Significance
Prokaryotic and eukaryotic organisms use analogous pathways to synthesize protein cofactors called iron–sulfur clusters. An unexplained difference between pathways is the functional requirements of the respective cysteine desulfurases. In eukaryotes, the cysteine desulfurase NFS1 requires additional accessory subunits for function. The lack of structural information has limited mechanistic insight into the role of these accessory proteins in mitochondrial Fe–S cluster biosynthesis. Here we determined crystallographic and electron microscopic structures of the NFS1–ISD11–ACP subcomplex. These results reveal an unexpected cysteine desulfurase architecture that reconciles mechanistic differences between the prokaryotic and eukaryotic systems, reveals the basis of control of iron–sulfur cluster assembly through fatty acid synthesis, and serves as a structural foundation for investigating human diseases related to iron–sulfur cluster assembly.
Keywords: LYR, ACP, iron–sulfur cluster, PLP, frataxin
Abstract
In eukaryotes, sulfur is mobilized for incorporation into multiple biosynthetic pathways by a cysteine desulfurase complex that consists of a catalytic subunit (NFS1), LYR protein (ISD11), and acyl carrier protein (ACP). This NFS1–ISD11–ACP (SDA) complex forms the core of the iron–sulfur (Fe–S) assembly complex and associates with assembly proteins ISCU2, frataxin (FXN), and ferredoxin to synthesize Fe–S clusters. Here we present crystallographic and electron microscopic structures of the SDA complex coupled to enzyme kinetic and cell-based studies to provide structure-function properties of a mitochondrial cysteine desulfurase. Unlike prokaryotic cysteine desulfurases, the SDA structure adopts an unexpected architecture in which a pair of ISD11 subunits form the dimeric core of the SDA complex, which clarifies the critical role of ISD11 in eukaryotic assemblies. The different quaternary structure results in an incompletely formed substrate channel and solvent-exposed pyridoxal 5′-phosphate cofactor and provides a rationale for the allosteric activator function of FXN in eukaryotic systems. The structure also reveals the 4′-phosphopantetheine–conjugated acyl-group of ACP occupies the hydrophobic core of ISD11, explaining the basis of ACP stabilization. The unexpected architecture for the SDA complex provides a framework for understanding interactions with acceptor proteins for sulfur-containing biosynthetic pathways, elucidating mechanistic details of eukaryotic Fe–S cluster biosynthesis, and clarifying how defects in Fe–S cluster assembly lead to diseases such as Friedreich’s ataxia. Moreover, our results support a lock-and-key model in which LYR proteins associate with acyl-ACP as a mechanism for fatty acid biosynthesis to coordinate the expression, Fe–S cofactor maturation, and activity of the respiratory complexes.
Iron–sulfur (Fe–S) clusters are protein cofactors and are required for critical biological processes such as oxidative respiration, nitrogen fixation, and photosynthesis. The iron–sulfur cluster (ISC) biosynthetic pathway, which is found in most prokaryotes and in the mitochondrial matrix of eukaryotes, is responsible for the synthesis of Fe–S clusters and distribution of these cofactors to the appropriate target proteins. Despite the homology between analogous components of the prokaryotic and eukaryotic ISC pathways, there are key unexplained differences, such as the requirement of the LYR protein ISD11 and acyl carrier protein (ACP) for function in eukaryotic but not prokaryotic ISC systems (1, 2).
Mitochondrial LYR proteins are members of a recently identified superfamily that are characterized by their small size (10–22 kDa), high positive charge, invariant Phe residue, and eponymous Leu–Tyr–Arg (LYR) motif near their N terminus (3). LYR proteins function as subunits or assembly factors for respiratory complexes I, II, III, and V. Human LYRM4, also known as ISD11, is critical for the function of the Fe–S assembly complex (4–9), whereas LYRM8, a key maturation factor for mitochondrial complex II, interacts with the HSC20 chaperone (10), which is important for Fe–S cluster delivery to apo targets. Despite their vital roles in Fe–S cluster cofactor biogenesis and oxidative respiration, structure-function details for these LYR proteins are poorly understood.
An emerging theme for LYR proteins is their interaction with ACP (3, 11). ACP operates in the mitochondrial fatty acid synthesis (mtFAS) pathway to synthesize fatty acids using a 4′-phosphopantethiene (4′-PPT) prosthetic group covalently attached to a serine residue on ACP. Acyl intermediates are linked by a thioester bond and shuttled between fatty acid biosynthetic enzymes to generate medium and long chain fatty acids (12–15). One of the best known functions of the mtFAS pathway is to generate octanoyl-ACP, which is required for lipoic acid biosynthesis. However, mitochondrial ACP also functions as a required subunit for respiratory complex I and is predominately associated with the long-chain fatty acid 3-hydroxytetradecanoate (16–23). More recently, ACP has been identified as an essential functional component of the eukaryotic Fe–S cluster biosynthetic complex (1). Currently, there are no X-ray crystal structures that detail interactions between ACP and LYR proteins that provide insight into the eukaryotic adaption of ACP for these moonlighting functions.
In eukaryotes, Fe–S clusters are synthesized by a multicomponent assembly complex (24–26). At the center of the complex, the pyridoxal 5′-phosphate (PLP)-dependent cysteine desulfurase NFS1 forms a tight complex with ISD11 (4, 8, 9, 27). The NFS1–ISD11 complex catalyzes the conversion of l-cysteine to l-alanine and generates a persulfide intermediate on a cysteine of the mobile S-transfer loop of NFS1 (S-loop) (24, 28). The terminal sulfur of this intermediate is transferred to the scaffold protein ISCU2, where it is combined with ferrous iron and electrons, from a ferredoxin (29, 30), to form Fe–S clusters. In humans, a low-activity Fe–S assembly complex consisting of NFS1–ISD11 and the catalytic subunit ISCU2 can be activated by binding of the allosteric activator frataxin (FXN) (25). NFS1 also provides sulfur for other processes including tRNA synthesis (31) and molybdenum cofactor biosynthesis (27). Importantly, clinical mutations of NFS1 (R72Q) (32) and ISD11 (R68L) (33) result in deficiencies in multiple oxidative respiratory complexes, whereas mutations of ISCU and FXN result in myopathy with exercise intolerance (34) and Friedreich’s ataxia (35), respectively. The lack of structural data for eukaryotic cysteine desulfurase and Fe–S assembly complexes has limited the mechanistic understanding of these critical biosynthetic processes, the role of the essential proteins ISD11 and ACP, and insight into how clinical mutants compromise function.
Results
Structure of the Cysteine Desulfurase Subcomplex for Eukaryotic Fe–S Cluster Biosynthesis.
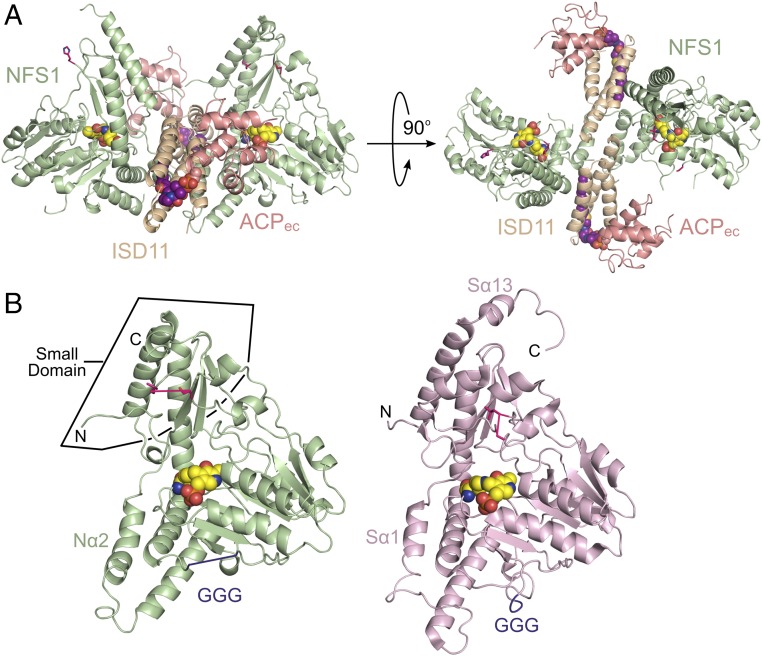
We identified recombinant coexpression conditions that allowed the purification of a complex between human NFS1–ISD11 and native Escherichia coli ACP (ACPec, 44% identical to human mitochondrial ACP) (SI Appendix, Fig. S1), consistent with a recent report (36). An X-ray crystal structure of the NFS1–ISD11–ACPec (SDAec) complex was determined using molecular replacement–single wavelength anomalous dispersion (MR-SAD) and refined to a resolution of 3.09 Å (Rwork/Rfree of 21.2/25.9%) with excellent geometry (SI Appendix, Table S1). The SDAec structure exhibited an overall α2β2γ2 assembly (Fig. 1A) with two α2β2γ2 assemblies in the asymmetric unit. The NFS1 fold is highly similar to E. coli IscS (Fig. 1B) (37) and shares 60% sequence identity (SI Appendix, Fig. S2). NFS1, like IscS, contains a larger domain that includes a PLP active site and a smaller domain that likely interacts with ISCU2. The primary differences between NFS1 and IscS include a small extension in NFS1 α-helix 2 (Nα2), larger disordered region for the mobile S loop, and disorder in a triple glycine-containing segment (GGG motif; see Subunit Orientation and Assembly of SDAec). In addition, some subunits display a different conformation for the equivalent C-terminal α-helix that is used by IscS (Sα13) to interact with IscU (38, 39). Surprisingly, this mitochondrial cysteine desulfurase crystal structure also revealed an ACP–lipid–ISD11 motif and a fundamentally different oligomeric architecture for NFS1 compared with its bacterial orthologs.
Fig. 1.
The architecture of a mitochondrial cysteine desulfurase. (A) Ribbon diagram of the SDAec structural architecture displayed with NFS1 (light green), ISD11 (wheat), and ACPec (salmon) in two orthogonal orientations. The K258-PLP (yellow) and lipid-bound 4’-PPT (magenta) cofactors are shown as spheres. (B) NFS1 and IscS (light pink; PDB ID code 3LVM) subunits displayed in similar orientations. Residues connecting disordered mobile S loop are shown in hot pink, whereas those connecting the GGG loop are in plum. The rmsd for 277 NFS1 (chain A) and IscS (chain A) Cα atoms is 0.84 Å.
Identification of the ACP–Lipid–ISD11 Motif.
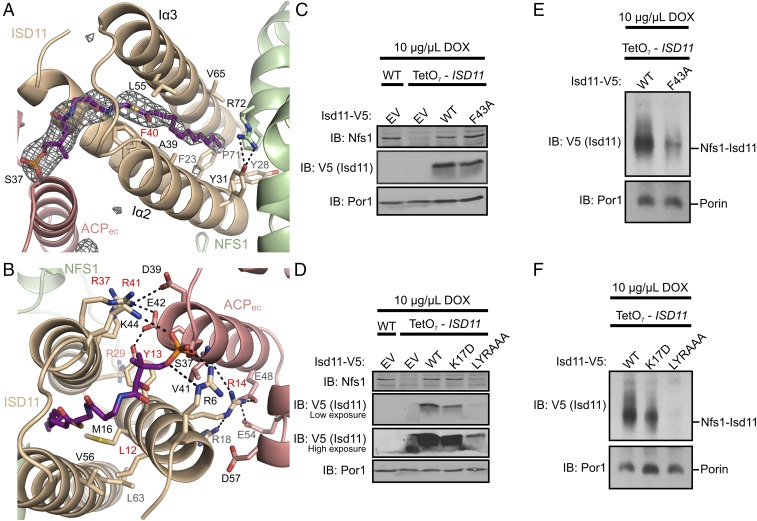
The electron density revealed a 3-helix bundle fold for ISD11 that included most of the highly conserved residues (SI Appendix, Fig. S3 and Table S2). ACPec displays a conformation similar to both uncomplexed bacterial ACP (40) and human ACP structures (SI Appendix, Fig. S4) with large B-factors (SI Appendix, Table S2), consistent with the reported high flexibility and dynamic properties of ACP (41). ACPec interacts with ISD11 through two distinct interfaces. The first interface is mediated by a 4′-PPT–congujated fatty acid covalently attached to ACPec, which is threaded into a groove on ISD11 (Fig. 2A). To generate this groove and accommodate the fatty acid, ISD11 adopts an unusual 3-helix bundle conformation in the SDAec structure that lacks a traditional hydrophobic core and contains remarkably few side-chain interactions between ISD11 α-helices Iα2 and Iα3. At least 12 carbons in the acyl-chain (dodecyl-ACP) were evident in the electron density (Fig. 2A), but longer acyl-chains could also be accommodated in the ISD11 core. In fact, we determined recombinant SDAec contains primarily a 16-carbon acyl-chain by performing gas chromatography–mass spectrometry (GC-MS) on isolated fatty-acid methyl esters (FAMEs) that were obtained from SDAec through a transesterification procedure (SI Appendix, Fig. S5). The aliphatic portion of the 4′-PPT cofactor is stabilized by hydrophobic interactions contributed by invariant residues on ISD11 including F40, which packs against the lipid cofactor, and F23, which is near the tip of the acyl-chain (Fig. 2A). The functional importance of F40 was verified using a chromosomally ISD11-depleted Saccharomyces cerevisiae strain covered with an ISD11 plasmid to test mutants. S. cerevisiae mitochondria with Isd11 containing the F43A variant (equivalent to human F40A) showed comparable expression levels to native Isd11 but decreased amounts of the Nfs1–Isd11 complex (Fig. 2 C and E). A similar result is observed for mitochondria either depleted in Acp1 (yeast ACP homolog) or with an Acp1 variant incapable of attaching the 4′-PPT prosthetic group (1).
Fig. 2.
ISD11–acyl-ACP interactions are critical for the formation of the SDAec complex. (A) Simulated annealing–omit mFo – DFc map (gray mesh) contoured to 3.0σ revealed a lipid-bound 4′-PPT inserted into the hydrophobic core of ISD11. The map was displayed with a 5 Å region padding. (B) ACPec interacts with ISD11 using electrostatic contacts and interactions with the LYR motif. Residues labeled in red in A and B were targeted for mutagenesis experiments in S. cerevisiae. Purified mitochondria from the indicated strains (WT or TetO7–ISD11) were either resolved by SDS/PAGE (C and D) or solubilized in 1% digitonin and resolved by BN-PAGE (E and F). Cells were grown for 18 h in the presence 10 μg/mL DOX. The indicated proteins and protein complexes were assessed by immunoblot. Low and high exposure indicates the intensity of light used to scan the blot. LYRAAA is the triple alanine variant (L15A Y16A K17A) of the LYR motif. EV is empty vector control. Porin is included as a loading control. IB, immunoblot.
Conserved residues on ISD11 have key roles in stabilizing the 3-helix bundle by providing a second, primarily electrostatic, interface with ACPec. The phosphate linker of the 4′-PPT is stabilized in the SDAec structure through interactions with side chains of conserved R6 and K44 residues on ISD11 (Fig. 2B). R29 interacts with backbone carbonyls of the F23-containing loop of ISD11 (SI Appendix, Fig. S6A), whereas R37 and R41 form salt bridges with ACPec residues E42 and D39, respectively (Fig. 2B). The residues of the characteristic ISD11 LYR motif contribute L12 hydrophobic interactions to an interface between ISD11 helices, Y13 forms a hydrogen bond to ACPec E42, and R14 forms a salt bridge with ACPec E48 (Fig. 2B). S. cerevisiae mitochondria containing R32D, R40D, and R44D ISD11 point mutations (equivalent to human ISD11 R29D, R37D, and R41D, respectively) or substitution of the LYR motif had decreased levels of Isd11 and loss of the Nfs1–Isd11 complex (Fig. 2 D and F and SI Appendix, Fig. S6 B and C). These results are consistent with a role for these residues in Isd11 stability and/or association with Acp1 (1). The residues on ACPec that interact with ISD11 in the SDAec structure are also conserved between E. coli and human mitochondrial ACP (SI Appendix, Fig. S4B) and suggest similar interactions will likely contribute to the interface between NFS1–ISD11 and human ACP. Together, these results provide insight into the unexpected role of ACP in eukaryotic Fe–S cluster biosynthesis by identifying ACP–lipid interactions that promote the stability of ISD11 and function of the Fe–S assembly complex.
Subunit Orientation and Assembly of SDAec.
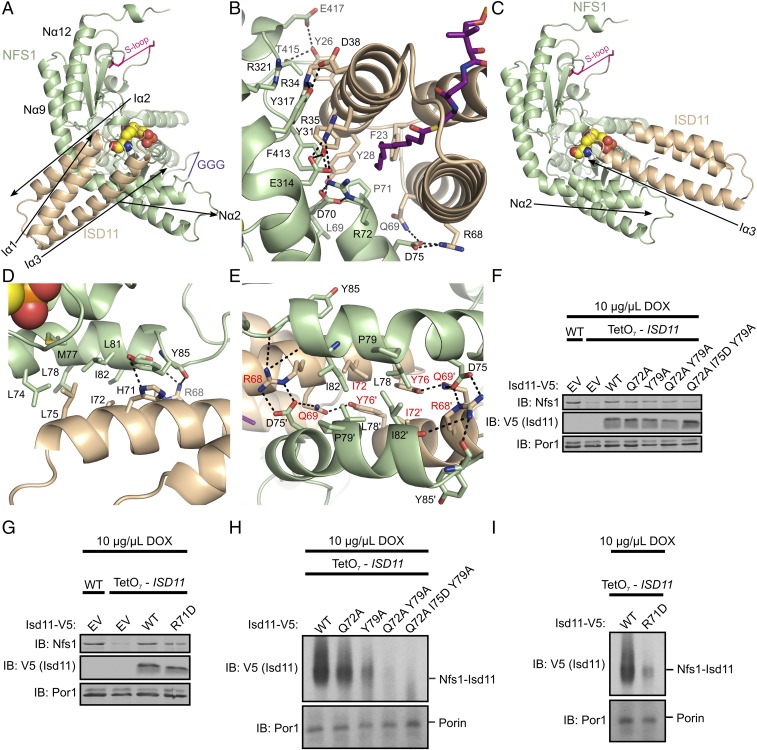
ISD11 forms two different interfaces with NFS1 that stabilize the SDAec subcomplex. In the first and larger interface, the Iα2 helix and acyl-chain are positioned in a pocket that is formed between the Nα2, Nα9, and Nα12 helices of NFS1 (Fig. 3A). The NFS1 residues in this ISD11-binding pocket are highly conserved in eukaryotes; whereas the analogous residues for bacterial IscS are more variable (SI Appendix, Fig. S7). Notable interactions in this interface include NFS1 residue R72, which forms a hydrogen bond with invariant Y31 on ISD11, as well as the highly conserved NFS1 P71 and ISD11 F23, and appear to cap the end of the acyl-chain (Fig. 3B). Interestingly, the R72Q NFS1 variant is associated with a Fe–S cluster disease, infantile mitochondrial complex II/III deficiency (32), suggesting that these interactions are functionally important. In the second, smaller, interface, the adjacent Iα3 helix of ISD11 lays across the Nα2 helix in an antiparallel orientation (Fig. 3C). In contrast to IscS, the NFS1 residues in this second interface with ISD11 are often conserved (SI Appendix, Fig. S7). A hydrophobic pocket is generated by residues L75 and I72 of ISD11 and L74, M77, L78, L81, and I82 of NFS1 (Fig. 3D). This pocket is bridged by hydrogen bonding residues on ISD11 to residues and backbone carbonyls on a loop between NFS1 Nα2 and Nα3.
Fig. 3.
Two distinct NFS1–ISD11 interfaces are important for forming the SDAec architecture. (A) Global overview of the primary NFS1–ISD11 interface (ACP and lipid are omitted for clarity). ISD11 packs against Nα9, Nα12, and Nα2 helices of NFS1. (B) Specific interactions of the primary NFS1–ISD11 interface (ACP omitted for clarity). The acylated 4′-PPT is shown in purple. (C) Global overview of the secondary NFS1–ISD11 interface. ISD11 Iα3 has an antiparallel orientation compared with NFS1 Nα2. (D) Specific interactions of the second NFS1–ISD11 interface. The PLP is shown in spheres. (E) The 4-helix bundle core of the SDAec complex. Residues labeled in red were tested in vivo using analogous substitution in S. cerevisiae. Arrows show the polypeptide direction for the α-helices. Purified mitochondria from the indicated strains were solubilized from cells similar to Fig. 2 and resolved by SDS/PAGE (F and G) and BN-PAGE (H and I). EV, empty vector; IB, immunoblot.
Interestingly, when two SDAec α1β1γ1 assemblies come together to form the α2β2γ2 architecture (Fig. 1A), the NFS1–ISD11 interfaces generate an unusual, interlocking 4-helix bundle core made up of two copies of the ISD11 Iα3 helices and two copies of the NFS1 Nα2 helices (Fig. 3E). A cluster of hydrophobic residues consisting of ISD11 (I72) and NFS1 (L78 and I82) from each of the two α1β1γ1 assemblies stabilizes the 4-helix bundle. The interactions between the four subunits appear to be facilitated by an extension of Nα2 (compared with Sα1 of IscS; Fig. 1B and SI Appendix, Fig. S2) and a kink induced by conserved P79. Moreover, hydrogen bonds across the ISD11–ISD11 interface between side chains of Q69 and Y76, and salt bridge interactions between ISD11 R68 and NFS1 D75 appear to further stabilize the SDAec complex (Fig. 3E). The importance of these interactions was tested through the introduction of single Y79A, double Q72A Y79A, and triple Q72A I75D Y79A (equivalent to human Q69A I72D Y76A) ISD11 point mutants along with the R71D ISD11 substitution (equivalent to human R68D) into S. cerevisiae. Isolated mitochondria showed Isd11 variants exhibit comparable expression levels to native Isd11 but decreased amounts of Nfs1–Isd11 complex (Fig. 3 F–I). These cell-based results coupled to the sequence conservation for interfacial residues (SI Appendix, Figs. S2 and S3), previous pull-down experiments (5), and involvement of variant ISD11 (R68L) with a mitochondrial genetic disorder (33) support the functional importance of these core SDAec interactions and explain the requirement of ISD11 for eukaryotic Fe–S cluster biosynthesis (4, 8, 9).
The positions of the two NFS1 subunits in the SDAec complex are fundamentally different compared with previous cysteine desulfurase structures. The limited, hydrophobic interactions between the Nα2 helices are the only contacts between NFS1 subunits (122 Å2 of buried surface area) in the SDAec complex. This observation is in stark contrast to all previously determined structures of cysteine desulfurases, which exhibit common homodimeric architectures (SI Appendix, Fig. S8) and extensive buried surface area (2,351 Å2 for E. coli IscS) (37, 42). The first NFS1–ISD11 interface in the SDAec complex would not exclude a bacterial-like dimer, possibly explaining the recent discovery that ISD11 can interact but not influence the activity of bacterial IscS (43). However, the second NFS1–ISD11 interface in the SDAec complex is not compatible with the IscS dimer. Overlay of an IscS molecule from the bacterial dimer with a NFS1 subunit reveals the second IscS molecule would spatially overlap with ISD11 (SI Appendix, Fig. S8). Interestingly, NFS1 and IscS use interactions on orthogonal faces of the equivalent helices (Nα2, Sα1) to generate the different oligomeric arrangements.
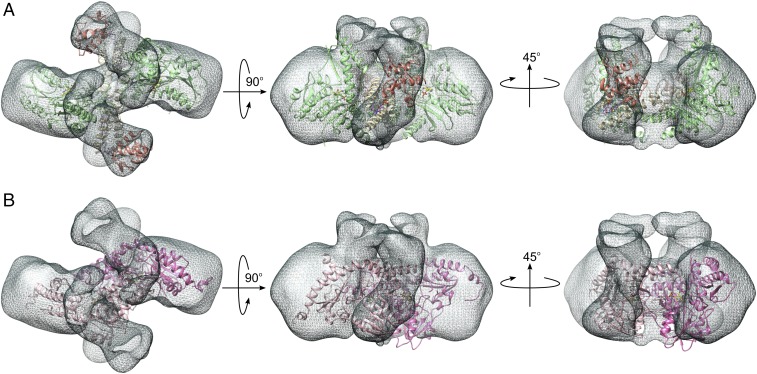
To further evaluate the protein assembly architecture of the complex, we used negative stain electron microscopy (EM) (Fig. 4 and SI Appendix, Fig. S9A). A total of 128 class averages were generated from the data (SI Appendix, Fig. S9B), 36 of which were used in an ab initio reconstruction (SI Appendix, Fig. S9C). This reconstruction was refined using 11,481 particles to yield a structure of the SDAec complex to ∼15 Å resolution (SI Appendix, Fig. S9 C–E). This reconstruction matched well with the overall arrangement of the SDAec crystal structure (correlation coefficient, 0.70) (Fig. 4A and SI Appendix, Fig. S9F). On the other hand, the 3D EM reconstruction was incompatible with the canonical E. coli IscS dimer (Fig. 4B and SI Appendix, Fig. S9G; correlation coefficient, 0.56). Overall, the crystal and EM structures for the SDAec complex reveal a rare situation in which orthologs with high sequence identity and the same protein fold exhibit distinct oligomeric architectures that result in dramatically different functional properties (see Biophysical and Functional Properties of SDAec Complex).
Fig. 4.
Single particle reconstruction of the SDAec complex with EM supports the crystallographic architecture. (A) SDAec was reconstructed from EM images of negatively stained specimens (isosurface contoured with dark gray mesh). The SDAec crystal structure (chains C, D, E, F, I, and L) fit well into the EM density map. Subunits are colored as in Fig. 1. (B) The 3D EM reconstruction does not match well with the architecture of the E. coli IscS structure (subunits shown in light and dark pink).
Biophysical and Functional Properties of SDAec Complex.
Crystallographic and EM structural analysis revealed that adaptor proteins (ISD11 and ACP) incorporated into the eukaryotic system support an unprecedented structural framework for the cysteine desulfurase reaction and Fe–S cluster biosynthesis. Incorporation of ISD11–ACPec generates a SDAec complex less prone to aggregation with increased solubility compared with the NFS1–ISD11 complex, consistent with the recently discovered Acp1 requirement for Nfs1–Isd11 stability and function in S. cerevisiae (1). In fact, the SDAec complex elutes from an analytical size-exclusion column with a mass consistent with a (SDAec)2 stoichiometry with no evidence of monomeric or oligomeric species at the concentration analyzed (SI Appendix, Fig. S10). Steady-state kinetic analysis on SDAec revealed a significantly lower KM for l-cysteine (Table 1) in comparison with the previously characterized complex that evidently lacked ACPec (25, 27, 33) and exhibited similar FXN binding and kcat activation phenomena (Table 1) that are hallmarks of mitochondrial cysteine desulfurases (25). Moreover, we found that SDAec variants reproduce the kinetic profiles of previously identified NFS1 (RRR/AAA is analogous to human R272A R275A R277A) (44) and ISD11 (R68L) (33) substitutions that lack FXN-based activation (SI Appendix, Fig. S11). Interestingly, these results indicate that NFS1 requires the association of four additional functional proteins (ISD11, ACPec, ISCU2, and FXN) to mimic the stability and steady-state kinetic parameters of the bacterial cysteine desulfurase ortholog IscS.
Table 1.
Michaelis–Menten kinetic parameters for cysteine turnover by Fe–S cluster assembly complexes
| Complex | KM, µM | kcat, min−1 | kcat/KM, M−1⋅s−1 | Source |
| NFS1–ISD11 (SD) | 340 ± 60 | 1.9 ± 0.1 | 93 ± 20 | 25 |
| SD + FXN (SDF) | 330 ± 60 | 1.7 ± 0.1 | 86 ± 20 | 25 |
| SD + ISCU2 (SDU) | 590 ± 50 | 0.89 ± 0.04 | 25 ± 2 | 25 |
| SDAec | 1.3 ± 0.3 | 0.60 ± 0.04 | 8,000 ± 2,000 | This work |
| SDAec + ISCU2 (SDAecU) | 0.82 ± 0.3 | 0.62 ± 0.11 | 13,000 ± 5,000 | This work |
| SDUF | 11 ± 3 | 5.2 ± 0.4 | 7,900 ± 2,000 | 25 |
| SDAecUF | 10.1 ± 0.2 | 11.6 ± 0.9 | 19,100 ± 2,000 | This work |
| IscS | 17 ± 2 | 7.5 ± 0.1 | 7,400 ± 900 | 49 |
The active sites were compared to understand the lower cysteine turnover rates for eukaryotic NFS1 complexes compared with prokaryotic IscS. Inspection of the NFS1 active site revealed that most of the interactions with PLP are similar to IscS (SI Appendix, Fig. S12). One notable difference between the NFS1 and IscS active sites is the substrate channel; NFS1 has a shallow substrate-binding pocket and solvent-exposed PLP (SI Appendix, Fig. S13). In contrast, one subunit of the IscS dimer contributes to the active site of the other subunit via interactions from the N35–S40 loop and a second loop that contains the GGG motif and T243, which forms a hydrogen bond to the phosphate of the PLP cofactor (SI Appendix, Figs. S12 and S13 B and C). These contributions to the active site are precluded by the different oligomeric architecture for the eukaryotic complex and may explain the low activity for the SDAec complex as well as the activator requirement in the eukaryotic system.
The SDAec crystal structure reveals a connection between the acylated 4′-PPT and PLP cofactors in which changes in the ACP cofactor could propagate to the NFS1 active site. Although ACPec does not directly interact with NFS1, the acyl-chain attached to 4′-PPT is threaded into the ISD11 hydrophobic core and is ∼18 Å from the PLP (SI Appendix, Fig. S14). The PLP is covalently attached to K258 and anchored by interactions between the phosphate moiety and the backbone amides and side chains of T128, S255, and H257, which are invariant residues in eukaryotic cysteine desulfurases. Interestingly, the backbone carbonyl of H257 forms a hydrogen bond to the side chain of T67 of the N-terminal loop that, in turn, packs against ISD11. The N terminus and additional adjacent hydrogen-bonding residues appear to function as a relay between the acyl-chain and PLP. Moreover, NFS1 residue R412, which is downstream of the presumed cysteine interacting residue R407, forms backbone interactions with ISD11 R34 and NFS1 Y260 and participates in a salt bridge network with NFS1 residues D70 and the clinical mutant R72. These invariant residues provide a possible link between the substrate-binding site and the acyl-chain that may contribute to the altered KM for cysteine in the presence of ACPec (Table 1).
Discussion
Eukaryotes synthesize Fe–S clusters in the mitochondrial matrix using a biosynthetic pathway that contains the same basic components as the prokaryotic ISC system. However, key, unexplained functional differences exist for the Fe–S assembly complexes that center on the respective cysteine desulfurases. First, two additional proteins, ISD11 and ACP, are required for the stabilization and function of NFS1 in the eukaryotic system (1, 4, 5, 8, 9, 27). In contrast, there is no evidence for a role of ACP in prokaryotic Fe–S cluster assembly, and the ACP interactions appear limited to a disulfide bond that would inactivate IscS (45, 46). Second, despite sharing 60% sequence identity with IscS, NFS1 is inactive on its own (27, 33). Third, in vitro assays reveal that FXN stimulates the cysteine desulfurase and Fe–S cluster biosynthetic activities in the eukaryotic system (25, 33, 44, 47–51), whereas the prokaryotic FXN homolog CyaY inhibits Fe–S cluster assembly on the scaffold protein IscU (49, 52). Unexpectedly, the cysteine desulfurases and not the FXN homolog control the mode of activation/inhibition in these assays for the respective systems (49). Despite these functional differences, analogous residues for the eukaryotic (SI Appendix, Fig. S11) (44) and prokaryotic (38) cysteine desulfurases appear to contribute to the binding of FXN homologs. Lastly, the electron donor for cluster assembly, ferredoxin (FDX2), appears to bind simultaneously with the FXN homolog in yeast (30) but to compete for binding with CyaY in the prokaryotic system (53, 54). To help explain these functional differences, we determined crystallographic and EM structures of the SDAec complex and demonstrated that the SDAec complex has a fundamentally different architecture compared with all previously determined cysteine desulfurase structures.
The unexpected architecture and incompletely formed substrate-binding channel for the SDAec complex provide a rationale for the differences in prokaryotic and eukaryotic cysteine desulfurase function and their interactions with accessory proteins for Fe–S cluster biosynthesis. Although the residues that interact directly with and tune the chemistry of the PLP cofactor are largely the same (SI Appendix, Fig. S12), the different quaternary structure of SDAec results in the loss of intersubunit interactions that line a tunnel to the active site in IscS (SI Appendix, Fig. S13). We hypothesize that FXN functions as an allosteric activator by promoting a conformation that replaces these interactions. In doing so, FXN guides the NFS1 sulfur-transfer loop trajectory, increasing the efficiency and rate of sulfur transfer chemistry, which results in the observed FXN-based stimulation in the cysteine desulfurase, interprotein sulfur transfer, and Fe–S cluster assembly kinetics (25, 30, 44, 47, 48, 50, 51, 55, 56). Moreover, the cup-like architecture for the SDAec complex, in contrast to the prokaryotic system, results in the two active sites facing one another and opens up the prospect of subunit interactions across the dimeric interface. A model (SI Appendix, Fig. S15) positions the ISCU2 subunits adjacent to one another in the complex and suggests the possibility that [2Fe–2S]–ISCU2 units may be able to reductively couple their clusters to form a [4Fe–4S]–ISCU2 species, consistent with uncomplexed bacterial IscU (57) and a previous report on the mammalian Fe–S assembly complex (56). Overall, the SDAec complex provides a framework for comprehending the differences in function compared with the prokaryotic system and understanding the binding of accessory proteins to the eukaryotic cysteine desulfurase.
We propose the cysteine desulfurase architecture provides a mechanism to integrate activity control elements into the eukaryotic system. In our monomer-based association model (SI Appendix, Fig. S16), a heterotrimeric SDA species is first formed from interactions of monomeric NFS1 with ISD11–ACP. In the second stage, the heterotrimeric SDA species dimerize to form the stable 4-helix bundle core of the SDAec architecture. Consistent with this model, NFS1–NFS1 interactions appear to be significantly weaker than IscS–IscS interactions and result in a population of monomeric NFS1 (8, 33, 58), which has a tendency to aggregate in the absence of either member of the ISD11–ACP pair (1, 8). In contrast, the SDAec dimeric architecture is the predominant species that elutes from a size exclusion column (SI Appendix, Fig. S10) and is the major species present in EM samples (Fig. 4 and SI Appendix, Fig. S9), which are conducted at submicromolar concentrations similar to Fe–S assembly protein levels measured in mitochondria (59–61). Additional EM studies of a much larger and functionally distinct oligomeric form of the eukaryotic Fe–S assembly complex are also consistent with an NFS1 quaternary structure different from IscS (62). Another PLP-containing enzyme, ornithine decarboxylase, also uses quaternary structure differences to modulate activity (63). Overall, we propose that eukaryotes have adopted a cysteine desulfurase architecture that allows FXN and ACP to control Fe–S cluster biosynthesis.
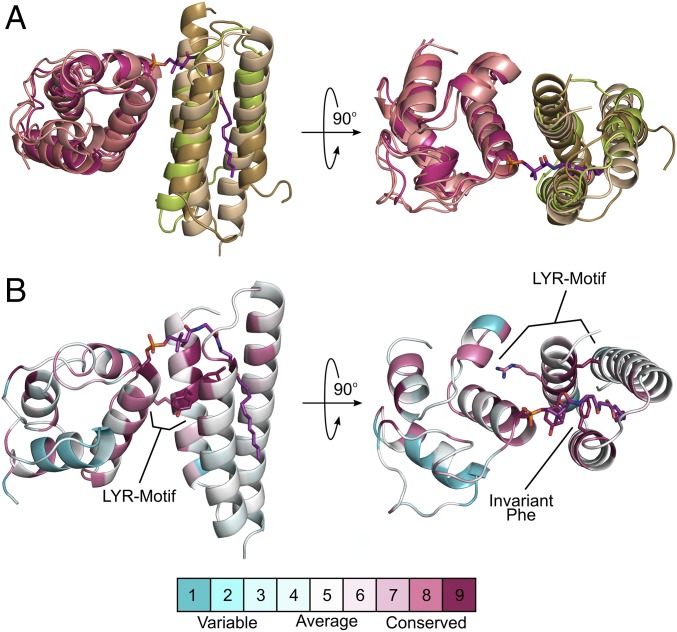
In addition, the SDAec structure provides another portrait of LYR–ACP interactions. Although the incorporation of ACP is a recently discovered component for the eukaryotic Fe–S cluster assembly machinery, its inclusion follows an emerging paradigm for interactions between mitochondrial ACP and LYR proteins (20). Previous studies demonstrate that ACP, in addition to its central role in mtFAS, forms a complex with LYRM3 and LYRM6 of Respiratory Complex I (17, 18, 21–23). Similar to the SDAec complex, the LYR motif and invariant Phe residue are required to anchor ACP to LYRM6 and to produce a functional complex (20). Overlay of ISD11–ACP with previously determined LYR–ACP structures reveals that all three LYR proteins have the same relative orientation to ACP (Fig. 5A). Mapping the sequence conservation of all 11 human LYR proteins, which include components implicated in Fe–S cluster insertion (10) and assembly factors of respiratory complexes II (64), III (65), and V (66), onto ISD11 reveals a common interface and invariant Phe that could be used to interact with ACP (Fig. 5B). Recent affinity capture MS studies further support this hypothesis by providing evidence that ACP interacts with at least 7 of the 11 human LYR proteins (67, 68), including LYR proteins that also interact with HSC20 and are implicated in the Fe–S cluster delivery mechanism (10, 69). These results suggest that the LYR superfamily forms lock-and-key interactions with ACP-associated 4′-PPT–conjugated fatty acids that influence or control their maturation and function.
Fig. 5.
LYR–ACP interactions identified in the SDAec complex appear conserved for the LYR superfamily of proteins. (A) Structural overlay of ISD11–ACP (wheat and salmon) with LYRM3–ACP (limon and deep salmon) and LYRM6–ACP (sand and warm pink) from bovine complex I (PDB ID code 4UQ8). (B) Sequence conservation of the 11 human LYR proteins mapped onto ISD11–ACP of the SDAec structure. Similar sequence conservation of ACPec and human mitochondrial ACP is also mapped to ISD11–ACP. Consurf scores, indicating relative conservation, are displayed in the scale bar.
The connection between ACP and Fe–S cluster biosynthesis is consistent with a previously proposed model in which mtFAS plays a regulatory role in oxidative respiration (14, 15). Experimental evidence supports a role of ACP-associated fatty acids in the processing of mitochondrial RNA and expression of the respiratory complexes (14, 70), synthesis of Fe–S cluster cofactors (1), and achieving the mature protein assembly and active conformation for the respiratory complexes (3, 23). Thus, it appears from our results and the work of others that the evolutionarily conserved pathways of Fe–S cluster biosynthesis, oxidative phosphorylation, and mtFAS have been connected by LYR proteins and their respective acyl-ACP associations. The mechanistic role for the acyl-chain is still unclear; however, it is possible that acyl-ACP acts as a metabolic sensor for the mitochondria by reporting the abundance of acetyl-CoA, as proposed previously (15), and allowing mtFAS to coordinate the expression, Fe–S cofactor assembly, and activity of the respiratory complexes. Overall, the cross-communication between biosynthetic and primary metabolic pathways through the utilization of LYR-acyl-ACP interactions is a potential and exciting avenue of regulation that warrants further investigation.
Materials and Methods
Protein Expression and Purification.
Plasmids containing the genes for NFS1 (pET-15b) and the natural S11A variant (71) of ISD11 (pACYDuet-1) were transformed into BL21(DE3) cells (25) and expressed using autoinduction media (72). Cultures were grown to an OD600 of 1–2 at 37 °C, the temperature was reduced to 18 °C, and the cells were collected after overnight growth. The cell pellet was resuspended in buffer A [50 mM Hepes, 500 mM NaCl, 5 mM imidazole, and 5% (vol/vol) glycerol, pH 7.8], and the cells were ruptured with six rounds of sonication. The cell lysate was combined with a buffered PLP solution (final concentration, 100 μM) and brought into an anaerobic Mbraun glovebox (∼12 °C, <1 ppm O2 monitored by a Teledyne model 310 analyzer). The lysate was loaded onto a 5 mL Ni-NTA (GE Healthcare Life Sciences) column and eluted using a linear gradient of buffer A and buffer B [50 mM Hepes, 250 mM NaCl, 500 mM imidazole, and 5% (vol/vol) glycerol, pH 7.8]. The resulting yellow fractions were pooled, diluted with buffer C (50 mM Hepes and 10% glycerol, pH 7.5), and combined with EDTA, DTT, and PLP at final concentrations of 2 mM, 5 mM, and 100 μM, respectively. The sample was loaded onto a 27-mL cation exchange (POROS 50HS, Applied Biosystems) column and eluted using a linear gradient of buffers C and D [50 mM Hepes, 1 M NaCl, and 10% (vol/vol) glycerol, pH 7.5]. Yellow fractions were pooled and combined with 2 mM EDTA, 5 mM DTT, and 100 μM PLP (final concentrations). The sample was then concentrated under argon using an amicon (10 kDa cutoff) and loaded onto a HiPrep 26/60 Sephacryl S300 HR (GE Healthcare Life Sciences) column equilibrated in buffer E [50 mM Hepes, 250 mM NaCl, and 10% (vol/vol) glycerol, pH 7.5]. The SDAec fractions were pooled, concentrated using the amicon to 280 μM (λmax = 420 nm, ε = 10.9 mM−1·cm−1), frozen in liquid nitrogen, and stored at –80 °C. For selenomethionine (SeMet)-labeled protein, proteins were expressed using a PASM-5052 medium as described by Studier (72), and the same purification strategy described above was used. SeMet containing proteins was concentrated to 215 μM, frozen in liquid nitrogen, and stored in a –80 °C freezer until further use. Site-directed mutants of NFS1 and ISD11 were constructed following the QuikChange protocol (Agilent). The resulting variants were expressed and purified as described above except that the cation column was skipped for the RRR/AAA variant. ISCU2 and FXN were expressed and purified as previously described (25). ISCU2 and FXN concentrations were determined using extinction coefficients at 280 nm of 8,250 and 26,030 M–1·cm–1, respectively.
Crystallization and X-Ray Data Collection.
SDAec aliquots, which were stored in buffer E, were thawed and diluted to 177 μM with 50 mM Hepes, 150 mM NaCl, pH 8.0, for crystallization. Optimized crystallization conditions were identified using the hanging drop vapor diffusion method in which 2 μL of 177 μM SDAec was combined with 2 μL of a well solution produced by mixing a 450-μL crystallization solution of 0.1 M Citrate BIS–Tris propane (CBTP), pH 6.4, 0.3 M CsCl, 0.2 M allylglycine, 5 mM Tris(2-carboxyethyl)phosphine (TCEP), and 8% (wt/vol) PEG 3350 with 50 μL of 40% (vol/vol) acetone. Single crystals grew within 1 wk at 22 °C. SeMet-labeled SDAec crystals were grown under similar conditions with a modified crystallization solution [0.1 M CBTP, pH 6.4, 0.3 M CsCl, 0.2 M allyglycine, 6 mM TCEP, and 4% (wt/vol) PEG 3350], which was also mixed with a 40% (vol/vol) acetone solution to provide a final well solution containing 4% (vol/vol) acetone. Crystals were harvested and cryoprotected by removing most of the liquor surrounding the crystal and gradually increasing the PEG 400 concentration in 5% (vol/vol) increments (30-s soaks) until the final concentration reached 25% (vol/vol). Crystals were then removed from the mother liquor/cryoprotection mixture using 0.3–0.4-mm loops (Hampton Research) and plunged into liquid nitrogen. All data sets were collected at SSRL beamline 7–1 (ADSC Q315R) at 100 K. A wavelength of 1.12709 Å was used for native data collection, and a wavelength of 0.9776 Å was used for SeMet data collection. The selenium peak for the SeMet containing crystals was determined by an X-ray fluorescence scan. Diffraction data were integrated, scaled, and merged using HKL2000 (73). Statistics are in SI Appendix, Table S1.
Crystallographic Structure Determination and Refinement.
Initial phases were determined by the MR-SAD method using Phaser-EP (74) in Phenix (75) using a monomer of IscS [Protein Data Bank (PDB) ID code 3LVM] (38) as a search model. A total of 46 SeMet sites were found with an initial figure-of-merit (FOM) of 0.47 at a resolution of 3.72 Å. Phenix fast chain tracing was used to build an initial model that was improved by the Phenix autobuild GUI. Phases were then extended to the native resolution using RESOLVE (76) for further model building and refinement. Phenix.refine (77) was used to randomly select the Rfree set, which corresponded to 3.4% of the total reflections and perform initial rounds of rigid body and xyz-coordinate refinement using 3LVM as a reference model to generate dihedral restraints (78). A partially sequenced NFS1–ISD11 pair was built using Coot (79) and subsequently used for MR in Phaser against the native dataset to generate four partially sequenced NFS1–ISD11 pairs in the asymmetric unit. Iterative rounds of model building and refinement were performed in Coot (79) and phenix.refine, respectively. Anomalous difference maps were used to aid in the sequencing of ISD11 and ACP (SI Appendix, Fig. S17). Due to high B factors in ACP regions, a high-resolution model of apo-ACP (1T8K.pdb) (40) was docked into the electron density for refinement. Ligand restraints from the Phenix library for the lys-PLP (LLP.pdb) cofactor were used during refinement. The acylated 4′-PPT cofactor was built using JLigand (80), and restraints were generated using ReadySet and eLBOW (81). The initial Rfree reflections selected included reflections between 3.09 Å and 3.07 Å corresponding to the edges of the detector. Thus, the Rfree set was truncated to 3.09 Å, and 26 reflections were added to the set using Phenix to generate a set corresponding to 2,000 reflections (3.4%) within refinement resolution. The B factors were reset to 103 Å2 (Wilson B factor calculated by Xtriage), and several rounds of Cartesian-simulated annealing were used to remove bias from the addition of these reflections by refining to convergence (82). The final model was refined by applying xyz-coordinate, group B factor (one per residue), TLS (one group per chain), and individual B-factor refinement. Stereochemistry, secondary structure, ADP, and torsion-angle NCS restraints were applied. Figures were generated in PyMOL (83). Data refinement statistics are listed in SI Appendix, Table S1. Chain details are listed in SI Appendix, Table S2. The final model was evaluated using Molprobity (84).
Acyl-Chain Isolation.
The acyl-chain associated with ACP in SDAec recombinant material was identified by transesterification to FAMEs (85) and GC-MS. Briefly, 1 mL of 110–150 μM SDAec was prepared by thawing frozen aliquots and diluting them with anhydrous methanol. A 10-mL solution of 10% (vol/vol) H2SO4 in anhydrous methanol was added to the 1 mL protein sample in a 25-mL screw cap vial. The sample was mixed vigorously and brought to reflux in a water bath (70–80 °C). Anhydrous sodium sulfate (1–1.5 g) was added to the mixture, incubated under reflux conditions for 2 h, and then cooled to room temperature. A small amount of anhydrous methanol was added to resuspend the white precipitate. Approximately 15 mL of hexane (mixture of isomers; Sigma-Aldrich) was added to this suspension. For best results and to maximize FAME extraction, the phases were allowed to mix vigorously on a shaker at 37 °C overnight. The organic phase was then extracted the following morning, transferred to a 15-mL glass screw cap vial, and evaporated under N2. Two additional 15-mL extractions with hexane were conducted with vigorous mixing at 37 °C for 1 h each. After all of the hexane had been evaporated, small portions of hexane were used to rinse the vial for transfer of the isolated product to a small 1-mL screw cap vial. The solvent was completely evaporated from the 1-mL vial under N2, and finally, 100 μL of hexane was added to the vial to prepare the sample for GC-MS analysis.
GC-MS Analysis of FAMEs.
GC-MS was performed on an Ultra GC/DSQ (ThermoElectron) using electron impact ionization (EI). An injection volume of 1 μL, inlet temperature of 225 °C, and Rxi-5ms (60 m length, 0.25 mm i.d., 0.25 μm film thickness; Restek) column were used. Helium was used as the carrier gas at a constant flow of 1.5 mL/min. For sample analysis, the oven temperature was initially held at 50 °C for 5 min, increased to 320 °C at a rate of 20 °C/min, and then held at 320 °C for 5 min. All standards were prepared at a concentration of 1 mg/mL in hexanes. Identification of standards and samples were based on both retention time and EI fragmentation.
Yeast Strains and Growth Conditions.
S. cerevisiae R1158 (BY4741 derivative; MATa, URA3::CMV-tTA, his3Δ1 leu2Δ0 met15Δ0) was used as the wild-type strain. Mutants were generated using standard methods by homologous recombination of PCR-amplified fragments. Yeast transformations were performed by the standard lithium acetate method, and transformed cells were recovered and grown in synthetic complete glucose (SD) medium lacking the appropriate amino acid(s) for selection purposes. S. cerevisiae cells were grown in YPA (rich media containing yeast extract, peptone, and adenine) or synthetic minimal medium supplemented with 2% glucose, 2% raffinose, or 2% glycerol. ISD11 mutants were tested with an ISD11 knockdown strain (R1158 plus isd11::KanRTetO7-CYC1TATA-ISD11) and a plasmid (ISD11-V5) previously constructed (1). To suppress ISD11 expression from the TetO7–ISD11 allele, overnight cultures were used to inoculate synthetic media containing either 2% glucose or 2% raffinose and 10 μg/mL doxycycline (DOX) to an approximate OD600 of 0.05 and were incubated for 16–24 h as indicated.
Isolation of Yeast Mitochondria and Analysis of Steady-State Protein Levels.
Cell pellets were washed once with ddH2O and incubated in TD buffer (100 mM Tris–SO4, pH 9.4 and 100 mM DTT) for 15 min at 30 °C. Spheroplasts were obtained by incubating cells in SP buffer (1.2 M sorbitol and 20 mM potassium phosphate, pH 7.4) supplemented with 0.3 mg/mL lyticase for 1 h at 30 °C to remove the cell wall. Spheroplasts were gently washed once and homogenized in ice-cold SEH buffer (0.6 M sorbitol, 20 mM Hepes-KOH, pH 7.4, 2 mM MgCl2, 1 mM EGTA) using a Dounce homogenizer applied with 30–40 strokes. Crude mitochondria were then isolated by differential centrifugation (1,500 × g for 5 min, 4,500 × g for 5 min, 10,000 × g for 10 min). Yeast mitochondria were solubilized in Laemmli buffer. Samples were resolved by SDS/PAGE and assessed by immunoblot. Blue native polyacrylamide gel electrophoresis (BN-PAGE) was performed as described previously (86). Mitochondria were resuspended in lysis buffer (Invitrogen) and solubilized with 1% digitonin. Lysates were resolved on a 4–16% gradient native gel (Invitrogen). The steady-state levels of each indicated complex was assessed by immunoblot. Porin was used as a loading control.
Single-Particle EM of the SDAec Complex.
The SDAec complex (5.0 μg/μL) was diluted to 15 ng/μL in 50 mM Hepes, pH 7.2, 200 mM NaCl, and 1 mM TCEP. A 5-μL sample was applied to a freshly glow-discharged carbon film supported by a 300 mesh copper EM grid (Electron Microscopy Sciences). After ∼1 min the protein solution was blotted from the grid and replaced with 5 μL 1% uranyl acetate. The blot and stain wash were repeated two more times. The final drop of stain was allowed to stand on the grid for ∼1 min before blotting slowly from the grid, leaving a thin film of stain that was air-dried.
SerialEM (87) was used to operate a Tecnai F30 microscope (FEI) at 300 kV equipped with a Falcon II direct electron detector (FEI) at a magnification of 59,000×, resulting in a pixel size of 1.833 Å at the specimen level, and acquire 109 images of the specimen with exposures of 42 e−/Å2 (SI Appendix, Fig. S9A). Defocus of each image was estimated with CTFFIND4 (88). In RELION-1.4 (89), an initial set of 4,167 particles were manually selected and windowed from 17 images, and a preliminary set of class averages were generated. Three averages with well-defined but dissimilar features were used to automatically select particles from all 109 images in RELION, the selections were visually assessed, and 21,973 particles were windowed. After an initial sorting procedure in RELION (90), 21,276 particles remained and these were further cleaned by 2D classification, yielding 128 class averages (SI Appendix, Fig. S9B). Sixty-three classes of incomplete complexes roughly the size of an NFS1 monomer and 20 classes of poorly defined aggregates or particles with encroaching neighbors were removed with 11,822 particles remaining (SI Appendix, Fig. S9B). In total, 36 class averages were input to e2initialmodel.py (EMAN2 version 2.12) to generate initial reconstructions with imposed C2 symmetry (91) (SI Appendix, Fig. S9C). Among the resulting initial models, a 3D map with projections that matched well with the averages was selected to initiate 3D refinement with 11,481 particles that remained following an additional round of 2D classification. The 3D classification within RELION into four classes failed to further separate the particles into meaningfully different groups, and therefore, the 11,481 particles were considered to be structurally homogenous and without further cleaning were used for refinement of the initial reconstruction in RELION with a strict high-resolution cutoff of 12 Å to prevent overfitting (SI Appendix, Fig. S9D). Fourier shell correlation calculated within RELION from independently refined maps of half of the data indicates that the final resolution of the map is ∼15 Å (SI Appendix, Fig. S9E). The final reconstruction was fit with atomic models of the SDAec crystal structure or an E. coli IscS dimer (PDB ID code 3LVM) (38) and was rendered with UCSF Chimera (92). Difference maps (SI Appendix, Fig. S9 F and G) and correlation coefficients were calculated with the “measure correlation” and “vop subtract” commands in Chimera.
Analytical Size Exclusion Chromatography.
Standards (Sigma Aldrich) including albumin (2.5 mg, 66 kDa), apoferritin (2.5 mg, 440 kDa), thyroglobulin (2 mg, 669 kDa), alcohol dehydrogenase (1.25 mg, 150 kDa), β-amylase (1 mg, 200 kDa), and carbonic anhydrase (1.25 mg, 29 kDa) were dissolved in 500 μL of 50 mM Hepes, 250 mM NaCl, pH 7.2. Approximately 250 μL of this standard mixture was loaded into a 500-μL sample loop that contained the same buffer as mentioned above. The entire sample loop (500 μL) was injected onto a Superdex 200 10/300 GL (S200, GE Healthcare Life Sciences) analytical size exclusion column. This procedure was conducted in triplicate. To determine the void volume of the column, 1 mg of blue dextran (Sigma Aldrich) was dissolved in 500 μL of 50 mM Hepes, 250 mM NaCl, pH 7.2. The entire sample was loaded into the sample loop and injected onto the S200 column. This experiment was conducted in duplicate providing a void volume (Vo) of 7.26 ± 0.10 mL. The factory provided column volume was 24 mL. The Kav for each standard was then determined using the following equation:
Samples of 4–10 μM SDAec were prepared by diluting frozen protein stock in the same buffer as mentioned above. Volumes of 500 µL were loaded into a 500-μL sample loop and the entire sample loop injected onto the S200 column. The Kav for the elution peak was determined as described above, and the molecular mass was estimated using a plot of the Kav for the standards versus log of molecular mass. An average of five runs provided a molecular mass of 123 ± 7 kDa. Protein was monitored at 280 nm.
Cysteine Desulfurase Activity Measurements.
The methylene blue assay for cysteine turnover was conducted as previously described (25) with minor modifications. Briefly, 800-μL reactions containing 0.5 μM SDA, 4 mM d,l-DTT, and 10 μM PLP in 50 mM Hepes, 250 mM NaCl, pH 7.5, were incubated for 15 min at 37 °C with and without three equivalents of the designated accessory subunit/subunits (Table 1). Reactions were initiated with up to 600 μM l-cysteine and were quenched as previously described after 6 min. The absorbance at 670 nm was converted to the concentration of sulfide using a standard curve generated from a sodium sulfide standard. Kinetic experiments were conducted in at least triplicate and fit to the Michaelis–Menten equation with KaleidaGraph (Synergy software). The errors in Table 1 represent errors in the fit.
Sequence and Structural Alignments.
The cysteine desulfurases, ISD11, and ACP were aligned using Clustal Omega default parameters (93) and displayed with Boxshade. Mitochondrial targeting sequences were truncated based on the Uniprot database. Alignments of human mitochondrial ACP and ACPec were conducted in Clustal Omega using default alignment parameters (93). All 11 human LYR proteins were aligned in a similar fashion. The sequence of LYRM6 was manually shifted, as previously described (3), using MEGA6 (94). The results of the alignment were visualized using the ConSurf server with default parameters (95). For the alignment of human and E. coli ACP (Fig. 5B), identical residues are shaded magenta and different residues are shaded based on the similarity of the amino acids. Structures were aligned in Pymol.
Supplementary Material
Acknowledgments
We thank Chris Putnam and Jennifer Bridwell-Rabb for helpful discussions, Chi-Lin Tsai for the initial crystallization conditions, and Pingwei Li and Mark F. Mabanglo for helpful experimental suggestions. We are grateful to Mike Rigney for management of the Electron Microscopy Facility at Brandeis University, where the EM data were collected; James Sacchettini for use of X-ray diffraction equipment; and the TAMU Chemistry Mass Spectrometry facility. This work was supported in part by NIH Grant R01GM110755 (to J.R. and D.R.W.), Robert A. Welch Grant A-1647 (to D.P.B.), NIH Grant R01GM096100 (to D.P.B.), and NSF Grant CHE 1508269 (to D.P.B.). Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). C.L.D and J.R. are Howard Hughes Medical Institute Investigators. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5USR).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702849114/-/DCSupplemental.
References
- 1.Van Vranken JG, et al. The mitochondrial acyl carrier protein (ACP) coordinates mitochondrial fatty acid synthesis with iron sulfur cluster biogenesis. eLife. 2016;5:e17828. doi: 10.7554/eLife.17828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roche B, et al. Iron/sulfur proteins biogenesis in prokaryotes: Formation, regulation and diversity. Biochim Biophys Acta. 2013;1827:455–469. doi: 10.1016/j.bbabio.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 3.Angerer H. Eukaryotic LYR proteins interact with mitochondrial protein complexes. Biology (Basel) 2015;4:133–150. doi: 10.3390/biology4010133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiedemann N, et al. Essential role of Isd11 in mitochondrial iron-sulfur cluster synthesis on Isu scaffold proteins. EMBO J. 2006;25:184–195. doi: 10.1038/sj.emboj.7600906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saha PP, Srivastava S, Kumar S K P, Sinha D, D’Silva P. Mapping key residues of ISD11 critical for NFS1-ISD11 subcomplex stability: Implications in the development of mitochondrial disorder, COXPD19. J Biol Chem. 2015;290:25876–25890. doi: 10.1074/jbc.M115.678508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terali K, Beavil RL, Pickersgill RW, van der Giezen M. The effect of the adaptor protein Isd11 on the quaternary structure of the eukaryotic cysteine desulphurase Nfs1. Biochem Biophys Res Commun. 2013;440:235–240. doi: 10.1016/j.bbrc.2013.09.039. [DOI] [PubMed] [Google Scholar]
- 7.Pandey A, Golla R, Yoon H, Dancis A, Pain D. Persulfide formation on mitochondrial cysteine desulfurase: Enzyme activation by a eukaryote-specific interacting protein and Fe-S cluster synthesis. Biochem J. 2012;448:171–187. doi: 10.1042/BJ20120951. [DOI] [PubMed] [Google Scholar]
- 8.Adam AC, Bornhövd C, Prokisch H, Neupert W, Hell K. The Nfs1 interacting protein Isd11 has an essential role in Fe/S cluster biogenesis in mitochondria. EMBO J. 2006;25:174–183. doi: 10.1038/sj.emboj.7600905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi Y, Ghosh MC, Tong WH, Rouault TA. Human ISD11 is essential for both iron-sulfur cluster assembly and maintenance of normal cellular iron homeostasis. Hum Mol Genet. 2009;18:3014–3025. doi: 10.1093/hmg/ddp239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maio N, et al. Cochaperone binding to LYR motifs confers specificity of iron sulfur cluster delivery. Cell Metab. 2014;19:445–457. doi: 10.1016/j.cmet.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 12.Brody S, Oh C, Hoja U, Schweizer E. Mitochondrial acyl carrier protein is involved in lipoic acid synthesis in Saccharomyces cerevisiae. FEBS Lett. 1997;408:217–220. doi: 10.1016/s0014-5793(97)00428-6. [DOI] [PubMed] [Google Scholar]
- 13.Hiltunen JK, et al. Mitochondrial fatty acid synthesis type II: More than just fatty acids. J Biol Chem. 2009;284:9011–9015. doi: 10.1074/jbc.R800068200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kursu VA, et al. Defects in mitochondrial fatty acid synthesis result in failure of multiple aspects of mitochondrial biogenesis in Saccharomyces cerevisiae. Mol Microbiol. 2013;90:824–840. doi: 10.1111/mmi.12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiltunen JK, et al. Mitochondrial fatty acid synthesis and respiration. Biochim Biophys Acta. 2010;1797:1195–1202. doi: 10.1016/j.bbabio.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Brody S, Mikolajczyk S. Neurospora mitochondria contain an acyl-carrier protein. Eur J Biochem. 1988;173:353–359. doi: 10.1111/j.1432-1033.1988.tb14005.x. [DOI] [PubMed] [Google Scholar]
- 17.Runswick MJ, Fearnley IM, Skehel JM, Walker JE. Presence of an acyl carrier protein in NADH:ubiquinone oxidoreductase from bovine heart mitochondria. FEBS Lett. 1991;286:121–124. doi: 10.1016/0014-5793(91)80955-3. [DOI] [PubMed] [Google Scholar]
- 18.Sackmann U, Zensen R, Röhlen D, Jahnke U, Weiss H. The acyl-carrier protein in Neurospora crassa mitochondria is a subunit of NADH:ubiquinone reductase (complex I) Eur J Biochem. 1991;200:463–469. doi: 10.1111/j.1432-1033.1991.tb16205.x. [DOI] [PubMed] [Google Scholar]
- 19.Carroll J, Fearnley IM, Shannon RJ, Hirst J, Walker JE. Analysis of the subunit composition of complex I from bovine heart mitochondria. Mol Cell Proteomics. 2003;2:117–126. doi: 10.1074/mcp.M300014-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Angerer H, et al. The LYR protein subunit NB4M/NDUFA6 of mitochondrial complex I anchors an acyl carrier protein and is essential for catalytic activity. Proc Natl Acad Sci USA. 2014;111:5207–5212. doi: 10.1073/pnas.1322438111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vinothkumar KR, Zhu J, Hirst J. Architecture of mammalian respiratory complex I. Nature. 2014;515:80–84. doi: 10.1038/nature13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu J, et al. Structure of subcomplex Iβ of mammalian respiratory complex I leads to new supernumerary subunit assignments. Proc Natl Acad Sci USA. 2015;112:12087–12092. doi: 10.1073/pnas.1510577112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fiedorczuk K, et al. Atomic structure of the entire mammalian mitochondrial complex I. Nature. 2016;538:406–410. doi: 10.1038/nature19794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lill R, Mühlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: Mechanisms, connected processes, and diseases. Annu Rev Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- 25.Tsai CL, Barondeau DP. Human frataxin is an allosteric switch that activates the Fe-S cluster biosynthetic complex. Biochemistry. 2010;49:9132–9139. doi: 10.1021/bi1013062. [DOI] [PubMed] [Google Scholar]
- 26.Schmucker S, et al. Mammalian frataxin: An essential function for cellular viability through an interaction with a preformed ISCU/NFS1/ISD11 iron-sulfur assembly complex. PLoS One. 2011;6:e16199. doi: 10.1371/journal.pone.0016199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marelja Z, Stöcklein W, Nimtz M, Leimkühler S. A novel role for human Nfs1 in the cytoplasm: Nfs1 acts as a sulfur donor for MOCS3, a protein involved in molybdenum cofactor biosynthesis. J Biol Chem. 2008;283:25178–25185. doi: 10.1074/jbc.M804064200. [DOI] [PubMed] [Google Scholar]
- 28.Rouault TA. Mammalian iron-sulphur proteins: Novel insights into biogenesis and function. Nat Rev Mol Cell Biol. 2015;16:45–55. doi: 10.1038/nrm3909. [DOI] [PubMed] [Google Scholar]
- 29.Shi Y, Ghosh M, Kovtunovych G, Crooks DR, Rouault TA. Both human ferredoxins 1 and 2 and ferredoxin reductase are important for iron-sulfur cluster biogenesis. Biochim Biophys Acta. 2012;1823:484–492. doi: 10.1016/j.bbamcr.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webert H, et al. Functional reconstitution of mitochondrial Fe/S cluster synthesis on Isu1 reveals the involvement of ferredoxin. Nat Commun. 2014;5:5013. doi: 10.1038/ncomms6013. [DOI] [PubMed] [Google Scholar]
- 31.Nakai Y, et al. Yeast Nfs1p is involved in thio-modification of both mitochondrial and cytoplasmic tRNAs. J Biol Chem. 2004;279:12363–12368. doi: 10.1074/jbc.M312448200. [DOI] [PubMed] [Google Scholar]
- 32.Farhan SM, et al. Exome sequencing identifies NFS1 deficiency in a novel Fe-S cluster disease, infantile mitochondrial complex II/III deficiency. Mol Genet Genomic Med. 2014;2:73–80. doi: 10.1002/mgg3.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim SC, et al. Mutations in LYRM4, encoding iron-sulfur cluster biogenesis factor ISD11, cause deficiency of multiple respiratory chain complexes. Hum Mol Genet. 2013;22:4460–4473. doi: 10.1093/hmg/ddt295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olsson A, Lind L, Thornell LE, Holmberg M. Myopathy with lactic acidosis is linked to chromosome 12q23.3-24.11 and caused by an intron mutation in the ISCU gene resulting in a splicing defect. Hum Mol Genet. 2008;17:1666–1672. doi: 10.1093/hmg/ddn057. [DOI] [PubMed] [Google Scholar]
- 35.Campuzano V, et al. Friedreich’s ataxia: Autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 36.Cai K, Frederick RO, Tonelli M, Markley JL. Mitochondrial cysteine desulfurase and ISD11 coexpressed in Escherichia coli yield complex containing acyl carrier protein. ACS Chem Biol. 2017;12:918–921. doi: 10.1021/acschembio.6b01005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cupp-Vickery JR, Urbina H, Vickery LE. Crystal structure of IscS, a cysteine desulfurase from Escherichia coli. J Mol Biol. 2003;330:1049–1059. doi: 10.1016/s0022-2836(03)00690-9. [DOI] [PubMed] [Google Scholar]
- 38.Shi R, et al. Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions. PLoS Biol. 2010;8:e1000354. doi: 10.1371/journal.pbio.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marinoni EN, et al. (IscS-IscU)2 complex structures provide insights into Fe2S2 biogenesis and transfer. Angew Chem Int Ed Engl. 2012;51:5439–5442. doi: 10.1002/anie.201201708. [DOI] [PubMed] [Google Scholar]
- 40.Qiu X, Janson CA. Structure of apo acyl carrier protein and a proposal to engineer protein crystallization through metal ions. Acta Crystallogr D Biol Crystallogr. 2004;60:1545–1554. doi: 10.1107/S0907444904015422. [DOI] [PubMed] [Google Scholar]
- 41.Byers DM, Gong H. Acyl carrier protein: Structure-function relationships in a conserved multifunctional protein family. Biochem Cell Biol. 2007;85:649–662. doi: 10.1139/o07-109. [DOI] [PubMed] [Google Scholar]
- 42.Kaiser JT, et al. Crystal structure of a NifS-like protein from Thermotoga maritima: Implications for iron sulphur cluster assembly. J Mol Biol. 2000;297:451–464. doi: 10.1006/jmbi.2000.3581. [DOI] [PubMed] [Google Scholar]
- 43.Yan R, et al. The eukaryotic-specific ISD11 is a complex-orphan protein with ability to bind the prokaryotic IscS. PLoS One. 2016;11:e0157895. doi: 10.1371/journal.pone.0157895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manicki M, et al. Overlapping binding sites of the frataxin homologue assembly factor and the heat shock protein 70 transfer factor on the Isu iron-sulfur cluster scaffold protein. J Biol Chem. 2014;289:30268–30278. doi: 10.1074/jbc.M114.596726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gully D, Moinier D, Loiseau L, Bouveret E. New partners of acyl carrier protein detected in Escherichia coli by tandem affinity purification. FEBS Lett. 2003;548:90–96. doi: 10.1016/s0014-5793(03)00746-4. [DOI] [PubMed] [Google Scholar]
- 46.Flint DH. Escherichia coli contains a protein that is homologous in function and N-terminal sequence to the protein encoded by the nifS gene of Azotobacter vinelandii and that can participate in the synthesis of the Fe-S cluster of dihydroxy-acid dehydratase. J Biol Chem. 1996;271:16068–16074. [PubMed] [Google Scholar]
- 47.Fox NG, Das D, Chakrabarti M, Lindahl PA, Barondeau DP. Frataxin accelerates [2Fe-2S] cluster formation on the human Fe-S assembly complex. Biochemistry. 2015;54:3880–3889. doi: 10.1021/bi5014497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bridwell-Rabb J, Fox NG, Tsai CL, Winn AM, Barondeau DP. Human frataxin activates Fe-S cluster biosynthesis by facilitating sulfur transfer chemistry. Biochemistry. 2014;53:4904–4913. doi: 10.1021/bi500532e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bridwell-Rabb J, Iannuzzi C, Pastore A, Barondeau DP. Effector role reversal during evolution: The case of frataxin in Fe-S cluster biosynthesis. Biochemistry. 2012;51:2506–2514. doi: 10.1021/bi201628j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pandey A, et al. Frataxin directly stimulates mitochondrial cysteine desulfurase by exposing substrate-binding sites, and a mutant Fe-S cluster scaffold protein with frataxin-bypassing ability acts similarly. J Biol Chem. 2013;288:36773–36786. doi: 10.1074/jbc.M113.525857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parent A, et al. Mammalian frataxin directly enhances sulfur transfer of NFS1 persulfide to both ISCU and free thiols. Nat Commun. 2015;6:5686. doi: 10.1038/ncomms6686. [DOI] [PubMed] [Google Scholar]
- 52.Adinolfi S, et al. Bacterial frataxin CyaY is the gatekeeper of iron-sulfur cluster formation catalyzed by IscS. Nat Struct Mol Biol. 2009;16:390–396. doi: 10.1038/nsmb.1579. [DOI] [PubMed] [Google Scholar]
- 53.Yan R, et al. Ferredoxin competes with bacterial frataxin in binding to the desulfurase IscS. J Biol Chem. 2013;288:24777–24787. doi: 10.1074/jbc.M113.480327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim JH, Frederick RO, Reinen NM, Troupis AT, Markley JL. [2Fe-2S]-ferredoxin binds directly to cysteine desulfurase and supplies an electron for iron-sulfur cluster assembly but is displaced by the scaffold protein or bacterial frataxin. J Am Chem Soc. 2013;135:8117–8120. doi: 10.1021/ja401950a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turowski VR, Busi MV, Gomez-Casati DF. Structural and functional studies of the mitochondrial cysteine desulfurase from Arabidopsis thaliana. Mol Plant. 2012;5:1001–1010. doi: 10.1093/mp/sss037. [DOI] [PubMed] [Google Scholar]
- 56.Colin F, et al. Mammalian frataxin controls sulfur production and iron entry during de novo Fe4S4 cluster assembly. J Am Chem Soc. 2013;135:733–740. doi: 10.1021/ja308736e. [DOI] [PubMed] [Google Scholar]
- 57.Agar JN, et al. IscU as a scaffold for iron-sulfur cluster biosynthesis: Sequential assembly of [2Fe-2S] and [4Fe-4S] clusters in IscU. Biochemistry. 2000;39:7856–7862. doi: 10.1021/bi000931n. [DOI] [PubMed] [Google Scholar]
- 58.Friemel M, Marelja Z, Li K, Leimkuhler S. The N-terminus of iron–sulfur cluster assembly factor ISD11 is crucial for subcellular targeting and interactions with l-cysteine desulfurase NFS1. Biochemistry. 2017;56:1797–1808. doi: 10.1021/acs.biochem.6b01239. [DOI] [PubMed] [Google Scholar]
- 59.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 60.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 61.Hudder BN, et al. Electron paramagnetic resonance and Mössbauer spectroscopy of intact mitochondria from respiring Saccharomyces cerevisiae. J Biol Inorg Chem. 2007;12:1029–1053. doi: 10.1007/s00775-007-0275-1. [DOI] [PubMed] [Google Scholar]
- 62.Gakh O, et al. Architecture of the human mitochondrial iron-sulfur cluster assembly machinery. J Biol Chem. 2016;291:21296–21321. doi: 10.1074/jbc.M116.738542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu HY, et al. Structural basis of antizyme-mediated regulation of polyamine homeostasis. Proc Natl Acad Sci USA. 2015;112:11229–11234. doi: 10.1073/pnas.1508187112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Na U, et al. The LYR factors SDHAF1 and SDHAF3 mediate maturation of the iron-sulfur subunit of succinate dehydrogenase. Cell Metab. 2014;20:253–266. doi: 10.1016/j.cmet.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Atkinson A, et al. The LYR protein Mzm1 functions in the insertion of the Rieske Fe/S protein in yeast mitochondria. Mol Cell Biol. 2011;31:3988–3996. doi: 10.1128/MCB.05673-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lefebvre-Legendre L, et al. Identification of a nuclear gene (FMC1) required for the assembly/stability of yeast mitochondrial F(1)-ATPase in heat stress conditions. J Biol Chem. 2001;276:6789–6796. doi: 10.1074/jbc.M009557200. [DOI] [PubMed] [Google Scholar]
- 67.Huttlin EL, et al. The BioPlex network: A systematic exploration of the human interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Floyd BJ, et al. Mitochondrial protein interaction mapping identifies regulators of respiratory chain function. Mol Cell. 2016;63:621–632. doi: 10.1016/j.molcel.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maio N, Kim KS, Singh A, Rouault TA. A single adaptable cochaperone-scaffold complex delivers nascent iron-sulfur clusters to mammalian respiratory chain complexes I-III. Cell Metab. 2017;25:945–953.e6. doi: 10.1016/j.cmet.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 70.Schonauer MS, Kastaniotis AJ, Hiltunen JK, Dieckmann CL. Intersection of RNA processing and the type II fatty acid synthesis pathway in yeast mitochondria. Mol Cell Biol. 2008;28:6646–6657. doi: 10.1128/MCB.01162-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gerhard DS, et al. MGC Project Team The status, quality, and expansion of the NIH full-length cDNA project: The Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 73.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromol Crystallogr Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 74.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Terwilliger T. SOLVE and RESOLVE: Automated structure solution, density modification and model building. J Synchrotron Radiat. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 77.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Headd JJ, et al. Use of knowledge-based restraints in phenix.refine to improve macromolecular refinement at low resolution. Acta Crystallogr D Biol Crystallogr. 2012;68:381–390. doi: 10.1107/S0907444911047834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lebedev AA, et al. JLigand: a graphical tool for the CCP4 template-restraint library. Acta Crystallogr D Biol Crystallogr. 2012;68:431–440. doi: 10.1107/S090744491200251X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moriarty NW, Grosse-Kunstleve RW, Adams PD. electronic Ligand Builder and Optimization Workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Crystallogr D Biol Crystallogr. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brünger AT. Assessment of phase accuracy by cross validation: The free R value. Methods and applications. Acta Crystallogr D Biol Crystallogr. 1993;49:24–36. doi: 10.1107/S0907444992007352. [DOI] [PubMed] [Google Scholar]
- 83. Schrödinger, LLC (2015) The PyMOL Molecular Graphics System (Schrödinger, LLC, New York), Version 1.3.
- 84.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35(Web Server issue):W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Christie WW. Preparation of ester derivatives of fatty acids for chromatographic analysis. In: Christie WW, editor. Advances in Lipid Methodology - Two. Oily Press; Dundee, Scotland: 1993. pp. 69–111. [Google Scholar]
- 86.Wittig I, Braun HP, Schägger H. Blue native PAGE. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 87.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 88.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Scheres SH. Semi-automated selection of cryo-EM particles in RELION-1.3. J Struct Biol. 2015;189:114–122. doi: 10.1016/j.jsb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 92.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 93.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Armon A, Graur D, Ben-Tal N. ConSurf: An algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J Mol Biol. 2001;307:447–463. doi: 10.1006/jmbi.2000.4474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.