Significance
Mosquitoes are important insects because several species transmit pathogens as adults that cause disease in humans and other vertebrates. One approach for control is preventing immature mosquitoes from developing into adults. Immature-stage mosquitoes require gut bacteria to develop, but the mechanisms underlying this dependence are unknown. Here, we identify cytochrome bd oxidase as a bacterial product involved in mosquito development. We also show that bacteria-mediated reduction of oxygen levels in the digestive tract of larvae serves as a signal for molting. These findings provide the first evidence that aerobic respiration by bacteria plays an essential role in mosquito development. This information can also potentially be used to develop tools for disabling the growth of larval mosquitoes into adults.
Keywords: insect, growth, bacteria, microbiota, hypoxia
Abstract
Mosquitoes host communities of microbes in their digestive tract that consist primarily of bacteria. We previously reported that several mosquito species, including Aedes aegypti, do not develop beyond the first instar when fed a nutritionally complete diet in the absence of a gut microbiota. In contrast, several species of bacteria, including Escherichia coli, rescue development of axenic larvae into adults. The molecular mechanisms underlying bacteria-dependent growth are unknown. Here, we designed a genetic screen around E. coli that identified high-affinity cytochrome bd oxidase as an essential bacterial gene product for mosquito growth. Bioassays showed that bacteria in nonsterile larvae and gnotobiotic larvae inoculated with wild-type E. coli reduced midgut oxygen levels below 5%, whereas larvae inoculated with E. coli mutants defective for cytochrome bd oxidase did not. Experiments further supported that hypoxia leads to growth and ecdysone-induced molting. Altogether, our results identify aerobic respiration by bacteria as a previously unknown but essential process for mosquito development.
Gut-dwelling microbes are of interest because of their potential effects on growth, development, and survival of animal hosts, including mosquitoes (1, 2). All mosquitoes are aquatic during their juvenile stages, with larvae feeding primarily on detritus and small organisms (3). As adults, most female mosquitoes blood-feed on vertebrates, which creates opportunities for transmission of pathogens that cause disease in humans and other species (4). Mosquitoes contain no microbes in their digestive tract after hatching from eggs, which results in larvae acquiring a gut microbiota anew each generation from the environment (5, 6). The majority of these community members are gram-negative aerobic and facultatively anaerobic bacteria (7–12). Some, but not all, community members are also transstadially transmitted to adults (9, 11). Prior studies show that gut microbes in adult females can affect vector competence and pathogen transmission to vertebrates (13, 14). They also indicate that gut bacteria strongly affect development (6). The absence of a tractable genetic model, however, has limited understanding of the molecular interactions that underlie bacteria-dependent growth of larvae into adults.
We recently established Escherichia coli K-12 BW25113 as a model for this purpose in conjunction with the mosquito Aedes aegypti. University of Georgia strain (UGAL) Ae. aegypti larvae reared under conventional (nonsterile) laboratory conditions contain a relatively simple community of ∼100 bacterial species if fed a nutritionally complete diet (9). Larvae also molt through four instars before pupating and emerging as adults (15). Axenic larvae with no gut microbiota die as first instars without molting but are rescued if inoculated with the community of bacteria present in a conventional culture (9). Several bacteria from this community and E. coli K-12, which is not a community member, also individually colonize the gut to produce gnotobiotic larvae that exhibit no differences in development time, adult size, or fecundity relative to conventionally reared larvae (9, 16). Field-collected Ae. aegypti and several other species of mosquitoes contain communities of gut bacteria that differ from laboratory-reared Ae. aegypti but exhibit the same defects under axenic conditions as UGAL Ae. aegypti (9, 17). Their development is also rescued if inoculated with only E. coli (9, 17).
Taken together, several mosquito species die as first instars when fed a nutritionally complete diet in the absence of a gut microbiota but develop into adults if living bacteria are present. Prior findings, however, argue against bacteria solely being a food source or provisioning nutrients essential for growth because axenic larvae do not develop beyond the first instar if fed dead bacteria, a standard diet with dead bacteria, or a standard diet that has been preconditioned by living bacteria before feeding (9). They also argue against Ae. aegypti requiring a particular species or community of bacteria because gnotobiotic larvae develop normally when singly colonized by several species, including E. coli (9, 16, 17). In this study, we used E. coli to gain insights into what bacteria provide that Ae. aegypti larvae require to grow. Our results identified cytochrome bd oxidase as a critical gene product. Functional data further indicated that bacteria reduce gut oxygen levels, which serves as a signal for growth and molting.
Results
Screening an E. coli Single-Gene Knockout Library Identifies Several Mutants That Adversely Affect Larval Growth.
Within each instar, insect larvae grow by feeding until they achieve a critical size (18). Endocrine events associated with critical size then stimulate a rise in titer of the steroid hormone 20-hydroxyecdysone (20E), which induces molting (18). Under standardized conditions, conventionally reared Ae. aegypti larvae and gnotobiotic larvae inoculated with wild-type E. coli K-12 exhibit no differences in when they achieve critical size (12–16 h posthatching) and molt to the second instar (∼24 h posthatching) (15). Conventional and gnotobiotic larvae also synchronously molt to the third instar and fourth instar at 48 h and 72 h posthatching, respectively, before pupating. In contrast, axenic first instars feed but never reach critical size and die without molting (15). We used this information and an E. coli K-12 knockout library of 3,985 single genes (Keio collection) (19) in a forward genetic screen to identify mutants defective in stimulating molting. After inoculating newly hatched axenic larvae with each mutant, the majority (97%) had little or no adverse effect, with most larvae molting by 24 h and all larvae molting by 48 h. However, 131 mutants were classified as defective because larvae either required longer than 48 h to molt or never molted.
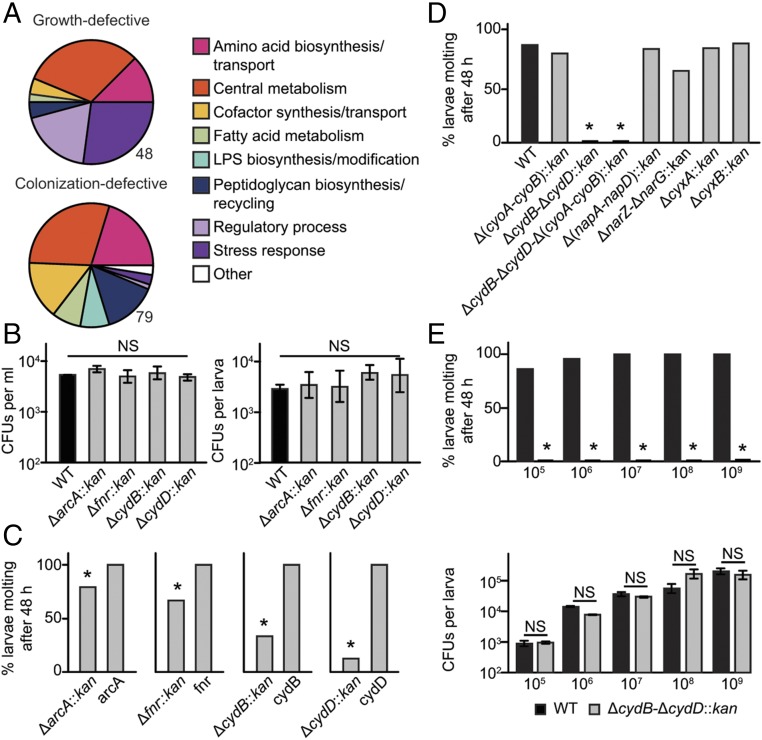
We subclassified 48 of these mutants as growth-defective because plate assays detected no bacteria at 24 h postinoculation in the culture environment, which consisted of water containing larval rearing diet (SI Appendix, Table S1). We subclassified 79 others as colonization-defective because viable bacteria were detected in the culture environment at 24 h postinoculation but were not detected in homogenates of larvae (SI Appendix, Table S1). The absence of viable bacteria in larvae suggested defects in the ability of these mutants to survive in the gut, which resulted in few or no larvae molting within 48 h. However, for many of these mutants, we determined that the proportion of larvae molting within 48 h increased if cultures were inoculated with a larger number of bacteria (SI Appendix, Table S1). Thus, defects for colonization-defective mutants were conditional, with starting abundance likely affecting the number of viable bacteria that persisted in the culture environment and/or the larval gut. A large proportion of the growth- and colonization-defective mutants lacked genes with roles in four broad gene ontology categories: central metabolism, amino acid biosynthesis/transport, peptidoglycan recycling, or stress responses that could slow or otherwise adversely affect bacterial growth under the culture conditions used to rear mosquitoes (Fig. 1A and SI Appendix, Table S1).
Fig. 1.
Multiple E. coli mutants affect growth and molting of Ae. aegypti larvae. (A) Functional clustering of the 48 growth-defective and 79 colonization-defective single-gene deletion mutants. Pie charts show the gene ontology (GO) categories to which deleted genes in the mutants belonged. Genes that fell into multiple GO categories are grouped together in the category designated as “Other.” (B) Abundance of the four rescue-defective mutants and wild-type (WT) E. coli in cultures (Left) and larvae (Right) at 24 h postinoculation. A minimum of four replicates were assayed per treatment. Each bar indicates mean ± SE colony-forming units. ANOVA detected no differences between treatments for either water (F4,14 = 1.385, P = 0.289) or larvae (F4,14 = 1.536, P = 0.245). (C) Percentage of first instars that molted when inoculated with each rescue-defective mutant vs. the same mutant transformed with an expression plasmid containing the deleted gene. An asterisk (*) indicates molting significantly differed between the two treatments, with all rescued mutants stimulating 100% of larvae to molt (P < 0.05, Fisher’s exact test). At least 24 larvae were assayed per treatment. (D) Percentage of first-instar larvae molting to the second instar when inoculated with WT E. coli or the mutants Δ(cyoA-cyoB)::kan, ΔcydB-ΔcydD::kan, ΔcydB-ΔcydD-Δ(cyoA-cyoB)::kan, Δ(napA-napD)::kan, ΔnarZ-ΔnarG::kan, ΔcyxA::kan, and ΔcyxB::kan. An asterisk (*) indicates a significant difference for a given mutant relative to the WT positive control (P < 0.0001, Fisher’s exact test). At least 72 larvae were assayed per treatment. (E, Upper) Percentage of first instars molting to the second instar when cultures were inoculated with 105–109 cfu of WT vs. ΔcydB-ΔcydD::kan E. coli. An asterisk (*) indicates a significant difference between treatments (P < 0.0001, Fisher’s exact test). At least 72 larvae were assayed per treatment. (E, Lower) Mean ± SE colony-forming units present per larva at 24 h postinoculation for the cultures shown above. For each inoculation amount, t tests detected no significant difference (NS) in colony-forming units per larva between treatments (P > 0.05). A minimum of four individuals were assayed per treatment.
The last four defective mutants (ΔarcA::kan, Δfnr::kan, ΔcydB::kan, and ΔcydD::kan) were of greatest interest because each was similarly abundant in cultures and first instars as wild-type E. coli at 24 h postinoculation, but the proportion of larvae that molted by 48 h was significantly lower than for wild-type E. coli (Fig. 1 B and C and SI Appendix, Table S1). We subclassified these mutants as rescue-defective. Notably, each had functions in respiration, with cydB and cydD encoding products required for assembly of cytochrome bd oxidase, a terminal enzyme in the aerobic electron transport chain (20, 21), and arcA and fnr encoding regulators that mediate expression of many genes, including cydB and cydD, that respond to aerobic vs. anaerobic conditions (22).
The Cytochrome bd Oxidase Respiratory Pathway Affects Larval Growth and Molting.
We assessed whether the molting defects associated with ΔarcA::kan, Δfnr::kan, ΔcydB::kan, and ΔcydD::kan E. coli were rescued by in trans provision of each gene on a plasmid. Results indicated they were, which further suggested E. coli respiration directly affected larval growth and molting (Fig. 1C). However, regulators like ArcA and Fnr mediate expression of other terminal oxidoreductases besides cytochrome bd oxidase, which, together, provide E. coli with respiratory flexibility as a facultative anaerobe. Under ambient (normoxic) aerobic conditions (21% O2), E. coli predominantly uses low-affinity cytochrome bo3 oxidase encoded by the cyoABCDE operon (23). However, under lower (hypoxic) oxygen conditions (10–3% O2), E. coli shifts to high-affinity cytochrome bd oxidase produced from the cydAB and cydDC genes (23, 24). A third enzyme, cytochrome bd oxidase II (encoded by cyxAB), has also been suggested to function under very low oxygen conditions (25). In contrast, anaerobic respiration in the vertebrate gut involves the reduction of nitrate by primary nitrate reductase encoded by the narGHJI operon, secondary nitrate reductase encoded by the narZYWV operon, and periplasmic nitrate reductase encoded by the napFDAGHBC operon (26–29).
We therefore examined whether multiple E. coli respiratory pathways affect growth and molting of Ae. aegypti first instars. For aerobic respiration, we produced a double mutant that deleted the genes encoding subunits I and II of cytochrome bo3 oxidase [Δ(cyoA-cyoB)::kan], a double mutant that deleted the gene for subunit II of cytochrome bd oxidase and the gene for a transporter subunit required for assembly in the membrane (ΔcydB-ΔcydD::kan) (20, 21), and a quadruple mutant defective for both enzymes [ΔcydB-ΔcydD-Δ(cyoA-cyoB)::kan]. We also assayed ΔcyxA::kan and ΔcyxB::kan from the Keio collection to assess whether defective cytochrome bd oxidase II (25) affected larvae. For anaerobic respiration, we generated a ΔnarZ-ΔnarG::kan double mutant to eliminate genes encoding subunits for both primary and secondary nitrate reductase, whereas periplasmic nitrate reductase was eliminated by generating a double mutant [Δ(napA-napD)::kan] lacking genes for the assembly protein and large reductase subunit (28). Only larvae inoculated with the mutants defective for cytochrome bd oxidase [ΔcydB-ΔcydD::kan or ΔcydB-ΔcydD-Δ(cyoA-cyoB)::kan] failed to molt (Fig. 1D). Increasing the number of ΔcydB-ΔcydD::kan bacteria used to inoculate cultures up to 109 per milliliter did not alter this outcome, whereas comparison with controls inoculated with wild-type E. coli showed no differences in the abundance of viable bacteria per larva (Fig. 1E). Prior results had shown that the abundance and distribution of bacteria in conventional and gnotobiotic larvae inoculated with wild-type E. coli were indistinguishable, with all bacterial cells in the midgut residing in the endoperitrophic space formed by the peritrophic matrix (15). Inoculating larvae with same starting density of bacteria (106 per milliliter) showed that the distribution of ΔcydB-ΔcydD::kan E. coli in the guts of larvae at 12 h posthatching was also indistinguishable from conventional larvae and wild-type E. coli gnotobiotic larvae (SI Appendix, Fig. S1). Loss of cytochrome bd oxidase therefore perturbed growth of Ae. aegypti first instars but had no effect on E. coli abundance and distribution in the gut.
Cytochrome bd Oxidase-Dependent Respiration Reduces Gut Oxygen Levels in Precritical Size Larvae.
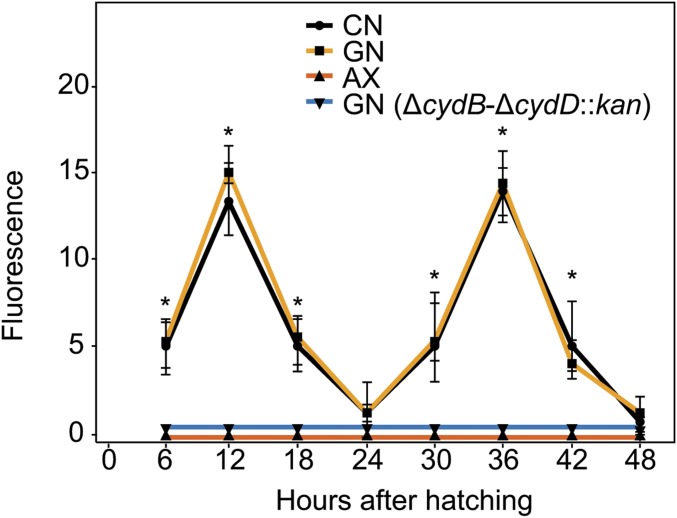
We assessed whether aerobic respiration by bacteria affects oxygen availability in the mosquito gut using a hypoxia marker (Image-iT Hypoxia Reagent; Life Technologies) that fluoresces with increasing intensity as atmospheric oxygen falls below 5%. For conventional and gnotobiotic first instars inoculated with wild-type E. coli, Image iT fluorescence increased up to 12 h posthatching but then declined after larvae achieved a critical size until molting at 24 h (Fig. 2 and SI Appendix, Fig. S2). Second instars exhibited the same pattern, with fluorescence increasing until larvae achieved a critical size (∼40 h) and then declining until larvae molted to the third instar at 48 h (Fig. 2 and SI Appendix, Fig. S2). In contrast, axenic and gnotobiotic larvae inoculated with ΔcydB-ΔcydD::kan E. coli, which remained first instars, exhibited no Image iT fluorescence over 48 h (Fig. 2 and SI Appendix, Fig. S3). Detection of a gut hypoxia signal in conventional larvae and wild-type E. coli gnotobiotic larvae, but the absence of a gut hypoxia signal in axenic larvae, supported a role for aerobic respiration by bacteria in lowering gut oxygen levels. The absence of a hypoxia signal in larvae inoculated with ΔcydB-ΔcydD::kan E. coli further implicated cytochrome bd oxidase in this response.
Fig. 2.
Quantitation of Image iT fluorescence in the midguts of conventional larvae (CN), gnotobiotic larvae inoculated with WT E. coli (GN), axenic larvae (AX), and gnotobiotic larvae inoculated with ΔcydB-ΔcydD::kan E. coli [GN (ΔcydB-ΔcydD::kan)]. Individual larvae for each treatment were examined by confocal microscopy from 6 to 48 h posthatching. Fluorescence intensity in the midgut was measured in 10 larvae per treatment and time point using the Pixel Intensity plug-in from ImageJ software (NIH). Mean pixel intensity values ± SD are presented. An asterisk (*) at a given time point indicates that the pixel intensity significantly differed for the CN and GN treatments relative to the AX and GN (ΔcydB-ΔcydD::kan) treatments (ANOVA followed by a post hoc Tukey–Kramer honest significant difference test, P < 0.05). Corresponding confocal images are shown in SI Appendix, Figs. S2 and S3.
Reduced Viability of Bacteria Correlates with Increased Oxygenation of the Gut in Postcritical Size Larvae.
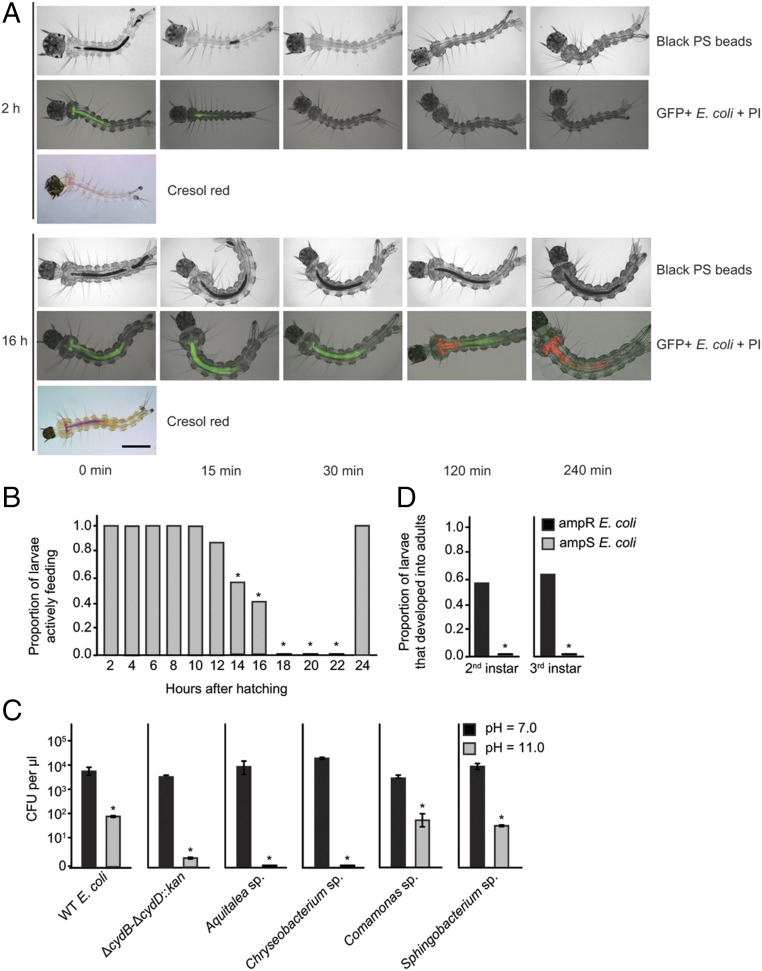
Unclear from the preceding results was why gut oxygen levels in conventional and gnotobiotic larvae inoculated with wild-type E. coli were below 5% before achieving critical size but above 5% after achieving critical size. To further examine why oxygen levels fluctuated in this manner, we fed first instars at 2 h posthatching (precritical size) a standard rearing diet containing black polystyrene microspheres and E. coli K-12 expressing green fluorescent protein (GFP+) (Fig. 3A). We then transferred larvae to culture wells containing only water and propidium iodide (PI) to monitor rates of excretion and the viability of ingested bacteria. Results showed that larvae excreted ingested food and bacteria within 30 min, whereas the absence of PI staining indicated most, if not all, bacteria in the midgut were viable before being excreted (Fig. 3A). We then examined larvae at 16 h posthatching, which were at the cusp of critical size. Many of these larvae fed but thereafter excreted little or no food and bacteria over a 240-min (4-h) observation period (Fig. 3A). The viability of the bacteria in the midgut also markedly declined, as evidenced by PI staining over the length of the midgut at 240 min (Fig. 3A). The pH of the water larvae were in was near neutral (pH 6.9), but the pH in the lumen of the anterior to near posterior midgut was above 10 in both 2-h and 16-h posthatching larvae (Fig. 3A). This high midgut pH was consistent with earlier data showing that fourth instar Ae. aegypti maintain a gut lumen alkalization profile and exhibit an anterior midgut pH of 11 (30, 31). The same assays conducted at 2-h intervals showed that larvae avidly fed and excreted beads plus bacteria within 30 min of ingestion up to 10 h posthatching (Fig. 3B). In contrast, larvae older than 16 h excreted little or no food and bacteria until molting (Fig. 3B). We thus concluded that gut oxygen levels in precritical size larvae fall below 5% due to aerobic respiration by viable bacteria but rise above 5% in postcritical size larvae because viability of bacteria declines.
Fig. 3.
Pre- and postcritical size larvae exhibit differences in food excretion and viability of gut bacteria. (A) Images show 2-h posthatching (precritical size) or 16-h posthatching (postcritical size) first instars that were monitored over a 240-min observation period. The head of each larva is oriented to the left (anterior). (Upper) Black polystyrene (PS) beads in the midgut (Mg) and hindgut (Hg) over the 240-min observation period. The Mg and Hg are filled with beads at 0 min in 2-h and 16-h larvae. All beads have been excreted by 30 min in 2-h larvae, whereas most beads remain present after 240 min in 16-h larvae. (Middle) E. coli expressing GFP+ and PI staining of bacteria. The Mg is filled with GFP+ bacteria (green) at 0 min in 2-h and 16-h larvae. No PI staining (red) is visible, indicating bacteria are viable. All bacteria have been excreted by 30 min in 2-h larvae, whereas bacteria remain present after 240 min in 16-h larvae. Bacteria in the anterior Mg are stained by PI at 120 min, whereas bacteria in both the anterior and posterior Mg are stained at 240 min. (Lower) Two-hour and 16-h larvae at 0 min after ingestion of the pH indicator cresol red. The magenta color in the anterior Mg extending to the posterior Mg indicates a strongly alkaline pH (>10). (Scale bar, 200 μm.) (B) Proportion of larvae from 2 to 24 h posthatching that excreted all PS beads and bacteria within 30 min. An asterisk (*) indicates time points when the proportion of larvae that have excreted beads in 30 min was significantly lower than the 2-h time point (P < 0.0001, Fisher’s exact test). At least 29 larvae were assayed per time point. (C) Sensitivity of WT E. coli, ΔcydB-ΔcydD::kan E. coli, and four abundant bacterial species present in conventionally reared larvae (Aquitalea sp., Chryseobacterium sp., Comamonas sp., and Sphingobacterium sp.) to culture at pH 7 vs. pH 11. Bacteria were incubated for 2 h in neutral or alkaline LB K medium and then plated on neutral LB agar to determine the number of colony-forming units per microliter. For each species, an asterisk (*) indicates a significant difference between treatments (t test, P < 0.01). Three replicate cultures were tested per species and pH treatment. (D) Proportion of larvae that molted and successfully developed into adults when inoculated with ampicillin-susceptible (ampS) or ampicillin-resistant (ampR) WT E. coli and subsequently treated with ampicillin. Larvae were treated with the antibiotic immediately after molting to the second or third instar. An asterisk (*) indicates a significant difference between treatments (P < 0.0001, Fisher’s exact test). At least 60 larvae were assayed per instar and treatment.
Most nonextremophilic bacteria grow over a range of external pH values (5.5–9.0) but exhibit rapid loss of viability under higher and lower pH conditions (32). We therefore reasoned that the decline in viability of bacteria in postcritical size larvae is due, at least in part, to longer exposure to high midgut pH. We directly tested the effects of high pH on viability of bacteria by incubating four of the most abundant species in conventionally reared Ae. aegypti (9) plus wild-type E. coli for 2 h in Luria broth (LB) K at pH 11. Results showed that the viability of each species declined 90% or more relative to bacteria cultured at pH 7 (Fig. 3C). We also assessed whether Ae. aegypti required bacteria for growth and molting after the first instar by inoculating axenic larvae with wild-type E. coli that were susceptible or resistant to ampicillin. Larvae were then treated with ampicillin immediately after molting to the second or third instar. Plate assays confirmed that ampicillin rapidly eliminated susceptible, but not resistant, bacteria from second and third instars (SI Appendix, Fig. S4). In turn, no ampicillin-treated second and third instars inoculated with susceptible bacteria ever molted, whereas most ampicillin-treated larvae inoculated with resistant bacteria molted to the next instar and completed development into adults (Fig. 3D). This outcome strongly suggested that bacteria-mediated gut hypoxia likely functions as a signal for growth and molting of Ae. aegypti in all instars.
An Oral Inhibitor of Prolyl Hydroxylases Stimulates Molting.
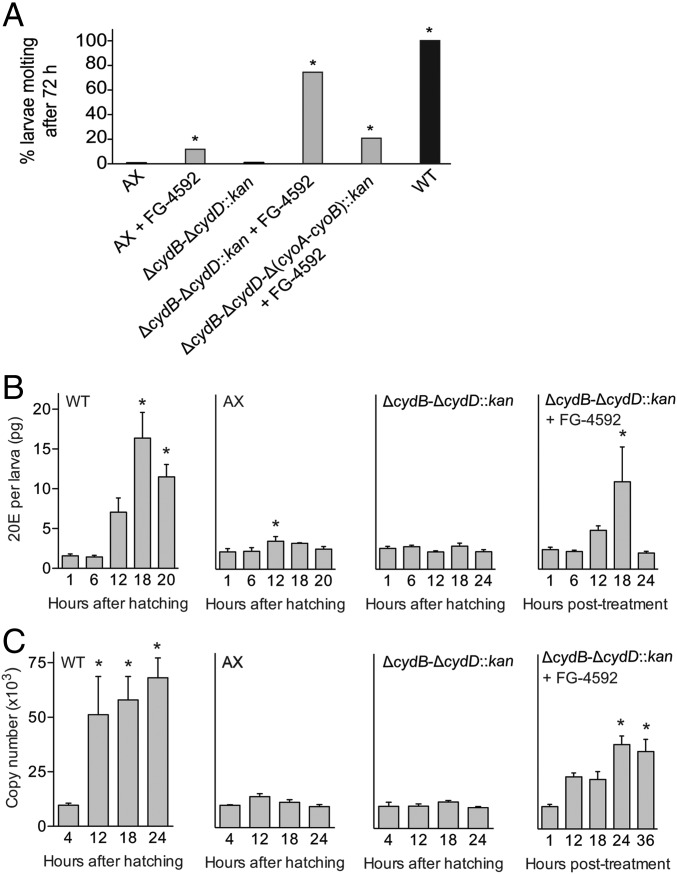
Cellular responses to hypoxia are primarily mediated by conserved α/β heterodimeric hypoxia-inducible transcription factors (HIFs) (33, 34). The HIF-β subunit is constitutively present, whereas specific prolyl hydroxylases (PHDs) hydroxylate HIF-α under normoxia, which targets it for degradation (33, 34). In contrast, PHD hydroxylation does not occur under hypoxia, which stabilizes HIF-α/β to activate hypoxia-responsive genes (33). FG-4592 is an oral inhibitor of HIF PHDs that stabilizes HIF-α/β and triggers a hypoxia transcriptional program under normoxic conditions (35–37). Transcriptional profiling indicated that Ae. aegypti orthologs of HIF-α (sima-1 and sima-2), HIF-β (tango), and PHD (fatiga) were expressed in conventional and gnotobiotic larvae inoculated with wild-type E. coli, as well as in axenic first instars (SI Appendix, Fig. S5). We therefore hypothesized that FG-4592 would mimic the effects of wild-type bacteria if hypoxia stimulates growth and molting through HIF signaling. FG-4592 treatment at 12 h posthatching significantly, but only modestly, increased the percentage of axenic larvae and gnotobiotic larvae inoculated with ΔcydB-ΔcydD-Δ(cyoA-cyoB)::kan E. coli that molted (Fig. 4A). However, it greatly increased the proportion of gnotobiotic larvae inoculated with ΔcydB-ΔcydD::kan E. coli that molted to levels approaching larvae inoculated with wild-type E. coli (Fig. 4A).
Fig. 4.
FG-4592 stimulates molting of gnotobiotic larvae inoculated with ΔcydB-ΔcydD::kan E. coli. (A) Percentage of axenic (AX) and gnotobiotic larvae inoculated with ΔcydB-ΔcydD::kan, ΔcydB-ΔcydD-Δ(cyoA-cyoB)::kan, or wild-type (WT) E. coli that molted to the second instar in the presence and absence of FG-4592. At least 40 larvae were assayed per treatment. An asterisk (*) indicates a given treatment significantly differed from untreated AX larvae (negative control) (P < 0.01, Fisher’s exact test). (B) Mean 20E titers (±SE) in gnotobiotic first instars inoculated with WT E. coli (WT), axenic first instars (AX), gnotobiotic first instars inoculated with ΔcydB-ΔcydD::kan E. coli (ΔcydB-ΔcydD::kan), and gnotobiotic first instars inoculated with ΔcydB-ΔcydD::kan E. coli treated with FG-4592 (ΔcydB-ΔcydD::kan + FG-4592). Titers were measured from 1 to 24 h posthatching for the WT, AX, and ΔcydB-ΔcydD::kan treatments. FG-4592 treatment began at 12 h posthatching, with titers measured from 1 to 24 h after treatment began. For each treatment, an asterisk (*) indicates the time point significantly differed from the 1-h time point (P < 0.05, ANOVA followed by a post hoc Dunnett’s test). A minimum of four larvae were analyzed per treatment and time point. Methods used for titer determination are discussed in SI Appendix, Supplemental Experimental Procedures. (C) Transcript abundance of the Ae. aegypti E74B gene in the WT, AX, ΔcydB-ΔcydD::kan, and ΔcydB-ΔcydD::kan + FG-4592 treatments. Larvae were collected from 4 to 24 h posthatching or 4–36 h posttreatment with FG-4592, followed by extraction of total RNA and RT-quantitative PCR analysis (SI Appendix, Supplemental Experimental Procedures). The bars in each graph show the copy number of each gene (±SE) per 500 ng of total RNA. For each treatment, an asterisk (*) indicates the time point significantly differed from the 4-h time point (P < 0.05, ANOVA followed by a post hoc Dunnett’s test). A minimum of four independent biological replicates were analyzed per treatment and time point.
Because insects molt in response to 20E, we assessed whether FG-4592 directly affected this signal. Because 20E titers had not been examined previously in Ae. aegypti first instars, we first compared gnotobiotic larvae inoculated with wild-type E. coli with axenic larvae. Gnotobiotic larvae exhibited a midinstar increase in 20E titer that preceded molting, whereas nonmolting axenic larvae exhibited little change in titer (Fig. 4B). We then examined gnotobiotic larvae inoculated with ΔcydB-ΔcydD::kan E. coli. No change in 20E titer was detected in untreated larvae, whereas larvae treated with FG-4592 exhibited an increase at 18 h posttreatment that preceded molting between 24 and 36 h (Fig. 4B). E74B is a transcription factor that has classically been used as a molecular marker for ecdysteroid signaling because expression rapidly up-regulates in response to an increase in 20E (38, 39). Transcript abundance of E74B increased with 20E titer in larvae inoculated with wild-type E. coli and in larvae inoculated with ΔcydB-ΔcydD::kan E. coli that were treated with FG-4592 (Fig. 4C). In contrast, copy number did not significantly change in axenic larvae or untreated ΔcydB-ΔcydD::kan-inoculated larvae (Fig. 4C). These results collectively indicated that FG-4592 mimicked the effects of wild-type E. coli by inducing larvae inoculated with ΔcydB-ΔcydD::kan to molt via activation of 20E release and downstream signaling.
Neither Environmental Hypoxia Nor Bacterial Fermentation Products Induce Normal Molting.
Given evidence that bacteria-induced gut hypoxia stimulated molting, we asked whether reducing oxygen levels in the environment had the same effect by incubating 16-h axenic larvae and gnotobiotic larvae inoculated with ΔcydB-ΔcydD::kan E. coli for 2 h in 21% to less than 1% atmospheric oxygen. The percentage of axenic and gnotobiotic larvae that initiated a molt increased to 42% and 55%, respectively, at 2.5% oxygen (SI Appendix, Fig. S6). However, larvae died without ecdysing, which indicated environmental hypoxia did not fully mimic the effects of gut hypoxia. Detritus-feeding insects like mosquito larvae, herbivores, and other insects that consume carbohydrate-rich diets also host bacteria that can produce fermentation products, including acetate, formate, lactate, and butyrate (1). These products have also been shown to affect the development of vertebrates and invertebrates, including Drosophila under nutrient-poor conditions (1, 2, 40). However, adding these products to cultures of axenic or gnotobiotic first instars inoculated with ΔcydB-cydD::kan E. coli resulted in no larvae initiating or completing a molt (SI Appendix, Fig. S7).
Discussion
Most insects that require symbionts for survival live on nutrient-poor diets where microbes provision essential resources and/or facilitate digestion (1). They also involve obligate associations where particular species of microbes are directly transferred from parents to offspring or between individuals of the same insect species (1, 41). Mosquito larvae can experience resource limitation in the field (42). However, they present the paradoxical situation of hosting highly variable communities of environmentally acquired microbes, yet fail to develop under axenic conditions when provided a nutritionally complete diet. We initially speculated that ingested microbes are simply digested and used as a source of nutrients. It was also plausible that multiple bacteria share traits that allow them to facilitate digestion or provide nutrients mosquitoes require. On the other hand, previous experiments strongly suggested these explanations are not why conventional and gnotobiotic larvae develop very similarly when fed a nutritionally complete diet, whereas axenic larvae die as first instars. We therefore hypothesized that microbes produce a cue in the gut that mosquitoes require to grow and molt. Our genetic screen of E. coli identified cytochrome bd oxidase as an important product for larval molting. Our results further suggest that (i) this enzyme is required for reducing midgut oxygen levels below 5%, (ii) hypoxia functions as a developmental signal, and (iii) hypoxia-initiated signaling triggers 20E release.
That multiple species of bacteria rescue development of Ae. aegypti is consistent with the widespread distribution of the cytochrome bd respiratory pathway, including its presence in most gram-negative heterotrophs (43, 44). It is also possible that aerobic respiration by nonbacterial members of the gut community, such as fungi, could contribute to producing a hypoxia signal in the midgut. Larvae inoculated over a range of densities with ΔcydB-ΔcydD::kan E. coli did not molt under the standardized conditions used in this study. However, FG-4592 induced a much stronger molting response in larvae colonized with this mutant than in axenic larvae or larvae colonized by ΔcydB-ΔcydD-Δ(cyoA-cyoB)::kan E. coli. Previous transcriptome data showed that axenic larvae exhibit pronounced alterations in expression of several genes with functions in nutrient acquisition and assimilation (15). Thus, ΔcydB-ΔcydD::kan E. coli and other bacteria may promote nutrient assimilation and growth through other processes besides inducing a hypoxia response. Alternatively, low-affinity cytochrome bo3 oxidase activity in ΔcydB-ΔcydD::kan E. coli could produce an intermediate level of hypoxia in the gut that promotes nutrient acquisition and/or assimilation. Our results showing that FG-4592 induces a weaker molting response in gnotobiotic larvae inoculated with ΔcydB-ΔcydD-Δ(cyoA-cyoB)::kan E. coli than with ΔcydB-ΔcydD::kan E. coli lends support for the latter suggestion.
The literature distinguishes between microbes that establish residency in the digestive tract (autochthonous) vs. microbes that are transiently present (allochthonous). In vertebrates, resident community members are usually more closely associated with the gut epithelium, whereas transient community members are located in the lumen with food (45). The digestive tract of insect larvae, including mosquitoes, differs from vertebrates in two ways that affect the gut microbiota. First, the foregut and hindgut are lined by cuticle, whereas epithelial cells secrete a peritrophic matrix that lines the midgut (6). Together, these barriers prevent many microbes from directly contacting gut epithelia. Second, molting results in shedding and replacement of these barriers and is associated with shifts in feeding activity. All midgut bacteria in conventional and gnotobiotic Ae. aegypti larvae reside inside the endoperitrophic space (15), and results of the current study show greatly reduced viability of these bacteria in postcritical size larvae. These results support the conclusion that aerobic respiration by bacteria passing through the gut reduces oxygen levels in precritical size larvae but retention and pH-mediated mortality of bacteria in postcritical size larvae cause oxygen levels to rise. We further hypothesize these shifts in gut oxygen levels potentially affect signaling by 20E, insulin-like peptides, and other factors that regulate insect growth and molting (18, 46). Prior results indicate that the larval microbiota of mosquitoes consists of a subset of the species in their aquatic environment (8–12). This study further refines this view by showing that most community members do not establish residency but, instead, are continuously acquired and lost between instars. Gut alkalization has also primarily been viewed as an adaptation by herbivores for digesting dietary protein and lowering binding tannins (47). However, our results suggest feeding, coupled with high midgut pH, plays a role in the phasic regulation of bacteria-mediated hypoxia.
Gut hypoxia likely serves as a cue for growth and molting in other mosquitoes, given that axenic larvae from several species exhibit the same inability to grow and molt as Ae. aegypti (9, 16, 17). Microbe-mediated gut hypoxia could also play an important regulatory role in other aquatic insects. In contrast, it is unclear whether gut microbes have such roles in terrestrial species. Oxygen supply has been implicated in regulating body size of terrestrial insects through interaction with the tracheal system (48). Unlike mosquitoes, however, gut microbes are not essential for survival of terrestrial species like Drosophila melanogaster because axenic cultures can be maintained over multiple generations when fed a nutritionally complete diet (49). Only under conditions of low nutrient availability do axenic Drosophila larvae exhibit delays in development (40, 49–53). One study links restoration of normal development under nutrient-poor conditions to acetate production by a gut community member (40), whereas another implicates a second community member in digestion and nutrient assimilation (52, 53). Our results do not support a role for acetate or other fermentation products in promoting growth of Ae. aegypti, but do suggest a role for gut bacteria in nutrient acquisition and/or assimilation. In summary, our results identify bacteria-mediated gut hypoxia as a cue for mosquito development. Our results also identify a prominent group of insects that do not form coevolved associations with their microbiota, yet require gut microbes for survival (54, 55).
Materials and Methods
Mosquitoes and Bacteria.
UGAL Ae. aegypti larvae were reared on a nutritionally complete diet consisting of rat chow, lactalbumin, and inactive torula yeast (1:1:1) sterilized by γ-irradiation (9, 56). Axenic larvae hatched from eggs that were surface-sterilized as described elsewhere (9). The K-12 single-gene knockout library (Keio collection) (19) and select ASKA (a complete set of E. coli K-12 ORF archive) strains (57) containing plasmids with genes of interest were obtained from the Nara Institute of Science and Technology. Double- and quadruple-gene deletions were constructed by either P1 phage transduction (58) or the phage-λ red recombinase method (59) (SI Appendix, Tables S2 and S3). Cells were grown in LB at 37 °C with 25 μg/mL kanamycin (Keio mutants) or 30 μg/mL chloramphenicol plus 50 μM isopropyl beta-d-thiogalactoside (IPTG) (ASKA clones).
Forward Genetic Screen and Growth Assays.
Axenic larvae were individually placed in wells of 24-well culture plates (Corning) containing 1 mL of sterile water and 100 μg of sterilized diet. Wild-type E. coli and mutants from the Keio collection were grown to midlog phase (OD600 ≈ 0.5–0.8), pelleted, resuspended in water, and inoculated at 106 cfu per well. A minimum of 30 larvae were bioassayed per mutant. Mutants where fewer than 100% of larvae molted after 48 h were further assessed by collecting 100 μL of water from each well and homogenizing the larva after surface sterilization in PBS. Water and larval samples were then added to LB agar plates containing kanamycin, followed by colony counts after 24 h at 37 °C. Plasmids were isolated from ASKA collection clones and transformed into Keio mutants that were subclassified as rescue-defective. Transformed E. coli were grown to midlog phase with IPTG (50 μM) and added to culture wells containing axenic larvae as described above, followed by recording the proportion of larvae that molted within 48 h. Bioassays with double and quadruple mutants used slightly modified methods that standardized cell growth and viability so that they were comparable to wild-type E. coli K-12. In brief, each mutant was grown in LB to midlog phase, followed by pelleting and suspension in M9 minimal medium containing sterilized diet (100 μg/mL) at 106 cfu/mL. One milliliter of this suspension containing an axenic larva was then added per well. To ensure that viable cell counts per well remained consistent between treatments, larvae were rinsed and moved to new wells containing a freshly prepared suspension every 4–48 h.
Gut Hypoxia and pH.
Conventional, axenic, and gnotobiotic larvae inoculated with either wild-type or ΔcydB-ΔcydD::kan E. coli were incubated in water containing rearing diet plus 1 μM Image-iT Hypoxia Reagent (Life Technologies) at specific intervals posthatching. Larvae were then moved to 4 °C for 10 min to slow movement, followed by slide mounting and imaging using a Zeiss LSM 710 confocal microscope. Images were processed using Adobe Photoshop CS6. The very small size of first instars precluded the use of a microelectrode to measure pH in the gut. We therefore estimated gut pH by adding 2.5% cresol red (wt/vol) to cultures for 30 min, followed by image acquisition using a Leica MZFLIII stereomicroscope, Canon EOS digital SLR camera, and Leica Application Suite software. Images were then processed using Adobe Photoshop CS6.
Excretion, Viability, and Antibiotic Clearance Assays.
Conventional first instars were fed 0.1% black polystyrene microspheres (Polysciences, Inc.) or 106 cfu of E. coli K-12 expressing GFP in 24-well plates containing standard diet at 2-h intervals for 45 min. Larvae were then rinsed three times in water and moved to new wells containing water and diet only. PI (1 μg/mL) was added to water containing larvae fed GFP+ E. coli to distinguish viable from nonviable bacteria in the gut. Movement of black microspheres and GFP+ E. coli through the gut was monitored at 15-min intervals for 4 h. Images were captured using either a Leica stereomicroscope or epifluorescent microscope fitted with digital cameras, followed by processing using Adobe Photoshop CS6. Plates containing axenic larvae were inoculated with 106 cfu of either ampicillin-susceptible or ampicillin-resistant E. coli K-12 and maintained as described above. Larvae were then treated with 25 mg/mL ampicillin (Sigma) immediately after molting to the second and third instars. The proportion of larvae that developed into adults was then recorded, and cohorts of larvae were also homogenized 24 h posttreatment and cultured on LB plates at 37 °C for 24 h to determine the number of colony-forming units per individual.
FG-4592, Environmental Hypoxia, and Fermentation Product Assays.
Gnotobiotic larvae inoculated with ΔcydB-ΔcydD::kan were reared as described above, followed by addition of 1 μM FG-4592 (Selleckchem) to cultures at 12 h posthatching. Larvae were then monitored to determine the proportion that molted to the second instar. Untreated larvae served as a negative control, whereas gnotobiotic larvae inoculated with wild-type E. coli K-12 served as a positive control. Other larvae were similarly treated by addition of 10 μM or 1 mM sodium acetate (Sigma), sodium butyrate (Sigma), or sodium formate (Sigma), or 0.2–2% (vol/vol) lactic acid. The 20E titers in axenic larvae and gnotobiotic larvae inoculated with wild-type or ΔcydB-ΔcydD::kan E. coli were determined by enzyme immunoassay (60). Transcript abundance of HIF-α (sima-1 and sima-2), HIF-β (tango), PHD (fatiga) and E74B was determined by reverse transcriptase-quantitative PCR analysis. Environmental hypoxia assays were conducted by placing axenic or gnotobiotic larvae inoculated with ΔcydB-ΔcydD::kan E. coli in 30-mL sterile glass culture tubes containing 1 mL of M9 medium. Tubes were made anaerobic by purging with oxygen-free nitrogen gas for 45 s to displace dissolved oxygen. Ambient air was then injected into anaerobic tubes using a sterile syringe to create atmospheric oxygen levels that ranged from 21 to <1%. After 2 h, larvae were individually moved to culture wells containing 1 mL of M9 medium and 100 μg of sterilized diet. The proportion of larvae that initiated a molt was then determined, with axenic and gnotobiotic larvae inoculated with wild-type E. coli in 21% oxygen serving as the negative and positive controls, respectively.
Additional methods are provided in SI Appendix, which is available online.
Supplementary Material
Acknowledgments
We thank J. A. Johnson for assistance with microscopy, B. Whitman for assistance with environmental hypoxia assays, V. F. Maples and S. R. Kushner for advice in designing gene knockout constructs, and B. Boyd for discussions. A. Elliot and S. R. Robertson assisted with rearing. This work was supported by an Achievement Rewards for College Scientists (ARCS) Foundation Scholarship (to K.L.C.), National Science Foundation Graduate Research Fellowship 038550-04 (to K.L.C.), and NIH Grants R01AI106892 and T32GM007103 (to M.R.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702983114/-/DCSupplemental.
References
- 1.Engel P, Moran NA. The gut microbiota of insects–Diversity in structure and function. FEMS Microbiol Rev. 2013;37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 2.Sommer F, Bäckhed F. The gut microbiota–Masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 3.Merritt RW, Dadd RH, Walker ED. Feeding behavior, natural food, and nutritional relationships of larval mosquitoes. Annu Rev Entomol. 1992;37:349–376. doi: 10.1146/annurev.en.37.010192.002025. [DOI] [PubMed] [Google Scholar]
- 4.Clements AN. The Biology of Mosquitoes, Development, Nutrition, and Reproduction. Vol 1 Chapman & Hall; New York: 1992. [Google Scholar]
- 5.Minard G, Mavingui P, Moro CV. Diversity and function of bacterial microbiota in the mosquito holobiont. Parasit Vectors. 2013;6:146. doi: 10.1186/1756-3305-6-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strand MR. The gut microbiota of mosquitoes: Diversity and function. In: Wikel SK, Aksoy S, Dimopoulos G, editors. Arthropod Vector: Controller of Disease Transmission. Vol 1. Academic Press; London: 2017. pp. 185–199. [Google Scholar]
- 7.Wang Y, Gilbreath TM, 3rd, Kukutla P, Yan G, Xu J. Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS One. 2011;6:e24767. doi: 10.1371/journal.pone.0024767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boissière A, et al. Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog. 2012;8:e1002742. doi: 10.1371/journal.ppat.1002742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coon KL, Vogel KJ, Brown MR, Strand MR. Mosquitoes rely on their gut microbiota for development. Mol Ecol. 2014;23:2727–2739. doi: 10.1111/mec.12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gimonneau G, et al. Composition of Anopheles coluzzii and Anopheles gambiae microbiota from larval to adult stages. Infect Genet Evol. 2014;28:715–724. doi: 10.1016/j.meegid.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 11.Duguma D, et al. Developmental succession of the microbiome of Culex mosquitoes. BMC Microbiol. 2015;15:140. doi: 10.1186/s12866-015-0475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muturi EJ, Bara JJ, Rooney AP, Hansen AK. Midgut fungal and bacterial microbiota of Aedes triseriatus and Aedes japonicus shift in response to La Crosse virus infection. Mol Ecol. 2016;25:4075–4090. doi: 10.1111/mec.13741. [DOI] [PubMed] [Google Scholar]
- 13.Hegde S, Rasgon JL, Hughes GL. The microbiome modulates arbovirus transmission in mosquitoes. Curr Opin Virol. 2015;15:97–102. doi: 10.1016/j.coviro.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Tol S, Dimopoulos G. Influences of the mosquito microbiota on vector competence. Adv Insect Physiol. 2016;51:243–291. [Google Scholar]
- 15.Vogel KJ, Valzania L, Coon KL, Brown MR, Strand MR. Transcriptome sequencing reveals large-scale changes in axenic Aedes aegypti larvae. PLoS Negl Trop Dis. 2017;11:e0005273. doi: 10.1371/journal.pntd.0005273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coon KL, Brown MR, Strand MR. Gut bacteria differentially affect egg production in the anautogenous mosquito Aedes aegypti and facultatively autogenous mosquito Aedes atropalpus (Diptera: Culicidae) Parasit Vectors. 2016;9:375. doi: 10.1186/s13071-016-1660-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coon KL, Brown MR, Strand MR. Mosquitoes host communities of bacteria that are essential for development but vary greatly between local habitats. Mol Ecol. 2016;25:5806–5826. doi: 10.1111/mec.13877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nijhout HF, et al. The developmental control of size in insects. Wiley Interdiscip Rev Dev Biol. 2014;3:113–134. doi: 10.1002/wdev.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green GN, Gennis RB. Isolation and characterization of an Escherichia coli mutant lacking cytochrome d terminal oxidase. J Bacteriol. 1983;154:1269–1275. doi: 10.1128/jb.154.3.1269-1275.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poole RK, Gibson F, Wu G. The cydD gene product, component of a heterodimeric ABC transporter, is required for assembly of periplasmic cytochrome c and of cytochrome bd in Escherichia coli. FEMS Microbiol Lett. 1994;117:217–223. doi: 10.1111/j.1574-6968.1994.tb06768.x. [DOI] [PubMed] [Google Scholar]
- 22.Lynch AS, Lin EC. Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: Characterization of DNA binding at target promoters. J Bacteriol. 1996;178:6238–6249. doi: 10.1128/jb.178.21.6238-6249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cotter PA, Gunsalus RP. Contribution of the fnr and arcA gene products in coordinate regulation of cytochrome o and d oxidase (cyoABCDE and cydAB) genes in Escherichia coli. FEMS Microbiol Lett. 1992;70:31–36. doi: 10.1016/0378-1097(92)90558-6. [DOI] [PubMed] [Google Scholar]
- 24.Fu HA, Iuchi S, Lin EC. The requirement of ArcA and Fnr for peak expression of the cyd operon in Escherichia coli under microaerobic conditions. Mol Gen Genet. 1991;226:209–213. doi: 10.1007/BF00273605. [DOI] [PubMed] [Google Scholar]
- 25.Brøndsted L, Atlung T. Effect of growth conditions on expression of the acid phosphatase (cyx-appA) operon and the appY gene, which encodes a transcriptional activator of Escherichia coli. J Bacteriol. 1996;178:1556–1564. doi: 10.1128/jb.178.6.1556-1564.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart V. Nitrate regulation of anaerobic respiratory gene expression in Escherichia coli. Mol Microbiol. 1993;9:425–434. doi: 10.1111/j.1365-2958.1993.tb01704.x. [DOI] [PubMed] [Google Scholar]
- 27.Rivera-Chávez F, et al. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe. 2016;19:443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart V, Lu Y, Darwin AJ. Periplasmic nitrate reductase (NapABC enzyme) supports anaerobic respiration by Escherichia coli K-12. J Bacteriol. 2002;184:1314–1323. doi: 10.1128/JB.184.5.1314-1323.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tiso M, Schechter AN. Nitrate reduction to nitrite, nitric oxide and ammonia by gut bacteria under physiological conditions. PLoS One. 2015;10:e0119712. doi: 10.1371/journal.pone.0119712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boudko DY, Moroz LL, Harvey WR, Linser PJ. Alkalinization by chloride/bicarbonate pathway in larval mosquito midgut. Proc Natl Acad Sci USA. 2001;98:15354–15359. doi: 10.1073/pnas.261253998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corena M, et al. Carbonic anhydrase in the midgut of larval Aedes aegypti: Cloning, localization and inhibition. J Exp Biol. 2002;205:591–602. doi: 10.1242/jeb.205.5.591. [DOI] [PubMed] [Google Scholar]
- 32.Padan E, Bibi E, Ito M, Krulwich TA. Alkaline pH homeostasis in bacteria: New insights. Biochim Biophys Acta. 2005;1717:67–88. doi: 10.1016/j.bbamem.2005.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Semenza GL. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci STKE. 2007;2007:cm8. doi: 10.1126/stke.4072007cm8. [DOI] [PubMed] [Google Scholar]
- 34.Dekanty A, et al. Drosophila genome-wide RNAi screen identifies multiple regulators of HIF-dependent transcription in hypoxia. PLoS Genet. 2010;6:e1000994. doi: 10.1371/journal.pgen.1000994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabinowitz MH. Inhibition of hypoxia-inducible factor prolyl hydroxylase domain oxygen sensors: Tricking the body into mounting orchestrated survival and repair responses. J Med Chem. 2013;56:9369–9402. doi: 10.1021/jm400386j. [DOI] [PubMed] [Google Scholar]
- 36.Flück K, Fandrey J. Oxygen sensing in intestinal mucosal inflammation. Pflugers Arch. 2016;468:77–84. doi: 10.1007/s00424-015-1722-4. [DOI] [PubMed] [Google Scholar]
- 37.Jain IH, et al. Hypoxia as a therapy for mitochondrial disease. Science. 2016;352:54–61. doi: 10.1126/science.aad9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karim FD, Thummel CS. Ecdysone coordinates the timing and amounts of E74A and E74B transcription in Drosophila. Genes Dev. 1991;5:1067–1079. doi: 10.1101/gad.5.6.1067. [DOI] [PubMed] [Google Scholar]
- 39.Sun G, Zhu J, Li C, Tu Z, Raikhel AS. Two isoforms of the early E74 gene, an Ets transcription factor homologue, are implicated in the ecdysteroid hierarchy governing vitellogenesis of the mosquito, Aedes aegypti. Mol Cell Endocrinol. 2002;190:147–157. doi: 10.1016/s0303-7207(01)00726-2. [DOI] [PubMed] [Google Scholar]
- 40.Shin SC, et al. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science. 2011;334:670–674. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- 41.Wilson ACC, Duncan RP. Signatures of host/symbiont genome coevolution in insect nutritional endosymbioses. Proc Natl Acad Sci USA. 2015;112:10255–10261. doi: 10.1073/pnas.1423305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rowbottom R, et al. Resource limitation, controphic ostracod density and larval mosquito development. PLoS One. 2015;10:e0142472. doi: 10.1371/journal.pone.0142472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jünemann S. Cytochrome bd terminal oxidase. Biochim Biophys Acta. 1997;1321:107–127. doi: 10.1016/s0005-2728(97)00046-7. [DOI] [PubMed] [Google Scholar]
- 44.Hao W, Golding GB. Asymmetrical evolution of cytochrome bd subunits. J Mol Evol. 2006;62:132–142. doi: 10.1007/s00239-005-0005-7. [DOI] [PubMed] [Google Scholar]
- 45.Nava GM, Stappenbeck TS. Diversity of the autochthonous colonic microbiota. Gut Microbes. 2011;2:99–104. doi: 10.4161/gmic.2.2.15416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baker KD, Thummel CS. Diabetic larvae and obese flies-emerging studies of metabolism in Drosophila. Cell Metab. 2007;6:257–266. doi: 10.1016/j.cmet.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barbehenn RV, Peter Constabel C. Tannins in plant-herbivore interactions. Phytochemistry. 2011;72:1551–1565. doi: 10.1016/j.phytochem.2011.01.040. [DOI] [PubMed] [Google Scholar]
- 48.Callier V, Nijhout HF. Control of body size by oxygen supply reveals size-dependent and size-independent mechanisms of molting and metamorphosis. Proc Natl Acad Sci USA. 2011;108:14664–14669. doi: 10.1073/pnas.1106556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bakula M. The persistence of a microbial flora during postembryogenesis of Drosophila melanogaster. J Invertebr Pathol. 1969;14:365–374. doi: 10.1016/0022-2011(69)90163-3. [DOI] [PubMed] [Google Scholar]
- 50.Broderick NA, Buchon N, Lemaitre B. Microbiota-induced changes in Drosophila melanogaster host gene expression and gut morphology. MBio. 2014;5:e01117–e14. doi: 10.1128/mBio.01117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ridley EV, Wong AC, Westmiller S, Douglas AE. Impact of the resident microbiota on the nutritional phenotype of Drosophila melanogaster. PLoS One. 2012;7:e36765. doi: 10.1371/journal.pone.0036765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Storelli G, et al. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 2011;14:403–414. doi: 10.1016/j.cmet.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 53.Erkosar B, et al. Pathogen virulence impedes mutualist-mediated enhancement of host juvenile growth via inhibition of protein digestion. Cell Host Microbe. 2015;18:445–455. doi: 10.1016/j.chom.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zilber-Rosenberg I, Rosenberg E. Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution. FEMS Microbiol Rev. 2008;32:723–735. doi: 10.1111/j.1574-6976.2008.00123.x. [DOI] [PubMed] [Google Scholar]
- 55.Moran NA, Sloan DB. The hologenome concept: Helpful or hollow? PLoS Biol. 2015;13:e1002311. doi: 10.1371/journal.pbio.1002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Riehle MA, Brown MR. Insulin receptor expression during development and a reproductive cycle in the ovary of the mosquito Aedes aegypti. Cell Tissue Res. 2002;308:409–420. doi: 10.1007/s00441-002-0561-8. [DOI] [PubMed] [Google Scholar]
- 57.Kitagawa M, et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 58.Purdy GE, Fisher CR, Payne SM. IcsA surface presentation in Shigella flexneri requires the periplasmic chaperones DegP, Skp, and SurA. J Bacteriol. 2007;189:5566–5573. doi: 10.1128/JB.00483-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kingan TG. A competitive enzyme-linked immunosorbent assay: Applications in the assay of peptides, steroids, and cyclic nucleotides. Anal Biochem. 1989;183:283–289. doi: 10.1016/0003-2697(89)90481-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.