Significance
Human cytosolic sulfotransferases (SULTs) regulate hundreds of signaling small molecules, yet little is known regarding their small-molecule regulation. Members of the SULT1 family harbor a conserved allosteric site that we hypothesize allows independent regulation of sulfonation in the 11 metabolic areas in which these isoforms operate. This hypothesis is validated using SULT1A3, which sulfonates and inactivates monoamine neurotransmitters. During validation it was discovered that tetrahydrobiopterin (THB), an essential cofactor in monoamine neurotransmitter biosynthesis, allosterically inhibits SULT1A3 with high affinity (Ki, 23 nM) and isozyme selectivity. Monoamine neurotransmitter metabolism shapes human behavior and social interactions and is a therapeutic target. These findings provide a paradigm for regulating sulfonation and a target for controlling neurotransmitter activity.
Keywords: sulfotransferase, tetrahydrobiopterin, neurotransmitter, allostery, evolution
Abstract
Monoamine neurotransmitters are among the hundreds of signaling small molecules whose target interactions are switched “on” and “off” via transfer of the sulfuryl-moiety (–SO3) from PAPS (3′-phosphoadenosine 5′-phosphosulfate) to the hydroxyls and amines of their scaffolds. These transfer reactions are catalyzed by a small family of broad-specificity enzymes—the human cytosolic sulfotransferases (SULTs). The first structure of a SULT allosteric-binding site (that of SULT1A1) has recently come to light. The site is conserved among SULT1 family members and is promiscuous—it binds catechins, a naturally occurring family of flavanols. Here, the catechin-binding site of SULT1A3, which sulfonates monoamine neurotransmitters, is modeled on that of 1A1 and used to screen in silico for endogenous metabolite 1A3 allosteres. Screening predicted a single high-affinity allostere, tetrahydrobiopterin (THB), an essential cofactor in monoamine neurotransmitter biosynthesis. THB is shown to bind and inhibit SULT1A3 with high affinity, 23 (±2) nM, and to bind weakly, if at all, to the four other major SULTs found in brain and liver. The structure of the THB-bound binding site is determined and confirms that THB binds the catechin site. A structural comparison of SULT1A3 with SULT1A1 (its immediate evolutionary progenitor) reveals how SULT1A3 acquired high affinity for THB and that the majority of residue changes needed to transform 1A1 into 1A3 are clustered at the allosteric and active sites. Finally, sequence records reveal that the coevolution of these sites played an essential role in the evolution of simian neurotransmitter metabolism.
Human cytosolic sulfotransferases (SULTs) regulate the activities of thousands of endogenous small-molecule metabolites and xenobiotics via transfer of the sulfuryl-moiety (–SO3) to and from the hydroxyl- and amine-moieties of these acceptors. The 13 full-length SULT isoforms encoded in the human genome are expressed in tissue- and developmentally specific patterns (1–3). SULT substrate specificities are typically broad, overlapping, and centered on different areas of metabolism. The diversity of function across SULT isoforms results in a remarkably broad range of metabolic functions including potent regulation of steroids (4), thyroid (5) and peptide hormones (6), oxysterols (7), pheromones (8), selectins (9), and neurotransmitters (10, 11).
Although recent work has deepened our understanding of SULT small-molecule allosteric regulation, the topic remains largely unexplored (12–16). The catechin-binding site is the most well-characterized SULT allosteric site (12). Catechins, a complex biomorphic family of SULT allosteric inhibitors, are found at high levels in tea leaves, cocoa, and coffee (17–19). The binding site is promiscuous in that it binds numerous catechins (15, 20) and related structures. The structure of the SULT1A1 catechin-binding site, the only published SULT allosteric-site structure, has recently come to light (12). The signature elements of the site are found in each of the 11 members of the SULT1 subfamily, three of which are known to be inhibited, with varying sensitivity, by catechins (12, 14, 20). The conservation, variable catechin responses, and promiscuity of these sites imply not only that endogenous metabolite allosteres exist (though none have been identified) but that nature may have solved the problem of how to independently regulate sulfonation in the various metabolic domains in which SULTs operate by “tuning” the binding properties of the allosteric sites, through adaptive selection, toward metabolites that lie within the domain of a particular isoform.
To test whether SULT catechin sites bind endogenous metabolites, an in silico model of the catechin-binding site of SULT1A3, which sulfonates monoamine neurotransmitters (i.e., dopamine, epinephrine, serotonin), was constructed and used in docking studies to screen monoamine neurotransmitter metabolites. The screen identified a single putative allostere, tetrahydrobiopterin (THB), an essential cofactor in the biosynthesis of all monoamine neurotransmitters. THB participates in the O2-cleavage reaction of aromatic amino acid hydroxylases, which rate-limit monoamine neurotransmitter biosynthesis. The sulfonation of such transmitters is extensive and tissue-dependent (11, 21–24). When tested, THB proved a potent (Ki, 23 nM) and highly selective allosteric inhibitor of SULT1A3. The biological relevance of the THB allosteric site is underscored by the fact that the allosteric and active-site specificities have coevolved over the last ∼60 My of evolution. Monoamine neurotransmitter metabolism shapes the behavior and social structures of humans and other primates (25). Although numerous therapeutics target neurotransmitter metabolism (26, 27), none do so by inhibiting sulfonation. The structure of the THB-binding site, presented here, provides an isozyme-specific means of controlling neurotransmitter activity.
Results and Discussion
Discovering the Allostere.
The complexity and biomorphic character of the catechin family and the promiscuity of the SULT1A3 catechin site suggested that it might also bind endogenous metabolite allosteres. To explore this possibility, the structure of the SULT1A3 site was modeled on the SULT1A1 site, and both templates were used in an in silico docking study to screen metabolite allosteres. The SULT1A1 control template helps gauge the specificity of candidate SULT1A3 allosteres. Given the high catalytic efficiency of SULT1A3 toward monoamine neurotranmitters, the screen was limited to the 115 human small-molecule metabolites associated with neurotransmitter metabolism found in the Small Molecule Pathways Database (SMPDB) (28). Screening was performed with the Genetic Optimization for Ligand Docking (GOLD) program (Materials and Methods).
The screens predicted that of the 115 metabolites tested, only THB, an essential cofactor in monoamine neurotransmitter biosynthesis, would bind SULT1A3 with high affinity (44 nM). The studies further predicted that THB would bind SULT1A3 far more tightly (230-fold) than SULT1A1. The compound whose predicted affinity was closest to that of THB is dihydrobiopterin (DHB), the oxidized form of THB produced during neurotransmitter biosynthesis. The affinity of DHB is predicted to be ∼130-fold weaker than that of THB.
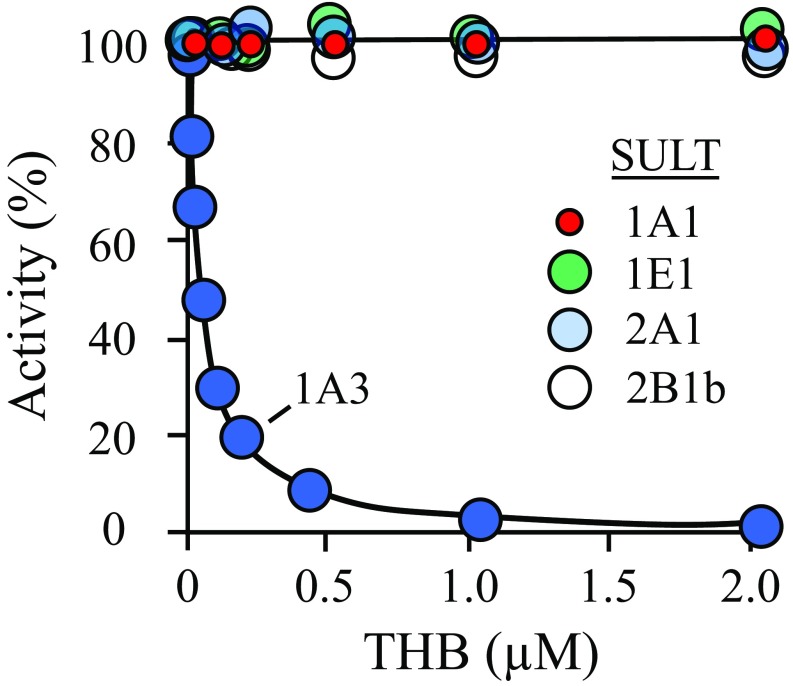
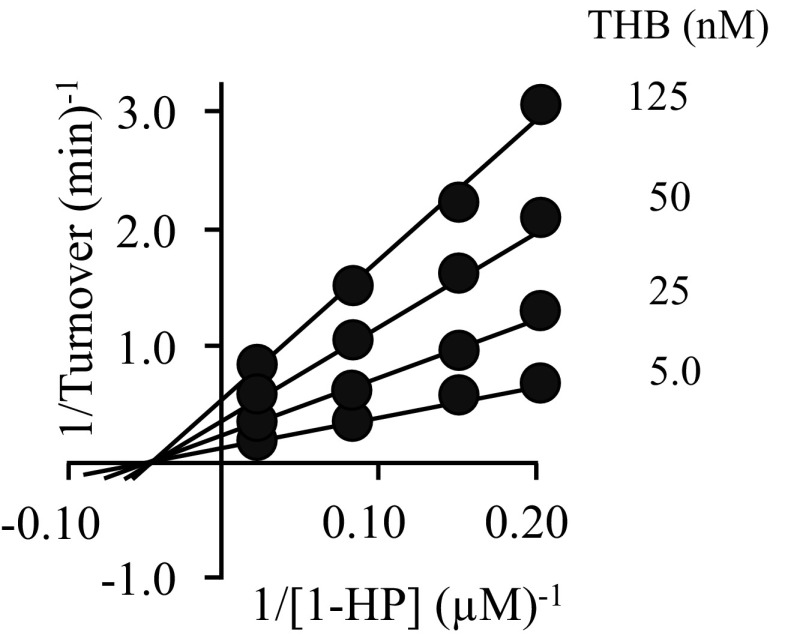
To test the binding and specificity predictions of the docking studies and whether, like catechins, THB functions as an inhibitor, THB was tested in initial-rate studies as an inhibitor of SULT1A3 and the four other SULTs that, together with SULT1A3, constitute the major SULT isoforms found in human brain (29, 30) and liver (1, 31) (i.e., SULTs 1A1, 1E1, 2A1, and 2B1b). The results (Fig. 1) reveal that THB binds and inhibits SULT1A3 with high affinity, Ki 23 (±2) nM, and appears to completely suppress SULT1A3 turnover at saturation. None of the other major brain and liver SULTs are inhibited at concentrations needed to saturate SULT1A3 (Fig. 1), and increasing THB further, to 200 μM, did not inhibit these SULTs. Thus, THB proved to be a remarkably potent and specific inhibitor of SULT1A3. The allosteric nature of THB inhibition was confirmed in an initial-rate study, which revealed that THB is a noncompetitive inhibitor versus 1-HP (Fig. S1).
Fig. 1.
THB inhibits SULT1A3 with high affinity and selectivity. SULT initial rates are plotted as a function of THB concentration. Initial rates are plotted as percent of activity at [THB] = 0. Reaction progress was monitored via a sulfonation-dependent change in 1-HP fluorescence [λex 325 nm, λex 375 nm (71)]. Less than 2% of the concentration-limiting substrate consumed at the reaction endpoints was converted to product during initial-rate measurements. Each point is the average of three independent determinations. The SULT1A3 inhibition constant, 23 ± 2 nM, was obtained by least-squares fitting using a single site per subunit model. The line through the SULT1A3 data is the behavior predicted by the best fit model. Conditions: SULT (25 nM, dimer), 1-HP (400 nM for SULT1A3, 20 × Km; 400 nM for SULT1A1, 20 × Km; 800 nM for SULT1E1, 20 × Km; 2.5 μM for SULT2A1, 20 × Km; 3.2 μM for SULT2B1b, 20 × Km), PAPS (6.0 μM, ∼20 × Km), DTT (5.0 mM), KPO4 (50 mM), pH 7.5, 25 ± 2 °C.
Fig. S1.
THB inhibition of SULT1A3 is noncompetitive. Reactions were initiated by addition of PAPS at saturation (500 μM, 1,500 × Km), 1-HP concentrations varied from 0.2 to 5 × Km (5–100 nM) in equal increments in double-reciprocal space, and THB concentrations are as indicated. Reaction conditions were as follows: SULT1A1 (10 nM, dimer), KPO4 (50 mM), pH 7.5, 25 ± 2 °C. Reaction progress was monitored by the fluorescence change associated with 1-HP sulfonation (λex 325 nm, λex 370 nm). Less than 5% of 1-HP formed at reaction endpoints was consumed during the measurements. Each point represents the average of two independent determinations. The lines through the points represent the behavior predicted by a global (1/v4)-weighted least-squares fit to a noncompetitive inhibition model. The predicted Ki of THB is 29 ± 3 µM.
THB appears to function both as an essential cofactor in monoamine neurotransmitter biosynthesis and as an allostere to inhibit the enzyme that inactivates these same transmitters. THB levels found in cells and tissues are consistent with an in vivo role for THB in regulating sulfonation. Examples include the following: (i) THB levels are near its Ki (23 nM) in human cerebrospinal fluid, erythrocytes, and plasma (∼15 nM) (32). (ii) THB levels in dopaminergic neurons appear to go from subsaturating in the cell body to well above saturation (∼100 μM) in neuronal termini (33)—a scenario that suggests that cytosolic dopamine, which diffuses into the cell body from leaky vesicles located in termini, might only be sulfonated once a suitable point in the THB concentration gradient is reached. (iii) THB levels are potently down-regulated by cytokines in a variety of cell types and tissues that express SULT1A3 (34–36). THB levels in normal-patient platelets are ∼200 nM (8.7 Ki THB) (36, 37) and expected to decrease significantly upon cytokine stimulation (36). (iv) SULT1A3 is up-regulated in neurons in response to dopamine (30) and prevents toxicity by suppressing dopamine free-radical formation (38). Exogenous THB is known to induce apoptosis via free-radical mechanisms only in dopaminergic cell lines (39) and may do so by preventing dopamine sulfonation.
The THB Binding-Site Structure.
The spin-label triangulation methodology.
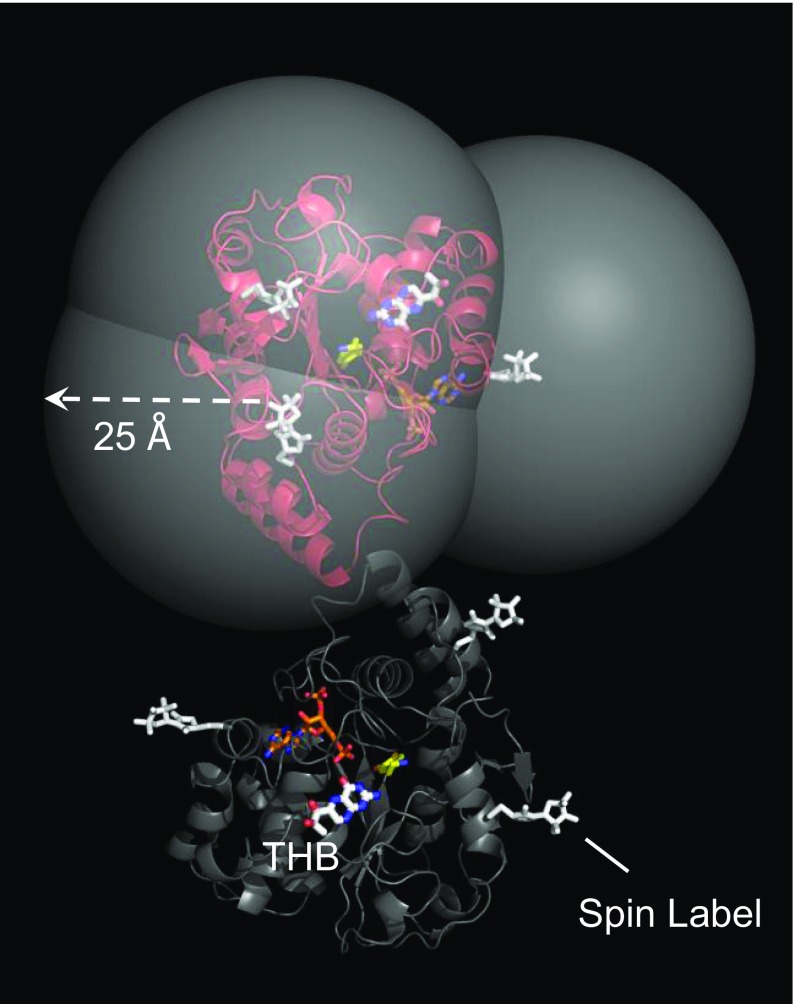
We have recently developed a spin-label method for determining the structures of SULT ligand-binding sites (12). The technique does not require X-ray crystallography or high-resolution multidimensional NMR and has no protein molecular weight limitations. The method involves identifying six solvent-accessible positions in a SULT scaffold at which cysteine can be inserted and spin-labeled without affecting catalysis. As a set, the individual cys constructs allow the entire surface of the enzyme to be “coated” in a paramagnetic field of sufficient strength to detect its effects on the 1D solution NMR spectrum of a ligand regardless of where it binds. Paramagnetic effects are detectible when a ligand docks within ∼25 Å of a given spin label and exchanges between protein and bulk solvent at rates comparable to or faster than the Larmor frequency difference between free- and bound-ligand NMR peaks. Distances between the spin label and ligand protons are calculated from the magnitudes of the paramagnetic effects. In most cases, effects can be detected from three different spin labels; hence, each ligand proton is separately positioned by triangulation, which allows the ligand to be oriented with respect to enzyme surface. A final, refined structure of the ligand-bound binding site is obtained using NMR distance-constrained molecular-dynamics docking.
The three SULT1A3 cys-insert mutants used in the current study were the same as those used to determine the catechin-site structure of SULT1A1. A model of SULT1A3 with spin labels attached at these positions is shown in Fig. 2. The identical subunits of the enzyme dimer are shown in red and blue. The 25-Å radii of the large, semitransparent spheres (which are centered on nitroxide oxygens) correspond to the approximate maximum distance over which proton/spin-label interactions are detectable. THB was positioned at the catechin site based on predictions from the docking screens described above. As is evident, the putative docking site is coated in a detectible paramagnetic field from each spin label.
Fig. 2.
The SULT1A3 spin-label constructs. The subunits of SULT1A3 dimer are in red and gray. THB is labeled and is positioned at the catechin-binding site on the basis of screening studies (Results and Discussion). The spin labels (white) are positioned such that their paramagnetic fields can perturb the solution NMR spectra of allosteres that bind the catechin site without affecting the enzyme’s initial-rate and catechin-inhibition parameters. The carbon atoms of dopamine and PAP are yellow and orange, respectively. The semitransparent spheres center on the nitroxyl-oxygen of the spin labels, and their radii are set to the approximate maximum distance over which ligand/spin label interactions can be detected (i.e., ∼25 Å). Unlike the figure, each experimental construct has spin label attached at a single position. The design allows allostere protons to be positioned by triangulation from three spin labels.
Building and testing cys constructs.
Three single-cys insertion mutants were constructed from a SULT1A3 coding region (Materials and Methods). Each construct was covalently labeled to >98% with spin label [TEMPO (2,2,6,6-tetramethylpiperidin-1-oxyl)] or N-cyclohexylmaleimide—a diamagnetic homolog in which the spin-label Proxyl (2,2,5,5-tetramethyl-1-pyrrolidinyloxy) moiety is replaced by a cyclohexyl-group. The diamagnetic constructs are used as controls in the NMR distance determinations (Distance Measurements). The initial-rate parameters of the labeled constructs were determined and were similar to those of the wild-type enzyme (Table 1 and Materials and Methods).
Table 1.
Initial-rate parameters for WT and spin-labeled SULT1A3 mutants
| Enzyme | kcat, min−1 | Km, nM | Ki THB, nM |
| WT | 120 (17) | 22 (3.1) | 23 (1.5) |
| 26* | 110 (15) | 25 (3.5) | 25 (2.7) |
| 151 | 115 (16) | 26 (2.2) | 20 (2.2) |
| 234 | 109 (15) | 24 (3.2) | 22 (1.6) |
Values in parentheses indicate SD.
Cys residue at which spin label is attached.
Distance measurements.
Methods for determining distances between the protons of bound ligands and covalently attached spin labels are well established (40, 41). In favorable cases (when ligand exchange is comparable to or greater than the Larmor frequency difference of the free and bound proton), the effects of the protein environment on ligand–proton relaxation can be detected in the ligand’s solution spectrum. In such cases, observed transverse relaxation rates, R2obs, can be calculated from solution-phase linewidths, which depend linearly on the fraction of bound ligand, FB, according to Eq. 1 (42):
| [1] |
R2B and R2F are the transverse relaxation rates for bound and free protons, and R2ex is the chemical exchange contribution to the relaxation.
The contribution of the electron to the transverse relaxation of the bound proton is required to calculate distances (41). To isolate the electron’s contribution from those of other factors, two R2 obs versus FB plots are created, using the para- and diamagnetic enzyme constructs (43). The electron’s contribution is given by the difference in slopes of the plots (41, 42).
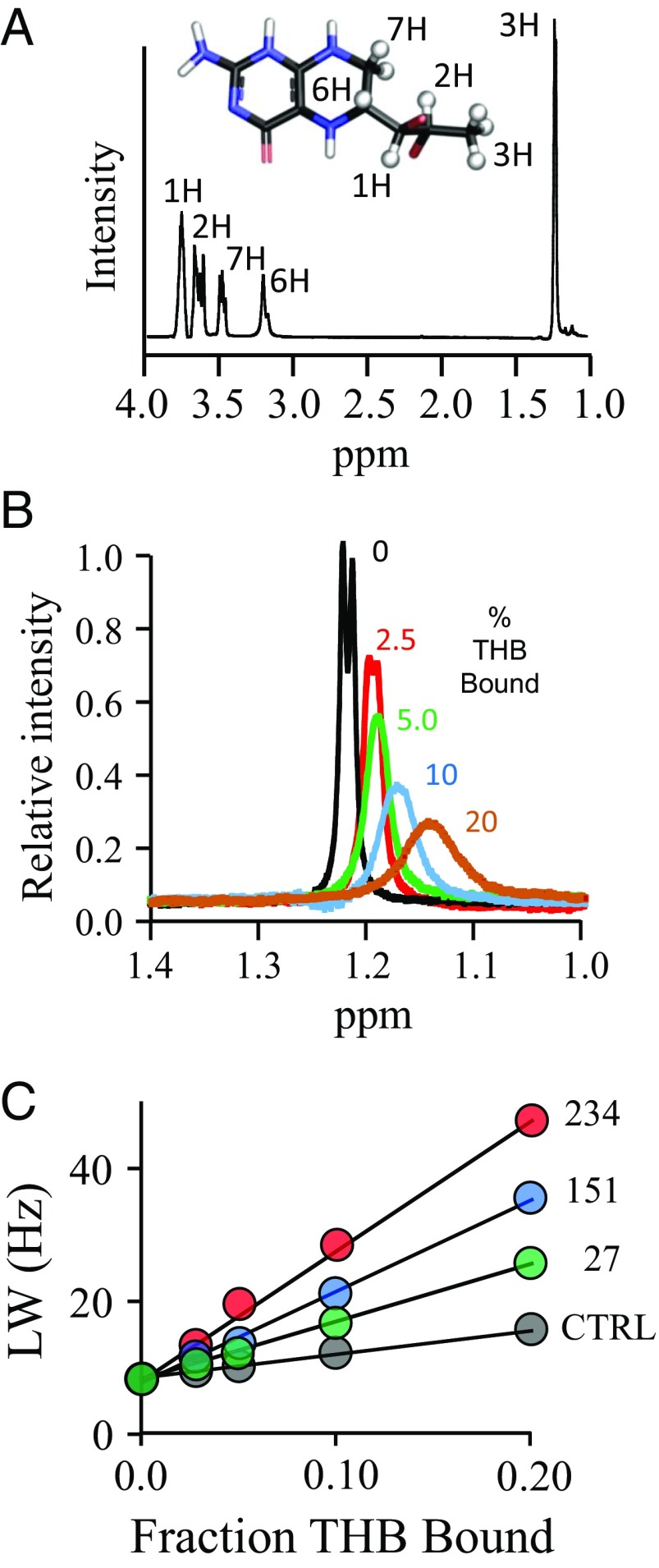
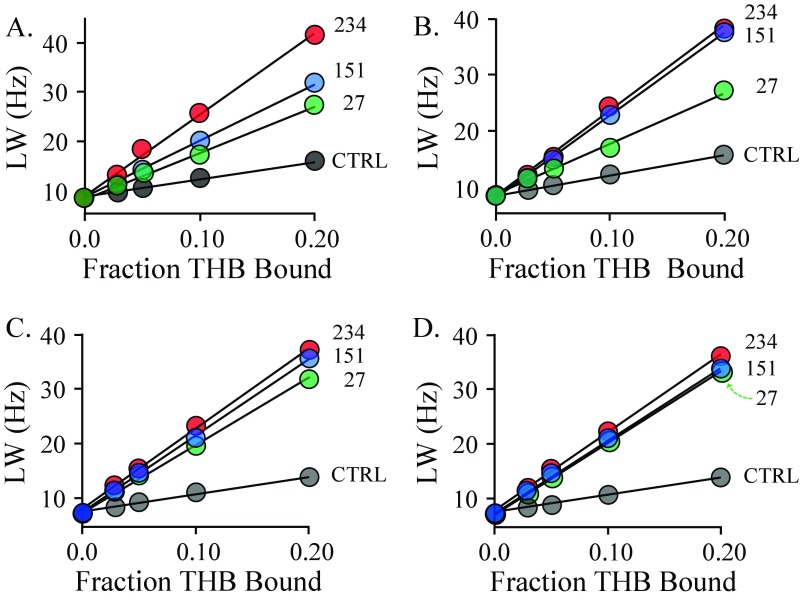
The structure and solution spectrum of THB are given in Fig. 3A, and the effects of the spin-labeled K234C construct on the H3 proton peak of THB are shown in Fig. 3B. A representative set of para- and diamagnetic R2 obs versus FB plots are given in Fig. 3C. Twelve such plot pairs were constructed; the remaining 11 pairs are presented in Fig. S2.
Fig. 3.
The NMR measurements. (A) The structure and 300-MHz 1H-NMR spectrum of THB. The THB protons used to construct the allosteric binding-site structure are depicted as white spheres and are labeled in the spectrum and structure. Peak assignments were determined previously (80). Conditions: THB (100 µM), KPO4 (50 mM), DTTdeuterated (5.0 mM), D2O (>98%), pH 7.4, 25 ± 1 °C. (B) Spin-label effects on the peak of the H3 protons of THB. The solution 1H-NMR spectrum (600 MHz) of the H3 peak of THB is shown as a function of the percent of THB bound to spin-labeled C234-SULT1A3. Conditions: THB [100 μM (brown), 200 μM (blue and black), 400 μM (green), and 800 μM (red)], spin-labeled C234-SULT1A3 [20 µM monomer or 0 µM (black)], PAP (500 µM, 17 × Kd), KPO4 (50 mM), DTTdeuterated (5.0 mM), D2O (>98%), pH 7.4, 25 ± 1 °C. The enzyme is saturated (≥4,400 Kd) at all THB concentrations (Kd THB = 23 nM). Peak amplitudes are normalized to THB concentration. (C) Line width versus fraction THB-bound plots. The effects of paramagnetic and diamagnetic C234-SULT1A3 constructs on the line width on the H3 proton peak of THB are plotted as a function of fraction of enzyme-bound THB. Conditions are described in B.
Fig. S2.
The line width versus fraction bound of the proton peaks of THB. The line width of H1 (A), H2 (B), H6 (C), and H7 (D) proton peaks as a function of fraction THB bound. Conditions: THB (150, 300, 600, and 1,200 µM), spin-labeled SULT1A3 [control (black), 27 (green), 151 (blue), and 234 (red), 30 µM monomer], PAP (500 µM, 17 × Kd), deuterated DTT (5.0 mM), KPO4 (50 mM), pH 7.4, 25 ± 1 °C. The enzyme is >99% saturated at all THB concentrations (Kd THB = 25 nM).
Refining the structure.
The final structure was obtained by distance-constrained molecular dynamics docking of the THB. Each NMR-determined distance represents a vector between the time-averaged position of the spin-label oxygen (calculated using GROMACS) and a given proton. Three such vectors are associated with each proton, each originates at the oxygen of a different spin label, and all three intersect at the proton. The errors associated with these three measurements constitute an ellipsoid that centers on the proton and whose axis magnitudes are given by the SEs (±1 σ) of the NMR measurements, which are compiled in Table 2. Docking is constrained by applying a 50 kj⋅mole−1⋅Å−1 restoring force [using distance_restraints, GROMACS (44, 45)] that drives the proton toward the ellipsoid center if any part of its van der Waals surface lies outside the ellipsoid; the restoring force inside the ellipsoid is 0. As is appropriate for NMR spin–spin interaction measurements (42, 46, 47), distance_restraints was parameterized to use time-averaged (1/r6)-weighted restraints. The motions of all five THB protons (H1, H2, H3, H6, and H7) were constrained simultaneously during docking. Docking was repeated 10 times, identical structures were achieved in each case, and the structure did not change once the distance constraints were removed.
Table 2.
Proton to spin-label distances (Å)
| Proton | Spin-label attachment residue | ||
| 27 | 151 | 234 | |
| H1 | 18 (14–21) | 19 (16–22) | 16 (14–20) |
| H2 | 20 (18–24) | 22 (19–25) | 14 (11–16) |
| H3 | 25 (21–28) | 24 (19–27) | 12 (10–15) |
| H6 | 17 (15–22) | 18 (14–20) | 18 (14–20) |
| H7 | 17 (14–21) | 19 (15–22) | 18 (13–20) |
Values in parentheses indicate 95% CI.
The refined structure.
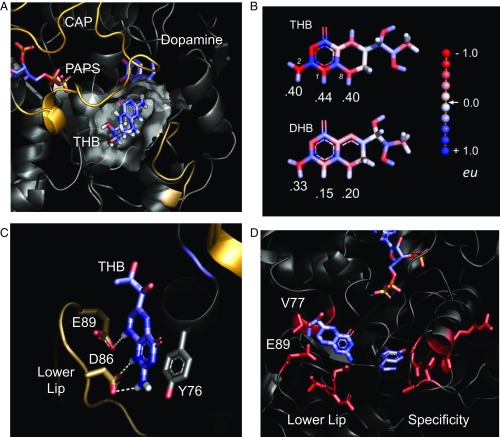
The MD-refined structure of THB bound to the catechin-binding site of SULT1A3 is shown in Fig. 4A [Protein Data Bank (PDB) coordinates are available at www.modelarchive.org, accession no. 3zswijvneac5ubl7f]. SULT1A3 harbors a conserved ∼30-residue active-site cap (shown in light orange) that opens and closes in response to nucleotide binding and a smaller “lower lip” segment (shown in beige) that together with the cap form a molecular pore, or sieve, through which acceptors must pass to enter the active site. The catechin-binding site of SULT1A3, like that of 1A1, is adjacent to the pore and sandwiched between the cap and lower lip. To validate the structure, three residues predicted to be in direct contact with THB (see labeled residues in Fig. 4B) were mutated, and the effects of the mutation on THB binding and inhibition and initial-rate turnover were tested. The results, compiled in Table 3, reveal that although the mutations did not significantly affect the catalytic integrity of the enzyme, neither THB binding nor inhibition could be detected at concentrations as high as 200 µM.
Fig. 4.
The THB-bound catechin-binding site of SULT1A3. (A) THB bound to the SULT1A3 allosteric-binding site. THB is sandwiched between the active-site cap (gold) and lower lip (sand) of the SULT1A3. The transparent surface of the binding pocket was calculated using Hollow. The donor (PAPS) and acceptor (dopamine) are labeled. (B) Residues used to verify the structure. Mutations at these positions prevent THB binding and inhibition but do not alter the initial-rate parameters of the enzyme (Table 3). (C) THB and DHB charge distributions. Distributions were calculated with Automated Topology Builder (49), which uses the B3L YP/631G* basis set. Charges are color-coded in electron units according the scale shown on the right. The protons that interact with D78 and E81 are labeled with their charges, and the nitrogen atoms bound to the protons are numbered (i.e., 1, 2, and 8). (D) Residues responsible for the allosteric and active-site specificity of SULT1A3. The sequences of SULT1A3 and SULT1A1 (the evolutionary progenitor of SULT1A3) differ at 21 positions, 11 of which, shown in red, cluster in two regions. Changes in the lower lip result in high affinity and specificity for THB; changes in the specificity region shift the substrate specificity from that of SULT1A1 to SULT1A3.
Table 3.
Initial-rate and THB binding and inhibition parameters for WT and mutant SULT1A3
| Enzyme | kcat, min−1* | Km 1-HP, nM* | Ki THB, nM* | Kd THB, nM† |
| WT | 120 (17)‡ | 24 (3.8) | 25 (2.7) | 23 (2.3) |
| D86A | 97 (18) | 27 (3.6) | ND‡ | ND‡ |
| E89I | 110 (11) | 22 (2.1) | ND | ND |
| Y76A | 107 (19) | 25 (3.0) | ND | ND |
Values in parentheses indicate SD.
Initial-rate protocols are described in Materials and Methods.
Binding-study protocols are described in Materials and Methods.
Inhibition and binding were not detected.
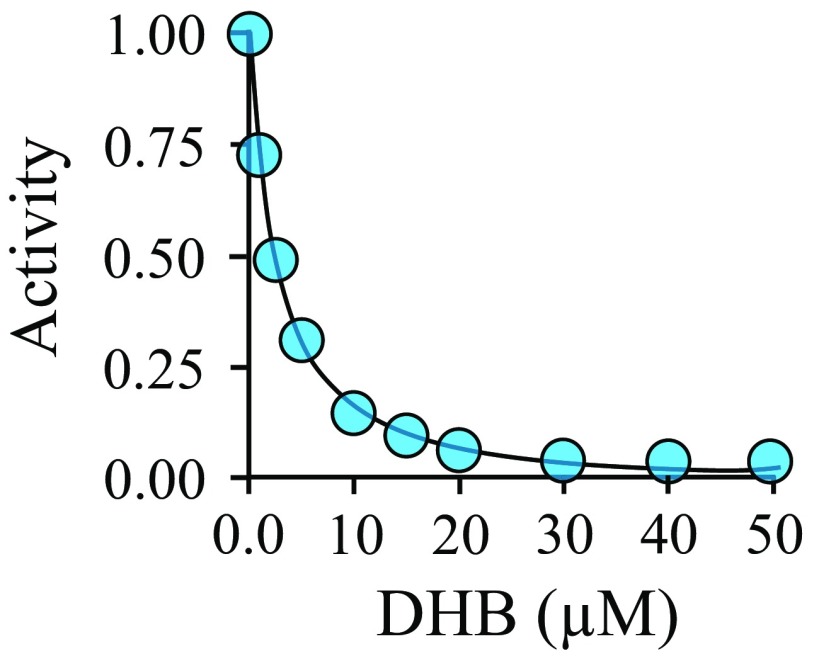
When bound, THB directly contacts six residues: P74, Y76, V77, D86, E89, and V243. Two (D86 and E89) are unique to SULT1A3; the remainder are also found in the catechin site of SULT1A1. The SULT1A3-specific residues are negatively charged and contact THB at the three hydrogen atoms that are linearly aligned along the N2–N1–N8 edge of its 2-amino-pterin ring (Fig. 4C). The charge distribution of THB, predicted by Automated Topology Builder (48, 49) using the B3L YP/631G* basis set, reveals that these three hydrogen atoms carry significant positive charge. Thus, their interactions with D86 and E89 are likely to contribute substantially to the site’s THB selectivity. The conversion of THB to DHB during catecholamine biosynthesis results in a subtle structural change and extends the resonance of the pterin ring, which delocalizes and thus diminishes charge at the N1–N2–N8 protons (Fig. 4C). This decrease in charge density is expected to weaken interactions with D86 and E89. The affinity of DHB was determined, by fluorescence titration, and found to be 130-fold weaker than that of THB (Fig. S3)—a value that agrees remarkably well with the ∼130-fold change predicted by GROMACS docking studies. The D86 and E89 ionic interactions engender both THB specificity and the ability to respond selectively to amino acid hydroxylase substrates, which report the cellular demand for neurotransmitter synthesis.
Fig. S3.
Inhibition of human SULT1A3 by DHB. The inhibition of DHB to SULT1A3 was studied using 1-HP. The reaction was monitored by the intrinsic fluorescence change of 1-HP sulfonate (λex 325 nm, λem 375 nm). The rates of 1-HP sulfonation in the presence of DHB were normalized to those without DHB and plotted vs. [DHB]. The reaction conditions were as follows: enzyme (10 nM, monomer), 1-HP (6.0 µM, 20 × Km), PAPS (6.0 µM, 20 × Km), DTT (5.0 mM), KPO4 (50 mM), pH 7.5, 25 ± 2 °C. The 1-HP was consumed less than 7.0% for all conditions. Each point represents the average of two independent determinations. The line through the points represents the best fit to data assuming one binding site per subunit. The Ki of DHB is 2.3 ± 0.1 µM.
The metabolic imperative of the THB-binding site.
If THB allosterism is essential to the proper functioning of neurotransmitter metabolism, one might expect that the allosteric and active sites, whose interdependent functions are coupled at different points in the same metabolic pathway, evolve as a single unit of metabolic circuitry until a more evolutionarily advantageous solution could be found. To evaluate the conservation and coevolution of these two sites, their evolutionary lineage was traced through the 14 SULT1A3 sequences available in the NCBI database.
SULT1A3 made its evolutionary debut at some point in the ∼10 My interval between the separation of monkeys (whose genomes encode the allosteric and active sites of SULT1A3; Fig. 4D) and lemurs (which encode neither) and the separation of old-world monkeys and apes. The interval begins 56–66 Mya (50)—a time that immediately follows the last mass extinction and is associated with a burst in mammalian speciation caused by a lack of competition in the many niches vacated by the extinction (51).
The immediate evolutionary progenitor of SULT1A3 is SULT1A1 (52)—their primary sequences are 295 residues in length, 92.9% identical, and 98.6% conserved. Eleven of the 21 nonidentical residues are clustered in two regions (Fig. 4D): the lower lip region of the THB allosteric site (seven residues), and the catechol-binding region of the active site (four residues), which is largely responsible for changing the substrate specificity of 1A1 to that of 1A3 (53). The remaining 10 residues are scattered throughout the scaffold and have not been assigned function. The active-site cluster is perfectly conserved in all 14 SULT1A3 sequences; however, the amino acids that line the SULT1A3 catechin-binding site drift slightly. Two of the six residues that directly contact THB (V77 and E89 in the human isoform) have drifted either to I77/D89 or M77/E89. To assess whether these conserved substitutions alter THB binding, they were inserted, using PCR mutagenesis, into the human SULT1A3, and the THB affinities of the hybrids were determined. The hybrids’ affinities were within a factor of 1.7 from that of the native construct (Table 4). Hence, the allosteric site has drifted only so far as to maintain its THB-binding properties. It appears that species that have lost either function have not survived the challenges of Darwinian selection to the current day—only those that retain the ability to allosterically down-regulate SULT1A3 turnover in response to THB remain.
Table 4.
THB inhibition of primate SULT1A3 mutants
| Enzyme | Ki, nM* |
| WT | 25 (2.6) |
| V77M | 32 (3.1) |
| E89D, V77I | 43 (5.0) |
Values in parentheses indicate SD.
Initial-rate protocol is described in Materials and Methods.
Conclusions
The central hypothesis of this manuscript—that the so-called catechin allosteric site of SULT1A3 can be used by endogenous metabolites—has been proven. THB, an essential metabolite in the synthesis of monoamine neurotransmitters, binds the SULT1A3 catechin site with high and physiologically relevant affinity (23 nM) and has no detectible affinity for four other major SULT isoforms. The binding site is highly selective for THB over DHB, its conjugate product, and has thus evolved to respond to the substrate side of the rate-limiting step in the biosynthesis of such transmitters. These findings suggest a classical feedback mechanism in which increased cellular demand for transmitters, which increase THB levels (54), results in allosteric inhibition of the enzyme that would otherwise have inactivated the transmitter and shorted the supply/demand circuitry.
The structure of the THB-bound binding site of SULT1A3 was determined and confirms that THB binds the catechin site of SULT1A3. THB is sandwiched between the active-site cap and lower lip of the enzyme and is adjacent to the pore through which acceptors must pass to enter the active site. The sequences of SULT1A3 and SULT1A1 differ by 21 amino acids, 11 of which are clustered into two spatially separate, functionally related regions—the allosteric and active sites—that have coevolved since the evolutionary inception of SULT1A3, ∼60 Mya (50).
Monoamine neurotransmitters are inextricably linked to brain activities ranging from executive functions that provide cognitive flexibility (55) to primitive functions that underlie reward seeking and recognition (56). Much of human emotion and behavior is determined by monoamine neurotransmitters levels in brain neuronal synapses, and modulating these levels is a major goal of the pharmaceutical industry (57). Classical therapeutic strategies aim to increase neurotransmitter levels either by preventing transmitter reuptake from synapses (57) or by inhibiting enzymes that inactivate neurotransmitters, particularly monoamine oxidases (MAOs) (58) and catechol-O-methyl transferases (COMTs) (59). MAOs attach to mitochondrial outer membranes with their active sites facing the cytosol, and COMTs are embedded in the outer membranes of pre- and postsynaptic neurons with their catalytic domains oriented extracellularly (60). Thus, these catalysts operate in spatially separate tissue compartments—cytosols and extracellular fluids.
SULT1A3 is, like MAO, located in neuronal cytosols (29) and inactivates catecholamines. Recent work reveals that sulfoconjugates predominate among serotonin metabolites in fluids extracted from brain cortices of living humans and are major metabolites in brain-ventricle and spinal fluids (61). Studies using subsections of rat brain reveal that MAO inhibition routes dopamine metabolism substantially through the sulfonation pathway and demonstrate the interactivity of the MAO and SULT systems (62, 63). These findings strongly recommend SULT1A3 as a candidate for controlling neurotransmitter activity, particularly when used in conjunction with MAO inhibitors. It is notable that nearly half of patients with neurocognitive disorders are not fully responsive to classical therapy but can be treated successfully with combination therapy (56, 64). To our knowledge, SULTs inhibitors have not been tested as therapeutics. The THB-binding site offers a structurally well-defined, isozyme-specific, allosteric target for developing SULT1A3 inhibitors that can be used to control neurotransmitter activity.
We are just beginning to appreciate the importance of small-molecule SULT allostery. With the exception of the current work, there appear to have been no studies aimed specifically at identifying SULT allosteres—such compounds have largely been discovered serendipitously during screening studies. In addition to allosteric inhibitors, activators and specificity modulators exist (65). As structures of these sites become available, they too can be used not only to identify new allosteres and deepen our understanding of SULT metabolism but also as molecular templates for the design of isoform-specific SULT allosteres—unique tools with which to probe and control SULT biology (66).
Materials and Methods
The materials and sources used in this study are as follows: DTT, deuterated DTT, 5,5′-dithiobis-2-nitrobenzoic acid (DNTB), ethylenediamine-tetraacetic acid (EDTA), l-glutathione (reduced), 1-hydroxypyrene (1-HP), imidazole, isopropyl-thio-β-d-galacto-pyranoside (IPTG), LB media, lysozyme, β-mercaptoethanol, 3-maleimido-PROXYL, N-cyclohexylmaleimide, pepstatin A, and potasium phosphate at the highest grade available from Sigma. Ampicillin, Hepes, KOH, KCl, MgCl2, and phenylmethylsulfonyl fluoride (PMSF) were purchased from Fisher Scientific. Glutathione- and nickel-chelating resins were obtained from GE Healthcare. Competent Escherichia coli [BL21(DE3)] was purchased from Novagen. PAPS and PAP were synthesized and purified according to previously published protocols (67).
Computer and Software.
Molecular dynamics simulations were performed using a Parallel Quantum Solutions QS32-2670C-XS8 computer. PQS Molecular Builder was purchased from Parallel Quantum Solutions. A GOLD license was obtained from the Cambridge Crystallographic Data Center. The source code for GROningen MAchine for Chemical Simulation (GROMACS) 4.5 was downloaded from www.GROMACS.org under the GROMCAS General Public License (GPL).
The SULT1A3 Constructs.
The SULT1A3 expression plasmid consists of the SULT1A3 coding region inserted into a triple-tag pGEX-6P expression vector containing an (N-terminal)-His/GST/MBP tag (68, 69). The cys-insertion mutants used for regio-specific attachment of maleimide-based labels were constructed as follows: Three single-cys mutants were created by inserting cys into the wild-type SULT 1A3 scaffold, which was not DTNB reactive, at residues E26, E151, and K234. All mutagenesis projects began with the SULT1A3 expression-plasmid and used standard PCR mutagenesis protocols (70).
The mutants used to confirm the structure of the THB binding (Y76A, D86A, and E89I) were constructed by mutating the wild-type SULT1A3 coding region.
Protein Purification.
SULT expression and purification were performed using protocols that have been described in detail previously (68, 69). The proteins (SULTs 1A3, 1A1, 1E1, 2A1, and 2B1b) are >95% pure as judged by Coomassie-blue staining of SDS/PAGE. Protein concentrations are determined by UV absorbance (Ɛ280 SULT1A3 = 53.9 mM−1⋅cm−1, Ɛ280 SULT1A1 = 53.9 mM−1⋅cm−1, Ɛ280 SULT1E1 = 61.1 mM−1⋅cm−1, Ɛ280 SULT2A1 = 79.5 mM−1⋅cm−1, and Ɛ280 SULT2B1b = 67.6 mM−1⋅cm−1). The final protein is flash-frozen and stored at –80 °C.
Covalent Tagging.
Labels (N-cyclohexylmaleimide or 3-maleimido-PROXYL) were added (20-fold excess over reactive cys) to a solution containing enzyme (50 μM, monomer), PAP (0.50 mM), KPO4 (50 mM), pH 7.4, 25 ± 2 °C. PAP was added to enhance enzyme stability (43). Reactions were monitored using DTNB to measure unreacted cysteine and were considered complete when >98% of the cysteine had reacted (∼3 h). Following completion, the reaction mixtures were dialyzed against PAP (0.50 mM), KPO4 (50 mM), pH 7.4, D2O (>95%), 4 ± 2 °C.
Initial-Rate Studies of Labeled Constructs and THB Binding-Site Mutants.
Initial-rate and THB-inhibition parameters were determined for the labeled constructs and THB binding-site mutants as previously described (14). Briefly, reactions are initiated by addition of PAPS (0.50 mM, 17 × Km) to a solution containing enzyme (25 nM), 1-HP (0.40 µM, ∼20 × Km), DTT (5.0 mM), and KPO4 (50 mM), pH 7.5, 25 ± 2 °C. Reaction progress was monitored via sulfonation-dependent change in 1-HP fluorescence [λex 325 nm, λex 370 nm (71)], and Km and Vmax are extracted from the data by progress-curve analysis (72). THB-inhibition studies used identical conditions. Ki was obtained from weighted least-squares fitting of initial rate versus [THB] data plotted in double reciprocal space (14). Inhibition by THB for the mutants used to verify the position of the THB-binding pocket (Y76A, D86A, E89I) was not detected at THB concentrations as high as 200 µM.
NMR Measurements.
Spectra were collected, using a Bruker DRX600 spectrometer equipped with a TCI H/F-cryogenic probe at 298 K, at the following THB concentrations: 0, 5, 10, 20, and 40× (active site). Peak widths for each THB proton used in the structure determination were obtained using NMRdraw (Lorentzian peak shape) (73). At each THB concentration, a 1D proton spectrum was collected from 0 to 14 ppm, using a 0.96 acquisition time and presaturating water pulse. Each spectrum was the time average of 512 scans, with a 1.5-s delay between each scan.
Screening for Human-Metabolite SULT1A3 Allosteres (GOLD Modeling).
SULT1A3 and SULT1A1 models were constructed using available crystal structures, 2A3R (74) and 2D06 (75), respectively, as described previously (72). In silico docking was performed using GOLD (76–78). The program was configured to use flexible side chains, 2,500 generations, and an initial population of 100 random positions. Docking was limited to a 15-Å radius around the center of the catechin-binding site. Simulations were performed three times for each compound. GOLD scores (which are indicative of binding affinity) were calibrated by constructing standard curves for SULT1A1 and 1A3 that correlate the GOLD scores and experimentally determined affinities of epigallocatechin gallate, epigallocatechin, epicatechin, and quercetin for each isoform.
Molecular Dynamics Modeling.
SWISS-MODEL (79) was used to create a ligand-free model of SULT1A3 from the SULT1A3·PAP⋅Dopamine (PDB ID code 2A3R) structure (74), which was missing 27 atoms. The model was protonated at pH 7.4 and energy-minimized in GROMACS (45). GROMAS57 energy-parameter files were created for THB, PAPS, and spin-labeled cysteines using Automated Topology Builder (49). The GROMAS57 field was modified in GROMACS to allow the program to recognize the spin-labeled cysteine as a canonical residue. Spin-labeled cysteines were inserted by replacing residues E26, E151, and K234; PAPS was positioned at the active site using GOLD (76–78); and the system was equilibrated using GROMACS (100 ps increments) to the following simulated condition: 298 K, NaCl (50 mM), pH 7.4. Once equilibrated, THB was positioned randomly in a simulated box of water (52 × 52 × 52 Å) containing the spin-labeled SULT1A3∙PAPS construct and then docked in GROMACS using the NMR distance constraints (Results and Discussion, Refining the Structure). The same structure was obtained each of the 10 times docking was repeated.
Equilibrium Binding Studies.
The binding of ligands to SULT1A3 was monitored via changes in the intrinsic fluorescence of the enzyme (λex 290 nm, λem 370 nm). Typical conditions were as follows: SULT (∼25 nM, dimer), PAP (0 or 0.50 mM, 17 × Kd low affinity), DTT (5.0 mM), KPO4 (50 mM), pH 7.5, 25 ± 2 °C. Titrations were performed in triplicate. Data were averaged and least-squares fit using a model that assumes a single binding site per monomer. THB concentration was varied from 0.10 to 20 × Kd. THB binding was not observed for the THB binding-site mutants (Y76A, D86A, E89I) at THB concentrations as high as 200 µM.
Acknowledgments
This work was supported by National Institutes of Health Grants GM106158 and GM121849.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704500114/-/DCSupplemental.
References
- 1.Riches Z, Stanley EL, Bloomer JC, Coughtrie MW. Quantitative evaluation of the expression and activity of five major sulfotransferases (SULTs) in human tissues: The SULT “pie”. Drug Metab Dispos. 2009;37:2255–2261. doi: 10.1124/dmd.109.028399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanchard RL, Freimuth RR, Buck J, Weinshilboum RM, Coughtrie MW. A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenetics. 2004;14:199–211. doi: 10.1097/00008571-200403000-00009. [DOI] [PubMed] [Google Scholar]
- 3.Nowell S, Falany CN. Pharmacogenetics of human cytosolic sulfotransferases. Oncogene. 2006;25:1673–1678. doi: 10.1038/sj.onc.1209376. [DOI] [PubMed] [Google Scholar]
- 4.Falany JL, Macrina N, Falany CN. Regulation of MCF-7 breast cancer cell growth by beta-estradiol sulfation. Breast Cancer Res Treat. 2002;74:167–176. doi: 10.1023/a:1016147004188. [DOI] [PubMed] [Google Scholar]
- 5.Visser TJ. Role of sulfation in thyroid hormone metabolism. Chem Biol Interact. 1994;92:293–303. doi: 10.1016/0009-2797(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 6.Matsubayashi Y, Sakagami Y. Peptide hormones in plants. Annu Rev Plant Biol. 2006;57:649–674. doi: 10.1146/annurev.arplant.56.032604.144204. [DOI] [PubMed] [Google Scholar]
- 7.Cook IT, Duniac-Dmuchowski Z, Kocarek TA, Runge-Morris M, Falany CN. 24-Hydroxycholesterol sulfation by human cytosolic sulfotransferases: Formation of monosulfates and disulfates, molecular modeling, sulfatase sensitivity and inhibition of LXR activation. Drug Metab Dispos. 2009;37:2069–2078. doi: 10.1124/dmd.108.025759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stowers L, Logan DW. Sexual dimorphism in olfactory signaling. Curr Opin Neurobiol. 2010;20:770–775. doi: 10.1016/j.conb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bistrup A, et al. Sulfotransferases of two specificities function in the reconstitution of high endothelial cell ligands for L-selectin. J Cell Biol. 1999;145:899–910. doi: 10.1083/jcb.145.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisenhofer G, Coughtrie MW, Goldstein DS. Dopamine sulphate: An enigma resolved. Clin Exp Pharmacol Physiol Suppl. 1999;26:S41–S53. [PubMed] [Google Scholar]
- 11.Cuche JL, et al. 1982. Plasma free and sulfoconjugated catecholamines in healthy men. Eur Heart J 3 Suppl C:3–8.
- 12.Cook I, Wang T, Girvin M, Leyh TS. The structure of the catechin-binding site of human sulfotransferase 1A1. Proc Natl Acad Sci USA. 2016;113:14312–14317. doi: 10.1073/pnas.1613913113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang T, Cook I, Leyh TS. Isozyme specific allosteric regulation of human sulfotransferase 1A1. Biochemistry. 2016;55:4036–4046. doi: 10.1021/acs.biochem.6b00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cook I, Wang T, Falany CN, Leyh TS. The allosteric binding sites of sulfotransferase 1A1. Drug Metab Dispos. 2015;43:418–423. doi: 10.1124/dmd.114.061887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coughtrie MW, Johnston LE. Interactions between dietary chemicals and human sulfotransferases-molecular mechanisms and clinical significance. Drug Metab Dispos. 2001;29:522–528. [PubMed] [Google Scholar]
- 16.Vietri M, De Santi C, Pietrabissa A, Mosca F, Pacifici GM. Inhibition of human liver phenol sulfotransferase by nonsteroidal anti-inflammatory drugs. Eur J Clin Pharmacol. 2000;56:81–87. doi: 10.1007/s002280050725. [DOI] [PubMed] [Google Scholar]
- 17.Sabhapondit S, Karak T, Bhuyan LP, Goswami BC, Hazarika M. Diversity of catechin in northeast Indian tea cultivars. Sci World J. 2012;2012:485193. doi: 10.1100/2012/485193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arts IC, van De Putte B, Hollman PC. Catechin contents of foods commonly consumed in The Netherlands. 2. Tea, wine, fruit juices, and chocolate milk. J Agric Food Chem. 2000;48:1752–1757. doi: 10.1021/jf000026+. [DOI] [PubMed] [Google Scholar]
- 19.Cordero-Herrera I, Martín MÁ, Goya L, Ramos S. Cocoa flavonoids attenuate high glucose-induced insulin signalling blockade and modulate glucose uptake and production in human HepG2 cells. Food Chem Toxicol. 2014;64:10–19. doi: 10.1016/j.fct.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 20.Pacifici GM. Inhibition of human liver and duodenum sulfotransferases by drugs and dietary chemicals: A review of the literature. Int J Clin Pharmacol Ther. 2004;42:488–495. doi: 10.5414/cpp42488. [DOI] [PubMed] [Google Scholar]
- 21.Racz K, Buu NT, Kuchel O. Regional distribution of free and sulfoconjugated catecholamines in the bovine adrenal cortex and medulla. Can J Physiol Pharmacol. 1984;62:622–626. doi: 10.1139/y84-100. [DOI] [PubMed] [Google Scholar]
- 22.Kuchel O, Hausser C, Buu NT, Tenneson S. CSF sulfoconjugated catecholamines in man: Their relationship with plasma catecholamines. J Neural Transm. 1985;62:91–97. doi: 10.1007/BF01260418. [DOI] [PubMed] [Google Scholar]
- 23.Davidson L, Vandongen R, Beilin LJ, Arkwright PD. Free and sulfate-conjugated catecholamines during exercise in man. J Clin Endocrinol Metab. 1984;58:415–418. doi: 10.1210/jcem-58-3-415. [DOI] [PubMed] [Google Scholar]
- 24.Yoshizumi M, et al. Physiological significance of plasma sulfoconjugated dopamine: Experimental and clinical studies. Hypertens Res. 1995;18:S101–S106. doi: 10.1291/hypres.18.supplementi_s101. [DOI] [PubMed] [Google Scholar]
- 25.Bergey CM, Phillips-Conroy JE, Disotell TR, Jolly CJ. Dopamine pathway is highly diverged in primate species that differ markedly in social behavior. Proc Natl Acad Sci USA. 2016;113:6178–6181. doi: 10.1073/pnas.1525530113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Youdim MB, Edmondson D, Tipton KF. The therapeutic potential of monoamine oxidase inhibitors. Nat Rev Neurosci. 2006;7:295–309. doi: 10.1038/nrn1883. [DOI] [PubMed] [Google Scholar]
- 27.Scott LJ. Opicapone: A review in Parkinson’s disease. Drugs. 2016;76:1293–1300. doi: 10.1007/s40265-016-0623-y. [DOI] [PubMed] [Google Scholar]
- 28.Jewison T, et al. SMPDB 2.0: Big improvements to the Small Molecule Pathway Database. Nucleic Acids Res. 2014;42:D478–D484. doi: 10.1093/nar/gkt1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salman ED, Kadlubar SA, Falany CN. Expression and localization of cytosolic sulfotransferase (SULT) 1A1 and SULT1A3 in normal human brain. Drug Metab Dispos. 2009;37:706–709. doi: 10.1124/dmd.108.025767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sidharthan NP, Minchin RF, Butcher NJ. Cytosolic sulfotransferase 1A3 is induced by dopamine and protects neuronal cells from dopamine toxicity: Role of D1 receptor-N-methyl-D-aspartate receptor coupling. J Biol Chem. 2013;288:34364–34374. doi: 10.1074/jbc.M113.493239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teubner W, Meinl W, Florian S, Kretzschmar M, Glatt H. Identification and localization of soluble sulfotransferases in the human gastrointestinal tract. Biochem J. 2007;404:207–215. doi: 10.1042/BJ20061431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duch DS, Bowers SW, Woolf JH, Nichol CA. Biopterin cofactor biosynthesis: GTP cyclohydrolase, neopterin and biopterin in tissues and body fluids of mammalian species. Life Sci. 1984;35:1895–1901. doi: 10.1016/0024-3205(84)90541-1. [DOI] [PubMed] [Google Scholar]
- 33.Levine RA, Miller LP, Lovenberg W. Tetrahydrobiopterin in striatum: Localization in dopamine nerve terminals and role in catecholamine synthesis. Science. 1981;214:919–921. doi: 10.1126/science.6117945. [DOI] [PubMed] [Google Scholar]
- 34.Werner-Felmayer G, et al. Pteridine biosynthesis in human endothelial cells. Impact on nitric oxide-mediated formation of cyclic GMP. J Biol Chem. 1993;268:1842–1846. [PubMed] [Google Scholar]
- 35.Simmons WW, Ungureanu-Longrois D, Smith GK, Smith TW, Kelly RA. Glucocorticoids regulate inducible nitric oxide synthase by inhibiting tetrahydrobiopterin synthesis and L-arginine transport. J Biol Chem. 1996;271:23928–23937. doi: 10.1074/jbc.271.39.23928. [DOI] [PubMed] [Google Scholar]
- 36.Franscini N, et al. Functional tetrahydrobiopterin synthesis in human platelets. Circulation. 2004;110:186–192. doi: 10.1161/01.CIR.0000134281.82972.57. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe E, Sasakawa S. Changes of platelet cell volumes in hypotonic solution. Thromb Res. 1983;31:13–21. doi: 10.1016/0049-3848(83)90003-8. [DOI] [PubMed] [Google Scholar]
- 38.Cavalieri E, Rogan E, Chakravarti D. The role of endogenous catechol quinones in the initiation of cancer and neurodegenerative diseases. Methods Enzymol. 2004;382:293–319. doi: 10.1016/S0076-6879(04)82017-2. [DOI] [PubMed] [Google Scholar]
- 39.Choi HJ, Kim SW, Lee SY, Hwang O. Dopamine-dependent cytotoxicity of tetrahydrobiopterin: A possible mechanism for selective neurodegeneration in Parkinson’s disease. J Neurochem. 2003;86:143–152. doi: 10.1046/j.1471-4159.2003.01808.x. [DOI] [PubMed] [Google Scholar]
- 40.Guan JY, et al. Small-molecule binding sites on proteins established by paramagnetic NMR spectroscopy. J Am Chem Soc. 2013;135:5859–5868. doi: 10.1021/ja401323m. [DOI] [PubMed] [Google Scholar]
- 41.Gochin M, Zhou G, Phillips AH. Paramagnetic relaxation assisted docking of a small indole compound in the HIV-1 gp41 hydrophobic pocket. ACS Chem Biol. 2011;6:267–274. doi: 10.1021/cb100368d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Solomon I. Relaxation processes in a system of two spins. Phys Rev. 1955;99:559–566. [Google Scholar]
- 43.Girvin ME, Fillingame RH. Determination of local protein structure by spin label difference 2D NMR: The region neighboring Asp61 of subunit c of the F1F0 ATP synthase. Biochemistry. 1995;34:1635–1645. doi: 10.1021/bi00005a020. [DOI] [PubMed] [Google Scholar]
- 44.Van Der Spoel D, et al. GROMACS: Fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 45.Berendsen HJC, Vanderspoel D, Vandrunen R. Gromacs - A message-passing parallel molecular-dynamics implementation. Comput Phys Commun. 1995;91:43–56. [Google Scholar]
- 46.Schmitz U, Ulyanov NB, Kumar A, James TL. Molecular dynamics with weighted time-averaged restraints for a DNA octamer. Dynamic interpretation of nuclear magnetic resonance data. J Mol Biol. 1993;234:373–389. doi: 10.1006/jmbi.1993.1593. [DOI] [PubMed] [Google Scholar]
- 47.Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 48.Koziara KB, Stroet M, Malde AK, Mark AE. Testing and validation of the Automated Topology Builder (ATB) version 2.0: Prediction of hydration free enthalpies. J Comput Aided Mol Des. 2014;28:221–233. doi: 10.1007/s10822-014-9713-7. [DOI] [PubMed] [Google Scholar]
- 49.Malde AK, et al. An automated force field topology builder (ATB) and repository: Version 1.0. J Chem Theory Comput. 2011;7:4026–4037. doi: 10.1021/ct200196m. [DOI] [PubMed] [Google Scholar]
- 50.Benton MJ, et al. Constraints on the timescale of animal evolutionary history. Palaeontol Electronica. 2015;18:1–107. [Google Scholar]
- 51.Feduccia A. Explosive evolution in tertiary birds and mammals. Science. 1995;267:637–638. doi: 10.1126/science.267.5198.637. [DOI] [PubMed] [Google Scholar]
- 52.Bradley ME, Benner SA. Phylogenomic approaches to common problems encountered in the analysis of low copy repeats: The sulfotransferase 1A gene family example. BMC Evol Biol. 2005;5:22. doi: 10.1186/1471-2148-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dajani R, Hood AM, Coughtrie MW. A single amino acid, glu146, governs the substrate specificity of a human dopamine sulfotransferase, SULT1A3. Mol Pharmacol. 1998;54:942–948. doi: 10.1124/mol.54.6.942. [DOI] [PubMed] [Google Scholar]
- 54.Hattori Y, Oka M, Kasai K, Nakanishi N, Shimoda SI. Lipopolysaccharide treatment in vivo induces tissue expression of GTP cyclohydrolase I mRNA. FEBS Lett. 1995;368:336–338. doi: 10.1016/0014-5793(95)00689-7. [DOI] [PubMed] [Google Scholar]
- 55.Mansouri FA, Tanaka K, Buckley MJ. Conflict-induced behavioural adjustment: A clue to the executive functions of the prefrontal cortex. Nat Rev Neurosci. 2009;10:141–152. doi: 10.1038/nrn2538. [DOI] [PubMed] [Google Scholar]
- 56.Russo SJ, Nestler EJ. The brain reward circuitry in mood disorders. Nat Rev Neurosci. 2013;14:609–625. doi: 10.1038/nrn3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berton O, Nestler EJ. New approaches to antidepressant drug discovery: Beyond monoamines. Nat Rev Neurosci. 2006;7:137–151. doi: 10.1038/nrn1846. [DOI] [PubMed] [Google Scholar]
- 58.Riederer P, Müller T. Use of monoamine oxidase inhibitors in chronic neurodegeneration. Expert Opin Drug Metab Toxicol. 2017;13:233–240. doi: 10.1080/17425255.2017.1273901. [DOI] [PubMed] [Google Scholar]
- 59.Männistö PT, Kaakkola S. Catechol-O-methyltransferase (COMT): Biochemistry, molecular biology, pharmacology, and clinical efficacy of the new selective COMT inhibitors. Pharmacol Rev. 1999;51:593–628. [PubMed] [Google Scholar]
- 60.Chen J, et al. Orientation and cellular distribution of membrane-bound catechol-O-methyltransferase in cortical neurons: Implications for drug development. J Biol Chem. 2011;286:34752–34760. doi: 10.1074/jbc.M111.262790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suominen T, et al. Determination of serotonin and dopamine metabolites in human brain microdialysis and cerebrospinal fluid samples by UPLC-MS/MS: Discovery of intact glucuronide and sulfate conjugates. PLoS One. 2013;8:e68007. doi: 10.1371/journal.pone.0068007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buu NT. Relationship between catechol-O-methyltransferase and phenolsulfotransferase in the metabolism of dopamine in the rat brain. J Neurochem. 1985;45:1612–1619. doi: 10.1111/j.1471-4159.1985.tb07234.x. [DOI] [PubMed] [Google Scholar]
- 63.Buu NT. Dopamine sulfoconjugation in the rat brain: Regulation by monoamine oxidase. J Neurochem. 1985;45:470–476. doi: 10.1111/j.1471-4159.1985.tb04012.x. [DOI] [PubMed] [Google Scholar]
- 64.Thomas SJ, Shin M, McInnis MG, Bostwick JR. Combination therapy with monoamine oxidase inhibitors and other antidepressants or stimulants: Strategies for the management of treatment-resistant depression. Pharmacotherapy. 2015;35:433–449. doi: 10.1002/phar.1576. [DOI] [PubMed] [Google Scholar]
- 65.Wang LQ, James MO. Sulfotransferase 2A1 forms estradiol-17-sulfate and celecoxib switches the dominant product from estradiol-3-sulfate to estradiol-17-sulfate. J Steroid Biochem Mol Biol. 2005;96:367–374. doi: 10.1016/j.jsbmb.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 66.Ho BK, Gruswitz F. HOLLOW: Generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct Biol. 2008;8:49. doi: 10.1186/1472-6807-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang T, Cook I, Leyh TS. 3′-Phosphoadenosine 5′-phosphosulfate allosterically regulates sulfotransferase turnover. Biochemistry. 2014;53:6893–6900. doi: 10.1021/bi501120p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang T, Cook I, Leyh TS. Design and interpretation of human sulfotransferase 1A1 assays. Drug Metab Dispos. 2016;44:481–484. doi: 10.1124/dmd.115.068205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cook I, et al. The gate that governs sulfotransferase selectivity. Biochemistry. 2013;52:415–424. doi: 10.1021/bi301492j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Andreassi JL, 2nd, Leyh TS. Molecular functions of conserved aspects of the GHMP kinase family. Biochemistry. 2004;43:14594–14601. doi: 10.1021/bi048963o. [DOI] [PubMed] [Google Scholar]
- 71.Ma B, Shou M, Schrag ML. Solvent effect on cDNA-expressed human sulfotransferase (SULT) activities in vitro. Drug Metab Dispos. 2003;31:1300–1305. doi: 10.1124/dmd.31.11.1300. [DOI] [PubMed] [Google Scholar]
- 72.Cook I, Wang T, Falany CN, Leyh TS. High accuracy in silico sulfotransferase models. J Biol Chem. 2013;288:34494–34501. doi: 10.1074/jbc.M113.510974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 74.Lu JH, et al. Crystal structure of human sulfotransferase SULT1A3 in complex with dopamine and 3′-phosphoadenosine 5′-phosphate. Biochem Biophys Res Commun. 2005;335:417–423. doi: 10.1016/j.bbrc.2005.07.091. [DOI] [PubMed] [Google Scholar]
- 75.Gamage NU, Tsvetanov S, Duggleby RG, McManus ME, Martin JL. The structure of human SULT1A1 crystallized with estradiol. An insight into active site plasticity and substrate inhibition with multi-ring substrates. J Biol Chem. 2005;280:41482–41486. doi: 10.1074/jbc.M508289200. [DOI] [PubMed] [Google Scholar]
- 76.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein-ligand docking using GOLD. Proteins. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 77.Verdonk ML, et al. Virtual screening using protein-ligand docking: Avoiding artificial enrichment. J Chem Inf Comput Sci. 2004;44:793–806. doi: 10.1021/ci034289q. [DOI] [PubMed] [Google Scholar]
- 78.Verdonk ML, et al. Modeling water molecules in protein-ligand docking using GOLD. J Med Chem. 2005;48:6504–6515. doi: 10.1021/jm050543p. [DOI] [PubMed] [Google Scholar]
- 79.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 80.Peres I, et al. NMR structural analysis of epigallocatechin gallate loaded polysaccharide nanoparticles. Carbohydr Polym. 2010;82:861–866. [Google Scholar]