Significance
In contrast to diffraction-limited microscopy, superresolution microscopy highly depends on the used fluorescent label. However, introducing a new label with suitable dynamics is not always straightforward. Here we describe how mCherry, a frequently used fluorescent protein in conventional microscopy, can be used for superresolution microscopy via a new caging mechanism involving the addition of β-mercaptoethanol. Moreover, we investigate the structural mechanism behind this chemical caging, using X-ray crystallography, NMR spectroscopy, and ab initio quantum mechanical calculations. These show that the mechanism is twofold: β-mercaptoethanol adds covalently to the protein’s chromophore, whereas it also acts as a reducing agent for the chromophore.
Keywords: fluorescent proteins, mCherry, localization microscopy, β-mercaptoethanol, photoactivation
Abstract
Fluorophores with dynamic or controllable fluorescence emission have become essential tools for advanced imaging, such as superresolution imaging. These applications have driven the continuing development of photoactivatable or photoconvertible labels, including genetically encoded fluorescent proteins. These new probes work well but require the introduction of new labels that may interfere with the proper functioning of existing constructs and therefore require extensive functional characterization. In this work we show that the widely used red fluorescent protein mCherry can be brought to a purely chemically induced blue-fluorescent state by incubation with β-mercaptoethanol (βME). The molecules can be recovered to the red fluorescent state by washing out the βME or through irradiation with violet light, with up to 80% total recovery. We show that this can be used to perform single-molecule localization microscopy (SMLM) on cells expressing mCherry, which renders this approach applicable to a very wide range of existing constructs. We performed a detailed investigation of the mechanism underlying these dynamics, using X-ray crystallography, NMR spectroscopy, and ab initio quantum-mechanical calculations. We find that the βME-induced fluorescence quenching of mCherry occurs both via the direct addition of βME to the chromophore and through βME-mediated reduction of the chromophore. These results not only offer a strategy to expand SMLM imaging to a broad range of available biological models, but also present unique insights into the chemistry and functioning of a highly important class of fluorophores.
Fluorescent proteins have greatly advanced the study of intracellular organization and dynamics (1–3). For example, the discovery of red fluorescent proteins (RFPs) has enabled multicolor imaging when combined with the original green fluorescent protein (GFP), whereas the development of photoconvertible proteins whose fluorescence emission can be either activated or altered has enabled studying protein dynamics and turnover, as well as diffraction-unlimited imaging through a range of different approaches (1, 3–5). Single-molecule localization microscopy [SMLM (6)], for example, depends on the semicontrolled switching of fluorophores between dark states and fluorescent states, which facilitates repetitive sparse sampling and nanometric localization of individual fluorophores. In contrast to synthetic fluorophores, where blinking is often induced by transient interactions between the dye and buffer components such as reducing agents (7), blinking in FPs is typically achieved through the use of photoactivatable variants whose emission spectrum changes upon exposure to specific wavelengths, which is largely independent of buffer conditions. This change can be irreversible, such as fluorescence activation (8, 9) or green to red photoconversion (10–12), or reversible on/off switching (13, 14). For example, photoactivatable GFP (PA-GFP) switches from dark to green fluorescent upon exposure to violet light, whereas mEos switches from green to red fluorescence in the same conditions (3).
The wide applicability of these types of “smart labels” has sparked a broad interest in development of new probes and in understanding the underlying structural mechanisms. In the case of fluorescent proteins, these fluorescence dynamics typically result from decarboxylation of a glutamate residue in the protein environment, extension of the conjugated system of the chromophore with a histidine residue, or cis/trans isomerization of the chromophore coupled to a change in protonation state (15). However, despite intensive efforts, rational fluorescent protein design has remained difficult due to the high structural complexity of these probes, the complex and subtle relation between the structural and spectroscopic properties, and the fact that these processes are difficult to synchronize at the ensemble level.
The downside to using engineered photoactivatible labels is that these require the construction and introduction of new labeling constructs, which can be costly and laborious. Introducing different types of FPs can also lead to perturbations of the cellular biology, requiring extensive validation efforts. Finally, for many photoactivatable FPs the activation efficiency is only around 50% (16), leaving half of the fluorophores undetected. For this reason, several investigators have explored the use of conventional FPs, such as GFP and YFP, for subdiffraction imaging, because they will blink either spontaneously or in specific conditions (17, 18). Recently, also SMLM with a conventional red FP has been achieved by exploiting a light-induced dark state of mCherry in the presence of the reducing agent β-mercaptoethylamine (19). Unfortunately, using a light-induced dark state can lead to premature photodestruction of the fluorophores before the start of the single-molecule image acquisition, resulting in suboptimal SMLM images.
Here we show that mCherry can be brought to a purely chemically induced dark state from which up to 80% of the fluorophores can be recovered to the fluorescent state. Incubating with β-mercaptoethanol (βME) quenches the fluorescence peak at 610 nm and introduces a fluorescence peak at 460 nm, which can be reversed by illumination with violet light or by βME washout. We find that this chemical caging can be used for mCherry-based SMLM, which should be widely applicable, given the many existing mCherry fusion proteins. Spectroscopic characterization provided insights into the kinetics of this process, whereas X-ray crystallography, NMR spectroscopy, and ab initio calculations demonstrate a unique mechanism in which βME can convert mCherry to a blue fluorescent protein both by reducing the chromophore and by covalent addition to the Cβ of the chromophore’s tyrosine. This discovery brings insights into the chemistry and functioning of a highly important class of fluorophores.
Results and Discussion
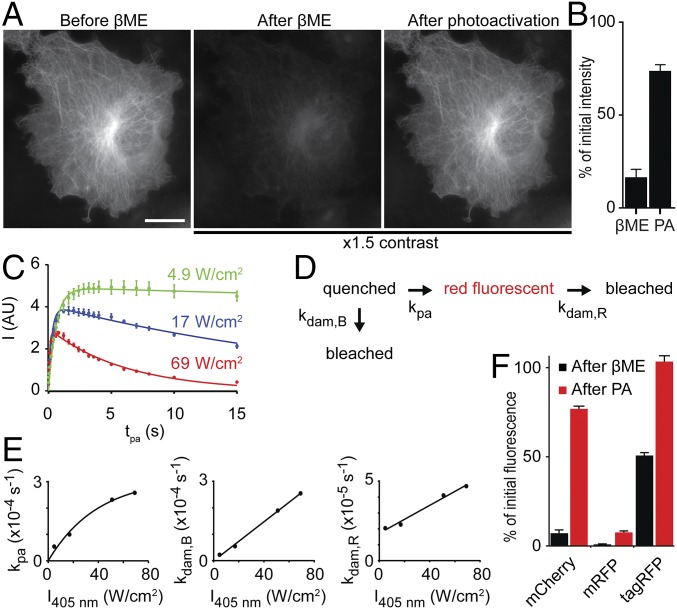
The effect of βME on RFPs was first observed in fixed COS-7 cells expressing mCherry-tubulin. The bright red fluorescence in these cells was largely quenched after addition of 286 mM (2% vol/vol) βME (Fig. 1A), but could be recovered with violet light from a mercury lamp. After optimization, we could recover 73% ± 4% (average ± SEM) of the initial red fluorescence (Fig. 1B), demonstrating that mCherry-tubulin can be chemically caged by βME and subsequently photoactivated to recover the red fluorescent state. To examine how uncaging efficiency depends on the intensity of activation light, purified mCherry was nonspecifically adsorbed onto glass coverslips, quenched by incubation with 286 mM βME in PBS, and subsequently activated by different exposure periods (0–15 s) at different intensities of 405 nm laser light (Fig. 1C). At the lowest intensity, we observed that uncaging first increased and then saturated with increasing exposure time. At higher intensities, longer exposures resulted in less fluorescence and also the maximum achievable fluorescence intensity was lower at increasing activation intensities.
Fig. 1.
RFPs can be caged by βME and uncaged by violet light. (A) Widefield images of a fixed COS-7 cell transfected with mCherry-tubulin, showing the initial fluorescence signal (Left), the fluorescence signal after adding 286 mM βME (Center), and the fluorescence signal after photoactivation with violet light (Right). The image contrast in Center and Right is 1.5 times that in Left. (Scale bar, 20 μm.) (B) Quantification of the fluorescence signal of widefield images of COS-7 cells transfected with mCherry-tubulin after adding βME and after photoactivation as percentage of the initial fluorescence. The fluorescence signal after photoactivation is 73% ± 4% of the initial signal (average ± SEM of 10 cells). (C) Fluorescence intensity of purified mCherry molecules in the presence of 286 mM βME after photoactivation, as a function of photoactivation exposure times for three 405-nm laser intensities. Solid lines indicate fits with a model incorporating both photoactivation and photodamage. (SI Methods, n = 6 measurements per time point). (D) Schematic of the model used for fitting data in E. (E) Rates of photoactivation and photodamage as function of 405-nm laser intensity, obtained from the fits described in C. Solid lines represent exponential (kpa) or linear (kdam,R and kdam,B) fits. (F) Quantification of the fluorescence signal of purified mCherry, mRFP, and TagRFP molecules from TIRF imaging. Black and red bars indicate the percentage of initial fluorescence after adding 286 mM βME and after photoactivation with low 405-nm laser intensity, respectively. n = 4, 2, and 2 samples and n = 40, 24, and 20 measurements for mCherry, mRFP, and tagRFP, respectively.
The data were fitted with a model describing a photoinduced transition from a caged to a fluorescent state and photodamaging pathways from both the caged state and the fluorescent state, with rates kpa, kdam,B, and kdam,R, respectively (Fig. 1D). As expected, the photoactivation rate and the photodamage rates increased with increasing 405-nm laser intensity (Fig. 1E). The linear fit of kdam,B approximately went through the origin, implying no photodamage without 405 nm light. In contrast, the fit of kdam,R had an offset, which can be explained by recaging of photoactivated molecules. The photoactivation rate also increased with laser intensity, but to a lesser extent at higher laser powers. Together, these results explain why the amount of molecules returning to the fluorescent state is highest when low photoactivation intensities are used. Using these activation settings, we found that mCherry, mRFP, and tagRFP were caged by βME to 7.1% ± 0.4%, 1.3% ± 0.5%, and 50.5% ± 0.2% of the original intensity, respectively, whereas the return percentages using violet illumination were 77% ± 2%, 10.4% ± 2.8%, and 103% ± 3% (mean ± SEM, Fig. 1F).
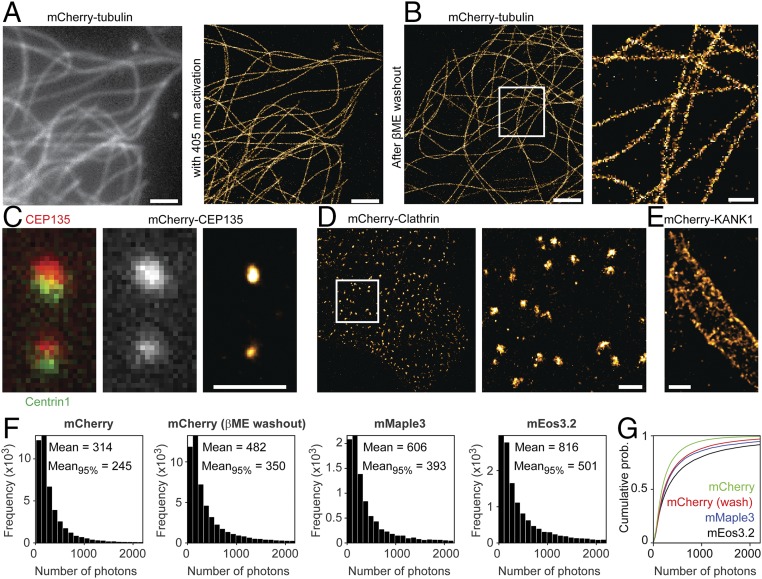
We next tested whether the observed photoactivation of chemically caged mCherry could be used for SMLM. Indeed, mCherry-based localization microscopy of fixed cells expressing mCherry-tubulin in a buffer containing 143 mM βME resulted in an improved resolution with a high density of emitters contributing to the superresolution reconstruction. mCherry molecules could either be photoactivated in the presence of βME (Fig. 2A) or return spontaneously after washout of βME during acquisition of the first thousands of frames (Fig. 2B). In both cases, the average localization accuracy was 11 nm. Other mCherry-labeled structures could also be better resolved using this approach, for example highlighting the centriole using mCherry-CEP135, clathrin-coated pits and lattices (20) using mCherry-Clathrin Light Chain (21), and cortical microtubule stabilization complexes using mCherry-KANK1 (22) (Fig. 2 C and D). Thus, βME-based caging combined with either 405-nm reactivation or βME washout enables mCherry-based localization microscopy.
Fig. 2.
Gradual uncaging enables superresolution imaging. (A) Widefield image of a COS-7 cell transfected with mCherry-tubulin (Left) and SMLM reconstruction of the same area (Right). SMLM was performed after addition of 143 mM βME. (Scale bars, 2 µm.) (B) SMLM reconstruction (Left) and zoom-in (Right) obtained by imaging after addition and washout of 286 mM βME, with no 405-nm activation during the first ∼3,000 frames. [Scale bars, 2 µm (Left) and 500 nm (Right).] (C) (Left and Center) Zoom-in of HeLa cell coexpressing GFP-Centrin-1 (Left) and mCherry-CEP135 (Center). (C, Right) SMLM of the same area. SMLM was performed after addition of 143 mM βME. (Scale bar, 1 µm.) (D and E) SMLM reconstructions of HeLa cells expressing mCherry-Clathrin Light Chain1 (D, Left) and zoom-in (D, Right) or mCherry-KANK1 (E). SMLM was performed after addition of 143 mM βME. A median filter of pixel size 2 was applied. (Scale bars, 1 µm.) (F) Photon number measurements of mCherry, obtained in the presence of βME (A) or after βME washout (B), compared with mMaple3 and mEos3.2. All constructs were fused to tubulin. For calculating the indicated mean photon numbers, also counts outside the histogram (i.e., counts >2,200) were included. Mean95% indicates the average photon count of the dimmest 95% blinking events for each type of sample. (G) Cumulative probability of photon counts for all four situations.
Interestingly, the photon counts per mCherry blinking event obtained after washout of βME were 54% higher than those obtained using photoactivation in the presence of βME (Fig. 2F). This could be caused by photodamage induced by the activation laser (Fig. 1 C–E) or by rapid βME-induced recaging. The high emitter densities in our images could suggest that fluorophores have multiple emission episodes. For example, we observed over 2,100 blinking events in the upper centriole in Fig. 2C, whereas the copy number of CEP135 per centriole has been estimated to be 300–500 (23). To compare the photon counts of mCherry with well-established photoconvertible fluorophores, we created tubulin constructs fused to mEos3.2 (24) or mMaple3 (25). Whereas the histograms of mCherry photon counts upon βME washout are very similar to the histograms of mEos3.2 and mMaple3, the average photon counts for mCherry are 40% and 20% lower, respectively. Closer inspection revealed that the higher average values of mEos3.2 and mMaple3 were caused by a small fraction of long-lived emission events. Excluding the brightest 5% of all emission events for all fluorophores resulted in photon counts of mCherry that were 70% and 89% of mEos3.2 and mMaple3, respectively. In addition, we found that the contrast ratio between the on-state and off-state fluorescence was also very similar for these fluorophores (43 ± 19, 45 ± 13, and 40 ± 15 for mCherry, mEos3.2, and mMaple3, respectively). Thus, for the vast majority of molecules, mCherry performs comparably to mMaple3 and slightly worse than mEos3.2.
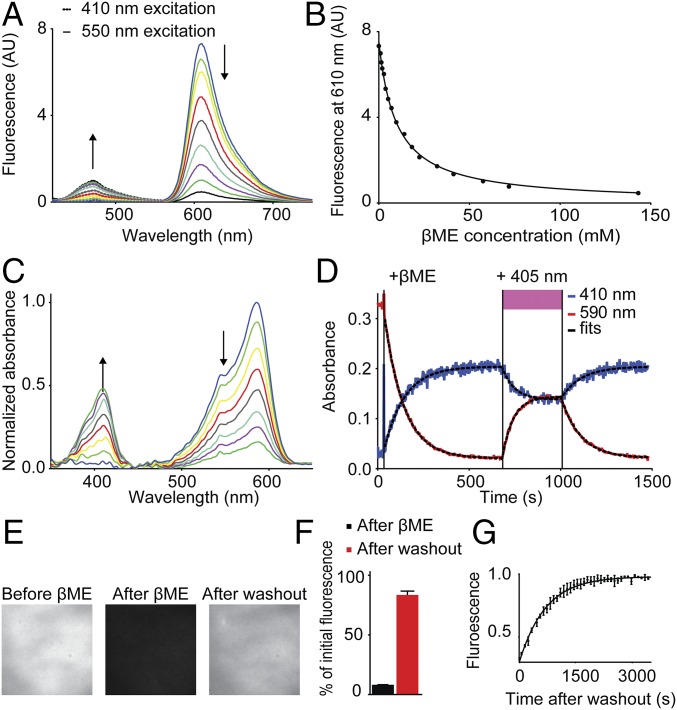
During photoactivation of mCherry-tubulin expressing and βME-quenched cells, we noted a faint blue fluorescence. To investigate this further, we recorded the fluorescence emission spectrum of purified mCherry in the presence of increasing βME concentrations and found that addition of βME reduced the emission peak at 610 nm, whereas a peak at 460 nm appeared, indicating blue fluorescence (Fig. 3A). The intensity decrease at 610 nm upon addition of βME and as a function of the βME concentration followed the expected behavior for a reversible bimolecular reaction (I = I0/(1 + Keq[βME])), yielding a Keq of 0.10 mM−1 (Fig. 3B). In addition to the emission spectrum, the absorbance spectrum of mCherry was also strongly altered upon addition of 286 mM βME. The absorbance peak at 590 nm decreased over time and a peak at 410 nm appeared (Fig. 3 C and D), which could be reversed by photoactivation with a 405-nm laser. These results indicate that βME converts mCherry molecules from a red fluorescent state to a blue fluorescent photoconvertible state with absorption maximum at 410 nm and fluorescence emission maximum at 460 nm.
Fig. 3.
βME transforms mCherry into a blue fluorescent protein. (A) Fluorescence spectra of in vitro mCherry molecules incubated with various βME concentrations, excited with 410 nm light (dotted lines) and 550 nm light (solid lines). Arrows indicate increasing βME concentration. (B) Quantification of mCherry fluorescence intensity at 610 nm as function of βME concentration. Solid line represents a fit of the fluorescence intensity according to an equation for a reversible bimolecular reaction I = I0 /(1 + Keq⋅[βME]), yielding Keq = 0.10 mM−1 (N = 1, n = 4). (C) Absorbance spectra of in vitro mCherry molecules over time after addition of 286 mM βME. Arrows indicate increasing time. (D) Absorbance of purified mCherry molecules at 410 nm (black line) and 590 nm (red line) as a function of time. Addition of 286 mM βME and exposure to 405 nm light are indicated. Dotted black lines represent fits for one phase decay: I = Plateau + (I0 − Plateau)·exp(−t/τ). Values for τ are 111 s−1 and 106 s−1 after βME, 65 s−1 and 60 s−1 during 405 nm illumination, and 109 s−1 and 108 s−1 after 405 nm illumination, for absorbance at 410 nm and 590 nm, respectively. (E) Fluorescence images of purified mCherry molecules on a coverslip before addition of 286 mM βME, after addition of βME, and 1.5 h after washout of βME. (F) Quantification of the fluorescence signal after addition of 286 mM βME (8% ± 0.1%) and 1.5 h after washout (83% ± 3%) (mean ± SEM; N = 2, n = 44 regions). (G) Kinetics of fluorescence return after βME washout. Fluorescence intensity is normalized to the final value. The solid line represents a fit for one phase dissociation of βME and mCherry, yielding τ = 7.0·102 s (95% confidence interval 6.6·102–7.4·102; N = 3).
To characterize the spontaneous return of mCherry to the red fluorescent state without photoactivation, coverslips with nonspecifically adsorbed mCherry molecules were incubated with 286 mM βME in PBS for 20 min and then washed extensively with PBS to remove the βME. Upon βME washout, the red fluorescence of mCherry returned over the course of 1 h, with an average recovery of 83% ± 3% of the initial intensity and a single-exponential time constant of 7.0·102 s (Fig. 3 E–G). In addition, we tested whether other fluorescent proteins could be caged using βME. Remarkably, βME caging could be observed only for mCherry and some other red fluorescent proteins like mRFP1, TagRFP-T, and monomeric dsRed, but not for FPs with other colors or red FPs such as tdTomato and mKate2 (Table S1).
Table S1.
Effect of 30–60 min incubation with 286 mM βME on the absorbance spectrum of different fluorescent proteins
| No caging effect | Small caging effect | Pronounced caging effect |
| Clover [GYG] | dsRed-express [QYG] | mCherry [MYG] |
| Dronpa [CYG] | mRFP1 [QYG] | dsRed monomer [QYG] |
| EGFP [TYG] | TagRFPT [MYG] | |
| mEos4b (green) [HYG] | mRuby2 [MYG] | |
| mKate2 [MYG] | ||
| mTFP1 [AYG] | ||
| mTFP0.7 [AYG] | ||
| Padron [CYG] | ||
| pcDronpa2 (green) [HYG] | ||
| rsEGFP [TYG] | ||
| rsEGFP2 [AYG] | ||
| rsGreen1 [TYG] | ||
| rsGreenF [TYG] | ||
| tdTomato [MYG] |
The chromophore amino acid composition is indicated in brackets.
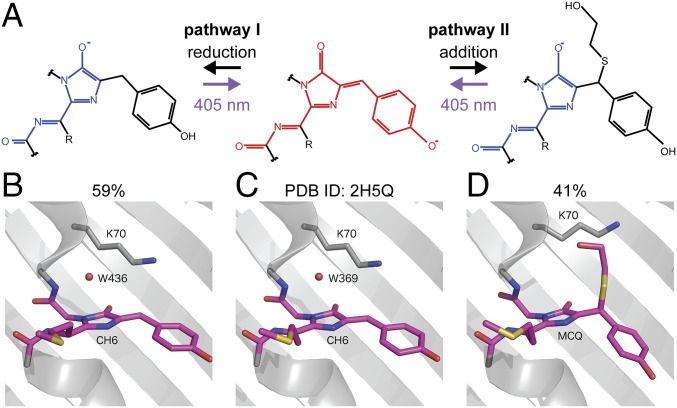
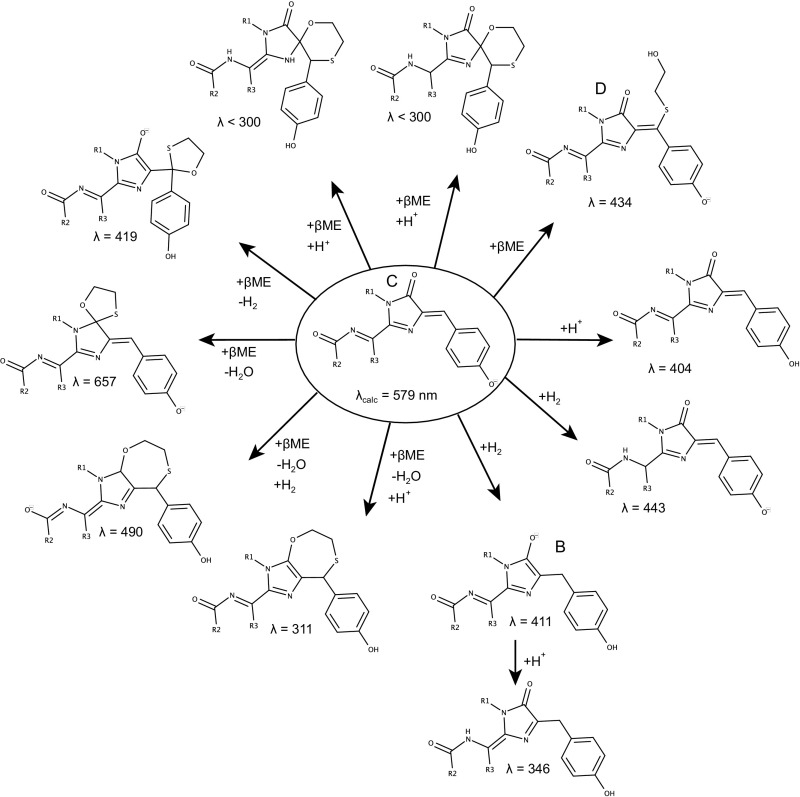
The βME-induced blue fluorescent state that we observed resembles the blue intermediate state reported in the maturation pathway of RFPs (26) and also matches the absorption and emission spectra reported for mTagBFP (27). Interestingly, exposure to violet light was found to accelerate the transition from the blue intermediate state to the final red state for dsRed (26), for mCherry-derived fluorescent timers, and for photoactivatable PAmCherry (9, 28). We therefore hypothesized that the addition of βME returns the RFP chromophore to a state similar to the blue intermediate state. The structure of the blue intermediate chromophore has been elucidated using X-ray crystallography (29) and ab initio quantum mechanics/molecular mechanics (QM/MM) calculations (30) and based on the reported differences in structure between the red fluorescent and blue fluorescent chromophore, we envisioned (at least) two ways for βME to interact with the chromophore to return it to the blue state (Fig. 4A). In the first mechanism, βME binds directly to the Cβ of the tyrosine, whereas in the second mechanism βME causes a chemical reduction of the chromophore without covalent addition. In both cases the conjugated system would be interrupted at the Cβ atom, resulting in the formation of an mTagBFP-type chromophore, with the shortening of the conjugated π system leading to the blue shift of the absorption and emission wavelengths.
Fig. 4.
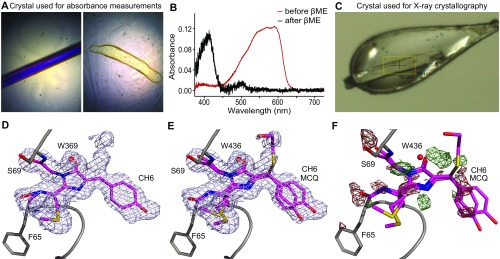
mCherry caging occurs through Michael addition of βME to the chromophore. (A) Two hypothesized pathways for βME-induced conversion of the RFP chromophore from the red fluorescent form to the blue fluorescent form. Pathway I is via reduction of the double bond between the Cα and Cβ of the tyrosine. In pathway II, βME is covalently attached to the Cβ of the tyrosine through a Michael addition, removing the double bond between the Cα and Cβ of the tyrosine. (B–D) Crystal structures of mCherry. C depicts the structure found in the PBD (ID: 2H5Q), whereas B and D depict the structures obtained after soaking a crystal in βME. The structure in B has 59% occupancy in the crystal vs. 41% occupancy in D.
To investigate the structural changes caused by βME, we crystallized mCherry and soaked it in a 2-M βME-containing solution just before flash freezing. Absorption measurements demonstrated that crystallized mCherry shows a similar blue shift in absorbance spectrum as noncrystallized mCherry and loses its red/purple color after soaking (Fig. S1 A–C). The crystal structure was determined with a resolution of 1.55 Å (Table S2) and displayed the typical β-barrel structure seen in other fluorescent proteins. Whereas most amino acid residues were well defined in the electron density maps, the region around the chromophore in the center of the barrel was initially less clear. We were able to model two different chromophore types in this electron density: a chromophore closely resembling that of untreated mCherry (with occupancy of 59%), and a variant in which βME adds to the Cβ of the tyrosine via its mercapto group (with occupancy of 41%) (Fig. 4 B–D). Although clear evidence for these two types was found, during structure refinement peaks in the 2Fo−Fc electron density map suggest the existence of alternative conformations of the free βME tail (Fig. S1 D and E).
Fig. S1.
(A) Color pictures of the mCherry crystal used for absorption measurements before (Left) and after (Right) soaking in βME. The odd shape of the crystal after soaking in βME is due to βME-induced dissolution because the absorption experiments were performed at room temperature (in contrast to the crystallography, where the crystal is flash-frozen after addition of βME, shown in C). (B) Absorption spectra of the crystals in A. The red line corresponds to the crystal before soaking in βME and the black line to the crystal after soaking in βME. The same absorbance shift is observed as for noncrystallized mCherry after soaking in βME. (C) Color picture of the (near) colorless mCherry crystal just before the start of the X-ray experiment. (D) The 2Fo−Fc map contoured at 0.5 rmsd together with the mCherry crystal structure as found in the PDB (ID: 2H5Q). Extra electron density is present near the Cβ of the chromophore’s tyrosine. (E) The 2Fo−Fc map contoured at 0.5 rmsd together with the two mCherry structures modeled in the electron density as described. (F) Fo−Fc difference map contoured at 3 rmsd, with positive peaks in green and negative peaks in red. The map shows that the model does not completely cover the electron density. Different cyclization reaction products can be fitted in the positive electron density, but cannot clearly be distinguished from each other.
Table S2.
Data collection and refinement statistics
| Space group | P 21 |
| Unit cell parameters | |
| a, b, c, Å | 48.783, 43.244, 61.234 |
| α, β, γ, ° | 90, 111.52, 90 |
| Resolution range, Å | 45.38–1.55 (1.58–1.55)* |
| Rmerge, % | 5.3 (49.9) |
| CC1/2, % | 99.9 (76.7) |
| <I/σ(I)> | 14.0 (2.1) |
| No. of reflections | 113,930 (5,098) |
| No. of unique reflections | 34,495 (1,686) |
| Multiplicity | 3.3 (3.0) |
| Completeness, % | 99.4 (99.0) |
| Rwork/Rfree†, % | 15.68/18.77 |
| rmsd from ideal | |
| Bond lengths, Å | 0.006 |
| Bond angles, ° | 1.269 |
| Average isotropic B factors, Å2 | |
| Main chain | 13.03 |
| Side chain | 17.81 |
| Water molecules | 28.98 |
| Ligands: BME, PGE, P6G | 35.84 |
| Ramachandran plot‡, % | |
| Residues in favored regions | 98.7 |
| Residues in allowed regions | 1.3 |
| Outliers | 0.0 |
| Rotamer‡ outliers | 1.0 |
Values in parentheses are for the highest-resolution shell.
Rfree is calculated using a random 5% of data excluded from the refinement.
Ramachandran and rotamer analysis was carried out using Molprobity.
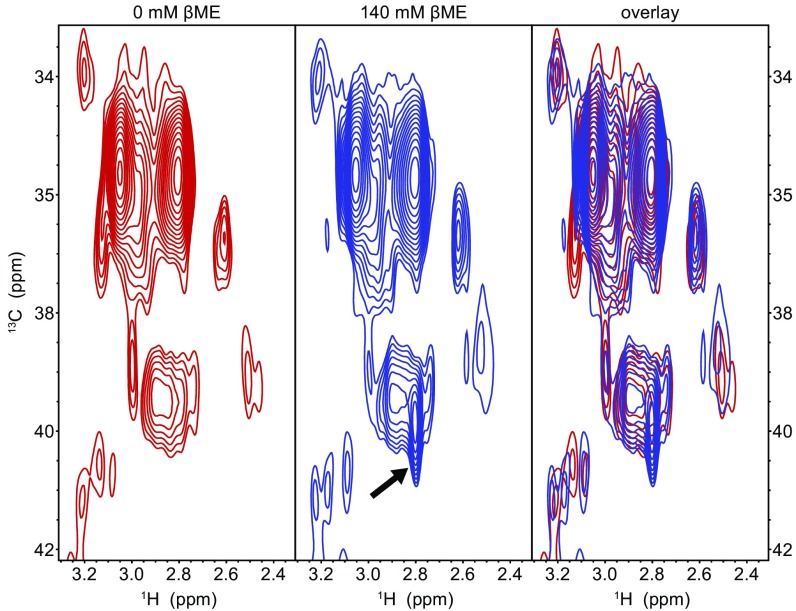
Whereas the crystal structure occupancies suggest that only about 41% of the mCherry molecules contain a βME adduct, the purple color of the crystal fully disappeared and the absorbance completely shifted to violet wavelengths upon addition of βME (Fig. S1 A and B). Although the partial occupancy could be caused by βME not having sufficient time before flash freezing, an image of the crystal on the goniometer head in the 100-K nitrogen stream just before X-ray data acquisition confirms that the crystal had fully lost its characteristic purple color (Fig. S1C). As a result, our data suggest that the remaining 59% does not correspond to the native mCherry chromophore, but rather to a chromophore in which the tyrosine Cβ is in the reduced state, albeit with nonoptimal bond angles for the Cβ sp3-hybridized carbon atom. Such deviations from model values are often observed in fluorescent protein chromophores. Further support for this model comes from our 2D [1H,13C] heteronuclear single quantum correlation (HSQC) measurements using 13C/15N tyrosine, methionine, and glycine-labeled mCherry (Fig. S2). Upon addition of βME, an extra peak emerged in the 13C-1H spectrum close to the characteristic Cβ resonance for tyrosine. This suggests that the double bond between the Cα and Cβ is converted to a single bond, resulting in blue-shifted absorption and emission. This peak most likely corresponds to the reduced state, because the covalent attachment of βME to the Cβ that was observed in the crystal structure is expected to induce a nontraceable shift away from the characteristic tyrosine Cβ resonance.
Fig. S2.
The 13C-1H HSQC spectra of purified mCherry molecules without βME (Left), with 143 mM βME (Center), and an overlay (Right). The arrow (Center) indicates an extra peak present in the spectrum in the presence of 143 mM βME at 39.9 ppm 13C and 2.8 ppm 1H, corresponding closely to the reported chemical shifts of the Cβ of tyrosine (39.3 ± 2.7 ppm 13C and 2.9 ± 0.5 ppm 1H; data from Biological Magnetic Resonance Data Bank). This indicates that the double bond between the Cα and Cβ could also be reduced after addition of βME, without the formation of a covalent adduct.
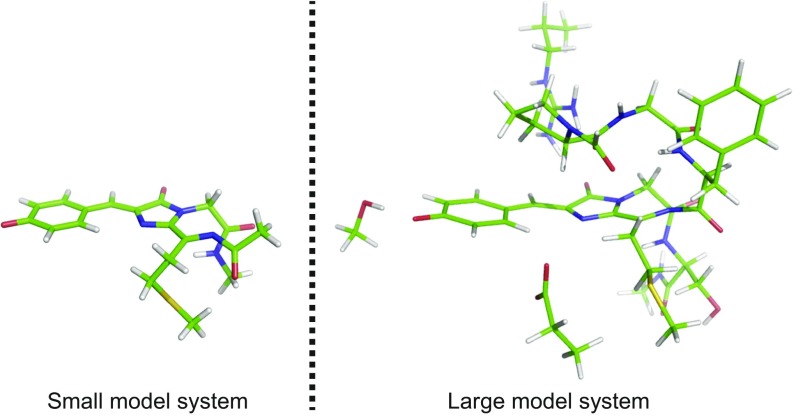
Despite the clear evidence for both reduction of and βME addition to the chromophore, the analyzed crystal structure did contain some peaks in the Fo−Fc difference electron density maps (Fig. S1F) that could not solely be explained by assuming conformational freedom for the free βME tail of the addition product. Moreover, we observed two conformations for lysine 70 and a lower occupancy of water molecule 436. Without these structural rearrangements, addition of βME would sterically not be possible, suggesting that the other possible enantiomer of the Michael addition cannot be accommodated by the β-barrel and cannot explain this residual electron density. Because we could not straightforwardly identify the missing structures or conformations, we next used quantum-chemical calculations on a model system of the chromophore (Fig. S3) to obtain detailed structures of possible βME-conjugated products (Fig. S4) and predict their absorption wavelengths (Table S3). These calculations were performed using coupled-cluster and time-dependent and time-independent density functional theory (SI Computation of Electronic States and Table S3).
Fig. S3.
Small and large model systems of chromophore C, the unmodified mCherry chromophore. Color code: green, C; white, H; blue, N; red, O; yellow, S.
Fig. S4.
Overview of the calculated chromophore variants and their RICC2/Def2TZVP absorption wavelength λ in nanometers. Structure names B, C, and D refer to the crystal structures shown in Fig. 4 B–D (reduced state, unreacted state, and linear adduct, respectively).
Table S3.
CAM-B3LYP/D3/6-31G(d,p) and RICC2/Def2TZVP excitation wavelengths in nanometers (and in kcal/mol) reported for the different chromophore variants
| Compound (total charge) | CAM-B3LYP/D3/6-31G(d,p) [*6-311++G(d,p)] | RICC2/Def2TZVP |
| C(−1) (Exp.: 49.4 kcal/mol; 587 nm) | 503 (56.8) | 579 (49.4) |
| C(−1) | 514 (55.6)* | — |
| C, large(−1) | 460 (62.2) | — |
| C-H+(0) | 391 (73.1) | 404 (70.8) |
| C-H2(−1) | 409 (69.9) | 443 (64.5) |
| C_spiro(−1) | 582 (49.1) | 657 (43.5) |
| B(−1) | 390 (73.3) | 411 (69.6) |
| B(−1) | 403 (70.9)* | — |
| B, large(−1) | 351 (81.5) | — |
| B-H+(0) | 334 (85.6) | 346 (82.7) |
| B_spiro(−1) | 392 (72.9) | 419 (68.2) |
| 7_ring(sp2)(0) | 292 (97.9) | 311 (91.9) |
| 7_ring(sp3)(−1) | 413 (69.2) | 490 (58.4) |
| D(−1) | 404 (70.8) | 434 (65.9) |
| D(−1) | 414 (69.1)* | — |
| D, large (0) | 372 (76.9)† [332 (86.1)] | — |
If not otherwise noted, the small model (and appropriate modifications thereof) of Fig. S3 has been used in the calculations. Where the larger model has been used, this is indicated using the word “large.”
Energies obtained with a larger basis set [6-311++G(d,p)] instead of 6-31G(d,p).
In this case the excitation energy for the structure optimized in PCM = water was chosen, because with PCM = diethylether (excitation energy reported in brackets) an artificial change in the protein environment occurs that influences the excitation energy significantly.
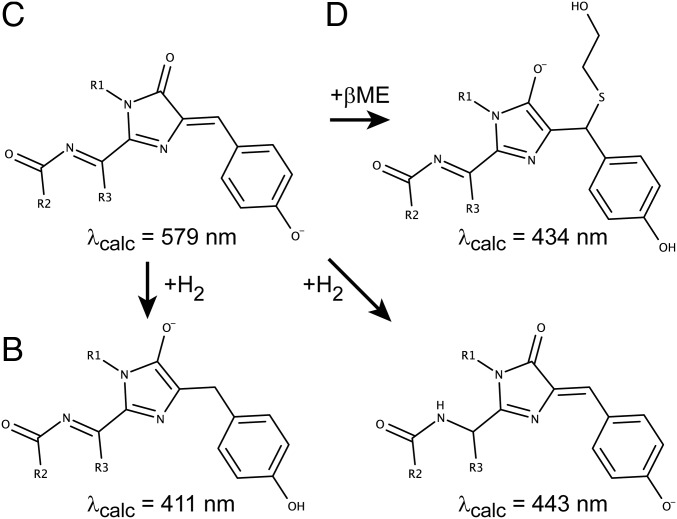
The absorption wavelengths for the mCherry chromophore matched very well with the experimental observation (579 nm vs. 587 nm experimentally). Of all tested structures featuring addition of βME, only two showed an absorption wavelength consistent with the experimental observations (434 nm and 419 nm calculated vs. 410 nm experimentally). The structure with calculated absorption at 434 nm corresponds with the formation of a linear adduct on the Cβ carbon atom of the chromophore’s tyrosine, whereas the other structure (419 nm) involves the formation of a five-membered spiro ring structure by double addition of βME at the same position. The X-ray data do not provide conclusive evidence for or disprove the presence of this structure. However, we consider it unlikely that this structure is formed because it would require an addition–oxidation–addition mechanism and the formation of acetals from βME is known to occur only under harsh conditions (31). Therefore, we propose that, of these structures, only the linear adduct (Fig. 5D) is formed. Of the structures that involve a reduction of the chromophore, both the structure initially suggested based on the NMR experiments (Fig. 5B) and an additional one involving reduction elsewhere in the chromophore lead to predicted absorption wavelengths consistent with experiment (411 nm and 443 nm, respectively). Our NMR data directly support the formation of the structure in Fig. 5B, but we do not have any direct data that prove or disprove the formation of the other reduced structure shown in Fig. 5. However, in our view the direct experimental support for the structures in Fig. 5 B and D and the high similarity of the absorption spectra to mTagBFP indicate that the structures proposed in Fig. 4 are most likely the chemical species formed in this process.
Fig. 5.
Quantum chemical calculations predict three possible reaction products. Three possible chromophore types can theoretically be formed after addition of βME, based on their RICC2/Def2TZVP absorption wavelength (indicated below each structure in nanometers). Structure names B, C, and D refer to the crystal structures shown in Fig. 4 B–D (reduced state, unreacted state, and linear adduct, respectively).
In summary, we have shown that mCherry can be chemically brought to a blue fluorescent state by βME and returned to the red fluorescent state by photoactivation with violet light or by βME washout. The crystallographic data unambiguously showed the appearance of electron density that can be explained only by the formation of a covalent bond at the Cβ atom of tyrosine 67, resulting in a blue fluorescent chromophore. Covalent addition of thiols has been previously reported for cyanine dyes (32), although in that case the addition required irradiation with light, whereas here quenching occurs in the absence of light. Other groups have observed a similar spontaneous addition of thiol compounds (33) and phosphine (34) to fluorophores, but to our knowledge this has not been previously shown for fluorescent proteins. We propose that the covalent modification of the chromophore occurs via a Michael addition through the SH group of βME. Reversibility of Michael additions has been shown for different addends and adducts (35, 36). For example, reversible Michael addition of a thiol to a fluorescent probe was shown to blue shift the absorbance and fluorescence spectra, allowing the measurement of glutathione concentration inside living cells (37).
We observed that βME can cage some, but not all RFPs. We believe that the ability and the magnitude of βME-induced caging depend on several factors, such as the allowance for βME to enter the β-barrel, the accessibility of the chromophore Cβ atom, the possibility for the lysine-70 conformational change upon Michael addition, and the possibility to change the Cβ hybridization following the Michael addition or reduction.
It is important to note that the additional covalent bond with βME was present in about 40% of mCherry molecules in the crystal, whereas the whole crystal changed color upon soaking in βME. This suggests that the quenching of the other 60% of the molecules is achieved by reduction of the tyrosine Cβ atom, consistent with the appearance of the characteristic tyrosine Cβ resonance in the NMR spectrum. Remarkably, irradiation with violet light could reverse both the reduction and the addition, given that the maximum recovery is about 80%. These mechanisms facilitate the use of conventional RFPs in SMLM. Here, the purely chemical transition to a dark state using βME decouples imaging laser power and duty ratio, which allows optimizing imaging conditions for single-molecule detection. This could be an advantage compared with using a light-induced dark state, as reported for mCherry in the presence of β-mercaptoethylamine (19). Given the many existing mCherry-based model systems, our strategy for mCherry-based SMLM using chemical caging followed by highly efficient photoactivation should be widely applicable.
SI Computation of Electronic States
Computational Methods.
First, we cut the chromophore variants as depicted in Fig. 4 B–D (subsequently in this section referred to as B, C, and D) out of the crystal structure such that the peptide bonds of the two amino acids that are covalently bound to the chromophore are included in our model system and the Cα atoms are replaced by methyl groups. Because our small model systems only vaguely resemble the protein-embedded chromophores, we also built larger model systems by including part of the protein environment (denoted as “large model system”). Here, we chose all amino acids that have at least one atom within 3 Å distance to any chromophore atom. If one of the backbone atoms is within the threshold distance, the whole amino acid is taken into account. On the other hand, if none of the backbone atoms is within the threshold distance, only the sidechain is taken into account. We capped the sidechains by adding a hydrogen to the Cβ atom and fixed the Cβ atom during structure optimizations. If the whole amino acid was taken into account, we included the peptide bonds of the two amino acids that are covalently bound to the specific amino acid and replaced the Cα atoms by methyl groups. We fixed the C, N, and O atoms of a capped peptide bond (excluding the ones inside the chromophore structure) during structure optimizations. As an example, in Fig. S3 the small and large model systems of chromophore C are shown.
We optimized the different chromophores B, C, and D and their derivatives in the small (and large) model system with B3LYP/D3/6–31G(d,p) (52), including empirical dispersion correction D3 (53) and continuum solvation by diethylether (which resembles the dielectric permittivity of the protein environment). The excitation energies were then obtained from CAM-B3LYP/D3/6-31G(d,p) single-point calculations, including the polarizable continuum model (PCM) (with solvent being diethylether) and RICC2/Def2TZVP single-point calculations on the optimized structures. For the first two methods we used Gaussian 09 (Gaussian Inc.) and for the latter one Turbomole/6.4 (54).
The average and maximum errors for absorption wavelengths with the RICC2 method used here have been found to be 0.29 eV and 1.25 eV (55). These errors relate to chromophores treated in isolation, whereas here we present predictions for the chromophores in a protein environment. As our calculations neglect the effect of the environment, this means that the error bar should be a little larger. On the other hand, the result for the native chromophore agrees well with experiment, suggesting favorable error cancellation. Accordingly, we believe that the presented calculated absorption wavelengths for a set of structurally related species should be accurate within a range of 50 nm.
Computational Results.
The TDDFT (CAM-B3LYP) and RICC2 vertical excitation energies calculated on the B3LYP/D3/6-31G(d,p)/PCM = diethylether optimized structures are reported in Table S3. In the main text and Fig. S4 we report only the RICC2/Def2TZVP excitation energies as they are shown to be more accurate (55). However, these results are based only on the small model systems and thus we compare here also the TDDFT results of the small and big model systems. The RICC2/Def2TZVP singlet–singlet excitation energy of C in the small model system amounts to 49.4 kcal/mol (579 nm), which is in good agreement with the experimental maximum absorption energy of 48.7 kcal/mol (587 nm). It is noteworthy that the experimental UV/VIS spectrum is not directly comparable, because the chromophore is embedded in the protein environment.
To gain some insight into the dependence of the excitation energies on the model size, we performed TDDFT(CAM-B3LYP) calculations on the small and the larger model system. It should be noted that even the larger model has some shortcomings and the agreement between the excitation energies obtained for these two model systems is acceptable, but not perfect (Table S3 shows the differences between the small and large model systems for B, C, and D, indicating discrepancies between 5 kcal/mol and 8 kcal/mol).
In comparison with the experimentally observed excitation energy, the CAM-B3LYP excitation energies for the small and the large model system are significantly too large. However, it should be noted that similarly large deviations of CAM-B3LYP excitation energies from experiment have been observed for fluorescent proteins (56–58). A benchmark over several fluorescent proteins (59) has shown that the overestimation of the excitation energy by CAM-B3LYP might be systematic independent of the conjugated system size, i.e., about 6.0 kcal/mol (0.26 eV) compared with the coupled-cluster (RICC2) reference result. Because RICC2 should yield the most reliable excitation energies, we mainly focus on the excitation energies obtained with this QM method. We also checked the basis set dependence of the CAM-B3LYP calculations and performed CAM-B3LYP/6-311++G(d,p) calculations on the B3LYP/6-31G(d,p) optimized structures. We observed deviations below 2.5 kcal/mol only for the excitation energies.
With RICC2, we observed that structures B and D have very similar, but 20.2-kcal/mol and 16.5-kcal/mol higher excitation energies compared with C. Thus, the relative difference in excitation energy between C and B/D resembles the experimentally observed one of 21.0 kcal/mol (from 587 nm to 410 nm), suggesting that either B or D or both are observed in the UV/VIS absorption spectrum. We also found that the protonated reduced chromophore B-H+ might well be formed after addition of βME and could explain the occurrence of absorption wavelength between about 350 nm and 450 nm, because the RICC2 excitation energy amounts to 346 nm. We also investigated the possibility of another reduced C chromophore with loss of the double bond between the chomophore methionine Cα and N, named C-H2, and found that the RICC2 excitation wavelength amounts to 443 nm (64.5 kcal/mol), which is in the area of the peak arising from the βME addition. Also the protonated C chromophore, C-H+, has been investigated and its RICC2 excitation energy lies within the 350- to 450-nm range. However, the pKa of mCherry is below 4.5, meaning that the chromophore is protonated only if the pH of the system is very low (39), which is not the case here.
Of the two possible βME adducts that contain a spiro 5 ring (at the tyrosine Cβ or the carbonyl C: B_spiro and C_spiro, respectively), only for B_spiro has an RICC2 excitation energy in the area of the experimental absorption spectrum been found.
The RICC2 excitation energies of the two investigated 6-ring structures (6_ring1 and 6_ring2) are lower than 300 nm, which is in the absorption wavelength area of many other structures in the protein and cannot explain the spectral peak around 400 nm. Furthermore, the excitation energy of the 7-membered ring structure with the original carbonyl carbon atom being sp3 hybridized, 7_ring(sp3), is slightly too low in energy whereas the excitation energy of the 7-membered ring with that carbon atom being sp2 hybrized, 7_ring(sp2), amounts to 91.9 kcal/mol (311 nm), which is too high in energy to explain the experimental absorption spectrum.
In summary, based on the calculated absorption wavelengths, we identified five different species that might contribute to the measured absorption wavelengths around 350–450 nm arising after addition of βME, which are the reduced chromophore B, the protonated reduced chomophore B-H+, the other reduced chromophore type C_H2, the spiro 5-ring structure B_spiro, and the Michael adduct D.
Materials and Methods
Microscopy.
All microscopy was performed on a Nikon Ti-E microscope equipped with a 100× Apo TIRF (N.A. 1.49) objective and a Perfect Focus System. Excitation was done either with a mercury lamp or via a custom illumination pathway with a 15-mW 405-nm diode laser (Power Technology) and a 100-mW 561-nm DPSS laser (Cobolt Jive). Fluorescence was detected using either an Andor DU-897D EMCDD with an additional 2.5× Optovar to achieve an effective pixel size of 64 nm or an Andor NEO 5.5 sCMOS with an effective pixel size of 65 nm. All components were controlled by Micromanager software (38). For intensity measurements as a function of photoactivation time an additional diffuser (Optotune LSR-C-3010) was inserted between a pair of lenses with focal distance of 75 mm in the illumination pathway for even illumination in TIRF mode. For SMLM, samples were continuously illuminated with 561 nm light in combination with increasing 405 nm intensity to maintain a constant number of molecules in the fluorescent state. Typically 5,000–15,000 frames were recorded with exposure times of 40–60 ms.
Crystallography.
mCherry crystals were grown in sitting drops containing 1 μL of protein solution (10 mg/mL), 1 μL of precipitant (0.2 M MgCl20.6H2O, 0.1 M Tris, pH 8.5, 25% PEG 4000), and 0.5 μL of βME (2 M), equilibrated against a 100-μL reservoir of precipitant. As the solution regained its color after several days, the crystals were soaked in a drop containing 1 μL PEG 400 (40%) and 1 μL βME (5 M) just before flash freezing in liquid nitrogen. Upon soaking, the crystals immediately lost their color and this absence of color remained until X-ray diffraction.
Data collection and refinement statistics can be found in Table S2. The structure was deposited in the PDB with accession code 5FHV. Images were created using the PyMOL Molecular Graphics System (Version 1.8; Schrödinger, LLC).
More experimental details and detailed information on the crystal structure determination and quantum-chemical calculations can be found in SI Methods.
SI Methods
Expression Constructs, Cell Culture, Transfection, and Fixation of Cells.
mCherry-α-Tubulin (39), vimentin-mMaple3 (25), mRFP1-Clathrin Light Chain A (21), and GFP-Centrin1 were obtained from Roger Tsien, University of California, San Diego, Xioawei Zhuang, Harvard University, Boston, Klemens Rottner, Technische Universität Braunschweig, Braunschweig, Germany, and Michel Bornens, Institut Curie, Paris, respectively, whereas mCherry-KANK1 (22) and pLVIP-β-tubulin-GFP (40) were obtained from Anna Akhmanova, Utrecht University, Utrecht, The Netherlands. mCherry-tubulin was subsequently subcloned into a β-actin vector (41). Full-length human CEP135 was amplified by PCR and inserted into an mCherry-C1 vector between XhoI and BamHI sites. mCherry-Clathrin Light Chain A was made from mRFP1-Clathrin by replacing RFP for mCherry, using the AgeI and XhoI restriction sites. mMaple3-α-Tubulin was cloned from the mCherry-α-Tubulin construct. mCherry was cut out by restriction digestion on the flanking HindIII and BglII sites and replaced by a restricted mMaple3 PCR product with an N-terminal HindIII site and a C-terminal 5-aa GS linker and BglII site. To generate β-Tubulin-mEos3.2, mEos3.2 (https://www.addgene.org/54525/) was PCR amplified to generate AgeI and NotI sites with which it was ligated into GW2-β-tubulin-eGFP. This GW2-β-tubulin-eGFP was obtained by restriction-based subcloning from pLVIP-β-tubulin-GFP (40), using NheI and XbaI.
COS-7 or HeLa cells were cultured in DMEM/Ham’s F-10 (50%/50%) containing 10% FCS and 1% penicillin/streptomycin at 37 °C and 5% CO2. Before transfection, COS-7 cells were plated on 18-mm glass coverslips for at least 2 d. COS-7 cells were transfected with Fugene6 (Roche) according to the manufacturer’s protocol.
For images of fixed cells before βME, after βME, and after photoactivation, cells were transfected for 2 d with mCherry-α-tubulin and then extracted and fixed using PEM80 with 0.3% Triton-X100 and 0.25% GA for 90 s and then PBS with 4% PFA for 20 min.
For SMLM imaging of mCherry-α-tubulin, after 2 d of transfection, the growth medium was exchanged with PBS containing 1 µM Taxol, preheated to 37 °C for 1 min, and then extracted with PEM80 (80 mM Pipes, 1 mM EGTA, and 5 mM MgCl, adjusted to pH 6.9) containing 1% Triton X-100 and 1 µM Taxol, preheated to 37 °C, for 2 min. The cells were then washed quickly three times with PEM80 and fixed with PBS containing 4% PFA and 0.25% GA for 20 min at room temperature. Samples were mounted in an imaging chamber filled with PBS, multiple positions were selected for SMLM imaging, and wide-field images at these positions were made. The PBS was then exchanged for PBS containing 143 mM βME, and SMLM imaging was started. When imaging mMaple3-α-Tubulin and β-Tubulin-mEos3.2, βME was omitted.
Protein Production.
mCherry and tagRFP were cloned by PCR from pmCherry-N1 (Clontech) and pTagRFP-N (Eurogene), respectively, and inserted into linearized pET-SUMO via TA ligation (Champion pET SUMO Expression System; Invitrogen). mRFP flanked by BamHI/XhoI was cloned by PCR, restricted, and inserted into a linearized pET28a-6×His vector. The identity of constructs was confirmed by sequencing analysis and the plasmids were retransformed into Escherichia coli BL21(DE3). Protein expression was induced for 3 h at 37 °C with 0.5 mM IPTG. Cells were pelleted by centrifugation at 4,600 × g. mCherry- and tagRFP-expressing cells were lysed in PBS containing 0.5% Triton X-100 and protease inhibitor mixture (Roche), sonicated for 15 min, and kept on ice for another 30 min. mRFP-expressing cells were lysed in PBS supplemented with protease inhibitor mixture (Roche) and lysozyme combined with five rounds of 1-min sonication. Bacterial extracts were prepared by centrifugation at 4,600 × g, at 4 °C for 30 min, and soluble protein was isolated with Probond resin (Invitrogen) according to a standard protocol. Purified protein was eluted from the beads by Imidazole gradient (150–300 mM), dialyzed into PBS, concentrated to 1 mg/mL, aliquoted, and stored at −80 °C.
For crystallization, a pRSetb plasmid containing the mCherry gene was transformed into JM109(DE3) E. coli cells (Promega). A single colony was inoculated in 1 L LB medium containing ampicillin. mCherry was expressed after 4 d at 20 °C. Cells were harvested by centrifugation at 5,000 × g for 10 min and resuspended in TN buffer (100 mM Tris, 300 mM NaCl, pH 7.4). Afterward, they were lysed using a French pressure cell, and the cellular debris was spin down for 20 min at 9,300 × g. Afterward, the cellular extract was loaded onto a HisTrap FF Crude column (GE Healthcare) coupled to an Aktra Prime system (GE Healthcare). Elution was performed using TN buffer supplemented with 500 mM imidazole. Then a size-exclusion chromatography was run on a HiLoad Superdex 200-pg 16/600 column (GE Healthcare) coupled to an Akta Purfier 10 system (GE Healthcare) with 0.1× TN buffer. Finally the protein was concentrated using a Vivaspin 10.000 MWCO column.
Measuring the Fluorescence Signal Return Percentage in Cells.
Coverslips were mounted in imaging chambers in PBS and positions for imaging were stored. A wide-field image was made per position first in PBS, then after incubation with 286 mM βME for 10 min, and finally after photoactivation for 5–10 s using a mercury lamp and a standard BFP2 filtercube (Chroma). Ten cells were analyzed and for each cell four regions were analyzed and averaged. The integrated fluorescence intensity was measured in nonsaturated parts of the images before adding βME, after incubation with 286 mM βME, and after photoactivation with violet light. Background intensity levels were determined from a part of the image outside of the cells and subtracted from all measurements. The fluorescence intensity before adding βME was set to 100%.
Measuring the Fluorescence Signal Return Percentage in Vitro.
For measurements of intensity as a function of photoactivation time, lane samples were created by attaching plasma-cleaned glass coverslips to microscope slides, using double-sided tape. Purified RFPs were diluted in PBS and nonspecifically adsorbed to the coverslips during 10 min incubation. After extensive washing with PBS to remove unbound molecules, 286 mM βME in PBS was added and the lanes were sealed. Imaging took place after ∼20 min. For each time point, the mean of the average intensity of six images was calculated. The data were fitted with a model describing photoactivation from the quenched state and photobleaching from both the quenched and red fluorescent state with rates kpa, kdam,B, and kdam,R, respectively,
where R is the amount of red fluorescent molecules after photoactivation and Q0 the amount of quenched molecules before photoactivation. Q0 is assumed constant for all photoactivation intensities and recaging was not included.
For washout experiments, the lane samples were washed extensively with PBS after 20 min incubation with βME and sealed with vacuum grease. Every ∼60 s an image was made and the average intensity calculated. The graph shows the mean ± SEM for n = 3 samples (Fig. 3G). The fitted model represents one phase dissociation of βME and mCherry:
To determine the return percentage, RFPs were nonspecifically adsorbed to plasma-cleaned 25-mm coverslips and mounted in imaging rings. In total five frames were made per position with an exposure time of 100 ms, using 561 nm laser light. Three images were made: before βME was added, one image after addition, and one image after photoactivation with 405 nm laser light. The percentages of fluorescence after adding βME and after photoactivation are relative to the last image before adding βME. n = 4, 2, and 2 samples and n = 40, 24, and 20 measurements for mCherry, mRFP, and tagRFP, respectively.
To test whether other FPs also experienced βME-induced caging, we measured the absorbance spectrum of several FPs in a Hepes/NaCl 50/30-mM buffer (pH 7.4) before and after the addition of 286 mM βME (30–60 min incubation time; Table S1).
SMLM Localization and Rendering Algorithms.
Single-molecule localization software was written in Java as an ImageJ plugin, called Detection of Molecules (DoM). Each image in an acquired stack was convoluted with the 2D Mexican hat kernel matching the microscope’s point spread function (PSF) size. The intensity histogram of the convolved image was fitted to a Gaussian distribution and used to calculate the threshold intensity value (mean value of the fit plus three SDs). The maximum intensity values within individual spots were chosen as initial positions for the peaks’ fitting performed on the original image. We used unweighted nonlinear least-squares fitting with a Levenberg–Marquardt algorithm to the assumed asymmetric 2D Gaussian PSF.
Only fits with a calculated width within ±30% of the measured PSF’s SD were accepted. Localizations within one pixel distance in a number of successive frames were considered to arise from the same molecule. In this case the weighted mean was calculated for each coordinate, where weights were equal to inverse squared localization precision. The resulting table with molecule coordinates and precision was used to render the final localization image with 10–20 nm pixel size. Each molecule was plotted as a 2D Gaussian of the integrated intensity equal to one and with SDs equal to the localization precision. Localization accuracies were estimated from the statistical uncertainties of the fitting parameters for the x and y positions ( and respectively) as The mode localization accuracies were determined from a histogram of all accuracies with 500 bins between 0 nm and 64 nm. Accuracies higher than 64 nm were excluded.
To calculate the number of photons per blinking event, the background-corrected integrated intensity was multiplied by the camera gain. The resulting number was divided by the chosen multiplication gain times the camera quantum efficiency in the spectral range of the fluorophore. When a blinking event lasted multiple frames, the photon counts were added for all frames.
To determine the contrast ratio between the on-state and off-state fluorescence for mCherry, mEos3.2, and mMaple3, we identified single-molecule photoactivation events in the superresolution recordings and determined the integrated, background-subtracted intensity of activated molecules in the first brightly fluorescent frame and in the second-to-last frame before bright emission. Background subtraction was performed by subtracting the integrated intensity of similarly sized adjacent regions. The obtained background values were consistent with the overall background values obtained at the end of recordings when all fluorophores had bleached. Because off-state fluorescence was barely detectable and because the SD of the background was considerable, values for off-state fluorescence were sometimes negative. We therefore averaged all on-state values and off-state values before calculating the contrast ratio. Uncertainties in the contrast ratio were determined by error propagation from the SEs of the off-state fluorescence and the on-state fluorescence. Thirty photoactivation events were analyzed per fluorophore.
Fluorescence Spectroscopy.
Fluorescence spectra as a function of βME percentage were recorded using a fluorescence spectrophotometer (Cary Eclipse; Agilent Technologies). Purified mCherry was diluted in PBS and pipetted in a 40-µL Quartz cuvette. βME concentration was increased from 0 mM to 143 mM. Four fluorescence spectra with both 550 nm and 410 nm excitation were recorded per βME concentration and averaged. Fluorescence intensity at 610 nm was fitted with an expression for a reversible bimolecular reaction:
Absorbance was measured over time, using a miniature deuterium-tungsten halogen lamp (Ocean Optics DT-MINI-2-GS) in line with a miniature spectrophotometer (Ocean Optics USB-4000). Purified mCherry was pipetted in a 1-mL cuvette and after the absorbance measurement was started 286 mM BME was added. Measurements were continued until the absorbance reached a steady state. An unfocused 405-nm diode laser (Coherent CUBE, 100 mW) was aimed from top to bottom through the cuvette to photoactivate the quenched mCherry molecules until again a steady state was reached. The photoactivation was then stopped and measurements continued for ∼6 min. The absorptions at both 410 nm and 590 nm are fitted with separate single-exponential functions for the three regimes.
NMR Spectroscopy.
The pET Sumo mCherry plasmid was transformed in BL21(DE3)Rosetta2, and a fresh colony was inoculated in 2 mL LB and cultured for 6 h at 37 °C. Cells were, after removal of the LB medium by centrifugation, transferred to 50 mL minimal medium (42), containing 4g/L 12C glucose and 1g/L 14N NH4Cl, and cultured overnight at 30 °C. Cells were transferred to 250 mL minimal medium containing 12C glucose and 14N NH4Cl supplemented with 100 mg/L of each amino acid. Glycine, Methionine and tyrosine (Cortecnet) were 13C/15N labeled; all of the other amino acids were not isotope enriched. mCherry expression was induced for 4 h at 30 °C by the addition of 0.5 mM IPTG, when the culture reached an OD600 of 0.7. Purification was performed as described in ref. 42, using metal affinity chromatography followed by sumo-tag removal by ULP1 according to instructions (Invitrogen). After overnight protease digestion, metal affinity chromatography was performed to separate the sumo tag from the essentially pure mCherry. The final sample was buffer exchanged by ultrafiltration to NMR buffer containing 50 mM NaPO4, 100 mM NaCl, pH 7.0, and 7.5% D2O at a final concentration of 100–200 µM mCherry.
Two-dimensional [1H,15N]- and [1H,13C]-HSQC spectra of mCherry were recorded at 298 K in the presence or absence of βME on a Bruker Avance 600-MHz spectrometer equipped with a TXI probe with z gradients. Spectra were processed using Topspin 2.1 and analysis was performed using Sparky (43).
Crystal Absorption Measurements.
Absorption spectra from single mCherry crystals were recorded using a home-built optical setup equipped with two objectives: a 10× Plan Apo objective (Nikon; N.A. 0.45) and a 10× Plan S objective (Olympus; N.A. 0.4). Depending on the working mode, the first objective was used to either focus white light on the crystal or illuminate the full field of view of the second objective, with the latter being responsible for the collection of the transmitted light and imaging of the sample surface. Transmitted light was split with a nonpolarizing beamsplitter and guided toward an optical fiber collimator and high-resolution color CMOS detector (DCC1645C), preceded by a tube lens. Imaging capability allowed for the precise beam positioning and collection of reliable reference absorption spectra (Ocean Optics DT-MINI-2-GS and USB4000). Both illumination and transmitted light were guided with a premium grade multimode optical fiber (Ocean Optics; QP600-1-UV-BX) with a core diameter of 600 µm. The crystallization buffer solution containing one mCherry crystal was sandwiched between two coverslips. Subsequently, the sample was placed between the objectives and mounted on the micrometer XYZ stage (Newport) that allowed for precise positioning within a travel range of 25 mm.
Crystal Structure Refinements.
X-ray diffraction data were collected on a Pilatus detector with a wavelength of 1.00 Å (12.4 keV) under a 100-K nitrogen stream at the X60DA beamline of the Swiss Light Source (SLS) at the Paul Scherrer Institute (PSI), Switzerland. The data were indexed and integrated using XDS v. January 22, 2015 (44) and scaled and merged using Aimless (45). The structure was solved by molecular replacement, using Phaser v. 2.5.6 (46) and the coordinates of mCherry (PDB ID: 2H5Q) as a model. Structure refinement was carried out using Phenix.refine v. 1.9 (47) and Coot v. 0.7.2 (48). After the first refinement cycle the original mCherry chromophore (PDB ligand ID: CH6) was modeled in the difference map. In subsequent refinement cycles, a modified version of the chromophore, being the Michael addition product of the chromophore and βME, was added to the model. The dictionary file of the CH6 chromophore was prepared using eLBOW (49). For the additional fragment in the other chromophore type (MCQ), an initial dictionary file was created using JLigand (50) and was manually changed using mean values of angles and dihedrals obtained from the Cambridge Crystallographic Database (CSD) (51). The occupancy factor of CH6 was coupled to that of K70 and water molecule 436 and converged to 59%, whereas the occupancy factor of MCQ was linked to that of the alternative conformation of K70 and converged to 41%. Water molecules were included in the model if they were in hydrogen bonding distance with chemically reasonable groups, appeared in Fo−Fc maps contoured at 3.0 σ, and had B factors less than 80 Å2.
Acknowledgments
We are grateful to Roger Tsien, Xioawei Zhuang, Klemens Rottner, and Anna Akhmanova for sharing constructs; to Kai Jiang, Roderick Tas, and Max Adrian for cloning constructs; and to Anne Janssen and York Ammon for technical assistance. We thank Wim Dehaen, Department of Chemistry, KU Leuven, for insightful discussion. This research was supported by the Dutch Technology Foundation (STW), which is part of the Netherlands Organisation for Scientific Research (NWO) Grant NWO-NANO 11421. Additional support came from the NWO Grant NWO-ALW-VIDI (to L.C.K.) and the European Research Council (ERC) Starting Grant 336291 (to L.C.K.). We also acknowledge support from the Research-Foundation Flanders (FWO) via Grants 1521915N, 1502314N, and 1525113N. M.M. is the recipient of a European Molecular Biology Organization (EMBO) Long-Term Fellowship (EMBO ALTF 88x4-2011), a Marie Curie IEF (FP7-PEOPLE-2011-IEF), and a DFG Emmy-Noether Grant Ml 1923/1-1. P.D. acknowledges support by the European Research Council via ERC Starting Grant 714688. E.D.Z. and V.G. thank the FWO Flanders for strategic basic research grants. B.K. received financial support from the Mobility Plus programme (1068/MOB/2013/0) founded by the Polish Ministry of Science and Higher Education. E.D.Z. and L.V.M. thank the staff of the beamline X60DA at the Swiss Light Source (Villigen, Switzerland) for their assistance with the data collection. M.B. gratefully acknowledges support from a fellowship from the Swiss National Science Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5FHV).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617280114/-/DCSupplemental.
References
- 1.Miyawaki A. Proteins on the move: Insights gained from fluorescent protein technologies. Nat Rev Mol Cell Biol. 2011;12:656–668. doi: 10.1038/nrm3199. [DOI] [PubMed] [Google Scholar]
- 2.Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 3.Shcherbakova DM, Sengupta P, Lippincott-Schwartz J, Verkhusha VV. Photocontrollable fluorescent proteins for superresolution imaging. Annu Rev Biophys. 2014;43:303–329. doi: 10.1146/annurev-biophys-051013-022836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adam V, Berardozzi R, Byrdin M, Bourgeois D. Phototransformable fluorescent proteins: Future challenges. Curr Opin Chem Biol. 2014;20:92–102. doi: 10.1016/j.cbpa.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 5.Dedecker P, De Schryver FC, Hofkens J. Fluorescent proteins: Shine on, you crazy diamond. J Am Chem Soc. 2013;135:2387–2402. doi: 10.1021/ja309768d. [DOI] [PubMed] [Google Scholar]
- 6.Patterson G, Davidson M, Manley S, Lippincott-Schwartz J. Superresolution imaging using single-molecule localization. Annu Rev Phys Chem. 2010;61:345–367. doi: 10.1146/annurev.physchem.012809.103444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van de Linde S, Sauer M. How to switch a fluorophore: From undesired blinking to controlled photoswitching. Chem Soc Rev. 2014;43:1076–1087. doi: 10.1039/c3cs60195a. [DOI] [PubMed] [Google Scholar]
- 8.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 9.Subach FV, et al. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat Methods. 2009;6:153–159. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci USA. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McEvoy AL, et al. mMaple: A photoconvertible fluorescent protein for use in multiple imaging modalities. PLoS One. 2012;7:e51314. doi: 10.1371/journal.pone.0051314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiedenmann J, et al. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc Natl Acad Sci USA. 2004;101:15905–15910. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 14.Duwé S, et al. Expression-enhanced fluorescent proteins based on enhanced green fluorescent protein for super-resolution microscopy. ACS Nano. 2015;9:9528–9541. doi: 10.1021/acsnano.5b04129. [DOI] [PubMed] [Google Scholar]
- 15.Shcherbakova DM, Verkhusha VV. Chromophore chemistry of fluorescent proteins controlled by light. Curr Opin Chem Biol. 2014;20:60–68. doi: 10.1016/j.cbpa.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durisic N, Laparra-Cuervo L, Sandoval-Álvarez A, Borbely JS, Lakadamyali M. Single-molecule evaluation of fluorescent protein photoactivation efficiency using an in vivo nanotemplate. Nat Methods. 2014;11:156–162. doi: 10.1038/nmeth.2784. [DOI] [PubMed] [Google Scholar]
- 17.Fölling J, et al. Fluorescence nanoscopy by ground-state depletion and single-molecule return. Nat Methods. 2008;5:943–945. doi: 10.1038/nmeth.1257. [DOI] [PubMed] [Google Scholar]
- 18.Biteen JS, et al. Super-resolution imaging in live Caulobacter crescentus cells using photoswitchable EYFP. Nat Methods. 2008;5:947–949. doi: 10.1038/NMETH.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winterflood CM, Ewers H. Single-molecule localization microscopy using mCherry. ChemPhysChem. 2014;15:3447–3451. doi: 10.1002/cphc.201402423. [DOI] [PubMed] [Google Scholar]
- 20.Grove J, et al. Flat clathrin lattices: stable features of the plasma membrane. Mol Biol Cell. 2014;25:3581–3594. doi: 10.1091/mbc.E14-06-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benesch S, et al. N-WASP deficiency impairs EGF internalization and actin assembly at clathrin-coated pits. J Cell Sci. 2005;118:3103–3115. doi: 10.1242/jcs.02444. [DOI] [PubMed] [Google Scholar]
- 22.Bouchet BP, et al. Talin-KANK1 interaction controls the recruitment of cortical microtubule stabilizing complexes to focal adhesions. eLife. 2016;5:e18124. doi: 10.7554/eLife.18124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bauer M, Cubizolles F, Schmidt A, Nigg EA. Quantitative analysis of human centrosome architecture by targeted proteomics and fluorescence imaging. EMBO J. 2016;35:2152–2166. doi: 10.15252/embj.201694462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang M, et al. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat Methods. 2012;9:727–729. doi: 10.1038/nmeth.2021. [DOI] [PubMed] [Google Scholar]
- 25.Wang S, Moffitt JR, Dempsey GT, Xie XS, Zhuang X. Characterization and development of photoactivatable fluorescent proteins for single-molecule-based superresolution imaging. Proc Natl Acad Sci USA. 2014;111:8452–8457. doi: 10.1073/pnas.1406593111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verkhusha VV, Chudakov DM, Gurskaya NG, Lukyanov S, Lukyanov KA. Common pathway for the red chromophore formation in fluorescent proteins and chromoproteins. Chem Biol. 2004;11:845–854. doi: 10.1016/j.chembiol.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 27.Subach OM, et al. Conversion of red fluorescent protein into a bright blue probe. Chem Biol. 2008;15:1116–1124. doi: 10.1016/j.chembiol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subach FV, et al. Photoactivation mechanism of PAmCherry based on crystal structures of the protein in the dark and fluorescent states. Proc Natl Acad Sci USA. 2009;106:21097–21102. doi: 10.1073/pnas.0909204106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subach OM, et al. Structural characterization of acylimine-containing blue and red chromophores in mTagBFP and TagRFP fluorescent proteins. Chem Biol. 2010;17:333–341. doi: 10.1016/j.chembiol.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bravaya KB, Subach OM, Korovina N, Verkhusha VV, Krylov AI. Insight into the common mechanism of the chromophore formation in the red fluorescent proteins: The elusive blue intermediate revealed. J Am Chem Soc. 2012;134:2807–2814. doi: 10.1021/ja2114568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Djerassi C, Gorman M. Studies in organic sulfur compounds. VI.1. Cyclic ethylene and trimethylene hemithioketals. J Am Chem Soc. 1953;75:3704–3708. [Google Scholar]
- 32.Dempsey GT, et al. Photoswitching mechanism of cyanine dyes. J Am Chem Soc. 2009;131:18192–18193. doi: 10.1021/ja904588g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng H, et al. Thiol reactive probes and chemosensors. Sensors. 2012;12:15907–15946. doi: 10.3390/s121115907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaughan JC, Dempsey GT, Sun E, Zhuang X. Phosphine quenching of cyanine dyes as a versatile tool for fluorescence microscopy. J Am Chem Soc. 2013;135:1197–1200. doi: 10.1021/ja3105279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Axel Castelli V, et al. Rates and equilibria of the Michael-type addition of benzenethiol to 2-cyclopenten-1-ones. J Org Chem. 1999;64:8122–8126. doi: 10.1021/jo9906882. [DOI] [PubMed] [Google Scholar]
- 36.Allen CFH, Happ GP. The thermal reversibility of the Michael reaction: I. Nitriles. Can J Chem. 1964;42:641–649. [Google Scholar]
- 37.Chen J, Jiang X, Carroll SL, Huang J, Wang J. Theoretical and experimental investigation of thermodynamics and kinetics of thiol-Michael addition reactions: A case study of reversible fluorescent probes for glutathione imaging in single cells. Org Lett. 2015;17:5978–5981. doi: 10.1021/acs.orglett.5b02910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using µManager. Curr Protoc Mol Biol. 2010;14:14.20.1–14.20.17. doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 40.Bouchet BP, et al. Mesenchymal cell invasion requires cooperative regulation of persistent microtubule growth by SLAIN2 and CLASP1. Dev Cell. 2016;39:708–723. doi: 10.1016/j.devcel.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kapitein LC, et al. NMDA receptor activation suppresses microtubule growth and spine entry. J Neurosci. 2011;31:8194–8209. doi: 10.1523/JNEUROSCI.6215-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Folkers GE, van Buuren BNM, Kaptein R. Expression screening, protein purification and NMR analysis of human protein domains for structural genomics. J Struct Funct Genomics. 2004;5:119–131. doi: 10.1023/B:JSFG.0000029200.66197.0c. [DOI] [PubMed] [Google Scholar]
- 43.Goddard T, Kneller D. Sparky 3. University of California; San Francisco: 2008. [Google Scholar]
- 44.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moriarty NW, Grosse-Kunstleve RW, Adams PD. Electronic Ligand Builder and Optimization Workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Crystallogr D Biol Crystallogr. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lebedev AA, et al. JLigand: A graphical tool for the CCP4 template-restraint library. Acta Crystallogr D Biol Crystallogr. 2012;68:431–440. doi: 10.1107/S090744491200251X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Groom CR, Bruno IJ, Lightfoot MP, Ward SC. The Cambridge Structural Database. Acta Crystallogr B Struct Sci Cryst Eng Mater. 2016;72:171–179. doi: 10.1107/S2052520616003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yanai T, Tew DP, Handy NC. A new hybrid exchange–correlation functional using the Coulomb-attenuating method (CAM-B3LYP) Chem Phys Lett. 2004;393:51–57. [Google Scholar]
- 53.Grimme S, Antony J, Ehrlich S, Krieg H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J Chem Phys. 2010;132:154104. doi: 10.1063/1.3382344. [DOI] [PubMed] [Google Scholar]
- 54.Ahlrichs R, Bar M, Haser M, Horn H, Kolmel C. Electronic-structure calculations on workstation computers - the program system Turbomole. Chem Phys Lett. 1989;162:165–169. [Google Scholar]
- 55.Schreiber M, Silva-Junior MR, Sauer SPA, Thiel W. Benchmarks for electronically excited states: CASPT2, CC2, CCSD, and CC3. J Chem Phys. 2008;128:134110. doi: 10.1063/1.2889385. [DOI] [PubMed] [Google Scholar]
- 56.List NH, Olsen JM, Rocha‐Rinza T, Christiansen O, Kongsted J. Performance of popular XC‐functionals for the description of excitation energies in GFP‐like chromophore models. Int J Quantum Chem. 2012;112:789–800. [Google Scholar]
- 57.Topol I, Collins J, Savitsky A, Nemukhin A. Computational strategy for tuning spectral properties of red fluorescent proteins. Biophys Chem. 2011;158:91–95. doi: 10.1016/j.bpc.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 58.Wanko M, García-Risueño P, Rubio A. Excited states of the green fluorescent protein chromophore: Performance of ab initio and semi-empirical methods. Physica Status Solidi B. 2012;249:392–400. [Google Scholar]
- 59.Surdhar PS, Armstrong DA. Reduction potentials and exchange reactions of thiyl radicals and disulfide anion radicals. J Phys Chem. 1987;91:6532–6537. [Google Scholar]