Significance
Innate immunity provides critical defense against pathogen invasion, and mutations in its cellular mechanisms have been implicated in autoimmunity, immune suppression, and other disease-producing conditions. However, knowledge of innate immunity pathways is largely biased toward model species. Thus, evolutionary interpretations suffer from large taxonomic gaps that ultimately weaken the strength of evolutionary inference. Our phylogenetic approach shows that the molecular machinery of the canonical TLR pathway was present in the last deuterostome ancestor before the rise of chordate lineages. Thus, TLR pathways with multiple gene–gene interactions have been conserved for more than 500 million years within vertebrates. Moreover, we provide evidence suggesting TLR3 may represent an ancient, evolutionarily conserved molecular interface for viral immune stimulation present across Deuterostomia.
Keywords: Deuterostomia, Toll-like receptors, molecular evolution, innate immunity, immunity evolution
Abstract
Animals have evolved an array of pattern-recognition receptor families essential for recognizing conserved molecular motifs characteristic of pathogenic microbes. One such family is the Toll-like receptors (TLRs). On pathogen binding, TLRs initiate specialized cytokine signaling catered to the class of invading pathogen. This signaling is pivotal for activating adaptive immunity in vertebrates, suggesting a close evolutionary relationship between innate and adaptive immune systems. Despite significant advances toward understanding TLR-facilitated immunity in vertebrates, knowledge of TLR pathway evolution in other deuterostomes is limited. By analyzing genomes and transcriptomes across 37 deuterostome taxa, we shed light on the evolution and diversity of TLR pathway signaling elements. Here, we show that the deuterostome ancestor possessed a molecular toolkit homologous to that which drives canonical MYD88-dependent TLR signaling in contemporary mammalian lineages. We also provide evidence that TLR3-facilitated antiviral signaling predates the origin of its TCAM1 dependence recognized in the vertebrates. SARM1, a negative regulator of TCAM1-dependent pathways in vertebrates, was also found to be present across all major deuterostome lineages despite the apparent absence of TCAM1 in invertebrate deuterostomes. Whether the presence of SARM1 is the result of its role in immunity regulation, neuron physiology, or a function of both is unclear. Additionally, Bayesian phylogenetic analyses corroborate several lineage-specific TLR gene expansions in urchins and cephalochordates. Importantly, our results underscore the need to sample across taxonomic groups to understand evolutionary patterns of the innate immunity foundation on which complex immunological novelties arose.
Innate immunity provides vital cellular and molecular defense against invading pathogens (1). Unlike immunological memory facilitated by jawed vertebrate immunoglobulins (2) and lamprey variable lymphocyte receptors (3), the molecular components of innate immunity do not recombine to diversify the breadth of defensive molecules (4). Thus, to provide substantial defense against a diversity of infectious agents with limited resources, the innate immune system exploits evolutionarily conserved pathogen-associated molecular patterns (PAMPs) (1, 4). PAMPs, such as Gram-negative lipopolysaccharide or viral dsRNA, often serve fundamental biological roles (4). Such structures are typically conserved over evolutionary time, providing targets for animal pattern-recognition receptors (5). Although there are many well-recognized families of innate immunity pattern-recognition receptors, Toll-like receptors (TLRs) evolved early in animals and have been extensively studied in model systems (6).
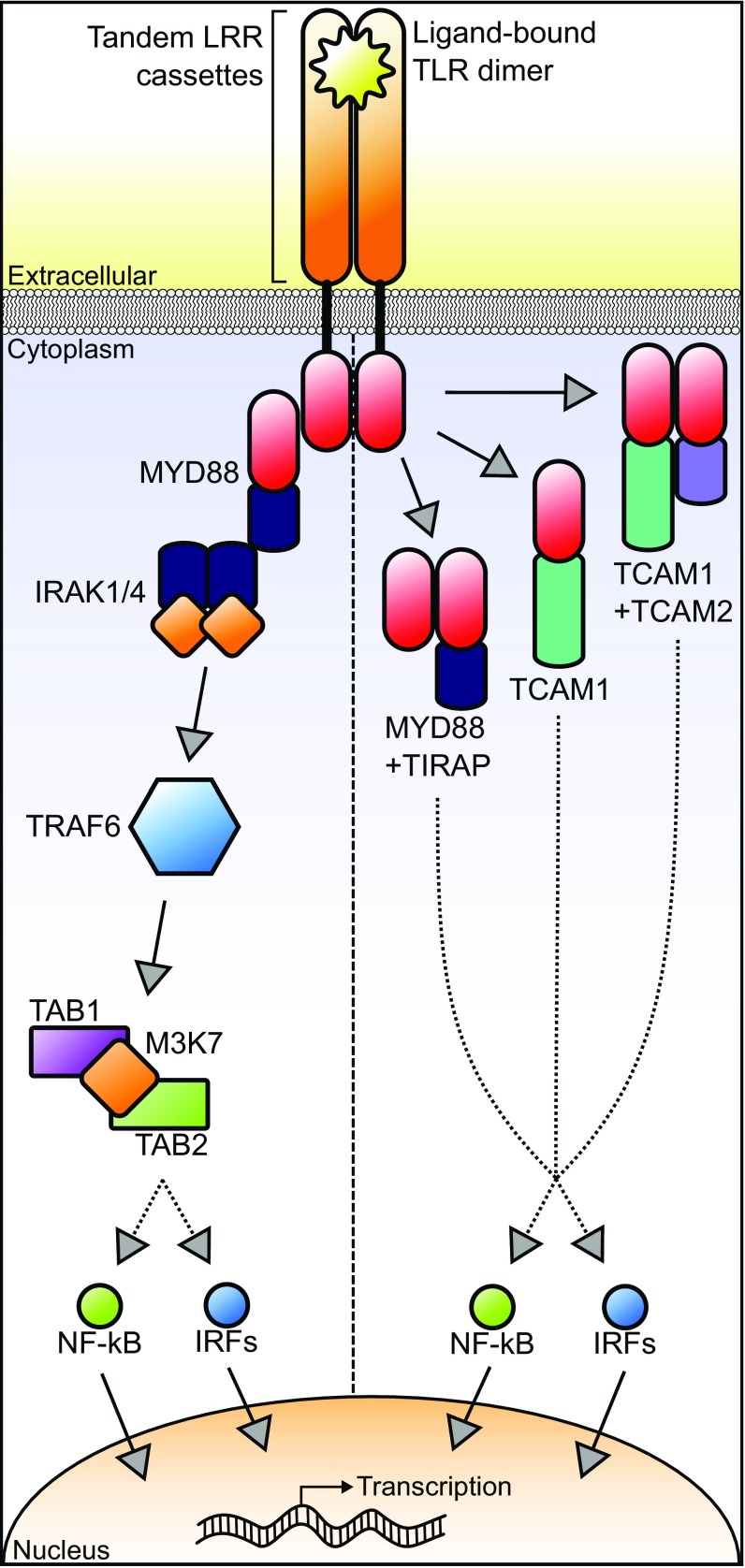
TLRs, named after the Toll protein in Drosophila melanogaster (7), are a group of type-I transmembrane glycoproteins localized to plasma membranes and endosomes (8). All TLRs possess three major regions: an extracellular domain of tandem leucine-rich repeats (LRRs), a transmembrane helix, and a cytoplasmic Toll/interleukin-1 receptor (TIR) domain. The breadth of TLR-facilitated immunity is determined by the ectodomain structure of LRRs and their associated glycosylated superstructure (8). Upon binding to PAMPs, TLRs dimerize, and a signal is transduced cytoplasmically via the TIR domain. Receptor dimers subsequently interact with cytoplasmic TIR domain-containing adaptor proteins (i.e., MYD88, TIRAP, TCAM1, and/or TCAM2) (9). Canonical signaling is mediated by MYD88 (Fig. 1) (10). This MYD88-dependent pathway proceeds through IRAK1/4, TRAF6, TAB1/2, and M3K7, terminating in activation of NF-κB for translocation to the nucleus where it acts as a transcription factor for a host of proinflammatory cytokines (10). This pathway rapidly provokes an inflammatory response and recruits additional phagocytic cells to confine and neutralize invading pathogens (11). Although TLRs were once thought to possess limited immune potential, research in jawed vertebrates has revealed that several TLRs possess pathogen-specific signaling pathways and are vital for activating adaptive immunity pathways (12).
Fig. 1.
Diagram of major TLR pathways. Upon ligand binding and receptor dimerization, TLRs interact with a TIR domain-containing adaptor protein. (Left) Canonically, TLR signal transduction occurs through the MYD88-dependent pathway. (Right) In some cases, such as with TLR3 and TLR4, TLRs require other TIR domain-containing adaptors to signal successfully for cytokine expression. SARM1, a TIR domain-containing negative regulator for TCAM1-dependent signaling pathways, is not shown. Red ellipses denote conserved TIR domains.
TLRs are functionally partitioned into two categories: those that are localized to host cell membranes and primarily recognize microbial cell membrane components (TLR1, 2, 4, 5, 6, and 10) and those that are localized to endosomes and recognize nucleic acids (TLR3, 7, 8, and 9) (10). TLR3, a vertebrate ortholog responsible for recognizing viral dsRNA, stimulates downstream signaling exclusively through the TIR domain-containing adaptor TCAM1 (9, 10). Independent of MYD88, TLR3 not only initiates downstream NF-κB activation but also initiates type-I IFN signaling that is fundamental to antiviral immunity (10). Concurrent with TLR3 activation and interaction with TCAM1, another TIR domain-containing protein, SARM1, increases in concentration and subsequently acts as a negative regulator for the TCAM1-dependent pathway (9). This negative-feedback loop is vital for efficient regulation of TLR signaling when overstimulation would be harmful to the host (10). The remaining two TIR domain-containing adaptors, TIRAP and TCAM2, are individually insufficient for TLR signal transduction. Instead, these proteins function as “sorting” proteins, with TIRAP promoting MYD88-dependent pathways and TCAM2 promoting TCAM1-dependent signaling pathways (9). In contrast to the earlier view which suggested that innate immunity acted merely as a molecular bridge to adaptive immunity (13), the presence of pathogen-specialized TLR-signaling pathways and their involvement in signaling immune responses indicates that innate immunity itself acts as a barrier to microbe pathogenesis.
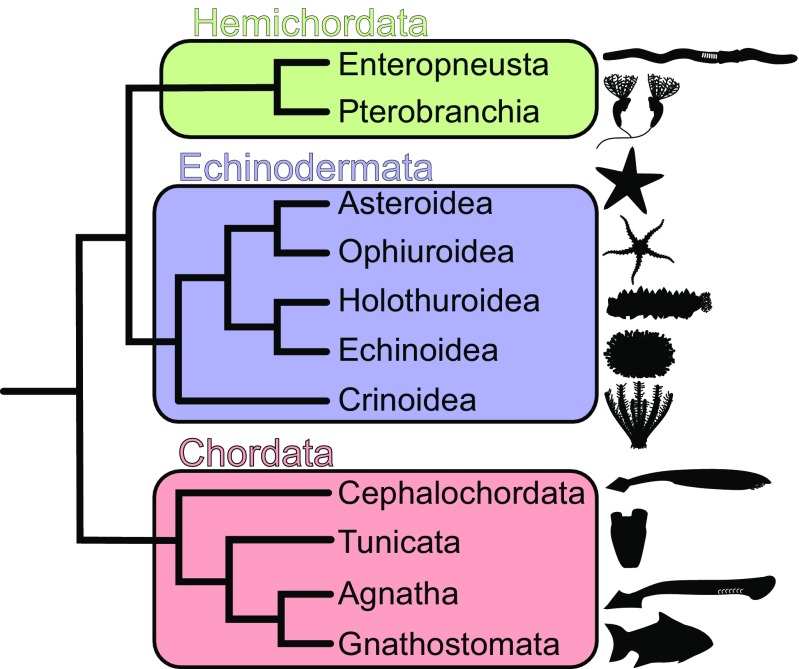
Jawed vertebrates possess ∼10 TLRs that have been functionally characterized. Far less is known about TLR diversity among other deuterostome groups. In addition to vertebrates, Deuterostomia comprises echinoderms (e.g., sea stars and urchins), hemichordates (acorn worms and pterobranchs), cephalochordates (lancelets), and tunicates (e.g., sea squirts) (Fig. 2). Genome surveys have revealed that the purple urchin Strongylocentrotus purpuratus and the lancelet Branchiostoma floridae have expanded repertoires of 253 and 72 TLRs, respectively (14–17). The difference between the size of TLR repertoires in these species and in jawed vertebrates appears to result from lineage-specific expansions of the majority of TLRs in S. purpuratus and B. floridae rather than from gene loss in jawed vertebrates (16, 17). How these TLR expansions affect the breadth of pathogen recognition has yet to be determined. In contrast, the tunicate Ciona intestinalis appears to possess only three TLRs, although C. intestinalis TLRs have broader PAMP recognition than those known in mammalian systems (18). Saccoglossus kowalevskii, an acorn worm hemichordate, has been reported to possess eight TLRs (17).
Fig. 2.
Deuterostome relationships as reported by recent phylogenomic studies (56). Echinoderms and hemichordates comprise the Ambulacraria, the sister group to Chordata.
In addition to TLRs, several immunity-related features appear to be evolutionarily and functionally conserved across deuterostome lineages; these features include pathogen-responsive phagocytic cell types, regulation of canonical cytokine homologs upon immune challenge, and differential regulation of TLR orthologs upon microbial challenge (Table 1). TLRs in the lancelet Branchiostoma belcheri have been shown to undergo obligatory MYD88 interactions to activate downstream NF-κB, consistent with observations in mammals (19). In the urchin S. purpuratus, gut epithelia have been shown to undergo stereotypical inflammatory responses in the presence of bacterial agents (20). This inflammatory response elicits specialization and migration of phagocytic cell types to regions of infection, mediated in part by TNFs and IL-17 signaling homologs (20). As a group, these conserved immune mechanisms suggest that the ancestor of all deuterostome possessed a common innate immunity toolkit with evolutionarily conserved function.
Table 1.
Functional conservation of immunity elements among invertebrate deuterostomes
| Chordata | Ambulacraria | |||
| Immunity feature | Tunicata | Cephalochordata | Echinodermata | Hemichordata |
| Phagocytes/coelomocytes | Present (57) | Present (58) | Present (20) | Present (58) |
| Cytokines and/or transcription factors expressed in infection | NF-κB, IL-1, TNFα (18, 57) | NF-κB, IL-17, IRFs (19, 29, 59) | NF-κB, TNF, IL-17 (20, 60) | Unknown |
| TLRs | ||||
| Subcellular expression | Cell membrane + endosomes (18) | Cell membrane and/or endosomes (19) | Unknown | Unknown |
| Cell/tissue expression | Pharynx and gut (18) | Epidermis, pharynx, and gut (19) | Coelomocytes and gut epithelium (17) | Unknown |
| PAMP-dependent regulation | Present (18) | Present (5) | Present (17) | Unknown |
| Molecular interactions | Activates NF-κB; induces TNFα expression (18) | Activates NF-κB via MYD88 (19) | Unknown | Unknown |
Although the origins and ancestral function of TLR signaling among animals are currently unclear, MYD88-facilitated TLR signaling is known to have been present in the bilaterian ancestor (6, 21). In contrast, virus-targeted TCAM1-dependent TLR signaling is known only from studies in select vertebrate taxa (e.g., mouse, human, and zebrafish) (9). As such, available evidence suggests that TCAM1-facilitated TLR signaling evolved in the vertebrate lineage at a similar time as the emergence of adaptive immunities (22). This hypothesis has been supported by the reported absence of a TCAM1 homolog among invertebrate model systems (9, 15, 16). However, the limited phylogenetic resolution provided by past comparisons between established vertebrate and invertebrate models is not adequate for an accurate understanding of TLR pathway evolution. In this study, we seek to illuminate the complement of TLR pathway components possessed in early deuterostomes and subsequent molecular innovation among contemporary lineages.
Results and Discussion
TLR-Signaling Adaptors and Their Associated Pathways.
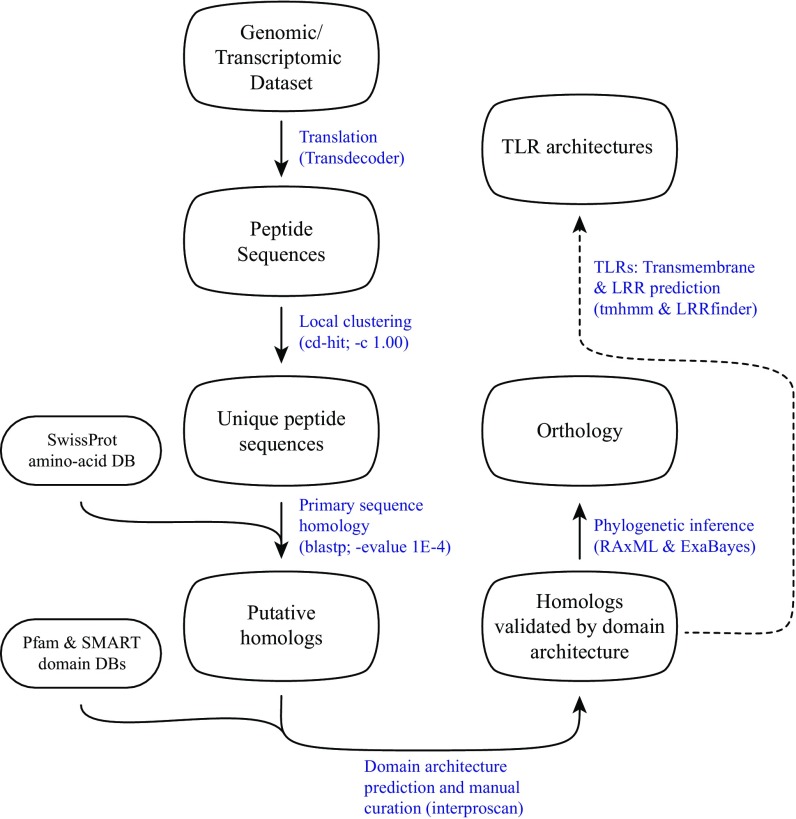
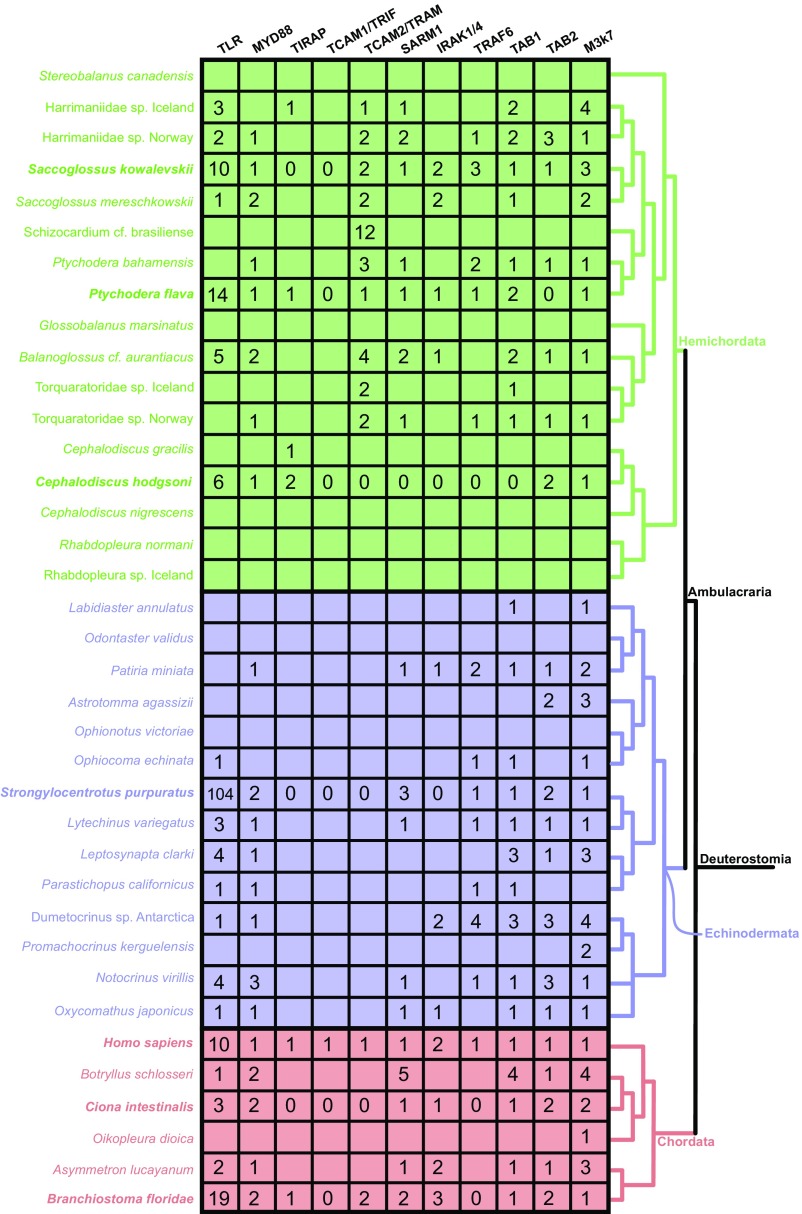
Our findings using bioinformatics tools (Fig. S1) to analyze genomic and transcriptomic data from 37 invertebrate deuterostome taxa, including humans (used as a genomic control) (Table S1) suggest the deuterostome ancestor possessed homologs to all canonical MYD88-dependent TLR-signaling components (Table 2). The presence of downstream TLR-signaling elements across all major deuterostome lineages may indicate conserved function of the pathway. The notion of conservation and a shared immunological ancestry among deuterostomes is supported by studies that show functional similarity between invertebrate deuterostomes and vertebrate lineages (Table 1). As mentioned previously, TLRs require MYD88 mediation to activate NF-κB in the lancelet (19), and TLR-mediated NF-κB activation also has been shown in the tunicate C. intestinalis (18). Homologs to several cytokines induced by the TLR pathway are expressed in immune-challenged invertebrates and have been implicated in inflammatory and immune responses in sea urchin embryos (20).
Fig. S1.
Diagram of the bioinformatic pipeline for homology identification. For details see Materials and Methods in the main text.
Table 2.
Presence of TLR pathway-signaling homologs in deuterostomes
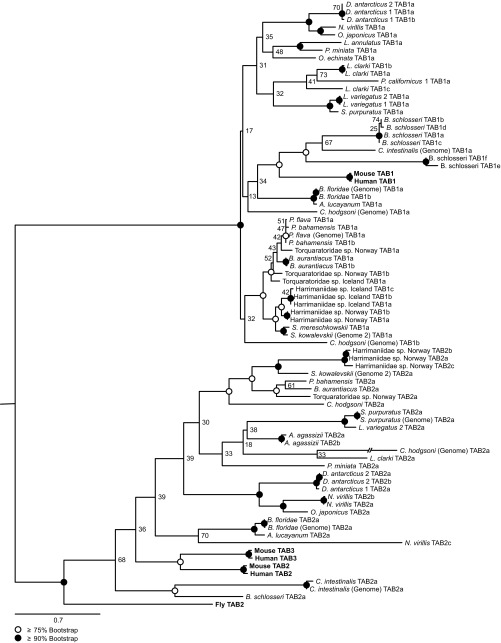
Although all other MYD88-dependent signaling mediators were identified by conserved domain architectures, invertebrate deuterostomes appear to lack a typical TAB2 signaling mediator (Table 2 and Fig. S2). TAB2, which facilitates coupling TRAF6 to the TAB1/M3K7 complex and NF-κB activation (Fig. 1), was frequently identifiable by primary sequence homology but could not be corroborated by assessment of typical domain architecture (except in the human control dataset). Specifically, each putative homolog identified lacked the N-terminal CUE domain essential in vertebrates for substantial activation of NF-κB (23). When placed in a phylogenetic framework including both known TABs and those identified in this study, TAB2 orthology was supported with high confidence (bootstrap = 100%) (Fig. S3), suggesting that TAB2’s CUE domain is a vertebrate-specific novelty. Because TAB2 homologs without the CUE domain were independently identified in 22 different datasets, the lack of the CUE domain is assumed to be correct and not the result of sequence assembly error. Interestingly, a functional TAB2 homolog that contains a CUE domain also has been reported in D. melanogaster (24). This finding suggests convergence regarding a CUE domain, perhaps selected for by molecular kinetics, between vertebrate and D. melanogaster TAB2s. Under a binary parsimony framework (i.e., presence vs. absence), a single deuterostome loss-and-regain event is equally parsimonious. Given the CUE domain’s potentially ancient eukaryotic origins [independent of TAB2 homology (25)], understanding the evolution of this domain will require much deeper taxon sampling within Metazoa.
Fig. S2.
Matrix containing the number of homologs identified per taxon. Taxa informed by both transcriptomic and genomic data are indicated in bold. Cells lacking values represent missing data for taxa informed solely by transcriptomic evidence. In contrast, gene absence is denoted by “0” only in taxa for which we analyzed genomic data. A cladogram depicting relationships among taxa is shown at the right of the matrix. Blocks/clades are colored by phylum: Hemichordata are green, Echinodermata are blue, and Chordata are red.
Fig. S3.
TAB1/2/3 gene-tree built using RAxML (54) rapid bootstrap analysis (1,000 replicates) and subsequent inference of best-fitting tree topology. Truncated TAB2s identified among invertebrate deuterostomes ally with human, mouse, and Drosophila TAB2/3s. Names in bold are sequences downloaded from public data repositories (Table S1).
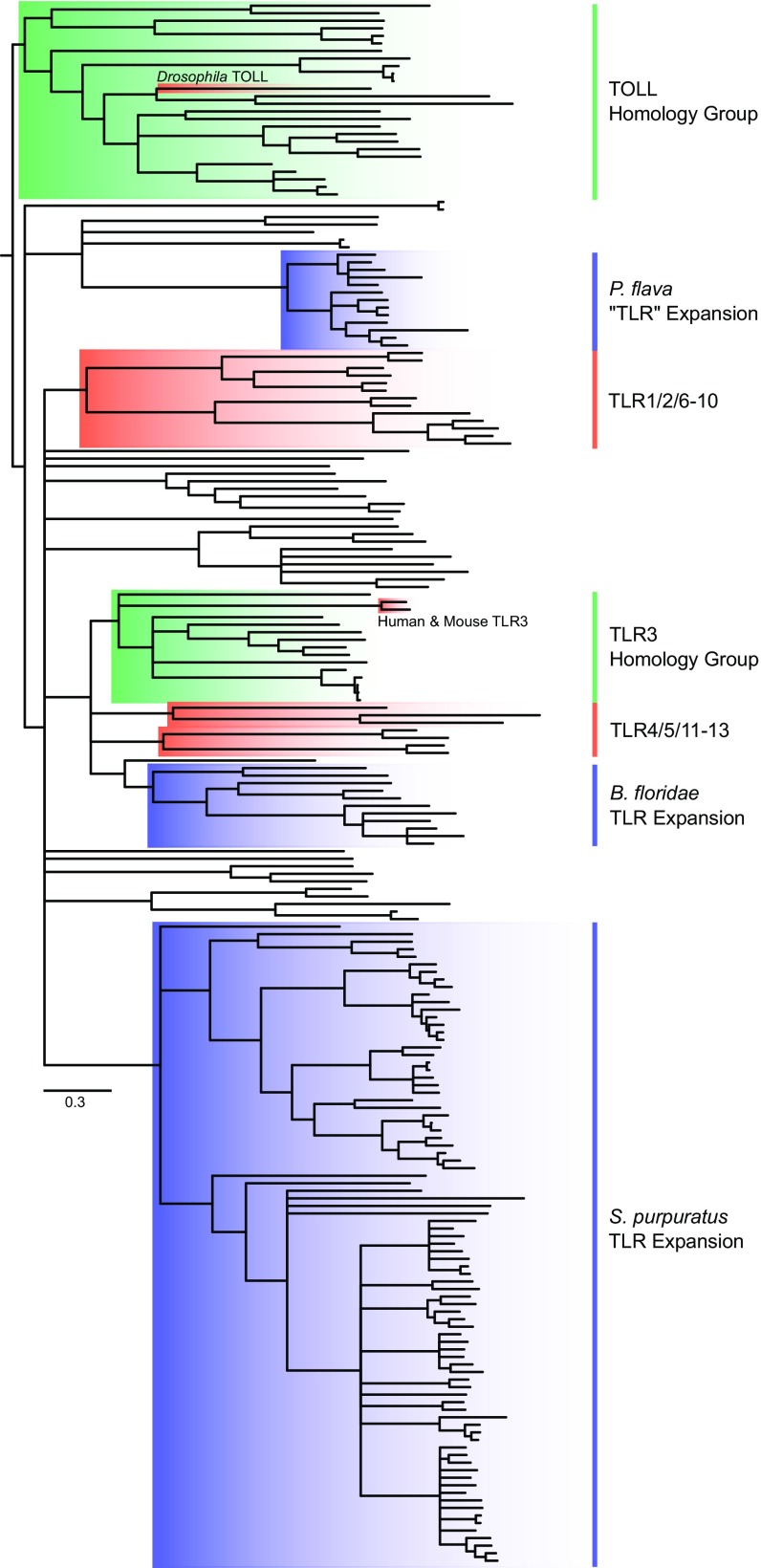
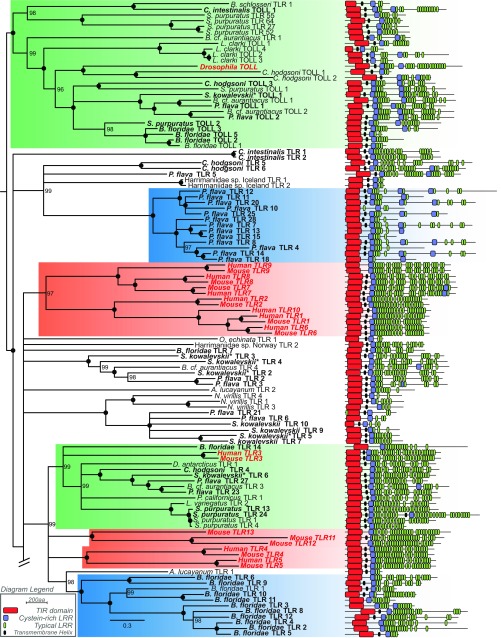
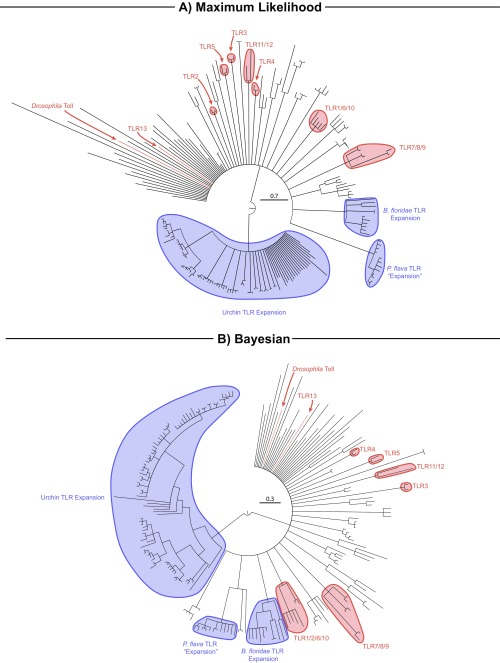
The second major pathway-defining protein, TCAM1, could not be identified in any genomic or transcriptomic dataset sampled herein aside from humans (Table 2 and Fig. S2). In mammals, TCAM1 is necessary for TLR3-facilitated antiviral cytokine signaling via interferons and is sufficient for TLR4 signal transduction (9, 10). Despite the absence of TLR3’s obligatory signaling adaptor, we found strong phylogenetic support (99% posterior probability) for TLR3 orthologs among invertebrate deuterostomes (Fig. 3 and Figs. S4 and S5). The sole exception was within tunicates, but the absence of TLR3 orthologs in tunicates is unsurprising, given their apparent TLR reduction (26). If the function of these TLR3 orthologs is conserved across taxa, the implication is that antiviral TLR3 signaling predates the origin of the TCAM1-dependent pathway. Molecular evidence suggests that the use of interferons (the primary antiviral cytokine family activated in TLR3-facilitated signaling) and their upstream transcriptional regulators (IFN regulatory factors; IRFs) for antiviral function are a vertebrate innovation (27, 28). Strikingly, IRFs from the lancelet B. belcheri have been shown to effectively recognize promoter regions of several human interferons (29) and are tightly regulated in response to dsRNA infection (19). These observations are consistent with the hypothesis that antiviral TLR3 signaling predates the emergence of TCAM1. Further molecular investigation will be required to determine the binding specificity of invertebrate TLR3 orthologs and their downstream signaling components. Such studies will be invaluable for understanding the ancestry and functional evolution of antiviral TLR signaling and for identifying subsequent vertebrate-specific molecular innovations.
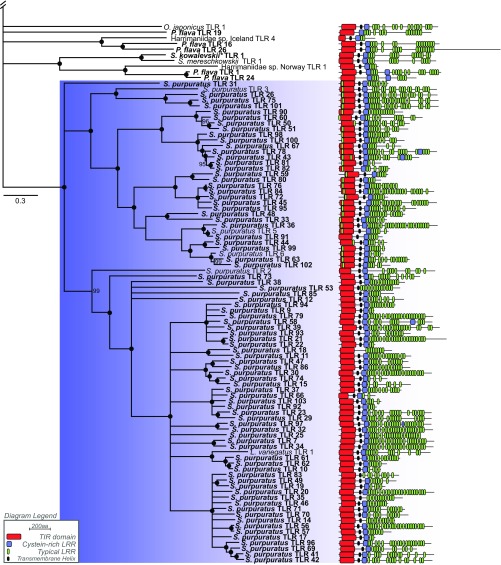
Fig. 3.
TLR gene tree from deuterostome taxa reconstructed with ExaBayes. Human and mouse TLRs, as well as Drosophila Toll, are included as positive controls and have been highlighted in red for orientation to known orthology groups. Tips have been removed for ease of interpretation; see Figs. S4 and S5 for more detail. All nodes have ≥95% posterior probability.
Fig. S4.
Detailed Bayesian gene-tree of deuterostome TLRs. Reference sequences from human, mouse, and Drosophila are highlighted in bold red font, and deuterostome sequences derived from genomic data are in bold font. The subtree contains Toll homology group, P. flava TLR expansion, reference TLR groups, the TLR3 homology group, and the B. floridae TLR expansion. Clades are colored as in Fig. 3. Black circles denote nodes with 100% posterior probability. Clades supported by less than 95% posterior probability have been collapsed. Asterisks indicate S. kowalevskii TLRs obtained from the revised S. kowalevskii genome (62).
Fig. S5.
Detailed Bayesian gene-tree of deuterostome TLRs. Reference sequences from human, mouse, and Drosophila are highlighted in bold red font, and deuterostome sequences derived from genomic data are in bold font. The subtree contains the S. purpuratus TLR expansion. Clades are colored as in Fig. 3. Black circles denote nodes with 100% posterior probability. Clades supported by less than 95% posterior probability have been collapsed. Asterisks indicate S. kowalevskii TLRs obtained from the revised S. kowalevskii genome (62).
The third TIR domain-containing adaptor, SARM1, acts in a negative-feedback loop for TCAM1-dependent pathways (e.g., TLR3 and TLR4), providing robust regulation where overexpression may yield deleterious effects. We found SARM1 to be present among all deuterostome phyla despite the apparent absence of TCAM1 (Table 2 and Fig. S2). Thus, SARM1’s function in TLR-signaling regulation may have existed before the origin of the TCAM1-dependent pathway in mammals. This conclusion is supported by research on the lancelet B. belcheri, in which SARM1 was found to play an inhibitory role for MYD88-dependent signaling rather than TCAM1-dependent signaling (30). SARM1 also has been shown to be central for a variety of neuronal processes [e.g., maintenance, behavior, and development (31)] and is a key player in injury-induced axon apoptosis in vertebrates (32). Additionally, SARM1 is implicated in embryological neuron development before its function in TLR-signaling regulation during B. belcheri ontogenesis (30). Thus, SARM1 homologs among invertebrate deuterostomes may be independent of TLR signaling but instead be a function of its role in neuron physiology/injury. Whether the original function of SARM1 was neuronal physiology/apoptosis, immunity regulation, or a coordination of these functions is still unclear.
The last two TIR domain-containing adaptors, TCAM2 and TIRAP, are individually insufficient for TLR signal transduction. Rather, TCAM2 and TIRAP facilitate the recruitment of TCAM1 and MYD88, respectively, providing downstream signaling specialization contingent upon the PAMP-bound TLR ortholog. Although our bioinformatics pipeline identified putative TCAM1 and TIRAP homologs among hemichordates and cephalochordates, orthology could not be supported when placed in a phylogenetic framework. These proteins may interact directly with TLRs and/or other TIR domain-containing adaptors; however, functional characterization will be required to elucidate their molecular signaling roles.
Lineage-Specific TLR Expansions.
Evidence for TLR expansions, as reported in echinoderms and lancelets (16, 17), could not be detected conclusively in hemichordates (Fig. 3 and Figs. S4 and S5). A possible exception appears to be the acorn worm Ptychodera flava, whose genome possesses 27 TLR homologs. However, a large group of P. flava’s TLRs (n = 13) possess multiple cysteine-rich LRR clusters (atypical even compared with Drosophila-like multicysteine cluster TLRs) and large regions that do not match any characterized protein domain (33, 34). When placed in a phylogenetic framework (Fig. 3 and Figs. S4 and S5), these TLRs form a monophyletic clade (100% posterior probability) closely related to homologs of more typical TLR structure. Their structural divergence from all other TLRs sampled in this study suggests that these TLRs may possess functional divergence specific to P. flava. In contrast, S. kowalevskii, for which a genome is also available (Table S1), possesses 13 TLRs.
Using genomic gene-model datasets coupled with transcriptomic evidence, we were able to identify only 104 TLRs in the urchin S. purpuratus and 19 in the lancelet B. floridae that could be confidently corroborated by domain architecture. Our estimates of TLR diversity in these two species are fewer than previously reported, possibly suggesting overly stringent homolog detection standards (see below). However, our methods identified all TLRs, signaling mediators, and gene variants present in the human gene-model dataset, a control for our bioinformatics pipeline. In an attempt to identify sources of error, we used the bioinformatics pipeline used by Buckley and Rast (17) on the most recent versions of the S. purpuratus and B. floridae genomes available at the time of this study (Table S1), but we were still unable to replicate the previously reported numbers of TLRs (253 and 72, respectively). Even when modifying detection methods in a way that would likely result in high false-discovery rates, we detected at most 185 and 39 TLRs in S. purpuratus and B. floridae, respectively. A large clade of S. purpuratus TLRs contained consistent overlapping domain signals from the cytoplasmic TIR domain and what appear to be cytoplasmic LRRs (Fig. S5). Although these overlapping domains may merely be domain-prediction artifacts, the S. purpuratus genome was the only dataset to show this predicted structure consistently. The function of expanded TLR repertoires in immune function or coopted for other roles remains unclear without further functional investigation.
With regard to S. purpuratus, the discrepancy between our results and previous reports (15, 17) can be attributed to two differences in the approach of bioinformatics workflows. First, our bioinformatics pipeline identifies unique peptide sequences that cannot be locally clustered by 100% identity, thus removing any translational redundancies and/or fragments of longer contigs (Materials and Methods, TLR Pathway Homolog Identification). This approach greatly reduces chances of including variants of the same TLR polypeptide that differ only by an N-terminal and/or C-terminal extension and thus cannot be confidently concluded to be unique TLRs. Second, extrinsic pseudogene prediction (15, 17) was not included in our analysis. If one combines the number of TLR pseudogenes identified in Buckley and Rast (17) with the number of TLRs identified through our overestimation pipeline, the results are consistent with previous estimates. Notably, however, we were able to reach this result only when deliberately overestimating the number of TLRs encoded in the S. purpuratus genome. We suspect that similar reasons led to discrepancies between our results and those previously reported for B. belcheri (17).
Conclusions
The last common deuterostome ancestor inherited a molecular toolkit sufficient for MYD88-dependent TLR signaling. However, unlike pathways in select model vertebrate taxa and Drosophila, conditions for complete signal transduction in the deuterostome ancestor likely lacked a typical TAB2 signaling mediator. TLR3 orthologs were recovered with strong phylogenetic support across invertebrate deuterostomes despite the apparent absence of its obligate signaling adaptor TCAM1, which is currently known only from vertebrates. Given our findings, coupled with functional evidence for evolutionarily and functionally conserved antiviral signaling mechanisms (29), we hypothesize that TLR3-facilitated antiviral cytokine signaling predates the origin of the TCAM1-dependent pathway. Considering the ubiquity of marine viruses (35) and their obligate virus–host interactions, extant deuterostome lineages may have inherited a TLR3-mediated antiviral defense from their most recent common ancestor, which was a marine organism. The TLR3-mediated antiviral defense would have been modified subsequently in vertebrates.
Materials and Methods
Data Acquisition and Assembly.
Datasets used for analyses, identified homologs, and their respective accession numbers are available in Table S1. cDNA downloaded as raw Illumina RNA-seq reads were digitally normalized via khmer (36) and assembled with Trinity using default parameters (37). Data generated on 454 sequencing platforms were assembled via 454’s NEWBLER. With regard to genomic data, only predicted and confirmed gene models were used for homolog identification. A total of 37 taxa were analyzed. Notably, transcriptomic datasets were evaluated only for the presence of homologs, and no conclusions were made on the absence of any particular homolog. Similarly, several transcriptomic datasets (such as the pterobranch hemichordates Cephalodiscus nigrescens and Rhabdopleura normani) had relatively low numbers of unique contigs and yielded no detectable homologs. Taxa included in this study were selected to represent phylogenetic depth and distribution representative of all major deuterostome groups while maintaining biocomputational feasibility by limiting vertebrate sampling.
TLR Pathway Homolog Identification.
TransDecoder (version 2.0.1) (38) was used on nucleotide sequences to identify putative ORFs and their associated protein sequences. Following TransDecoder’s log-likelihood scoring metric, high-scoring, small ORFs encapsulated by larger ORFs of the same reading frame were consolidated to avoid redundancy. Extracted amino acid sequences were additionally clustered by 100% identity using CD-HIT (version 4.6.1) (39) to control for translation redundancies and fragments. Following translation, amino acid sequences were queried against the Swiss-Prot database (downloaded May 2015) (40) using blastp (version 2.2.29+) (41) with an e-value cutoff of 1E-4. Sequences with best hits to target proteins were annotated for protein domain architecture using the SMART (33) and Pfam (34) databases included in InterProScan (version 5.17-56.0) (42). Domain architectures were cross-referenced with the putative homology for each sequence to control for nontarget or fragmented proteins. The absence of stop codons was not used as a criterion for removal of putative TLR genes because of the potential for extensive data loss; additionally, this type of filtering merely controls for C-terminal fragments. TMHMM (version 2.0c) (43) was used to predict TLR transmembrane helices; however, the absence of a transmembrane helix was not used as a criterion for TLR structural homology, because TLRs with known transmembrane helices (i.e., human and mouse TLR9) lacked confident transmembrane predictions with this software. The complete homolog identification bioinformatic pipeline can be found at https://github.com/mtassia/Homolog_identification and is diagramed in Fig. S1.
LRRs were additionally annotated with LRRfinder (44) to filter for particularly short fragments (see Table S2 for TLR domain architecture criteria) or TLR products of erroneous gene prediction. Because LRRfinder’s prediction confidence statistics were unreliable and appeared to prioritize overprediction rather than underprediction given its performance on human TLRs, we did not remove TLRs with predicted LRR coordinates lying within cytoplasmic regions (as was the case for a group of S. purpuratus TLRs). Hemichordate TLRs identified from S. kowalevskii and P. flava genomes were mapped back to their respective genomes using BWA-MEM (version 0.7.12) (45) to confirm gene models and check for overlap.
Phylogenetic Analyses.
To evaluate the orthology of recovered sequences, human and mouse TLR pathway protein sequences (in addition to Drosophila TOLL) were obtained from the Swiss-Prot database and used for phylogenetic inference. We used the E-INS-I alignment algorithm in MAFFT (version 7.215) (46), which optimizes alignments for sequences of multiple domains separated by hypervariable regions, to align complete TLR amino acid sequences. ProtTest (version 3.4) (47) was used to test and select best-fit amino acid substitution model using the Bayesian Inference Criterion (BIC). A gene-tree of TLRs was inferred with ExaBayes (version 1.4.2) (48). ExaBayes was executed with two parallel runs of four Metropolis-coupled chains analyzed for 1 × 106 generations (sampled every 500 generations) using a γ-distributed rate heterogeneity, empirical amino acid state frequencies, and a fixed amino acid substitution model (Whelan and Goldman; WAG) (49). All chains appeared to converge; convergence statistics are available in Table S3. Partitioning TLRs by cytoplasmic, transmembrane, and extracellular regions was tested using PartitionFinder (version 1.1.1) (50); however, partitioned analyses did not improve phylogenetic resolution. A majority-rule consensus tree was generated after discarding the first 25% sampled Markov chain Monte Carlo (MCMC) generations (250,000) as burn-in and was visualized with Mesquite (version 3.10) (51). All nodes with posterior probabilities less than 95% were collapsed. Two additional TLR phylogenetic analyses were inferred using only TIR domains to compare with previous results of TLR orthology inference (52, 53). TIR domain-only methods and results are available in the SI Text, and topologies are shown in Fig. S6. Aligning matrices of full TLR contigs provided greater nodal support and increased resolution relative to the TIR domain-only tree (Fig. 3 and Fig. S6). Non-TLR gene-trees were inferred using RAxML’s (version 8.0.23) (54) rapid bootstrap analysis with subsequent best-fit tree inference. Amino acid substitution matrices for non-TLR gene-trees were also inferred using ProtTest, and phylogenetic analyses were run with γ-distributed rate heterogeneity and empirical amino acid state frequencies. RAxML was preferred over ExaBayes for small gene-trees because of its accessibility when working with small datasets and short computation time.
Fig. S6.
TIR domain-only TLR amino acid trees. Topologies are consistent with those displayed in Fig. 3 and Figs. S4 and S5, although with considerably lower resolution and node support values. (A) Tree produced using RAxML; all nodes have ≥75% bootstrap support. (B) Tree produced using ExaBayes; all nodes have ≥95% posterior probability.
TLR Overestimation Pipeline.
Genome assemblies of B. floridae and S. purpuratus (Table S1) were analyzed in an attempt to detect TLR diversifications similar to previous findings (16, 17). Whole-genome scaffolds were translated in all ORFs using EMBOSS’ getorf (55) command with a minimum peptide length of 75 amino acids. Sequences were annotated for Pfam domains using HMMER with default detection thresholds. Polypeptides possessing TIR and LRRs were considered complete and subsequently were clustered by 100% identity by cd-hit to remove any sequences that might have been fragments of larger sequences in datasets. To provide the highest possible TLR counts, we did not use domain architecture to validate TLR homology in this reanalysis. Although this pipeline was assembled in an attempt to maximize overestimation of TLRs in a given genome, we still were unable to identify the magnitude of TLR diversity identified in previous studies (16, 17).
SI Text
Using the same base dataset, TIR domain amino acid sequences were extracted from TLRs, aligned, and placed in both a likelihood and Bayesian phylogenetic framework using the bioinformatic workflow described in Materials and Methods in the main text. Although both resulting likelihood and Bayesian TIR domain-only topologies (Fig. S6) were consistent with the topology generated using full TLR sequences (Fig. 3), support values in the TIR domain-only tree were low, and the resulting topology lacked the resolution yielded by using whole TLR alignment matrices. Despite being allowed to run for nearly double the number of generations as the full TLR Bayesian analysis, MCMC convergence statistics for the TIR domain-only matrix were reflective of less stationary sampling of parameter space relative to each MCMC chain compared with the full TLR analysis (asdsf = 1.11 and 0.60%, respectively) (Table S3). According to ExaBayes’ recommended convergence diagnostics, an asdsf value of 1.11% still falls within the “acceptable convergence” range, whereas a value of ≤1.00% is regarded as “excellent convergence” (48). We expect that orthology inference across 207 sequences using an amino acid matrix of only 246 sites is insufficient for phylogenetic inference at our target depth. Furthermore, by including a broad distribution of deuterostome taxa that more accurately represent deep deuterostome phylogeny, we are able to align complete TLR sequences more reliably than would have been possible when restricted to comparisons between model species (e.g., S. purpuratus vs. C. intestinalis vs. mouse).
Supplementary Material
Acknowledgments
The Auburn University Cellular and Molecular Biosciences Graduate Research Apprenticeship provided funding to M.G.T. and computational resources were made available by the Alabama Supercomputer Authority. This publication is Molette Biology Laboratory contribution 64 and Auburn University Marine Biology Program contribution 158. The findings and conclusions in the article are those of the authors and do not necessarily represent the findings and conclusions of the US Fish and Wildlife Service.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The datasets analyzed and homologs identified have been deposited in the NCBI database (www.ncbi.nlm.nih.gov) and figshare (www.figshare.com). Accession numbers are given in Table S1.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617722114/-/DCSupplemental.
References
- 1.Beutler B. Innate immunity: An overview. Mol Immunol. 2004;40:845–859. doi: 10.1016/j.molimm.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Rast JP, Litman GW. Towards understanding the evolutionary origins and early diversification of rearranging antigen receptors. Immunol Rev. 1998;166:79–86. doi: 10.1111/j.1600-065x.1998.tb01254.x. [DOI] [PubMed] [Google Scholar]
- 3.Pancer Z, et al. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature. 2004;430:174–180. doi: 10.1038/nature02740. [DOI] [PubMed] [Google Scholar]
- 4.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Dzik JM. The ancestry and cumulative evolution of immune reactions. Acta Biochim Pol. 2010;57:443–466. [PubMed] [Google Scholar]
- 6.Gauthier MEA, Du Pasquier L, Degnan BM. The genome of the sponge Amphimedon queenslandica provides new perspectives into the origin of Toll-like and interleukin 1 receptor pathways. Evol Dev. 2010;12:519–533. doi: 10.1111/j.1525-142X.2010.00436.x. [DOI] [PubMed] [Google Scholar]
- 7.Lemaitre B, Nicolas E, Michaut L, Reichhart J-M, Hoffmann JA. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 8.Botos I, Segal DM, Davies DR. The structural biology of Toll-like receptors. Structure. 2011;19:447–459. doi: 10.1016/j.str.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Neill LAJ, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 10.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 11.Sanjuan MA, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 12.Pasare C, Medzhitov R. Toll-like receptors: Linking innate and adaptive immunity. Microbes Infect. 2004;6:1382–1387. doi: 10.1016/j.micinf.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 13.O’Neill LA, Golenbock D, Bowie AG. The history of Toll-like receptors - redefining innate immunity. Nat Rev Immunol. 2013;13:453–460. doi: 10.1038/nri3446. [DOI] [PubMed] [Google Scholar]
- 14.Buckley KM, Rast JP. Diversity of animal immune receptors and the origins of recognition complexity in the deuterostomes. Dev Comp Immunol. 2015;49:179–189. doi: 10.1016/j.dci.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 15.Hibino T, et al. The immune gene repertoire encoded in the purple sea urchin genome. Dev Biol. 2006;300:349–365. doi: 10.1016/j.ydbio.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 16.Huang S, et al. Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity. Genome Res. 2008;18:1112–1126. doi: 10.1101/gr.069674.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buckley KM, Rast JP. Dynamic evolution of toll-like receptor multigene families in echinoderms. Front Immunol. 2012;3:136. doi: 10.3389/fimmu.2012.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasaki N, Ogasawara M, Sekiguchi T, Kusumoto S, Satake H. Toll-like receptors of the ascidian Ciona intestinalis: Prototypes with hybrid functionalities of vertebrate Toll-like receptors. J Biol Chem. 2009;284:27336–27343. doi: 10.1074/jbc.M109.032433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan S, Ruan J, Huang S, Chen S, Xu A. Amphioxus as a model for investigating evolution of the vertebrate immune system. Dev Comp Immunol. 2015;48:297–305. doi: 10.1016/j.dci.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 20.Ch Ho E, et al. Perturbation of gut bacteria induces a coordinated cellular immune response in the purple sea urchin larva. Immunol Cell Biol. 2016;94:861–874. doi: 10.1038/icb.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, et al. Characteristic and functional analysis of toll-like receptors (TLRs) in the lophotrocozoan, Crassostrea gigas, reveals ancient origin of TLR-mediated innate immunity. PLoS One. 2013;8:e76464. doi: 10.1371/journal.pone.0076464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leulier F, Lemaitre B. Toll-like receptors––taking an evolutionary approach. Nat Rev Genet. 2008;9:165–178. doi: 10.1038/nrg2303. [DOI] [PubMed] [Google Scholar]
- 23.Kanayama A, et al. TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 24.Zhuang ZH, et al. Drosophila TAB2 is required for the immune activation of JNK and NF-kappaB. Cell Signal. 2006;18:964–970. doi: 10.1016/j.cellsig.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 25.Ponting CP. Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic Acids Res. 2002;30:3643–3652. doi: 10.1093/nar/gkf498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu C, et al. Genes “waiting” for recruitment by the adaptive immune system: The insights from amphioxus. J Immunol. 2005;174:3493–3500. doi: 10.4049/jimmunol.174.6.3493. [DOI] [PubMed] [Google Scholar]
- 27.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 28.Nehyba J, Hrdlicková R, Bose HR. Dynamic evolution of immune system regulators: The history of the interferon regulatory factor family. Mol Biol Evol. 2009;26:2539–2550. doi: 10.1093/molbev/msp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan S, et al. Characterization of amphioxus IFN regulatory factor family reveals an archaic signaling framework for innate immune response. J Immunol. 2015;195:5657–5666. doi: 10.4049/jimmunol.1501927. [DOI] [PubMed] [Google Scholar]
- 30.Yuan S, et al. Amphioxus SARM involved in neural development may function as a suppressor of TLR signaling. J Immunol. 2010;184:6874–6881. doi: 10.4049/jimmunol.0903675. [DOI] [PubMed] [Google Scholar]
- 31.Lin C-W, Chen C-Y, Cheng S-J, Hu H-T, Hsueh Y-P. Sarm1 deficiency impairs synaptic function and leads to behavioral deficits, which can be ameliorated by an mGluR allosteric modulator. Front Cell Neurosci. 2014;8:87. doi: 10.3389/fncel.2014.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerdts J, Summers DW, Milbrandt J, DiAntonio A. Axon self-destruction: New links among SARM1, MAPKs, and NAD+ metabolism. Neuron. 2016;89:449–460. doi: 10.1016/j.neuron.2015.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Letunic I, Doerks T, Bork P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43:D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finn RD, et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suttle CA. Marine viruses––major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 36.Brown CT, Howe A, Zhang Q, Pyrkosz AB, Brom TH. A reference-free algorithm for computational normalization of shotgun sequencing data. arXiv. 2012;1203.4802v2:1–18. [Google Scholar]
- 37.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.UniProt Consortium UniProt: A hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Camacho C, et al. BLAST+: Architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones P, et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 44.Offord V, Coffey TJ, Werling D. LRRfinder: A web application for the identification of leucine-rich repeats and an integrative Toll-like receptor database. Dev Comp Immunol. 2010;34:1035–1041. doi: 10.1016/j.dci.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 45.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2:1-3.
- 46.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aberer AJ, Kobert K, Stamatakis A. ExaBayes: Massively parallel Bayesian tree inference for the whole-genome era. Mol Biol Evol. 2014;31:2553–2556. doi: 10.1093/molbev/msu236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 50.Lanfear R, Calcott B, Ho SYW, Guindon S. Partitionfinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- 51.Maddison W, Maddison D. 2016 Mesquite: A modular system for evolutionary analysis. Version 3.2. Available at mesquiteproject.org. Accessed March 30, 2017.
- 52.Roach JC, et al. The evolution of vertebrate Toll-like receptors. Proc Natl Acad Sci USA. 2005;102:9577–9582. doi: 10.1073/pnas.0502272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roach JM, Racioppi L, Jones CD, Masci AM. Phylogeny of Toll-like receptor signaling: Adapting the innate response. PLoS One. 2013;8:e54156. doi: 10.1371/journal.pone.0054156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rice P, Longden I, Bleasby A. EMBOSS: The European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 56.Cannon JT, et al. Phylogenomic resolution of the hemichordate and echinoderm clade. Curr Biol. 2014;24:2827–2832. doi: 10.1016/j.cub.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 57.Cima F, Franchi N, Ballarin L. In: Origin and Functions of Tunicate Hemocytes. Evolution of the Immune System. Malagoli D, editor. Academic; London: 2016. pp. 29–49. [Google Scholar]
- 58.Rhodes CP, Ratcliffe NA. Coelomocytes and defence reactions of the primitive chordates, Branchiostoma lanceolatum and Saccoglossus horsti. Dev Comp Immunol. 1983;7:695–698. [Google Scholar]
- 59.Wu B, Jin M, Gong J, Du X, Bai Z. Dynamic evolution of CIKS (TRAF3IP2/Act1) in metazoans. Dev Comp Immunol. 2011;35:1186–1192. doi: 10.1016/j.dci.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 60.Smith LC. Innate immune complexity in the purple sea urchin: Diversity of the sp185/333 system. Front Immunol. 2012;3:70. doi: 10.3389/fimmu.2012.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Terajima D, et al. Identification of candidate genes encoding the core components of the cell death machinery in the Ciona intestinalis genome. Cell Death Differ. 2003;10:749–753. doi: 10.1038/sj.cdd.4401223. [DOI] [PubMed] [Google Scholar]
- 62.Simakov O, et al. Hemichordate genomes and deuterostome origins. Nature. 2015;527:459–465. doi: 10.1038/nature16150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.