Significance
CREB-binding protein (CBP) and its paralog p300 play a vital role in regulating gene transcription. Through the enzymatic activity of their histone acetyltransferase (HAT) domain, CBP and p300 control the accessibility of genes in chromatin and activate transcription. They also function as transcriptional repressors following SUMOylation of the cell cycle regulatory domain 1 (CRD1) located N-terminal to the catalytic core. We present structural and biochemical results showing that the CBP bromodomain, CH2, and ZZ domains, which flank the acetyltransferase domain, regulate acetyltransferase activity and also promote SUMOylation of the adjacent CRD1 cell cycle regulatory domain. This study provides insights into the function of the catalytic core and the role of adjacent domains and a disordered regulatory loop in mediating CBP/p300 activity.
Keywords: acetyltransferase, transcriptional coactivator, X-ray structure, bromodomain, Ubc9
Abstract
The histone acetyl transferases CREB-binding protein (CBP) and its paralog p300 play a critical role in numerous cellular processes. Dysregulation of their catalytic activity is associated with several human diseases. Previous work has elucidated the regulatory mechanisms of p300 acetyltransferase activity, but it is not known whether CBP activity is controlled similarly. Here, we present the crystal structure of the CBP catalytic core encompassing the bromodomain (BRD), CH2 (comprising PHD and RING), HAT, and ZZ domains at 2.4-Å resolution. The BRD, PHD, and HAT domains form an integral structural unit to which the RING and ZZ domains are flexibly attached. The structure of the apo-CBP HAT domain is similar to that of acyl-CoA–bound p300 HAT complexes and shows that the acetyl-CoA binding site is stably formed in the absence of cofactor. The BRD, PHD, and ZZ domains interact with small ubiquitin-like modifier 1 (SUMO-1) and Ubc9, and function as an intramolecular E3 ligase for SUMOylation of the cell cycle regulatory domain 1 (CRD1) of CBP, which is located adjacent to the BRD. In vitro HAT assays suggest that the RING domain, the autoregulatory loop (AL) within the HAT domain, and the ZZ domain do not directly influence catalytic activity, whereas the BRD is essential for histone H3 acetylation in nucleosomal substrates. Several lysine residues in the intrinsically disordered AL are autoacetylated by the HAT domain. Upon autoacetylation, acetyl-K1596 (Ac-K1596) binds intramolecularly to the BRD, competing with histones for binding to the BRD and acting as a negative regulator that inhibits histone H3 acetylation.
CREB-binding protein (CBP) and the closely related E1A-binding protein p300 are transcriptional coactivators that regulate expression of genes involved in a plethora of cellular processes (1–3). CBP and p300, which constitute the unique KAT3 acetyltransferase family, perform their regulatory functions by catalyzing the transfer of an acetyl group to lysine residues in histones and transcription factors (4–8). Mutations that dysregulate their catalytic activity are associated with a number of human diseases including Rubinstein–Taybi syndrome, leukemia, and lymphoma (9–11).
CBP and p300 contain both intrinsically disordered regions and folded functional domains. The histone acetyltransferase (HAT) domain is flanked by other domains, including the bromodomain (BRD), cysteine/histidine-rich region 2 (CH2), and cysteine/histidine-rich region 3 (CH3), which have a potential role in regulation of the acetyltransferase activity and substrate specificity. The domain architecture of the BRD, CH2, HAT, and CH3 regions is highly conserved among higher organisms. The BRD of CBP/p300 binds promiscuously to acetylated lysines in histones H2A, H2B, H3, and H4 (12–14), and contributes to the function of the HAT domain in acetylating nucleosomes in native chromatin (15).
The CH2 region of CBP and p300 has been shown to influence HAT activity in in vitro assays and to reduce transcriptional activity on transfected reporter genes, although conflicting results have been reported (13, 16–19). An X-ray structure of the p300 catalytic core showed that the CH2 region adopts a unique fold consisting of a plant homeodomain (PHD) motif and an interleaved RING domain (13). Deletion or mutation of the p300 RING, or an E1242K mutation that disrupts the interaction between the CH2 domain and the HAT, enhanced the autoacetylation of K1499 of p300 and acetylation of K382 of p53 (13, 18). In contrast, deletion of the p300 RING impaired acetylation of K9 or K14 of histone H3 in cells (18), suggesting that the regulatory role of the RING domain may be substrate specific.
The HAT domains of p300 and CBP contain a lysine-rich autoregulatory loop (AL) that, when hypoacetylated, inhibits acetyltransferase activity by competing with positively charged substrates for binding to the active site (20). Hyperacetylation of the AL of p300 greatly enhances HAT activity, suggesting that autoacetylation may play a regulatory role (20). Despite 88% sequence identity of the HAT and flanking domains, CBP and p300 differ in their specificity and selectivity for substrates (21, 22). The molecular basis for the difference in activity of CBP and p300 is largely unknown, and additional studies are required to further our understanding of the regulation of the HAT activity of the KAT3 family proteins.
Although a body of data implicates the BRD and CH2 domains, located N-terminal to the HAT domain, in regulation of acetyltransferase activity, much less is known about the role of the C-terminal CH3 region in regulation of the CBP/p300 catalytic core (23). The CH3 region consists of the ZZ-type zinc finger (ZZ) and transcriptional adaptor zinc finger-2 (TAZ2) domains (24). The TAZ2 domain recruits many proteins to the coactivator complex via protein–protein interactions (2). The ZZ domain has been suggested to interact with small ubiquitin-like modifier 1 (SUMO-1) (25), but the functional role of this interaction is unknown. It is of note, however, that SUMOylation of the cell cycle regulatory domain 1 (CRD1) of CBP and p300, located N-terminal to the BRD, mediates transcriptional repression (26, 27). Investigation of SUMO-related activities of the CBP/p300 catalytic core is thus of considerable interest.
Earlier studies of the structure and regulation of the HAT domain were largely focused on p300, and much less information is available for CBP. To gain insights into the contributions made by proximal domains to HAT activity, we determined the crystal structure at 2.4-Å resolution of the CBP HAT core, including the flanking BRD, CH2, and ZZ domains, and investigated its acetyltransferase activity using in vitro assays. Unexpectedly, we discovered that the BRD, PHD, and ZZ domains interact with SUMO-1 and the SUMO E2 ligase Ubc9 and function synergistically to facilitate intramolecular SUMOylation of the CRD1 repression domain. We also identified an intramolecular interaction between the BRD and AL, in which the autoacetylated K1596 binds to the BRD in competition with nucleosomes and inhibits acetylation of histone H3. Overall, our studies provide insights into the role of flanking domains and the AL in modulating the acetyltransferase activity of the central core of CBP/p300 and reveal a previously unrecognized intramolecular SUMOylation activity that may play an important role in repression of CBP/p300-mediated transcriptional activity.
Results
Overall Structure of the CBP Core.
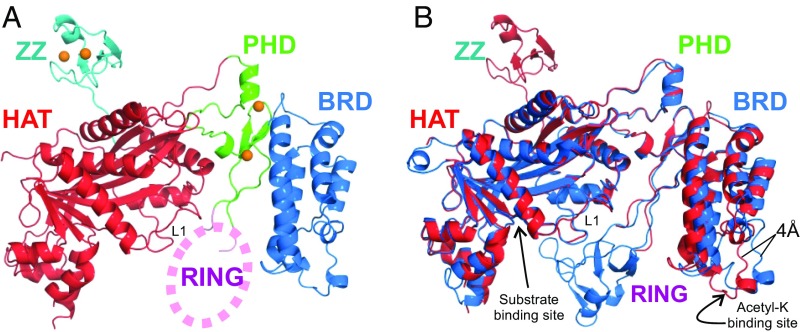
Previous X-ray structures of the p300 HAT domain (13, 28, 29) were determined from crystals of semisynthetic proteins or catalytically inactive mutants. In this study, we expressed the active wild-type mouse CBP catalytic core in Escherichia coli. Constructs containing the HAT domain were heavily acetylated in E. coli and were deacetylated using SIRT2 before crystallization. The AL, residues 1,557–1,619 of the HAT domain, was deleted because the corresponding region of p300 was reported to be flexible and inhibit crystallization (13, 28, 29). The crystal structure of the CBP core encompassing the BRD, CH2, HAT, and ZZ domains (abbreviated as BCHZ) was determined by molecular replacement, using the p300 structure (PDB ID code 4BHW) (13) as a search model, and refined to 2.4-Å resolution (Fig. 1, Fig. S1, and Table S1).
Fig. 1.
Crystal structure of the CBP catalytic core. (A) Cartoon representation of the structure of the BCHZ∆AL construct. The BRD, PHD, HAT, and ZZ domains are shown in blue, green, red, and cyan, respectively. Zinc atoms are shown as orange spheres. Electron density for the RING domain was largely absent, and its approximate location is indicated schematically by the pink oval. (B) Superposition of the CBP (red) and p300 (PDB ID code 4BHW, blue) structures showing that the acetyl-lysine binding site of the BRD is ∼4 Å closer to the HAT domain in CBP compared with p300. The acetyl-lysine binding site of the BRD and the substrate binding site of the HAT domain are indicated by arrows.
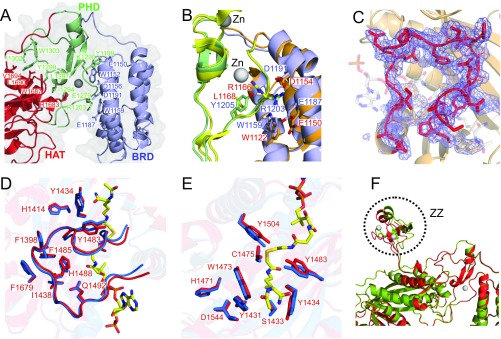
Fig. S1.
Details of the crystal structure of CBP BCHZ and comparison with p300. (A) Interfaces between BRD (blue) and PHD (green) domains and between the PHD and HAT (red) domains of CBP. The residues that participate in hydrophobic interactions and electrostatic interactions are labeled, and their side chains are shown as sticks. (B) Differences in side-chain packing in the interface between the BRD and PHD domains of CBP and p300 (PDB id: 4BHW). The CBP and p300 bromodomains are shown in pale blue and orange, respectively, whereas the PHD domains are pale green (CBP) and yellow (p300). The HAT domains are omitted for clarity. CBP side chains are labeled in blue, and p300 side chains in red. An electrostatic interaction occurs between R1203 and D1191/E1187 of CBP, whereas no corresponding interaction occurs between R1166 and E1150 in p300 because the side chain of R1166 is facing in the opposite direction. (C) Cartoon representation of the CBP HAT domain overlaid on an Fo–Fc map contoured at 1.5σ at the acetyl-CoA binding site. The backbone and side chains of the L1 loop are shown as red sticks. Note that no significant electron density is observed in the cofactor binding site, showing that the structure is that of the apo enzyme. As a point of reference, the expected position of bound cofactor, modeled from the Lys-CoA structure bound to the p300 catalytic core (4BHW), is indicated as transparent sticks. (D) Acetyl-CoA binding site, showing the L1 loops of apo-CBP (red) and Lys-CoA–bound p300 (3BIY; blue). Labeled residues participate in the hydrophobic interaction between the L1 loop and the core of the HAT domain. (E) Active site of apo-CBP (red) and Lys-CoA–bound p300 (3BIY; blue). Lys-CoA is shown as yellow sticks in D and E. (F) Ribbon representation showing the superposition of the two CBP molecules in the crystallographic asymmetric unit. The locations of the ZZ domain are indicated. Electron density is weak or missing for most of the RING domain residues of both CBP molecules.
Table S1.
Data collection and refinement statistics
| Data collection | BCHZΔAL |
| Space group | C2 |
| Cell dimensions | |
| a, b, c, Å | 214.2, 87.2, 115.0 |
| α, β, γ, ° | 90.0, 90.0, 109.7 |
| Resolution, Å | 50–2.4 |
| Rsym or Rmerge | 11.2 (100) |
| Rmeas | 12.0 (100) |
| Rpim | 6.0 (55.5) |
| I/σI | 12.7 (1.3) |
| CC1/2 | 83.7 (36.6) |
| Completeness, % | 99.8 (99.8) |
| Redundancy | 5.0 (4.9) |
| Refinement | |
| Resolution, Å | 50–2.4 |
| No. of reflections | 77,429 |
| Rwork/Rfree | 0.25/0.28 |
| No. of atoms | |
| Proteins | 8,742 |
| Water | 155 |
| Zinc | 8 |
| B factors | |
| Proteins | 57.2 |
| Water | 33.0 |
| Zinc | 53.9 |
| rms deviations | |
| Bond length, Å | 0.013 |
| Bond angles, ° | 1.067 |
The CBP BCHZ construct contains a modular core, similar to that of p300 (13), in which the BRD, PHD, and HAT domains are intimately packed through electrostatic and hydrophobic interactions (Fig. 1A and Fig. S1 A and B). The structures of the BRD, PHD, and HAT domains of CBP and p300 are very similar with a backbone rmsd of 0.5 Å (Fig. 1B). However, in contrast to the available p300 HAT domain structures, which all contain acyl-CoA variants bound in the active site (13, 28–30), the CBP HAT domain is in the ligand-free apo state (Fig. S1C). The L1 loop, which forms the Ac-CoA binding site, adopts an almost identical structure in the apo CBP and p300 HATs, showing that the cofactor binding site is preformed in the absence of Ac-CoA (Fig. S1D). Most of the active site residues, including the catalytic tyrosine (Y1504 in CBP, Y1467 in p300), adopt almost identical conformations in the apo CBP HAT and in the structure of the p300 HAT bound to the bisubstrate inhibitor Lys-CoA (Fig. S1E).
Electron density was observed for the zinc-binding CH2 domain in the p300 core, revealing a unique motif comprising a discontinuous PHD domain with a RING domain embedded in one of the loops (13). The p300 RING packs intimately against the HAT domain of a neighboring molecule in the lattice. In contrast, electron density for the CBP RING is weak and disordered and projects into a wide channel in the crystal lattice (Fig. 1A). Our CBP core construct contains the ZZ domain, which was omitted from the p300 core construct used for crystallization (13). The structure of the ZZ domain is very similar to that of the isolated domain determined previously by NMR (31). Except for the linker, the ZZ domain makes minimal contact with the HAT domain to which it is attached. However, its orientation is constrained by contacts to HAT and ZZ domains in neighboring molecules in the crystal lattice. The two CBP molecules per asymmetric unit of the crystal superimpose well, except for the ZZ domain, suggesting that ZZ is flexibly attached to the catalytic core (Fig. S1F).
A notable difference between the structures of the BRD–PHD–HAT modules of p300 and CBP is that the BRD is rotated in the CBP structure and brings the acetyl-lysine binding site ∼4 Å closer to the HAT domain (Fig. 1B). This difference, which persists in structures of the CBP core crystallized in different space groups, appears to arise from differing interactions in the interface between the BRD and PHD domains of CBP and p300 (SI Text and Fig. S1B). Although the structure of the substrate-binding site of the HAT domain is the same in CBP and p300, the proteins exhibit different selectivity in acetylating histones (21, 22). It is likely that the BRD plays a role in hyperacetylation by anchoring the substrate to the HAT core through an already acetylated lysine to facilitate acetylation at additional sites. Because the structures and acetyl-lysine binding activities (12–14) of the BRDs of CBP and p300 are almost identical, their differing substrate selectivity might be associated with differences in the distance between the acetyl-lysine binding site of the BRD and the substrate binding site of their HAT domains.
Contribution of Proximal Regions to HAT Activity.
To address the influence of the proximal domains on the catalytic activity of the HAT domain, four recombinant CBP core constructs (Fig. 2) were prepared for in vitro assays of histone H3 acetylation at K18 and K27 using HeLa cell nuclear extract as a substrate, autoacetylation of the CBP catalytic core, and acetylation of K382 in full-length p53. Acetylation of histone H3 and full-length p53 was observed for all CBP constructs. Enhanced CBP autoacetylation and p53 K382 acetylation were observed for the RING domain-deletion mutant (Fig. 3 A and B, lane 3), consistent with observations made for p300 (13), whereas RING deletion has no effect on histone H3 K18 and K27 acetylation levels (Fig. 3A, lane 3). To further investigate regulation by the RING, we tested acetylation of a recombinant 63-residue AL peptide and a C-terminal peptide from p53 (residues 361–393; p53C) as substrates. In contrast to the results obtained with full-length p53 and with the CBP catalytic core, the RING-deleted mutant did not enhance acetylation of either p53C or the isolated AL peptide (Fig. 3 B and C, lane 3). This differential regulation by the RING domain can be explained by the structure of the substrates. The RING domain is positioned close to the substrate entry channel where it may occlude access to the active site by bulky substrates such as the full-length p53 tetramer or the intact CBP/p300 core, which is autoacetylated on lysine residues in the AL and on K1536 (CBP) or K1499 (p300) located in a structured region of the HAT domain (18, 20). Deletion of the RING thus enhances acetylation of bulky substrates. In contrast, acetylation of short and flexible peptides that can easily reach the active site of the HAT domain, such as the p53C and AL peptides, are unaffected by RING deletion.
Fig. 2.
Schematic representation of the domain architecture of CBP. The recombinant protein constructs used in the HAT and SUMOylation assays are shown below.
Fig. 3.
The influence of proximal domains on CBP HAT activity. (A) In vitro HAT activity of recombinant CBP protein constructs. Acetylation of nucleosomal histone H3 in HeLa cell nuclear extract and of full-length p53 probed using anti-H3K18Ac/anti-H3K27Ac and anti-p53K382Ac antibodies, respectively. A Coomassie-stained gel for the CBP proteins is shown below. Other irrelevant bands from the nuclear extract also appear in this gel. (B) In vitro assays showing autoacetylation of the CBP protein constructs and acetylation of a recombinant 62-residue AL substrate, probed using an anti–acetyl-K antibody. A Coomassie-stained gel for the CBP proteins and the AL is shown below. (C) In vitro acetylation of a recombinant p53 C-terminal peptide (p53C, 31 aa). p53 acetylation was monitored by using an anti-p53K382Ac antibody. Coomassie-stained gels for the CBP proteins and the p53C peptide are shown below.
No differences were observed in acetylation of p53 or histone H3 in HeLa cell extracts by the BCHZ and BCH constructs (Fig. 3 A and C, compare lanes 2 and 4), implying that the ZZ domain does not directly influence the activity of the HAT domain. Although little difference in histone H3 acetylation was detected upon deletion of the AL (construct BCHZ∆AL), substantial reduction was observed in acetylation of full-length p53 but not the p53C peptide (Fig. 3 A and C, lanes 2 and 5). It was previously suggested that the AL is autoinhibitory, competing with substrates for binding in the active site and resulting in decreased acetyltransferase activity (20). However, deletion of the AL causes no change in acetylation of peptide substrates (histone H3 and the AL and p53C peptides) and greatly impairs acetylation of K382 in full-length p53. Similarly, deletion of the AL from the p300 HAT caused no change in autoacetylation of K1499 or acetylation of K9 or K14 of histone H3 (18).
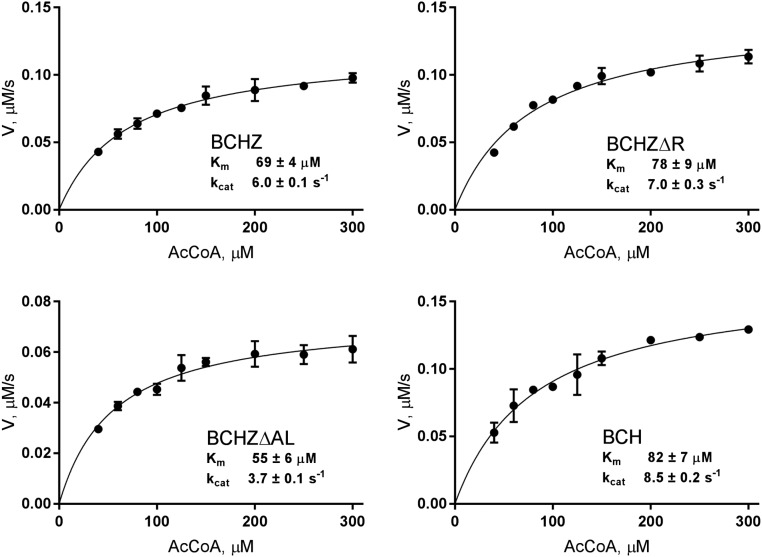
To assess HAT activity, acetylation kinetics were measured for the various CBP constructs using a histone H3 peptide as substrate. The assays show that the intrinsic catalytic activity of the HAT domain is not directly modulated by the neighboring RING, AL, or ZZ domains because kcat and Km (for AcCoA) are similar for all constructs (Fig. S2). Changes in acetylation level upon deletion of the RING or AL loop must therefore reflect differing interactions between the catalytic core and its substrates.
Fig. S2.
Kinetic analysis of the HAT activity of CBP constructs measured using a DTNB-coupled acetyltransferase assay (20) with a histone H3 peptide substrate. In each panel, filled circles are experimental data, and solid lines are the fitting results. The error bars represent the rmsd from duplicate experiments. Purified proteins were exhaustively deacetylated with SIRT2 and exchanged into fresh assay buffer containing 50 mM Hepes, pH 7.6, 150 mM NaCl, and 50 μg/mL BSA. Multiple reactions were set up for each construct, using 0.4 mM histone H3 peptide (residues 5–23) as substrate and a range of Ac-CoA concentrations from 40 to 300 μM in the assay buffer. Individual reactions containing 20 nM CBP were allowed to proceed for 10 min at 30 °C, and then quenched by the addition of an equal volume of quench buffer (6 M guanidinium-HCl in the assay buffer). The concentration of CoASH product was quantified by measuring the absorbance at 412 nm after adding an equal volume of DTNB solution (to a final concentration of 0.2 mM, in buffer containing 10 mM EDTA) to each reaction tube. Controls for CBP autoacetylation were performed in parallel by setting up reactions without histone H3 substrate. The contribution to the absorbance arising from autoacetylation was subtracted from the data points for each reaction. Average values from duplicate experiments were used to fit the initial rates to the Michaelis–Menten equation by nonlinear regression using the program Prism.
The PHD and ZZ Domain Interact with SUMO-1.
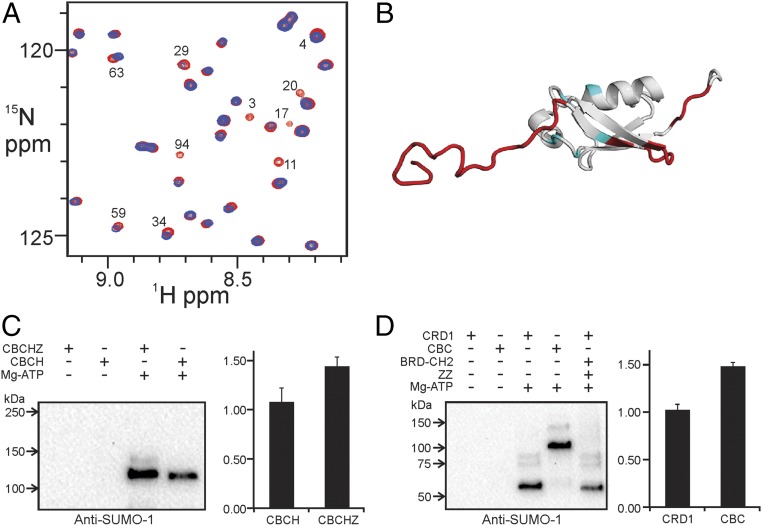
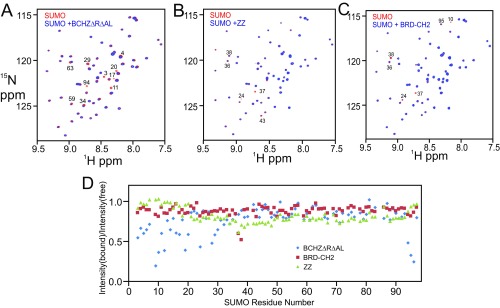
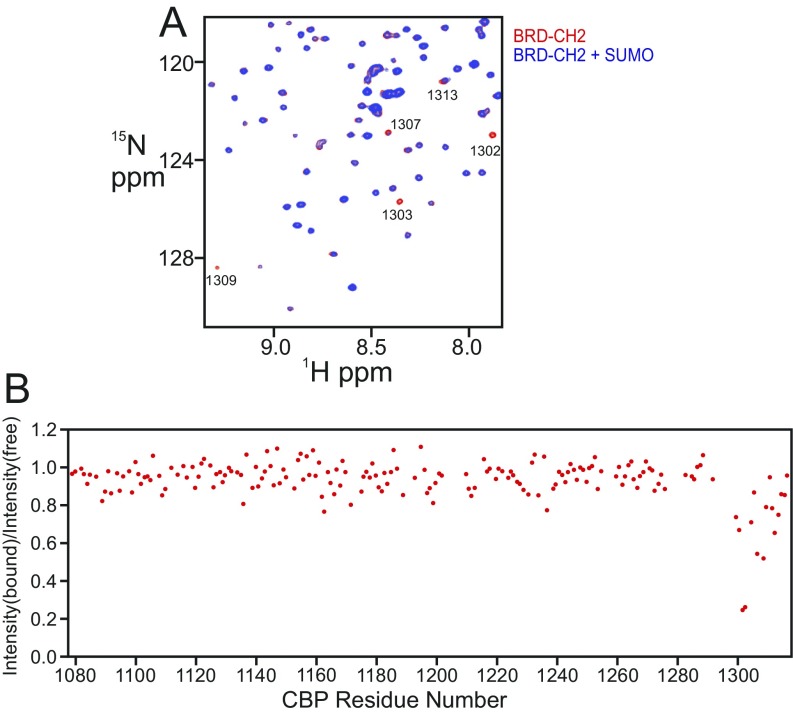
Although ZZ-type zinc fingers have been implicated in protein–protein interactions (24, 31), the biological function of the ZZ domain of CBP/p300 has yet to be determined. The discovery that the ZZ domain of the ubiquitin-ligase HERC2 binds SUMO-1 (32) prompted us to examine whether the CBP core can also bind SUMO-1. Several backbone amide cross peaks in the 1H–15N heteronuclear single quantum coherence (HSQC) spectrum of 15N-labeled SUMO-1 are selectively broadened upon addition of equimolar BCHZ∆R∆AL (Fig. 4A and Fig. S3A). The residues experiencing the greatest perturbation are located in the N- and C-terminal regions and β-sheet of SUMO-1, suggesting that these regions interact directly with CBP (Fig. 4B and Fig. S3D). While this manuscript was in preparation, Diehl et al. (25) reported that the ZZ domain of CBP/p300 interacts with SUMO-1. Surprisingly, the site through which SUMO-1 binds to the CBP core differs from that through which it binds to the isolated ZZ domain (Fig. 4B), suggesting that additional sites on the CBP core interact with SUMO-1. We therefore investigated the interactions of SUMO-1 with various CBP constructs. Addition of the isolated ZZ domain caused slight broadening of several cross peaks in the SUMO-1 1H–15N HSQC spectrum (Fig. S3B), in agreement with the previous finding that the ZZ domain has a SUMO-interacting motif (25). Specific broadening of SUMO-1 cross peaks also occurs upon addition of the BRD–CH2 construct (Fig. S3C), although to a lesser extent than for BCHZ∆R∆AL (Fig. S3 A and C), suggesting that the isolated BRD–CH2 cannot bind to SUMO-1 as tightly as BCHZ. Conversely, cross peaks in the HSQC spectrum of the isolated BRD–CH2 construct, assigned to residues in the C-terminal region of the PHD finger, are also broadened by interaction with SUMO-1 (Fig. S4). Taken together, it is clear that within the context of the CBP catalytic core, the PHD and ZZ domains function cooperatively to bind SUMO-1.
Fig. 4.
The CBP core interacts with SUMO-1. (A) Overlay of a region of the 1H–15N HSQC spectra of free 15N-labeled SUMO-1 (red) and in the presence of equimolar BCHZ∆R∆AL (blue). Cross peaks that are broadened or shifted are marked with residue numbers. (B) Mapping of the binding site (red) of the CBP core on the SUMO-1 structure (PDB ID code 1A5R). The binding site for the isolated ZZ domain, identified by Diehl et al. (25), is shown in blue. (C) In vitro SUMOylation of CRD1 repressor domain in CBCHZ and CBCH constructs. SUMOylated protein was detected using anti–SUMO-1 antibody (Left). Densitometric quantification (Right) was based on the average of five independent repeats and SEs are shown. (D) In vitro SUMOylation of GST–CRD1 and GST–CRD1–BRD–CH2. SUMOylated protein was detected by anti–SUMO-1 antibody (Left). Densitometric quantification (Right) was based on the average of five repeats, and SEs are shown.
Fig. S3.
Interaction of SUMO with domains of CBP. (A) Overlay of the 1H–15N HSQC spectra of 15N-labeled SUMO-1 free (red) and in the presence of equimolar BCHZ∆R∆AL (blue). This figure is an expansion of Fig. 3A in the main text. Cross peaks that are broadened or shifted are marked with residue numbers. (B) Overlay of the 1H–15N HSQC spectra of 15N-labeled SUMO-1 free (red) and in the presence of equimolar ZZ domain (blue). Cross peaks that exhibit broadening or chemical shift changes are marked by residue numbers. (C) Overlay of the 1H–15N HSQC spectra of 15N-labeled SUMO-1 free (red) and in the presence of equimolar BRD–CH2 (blue). Cross peaks that exhibit broadening or changes in chemical shift are indicated by residue numbers. (D) Fractional change in the intensity of cross peaks in the 1H–15N HSQC spectrum of SUMO-1 upon addition of equimolar BCHZ∆R∆AL (blue), BRD–CH2 (red), and ZZ (green).
Fig. S4.
(A) Overlay of the 1H–15N HSQC spectra of 15N-labeled BRD–CH2 free (red) and in the presence of equimolar SUMO-1 (blue). Peaks that are shifted or broadened are marked with residue numbers. (B) Plot of the ratio of cross-peak intensities in the 1H–15N HSQC spectra of BRD–CH2 in the presence and absence of SUMO-1 versus CBP residue number.
The BRD, PHD, and ZZ Domains Facilitate Intramolecular SUMOylation of CBP.
Because RING and PHD zinc fingers frequently function as E3–SUMO ligases (33, 34), we performed SUMOylation assays to determine whether the CH2 and ZZ domains might perform a similar function in the CBP core. CBP becomes SUMOylated at three lysines in the CRD1 domain, recruiting Daxx and histone deacetylases through the covalently attached SUMO-1 to negatively modulate the transcription of target genes (27). In vitro SUMOylation assays were performed for two CBP constructs, CBCHZ spanning residues from CRD1 to ZZ and CBCH spanning residues from CRD1 to HAT (Fig. 2). Increased SUMOylation was observed for CBCHZ (Fig. 4C), showing that the ZZ domain enhances CRD1 SUMOylation. GST–CRD1 and GST–CRD1–BRD–CH2 (CBC; Fig. 2) fusions were also assayed for SUMOylation in vitro. CRD1 SUMOylation was significantly increased for CBC, showing that the BRD–CH2 region also promotes intramolecular SUMOylation (Fig. 4D). The enhancement of CRD1 SUMOylation by ZZ and BRD–CH2 is consistent with the NMR results that show that the ZZ and PHD domains both interact with SUMO-1 (Fig. S3). This enhancement was not observed for the isolated CRD1 peptide when BRD–CH2 and ZZ domains were separately added to the reaction mixture (Fig. 4D, lane 5), showing that these domains promote SUMOylation only when they are covalently linked to the CRD1 domain.
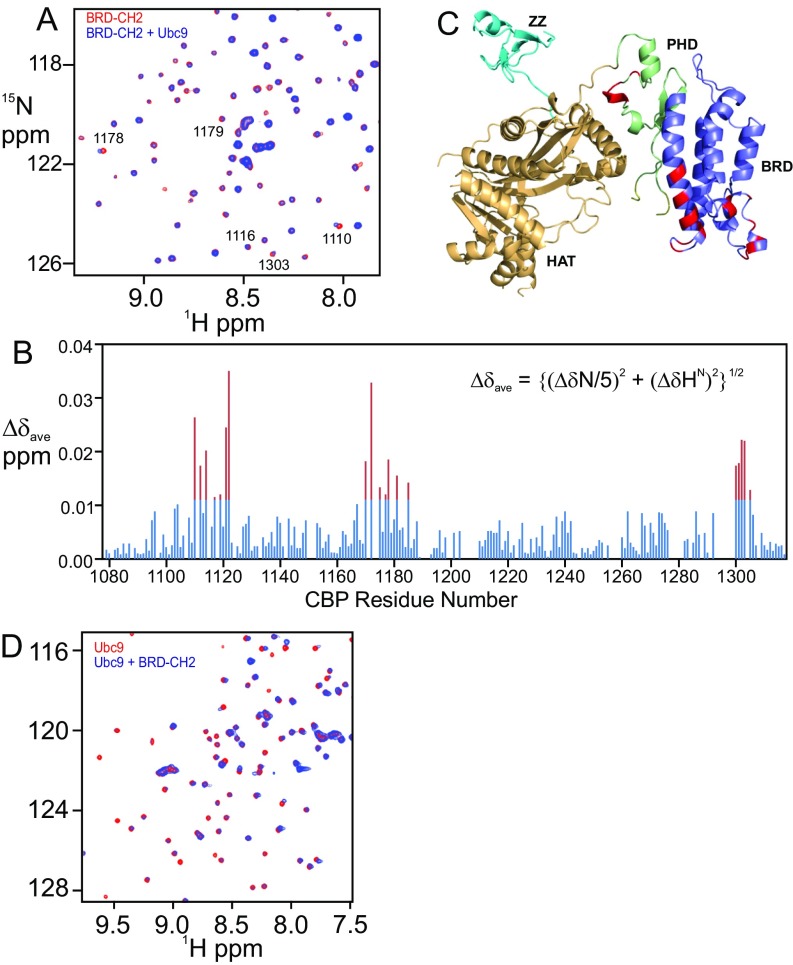
Because E3 ligases often recruit the E2∼SUMO-1 thioester conjugate to facilitate SUMOylation of substrate (35), we used NMR to probe for interactions with the SUMO E2 ligase Ubc9. Many cross peaks in the 1H–15N HSQC spectrum of BRD–CH2 were broadened and shifted upon addition of Ubc9, indicating direct protein–protein interaction (Fig. S5). The residues that showed the largest chemical shift changes are localized on two surfaces: one in the BRD and the other in the PHD (Fig. S5 B and C). Conversely, many cross peaks in the 1H–15N HSQC spectrum of Ubc9 were shifted or broadened upon addition of BRD–CH2, confirming the binding interaction (Fig. S5D). Taken together, the in vitro SUMOylation assays and NMR experiments strongly suggest that the BRD, PHD, and ZZ domains function synergistically, within the context of the intact CBP catalytic core, as an E3 ligase to promote intramolecular SUMOylation of the CRD1 domain.
Fig. S5.
BRD–CH2 interacts with Ubc9. (A) Overlay of the 1H–15N HSQC spectra of the 15N-labeled BRD–CH2 free (red) and after addition of equimolar Ubc9 (blue). Peaks that exhibit chemical shift perturbation are marked by residue numbers. (B) Plot of chemical shift differences (∆δave) of the backbone HN and N resonances of BRD–CH2 between free and Ubc9-bound forms (Bottom) versus CBP residue number. Red bars indicate shift changes greater than the average plus 1 SD. (C) Residues in the BRD–CH2 construct that undergo significant chemical shift changes upon addition of 1:1 Ubc9 mapped onto the structure of the CBP catalytic core. Perturbed residues (red) correspond to the red peaks in the histogram in B. (D) Overlay of the 1H–15N HSQC spectra of the 15N-labeled Ubc9 free (red) and in the presence of 1:1 BRD–CH2 (blue).
The BRD Interacts with the Acetylated AL.
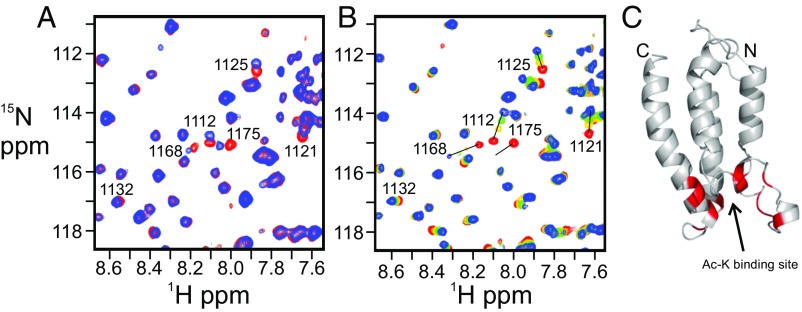
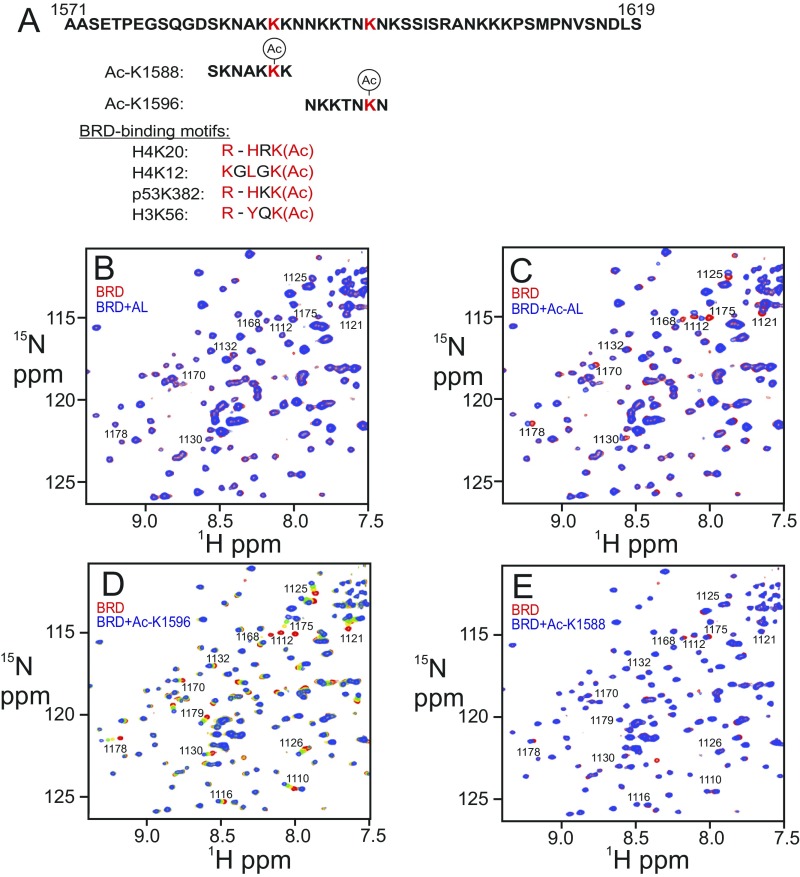
The 1H–15N HSQC spectrum of 15N-labeled BRD is perturbed by addition of an acetylated peptide corresponding to the AL (residues L1557 to S1619 of CBP), showing that the BRD binds to the acetylated AL (Fig. 5A and Fig. S6C); in contrast, no change was observed upon addition of the nonacetylated AL (Fig. S6B). The consensus binding motif for the CBP BRD consists of a hydrophobic or aromatic residue at position −2 and a lysine or arginine at position −3 or −4 from the acetylated lysine (36–38) (Fig. S6A). None of the lysines within the AL have hydrophobic or aromatic residues at position −2, but two (K1588 and K1596) have lysines at the −3 or −4 position (Fig. S6A). Addition of the seven-residue acetyl-K1596 (Ac-K1596) peptide (Fig. S6A) to the isolated BRD caused chemical shift changes and broadening of the same 1H–15N HSQC cross peaks of the BRD as are perturbed by binding of the acetylated 63-residue AL construct (Fig. 5 A and B). Addition of 10-fold excess of the Ac-K1596 peptide resulted in substantial changes in chemical shift of BRD cross peaks (Fig. 5B and Fig. S6D), associated with residues in the acetyl-lysine binding pocket (Fig. 5C). In contrast, addition of a 10-fold excess of the Ac-K1588 peptide caused little change in the 1H–15N HSQC spectrum of the BRD (Fig. S6E). Thus, the interaction between the BRD and the AL region of CBP appears to be specific and requires acetylation of K1596.
Fig. 5.
Interaction between the BRD and acetylated autoregulatory loop (AL) of CBP. (A) Overlay of a region of the 1H–15N HSQC spectra of the 15N-labeled BRD free (red) and in complex with the acetylated 63-residue AL peptide (blue). (B) Changes in the 1H–15N HSQC spectrum of the 15N-labeled BRD upon titration with the acetylated K1596 peptide at a molar ratio of 1:0 (red), 1:2.5 (yellow), 1:5 (green), and 1:10 (blue). Peaks that exhibit chemical shift perturbations are marked with residue numbers. Full versions of these spectral overlays can be found in Fig. S6 C and D. (C) Residues that experience chemical shift changes greater than average plus 1 SD upon addition of Ac-K1596 peptide are mapped (in red) onto the structure of the BRD of CBP.
Fig. S6.
Interaction between the BRD and acetylated AL of CBP. (A) Amino acid sequence of the AL and the two autoacetylation sites (red) chosen for testing BRD binding. Known CBP BRD-binding motifs are shown below. The acetylated lysine and residues at positions −2 and −3/−4 relative to the acetylated lysine are colored in red. (B) Overlay of the 1H–15N HSQC spectra of the free 15N-labeled BRD (red) and after addition of equimolar unacetylated AL (blue). (C) Overlay of the 1H–15N HSQC spectra of the free 15N-labeled BRD (red) and after addition of equimolar acetylated AL (blue). (D) Changes in the 1H–15N HSQC spectrum of the 15N-labeled BRD upon titration with the acetylated K1596 peptide at molar ratios of 1:0 (red), 1:2.5 (yellow), 1:5 (green), and 1:10 (blue). Peaks that exhibit chemical shift perturbations are marked with residue numbers. (E) Overlay of the 1H–15N HSQC spectra of the free 15N-labeled BRD (red) and after addition of 10-fold molar access of acetylated K1588 peptide (blue).
Negative Regulation of Histone Acetyltransferase Activity by Autoacetylation.
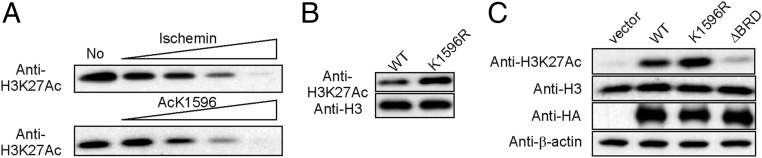
The CBP and p300 HATs acetylate multiple lysines on the four core histones (39), including H3K27Ac, which is an important histone mark associated with transcriptionally active genes (40–42). The CBP and p300 BRDs interact with hyperacetylated nucleosomes (43, 44), binding promiscuously to acetylated histone tails (12–14). To determine whether the CBP BRD is functionally important in histone acetylation, we monitored H3K27 acetylation in promiscuously acetylated nucleosomes in HeLa cell nuclear extract in the presence of ischemin, a specific CBP/p300 BRD inhibitor (45). Addition of increasing amounts of ischemin strongly attenuated acetylation of H3K27 by the CBP catalytic core (Fig. 6A), showing that interactions with the BRD are essential for H3K27 acetylation. A similar attenuation was observed upon addition of increasing amounts of the seven-residue Ac-K1596 peptide, although higher concentrations are required to reach the same level of inhibition as ischemin. These results suggest that acetylation of K1596 in the AL might down-regulate histone acetylation by competition for binding to the CBP BRD. Binding of Ac-K1596 to the BRD is likely to be enhanced within the intact CBP catalytic core due to its increased effective concentration. In contrast to our observations with ischemin, the bromodomain inhibitor I-CBP112 activates CBP/p300 toward acetylation of a subset of H3 and H4 residues (H3K18, H3K23, H4K5) within nucleosomal substrates, whereas a different inhibitor, CBP30, has no effect on histone acetylation (46). It is apparent that the role of the BRD in regulating acetylation of histones within intact nucleosomes is complex and that further work will be required to elucidate the molecular mechanisms that underlie the differing effects of the various bromodomain inhibitors.
Fig. 6.
Acetylation at K1596 negatively regulates the HAT activity. (A) Acetylation of nucleosomal H3K27 by the CBP BCHZ construct is impaired by the BRD inhibitor ischemin (Top) or the Ac-K1596 peptide (Bottom) in an in vitro HAT assay using nucleosomes in HeLa cell nuclear extract as substrate. Acetylation of H3K27 was probed using an anti-H3K27Ac antibody. The concentrations of ischemin were 12.6, 31.3, 62.5, and 125 µM in lanes 2–5, and the concentrations of Ac-K1596 motif were 62.5, 125, 250, and 500 µM in lanes 2–5. (B) In vitro HAT assays using nucleosomes in HeLa cell nuclear extract as substrate showing enhancement of histone H3K27 acetylation by the K1596R mutant BCHZ construct. Acetylation of H3K27 was probed using an anti-H3K27Ac antibody. (C) Acetylation of H3K27 in HEK293T cells that were transiently transfected with full-length CBP (wild-type, K1596R, or ∆BRD). CBP was expressed as an HA-tagged protein. Acetylation of H3K27 was probed using an anti-H3K27Ac antibody. The amount of expressed CBP was monitored using an anti-HA antibody, and β-actin was used as a loading control.
To establish whether negative regulation of HAT activity by the autoacetylated AL can occur in cis, we replaced K1596 in the BCHZ construct by arginine. In HAT assays using HeLa cell nuclear extract as substrate, the K1596R mutant exhibited substantially increased H3K27 acetylation compared with the self-acetylated wild-type BCHZ (Fig. 6B). The increased acetylation activity of K1596R can be attributed to the loss of negative regulatory interactions between the BRD and the AL. Negative regulation of the HAT activity was also observed for mutants of full-length CBP in a human cell line. Acetylation of H3K27 by CBP monitored after transient expression of wild-type, K1596R, or ∆BRD CBP in HEK293T cells showed that deletion of the BRD almost completely abrogates acetylation of H3K27 (Fig. 6C, lane 4). Furthermore, H3K27 was more highly acetylated by the K1596R mutant than by wild-type CBP (Fig. 6C, lane 3), lending support to a model in which self-acetylation of the AL has a negative regulatory function in mammalian cells.
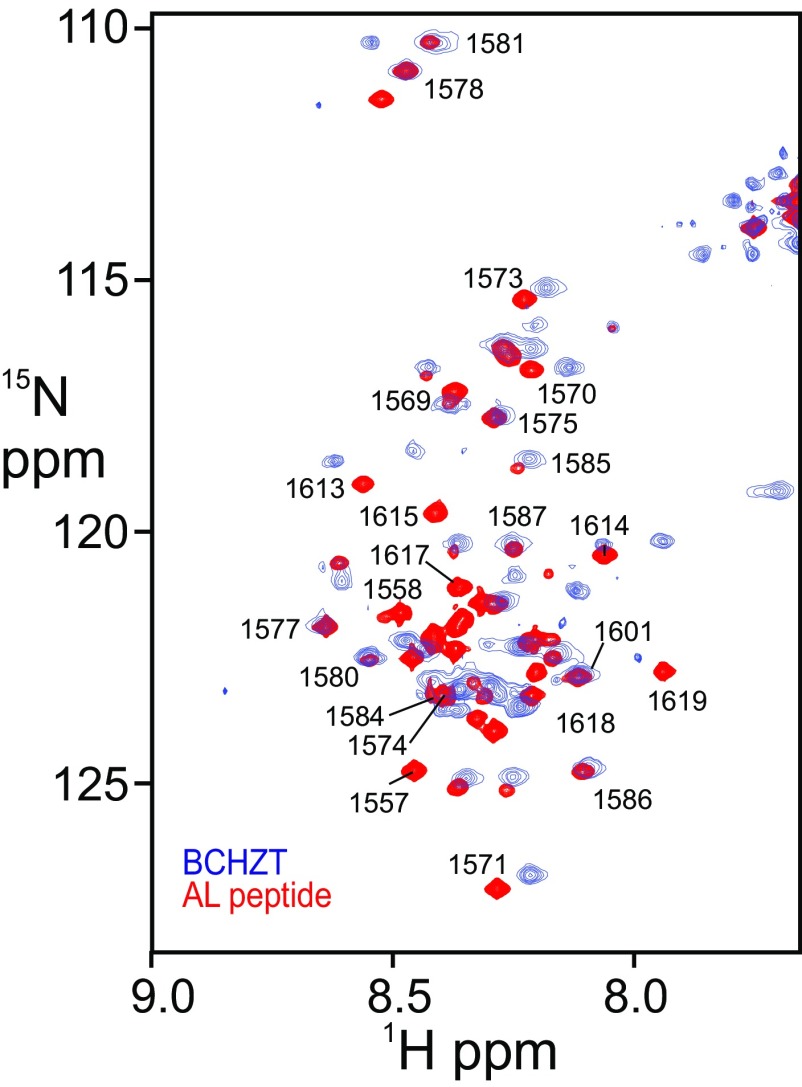
The AL, which is rich in charged and polar amino acids and highly deficient in bulky hydrophobic side chains, is predicted to be intrinsically disordered (47). Disorder of the AL, in a 93-kDa CBP construct containing the BRD, CH2, HAT, ZZ, and the adjacent TAZ2 domain (BCHZT), was confirmed by NMR (SI Text and Fig. S7). K1596 is located near the center of the AL region (Fig. S6A), which is long enough to allow Ac-K1596 to reach the binding site of the BRD.
Fig. S7.
Overlay of the 1H–15N HSQC spectra of BCHZT (blue, 93 kDa) and the recombinant 63-residue AL peptide (red). Assignments of dispersed cross peaks in the AL peptide spectrum are indicated by residue numbers.
Discussion
The regulation of histone acetylation can be achieved at several different levels, including the competing activity of HATs and HDACs and Ac-CoA metabolism (46, 47). The activity of HATs is also regulated by binding partners, associated protein domains, and autoacetylation (48). Taken together, the X-ray structures of the HAT and flanking domains of p300 (13) and CBP (this work) provide a comprehensive view of the central catalytic core of these essential transcriptional coactivators and insights into their regulation. The structures of the apo CBP HAT domain and the Lys-CoA–bound p300 HAT domain (13, 29) are nearly identical, showing that the active site and cofactor binding site are fully preformed in the absence of bound ligands. No significant structural changes are required to accommodate Ac-CoA, and the hydroxyl oxygen atom of the critical Y1504 (equivalent to Y1467 in p300), which protonates the CoA leaving group, is close (0.9 Å) to its position in Lys-CoA–bound p300 (29, 49).
The domains that flank the CBP and p300 HATs play an important role in regulation of their catalytic activity. Deletion of the CBP BRD strongly impairs histone H3K27 acetylation in HEK293 cells (Fig. 6C), and in vitro experiments using posttranslationally modified nucleosome arrays showed that deletion of the p300 BRD abrogated acetylation at all histone sites that were assayed (44). The BRD clearly plays an important regulatory role in recruiting CBP/p300 to hyperacetylated nucleosomes to activate CBP/p300-mediated acetylation programs. The PHD appears to be tightly packed against the BRD and HAT domains through a network of hydrophobic, hydrogen-bonding, and electrostatic interactions. Substitution of Y1205 in CBP by Leu in p300 (Fig. S1B) appears to alter the PHD–BRD interface and reposition the BRD active site at a larger distance from the HAT domain, which may account for the differing selectivity of CBP and p300 in acetylation of histone substrates (21, 22). The interface between the PHD and HAT domains is highly conserved in the CBP and p300 structures. The Rubinstein–Taybi syndrome mutation E1279K (E1278 in human CBP), which would disrupt this interface by breaking salt bridges to R1683 and R1684, impairs histone acetylation in vitro and transcriptional activation in cells (16).
The principal differences between the structures of the CBP and p300 catalytic cores are the presence of the ZZ domain in the CBP construct and the weak and diffuse nature of the electron density for the CBP RING domain (T1211–K1272). The diffuse electron density most likely reflects orientational disorder rather than internal flexibility of the RING domain; the elements of secondary structure identified in NMR studies of a CBP BRD–CH2 construct (12) correspond precisely to the locations of secondary structural elements in the X-ray structure of the p300 RING domain, suggesting that the CBP RING is stably folded. Electron density was also absent for most RING residues in a CBP BRD–CH2 construct, consistent with flexibility of the CBP RING in the absence of stabilizing lattice contacts (37). The bromodomain structures are nearly identical in the CBP core and the BRD–CH2 construct, but structural differences are observed in regions of the PHD that pack against the HAT. Although the ZZ and most likely the RING form well-folded globular domains, both appear to be linked to the catalytic core through flexible connections, as shown for the ZZ domain in Fig. S1F.
Deletion of the CBP or p300 RING domains enhances autoacetylation and acetylation of the p53 tetramer (13, 18) (Fig. 3 A and B). The RING domain appears to function as a gatekeeper, regulating access to the acetyltransferase active site by bulky substrates (13). Because the RING is attached to the catalytic core through a flexible linker, interactions or posttranslational modifications that modulate its position may play a regulatory role by controlling access of substrates to the active site of the HAT domain.
The CBP and p300 HAT domains contain a lysine-rich AL that functions as an autoinhibitor of HAT activity by competing with substrates for binding in the active site; inhibition is relieved by loop deletion or by hyperacetylation of the AL to displace it from the active site and allow access by other substrates (20). Inhibition is also relieved by enhancer RNAs, which bind directly to the AL to stimulate CBP acetyltransferase activity and promote gene expression (50). The present work reveals additional functions of the AL. First, deletion of the AL greatly impairs K382 acetylation of the p53 tetramer (Fig. 3A), suggesting that the loop may also play a role in substrate recruitment, perhaps, in the case of p53, by binding to the acidic grooves in the tetramerization domain located close to the site of acetylation. Second, following activation of the CBP HAT by autoacetylation of the disordered AL, Ac-K1596 binds in an intramolecular manner to its own bromodomain, competing with histones for binding to the CBP catalytic core. This intramolecular interaction acts as a negative regulator of histone acetylation and may promote dissociation of CBP from chromatin. Our results suggest a potential mechanism, involving intramolecular interaction between acetylated K1596 and the BRD, by which autoacetylation induces dissociation of CBP and p300 from the preinitiation complex, a requirement for transcription to begin (51).
Although the bromodomain of CBP/p300 performs a well-established function in acetyl-lysine recognition (12, 13, 52), the functions of the PHD, RING, and ZZ domains remain poorly understood. The observation that SUMO-1 interacts with the ZZ domain (ref. 25 and this work) and the PHD domain, and that the E2 ligase Ubc9 interacts with the BRD and PHD domains, suggests an intriguing possibility that the CBP/p300 core might function as a SUMO E3 ligase for intramolecular SUMOylation of the adjacent CRD1 domain. Indeed, in vitro assays confirmed that the BRD–CH2 region does promote SUMOylation of CRD1, that SUMOylation is enhanced when the ZZ domain is also present in the CBP core construct, and that SUMOylation is most efficient in cis, that is, when the CRD1, BRD–CH2, and ZZ domains are part of the same polypeptide chain (Fig. 4). Mechanistically, SUMO E3 ligases function to bring substrates and E2∼SUMO thioester into close proximity to promote SUMO transfer (35). In CBP, the substrate CRD1 domain is covalently attached to the E3 moiety, so recruiting and properly positioning the E2∼SUMO thioester through the E3 function can facilitate SUMOylation of the substrate. Intramolecular SUMO E3 ligase activity has also been observed in the KAP1 corepressor, where a PHD domain functions to promote SUMOylation of specific lysine residues in the adjacent bromodomain (53). PHD and RING domains frequently function as SUMO E3 ligases (33, 34, 54). Although the CBP core contains a RING finger, our NMR data show that the RING domain does not interact directly with either SUMO-1 or Ubc9. Instead, the BRD, PHD, and ZZ zinc finger function synergistically to endow E3 ligase activity on the CBP catalytic core.
The domains flanking the HAT domain play a pivotal role in regulation of acetyltransferase activity by directing the enzymatic activity of the core. Another important regulatory function of these domains is in control of SUMOylation of the cell cycle regulatory domain CRD1. These functions are intimately linked: competition between autoacetylation and SUMOylation of lysine residues in CRD1 determines whether CBP/p300 function as transcriptional activators or repressors in response to various signals.
Materials and Methods
Protein Preparation.
cDNA sequences from mouse CBP encoding residues 1,079–1,751 for BCHZ, 1,079–1,872 with a Y1504F mutation for BCHZT, 1,079–1,700 for BCH, 989–1,751 for CBCHZ, 989–1,700 for CBCH, 1,079–1,751 with a deletion of 1,211–1,272 for BCHZ∆R, 1,079–1,751 with a replacement of 1,557–1,618 by a 5-aa SGGSG linker for BCHZ∆AL, 1,079–1,751 with a deletion of 1,211–1,272 and a replacement of 1,557–1,618 by a 5-aa SGGSG linker for BCHZ∆R∆AL, 1,324–1,700 for HAT, and 1,557–1,619 for AL were cloned into the vector pHis parallel 2 (55) with a TEV-cleavable N-terminal His6 tag. The proteins were expressed in E. coli BL21(DE3)DNAY cells and induced by 1 mM IPTG at 15 °C. Cells were resuspended in buffer A (25 mM Tris⋅HCl, pH 8.0, 0.3 M NaCl, 2 mM DTT, and 50 µM ZnSO4) that contains 10 units of benzonase (Novagen) and EDTA-free protease inhibitor mixture (Roche) and sonicated. The soluble fraction after sonication was applied to cOmplete His-Tag purification resin (Roche) equilibrated with buffer A containing 5 mM imidazole. The proteins were eluted with buffer A containing 300 mM imidazole. The His6 tag was cleaved by TEV protease and removed from the protein solution by cOmplete His-Tag purification resin. All of the HAT-containing proteins expressed in E. coli were deacetylated with His6-tagged SIRT2 at 1:100 molar ratio in buffer A containing 1 mM NAD+ for 16 h at 4 °C. SIRT2 was removed from CBP constructs using cOmplete His-Tag purification resin. Proteins were further purified by size exclusion chromatography using Superdex 75 (GE Healthcare Life Sciences) equilibrated with buffer A. CBCHZ and CBCH were purified by ion exchange chromatography using Q-Sepharose (GE Healthcare) before size exclusion chromatography.
cDNA sequences from mouse CBP encoding residues 989–1,067 for CRD1 and 989–1,317 for CRD1–BRD–CH2, human SUMO-1, and human Ubc9 were cloned into the vector pGEX-4T-2 (GE Healthcare) with a thrombin-cleavable N-terminal GST tag. The proteins were expressed in E. coli BL21 cells and induced by 1 mM IPTG at 25 °C. Cells were resuspended in buffer B (25 mM Tris⋅HCl, pH 7.5, 0.1 M NaCl, 1 mM DTT, and 50 µM ZnSO4) containing 10 units of benzonase (Novagen) and EDTA-free protease inhibitor mixture (Roche) and sonicated. The soluble fraction after sonication was applied to glutathione-Sepharose 4B (Sigma-Aldrich) equilibrated with buffer. The proteins were eluted with 50 mM Tris⋅HCl, pH 8.0, and 10 mM reduced l-glutathione. CRD1 and CRD1–BRD–CH2 were further purified by ion exchange chromatography using SP-Sepharose (GE Healthcare) and then dialyzed against 25 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, 1 mM DTT. GST-tagged SUMO-1 and Ubc9 were cleaved using thrombin and then passed through a second glutathione-Sepharose 4B column to remove GST. Cleaved SUMO-1 and Ubc9 were further purified by size exclusion chromatography using Superdex 75 (GE Healthcare Life Sciences) equilibrated with buffer containing 25 mM Tris⋅HCl, pH 8.0, 100 mM NaCl, and 1 mM DTT.
Bromodomain-binding and NMR experiments were performed using the more stable BRDCH2 construct containing residues 1,079–1,317 of mouse CBP. The BRDCH2 protein was expressed in E. coli BL21 cells and purified by a combination of glutathione-Sepharose 4B (Sigma-Aldrich), thrombin cleavage, and Sephacryl S-100 (GE Healthcare Life Sciences), as described previously (12).
Crystallization and Structure Determination.
Purified BCHZ∆AL at 10 mg/mL in buffer A was crystallized using the sitting-drop vapor diffusion method. Crystals were grown by mixing 1 µL of protein and reservoir solution containing 0.3 M trisodium citrate, pH 7.0, and 20% (wt/vol) PEG3350 at 22 °C. The crystals were flash-cooled in liquid nitrogen using reservoir solution supplemented with 25% (vol/vol) ethylene glycol as cryoprotectant. Diffraction data were collected at the Advanced Photon Source (APS) on the General Medical Sciences and Cancer Institute Structural Biology Facility (GM/CA) CAT 23ID-D beamline and processed with HKL-2000 (56) in space group C2. The structure of BCHZ∆AL was solved by molecular replacement using Phaser (57) with the p300 core structure (PDB ID code 4BHW) as a search model. Model building and refinement were carried out using Coot (58) and Phenix (59), respectively.
NMR Spectroscopy.
15N-labeled proteins were prepared by growing E. coli cells in 15NH4Cl-supplemented minimal media. Two-dimensional 1H,15N-HSQC spectra were acquired for uniformly 15N-labeled proteins in NMR buffer (25 mM Tris⋅HCl, pH 7.0, 50 mM NaCl, 1 mM DTT in 90% H2O:10% 2H2O). Unless otherwise specified, NMR spectra were acquired at 30 °C on Bruker 600-, 750-, 800-, and 900-MHz spectrometers. All NMR data were processed and analyzed using NMRPipe (60) and Sparky (61). 1H,15N-HSQC spectra of 100 μM free SUMO-1 and its 1:1 complexes with the ZZ or BRD–CH2 domains were obtained in NMR buffer, as were spectra of 100 μM free BRD–CH2 and its 1:1 complex with SUMO-1. 1H,15N-HSQC spectra of 75 μM free SUMO-1 and the 1:1 complex with BCHZ∆R∆AL were recorded in 25 mM Tris⋅HCl, pH 7.0, 300 mM NaCl, 1 mM DTT in 90% H2O:10% 2H2O at 28 °C. 1H,15N-HSQC spectra of 100 μM free BRD–CH2 and the 1:1 complex with Ubc9 were obtained in 25 mM Tris⋅HCl, pH 7.0, 200 mM NaCl, 1 mM DTT in 90% H2O:10% 2H2O. 1H,15N-HSQC spectra of 100 µM free Ubc9 and the 1:1 complex with BRD–CH2 were obtained in 25 mM Tris⋅HCl, pH 7.0, 200 mM NaCl, 1 mM DTT in 90% H2O:10% 2H2O. 1H,15N-HSQC spectra of 100 µM AL peptide (63 aa) and 83 µM BCHZT were obtained in 25 mM Tris⋅HCl, pH 7.0, 200 mM NaCl, 1 mM DTT in 90% H2O:10% 2H2O. Interactions between the BRD and AL were investigated by monitoring chemical shift perturbations upon addition of equimolar unmodified or acetylated AL peptides to 200 µM 15N-BRDCH2. The peptides containing Ac-K1588 (SKNAK-acK-K) or Ac-K1596 (NKKTN-acK-N) were commercially prepared by Biomatik. The Ac-K1588 peptide was added to 200 µM 15N-BRDCH2 at 10:1 molar ratio. The Ac-K1596 peptide was titrated into 200 µM 15N-BRDCH2 at 2.5:1, 5:1, and 10:1 molar ratio.
The backbone resonances of BRD–CH2 were assigned previously (12). The backbone assignments of SUMO-1 were obtained from Biological Magnetic Resonance Data Bank (ID 6304) (62). The assignments of the dispersed cross peaks in the HSQC spectrum of the 63-residue AL peptide (Fig. S5D) were made using triple-resonance methods and 13C,15N-labeled peptide. Because of severe cross-peak overlap, most of the Glu and Lys resonances have not been assigned at this time.
In Vitro SUMOylation Assay.
In vitro SUMOylation assays were carried out using a commercial SUMOylation kit (Boston Biochem) following the manufacturer-provided protocol. The final CBCHZ, CBCH, CRD1, and CRD1–BRD–CH2 substrate concentration was 2 µM. For the control experiments, Mg-ATP was not added to the reaction mixture. To assess the function of BRD–CH2 and ZZ in trans SUMOylation, 5 µM BRD–CH2 and ZZ were added to a CRD1 sample. All SUMOylation reactions were incubated at 37 °C for 3 h and then quenched by adding 2× SDS-sampling buffer. SUMOylated proteins were detected by a Western blot using anti–SUMO-1 antibody (ENZO Life Science). Five independent SUMOylation assays were performed, and band intensities were quantified using densitometry.
In Vitro HAT Assay.
In vitro histone acetylation assays were performed on 25 μg of HeLa cell nuclear extract (Santa Cruz Biotechnology) in 50 mM Tris⋅HCl, pH 8.0, 100 mM NaCl, 1 mM DTT buffer supplemented with 1 mM Ac-CoA. The reaction was initiated by adding 0.5 µM purified CBP proteins (BCHZ, BCHZ∆R, BCHZ∆AL, and BCH, all of which were exhaustively deacetylated by incubation with SIRT2), and samples were incubated at 37 °C for 1 h. To test the acetyltransferase activity with the BRD inhibition, 12.6, 31.3, 62.5, or 125 µM ischemin and 62.5, 125, 250, or 500 µM Ac-K1596 peptide were added to reactions. Acetylation of H3, p53, and the AL was monitored by Western blots using anti-H3K27Ac antibody (Abcam), anti-p53K382Ac antibody (Abcam), and anti-acK antibody (Abcam), respectively. Ischemin was purchased from Calbiochem. All acetylation assays were performed in duplicate.
In-Cell HAT Assay.
Human HEK293T cells were obtained from ATCC and maintained in DMEM (Gibco) supplemented with 10% calf serum. The cells were transiently transfected with 1 µg of vector control or pcDNA3.1 plasmid constructs expressing HA-tagged wild-type or mutant CBP using Lipofectamine 2000 (Invitrogen). After 48 h, cells were harvested and lysed by the freeze-and-thaw method in 50 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, and 0.1% Triton X-100 supplemented with 1 mM PMSF and one Complete Protease Inhibitor tablet (EDTA-free; Roche), and then centrifuged at 13,000 × g for 10 min. The supernatant was used to check the expression level of CBP by Western blot using anti-HA.11 antibody (Covance). Anti–β-actin antibody (Abcam) was used for the loading control. The pellet containing the nucleus was further lysed by sonication in RIPA buffer (50 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 0.5% sodium deoxycholate, and 1% Nonidet P-40) followed by centrifugation at 13,000 × g for 10 min. The supernatant was assayed for acetylated histone H3K27 using anti-H3K27Ac antibody (Abcam), whereas anti-H3 antibody (Abcam) was used for the loading control. The in-cell assay was performed in duplicate with consistent results.
Structure Description
Structural Differences Between CBP and p300 Catalytic Cores.
Differences in the orientation of the BRD in the CBP and p300 catalytic cores appear to be associated with differences in packing interactions between the BRD and PHD domains. The side chain of R1203 in the PHD of CBP makes electrostatic and hydrogen-bonding interactions with E1187 and D1191 on the BRD (Fig. S1A). These interactions are buttressed by hydrogen bonding between the side chains of W1159 and D1191 in the BRD and the backbone NH and CO of G1202 in the PHD. In contrast, R1166 of p300, which corresponds to R1203 of CBP, does not interact with the conserved Glu and Asp side chains of the BRD, but is directed toward the HAT, a conformational change that is permitted by substitution of the bulky Y1205 side chain of CBP by the smaller L1168 (Fig. S1B). The resulting rotation of the p300 BRD away from the PHD disrupts the hydrogen bond between the tryptophan indole (W1122) and the glycine (G1165) carbonyl.
No significant electron density for an acetyl-CoA ligand was observed in the cofactor binding site (Fig. S1C), showing that our structure is that of the ligand-free (apo) CBP HAT domain. In contrast, all of the available p300 HAT domain structures contain Ac-CoA substrate or the analogs Lys-CoA, CoA, acetonyl-CoA, or acyl-CoA variants bound in the active site (13, 28–30). The structures of the apo and ligand-bound HAT domains are very similar, even in the Ac-CoA binding site. The CBP and p300 HAT domains have an unusually long L1 loop, which in the p300 structure makes extensive interactions with CoA ligands and with the core of the HAT domain (13, 28, 29). The L1 loop adopts an almost identical structure in the apo CBP HAT, showing that the cofactor binding site, a long channel that can accommodate the acetyl group and the pantetheine arm, is preformed in the absence of Ac-CoA (Fig. S1D). Most of the active-site residues, including the catalytic tyrosine (Y1504 in CBP, Y1467 in p300), adopt almost identical conformations in the apo CBP HAT and in the structure of the p300 HAT bound to the bisubstrate inhibitor Lys-CoA (Fig. S1E); no significant structural changes are required to accommodate either Ac-CoA or substrate.
Demonstration of Intrinsic Disorder in the AL.
Intramolecular inhibition of the bromodomain by Ac-K1596 requires that the AL be long enough and sufficiently flexible to extend ∼65–70 Å from the HAT domain to the active site of the BRD. The AL is rich in charged and polar amino acids and highly deficient in bulky hydrophobic side chains; residues 1,555–1,620 are predicted to be intrinsically disordered (63, 64). We verified that the AL is disordered and flexible in the context of the HAT domain by recording the 1H–15N HSQC spectrum of a 15N-labeled, catalytically inactive (Y1504F mutation) CBP construct containing the BRD, CH2, HAT, ZZ, and the adjacent TAZ2 domain (BCHZT) (Fig. S7). For proteins of the size of BCHZT (93 kDa), resonances from structured regions are not typically observable in the 1H–15N HSQC spectrum due to slow molecular tumbling, but the amide cross peaks of residues in disordered regions with fast dynamic motion can be observed. The 1H–15N HSQC spectrum of BCHZT shows about 50 backbone amide cross peaks, indicating that there are about 50 disordered and flexible residues in this HAT construct. Comparison of the BCHZT spectrum with that of the isolated AL peptide (63 aa) showed that the most of the BCHZT cross peaks overlay with peaks in the 1H–15N HSQC spectrum of the AL peptide (Fig. S7), indicating that most of the cross peaks observed in the spectrum of BCHZT belong to residues in the AL. Cross peaks arising from the N-terminal Glu-rich region of the AL (1,557–1,569) are not observed in the 1H–15N HSQC spectrum of BCHZT (Fig. S7), suggesting that this region may interact with the slowly tumbling globular domains. Because K1596 is located nearly at the center of the disordered AL loop, it could undergo interactions up to 80–100 Å from the site of attachment of AL to the HAT domain, assuming a contour length per amino acid of 4.0 Å (65). Thus, the AL is clearly long enough to allow Ac-K1596 to reach the binding site of the BRD.
Acknowledgments
We thank Gerard Kroon for expert assistance with NMR experiments and Euvel Manlapaz for technical assistance. We thank Drs. Peiqing Sun and Ian MacRae for access to cell culture facilities. This work was supported by NIH Grant CA096865 and by the Skaggs Institute for Chemical Biology. The GM/CA CAT 23-ID-D has been funded in whole or in part with federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104). Use of the APS was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5U7G).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703105114/-/DCSupplemental.
References
- 1.Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 2.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 3.Giordano A, Avantaggiati ML. p300 and CBP: Partners for life and death. J Cell Physiol. 1999;181:218–230. doi: 10.1002/(SICI)1097-4652(199911)181:2<218::AID-JCP4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 4.Arany Z, Sellers WR, Livingston DM, Eckner R. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell. 1994;77:799–800. doi: 10.1016/0092-8674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 5.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 6.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 7.Yuan LW, Giordano A. Acetyltransferase machinery conserved in p300/CBP-family proteins. Oncogene. 2002;21:2253–2260. doi: 10.1038/sj.onc.1205283. [DOI] [PubMed] [Google Scholar]
- 8.Urnov FD, Wolffe AP. Chromatin remodeling and transcriptional activation: The cast (in order of appearance) Oncogene. 2001;20:2991–3006. doi: 10.1038/sj.onc.1204323. [DOI] [PubMed] [Google Scholar]
- 9.Petrij F, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 10.Mullighan CG, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471:235–239. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasqualucci L, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park S, Martinez-Yamout MA, Dyson HJ, Wright PE. The CH2 domain of CBP/p300 is a novel zinc finger. FEBS Lett. 2013;587:2506–2511. doi: 10.1016/j.febslet.2013.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delvecchio M, Gaucher J, Aguilar-Gurrieri C, Ortega E, Panne D. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nat Struct Mol Biol. 2013;20:1040–1046. doi: 10.1038/nsmb.2642. [DOI] [PubMed] [Google Scholar]
- 14.Filippakopoulos P, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kraus WL, Manning ET, Kadonaga JT. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol Cell Biol. 1999;19:8123–8135. doi: 10.1128/mcb.19.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalkhoven E, et al. Loss of CBP acetyltransferase activity by PHD finger mutations in Rubinstein-Taybi syndrome. Hum Mol Genet. 2003;12:441–450. doi: 10.1093/hmg/ddg039. [DOI] [PubMed] [Google Scholar]
- 17.Kalkhoven E, Teunissen H, Houweling A, Verrijzer CP, Zantema A. The PHD type zinc finger is an integral part of the CBP acetyltransferase domain. Mol Cell Biol. 2002;22:1961–1970. doi: 10.1128/MCB.22.7.1961-1970.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rack JGM, et al. The PHD finger of p300 influences its ability to acetylate histone and non-histone targets. J Mol Biol. 2014;426:3960–3972. doi: 10.1016/j.jmb.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 19.Bordoli L, et al. Functional analysis of the p300 acetyltransferase domain: The PHD finger of p300 but not of CBP is dispensable for enzymatic activity. Nucleic Acids Res. 2001;29:4462–4471. doi: 10.1093/nar/29.21.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson PR, et al. Regulation of the p300 HAT domain via a novel activation loop. Nat Struct Mol Biol. 2004;11:308–315. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- 21.Henry RA, Kuo YM, Andrews AJ. Differences in specificity and selectivity between CBP and p300 acetylation of histone H3 and H3/H4. Biochemistry. 2013;52:5746–5759. doi: 10.1021/bi400684q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McManus KJ, Hendzel MJ. Quantitative analysis of CBP- and P300-induced histone acetylations in vivo using native chromatin. Mol Cell Biol. 2003;23:7611–7627. doi: 10.1128/MCB.23.21.7611-7627.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dancy BM, et al. Live-cell studies of p300/CBP histone acetyltransferase activity and inhibition. ChemBioChem. 2012;13:2113–2121. doi: 10.1002/cbic.201200381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ponting CP, Blake DJ, Davies KE, Kendrick-Jones J, Winder SJ. ZZ and TAZ: New putative zinc fingers in dystrophin and other proteins. Trends Biochem Sci. 1996;21:11–13. [PubMed] [Google Scholar]
- 25.Diehl C, et al. Structural analysis of a complex between small ubiquitin-like modifier 1 (SUMO1) and the ZZ domain of CREB-binding protein (CBP/p300) reveals a new interaction surface on SUMO. J Biol Chem. 2016;291:12658–12672. doi: 10.1074/jbc.M115.711325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Girdwood D, et al. P300 transcriptional repression is mediated by SUMO modification. Mol Cell. 2003;11:1043–1054. doi: 10.1016/s1097-2765(03)00141-2. [DOI] [PubMed] [Google Scholar]
- 27.Kuo HY, et al. SUMO modification negatively modulates the transcriptional activity of CREB-binding protein via the recruitment of Daxx. Proc Natl Acad Sci USA. 2005;102:16973–16978. doi: 10.1073/pnas.0504460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maksimoska J, Segura-Peña D, Cole PA, Marmorstein R. Structure of the p300 histone acetyltransferase bound to acetyl-coenzyme A and its analogues. Biochemistry. 2014;53:3415–3422. doi: 10.1021/bi500380f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, et al. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature. 2008;451:846–850. doi: 10.1038/nature06546. [DOI] [PubMed] [Google Scholar]
- 30.Kaczmarska Z, et al. Structure of p300 in complex with acyl-CoA variants. Nat Chem Biol. 2017;13:21–29. doi: 10.1038/nchembio.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Legge GB, et al. ZZ domain of CBP: An unusual zinc finger fold in a protein interaction module. J Mol Biol. 2004;343:1081–1093. doi: 10.1016/j.jmb.2004.08.087. [DOI] [PubMed] [Google Scholar]
- 32.Danielsen JR, et al. DNA damage-inducible SUMOylation of HERC2 promotes RNF8 binding via a novel SUMO-binding Zinc finger. J Cell Biol. 2012;197:179–187. doi: 10.1083/jcb.201106152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng J, Wysocka J. It takes a PHD to SUMO. Trends Biochem Sci. 2008;33:191–194. doi: 10.1016/j.tibs.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Hochstrasser M. SP-RING for SUMO: New functions bloom for a ubiquitin-like protein. Cell. 2001;107:5–8. doi: 10.1016/s0092-8674(01)00519-0. [DOI] [PubMed] [Google Scholar]
- 35.Gareau JR, Lima CD. The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng L, Zhang Q, Gerona-Navarro G, Moshkina N, Zhou MM. Structural basis of site-specific histone recognition by the bromodomains of human coactivators PCAF and CBP/p300. Structure. 2008;16:643–652. doi: 10.1016/j.str.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plotnikov AN, et al. Structural insights into acetylated-histone H4 recognition by the bromodomain-PHD finger module of human transcriptional coactivator CBP. Structure. 2014;22:353–360. doi: 10.1016/j.str.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Das C, et al. Binding of the histone chaperone ASF1 to the CBP bromodomain promotes histone acetylation. Proc Natl Acad Sci USA. 2014;111:E1072–E1081. doi: 10.1073/pnas.1319122111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schiltz RL, et al. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J Biol Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- 40.Pasini D, et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res. 2010;38:4958–4969. doi: 10.1093/nar/gkq244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tie F, et al. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ragvin A, et al. Nucleosome binding by the bromodomain and PHD finger of the transcriptional cofactor p300. J Mol Biol. 2004;337:773–788. doi: 10.1016/j.jmb.2004.01.051. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen UTT, et al. Accelerated chromatin biochemistry using DNA-barcoded nucleosome libraries. Nat Methods. 2014;11:834–840. doi: 10.1038/nmeth.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borah JC, et al. A small molecule binding to the coactivator CREB-binding protein blocks apoptosis in cardiomyocytes. Chem Biol. 2011;18:531–541. doi: 10.1016/j.chembiol.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zucconi BE, et al. Modulation of p300/CBP acetylation of nucleosomes by bromodomain ligand I-CBP112. Biochemistry. 2016;55:3727–3734. doi: 10.1021/acs.biochem.6b00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oates ME, et al. D2P2: Database of disordered protein predictions. Nucleic Acids Res. 2013;41:D508–D516. doi: 10.1093/nar/gks1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCullough CE, Marmorstein R. Molecular basis for histone acetyltransferase regulation by binding partners, associated domains, and autoacetylation. ACS Chem Biol. 2016;11:632–642. doi: 10.1021/acschembio.5b00841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X, et al. Catalytic mechanism of histone acetyltransferase p300: From the proton transfer to acetylation reaction. J Phys Chem B. 2014;118:2009–2019. doi: 10.1021/jp409778e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bose DA, et al. RNA binding to CBP stimulates histone acetylation and transcription. Cell. 2017;168:135–149.e22. doi: 10.1016/j.cell.2016.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Black JC, Choi JE, Lombardo SR, Carey M. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell. 2006;23:809–818. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 52.Mujtaba S, et al. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol Cell. 2004;13:251–263. doi: 10.1016/s1097-2765(03)00528-8. [DOI] [PubMed] [Google Scholar]
- 53.Ivanov AV, et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell. 2007;28:823–837. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jackson PK. A new RING for SUMO: Wrestling transcriptional responses into nuclear bodies with PIAS family E3 SUMO ligases. Genes Dev. 2001;15:3053–3058. doi: 10.1101/gad.955501. [DOI] [PubMed] [Google Scholar]
- 55.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 56.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 57.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 59.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 61.Goddard TD, Kneller DG. 2006. SPARKY 3 (University of California, San Francisco), Version 3.115.
- 62.Macauley MS, et al. Structural and dynamic independence of isopeptide-linked RanGAP1 and SUMO-1. J Biol Chem. 2004;279:49131–49137. doi: 10.1074/jbc.M408705200. [DOI] [PubMed] [Google Scholar]
- 63.Dosztányi Z, Csizmok V, Tompa P, Simon I. IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 64.Peng K, et al. Optimizing long intrinsic disorder predictors with protein evolutionary information. J Bioinform Comput Biol. 2005;3:35–60. doi: 10.1142/s0219720005000886. [DOI] [PubMed] [Google Scholar]
- 65.Ainavarapu SR, et al. Contour length and refolding rate of a small protein controlled by engineered disulfide bonds. Biophys J. 2007;92:225–233. doi: 10.1529/biophysj.106.091561. [DOI] [PMC free article] [PubMed] [Google Scholar]