Significance
Of the many receptors that were pharmacologically described during the 20th century, almost all were cloned by the end of the 1990s. A key exception is the σ2 receptor, a potential therapeutic target for diseases as diverse as schizophrenia, Alzheimer’s disease, and cancer. Despite the development of a rich pharmacopeia, the unknown molecular identity of the receptor has crippled biological investigation. Here, we identify the σ2 receptor as TMEM97, a membrane protein implicated in cancer and a binding partner of Niemann–Pick disease protein NPC1. Our results unite two fields of research, bringing the σ2 receptor into the modern age of biological inquiry and providing the TMEM97 field with a rich pool of ligands and pharmacological tools.
Keywords: sigma-2 receptor, sigma receptors, TMEM97, cholesterol regulation, NPC1
Abstract
The σ2 receptor is an enigmatic protein that has attracted significant attention because of its involvement in diseases as diverse as cancer and neurological disorders. Unlike virtually all other receptors of medical interest, it has eluded molecular cloning since its discovery, and the gene that codes for the receptor remains unknown, precluding the use of modern biological methods to study its function. Using a chemical biology approach, we purified the σ2 receptor from tissue, revealing its identity as TMEM97, an endoplasmic reticulum-resident transmembrane protein that regulates the sterol transporter NPC1. We show that TMEM97 possesses the full suite of molecular properties that define the σ2 receptor, and we identify Asp29 and Asp56 as essential for ligand recognition. Cloning the σ2 receptor resolves a longstanding mystery and will enable therapeutic targeting of this potential drug target.
Pharmacological approaches such as radioactive ligand-binding assays have enabled the identification and characterization of a plethora of cellular receptors for hormones, peptides, neurotransmitters, and other biologically active molecules (1). Among the receptors thus identified are the enigmatic σ receptors, which were first reported in 1976 and later classified into σ1 and σ2 subtypes (2, 3). Recently, the σ2 receptor has emerged as a potential therapeutic target, and compounds targeting it are now in clinical trials for the diagnosis of breast cancer (4) and for the treatment of Alzheimer’s disease (5–7) and schizophrenia (8). However, despite the increasingly apparent medical importance of the σ2 receptor, progress toward understanding its biological role has been stymied because the gene that encodes the receptor has never been identified.
The σ2 receptor was initially identified by radioligand-binding studies as a binding site with high affinity for di-o-tolylguanidine (DTG) (Ki = 21.2 nM) and haloperidol (Ki = 48.7 nM) (3) but with low affinity for (+)-benzomorphans (9). In contrast, the σ1 receptor shows high-affinity binding for all these ligands (10). The σ2 receptor is known to be an 18- to 21-kDa membrane protein (3), and ligand-binding studies have shown that it is highly expressed in the liver and kidney (11, 12) and in the CNS (13) as well as in several cancer cell lines (14) and proliferating tumors (15). These intriguing features and the potential medical applications of compounds that modulate σ2 have stimulated a number of investigations directed toward the development of σ2 receptor ligands, and numerous high-affinity and subtype-selective compounds have been developed as a result (12, 16–20).
In 1996, the cloning of the σ1 receptor revealed that it has no mammalian paralogs (21), and subsequent σ1-knockout mouse studies demonstrated that the pharmacologically similar σ2 subtype derives from an altogether different, unknown gene (22). In 2011, it was reported that the σ2 receptor-binding site resides within the progesterone receptor membrane component 1 (PGRMC1) protein complex (23). However, subsequent reports have shown that overexpression of PGRMC1 does not increase σ2 receptor binding (24, 25), and σ2-binding levels are unaffected by either siRNA knockdown or CRISPR-Cas9 knockout of PGRMC1 (25). Moreover, a recent report using fluorescent σ2 ligands also showed that binding levels were unaffected by whether PGRMC1 was overexpressed or knocked down (26), further confirming that the σ2 receptor and PGRMC1 are two distinct molecular entities. The identity of the gene encoding σ2 thus continues to elude discovery despite almost 30 years of effort, posing a major roadblock to understanding σ2 receptor biology and therapeutic potential. To address this problem, we sought to leverage chemistry being developed in our laboratories toward σ2 receptor ligand discovery to identify the σ2 receptor.
Results
Affinity Purification of σ2 from Calf Liver Tissue.
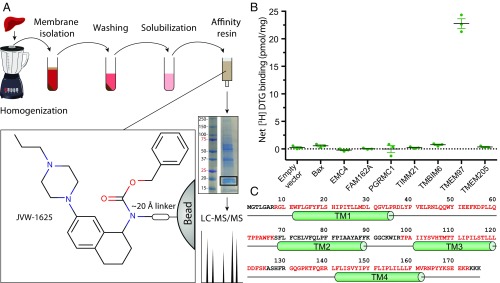
We first synthesized JVW-1625 (Ki = 16.6 nM), a σ2 receptor-binding ligand that was derived from the high-affinity σ2 ligand JVW-1601 (σ2 Ki = 19.6 nM) (SI Appendix, Fig. S1). We then selected a tissue source material (SI Appendix, Fig. S2A) and devised a protocol to extract σ2 efficiently from calf liver membranes in a functional form (SI Appendix, Fig. S2B). We next covalently coupled JVW-1625 to agarose beads to prepare an affinity chromatography resin (SI Appendix, Fig. S2C) that was used to capture σ2 extracted from calf liver membranes (Fig. 1). The resulting eluate was separated by SDS/PAGE and analyzed by mass spectrometry. From the many proteins identified by mass spectrometry, seven candidates were prioritized for validation on the basis of being membrane proteins of the expected molecular weight. These proteins along with PGRMC1 were overexpressed in HEK293 cells and were assayed for their ability to bind 3H DTG at a site competitive with haloperidol, a core feature of the pharmacological definition of the σ2 receptor (Fig. 1B). Among the candidate proteins tested, only the endoplasmic reticulum (ER)-resident membrane protein TMEM97 resulted in a significant increase of net 3H DTG binding (Fig. 1 B and C). As reported by others (24, 25), PGRMC1 overexpression did not lead to increased specific 3H DTG binding.
Fig. 1.
Purification and molecular cloning of the σ2 receptor. (A) A schematic of the purification of σ2 from calf liver. The Inset depicts the portion of the ligand that binds σ2. (B) Single-point 3H DTG-binding assay of membrane preparations from expi293 cells expressing LC-MS/MS hits, shown as means ± SEM for an experiment performed in triplicate. (C) The sequence of TMEM97. Red font indicates regions that were identified by LC-MS/MS. Green cylinders denote predicted transmembrane (TM) helices.
The Pharmacological Profile of TMEM97 Is Identical to That of the σ2 Receptor.
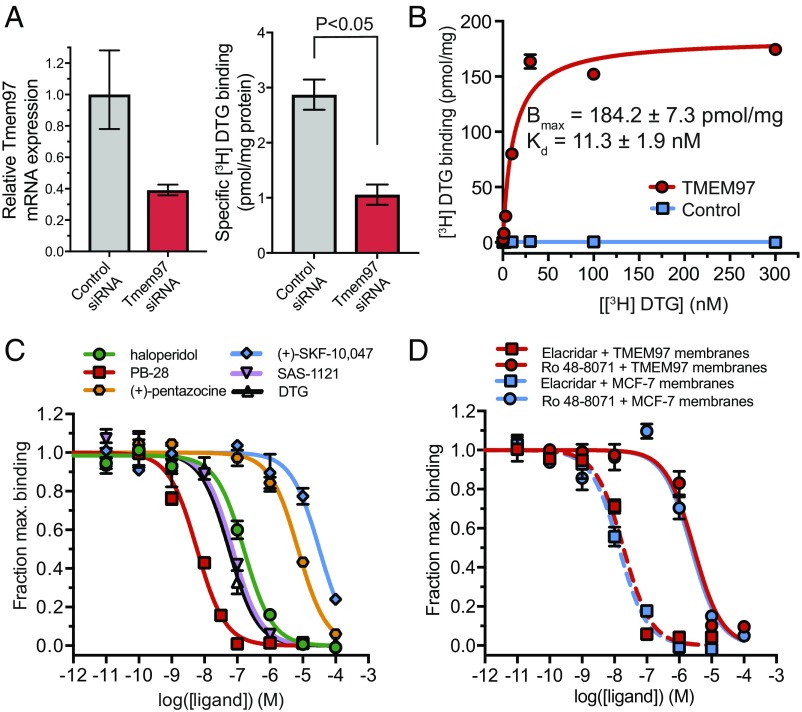
If TMEM97 is indeed the σ2 receptor, its expression should be required for σ2 binding activity. To test whether TMEM97 expression contributes to the σ2 site, we used siRNA knockdown to reduce mRNA expression of Tmem97 by ∼60% in PC-12 cells, a classical σ2 receptor cell line. This knockdown resulted in a nearly identical reduction in σ2 expression levels as measured by saturating 3H DTG binding (Fig. 2A). We then assessed whether TMEM97 possesses the same ligand-binding profile as the σ2 receptor. To do so, human TMEM97 was overexpressed in Sf9 insect cells, which lack an endogenous TMEM97 homolog (27) and show no appreciable 3H DTG binding. Expression of TMEM97 in these cells conferred saturable 3H DTG binding with an affinity of 11.3 nM, a value closely in line with literature values ranging from 17.9 to 37.6 nM (Fig. 2B) (3, 9, 14, 24). Competition binding experiments with a collection of σ2 ligands representing diverse chemical classes show that the affinity of each ligand for TMEM97 is consistent with previously reported σ2 receptor-binding affinities (Fig. 2C, Table 1, and SI Appendix, Fig. S3) (20).
Fig. 2.
Pharmacological validation of TMEM97 as the σ2 receptor. (A) PC-12 cells treated with Tmem97-targeted or control siRNA were tested to measure Tmem97 mRNA levels (Left) and σ2 expression by 3H DTG binding (Right). qPCR data are shown as mean ± SD. Radioligand binding data are shown as mean ± SEM and are representative of two independent experiments performed in triplicate. (B) 3H DTG saturation binding on Sf9 insect cell membranes expressing either TMEM97 (red circles) or the negative control β2 adrenergic receptor (blue squares). (C) 3H DTG competition curves in Sf9 insect cell membranes expressing TMEM97 against the known σ2 receptor ligands haloperidol (green circles), PB-28 (red squares), (+)-pentazocine (orange hexagons), (+)-SKF-10,047 (blue diamonds), SAS-1121 (purple triangles), and cold DTG (white triangles). (D) 3H DTG competition curves with the known TMEM97 ligands Elacridar (squares, dotted line) and Ro 48-8071 (circles, solid line) in Sf9 insect cell membranes expressing TMEM97 (red) and in MCF-7 cell membranes (blue). For all curves, data points are shown as mean ± SEM and are representative of two independent experiments performed in triplicate.
Table 1.
Radioligand-binding analysis
| Ligand | Ki for TMEM97 in Sf9 cells, nM | σ2 Ki, nM |
| DTG | 19.6 ± 3.0 | 21.2* |
| Haloperidol | 54.1 ± 6.7 | 48.7* |
| PB-28 | 2.0 ± 0.3 | 1.8† |
| (+)-pentazocine | 2,467 ± 436 | 1,402* – 3,890† |
| (+)-SKF 10,047 | >10,000 | No inhibition* |
| SAS-1121 | 25.2 ± 3.8 | 23.8‡ |
| Elacridar | 6.5 ± 1.1 | 4.7 ± 0.7 |
| Ro 48-8071 | 971 ± 228 | 817 ± 226 |
The inhibition constant (Ki) for various ligands binding to recombinant TMEM97 and to the σ2 receptor natively expressed in cell membranes. Affinities were measured as described in Methods and are reported from literature where indicated.
Data from Hellewell and Bowen, 1990 (3).
Data from Colabufo et al., 2004 (49).
Data from Sahn et al., 2017 (20).
TMEM97 Ligands Bind the σ2 Receptor.
Having demonstrated that TMEM97 expression is both necessary and sufficient for σ2 receptor-binding activity, we tested whether two recently reported TMEM97 ligands, Elacridar and Ro 48-8071 (28), could bind the σ2 receptor in a classical binding assay. The measured binding affinities of these compounds to Sf9 membranes overexpressing TMEM97 were identical to those measured in MCF-7 cell membranes, a classical σ2 receptor cell line (Fig. 2D and Table 1) (14). Hence, known σ2 ligands bind to TMEM97, and, conversely, known TMEM97 ligands bind to the σ2 receptor. In each case, ligand affinities for the σ2 receptor in a classical binding assay are identical to those for recombinant TMEM97. Collectively, these data lead us to conclude that TMEM97 is synonymous with the σ2 receptor.
Structural Insights into TMEM97 Ligand Recognition.
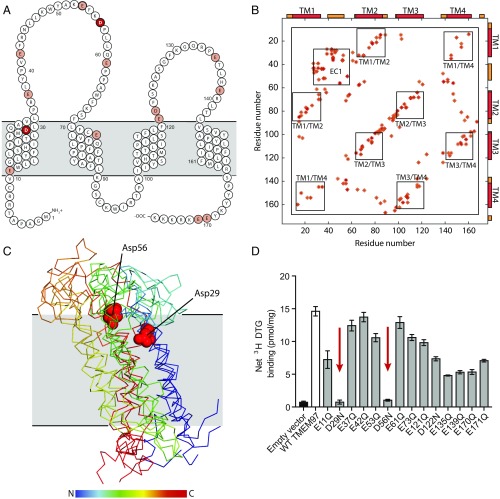
To map the ligand-binding site, we performed site-directed mutagenesis of all Glu and Asp residues, hypothesizing that one of these must be the counter ion to the basic amine found in all σ2 ligands. Mutation of two conserved aspartate residues (SI Appendix, Fig. S4), D29N and D56N, abolished all binding to 3H DTG (Fig. 3D and SI Appendix, Fig. S5). We then performed evolutionary coupling analysis (29) that revealed strong antiparallel correlations among predicted transmembrane helices (Fig. 3B). Using these data for ab initio structure prediction suggested that the protein likely possesses a four-helix bundle fold (Fig. 3C). The two Asp residues essential for binding are located in close proximity to one another in our predicted structural model (Fig. 3C). That two acidic groups are essential, rather than just one, is reminiscent of the σ1 receptor, in which both Asp126 and Glu172 are required for ligand binding (30); in the case of σ1 these two residues interact directly with each other as part of a hydrogen bond network that includes the basic amine of the ligand (30). Asp29 and Asp56 may play a similar role in ligand binding to the σ2 receptor.
Fig. 3.
Structural analysis of TMEM97 and mapping of the ligand-binding site. (A) A schematic of TMEM97 in the membrane. Acidic residues chosen for mutagenesis are colored pale red, with Asp29 and Asp56 highlighted in dark red. Membrane helices and TMEM97 topology are according to TMHMM server prediction. (B) Evolutionary coupling map of the top 140 pairs of human TMEM97. Transmembrane helices (TM) and the extracellular domain (EC) are indicated and boxed. (C) Molecular models of TMEM97 generated by evolutionary coupling analysis (29). The top four ranked models are presented. Models are rainbow colored from the N terminus to the C terminus as indicated. The location of the membrane plane was calculated by the PPM server (48) and is shown in gray. Asp29 and Asp56 are shown as red spheres. For clarity a single representative residue pair is presented. (D) Single-point 3H DTG-binding assay of membrane preparations from Expi293 cells expressing TMEM97 single-point mutants. Empty vector is shown as a black bar, wild-type TMEM97 is shows as a white bar, and point mutants are shown as gray bars. Positions 29 and 56, which show abolished binding, are indicated by red arrows. Data are shown as means ± SEM for an experiment performed in triplicate.
Discussion
In contrast to the hundreds of studies of σ2 receptor biology, comparatively little is known about TMEM97. Although its molecular function is not well understood, TMEM97 has attracted interest because of its involvement in cholesterol homeostasis (31). TMEM97 is a binding partner of the lysosomal cholesterol transporter NPC1, whose loss causes Niemann–Pick disease type C1, a fatal lysosomal storage disorder (32). The connection between the σ2 receptor and sterol biology is not entirely unforeseen, because pharmacological studies in the mid-1990s had suggested a possible connection between σ receptors and cholesterol metabolism (33). Moreover, TMEM97 has also been recently described as a highly ligandable protein because of its ability to bind multiple small-molecule chemotypes (28), a finding consistent with the relatively large number of σ2 ligands reported to date (12, 13, 34).
Additionally, the proposed role of σ2 receptor in cancer cell proliferation fits well with its identity as TMEM97. For many years, it has been known that the σ2 receptor is overexpressed in several rat and human cancer cell lines, and it has been reported that the receptor is particularly overexpressed in proliferating tumors (12, 14). Similarly, TMEM97, which is also known as “MAC30,” is overexpressed in epithelial, lung, colorectal, ovarian, and breast cancers (35–39). In some of these cancers, TMEM97 expression has been correlated with poor prognosis and metastasis (35, 37). Furthermore, a recent study has reported that RNA silencing of TMEM97 in human gastric cancer cells can inhibit cell growth (40), and pharmacological targeting of the σ2 receptor in human cancer cell lines has been shown to exert antiproliferative effects (12). Together, these data support the identity of the σ2 receptor as TMEM97, and the unification of σ2 receptor pharmacology and the emerging biology of TMEM97 may be useful for assessing the value of this receptor as a therapeutic target.
Revealing the identity of the σ2 receptor as TMEM97 makes it possible to apply parallel computational and mutagenic analysis to develop a better understanding of the molecular basis for the binding of σ2 ligands to their target (41). Sequence analysis indicates that TMEM97 consists of an “EXPERA” domain, which in humans is shared among TMEM97 and its distant homologs TM6SF1, TM6SF2, EBP, and EBPL (42). Like TMEM97, these genes are implicated in cholesterol biology, and one of them, TM6SF2, has been implicated in nonalcoholic fatty liver disease in humans (43). Like other EXPERA domains, TMEM97 is predicted to have a four-pass transmembrane topology with cytosolic N and C termini (42), the latter of which contains the predicted ER-retention sequence “KRKKK” (Fig. 3A).
The identification of the σ2 receptor as TMEM97 resolves a longstanding pharmacological mystery and opens the door to applying the full suite of modern molecular biology tools and techniques to mechanistic studies of this receptor. Furthermore, because TMEM97 appears to be involved in cholesterol homeostasis, a ready trove of ligands that had originally been identified as σ2 receptor binders may now be used to study pathologies associated with aberrant cholesterol trafficking. Similarly, other EXPERA domain proteins, such as TM6SF2, may be amenable to ligand discovery aided by the abundant collection of σ2 ligands developed to date. The molecular cloning of the σ2 receptor provides an opportunity to revisit the biology and pharmacology of this receptor with implications for all areas of σ2 research, ranging from cancer to neuroscience.
Methods
Purification of the σ2 Receptor from Calf Liver.
Frozen calf livers (Omaha Steaks) were thawed, cut into 1-cm cubes, and suspended in a buffer of 20 mM Hepes (pH 7.5), 2 mM magnesium chloride, and 1:100,000 (vol:vol) benzonase nuclease (Sigma Aldrich), supplemented with cOmplete Mini, EDTA-free Protease Inhibitor Mixture Tablets (Roche). Tissue was homogenized with a blender and then was centrifuged for 20 min at 50,000 × g. The supernatant was discarded, and the pelleted membranes were washed by resuspension with a glass Dounce tissue grinder in Hepes-buffered saline (HBS) [20 mM Hepes (pH 7.5), 150 mM NaCl]. Washing was repeated until the protein content in the supernatant was below detection (typically 5–10 washes). Membranes were washed further with HBS supplemented with 2 M urea and then with HBS supplemented with 0.5 M sodium chloride to remove peripheral membrane proteins.
To extract the receptor, membranes were homogenized with a glass Dounce tissue grinder in a 1:5 (vol/vol) solubilization buffer consisting of 150 mM NaCl, 20 mM Hepes (pH 7.5), 10% (vol/vol) glycerol, and 1% (wt/vol) lauryl maltose neopentyl glycol (LMNG) (Anatrace). Samples were stirred for 2 h at 4 °C and then were centrifuged as before for 20 min. The resulting supernatant was filtered with a glass microfiber filter (VWR). The filtered supernatant containing solubilized receptor was loaded by gravity flow onto 2-mL affinity resin made by coupling compound JVW-1625 at 100 μM density to Affi-gel 10 (Bio-Rad) according to the manufacturer’s instructions. The resin was washed with 50 mL of buffer containing 150 mM NaCl, 20 mM Hepes (pH 7.5), 1% glycerol, and 0.1% LMNG. The receptor was eluted with 50 mL of the same buffer supplemented with 100 μM DTG using a syringe pump over a period of 3 h and with the eluate flowing directly onto a 250-μL hydroxyapatite resin column. Receptor then was eluted from hydroxyapatite resin using 500 μL of a buffer containing 500 mM potassium chloride (pH 7.2), 25 mM NaCl, and 0.1% (wt/wt) LMNG. Proteins were precipitated by trichloroacetic acid and were resolved on SDS/PAGE. A segment of the gel corresponding to a mass range of 15–25 kDa was sent for LC-MS/MS analysis at the Harvard Medical School Taplin Mass Spectrometry Facility and at Harvard’s Faculty of Arts and Sciences Mass Spectrometry and Proteomics Resource Laboratory.
Recombinant Receptor Expression.
Eight selected hits from LC-MS/MS were cloned into a pTARGET vector (Promega), followed by a porcine teschovirus-1 2A skip peptide (ATNFSLLKQAGDVEENPGP) and by the fluorescent protein mCardinal (44) to assess transfection efficiency. Plasmids were transfected into Expi293 cells (Thermo Fisher) according to the manufacturer’s instructions. After 36 h, expression of each target protein was confirmed by flow cytometry analysis of mCardinal fluorescence levels. Receptor point mutants were generated by site-directed mutagenesis using KAPA polymerase (KAPA Biosystems), and resulting constructs were expressed in Expi293 cells as described above.
For insect cell expression, human TMEM97 was cloned into the vector pVL1392, and baculovirus was prepared using the BestBac system (Expression Systems) in accordance with the manufacturer’s instructions. For large-scale production, Sf9 insect cells were infected at a density of 4 × 106 cells/mL and then were shaken at 27 °C for 60 h before harvest.
Preparation of Cell Membranes from Cultured Cells.
Membranes were prepared from PC-12, MCF-7, Sf9, or Expi293 cells with a protocol based on that of Vilner et al. (14) In brief, adherent cells were washed with ice-cold PBS or HBS and were harvested with a cell scraper; suspension cells were simply pelleted. Cell pellets were suspended in 20 mM Hepes (pH 7.5), 2 mM magnesium chloride, and 1:100,000 (vol:vol) benzonase nuclease (Sigma Aldrich) and were supplemented with cOmplete Mini, EDTA-free Protease Inhibitor Mixture Tablets (Roche). Following Dounce homogenization, the cells were centrifuged at 50,000 × g for 20 min. The supernatant was discarded, and the membranes were washed one more time with cold 50 mM Tris (pH 8.0) containing one cOmplete Mini, EDTA-free Protease Inhibitor Mixture Tablet (Roche) per 50 mL buffer. The membranes were centrifuged once more at 50,000 × g for 20 min and then were resuspended in a variable volume of cold 50 mM Tris (pH 8.0) with the same protease inhibitor mixture. Protein content was assessed by DC protein assay (Bio-Rad) according to the manufacturer’s instructions. Membranes were aliquoted, flash frozen, and stored at −80 °C until use in the radioligand binding experiments described below.
Single-Point Radioligand-Binding Assays.
Membrane radioligand-binding assays were performed as described (45) with slight modifications. Briefly, samples were incubated with 10–30 nM 3H DTG (Perkin-Elmer) in 50 mM Tris (pH 8) buffer supplemented with either 1.8 μM (+)SKF-10,047 or 50 nM PD-144418, both potent and selective σ1 receptor ligands (46), to block σ1 sites. Nonspecific binding was measured by the addition of 2 μM haloperidol to otherwise identical conditions measured in parallel. For siRNA experiments in PC-12 cells, 30 nM 3H DTG was isotopically diluted with 270 nM cold DTG to ensure that total σ2 binding was assayed. Samples were incubated at room temperature with shaking for 1.5 h; then the reaction was terminated by the addition of ice-cold water. Samples then were applied to glass fiber filters (Merck Millipore) that had been pretreated with 0.3% (vol/vol) polyethylenimine. Filters were immediately washed twice with ice-cold water and then dried. Radioactivity was measured by liquid scintillation counting.
Radioligand-binding experiments on solubilized, detergent-extracted samples were done with slight modifications. The buffer in these assays contained 0.01% (wt/vol) LMNG, and incubation with 3H DTG was done without shaking. Bound radioligand was separated from unbound radioligand using a desalting column of G50 fine resin (GE Healthcare) with a separation buffer also containing 0.01% (wt/vol) LMNG.
3H DTG Saturation Binding in Cell Membranes.
3H DTG saturation binding to membranes was determined using an assay similar to that described by Chu and Ruoho (45). Briefly, membranes from infected Sf9 insect cells (2.5 μg total protein per reaction) or MCF-7 cells (15-30 μg total protein per reaction) were incubated in a 100-μL reaction buffered with 50 mM Tris (pH 8.0), containing 1.8 μM (+)SKF-10,047, and 0–30 nM 3H DTG. Concentrations of 100 and 300 nM DTG were assayed by isotopic dilution to minimize the use of 3H DTG. For each membrane type, a second curve that was otherwise identical save for the addition of 2 μM haloperidol was measured in parallel to determine nonspecific binding. Reactions were incubated at 37 °C for 90 min and then were terminated via filtration through a glass fiber filter using a Brandel cell harvester. After washing, filters were soaked in 5 mL Cytoscint scintillation fluid (MP Biomedicals) overnight, and scintillation was measured on a Beckman Coulter LS 6500 scintillation counter. Kd values were calculated using nonlinear regression tools from GraphPad Prism.
Competition Binding Assays in Cell Membranes.
3H DTG competition curves testing the binding of σ ligands haloperidol, DTG, PB-28, SAS-1121, (+)-pentazocine, and (+)-SKF-10,047 or the TMEM97 ligands Elacridar or Ro 48-8071 were performed as described by Chu and Ruoho (45), with slight modifications. Briefly, Sf9 insect membranes overexpressing TMEM97 (2.5 μg of total protein per reaction) or MCF-7 membranes (12–30 μg total protein per reaction) were incubated in a 100-μL reaction buffered with 50 mM Tris (pH 8.0), with 30 nM 3H DTG and eight concentrations (ranging from 10 to 100 μM) of the competing cold ligand. (+)SKF-10,047 (1.8 μM) was included to block σ1 receptor sites in all MCF-7 membrane-binding assays and in Sf9 membranes when testing TMEM97 ligands. Reactions were incubated for 90 min at 37 °C and then were terminated by filtration through a glass fiber filter using a Brandel cell harvester. Glass fiber filters were soaked in 0.3% polyethylenimine for at least 30 min at room temperature before harvesting. All reactions were performed in triplicate using a 96-well block. After the membranes were transferred to the filters and washed, the filters were soaked in 5 mL Cytoscint scintillation fluid overnight, and radioactivity was measured using a Beckman Coulter LS 6500 scintillation counter. Data were analyzed using GraphPad Prism software. Ki values were computed by directly fitting the data and using the experimentally determined probe Kd to calculate a Ki value, using the GraphPad Prism software. This process implicitly uses a Cheng–Prusoff correction, so no secondary correction was applied.
siRNA Knockdown of Tmem97.
A pair of siRNA oligos was designed against Rattus norvegicus Tmem97 mRNA using the Stealth RNAi tool available through Thermo Fisher Scientific. The sense strand for the siRNA was 5′-CAACCUGUUGCGGUGGUACUCUAAG-3′, and the antisense strand was 5′- CUUAGAGUACCACCGCAACAGGUUG-3′. As a control, we used the AllStars Negative Control siRNA from Qiagen.
For the transfection, 2.2 × 106 PC-12 cells suspended in 8.0 mL of DMEM with 10% FBS and 10 μg/mL gentamicin were placed in a 10-cm dish and immediately transfected with a 2.0-mL solution containing 20 μL of Lipofectamine RNAimax from Thermo Fisher Scientific and 10 nM of either the control or Tmem97 siRNA. After 24 h, the medium was replaced with fresh medium, and the cells were transfected again in the same way. Forty-eight hours after the first transfection, the cells were trypsinized and split 1:2 into two new 10-cm plates. On the fifth day after the first transfection, cells were harvested by trypsinization and centrifugation. Ten percent were set aside for RNA extraction, and 90% were used for binding analysis.
RNA extraction was done using the RNeasy kit from Qiagen according to the manufacturer’s instructions. The RNA was converted to cDNA using Invitrogen’s SuperScript II reverse transcriptase kit according to the manufacturer’s instructions. RNA was removed from the cDNA after reverse transcription by digestion using Escherichia coli RNase H.
Real-Time qPCR for Quantification of TMEM97 mRNA Levels in PC-12 Cells.
After preparation of the cDNA was complete, real-time qPCR was performed on a QuantStudio 6 qPCR instrument (Applied Biosystems) at the Harvard Medical School Center for Molecular Interactions using PowerUp SYBR Green Master Mix from Applied Biosystems Life Technologies. The qPCR was performed according to the recommendations of the master mix manufacturer, using a range of different template input concentrations and a final primer concentration of 250 nM. Primers for qPCR were designed using the National Center for Biotechnology Information (NCBI) primer design tools. The primers used for the forward and reverse primer for R. norvegicus Tmem97 were 5′-TACTTCGTCTCGCACATCCC-3′ and 5′-TTGCTGAACTCCTGCGGGTA-3′, respectively. R. norvegicus Actb was used as a reference gene, for which the forward and reverse primers were 5′-CCCGCGAGTACAACCTTCTTG-3′ and 5′-GTCATCCATGGCGAACTGGTG-3′, respectively. Fold differences in Tmem97 expression levels were calculated using the ΔΔCT method (47). All measurements were performed in triplicate.
Synthetic Chemistry.
See SI Appendix for details of synthetic chemistry.
Supplementary Material
Acknowledgments
We thank Bryan L. Roth, MD, PhD, the director of the National Institute of Mental Health (NIMH) Psychoactive Drug Screening Program (PDSP) at the University of North Carolina at Chapel Hill; Project Officer Jamie Driscoll at NIMH, Bethesda; Dr. Roberta Pascolutti for instructional assistance with siRNA oligo design and transfection; Dr. Dominique Burri, Dr. Bingqian Guo, and Dr. Kelly Arnett for instructional assistance with the qPCR experimental design; and the Harvard Medical School Center for Macromolecular Interactions for access to the QuantStudio 6 qPCR machine. Initial determination of the receptor-binding profiles of SAS-1121 and JVW-1601 was generously provided by NIMH PDSP Contract HHSN-271-2013-00017-C. Financial support was provided by NIH Grant 1DP5OD021345, the Vallee Scholars Program, the Harvard Brain Science Initiative Winthrop Fund, the Klingenstein–Simons Fellowship Program, Robert A. Welch Foundation Grant F-0652, the donors of the Alzheimer's Disease Research Program, the BrightFocus Foundation, and Dell Medical School’s Texas Health Catalyst Program. H.R.S. is supported by a National Science Foundation Graduate Research Fellowship.
Footnotes
Conflict of interest statement: The authors are inventors on a provisional patent application related to the work described in this article.
This article is a PNAS Direct Submission.
See Commentary on page 6888.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705154114/-/DCSupplemental.
References
- 1.Lefkowitz RJ. A brief history of G-protein coupled receptors (Nobel Lecture) Angew Chem Int Ed Engl. 2013;52:6366–6378. doi: 10.1002/anie.201301924. [DOI] [PubMed] [Google Scholar]
- 2.Martin WR, Eades CG, Thompson JA, Huppler RE, Gilbert PE. The effects of morphine- and nalorphine- like drugs in the nondependent and morphine-dependent chronic spinal dog. J Pharmacol Exp Ther. 1976;197:517–532. [PubMed] [Google Scholar]
- 3.Hellewell SB, Bowen WD. A sigma-like binding site in rat pheochromocytoma (PC12) cells: Decreased affinity for (+)-benzomorphans and lower molecular weight suggest a different sigma receptor form from that of guinea pig brain. Brain Res. 1990;527:244–253. doi: 10.1016/0006-8993(90)91143-5. [DOI] [PubMed] [Google Scholar]
- 4. Abramson Cancer Center of the University of Pennsylvania (2014) Imaging of in vivo sigma-2 receptor expression with 18F ISO-1 positron emission tomography (PET/CT) in primary breast cancer. Clinical trial NCT02762110. Available at https://clinicaltrials.gov/ct2/show/NCT02762110?term=NCT02762110&rank=1. Accessed November 2, 2016.
- 5. Cognition Therapeutics (2016) Clinical trial of CT1812 in mild to moderate Alzheimer's Disease. Clinical trial NCT02907567. Available at https://clinicaltrials.gov/ct2/show/NCT02907567?term=NCT02907567&rank=1. Accessed November 2, 2016.
- 6.Yi B, et al. Small molecule modulator of sigma 2 receptor is neuroprotective and reduces cognitive deficits and neuro-inflammation in experimental models of Alzheimer's disease. J Neurochem. 2017;140:561–575. doi: 10.1111/jnc.13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Izzo NJ, et al. Alzheimer's therapeutics targeting amyloid beta 1-42 oligomers I: Abeta 42 oligomer binding to specific neuronal receptors is displaced by drug candidates that improve cognitive deficits. PLoS One. 2014;9:e111898. doi: 10.1371/journal.pone.0111898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Minerva Neurosciences, Inc. (2014) Study to evaluate the efficacy, safety, and tolerability of MIN-101 in patients with negative symptoms of schizophrenia. Clinical trial 2014-004878-42. Available at https://www.clinicaltrialsregister.eu/ctr-search/trial/2014-004878-42/LV. Accessed November 2, 2016.
- 9.Walker JM, et al. Sigma receptors: Biology and function. Pharmacol Rev. 1990;42:355–402. [PubMed] [Google Scholar]
- 10.Chu UB, Ruoho AE. Biochemical pharmacology of the sigma-1 receptor. Mol Pharmacol. 2016;89:142–153. doi: 10.1124/mol.115.101170. [DOI] [PubMed] [Google Scholar]
- 11.Hellewell SB, et al. Rat liver and kidney contain high densities of sigma 1 and sigma 2 receptors: Characterization by ligand binding and photoaffinity labeling. Eur J Pharmacol. 1994;268:9–18. doi: 10.1016/0922-4106(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 12.Mach RH, Zeng C, Hawkins WG. The σ2 receptor: A novel protein for the imaging and treatment of cancer. J Med Chem. 2013;56:7137–7160. doi: 10.1021/jm301545c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo L, Zhen X. Sigma-2 receptor ligands: Neurobiological effects. Curr Med Chem. 2015;22:989–1003. doi: 10.2174/0929867322666150114163607. [DOI] [PubMed] [Google Scholar]
- 14.Vilner BJ, John CS, Bowen WD. Sigma-1 and sigma-2 receptors are expressed in a wide variety of human and rodent tumor cell lines. Cancer Res. 1995;55:408–413. [PubMed] [Google Scholar]
- 15.Wheeler KT, et al. Sigma-2 receptors as a biomarker of proliferation in solid tumours. Br J Cancer. 2000;82:1223–1232. doi: 10.1054/bjoc.1999.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai S, et al. Synthesis and structure-activity relationship studies of conformationally flexible tetrahydroisoquinolinyl triazole carboxamide and triazole substituted benzamide analogues as σ2 receptor ligands. J Med Chem. 2014;57:4239–4251. doi: 10.1021/jm5001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berardi F, et al. Exploring the importance of piperazine N-atoms for sigma(2) receptor affinity and activity in a series of analogs of 1-cyclohexyl-4-[3-(5-methoxy-1,2,3,4-tetrahydronaphthalen-1-yl)propyl]piperazine (PB28) J Med Chem. 2009;52:7817–7828. doi: 10.1021/jm9007505. [DOI] [PubMed] [Google Scholar]
- 18.Sahn JJ, Hodges TR, Chan JZ, Martin SF. Norbenzomorphan framework as a novel scaffold for generating sigma 2 receptor/PGRMC1 subtype-selective ligands. ChemMedChem. 2016;11:556–561. doi: 10.1002/cmdc.201500551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu ZW, et al. Synthesis and evaluation of tetrahydroindazole derivatives as sigma-2 receptor ligands. Bioorg Med Chem. 2015;23:1463–1471. doi: 10.1016/j.bmc.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 20.Sahn JJ, Hodges TR, Chan JZ, Martin SF. Norbenzomorphan scaffold: Chemical tool for modulating sigma receptor-subtype selectivity. ACS Med Chem Lett. 2017;8:455–460. doi: 10.1021/acsmedchemlett.7b00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanner M, et al. Purification, molecular cloning, and expression of the mammalian sigma1-binding site. Proc Natl Acad Sci USA. 1996;93:8072–8077. doi: 10.1073/pnas.93.15.8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langa F, et al. Generation and phenotypic analysis of sigma receptor type I (sigma 1) knockout mice. Eur J Neurosci. 2003;18:2188–2196. doi: 10.1046/j.1460-9568.2003.02950.x. [DOI] [PubMed] [Google Scholar]
- 23.Xu J, et al. Identification of the PGRMC1 protein complex as the putative sigma-2 receptor binding site. Nat Commun. 2011;2:380. doi: 10.1038/ncomms1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abate C, Niso M, Infantino V, Menga A, Berardi F. Elements in support of the ‘non-identity’ of the PGRMC1 protein with the σ2 receptor. Eur J Pharmacol. 2015;758:16–23. doi: 10.1016/j.ejphar.2015.03.067. [DOI] [PubMed] [Google Scholar]
- 25.Chu UB, et al. The sigma-2 receptor and progesterone receptor membrane component 1 are different binding sites derived from independent genes. EBioMedicine. 2015;2:1806–1813. doi: 10.1016/j.ebiom.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pati ML, et al. Sigma-2 receptor and progesterone receptor membrane component 1 (PGRMC1) are two different proteins: Proofs by fluorescent labeling and binding of sigma-2 receptor ligands to PGRMC1. Pharmacol Res. 2017;117:67–74. doi: 10.1016/j.phrs.2016.12.023. [DOI] [PubMed] [Google Scholar]
- 27.Kakumani PK, Malhotra P, Mukherjee SK, Bhatnagar RK. A draft genome assembly of the army worm, Spodoptera frugiperda. Genomics. 2014;104:134–143. doi: 10.1016/j.ygeno.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Niphakis MJ, et al. A global map of lipid-binding proteins and their ligandability in cells. Cell. 2015;161:1668–1680. doi: 10.1016/j.cell.2015.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hopf TA, et al. Sequence co-evolution gives 3D contacts and structures of protein complexes. eLife. 2014;3:3. doi: 10.7554/eLife.03430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt HR, et al. Crystal structure of the human σ1 receptor. Nature. 2016;532:527–530. doi: 10.1038/nature17391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartz F, et al. Identification of cholesterol-regulating genes by targeted RNAi screening. Cell Metab. 2009;10:63–75. doi: 10.1016/j.cmet.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Ebrahimi-Fakhari D, et al. Reduction of TMEM97 increases NPC1 protein levels and restores cholesterol trafficking in Niemann-pick type C1 disease cells. Hum Mol Genet. 2016;25:3588–3599. doi: 10.1093/hmg/ddw204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moebius FF, Striessnig J, Glossmann H. The mysteries of sigma receptors: New family members reveal a role in cholesterol synthesis. Trends Pharmacol Sci. 1997;18:67–70. doi: 10.1016/s0165-6147(96)01037-1. [DOI] [PubMed] [Google Scholar]
- 34.Huang YS, Lu HL, Zhang LJ, Wu Z. Sigma-2 receptor ligands and their perspectives in cancer diagnosis and therapy. Med Res Rev. 2014;34:532–566. doi: 10.1002/med.21297. [DOI] [PubMed] [Google Scholar]
- 35.Ding H, et al. Prognostic value of MAC30 expression in human pure squamous cell carcinomas of the lung. Asian Pac J Cancer Prev. 2016;17:2705–2710. [PubMed] [Google Scholar]
- 36.Zhao ZR, et al. Significance of mRNA and protein expression of MAC30 in progression of colorectal cancer. Chemotherapy. 2011;57:394–401. doi: 10.1159/000331716. [DOI] [PubMed] [Google Scholar]
- 37.Yan BY, et al. Overexpression of MAC30 in the cytoplasm of oral squamous cell carcinoma predicts nodal metastasis and poor differentiation. Chemotherapy. 2010;56:424–428. doi: 10.1159/000317582. [DOI] [PubMed] [Google Scholar]
- 38.Moparthi SB, et al. Expression of MAC30 protein is related to survival and biological variables in primary and metastatic colorectal cancers. Int J Oncol. 2007;30:91–95. [PubMed] [Google Scholar]
- 39.Wilcox CB, et al. Coordinate up-regulation of TMEM97 and cholesterol biosynthesis genes in normal ovarian surface epithelial cells treated with progesterone: Implications for pathogenesis of ovarian cancer. BMC Cancer. 2007;7:223. doi: 10.1186/1471-2407-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu XY, et al. Down-regulated MAC30 expression inhibits proliferation and mobility of human gastric cancer cells. Cell Physiol Biochem. 2014;33:1359–1368. doi: 10.1159/000358703. [DOI] [PubMed] [Google Scholar]
- 41.Laurini E, et al. A 3D-pharmacophore model for sigma2 receptors based on a series of substituted benzo[d]oxazol-2(3H)-one derivatives. Bioorg Med Chem Lett. 2010;20:2954–2957. doi: 10.1016/j.bmcl.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 42.Sanchez-Pulido L, Ponting CP. TM6SF2 and MAC30, new enzyme homologs in sterol metabolism and common metabolic disease. Front Genet. 2014;5:439. doi: 10.3389/fgene.2014.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kozlitina J, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46:352–356. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chu J, et al. Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein. Nat Methods. 2014;11:572–578. doi: 10.1038/nmeth.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chu UB, Ruoho AE. Sigma receptor binding assays. Curr Protoc Pharmac. 2015;71:1.34.1–1.34.21. doi: 10.1002/0471141755.ph0134s71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lever JR, et al. Relationship between cerebral sigma-1 receptor occupancy and attenuation of cocaine's motor stimulatory effects in mice by PD144418. J Pharmacol Exp Ther. 2014;351:153–163. doi: 10.1124/jpet.114.216671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 48.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–D376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colabufo NA, et al. Antiproliferative and cytotoxic effects of some sigma2 agonists and sigma1 antagonists in tumour cell lines. Naunyn Schmiedebergs Arch Pharmacol. 2004;370:106–113. doi: 10.1007/s00210-004-0961-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.