Significance
Escalating antibiotic resistance has stimulated interest in understanding the mechanisms of phosphoglycosyl transferases (PGTs) that initiate glycoconjugate biosynthesis and are implicated in bacterial survival and pathogenicity. This study provides compelling evidence that the action of PglC, a prototypic dual domain PGT, proceeds through a unique covalent phosphosugar enzyme intermediate. Thus, the reaction course differs significantly from the mechanism of polytopic PGTs, illustrating nature’s ability to catalyze a biologically critical phosphosugar transfer reaction by two fundamentally different mechanisms. Dual domain PGTs, despite their widespread distribution throughout bacteria, have not been extensively studied to date. This analysis of dual domain PGTs will fuel approaches targeted at the development of small molecule inhibitors of critical first steps in bacterial glycoconjugate biosynthesis.
Keywords: phosphoglycosyl transferase, membrane protein, dual domain PGT, covalent intermediate, glycoconjugate biosynthesis
Abstract
Phosphoglycosyl transferases (PGTs) are integral membrane proteins with diverse architectures that catalyze the formation of polyprenol diphosphate-linked glycans via phosphosugar transfer from a nucleotide diphosphate-sugar to a polyprenol phosphate. There are two PGT superfamilies that differ significantly in overall structure and topology. The polytopic PGT superfamily, represented by MraY and WecA, has been the subject of many studies because of its roles in peptidoglycan and O-antigen biosynthesis. In contrast, less is known about a second, extensive superfamily of PGTs that reveals a core structure with dual domain architecture featuring a C-terminal soluble globular domain and a predicted N-terminal membrane-associated domain. Representative members of this superfamily are the Campylobacter PglCs, which initiate N-linked glycoprotein biosynthesis and are implicated in virulence and pathogenicity. Despite the prevalence of dual domain PGTs, their mechanism of action is unknown. Here, we present the mechanistic analysis of PglC, a prototypic dual domain PGT from Campylobacter concisus. Using a luminescence-based assay, together with substrate labeling and kinetics-based approaches, complementary experiments were carried out that support a ping-pong mechanism involving a covalent phosphosugar intermediate for PglC. Significantly, mass spectrometry-based approaches identified Asp93, which is part of a highly conserved AspGlu dyad found in all dual domain PGTs, as the active-site nucleophile of the enzyme involved in the formation of the covalent adduct. The existence of a covalent phosphosugar intermediate provides strong support for a ping-pong mechanism of PglC, differing fundamentally from the ternary complex mechanisms of representative polytopic PGTs.
To advance progress toward strategies for combating the escalating problem of antibiotic resistance, there is increasing focus on the mechanisms of enzymes involved in bacterial survival and virulence. As such, the polyprenol phosphate (Pren-P) C-1-phosphoglycosyltransferases (PGTs) represent an important class of enzymes because of the critical roles they play in bacterial pathogenicity through catalysis of key steps in the biosynthesis of complex glycoconjugates. PGTs are integral membrane proteins that mediate transfer of a phosphosugar moiety from a soluble nucleotide diphosphate-sugar (NDP-sugar) donor to a Pren-P acceptor. Because PGTs function at the beginning of many diverse membrane-associated glycoconjugate assembly pathways, they are also referred to as “priming” or “initiating” glycosyl transferases (GTs) (1, 2) even though they are formally PGTs. For example, MraY (3, 4), the ubiquitous bacterial phospho-N-acetylmuramyl-pentapeptide-transferase, catalyzes formation of the first lipid-linked intermediate in peptidoglycan biosynthesis (5, 6). The Campylobacter jejuni PglC (7, 8) and the Neisseria gonorrhoeae PglB (9) enzymes catalyze the first steps in N- and O-linked glycoprotein biosynthesis, respectively, and WecA, in Gram-negative bacteria (3, 10), initiates the lipopolysaccharide O-antigen biosynthesis pathway (11). In addition, although far more abundant in bacteria because of the greater variety of glycoconjugates, PGTs are also important in eukaryotes. For example, the mammalian dolichol phosphate GlcNAc-1-P transferase (GPT) (12), which is a WecA homolog, is an essential eukaryotic PGT that functions at the beginning of the dolichol pathway for N-linked protein glycosylation (13).
There are two PGT superfamilies, which display distinct predicted structures and membrane topologies. PGTs including MraY, WecA, and GPT are polytopic membrane proteins with 10–11 predicted transmembrane α-helical domains that align key cytoplasmic loops for catalysis (14). To date, MraY is the only PGT with an experimentally determined X-ray crystal structure, providing a structural basis for enzyme function (15), as well as the opportunity to develop models for substrate and inhibitor binding (16, 17). Long-standing interest in the development of antibiotics has prompted mechanistic analysis of both MraY and WecA, which are the best studied of the polytopic PGTs. Ultimately, although studies on MraY initially suggested a substituted enzyme “ping-pong” mechanism (4, 18–20), recent investigations have provided evidence that both MraY and WecA follow a ternary complex mechanism (21, 22).
In contrast to the polytopic PGTs, less is known about the other superfamily, despite the prevalence of homologous sequences in many diverse bacterial glycoconjugate biosynthetic pathways (8). To better understand this superfamily, we carried out bioinformatics analysis on 15,000 sequences from the three related PGT families, which are mainly distinguished by auxiliary C- or N-terminal domains (1, 8). The least elaborated member of the superfamily is represented by PglC from the Campylobacter genus, which lacks appended domains. PglC catalyzes the reaction of a linear Pren-P (e.g., C55) with UDP-di-N-acetylbacillosamine (UDP-diNAcBac) to form the first membrane-associated intermediate (Pren-PP-diNAcBac) in the bacterial N-linked protein glycosylation (pgl) pathway. Extensive homology analysis reveals a deceptively simple “dual domain” architecture, in which the minimal functional PGT unit is represented by a small (ca. 180 residues) soluble globular C-terminal domain and a single predicted N-terminal membrane-inserted domain (ca. 20 residues) (1, 8). Using the aforementioned bioinformatics results as a guide, we mutated several highly conserved amino acids to identify key catalytic residues and applied conservation covariance analysis to generate a structural model of the C. jejuni PglC.
Despite the similarities in the overall reaction, the predicted structural and topological distinctions between the PGT enzyme superfamilies implied that the mechanisms could either differ or converge to a common catalytic path. As a bisubstrate enzyme, a PGT such as PglC can potentially follow a mechanism similar to the polytopic PGTs, involving the formation of a ternary complex (Fig. 1A) (21, 22). In this case, substrate binding and product release can either be ordered or random (23). Alternatively, the enzyme may operate through a ping-pong mechanism, involving, first, formation of an enzyme-substrate covalent intermediate and release of UMP, followed by nucleophilic attack on the covalent adduct by the Pren-P substrate to generate the Pren-PP–linked product in the second half of the reaction (Fig. 1B).
Fig. 1.
Potential mechanisms of PglC, a prototypic dual domain PGT. (A) The sequential mechanism involves formation of a noncovalent ternary complex featuring both substrates. (B) The ping-pong mechanism involves reaction of an active-site nucleophile with the α-phosphate of the NDP-sugar substrate in step 1 to generate a covalent enzyme-substrate intermediate with release of UMP. In step 2, the covalent enzyme intermediate undergoes nucleophilic attack by the second substrate to generate the product. [ENZ-NU represents a reactive nucleophilic functional group (NU) as part of the enzyme (ENZ) active site.]
Here we report mechanistic analysis of PglC, a prototypic member of the dual domain PGT superfamily. The studies reveal a stepwise ping-pong mechanism and the intermediacy of a covalent phosphosugar adduct. We anticipate that the mechanistic insight provided by this research will fuel further functional and structural analysis on this important class of glycoconjugate biosynthesis enzymes.
Results and Discussion
Recent research on the dual domain Campylobacter PGTs (8) forms the foundation for this study. Although the C. jejuni PglC has been the subject of the majority of biochemical studies (7, 8, 24), the Campylobacter concisus enzyme, which shares 72% sequence identity, has proven to be more tractable to overexpression, solubilization, and purification, which is critical for mechanistic studies (SI Appendix, Fig. S1). By applying a luminescent PGT assay (25), radioactivity and mass spectrometry-based approaches, and enzyme kinetic analysis, we provide insight into the mechanism of this dual domain PGT. The potential mechanisms of PglC (Fig. 1) can be distinguished by the timing of UMP release and the intermediacy of a covalent enzyme-substrate intermediate. Additional complementary information can be derived from tracer experiments (4, 21) and kinetic analysis (26).
Monitoring UMP Release in the Reaction of PglC with UDP-diNAcBac.
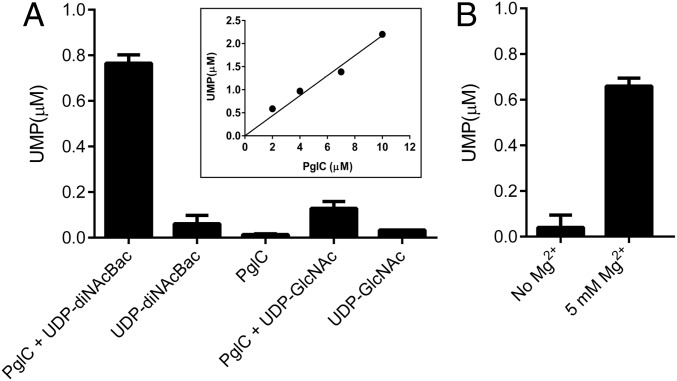
PglC catalyzes transfer of α-C1′-phospho-d-diNAcBac from the soluble UDP-diNAcBac donor to the membrane-associated Pren-P acceptor to afford Pren-PP-diNAcBac and UMP (7, 25). Therefore, measurement of UMP using a luminescence-based assay (UMP/CMP-Glo; Promega) (25) in the PglC reaction can be applied to assess the timing of UMP release in the absence of the Pren-P acceptor, as well as the overall enzyme activity. When 4 µM PglC was reacted with 80 µM UDP-diNAcBac in the absence of Pren-P, 0.75 µM UMP was generated in 20 min (Fig. 2A). The amount of UMP release was significantly above the lower limits of UMP detection measurable using the UMP-Glo reagent (∼0.05 µM) (25) and corresponded to conversion of 20% of the PglC. Release of UMP was proportionate to PglC concentration up to at least 10 µM (Fig. 2A, inset) and dependent on the presence of both PglC and UDP-diNAcBac (Fig. 2A). Further analysis showed that UMP release in the first half reaction, in the presence of 1 µM PglC and 80 µM UDP-diNAcBac, was complete in less than 5 s (SI Appendix, Fig. S2). The PglC reaction was also carried out in the presence of a noncognate NDP-sugar, UDP-GlcNAc, which is the preferred substrate for several other PGTs including WecA (10, 11, 27) and the avian GPT (26, 28). When 4 µM PglC was reacted with 80 µM UDP-GlcNAc, only a trace of UMP (0.13 µM) was detected (Fig. 2A). This shows that PglC accepts UDP-GlcNAc as a substrate, albeit with far poorer efficiency relative to UDP-diNAcBac, and underscores that the UMP release is substrate dependent and enzyme catalyzed.
Fig. 2.
UMP release in the reaction of PglC with UDP-diNAcBac. (A) In the absence of Und-P 0.75 µM, UMP was released in the reaction of 4 µM PglC with 80 µM UDP-diNAcBac in 20 min. Under similar conditions, control experiments afforded 0.01–0.13 µM UMP. (Inset) Correlation of UMP release with PglC concentration (2–10 µM). (B) In the absence of Mg2+, negligible UMP (0.04 µM) was produced. When the assay was supplemented with 5 mM Mg2+, 0.66 µM UMP was released. Assays were carried out in duplicate. Error bars represent mean ± SD.
The possible copurification of PglC with endogenous Pren-P (29) was rigorously assessed, as residual Pren-P could account for the observed UMP release in the reaction of PglC with UDP-diNAcBac, in the absence of exogenous Pren-P. Radioactivity-based assays were performed by reacting PglC with UDP-[3H]-diNAcBac (8, 30), wherein contamination with copurifying Pren-P in the PglC preparation would manifest as a transfer of radioactivity from UDP-[3H]-diNAcBac to Pren-P to generate the organic-soluble Pren-PP-[3H]-diNAcBac (7, 8). When 10 µM PglC was reacted with 65 µM UDP-[3H]-diNAcBac (5.4 mCi/mmol, corresponding to 12,000 dpm/nmol) and the reaction was extracted with chloroform-methanol, minimal radioactivity (317 ± 54 dpm) at a level comparable to that (315 ± 25 dpm) in the organic extract of a PglC-free control reaction was observed. The radioactivity in the organic extracts in both experiments was extremely low compared with the total radioactivity (2.9 nmol of 65 µM UDP-[3H]-diNAcBac, 34,800 dpm) supplied in the assay. Together, these results demonstrate that the PglC in the UMP release experiments did not contain residual endogenous Pren-P at a level that could obscure the results. In a complementary experiment, UMP-Glo was used to quantify the UMP produced in a similar reaction that also contained 10 µM PglC and 80 µM UDP-diNAcBac. In this case, 2.2 µM UMP was detected in the reaction. If an equivalent amount of P-diNAcBac had been transferred to Pren-P in the radioactivity-based assay, this would have resulted in the transfer of radioactivity (918 dpm) to the organic extract. Together, these experiments ruled out the presence of confounding levels of endogenous Pren-P in the purified PglC.
Validation of enzyme homogeneity is critical. Previously, mechanistic studies on MraY suggested that this polytopic PGT followed a ping-pong mechanism involving a covalent enzyme–substrate intermediate (4, 18, 19). This was based on evidence derived from tracer studies showing exchange of radiolabeled UMP into the NDP-sugar substrate in the absence of an exogenous Pren-P (18), kinetic analysis (19), and the apparent detection of an enzyme–substrate covalent intermediate (4). However, these studies were carried out using crude or partially purified enzyme preparations, which would have suffered from contamination with endogenous Pren-P, thus complicating the analyses. Recently, isotope exchange studies performed on purified WecA and MraY support a one-step, ternary complex mechanism (21, 22).
UMP release in the first half of the reaction (Fig. 1B) occurs in the absence of Pren-P, and thus suggests the intermediacy of a covalent enzyme–substrate intermediate. This result represents a significant departure from the known mechanisms of the polytopic PGT superfamily such as MraY, WecA, and GPT, which follow a one-step ternary complex mechanism that does not involve a covalent enzyme intermediate (21, 22, 26).
Essentiality of Mg2+ in UMP Release in the Reaction of PglC with UDP-diNAcBac.
Mg2+ is an essential cofactor in polytopic PGTs such MraY (4, 31), WecA (10, 32), and GPT (26, 28), and in dual domain PGTs such as WbaP (33) and PglC (7). Our recent sequence alignment and mutagenesis analyses on dual domain PGTs (8) highlight an adjacent, highly conserved Asp-Glu dyad (D92/E93 in C. jejuni and D93/E94 in C. concisus) as possible Mg2+ coordinating residues that might serve a key function in the dual domain PGTs. Interestingly, previous biochemical studies on other PGT mutants have also identified adjacent conserved acidic residues in the dual domain WbaP (D382/E383) and the polytopic WecA (D90/D91), and these residues have been implicated in coordinating to Mg2+ (32, 33).
As proposed for MraY and WecA (4, 21), the essential Mg2+ in PglC may coordinate to the diphosphate of the NDP-sugar substrate and to acidic residues. In this case, Mg2+ coordination would neutralize the electrostatic repulsion between the negatively charged phosphate and acidic residues, as observed in the catalysis of phosphotransferase enzymes of the HAD superfamily (34). Moreover, coordination of Mg2+ to the diphosphate group could assist in substrate orientation for catalysis and also increase the electrophilicity of the α-phosphate in the NDP-sugar by polarizing the P–O bond for the subsequent nucleophilic attack. Such effects would be reminiscent of the role of Mg2+ in enzymes such as phosphoenolpyruvate carboxykinase (35, 36) and DNA gyrase B (37).
We investigated the essentiality of Mg2+ in the PglC reaction in the absence of Pren-P and in the presence of UDP-diNAcBac to establish the catalytic relevance of UMP release as an “on path” process. PglC assays with UDP-diNAcBac were carried out with apoenzyme and Mg2+-free buffers. Under these conditions, when assays were carried out using 4 µM PglC and 80 µM UDP-diNAcBac, negligible UMP release was detected using UMP/CMP-Glo (Fig. 2B), confirming that UMP release was Mg2+-dependent. On addition of 5 mM Mg2+, ∼90% of the enzyme activity was recovered (Fig. 2B). Therefore, Mg2+ plays an indispensable role in the half reaction of PglC with UDP-diNAcBac (Fig. 1B).
Structural analysis of PglC in complex with cofactors and ligands would be necessary to define the mechanistic course of the PglC reaction. However, based on precedence with other Mg2+-dependent phosphoryl transfer enzymes, it is likely that the Mg2+ plays a role in activating the UDP-sugar for α-phosphate nucleophilic attack by the conserved Asp in the Asp-Glu dyad and that the adjacent Glu residue may assist in UMP release and Pren-P attack to complete the reaction.
Radioactivity-Based UMP-Exchange Analysis.
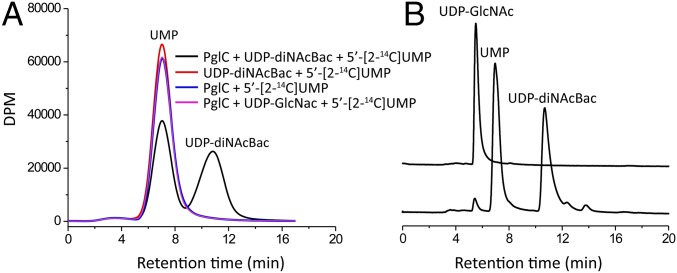
Tracer studies, previously used to probe the mechanisms of MraY and WecA (18–21), were also applied to PglC. Specifically, we examined the reaction of PglC with UDP-diNAcBac in the presence 5′-[2-14C] UMP and absence of Pren-P. Because of the reversible nature of this step, exogenous UMP should be incorporated into UDP-diNAcBac. To probe exchange, 10 µM PglC was reacted with 100 µM UDP-diNAcBac in the presence of 100 µM 5′-[2-14C] UMP (50 mCi/mmol, corresponding to 111,000 dpm/nmol) in the assay. After 1 h, the components of the reaction were separated by RP-HPLC, and the fractions were quantified by scintillation counting. This analysis showed exchange of ∼50% radioactivity of 5′-[2-14C] UMP with the UMP moiety of the UDP-diNAcBac substrate (Fig. 3A). In the absence of PglC, no radioactivity exchange was observed, confirming that the UMP exchange was enzyme-dependent. Furthermore, incubation of PglC with UDP-GlcNAc in the presence of 5′-[2-14C] UMP did not reveal significant incorporation of 5′-[2-14C] UMP into UDP-GlcNAc (Fig. 3A). These studies demonstrate that PglC catalyzes exchange of externally added UMP into UDP-diNAcBac in a substrate-specific manner in the absence of Pren-P. Overall, these results diverge from the recent reports on similar tracer experiments carried out on the polytopic PGTs, which showed that Pren-P was indispensable for UMP exchange reactions (21).
Fig. 3.
Radioactivity-based UMP-exchange assays. (A) Incubation of 10 µM PglC with 100 µM UDP-diNAcBac in the presence of exogenous 100 µM 5′-[2-14C] UMP led to incorporation of ∼50% radioactivity of 5′-[2-14C] UMP into UDP-diNAcBac (black). Control reactions in the absence of the enzyme (red) or with 100 µM UDP-GlcNAc (magenta) instead of UDP-diNAcBac did not show radioactivity incorporation. (B) HPLC traces of UDP-GlcNAc, UMP, and UDP-diNAcBac.
LC-MS/MS Analysis of the Covalent Enzyme-Substrate Intermediate.
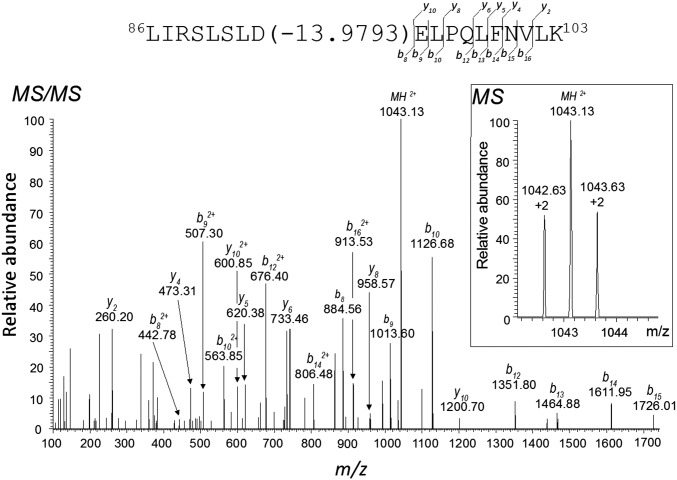
Complementary experiments provide evidence supporting a ping-pong mechanism for PglC catalysis; however, a unique and unequivocal feature of such a mechanism is the formation of a covalent enzyme-substrate intermediate. On the basis of strong precedent from mechanistic studies on enzymes such as phosphotransferases (38, 39) and P-type ATPases (40), wherein the formation of a covalent aspartyl-phosphate has been demonstrated, we hypothesized that a conserved acidic residue of PglC might serve as an active-site nucleophile in a chemically analogous step. In this case, nucleophilic attack on the α-phosphate of UDP-diNAcBac would afford an acyl phosphosugar-enzyme intermediate. However, although acyl phosphate intermediates are widely recognized, the corresponding acyl phosphosugar intermediate is unprecedented and its stability is unknown. To investigate intermediacy of a covalent adduct, MS-based experiments were carried out. Initial studies involved incubation of PglC with UDP-diNAcBac and direct analysis of the reaction mixture by LC-MS. However, this approach was unsuccessful for direct detection of a covalent intermediate, potentially as a result of lability under the experimental conditions. Therefore, a chemical trapping approach, following protocols used in the study of phosphotransferases (38) was adopted. In this method, PglC was incubated with UDP-diNAcBac, followed by treatment of the sample with sodium borohydride (NaBH4). In this case, the acyl phosphosugar intermediate, if generated, would be reduced to the corresponding protein-based alcohol, which would be amenable to protease digestion and MS analysis. Experiments were carried out by reacting PglC with UDP-diNAcBac, recovering the protein by precipitation, followed by NaBH4 treatment. LC-MS/MS analysis of the LysC digested samples revealed a modified peptide, 86LIRSLSLDELPQLFNVIK103, with a molecular weight decreased by 13.9793 Da (Fig. 4) relative to the mass of the native peptide. This observation supports reduction of either an aspartyl or glutamyl phosphosugar derivative to the corresponding alcohol in the unique PglC peptide. Subsequent fragmentation of the modified peptide and analysis of the resulting b and y ions (Fig. 4; SI Appendix, Table S1) allowed identification of D93 as the modified residue. This result strongly supports D93 as the active-site nucleophile that reacts with UDP-diNAcBac to generate an acyl phosphosugar enzyme intermediate, which is reduced by NaBH4 to generate homoserine. This finding was consistent among various samples generated by incubating PglC with different amounts of UDP-diNAcBac. MS-based analysis of samples that had not been exposed to the UDP-sugar did not exhibit modification of the 86LIRSLSLDELPQLFNVIK103 peptide (SI Appendix, Fig. S3 and Table S2).
Fig. 4.
Mass spectrometric analysis of the covalent enzyme–substrate intermediate. Analysis was performed on a LysC digest of PglC that had been reacted with UDP-diNAcBac and then treated with NaBH4. The PglC-derived peptide 86LIRSLSLDELPQLFNVLK103 was identified as the modified peptide with a mass reduction of 13.9793 Da, which was determined from the observed mass of the peptide 1,043.13 Da (Inset with isotopic masses, z = +2). The MS/MS spectrum of the peptide exhibited several b and y ions, characteristic of D93 modification in the peptide.
The MS analysis also showed a peak at a later retention time characterized by a mixed MS/MS spectrum (SI Appendix, Fig. S4). The peptide-mixture was an order of magnitude less abundant relative to the aforementioned modified peptide (SI Appendix, Table S3). Interestingly, one of the peptides in the mixture was also the 86LIRSLSLDELPQLFNVIK103 peptide with the same modification on D93. The second peptide was a similar species, 86LIRSLSLDELPQLFNVIK103, but with modification on E94. This observation suggests that in a minor population of the enzyme, phosphosugar migration may have occurred between adjacent acidic residues before NaBH4 reduction during sample processing.
Support for a Ping-Pong Mechanism from Kinetic Analysis.
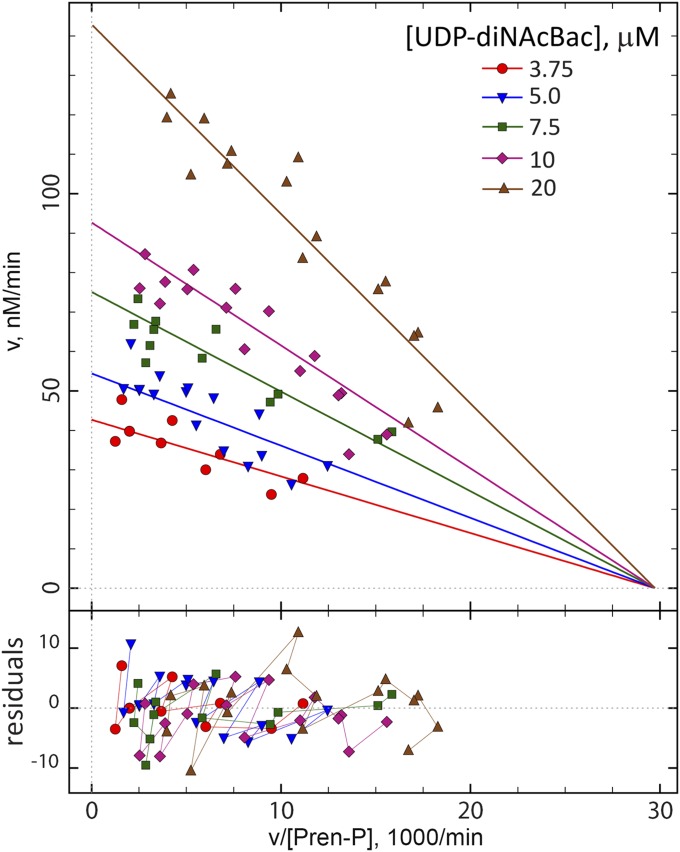
Kinetic experiments were performed to assess the initial rate of PglC-catalyzed reactions as a function of each of the substrates. For each reaction, the rate was measured by quantifying UMP production using UMP-Glo (25) (SI Appendix, Table S4). The initial reaction rates were fit to a variety of alternate kinetic mechanisms (SI Appendix, Methods, Figs. S5 and S6, and Tables S5 and S6 for details of the mathematical and statistical procedures). Based on a combination of statistical model selection measures (41–43), including in particular the nonsymmetrical confidence interval estimation (profile-t) method (44–46) (SI Appendix, Fig. S7), the ping-pong model was selected as the most plausible candidate. Adherence to the ping-pong mechanism is graphically illustrated in Fig. 5 by a set of intersecting straight lines in the Eadie-Hofstee plot. Together, the analysis suggests that PglC catalysis very likely occurs through a ping-pong mechanism. The steady-state kinetic parameters obtained from the global fitting are kcat = 1,540 ± 120 min−1 (∼26 s−1); Km(Und-P) = 9.9 ± 1.1 µM; Km(UDP-diNAcBac) = 22.9 ± 2.5 µM (kcat/Km(Und-P) = 2.59 × 106 M−1⋅s−1 and kcat/Km(UDP-diNAcBac) = 1.12 × 106 M−1⋅s−1). Kinetic analyses were performed by using the software package DynaFit (47, 48).
Fig. 5.
Initial rate kinetic analysis of PglC. Initial rates (v, vertical axis) were fit to alternative kinetic mechanisms (SI Appendix, Methods). The best-fit theoretical curves corresponding to the ping-pong kinetic mechanism are shown in the Eadie-Hofstee coordinates [horizontal axis: initial rate v divided by the substrate concentrations (Pren-P)]. The values on the x-axis are multiplied by 1,000. Symbols and best-fit lines correspond to fixed concentrations of UDP-diNAcBac. The intersection point on the horizontal axis illustrates the involvement of the ping-pong mechanism.
Conclusions
PGT enzymes catalyze the first membrane-committed step in biosynthetic pathways of diverse glycoconjugates. The complex biomolecules produced in these pathways play critical roles in eukaryotic biology and bacterial survival, communication, and pathogenesis. The current studies reveal that the mechanism of PglC, a prototypic dual domain PGT, differs significantly from that of the polytopic PGTs that have been described to date. For example, MraY and WecA, members of different families of the polytopic PGTs, catalyze phosphoglycosyl transfer via the intermediacy of a ternary complex (21, 22). In contrast, our studies, involving several complementary lines of evidence, support a ping-pong mechanism with the intermediacy of a covalent acyl phosphosugar enzyme derivative at Asp93 in the C. concisus PglC. Asp93 is essential for catalysis and features in a highly conserved AspGlu dyad found in thousands of dual domain PGTs, including all three identified subfamilies (8). In addition, we provide clear evidence for the catalytic relevance of a discrete first step in the PGT reaction by quantifying UMP release and monitoring exchange of exogenous UMP with the UDP-sugar in the presence and absence of native substrate, substrate analogs, and the Mg2+ cofactor.
Understanding the mechanism of dual domain PGTs has fundamental and practical implications. Questions certainly emerge as to why the PGT superfamilies evolved independently and whether the existence of PGTs with topologically different architectures might provide an advantage. Now, experiments can be designed to probe whether the identity of the PGT might facilitate recruitment or “sorting” of enzymes and substrates into specific pathways, providing an advantage with respect to pathway efficiency and fidelity (49). For example, many bacteria express polytopic WecA and MraY homologs that catalyze biosynthesis of Pren-PP-linked glycans on the cytosolic face of the inner membrane, yet in those same bacteria, dual domain PGTs, which are involved in multistep capsular polysaccharide or glycoprotein biosynthesis, occur in the same cellular location. It is also intriguing that the major biosynthetic pathway in eukaryotes to use a PGT is the dolichol pathway for N-linked protein glycosylation, and this pathway exclusively exploits the polytopic PGTs, which are WecA homologs. Indeed, the dual domain PGTs seem to be found exclusively in bacteria (1, 8).
Knowledge of the mechanism of the dual domain PGTs will also add important insight into inhibitor development, particularly because these enzymes are as yet structurally uncharacterized. Pathway-specific dual domain PGT inhibitors would represent valuable tools for unraveling key glycoconjugate biosynthesis pathways and potential leads toward the development of therapeutic agents to mitigate the virulence of microbial pathogens.
In conclusion, these studies provide an intriguing example of the evolution of alternative solutions to challenges in catalysis. A challenge that is, in this case, constrained as the enzymes initiate sequential biochemical pathways that are localized at the membrane interface. Evolutionary pressures, including originating from different protein scaffolds, may have led to the adoption of different PGT reaction mechanisms in structurally and topologically distinct enzyme superfamilies.
Materials and Methods
Materials.
All standard chemical and biochemical reagents were obtained at the highest purity possible. The 5′-[2-14C] UMP (Cat. ARC 1230) was purchased from American Radiolabeled Chemicals, Inc. Ultrapure UDP-GlcNAc (Cat. V7071) and the UMP/CMP-Glo glycosyl transferase assay reagent were obtained from Promega.
Expression and Purification of PglC from C. concisus.
PglC was expressed and purified as described in the SI Appendix (SI Appendix, Methods and Fig. S1).
Measurement of UMP Using UMP-Glo.
UMP/CMP-Glo (Promega) was used for measuring UMP, as described previously (25). Briefly, at the end of the reaction, a 15-µL aliquot was quenched with the UMP-Glo reagent, mixed gently, and transferred to a 96-well plate (Corning; white, flat bottom, nonbinding surface, half area). A SynergyH1 multimode plate reader (Biotek) was used to measure luminescence. The 96-well plate was shaken inside the plate reader chamber at 237 cpm at 25 °C in the double orbital mode for 16 min, followed by 44 min incubation at the same temperature, after which time the luminescence was recorded (gain: 200, integration time: 0.5 s). Conversion of luminescence to UMP concentration was carried out using a standard curve (25).
UMP Release in the Reaction of PglC with UDP-diNAcBac.
Assays were performed with 4 µM PglC and 80 µM UDP-diNAcBac at room temperature for 20 min in assay buffer containing 50 mM Hepes at pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.1% Triton X-100, and 10% DMSO. PglC was varied from 2 to 10 µM for measuring the concentration dependence of UMP release. A similar assay with 1 µM PglC and 80 µM UDP-diNAcBac was carried out with quenching at shorter times (5, 15, 30, and 60 s) (SI Appendix, Fig. S2). Control assays were performed in the absence of PglC and UDP-diNAcBac, and also in the presence of 4 µM PglC and 80 µM of UDP-GlcNAc.
Radioactivity-Based Assays to Investigate the Presence of Pren-P in Purified PglC.
Assays were performed in quadruplicate in 45 µL volume by reacting 10 µM PglC with 65 µM UDP-[3H]-diNAcBac (5.4 mCi/mmol, corresponding to 12,000 dpm/nmol). The [3H] label was incorporated in the C4′-N-acetyl moiety as described previously (30). Control experiments were carried out in the absence of PglC. Reactions were incubated for 20 min at room temperature and then quenched with 1 mL CHCl3:MeOH (2:1). The lower organic layer was washed thoroughly with 6 × 400 µL PSUP (Pure Solvent Upper Phase composed of 235 mL H2O, 240 mL MeOH, 15 mL CHCl3, and 1.83 g KCl). The resulting organic and aqueous layers were combined with 5 mL OptiFluor (PerkinElmer) and 5 mL EcoLite (MP Biomedicals) scintillation fluids, respectively, and radioactivity was measured using scintillation counting (Beckman Coulter LS 6500). The low level of radioactivity observed in the organic extracts of both reactions is ascribed to the partial solubility of UDP-[3H]-diNAcBac in the organic phase.
Measuring the Effect of Mg2+ on UMP Release.
Assays were performed using PglC that had been treated with Chelex 100 resin (Bio-Rad, Cat. 143-2832, sodium form) to remove divalent metal ions from the enzyme. A sample of 70 µM PglC (200 µl) was treated twice with 14 mg resin for 10 min at 4 °C, followed by centrifugation at 2,000 × g for 2 min to separate the resin. In addition, the assay buffer (50 mM Hepes at pH 7.5, 150 mM NaCl, 0.1% Triton X-100, and 10% DMSO) and UDP-diNAcBac solution were treated with Chelex. Assays were performed in a 15-µL volume at room temperature for 20 min by reacting 4 µM PglC with 80 µMUDP-diNAcBac in the presence of 0.5 mM EDTA to remove any residual Mg2+ from the reaction. After the reaction, samples were boiled for 10 min to inactivate PglC, cooled to room temperature, and centrifuged at 14,000 × g for 10 min. The resulting solutions were then supplemented with 0.5 mM Mg2+ to saturate the EDTA before addition of the UMP-Glo reagent to the solutions for UMP measurement. Control assays were also performed with the addition of 5 mM Mg2+.
Radioactivity-Based UMP Exchange Assays.
Assays were performed in 25 µL volume by reacting 10 µM PglC with 100 µM UDP-diNAcBac in the presence of 100 µM 5′-[2-14C] UMP (50 mCi/mmol) for 1 h at room temperature. The samples were then boiled for 10 min, followed by centrifugation at 14,000 × g for 10 min to remove the precipitated protein. Separation of a 10-µL aliquot of the reaction was carried out by RP-HPLC (C18 column, YMC-Pack-ODS-A, 250 × 4.6 nm I.D.; Cat. AA12S05-2546WT) on a Waters 600 system coupled to a UV detector. A buffer containing 50 mM ammonium formate at pH 4.2 was used as the eluent after a previously reported protocol (21) at an isocratic flow (1 mL/min). Elution fractions were combined with EcoLite liquid scintillation mixture (5 mL mixture/mL fraction), and radioactivity was measured using scintillation counting. Control experiments were performed by systematically eliminating PglC or UDP-diNAcBac from the assay. Control assays were also performed with 10 µM PglC and 100 µM UDP-GlcNAc in the presence of 100 µM 5′-[2-14C] UMP.
LC-MS Analysis of PglC Reaction with UDP-diNAcBac.
Assays were performed by incubating varying amounts of PglC (4–70 µM) with 100 µMUDP-diNAcBac in a 25-µL reaction for various times (5 s–20 min) at room temperature. After the reaction, the samples were analyzed by LC-MS, which involves elution of the sample through a C18 column using a water (0.1% TFA): acetonitrile (0.1% TFA) gradient.
LC-MS/MS Analysis of the Covalent Enzyme-Substrate Intermediate in the PglC Reaction.
Assays were performed by reacting 100 µL of 70 µM PglC (160 µg) with various concentrations of UDP-diNAcBac (70, 140, and 280 µM) at room temperature for 20 min. A control experiment was also performed in the absence of UDP-diNAcBac. Assays were quenched with ice-cold trichloroacetic acid (to 16%) and kept on ice for 20 min to precipitate the protein. To complete precipitation, the samples were further incubated at 30 °C for 10 min, followed by incubation on ice for 5 min. The precipitate was separated by centrifugation (10,000 × g for 10 min) at 4 °C, washed with 3 × 400 µL 10 mM ice-cold HCl, dried under vacuum, and dissolved in 20 µL DMSO. To this solution, 15 µL 0.1 M NaBH4 in DMSO was added and incubated at 30 °C for 10 min, followed by addition of 1 mL ice-cold perchloric acid (0.44 M). The mixture was incubated on ice for 30 min, followed by centrifugation at 10,000 × g for 20 min at 4 °C. The resulting precipitate was washed with 3 × 400 µL 10 mM ice-cold HCl, dried under vacuum, and resuspended in 50-µL buffer containing 25 mM Tris at pH 8, 1 mM EDTA. Methods for LysC digestion and LC-MS/MS analysis are detailed in the SI Appendix, Methods.
PglC Kinetics.
The UMP-Glo assay was used to explore the kinetics of the PglC reaction. Assays were performed at room temperature for 15 min in 15 µL volume. A standard PglC assay contained 0.2 nM PglC in assay buffer containing 50 mM Hepes at pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.1% Triton X-100, and 10% DMSO and combinations of various concentrations UDP-diNAcBac (2.5–20 µM) and Und-P (3.75–30 µM). Assays were carried out in duplicate.
Supplementary Material
Acknowledgments
We thank Prof. Karen Allen, Dr. Vinita Lukose, and Sonya Entova for valuable discussions; Dr. Amanda Del Rosario (MIT Koch Institute) for performing mass spectrometry experiments and analyzing the data; and Dr. Hicham Zegzouti (Promega) for providing the UMP/CMP-Glo Reagent. This research was supported by NIH GM-039334 (to B.I.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703397114/-/DCSupplemental.
References
- 1.Hug I, Feldman MF. Analogies and homologies in lipopolysaccharide and glycoprotein biosynthesis in bacteria. Glycobiology. 2011;21:138–151. doi: 10.1093/glycob/cwq148. [DOI] [PubMed] [Google Scholar]
- 2.Linton D, et al. Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol Microbiol. 2005;55:1695–1703. doi: 10.1111/j.1365-2958.2005.04519.x. [DOI] [PubMed] [Google Scholar]
- 3.Price NP, Momany FA. Modeling bacterial UDP-HexNAc: Polyprenol-P HexNAc-1-P transferases. Glycobiology. 2005;15:29R–42R. doi: 10.1093/glycob/cwi065. [DOI] [PubMed] [Google Scholar]
- 4.Lloyd AJ, Brandish PE, Gilbey AM, Bugg TD. Phospho-N-acetyl-muramyl-pentapeptide translocase from Escherichia coli: Catalytic role of conserved aspartic acid residues. J Bacteriol. 2004;186:1747–1757. doi: 10.1128/JB.186.6.1747-1757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Heijenoort J. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat Prod Rep. 2001;18:503–519. doi: 10.1039/a804532a. [DOI] [PubMed] [Google Scholar]
- 6.Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 7.Glover KJ, Weerapana E, Chen MM, Imperiali B. Direct biochemical evidence for the utilization of UDP-bacillosamine by PglC, an essential glycosyl-1-phosphate transferase in the Campylobacter jejuni N-linked glycosylation pathway. Biochemistry. 2006;45:5343–5350. doi: 10.1021/bi0602056. [DOI] [PubMed] [Google Scholar]
- 8.Lukose V, et al. Conservation and covariance in small bacterial phosphoglycosyltransferases identify the functional catalytic core. Biochemistry. 2015;54:7326–7334. doi: 10.1021/acs.biochem.5b01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartley MD, et al. Biochemical characterization of the O-linked glycosylation pathway in Neisseria gonorrhoeae responsible for biosynthesis of protein glycans containing N,N′-diacetylbacillosamine. Biochemistry. 2011;50:4936–4948. doi: 10.1021/bi2003372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Dabbagh B, Mengin-Lecreulx D, Bouhss A. Purification and characterization of the bacterial UDP-GlcNAc:undecaprenyl-phosphate GlcNAc-1-phosphate transferase WecA. J Bacteriol. 2008;190:7141–7146. doi: 10.1128/JB.00676-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehrer J, Vigeant KA, Tatar LD, Valvano MA. Functional characterization and membrane topology of Escherichia coli WecA, a sugar-phosphate transferase initiating the biosynthesis of enterobacterial common antigen and O-antigen lipopolysaccharide. J Bacteriol. 2007;189:2618–2628. doi: 10.1128/JB.01905-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckert V, et al. Cloning and functional expression of the human GlcNAc-1-P transferase, the enzyme for the committed step of the dolichol cycle, by heterologous complementation in Saccharomyces cerevisiae. Glycobiology. 1998;8:77–85. doi: 10.1093/glycob/8.1.77. [DOI] [PubMed] [Google Scholar]
- 13.Burda P, Aebi M. The dolichol pathway of N-linked glycosylation. Biochim Biophys Acta. 1999;1426:239–257. doi: 10.1016/s0304-4165(98)00127-5. [DOI] [PubMed] [Google Scholar]
- 14.Anderson MS, Eveland SS, Price NP. Conserved cytoplasmic motifs that distinguish sub-groups of the polyprenol phosphate:N-acetylhexosamine-1-phosphate transferase family. FEMS Microbiol Lett. 2000;191:169–175. doi: 10.1111/j.1574-6968.2000.tb09335.x. [DOI] [PubMed] [Google Scholar]
- 15.Chung BC, et al. Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis. Science. 2013;341:1012–1016. doi: 10.1126/science.1236501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen KT, et al. Structural investigation of Park’s nucleotide on bacterial translocase MraY: Discovery of unexpected MraY inhibitors. Sci Rep. 2016;6:31579. doi: 10.1038/srep31579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bugg TD, Lloyd AJ, Roper DI. Phospho-MurNAc-pentapeptide translocase (MraY) as a target for antibacterial agents and antibacterial proteins. Infect Disord Drug Targets. 2006;6:85–106. doi: 10.2174/187152606784112128. [DOI] [PubMed] [Google Scholar]
- 18.Struve WG, Sinha RK, Neuhaus FC. On the initial stage in peptidoglycan synthesis. Phospho-N-acetylmuramyl-pentapeptide translocase (uridine monophosphate) Biochemistry. 1966;5:82–93. doi: 10.1021/bi00865a012. [DOI] [PubMed] [Google Scholar]
- 19.Heydanek MG, Jr, Struve WG, Neuhaus FC. On the initial stage in peptidoglycan synthesis. 3. Kinetics and uncoupling of phospho-N-acetylmuramyl-pentapeptide translocase (uridine 5′-phosphate) Biochemistry. 1969;8:1214–1221. doi: 10.1021/bi00831a056. [DOI] [PubMed] [Google Scholar]
- 20.Heydanek MG, Jr, Neuhaus FC. The initial stage in peptidoglycan synthesis. IV. Solubilization of phospho-N-acetylmuramyl-pentapeptide translocase. Biochemistry. 1969;8:1474–1481. doi: 10.1021/bi00832a024. [DOI] [PubMed] [Google Scholar]
- 21.Al-Dabbagh B, et al. Catalytic mechanism of MraY and WecA, two paralogues of the polyprenyl-phosphate N-acetylhexosamine 1-phosphate transferase superfamily. Biochimie. 2016;127:249–257. doi: 10.1016/j.biochi.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, et al. New insight into the catalytic mechanism of bacterial MraY from enzyme kinetics and docking studies. J Biol Chem. 2016;291:15057–15068. doi: 10.1074/jbc.M116.717884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cornish-Bowden A. Fundamentals of enzyme kinetics. Two-substrate reactions. Butterworth & Co; London: 1979. pp. 99–129. [Google Scholar]
- 24.Walvoort MT, Lukose V, Imperiali B. A modular approach to phosphoglycosyltransferase inhibitors inspired by nucleoside antibiotics. Chemistry. 2016;22:3856–3864. doi: 10.1002/chem.201503986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das D, Walvoort MT, Lukose V, Imperiali B. A rapid and efficient luminescence-based method for assaying phosphoglycosyltransferase enzymes. Sci Rep. 2016;6:33412. doi: 10.1038/srep33412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller RK, Boon DY, Crum FC. N-Acetylglucosamine-1-phosphate transferase from hen oviduct: Solubilization, characterization, and inhibition by tunicamycin. Biochemistry. 1979;18:3946–3952. doi: 10.1021/bi00585a016. [DOI] [PubMed] [Google Scholar]
- 27.Meier-Dieter U, Starman R, Barr K, Mayer H, Rick PD. Biosynthesis of enterobacterial common antigen in Escherichia coli. Biochemical characterization of Tn10 insertion mutants defective in enterobacterial common antigen synthesis. J Biol Chem. 1990;265:13490–13497. [PubMed] [Google Scholar]
- 28.Kaushal GP, Elbein AD. Purification and properties of UDP-GlcNAc:dolichyl-phosphate GlcNAc-1-phosphate transferase. Activation and inhibition of the enzyme. J Biol Chem. 1985;260:16303–16309. [PubMed] [Google Scholar]
- 29.Barreteau H, et al. Quantitative high-performance liquid chromatography analysis of the pool levels of undecaprenyl phosphate and its derivatives in bacterial membranes. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:213–220. doi: 10.1016/j.jchromb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 30.Olivier NB, Chen MM, Behr JR, Imperiali B. In vitro biosynthesis of UDP-N,N′-diacetylbacillosamine by enzymes of the Campylobacter jejuni general protein glycosylation system. Biochemistry. 2006;45:13659–13669. doi: 10.1021/bi061456h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouhss A, Crouvoisier M, Blanot D, Mengin-Lecreulx D. Purification and characterization of the bacterial MraY translocase catalyzing the first membrane step of peptidoglycan biosynthesis. J Biol Chem. 2004;279:29974–29980. doi: 10.1074/jbc.M314165200. [DOI] [PubMed] [Google Scholar]
- 32.Amer AO, Valvano MA. Conserved aspartic acids are essential for the enzymic activity of the WecA protein initiating the biosynthesis of O-specific lipopolysaccharide and enterobacterial common antigen in Escherichia coli. Microbiology. 2002;148:571–582. doi: 10.1099/00221287-148-2-571. [DOI] [PubMed] [Google Scholar]
- 33.Patel KB, Furlong SE, Valvano MA. Functional analysis of the C-terminal domain of the WbaP protein that mediates initiation of O antigen synthesis in Salmonella enterica. Glycobiology. 2010;20:1389–1401. doi: 10.1093/glycob/cwq104. [DOI] [PubMed] [Google Scholar]
- 34.Allen KN, Dunaway-Mariano D. Phosphoryl group transfer: Evolution of a catalytic scaffold. Trends Biochem Sci. 2004;29:495–503. doi: 10.1016/j.tibs.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 35.Sudom A, et al. Mechanisms of activation of phosphoenolpyruvate carboxykinase from Escherichia coli by Ca2+ and of desensitization by trypsin. J Bacteriol. 2003;185:4233–4242. doi: 10.1128/JB.185.14.4233-4242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tari LW, Matte A, Goldie H, Delbaere LT. Mg(2+)-Mn2+ clusters in enzyme-catalyzed phosphoryl-transfer reactions. Nat Struct Biol. 1997;4:990–994. doi: 10.1038/nsb1297-990. [DOI] [PubMed] [Google Scholar]
- 37.Wigley DB, Davies GJ, Dodson EJ, Maxwell A, Dodson G. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature. 1991;351:624–629. doi: 10.1038/351624a0. [DOI] [PubMed] [Google Scholar]
- 38.Collet JF, Stroobant V, Van Schaftingen E. Evidence for phosphotransferases phosphorylated on aspartate residue in N-terminal DXDX(T/V) motif. Methods Enzymol. 2002;354:177–188. doi: 10.1016/s0076-6879(02)54014-3. [DOI] [PubMed] [Google Scholar]
- 39.Collet JF, Stroobant V, Pirard M, Delpierre G, Van Schaftingen E. A new class of phosphotransferases phosphorylated on an aspartate residue in an amino-terminal DXDX(T/V) motif. J Biol Chem. 1998;273:14107–14112. doi: 10.1074/jbc.273.23.14107. [DOI] [PubMed] [Google Scholar]
- 40.Post RL, Kume S. Evidence for an aspartyl phosphate residue at the active site of sodium and potassium ion transport adenosine triphosphatase. J Biol Chem. 1973;248:6993–7000. [PubMed] [Google Scholar]
- 41.Myung JI, Pitt MA. Model comparison methods. Methods Enzymol. 2004;383:351–366. doi: 10.1016/S0076-6879(04)83014-3. [DOI] [PubMed] [Google Scholar]
- 42.Myung JI, Tang Y, Pitt MA. Evaluation and comparison of computational models. Methods Enzymol. 2009;454:287–304. doi: 10.1016/S0076-6879(08)03811-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burnham KB, Anderson DR. Model selection and multimodel inference: A practical information-theoretic approach. 2nd Ed Springer-Verlag; New York: 2002. [Google Scholar]
- 44.Bates DM, Watts DG. Nonlinear regression analysis and its applications. Wiley; New York: 1988. [Google Scholar]
- 45.Brooks I, Watts DG, Soneson KK, Hensley P. Determining confidence intervals for parameters derived from analysis of equilibrium analytical ultracentrifugation data. Methods Enzymol. 1994;240:459–478. doi: 10.1016/s0076-6879(94)40060-1. [DOI] [PubMed] [Google Scholar]
- 46.Watts DG. Parameter estimates from nonlinear models. Methods Enzymol. 1994;240:23–36. doi: 10.1016/s0076-6879(94)40041-5. [DOI] [PubMed] [Google Scholar]
- 47.Kuzmic P. DynaFit––a software package for enzymology. Methods Enzymol. 2009;467:247–280. doi: 10.1016/S0076-6879(09)67010-5. [DOI] [PubMed] [Google Scholar]
- 48.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: Application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 49.Kain J, He GG, Losick R. Polar localization and compartmentalization of ClpP proteases during growth and sporulation in Bacillus subtilis. J Bacteriol. 2008;190:6749–6757. doi: 10.1128/JB.00589-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.