Significance
Lanthipeptides are a large family of microbial natural products of ribosomal origin. Prochlorosins are a group of unusually diverse lanthipeptides found in strains of the marine cyanobacteria Prochlorococcus and Synechococcus—the most abundant photosynthetic microorganisms on Earth. By analyzing the prochlorosin biosynthesis genes from cultured strains and wild cyanobacteria, we show that the global collective of these microorganisms has evolved thousands of structurally distinct lanthipeptides via a process of evolutionary radiation favoring the sustained emergence of new structures over refinement of an existing one. The evolutionary history of prochlorosins suggests a fundamentally different structure-to-function relationship compared with other lanthipeptides and opens the question of how structural diversification contributes to their function and mode of action in the marine environment.
Keywords: lanthipeptides, prochlorosin, RiPPs, Prochlorococcus, Synechococcus
Abstract
Lanthipeptides are ribosomally derived peptide secondary metabolites that undergo extensive posttranslational modification. Prochlorosins are a group of lanthipeptides produced by certain strains of the ubiquitous marine picocyanobacteria Prochlorococcus and Synechococcus. Unlike other lanthipeptide-producing bacteria, picocyanobacteria use an unprecedented mechanism of substrate promiscuity for the production of numerous and diverse lanthipeptides using a single lanthionine synthetase. Through a cross-scale analysis of prochlorosin biosynthesis genes—from genomes to oceanic populations—we show that marine picocyanobacteria have the collective capacity to encode thousands of different cyclic peptides, few of which would display similar ring topologies. To understand how this extensive structural diversity arises, we used deep sequencing of wild populations to reveal genetic variation patterns in prochlorosin genes. We present evidence that structural variability among prochlorosins is the result of a diversifying selection process that favors large, rather than small, sequence changes in the precursor peptide genes. This mode of molecular evolution disregards any conservation of the ancestral structure and enables the emergence of extensively different cyclic peptides through short mutational paths based on indels. Contrary to its fast-evolving peptide substrates, the prochlorosin lanthionine synthetase evolves under a strong purifying selection, indicating that the diversification of prochlorosins is not constrained by commensurate changes in the biosynthetic enzyme. This evolutionary interplay between the prochlorosin peptide substrates and the lanthionine synthetase suggests that structure diversification, rather than structure refinement, is the driving force behind the creation of new prochlorosin structures and represents an intriguing mechanism by which natural product diversity arises.
Microbial secondary metabolism produces a wealth of small molecules, referred to as natural products. These metabolites are fundamental for the function of microbial communities because they play diverse roles in mediating both biotic and abiotic interactions (1, 2). Studies on the diversity of secondary metabolite biosynthetic pathways in the human gut (3), soil (4), and marine sediments (5) indicate that the production of structurally diverse natural products is an integral feature of microbial communities. The oligotrophic ocean is the largest ecosystem on Earth, yet little is known about secondary metabolite production in planktonic marine microbial communities in part because their dilute habitat presents an unconventional stage for the action of these types of often-secreted compounds.
A survey of secondary metabolite pathways in sequenced genomes of Prochlorococcus and Synechococcus, the most abundant phytoplankton groups in the ocean, revealed the presence of a lanthipeptide biosynthesis pathway (6). The compounds produced by this pathway were named prochlorosins and are the first natural products identified in marine picocyanobacteria (6). The discovery of these compounds in these globally distributed and diverse microorganisms—emerging models for integrative systems biology (7, 8)—provides a unique opportunity to study the diversity and evolution of secondary metabolites in the natural environment.
Lanthipeptides are a class of ribosomally synthesized and posttranslationally modified peptides (RiPPs) characterized by the presence of intramolecular thioether cross-links that render them into complex polycyclic structures. The biosynthesis of lanthipeptides generally involves the synthesis of a precursor peptide composed of an N-terminal leader peptide and a C-terminal core region. The latter undergoes posttranslational modifications performed by a dedicated lanthionine synthetase that dehydrates select Ser and Thr residues and catalyzes the intramolecular addition of Cys thiols to the resulting unsaturated amino acids, forming lanthionine and methyl-lanthionine bridges, respectively. Following modification, the precursor peptide is often trafficked to the cell membrane where an ABC transporter with a C39 protease domain cleaves the leader peptide and releases the modified core peptide to the extracellular environment (9). Although the vast majority of known lanthipeptides are bactericidal (9, 10), some can act as signaling molecules (11, 12) or morphogenetic peptides (13). The function of prochlorosins, as well as the products of several other recently discovered lanthipeptide pathways in bacteria (14), remains unknown.
The lanthionine synthetase in most lanthipeptide-producing bacteria modifies only one cognate precursor peptide, producing a single product (15). Lanthipeptide-encoding strains of Prochlorococcus and Synechococcus, on the other hand, can produce multiple different lanthipeptides from distinct gene-derived precursors [called prochlorosin precursor peptides (ProcA) in this study] using a single highly substrate-tolerant prochlorosin lanthionine synthetase, ProcM (6) (Fig. 1A). Biochemical characterization of a ProcM from a Prochlorococcus strain (MIT9313) that encodes 29 different procA precursor peptide genes showed that this ProcM can catalyze the dehydration and cyclization of all 18 prochlorosin precursor peptide substrates tested (6) using a distinct substrate-tolerant catalysis mechanism (16).
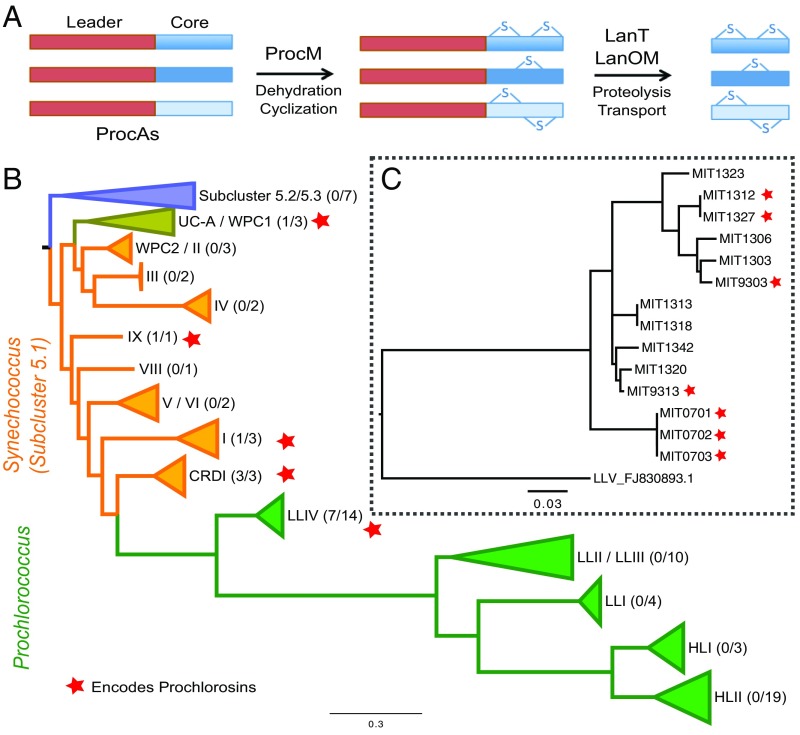
Fig. 1.
The prochlorosin biosynthesis pathway and its distribution among picocyanobacterial lineages. (A) ProcAs, which contain a conserved leader region (red) and hypervariable core region (blue shades), become mature products, first through the action of ProcM, which catalyzes the cyclization of the core peptide, and then by coupled cleavage of the leader peptide at a conserved Gly-Gly site by a membrane-associated ABC transporter with a C39 proteolytic domain (LanT). The transport across the cell membranes is thought to be the combined action of the LanT and a putative TolC-like outer membrane transporter, LanOM. (B) Schematic of a phylogenetic tree showing the principal clades of marine picocyanobacteria (38) showing the clades that contain a strain that encodes prochlorosin biosynthesis genes (red stars). Parentheses to the right of the clades indicate the fraction of sequenced genomes within each clade that encode the prochlorosin biosynthesis pathway. (C) Phylogenetic tree of the low-light–adapted IV (LLIV) clade of Prochlorococcus based on ITS rDNA sequence diversity (SI Appendix).
Sequence analysis of the 29 prochlorosin precursor peptides from Prochlorococcus MIT9313 revealed two features that make them unique among lanthipeptides (6). First, their core regions are highly dissimilar, and none have Ser/Thr or Cys residues in positions that would result in ring topologies similar to other known lanthipeptides, or to each other. Second, in contrast to the core regions, the sequence of the leader peptide is remarkably conserved. This unprecedented hypervariability in the prochlorosin core peptide observed in a single genome raises questions about the extent of the structural diversity of these compounds across marine picocyanobacterial lineages and, more broadly, about how widespread and diverse these natural products are along oceanic environmental gradients. Additionally, the hypervariability poses questions about what evolutionary mechanisms would enable the generation of such extreme diversity in prochlorosin precursor peptide genes.
Here we analyzed the prochlorosin biosynthesis pathway in genomes of previously and newly sequenced strains of Prochlorococcus and Synechococcus to better understand the distribution of this trait among lineages in these two genera and the degree of prochlorosin diversity within each group. To investigate how lanthipeptides evolve in the open ocean environment, we used a biogeographic approach that characterizes the distribution and abundance of the prochlorosin trait in different oceanic regions and that uncovers the extent of lanthipeptide structural diversity in wild picocyanobacterial populations. We examined fine-scale genetic variation in prochlorosin precursor peptide genes derived from these wild populations to begin to explore the evolutionary mechanisms responsible for this extensive diversity. Through this cross-scale analysis—from genomes to populations—we present evidence consistent with the hypothesis that structural diversity, instead of structural constraint, drives the evolution of lanthipeptides in marine picocyanobacteria, an unusual evolutionary scenario for the creation of complex polycyclic peptide natural products.
Results and Discussion
Phylogenomic Analysis of the Prochlorosin Biosynthesis Pathway.
To examine the phylogenetic distribution of the prochlorosin trait in cultured strains of Prochlorococcus and Synechococcus, we first searched for homologs of the prochlorosin lanthionine synthetase gene (procM) and the prochlorosin precursor peptide genes (procA) in 50 Prochlorococcus and 26 Synechococcus genomes. This set of genomes includes nine Prochlorococcus and three Synechococcus strains that are described in this work (SI Appendix, Table S1). Prochlorosin genes appear in 13 of these 76 genomes and have a restricted phylogenetic distribution. Among Synechococcus strains, the prochlorosin trait is present in four distantly related clades (I, IX, UC-A, and CRD1) of the 5.1 marine subcluster (Fig. 1B and SI Appendix, Fig. S1). Among Prochlorococcus strains, the trait is found only in the deeply branching low-light–adapted LLIV clade (Fig. 1B). Within the LLIV clade, 7 of the 14 strains available harbor prochlorosin genes (Fig. 1C and SI Appendix, Fig. S2). This sporadic occurrence of the trait among closely related strains is also evident within Synechococcus clades (Fig. 1B and SI Appendix, Fig. S1), indicating that lanthipeptide production is not a stably maintained trait within picocyanobacterial lineages. To refine the set of genomes used for the analysis of the prochlorosin biosynthesis pathway, we removed strains that were identical in their internal transcribed spacer (ITS) ribosomal DNA sequence and displayed more than 99% overall genome similarity, which in all cases had identical procA genes (SI Appendix, Figs. S3 and S4). This left four Prochlorococcus (MIT9313, MIT9303, MIT0701, and MIT1327) and five Synechococcus (MITS9509, MITS9508, RS9916, WH8016, and KORDI-100) strains for further analysis (SI Appendix, Table S1).
These nine genomes display flexibility in the genetic organization of the three principal components of the prochlorosin pathway: the precursor peptide genes (procA), the lanthionine synthetase gene (procM), and the genes that encode the inner and outer membrane transporters (lanT, lanOM) (SI Appendix, Fig. S5). Although there is a single procM gene in each genome, there is great variation in the number and genomic location of the procA genes, some of which can be present in tandem arrays of multiple genes (SI Appendix, Table S2). In the set of four Prochlorococcus strains, the number of procA genes varies between 9 and 29. Among Synechococcus strains, which span a larger genetic distance, the procA gene number varies from 1 to 80—an unprecedented expansion of lanthipeptide precursor peptide genes (SI Appendix, Table S2). In total, these nine genomes encode 181 procA genes. Similarly to the procA genes, the procM and transporter genes are not necessarily located in the same genomic locus in all strains. Although strains MIT1327, MITS9509, and RS9916 have their procM, lanT, and lanOM genes present in a single gene cluster, the other strains have these genes scattered across multiple genomic loci, suggesting genetic mobility and modularity within components of the prochlorosin pathway (SI Appendix, Fig. S5). These architectures contrast with the genetic organization of lanthipeptide biosynthesis pathways in other bacteria and with canonical peptide-based secondary metabolite pathways in which components for biosynthesis are usually found in a gene cluster that behaves as a single evolutionary unit (17). Flexibility in the configuration of the genetic components of the prochlorosin pathway implies an uncommon mode of evolution for this family of lanthipeptides.
Prochlorosin Precursor Peptide Gene Diversity in Picocyanobacterial Genomes.
We assessed the potential diversity of lanthipeptide structures encoded in this set of nine strains by examining the amino acid sequence similarity of the leader and core peptide regions of the prochlorosin precursor peptides. Strikingly, the core region—the substrate for posttranslational modifications—of all but 2 of the 181 precursor peptides is different, with 98.4% of all possible pairs displaying less than 30% protein identity to each other (Fig. 2A). Despite their massive sequence diversity, these core peptides are not random protein sequences; their amino acid composition is dominated by Gly, a small flexible amino acid, and the amino acids required for cyclization (Cys, Thr, and Ser), with Gly, Cys, and Thr residues enriched relative to the total proteome of their host organisms (Fig. 2B). Based on the co-occurrence of Cys/Ser or Cys/Thr residues, 91% of the precursor peptides identified have the potential to form a (methyl)lanthionine-containing cyclic peptide.
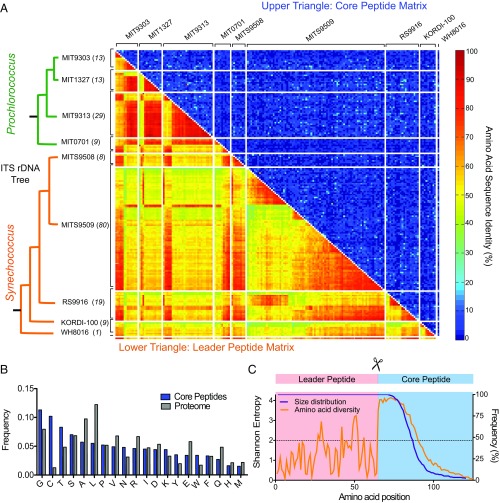
Fig. 2.
Genetic variation of prochlorosin precursor peptides. (A) Percentage of amino acid identity for each pairwise comparison in a multiple sequence alignment of the leader peptide region (Lower triangle) and the core peptide region (Upper triangle) of the 181 prochlorosin precursor peptides found in nine nonclonal lanthipeptide-encoding strains of Prochlorococcus and Synechococcus. The precursor peptides in the matrix were grouped first by the phylogenetic relationship of the strain of origin (based on ITS ribosomal DNA, at left) and then by the intragenome amino acid identity of their leader peptide region. Only one pair of prochlorosins originating in MIT9313 and MIT1327 share 100% identity in their core region; their leader peptide region is 98% identical. (B) Amino acid composition of the prochlorosin core peptides (weighted average of frequency) found in Prochlorococcus and Synechococcus genomes compared with the general proteome composition of these strains. Gly, a flexible small amino acid, along with amino acids necessary for the formation of ring structures (Cys, Ser, and Thr), are the most frequent residues in the prochlorosin core peptides; all but Ser are found at a higher frequency in prochlorosin core peptides than in the general proteome. (C) Size distribution (purple line, right y axis) and amino acid diversity by Shannon entropy at each position (orange line, left y axis) of prochlorosin precursor peptides. Positions within the leader peptide region correspond to the consensus of the multiple sequence alignment of 181 leader peptides. Because of the difficulty in producing a meaningful multiple alignment of the core region, the positions for this region correspond to stacked sequences that use the Gly-Gly motif as a starting reference. Shannon entropy values higher than 2 are considered variable (dashed line); the maximum theoretical value of the Shannon entropy for 20 amino acids is 4.322 and signifies that all residues are equally represented in that position (39).
The length of the precursor peptide also varies greatly among the sequences, and the amino acid diversity within the peptide increases dramatically immediately after the end of the leader peptide, indicating no preference for a particular amino acid at any given position in the core peptide following the Gly-Gly motif of proteolytic cleavage (Fig. 2C). Consequently, these core peptides would result in the formation of lanthipeptides with distinct ring topologies. In contrast to the core region, the leader peptide region exhibits a high degree of inter- and intragenome conservation, consistent with its functional role in directing the biosynthesis of prochlorosins. Interestingly, although the molecular determinants for ProcM activity are located only in the final third of the C-terminal end of the ProcA leader peptide (18), the high degree of conservation of the N-terminal end of the leader peptide across distant lineages suggests that this region may be important for additional steps of the biosynthesis process, perhaps transport of prochlorosins.
When performing a BLAST search for procA homologs in our 76 genomes, we found a total of 53 procA pseudogenes, i.e., prochlorosin precursor peptide genes containing a mutation (early stop codon, frameshifts, or elimination of the start codon) that prevents the formation of a functional prochlorosin ORF. The procA pseudogenes were detected in seven of the nine prochlorosin-encoding strains and also in four strains of the LLIV clade of Prochlorococcus that do not encode the prochlorosin biosynthesis pathway (SI Appendix, Tables S1 and S3). After correction of mutations, we found that, as in intact procA genes, most of the core regions of the procA pseudogenes have no sequence similarity to other procA genes or procA pseudogenes (SI Appendix, Fig. S6 A and B). Also, the putative core regions of the procA pseudogenes have a similar amino acid composition to the intact procA genes and are also enriched in Gly, Cys, Ser, and Thr residues, indicating that they could have encoded substrates for the creation of prochlorosins before the deactivating mutations occurred (SI Appendix, Fig. S6C). Despite the lack of similarity between their core regions, phylogenetic analysis of the leader peptide region of the intact and pseudoprocA genes indicates that they are often closely related (SI Appendix, Fig. S7), suggesting that many of the pseudogenization events are recent. The high frequency of procA pseudogenes in these genomes suggests a high turnover rate of paralogous genes, indicating that the prochlorosin biosynthesis pathway is a dynamic trait involving sustained expansion, diversification, and elimination of procA genes.
The 180 different procA genes that we have identified in only nine closely related marine picocyanobacteria strains is double the ∼90 other lanthipeptides that have been described from vastly different branches of the bacterial domain (15). This impressive diversity led us to wonder how many lanthipeptide structures have evolved within the enormous genetic diversity embodied in Prochlorococcus and Synechococcus populations on a global scale and how these metabolites are distributed along environmental gradients in the oceans.
Diversity and Distribution of Prochlorosin Precursor Peptide Genes in Wild Populations.
An initial survey of prochlorosin biosynthesis genes in metagenomic databases of surface ocean samples revealed the presence of the lanthipeptide production trait in wild picocyanobacterial populations (6). To obtain a comprehensive picture of the biogeography of the prochlorosin production trait, we investigated ocean regions where gradients of light, temperature, and nutrient concentrations are known to shape the relative abundance of Prochlorococcus ecotypes and Synechococcus groups (19, 20). Using quantitative PCR targeting highly conserved regions of the prochlorosin precursor peptide gene, we measured the abundance of procA genes in samples collected at Station ALOHA in the North Pacific Subtropical Gyre and along 16 stations spanning an Atlantic Meridional Transect (AMT) (Fig. 3A). The prochlorosin trait is widespread, but patchily distributed along the transect (Fig. 3B); procA genes have distinctive depth-distribution patterns and vary in abundance across the transects. These patterns are likely influenced by the current restriction of the trait, within Prochlorococcus, to the LLIV clade; members of this clade are adapted to low-light conditions and typically found deeper in the water column (19, 20).
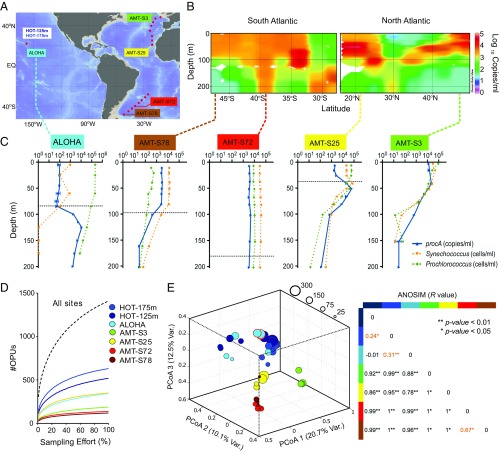
Fig. 3.
Biogeography and diversity of prochlorosin precursor peptides in the Atlantic and Pacific oceans. (A) Oceanic sites sampled in this study. Red dots denote sampling locations: Station ALOHA in the North Pacific and the Atlantic Meridional Transect (AMT13) in the Atlantic. Colored labels correspond to stations shown as depth profiles in C and analyzed by deep amplicon sequencing of the prochlorosin population in D and E. (B) Depth distribution of procA genes within the upper 250 m of the water column along the AMT13 transect. Color-scale indicates the logarithm of concentration in copies/mL as determined by qPCR targeting conserved sequences of the leader peptide region. (C) Depth profiles showing abundance of procA genes (blue) and flow cytometry cell counts of total Prochlorococcus (green) and Synechococcus (orange) at five oceanic sites. Dashed lines indicate the depth of the surface mixed layer, as determined by potential density differential from surface values by >0.125 kg⋅m−3. Mixed layer data are not available for AMT-S3, but from the shape of the profiles it can be presumed to be about 50 m. (D) OPU rarefaction curves for each of the five oceanic sites: AMT-S3 (9, 54, 79, and 104 m), AMT-S25 (17, 53, 83, and 103 m), AMT-S72 (9, 53, 74, and 154 m), AMT-S78 (10, 50, 80, and 160 m), Station ALOHA (25, 85, 100, 125, and 175 m), and two depths from nine monthly time-series samplings at Station ALOHA between June 2005 and February 2006 (HOT-125 m and HOT-175 m). (E) OPU clustering on the basis of a Jaccard dissimilarity matrix (visualized by principal coordinates analysis). Circle diameters are proportional to the richness of OPUs in each sample. We included a technical replicate (same DNA extraction) and a biological replicate (different DNA extraction from the same water sample) of the ALOHA H175 125-m sample as controls for the clustering, outlined in red. ANOSIM R = 1 (complete dissimilarity), R = 0 (complete evenness), and R < 0 (dissimilarity is greater within groups than between groups). Values highlighted in orange correspond to site comparisons for which there was overlap in their OPU populations.
The relationship between the abundance of procA genes and cells belonging to the LLIV clade is best illustrated by procA distribution patterns at two subtropical locations (Station ALOHA and AMT-S25, Fig. 3C), where the surface mixed layer is dominated by high-light–adapted ecotypes of Prochlorococcus (not known to encode prochlorosins) and where Synechococcus is present in low numbers relative to Prochlorococcus (19, 21). In these regions, the abundance of procA genes is relatively low throughout the surface mixed layer but increases in deeper waters where procA-encoding LLIV Prochlorococcus are known to thrive (Fig. 3C) (19). In contrast, at low or high latitudes of the Atlantic Ocean (i.e., AMT-S78 and AMT-S3, respectively) where Synechococcus dominates and low-light–adapted Prochlorococcus are relatively rare (20), procA abundances track Synechococcus abundances and are low at the base of the euphotic zone. The only instance where procA genes were found equally distributed throughout the euphotic zone was when a deep-mixing event homogenized the distribution of Prochlorococcus and Synechococcus populations in the water column (i.e., AMT-S72, Fig. 3C). We conclude that prochlorosin production is widespread in the oceans and constitutes an integral component of planktonic cyanobacterial populations throughout the oligotrophic ocean.
Having identified picocyanobacterial populations that harbor the prochlorosin trait, we next undertook analyses to better understand how selective pressures on the prochlorosin trait shape the diversity of prochlorosin populations (i.e., the total collection of different lanthipeptide structures encoded by the picocyanobacterial population at a location) across different oceanic sites. To obtain most of the coding sequence of the procA gene and predict its prochlorosin product, we used the highly conserved 5′ region of the leader peptide and a moderately conserved 3′ intergenic region downstream of the procA gene as primer-anchoring sites for amplification (SI Appendix, Fig. S8). Evaluation of this set of primers on a mock seawater sample containing cells from prochlorosin-encoding Prochlorococcus and Synechococcus strains suggests that ∼25% of the potential procA genes can be recovered by this method (SI Appendix, Supplemental Materials and Methods and Table S6). Thus, although this set of primers likely will not amplify all procA genes at a given location, it places a lower bound on their diversity and facilitates comparisons among prochlorosin populations from different natural environments. For this analysis, we selected four to five depths from each of five locations displaying distinct patterns in the distribution of procA genes (Fig. 3C) and nine samples collected monthly (June 2005 to February 2006) from 125 m and 175 m at Station ALOHA for the Hawaii Ocean Time-Series (HOT) program (19, 22). The procA genes from these 39 environmental samples were deeply sequenced and quality-filtered (Materials and Methods). To determine the total number of different prochlorosin precursor peptides in the dataset, procA ORFs were predicted from the cleaned reads from all samples and clustered at a genetic distance of 3% to account for possible sequencing errors. Each unique 97% DNA identity cluster (excluding singletons) was defined as an operational prochlorosin unit (OPU), representing a unique prochlorosin precursor peptide gene from which the sequence information of the leader and core peptide regions could be obtained. After clustering 1.6 million procA ORF sequences, a total of 1,697 OPUs were identified, an impressive number considering it represents a lower bound estimate of prochlorosin diversity due to the constraints on conserved priming sites mentioned above and the conservative clustering procedure.
To determine how similar the prochlorosin populations are from different oceanic regions, we mapped the procA ORF sequences from each site to the total OPU set to create an OPU composition matrix. Rarefaction analysis shows that the number of OPUs sampled globally does not tend toward an asymptote, but those for each site are closer to saturation (Fig. 3D). Suspecting that this reflects variation in the composition of OPU populations between sampling sites, we constructed a dissimilarity matrix based on the OPU composition at each site and analyzed it using principal coordinates analysis (Materials and Methods). We found that prochlorosin populations primarily cluster by geographic location (i.e., all depths from the same station formed close groups) (Fig. 3E). Furthermore, analysis of similarities (ANOSIM) between geographic locations indicates that there is little overlap between the OPU populations, demonstrating that different picocyanobacterial populations encode distinct sets of lanthipeptide genes (Fig. 3E). The only instance in which two different sampling sites showed a significant overlap between their prochlorosin populations was for two stations in the South Atlantic (AMT-S72 and AMT-S78, ANOSIM, R = 0.67; P value <0.05), both of which were heavily dominated by Synechococcus and under a similar deep-mixing regime (Fig. 3C). Among the time-series samples from Station ALOHA for HOT, there was a significant overlap between the OPU populations for the majority of the monthly samples from the 125-m and 175-m depths (ANOSIM, R = 0.24; P value < 0.05). However, despite the overall cohesion of prochlorosin populations from Station ALOHA, we observed a few samples distant from the main cluster (25-, 85-, and 100-m depths and time-series samples from 3 mo in the fall and 1 mo in the winter at 125 m), indicating that environmental gradients along the water column and seasonal changes can also influence the composition of prochlorosin populations at this site.
That picocyanobacterial populations from different oceanic environments harbor significantly different sets of prochlorosin precursor peptide genes suggests that selection pressures operating at each site drive the evolution of divergent lanthipeptide structures within this group of closely related microorganisms. This led us to wonder about the full extent of the structural differences between prochlorosins in these natural populations and how this diversity might originate.
Hypervariability of Prochlorosin Structures in Wild Populations.
To explore the scope of structural variability among prochlorosins observed in wild populations, we examined in detail the sequence information of the leader and core peptide regions of the 1,697 OPUs identified. At the nucleotide level, the majority of the OPUs share only 40–70% identity, indicating extensive sequence diversity in the wild (Fig. 4A). At the protein level, the identity of the core peptide region among OPUs is less than 30% for most (98.1%) of the sequence comparisons, whereas the leader peptide is relatively more conserved with nearly half of the sequence comparisons showing greater than 60% pairwise amino acid identity (Fig. 4B). Moreover, in these sequences the vast majority of OPUs that display high identity between their leader peptides have low identity between their core peptide regions (Fig. 4C). This metagenomic analysis, which enables a sampling scope far exceeding the limitations of cultured genomes, indicates that prochlorosins with similar core peptides are rare in the natural environment, consistent with our observations using a limited number of cultured isolates.
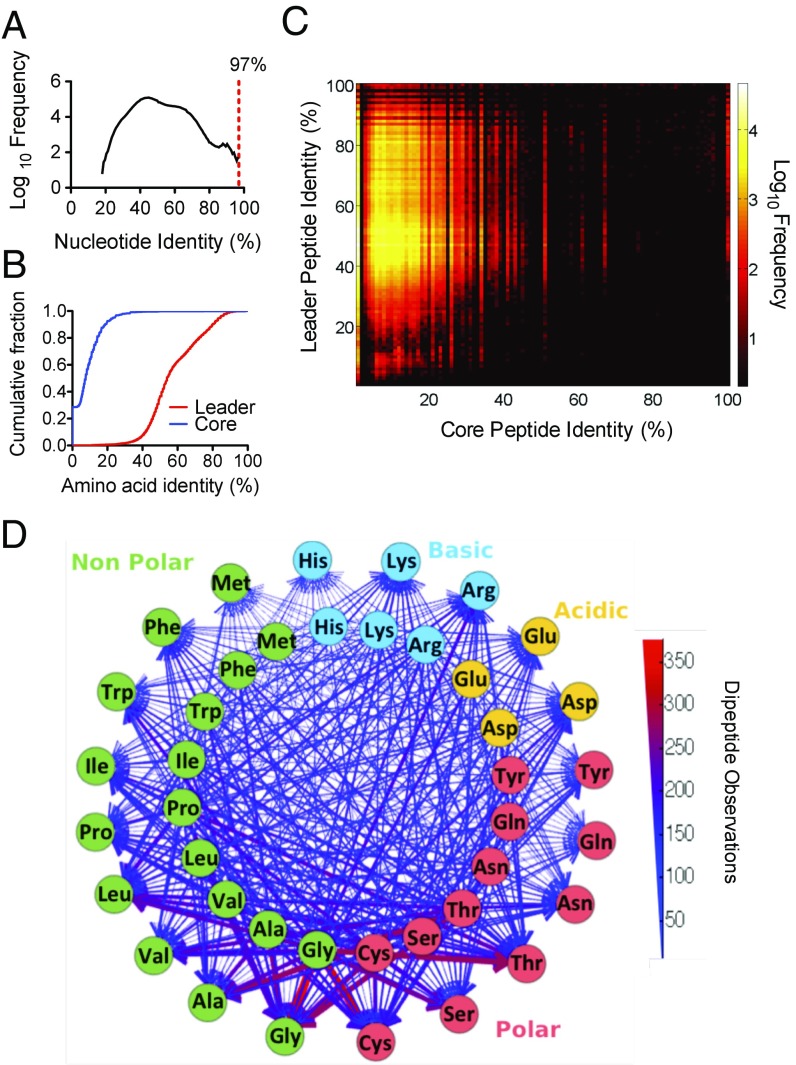
Fig. 4.
The sequence diversity of OPUs from wild populations. (A) Frequency distribution of nucleotide identities among pairwise comparisons in a multiple alignment of all OPUs. The red dashed line represents the 3% distance cutoff used in the procA ORF clustering. (B) Cumulative distribution function of the amino acid identity of the leader and core peptide regions in the dataset. (C) Amino acid identity matrix heat map with every pairwise OPU comparison in a multiple sequence alignment for the core peptide and leader peptide regions. (D) Dipeptide composition of the prochlorosin core peptides identified in the OPUs. Inner and outer circles correspond to the N-terminal and C-terminal amino acid positions, respectively. Lines connecting amino acids represent the dipeptide observations. Color and width of the lines are weighted for the number of dipeptide observations in the dataset.
To investigate whether the core regions within this extensively diverse set of OPUs from wild cells possess structural properties that could actually result in a cyclic peptide, we first determined the co-occurrence of Cys/Ser and Cys/Thr pairs in the core regions and found that 81% of the OPUs could result in a lanthipeptide with at least one ring. We next calculated the dipeptide composition of all core regions in the set of OPUs to identify the amino acid associations that might represent underlying structural commonalities. The most frequent dipeptides were a polar amino acid involved in cyclization (Cys, Ser, or Thr) associated with a small side-chain–containing nonpolar amino acid (Gly or Ala) (Fig. 4D) (23–25). This dipeptide signature is also observed in core regions of prochlorosin precursor peptides from genomes of cultured strains and is different from the dipeptide frequency observed for the general proteome of prochlorosin-encoding strains (SI Appendix, Fig. S9). Thus, despite the lack of sequence conservation between most of the OPU core peptides, the OPUs contain the general structural signatures of cyclic peptides. This metagenomic survey suggests that natural picocyanobacterial populations collectively have the potential to produce thousands of structurally different lanthipeptides.
Molecular Diversification of Prochlorosin Precursor Peptides.
What are the underlying evolutionary mechanisms that might generate this expansive diversity? Analysis of the procA genes found in the set of cultured strains alone cannot provide a clear picture of how the expansion and diversification of these genes takes place because the variation in the core region of the procA gene is so extreme that meaningful multiple sequence alignments cannot be obtained for the 3′ end of the gene. This extreme variation precludes phylogenetic reconstruction for the entire precursor peptide. Using only the leader peptide region, however, one can generate a partial reconstruction of the phylogeny of these genes (SI Appendix, Fig. S10). Prochlorococcus leader peptides generally group into a distinct branch separate from most Synechococcus leader peptides, consistent with the phylogeny of these two genera. These extreme differences in the degree of genetic variability between the leader and core peptide regions suggest an atypical molecular evolution mechanism for the procA gene. To gain insight into the evolutionary mechanisms that could generate this abnormal diversity, we analyzed the genetic variation of the prochlorosin precursor peptide in the OPUs obtained from wild populations.
To explore possible early steps in the diversification of the core peptide region of the procA gene, and to characterize the types of molecular changes that give rise to new lanthipeptide structures, we searched for OPUs that share 100% nucleotide identity in their leader peptide region, allowing us to identify groups of genes that might have undergone recent changes in their core peptide region. We identified 156 such groups of OPUs with identical leader peptides, 7 of which contained more than 7 OPUs. The core peptide regions within these groups were 0–88% identical—a striking range of variation considering that they have identical leader peptides (Fig. 5A). To further explore variation at the 3′ end of the procA gene in these groups, we recovered sequence information from the 3′ intergenic region downstream of the procA ORFs that had previously been removed as part of the read processing and clustering procedure (Materials and Methods). Multiple sequence alignments using the full length of the procA genes revealed that the 3′ intergenic region downstream of the ORF is highly conserved within these sets, further suggesting that the changes within the core peptide region of these procA genes reflect recent events (Fig. 5B).
Fig. 5.
Genetic variation and molecular diversification of procA genes. (A) Amino acid identity (as percentage of aligned sequence ignoring indels) matrix of the core peptide regions (Upper triangle) of seven groups (G1–7) of OPUs selected from environmental sequences that display complete conservation of their leader peptide sequence (Lower triangle). (B) Variation in the procA gene region of the G3 group of OPUs. DNA multiple sequence alignment of the procA genes indicates the presence of deletion events that occurred on the OPU116 ancestor. The predicted core peptide regions resulting from the diversification of this gene region are presented below the alignment. Cys, Ser, and Thr are highlighted to note that, given the positions of these residues, each peptide would display a different ring topology. Numbers on the left correspond to OPU designations.
Using sequence information from the entire procA gene, it is possible to examine the molecular changes that gave rise to new procA variants in these closely related OPU groups. For example, examination of the nine procA genes in the G3 group of recently diverged OPUs revealed that OPU116 is the likely ancestor of eight other OPUs, which all arose as the result of individual deletion events in various parts of the 3′ end of the gene (Fig. 5B). Five of these genes suffered deletion events that included several nucleotides outside of the coding region of the original OPU116. Compared with their ancestor OPU116, all of these deletions resulted in procA ORFs with core regions displaying large differences in their sequence composition, but that still contain Cys, Ser, and Thr residues in different positions that could serve as substrates for the creation of lanthionine bridges (Fig. 5B). Thus, in a very short mutational path a single procA gene region rapidly diversified into eight prochlorosin precursor peptide genes, the products of which would display completely different ring topologies.
The sequence variation patterns in the other groups of closely related OPUs were also dominated by multiple insertion-deletion (indel) events of variable lengths, indicating that the majority of polymorphisms in the core peptide region of recently diverged procA genes resulted in large changes in the amino acid sequence of the final product (SI Appendix, Fig. S11). This pattern of variation implies that the diversity in the core peptide region of prochlorosins is not likely to have resulted from a slow multistep exploration of the sequence space around a particular ring topology, as for variants in other lanthipeptide groups (26), but rather from a single-step diversification process that rapidly explores very large changes in sequence composition. In contrast, in groups of OPUs with 100% nucleotide identity in their core peptide region, variations in the leader peptide region were dominated by single-nucleotide substitutions (SI Appendix, Fig. S12).
These contrasting variation patterns indicate that the leader and core regions of the prochlorosin precursor peptide gene are undergoing distinct modes of molecular evolution. The leader peptide, the function of which is to direct biosynthesis, follows the molecular evolution pattern of most proteins where large sequence changes are thought to be detrimental to the protein structure, and thus only small changes in the exploration of the sequence space are permitted. Conversely, the core peptide evolves mainly by the action of large sequence polymorphisms that greatly affect the overall sequence of the core peptide region, disregarding any conservation of the ancestral structure.
Structural Diversity Is Under Selection in Picocyanobacterial Lanthipeptides.
In the absence of selection, a diversification process of procA genes that is driven by large indel events is expected to create random peptides. In a simulation of a neutral evolution process of 100 procA genes driven by indel mutations in the 3′ end of the gene, the resulting population of potentially cyclic peptides would not exceed 50%, and the expected amino acid composition of the pool of core peptides tends to be more similar to the general proteome composition of marine picocyanobacteria (SI Appendix, Fig. S13). Therefore, the fact that 91% of the prochlorosin core peptides identified in the genomes and 81% of the core peptides in the OPUs contain residues and structural features that favor the formation of a cyclic product indicates that prochlorosin core peptide diversity is not likely the result of genetic drift. In addition, as evidenced by the complete diversity (no core peptide is highly similar to any other) of procA genes found within genomes of cultured strains, selection also seems to allow only the maintenance of lanthipeptide variants that display a different ring topology from the rest of the prochlorosins in the genome. Also, the prevalence of procA pseudogenes in prochlorosin-encoding cells supports the notion of a constant selective pressure on the prochlorosin trait that inactivates procA genes that are no longer advantageous, thus relieving the cell from having to produce such peptides. In turn, this observation also suggests that the extant prochlorosins have a functional role.
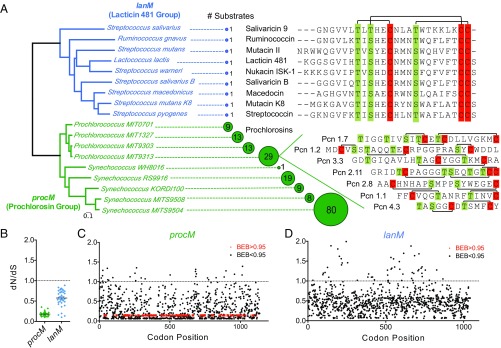
Additional evidence for the selection of traits in the prochlorosin biosynthesis pathway that favor structural diversification comes from an analysis of the evolutionary interplay between lanthionine synthetases and their peptide substrates. Does the rapid diversification of the prochlorosin core peptide substrates demand a fast evolution of the lanthionine synthetase? To explore this, we compared the patterns of natural selection on LanM enzymes from the lacticin 481 group, which modify a single precursor peptide substrate, the putative ring topologies of which are conserved (26, 27), to patterns of selection on ProcM enzymes within individual strains of picocyanobacteria harboring multiple precursor peptides of different ring topologies (Fig. 6A). Genes from the ProcM clade display a very low ratio of the rate of nonsynonymous substitutions to the rate of synonymous substitutions (dN/dS) compared with the enzymes of the lacticin 481 (LanM) clade, which is indicative of negative selection operating on the former group (Fig. 6B). In the case of the lanthionine synthetases from the ProcM clade, even when their peptide substrates rapidly expand and diversify within a genome, about 25% of the codons in the ProcM enzyme evolve under a strong purifying selection (Fig. 6C). This level of conservation is not observed in members of the lacticin 481 group, the substrates of which experience less drastic changes than the prochlorosins (Fig. 6D). This interplay between enzyme and substrates suggests that the prochlorosin lanthionine synthetase is evolutionarily locked in a state that favors maintaining its substrate promiscuity. The ProcM sequence analysis further suggests that the evolution of new prochlorosin structures is not constrained by a need for large changes in the lanthionine synthetase. Outstanding examples of this are the Prochlorococcus strains MIT9313 and MIT9303, which encode vastly different sets of prochlorosin precursor peptides, yet harbor procM genes that are 96% identical.
Fig. 6.
Evolutionary history of two lineages of type II lanthionine synthetases. (A) Maximum-likelihood phylogeny of the procM genes of marine picocyanobacteria (green) and lanM genes of the lacticin 481 group found in Firmicutes (blue). The diameter of each circle is proportional to the number of cognate prochlorosin substrates (ProcA) of each ProcM enzyme. Sequence alignments of core peptide regions of members of the lacticin 481 group show the conservation of ring topology (black lines above sequence) among variants. Ring topologies for the 7 (of 29) prochlorosins from Prochlorococcus MIT9313 that have been determined are presented as an example of the extensive structural diversity in this group (6, 40). (B) Average value of the ratio between dS and dN substitution rates for every pairwise intraclade comparison for the entire length of the procM and lanM genes. (C and D) Codon-by-codon analysis of the synonymous and nonsynonymous substitution rates along lanthionine synthetase genes of the prochlorosin group (procM: C) and the lacticin 481 group (lanM: D). Significant dN/dS values are shown in red (BEB values greater than 0.95) indicating codons under purifying selection.
Conclusion.
This cross-scale analysis of prochlorosin precursor peptide genes reveals that the hypervariability of prochlorosins is a feature that occurs across multiple scales of complexity. At the genome level, all of the procA genes found within a single genome are different (Fig. 2). Between genomes, prochlorosin-encoding genomes from several picocyanobacterial lineages harbor distinctive sets of procA genes (Fig. 2). In the global ocean, geographically distinct marine picocyanobacterial populations contain largely dissimilar collections of procA genes (Fig. 3). Collectively, these findings indicate that lanthipeptides in marine picocyanobacteria are undergoing a rapid evolutionary process of structural diversification. It appears that the prochlorosin biosynthesis pathway has evolved such that it can generate a suite of structurally diverse natural products by using the combination of a substrate-tolerant lanthionine synthetase and a collection of precursor peptide substrates encoded in a highly dynamic family of multicopy genes poised for rapid expansion and diversification.
This type of diversity-prone selection contrasts with the evolutionary path of other lanthipeptides such as lantibiotics, which display antimicrobial activity (15). Lantibiotic biosynthesis pathways have generally evolved to be efficient in the creation of one cyclic peptide with a defined ring topology that endows the molecule with its bioactivity; the ring topology resulting from posttranslational modifications is thought to be the major determinant of the bioactivity. For example, in lantibiotics of the nisin group or the lacticin 481 group, the targets of which are specific lipid components of Gram-positive bacteria, variations found within their core peptide sequences are composed mainly of small amino acid changes that do not alter the structure of the principal rings, thereby preserving the antimicrobial activity (27, 28). Consequently, lantibiotics follow the evolutionary pattern of a typical protein with an established structure–function relationship, where structural similarity is maintained through negative selection. The case of prochlorosins is different: the predominance of prochlorosins that would display extensively different ring topologies, and the high frequency of large sequence polymorphisms that tend to change the structure of the core peptide, suggest that the potential bioactivity of the prochlorosins does not have a relationship with a particular ring topology. Rather, the evolutionary history of prochlorosins suggests that the selective advantage of the trait does not rely solely on the function afforded by the structure of a single peptide, but rather on the plasticity in the production of a suite of cyclic peptides with diverse ring topologies and their sustained diversification. Previous examples of pathways resulting in many structurally diverse products have been interpreted by the “screening hypothesis” (29), but, in the absence of definitive information on the function of prochlorosins, we believe it is premature to do so for the prochlorosins.
Aside from lanthipeptides, other RiPP families are known to display hypervariability in the core region of their precursor peptide substrates (30). Cyanobactins, for example, are a group of structurally diverse RiPPs, the biosynthetic enzymes of which also have remarkable substrate promiscuity in the creation of these small cyclic peptides (31). Similar to prochlorosins, some cyanobactin variants arise from duplication and diversification events in their precursor peptide genes (32). However, in contrast to prochlorosins, natural genetic variation patterns found in precursor peptides of the patellamide family of cyanobactins from tunicate-associated cyanobacteria suggest that their evolutionary diversification occurs through small stepwise changes, somewhat constrained by the maintenance of conserved residues and the overall length of the peptide (33). Not all families of cyanobactins, however, display marked similarities between closely related variants of precursor peptide genes. For example, analysis of the genome of Planktothrix agardhii (34) reveals that the prenylagaramide biosynthetic gene cluster harbors six precursor peptide genes that display very low similarity in their core peptide region, suggesting that variants within this family could have arisen through an evolutionary process similar to that of prochlorosins. Furthermore, genome mining of other RiPPs such as the thiazole/oxazole-modified microcin family revealed the presence of cyclic peptide biosynthetic gene clusters in bacteria other than cyanobacteria. These clusters resemble the prochlorosin pathway in terms of the multiplicity of precursor peptide genes per genome and their striking core peptide sequence diversity, suggesting that evolutionary diversification of natural products might occur in other branches of the bacterial phylogeny (25).
Materials and Methods
Detailed materials and methods for Prochlorococcus isolations, genome sequencing, qPCR detection of procA genes, amplicon library construction, and sequence processing and analysis are provided in SI Appendix. For the clustering of procA amplicons, a total of 1.6 million procA ORF sequences were predicted from the pool of cleaned reads, for an average of 41,000 per sample. These reads were then used to create 3% similarity clusters using the cluster_otu function of USEARCH (35). The resulting 3% similarity clusters were mapped back to the original procA ORF reads from each one of the sites sampled using USEARCH with a DNA identity threshold of 0.97. The resulting nonsingleton clusters from this mapping are considered to be OPUs (Dataset S1). For the comparative analysis of OPU populations, we constructed a dissimilarity matrix based on the OPU composition at each site using the Jaccard distance and used principal coordinates analysis to represent the similarity between samples in ordination space. In addition, ANOSIM was used to assess the statistical significance of the observed Jaccard distances between groups of samples from different locations. Codon-specific dN/dS analysis of lanthionine synthetase genes of the procM and lacticin 481 lanM groups was calculated using codeml from the PAML package (36, 37) implemented with a user-provided maximum-likelihood tree and the M8 Model (beta&ω > 1). Selection indices for each codon were considered significant when Bayes Empirical Bayes (BEB) probabilities were BEB ≥ 0.95.
Supplementary Material
Acknowledgments
We thank the crew and the science party that participated in the Hawaii Ocean Experiment-Phosphorus Rally (HOE-PhoR) expedition for assistance with sampling. The work was supported in part by the following grants to S.W.C.: Grant GBMF495 from the Gordon and Betty Moore Foundation; National Science Foundation Science and Technology Center for Microbial Oceanography Research and Education (C-MORE); and grants from the Simons Foundation [Simons Collaboration on Ocean Processes and Ecology (SCOPE) Award 329108-SWC; Life Sciences 337262]. A.C.-R. was supported by a Howard Hughes Medical Institute international student research fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Genomes described in this work have been deposited in the National Center for Biotechnology Information databases and their accession numbers are listed in SI Appendix, Table S4. Sequence data related to the metagenomic procA genes have been deposited in the Sequence Read Archive (SRA) database, https://www.ncbi.nlm.nih.gov/sra (BioProject ID: PRJNA387049).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700990114/-/DCSupplemental.
References
- 1.Romero D, Traxler MF, López D, Kolter R. Antibiotics as signal molecules. Chem Rev. 2011;111:5492–5505. doi: 10.1021/cr2000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Traxler MF, Kolter R. Natural products in soil microbe interactions and evolution. Nat Prod Rep. 2015;32:956–970. doi: 10.1039/c5np00013k. [DOI] [PubMed] [Google Scholar]
- 3.Donia MS, et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158:1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charlop-Powers Z, Owen JG, Reddy BV, Ternei MA, Brady SF. Chemical-biogeographic survey of secondary metabolism in soil. Proc Natl Acad Sci USA. 2014;111:3757–3762. doi: 10.1073/pnas.1318021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ziemert N, et al. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc Natl Acad Sci USA. 2014;111:E1130–E1139. doi: 10.1073/pnas.1324161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li B, et al. Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc Natl Acad Sci USA. 2010;107:10430–10435. doi: 10.1073/pnas.0913677107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scanlan DJ, et al. Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev. 2009;73:249–299. doi: 10.1128/MMBR.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biller SJ, Berube PM, Lindell D, Chisholm SW. Prochlorococcus: The structure and function of collective diversity. Nat Rev Microbiol. 2015;13:13–27. doi: 10.1038/nrmicro3378. [DOI] [PubMed] [Google Scholar]
- 9.Willey JM, van der Donk WA. Lantibiotics: Peptides of diverse structure and function. Annu Rev Microbiol. 2007;61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- 10.Chatterjee C, Paul M, Xie L, van der Donk WA. Biosynthesis and mode of action of lantibiotics. Chem Rev. 2005;105:633–684. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- 11.Kuipers OP, Beerthuyzen MM, de Ruyter PG, Luesink EJ, de Vos WM. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J Biol Chem. 1995;270:27299–27304. doi: 10.1074/jbc.270.45.27299. [DOI] [PubMed] [Google Scholar]
- 12.Schmitz S, Hoffmann A, Szekat C, Rudd B, Bierbaum G. The lantibiotic mersacidin is an autoinducing peptide. Appl Environ Microbiol. 2006;72:7270–7277. doi: 10.1128/AEM.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willey JM, Willems A, Kodani S, Nodwell JR. Morphogenetic surfactants and their role in the formation of aerial hyphae in Streptomyces coelicolor. Mol Microbiol. 2006;59:731–742. doi: 10.1111/j.1365-2958.2005.05018.x. [DOI] [PubMed] [Google Scholar]
- 14.Skinnider MA, et al. Genomic charting of ribosomally synthesized natural product chemical space facilitates targeted mining. Proc Natl Acad Sci USA. 2016;113:E6343–E6351. doi: 10.1073/pnas.1609014113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knerr PJ, van der Donk WA. Discovery, biosynthesis, and engineering of lantipeptides. Annu Rev Biochem. 2012;81:479–505. doi: 10.1146/annurev-biochem-060110-113521. [DOI] [PubMed] [Google Scholar]
- 16.Thibodeaux CJ, Ha T, van der Donk WA. A price to pay for relaxed substrate specificity: A comparative kinetic analysis of the class II lanthipeptide synthetases ProcM and HalM2. J Am Chem Soc. 2014;136:17513–17529. doi: 10.1021/ja5089452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischbach MA, Walsh CT, Clardy J. The evolution of gene collectives: How natural selection drives chemical innovation. Proc Natl Acad Sci USA. 2008;105:4601–4608. doi: 10.1073/pnas.0709132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Q, Yang X, Wang H, van der Donk WA. High divergence of the precursor peptides in combinatorial lanthipeptide biosynthesis. ACS Chem Biol. 2014;9:2686–2694. doi: 10.1021/cb500622c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malmstrom RR, et al. Temporal dynamics of Prochlorococcus ecotypes in the Atlantic and Pacific oceans. ISME J. 2010;4:1252–1264. doi: 10.1038/ismej.2010.60. [DOI] [PubMed] [Google Scholar]
- 20.Johnson ZI, et al. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science. 2006;311:1737–1740. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- 21.Zwirglmaier K, et al. Basin-scale distribution patterns of picocyanobacterial lineages in the Atlantic Ocean. Environ Microbiol. 2007;9:1278–1290. doi: 10.1111/j.1462-2920.2007.01246.x. [DOI] [PubMed] [Google Scholar]
- 22.Karl DM, Church MJ. Microbial oceanography and the Hawaii Ocean time-series programme. Nat Rev Microbiol. 2014;12:699–713. doi: 10.1038/nrmicro3333. [DOI] [PubMed] [Google Scholar]
- 23.Ennahar S, Sashihara T, Sonomoto K, Ishizaki A. Class IIa bacteriocins: Biosynthesis, structure and activity. FEMS Microbiol Rev. 2000;24:85–106. doi: 10.1111/j.1574-6976.2000.tb00534.x. [DOI] [PubMed] [Google Scholar]
- 24.Jack RW, Tagg JR, Ray B. Bacteriocins of gram-positive bacteria. Microbiol Rev. 1995;59:171–200. doi: 10.1128/mr.59.2.171-200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haft DH, Basu MK, Mitchell DA. Expansion of ribosomally produced natural products: A nitrile hydratase- and Nif11-related precursor family. BMC Biol. 2010;8:70. doi: 10.1186/1741-7007-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q, Yu Y, Vélasquez JE, van der Donk WA. Evolution of lanthipeptide synthetases. Proc Natl Acad Sci USA. 2012;109:18361–18366. doi: 10.1073/pnas.1210393109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dufour A, Hindré T, Haras D, Le Pennec JP. The biology of lantibiotics from the lacticin 481 group is coming of age. FEMS Microbiol Rev. 2007;31:134–167. doi: 10.1111/j.1574-6976.2006.00045.x. [DOI] [PubMed] [Google Scholar]
- 28.Cotter PD, Hill C, Ross RP. Bacterial lantibiotics: Strategies to improve therapeutic potential. Curr Protein Pept Sci. 2005;6:61–75. doi: 10.2174/1389203053027584. [DOI] [PubMed] [Google Scholar]
- 29.Firn RD, Jones CG. Natural products: A simple model to explain chemical diversity. Nat Prod Rep. 2003;20:382–391. doi: 10.1039/b208815k. [DOI] [PubMed] [Google Scholar]
- 30.Arnison PG, et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donia MS, et al. Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat Chem Biol. 2006;2:729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 32.Lin Z, Torres JP, Tianero MD, Kwan JC, Schmidt EW. Origin of chemical diversity in prochloron-tunicate symbiosis. Appl Environ Microbiol. 2016;82:3450–3460. doi: 10.1128/AEM.00860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt EW, Donia MS, McIntosh JA, Fricke WF, Ravel J. Origin and variation of tunicate secondary metabolites. J Nat Prod. 2012;75:295–304. doi: 10.1021/np200665k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donia MS, Schmidt EW. Linking chemistry and genetics in the growing cyanobactin natural products family. Chem Biol. 2011;18:508–519. doi: 10.1016/j.chembiol.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 36.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 37.Xu B, Yang Z. PAMLX: A graphical user interface for PAML. Mol Biol Evol. 2013;30:2723–2724. doi: 10.1093/molbev/mst179. [DOI] [PubMed] [Google Scholar]
- 38.Scanlan DJ. Marine picocyanobacteria. In: Whitton BA, editor. Ecology of Cyanobacteria II: Their Diversity in Space and Time. Springer Netherlands; Dordrecht: 2012. pp. 503–533. [Google Scholar]
- 39.Litwin S, Jores R. Shannon information as a measure of amino acid diversity. In: Perelson A, Weisbuch G, editors. Theoretical and Experimental Insights into Immunology. Vol 66. Springer; Paris: 1992. pp. 289–296. [Google Scholar]
- 40.Zhang Q, et al. Structural investigation of ribosomally synthesized natural products by hypothetical structure enumeration and evaluation using tandem MS. Proc Natl Acad Sci USA. 2014;111:12031–12036. doi: 10.1073/pnas.1406418111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.