mortality follow-up. J Natl Med Assoc. 2008;100(5):565–71. doi: 10.1016/s0027-9684(15)31304-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Church TS, et al. Metabolic syndrome and diabetes, alone and in combination, as predictors of cardiovascular disease mortality among men. Diabetes Care. 2009;32(7):1289–94. doi: 10.2337/dc08-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1998;15(7):539–53. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 15.Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, And Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III) Jama. 2001;285(19):2486–97. doi: 10.1001/jama.285.19.2486. [DOI] [PubMed] [Google Scholar]
- 16.Alberti KG, Zimmet P, Shaw J. The metabolic syndrome--a new worldwide definition. Lancet. 2005;366(9491):1059–62. doi: 10.1016/S0140-6736(05)67402-8. [DOI] [PubMed] [Google Scholar]
- 17.Grundy SM, et al. Diagnosis and management of the metabolic syndrome: an American Heart Association/National Heart, Lung, and Blood Institute Scientific Statement. Circulation. 2005;112(17):2735–52. doi: 10.1161/CIRCULATIONAHA.105.169404. [DOI] [PubMed] [Google Scholar]
- 18.Alberti KG, et al. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120(16):1640–5. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- 19.Ervin RB. Prevalence of metabolic syndrome among adults 20 years of age and over, by sex, age, race and ethnicity, and body mass index: United States, 2003–2006. Natl Health Stat Report. 2009;(13):1–7. [PubMed] [Google Scholar]
- 20.Brown TM, et al. Variations in prevalent cardiovascular disease and future risk by metabolic syndrome classification in the REasons for Geographic and Racial Differences in Stroke (REGARDS) study. Am Heart J. 2010;159(3):385–91. doi: 10.1016/j.ahj.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voulgari C, et al. The impact of metabolic syndrome on left ventricular myocardial performance. Diabetes Metab Res Rev. 2010;26(2):121–7. doi: 10.1002/dmrr.1063. [DOI] [PubMed] [Google Scholar]
- 22.Ballantyne CM, et al. Metabolic syndrome risk for cardiovascular disease and diabetes in the ARIC study. Int J Obes (Lond) 2008;32(Suppl 2):S21–4. doi: 10.1038/ijo.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ninomiya JK, et al. Association of the metabolic syndrome with history of myocardial infarction and stroke in the Third National Health and Nutrition Examination Survey. Circulation. 2004;109(1):42–6. doi: 10.1161/01.CIR.0000108926.04022.0C. [DOI] [PubMed] [Google Scholar]
- 24.Lakka HM, et al. The metabolic syndrome and total and cardiovascular disease mortality in middle-aged men. Jama. 2002;288(21):2709–16. doi: 10.1001/jama.288.21.2709. [DOI] [PubMed] [Google Scholar]
- 25.Rankinen T, et al. The human obesity gene map: the 2005 update. Obesity (Silver Spring) 2006;14(4):529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- 26.Lyon HN, Hirschhorn JN. Genetics of common forms of obesity: a brief overview. Am J Clin Nutr. 2005;82(1 Suppl):215S–217S. doi: 10.1093/ajcn/82.1.215S. [DOI] [PubMed] [Google Scholar]
- 27.Loos RJ, Bouchard C. Obesity--is it a genetic disorder? J Intern Med. 2003;254(5):401–25. doi: 10.1046/j.1365-2796.2003.01242.x. [DOI] [PubMed] [Google Scholar]
- 28.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–94. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scuteri A, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3(7):e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loos RJ, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40(6):768–75. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chambers JC, et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet. 2008;40(6):716–8. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- 32.Heard-Costa NL, et al. NRXN3 is a novel locus for waist circumference: a genome-wide association study from the CHARGE Consortium. PLoS Genet. 2009;5(6):e1000539. doi: 10.1371/journal.pgen.1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyre D, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41(2):157–9. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 34.Thorleifsson G, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41(1):18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 35.Willer CJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41(1):25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cotsapas C, et al. Common body mass index-associated variants confer risk of extreme obesity. Hum Mol Genet. 2009;18(18):3502–7. doi: 10.1093/hmg/ddp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindgren CM, et al. Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS Genet. 2009;5(6):e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manolio TA, et al. New models of collaboration in genome-wide association studies: the Genetic Association Information Network. Nat Genet. 2007;39(9):1045–51. doi: 10.1038/ng2127. [DOI] [PubMed] [Google Scholar]
- 39.McCarthy MI, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9(5):356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 40.Argyropoulos G. Genetics of the metabolic syndrome. In: Kumar S, O’Rahilly S, editors. Insulin Resistance. John Wiley & Sons, Ltd; West Essex, England: 2005. pp. 401–450. [Google Scholar]
- 41.Perusse L, et al. Genetic and environmental determinants of serum lipids and lipoproteins in French Canadian families. Arteriosclerosis. 1989;9(3):308–18. doi: 10.1161/01.atv.9.3.308. [DOI] [PubMed] [Google Scholar]
- 42.Katzmarzyk PT, et al. Familial aggregation of seven-year changes in blood pressure in Canada. Can J Cardiol. 2001;17(12):1267–74. [PubMed] [Google Scholar]
- 43.An P, et al. Familial aggregation of resting blood pressure and heart rate in a sedentary population: the HERITAGE Family Study. Health, Risk Factors, Exercise Training, and Genetics. Am J Hypertens. 1999;12(3):264–70. doi: 10.1016/s0895-7061(98)00261-1. [DOI] [PubMed] [Google Scholar]
- 44.Rice T, et al. Genome-wide linkage analysis of systolic and diastolic blood pressure: the Quebec Family Study. Circulation. 2000;102(16):1956–63. doi: 10.1161/01.cir.102.16.1956. [DOI] [PubMed] [Google Scholar]
- 45.Lehtovirta M, et al. Insulin sensitivity and insulin secretion in monozygotic and dizygotic twins. Diabetologia. 2000;43(3):285–93. doi: 10.1007/s001250050046. [DOI] [PubMed] [Google Scholar]
- 46.Mayer EJ, et al. Genetic and environmental influences on insulin levels and the insulin resistance syndrome: an analysis of women twins. Am J Epidemiol. 1996;143(4):323–32. doi: 10.1093/oxfordjournals.aje.a008746. [DOI] [PubMed] [Google Scholar]
- 47.Carmelli D, Cardon LR, Fabsitz R. Clustering of hypertension, diabetes, and obesity in adult male twins: same genes or same environments? Am J Hum Genet. 1994;55(3):566–73. [PMC free article] [PubMed] [Google Scholar]
- 48.Mitchell BD, et al. Genetic analysis of the IRS. Pleiotropic effects of genes influencing insulin levels on lipoprotein and obesity measures. Arterioscler Thromb Vasc Biol. 1996;16(2):281–8. doi: 10.1161/01.atv.16.2.281. [DOI] [PubMed] [Google Scholar]
- 49.Kissebah AH, et al. Quantitative trait loci on chromosomes 3 and 17 influence phenotypes of the metabolic syndrome. Proc Natl Acad Sci U S A. 2000;97(26):14478–83. doi: 10.1073/pnas.97.26.14478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liese AD, et al. Familial components of the multiple metabolic syndrome: the ARIC Study. Diabetologia. 1997;40(8):963–970. doi: 10.1007/s001250050775. [DOI] [PubMed] [Google Scholar]
- 51.Hong Y, et al. Familial clustering of insulin and abdominal visceral fat: the HERITAGE Family Study. J Clin Endocrinol Metab. 1998;83(12):4239–45. doi: 10.1210/jcem.83.12.5312. [DOI] [PubMed] [Google Scholar]
- 52.Lehman DM, et al. Bivariate linkage analysis of the insulin resistance syndrome phenotypes on chromosome 7q. Hum Biol. 2005;77(2):231–46. doi: 10.1353/hub.2005.0040. [DOI] [PubMed] [Google Scholar]
- 53.Arya R, et al. Evidence for bivariate linkage of obesity and HDL-C levels in the Framingham Heart Study. BMC Genet. 2003;4(Suppl 1):S52. doi: 10.1186/1471-2156-4-S1-S52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turner ST, et al. Multivariate linkage analysis of blood pressure and body mass index. Genet Epidemiol. 2004;27(1):64–73. doi: 10.1002/gepi.20002. [DOI] [PubMed] [Google Scholar]
- 55.Loos RJ, et al. Genome-wide linkage scan for the metabolic syndrome in the HERITAGE Family Study. J Clin Endocrinol Metab. 2003;88(12):5935–43. doi: 10.1210/jc.2003-030553. [DOI] [PubMed] [Google Scholar]
- 56.Kraja AT, et al. Quantitative trait loci for metabolic syndrome in the Hypertension Genetic Epidemiology Network study. Obes Res. 2005;13(11):1885–90. doi: 10.1038/oby.2005.231. [DOI] [PubMed] [Google Scholar]
- 57.Kraja AT, et al. Two major QTLs and several others relate to factors of metabolic syndrome in the family blood pressure program. Hypertension. 2005;46(4):751–7. doi: 10.1161/01.HYP.0000184249.20016.bb. [DOI] [PubMed] [Google Scholar]
- 58.Park YM, et al. Longitudinal trends in the association of metabolic syndrome with 550 k single-nucleotide polymorphisms in the Framingham Heart Study. BMC Proc. 2009;3(Suppl 7):S116. doi: 10.1186/1753-6561-3-s7-s116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pirruccello J, Kathiresan S. Genetics of lipid disorders. Curr Opin Cardiol. 2010 doi: 10.1097/HCO.0b013e328338574d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(2):105–16. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40(5):638–45. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kathiresan S, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41(1):56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grundy SM. Obesity, metabolic syndrome, and cardiovascular disease. J Clin Endocrinol Metab. 2004;89(6):2595–600. doi: 10.1210/jc.2004-0372. [DOI] [PubMed] [Google Scholar]
- 64.Caprio S. Insulin resistance in childhood obesity. J Pediatr Endocrinol Metab. 2002;15(Suppl 1):487–92. [PubMed] [Google Scholar]
- 65.Arslanian S. Type 2 diabetes in children: clinical aspects and risk factors. Horm Res. 2002;57(Suppl 1):19–28. doi: 10.1159/000053308. [DOI] [PubMed] [Google Scholar]
- 66.Berenson GS, et al. Association between multiple cardiovascular risk factors and atherosclerosis in children and young adults. The Bogalusa Heart Study. N Engl J Med. 1998;338(23):1650–6. doi: 10.1056/NEJM199806043382302. [DOI] [PubMed] [Google Scholar]
- 67.Goran MI, Gower BA. Abdominal obesity and cardiovascular risk in children. Coron Artery Dis. 1998;9(8):483–7. doi: 10.1097/00019501-199809080-00003. [DOI] [PubMed] [Google Scholar]
- 68.Must A, et al. Long-term morbidity and mortality of overweight adolescents. A follow-up of the Harvard Growth Study of 1922 to 1935. N Engl J Med. 1992;327(19):1350–5. doi: 10.1056/NEJM199211053271904. [DOI] [PubMed] [Google Scholar]
- 69.Steinberger J, Daniels SR. Obesity, insulin resistance, diabetes, and cardiovascular risk in children: an American Heart Association scientific statement from the Atherosclerosis, Hypertension, and Obesity in the Young Committee (Council on Cardiovascular Disease in the Young) and the Diabetes Committee (Council on Nutrition, Physical Activity, and Metabolism) Circulation. 2003;107(10):1448–53. doi: 10.1161/01.cir.0000060923.07573.f2. [DOI] [PubMed] [Google Scholar]
- 70.Biro FM, Wien M. Childhood obesity and adult morbidities. Am J Clin Nutr. 2010 doi: 10.3945/ajcn.2010.28701B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weiss R, et al. Obesity and the metabolic syndrome in children and adolescents. N Engl J Med. 2004;350(23):2362–74. doi: 10.1056/NEJMoa031049. [DOI] [PubMed] [Google Scholar]
- 72.Reaven GM. Banting lecture 1988. Role of insulin resistance in human disease. Diabetes. 1988;37(12):1595–607. doi: 10.2337/diab.37.12.1595. [DOI] [PubMed] [Google Scholar]
- 73.Haffner SM, et al. Prospective analysis of the insulin-resistance syndrome (syndrome X) Diabetes. 1992;41(6):715–22. doi: 10.2337/diab.41.6.715. [DOI] [PubMed] [Google Scholar]
- 74.Liese AD, et al. Development of the multiple metabolic syndrome in the ARIC cohort: joint contribution of insulin, BMI, and WHR. Atherosclerosis risk in communities. Ann Epidemiol. 1997;7(6):407–16. doi: 10.1016/s1047-2797(97)00047-1. [DOI] [PubMed] [Google Scholar]
- 75.Haffner SM, et al. Is leptin concentration associated with the insulin resistance syndrome in nondiabetic men? Obes Res. 1999;7(2):164–9. doi: 10.1002/j.1550-8528.1999.tb00698.x. [DOI] [PubMed] [Google Scholar]
- 76.Alexander CM, Landsman PB, Grundy SM. The influence of age and body mass index on the metabolic syndrome and its components. Diabetes Obes Metab. 2008;10(3):246–50. doi: 10.1111/j.1463-1326.2006.00695.x. [DOI] [PubMed] [Google Scholar]
- 77.Chang C, et al. Metabolic syndrome phenotype in very obese women. Metab Syndr Relat Disord. 2007;5(1):3–12. doi: 10.1089/met.2006.0016. [DOI] [PubMed] [Google Scholar]
- 78.Grundy SM. Metabolic complications of obesity. Endocrine. 2000;13(2):155–65. doi: 10.1385/ENDO:13:2:155. [DOI] [PubMed] [Google Scholar]
- 79.Grundy SM. Obesity, metabolic syndrome, and coronary atherosclerosis. Circulation. 2002;105(23):2696–8. doi: 10.1161/01.cir.0000020650.86137.84. [DOI] [PubMed] [Google Scholar]
- 80.Grundy SM. What is the contribution of obesity to the metabolic syndrome? Endocrinol Metab Clin North Am. 2004;33(2):267–82. doi: 10.1016/j.ecl.2004.03.001. table of contents. [DOI] [PubMed] [Google Scholar]
- 81.Grundy SM. Metabolic syndrome: a multiplex cardiovascular risk factor. J Clin Endocrinol Metab. 2007;92(2):399–404. doi: 10.1210/jc.2006-0513. [DOI] [PubMed] [Google Scholar]
- 82.Grundy SM. Metabolic syndrome pandemic. Arterioscler Thromb Vasc Biol. 2008;28(4):629–36. doi: 10.1161/ATVBAHA.107.151092. [DOI] [PubMed] [Google Scholar]
- 83.Haffner S, Taegtmeyer H. Epidemic obesity and the metabolic syndrome. Circulation. 2003;108(13):1541–5. doi: 10.1161/01.CIR.0000088845.17586.EC. [DOI] [PubMed] [Google Scholar]
- 84.Haffner SM. Obesity and the metabolic syndrome: the San Antonio Heart Study. Br J Nutr. 2000;83(Suppl 1):S67–70. doi: 10.1017/s0007114500000970. [DOI] [PubMed] [Google Scholar]
- 85.Haffner SM. Relationship of metabolic risk factors and development of cardiovascular disease and diabetes. Obesity (Silver Spring) 2006;14(Suppl 3):121S–127S. doi: 10.1038/oby.2006.291. [DOI] [PubMed] [Google Scholar]
- 86.Lorenzo C, et al. Is waist circumference an essential component of the metabolic syndrome? Diabetes Care. 2007;30(8):2141–2. doi: 10.2337/dc06-2627. [DOI] [PubMed] [Google Scholar]
- 87.Pladevall M, et al. A single factor underlies the metabolic syndrome: a confirmatory factor analysis. Diabetes Care. 2006;29(1):113–22. doi: 10.2337/diacare.29.1.113. [DOI] [PubMed] [Google Scholar]
- 88.Stancakova A, et al. Changes in insulin sensitivity and insulin release in relation to glycemia and glucose tolerance in 6,414 Finnish men. Diabetes. 2009;58(5):1212–21. doi: 10.2337/db08-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vega GL, et al. Influence of body fat content and distribution on variation in metabolic risk. J Clin Endocrinol Metab. 2006;91(11):4459–66. doi: 10.1210/jc.2006-0814. [DOI] [PubMed] [Google Scholar]
- 90.Zhao D, et al. Ten-year cardiovascular disease risk of metabolic syndrome without central obesity in middle-aged chinese. Am J Cardiol. 2007;100(5):835–9. doi: 10.1016/j.amjcard.2007.03.103. [DOI] [PubMed] [Google Scholar]
- 91.Han TS, et al. Analysis of obesity and hyperinsulinemia in the development of metabolic syndrome: San Antonio Heart Study. Obes Res. 2002;10(9):923–31. doi: 10.1038/oby.2002.126. [DOI] [PubMed] [Google Scholar]
- 92.Ascaso JF, et al. Abdominal obesity, insulin resistance, and metabolic syndrome in a southern European population. Eur J Intern Med. 2003;14(2):101–106. doi: 10.1016/S0953-6205(03)00022-0. [DOI] [PubMed] [Google Scholar]
- 93.Kraja AT, et al. An evaluation of the metabolic syndrome in the HyperGEN study. Nutr Metab (Lond) 2005;2(1):2. doi: 10.1186/1743-7075-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maison P, et al. Do different dimensions of the metabolic syndrome change together over time? Evidence supporting obesity as the central feature. Diabetes Care. 2001;24(10):1758–63. doi: 10.2337/diacare.24.10.1758. [DOI] [PubMed] [Google Scholar]
- 95.Meigs JB, et al. Risk variable clustering in the insulin resistance syndrome. The Framingham Offspring Study. Diabetes. 1997;46(10):1594–600. doi: 10.2337/diacare.46.10.1594. [DOI] [PubMed] [Google Scholar]
- 96.Arnlov J, et al. Impact of body mass index and the metabolic syndrome on the risk of cardiovascular disease and death in middle-aged men. Circulation. 2010;121(2):230–6. doi: 10.1161/CIRCULATIONAHA.109.887521. [DOI] [PubMed] [Google Scholar]
- 97.Gade W, et al. Beyond obesity: the diagnosis and pathophysiology of metabolic syndrome. Clin Lab Sci. 2010;23(1):51–61. quiz 62–5. [PubMed] [Google Scholar]
- 98.Abbasi F, et al. Relationship between obesity, insulin resistance, and coronary heart disease risk. J Am Coll Cardiol. 2002;40(5):937–43. doi: 10.1016/s0735-1097(02)02051-x. [DOI] [PubMed] [Google Scholar]
- 99.Libby P, Ridker PM, Maseri A. Inflammation and atherosclerosis. Circulation. 2002;105(9):1135–43. doi: 10.1161/hc0902.104353. [DOI] [PubMed] [Google Scholar]
- 100.Nishimura S, Manabe I, Nagai R. Adipose tissue inflammation in obesity and metabolic syndrome. Discov Med. 2009;8(41):55–60. [PubMed] [Google Scholar]
- 101.Kopp W. High-insulinogenic nutrition--an etiologic factor for obesity and the metabolic syndrome? Metabolism. 2003;52(7):840–4. doi: 10.1016/s0026-0495(02)05294-0. [DOI] [PubMed] [Google Scholar]
- 102.Le Stunff C, Bougneres P. Early changes in postprandial insulin secretion, not in insulin sensitivity, characterize juvenile obesity. Diabetes. 1994;43(5):696–702. doi: 10.2337/diab.43.5.696. [DOI] [PubMed] [Google Scholar]
- 103.Sigal RJ, et al. Acute postchallenge hyperinsulinemia predicts weight gain: a prospective study. Diabetes. 1997;46(6):1025–9. doi: 10.2337/diab.46.6.1025. [DOI] [PubMed] [Google Scholar]
- 104.Djousse L, et al. Diet and Metabolic Syndrome. Endocr Metab Immune Disord Drug Targets. 2010 doi: 10.2174/187153010791213056. [DOI] [PubMed] [Google Scholar]
- 105.McGarry JD. Banting lecture 2001: dysregulation of fatty acid metabolism in the etiology of type 2 diabetes. Diabetes. 2002;51(1):7–18. doi: 10.2337/diabetes.51.1.7. [DOI] [PubMed] [Google Scholar]
- 106.Carr MC, Brunzell JD. Abdominal obesity and dyslipidemia in the metabolic syndrome: importance of type 2 diabetes and familial combined hyperlipidemia in coronary artery disease risk. J Clin Endocrinol Metab. 2004;89(6):2601–7. doi: 10.1210/jc.2004-0432. [DOI] [PubMed] [Google Scholar]
- 107.Hopkins PN, et al. Coronary artery disease risk in familial combined hyperlipidemia and familial hypertriglyceridemia: a case-control comparison from the National Heart, Lung, and Blood Institute Family Heart Study. Circulation. 2003;108(5):519–23. doi: 10.1161/01.CIR.0000081777.17879.85. [DOI] [PubMed] [Google Scholar]
- 108.Hunt SC, et al. Apolipoprotein, low density lipoprotein subfraction, and insulin associations with familial combined hyperlipidemia. Study of Utah patients with familial dyslipidemic hypertension. Arteriosclerosis. 1989;9(3):335–44. doi: 10.1161/01.atv.9.3.335. [DOI] [PubMed] [Google Scholar]
- 109.Soverini V, et al. Metabolic syndrome and insulin resistance in subjects with morbid obesity. Obes Surg. 2010;20(3):295–301. doi: 10.1007/s11695-009-9999-z. [DOI] [PubMed] [Google Scholar]
- 110.Nisoli E, Carruba MO. Emerging aspects of pharmacotherapy for obesity and metabolic syndrome. Pharmacol Res. 2004;50(5):453–69. doi: 10.1016/j.phrs.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 111.Speakman JR. Thrifty genes for obesity and the metabolic syndrome--time to call off the search? Diab Vasc Dis Res. 2006;3(1):7–11. doi: 10.3132/dvdr.2006.010. [DOI] [PubMed] [Google Scholar]
- 112.Bosello O, Zamboni M. Visceral obesity and metabolic syndrome. Obes Rev. 2000;1(1):47–56. doi: 10.1046/j.1467-789x.2000.00008.x. [DOI] [PubMed] [Google Scholar]
- 113.Zoico E, et al. Adipose tissue infiltration in skeletal muscle of healthy elderly men: relationships with body composition, insulin resistance, and inflammation at the systemic and tissue level. J Gerontol A Biol Sci Med Sci. 2010;65(3):295–9. doi: 10.1093/gerona/glp155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gustafson B, et al. Inflamed adipose tissue: a culprit underlying the metabolic syndrome and atherosclerosis. Arterioscler Thromb Vasc Biol. 2007;27(11):2276–83. doi: 10.1161/ATVBAHA.107.147835. [DOI] [PubMed] [Google Scholar]
- 115.Iyer A, et al. Inflammatory lipid mediators in adipocyte function and obesity. Nat Rev Endocrinol. 2010;6(2):71–82. doi: 10.1038/nrendo.2009.264. [DOI] [PubMed] [Google Scholar]
- 116.Kraja AT, et al. Do inflammation and procoagulation biomarkers contribute to the metabolic syndrome cluster? Nutr Metab (Lond) 2007;4:28. doi: 10.1186/1743-7075-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mertens I, et al. Among inflammation and coagulation markers, PAI-1 is a true component of the metabolic syndrome. Int J Obes (Lond) 2006;30(8):1308–14. doi: 10.1038/sj.ijo.0803189. [DOI] [PubMed] [Google Scholar]
- 118.Teran-Garcia M, Bouchard C. Genetics of the metabolic syndrome. Appl Physiol Nutr Metab. 2007;32(1):89–114. doi: 10.1139/h06-102. [DOI] [PubMed] [Google Scholar]
- 119.North KE, et al. Linkage analysis of factors underlying insulin resistance: Strong Heart Family Study. Obes Res. 2005;13(11):1877–84. doi: 10.1038/oby.2005.230. [DOI] [PubMed] [Google Scholar]
- 120.Kullo IJ, et al. Pleiotropic genetic effects contribute to the correlation between HDL cholesterol, triglycerides, and LDL particle size in hypertensive sibships. Am J Hypertens. 2005;18(1):99–103. doi: 10.1016/j.amjhyper.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 121.Edwards KL, et al. Pleiotropic genetic effects on LDL size, plasma triglyceride, and HDL cholesterol in families. Arterioscler Thromb Vasc Biol. 1999;19(10):2456–64. doi: 10.1161/01.atv.19.10.2456. [DOI] [PubMed] [Google Scholar]
- 122.Kondo H, et al. Association of adiponectin mutation with type 2 diabetes: a candidate gene for the insulin resistance syndrome. Diabetes. 2002;51(7):2325–8. doi: 10.2337/diabetes.51.7.2325. [DOI] [PubMed] [Google Scholar]
- 123.Ohashi K, et al. Adiponectin I164T mutation is associated with the metabolic syndrome and coronary artery disease. J Am Coll Cardiol. 2004;43(7):1195–200. doi: 10.1016/j.jacc.2003.10.049. [DOI] [PubMed] [Google Scholar]
- 124.Lin RC, et al. Association of obesity, but not diabetes or hypertension, with glucocorticoid receptor N363S variant. Obes Res. 2003;11(6):802–8. doi: 10.1038/oby.2003.111. [DOI] [PubMed] [Google Scholar]
- 125.Rosmond R, Holm G. A 5-year follow-up study of 3 polymorphisms in the human glucocorticoid receptor gene in relation to obesity, hypertension, and diabetes. J Cardiometab Syndr. 2008;3(3):132–5. doi: 10.1111/j.1559-4572.2008.00008.x. [DOI] [PubMed] [Google Scholar]
- 126.Scott LJ, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–5. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zeggini E, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(5829):1336–41. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sabatti C, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41(1):35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Aulchenko YS, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41(1):47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kathiresan S, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40(2):189–97. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Willer CJ, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40(2):161–9. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wallace C, et al. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am J Hum Genet. 2008;82(1):139–49. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Assimes TL, et al. Susceptibility locus for clinical and subclinical coronary artery disease at chromosome 9p21 in the multi-ethnic ADVANCE study. Hum Mol Genet. 2008;17(15):2320–8. doi: 10.1093/hmg/ddn132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Helgadottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316(5830):1491–3. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 135.McCarthy JJ. Gene by sex interaction in the etiology of coronary heart disease and the preceding metabolic syndrome. Nutr Metab Cardiovasc Dis. 2007;17(2):153–61. doi: 10.1016/j.numecd.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 136.Pollex RL, Hegele RA. Genetic determinants of the metabolic syndrome. Nat Clin Pract Cardiovasc Med. 2006;3(9):482–9. doi: 10.1038/ncpcardio0638. [DOI] [PubMed] [Google Scholar]
- 137.Williams RR, et al. NHLBI family blood pressure program: methodology and recruitment in the HyperGEN network. Hypertension genetic epidemiology network. Ann Epidemiol. 2000;10(6):389–400. doi: 10.1016/s1047-2797(00)00063-6. [DOI] [PubMed] [Google Scholar]
- 138.Province MA, et al. Association between the alpha-adducin gene and hypertension in the HyperGEN Study. Am J Hypertens. 2000;13(6 Pt 1):710–8. doi: 10.1016/s0895-7061(99)00282-4. [DOI] [PubMed] [Google Scholar]
- 139.Lukaski H. Use of bioelectric impedance analysis to assess human body composition. In: Livingston G, editor. Nutritional Status Assessment of the Individual. Food and Nutrition Press, Inc; Conn: 1989. [Google Scholar]
- 140.Kraja AT, et al. An evaluation of the metabolic syndrome in a large multi-ethnic study: the Family Blood Pressure Program. Nutr Metab (Lond) 2005;2:17. doi: 10.1186/1743-7075-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Parikh NI, et al. Visceral and subcutaneous adiposity and brachial artery vasodilator function. Obesity (Silver Spring) 2009;17(11):2054–9. doi: 10.1038/oby.2009.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kannel W. Obesity as a force of morbidity. In: Heald F, editor. Adolescent Nutrition and Growth. Appleton-Century-Crofts; New York: 1969. pp. 51–71. [Google Scholar]
- 143.Kraja AT, et al. QTLs of factors of the metabolic syndrome and echocardiographic phenotypes: the hypertension genetic epidemiology network study. BMC Med Genet. 2008;9:103. doi: 10.1186/1471-2350-9-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Huang P, et al. Factor relationships of metabolic syndrome and echocardiographic phenotypes in the HyperGEN study. J Hypertens. 2008;26(7):1360–6. doi: 10.1097/HJH.0b013e3282ffdc80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Huang ZY, et al. T-cadherin-mediated cell growth regulation involves G2 phase arrest and requires p21(CIP1/WAF1) expression. Mol Cell Biol. 2003;23(2):566–78. doi: 10.1128/MCB.23.2.566-578.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Philippova M, et al. Atypical GPI-anchored T-cadherin stimulates angiogenesis in vitro and in vivo. Arterioscler Thromb Vasc Biol. 2006;26(10):2222–30. doi: 10.1161/01.ATV.0000238356.20565.92. [DOI] [PubMed] [Google Scholar]
- 147.Kipmen-Korgun D, et al. T-cadherin mediates low-density lipoprotein-initiated cell proliferation via the Ca(2+)-tyrosine kinase-Erk1/2 pathway. J Cardiovasc Pharmacol. 2005;45(5):418–30. doi: 10.1097/01.fjc.0000157458.91433.86. [DOI] [PubMed] [Google Scholar]
- 148.Org E, et al. Genome-wide scan identifies CDH13 as a novel susceptibility locus contributing to blood pressure determination in two European populations. Hum Mol Genet. 2009;18(12):2288–96. doi: 10.1093/hmg/ddp135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hug C, et al. T-cadherin is a receptor for hexameric and high-molecular-weight forms of Acrp30/adiponectin. Proc Natl Acad Sci U S A. 2004;101(28):10308–13. doi: 10.1073/pnas.0403382101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yamauchi T, et al. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature. 2003;423(6941):762–9. doi: 10.1038/nature01705. [DOI] [PubMed] [Google Scholar]
- 151.Dolley G, et al. Promoter adiponectin polymorphisms and waist/hip ratio variation in a prospective French adults study. Int J Obes (Lond) 2008;32(4):669–75. doi: 10.1038/sj.ijo.0803773. [DOI] [PubMed] [Google Scholar]
- 152.Guerre-Millo M. Adiponectin: an update. Diabetes Metab. 2008;34(1):12–8. doi: 10.1016/j.diabet.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 153.Halvatsiotis I, et al. Genetic variation in the adiponectin receptor 2 (ADIPOR2) gene is associated with coronary artery disease and increased ADIPOR2 expression in peripheral monocytes. Cardiovasc Diabetol. 2010;9:10. doi: 10.1186/1475-2840-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kotronen A, et al. Genetic variation in the ADIPOR2 gene is associated with liver fat content and its surrogate markers in three independent cohorts. Eur J Endocrinol. 2009;160(4):593–602. doi: 10.1530/EJE-08-0900. [DOI] [PubMed] [Google Scholar]
- 155.Ling H, et al. Genome-wide linkage and association analyses to identify genes influencing adiponectin levels: the GEMS Study. Obesity (Silver Spring) 2009;17(4):737–44. doi: 10.1038/oby.2008.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Takeuchi T, et al. Adiponectin receptors, with special focus on the role of the third receptor, T-cadherin, in vascular disease. Med Mol Morphol. 2007;40(3):115–20. doi: 10.1007/s00795-007-0364-9. [DOI] [PubMed] [Google Scholar]
- 157.Vasseur F, Meyre D, Froguel P. Adiponectin, type 2 diabetes and the metabolic syndrome: lessons from human genetic studies. Expert Rev Mol Med. 2006;8(27):1–12. doi: 10.1017/S1462399406000147. [DOI] [PubMed] [Google Scholar]
- 158.Vaxillaire M, et al. Genetic analysis of ADIPOR1 and ADIPOR2 candidate polymorphisms for type 2 diabetes in the Caucasian population. Diabetes. 2006;55(3):856–61. doi: 10.2337/diabetes.55.03.06.db05-0665. [DOI] [PubMed] [Google Scholar]
- 159.Hebbard LW, et al. T-cadherin supports angiogenesis and adiponectin association with the vasculature in a mouse mammary tumor model. Cancer Res. 2008;68(5):1407–16. doi: 10.1158/0008-5472.CAN-07-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kadowaki T, et al. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J Clin Invest. 2006;116(7):1784–92. doi: 10.1172/JCI29126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nishida M, Funahashi T, Shimomura I. Pathophysiological significance of adiponectin. Med Mol Morphol. 2007;40(2):55–67. doi: 10.1007/s00795-007-0366-7. [DOI] [PubMed] [Google Scholar]
- 162.Philippova M, et al. A guide and guard: the many faces of T-cadherin. Cell Signal. 2009;21(7):1035–44. doi: 10.1016/j.cellsig.2009.01.035. [DOI] [PubMed] [Google Scholar]
- 163.Joy T, et al. Genetics of metabolic syndrome. Curr Diab Rep. 2008;8(2):141–8. doi: 10.1007/s11892-008-0025-y. [DOI] [PubMed] [Google Scholar]
- 164.Lawson HA, Cheverud JM. Metabolic syndrome components in murine models. Endocr Metab Immune Disord Drug Targets. 2010;10(1):25–40. doi: 10.2174/187153010790827948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Su Z, et al. Candidate genes for obesity revealed from a C57BL/6J x 129S1/SvImJ intercross. Int J Obes (Lond) 2008;32(7):1180–9. doi: 10.1038/ijo.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hinney A, Hebebrand J. Polygenic Obesity in Humans. Obes Facts. 2008;1(1):35–42. doi: 10.1159/000113935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hinney A, Vogel CI, Hebebrand J. From monogenic to polygenic obesity: recent advances. Eur Child Adolesc Psychiatry. 2010;19(3):297–310. doi: 10.1007/s00787-010-0096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tonjes A, et al. Association of FTO variants with BMI and fat mass in the self-contained population of Sorbs in Germany. Eur J Hum Genet. 2010;18(1):104–10. doi: 10.1038/ejhg.2009.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Johansson A, et al. Linkage and genome-wide association analysis of obesity-related phenotypes: association of weight with the MGAT1 gene. Obesity (Silver Spring) 2010;18(4):803–8. doi: 10.1038/oby.2009.359. [DOI] [PubMed] [Google Scholar]
- 170.Fox CS, et al. Genome-wide association to body mass index and waist circumference: the Framingham Heart Study 100K project. BMC Med Genet. 2007;8(Suppl 1):S18. doi: 10.1186/1471-2350-8-S1-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cho YS, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41(5):527–34. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 172.Wang Y, et al. From the Cover: Whole-genome association study identifies STK39 as a hypertension susceptibility gene. Proc Natl Acad Sci U S A. 2009;106(1):226–31. doi: 10.1073/pnas.0808358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Levy D, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009 doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Newton-Cheh C, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009 doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kooner JS, et al. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat Genet. 2008;40(2):149–51. doi: 10.1038/ng.2007.61. [DOI] [PubMed] [Google Scholar]
- 176.Hiura Y, et al. Identification of genetic markers associated with high-density lipoprotein-cholesterol by genome-wide screening in a Japanese population: the Suita study. Circ J. 2009;73(6):1119–26. doi: 10.1253/circj.cj-08-1101. [DOI] [PubMed] [Google Scholar]
- 177.Burkhardt R, et al. Common SNPs in HMGCR in micronesians and whites associated with LDL-cholesterol levels affect alternative splicing of exon13. Arterioscler Thromb Vasc Biol. 2008;28(11):2078–84. doi: 10.1161/ATVBAHA.108.172288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pollin TI, et al. A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection. Science. 2008;322(5908):1702–5. doi: 10.1126/science.1161524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sandhu MS, et al. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 2008;371(9611):483–91. doi: 10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zemunik T, et al. Genome-wide association study of biochemical traits in Korcula Island, Croatia. Croat Med J. 2009;50(1):23–33. doi: 10.3325/cmj.2009.50.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Heid IM, et al. Clear detection of ADIPOQ locus as the major gene for plasma adiponectin: results of genome-wide association analyses including 4659 European individuals. Atherosclerosis. 2010;208(2):412–20. doi: 10.1016/j.atherosclerosis.2009.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Richards JB, et al. A genome-wide association study reveals variants in ARL15 that influence adiponectin levels. PLoS Genet. 2009;5(12):e1000768. doi: 10.1371/journal.pgen.1000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Danik JS, et al. Novel loci, including those related to Crohn disease, psoriasis, and inflammation, identified in a genome-wide association study of fibrinogen in 17 686 women: the Women’s Genome Health Study. Circ Cardiovasc Genet. 2009;2(2):134–41. doi: 10.1161/CIRCGENETICS.108.825273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Dehghan A, et al. Association of novel genetic Loci with circulating fibrinogen levels: a genome-wide association study in 6 population-based cohorts. Circ Cardiovasc Genet. 2009;2(2):125–33. doi: 10.1161/CIRCGENETICS.108.825224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Elliott P, et al. Genetic Loci associated with C-reactive protein levels and risk of coronary heart disease. Jama. 2009;302(1):37–48. doi: 10.1001/jama.2009.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Reiner AP, et al. Polymorphisms of the HNF1A gene encoding hepatocyte nuclear factor-1 alpha are associated with C-reactive protein. Am J Hum Genet. 2008;82(5):1193–201. doi: 10.1016/j.ajhg.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chen WM, et al. Variations in the G6PC2/ABCB11 genomic region are associated with fasting glucose levels. J Clin Invest. 2008;118(7):2620–8. doi: 10.1172/JCI34566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bouatia-Naji N, et al. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet. 2009;41(1):89–94. doi: 10.1038/ng.277. [DOI] [PubMed] [Google Scholar]
- 189.Chambers JC, et al. Common genetic variation near melatonin receptor MTNR1B contributes to raised plasma glucose and increased risk of type 2 diabetes among Indian Asians and European Caucasians. Diabetes. 2009;58(11):2703–8. doi: 10.2337/db08-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Takeuchi F, et al. Confirmation of multiple risk Loci and genetic impacts by a genome-wide association study of type 2 diabetes in the Japanese population. Diabetes. 2009;58(7):1690–9. doi: 10.2337/db08-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Steinthorsdottir V, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39(6):770–5. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 192.Sladek R, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445(7130):881–5. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 193.Yasuda K, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet. 2008;40(9):1092–7. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- 194.Unoki H, et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet. 2008;40(9):1098–102. doi: 10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- 195.Meigs JB, et al. Genome-wide association with diabetes-related traits in the Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S16. doi: 10.1186/1471-2350-8-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Tsai FJ, et al. A genome-wide association study identifies susceptibility variants for type 2 diabetes in Han Chinese. PLoS Genet. 2010;6(2):e1000847. doi: 10.1371/journal.pgen.1000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Salonen JT, et al. Type 2 diabetes whole-genome association study in four populations: the DiaGen consortium. Am J Hum Genet. 2007;81(2):338–45. doi: 10.1086/520599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Timpson NJ, et al. Adiposity-related heterogeneity in patterns of type 2 diabetes susceptibility observed in genome-wide association data. Diabetes. 2009;58(2):505–10. doi: 10.2337/db08-0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Prokopenko I, et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009;41(1):77–81. doi: 10.1038/ng.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Saxena R, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–6. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 202.Hanson RL, et al. A search for variants associated with young-onset type 2 diabetes in American Indians in a 100K genotyping array. Diabetes. 2007;56(12):3045–52. doi: 10.2337/db07-0462. [DOI] [PubMed] [Google Scholar]
- 203.Hayes MG, et al. Identification of type 2 diabetes genes in Mexican Americans through genome-wide association studies. Diabetes. 2007;56(12):3033–44. doi: 10.2337/db07-0482. [DOI] [PubMed] [Google Scholar]
- 204.Rampersaud E, et al. Identification of novel candidate genes for type 2 diabetes from a genome-wide association scan in the Old Order Amish: evidence for replication from diabetes-related quantitative traits and from independent populations. Diabetes. 2007;56(12):3053–62. doi: 10.2337/db07-0457. [DOI] [PubMed] [Google Scholar]
- 205.Hindorff LA, et al. A catalog of published genome-wide associaiton studies. [cited 2010 April 5]; Available from: www.genome.gov/gwastudies.
- 206.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106(23):9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Kraja A, Vaidya D, Pankow JS. A bivariate genome-wide approach to metabolic syndrome: STAMPEED Consortium. Annals of Internal Medicine. doi: 10.2337/db10-1011. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Mardis ER. New strategies and emerging technologies for massively parallel sequencing: applications in medical research. Genome Med. 2009;1(4):40. doi: 10.1186/gm40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Andriole KP, Khorasani R. Cloud computing: what is it and could it be useful? J Am Coll Radiol. 2010;7(4):252–4. doi: 10.1016/j.jacr.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 211.Joy T, Hegele RA. Genetics of metabolic syndrome: is there a role for phenomics? Curr Atheroscler Rep. 2008;10(3):201–8. doi: 10.1007/s11883-008-0032-0. [DOI] [PubMed] [Google Scholar]
- 212.Lynn FC. Meta-regulation: microRNA regulation of glucose and lipid metabolism. Trends Endocrinol Metab. 2009;20(9):452–9. doi: 10.1016/j.tem.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 213.Kim M, et al. DNA methylation as a biomarker for cardiovascular disease risk. PLoS One. 2010;5(3):e9692. doi: 10.1371/journal.pone.0009692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Heid IM, et al. Genome-wide association analysis of high-density lipoprotein cholesterol in the population-based KORA study sheds new light on intergenic regions. Circ Cardiovasc Genet. 2008;1(1):10–20. doi: 10.1161/CIRCGENETICS.108.776708. [DOI] [PubMed] [Google Scholar]
- 215.Ridker PM, et al. Polymorphism in the CETP gene region, HDL cholesterol, and risk of future myocardial infarction: Genomewide analysis among 18 245 initially healthy women from the Women’s Genome Health Study. Circ Cardiovasc Genet. 2009;2(1):26–33. doi: 10.1161/CIRCGENETICS.108.817304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bouatia-Naji N, et al. A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science. 2008;320(5879):1085–8. doi: 10.1126/science.1156849. [DOI] [PubMed] [Google Scholar]
- 217.Rung J, et al. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat Genet. 2009;41(10):1110–5. doi: 10.1038/ng.443. [DOI] [PubMed] [Google Scholar]