Significance
Mucosa-associated invariant T (MAIT) cells are a large subset of unconventional T cells in humans, recognizing microbial riboflavin metabolites presented by the monomorphic MR1 molecule. The extraordinary level of conservation of MR1 and the limited diversity of riboflavin-derived antigens have suggested that MAIT cells are homogeneous, and their functional specialization has not been thoroughly investigated. Here, we show that MAIT cell responses against two distinct riboflavin biosynthesis-competent microorganisms display microbe-specific response patterns with multiple layers of heterogeneity. Furthermore, a set of natural killer cell-associated receptors define a subset with enhanced capacity to respond to innate cytokine stimulus. Thus, MAIT cells harbor multiple layers of functional heterogeneity and can adapt their antimicrobial responses to the type of microbial stimuli.
Keywords: T cells, MAIT cells, immunity, MR1, microbial immunity
Abstract
Mucosa-associated invariant T (MAIT) cells are a large innate-like T-cell subset in humans defined by invariant TCR Vα7.2 use and expression of CD161. MAIT cells recognize microbial riboflavin metabolites of bacterial or fungal origin presented by the monomorphic MR1 molecule. The extraordinary level of evolutionary conservation of MR1 and the limited known diversity of riboflavin metabolite antigens have suggested that MAIT cells are relatively homogeneous and uniform in responses against diverse microbes carrying the riboflavin biosynthesis pathway. The ability of MAIT cells to exhibit microbe-specific functional specialization has not been thoroughly investigated. Here, we found that MAIT cell responses against Escherichia coli and Candida albicans displayed microbe-specific polyfunctional response profiles, antigen sensitivity, and response magnitudes. MAIT cell effector responses against E. coli and C. albicans displayed differential MR1 dependency and TCR β-chain bias, consistent with possible divergent antigen subspecificities between these bacterial and fungal organisms. Finally, although the MAIT cell immunoproteome was overall relatively homogenous and consistent with an effector memory-like profile, it still revealed diversity in a set of natural killer cell-associated receptors. Among these, CD56, CD84, and CD94 defined a subset with higher expression of the transcription factors promyelocytic leukemia zinc finger (PLZF), eomesodermin, and T-bet and enhanced capacity to respond to IL-12 and IL-18 stimulation. Thus, the conserved and innate-like MAIT cells harbor multiple layers of functional heterogeneity as they respond to bacterial or fungal organisms or innate cytokines and adapt their antimicrobial response patterns in a stimulus-specific manner.
Mucosa-associated invariant T (MAIT) cells are an evolutionarily conserved subset of unconventional T cells, highly abundant in human mucosal tissues as well as in peripheral blood and the liver (1–3). Human MAIT cells express a semi-invariant αβ T-cell receptor (TCR) with the Vα7.2 segment coupled with restricted Jα segments and limited Vβ repertoires (3–8). Coexpression of the Vα7.2 TCR and high levels of the C-type lectin CD161 or the interleukin (IL)-18 receptor α subunit (IL-18Rα) defines the major MAIT cell population in healthy adult humans (2, 9). MAIT cells display a transcriptional profile characterized by the expression of promyelocytic leukemia zinc finger (PLZF or zinc finger and BTB domain containing 16, ZBTB16) and retinoid-related orphan receptor (ROR) γt (1, 2, 10). They also express eomesodermin (Eomes) and T box transcription factor 21 (TBX21 or T-bet) (11, 12), reciprocally expressed in effector and memory CD8+ T cells (13), and Helios (or IKAROS family zinc finger 2, IKZF2) (11, 12), involved in T-cell activation and proliferation (14).
MAIT cells recognize microbial vitamin B2 metabolite antigens from a wide range of bacteria and fungi presented by the major histocompatibility complex (MHC) class I-related (MR) 1 molecules (15, 16). MAIT cells activated by such antigens display an innate-like response pattern and rapidly release cytokines, including IFNγ, TNF, IL-17, and IL-22 (1, 9, 11, 17), and mediate cytolytic function against bacterially infected cells (12, 18, 19). These response patterns and functions contribute to their involvement and protective role against bacterial infections in animal models as well as humans (9, 20–30). In addition, high expression of receptors for IL-18 and IL-12 (9, 31, 32) provides MAIT cells with the capacity to respond to antigen-presenting cell (APC)-derived cytokines, recently shown to be important for TCR-mediated activation (33, 34), as well as to MR1-independent innate responses (35, 36). Given that the activating antigens identified thus far are of bacterial or fungal origin, MR1-independent responses are probably important for the involvement of MAIT cells in viral immunopathogenesis in diseases caused by HIV, HCV, dengue virus, and influenza virus (37–40). In addition, MR1-dependent responses might play a role during viral infections when the mucosal barriers are compromised and microbial translocation occurs (41).
The extraordinary level of MR1 evolutionary conservation and the limited set of riboflavin-derived antigens known to date have favored the idea that MAIT cells are relatively homogenous in their responses and effector functions. However, although the MAIT cell TCR repertoire is limited, the variability in the β-chain adds some diversity to the MAIT cell compartment (3–8). The phenotypic heterogeneity among MAIT cells is incompletely understood, and functionally diverse subsets may exist. It is also possible that distinct microbial characteristics may influence the quality of MAIT cell responses. In this study, we investigated the extent to which there is functional heterogeneity and diversity in the human MAIT cell compartment, by extensive characterization of their surface immuno-proteome and their responses against the two distinct microbes Escherichia coli and Candida albicans as well as against innate cytokine stimulation. The findings indicate that MAIT cells encompass multiple layers of functional heterogeneity at both the single cell level and population level that allow distinct response profiles against different microbes and in response to innate cytokine triggering. These findings indicate that MAIT cells can adapt their response patterns to the microbial challenge at hand and play diverse roles in immune responses against different types of bacteria, fungi, and viruses.
Results
Differential Magnitude and Sensitivity in MAIT Cell Recognition of E. coli and C. albicans.
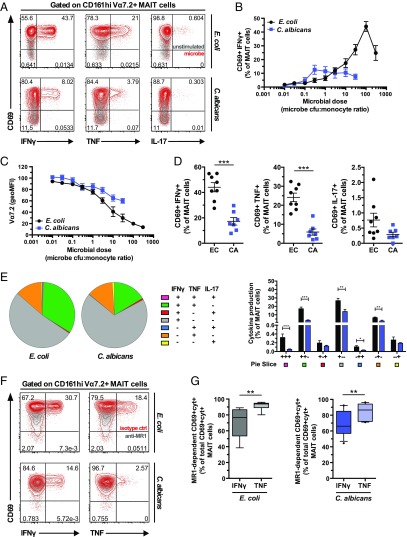
To investigate if CD161hi Vα7.2+ MAIT cells may respond differently to distinct microbes carrying the riboflavin biosynthesis pathway, we compared responses to a nonpathogenic strain of E. coli and the opportunistic fungal pathogen C. albicans. Monocytes were fed mildly formaldehyde-fixed microbes, and purified Vα7.2+ cells were added and cultured for 24 h and then intracellularly stained for IFNγ, TNF, and IL-17 (Fig. 1A) (42). We first determined the microbial dose requirement, measured as the microbe colony-forming unit (cfu) to monocyte ratio at which the MAIT cell activation peaked, using the coexpression of CD69 and IFNγ as readout (Fig. 1A). MAIT cell activation peaked at the ratios of 100 E. coli cfu and 0.3 C. albicans cfu per monocyte, whereas the response magnitude was higher against E. coli compared with C. albicans (Fig. 1B). Interestingly, E. coli induced strong dose-dependent down-regulation of the MAIT cell TCR, as the surface Vα7.2 staining decreased to levels as low as 20% of the unstimulated control (Fig. 1C). This effect was less pronounced in MAIT cells stimulated with C. albicans (Fig. 1C). These patterns reveal differential magnitude and sensitivity in MAIT cell responses against two distinct microbes.
Fig. 1.
Distinct polyfunctional profile and MR1 dependency in the MAIT cell response against E. coli and C. albicans. (A) MAIT cells were stimulated with E. coli or C. albicans-fed monocytes for 24 h and in the presence of anti-CD28 mAb. Representative example of the CD69 and IFNγ, TNF, or IL-17 expression by MAIT cells after stimulation with the microbes (red) or at baseline conditions (gray). (B) Frequency of CD69+IFNγ+ MAIT cells after stimulation with different doses of E. coli (0.01–300 cfu per monocyte) and C. albicans (0.01–30 cfu per monocyte). (C) Down-regulation of the TCR Vα7.2 segment upon stimulation with E. coli or C. albicans, represented as the Vα7.2 geometric MFI of the stimulated CD161hi cells normalized to the one of the unstimulated control. (D) Frequency of MAIT cells expressing IFNγ, TNF, or IL-17. The microbial dose selected for each donor was the one inducing the highest frequency of CD69+IFNγ+ MAIT cells. (E) Polyfunctional profile of the MAIT cells responding to the microbes. The pie charts and the bar plot show the frequency of MAIT cells expressing CD69 and all different combinations of cytokines. (F) MAIT cells were stimulated for 18 h with E. coli-fed monocytes or 24 h with C. albicans-fed monocytes in the presence of anti-CD28 mAb and MR1-blocking mAb or isotype ctrl. Representative example of the expression of CD69 and IFNγ or TNF by MAIT cells after stimulation with the microbes and in the presence of MR1-blocking mAb (gray) or isotype ctrl (red). (G) Proportion of MR1-dependent IFNγ and TNF expression out of the total IFNγ and TNF expression by MAIT cells upon stimulation with E. coli or C. albicans. Graphs show the mean ± SEM (B–D) and mean + SEM (E, bar plot), and the box and whisker plots show the median, the 10th and 90th percentiles, and the interquartile range (G). Data are from 8 and 4–7 donors for E. coli and C. albicans, respectively (A–E) and from 8 and 9 donors for E. coli and 11 donors for C. albicans for IFNγ and TNF production, respectively (F and G). The unpaired t test or Mann–Whitney test was used (D, E, and G) to detect significant differences across multiple, unpaired samples. CA, C. albicans; cyt, cytokine; EC, E. coli. *P < 0.05; **P < 0.01; ***P < 0.001.
Distinct Polyfunctional Response Profiles of MAIT Cells Against E. coli and C. albicans.
Next, we investigated the cytokine profile of MAIT cells responding at the optimal microbial doses. E. coli induced robust and significantly higher frequencies of CD69+IFNγ+-producing MAIT cells than C. albicans (P = 0.0006) (Fig. 1D). E. coli also stimulated high levels of TNF, whereas the frequency of CD69+TNF+ MAIT cells was significantly lower for C. albicans (P = 0.0003) (Fig. 1D). IL-17 production was much lower than IFNγ or TNF and trended toward higher levels in response to E. coli (P = 0.1197) (Fig. 1D). The dominant MAIT cell cytokine response profile was monofunctional IFNγ (Fig. 1E). Nevertheless, E. coli induced significantly higher levels of IFNγ/TNF/IL-17 triple-producing MAIT cells than did C. albicans (P = 0.0006), suggesting that MAIT cell responses to E. coli are more polyfunctional. Furthermore, E. coli induced higher levels of TNF/IL-17 double-producing MAIT cells than did C. albicans (P = 0.0205) (Fig. 1E), suggesting that the response to E. coli might be more skewed toward a Th17-like response. These results suggest that MAIT cells display significant cytokine profile heterogeneity in response to different microbes.
We next investigated the importance of the TCR–MR1 interaction for the MAIT cell response against the two microbes. Anti-MR1 mAb blocking was used to determine the proportion of MR1-dependent CD69+IFNγ+ or CD69+TNF+ MAIT cells out of the total CD69+IFNγ+ or CD69+TNF+ MAIT cells (Fig. 1F). The MAIT cell IFNγ production induced by E. coli and C. albicans was predominantly MR1-dependent, although the MR1 dependency was not complete (mean MR1 dependency ± SD = 70.9% ± 18.4% and 67.9% ± 15.7% for E. coli and C. albicans, respectively) (Fig. 1G). This is consistent with the notion that MAIT cells also respond to cytokines produced by the monocytes after microbial exposure. Interestingly, however, anti-MR1 blocking strongly reduced MAIT cell TNF production (mean MR1 dependency ± SD = 91.6% ± 5.1% and 84.0% ± 10.2% for E. coli and C. albicans, respectively). For both E. coli and C. albicans, TNF production was significantly more MR1-dependent than that of IFNγ (P = 0.0025 and P = 0.0083, respectively) (Fig. 1G). These results indicate that production of the proinflammatory cytokine TNF by MAIT cells is primarily controlled by TCR engagement.
TCR β-Chain Influence on MR1-Restricted MAIT Cell Responses.
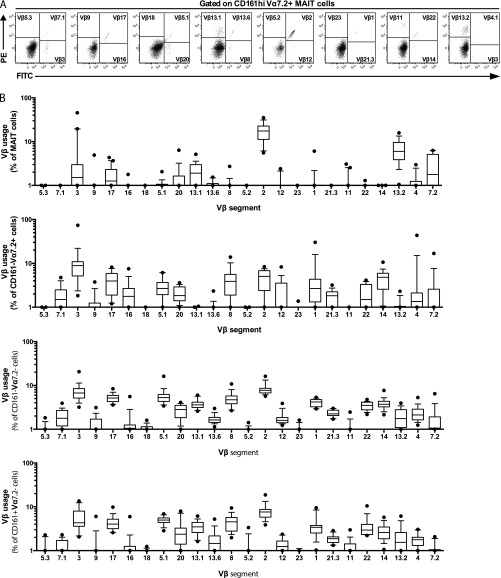
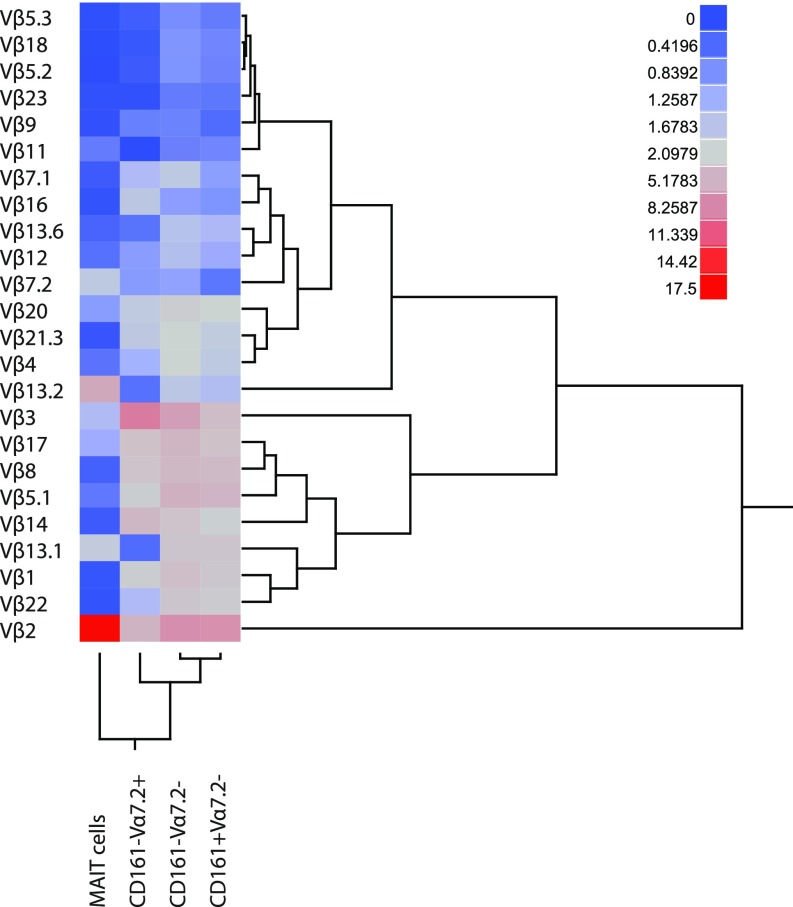
To investigate the influence of the TCR β-chain in the MAIT cell response against the different microbes, we initially determined by flow cytometry which Vβ segments were frequently expressed by MAIT cells (Fig. S1A). Out of the 24 Vβ segments screened, Vβ2 and Vβ13.2 covered the majority of the MAIT cell Vβ repertoire (Fig. 2A and Fig. S1B), as previously described (5, 7, 8). This contrasted with the other three T-cell populations defined by CD161 and Vα7.2—CD161–Vα7.2+, CD161–Vα7.2–, and CD161+Vα7.2– cells—which displayed more diverse Vβ use that was similar between the populations (Fig. 2A and Fig. S1B). Clustering analysis of the Vβ repertoire confirmed that MAIT cells were distinct from the other T-cell populations (Fig. S2). Although Vβ2 and Vβ13.2 were predominantly expressed by MAIT cells, other Vβ segments were expressed to a lesser extent, and these included Vβ3, 7.2, 8, 13.1, 13.6, 17, and 20 (Fig. 2A and Fig. S1B). Although the Vβ use patterns were relatively similar between donors, the frequency of Vβ2, 3, 7.2, and 13.2 displayed significant interdonor variability (Fig. 2B). Thus, the MAIT cell TCR Vβ repertoire measured in this way is overall similar between individuals but still displays some differences that may reflect the antigenic experience of MAIT cells in vivo.
Fig. S1.
TCR Vβ use in MAIT cells and other T cells. (A) Representative example from 16 to 20 donors of the gating strategy to identify each Vβ segment on MAIT cells. (B) Frequency of MAIT, CD161–Vα7.2+, CD161–Vα7.2–, and CD161+Vα7.2– cells expressing each Vβ segment. Box and whisker plots show the median, the 10th and 90th percentiles, and the interquartile range. Data are from 16 to 20 donors.
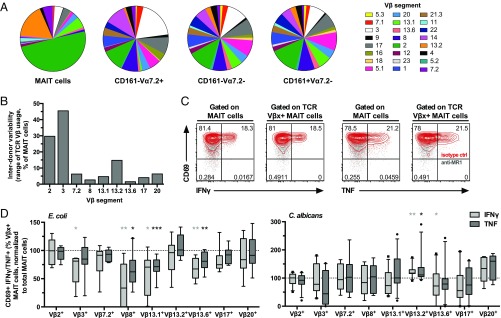
Fig. 2.
TCR Vβ use in MAIT cells influences the response to microbial stimulus. (A) Distribution of the 24 Vβ segments in the CD161hi Vα7.2+ MAIT, CD161–Vα7.2+, CD161–Vα7.2–, and CD161+Vα7.2– populations. Each pie slice represents the median frequency of cells expressing each TCR Vβ segment. (B) Range of expression (maximum minus minimum frequency of MAIT cells expressing each Vβ segment) of the nine Vβ segments selected for further functional experiments. (C) Representative example of the strategy used to investigate the influence of the Vβ segment in MAIT cell responses. The MR1-dependent expression of IFNγ and TNF in each Vβ subpopulation was calculated and compared with that of the total MAIT cell population. (D) Frequency of MR1-dependent CD69+IFNγ+ or CD69+TNF+ CD8+ and CD8–CD4– MAIT cells defined by each Vβ segment normalized to that of the total CD8+ and CD8–CD4– MAIT cells, after stimulation with E. coli or C. albicans-fed monocytes for 18 h or 24 h, respectively, and in the presence of anti-CD28 mAb. Due to fluorochrome overlap in the anti-Vβ antibodies, the staining for each TCR Vβ segment was done separately (nine different samples per stimulation and per donor) and the Wilcoxon’s test or paired t test was therefore used to detect significant differences between the paired samples (total vs. Vβx+ MAIT cells from the same sample). Box and whisker plots show the median, the 10th and 90th percentiles, and the interquartile range. Data are from 16 to 20 donors (A and B) and from 6 to 9 and 8–11 donors (C and D, for E. coli and C. albicans, respectively). *P < 0.05; **P < 0.01; ***P < 0.001.
Fig. S2.
MAIT cells cluster separately from other T cells based on the TCR Vβ repertoire. Heat map and dendrogram based on the median frequency of cells expressing each Vβ segment. Data are from 16 to 20 donors.
The nine most frequently represented Vβ segments were selected for subsequent E. coli and C. albicans-mediated MAIT cell activation experiments, where the frequencies of MR1-dependent CD69+IFNγ+ and CD69+TNF+ MAIT cells were calculated and compared between the total MAIT cell population and the MAIT cell population defined by each of the Vβ segments (Fig. 2C). This high-throughput analysis approach allows assessment of Vβ influence on responsiveness in a large number of individual MAIT cells. Interestingly, MAIT cells expressing Vβ8, 13.1, and 13.6 were hyporesponsive to E. coli-fed monocytes, as they produced significantly less IFNγ and TNF than the total MAIT cells (P = 0.0078, P = 0.0156, and P = 0.0056 for IFNγ and P = 0.0117, P = 0.0008, and P = 0.0056 for TNF for Vβ8, 13.1, and 13.6, respectively) (Fig. 2D and Fig. S3A). This bias was only observed for the MR1-dependent response, as no difference was detected in the MR1-independent IFNγ production. Interestingly, the MAIT cell response against C. albicans-fed monocytes had a different TCR Vβ bias, where Vβ13.2+ MAIT cells displayed slightly higher MR1-dependent IFNγ and TNF compared with the total MAIT cells (P = 0.0049 and P = 0.0177 for IFNγ and TNF, respectively) (Fig. 2D and Fig. S3B). Taken together, these results indicate that the MAIT cell TCR Vβ use can influence the ability to respond to an MR1-presented antigen and suggest that this bias is to some extent dependent on the specific microbe.
Fig. S3.
The MAIT cell response against microbial stimuli is influenced by the TCR Vβ use. Frequency of MR1-dependent CD69+TNF+ (Upper) and MR1-dependent CD69+IFNγ+ (Lower) MAIT cells upon stimulation with (A) E. coli-fed monocytes or (B) C. albicans-fed monocytes. Lines represent individual donors. The Wilcoxon’s test or paired t test was used to detect significant differences between the paired samples. *P < 0.05; **P < 0.01; ***P < 0.001.
Rare Vβ Segments Are Associated with Lower Proliferative Capacity of MAIT Cells.
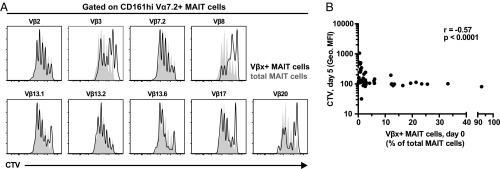
We next investigated whether MAIT cells expressing different TCR β-chains also differ in their capacity to proliferate in vitro. MAIT cells in cell trace violet (CTV)-labeled PBMC samples were cultured for 5 d in the presence of E. coli and IL-2 and then stained for the nine most frequently used Vβ segments (Fig. 3A). Interestingly, the input frequency of Vβ-defined MAIT cell subpopulations in the fresh PBMC sample was inversely correlated with the CTV geometric MFI in these subpopulations at day 5 (Spearman’s r = –0.57, P < 0.0001) (Fig. 3B). Thus, the initially less abundant Vβ-defined MAIT cell subpopulations ex vivo were also less proliferative in vitro.
Fig. 3.
Rare TCR Vβ segments are associated with lower proliferative capacity of MAIT cells in vitro. MAIT cells in the PBMC sample were labeled with CTV and cultured for 5 d in the presence of E. coli (bacterial dose of 10), IL-2, and anti-CD28 mAb. (A) Representative example of the CTV dilution in Vβx+ (black histogram) and total (solid gray) CD8+ and CD8–CD4– MAIT cells after 5 d in culture. (B) Correlation between the frequency of Vβx+CD8+ and CD8–CD4– MAIT cells in the initial PBMC sample and the geometric MFI of the CTV staining in Vβx+CD8+ and CD8–CD4– MAIT cells normalized to that of total CD8+ and CD8–CD4– MAIT cells after 5 d of proliferation. Correlation was calculated using the Spearman’s test. Data are from five to seven donors.
MAIT Cells Have Distinct Surface Immuno-Proteome and Transcription Factor Profiles.
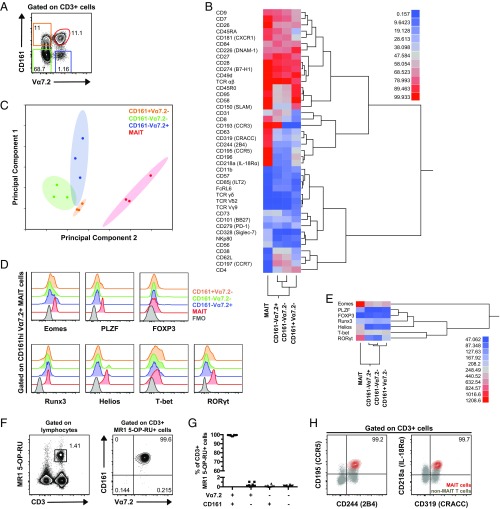
With the aim to investigate associations between phenotypic characteristics and function among MAIT cells, we next performed a broad surface immuno-receptor profiling of MAIT cells in comparison with the other three T-cell populations defined by Vα7.2 and CD161 (Fig. 4A). PBMCs from healthy individuals were prestained for CD161 and Vα7.2 (Fig. S4A) and then screened by flow cytometry for a panel of 332 immuno-receptor surface proteins. Based on this dataset, the T-cell populations lacking CD161 clustered together in the hierarchical clustering analysis, followed by the CD161+Vα7.2– cells, whereas the MAIT cells clustered separately (Fig. 4B). CD63, CD319, CD244, CD195, CD196, and CD218 were highly expressed on MAIT cells and contributed to the separation between MAIT cells and the other T-cell populations. MAIT cells further expressed CD328, NKp80, and CD56 at intermediate levels (32–42% of cells), whereas the expression of these receptors in other T cells was low. Interestingly, Vα7.2+ cells shared high (>95%) and intermediate (approx. 40%) expression of CD193 and CD73, respectively, whereas CD161+ T cells shared relatively high expression of CD45RO, CD95, CD58, and CD150. The total CD161– cell population shared intermediate expression (45–72%) of CD38, CD62L, and CD197 (Fig. 4B and Table S1). Dimensionality reduction by principal component analysis (PCA) integrates the expression of all data points into principal components, which cumulatively explain the variability within the dataset. In the present dataset, the first two principal components accounted for ∼60% of the total variability (Fig. S4B). The PCA indicated that the MAIT cells represent a distinct cell population compared with the other T-cell subsets defined by Vα7.2 and CD161 (Fig. 4C), in agreement with the hierarchical clustering analysis.
Fig. 4.
Surface immuno-proteome and transcription factor analysis of human MAIT cells. (A) Representative flow cytometry identification of the CD161hi Vα7.2+ MAIT, CD161–Vα7.2+, CD161–Vα7.2–, and CD161+Vα7.2– cell populations. (B) Heat map and dendrogram based on the mean frequency of cells expressing a restricted group of proteins from the surface immuno-proteome dataset. The proteins for which the range of expression was equal or higher than 10% between any two T-cell populations were included. (C) PCA on the whole surface immuno-proteome dataset with the four T-cell populations plotted against principal components 1 and 2. (D) Representative example of the expression of Eomes, PLZF, FOXP3, Runx3, Helios, T-bet, and RORγt in CD161hi Vα7.2+ MAIT (red), CD161–Vα7.2+ (blue), CD161–Vα7.2– (green), and CD161+Vα7.2– cells (orange). The fluorescence minus one (FMO) control (black) is also shown. (E) Heat map and dendrogram based on the geometric MFI of the transcription factor stainings in MAIT, CD161–Vα7.2+, CD161–Vα7.2–, and CD161+Vα7.2– cells. (F) Representative example of the CD3 and MR1 5-OP-RU tetramer staining in lymphocytes and CD161 and Vα7.2 expression in CD3+ MR1 5-OP-RU+ cells. (G) Frequency of CD161+Vα7.2+, CD161+Vα7.2–, CD161–Vα7.2+, and CD161–Vα7.2– within the CD3+ MR1 5-OP-RU+ cell population in PBMCs from 10 healthy donors. (H) Representative example of the expression of CD195, CD244, CD218a, and CD319 on CD161hi MR1 5-OP-RU+ MAIT cells (red) and non-MAIT T cells (gray). Data are from three donors (A–C), eight donors for all transcription factors except for Runx3 (three donors) and FOXP3 (five donors) (D and E), and 10 donors (F–H).
Fig. S4.
Identification of cell populations. (A) Gating strategy representative of three donors to identify MAIT, CD161–Vα7.2+, CD161–Vα7.2–, and CD161+Vα7.2– cells by flow cytometry in the immuno-proteome dataset. (B) Percent variability explained by each principal component and cumulatively. (C) Representative example from 10 donors of MR1 5-OP-RU and MR1 6-FP tetramer staining in CD161hi Vα7.2+ MAIT (red), CD161+ Vα7.2– (orange), and CD161–Vα7.2+ (blue) cells.
Next, the intracellular expression levels of several transcription factors in peripheral blood MAIT cells were compared with those in CD161–Vα7.2+, CD161–Vα7.2–, and CD161+Vα7.2– cells (Fig. 4D). This analysis included Eomes, PLZF, forkhead box P3 (FOXP3), runt-related transcription factor 3 (Runx3), Helios, T-bet, and RORγt. The T-cell populations clustered similarly based on transcription factor expression levels (geometric MFI) (Fig. 4E), as in the surface immuno-proteome (Fig. 4B) and Vβ repertoire analysis (Fig. S2). In addition to the defining expression of PLZF, high expression of Eomes and RORγt and intermediate expression of Helios distinguished MAIT cells from the other T-cell populations (Fig. 4E and Table S2). Taken together, these data indicate that MAIT cells have distinctive phenotypic and transcriptional characteristics that set them apart from the broader T-cell compartment.
The peripheral blood MAIT cell population, as defined in these studies by the expression of Vα7.2 and CD161, stained with the MR1 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil (5-OP-RU) tetramer and not with the MR1 6-formyl pterin (6-FP) negative control tetramer (Fig. S4C). Conversely, the MR1 5-OP-RU+ cell population within total CD3+ cells was from 98.2–99.7% composed of CD161hi Vα7.2+ cells (Fig. 4F), with minor CD161–Vα7.2+, CD161+Vα7.2–, and CD161–Vα7.2– cell populations (Fig. 4G). Furthermore, the high expression of CD195, CD244, CD218a, and CD319 was reproduced when MAIT cells were identified by MR1 5-OP-RU tetramer (Fig. 4H). Thus, the CD161hi Vα7.2+ T-cell phenotype accurately identifies the major MR1-restricted MAIT cell population in peripheral blood and allows consistent phenotypic characterization of these cells.
MAIT Cells Are Effector Memory-Like T Cells with Pronounced Phenotypic Heterogeneity Primarily in Receptors Associated with Innate Immunity.
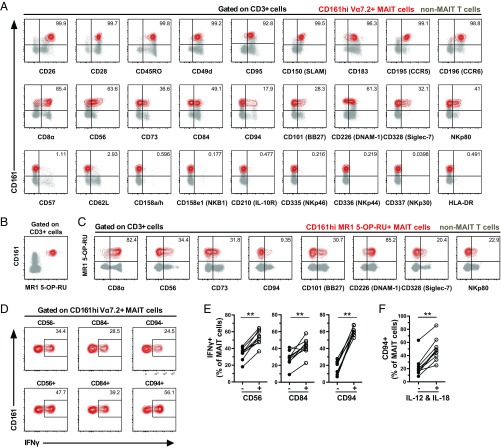
The surface immuno-proteome dataset was next revisited to extensively characterize the MAIT cell population surface receptor repertoire (Fig. 5A and Table S1). MAIT cells were relatively homogeneous in their surface phenotype and were distinctly positive for a subset of 67 surface proteins (>90% of cells). On the other hand, 235 surface proteins were not expressed or expressed in small minority populations (<10% of MAIT cells). In contrast, 21 surface proteins displayed intermediate expression frequency, and many of these were associated with innate cellular immune functions.
Fig. 5.
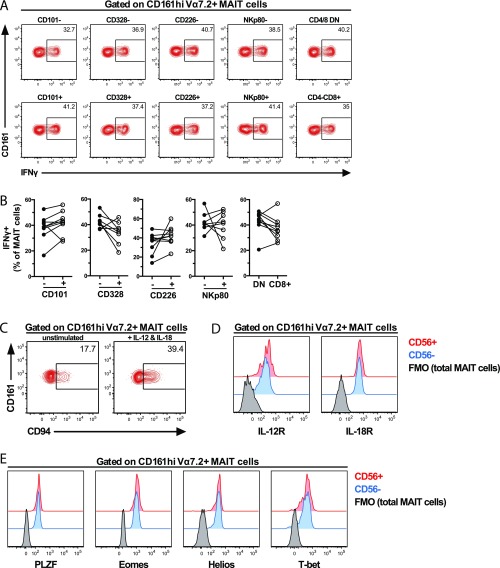
MAIT cells expressing innate receptors respond stronger to IL-12 and IL-18 stimulation. (A) Representative example of immuno-proteome receptors with high (>90%, Upper), low (<10%, Lower), and intermediate (between 10% and 90%, Middle) expression on CD161hi Vα7.2+ MAIT cells (red). (B) Representative example of the identification of CD161hi MR1 5-OP-RU+ MAIT cell population (red) and non-MAIT T cells (gray) by flow cytometry. (C) Representative example of immuno-proteome receptors with intermediate expression on CD161hi MR1 5-OP-RU+ MAIT cells (red) and non-MAIT T cells (gray). (D) MAIT cells in the PBMC sample were stimulated for 24 h with IL-12 and IL-18. Representative example of the production of IFNγ in the MAIT cell subsets defined by the expression of CD56, CD84, and CD94. (E) Frequency of MAIT cells expressing IFNγ after 24 h stimulation with IL-12 and IL-18. MAIT cells were divided into groups expressing or not expressing CD56, CD84, and CD94. (F) Frequency of CD94+ MAIT cells, at baseline conditions and after IL-12 and IL-18 stimulation. Data are from 3 donors (A), 10 donors (B and C), and 9 donors (D–F). Lines represent individual donors. The Wilcoxon’s test or paired t test was used to detect significant differences between the paired samples. **P < 0.01.
It is worthwhile to consider these data in some detail and compare them with the overall T-cell compartment. First, MAIT cells displayed a distinct homing receptor pattern with expression of the inflammatory chemokine receptors CXCR3 (CD183), CCR5 (CD195), and CXCR1 (CD181) as well as CCR3 (CD193), CXCR4 (CD184), and CCR6 (CD196), homing receptors to hematopoietic organs and the gut, respectively. Expression levels of CXCR2 (CD182), CXCR7, and the lymph node homing receptor CCR7 (CD197) were very low. MAIT cells highly expressed IL-18Rα (CD218a), which can activate TCR-independent effector functions, as previously reported (36). The bright expression of CD45RO and the combination of high expression of CD127 (IL-7Rα) and low expression of CD62L identified MAIT cells as effector memory-like T cells.
The costimulatory molecules CD26, CD27, CD28, CD150 (signaling lymphocyte activation molecule family 1, SLAMF1), and CD352 [SLAMF6 or natural killer (NK)-, T-, and B-cell antigen, NTB-A] were expressed by 80–100% of MAIT cells. MAIT cells also expressed members of the tetraspanin family of costimulatory molecules CD53, CD63, CD81, and CD9, whereas CD231 (TALLA-1) was not detected. Apart from SLAMF1 and SLAMF6, MAIT cells also expressed other members of the SLAM family of proteins, broadly involved in the regulation of the activity of NK cells and T cells. These included SLAMF2 (CD48), SLAMF4 (CD244 or 2B4), SLAMF7 (CD319 or CRACC), and SLAMF5 (CD84). In contrast to these aforementioned NK cell-associated molecules, MAIT cells did not express the activating natural cytotoxicity receptors NKp46 (CD335), NKp44 (CD336), and NKp30 (CD337), whereas NKp80 was expressed by approximately a third of the MAIT cell population. The immuno-proteome dataset also revealed relatively high expression of CD95 (Fas), CD172γ (SIRPγ), CD52, and CD43. The expression of these proapoptotic molecules at baseline conditions occurred despite low expression of late activation markers such as CD38 and HLA-DR. Interestingly, relatively small proportions of MAIT cells expressed proteins involved in inhibition of TCR-mediated activation and proliferation, including PD-1 (CD279) and siglec-9, CD35, and siglec-7 (CD328). The intermediate expression levels of these receptors were reproduced when MAIT cells were defined by MR1 5-OP-RU tetramer staining (Fig. 5 B and C), consistent with the high degree of overlap between this population and the one identified by CD161 and Vα7.2 (Fig. 4 F and G). Altogether, these data indicate that MAIT cells are relatively uniform in their effector memory-like inflammation-homing profile, with pronounced phenotypic heterogeneity primarily in receptors associated with innate immune cell functions.
MAIT Cells Expressing CD56, CD84, and CD94 Have Enhanced Capacity to Respond to Innate Cytokine Stimuli.
Several of the surface proteins with heterogeneous expression on MAIT cells are known to be associated with NK cell function, including CD56, CD84, CD101, CD94, CD328 (Siglec-7), CD226 (DNAM-1), and NKp80. We next investigated if expression of these receptors on MAIT cells was associated with responsiveness in vitro to TCR-independent innate stimulus with IL-12 and IL-18. MAIT cells in PBMC samples were stimulated for 24 h with IL-12 and IL-18 and stained for the aforementioned markers as well as IFNγ (Fig. 5D and Fig. S5A). MAIT cells expressing CD56 or CD84, but not CD101, CD328, CD226, NKp80, or CD8, produced significantly more IFNγ in response to IL-12 and IL-18 stimulation than their negative counterparts (P = 0.0039 and P = 0.0013 for CD56 and CD84, respectively) (Fig. 5E and Fig. S5B). CD94 was up-regulated on MAIT cells upon cytokine stimulation (P = 0.0039) (Fig. S5C and Fig. 5F), and CD94+ MAIT cells produced more IFNγ than their CD94– counterparts (P = 0.0039) (Fig. 5E).
Fig. S5.
MAIT cell subset responses to innate cytokine stimulus and further characterization of CD56+ and CD56– MAIT cells. (A) MAIT cells in the PBMC sample were stimulated with IL-12 and IL-18 for 24 h. Representative example of IFNγ production in the MAIT cell subsets defined by the expression of CD101, CD328, CD226, NKp80, and CD8α. (B) Frequency of MAIT cells expressing IFNγ after 24-h stimulation with IL-12 and IL-18. MAIT cells were divided into groups expressing or not CD101, CD328, CD226, and NKp80, as well as in CD4–CD8– double-negative (DN) and CD4–CD8+ MAIT cells. Lines represent individual donors. Representative example of the expression of (C) CD94 on MAIT cells before and after 24-h stimulation with IL-12 and IL-18 and of (D) IL-12R and IL-18R and (E) Eomes, PLZF, Helios, and T-bet in resting CD56+ (red) and CD56– (blue) MAIT cells. The fluorescence minus one (FMO) control in total MAIT cells (black) is also shown. Data are from (A–C) 8–9 donors and (D and E) 9 donors, except for PLZF (8 donors). Lines represent individual donors. The Wilcoxon’s test or paired t test was used to detect significant differences between the paired samples.
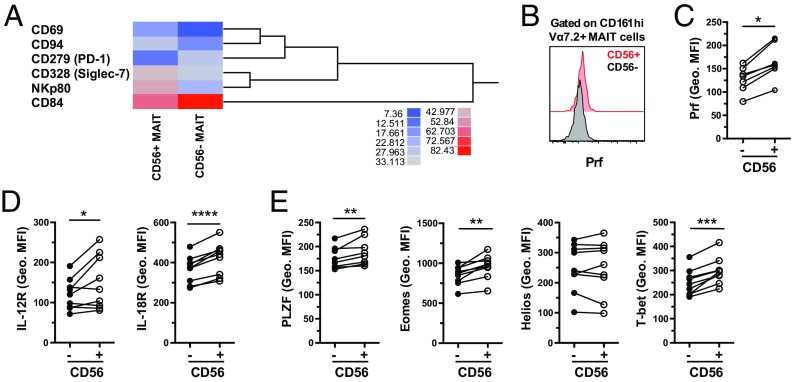
We next investigated in more detail the CD56+ and CD56– MAIT cell subsets with regard to their surface phenotype and transcription factor expression. The two subsets were overall very similar, with the surface immuno-proteome dataset revealing differences in the expression of only six receptors (Fig. 6A). Interestingly, CD56+ MAIT cells expressed higher levels of the NK cell-associated receptors CD94, CD328, and NKp80 but lower levels of CD84. Also, CD56+ MAIT cells expressed lower levels of CD279 despite higher expression of CD69 (Fig. 6A). At resting state, CD56+ MAIT cells expressed more perforin than did CD56– MAIT cells (P = 0.0156) (Fig. 6 B and C) and, interestingly, also expressed significantly higher levels of IL-12R (P = 0.0325) and IL-18R (P < 0.0001) (Fig. 6D and Fig. S5D) as well as of the transcription factors PLZF (P = 0.0084), Eomes (P = 0.0020), and T-bet (P = 0.0001) (Fig. 6E and Fig. S5E). Together, these data demonstrate heterogeneity among MAIT cells in the expression of innate immune receptors, some of which are associated with an increased capacity of MAIT cells to respond to TCR-independent innate cytokine stimuli.
Fig. 6.
Surface phenotype and transcription factor analyses of CD56+ and CD56– MAIT cells. (A) Heat map and dendrogram based on the mean frequency of cells expressing a restricted group of proteins from the surface immuno-proteome dataset. The proteins for which the difference in expression between CD56+ and CD56– MAIT cells was equal or higher than 10% were included. (B) Representative example of the expression of perforin and (C) geometric MFI of the perforin staining, in resting CD56– and CD56+ MAIT cells. Geometric MFI of the staining of (D) IL-12R and IL-18R and (E) PLZF, Eomes, Helios, and T-bet in MAIT cells at baseline conditions. Data are from 3 donors (A), 7 donors (B and C), and 8–9 donors (D and E). Lines represent individual donors. The Wilcoxon’s test or paired t test was used to detect significant differences between the paired samples. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Discussion
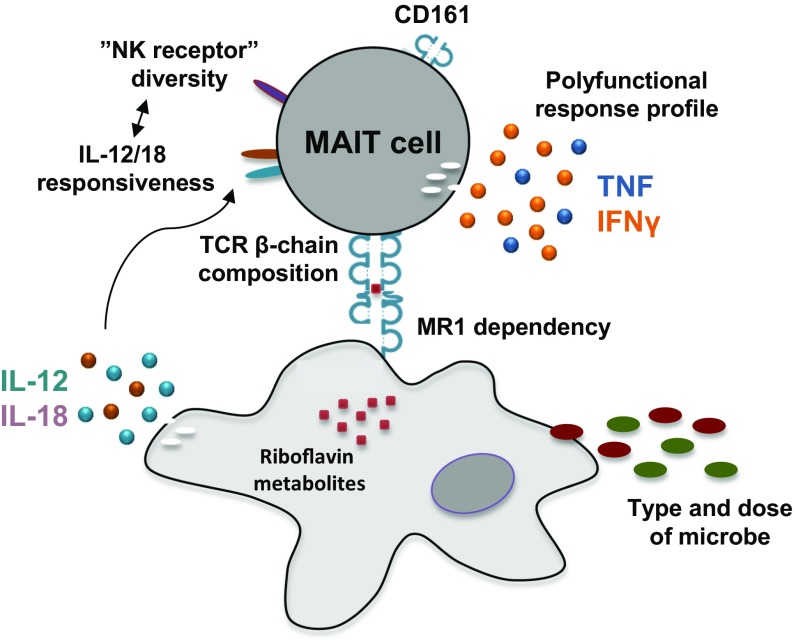
MAIT cells are unconventional innate-like T cells with the capacity to rapidly respond against cells presenting microbial riboflavin metabolite antigens on MR1 molecules. The notion that these cells may have relatively uniform response patterns to different microbes has been favored by the high degree of evolutionary conservation of MR1 and the limited set of activating antigens discovered to date. In this study, we found that MAIT cells displayed microbe-specific polyfunctional response profiles, antigen sensitivity, MR1 dependency, and response magnitudes. Responses against E. coli and C. albicans displayed clear, albeit modest, TCR β-chain bias, indicative of possible divergent antigen subspecificities. Finally, the MAIT cell immuno-proteome was overall relatively homogenous but still showed diversity in some NK cell-associated receptors. In particular, CD56, CD84, and CD94 defined a subset with higher expression of PLZF, Eomes, and T-bet and enhanced capacity to respond to IL-12 and IL-18. Thus, MAIT cells harbor significant functional heterogeneity as they respond to different bacteria, fungi, or innate cytokines (Fig. 7).
Fig. 7.
Factors contributing to the heterogeneity in MAIT cell responses to microbial antigens and innate cytokine stimuli.
To better understand MAIT cell response characteristics against different types of microbes, we dissected the in vitro responses against E. coli and C. albicans. The response of MAIT cells against E. coli was characterized by robust production of IFNγ as well as TNF and may reflect the effector functions of MAIT cells upon encounter of potentially pathogenic microbes. E. coli induced strong TCR down-regulation, consistent with this being a potent MAIT cell-activating microbe. In contrast, the response of MAIT cells to C. albicans was less robust with weaker TCR down-regulation. It is tempting to speculate that differences in the microbial riboflavin biosynthesis pathways, such as the potential production of MR1 ligands with differential capacity to activate MAIT cells (43), may influence these differences in MAIT cell responses to the two highly divergent microbes. In the responses against both E. coli and C. albicans, IFNγ production was predominantly but not exclusively dependent on MR1, whereas TNF production was strongly MR1-dependent, suggesting differential regulation of these cytokines in MAIT cells. This may be explained by the contributions of cytokines such as IL-12 and IL-18 previously shown to contribute to the total IFNγ response by MAIT cells (35, 36). The requirement of the TCR–MR1 interaction for the production of TNF is consistent with the notion that the production of highly proinflammatory cytokines may be tightly regulated to avoid inflammatory responses against commensal microbes that may not be actively producing antigenic vitamin B metabolites in the state of homeostasis. On the other hand, the difference between the E. coli- and C. albicans-triggered cytokine profiles indicates that MAIT cells can respond differently to antigenic microbes, despite the highly conserved nature of MR1. Altogether, these findings suggest that MAIT cells can adapt their response patterns to suit different microbial challenges.
The immuno-proteome analysis confirmed that MAIT cells are effector memory-like T cells with a distinct homing receptor profile indicating a capacity to migrate to mucosal tissues as well as sites of infection. Detailed inspection of the surface immuno-proteome revealed that the expression of receptors associated with innate immunity and NK cells, including CD56, CD84, CD94, CD101, CD226, CD328, and NKp80, was heterogeneous within the MAIT cell population. MAIT cells expressing CD56, CD84, and CD94 responded to IL-12 and IL-18 with higher IFNγ production than their negative counterparts. MAIT cells also strongly up-regulated CD94 upon stimulation, similar to that previously reported in conventional CD8+ T cells (44). Interestingly, CD56+ MAIT cells displayed higher expression of IL-12R and IL-18R as well as of the transcription factors Eomes and T-bet, and this may explain the higher responsiveness. Thus, CD56 identifies a MAIT cell subset more responsive to IL-12 and IL-18, which may be important in MR1-independent MAIT cell responses during inflammation and viral infections.
To further dissect the levels of heterogeneity in the MR1-dependent response against microbes, we explored the possibility that the MAIT cell Vβ repertoire may influence antigen recognition and responses. Here, we took advantage of the finding that the TNF response by MAIT cells was strongly MR1-dependent and residual MR1-independent contributions could be eliminated by subtracting the residual response upon MR1 blocking. MAIT cells expressing Vβ8, Vβ13.1, and Vβ13.6 were hyporesponsive to E. coli, whereas Vβ13.2-expressing MAIT cells were hyperresponsive to C. albicans. This suggests that the Vβ segment influences the functional outcome of the TCR–MR1 interaction, consistent with previous studies (45, 46). Another study, however, indicated that single mutations in the Vβ or CDR3β loop did not affect MR1 recognition (47). Eckle et al. (48) reported that TCRs using Vβ13.3 and Vβ13.5 displayed similar affinity to MR1, whereas Vβ13.3+ TCRs with different CDR3β loops bound MR1 with different affinities. However, the number of TCRs used in these previous studies was limited, and the functional responses were evaluated using TCR-transduced SKW3 cells and C1R.MR1 cells preincubated with synthetic MR1 ligands (48). In contrast, our experimental approach evaluated a large number of responding MAIT cells and thus a large number of different TCRs and allowed detection of TCR Vβ bias in responses that would have gone undetected in lower throughput approaches. Gold et al. (49) evaluated the TCR Jα and Vβ repertoires of responding CD4–TCRγδ–CD8+TRAV1-2+TNF+ cells in response to Mycobacterium smegmatis, Salmonella typhimurium, and C. albicans and reported diversity across donors and microbes in the repertoires of responding cells. Altogether, the present results together with previously published data suggest that the TCR β-chain has some limited influence, which can be positive or negative, on antigen recognition by MAIT cells and contributes to the diversity in responses against different microbes.
The differential influence of the Vβ use in response to E. coli and C. albicans is compatible with the notion that there may be diversity in the antigens presented by MR1. Furthermore, individual MAIT cell clones may contribute differentially to the MR1-restricted response to different antigenic microbes. Interestingly, our data from in vitro proliferation experiments support the possibility that the TCR Vβ repertoire may be shaped by interaction with commonly encountered microbes, where responsive TCRs may expand at the expense of less responsive ones. This notion would be consistent with both the Vβ bias in anti-E. coli effector responses and with the poor proliferative response from MAIT cells carrying rare Vβ segments. Also in support of this hypothesis, the Vβ repertoire of CD161++CD8α+ T cells in cord blood was previously reported to be diverse with no Vβ2 and Vβ13.2 dominance (8).
Our investigation regarding the relationship between MAIT cells and the other T-cell subsets defined by CD161 and Vα7.2 indicates that MAIT cells are distinct based on an extensive set of immunologically relevant surface proteins, a group of seven transcription factors, and a panel of 24 TCR Vβ segments. These findings are supported by those obtained by Fergusson et al. (4) with CD161++CD8+ T cells, where these cells were distinct from CD161+CD8+ and CD161–CD8+ T cells based on the expression of 23 surface molecules. In the study by Fergusson et al. (4), however, different cell types expressing CD161 were reported to share a common transcriptional signature. The unique transcription factor profile of MAIT cells is consistent with our previous study (12), where the expression of PLZF, RORγt, T-bet, Eomes, and Helios was found to be different from that of conventional CD4+ and CD8+ T cells. We furthermore confirmed that MAIT cells displayed a Vβ repertoire distinct from the diverse repertoires of other T-cell populations defined by CD161 and Vα7.2. This is in agreement with previous reports based on TCR sequencing (4, 5). Altogether, our data support the distinctiveness of the MAIT cell population in relation to other T-cell subsets. Stainings with the MR1 5-OP-RU tetramer showed that a vast majority, around 99%, of the MR1 5-OP-RU+ T cells in healthy donor PBMCs coexpressed CD161 and Vα7.2, confirming the CD161hi Vα7.2+ identity of the major MR1-restricted MAIT cell population. Nevertheless, minor populations of MR1-restricted cells lacking either CD161 or Vα7.2 can be identified, a pattern consistent with previous studies (50, 51).
In summary, the findings in this study show that the MAIT cell population harbors multiple layers of heterogeneity in response to microbial antigens and innate cytokine stimuli. This opens the possibility that MAIT cells can adapt their response patterns to the microbial challenge at hand and play different roles in immune responses against different types of bacteria, fungi, and viruses.
Materials and Methods
Cell Isolation.
Peripheral blood was collected from healthy individuals recruited at the Blood Transfusion Clinic at the Karolinska University Hospital Huddinge. Written informed consent was obtained from all of the donors in accordance with study protocols conforming to the provisions of the Declaration of Helsinki and approved by the Regional Ethics Review Board in Stockholm. PBMCs were isolated from peripheral blood by Ficoll–Hypaque density gradient centrifugation (Lymphoprep, Axis-Shield). After isolation, PBMCs were either rested overnight in RPMI-1640 medium supplemented with 25 mM Hepes, 2 mM l-glutamine (all from Thermo Fisher Scientific), 10% FBS (Sigma-Aldrich), 50 μg/mL gentamicin (Life Technologies), and 100 μg/mL normocin (InvivoGen) (complete medium) or immediately used for Vα7.2+ cell isolation, depending on the type of assay to be carried out. Vα7.2+ cells were isolated from PBMCs using anti-Vα7.2 PE- or APC-conjugated mAb (Biolegend), followed by positive selection with magnetic-activated cell sorting (MACS) anti-PE or anti-APC microbeads, respectively (Miltenyi Biotec), as per the manufacturer’s instructions. Monocytes were isolated from peripheral blood by negative selection using the RosetteSep human monocyte enrichment cocktail (STEMCELL Technologies).
Microbes.
The E. coli strain D21 and C. albicans (ATCC 10231) were cultured overnight and for 5 d, respectively, at 37 °C in Luria–Bertani broth. Live microbes were counted by the standard plate counting method on appropriate culture media, and counts were expressed as cfu/mL. The microbes were then stored at –80 °C in 50% glycerol/50% FCS.
MAIT Cell Activation Assays.
Microbes were washed once in PBS, fixed in 1% formaldehyde for 3 min, and extensively washed in PBS before feeding to monocytes. Contrarily to E. coli, C. albicans was not vortexed for the first 60 s and last 30 s of fixation and was only washed once after fixation to prevent cell damage and loss. The activation assay was performed as previously described (42). Cells were cultured at the Vα7.2+ cell:monocyte ratio of 2:1 for 18 h or 24 h in the presence of fixed microbes at the indicated dose and 1.25 μg/mL anti-CD28 mAb (L293, BD Biosciences). In the 18-h experiments and before adding the Vα7.2+ cells, medium was washed away and fresh complete medium was added with anti-MR1 mAb (26.5, Biolegend) or IgG2a isotype control (ctrl) (MOPC-173, Biolegend) at the final concentration of 20 μg/mL. In selected experiments, PBMCs were stimulated with IL-12 p70 (10 ng/mL, Peprotech) and IL-18 (100 ng/mL, Medical & Biological Laboratories) for 24 h. Stimulation of Vα7.2+ cells or PBMCs for 6 h with PMA/ionomycin (Leukocyte Activation Cocktail with Golgi Plug, BD Biosciences) and in the presence of monensin was included in all experiments as the positive control. The frequency of CD69+cytokine+ MAIT cells was calculated by subtracting the residual frequency of unstimulated CD69+cytokine+ MAIT cells to the frequency of stimulated CD69+cytokine+ MAIT cells. In selected experiments, the frequency of MR1-dependent CD69+cytokine+ MAIT cells was determined by subtracting the frequency of stimulated CD69+cytokine+ MAIT cells in the presence of anti-MR1 mAb from the frequency of stimulated CD69+cytokine+ MAIT cells in the presence of IgG2a isotype ctrl.
MAIT Cell Proliferation Assay.
The proliferation experiments were performed as previously described (42) with minor modifications. Briefly, PBMCs were stained with 1.25 μM CTV (Thermo Fisher Scientific Life Sciences), as per the manufacturer’s instructions. CTV-labeled PBMCs (106 per well) were then cultured for 5 d in complete medium with fixed E. coli (10 E. coli cfu per PBMC) and in the presence of 1.25 μg/mL anti-CD28 mAb and 20 μg/mL anti-MR1 mAb or IgG2a isotype ctrl. Recombinant human IL-2 (Peprotech) was added at the final concentration of 100 IU/mL after 24 h and was replenished 48 h later. Anti-MR1 mAb and IgG2a isotype ctrl were replenished at 10 μg/mL 72 h after the first addition.
Flow Cytometry.
Cell surface and intracellular stainings as well as stainings using the LEGENDScreen kit were performed as previously described (12, 17, 39, 52). Stainings with CD195 (CCR5) mAb and the MR1 5-OP-RU and MR1 6-FP tetramers conjugated with PE were performed for 40 min at room temperature (15) before surface staining with other mAbs for 20 min at 4 °C. The antibodies used are listed in Table S3. Samples were acquired on an LSRFortessa flow cytometer (BD Biosciences) equipped with 355-, 405-, 488-, 561-, and 639-nm lasers. Compensation was performed using single-stained polystyrene beads (BD Biosciences) and the compensation platform in the FlowJo software v. 9.9 (TreeStar). Clustering analyses and PCA were performed using the JMP software v. 12 (SAS Institute Inc.), and the polyfunctionality analyses were performed using the SPICE software v. 5.3 (NIH).
Statistical Analyses.
Statistical analyses were performed using Prism software v.6 (GraphPad). Datasets were first assessed for normality of the data distribution. Statistically significant differences between samples were determined as appropriate using the unpaired t test or Mann–Whitney’s test for unpaired samples and the paired t test or Wilcoxon’s signed-rank test for paired samples. Correlations were assessed using the Spearman’s rank correlation. Two-sided P values < 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Caroline Boulouis, Salah Zangenah, and David Malone for technical assistance. The MR1 tetramer technology was developed jointly by Dr. James McCluskey, Dr. Jamie Rossjohn, and Dr. David Fairlie, and the material was produced by the NIH Tetramer Core Facility as permitted to be distributed by the University of Melbourne. This research was supported by Swedish Research Council Grant K2012-56X-14449-11-5, Swedish Cancer Society Grant CAN 2014/879, Petrus and Augusta Hedlund Foundation Grant M-2016-0443, and National Institutes of Health Grant R01DK108350 (to J.K.S.). Further support came from the Clas Groschinsky Memorial Fund, Swedish Medical Doctors Against AIDS Foundation Grant FOb2013-0005, the Swedish Society of Medicine, the Jonas Söderquist Foundation for Virology and Immunology, Swedish Research Council Grant 2015-00174, and Marie Skłodowska Curie Actions, Cofund, Project INCA 600398 (to E.L.). J.D. was supported by Fundação para a Ciência e a Tecnologia Doctoral Fellowship SFRH/BD/85290/2012, through program QREN-POPH-typology 4.1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705759114/-/DCSupplemental.
References
- 1.Dusseaux M, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. 2011;117:1250–1259. doi: 10.1182/blood-2010-08-303339. [DOI] [PubMed] [Google Scholar]
- 2.Martin E, et al. Stepwise development of MAIT cells in mouse and human. PLoS Biol. 2009;7:e54. doi: 10.1371/journal.pbio.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Treiner E, et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 2003;422:164–169. doi: 10.1038/nature01433. [DOI] [PubMed] [Google Scholar]
- 4.Fergusson JR, et al. CD161 defines a transcriptional and functional phenotype across distinct human T cell lineages. Cell Reports. 2014;9:1075–1088. doi: 10.1016/j.celrep.2014.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lepore M, et al. Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRβ repertoire. Nat Commun. 2014;5:3866. doi: 10.1038/ncomms4866. [DOI] [PubMed] [Google Scholar]
- 6.Reantragoon R, et al. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J Exp Med. 2013;210:2305–2320. doi: 10.1084/jem.20130958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tilloy F, et al. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J Exp Med. 1999;189:1907–1921. doi: 10.1084/jem.189.12.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker LJ, et al. Human MAIT and CD8αα cells develop from a pool of type-17 precommitted CD8+ T cells. Blood. 2012;119:422–433. doi: 10.1182/blood-2011-05-353789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le Bourhis L, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol. 2010;11:701–708. doi: 10.1038/ni.1890. [DOI] [PubMed] [Google Scholar]
- 10.Savage AK, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbs A, et al. MAIT cells reside in the female genital mucosa and are biased towards IL-17 and IL-22 production in response to bacterial stimulation. Mucosal Immunol. 2017;10:35–45. doi: 10.1038/mi.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leeansyah E, et al. Arming of MAIT cell cytolytic antimicrobial activity is induced by IL-7 and defective in HIV-1 infection. PLoS Pathog. 2015;11:e1005072. doi: 10.1371/journal.ppat.1005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012;12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akimova T, Beier UH, Wang L, Levine MH, Hancock WW. Helios expression is a marker of T cell activation and proliferation. PLoS One. 2011;6:e24226. doi: 10.1371/journal.pone.0024226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corbett AJ, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 2014;509:361–365. doi: 10.1038/nature13160. [DOI] [PubMed] [Google Scholar]
- 16.Kjer-Nielsen L, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012;491:717–723. doi: 10.1038/nature11605. [DOI] [PubMed] [Google Scholar]
- 17.Leeansyah E, Loh L, Nixon DF, Sandberg JK. Acquisition of innate-like microbial reactivity in mucosal tissues during human fetal MAIT-cell development. Nat Commun. 2014;5:3143. doi: 10.1038/ncomms4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurioka A, et al. MAIT cells are licensed through granzyme exchange to kill bacterially sensitized targets. Mucosal Immunol. 2015;8:429–440. doi: 10.1038/mi.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Bourhis L, et al. MAIT cells detect and efficiently lyse bacterially-infected epithelial cells. PLoS Pathog. 2013;9:e1003681. doi: 10.1371/journal.ppat.1003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Booth JS, et al. Mucosal-associated invariant T cells in the human gastric mucosa and blood: Role in Helicobacter pylori infection. Front Immunol. 2015;6:466. doi: 10.3389/fimmu.2015.00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z, et al. Mucosal-associated invariant T-cell activation and accumulation after in vivo infection depends on microbial riboflavin synthesis and co-stimulatory signals. Mucosal Immunol. 2017;10:58–68. doi: 10.1038/mi.2016.39. [DOI] [PubMed] [Google Scholar]
- 22.Chua WJ, et al. Polyclonal mucosa-associated invariant T cells have unique innate functions in bacterial infection. Infect Immun. 2012;80:3256–3267. doi: 10.1128/IAI.00279-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Georgel P, Radosavljevic M, Macquin C, Bahram S. The non-conventional MHC class I MR1 molecule controls infection by Klebsiella pneumoniae in mice. Mol Immunol. 2011;48:769–775. doi: 10.1016/j.molimm.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Gold MC, et al. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. 2010;8:e1000407. doi: 10.1371/journal.pbio.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimaldi D, et al. Specific MAIT cell behaviour among innate-like T lymphocytes in critically ill patients with severe infections. Intensive Care Med. 2014;40:192–201. doi: 10.1007/s00134-013-3163-x. [DOI] [PubMed] [Google Scholar]
- 26.Leung DT, et al. Circulating mucosal associated invariant T cells are activated in Vibrio cholerae O1 infection and associated with lipopolysaccharide antibody responses. PLoS Negl Trop Dis. 2014;8:e3076. doi: 10.1371/journal.pntd.0003076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meierovics A, Yankelevich WJ, Cowley SC. MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc Natl Acad Sci USA. 2013;110:E3119–E3128. doi: 10.1073/pnas.1302799110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meierovics AI, Cowley SC. MAIT cells promote inflammatory monocyte differentiation into dendritic cells during pulmonary intracellular infection. J Exp Med. 2016;213:2793–2809. doi: 10.1084/jem.20160637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okada S, et al. Immunodeficiencies. Impairment of immunity to Candida and Mycobacterium in humans with bi-allelic RORC mutations. Science. 2015;349:606–613. doi: 10.1126/science.aaa4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith DJ, Hill GR, Bell SC, Reid DW. Reduced mucosal associated invariant T-cells are associated with increased disease severity and Pseudomonas aeruginosa infection in cystic fibrosis. PLoS One. 2014;9:e109891. doi: 10.1371/journal.pone.0109891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeffery HC, et al. Biliary epithelium and liver B cells exposed to bacteria activate intrahepatic MAIT cells through MR1. J Hepatol. 2016;64:1118–1127. doi: 10.1016/j.jhep.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson RP, et al. STAT3 is a critical cell-intrinsic regulator of human unconventional T cell numbers and function. J Exp Med. 2015;212:855–864. doi: 10.1084/jem.20141992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slichter CK, et al. Distinct activation thresholds of human conventional and innate-like memory T cells. JCI Insight. 2016;1:e86292. doi: 10.1172/jci.insight.86292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ussher JE, et al. TLR signaling in human antigen-presenting cells regulates MR1-dependent activation of MAIT cells. Eur J Immunol. 2016;46:1600–1614. doi: 10.1002/eji.201545969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loh L, et al. Human mucosal-associated invariant T cells contribute to antiviral influenza immunity via IL-18-dependent activation. Proc Natl Acad Sci USA. 2016;113:10133–10138. doi: 10.1073/pnas.1610750113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ussher JE, et al. CD161++ CD8+ T cells, including the MAIT cell subset, are specifically activated by IL-12+IL-18 in a TCR-independent manner. Eur J Immunol. 2014;44:195–203. doi: 10.1002/eji.201343509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cosgrove C, et al. Early and nonreversible decrease of CD161++ /MAIT cells in HIV infection. Blood. 2013;121:951–961. doi: 10.1182/blood-2012-06-436436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hengst J, et al. Nonreversible MAIT cell-dysfunction in chronic hepatitis C virus infection despite successful interferon-free therapy. Eur J Immunol. 2016;46:2204–2210. doi: 10.1002/eji.201646447. [DOI] [PubMed] [Google Scholar]
- 39.Leeansyah E, et al. Activation, exhaustion, and persistent decline of the antimicrobial MR1-restricted MAIT-cell population in chronic HIV-1 infection. Blood. 2013;121:1124–1135. doi: 10.1182/blood-2012-07-445429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Wilgenburg B, et al. STOP-HCV consortium MAIT cells are activated during human viral infections. Nat Commun. 2016;7:11653. doi: 10.1038/ncomms11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandberg JK, Dias J, Shacklett BL, Leeansyah E. Will loss of your MAITs weaken your HAART [corrected]? AIDS. 2013;27:2501–2504. doi: 10.1097/QAD.0b013e3283620726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dias J, Sobkowiak MJ, Sandberg JK, Leeansyah E. Human MAIT-cell responses to Escherichia coli: Activation, cytokine production, proliferation, and cytotoxicity. J Leukoc Biol. 2016;100:233–240. doi: 10.1189/jlb.4TA0815-391RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soudais C, et al. In vitro and in vivo analysis of the gram-negative bacteria-derived riboflavin precursor derivatives activating mouse MAIT cells. J Immunol. 2015;194:4641–4649. doi: 10.4049/jimmunol.1403224. [DOI] [PubMed] [Google Scholar]
- 44.Derre L, et al. Expression of CD94/NKG2-A on human T lymphocytes is induced by IL-12: Implications for adoptive immunotherapy. J Immunol. 2002;168:4864–4870. doi: 10.4049/jimmunol.168.10.4864. [DOI] [PubMed] [Google Scholar]
- 45.López-Sagaseta J, et al. The molecular basis for Mucosal-Associated Invariant T cell recognition of MR1 proteins. Proc Natl Acad Sci USA. 2013;110:E1771–E1778. doi: 10.1073/pnas.1222678110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.López-Sagaseta J, et al. MAIT recognition of a stimulatory bacterial antigen bound to MR1. J Immunol. 2013;191:5268–5277. doi: 10.4049/jimmunol.1301958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reantragoon R, et al. Structural insight into MR1-mediated recognition of the mucosal associated invariant T cell receptor. J Exp Med. 2012;209:761–774. doi: 10.1084/jem.20112095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eckle SB, et al. A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J Exp Med. 2014;211:1585–1600. doi: 10.1084/jem.20140484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gold MC, et al. MR1-restricted MAIT cells display ligand discrimination and pathogen selectivity through distinct T cell receptor usage. J Exp Med. 2014;211:1601–1610. doi: 10.1084/jem.20140507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gherardin NA, et al. Diversity of T cells restricted by the MHC class I-related molecule MR1 facilitates differential antigen recognition. Immunity. 2016;44:32–45. doi: 10.1016/j.immuni.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 51.Meermeier EW, et al. Human TRAV1-2-negative MR1-restricted T cells detect S. pyogenes and alternatives to MAIT riboflavin-based antigens. Nat Commun. 2016;7:12506. doi: 10.1038/ncomms12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dias J, Sandberg JK, Leeansyah E. Extensive phenotypic analysis, transcription factor profiling, and effector cytokine production of human MAIT cells by flow cytometry. Methods Mol Biol. 2017;1514:241–256. doi: 10.1007/978-1-4939-6548-9_17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.