Significance
BRD4 plays a vital role in cellular growth control. Because BRD4 is dysregulated in a wide range of aggressive malignancies, it is being increasingly implicated as a major driver of oncogenic growth and a novel target for cancer therapy. However, how BRD4 is regulated to maintain its normal function in healthy cells and how alteration of this process leads to cancer remain poorly understood. We discovered that BRD4 is hyperphosphorylated in cancers and that this hyperphosphorylation may be a general mechanism to support its oncogenic activities. Our study shows how dysregulation of BRD4 function could lead to tumorigenesis. Our discovery also provides the rationale for investigating how cellular signaling pathways modulate BRD4 phosphorylation to control its function during cancer development.
Keywords: bromodomain-containing protein 4, NUT midline carcinoma, CDK9, cellular transformation, cancer
Abstract
The epigenetic reader BRD4 plays a vital role in transcriptional regulation, cellular growth control, and cell-cycle progression. Dysregulation of BRD4 function has been implicated in the pathogenesis of a wide range of cancers. However, how BRD4 is regulated to maintain its normal function in healthy cells and how alteration of this process leads to cancer remain poorly understood. In this study, we discovered that BRD4 is hyperphosphorylated in NUT midline carcinoma and identified CDK9 as a potential kinase mediating BRD4 hyperphosphorylation. Disruption of BRD4 hyperphosphorylation using both chemical and molecular inhibitors led to the repression of BRD4 downstream oncogenes and abrogation of cellular transformation. BRD4 hyperphosphorylation is also observed in other cancers displaying enhanced BRD4 oncogenic activity. Our study revealed a mechanism that may regulate BRD4 biological function through phosphorylation, which, when dysregulated, could lead to oncogenesis. Our finding points to strategies to target the aberrant BRD4 signaling specifically for cancer intervention.
BRD4 (bromodomain-containing protein 4) is a member of the bromodomain and extraterminal (BET) family. The tandem bromodomains (BD1 and BD2) of BRD4 specifically recognize acetylated histones H3 and H4 on chromatin (1). Through this interaction, BRD4 actively recruits P-TEFb (positive transcription elongation factor b), mediators and other transcriptional activators to facilitate gene activation (2–4). P-TEFb is a heterodimer of CDK9 and cyclin T1, which together phosphorylate serine 5 in the C-terminal domain of RNA polymerase II (RNAPII) to support transcription elongation (5–7). Therefore, BRD4 functions as an epigenetic reader that plays a central role in transcriptional regulation, cellular growth control, and cell-cycle progression (2, 8, 9).
Dysregulation of BRD4 function has been implicated in the pathogenesis of a wide range of cancers including acute myeloid leukemia (AML), multiple myeloma, Burkitt’s lymphoma, diffuse large B-cell lymphoma, breast cancer, colon cancer, and ovarian carcinoma (4, 10–14). It is also the target of a genetic translocation between chromosomes 15 and 19, denoted as t(15;19), which results in the formation of a novel fusion oncogene BRD4-NUT that accounts for the highly lethal NUT (nuclear protein in testis) midline carcinoma (NMC) (15).
Although BRD4 has emerged as a critical therapeutic target for a wide variety of cancers (4, 10–14), the mechanisms that regulate BRD4 function have not been clearly elucidated. How alteration of BRD4 function leads to cancer development remains largely unknown. BRD4 has been shown to enrich disproportionately at a subset of key oncogenic and lineage-specific genes such as c-MYC and selectively stimulates their expression to drive cellular proliferation in cancers (4, 14, 16–18). Blocking bromodomain binding to acetylated histones with BET inhibitors, including (+)-JQ1 (19) and I-BET (20), specifically down-regulate these oncogenes (4, 16, 17). Addiction of the tumor cells to high-level expression of these oncogenes provides the basis for using BET inhibitors to abrogate BRD4 function for treating these cancers (4, 14, 16, 17). Multiple BET inhibitors have thus entered clinical trials. Early clinical trials have shown promising results, especially for hematological malignancies (21), highlighting the potential of targeting BRD4 in anticancer treatment. However, resistance to BET inhibitors has also emerged (22, 23), revealing the therapeutic limitations of BET inhibitors and the complexity of BRD4 regulation mechanisms. More importantly, we and others have shown that BRD4 also plays an important role in noncancerous systems such as mouse embryonic stem cells, preimplantation embryos, and keratinocyte differentiation (9, 24–26). There are growing concerns regarding the consequences of disrupting BRD4 function in the normal cells by using BET inhibitors (27). Therefore, it is critical to elucidate the molecular mechanisms that regulate BRD4’s biological function in both normal and disease settings so that therapeutic interventions can be developed to switch off the oncogenic activity of BRD4 specifically in cancer cells while sparing the normal BRD4 function in healthy cells.
NMC is a highly lethal tumor typically caused by translocation t(15;19). The translocation breakpoint splits BRD4 in half, resulting in the in-frame fusion of BRD4 bromodomains and extraterminal domain with nearly the entire sequence of the NUT gene (15, 28). NMCs represent the most lethal subset of squamous cell carcinomas (15). They metastasize rapidly and are extremely aggressive; patients have a median survival of <7 mo (15). Translocation t(15;19), which causes the formation of the BRD4-NUT fusion oncogene in NMC (15), has been described in pediatric head and neck tumors as well as in lung cancers (29). All NMCs carry an intact BRD4 locus and simultaneously express BRD4 and the BRD4-NUT fusion oncogene (30, 31), providing a unique tumor model to investigate how alteration of BRD4 function by oncogenic mutation leads to cancer. The BRD4-NUT fusion oncoprotein is also tethered to acetylated chromatin by the bromodomains (31, 32). It causes malignancy by blocking NMC differentiation while driving tumor growth (15, 19). We and others demonstrated that BRD4-NUT stimulates BRD4 transcription function to activate specifically the expression of oncogenes such as SOX2 (28, 30) and c-MYC (18), which collectively drive the potent NMC transforming activity. However, the molecular mechanisms by which BRD4-NUT modulates BRD4 function to induce such highly aggressive carcinomas remain to be elucidated.
In this study, we found that BRD4 is hyperphosphorylated in NMC tumors and that this hyperphosphorylation is linked to its ability to drive oncogene expression and cellular transformation. We found that BRD4 is hyperphosphorylated in other BRD4-associated cancers as well. Our study revealed a cellular mechanism that could regulate BRD4’s biological function through phosphorylation, which, when dysregulated could lead to oncogenesis.
Results
BRD4 Is Hyperphosphorylated in NMC Tumors.
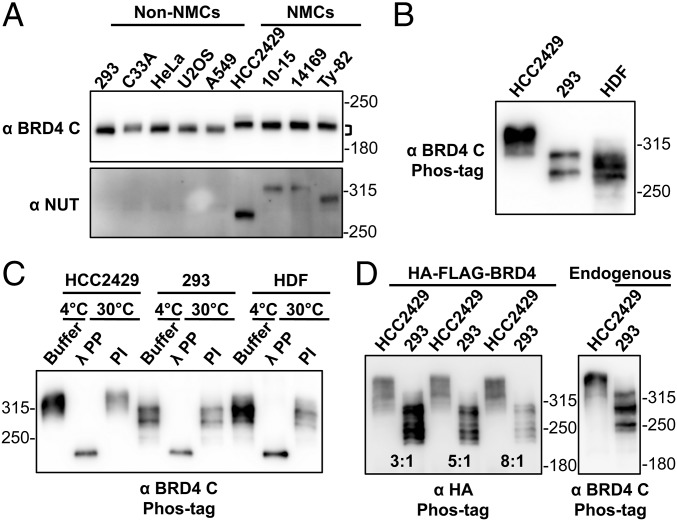
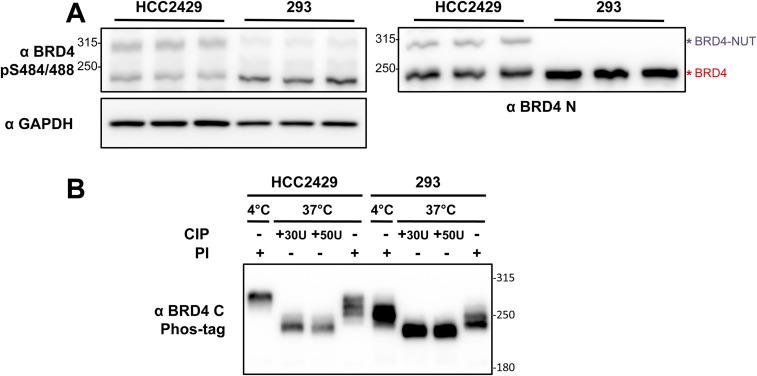
From our previous NMC studies (28, 30, 31), we observed that BRD4 isolated from NMC cells, including HCC2429, 10-15, 14169, and Ty-82 cells, migrates more slowly in SDS/PAGE than do a number of non–NMC cells, such as HEK293, C33A, HeLa, U2OS, and A549 cells (Fig. 1A). Because both types of cells carry the wild-type BRD4 gene, our observation indicates that BRD4 has different posttranslational modification(s) in NMC and non–NMC cells. Therefore, we investigated the BRD4 phosphorylation status in these cells. Whole-cell lysates isolated from both HCC2429 and HEK293 cells were reactive to a phospho-BRD4 antibody, α-pS484/488 (Fig. S1A), which was raised against diphosphorylated S484 and S488 residues of BRD4 (33). This result suggested that BRD4 is phosphorylated in both cell types. To understand the different mobility shift of BRD4 in these cell types, we analyzed BRD4 proteins from HCC2429 and HEK293 cells and from noncancerous cells such as human dermal fibroblasts (HDFs) on an acrylamide gel containing the chemical Phos-tag that binds specifically to phosphorylated proteins and selectively retards their migration during electrophoresis (34). Remarkably, BRD4 isolated from HCC2429 cells migrates to a much higher position than BRD4 isolated from HEK293 cells or from HDFs (Fig. 1B). More importantly, after dephosphorylation treatment with lambda protein phosphatase (λPP) or calf intestinal alkaline phosphatase, BRD4 proteins isolated from HCC2429 and HEK293 cells and from HDFs migrated to the same position in Phos-tag gel, which was much lower the position of untreated BRD4 from any of the cell types (Fig. 1C and Fig. S1B). This mobility shift was reversed when phosphatase inhibitors (PIs) were added to the phosphatase reaction. These studies demonstrated that BRD4 is phosphorylated in all three cell types, but the phosphorylation in HCC2429 NMC cells is clearly different, most likely from hyperphosphorylation.
Fig. 1.
BRD4 is hyperphosphorylated in NMCs. (A) Whole-cell lysates of non–NMCs and NMCs were resolved on SDS/PAGE and immunoblotted with BRD4 C-terminal domain (BRD4 C) and NUT antibodies. Brackets mark the difference in the position of NMC and non–NMC BRD4. (B) Whole-cell lysates of HCC2429 and HEK293 cells and HDFs were resolved on a Phos-tag gel and immunoblotted with BRD4 C antibody. Molecular weight markers are shown on the right. However, as indicated by the manufacturer, the molecular weight markers are frequently distorted during Phos-tag gel electrophoresis, and therefore they can be used only as rough estimates of the molecular weights. (C) BRD4 immunoprecipitated from HCC2429 and HEK293 cells and HDFs was mixed with buffer, λPP, or PIs. Samples were incubated at 4 °C or 30 °C as indicated. Proteins then were analyzed on Phos-tag gel and immunoblotted with BRD4 C antibody. (D) HCC2429 and HEK293 cells were transfected with a construct expressing HA-FLAG–tagged BRD4. At 2 d posttransfection, whole-cell lysates were analyzed on Phos-tag gel and immunoblotted with HA antibody. Because of the different transfection efficiencies of the two cell lines, the lysates of HCC2429 and HEK293 were loaded at the ratios of 3:1, 5:1, and 8:1 as indicated. Lysates from untransfected HCC2429 and HEK293 cells were also loaded on the same Phos-tag gel to show the endogenous BRD4 controls. The samples were immunoblotted with BRD4 C antibody. See also Fig. S1.
Fig. S1.
BRD4 is hyperphosphorylated in NMCs. (A) Whole-cell lysates from HCC2429 and HEK293 cells were resolved on SDS/PAGE and immunoblotted with the indicated antibodies. Samples isolated from three independent experiments were examined. (B) Whole-cell lysates from HCC2429 or HEK293 cells were treated with calf intestinal alkaline phosphatase (CIP) or PIs. Samples with 100 μg of protein in a volume of 80 μL were incubated at 4 °C or 37 °C for 30 min as indicated. Proteins then were analyzed on a 4 μM Phos-tag gel and immunoblotted with BRD4 C antibody.
That the dephosphorylated BRD4 proteins in HCC2429 and HEK293 cells and HDFs migrated to the same position in Phos-tag gel also confirmed that the unphosphorylated BRD4 proteins in these three cell types have the same molecular weight. The result suggested that the unique cellular environment in HCC2429 NMC cells might induce BRD4 hyperphosphorylation. To test this possibility, we transfected a construct expressing HA and FLAG dual-tagged BRD4 into HCC2429 and HEK293 cells. At 2 d posttransfection, the whole-cell lysates were analyzed on a Phos-tag gel (Fig. 1D). As observed with the endogenous BRD4, HA-FLAG-BRD4 expressed in HCC2429 cells migrated to a much higher position than the same protein expressed in HEK293 cells (Fig. 1D). Together, our study demonstrated that BRD4 is hyperphosphorylated in NMC cells.
CDK9 Is a Potential Kinase That Mediates BRD4 Hyperphosphorylation.
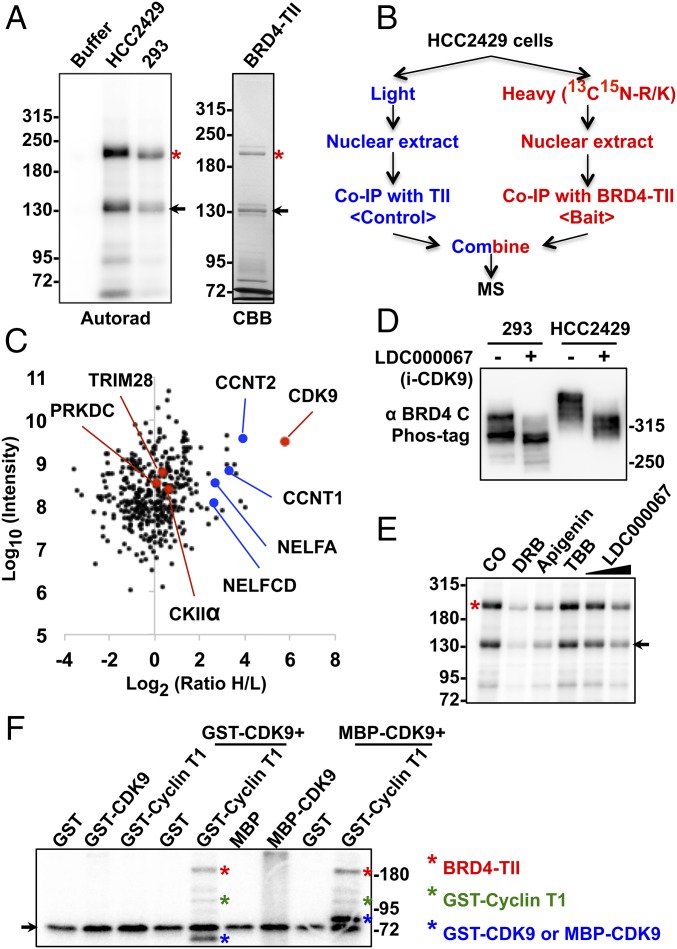
We then performed an in vitro kinase assay to confirm that BRD4 is hyperphosphorylated in NMC cells compared with HEK293 cells. Recombinant BRD4-TII protein, in which BRD4 is fused to two IgG-binding domains of protein A through a tobacco etch virus (TEV) protease cleavage site, was expressed in Escherichia coli and affinity purified on IgG beads. The BRD4-TII beads were incubated with an equal amount of nuclear proteins isolated from HCC2429 or HEK293 cells to immunoprecipitate the kinases for BRD4. The immunocomplexes captured on the beads were then subjected to an in vitro kinase assay (Fig. 2A). Compared with the buffer control, BRD4-TII was phosphorylated in both reactions using either HCC2429 or HEK293 nuclear extract, but much more [γ-32P]ATP was incorporated into BRD4-TII incubated with the HCC2429 nuclear extract (Fig. 2A). Thus, this in vitro system was able to mimic the situation in the cells and confirmed that a higher level of BRD4 phosphorylation was induced by the cellular factors from NMC cells.
Fig. 2.
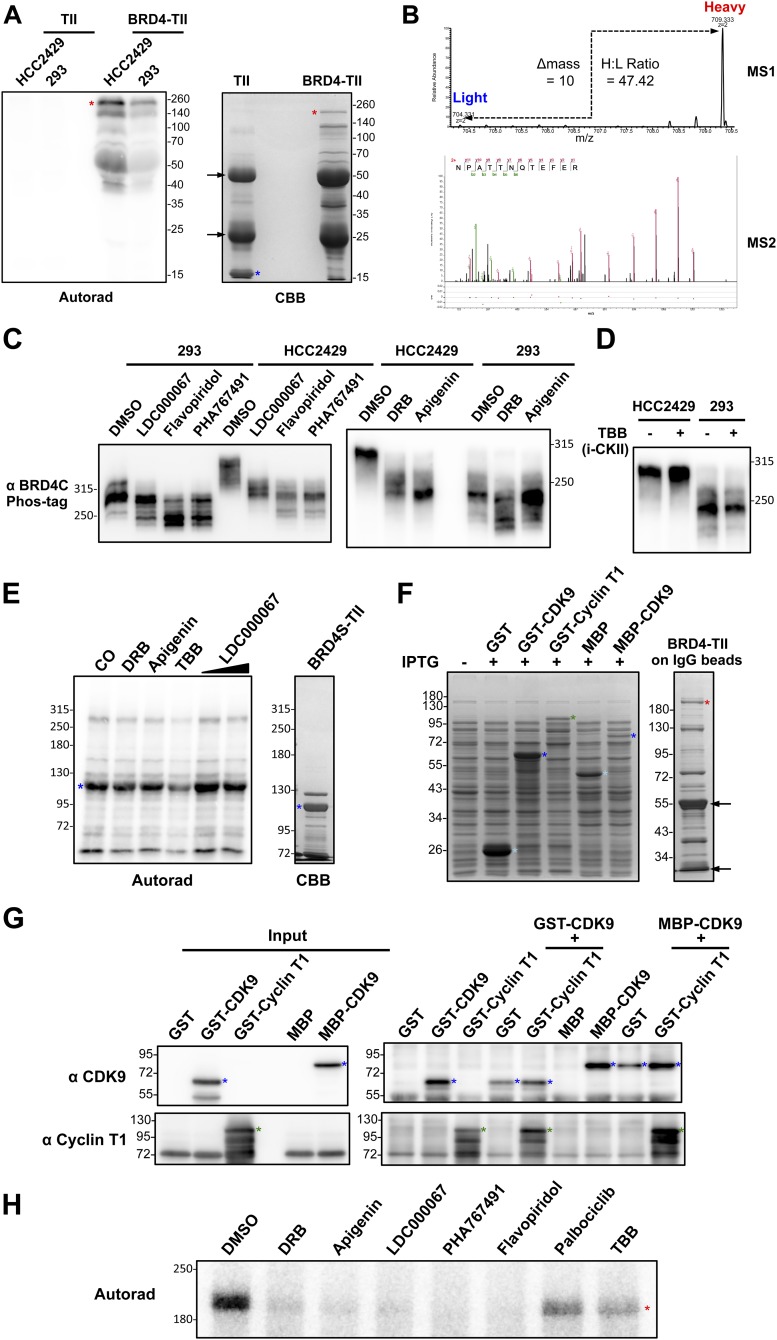
CDK9 contributes to BRD4 hyperphosphorylation in NMC. (A) BRD4-TII expressed in E. coli was affinity purified on IgG beads. The beads were either kept in buffer or coimmunoprecipitated with equal amounts of nuclear proteins from HCC2429 or HEK293 cells. After washing, beads were subjected to kinase assay. BRD4-TII samples then were analyzed on SDS/PAGE and visualized by autoradiography (Autorad). BRD4-TII extracted from the beads before kinase assay was analyzed on SDS/PAGE and stained with Coomassie Brilliant Blue (CBB). Asterisks indicate the full-length BRD4-TII. Arrows mark a BRD4-TII fragment purified from E. coli. (B) Flow diagram of the SILAC experiment for identifying BRD4-associated proteins. HCC2429 cells were cultured in light medium (with normal arginine and lysine) or in heavy medium (with 13C- and 15N-labeled arginine and lysine). Nuclear extracts from light-labeled cells were coimmunoprecipitated with beads containing cross-linked TII tag protein, which served as the background control. Nuclear extracts from heavy-labeled cells were coimmunoprecipitated with beads with cross-linked BRD4-TII. After extensive washing, the two samples were mixed at a 1:1 ratio and subjected to mass spectrometry. (C) Plot showing the normalized SILAC ratio intensity. Shown is the SILAC H/L ratio versus the corresponding protein intensity distribution. Red dots indicate BRD4-associated kinases identified with log2(ratio H/L)>0. Blue dots highlight cyclin T1, cyclin T2, and components of the NELF complex. PRKDC encodes the catalytic subunit of the DNA-dependent protein kinase (DNA-PK). The total SILAC data are shown in Dataset S1. (D) HEK293 and HCC2429 cells were treated with DMSO or 5 μM LDC000067 for 1 h. Whole-cell lysates were analyzed on Phos-tag gel and immunoblotted with BRD4 C antibody. (E) BRD4-TII expressed in E. coli was affinity purified on IgG beads and used to coimmunoprecipitate nuclear proteins from HCC2429 cells. Equal amounts of immunocomplexes were subjected to in vitro kinase assay in the presence of DMSO, 50 μM DRB, 50 μM apigenin, 50 μM TBB, or 2 μM or 10 μM LDC000067. The samples then were analyzed on SDS/PAGE and visualized by autoradiography. The asterisk indicates the full-length BRD4-TII. The arrow marks a BRD4-TII fragment purified from E. coli. (F) BRD4-TII expressed in E. coli was used to coimmunoprecipitate GST, GST-CDK9, GST-cyclin T1, MBP (Maltose-binding protein), or MBP-CDK9 from E. coli lysates as indicated. The immunocomplexes were subjected to in vitro kinase assay. The samples were resolved on SDS/PAGE and visualized by autoradiography. Phosphorylated BRD4-TII, cyclin T1, and CDK9 are marked with red, green, and blue asterisks, respectively. The arrow marks a nonspecific band present in all the samples. See also Fig. S2, Table S1, and Dataset S1.
Kinases often bind directly to their substrates (35). The data shown in Fig. 2A also suggest that BRD4 kinases can be isolated using the BRD4-TII beads. We then used SILAC (stable isotope labeling with amino acids in cell culture)-based proteomic analysis to identify the BRD4-interacting kinases that contribute to its hyperphosphorylation in NMC cells (Fig. 2B). BRD4-TII was used as the bait protein, and TII was used as the tag control (Fig. 2B and Fig. S2A). An in vitro kinase assay confirmed that TII did not pull down any detectable kinase activity, but BRD4-TII coimmunoprecipitated more kinase activity from HCC2429 cells than from HEK293 cells (Fig. S2A). HCC2429 cells then were cultured either in normal (light) medium or medium containing heavy isotope (13C15N)-labeled arginine and lysine. Nuclear extracts from light-labeled cells were coimmunoprecipitated with TII tag protein immobilized on IgG beads, and the nuclear extracts from heavy-labeled cells were coimmunoprecipitated with BRD4-TII beads (Fig. 2B). After extensive washing to remove unbound proteins, the two samples were combined at a 1:1 ratio and subjected to mass spectrometry. Following the principles of SILAC technology (36), the proteins with a heavy-to-light (H/L) ratio of more than 1 as determined by mass spectrometry were identified as BRD4-associated proteins (Fig. 2C). From this study, several protein kinases, including CDK9, the kinase component of P-TEFb, PRKDC, CKIIα, and TRIM28 (37), were identified as BRD4-interacting proteins in NMCs (Fig. 2C and Fig. S2B). Among these, CDK9 was found to be the most abundant BRD4-associated kinase with the highest H/L ratio (Fig. 2C and Fig. S2B); the levels of the other three kinases were much lower in the BRD4-binding proteins (Fig. 2C).
Fig. S2.
CDK9 contributes to BRD4 hyperphosphorylation in NMC. (A) The TII tag control and BRD4-TII expressed in E. coli were affinity purified on IgG beads. The beads were used to coimmunoprecipitate equal amounts of nuclear proteins from HCC2429 or HEK293 cells. After washing, the beads were subjected to kinase assay. Phosphorylated proteins then were analyzed on SDS/PAGE and visualized by autoradiography (Autorad). TII and BRD4-TII extracted from the beads before the kinase assay were analyzed on SDS/PAGE and stained with Coomassie Brilliant Blue (CBB). Red asterisks indicate the full-length BRD4-TII; the blue asterisk indicates the TII tag control; arrows indicate the heavy chain and light chain from IgG beads. (B) The MS1 spectrum shows a SILAC peptide pair identified for CDK9. The peaks at m/z 709.333 and 704.331 correspond to the peptides coimmunoprecipitated with BRD4-TII (bait) and TII (control), respectively, exhibiting a SILAC H/L ratio of 47.42. The precursor ion was isolated, fragmented, and analyzed in the MS2. The MS2 spectrum shows the identification of the peptide sequence. (C) HEK293 and HCC2429 cells were treated with DMSO, 10 μM LDC000067, 2 μM flavopiridol, or 10 μM PHA767491 for 1 h or were treated with DMSO, 50 μM DRB, or 50 μM apigenin for 4 h. Whole-cell lysates were analyzed on a 10-μM (Left) or 4-μM (Right) Phos-tag gel and were immunoblotted with BRD4 C antibody. (D) HCC2429 and HEK293 cells were treated with DMSO or 50 μM TBB for 4 h. Whole-cell lysates were analyzed on a 4-μM Phos-tag gel and were immunoblotted with BRD4 C antibody. (E) BRD4S-TII expressed in E. coli was affinity purified on IgG beads and used to coimmunoprecipitate nuclear proteins from HCC2429 cells. The immunocomplexes were subjected to an in vitro kinase assay in the presence of DMSO, 50 μM DRB, 50 μM apigenin, 50 μM TBB, or 2 μM or 10 μM LDC000067. The samples then were analyzed on SDS/PAGE and visualized by autoradiography (Autorad). BRD4S-TII extracted from the beads before the kinase assay was analyzed on SDS/PAGE and stained with CBB. The asterisk indicates the full-length BRD4S-TII. (F, Left) After induction of GST, GST-CDK9, GST-cyclin T1, MBP, and MBP-CDK9 expression in E. coli, proteins extracted from E. coli were analyzed on SDS/PAGE and stained with CBB. (Right) BRD4-TII expressed in E. coli was affinity purified on IgG beads, analyzed on SDS/PAGE, and stained with CBB. Light blue asterisks indicate GST or MBP tag control. Blue asterisks indicate GST-CDK9 or MBP-CDK9. The green asterisk indicates GST-cyclin T1. The red asterisk indicates the full-length BRD4-TII. Arrows indicate the heavy chain and light chain from IgG beads. (G) BRD4-TII expressed in E. coli was affinity purified on IgG beads and used to coimmunoprecipitate GST, GST-CDK9, GST-cyclin T1, MBP, or MBP-CDK9 from E. coli lysates as indicated. The immunocomplexes extracted from the beads were resolved on SDS/PAGE and immunoblotted with CDK9 or cyclin T1 antibodies. Blue asterisks indicate GST-CDK9 or MBP-CDK9. Green asterisks indicate GST-cyclin T1. (H) BRD4-TII expressed in E. coli was affinity purified on IgG beads and used to coimmunoprecipitate GST-cyclin T1 and MBP-CDK9 from E. coli lysates. The immunocomplexes were subjected to an in vitro kinase assay in the presence of DMSO, 50 μM DRB, 50 μM apigenin, 10 μM LDC000067, 10 μM PHA767491, 2 μM flavopiridol, 10 μM palbociclib, or 50 μM TBB. The samples were analyzed on SDS/PAGE and visualized by autoradiography (Autorad). The asterisk indicates the full-length BRD4-TII.
In parallel with the proteomic study, we also performed a minicompound screening to determine the effect of numerous drugs on BRD4 hyperphosphorylation. Among all the kinase inhibitors tested, only the known CDK9 inhibitors could block BRD4 hyperphosphorylation in NMC cells (Table S1). In a Phos-tag gel phosphorylation assay, all three CDK9 inhibitors tested—LDC000067, flavopiridol, and PHA767491—showed a significant inhibition of BRD4 hyperphosphorylation in HCC2429 NMC cells (Fig. 2D and Fig. S2C). In addition, two other less specific CDK9 inhibitors, 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) (38) and apigenin (39), also reduced BRD4 hyperphosphorylation in HCC2429 cells (Fig. S2C). All the inhibitors tested also moderately inhibited BRD4 phosphorylation in HEK293 cells (Fig. 2D and Fig. S2C). In contrast, treatment with a casein kinase II-specific inhibitor, 4,5,6,7-tetrabromobenzotriazole (TBB), which has been shown to inhibit BRD4 phosphorylation (33), did not affect BRD4 phosphorylation in either HCC2429 NMC cells or HEK293 cells in the Phos-tag gel assay (Fig. S2D).
Table S1.
Drugs screened for the effect on BRD4 hyperphosphorylation
| Name | Target | Effect on BRD4 hyperphosphorylation |
| Ibrutinib | BTK | No effect |
| Idelalisib | PI3K (p110δ) | No effect |
| Trametinib | MEK1/2 | No effect |
| TBB | CKII | No effect |
| LDC000067 | CDK9 | Inhibits BRD4 hyperphosphorylation in HCC2429 |
| PHA767491 | CDK9/CDC7 | Inhibits BRD4 hyperphosphorylation in HCC2429 |
| Flavopiridol | CDKs | Inhibits BRD4 hyperphosphorylation in HCC2429 |
| DRB | CDK9 and many other kinases | Inhibits BRD4 hyperphosphorylation in HCC2429 |
| Apigenin | CDK9 and many other kinases | Inhibits BRD4 hyperphosphorylation in HCC2429 |
| Wortmannin | PI3K/ATM | No effect |
| Danusertib | Aurora A/B/C | No effect |
| Sotrastaurin | pan-PKC | No effect |
| MK-2206 | Akt1/2/3 | No effect |
| KU-55933 | ATM | No effect |
| D4476 | CKI | No effect |
| CHIR99021 | GSK-3α/β | No effect |
| SRPIN340 | SRPK1 | No effect |
| NU7441 | DNA-PK | No effect |
| Palbociclib | CDK4/6 | No effect |
| 3-Deazaneplanocin A (DZNep) | EZH2 | No effect |
| BIX01294 | G9a | No effect |
| GSK343 | EZH2 | No effect |
| CPI-169 | EZH2 | No effect |
| C646 | p300 | Inhibits BRD4 hyperphosphorylation in HCC2429 |
| (+)-JQ1 | BRD4 and other BET proteins | Reduces BRD4 hyperphosphorylation in HCC2429 |
| HMBA | Releases P-TEFb, induces HEXIM1, also targets p300 and BRD4 | Reduces BRD4 hyperphosphorylation in HCC2429 |
Notably, BRD4 from the NMC cells treated with these CDK9 inhibitors migrated to the same position as the less phosphorylated BRD4 in the untreated HEK293 cells (Fig. 2D and Fig. S2C) but not to the lower position of completely dephosphorylated BRD4 as shown in Fig. 1C. This result suggested that other kinase(s) besides CDK9 could be involved in BRD4 phosphorylation. In addition, the CDK9 inhibitors were able to block BRD4 hyperphosphorylation efficiently in NMC cells but inhibited BRD4 phosphorylation in HEK293 cells to a lesser degree (Fig. 2D and Fig. S2C). These results indicated that an NMC-specific mechanism might allow CDK9 to hyperphosphorylate BRD4 in these cells.
In the in vitro kinase assay using nuclear extract of NMC cells to phosphorylate recombinant BRD4, the CDK9 kinase inhibitors also dramatically inhibited BRD4 phosphorylation, whereas TBB had no appreciable effect (Fig. 2E). However, TBB was able to inhibit in vitro phosphorylation of the short isoform of BRD4, BRD4S (2), which contains the N-terminal bromodomains and the extraterminal domain (Fig. S2E). Together, these results suggest that CDK9 plays a critical role in BRD4 hyperphosphorylation in NMC cells. Therefore, we focused on CDK9 in the rest of this study.
In a previous study, P-TEFb purified from mammalian cells was shown to phosphorylate BRD4 protein isolated from Sf9 insect cells in vitro (40). To test whether CDK9 could directly phosphorylate BRD4, recombinant BRD4-TII expressed in E. coli and immunoprecipitated on IgG beads was used to coimmunoprecipitate CDK9 and cyclin T1, which also were expressed in E. coli (Fig. S2F). Western blotting showed that BRD4-TII could pull down CDK9 and cyclin T1 efficiently, individually or in complex (Fig. S2G). Interestingly, only the immunocomplexes containing both CDK9 and cyclin T1 demonstrated strong kinase activity for BRD4 in the in vitro phosphorylation assay (Fig. 2F). The results provided direct evidence that CDK9 in the P-TEFb complex functions as a BRD4 kinase. Various CDK9 inhibitors also dramatically inhibited BRD4 phosphorylation in this in vitro phosphorylation assay, whereas TBB and palbociclib, a selective inhibitor of CDK4 and CDK6, had moderate effects on BRD4 phosphorylation (Fig. S2H). This in vitro experiment using all recombinant proteins further confirmed the specific role of CDK9 in BRD4 hyperphosphorylation.
The Molecular Mechanism Underlying BRD4 Hyperphosphorylation in NMC.
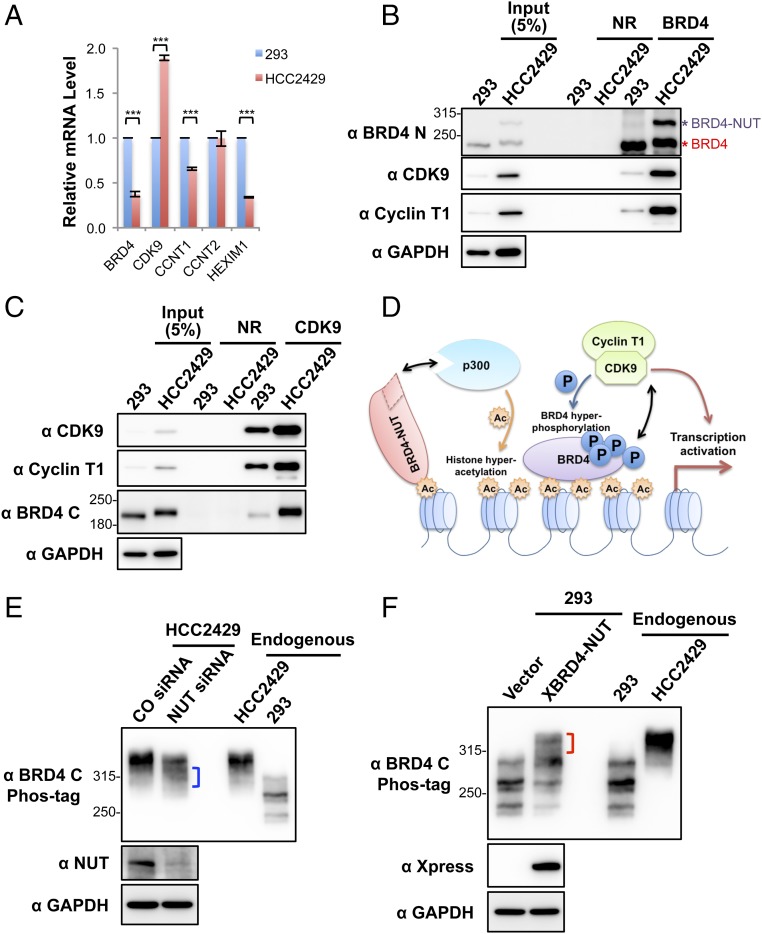
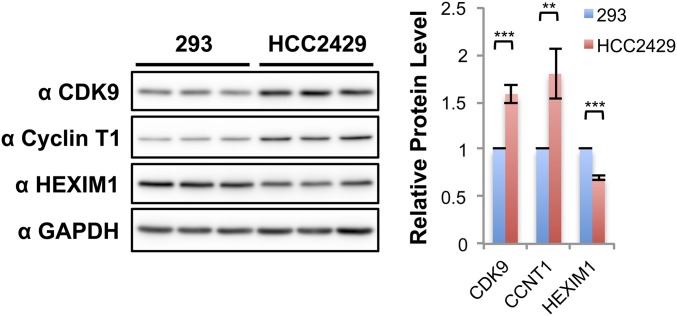
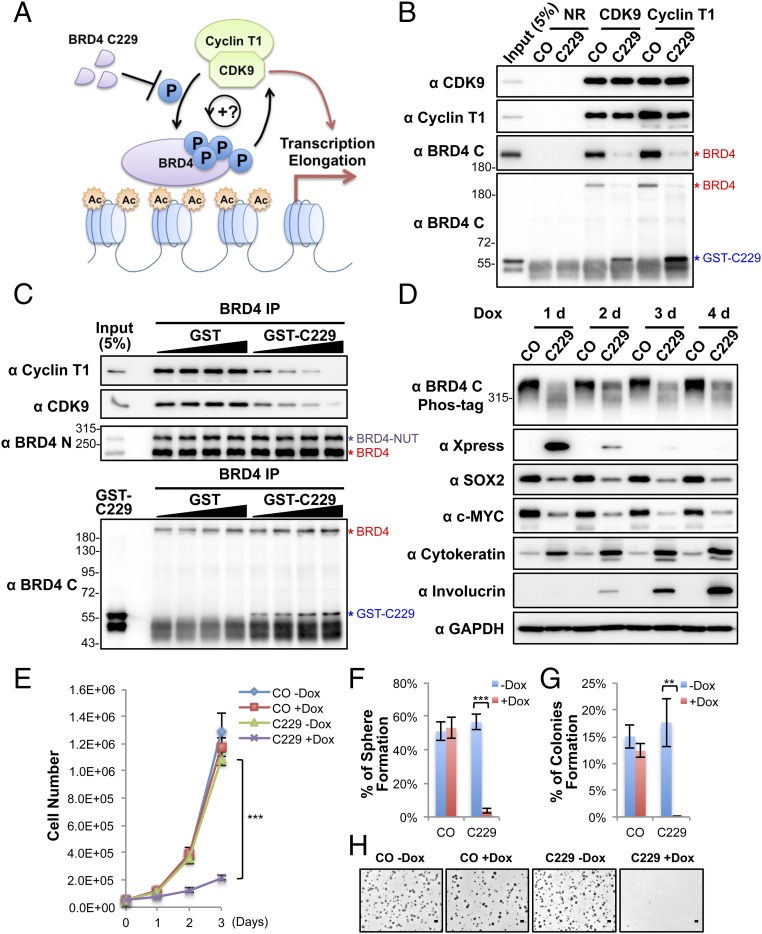
We then set out to understand the mechanism by which CDK9 leads to BRD4 hyperphosphorylation in NMCs. We first examined the expression level of relevant genes in HCC2429 NMC cells and compared them with HEK293 cells by RT-qPCR (Fig. 3A). Because NMC cells carry only one copy of the intact BRD4 gene, it was not surprising that the BRD4 transcript level was about half that observed in HEK293 cells (Fig. 3A). In contrast, the CDK9 transcript level was nearly doubled in HCC2429 cells compared with HEK293 cells (Fig. 3A). The expression levels of cyclin T1 and cyclin T2 were comparable in HCC2429 and HEK293 cells. Hexamethylene bisacetamide (HMBA)-inducible protein 1 (HEXIM1) is a P-TEFb inhibitor that binds to cyclin T1 and sequesters the P-TEFb complex into an inhibitory complex containing the 7SK small nuclear ribonucleoprotein (41). Interestingly, the level of HEXIM1 was significantly lower in HCC2429 cells (Fig. 3A). Western blotting analysis of these proteins in the two cell types confirmed the higher level of CDK9 and lower level of HEXIM1 in HCC2429 cells (Fig. S3). In addition, we consistently detected a higher level of cyclin T1 protein in HCC2429 cells than in HEK293 cells (Fig. 3 B and C and Fig. S3). Therefore, the higher level of P-TEFb and the reduced level of the P-TEFb inhibitor HEXIM1 may contribute to BRD4 hyperphosphorylation in NMC cells.
Fig. 3.
Enrichment of CDK9 in BRD4–NUT–induced hyperacetylated chromatin foci causes BRD4 hyperphosphorylation. (A) Relative mRNA levels in HEK293 and HCC2429 cells were measured by RT-qPCR and normalized to GAPDH mRNA levels. The values for HEK293 mRNA levels were set as 1. Values represent the average of three independent experiments; error bars indicate SD. ***P < 0.001. (B) Nuclear extracts of HEK293 and HCC2429 cells were immunoprecipitated with normal rabbit (NR) or Brd4 N-terminal domain (Brd4 N) antibodies. The samples were immunoblotted with the indicated antibodies. (C) Nuclear extracts of HEK293 and HCC2429 cells were immunoprecipitated with normal rabbit (NR) or CDK9 antibodies. The samples were immunoblotted with the indicated antibodies. (D) The mechanisms underlying BRD4 hyperphosphorylation in NMC. BRD4-NUT recruits and activates p300 to induce histone hyperacetylation in discrete chromatin foci in NMC. This hyperacetylation leads to the sequestration and enrichment of BRD4 and associated P-TEFb in the hyperacetylated chromatin domains, leading to BRD4 hyperphosphorylation. (E) HCC2429 cells were transfected with control (CO) or NUT siRNA. At 30 h posttransfection, whole-cell lysates were analyzed on Phos-tag or SDS/PAGE gels and were immunoblotted with the indicated antibodies. The bracket marks the BRD4 bands with reduced levels of phosphorylation. (F) HEK293 cells were transfected with the empty vector control or a construct expressing Xpress-tagged BRD4-NUT (XBRD4-NUT). At 2 d posttransfection, whole-cell lysates were analyzed on Phos-tag or SDS/PAGE gels and were immunoblotted with the indicated antibodies. The bracket marks the hyperphosphorylated BRD4 bands. In both E and F, lysates from untransfected HCC2429 and HEK293 cells were also loaded on the same Phos-tag gel to show the endogenous BRD4. See also Fig. S3.
Fig. S3.
The protein levels of the P-TEFb components and inhibitor in HEK293 and HCC2429 cells. Whole-cell lysates from HEK293 and HCC2429 cells were resolved on SDS/PAGE and immunoblotted with the indicated antibodies. Samples isolated from three independent experiments were examined. The intensity of the protein bands was analyzed using ImageJ, and the values of HEK293 levels were set as 1. Shown are the relative protein levels of CDK9, cyclin T1, and HEXIM1 normalized with GAPDH. Values represent the average of three independent experiments; error bars indicate SD. **P < 0.01, ***P < 0.001.
We also examined the BRD4–P-TEFb interaction in HCC2429 and HEK293 cells. An antibody specific for the N terminus of BRD4 pulled down a similar amount of BRD4 from HCC2429 and HEK293 cells, but the amount of CDK9 and cyclin T1 coimmunoprecipitated with BRD4 was dramatically greater in HCC2429 than in HEK293 cells (Fig. 3B). In the reciprocal coimmunoprecipitation experiment, a CDK9 antibody also pulled down a significantly increased level of BRD4 from HCC2429 cells compared with HEK293 cells (Fig. 3C). Although these coimmunoprecipitation results suggested an increased interaction between P-TEFb and BRD4 in HCC2429 NMC cells compared with HEK293 cells, this increase might also be caused in part by the higher levels of CDK9 and cyclin T1 detected in HCC2429 cells (Fig. 3 B and C).
We also examined whether other cellular factors such as local kinase and substrate concentrations might contribute to BRD4 hyperphosphorylation in NMC cells. In previous studies (28, 30, 31), we showed that in NMC cells BRD4-NUT localizes to discrete chromatin foci containing hyperacetylated histones and recruits histone acetyltransferases (HATs), such as p300, to stimulate more histone hyperacetylation in these chromatin foci. The hyperacetylated histones provide binding sites for the accumulation of the BRD4 and P-TEFb complexes (Fig. 3D). We also have shown that P-TEFb is enriched in these hyperacetylated chromatin foci (31). We therefore speculated that BRD4 enrichment and associated P-TEFb in these hyperacetylated BRD4-NUT foci might increase the local concentrations of both the kinase and substrate, thus leading to BRD4 hyperphosphorylation in NMC cells (Fig. 3D). BRD4-NUT knockdown in NMC cells disperses the hyperacetylated chromatin foci, causing BRD4 to be released from the punctate chromatin foci and to diffuse throughout chromatin (31). Importantly, BRD4-NUT knockdown in HCC2429 NMC cells also caused a partial reduction of BRD4 hyperphosphorylation (Fig. 3E). Conversely, BRD4-NUT expressed ectopically in non–NMC cells induces the formation of histone-hyperacetylated nuclear foci in non–NMC cells and recruits BRD4 to punctate chromatin foci (31). We found that in HEK293 cells transfected with a BRD4-NUT expression construct a portion of the endogenous BRD4 also became hyperphosphorylated and migrated to the same position as BRD4 isolated from HCC2429 NMC cells (Fig. 3F). These studies supported the role of BRD4-NUT in promoting BRD4 hyperphosphorylation.
We also demonstrated that BRD4-NUT enriched in the hyperacetylated chromatin domain strongly stimulates the abnormal activation of the stem cell marker SOX2 to support the aberrant stem cell-like proliferation and highly aggressive transforming activity of NMC (30). Inhibition of BRD4 hyperphosphorylation in NMC cells by BRD4-NUT knockdown also correlates with SOX2 repression (30), suggesting that BRD4 hyperphosphorylation induced by BRD4-NUT might contribute to the oncogene transactivation that supports NMC transformation.
Inhibition of BRD4 Hyperphosphorylation by Chemical Compounds Represses Oncogene Expression and Cellular Transformation.
Because we discovered that BRD4 recruitment to hyperacetylated chromatin foci stimulates its hyperphosphorylation, we tested whether blocking the formation of these foci in NMC cells could block BRD4 hyperphosphorylation and downstream oncogene expression. Both BRD4-NUT knockdown and overexpression provided evidence that BRD4–NUT–induced hyperacetylated chromatin foci are critical for triggering BRD4 hyperphosphorylation in NMC cells. However, the low transfection efficiency for BRD4-NUT siRNA and expression construct resulted in very moderate changes in BRD4 hyperphosphorylation in the transfected cells (Fig. 3 E and F). We therefore used the chemical approach using three different compounds to examine further whether blocking BRD4–NUT–induced HAT recruitment and histone hyperacetylation prevents BRD4 hyperphosphorylation in NMC cells (Fig. 4).
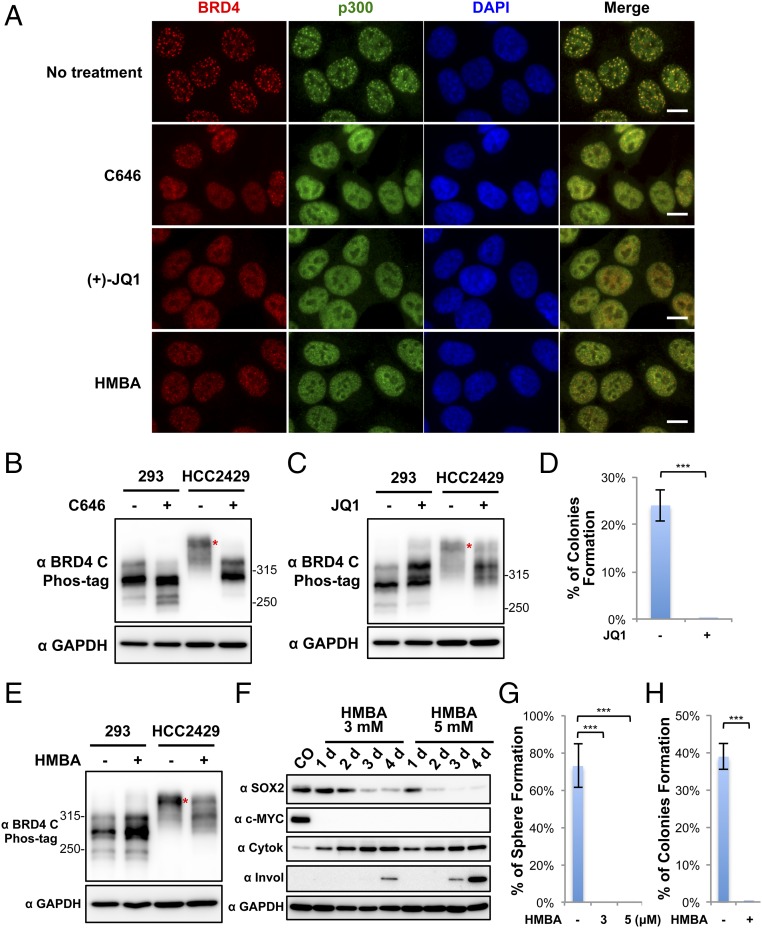
Fig. 4.
Dispersion of BRD4-NUT foci reduces BRD4 hyperphosphorylation, inhibits downstream oncogene expression, and abolishes NMC transformation. (A) HCC2429 cells were treated with 20 μM C646 for 1 h, 1 μM (+)-JQ1 for 1 h, or 5 mM HMBA for 12 h. Untreated control cells and treated cells were fixed and immunostained with BRD4 C and p300 antibodies and were counterstained with DAPI. (Scale bars: 10 μm.) (B) HEK293 and HCC2429 cells were treated with DMSO or 20 μM C646 for 1 h. Whole-cell lysates were analyzed on Phos-tag or SDS/PAGE gel and were immunoblotted with the indicated antibodies. (C) HEK293 and HCC2429 cells were treated with 1 μM (−)-JQ1 or (+)-JQ1 for 1.5 h. Whole-cell lysates were analyzed on Phos-tag or SDS/PAGE gel and were immunoblotted with the indicated antibodies. (D) HCC2429 cells were seeded in the presence of 100 nM (−)-JQ1 or (+)-JQ1 for the soft agar assay. The percentage of cells forming colonies was calculated from 8,000 seeded cells. Values represent the average of three independent experiments; error bars indicate SD. (E) HEK293 and HCC2429 cells were treated with or without 5 mM HMBA for 12 h. Whole-cell lysates were analyzed on Phos-tag or SDS/PAGE gel and were immunoblotted with the indicated antibodies. (F) HCC2429 cells were treated with 3 mM or 5 mM HMBA. Untreated control (CO) or treated cells were harvested at the indicated times. Whole-cell lysates were immunoblotted with the indicated antibodies. (G) HCC2429 cells were seeded in the presence or absence of HMBA at the indicated concentration for the sphere-formation assay. The percentage of cells forming spheres was calculated from 80 seeded cells. Values represent the average of three independent experiments; error bars indicate SD. (H) HCC2429 cells were seeded in the absence or presence of 3 mM HMBA for the soft agar assay. The percentage of cells forming colonies was calculated from 8,000 seeded cells. Values represent the average of three independent experiments; error bars indicate SD. ***P < 0.001. See also Fig. S4.
We first started by disrupting the BRD4–NUT/p300–induced hyperacetylated chromatin foci (Figs. 3D and 4A). In the untreated HCC2429 cells, both BRD4 and p300 were sequestered in the hyperacetylated, distinct chromatin foci (Fig. 4A). Treatment of HCC2429 cells with the p300 inhibitor C646 (30) for only 1 h efficiently dispersed the hyperacetylated BRD4-NUT chromatin foci in all the treated cells (Fig. 4A). Remarkably, C646 treatment also resulted in a significant reduction of BRD4 hyperphosphorylation in HCC2429 cells without dramatically affecting the BRD4 phosphorylation level in HEK293 cells (Fig. 4B). We, and others, have demonstrated that BRD4-NUT and BRD4 are recruited to induce abnormal activation of oncogenes such as SOX2 (30) and c-MYC (18) within the hyperacetylated chromatin domains in NMC. By inhibiting p300 activity with C646, we observed a significant repression of SOX2 (30). Both the mRNA and protein levels of c-MYC were also reduced in C646-treated NMC cells (Fig. S4). These results indicate that BRD4–NUT–recruited p300 plays an important role in BRD4 hyperphosphorylation, which correlates with downstream oncogene expression.
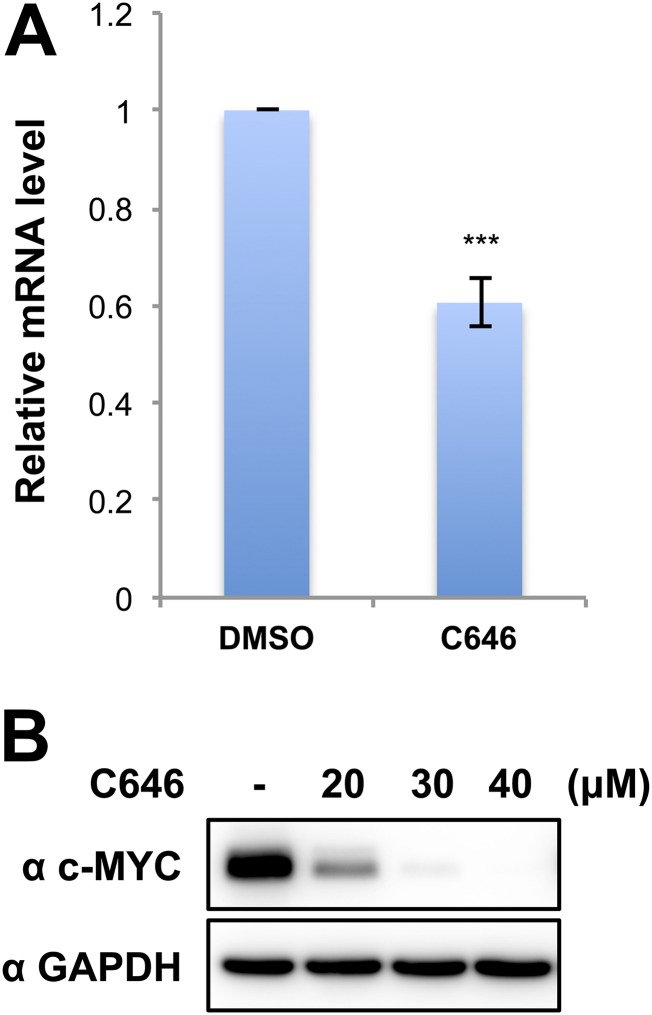
Fig. S4.
The p300 inhibitor C646 represses c-MYC expression in HCC2429 cells. (A) c-MYC mRNA levels in HCC2429 cells treated with DMSO or 40 μM C646 for 4 h were measured by RT-qPCR and normalized to GAPDH mRNA levels. The value for the c-MYC mRNA level in DMSO control cells was set as 1. Values represent the average of three independent experiments; error bars indicate SD. ***P < 0.001. (B) HCC2429 cells were treated with DMSO or with 20, 30, or 40 μM C646 for 6 h. Whole-cell lysates were analyzed on SDS/PAGE and immunoblotted with indicated antibodies.
We also reasoned that if BRD4 binding to the hyperacetylated chromatin foci is critical for enriching P-TEFb to stimulate BRD4 hyperphosphorylation (Fig. 3D), dissociation of BRD4 from acetylated histones by the BET inhibitor (+)-JQ1 (19) might also inhibit its hyperphosphorylation by dispersing the foci containing both highly concentrated CDK9 kinase and BRD4 substrate. Indeed, HCC2429 cells treated with (+)-JQ1 showed significantly reduced BRD4 hyperphosphorylation compared with the cells treated with the inactive stereoisomer (−)-JQ1 (Fig. 4C). In contrast, the BRD4 phosphorylation level in HEK293 cells was not reduced but instead was slightly increased by (+)-JQ1 treatment (Fig. 4C), indicating that it might be regulated through a different mechanism, such as BET inhibitor-induced adaptive kinome reprogramming (42). The (+)-JQ1 treatment of NMC cells also led to SOX2 repression, abrogation of the aberrant stem cell-like proliferation, and complete disruption of colony formation in soft agar plates by cancer cells (Fig. 4D) (30). Together, these results provided additional evidence that BRD4 hyperphosphorylation is linked to its downstream oncogene expression during NMC cellular transformation.
Another chemical compound, HMBA, has been used to induce cancer cell differentiation for treating a number of cancers (43). The mechanism of HMBA action involves inducing the expression of the P-TEFb inhibitor HEXIM1 (41). In addition, HMBA also acts as a selective bromodomain inhibitor and displaces BET proteins from chromatin (43). We therefore tested this early anticancer agent in NMC cells. HCC2429 cells treated with HMBA indeed showed diffusion of BRD4 away from the punctate foci observed in untreated cells, indicating that HMBA could dissociate BRD4 from hyperacetylated chromatin domain (Fig. 4A). HMBA also efficiently decreased BRD4 hyperphosphorylation in HCC2429 cells without dramatically affecting the BRD4 phosphorylation level in HEK293 cells (Fig. 4E). This effect of HMBA may be attributable to its ability to induce HEXIM1, which then can negatively regulate the CDK9/cyclin T1 complex (41). More importantly, HMBA treatment also led to a dramatic repression of the BRD4 downstream oncogenes SOX2 and c-MYC while inducing the cellular differentiation markers cytokeratin and involucrin (Fig. 4F). As we reported previously (30), NMC cells can grow into stem cell-like spheres in culture dishes and form colonies in soft agar. We therefore performed sphere-formation and soft agar transformation assays to determine if HMBA inhibits NMC proliferation and cellular transformation. Unlike the untreated NMC cells, HMBA-treated NMC cells could no longer grow into cancer stem cell-like spheres or as transformed colonies in soft agar plates (Fig. 4 G and H).
In summary, three different chemical compounds used in this study, with different mechanisms of action for blocking BRD4 recruitment to the hyperacetylated chromatin foci, were all able to inhibit BRD4 hyperphosphorylation. We also demonstrated a correlation between BRD4 hyperphosphorylation and its ability to drive downstream oncogenes that directly contribute to NMC tumorigenesis. This study further indicates that BRD4 hyperphosphorylation may be an important factor for sustaining target oncogene expression and cellular transformation.
A Dominant Negative Inhibitor of BRD4–CDK9 Interaction Abrogates BRD4 Hyperphosphorylation, Oncogene Expression, and Cellular Transformation in NMC.
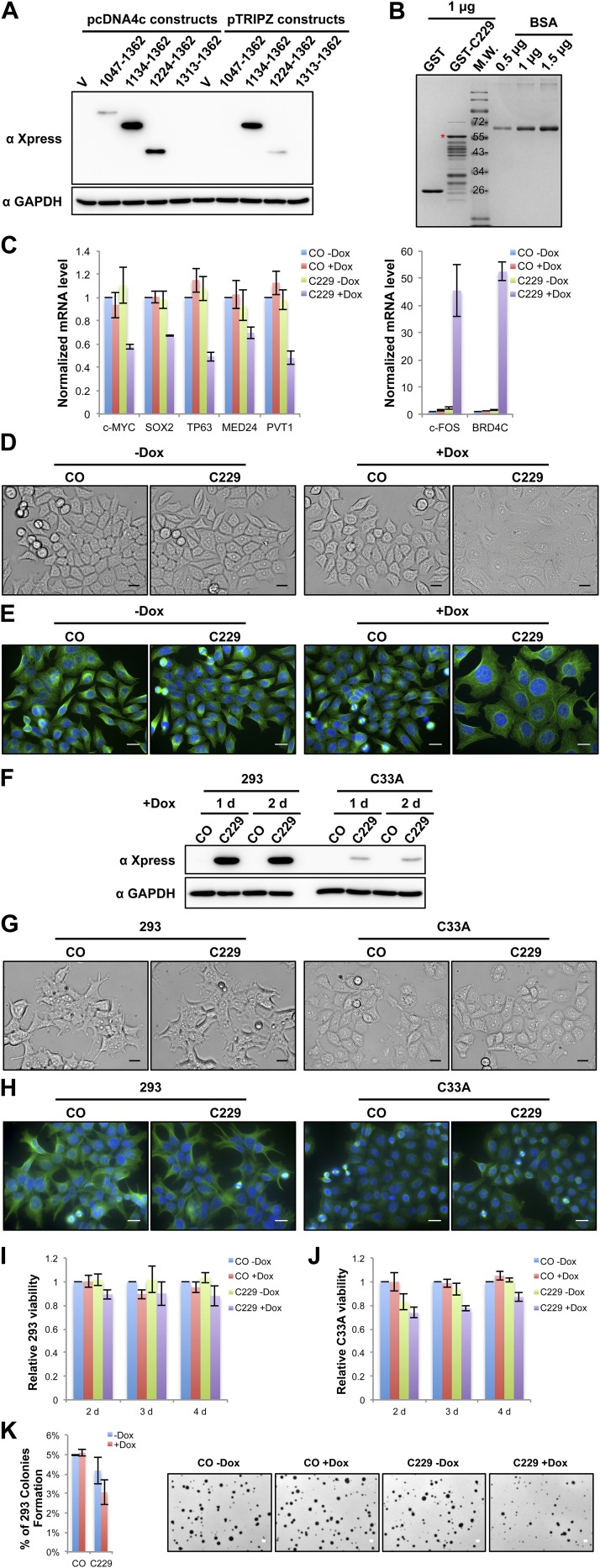
Because CDK9 binds BRD4 through its C-terminal domain (44), we tested if the BRD4 C-terminal domain could be used as a dominant negative inhibitor (DNI) to block the BRD4 and CDK9 interaction competitively and to inhibit BRD4 hyperphosphorylation (Fig. 5A). A set of constructs encoding varied lengths of the BRD4 C-terminal domains fused to the Xpress tag were transfected into HCC2429 cells. The fragment spanning amino acids 1134–1362 showed the highest expression level in HCC2429 cells when expressed from two different vectors (Fig. S5A). Therefore, it was chosen for the rest of this study and was named “C229.” We first tested the effect of C229 on the P-TEFb–BRD4 interaction by performing coimmunoprecipitation. Because C229 was degraded very quickly in the HCC2429 cell lysates used for coimmunoprecipitation, we performed the coimmunoprecipitation using HCC2429 cell lysates supplemented with or without recombinant GST-C229 expressed in E. coli (Fig. S5B). Both CDK9 and cyclin T1 antibodies coimmunoprecipitated BRD4 in the absence of the C229, but the antibodies pulled down only C229 when it was added into the reaction (Fig. 5B). C229 also caused a dose-dependent reduction of P-TEFb coimmunoprecipitated with BRD4 antibody (Fig. 5C). NMC cells then were transduced with lentiviruses encoding the doxycycline (Dox)-inducible C229 to test how blocking the P-TEFb–BRD4 interaction affects BRD4 hyperphosphorylation and downstream gene expression. Remarkably, expression of C229 in NMC cells effectively inhibited BRD4 hyperphosphorylation and repressed its oncogenic target genes SOX2 and MYC (Fig. 5D and Fig. S5C). On the other hand, C229 expression led to significant induction of NMC cell differentiation, as indicated by the robustly increased differentiation markers cytokeratin, involucrin, and c-FOS (Fig. 5D and Fig. S5C). The C229-expressing HCC2429 cells also showed a clearly differentiated morphology, with nearly all the cells becoming flat and enlarged (Fig. S5 D and E). C229 expression also significantly inhibited the growth of NMC tumor cells (Fig. 5E). The induced C229 expression diminished the ability of NMC cells to grow as stem cell-like spheres and soft agar colonies, abolishing NMC transformation (Fig. 5 F–H). In contrast, C229 expression in HEK293 and C33A cells, which do not show BRD4 hyperphosphorylation, did not significantly repress cellular proliferation, differentiation, or transformation (Fig. S5 F–K). Although C229 expression in HEK293 cells did not dramatically change the number of colonies grown in soft agar, the size of these colonies was reduced (Fig. S5K). The underlying mechanism remains to be investigated. Thus, the effect of C229 on BRD4 hyperphosphorylation and cellular transformation appeared to be specific to NMC. Taken together, our results showed that C229 functions as a DNI to block the P-TEFb–BRD4 interaction, inhibiting BRD4 hyperphosphorylation, oncogene expression, and NMC transformation.
Fig. 5.
Blocking the BRD4–CDK9 interaction reduces BRD4 hyperphosphorylation, inhibits downstream oncogene expression, and abolishes cellular transformation. (A) BRD4–C229 functions as a DNI to block the BRD4–CDK9 interaction. (B) Nuclear extracts of HCC2429 cells were coimmunoprecipitated with normal rabbit (NR), CDK9, or cyclin T1 antibodies in the presence of 1.3 μg purified GST control (CO) or GST–C229. The immunocomplexes were immunoblotted with the indicated antibodies. (C) Nuclear extracts of HCC2429 cells were coimmunoprecipitated with normal rabbit (NR) or BRD4 N antibodies in the presence of 2, 4, 8, or 16 μg purified GST or GST–C229. The immunocomplexes were immunoblotted with the indicated antibodies. (D) HCC2429 cells carrying a vector control (CO) or an inducible Xpress-C229 construct were treated with Dox and harvested as indicated. Cell lysates were analyzed on Phos-tag or SDS/PAGE gels and blotted with the indicated antibodies. (E) Growth curves of HCC2429 control (CO) or C229 inducible cells after treating with or without 1 μg/mL Dox. (F). HCC2429 control (CO) or C229 inducible cells were seeded in the presence or absence of 1 μg/mL Dox for the sphere-forming assay. The percentage of cells forming spheres was calculated from 240 seeded cells. Values represent the average of four independent experiments; error bars indicate SD. (G) HCC2429 control (CO) or C229 inducible cells were seeded in the presence or absence of 1 μg/mL Dox for the soft agar assay. The percentage of cells forming colonies was calculated from 12,000 seeded cells. Values represent the average of three independent experiments; error bars indicate SD. (H) Representative images of soft agar plates for the experiments shown in G. (Scale bar: 1 mm.) **P < 0.01; ***P < 0.001. See also Fig. S5.
Fig. S5.
Blocking the BRD4–CDK9 interaction specifically reduces BRD4 hyperphosphorylation, inhibits downstream oncogene expression, induces differentiation, and abolishes cellular transformation in NMC cells. (A) HCC2429 cells were transfected with the vector control (V) or the various Xpress-tagged BRD4 C-terminal domain constructs. At 1 d posttransfection, whole-cell lysates were analyzed on SDS/PAGE and immunoblotted with Xpress and GAPDH antibodies. (B) GST control and GST-BRD4-C229 purified proteins from E. coli were analyzed on SDS/PAGE and stained with CBB. Varying amounts of BSA were also loaded in the same gel to estimate the abundance of the purified proteins. (C) HCC2429 control (CO) or C229 inducible cells were treated with or without 1 μg/mL Dox for 1 d. Relative mRNA levels were measured by RT-qPCR and normalized to GAPDH mRNA levels. The values for mRNA levels from control cells without Dox treatment were set as 1. Values represent the average of three independent experiments; error bars indicate SD. (D) HCC2429 control (CO) or C229 inducible cells were treated with or without 1 μg/mL Dox for 1 d. Shown are the bright-field images of the cells captured under a microscope. (Scale bars: 20 μm.) (E) HCC2429 control (CO) or C229 inducible cells were treated with or without 1 μg/mL Dox for 1 d. Cells were fixed and immunostained with β-tubulin antibody and were counterstained with DAPI. (Scale bars: 20 μm.) (F) HEK293 and C33A cells stably carrying a vector control (CO) or the inducible C229 expression construct were treated with 1 μg/mL Dox and harvested as indicated. Whole-cell lysates were analyzed on SDS/PAGE and immunoblotted with Xpress and GAPDH antibodies. (G) HEK293 and C33A control (CO) or C229 inducible cells were treated with 1 μg/mL Dox for 1 d. Shown are the bright-field images of the cells captured under a microscope. (Scale bars: 20 μm.) (H) HEK293 and C33A control (CO) or C229 inducible cells were treated with 1 μg/mL Dox for 1 d. Cells were fixed and immunostained with β-tubulin antibody and counterstained with DAPI. (Scale bars: 20 μm.) (I) HEK293 control (CO) or C229 inducible cells were treated or not treated with 1 μg/mL Dox. Cell viability was measured using the CellTiter-Glo assay at the indicated time points. Relative cell viability was calculated by setting the values for the control cells without Dox treatment as 1. Values represent the average of three independent experiments; error bars indicate SD. (J) C33A control (CO) or C229 inducible cells were treated or not treated with 1 μg/mL Dox. Cell viability was measured using the CellTiter-Glo assay at the indicated time points. Relative cell viability was calculated by setting the values for the control cells without Dox treatment as 1. Values represent the average of three independent experiments; error bars indicate SD. (K, Left) HEK293 control (CO) or C229 inducible cells were seeded in the presence or absence of 1 μg/mL Dox for the soft agar assay. The percentage of cells forming colonies was calculated from 12,000 seeded cells. Values represent the average of three independent experiments; error bars indicate SD. (Right) Representative images of soft agar plates for this experiment. (Scale bars: 1 mm.)
BRD4 Is Also Hyperphosphorylated in Non–NMC Cancers Associated with High BRD4 Oncogenic Activity.
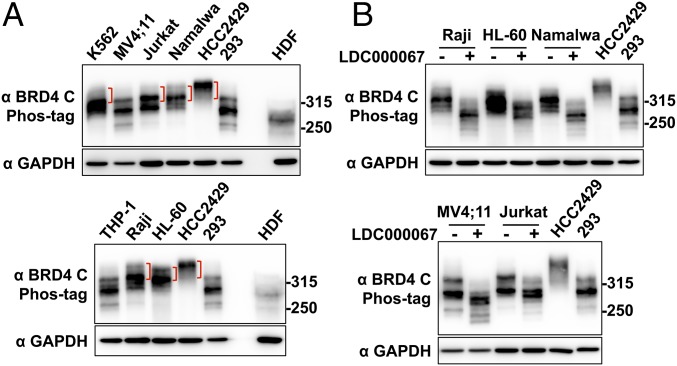
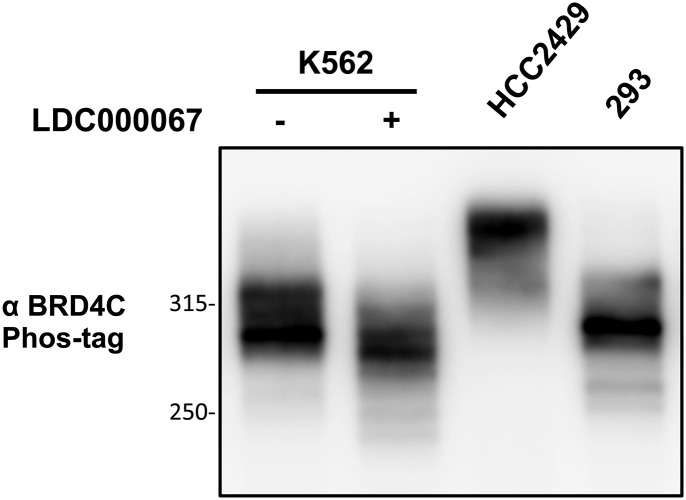
Our study suggested that BRD4 hyperphosphorylation is linked to oncogene expression and cellular transformation in NMC tumors. Because BRD4 is a key therapeutic target that drives oncogene expression in multiple cancers (4, 14), we decided to examine if BRD4 is also hyperphosphorylated in other cancer cells that have demonstrated enhanced BRD4 oncogenic activity. Activation of the BRD4-dependent core transcriptional program is the key oncogenic driver for various leukemia diseases (10, 45, 46). BRD4 is also implicated in the pathogenesis of Burkitt’s lymphoma (12). We therefore examined the BRD4 phosphorylation status in a number of leukemia and lymphoma cell lines, including K562, MV4;11, Jurkat, THP-1, HL-60, Namalwa, and Raji cells (Fig. 6). Using the Phos-tag gel assay, we compared the BRD4 phosphorylation status in these cancer cells and in normal cells such as HDFs, as well as in HCC2429 and HEK293 cells. BRD4 in the normal HDFs migrated to the lowest position in Phos-tag gel, showing very little signal for phosphorylated BRD4 bands (Fig. 6A). BRD4 is phosphorylated to some degree in HEK293 cells but shows robust hyperphosphorylation in HCC2429 cells (Fig. 6A). We detected various degrees of BRD4 hyperphosphorylation in K562, Jurkat, Namalwa, Raji, and HL-60 cells, because some of the BRD4 isolated from these cancer cells migrated to a higher position in the Phos-tag gel than BRD4 isolated from HDFs and HEK293 cells (Fig. 6A). On the other hand, MV4;11 and THP-1 cells showed much less hyperphosphorylated BRD4 (Fig. 6A). Notably, treatment with the CDK9 inhibitor LDC000067 dramatically inhibited BRD4 hyperphosphorylation in these non–NMC cancer cells (Fig. 6B and Fig. S6). These results showed that, in addition to NMC cells, BRD4 is also hyperphosphorylated in some other cancer cells displaying high levels of BRD4 oncogenic function and that CDK9 may be a key kinase that contributes to BRD4 hyperphosphorylation.
Fig. 6.
BRD4 is also hyperphosphorylated in non–NMC cancer cells associated with BRD4 dysfunction. (A) Whole-cell lysates from various leukemia and lymphoma cells were analyzed on a Phos-tag gel and compared with the samples from HCC2429 and HEK293 cells and HDFs. The samples were immunoblotted with BRD4 C antibody. Brackets mark the hyperphosphorylated BRD4 bands. The samples were also analyzed on SDS/PAGE and immunoblotted with GAPDH. Because the BRD4 protein level in HDFs is extremely low, seven times more HDF protein sample was loaded in the Phos-tag gel than in the SDS/PAGE for the GAPDH blot. The images shown are representative of three independent experiments. (B) Leukemia and lymphoma cells were treated with DMSO or 10 μM LDC000067 for 2 h. Whole-cell lysates were analyzed on Phos-tag or SDS/PAGE gel and were immunoblotted with the indicated antibodies. Lysates from untreated HCC2429 and HEK293 cells were also loaded on the same Phos-tag gel to show the endogenous BRD4. See also Fig. S6.
Fig. S6.
BRD4 is also hyperphosphorylated in non–NMC cancer cells. K562 cells were treated with DMSO or 10 μM LDC000067 for 2 h. Whole-cell lysates were analyzed on a 10-μM Phos-tag gel and were immunoblotted with BRD4 C antibody. Lysates from untreated HCC2429 and HEK293 cells were also loaded on the same Phos-tag gel to show the endogenous BRD4.
Discussion
BRD4 is dysregulated in a range of aggressive malignancies (4, 10–12, 14, 16, 17) and is being increasingly implicated as a major driver of oncogenic growth. Hence, it is under intense investigation as a target for cancer therapy. BRD4 also plays a fundamental role in the regulation of transcription, cell-cycle progression, and cellular proliferation. However, how BRD4 activity is regulated to maintain its normal function in healthy cells and how dysregulation of this process leads to cancer remain poorly understood. Therefore, it has been challenging to develop anticancer strategies that specifically target the aberrant BRD4 oncogenic activity in cancer cells without affecting the normal BRD4 function in healthy cells.
In this study, we tackled the important question regarding the mechanistic regulation of BRD4 function during cancer development. We discovered that BRD4 is hyperphosphorylated in NMC tumors and identified CDK9 as a potential kinase that contributes to BRD4 hyperphosphorylation in NMC (Figs. 1 and 2). High CDK9 expression levels and robust P-TEFb–BRD4 interaction in NMC may contribute to BRD4 hyperphosphorylation (Fig. 3). Also, in NMC cells, the BRD4 and P-TEFb complexes are enriched at the BRD4–NUT–induced histone-hyperacetylated chromatin domain (31), thus stimulating BRD4 hyperphosphorylation (Fig. 3D). BRD4-NUT knockdown, disrupting the histone-hyperacetylated chromatin foci, inhibited BRD4 hyperphosphorylation, whereas induction of histone-hyperacetylated chromatin domain by expressing BRD4-NUT in non–NMC cells stimulated BRD4 hyperphosphorylation (Fig. 3). Using both chemical and molecular inhibitors to disperse BRD4–NUT–induced hyperphosphorylated chromatin foci or to disrupt the P-TEFb–BRD4 interaction in NMC cells abrogated BRD4 hyperphosphorylation, downstream oncogene expression, and NMC cellular transformation (Figs. 3–5). Therefore, our study suggested that BRD4 hyperphosphorylation is linked to its ability to drive downstream oncogene expression and cellular transformation in NMC tumors.
Up-regulation of a specific set of oncogenic target genes driven by BRD4 has been observed in a variety of cancers (4, 14). Therefore, we compared the BRD4 phosphorylation status in these BRD4-associated cancers with that in normal cells. We discovered that BRD4 is also hyperphosphorylated in a number of non–NMC cancers associated with enhanced BRD4 oncogenic transcription activities (Fig. 6), suggesting that this hyperphosphorylation may be a general mechanism that supports BRD4 oncogenic function. In line with this notion, the chemical compounds that we found to cause reduction of BRD4 hyperphosphorylation could significantly repress the growth of both NMC and non–NMC tumor growth in vivo, with several of them demonstrating potent antitumor activities in clinical trials (Table 1). Therefore, it is tempting to speculate that these compounds may function, at least in part, by blocking BRD4 hyperphosphorylation.
Table 1.
Summary of in vivo studies using compounds that inhibit BRD4 hyperphosphorylation
| Compound | Target | In vivo studies and references |
| Flavopiridol | CDK1/2/4/6/7/9 | Flavopiridol significantly inhibits tumor growth in one of the two NMC xenograft lines (47). |
| Flavopiridol prolongs survival in murine transplanted models of MLL-ENL–transformed cells (48). | ||
| Flavopiridol has potent antitumor activity in human leukemia and lymphoma xenografts (49). | ||
| Flavopiridol has modest to significant activities in relapsed CLL in clinical trials (50, 51). | ||
| PHA-767491 | CDC7/CDK9 | PHA-767491 has antitumor activity in rodent models of human leukemia, human colon carcinoma, and DMBA-induced mammary carcinomas (52). |
| C646 | p300 | C646 suppresses in vivo growth of transplanted AE9a leukemia blasts in mice (53). |
| C646 suppresses tumor growth in a lung cancer xenograft and a leukemia xenograft (54). | ||
| (+)-JQ1 | BRD4/BRD4-NUT | JQ1 promotes tumor regression and prolongs survival in murine models of NMC (19). Therapeutic effects for JQ1 were observed in mouse models of hematological malignancies, such as acute myeloid leukemia, multiple myeloma, and lymphoma, and in some solid tumors, such as neuroblastoma, breast cancer, and prostate cancer (10–14, 55, 56). |
| OTX015/MK-8628 (A JQ1 analog) | BRD4/BRD4-NUT | Among four patients with advanced-stage NMC treated with OTX015/MK-8628, two patients responded rapidly with tumor regression and symptomatic relief, and a third had meaningful disease stabilization with a minor metabolic response (57). |
| HMBA | Releases CDK9 from its inactive complex; induces HEXIM1; targets p300 and BRD4 (43) | HMBA induces a sharp decrease in tumorigenicity in a murine model of human colon cancer (58). |
| HMBA slightly promotes tumor regression and prolongs survival in murine models of human oral squamous cell carcinoma (59). | ||
| A phase II clinical trial was conducted using HMBA to treat patients with MDS or AML. Of the 41 patients in the trial, HMBA induced a complete remission in three patients and a partial remission in six patients (60). |
AE9a, AML1-ETO 9a fusion gene; AML, acute myelogenous leukemia; CLL, chronic lymphocytic leukemia; DMBA, 7,12-dimethylbenz(a)anthracene; MDS, myelodysplastic syndrome; MLL-ENL, mixed-lineage leukemia (MLL) oncogenic fusion caused by the translocation between chromosomes 11 and 19 (also called eleven nighteen leukemia, ENL); NMC, NUT midline carcinoma.
Our study suggests that CDK9 is one of the major kinases that potentially could phosphorylate BRD4 in NMCs. In NMCs, BRD4–NUT–induced enrichment of CDK9 at the hyperacetylated chromatin foci may lead to BRD4 hyperphosphorylation. In addition, p300 recruited by BRD4-NUT to the hyperacetylated chromatin foci also has the ability to activate CDK9 (61), and this activation could contribute to BRD4 hyperphosphorylation in NMC cells. Treating additional BRD4-associated cancers with a CDK9 inhibitor also blocked BRD4 hyperphosphorylation in these cancer cells (Fig. 6B), suggesting that CDK9 could also mediate BRD4 hyperphosphorylation in these cancers. However, other cellular kinases and protein phosphatases may also regulate BRD4 phosphorylation under different physiological conditions. For example, from the SILAC BRD4 proteomic study described above, we identified a number of kinases and protein phosphatases as BRD4-binding partners (Fig. 2 and Dataset S1). Protein phosphatases such as PP2A frequently have been found to be genetically mutated or functionally inactivated in many cancers and therefore have become attractive targets for anticancer therapy (62). In fact, PP2A has been identified as a BRD4 serine phosphatase in triple-negative breast cancer (23). Building on our finding and these recent studies, we reason that coordinated actions of multiple cellular kinases and phosphatases provide a tightly controlled molecular mechanism to regulate BRD4 phosphorylation and function in normal cells and that abnormal activation of these kinases or inhibition of phosphatases may lead to aberrant BRD4 phosphorylation, thus inducing BRD4 activity in oncogenes to activate tumorigenesis in NMC and other cancers. Further studies are critical to identify the additional kinases and phosphatases that regulate BRD4 phosphorylation and/or function. Identification of these BRD4 regulatory factors may allow us to examine how dysregulation of the relevant signaling pathways contributes to BRD4 hyperphosphorylation in BRD4-associated cancers.
So far, our study showed only that BRD4 hyperphosphorylation correlates with downstream oncogene expression and cellular transformation. Whether phosphorylation of BRD4 itself is the key event driving cellular transformation or whether the hyperphosphorylated BRD4 recruits P-TEFb to stimulate downstream oncogene expression is unclear and remains an important question for future studies. To prove definitively that BRD4 hyperphosphorylation supports its ability to activate the oncogenes that drive cellular transformation, we will need to identify BRD4 residues that are hyperphosphorylated in cancers and test if mutagenesis of BRD4 hyperphosphorylation sites abolishes cellular transformation. Compared with normal cells, BRD4 is hyperphosphorylated in a number of BRD4-associated cancers (Fig. 6). Also, the wide range of BRD4 mobility shifts observed in these cancers suggested that BRD4 is phosphorylated at multiple sites. Indeed, using the KinasePhos 2.0 web server (63), we analyzed the BRD4 protein sequence and identified more than 40 consensus phosphorylation sites for kinases that potentially could phosphorylate BRD4. As described above, BRD4 also could be phosphorylated by different kinases at different sites in various cancers. The complexity of BRD4 phosphorylation pattern prevents us from mutating a specific residue of BRD4 to test its function in cellular transformation. Our current effort focuses on using quantitative mass spectrometry to identify systematically the BRD4 residues differentially phosphorylated in various cancers. Identifying the BRD4 hyperphosphorylation sites that are uniquely present in the relevant cancers will allow us to develop phosphospecific antibodies that recognize hyperphosphorylated BRD4 as new diagnostic markers to detect abnormal BRD4 function in related cancers. The BRD4 phosphorylation patterns will also be critical for future mechanistic studies to elucidate how BRD4 phosphorylation may dynamically regulate its function in both normal and cancer cells.
In summary, our study uncovered a mechanism by which BRD4 function could be regulated by cellular kinases and signaling pathways, providing insights into how dysregulation of this important epigenetic reader could lead to tumorigenesis. AML, NMC, and other BRD4-associated cancers are often resistant to conventional chemotherapy (15, 64). Therefore, to develop novel therapeutic approaches, it is imperative to determine the oncogenic mechanisms of these cancers. BRD4 and other BET proteins provide ideal “druggable” targets for epigenetic therapy. Elucidation of the mechanism underlying BRD4 hyperphosphorylation will reveal novel targets, allowing the design of more effective anticancer therapeutic strategies to modulate BRD4 phosphorylation specifically, thereby turning off its oncogenic activity in the relevant cancers while sparing the normal BRD4 functions described in previous studies from our group and others (9, 24–26). Our studies also open opportunities for developing BET inhibitors and kinase inhibitors as combinatorial treatment approaches for BRD4-associated cancers.
Materials and Methods
In vitro kinase and phosphatase assays, Western blot analyses, immunoprecipitation, immunofluorescent staining, RT-qPCR, soft agar growth and sphere-formation assays, and SILAC studies were performed using standard methods. Phos-tag gel analysis was performed using 5% PAGE gel containing 10 μM Phos-tag acrylamide AAL-107 (Wako Chemicals) and 40 μM MnCl2 following the manufacturer’s instructions. A full description of reagents and methods used in this study can be found in SI Materials and Methods.
SI Materials and Methods
Recombinant Plasmid Constructs.
pOZN-BRD4 (encoding HA-FLAG-BRD4), pcDNA4c-BRD4-NUT (encoding Xpress-BRD4-NUT), and pGEX-6p-1-BRD4 1134–1362 (encoding GST-C229) have been described in our previous study (30, 65, 66). pET23a(+)-TII, encoding two IgG-binding domains of protein A fused to a TEV protease cleavage site, and hence named “TII tag,” was generated by cloning the PCR-amplified TII fragments from pTAPC2 (67) into the NotI and XhoI sites of the pET23a(+) vector. pET23a(+)-BRD4-TII, expressing BRD4-TII, was generated by further cloning the PCR-amplified BRD4-coding sequence from pcDNA4c-BRD4 (66) into the NdeI and NotI sites of pET23a(+)-TII. For the pMAL-c4X-CDK9 construct, the CDK9 ORF was PCR amplified and subcloned into the EcoRI and SalI sites of pMAL-c4X. For pGEX-6p-1-CDK9 construction, the CDK9 ORF was PCR amplified and subcloned into the EcoRI and NotI sites of pGEX-6p-1. Similarly, the cyclin T1 ORF was PCR amplified and subcloned into the EcoRI and NotI sites of pGEX-6p-1 to make the pGEX-6p-1-cyclin T1 construct. In order to generate a lentivirus construct for preparing C229 stable cells, firstly, the nuclear localization signal (NLS) coding sequence was inserted into the BamHI site of pcDNA4c-hBRD4 1134–1362 (66) using annealed products of two paired primers, BamHI-NLS F (5′-/5Phos/GAT CCC CTA AGA AGA AGA GGA AGG TTG-3′) and BamHI-NLS R (5′-/5Phos/GAT CCA ACC TTC CTC TTC TTC TTA GGG-3′), then the entire coding sequence for the Xpress-NLS-C229 was PCR amplified with pcDNA4C-specific primers and subcloned into pTRIPZ vector (Open Biosystems) using the AgeI and MluI sites. All plasmid constructs were verified by DNA sequencing.
Cell Culture, Transfection, and Stable Cell Line Construction.
HCC2429 (a gift from Thao P. Dang, Vanderbilt University, Nashville, TN), Ty-82 (Japanese Collection of Research Bioresources Cell Bank), 10-15, 14169, and A549 cells were maintained in RPMI 1640 medium (Invitrogen) with 10% FBS (HyClone) and 1% penicillin/streptomycin (P/S, Invitrogen). The 10-15 and 14169 cell lines were provided by Christopher A. French, Harvard Medical School, Boston. HEK293, C33A, and HeLa cells were maintained in DMEM (Invitrogen) with 10% FBS and 1% P/S. U2OS cells were maintained in McCoy’s 5A medium (Invitrogen) with 10% FBS and 1% P/S. K562, MV4;11, Jurkat, Namalwa, and THP-1 cells were grown in suspension culture using RPMI 1640 medium with 10% FBS and 1% P/S. HL-60 cells were maintained in RPMI 1640 medium with 20% FBS and 1% P/S. The adherent subpopulation of Raji cells was isolated from suspension culture (68) and maintained in adherent culture using RPMI 1640 medium with 10% FBS and 1% P/S for at least four passages. Jurkat, Namalwa, and Raji cells were provided by Erle S. Robertson, University of Pennsylvania, Philadelphia. MV4;11 and THP-1 cells were provided by Martin P. Carroll, University of Pennsylvania. HDFs were prepared as described in our previous study (69) and were maintained in DMEM with 10% FBS and 1% P/S.
HCC2429 and HEK293 cells were transiently transfected using Lipofectamine 2000 reagent (Invitrogen) following the manufacturer’s instructions. The nontarget control and NUT siRNAs were purchased from Dharmacon. For siRNA knockdown, HCC2429 cells were transfected using DharmaFECT reagents (GE Dharmacon).
HCC2429 control (CO) and C229 stable cells were prepared as described previously (30). To package the C229 inducible lentiviruses, HEK293T cells were cultured to 95% confluence. pTRIPZ-C229 was transfected into HEK293T cells together with psPAX2 and pMD.2G using Lipofectamine 2000 (Invitrogen). At 6 h posttransfection, the culture medium was changed to fresh medium. Twenty-four hours later, lentiviruses in the supernatant were harvested and filtered through a 0.45-μm syringe filter. HCC2429 cells were transduced with filtered viral supernatant supplemented with 6 μg/mL Polybrene (Sigma). Starting on day 2 after transduction, cells were selected using 0.6 µg/mL puromycin for 4 d. A control cell line stably transduced with lentiviruses carrying the pTRIPZ vector containing an inducible RFP gene was generated using the same method. Media for HEK293T and HCC2429 used in lentivirus packaging, transduction, selection, and stable cell culture process were supplemented with Tet System Approved FBS (Clontech).
Phos-Tag Gel Technique.
To prepare whole-cell lysates for the Phos-tag gel assay, cells were washed once with 1× TBS [50 mM Tris⋅HCl (pH 7.5), 150 mM NaCl] buffer and then were lysed in Phos-tag lysis buffer containing 20 mM Tris⋅HCl (pH 7.5), 100 mM NaCl, 1 mM DTT, 1 mM PMSF, 1 μg/mL aprotinin, 1 μg/mL leupeptin, 1 μg/mL pepstatin and supplemented with PhosSTOP phosphatase inhibitors (Roche). After sonication on ice at 10 W for 30–60 s, whole-cell lysates were centrifuged at 16,000 × g for 15 min at 4 °C to remove the pellets. The protein concentration was determined using the Bradford assay. The lysates then were boiled in SDS/PAGE sample buffer. The protein samples were resolved on 5% PAGE gel containing 10 μM Phos-tag acrylamide AAL-107 (Wako Chemicals) and 40 μM MnCl2 following the manufacturer’s instructions. The Phos-tag gels were washed with transfer buffer containing 4 mM EDTA for 20 min and then were washed with transfer buffer containing 0.1% SDS for 5 min before transfer onto PVDF membranes. The membranes then were immunoblotted with specific primary antibodies as indicated in the figures.
In Vitro Kinases Assay.
In vitro kinase assays using BRD4-TII and associate proteins purified on IgG beads were performed in 20 μL kinase buffer containing 50 mM Tris (pH 7.5), 1 mM DTT, 1 mM MnCl2, 5 mM MgCl2, 100 μM cold ATP, and 10 μCi [γ-32P]ATP (3,000 Ci/mM) supplemented with protease inhibitors and phosphatase inhibitors. The assay samples were incubated at 30 °C for 1 h with shaking. The beads were boiled in 2× SDS/PAGE sample buffer. The proteins were resolved on SDS/PAGE. The dried gels were exposed to a phosphor screen and detected using a Typhoon FLA 7000 imager (GE Healthcare).
SILAC.
HCC2429 cells were cultured in 10% dialyzed FBS (Pierce) in RPMI 1640 medium (Pierce) supplemented with either l-arginine (Sigma), l-lysine monohydrochloride (Sigma), and l-Proline (Sigma) in the light medium or with (13C15N)-labeled l-arginine monohydrochloride (Silantes), (13C15N)-labeled l-lysine monohydrochloride (Silantes), and l-proline (Sigma) in the heavy medium to reach final concentrations of 200 mg/L, 40 mg/L, and 100 mg/L for the three amino acids, respectively. After three passages, mass spectrometry was performed to confirm that HCC2429 cells cultured in the heavy medium were efficiently labeled with the heavy isotopes. The light-labeled and heavy-labeled cells were lysed, and the nuclear extracts were prepared as described previously (70). Nuclear extracts from light-labeled cells were immunoprecipitated with beads containing the cross-linked TII tag protein, and nuclear extracts from heavy-labeled cells were immunoprecipitated with beads containing the cross-linked BRD4-TII. After rotating in the cold room for 4 h, the beads were washed three times with KCl wash buffer [20 mM Tris⋅HCl (pH 7.5), 100 mM KCl, 10% glycerol, 0.1% Tween-20 supplemented with protease inhibitors and phosphatase inhibitors] and three times with KCl wash buffer without Tween-20. In the last wash, the light and heavy samples were combined at a 1:1 ratio. The binding proteins were eluted with 2% SDS in 0.1 M Tris⋅HCl (pH 7.5) with heating at 95 °C for 10 min.
Protein Digestion, LC-MS/MS Analysis, and Data Analysis.
An approximately fivefold volume of prechilled acetone was added to the protein solution and incubated overnight at −20 °C. The protein pellet was reconstituted in 20 μL urea lysis buffer [8 M urea, 50 mM Tris⋅HCl (pH 8.3), 50 mM NaCl] and was reduced using 10 mM DTT (Invitrogen) and alkylated using 50 mM iodoacetamide (Bio-Rad). Urea was diluted to 1.5 M using 50 mM Tris⋅HCl (pH 8.3), and trypsin (Promega) was added at a ratio of 1:20 (wt/wt) for digestion overnight at room temperature. Peptides were desalted using Sep-Pak C18 cartridges (Waters) as described previously (71) and then were dried by Vacufuge Plus Model 5305 Speed Vac (Eppendorf). The dried peptides were reconstituted in 0.1% formic acid and were loaded onto a 75-μm i.d. × 20-cm fused silica capillary column packed with Reprosil-Pur C18-AQ resin (3 μm; Dr. Maisch GmbH) and resolved using an EASY-nLC 1000 HPLC system (Thermo Scientific) coupled in-line with a Q-Exactive mass spectrometer (Thermo Scientific). The HPLC gradient consisted of 5–30% solvent B (solvent A = 0.1% formic acid in water; solvent B = 0.1% formic acid in acetonitrile) for 160 min, followed by 30–95% solvent B for 10 min, and then held at 95% solvent B for 10 min, with a constant flow rate of 300 nL/min applied throughout. Full MS scans (m/z 350–1600) were performed at a resolution of 70,000 (at 200 m/z), and the 12 most intense ions were selected for MS/MS performed with high-energy collision dissociation with normalized collision energy of 24 at a resolution of 17,500 (at 200 m/z). Automatic gain control targets of full MS and MS/MS scans were 1 × 106 and 1 × 105, respectively. Unassigned charge states and singly charged species were rejected, dynamic exclusion was set at 20 s, and lock mass calibration was implemented using polysiloxane ions 371.10123 and 445.12000.
MaxQuant (version 1.5.3.30) was used to search the UniProtKB/Swiss-Prot/TrEMBL Human Reference Proteome database [9606] (retrieved on 21 December, 2015; containing 88,951 entries including isoforms) for peptide and protein identification and quantification (72, 73). Cysteine carbamidomethylation was set as a fixed modification; methionine oxidation and protein N-terminal acetylation were set as variable modifications. Trypsin was specified as the digestive enzyme. Minimal peptide length was set to seven amino acids, and a maximum of two missed cleavages was allowed. The false-discovery rate was set to 0.01 for peptide-spectrum matches and protein identifications. Protein grouping was enabled. Peptide identification was performed with an allowed precursor mass deviation up to 4.5 ppm after time-dependent mass calibration and an allowed mass deviation of 20 ppm for the contaminant database search (preceding the main search). Fragment ion tolerance was set to 50 ppm. Protein identification required at least one unique or razor peptide per protein group. Protein tables were filtered to eliminate the identifications from the reverse database identified only by site and common contaminants.
After the H/L protein ratios were retrieved, data were log2-transformed and normalized by subtracting the median of the distribution.
Chemical Inhibitors.
LDC000067 (Selleck Chemicals), Flavopiridol (Santa Cruz), PHA767491 (Santa Cruz), (−)-JQ1 (BPS Bioscience), and (+)-JQ1 (BPS Bioscience) powders were dissolved in DMSO to a stock concentration of 10 mM. DRB (Santa Cruz), apigenin (Santa Cruz), and TBB (Santa Cruz) powders were dissolved in DMSO to prepare 100-mM stock solutions. C646 (Sigma) powder was dissolved in DMSO to reach a stock concentration of 25 mM. HMBA (Sigma) powder was dissolved in distilled water to prepare a 0.5-M stock solution. After filtered with a 0.2-μm syringe filter, the HMBA stock solution was stored as small aliquots at −20 °C. All other stock solutions were stored as small aliquots at −80 °C.
Western Blot Analyses.
Western blot analyses were performed as described previously (30). The primary antibodies used in the Western blot analyses include anti-BRD4 N (targeting BRD4 156–284; 1:40,000); anti-BRD4 C (targeting BRD4 1313–1362; 1:40,000); anti-NUT (targeting NUT 300–700; 1:20,000); anti-CDK9 (1:500; sc13130; Santa Cruz); anti-cyclin T1 (1:500; sc271575; Santa Cruz); anti-SOX2 (1:2,000; MAB2018; R&D); anti–c-MYC (1:2,000; 9402S; Cell Signaling Technology); anti-cytokeratin (1:4,000; M3515; Dako); anti-involucrin (1:2,000; I9018; Sigma); anti-GAPDH (1:4,000; G8140-01; US Biological); anti-Xpress (1:5,000; R910-25l; Invitrogen); and anti-HEXIM1 (1:2,000; 9064S; Cell Signaling Technology). HRP-linked anti-rabbit IgG (1:3,000; 7074S; Cell Signaling Technology) and HRP-linked anti-mouse IgG (1:3,000; 7076S; Cell Signaling Technology) were used as secondary antibodies. Western blots were developed using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific), and images were captured using an Amersham Imager 600 (GE Healthcare).
Immunoprecipitation.
For preparing nuclear extracts, cells were first gently resuspended in buffer A [20 mM Hepes (pH 7.5), 10 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.4% Nonidet P-40 supplemented with protease inhibitors and phosphatase inhibitors]. After 10-min incubation on ice, nuclear pellets were centrifuged at 1,000 × g for 5 min at 4 °C. The nuclear pellets were resuspended in buffer B [20 mM Hepes (pH 7.5), 450 mM NaCl supplemented with protease inhibitors and phosphatase inhibitors] and subjected to sonication on ice at 10 W for 60 s. After rotating in the cold room for 20 min, the nuclear extracts were centrifuged at 20,000 × g for 5 min at 4 °C. The nuclear extracts were diluted with two volumes of buffer B without NaCl to reach the final salt concentration of ∼150 mM. The samples were centrifuged twice at 20,000 × g for 15 min at 4 °C. The supernatants were collected, and the protein concentrations were determined using Bradford assay. A total of 600 μg of nuclear extracts were immunoprecipitated using protein A beads (preblocked with 1% BSA in PBS at 4 °C for 1 h) mixed with 2 μg of either normal rabbit IgG or the antibodies indicated in the figures for 4 h at 4 °C. The beads were washed three times with KCl wash buffer [20 mM Tris⋅HCl (pH 7.5), 150 mM KCl, 10% glycerol, 0.1% Tween-20 supplemented with protease inhibitors and phosphatase inhibitors] and eluted by boiling in 2× SDS/PAGE sample buffer. Antibodies used in the coimmunoprecipitation experiments include anti-BRD4 N, anti-CDK9 (sc8338; Santa Cruz), and anti-cyclin T1 (sc10750; Santa Cruz).
In Vitro Phosphatase Assay.
The nuclear extracts (1.2 mg) isolated from HCC2429 and HEK293 cells and HDFs as described in the Immunoprecipitation section above were immunoprecipitated with preblocked protein A beads and 2 μg BRD4C antibody for 2 h. The beads were washed twice with KCl wash buffer, once quickly with 1 M NaCl in 20 mM Tris⋅HCl (pH 7.5), and then twice with KCl wash buffer. After washing, the beads were divided into three aliquots and mixed with buffer or with buffer supplemented with λPP (New England BioLabs) or PIs. Samples mixed with λPP and PIs were incubated at 30 °C for 30 min with shaking every 5 min. After the assay, the beads were boiled in 2× SDS/PAGE sample buffer.
Immunofluorescent Staining.
Cells cultured on coverslips were fixed with 3% paraformaldehyde in PBS for 20 min. Immunofluorescent staining was performed as previously described (31). Anti-BRD4C and anti-p300 (1:300; 05-257; Millipore) were used as primary antibodies. Alexa Fluor 594 goat anti-rabbit IgG (1:1,000; A11012; Invitrogen) and Alexa Fluor 488 goat anti-mouse IgG (1:500; A11001; Invitrogen) were used as secondary antibodies. All immunofluorescent images were captured using an inverted fluorescence microscope (IX81; Olympus) as described previously (65). The scale bars were added using ImageJ software.
RT-qPCR.
Total RNA was isolated using the NucleoSpin RNA II Kit (Macherey-Nagel) following the manufacturer’s instructions. Reverse transcription was performed using a 20-μL reaction mixture containing 350 ng of total RNA, oligo(dT) primer (Invitrogen), dNTP (Invitrogen), and M-MLV reverse transcriptase (Invitrogen). Real-time qPCR was performed using a CFX96 real-time PCR detection system (Bio-Rad) with IQ SYBR Green supermix (Bio-Rad). The mRNA level of each gene was normalized to the GAPDH mRNA level. Primer sequences were as follows: HBRD4IF3.5, 5′-CCAGAGCAGAACGCACCGC-3′; HBRD4IR3.8, 5′-CTCCAGCGCATCCTCGTCC-3′; CDK9 RT1 F, 5′-GTGCTGATGGAAAACGAGAA-3′; CDK 9RT1 R, 5′-ATAGGGGGAAGCTTTGGTTC-3′; CCNT1 RT1 F, 5′-TCCTTCACACAGTTCCCTGG-3′; CCNT1 RT1 R, 5′-GCTACCTTGATGACATGTTCCA-3′; CCNT2 RT1 F, 5′-GCGTCTCAATGTCTCTCAGC-3′; CCNT2 RT1 R, 5′-TGCAGCCAAAAATAATGCAG-3′; HEXIM1 RT1 F, 5′-GGGCAAAGGGGACTTTTTAC-3′; and HEXIM1 RT1 R, 5′-TTACGAAACCAACCAAAGCC-3′ (where F is forward, and R is reverse).
Soft Agar Assay.
The soft agar growth assay was performed as described previously (30). The bottom layer consisted of 0.5% agar in the medium solution containing RPMI 1640 medium with 10% FBS, 1% P/S, and JQ1, HMBA, or Dox at the working concentrations indicated in the figure legends. The middle layer was composed of 0.33% agar in the same medium solution mixed with the cells and the chemicals at the working concentrations shown in the figure legends. After overnight culture, 0.5% agar/medium solution containing each drug at twice the working concentration was poured as the top layer of the agar plate. In the soft agar experiment using Dox-induced BRD4 C229 or the control HCC2429 stable cells, fresh medium with Dox was added to the top of the agar to replace the old medium every 3 d. Two weeks later, the colonies were stained with 0.005% crystal violet in PBS. The number of colonies was quantified using ImageJ software.
Sphere-Formation Assay.
The sphere-formation assay was performed as described previously (30).
Statistical Analyses.
Statistical analysis was performed using the unpaired t test of GraphPad Prism software (version 5.0) to compare the data from the control and experimental groups. A two-tailed P value of <0.05 was considered statistically significant. ***P < 0.001, **P < 0.01.
Supplementary Material
Acknowledgments
We thank Dr. Junpeng Yan (Case Western Reserve University) for providing plasmids; Drs. Martin P. Carroll (University of Pennsylvania), Erle S. Robertson (University of Pennsylvania), Thao P. Dang (Vanderbilt University), and Christopher A. French (Harvard Medical School) for providing cell lines; Dr. Chao-Xing Yuan and Anne Lehman for technical support; and the members of our laboratories for helpful discussion. This work was supported by NIH Grants R01CA148768, R01CA142723, R01CA187718, R01GM110174, P01CA196539, and R01CA103867; Cancer Prevention Research Institute of Texas Grants RP110471 and RP140367; and Welch Foundation Grant I-1805.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703071114/-/DCSupplemental.
References
- 1.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci USA. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 3.Rahman S, et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol. 2011;31:2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lovén J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 6.Czudnochowski N, Bösken CA, Geyer M. Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat Commun. 2012;3:842. doi: 10.1038/ncomms1846. [DOI] [PubMed] [Google Scholar]
- 7.Yang Z, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 8.Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol. 2008;28:967–976. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Houzelstein D, et al. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol Cell Biol. 2002;22:3794–3802. doi: 10.1128/MCB.22.11.3794-3802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mertz JA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci USA. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puissant A, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013;3:308–323. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.French CA. Pathogenesis of NUT midline carcinoma. Annu Rev Pathol. 2012;7:247–265. doi: 10.1146/annurev-pathol-011811-132438. [DOI] [PubMed] [Google Scholar]
- 16.Chapuy B, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dawson MA, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grayson AR, et al. MYC, a downstream target of BRD-NUT, is necessary and sufficient for the blockade of differentiation in NUT midline carcinoma. Oncogene. 2014;33:1736–1742. doi: 10.1038/onc.2013.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicodeme E, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berthon C, et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: A dose-escalation, phase 1 study. Lancet Haematol. 2016;3:e186–e195. doi: 10.1016/S2352-3026(15)00247-1. [DOI] [PubMed] [Google Scholar]
- 22.Fong CY, et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature. 2015;525:538–542. doi: 10.1038/nature14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shu S, et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature. 2016;529:413–417. doi: 10.1038/nature16508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu W, et al. BRD4 regulates Nanog expression in mouse embryonic stem cells and preimplantation embryos. Cell Death Differ. 2014;21:1950–1960. doi: 10.1038/cdd.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu S-Y, et al. BRD4 phosphorylation regulates HPV E2-mediated viral transcription, origin replication, and cellular MMP-9 expression. Cell Reports. 2016;16:1733–1748. doi: 10.1016/j.celrep.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolden JE, et al. Inducible in vivo silencing of Brd4 identifies potential toxicities of sustained BET protein inhibition. Cell Reports. 2014;8:1919–1929. doi: 10.1016/j.celrep.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andrieu G, Belkina AC, Denis GV. Clinical trials for BET inhibitors run ahead of the science. Drug Discov Today Technol. 2016;19:45–50. doi: 10.1016/j.ddtec.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang R, You J. Mechanistic analysis of the role of bromodomain-containing protein 4 (BRD4) in BRD4-NUT oncoprotein-induced transcriptional activation. J Biol Chem. 2015;290:2744–2758. doi: 10.1074/jbc.M114.600759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haruki N, et al. Cloned fusion product from a rare t(15;19)(q13.2;p13.1) inhibit S phase in vitro. J Med Genet. 2005;42:558–564. doi: 10.1136/jmg.2004.029686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang R, et al. Activation of SOX2 expression by BRD4-NUT oncogenic fusion drives neoplastic transformation in NUT midline carcinoma. Cancer Res. 2014;74:3332–3343. doi: 10.1158/0008-5472.CAN-13-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan J, Diaz J, Jiao J, Wang R, You J. Perturbation of BRD4 protein function by BRD4-NUT protein abrogates cellular differentiation in NUT midline carcinoma. J Biol Chem. 2011;286:27663–27675. doi: 10.1074/jbc.M111.246975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reynoird N, et al. Oncogenesis by sequestration of CBP/p300 in transcriptionally inactive hyperacetylated chromatin domains. EMBO J. 2010;29:2943–2952. doi: 10.1038/emboj.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu SY, Lee AY, Lai HT, Zhang H, Chiang CM. Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol Cell. 2013;49:843–857. doi: 10.1016/j.molcel.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinoshita E, Kinoshita-Kikuta E, Koike T. Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nat Protoc. 2009;4:1513–1521. doi: 10.1038/nprot.2009.154. [DOI] [PubMed] [Google Scholar]
- 35.Fernandes N, et al. DNA damage-induced association of ATM with its target proteins requires a protein interaction domain in the N terminus of ATM. J Biol Chem. 2005;280:15158–15164. doi: 10.1074/jbc.M412065200. [DOI] [PubMed] [Google Scholar]
- 36.Chen X, Wei S, Ji Y, Guo X, Yang F. Quantitative proteomics using SILAC: Principles, applications, and developments. Proteomics. 2015;15:3175–3192. doi: 10.1002/pmic.201500108. [DOI] [PubMed] [Google Scholar]
- 37.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 38.Wang S, Fischer PM. Cyclin-dependent kinase 9: A key transcriptional regulator and potential drug target in oncology, virology and cardiology. Trends Pharmacol Sci. 2008;29:302–313. doi: 10.1016/j.tips.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 39.Polier G, et al. Wogonin and related natural flavones are inhibitors of CDK9 that induce apoptosis in cancer cells by transcriptional suppression of Mcl-1. Cell Death Dis. 2011;2:e182. doi: 10.1038/cddis.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Devaiah BN, Singer DS. Cross-talk among RNA polymerase II kinases modulates C-terminal domain phosphorylation. J Biol Chem. 2012;287:38755–38766. doi: 10.1074/jbc.M112.412015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yik JH, et al. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 42.Kurimchak AM, et al. Resistance to BET bromodomain inhibitors is mediated by kinome reprogramming in ovarian cancer. Cell Reports. 2016;16:1273–1286. doi: 10.1016/j.celrep.2016.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nilsson LM, et al. Cancer differentiating agent hexamethylene bisacetamide inhibits BET bromodomain proteins. Cancer Res. 2016;76:2376–2383. doi: 10.1158/0008-5472.CAN-15-2721. [DOI] [PubMed] [Google Scholar]
- 44.Yan J, Li Q, Lievens S, Tavernier J, You J. Abrogation of the Brd4-positive transcription elongation factor B complex by papillomavirus E2 protein contributes to viral oncogene repression. J Virol. 2010;84:76–87. doi: 10.1128/JVI.01647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valent P, Zuber J. BRD4: A BET(ter) target for the treatment of AML? Cell Cycle. 2014;13:689–690. doi: 10.4161/cc.27859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knoechel B, et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet. 2014;46:364–370. doi: 10.1038/ng.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beesley AH, et al. Comparative drug screening in NUT midline carcinoma. Br J Cancer. 2014;110:1189–1198. doi: 10.1038/bjc.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia-Cuellar MP, et al. Efficacy of cyclin-dependent-kinase 9 inhibitors in a murine model of mixed-lineage leukemia. Leukemia. 2014;28:1427–1435. doi: 10.1038/leu.2014.40. [DOI] [PubMed] [Google Scholar]
- 49.Arguello F, et al. Flavopiridol induces apoptosis of normal lymphoid cells, causes immunosuppression, and has potent antitumor activity In vivo against human leukemia and lymphoma xenografts. Blood. 1998;91:2482–2490. [PubMed] [Google Scholar]
- 50.Byrd JC, et al. Cancer and Leukemia Group B Treatment of relapsed chronic lymphocytic leukemia by 72-hour continuous infusion or 1-hour bolus infusion of flavopiridol: Results from Cancer and Leukemia Group B study 19805. Clin Cancer Res. 2005;11:4176–4181. doi: 10.1158/1078-0432.CCR-04-2276. [DOI] [PubMed] [Google Scholar]
- 51.Lin TS, et al. Phase II study of flavopiridol in relapsed chronic lymphocytic leukemia demonstrating high response rates in genetically high-risk disease. J Clin Oncol. 2009;27:6012–6018. doi: 10.1200/JCO.2009.22.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Montagnoli A, et al. A Cdc7 kinase inhibitor restricts initiation of DNA replication and has antitumor activity. Nat Chem Biol. 2008;4:357–365. doi: 10.1038/nchembio.90. [DOI] [PubMed] [Google Scholar]
- 53.Gao XN, et al. A histone acetyltransferase p300 inhibitor C646 induces cell cycle arrest and apoptosis selectively in AML1-ETO-positive AML cells. PLoS One. 2013;8:e55481. doi: 10.1371/journal.pone.0055481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ogiwara H, et al. Targeting p300 addiction in CBP-deficient cancers causes synthetic lethality by apoptotic cell death due to abrogation of MYC expression. Cancer Discov. 2016;6:430–445. doi: 10.1158/2159-8290.CD-15-0754. [DOI] [PubMed] [Google Scholar]
- 55.Shi J, et al. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell. 2014;25:210–225. doi: 10.1016/j.ccr.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Asangani IA, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stathis A, et al. Clinical response of carcinomas harboring the BRD4–NUT oncoprotein to the targeted bromodomain inhibitor OTX015/MK-8628. Cancer Discov. 2016;6:492–500. doi: 10.1158/2159-8290.CD-15-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schroy P, Winawer S, Friedman E. Effect on in vivo tumorigenicity of lengthy exposure of human colon cancer cells to the differentiation agent hexamethylene bisacetamide. Cancer Lett. 1989;48:53–58. doi: 10.1016/0304-3835(89)90202-4. [DOI] [PubMed] [Google Scholar]
- 59.Naito S, et al. Enhancement of antitumor activity of herpes simplex virus gamma(1)34.5-deficient mutant for oral squamous cell carcinoma cells by hexamethylene bisacetamide. Cancer Gene Ther. 2006;13:780–791. doi: 10.1038/sj.cgt.7700957. [DOI] [PubMed] [Google Scholar]
- 60.Andreeff M, et al. Hexamethylene bisacetamide in myelodysplastic syndrome and acute myelogenous leukemia: A phase II clinical trial with a differentiation-inducing agent. Blood. 1992;80:2604–2609. [PubMed] [Google Scholar]
- 61.Fu J, Yoon H-G, Qin J, Wong J. Regulation of P-TEFb elongation complex activity by CDK9 acetylation. Mol Cell Biol. 2007;27:4641–4651. doi: 10.1128/MCB.00857-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sangodkar J, et al. All roads lead to PP2A: Exploiting the therapeutic potential of this phosphatase. FEBS J. 2016;283:1004–1024. doi: 10.1111/febs.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wong YH, et al. KinasePhos 2.0: A web server for identifying protein kinase-specific phosphorylation sites based on sequences and coupling patterns. Nucleic Acids Res. 2007;35:W588–594. doi: 10.1093/nar/gkm322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O’Brien JA, Rizzieri DA. Leukemic stem cells: A review. Cancer Invest. 2013;31:215–220. doi: 10.3109/07357907.2012.700986. [DOI] [PubMed] [Google Scholar]
- 65.Wang R, Li Q, Helfer CM, Jiao J, You J. Bromodomain protein Brd4 associated with acetylated chromatin is important for maintenance of higher-order chromatin structure. J Biol Chem. 2012;287:10738–10752. doi: 10.1074/jbc.M111.323493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.You J, Croyle JL, Nishimura A, Ozato K, Howley PM. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell. 2004;117:349–360. doi: 10.1016/s0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- 67.Li Q, et al. A modified mammalian tandem affinity purification procedure to prepare functional polycystin-2 channel. FEBS Lett. 2004;576:231–236. doi: 10.1016/j.febslet.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 68.Nyormoi O, Sinclair JH, Klein G. Isolation and characterization of an adherent, 8-azaguanine resistant variant of the Burkitt lymphoma cell line, Raji. Exp Cell Res. 1973;82:241–251. doi: 10.1016/0014-4827(73)90337-6. [DOI] [PubMed] [Google Scholar]
- 69.Liu W, et al. Identifying the target cells and mechanisms of Merkel cell polyomavirus infection. Cell Host Microbe. 2016;19:775–787. doi: 10.1016/j.chom.2016.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Villén J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc. 2008;3:1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 73.Cox J, et al. Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.