Significance
Sex chromosomes can display divergent evolution, as seen in humans, in which the Y chromosome underlying maleness is smaller and contains much less information than the X chromosome. The differentiation between sex chromosomes can occur stepwise along their length, which is thought to result from the successive beneficial linkage of genes with different phenotype optima in the two sexes to sex-determining genes. However, there is little evidence to support this hypothesis. Here, we recovered ancestral chromosome structures and gathered evidence for stepwise differentiation between fungal mating-type chromosomes despite the absence of male/female roles. Our results suggest that the analogous features of sex chromosomes may not be due to differences in selection between males and females.
Keywords: evolutionary strata, chromosomal rearrangements, fungi, genomic degeneration, mating-type chromosomes
Abstract
Sex chromosomes can display successive steps of recombination suppression known as “evolutionary strata,” which are thought to result from the successive linkage of sexually antagonistic genes to sex-determining genes. However, there is little evidence to support this explanation. Here we investigate whether evolutionary strata can evolve without sexual antagonism using fungi that display suppressed recombination extending beyond loci determining mating compatibility despite lack of male/female roles associated with their mating types. By comparing full-length chromosome assemblies from five anther-smut fungi with or without recombination suppression in their mating-type chromosomes, we inferred the ancestral gene order and derived chromosomal arrangements in this group. This approach shed light on the chromosomal fusion underlying the linkage of mating-type loci in fungi and provided evidence for multiple clearly resolved evolutionary strata over a range of ages (0.9–2.1 million years) in mating-type chromosomes. Several evolutionary strata did not include genes involved in mating-type determination. The existence of strata devoid of mating-type genes, despite the lack of sexual antagonism, calls for a unified theory of sex-related chromosome evolution, incorporating, for example, the influence of partially linked deleterious mutations and the maintenance of neutral rearrangement polymorphism due to balancing selection on sexes and mating types.
Chromosomes carrying genes controlling mating compatibility may display suppressed recombination, as seen in sex chromosomes in some animals and plants (1, 2). When recombination is suppressed and one of the sex chromosomes is always in a heterozygous state (e.g., the Y chromosome in humans), extensive rearrangements of that chromosome typically occur. Our understanding of how sex chromosomes evolve remains incomplete, with many unanswered questions (3), including the reasons why these chromosomes display such large regions of suppressed recombination, extending well beyond the sex-determining genes. Sex chromosomes may bear discrete “evolutionary strata” of differentiation between alleles, with divergence decreasing with distance from sex-determining genes in the ancestral gene order, as inferred from the nonrearranged sex chromosome (1). It is generally accepted that such strata result from successive rounds of selection favoring the linkage of sexually antagonistic genes (with alleles beneficial in one sex but detrimental in the other) and sex-determining loci (1, 4). However, there is no definitive evidence to support a prominent role for sexually antagonistic genes in the formation of evolutionary strata in sex chromosomes (3). Other hypotheses have been put forward, including the permanent sheltering of deleterious alleles, which may accumulate in the margins of nonrecombining regions, provided that recombination rates are low in these regions. Another hypothesis is the fixation of neutral rearrangements by drift in one of the two sex chromosomes with automatic recombination arrest and the maintenance of a polymorphic state due to balancing selection on sexes (2, 5–8) (SI Appendix, SI Text and Fig. S1).
Recombination suppression has been documented in fungal mating-type chromosomes (9, 10), which carry key loci determining mating compatibility. In basidiomycetes, mating type is typically controlled by two loci: (i) the pheromone receptor (PR) locus, which carries one gene encoding a mating-type–specific PR and one to several genes encoding pheromones, and (ii) the homeodomain gene (HD) locus, encoding two homeodomain-type transcription factors controlling postmating growth. Recombination suppression ensures full linkage of the mating-type genes within each of these two loci, which is required for correct mating-type determination (11), and typically does not extend beyond the mating-type genes (12, 13). Recombination suppression in fungal mating-type chromosomes can further link the two mating-type loci, HD and PR, to each other and/or the centromere (9, 14–18). Such linkage is beneficial in species with selfing-based mating systems, as it increases the odds of compatibility between gametes from a diploid parent (16, 19, 20) (SI Appendix, SI Text and Fig. S2). It has been repeatedly suggested that some fungal mating-type chromosomes have additional evolutionary strata extending the suppression of recombination beyond mating-type genes (9, 18, 21, 22) (SI Appendix, SI Text). However, no evidence has been provided of multiple discrete regions in which divergence between fungal mating-type–associated alleles decreases with increasing distance from sexual compatibility loci. Uncertainty about fungal evolutionary strata persists due to difficulties in inferring the ancestral gene order, which is essential for reconstructions of the evolution of mating-type chromosomes. If fungi have evolutionary strata not involving the linkage of mating-type genes, their presence cannot be explained by sexually antagonistic selection, as fungal mating types are not associated with male/female functions. Moreover, there are virtually no differences in ecological or life-history traits between fungal mating types (23), making it unlikely that there is any analogous mating-type–antagonistic selection improving the function of one mating type while having deleterious effects on the function of the other.
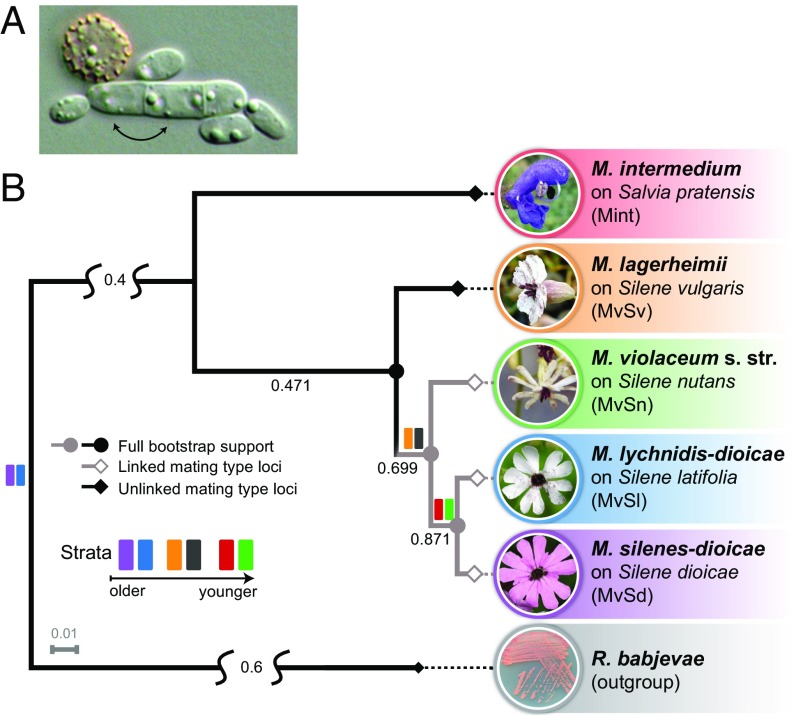
We investigated the existence of evolutionary strata in Microbotryum lychnidis-dioicae, a castrating anther-smut fungus. This species is particularly suited for determining whether the stepwise evolution of suppressed recombination can occur without sexual antagonism. It has two alternative mating-type chromosomes with a large region of suppressed recombination (10, 17, 24, 25), complete absence of male/female function, and virtually no opportunity for mating-type–antagonistic selection. Indeed, almost all matings occur within the tetrad (26–28) with the isogamous gametes fusing rapidly after meiosis without mitotic proliferation in a free haploid stage (Fig. 1A). There has been much debate about the existence of evolutionary strata in the M. lychnidis-dioicae mating-type chromosomes (17, 21, 22, 25) (SI Appendix, SI Text). Genes with different levels of synonymous divergence between alleles linked to the two mating types (called a1 and a2) are widely dispersed over 90% of the chromosome length, suggesting that, if there were any evolutionary strata, they have been erased by massive rearrangements (17). As in all fungi with distinct mating types, and in contrast to plant and animal sex chromosomes, M. lychnidis-dioicae mating-type chromosomes are always heterozygous in the diploid stage, so neither conserves the ancestral gene order (17). This precludes the typical use of the nonrearranged chromosome for the detection of evolutionary strata (1). The recombining regions in M. lychnidis-dioicae, such as the autosomes and the pseudoautosomal regions (PARs), are, however, highly collinear (17) (SI Appendix, Fig. S3), suggesting that the ancestral gene order should be retained in mating-type chromosomes undergoing recombination, even in selfing species. We use the term “autosome” as a contrast to “dimorphic” chromosomes, as originally intended (29, 30), rather than in opposition to sex chromosomes per se. Similarly, PAR is used to name the genomic regions at the edges of the mating-type chromosomes that are collinear and recombine like autosomes.
Fig. 1.
Anther-smut fungi. (A) M. lychnidis-dioicae tetrad with a cell in the spore and three cells in a horizontal promycelium; mating occurs rapidly after meiosis within the tetrad (black arrows). Some haploid cells are budding, but almost 100% of matings occur within the promycelium. (B) Microbotryomycete phylogenetic tree based on 4,000 orthologous genes, including the studied Microbotryum species (shown in the anthers of their host plants) and the outgroup Rhodotorula babjevae. Branch color and symbol indicate linked (gray branches and open diamonds) or unlinked (black branches and closed diamonds) mating-type loci. The closed circles indicate full bootstrap support. Internode certainty with no conflict bipartitions is given below the branches (i.e., the normalized frequency of the most frequent bipartition across gene genealogies relative to the summed frequencies of the two most frequent bipartitions), indicating good support for the nodes. Relative tree certainty of the tree was 0.68. Colored rectangles along the branches indicate the evolution of the various evolutionary strata (see Fig. 3).
We compiled chromosome-length genome assemblies for five Microbotryum species and adopted an approach in which species carrying mostly recombining mating-type chromosomes were identified and used to recover the ancestral chromosomal state and gene order. Using this method, we were able to reconstruct the rearrangements leading to the current nonrecombining mating-type chromosomes in the selfing M. lychnidis-dioicae. One important step involved chromosomal rearrangements and fusion of the two separate ancestral mating-type chromosomes, linking the two mating-type loci in the same chromosome, which is beneficial under selfing (although not required) (SI Appendix, Fig. S2) (16). This finding sheds light on the genomic evolutionary steps leading to changes in breeding systems in fungi. We also provide compelling evidence for multiple, clearly differentiated and ancestrally adjacent evolutionary strata in fungal mating-type chromosomes with several strata devoid of genes controlling mating types. The existence of successive extensions of the region of recombination suppression beyond the linkage of mating-type genes in fungi without male/female functions indicates that evolutionary strata can occur without sexual antagonism. This finding has profound implications for our understanding of the evolutionary genomics of sexual eukaryotes across three kingdoms, highlighting the need for more serious consideration of alternative theoretical models of sex-related chromosome evolution beyond sexually antagonistic processes.
Results and Discussion
Ancestral Gene Order in Microbotryum Mating-Type Chromosomes.
We obtained high-quality assemblies of haploid genomes for two Microbotryum species expected to have retained the ancestral gene order: Microbotryum intermedium, a distant relative of M. lychnidis-dioicae (Fig. 1B) (31, 32), and Microbotryum lagerheimii, a species known to have unlinked HD and PR mating-type loci (32). As hypothesized, M. intermedium was found to have the PR and HD loci in two different chromosomes that were highly collinear with the mating-type chromosomes of M. lagerheimii (Fig. 2A). The collinearity between these distantly related species indicated a high degree of gene order conservation across multiple speciation events (31), therefore closely reflecting the ancestral state in this clade. The ancestral gene order has probably been maintained by regular recombination, as also indicated by the collinearity between the a1 and a2 mating-type chromosomes in M. lagerheimii (SI Appendix, Fig. S4), typical of recombining genomic regions (SI Appendix, Fig. S3).
Fig. 2.
Comparison of gene order across Microbotryum mating-type chromosomes. The mating-type HD and PR genes are indicated by small black and gray circles, respectively. Blue and orange lines link regions with collinearity extending over 2 kb, the latter corresponding to inversions. Areas without links correspond to highly rearranged regions. Yellow regions on the outer track indicate centromere-specific repeats (17). (A) Comparison of gene order between the mating-type chromosomes of M. intermedium (Left) and M. lagerheimii (Right). The darkest contig on the Right corresponds to MC16, as discussed in SI Appendix, SI Text. (B) Comparison of a1 (light contig) and a2 (dark contig) M. lychnidis-doicae mating-type chromosomes. The red and green genes in Fig. 4B are indicated in the a1 genome.
Ancient Linkage of Mating-Type Genes at Each of the HD and PR Loci.
We found footprints of ancient recombination suppression linking the essential pairs of interacting mating-type genes at each of the HD and PR loci, as is typical and ancestral in basidiomycete fungi (13). The nucleotide sequences of the a1 and a2 alleles of the PR gene were too differentiated to be aligned (Figs. 3A and 4) and a previous study inferred that these alleles had been diverging for about 370 million years (33). Recombination suppression linking the two HD mating-type genes was also very old (Fig. 3B), as shown by the second-highest level of synonymous divergence (dS) between the a1 and a2 mating types at these genes in both M. lagerheimii and M. lychnidis-dioicae (Fig. 4) and by gene genealogies (34).
Fig. 3.
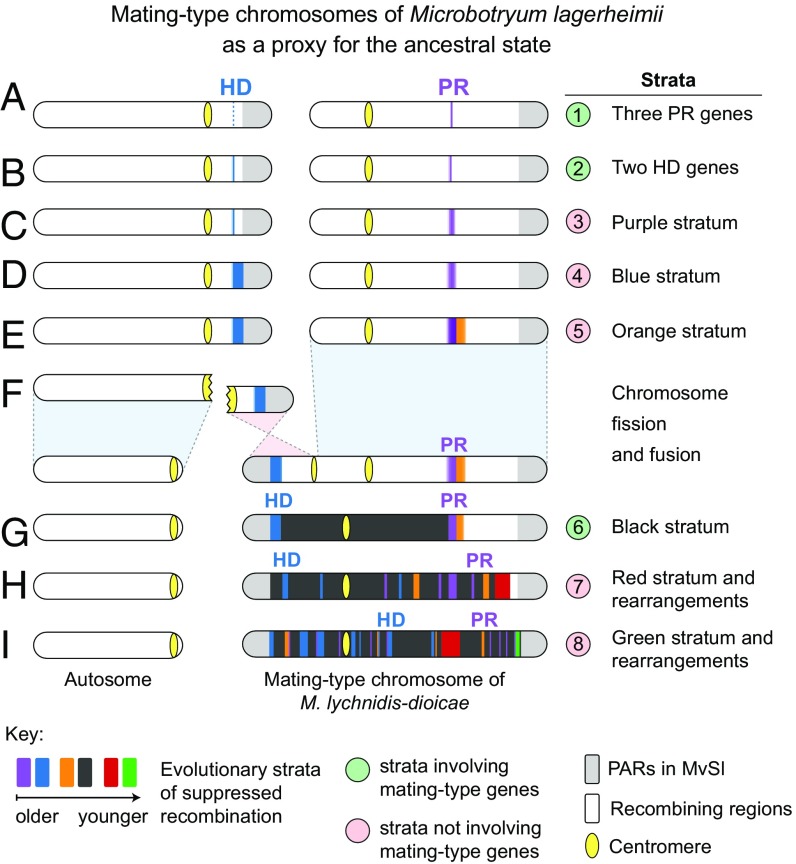
Scenario of mating-type chromosome evolution in the genus Microbotryum. Strata involving the linkage of mating-type genes are numbered in green circles, whereas strata devoid of mating-type genes are numbered in red circles. (A) Ancestral state with separate HD and PR chromosomes and suppressed recombination restricted to the PR mating-type genes. (B) Suppressed recombination at all mating-type genes (as typically seen in basidiomycete fungi, with linked PR and pheromone genes at one locus and the two linked HD genes linked at a second locus). (C) Extension of suppressed recombination to purple nonmating-type genes. (D) Extension of suppressed recombination to blue nonmating-type genes. (E) Extension of suppressed recombination distal to the purple stratum, forming the orange stratum, devoid of mating-type genes. (F) Fission of the HD chromosome and fusion of one arm with the PR chromosome. (G) Complete linkage between the HD and PR mating-type loci, forming the black stratum. (H) Extension of suppressed recombination to nonmating-type genes forming the red stratum and rearrangements shuffling older evolutionary strata. (I) Further extension of suppressed recombination forming the green stratum and further rearrangements, leading to the current M. lychnidis-dioicae (MvSl) mating-type chromosome with the red stratum located between the PR and HD genes.
Fig. 4.
Synonymous divergence (dS) between alleles associated with the a1 and a2 mating types along mating-type chromosomes, as found in the PacBio-sequenced diploid individual in each Microbotryum species. Per-gene mean dS values and SE values. The locations of the mating-type genes (PR and HD) are indicated. The divergence between the a1 and a2 PR alleles was too extensive (33) and could not be computed. It is plotted as an “unalignable” (Un) open purple circle. The M. lagerheimii centromeres are shown in yellow. (A) Synonymous divergence for all single-copy genes common to the M. lagerheimii a1 and a2 mating-type chromosomes plotted according to the genomic coordinates of its a1 mating-type chromosomes. The HD chromosome is displayed on the left and the PR chromosome on the right. Genes with within-individual a1–a2 ds > 0 are shown in blue around HD and purple around PR. The mating-type loci are partially linked to the centromere in M. lagerheimii (32). (B) Synonymous divergence for all single-copy genes common to the M. lychnidis-dioicae mating-type chromosomes and M. lagerheimii, plotted in the ancestral gene order, as inferred from the genomic coordinates of the M. lagerheimii a1 mating-type chromosomes. The limits of the M. lagerheimii PR and HD mating-type chromosomes are indicated, together with the positions of their centromeres. Blue and purple genes correspond to genes orthologous to the blue and purple regions in A. Other evolutionary strata are indicated in orange, black, red, and green. (Inset) Magnification of the youngest strata. The gray gene with nonzero dS value at the right of the green stratum was located within the collinear PAR in Fig. 2B and was separated from the green genes by a small noncollinear region. The PARs in M. lychnidis-dioicae are shown in gray.
Chromosomal Fusion Led to the Large Nonrecombining Region Linking HD and PR Mating-Type Genes in M. lychnidis-dioicae.
A more recent recombination suppression event involved the linkage of the HD and PR mating-type loci to each other (Fig. 3), which is favorable under selfing (15, 16) (SI Appendix, Fig. S2), the predominant mating system in Microbotryum (26, 28). Genome comparisons indicated that the M. lychnidis-dioicae mating-type chromosome was formed by the fusion of the complete ancestral PR chromosome with one arm of the ancestral chromosome carrying the HD locus (Fig. 3 D–F and SI Appendix, Fig. S5A). The remaining ancestral HD chromosome arm became an autosomal component in M. lychnidis-dioicae (Fig. 3 D–F and SI Appendix, Fig. S5A). Based on the inferred ancestral state, the two mating-type loci were initially some 800 kb apart following the fusion event and the fused chromosome was about 2 Mb long. The subsequent cessation of recombination between the PR and HD mating-type loci led to extensive rearrangements (SI Appendix, Fig. S3) and divergence between alleles associated with the a1 and a2 mating types in this region (in black in Fig. 4B and Fig. 3 G–I). Conversely, the chromosome regions ancestrally distal to the PR and HD loci largely constituted PARs that remained collinear (SI Appendix, Fig. S1), with a within-individual dS of 0 (gray in Fig. 4B), as expected for recombining regions in organisms with high selfing rates.
For confirmation of the complete and ancient nature of recombination suppression in the region ancestrally between the HD and PR loci (black in Fig. 4B), we obtained and analyzed high-quality genome assemblies of M. silenes-dioicae and M. violaceum s. str., two species closely related to M. lychnidis-dioicae (Fig. 1B). All three species had homologous mating-type chromosomes (SI Appendix, Fig. S5 B and C), each including the large black nonrecombining region ancestrally located between PR and HD, with a high degree of rearrangement between mating types and between species (SI Appendix, Fig. S5). Most of the genes (66%) in the black region displayed trans-specific polymorphism across M. lychnidis-dioicae and M. silenes-dioicae, with alleles associated with the a1 mating type of both species branching together rather than clustering with the allele associated with the a2 mating type from the same species. Trans-specific polymorphism also extended to the divergence from M. violaceum s. str. in 20% of the genes (SI Appendix, Figs. S6 and S7), which were dispersed along the ancestral gene order in the black stratum (corresponding to the black points with the highest dS values in Fig. 4B). Such old trans-specific polymorphism associated with mating type could not have persisted without an ancient and complete cessation of recombination preceding the divergence of these three species, as even very low levels of recombination would have had time to break down the association between alleles at genes in the black stratum and mating-type genes. The less marked or absent trans-specific polymorphism for some of the genes within the black stratum probably results from occasional localized gene conversion events, as demonstrated for mating-type chromosomes in Microbotryum (35) and other fungi (36, 37). Trans-specific polymorphism in the genealogies of genes in the black stratum never extended to M. lagerheimii, consistent with the lack of linkage between the PR and HD mating-type loci in this species. When calibrated by the previous estimate for the date of the speciation event between M. lychnidis-dioicae and M. silenes-dioicae to 420,000 years ago (38), the divergence between a1- and a2- associated alleles in the black region led to an estimate for the date of linkage between the HD and PR loci of about 1.3 million years ago (95% confidence interval between 1.1 and 1.6 million years) (SI Appendix, SI Text and Table S1).
Evolutionary Strata Extending Suppressed Recombination Around Each of the PR and HD Mating-Type Loci.
We detected further independent evolutionary strata, extending recombination suppression to nonmating-type genes in several successive steps (Fig. 3 C and D and SI Appendix, Table S1). Three strata (blue, purple, and orange strata, Figs. 1, 3, and 4), devoid of mating-type genes, evolved independently after the recombination suppression events at each of the PR and HD mating-type loci but before the HD and PR linkage via chromosome fusion (black stratum, Figs. 1, 3, and 4). Indeed, M. lagerheimii, which has unlinked HD and PR mating-type loci and therefore no black stratum, showed high divergence levels between alleles associated with the a1 mating type and those associated with the a2 mating type for 14 HD-proximal (blue) genes and 31 PR-proximal (purple) genes, none of which are involved in mating-type determination (Fig. 4A and SI Appendix, Tables S1, S2, and S3). These findings are in stark contrast with the near-zero within-individual dS values along the rest of the mating-type chromosomes in this species. These same genes ancestrally located near the PR and HD loci also had the next-highest level of divergence after the mating-type genes themselves in M. lychnidis-dioicae, M. silenes-dioicae, and M. violaceum s. str. (purple and blue in Fig. 4B; SI Appendix, Tables S1 and S3 and Fig. S8). Most of these genes also displayed high levels of synonymous divergence between mating types in three other Microbotryomycetes (SI Appendix, Table S2 and figure 2B in ref. 39) and were also localized within noncollinear regions in these distant outgroups (figure 1A in ref. 39). These results suggest that recombination suppression linking nonmating-type genes to key mating-type determinants is very old and occurred before these Microbotryomycetes species diverged (Fig. 1B).
Further supporting this inference, all genes in the purple stratum displayed trans-specific polymorphism with 50% displaying trans-specific polymorphism extending up to the divergence of M. lychnidis-dioicae and M. lagerheimii and 12.5% back to the root of the Microbotryomycetes (SI Appendix, Figs. S6 and S7). This confirms that a set of genes has been linked to the mating-type genes since long before the black stratum evolved, at least before the split between the common ancestor of Microbotryum and these outgroups. Indeed, such trans-specific polymorphisms separating alleles associated with the a1 and a2 mating types across species cannot be maintained without a full recombination suppression preceding all those speciation events. Additional strong evidence for an ancient and complete recombination cessation in the purple stratum is provided by the rearrangements between mating types present in M. lagerheimii, with the fully linked PR and pheromone genes of the a1 genome located at opposite edges of the purple region (SI Appendix, Fig. S4). Recombination was suppressed in the purple stratum about 2.1 million years ago (95% confidence interval between 1.6 and 2.7 million years), as estimated based on the divergence between alleles associated with the alternative mating types (SI Appendix, SI Text and Table S1). This date confirms that the suppression of recombination predates the black stratum. We also found trans-specific polymorphism in the blue stratum in 75% of the genes (SI Appendix, Fig. S7). Recombination suppression in the purple and blue nonmating-type genes was more recent than that between the genes at each of the HD and PR mating-type loci, as shown by the lower dS than for the mating-type genes and by the ancestral situation in basidiomycetes, with only mating-type genes linked at each of the PR and HD loci. Furthermore, the estimated dates of recombination suppression and trans-specifc polymorphism in the purple stratum were much more recent than those for the PR gene (SI Appendix, Table S1 and Fig. S7) (33). The blue stratum may be more recent than the purple stratum, as suggested by its lower dS values and less deep trans-specific polymorphism. However, even if equally ancient, the blue and purple strata constitute independent events of recombination suppression, given that they evolved in separate chromosomes, as seen in M. lagerheimii (Figs. 3 and 4).
We found another independent stratum ancestrally located distal to the purple stratum, toward the PARs (orange in Fig. 4B), with high dS values in M. lychnidis-dioicae, M. silenes-dioicae, and M. violaceum s. str. (Fig. 4 and SI Appendix, Fig. S8). Genes in the orange stratum were not involved in mating-type determination and also displayed deep trans-specific polymorphism, indicating an ancient complete suppression of recombination (SI Appendix, Figs. S6 and S7). The orange stratum most likely evolved before the black stratum linking the HD and PR mating-type genes (Fig. 3E), as it showed a much higher mean dS value (Figs. 4B and SI Appendix, Table S1). The orange stratum genes had zero dS in M. lagerheimii (Fig. 4 and SI Appendix, Fig. S8), indicating that recombination suppression in this region evolved after the divergence with this species and therefore after the purple stratum (Fig. 3). Consistent with this inference, the genes of the orange stratum had lower dS values than those of the purple stratum.
Additional, Younger Evolutionary Strata Evolved After the Linkage Between HD and PR Mating-Type Loci in M. lychnidis-dioicae.
We found evidence for two additional and younger evolutionary strata in M. lychnidis-dioicae. These strata did not involve mating-type genes either and arose much later than the linkage between HD and PR mating-type genes (red and green in Fig. 4B and SI Appendix, Tables S1 and S3). The red stratum contains genes that were ancestrally located distal to the orange stratum in the PAR and displayed intermediate dS values in M. lychnidis-dioicae (SI Appendix, Table S1 and Fig. 4B, Inset). These findings indicate a further distal expansion of recombination suppression, after the evolution of the black stratum, in the lineage leading to M. lychnidis-dioicae (Fig. 3 H and I). The red stratum is currently located between the HD and PR loci in M. lychnidis-dioicae (Fig. 2B), where recombination has been shown to be completely suppressed using segregation analyses (10, 24). This additional recombination suppression step occurred before the divergence of M. lychnidis-dioicae and M. silenes-dioicae, as shown by the nonzero within-individual dS values found in M. silenes-dioicae (SI Appendix, Fig. S8A) and the trans-specific polymorphism observed in almost 30% of the genes across these species (SI Appendix, Fig. S7). In M. silenes-dioicae, the red stratum genes have remained in close proximity but are separated from the PAR by a small noncollinear region in M. silenes-dioicae (SI Appendix, Fig. S5E). In contrast, the trans-specific polymorphism in the red stratum genes never extended to M. violaceum s. str. (SI Appendix, Fig. S7). In this later species, the red stratum genes were located in the PAR and had zero dS values within the sequenced diploid strain (SI Appendix, Figs. S5 B and D and S7), demonstrating the regular occurrence of recombination. This indicates that the red stratum, which contains no mating-type genes, was formed more recently than the black stratum, in the lineage leading to M. lychnidis-dioicae and M. silenes-dioicae, after its divergence with M. violaceum s. str. (Fig. 1). We estimated the date of recombination suppression in the red stratum at about 0.9 millions years ago (95% confidence interval between 0.7 and 1.1 million years), which is much more recent than the black stratum linking the HD and PR loci (SI Appendix, SI Text and Table S1).
We found yet another small, very recent putative evolutionary stratum, also common only to M. lychnidis-dioicae and M. silenes-dioicae (green in Figs. 2B and 4B). In both species, this green stratum was located close to the PAR, but was separated from it by a region with an inversion between a1 and a2 (Fig. 2B and SI Appendix, Fig. S5E), and the genes had low but nonzero dS values. These genes probably correspond to a very recent expansion of the nonrecombining region after the formation of the red stratum (Fig. 3I), as suggested by their lower dS values (Fig. 4B and SI Appendix, Table S1), distinctive current location in the M. lychnidis-dioicae genome, and lack of trans-specific polymorphism (SI Appendix, Fig. S7). Alternatively, the red and green strata may have been generated in a single event, followed by physical separation due to rearrangements.
We further confirmed recombination suppression in the red and green strata using multiple available resequenced genomes for M. lychnidis-dioicae and M. silenes-dioicae (22, 40). Gene genealogies revealed that the alleles associated with the a1 and a2 mating types formed two different clades for at least one species in all of the gene genealogies, and trans-specific polymorphism was observed across all individuals in 23% of cases (SI Appendix, Fig. S9). These patterns can be explained only by full linkage to mating type before the two species split. Furthermore, the mean levels of polymorphism per mating type and per species were significantly lower in all of the evolutionary strata than in the PARs (Student’s t tests, P < 0.05) (SI Appendix, Fig. S10), as expected in regions without recombination due to the lower effective population size. These findings indicated that the nonzero dS in the evolutionary strata within the sequenced individuals was due to recombination suppression rather than elevated substitution rates. Maximum-likelihood Hudson–Kreitman–Agade tests further supported that the higher dS levels in the purple, blue, orange, black, and red strata compared with the PARs in M. lychnidis-dioicae and M. silenes-dioicae were due to balancing selection (because of linkage to mating type) rather than to elevated mutation rates (SI Appendix, SI Text).
Conclusion.
We found multiple evolutionary strata in Microbotryum mating-type chromosomes, several of which included no genes related to mating-type determination (SI Appendix, Tables S2 and S3). Only the two oldest strata (linking pairs of mating-type genes within each of the separate PR and HD loci, respectively) and the black stratum (linking the HD and PR loci together) involved mating-type genes. The linkage between HD and PR genes probably evolved because it increased the likelihood of compatibility between gametes in a selfing system (16) (SI Appendix, SI Text and Fig. S2). Mating-type loci linkage has been reported in several fungi (9, 14, 41, 42), and our study provides clues to the chromosomal rearrangements underlying this phenomenon. The five other M. lychnidis-dioicae evolutionary strata (purple, blue, orange, red, and green strata) did not involve mating-type genes. As this species has no male/female traits, our results indicate that sexual antagonism is insufficient to account for the occurrence of these evolutionary strata. Furthermore, given the M. lychnidis-dioicae life cycle, the existence of ecological differences between mating types that would enhance alternative mating-type functions is highly unlikely. Alternative hypotheses should therefore be considered, including the capture and shelter of deleterious alleles in a permanently heterozygous state, the fixation of neutral rearrangements by drift in one gametolog (see SI Appendix, SI Text and Fig. S1 for more details concerning these hypotheses). These general mechanisms apply to both sex and mating-type chromosomes across the plant, animal, and fungal kingdoms. Our results thus call for an expansion of theories concerning the evolution of eukaryotic sex-related chromosomes to forces beyond sexual antagonism.
Materials and Methods
Haploid genomes were sequenced with P6/C4 Pacific Biosciences SMRT technology (University of California, San Diego IGM Genomics Facility) (SI Appendix, Table S4). Orthologs were identified by orthomcl (43). Trees were inferred with RAxML (44). Strata divergence times were inferred with BEAST2 (45). More details about the materials and methods used are provided in SI Appendix, SI Text.
Supplementary Material
Acknowledgments
We thank Stéphanie Le Prieur, Alodie Snirc, Cécile Fairhead, and Gilles Deparis for help with DNA extraction. This work was supported by European Research Council GenomeFun Grant 309403 and a grant from Institut Diversité Ecologie et Evolution du Vivant (to T.G.); NSF Grant DEB-1115765 and NIH Grant R15GM119092 (to M.E.H.); the Marie Curie European Grant 701646 (to S.B.); and a postdoctoral fellowship (SFRH/BPD/79198/2011) from Fundação para a Ciência e a Tecnologia, Portugal (to M.A.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The genome assemblies are available at the European Nucleotide Archive. Accession numbers are as follows: PRJEB12080, ERS459551, and ERZ250722 for Microbotryum lychnidis-dioicae 1064 a2; PRJEB12080, ERS1013679, and ERZ250721 for M. lychnidis-dioicae 1064 a1; PRJEB12080, ERS1013678, and ERZ250720 for Microbotryum lagerheimii 1253 a2; PRJEB12080, ERS1013677, and ERZ250719 for M. lagerheimii 1253 a1; PRJEB12080, ERS1013672, and ERZ250714 for Microbotryum violaceum s. str. 1249 a2; PRJEB12080, ERS1013671, and ERZ250713 for M. violaceum s. str. 1249 a1; PRJEB16471 and ERZ369313 for Microbotryum intermedium; PRJEB16741, ERS1436593, and ERZ370138 for Microbotryum silenes-dioicae 1303 a2; and PRJEB16741, ERS1436592, and ERZ369313 for M. silenes-dioicae 1303 a1.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701658114/-/DCSupplemental.
References
- 1.Bergero R, Charlesworth D. The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol. 2009;24:94–102. doi: 10.1016/j.tree.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 2.Charlesworth D. The status of supergenes in the 21st century: Recombination suppression in Batesian mimicry and sex chromosomes and other complex adaptations. Evol Appl. 2015;9:74–90. doi: 10.1111/eva.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wright AE, Dean R, Zimmer F, Mank JE. How to make a sex chromosome. Nat Commun. 2016;7:12087. doi: 10.1038/ncomms12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charlesworth B. The evolution of sex chromosomes. Science. 1991;251:1030–1033. doi: 10.1126/science.1998119. [DOI] [PubMed] [Google Scholar]
- 5.Ironside JE. No amicable divorce? Challenging the notion that sexual antagonism drives sex chromosome evolution. BioEssays. 2010;32:718–726. doi: 10.1002/bies.200900124. [DOI] [PubMed] [Google Scholar]
- 6.Charlesworth B, Wall JD. Inbreeding, heterozygote advantage and the evolution of neo-X and neo-Y sex chromosomes. Proc Biol Sci. 1999;266:51–56. [Google Scholar]
- 7.Antonovics J, Abrams JY. Intratetrad mating and the evolution of linkage relationships. Evolution. 2004;58:702–709. doi: 10.1111/j.0014-3820.2004.tb00403.x. [DOI] [PubMed] [Google Scholar]
- 8.Johnson LJ, Antonovics J, Hood ME. The evolution of intratetrad mating rates. Evolution. 2005;59:2525–2532. [PubMed] [Google Scholar]
- 9.Menkis A, Jacobson DJ, Gustafsson T, Johannesson H. The mating-type chromosome in the filamentous ascomycete Neurospora tetrasperma represents a model for early evolution of sex chromosomes. PLoS Genet. 2008;4:e1000030. doi: 10.1371/journal.pgen.1000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hood ME. Dimorphic mating-type chromosomes in the fungus Microbotryum violaceum. Genetics. 2002;160:457–461. doi: 10.1093/genetics/160.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kües U. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol Mol Biol Rev. 2000;64:316–353. doi: 10.1128/mmbr.64.2.316-353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coelho MA, Bakkeren G, Sun S, Hood ME, Giraud T. Fungal sex: The basidiomycota. Microbiol Spectr. 2017 doi: 10.1128/microbiolspec.FUNK-0046-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kües U. From two to many: Multiple mating types in Basidiomycetes. Fungal Biol Rev. 2015;29:126–166. [Google Scholar]
- 14.Bakkeren G, Kronstad JW. Linkage of mating-type loci distinguishes bipolar from tetrapolar mating in basidiomycetous smut fungi. Proc Natl Acad Sci USA. 1994;91:7085–7089. doi: 10.1073/pnas.91.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Idnurm A, Hood ME, Johannesson H, Giraud T. Contrasted patterns in mating-type chromosomes in fungi: Hotspots versus coldspots of recombination. Fungal Biol Rev. 2015;29:220–229. doi: 10.1016/j.fbr.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nieuwenhuis BPS, et al. Evolution of uni- and bifactorial sexual compatibility systems in fungi. Heredity (Edinb) 2013;111:445–455. doi: 10.1038/hdy.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Badouin H, et al. Chaos of rearrangements in the mating-type chromosomes of the anther-smut fungus Microbotryum lychnidis-dioicae. Genetics. 2015;200:1275–1284. doi: 10.1534/genetics.115.177709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraser JA, et al. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2004;2:e384. doi: 10.1371/journal.pbio.0020384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.James TY. Why mushrooms have evolved to be so promiscuous: Insights from evolutionary and ecological patterns. Fungal Biol Rev. 2015;29:167–178. [Google Scholar]
- 20.Giraud T, Yockteng R, López-Villavicencio M, Refrégier G, Hood ME. Mating system of the anther smut fungus Microbotryum violaceum: Selfing under heterothallism. Eukaryot Cell. 2008;7:765–775. doi: 10.1128/EC.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Votintseva AA, Filatov DA. Evolutionary strata in a small mating-type-specific region of the smut fungus Microbotryum violaceum. Genetics. 2009;182:1391–1396. doi: 10.1534/genetics.109.103192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whittle CA, Votintseva A, Ridout K, Filatov DA. Recent and massive expansion of the mating-type-specific region in the smut fungus Microbotryum. Genetics. 2015;199:809–816. doi: 10.1534/genetics.114.171702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Billiard S, et al. Having sex, yes, but with whom? Inferences from fungi on the evolution of anisogamy and mating types. Biol Rev Camb Philos Soc. 2011;86:421–442. doi: 10.1111/j.1469-185X.2010.00153.x. [DOI] [PubMed] [Google Scholar]
- 24.Hood ME, Antonovics J, Koskella B. Shared forces of sex chromosome evolution in haploid-mating and diploid-mating organisms: Microbotryum violaceum and other model organisms. Genetics. 2004;168:141–146. doi: 10.1534/genetics.104.029900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hood ME, Petit E, Giraud T. Extensive divergence between mating-type chromosomes of the anther-smut fungus. Genetics. 2013;193:309–315. doi: 10.1534/genetics.112.146266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hood ME, Antonovics J. Intratetrad mating, heterozygosity, and the maintenance of deleterious alleles in Microbotryum violaceum (=Ustilago violacea) Heredity (Edinb) 2000;85:231–241. doi: 10.1046/j.1365-2540.2000.00748.x. [DOI] [PubMed] [Google Scholar]
- 27.Hood ME, Antonovics J. Mating within the meiotic tetrad and the maintenance of genomic heterozygosity. Genetics. 2004;166:1751–1759. doi: 10.1534/genetics.166.4.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giraud T, Jonot O, Shykoff JA. Selfing propensity under choice conditions in a parasitic fungus, Microbotryum violaceum, and parameters influencing infection success in artificial inoculations. Int J Plant Sci. 2005;166:649–657. [Google Scholar]
- 29.Montgomery T. Are particular chromosomes sex determinants? Biol Bull. 1911;19:1–17. [Google Scholar]
- 30.Montgomery TH., Jr The terminology of aberrant chromosomes and their behavior in certain Hemiptera. Science. 1906;23:36–38. doi: 10.1126/science.23.575.36. [DOI] [PubMed] [Google Scholar]
- 31.Kemler M, Göker M, Oberwinkler F, Begerow D. Implications of molecular characters for the phylogeny of the Microbotryaceae (Basidiomycota: Urediniomycetes) BMC Evol Biol. 2006;6:35. doi: 10.1186/1471-2148-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hood ME, Scott M, Hwang M. Breaking linkage between mating compatibility factors: Tetrapolarity in Microbotryum. Evolution. 2015;69:2561–2572. doi: 10.1111/evo.12765. [DOI] [PubMed] [Google Scholar]
- 33.Devier B, Aguileta G, Hood ME, Giraud T. Ancient trans-specific polymorphism at pheromone receptor genes in basidiomycetes. Genetics. 2009;181:209–223. doi: 10.1534/genetics.108.093708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petit E, et al. Linkage to the mating-type locus across the genus Microbotryum: Insights into nonrecombining chromosomes. Evolution. 2012;66:3519–3533. doi: 10.1111/j.1558-5646.2012.01703.x. [DOI] [PubMed] [Google Scholar]
- 35.Fontanillas E, et al. Degeneration of the nonrecombining regions in the mating-type chromosomes of the anther-smut fungi. Mol Biol Evol. 2015;32:928–943. doi: 10.1093/molbev/msu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menkis A, Whittle CA, Johannesson H. Gene genealogies indicates abundant gene conversions and independent evolutionary histories of the mating-type chromosomes in the evolutionary history of Neurospora tetrasperma. BMC Evol Biol. 2010;10:234. doi: 10.1186/1471-2148-10-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun S, Hsueh Y-P, Heitman J. Gene conversion occurs within the mating-type locus of Cryptococcus neoformans during sexual reproduction. PLoS Genet. 2012;8:e1002810. doi: 10.1371/journal.pgen.1002810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gladieux P, et al. Maintenance of fungal pathogen species that are specialized to different hosts: Allopatric divergence and introgression through secondary contact. Mol Biol Evol. 2011;28:459–471. doi: 10.1093/molbev/msq235. [DOI] [PubMed] [Google Scholar]
- 39.Maia TM, et al. Evolution of mating systems in Basidiomycetes and the genetic architecture underlying mating-type determination in the yeast Leucosporidium scottii. Genetics. 2015;201:75–89. doi: 10.1534/genetics.115.177717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Badouin H, et al. Widespread selective sweeps throughout the genome of model plant pathogenic fungi and identification of effector candidates. Mol Ecol. 2017;26:2041–2062. doi: 10.1111/mec.13976. [DOI] [PubMed] [Google Scholar]
- 41.Fraser JA, Heitman J. Chromosomal sex-determining regions in animals, plants and fungi. Curr Opin Genet Dev. 2005;15:645–651. doi: 10.1016/j.gde.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 42.Grognet P, et al. Maintaining two mating types: Structure of the mating type locus and its role in heterokaryosis in Podospora anserina. Genetics. 2014;197:421–432. doi: 10.1534/genetics.113.159988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 45.Bouckaert R, et al. BEAST 2: A software platform for Bayesian evolutionary analysis. PLOS Comput Biol. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.