Significance
Male ejaculates exhibit remarkable diversity, including variation in their spatial and temporal molecular composition. This complexity suggests that ejaculates provide functions far beyond the delivery of sperm. Here, we investigated the molecular and functional specificity of the butterfly spermatophore, a structurally complex ejaculate. We discovered that its two distinct parts originate from separate regions of the male reproductive tract, are transferred sequentially during mating, and seem to be the result of a complex evolutionary history. We also highlight a large and previously unrecognized female contribution to the spermatophore, which calls into question traditional characterizations of females as passive recipients of these male ejaculates.
Keywords: seminal fluid proteins, reproductive physiology, ejaculate, spermatophore
Abstract
Male ejaculates are often structurally complex, and this complexity is likely to influence key reproductive interactions between males and females. However, despite its potential evolutionary significance, the molecular underpinnings of ejaculate structural complexity have received little empirical attention. To address this knowledge gap, we sought to understand the biochemical and functional properties of the structurally complex ejaculates of Pieris rapae butterflies. Males in this species produce large ejaculates called spermatophores composed of an outer envelope, an inner matrix, and a bolus of sperm. Females are thought to benefit from the nutrition contained in the soluble inner matrix through increases in longevity and fecundity. However, the indigestible outer envelope of the spermatophore delays female remating, allowing males to monopolize paternity for longer. Here, we show that these two nonsperm-containing spermatophore regions, the inner matrix and the outer envelope, differ in their protein composition and functional properties. We also reveal how these divergent protein mixtures are separately stored in the male reproductive tract and sequentially transferred to the female reproductive tract during spermatophore assembly. Intriguingly, we discovered large quantities of female-derived proteases in both spermatophore regions shortly after mating, which may contribute to spermatophore digestion and hence, female control over remating rate. Finally, we report evidence of past selection on these spermatophore proteins and female proteases, indicating a complex evolutionary history. Our findings illustrate how structural complexity of ejaculates may allow functionally and/or spatially associated suites of proteins to respond rapidly to divergent selective pressures, such as sexual conflict or reproductive cooperation.
Ejaculates provide important functions far beyond the simple delivery of sperm, playing roles in sperm survival, sperm storage, and the manipulation of female reproductive physiology (1). Understanding these reproductive functions and the biochemical compounds associated with them has been an area of active, fruitful investigation for the past several decades. In Drosophila, for example, substances in the male ejaculate have been shown to increase oviposition and reduce female receptivity to remating (2–5). Similarly, substances in llama and alpaca ejaculates have been shown to induce ovulation (6). However, one aspect of ejaculate biology that remains less well-understood is how these different roles are reflected in spatial and structural adaptations of ejaculates themselves. Observations of structural diversity in ejaculates across animal groups suggest that males may position reproductively important compounds where their functions can be used to best advantage. For example, in many primates and rodents, specific seminal fluid proteins (SFPs) form a copulatory plug inside the female reproductive tract that can inhibit subsequent mates from siring offspring (7–9). These proteins are transferred after the bolus of sperm so that they block the female reproductive tract but do not inhibit the male’s own sperm. In other taxonomic groups as diverse as insects, squid, and salamanders, male ejaculates form complex structures called spermatophores, which are likely to serve a variety of functions, including providing manipulative and/or nutritional substances to mates (10, 11).
In insects, spermatophores exhibit remarkable diversity in shape and structural complexity (12, 13). For example, many orthopteran (e.g., katydid) spermatophores are composed of two distinct parts: a gelatinous spermatophylax and a separate sperm-containing ampulla. During mating, orthopteran females eat the externally deposited spermatophylax first before moving to consumption of the ampulla (14–16). Larger and/or more phagostimulating spermatophylaxes favor males by extending the time that their sperm have to migrate out of the ampulla and into the female reproductive tract, where they can fertilize ova (17). However, whether the spermatophylax provides any nutritive value and can, therefore, be considered parental or mating investment or simply acts to manipulate female control over insemination by distracting the female through hijacking of her taste receptors [e.g., the “candymaker hypothesis” (18)] remains the subject of ongoing inquiry (17). Parsing between these alternative evolutionary scenarios requires an understanding of not only the identities and functions of male reproductive compounds but also, their spatial placement within the ejaculate.
Other even more complex and poorly understood examples exist, such as the elaborate biochemical choreography of ejaculate transfer in the rove beetle Aleochara curtula. In this species, spermatophore formation begins with male secretion of a tube-like structure inside the female spermatheca. This ejaculate tube then serves as a guide for the elongation of a second interior tube, creating a bilayer structure. The second tube then extends until it reaches the apical bulb of the spermatheca, where it inflates to form a balloon that ultimately bursts to release the sperm (19–21). How this sequence of events is enacted at the biochemical level remains unknown, and the adaptive significance of such complexity is likewise a mystery. However, these examples and others highlight the trove of uncharacterized molecular mechanisms and evolutionary strategies currently hidden within the structural complexity of male ejaculates.
We studied the molecular basis and functional significance of ejaculate complexity in the butterfly Pieris rapae. Lepidopteran spermatophores are internally deposited ejaculate structures that play a number of important reproductive roles. In many species, females use nutrition from the spermatophore to support egg production and somatic maintenance (22, 23). Thus, spermatophores may act as paternal investment by increasing the quality and/or number of offspring that a male sires with a given female (24). This nutritional exchange represents a cooperative relationship between the sexes. However, spermatophores may also be used to manipulate female remating rate in species in which females are unwilling to remate until they have digested a spermatophore (12, 24). Because of widespread last male sperm precedence in Lepidoptera (25–29), males benefit when their spermatophore delays female remating, thereby increasing the amount of time in which his sperm are preferentially used to fertilize her eggs. However, this delay may come at a cost to females, because it prevents their access to the nutritive ejaculates of future partners (30–32). Thus, interactions between the spermatophore and the female reproductive tract may play an important role in mediating sexual conflict over female remating rate.
In the Cabbage White butterfly, P. rapae, males transfer large spermatophores with three distinct regions—a tough outer envelope, a soft inner matrix, and a bolus of sperm at the spermatophore base (33). This complex tripartite ejaculate is secreted by the male aedeagus into a specialized organ in the female reproductive tract called the bursa copulatrix (34). After mating, sperm rapidly migrate to the sperm storage organ in the female, called the spermatheca, and the bursa copulatrix is left to digest and absorb the nutritional content of the spermatophore (35). This process of digestion involves both mechanical and chemical digestion. Muscular contractions of the bursal wall animate a hardened, toothed device called the signum, which bores a hole through the tough outer envelope (34, 36, 37). Chemical breakdown of the spermatophore is then accomplished through a mixture of enzymes, including as many as nine proteases, which the female secretes in concentrations dramatically surpassing those found in the larval gut (38).
Although our knowledge of bursal physiology and function is increasingly sophisticated, our corresponding understanding of the biochemical complexity of the spermatophore remains rudimentary in this species and indeed, the Lepidoptera in general. For example, almost nothing is known about the biochemical basis of the outer spermatophore envelope, except that it is often indigestible, allowing it to persist in the bursa for the female’s entire lifespan. This indigestibility has led to the common practice of counting spermatophore envelopes in the bursae of field-caught females to quantify female remating rates in natural populations (39, 40). The biochemical composition of the spermatophore inner matrix is also poorly understood, except in a few cases where we know the relative proportions of bulk protein, lipid, and carbohydrates (41, 42). Thus, a critical first step in better understanding the functional significance of spermatophore complexity in the Lepidoptera is to characterize the underlying biochemical constituents and physical properties of these spermatophore regions.
In our study, we sought to decipher the structural complexity of the spermatophore in P. rapae, with an emphasis on its constituent proteins and their evolution. We focused on the envelope and the inner matrix, in light of their potential to shape female reproductive physiology through interactions with the bursa copulatrix. We reveal that the hard, indigestible outer envelope and the soft inner matrix are composed of different protein suites, consistent with their potential to impart distinct functions after copulation. Using a combination of proteomics, transcriptomics, and amino acid analyses, we identify two likely candidates for the structural proteins that impart digestion resistance to the spermatophore envelope. Then, using spatially explicit proteomic sampling of the spermatophore and male reproductive tract, we reveal that the distinct protein mixtures found in the envelope and inner matrix are stored in specific regions of the male reproductive tract and sequentially transferred to the female bursa during spermatophore assembly. In addition, we find evidence for high levels of incorporation of female-produced proteins, particularly proteases, into the spermatophore during or shortly after mating, suggesting an early involvement of female biochemical digestion in spermatophore processing. Lastly, we report that many of the identified spermatophore proteins appear to have evolved rapidly, and a number of the female proteases are the product of recent gene duplications, consistent with a history of adaptive evolution. Each of these findings—the distinct biochemistry and functional properties of the envelope and inner matrix, the presence of female proteases inside the spermatophore, and evidence of strong past selection on protein sequences—supports an emerging picture that this male ejaculate has been shaped by a complex evolutionary history. Furthermore, this history seems to have selected for specialized suites of proteins that impart specific functional attributes to the spermatophore, including digestion resistance to the spermatophore envelope and solubility to the inner matrix.
More generally, our study illustrates why the field of reproductive physiology should move beyond its empirical tendency to treat ejaculates as biochemically homogenous and strictly male-produced into exciting territory involving interactions between functionally integrated suites of male and female reproductive molecules. Such research is likely to change the way that we think about male–female reproductive interactions and open up avenues of inquiry into the specialized tissues that produce these unique properties of male ejaculates.
Results
Envelope and Inner Matrix Are Composed of Distinct Protein Suites Stored in Different Regions of the Male Reproductive Tract, Suggesting Their Sequential Transfer During Mating.
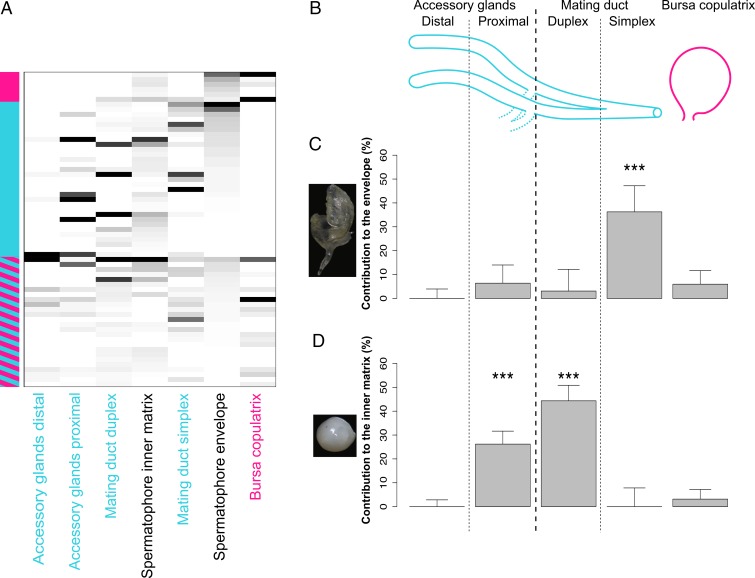
We first sought to identify the protein constituents of the different regions of the spermatophore and the male and female reproductive tissues that contribute them. We dissected out four morphologically distinct secretory glands/regions of the male reproductive tract (Fig. 1B) as well as the female bursa copulatrix. Accessory glands were separated into distal and proximal regions (defined relative to the genital opening of the male). We also separated the male mating duct into duplex and simplex sections based on morphology (Methods). From MS analysis of these nine tissues, we identified 341 proteins. Protein quantities were highly correlated between biological replicates after bioinformatic subtraction of hemolymph proteins (Methods, Fig. S1, and Table S1). The spermatophore envelope and inner matrix contained 63 proteins as major components (Fig. 1A and Table S2). By comparing these proteins with SFPs previously identified in Heliconius butterflies (42), we found that 10 of our proteins are homologous to SFPs from Heliconius erato and/or Heliconius melpomene (Table S3). Most of the spermatophore proteins found in P. rapae are enzymes (n = 27; 42.8%), including 11 proteases. The abundance and diversity of proteases are a recurring characteristic of ejaculates in insects and more divergent taxonomic groups (43–45). There were also many proteins of undetermined function. Thirteen proteins had no discernable molecular domain or homolog, and five had homologs only to uncharacterized proteins in Lepidoptera, prohibiting their functional annotation.
Fig. 1.
The spermatophore is a complex product of the male and female reproductive tracts and sequentially transferred during mating. (A) Abundances of 63 proteins identified between the inner matrix and the envelope of the spermatophore. Each row represents a protein, and each column represents a tissue. The vertical column on the left side of the heat map represents the origin of the gene product (blue for male and pink for female; a combination of pink and blue indicates that transcripts encoding the proteins are found in a combination of reproductive and nonreproductive tissues of both sexes). Abundance is represented by grayscale (black, highly abundant; white, absent from sample). (B) Simplified schematics of an unfolded P. rapae male reproductive tract and the female bursa copulatrix. Segments are not drawn to scale. (C) Contributions of male and female reproductive tissues to the envelope of the spermatophore. ***P < 0.001. (D) Contributions of male and female reproductive tissues to the inner matrix of the spermatophore.
Fig. S1.
Hemolymph subtraction reveals tissues cluster by type. (A) Before hemolymph subtraction, most tissues from separate samples clustered by sample and not type. All proteins found in the 2015 hemolymph sample were subtracted bioinformatically from the protein profile of each tissue (hemolymph proteins were removed, and the quantitative normal values were rescaled accordingly). (B) After hemolymph subtraction, all duplicate tissues were clustered by type. This analysis allowed subsequent merging of duplicate tissues, such that they were treated as replicates of one sample.
Table S1.
Hemolymph subtraction alters tissue correlations
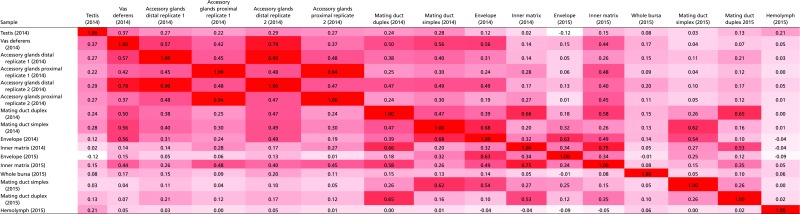 |
Before hemolymph subtraction, the proteomic relationship between many tissues was obscured by hemolymph contamination (Fig. S1). Proteins present in the 2015 hemolymph sample were removed from the protein profile of each tissue. After this subtraction, biological replicates were more strongly correlated and clustered together. Red shading indicates the strength of correlation, as measured by the correlation coefficient r.
Table S2.
Inventory of 63 proteins identified in the P. rapae spermatophore
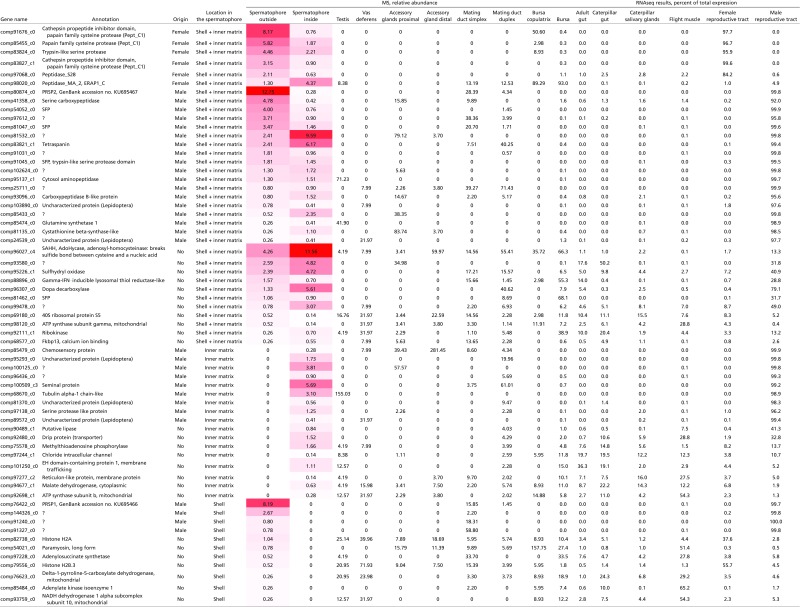 |
Red shading reflects the abundance of specific proteins in the spermatophore.
Table S3.
Homology search between P. rapae proteins and Heliconius melpomene and Heliconius erato proteins
| Gene name | Best Blast hits? | |||||||
| H. erato | E value | Identity, % | Positives, % | H. melpomene | E value | Identity, % | Positives, % | |
| comp91676_c0 | No | / | / | / | HACP057 | 1.00E-13 | 38 | 58 |
| comp85455_c0 | No | / | / | / | No | / | / | / |
| comp83824_c0 | HACP001 | 5.00E-27 | 33 | 52 | HACP001 | 3.00E-28 | 34 | 53 |
| comp83827_c1 | No | / | / | / | HACP057 | 1.00E-06 | 30 | 57 |
| comp97068_c0 | No | / | / | / | No | / | / | / |
| comp98020_c0 | No | / | / | / | No | / | / | / |
| comp80874_c0 | No | / | / | / | No | / | / | / |
| comp41358_c0 | No | / | / | / | No | / | / | / |
| comp54052_c0 | HACP004 | 6.00E-30 | 43 | 69 | HACP004 | 6.00E-28 | 52 | 74 |
| comp97612_c0 | No | / | / | / | No | / | / | / |
| comp81047_c0 | HACP026 | 1.00E-121 | 58 | 74 | HACP026 | 2.00E-124 | 59 | 75 |
| comp81532_c0 | No | / | / | / | No | / | / | / |
| comp83821_c1 | No | / | / | / | No | / | / | / |
| comp91031_c0 | No | / | / | / | HACP016 | 3.00E-05 | 41 | 58 |
| comp91045_c0 | HACP010 | 0 | 78 | 88 | HACP010 | 0 | 75 | 85 |
| comp102624_c0 | No | / | / | / | No | / | / | / |
| comp95137_c1 | No | / | / | / | No | / | / | / |
| comp25711_c0 | No | / | / | / | No | / | / | / |
| comp93096_c0 | No | / | / | / | No | / | / | / |
| comp103890_c0 | No | / | / | / | No | / | / | / |
| comp85433_c0 | No | / | / | / | No | / | / | / |
| comp85474_c0 | No | / | / | / | No | / | / | / |
| comp81135_c0 | No | / | / | / | No | / | / | / |
| comp24539_c0 | No | / | / | / | No | / | / | / |
| comp96027_c4 | No | / | / | / | No | / | / | / |
| comp93580_c0 | No | / | / | / | No | / | / | / |
| comp95226_c1 | No | / | / | / | No | / | / | / |
| comp88896_c0 | No | / | / | / | No | / | / | / |
| comp96307_c0 | No | / | / | / | No | / | / | / |
| comp81462_c0 | HACP057 | 2.00E-18 | 53 | 64 | HACP057 | 1.00E-30 | 52 | 67 |
| comp99478_c0 | No | / | / | / | No | / | / | / |
| comp69180_c0 | No | / | / | / | No | / | / | / |
| comp98120_c0 | No | / | / | / | No | / | / | / |
| comp92111_c1 | No | / | / | / | No | / | / | / |
| comp68577_c0 | No | / | / | / | No | / | / | / |
| comp85479_c0 | No | / | / | / | No | / | / | / |
| comp95293_c0 | No | / | / | / | No | / | / | / |
| comp100125_c0 | No | / | / | / | No | / | / | / |
| comp96436_c0 | No | / | / | / | No | / | / | / |
| comp100509_c3 | HACP011 | 5.00E-52 | 43 | 62 | HACP011 | 2.00E-53 | 43 | 62 |
| comp68670_c0 | No | / | / | / | No | / | / | / |
| comp81370_c0 | No | / | / | / | No | / | / | / |
| comp97138_c0 | HACP037 | 2.00E-100 | 74 | 88 | HACP037 | 4.00E-155 | 72 | 86 |
| comp89572_c0 | No | / | / | / | No | / | / | / |
| comp90489_c1 | No | / | / | / | No | / | / | / |
| comp92480_c0 | No | / | / | / | No | / | / | / |
| comp75578_c0 | No | / | / | / | No | / | / | / |
| comp97244_c1 | No | / | / | / | No | / | / | / |
| comp101250_c0 | No | / | / | / | No | / | / | / |
| comp97277_c2 | No | / | / | / | No | / | / | / |
| comp94677_c1 | No | / | / | / | No | / | / | / |
| comp92698_c1 | No | / | / | / | No | / | / | / |
| comp76422_c0 | No | / | / | / | No | / | / | / |
| comp144326_c0 | No | / | / | / | No | / | / | / |
| comp91240_c0 | No | / | / | / | No | / | / | / |
| comp91327_c0 | No | / | / | / | No | / | / | / |
| comp82738_c0 | No | / | / | / | No | / | / | / |
| comp54021_c0 | No | / | / | / | No | / | / | / |
| comp97228_c0 | No | / | / | / | No | / | / | / |
| comp79556_c0 | No | / | / | / | No | / | / | / |
| comp76623_c0 | No | / | / | / | No | / | / | / |
| comp85484_c0 | No | / | / | / | No | / | / | / |
| comp93759_c0 | No | / | / | / | No | / | / | / |
/ represents not applicable.
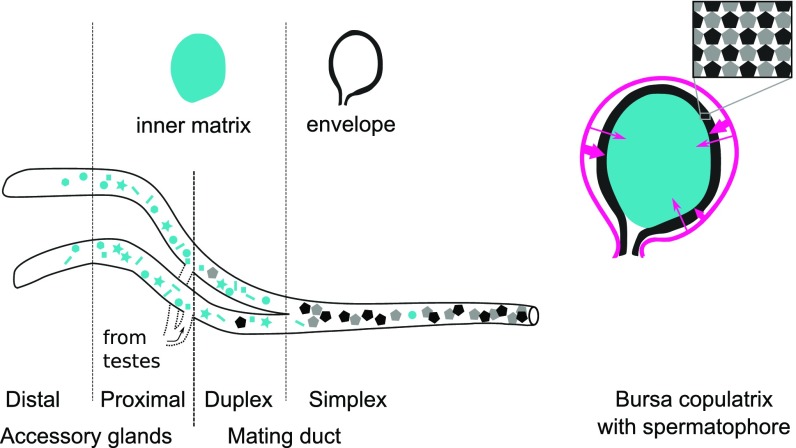
We then estimated the proportional contribution of each male and female reproductive tissue to the spermatophore. We used a likelihood model to estimate the weighted contribution of each individual tissue based on its quantitative protein profile (Methods). Data from testes and vas deferens showed a negligible contribution to the spermatophore, and these tissues were, therefore, excluded from subsequent analyses. The best fit model for the outer envelope showed the mating duct simplex to be the main contributor (36.2 ± 1.1%). As for the inner matrix, the mating duct duplex and the proximal accessory glands were the main contributors (44.3 ± 6.5% and 26.2 ± 5.4%, respectively) (Fig. 1 C and D). These contributions suggest that the spermatophore is assembled in a sequential process. The mating duct simplex is the most proximal to the male genital opening, and therefore, its secretion products (i.e., the envelope components) would be transferred first to the female. Secretion products from reproductive tract regions more distal to the genital opening, such as the mating duct duplex and the proximal accessory glands, would be transferred afterward to form the inner matrix. Thus, our data suggest that spermatophore formation begins with transfer of the envelope, which is then inflated by the inner matrix material (Fig. 2).
Fig. 2.
Mechanistic model of spermatophore formation. The male reproductive tract (MRT) and its different regions are represented on the left. The blue, black, and gray shapes in the MRT represent proteins. The pink arrows represent female proteases. For visual clarity, organs are not represented to scale. In a typical P. rapae male, the accessory glands (distal and proximal) measure around 20 mm, whereas the mating duct (duplex and simplex) can measure more than 110 mm. On the female side, the bursa copulatrix typically measures around 2.5 mm in diameter.
Large Amounts of Female Proteases Invade the Spermatophore During or Shortly After Mating.
By combining the proteomic results with our nine previously published tissue-specific transcriptomes (34), we determined the origins of spermatophore proteins and whether they came from females and/or males. As expected, most spermatophore proteins (n = 31; 49.2%) were highly specific to the male reproductive tract, whereas others (n = 26; 41.2%) are expressed in reproductive and nonreproductive organs of both sexes (Fig. 1A and Table S2). Surprisingly, we uncovered a large female contribution to the spermatophore. Five proteins found in large quantities in the spermatophore were identified as being solely expressed in the female reproductive tract, and one other is found predominantly in the bursa copulatrix. By abundance, these female-specific proteins represent 25% of the soluble protein content in the envelope and 10.7% of the inner matrix (Fig. 1A). Interestingly, all six of these female-produced proteins are proteases, each of which was previously identified as expressed in the bursa copulatrix (38). Because the spermatophores sampled here were collected 2 h after mating—the earliest time point at which the envelope is fully hardened—these proteases could have been incorporated either during the transfer of the ejaculate or shortly after the transfer was complete. This rapid incorporation of large amounts of female-derived proteases is consistent with our prior observations of the remarkably high concentrations of bursal proteases in virgin females (34, 38) and implies that females may already have an active role in spermatophore digestion even before breach of the indigestible envelope by the signum.
The Indigestible Portion of the Envelope Is Composed of Proline-Rich Proteins, a Hallmark of Extracellular Structures.
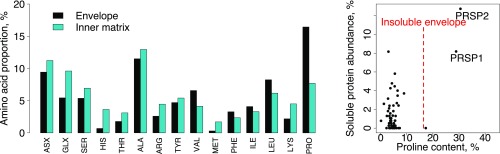
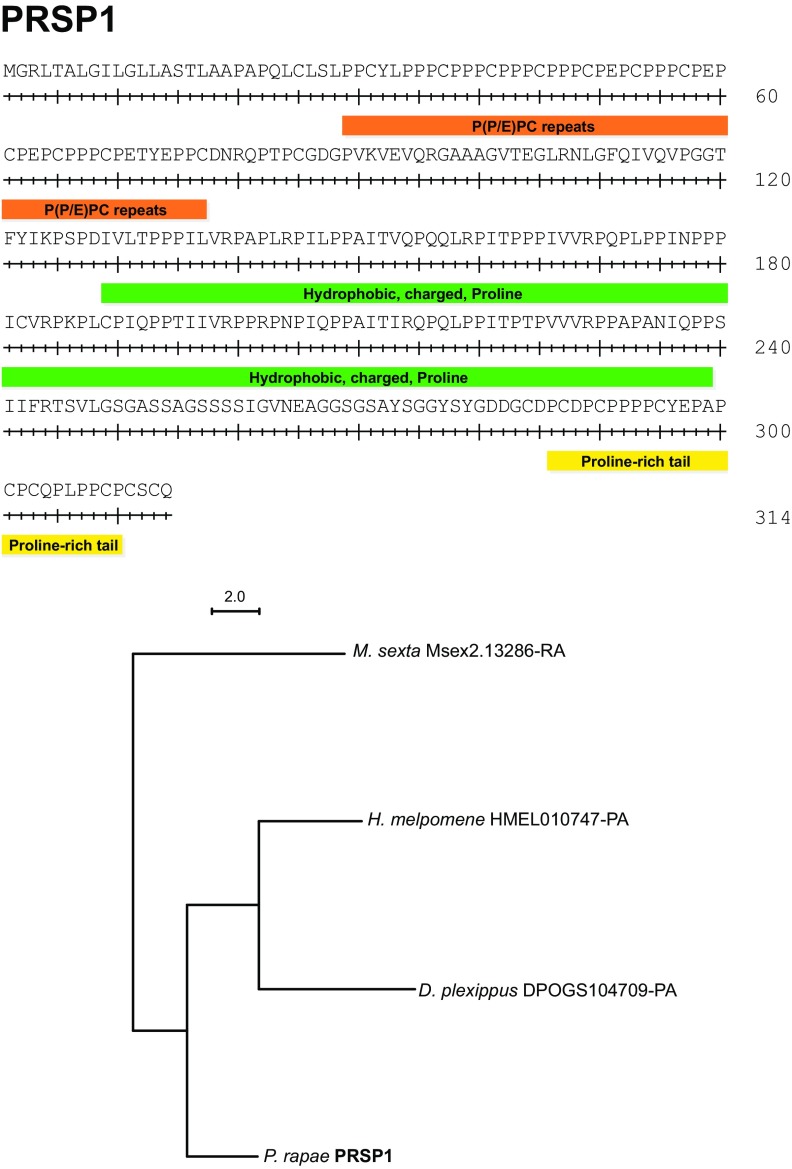
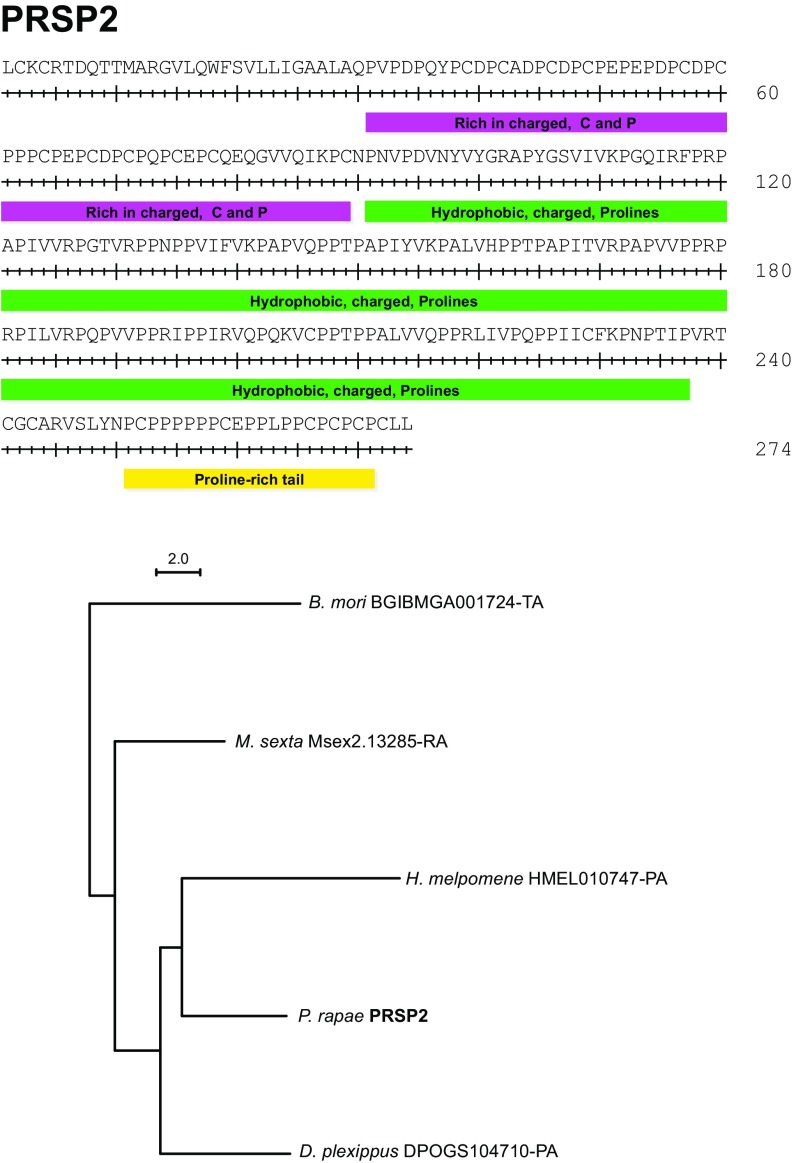
In P. rapae and other butterflies (46), spermatophore envelopes are never fully digested and remain in the bursa copulatrix for the female’s lifespan. This persistence implies that spermatophore envelopes are made of substances that the female is unable to digest. We found that spermatophore envelopes were also insoluble in vitro when incubated with detergents (SDS), proteases (proteinase K and trypsin), and reducing agents (DTT). Only strong oxidizing agents were able to completely dissolve envelopes (96% sulfuric acid or 8.25% sodium hypochlorite). These results suggest that covalent bonds, other than disulfide bonds, are involved in the polymerization of the envelope. To determine the probable composition of the envelope, we performed an amino acid composition analysis on both the envelope and the inner matrix. We determined that the proportion of protein content by dry weight in the insoluble envelope was 73.3%, whereas the proportion of total protein in the inner matrix was 48.8%. As such, the envelope is mainly composed of protein and not composed of alternative structural molecules, such as chitin, which has been previously hypothesized (47, 48). We then compared the amino acid composition of the envelope and inner matrix. Although most amino acids were present at roughly similar levels, there was a twofold higher proportion of proline in the envelope compared with the inner matrix (Fig. 3). We identified two proteins that could explain this high proportion of proline residues, which we named Proline-Rich Seminal Protein 1 (PRSP1) and PRSP2 (GenBank accession nos. KU695466 and KU695467). These two proteins have the highest proline content of all of the proteins that we identified by MS (Fig. 3 and Table S4). Moreover, they are highly abundant in the soluble portion of the envelope, accounting for almost 21% of soluble protein. In contrast, PRSP1 and PRSP2 are almost entirely absent from the spermatophore inner matrix. Although the PRSPs thus likely form the majority of the insoluble envelope structure, their amino acid profiles do not perfectly match the amino acid composition of the envelope. The envelope is then probably composed of a mixture of several proteins, including PRSPs. A search against the PFAM database failed to identify recognized domains in either PRSP. Furthermore, they seem to be specific to Lepidoptera, because we identified homologs in the postman butterfly (H. melpomene), the monarch butterfly (Danaus plexippus), the tobacco hornworm (Manduca sexta), and the silkworm moth (Bombyx mori) but not in more divergent taxonomic groups (e.g., Drosophila melanogaster). However, both PRSPs contain three distinct regions with unusual amino acid compositions and highly repetitive sequences (Figs. S2 and S3). They also possess two similar regions in their N-terminal ends: (i) a region rich in repeats of hydrophobic amino acids, charged amino acids, and proline and (ii) a C-terminal region rich in proline. Such repetitive and proline-rich regions have also been reported in a protein purified from the spermatophore of the yellow mealworm beetle Tenebrio molitor, aptly named spermatophorin (49), and other structural proteins, such as spider silk, the mammalian copulatory plug, collagens, and plant cell walls (50–53).
Fig. 3.
The spermatophore envelope is enriched in proline-rich proteins compared with the inner matrix. Relative proportions of each amino acid in proteins from the spermatophore inner matrix and the envelope were determined by amino acid composition analysis after digestion of both structures in hot HCl. Only two proteins present in the spermatophore (PRSP1 and PRSP2) have a proline content high enough to explain the overall proline content of the insoluble envelope (dashed red line). Each black dot represents 1 of 63 proteins identified in the spermatophore.
Table S4.
Amino acid composition of 63 proteins identified in the P. rapae spermatophore
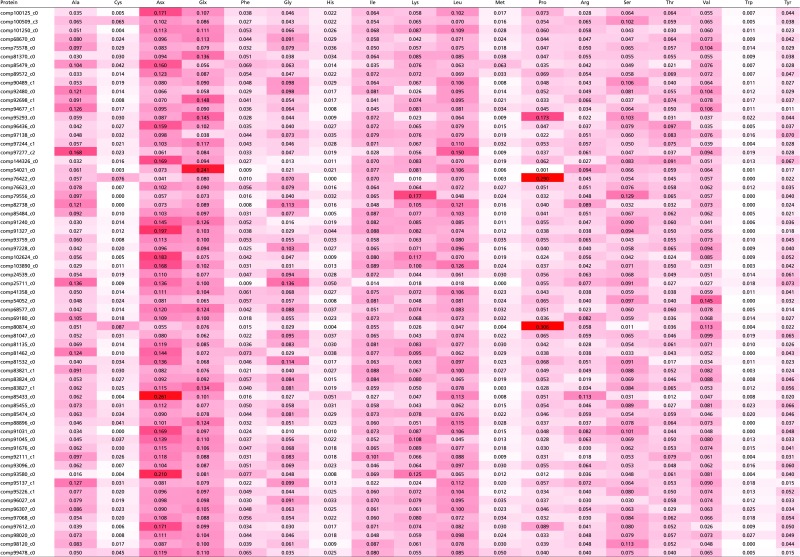 |
Red shading reflects the abundance of specific amino acids in each protein.
Fig. S2.
PRSP1 domains and phylogenetic analysis.
Fig. S3.
PRSP2 domains and phylogenetic analysis.
Male- and Female-Contributed Spermatophore Proteins Exhibit Rapid Evolutionary Divergence.
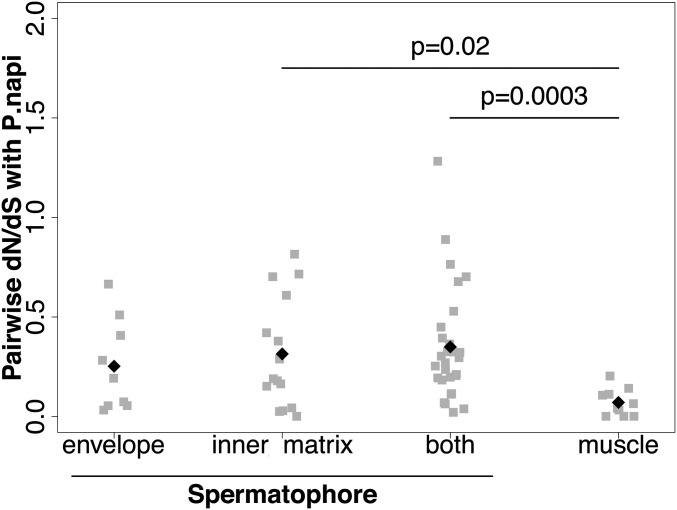
Because the spermatophore envelope and inner matrix are different in composition and thus, likely serve different functions, we compared their evolutionary rates to ask if selection has acted differently on them. We used sequences from P. rapae and a congener, Pieris napi, to estimate rates of evolution through their global dN/dS ratio. Although orthologs were clearly identified in more distant species of Lepidoptera, their divergence at synonymous sites was so high as to be saturated and could not be used. However, using pairwise estimates between P. rapae and P. napi, we determined that, as a class, genes encoding spermatophore proteins exhibit elevated dN/dS ratios, indicating rapid evolutionary rates compared with muscle genes expressed in the bursa copulatrix (Fig. 4). The latter were used for comparison, because these muscle genes, which are coexpressed with other muscle tissues throughout the body [e.g., flight muscle (34)], represent a gene set that is relatively conserved across species. This finding is largely consistent with reports of elevated evolutionary rates for ejaculate proteins in diverse taxonomic groups (42, 54–57). Among the male-specific genes, eight of them have discernable orthologs only in the close relative P. napi, whereas three others have no orthologs at all, suggesting either their rapid evolution or recent gene formation. Interestingly, the majority of the male-specific genes without an identified ortholog or with an ortholog just in P. napi (nine genes; 72.7%) are part of the spermatophore inner matrix (Table S5), suggesting that this region of the spermatophore may be a hotspot for the evolution of SFPs.
Fig. 4.
Genes encoding proteins from the spermatophore evolve faster than other genes. Pairwise dN/dS ratios between P. rapae and P. napi orthologs were calculated for genes found exclusively in the envelope or inner matrix, in both the envelope and the inner matrix, and for muscle genes. Muscle proteins are more conserved than spermatophore proteins and hence, exhibit lower dN/dS ratios.
Table S5.
Phylogenetic analysis of 63 proteins identified in the P. rapae spermatophore
| Gene identification | Origin | Duplications in P. rapae, including gene of interest | Lepidoptera specific? | Notes | Location | dN/dS |
| comp81047_c0 | Male | 2 | No | Shell + inner matrix | 0.1977 | |
| comp97138_c0 | Male | 1 | No | Inner matrix | 0.0435 | |
| comp81370_c0 | Male | 1 | Yes | Inner matrix | 0.4207 | |
| comp68670_c0 | Male | 1 | No | Inner matrix | 0.001 | |
| comp54052_c0 | Male | 1 | Yes | Shell + inner matrix | N/A | |
| comp91240_c0 | Male | Only one sequence | Shell | 0.5103 | ||
| comp81532_c0 | Male | Only one sequence | Shell + inner matrix | N/A | ||
| comp95293_c0 | Male | Only one sequence | Inner matrix | 0.609 | ||
| comp24539_c0 | Male | 1 | Yes | Shell + inner matrix | 0.0631 | |
| comp91031_c0 | Male | Only one sequence | Shell + inner matrix | 0.2111 | ||
| comp95137_c1 | Male | 3 | No | Shell + inner matrix | 0.2533 | |
| comp85479_c0 | Male | 15 | No | Inner matrix | 0.8153 | |
| comp89572_c0 | Male | 2 | Yes | Inner matrix | 0.0289 | |
| comp97612_c0 | Male | 2 | Yes | Shell + inner matrix | 0.6777 | |
| comp80874_c0 | Male | 1 | Yes | PRSP2 | Shell + inner matrix | 0.3031 |
| comp103890_c0 | Male | 1 | Yes | Shell + inner matrix | 0.3253 | |
| comp102624_c0 | Male | Only one sequence | Shell + inner matrix | N/A | ||
| comp85474_c0 | Male | 1 | No | Shell + inner matrix | 0.1119 | |
| comp100509_c3 | Male | 1 | Yes | Inner matrix | 0.7155 | |
| comp83821_c1 | Male | 2 | Yes | Shell + inner matrix | 0.5287 | |
| comp76422_c0 | Male | 1 | Yes | PRSP1 | Shell | 0.2825 |
| comp85433_c0 | Male | Only one sequence | Shell + inner matrix | N/A | ||
| comp41358_c0 | Male | 6 | No | Shell + inner matrix | 0.3641 | |
| comp91045_c0 | Male | 5 | Yes | Shell + inner matrix | N/A | |
| comp96436_c0 | Male | Only one sequence | Inner matrix | 0.7024 | ||
| comp93096_c0 | Male | 10 | No | Shell + inner matrix | 0.0691 | |
| comp25711_c0 | Male | Only one sequence | Shell + inner matrix | 1.2823 | ||
| comp144326_c0 | Male | Only one sequence | Shell | 0.4074 | ||
| comp100125_c0 | Male | Only one sequence | Inner matrix | 0.3786 | ||
| comp91327_c0 | Male | Only one sequence | Shell | 0.6649 | ||
| comp81135_c0 | Male | 2 | No | Shell + inner matrix | 0.236 | |
| comp98020_c0 | Female | 2 | No | Shell + inner matrix | 0.2053 | |
| comp97068_c0 | Female | 3 | No | Shell + inner matrix | 0.4491 | |
| comp83824_c0 | Female | 4 | No | Shell + inner matrix | 0.2953 | |
| comp91676_c0 | Female | 3 | No | Same phylogenetic tree | Shell + inner matrix | 0.3941 |
| comp83827_c1 | Female | 3 | No | Shell + inner matrix | 0.7021 | |
| comp85455_c0 | Female | 2 | No | Shell + inner matrix | N/A | |
| comp96307_c0 | Both sexes | 1 | No | Shell + inner matrix | 0.1151 | |
| comp99478_c0 | Both sexes | 2 | Yes | Shell + inner matrix | N/A | |
| comp95226_c1 | Both sexes | 1 | No | Shell + inner matrix | 0.1938 | |
| comp93580_c0 | Both sexes | Only one sequence | Shell + inner matrix | 0.2695 | ||
| comp81462_c0 | Both sexes | 5 | Yes | Shell + inner matrix | 0.7643 | |
| comp88896_c0 | Both sexes | 2 | No | Shell + inner matrix | 0.3223 | |
| comp96027_c4 | Both sexes | 1 | No | Shell + inner matrix | 0.0209 | |
| comp92111_c1 | Both sexes | 1 | No | Shell + inner matrix | 0.3242 | |
| comp69180_c0 | Both sexes | 1 | No | Shell + inner matrix | 0.0385 | |
| comp68577_c0 | Both sexes | 1 | No | Shell + inner matrix | 0.1831 | |
| comp98120_c0 | Both sexes | 1 | No | Shell + inner matrix | N/A | |
| comp90489_c1 | Both sexes | 4 | No | Inner matrix | 0.1795 | |
| comp92480_c0 | Both sexes | 1 | No | Inner matrix | N/A | |
| comp75578_c0 | Both sexes | 1 | No | Inner matrix | 0.1897 | |
| comp97244_c1 | Both sexes | 1 | No | Inner matrix | N/A | |
| comp101250_c0 | Both sexes | 1 | No | Inner matrix | 0.025 | |
| comp97277_c2 | Both sexes | 1 | No | Inner matrix | 0.1638 | |
| comp94677_c1 | Both sexes | 1 | No | Inner matrix | 0.2887 | |
| comp92698_c1 | Both sexes | 2 | No | Inner matrix | 0.1521 | |
| comp97228_c0 | Both sexes | 1 | No | Shell | 0.0542 | |
| comp76623_c0 | Both sexes | 1 | No | Shell | 0.0548 | |
| comp79556_c0 | Both sexes | 2 | No | Shell | N/A | |
| comp82738_c0 | Both sexes | 3 | No | Shell | N/A | |
| comp85484_c0 | Both sexes | 1 | No | Shell | 0.1923 | |
| comp54021_c0 | Both sexes | 1 | No | Shell | 0.0322 | |
| comp93759_c0 | Both sexes | 1 | No | Shell | 0.0737 |
N/A, not applicable.
In addition to sequence-level substitutions, evolutionary diversity can also arise from gene duplication. Accordingly, we estimated rates of gene duplication for spermatophore constituents through reconstructed phylogenetic trees within the Lepidoptera. We found that female-expressed genes with products that are found in the spermatophore are duplicated at a significantly higher rate than a set of genes randomly chosen from the P. rapae transcriptome (Table 1 and Table S5). In fact, all six female proteases found in the spermatophore exhibited evidence of butterfly-specific gene duplication. Three of the female proteases are found in the same phylogenetic tree and originated from recent duplication events, either specific to butterflies or even potentially specific to the Pieris genus (Fig. S4 and Table S6). In addition to their recent origin, these proteases appear to be diverging rapidly, exhibiting elevated dN/dS ratios (comp83827_c1 dN/dS = 0.70, comp91676_c0 dN/dS = 0.39) compared with genes encoding muscle proteins (dN/dS ∼ 0.07 ± 0.07) (Table S6). In contrast, spermatophore genes expressed in males or both sexes were not duplicated more often than random controls.
Table 1.
Proportion of duplicated genes encoding spermatophore and random control genes
| Site of expression | Proportion of duplicate genes |
| Spermatophore | |
| Male | 0.23 (n = 31) |
| Female | 1.00 (n = 6)* |
| Both sexes | 0.27 (n = 26) |
| Random controls | 0.14 (n = 22) |
Fisher’s exact test: female vs. male (P value = 0.0007), female vs. both sexes (P value = 0.002), and female vs. random controls (P value = 0.0002).
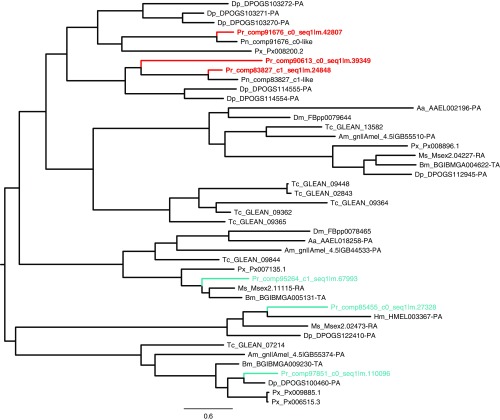
Fig. S4.
Phylogenetic tree of the papain proteases from the spermatophore. Proteases identified in the Cabbage White butterfly (Pieris rapae) spermatophore are indicated in red, whereas other proteases from the papain family are indicated in blue.
Table S6.
Protein identity between P. rapae papain proteases and their orthologs in Pieris napi
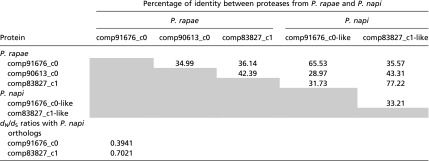 |
Grey shading indicates no value is reported since the identity matrix is symmetrical.
Discussion
Many animal groups produce ejaculates that form distinct structural regions within or outside the female reproductive tract (e.g., copulatory plugs and spermatophores), and these structures are thought to influence reproductive outcomes through their unique functional properties (10). We used the spermatophore of the butterfly P. rapae to characterize how a structurally complex ejaculate is formed, how it provides key reproductive functions, and how its protein components evolve between species. We first provided evidence that the spermatophore envelope and inner matrix are composed of distinct protein profiles (Fig. 1). This result shows that, in addition to being structurally complex, the lepidopteran spermatophore is also biochemically partitioned. This finding represents an advance in our understanding of ejaculate function, because it provides an important example of how biochemical heterogeneity can produce structural complexity. In terms of evolutionary dynamics, such heterogeneous ejaculates would allow the proteins from distinct regions to evolve more independently in response to different selective pressures, because they may occupy distinct “biochemical niches” during reproduction.
Additional analysis showed that the constituent proteins of each spermatophore region are stored in distinct subsections of the male reproductive tract. This finding supports a model of spermatophore assembly, in which the envelope material is transferred first followed by the inner matrix and then sperm (Fig. 2). It also raises the question of whether ejaculate proteins are expressed specifically in those sections or alternatively, if males slowly modify the transcriptional profile of their accessory glands to produce the divergent protein blends observed within the reproductive tract. This distinction has important implications for how males might strategically tailor their ejaculates to affect spermatophore composition and structure, perhaps in response to social experience or environment (1). For example, males may be unable to readily alter their relative investment in spermatophore envelopes if these proteins are already produced and stored in the male simplex well in advance of mating.
The focus of our empirical approach on structural complexity also drove more careful examination of particular morphological interfaces. Most notably, we were motivated to better understand the spermatophore envelope, which represents an important reproductive interface between male- and female-expressed proteins. We revealed that the indigestible portion of the envelope is a proteinaceous structure largely composed of two proline-rich proteins, PRSP1 and PRSP2. We also found that the envelope is insoluble, even under biologically extreme proteolytic conditions, but was dissolved by harsh oxidizers, which suggests that its structural toughness is the result of covalent bonding between the repetitive PRSP domains, similar to the structural underpinnings of plant cell walls (50).
Our examination of the physical origins of spermatophore constituents also led us to a surprising finding: females are far more involved in spermatophore formation than has been previously appreciated. We found large amounts of female-produced proteases throughout the spermatophore, making up 25% and 11% of soluble protein in the envelope and inner matrix, respectively. These proteases are expressed at extraordinary levels in virgin females (38), suggesting that females may concentrate them in the bursa and/or reproductive tract before mating in an attempt to incorporate them into the spermatophore either during formation or shortly thereafter. These proteases may then increase the rate of digestion of the spermatophore material, particularly the inner matrix, and may also assist the signum in breaching the outer envelope. This finding alters the decades-old interpretation of spermatophores as strictly male contributions. In fact, previous studies have often quantified spermatophore mass as a measure of male reproductive investment (30, 58). However, our work suggests that some nontrivial proportion of spermatophore mass is actually contributed by the female and may, therefore, have been misattributed as male investment in past work.
It is still unclear which selective pressures led to the evolution of the spermatophore structure. In such a male–female interface, several hypotheses are possible and are not all mutually exclusive. A first hypothesis is that the envelope could provide a simple mechanical function to keep ejaculate contents from leaving the female tract; however, females receive the spermatophore in a closed-end pouch—the bursa copulatrix—and leakage is not observed during the hour before the envelope hardens. A second hypothesis emphasizes the spermatophore as a cooperative gift to the female. In this scenario, the spermatophore envelope together with the female digestive traits (signum and the proteases) lead to optimal timing of release of the nutrients to be used by the female for her and her progeny’s benefit. A third hypothesis instead focuses on the opportunity for sexual conflict, in which the envelope’s resistance to digestion enables males to delay female remating and thereby, sire a larger proportion of her offspring. Under such sexual antagonism, spermatophore traits leading to slower digestion would be favored in social systems in which females have greater opportunity to mate with multiple males (i.e., polyandry) (47). Thus, sexual conflict over female remating rate could have played an important role in the origin and subsequent evolution of these unusual proline-rich envelope proteins. Consistent with this latter hypothesis, prior research in Heliconius butterflies has shown that polyandrous butterflies have thicker spermatophore envelopes than monandrous species (59). However, sexual conflict would also imply that females evolve to counter these delays. Again, prior research indicates that the signum, a mechanical device for breaching the spermatophore envelope, tends to be maintained in polyandrous Lepidoptera species and lost in monandrous lineages (60). The extreme proteolytic contribution of females also raises the possibility that these proteins are locked in a coevolutionary arms race with male-derived spermatophore proteins, driven by sexual conflict over female remating rate. Whether the evolutionary rates of these male and female proteins are driven by sexual conflict vs. cooperative coevolution is an exciting avenue for future research.
Finally, our focus on the biochemistry of spermatophore structure may offer inroads into the debate about spermatophores as paternal investment vs. mating effort (61–63). This debate hinges on the relative timing of fertilization vs. uptake of male resources by eggs. In species where a male fertilizes the same eggs that he provisions with ejaculate resources, the spermatophore can be considered paternal investment. However, males may often provision eggs that are destined to be fertilized by a subsequent male. Such “stepfather” situations (62) have led researchers to question whether spermatophores are indeed parental effort or should rather be considered simply mating effort. Here, we report that the outer envelope is enriched with indigestible PRSPs, which delay female access to the soluble inner matrix. This structural arrangement extends the time that a male can fertilize a female’s eggs, but it also delays her uptake of the potentially nutritive inner matrix. Thus, the structural strategy adopted by P. rapae males seems to separate the initiation of fertilization from the timing of resource uptake from the spermatophore. As such, spermatophores in this species may more frequently function as mating effort, particularly in populations where females mate readily again after spermatophore digestion. However, in populations with lower remating rates, spermatophore resources may also contribute to paternal investment. However, this scenario need not be more generally the case. Spermatophores with structural phenotypes that make egg-provisioning resources more immediately available could favor parental investment. Our approach would enable evidence for such strategies to be more readily identified.
In summary, we urge more careful consideration of the structural and biochemical complexity of male ejaculates. Such work is likely to uncover insights into reproductive interactions between the sexes and highlight previously unconsidered axes of reproductive adaptation (64, 65). How males and females collaborate and quarrel over ejaculate processing in both space and time may lead us to better understand the control of reproductive tissues and by extension, the origin of reproductive dysfunctions that play critical roles in infertility, individual fitness, and reproductive isolation during speciation.
Methods
Animal Stocks.
P. rapae individuals were sampled from a continuous laboratory colony established in 2012 from individuals collected in Rochester, Pennsylvania (40°44′45″ N, 80°9′45″ W) and supplemented periodically by individuals collected from the same site. Individuals were raised in insect growth chambers under controlled climatic conditions (24 °C, 60% relative humidity, 16-h light:8-h dark photoperiod). Larvae were reared on kale grown in greenhouses at the University of Pittsburgh.
Tissue Dissection.
Live animals had their head, thorax, and abdomen separated and immediately spread open in butterfly Ringers (66) to expose tissues of interest. To obtain a tissue-specific view of protein expression, we dissected several tissues from males and females: female bursa copulatrix (virgin), testes, accessory glands, vas deferens, and mating duct (Fig. 2). Beginning at the male genital opening, the mating duct simplex is the first gland/tissue followed by the mating duct duplex. In a typical P. rapae male, the whole mating duct is ∼10 cm. Moving distally, the accessory glands are next after the bifurcation of the vas deferens leading to the testes. Accessory glands were subdivided into proximal and distal segments based on appearance. When dissected into butterfly Ringers solution, the contents of the proximal segment became cloudy, whereas the distal segment remained transparent. The same phenomenon happened when dissecting out the mating duct, with the duplex becoming cloudy, whereas the simplex remained transparent. We, therefore, used both visual appearance and morphology to divide the mating duct into duplex (distal from the genital opening) and simplex (proximal from the genital opening). Spermatophores retrieved from females 2 h after mating—the time at which they are solidified and adopt their typical shape in the female reproductive tract—were carefully rinsed in PBS one time as a whole after their removal from the bursa copulatrix and dissected into two different parts: the envelope and the inner matrix. Tissues were stored at −20 °C in Laemmli buffer [4% SDS, 120 mM Tris⋅HCl, 0.02% (wt/vol) bromophenol blue] until use.
Hemolymph Collection.
Individuals were decapitated and placed into a 500-μL Eppendorf tube with a perforated bottom tip, which was inserted into a 1.5-mL Eppendorf tube. The Eppendorf tube was centrifuged at 1,000 × g for 10 min at 4 °C to obtain 3–5 μL clear hemolymph per individual. The hemolymph was stored in Laemmli buffer at −20 °C until use.
Protein Identification by MS.
Tissues were stored in butterfly Ringers before being homogenized with a pestle in Laemmli sample buffer. Protein was quantified using a Qubit fluorometer (Life Technologies), and equal amounts of protein for each tissue were run for a short time on SDS/PAGE until they migrated a few millimeters into a 12% resolving gel. Gel bands containing all proteins were excised from the gel and processed by the Biomedical Mass Spectrometry Center of the University of Pittsburgh.
In-Gel Trypsin Digestion.
In-gel trypsin digestion was carried out as previously described (67). Excised gel bands were washed with HPLC water and destained with 50% acetonitrile (ACN)/25 mM ammonium bicarbonate until no visible staining. Gel pieces were dehydrated with 100% ACN and reduced with 10 mM DTT at 56 °C for 1 h followed by alkylation with 55 mM iodoacetamide (IAA) at room temperature for 45 min in the dark. Gel pieces were then again dehydrated with 100% ACN to remove excess DTT and IAA, rehydrated with 20 ng/μL trypsin/25 mM ammonium bicarbonate, and digested overnight at 37 °C. The resultant tryptic peptides were extracted with 70% ACN/5% formic acid, vacuum dried, and reconstituted in 18 μL 0.1% formic acid.
Tandem MS.
Proteolytic peptides from gel-based trypsin digestion were analyzed by a nanoflow reverse-phased liquid chromatography tandem MS. Tryptic peptides were loaded onto a C18 column (PicoChip column packed with 10.5 cm Reprosil C18, 3 μm 120 Å chromatography media with a 75-μm i.d. column and a 15-μm tip; New Objective, Inc.) using a Dionex HPLC system (Dionex Ultimate 3000; ThermoFisher Scientific) operated with a double-split system to provide an in-column nanoflow rate (∼300 nL/min). Mobile phases used were 0.1% formic acid for the initial mobile phase A and 0.1% formic acid in ACN for the subsequent mobile phase B. Peptides were eluted off the column using a 52-min gradient (2–40% B in 42 min, 40–95% B in 1 min, 95% B for 1 min, 2% B for 8 min) and injected into a linear ion trap MS (LTQ-XL; ThermoFisher Scientific) through electrospray.
The LTQ XL was operated in a date-dependent MS/MS mode, in which each full MS spectrum [acquired at 30,000 automatic gain control (AGC) target, 50 ms maximum ion accumulation time, and precursor ion selection range of m/z 300–1,800] was followed by MS/MS scans of the five most abundant molecular ions determined from full MS scan (acquired based on the setting of 1,000 signal threshold, 10,000 AGC target, 100 ms maximum accumulation time, 2.0 Da isolation width, 30 ms activation time, and 35% normalized collision energy). Dynamic exclusion was enabled to minimize redundant selection of peptides previously selected by collision-induced dissociation.
Peptide Identification by Database Search.
MS/MS spectra were searched using the MASCOT search engine (Version 2.4.0; Matrix Science Ltd) against the previously published translated transcriptome of P. rapae (34). The following modifications were used: static modification of cysteine (carboxyamidomethylation, +57.05 Da) and variable modification of methionine (oxidation, +15.99Da). The mass tolerance was set at 1.4 Da for the precursor ions and 0.5 Da for the fragment ions. Peptide identifications were filtered using PeptideProphet and ProteinProphet algorithms with a protein threshold cutoff of 99% and peptide threshold cutoff of 95% implemented in Scaffold (Proteome Software). Proteins were identified with a 99% probability and by a minimum of two peptides. By using those thresholds, 341 proteins were identified across all samples, including 117 proteins that were identified by five peptides. Normalized spectral counts were used as a semiquantitative measure of protein abundance. Sex specificity was assessed by computing normalized expression values proportions for each gene from our previous transcriptomic study (34). The bursa copulatrix and the female reproductive tract were considered as female-specific tissues, whereas the male reproductive tract was considered male-specific. A gene was considered sex-specific when the mean of its expression in sex-specific tissues represented more than 85% of its total expression across all nine tissues sampled in the study.
Hemolymph Subtraction from MS Data and Merger of Proteins Isoforms.
Because of hemolymph infiltration during the dissection of tissues of interest, all proteins identified by MS in the hemolymph were removed. If multiple proteins encoded by the same gene were identified, the maximum value for each tissue type was kept to create only one entry per protein/gene. By subtracting hemolymph proteins and duplicates, 144 proteins remained in the dataset across all tissues, 63 being present in the spermatophore (Dataset S1). Data from testes and vas deferens showed a negligible contribution to the spermatophore and were not further considered.
Quantification of Tissue Contributions to the Spermatophore.
Contributions to the spermatophore from five male and female tissues—the distal and proximal accessory glands, duplex and simplex mating ducts, and the bursa copulatrix—were quantitatively estimated using a likelihood framework. The protein abundances of a spermatophore part (envelope or inner matrix) were represented as a row vector, s. The protein abundances of each tissue were represented in a matrix T, with rows as tissues and proteins as columns. The goal was to estimate a weight for each tissue to represent its proportional contribution to a finished spermatophore part. Weights were represented in a row vector w, with columns that correspond to the rows of T (tissues). Weights were not permitted to take negative values, because we do not expect tissues to subtract proteins from the spermatophore. The rows of T and s were normalized to have identical sums, so that weights reflect proportional contribution to the spermatophore. The predicted protein composition of a spermatophore part was modeled as the product of the relative weights on each tissue (w) and the protein abundances in the five contributing tissues (T):
The error term (ε) was modeled as the normal distribution with zero mean and SD as a freely estimated parameter. The likelihood function was defined as the probability of the residual values given the estimated error distribution. The five tissue weights and the SD of the error were jointly optimized by numerically maximizing the likelihood function through the bbmle package in R (Ben Bolker; R Development Core Team, McMaster University, Hamilton, Ontario, Canada). The marginal SEs for each tissue weight were used to calculate the probability of that weight being greater than zero. In a complementary approach, we estimated the weights using nonnegative least squares (NNLS) through the nnls package in R (Katharine M. Mullen, University of California, Los Angeles, and Ivo H. M. van Stokkum, Vrije Universiteit Amsterdam). NNLS returned the same weights as the maximum likelihood estimates; however, NNLS does not yield SEs.
Determination of the Amino Acid Composition of the Envelope and the Inner Matrix of the Spermatophore.
A pool of eight spermatophore envelopes was boiled in 4% SDS at 95 °C for 1 h and rinsed five times in deionized water to keep only the insoluble fraction. Pools of envelopes and corresponding inner matrices were then lyophilized and massed before submission to the Protein Chemistry Laboratory of Texas A&M University to determine their amino acid composition. Each sample was transferred to a hydrolysis tube, and 200 µL 6 N HCl and 50 nmol internal standards were added. The same procedure was performed on the assay blank: two 50-nmol standards and a human serum albumin control. All samples, standards, blank, and control were subjected to 22 h of hydrolysis at 100 °C in 200 µL 6 N HCl. At the end of the hydrolysis, 10 µL were taken from all hydrolysates, dried down, resuspended in 100 µL 0.4 M borate buffer for derivatization, and transferred to the Agilent G1367E autosampler for automated derivatization and loading. The Agilent 1260 HPLC analyzes all samples by precolumn derivatization of the amino acids with o-phthalaldehyde (OPA) and 9-fluoromethyl-chloroformate (FMOC). OPA reacts with primary amino acids, and FMOC reacts with secondary amino acids (proline). Both reagents react rapidly and quantitatively and give highly fluorescent and UV-absorbing isoindole derivatives. The derivatized amino acids are separated by reverse-phase HPLC and detected by UV absorbance. In this assay, asparagine and glutamine are deamidated to their respective acids. Results for those residues are then reported as Asx and Glx that combine the amounts for both the amide and the acid. Tryptophan and cysteine are destroyed by the acid hydrolysis and are not quantified. Additionally, the acid used for hydrolysis is typically contaminated with glycine residues, and therefore, the results for this amino acid are not reported. Nanomolar data were converted to micrograms to give an estimate of the protein mass in our samples. However, because not all amino acids were quantified, protein masses are underestimates.
Phylogenetic Analyses.
Homologous genes searches were performed with BLASTP using protein sequences identified by MS as queries against protein databases of the following species: six Lepidoptera species: P. rapae translated ORFs (our data), B. mori (silkworm.genomics.org.cn/; Bombyx_mori.Bmor1.21.pep.all.fa), D. plexippus (monarchbase.umassmed.edu/; Dp_geneset_OGS1_pep.fasta), H. melpomene (www.butterflygenome.org/; heliconius_melpomene_v1.1_primaryScaffs_Protein.faa), M. sexta (agripestbase.org/manduca/; Manduca_sexta_OGS2_20140407_proteins.fa), and Plutella xylostella (gigadb.org/dataset/100078; P.xylostella.pep.fasta); two Diptera: D. melanogaster (flybase.org/; dmel-all-translation-r5.57.fasta) and Aedes aegypti (Liverpool strain; https://www.vectorbase.org/; Aedes-aegypti-Liverpool_PEPTIDES_AaegL3.1.fa); and two additional insects, Apis mellifera (hymenopteragenome.org/beebase/; amel_OGSv3.2_pep.fa) and Tribolium castaneum (ftp://ftp.ncbi.nih.gov/genomes/Tribolium_castaneum/protein/; protein.fasta.gz). BLASTP outputs were then parsed to cluster homologous protein sequences together (E-value threshold of 1e−10). Multiple sequence alignments of clusters were performed using Clustal Omega (68), and phylogenetic trees were reconstructed using PhyML (69). Branch support values were computed using approximate likelihood ratio test (aLRT) statistics (70). Duplicates for our genes of interest were accounted for when P. rapae homologs were found in the same clade containing our focus gene and Lepidoptera sequences only. Control genes for the determination of the proportion of duplicated genes were chosen randomly within the whole transcriptome of P. rapae. Estimates of dN/dS were determined using pairwise alignments of P. rapae and P. napi sequences as an input for the codeml program (71). Control genes for the dN/dS estimates were muscle genes previously identified in the bursa copulatrix (34).
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KU695466.1, KU695467.1, MF319685, and MF319745).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707680114/-/DCSupplemental.
References
- 1.Perry JC, Sirot L, Wigby S. The seminal symphony: How to compose an ejaculate. Trends Ecol Evol. 2013;28:414–422. doi: 10.1016/j.tree.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kubli E, Bopp D. Sexual behavior: How sex peptide flips the postmating switch of female flies. Curr Biol. 2012;22:R520–R522. doi: 10.1016/j.cub.2012.04.058. [DOI] [PubMed] [Google Scholar]
- 3.Chapman T, et al. The sex peptide of Drosophila melanogaster: Female post-mating responses analyzed by using RNA interference. Proc Natl Acad Sci USA. 2003;100:9923–9928. doi: 10.1073/pnas.1631635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avila FW, Ravi Ram K, Bloch Qazi MC, Wolfner MF. Sex peptide is required for the efficient release of stored sperm in mated Drosophila females. Genetics. 2010;186:595–600. doi: 10.1534/genetics.110.119735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gioti A, et al. Sex peptide of Drosophila melanogaster males is a global regulator of reproductive processes in females. Proc Biol Sci. 2012;279:4423–4432. doi: 10.1098/rspb.2012.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams GP, Ratto MH, Huanca W, Singh J. Ovulation-inducing factor in the seminal plasma of alpacas and llamas. Biol Reprod. 2005;73:452–457. doi: 10.1095/biolreprod.105.040097. [DOI] [PubMed] [Google Scholar]
- 7.Dixson AL, Anderson MJ. Sexual selection, seminal coagulation and copulatory plug formation in primates. Folia Primatol (Basel) 2002;73:63–69. doi: 10.1159/000064784. [DOI] [PubMed] [Google Scholar]
- 8.Sutter A, Simmons LW, Lindholm AK, Firman RC. Function of copulatory plugs in house mice: Mating behavior and paternity outcomes of rival males. Behav Ecol. 2016;27:185–195. [Google Scholar]
- 9.Mangels R, et al. Genetic and phenotypic influences on copulatory plug survival in mice. Heredity (Edinb) 2015;115:496–502. doi: 10.1038/hdy.2015.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mann T. Spermatophores: Development, Structure, Biochemical Attributes and Role in the Transfer of Spermatozoa. Springer; New York: 1984. [Google Scholar]
- 11.South A, Lewis SM. The influence of male ejaculate quantity on female fitness: A meta-analysis. Biol Rev Camb Philos Soc. 2011;86:299–309. doi: 10.1111/j.1469-185X.2010.00145.x. [DOI] [PubMed] [Google Scholar]
- 12.Vahed K. The function of nuptial feeding in insects: A review of empirical studies. Biol Rev Camb Philos Soc. 1998;73:43–78. [Google Scholar]
- 13.Lewis S, South A. The evolution of animal nuptial gifts. Adv Study Behav. 2012;44:53–97. [Google Scholar]
- 14.Sakaluk SK. Male crickets feed females to ensure complete sperm transfer. Science. 1983;35:609–610. doi: 10.1126/science.223.4636.609. [DOI] [PubMed] [Google Scholar]
- 15.Gwynne DT, Bowen BJ, Codd CG. The function of the katydid spermatophore and its role in fecundity and insemination (Orthoptera: Tettigoniidae) Aust J Zool. 1984 doi: 10.1071/ZO9840015. [DOI] [Google Scholar]
- 16.Gwynne DT. The evolution of edible “sperm sacs” and other forms of courtship feeding in crickets, katydids and their kin (Orthoptera: Ensifera) In: Choe JC, Crespi BJ, editors. The Evolution of Mating Systems in Insects and Arachnids. Cambridge Univ Press; Cambridge, UK: 1997. pp. 110–129. [Google Scholar]
- 17.Gwynne DT. Sexual conflict over nuptial gifts in insects. Annu Rev Entomol. 2008;53:83–101. doi: 10.1146/annurev.ento.53.103106.093423. [DOI] [PubMed] [Google Scholar]
- 18.Warwick S, Vahed K, Raubenheimer D, Simpson SJ. Free amino acids as phagostimulants in cricket nuptial gifts: Support for the ‘Candymaker’ hypothesis. Biol Lett. 2009;5:194–196. doi: 10.1098/rsbl.2008.0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gack C, Peschke K. Spermathecal morphology, sperm transfer and a novel mechanism of sperm displacement in the rove beetle, Aleochara curtula (Coleoptera, Staphylinidae) Zoomorphology. 1994;114:227–237. [Google Scholar]
- 20.Gadzama NM, Happ GM. The structure and evacuation of the spermatophore of Tenebrio molitor L. (Coleoptera: Tenebrionidae) Tissue Cell. 1974;6:95–108. doi: 10.1016/0040-8166(74)90025-1. [DOI] [PubMed] [Google Scholar]
- 21.Forster M, Gack C, Peschke K. Morphology and function of the spermatophore in the rove beetle, Aleochara curtula (Coleoptera: Staphylinidae) Zoology. 1998;101:34–44. [Google Scholar]
- 22.Boggs CL. A general model of the role of male-donated nutrients in female insects’ reproduction. Am Nat. 1990;136:598–617. [Google Scholar]
- 23.Boggs CL, Gilbert LE. Male contribution to egg production in butterflies: Evidence for transfer of nutrients at mating. Science. 1979;206:83–84. doi: 10.1126/science.206.4414.83. [DOI] [PubMed] [Google Scholar]
- 24.Boggs CL. Male nuptial gifts: Phenotypic consequences and evolutionary implications. In: Leather SR, Hardie J, editors. Insect Reproduction. CRC; Cleveland: 1995. pp. 215–242. [Google Scholar]
- 25.Drummond BA. Multiple mating and sperm competition in the Lepidoptera. In: Smith RL, editor. Sperm Competition and the Evolution of Animal Mating Systems. Academic; New York: 1984. pp. 291–370. [Google Scholar]
- 26.Gwynne DT. Male mating effort, confidence of paternity, and insect sperm competition. In: Smith RL, editor. Sperm Competition and the Evolution of Animal Mating Systems. Academic; New York: 1984. pp. 117–149. [Google Scholar]
- 27.Ridley M. The incidence of sperm displacement in insects: Four conjectures, one corroboration. Biol J Linn Soc Lond. 1989;38:349–367. [Google Scholar]
- 28.LaMunyon CW, Eisner T. Spermatophore size as determinant of paternity in an arctiid moth (Utetheisa ornatrix) Proc Natl Acad Sci USA. 1994;91:7081–7084. doi: 10.1073/pnas.91.15.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svärd L, McNeil JN. Female benefit, male risk: Polyandry in the true armyworm Pseudaletia unipuncta. Behav Ecol Sociobiol. 1994;35:319–326. [Google Scholar]
- 30.Karlsson B. Nuptial gifts, resource budgets, and reproductive output in a polyandrous butterfly. Ecology. 1998;79:2931–2940. [Google Scholar]
- 31.Watanabe M, Ando S. Influence of mating frequency on lifetime fecundity in wild females of the small white Pieris rapae (Lepidoptera, Pieridae) Jap J Entomol. 1993;61:691–696. [Google Scholar]
- 32.Arnqvist G, Nilsson T. The evolution of polyandry: Multiple mating and female fitness in insects. Anim Behav. 2000;60:145–164. doi: 10.1006/anbe.2000.1446. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe M, Sato K. A spermatophore structured in the bursa copulatrix of the small white Pieris rapae (Lepidoptera, Pieridae) during copulation, and its sugar content. J Res Lepid. 1993;32:26–36. [Google Scholar]
- 34.Meslin C, et al. Digestive organ in the female reproductive tract borrows genes from multiple organ systems to adopt critical functions. Mol Biol Evol. 2015;32:1567–1580. doi: 10.1093/molbev/msv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rutowski RL, Gilchrist GW. Copulation in Colias eury theme (Lepidoptera: Pieridae): Patterns and frequency. J Zool. 1986;209:115–124. [Google Scholar]
- 36.Rogers SH, Wells H. The structure and function of the bursa copulatrix of the monarch butterfly (Danaus plexippus) J Morphol. 1984;180:213–221. doi: 10.1002/jmor.1051800305. [DOI] [PubMed] [Google Scholar]
- 37.Lincango P, Fernández G, Baixeras J. Microstructure and diversity of the bursa copulatrix wall in Tortricidae (Lepidoptera) Arthropod Struct Dev. 2013;42:247–256. doi: 10.1016/j.asd.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 38.Plakke MS, Deutsch AB, Meslin C, Clark NL, Morehouse NI. Dynamic digestive physiology of a female reproductive organ in a polyandrous butterfly. J Exp Biol. 2015;218:1548–1555. doi: 10.1242/jeb.118323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burns JM. Mating frequency in natural population of skippers and butterflies as determined by spermatophore counts. Proc Natl Acad Sci USA. 1968;61:852–859. doi: 10.1073/pnas.61.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ehrlich AH, Ehrlich PR. Reproductive strategies in the butterflies. I. Mating frequency, plugging, and egg number. J Kans Entomol Soc. 1978;51:666–697. [Google Scholar]
- 41.Marshall LD. Protein and lipid composition of Colias philodice and C. eurytheme spermatophores and their changes over time (Pieridae) J Res Lepid. 1985;24:21–30. [Google Scholar]
- 42.Walters JR, Harrison RG. Combined EST and proteomic analysis identifies rapidly evolving seminal fluid proteins in Heliconius butterflies. Mol Biol Evol. 2010;27:2000–2013. doi: 10.1093/molbev/msq092. [DOI] [PubMed] [Google Scholar]
- 43.Avila FW, Sirot LK, LaFlamme BA, Rubinstein CD, Wolfner MF. Insect seminal fluid proteins: Identification and function. Annu Rev Entomol. 2011;56:21–40. doi: 10.1146/annurev-ento-120709-144823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laflamme BA, Wolfner MF. Identification and function of proteolysis regulators in seminal fluid. Mol Reprod Dev. 2013;80:80–101. doi: 10.1002/mrd.22130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelleher ES, Clark NL, Markow TA. Diversity-enhancing selection acts on a female reproductive protease family in four subspecies of Drosophila mojavensis. Genetics. 2011;187:865–876. doi: 10.1534/genetics.110.124743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe M. Sperm Competition in Butterflies. Springer; Tokyo: 2016. [Google Scholar]
- 47.Cordero C. The evolutionary origin of signa in female Lepidoptera: Natural and sexual selection hypotheses. J Theor Biol. 2005;232:443–449. doi: 10.1016/j.jtbi.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 48.Callahan PS. Serial morphology as a technique for determination of reproductive patterns in the corn earworm, Heliothis zea (Boddie) Ann Entomol Soc Am. 1958;51:413–428. [Google Scholar]
- 49.Shinbo H, Yaginuma T, Happ GM. Purification and characterization of a proline-rich secretory protein that is a precursor to a structural protein of an insect spermatophore. J Biol Chem. 1987;262:4794–4799. [PubMed] [Google Scholar]
- 50.Kavi Kishor PB, Hima Kumari P, Sunita MSL, Sreenivasulu N. Role of proline in cell wall synthesis and plant development and its implications in plant ontogeny. Front Plant Sci. 2015;6:544. doi: 10.3389/fpls.2015.00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shoulders MD, Raines RT. Collagen structure and stability. Annu Rev Biochem. 2009;78:929–958. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Römer L, Scheibel T. The elaborate structure of spider silk: Structure and function of a natural high performance fiber. Prion. 2008;2:154–161. doi: 10.4161/pri.2.4.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harris SE, et al. Structural characterization of the rat seminal vesicle secretion II protein and gene. J Biol Chem. 1990;265:9896–9903. [PubMed] [Google Scholar]
- 54.Swanson WJ, Clark AG, Waldrip-Dail HM, Wolfner MF, Aquadro CF. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc Natl Acad Sci USA. 2001;98:7375–7379. doi: 10.1073/pnas.131568198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clark NL, Swanson WJ. Pervasive adaptive evolution in primate seminal proteins. PLoS Genet. 2005;1:e35. doi: 10.1371/journal.pgen.0010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andrés JA, Maroja LS, Bogdanowicz SM, Swanson WJ, Harrison RG. Molecular evolution of seminal proteins in field crickets. Mol Biol Evol. 2006;23:1574–1584. doi: 10.1093/molbev/msl020. [DOI] [PubMed] [Google Scholar]
- 57.Dean MD, et al. Proteomics and comparative genomic investigations reveal heterogeneity in evolutionary rate of male reproductive proteins in mice (Mus domesticus) Mol Biol Evol. 2009;26:1733–1743. doi: 10.1093/molbev/msp094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Svärd L, Wiklund C. Mass and production rate of ejaculates in relation to monandry/polyandry in butterflies. Behav Ecol Sociobiol. 1989;24:395–402. [Google Scholar]
- 59.Sánchez V, Cordero C. Sexual coevolution of spermatophore envelopes and female genital traits in butterflies: Evidence of male coercion? PeerJ. 2014;2:e247. doi: 10.7717/peerj.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sánchez V, Hernández-Baños BE, Cordero C. The evolution of a female genital trait widely distributed in the Lepidoptera: Comparative evidence for an effect of sexual coevolution. PLoS One. 2011;6:e22642. doi: 10.1371/journal.pone.0022642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simmons LW, Parker GA. Nuptial feeding in insects: Mating effort versus paternal investment. Ethology. 1989;81:332–343. [Google Scholar]
- 62.Wickler W. Stepfathers in insects and their pseudo-parental investment. Z Tierpsychol. 1985;69:72–78. [Google Scholar]
- 63.Wedell N. Mating effort or paternal investment? Incorporation rate and cost of male donations in the wartbiter. Behav Ecol Sociobiol. 1993;32:239–246. [Google Scholar]
- 64.Lewis SM, et al. Emerging issues in the evolution of animal nuptial gifts. Biol Lett. 2014;10:20140336. doi: 10.1098/rsbl.2014.0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Al-Wathiqui N, Fallon TR, South A, Weng JK, Lewis SM. Molecular characterization of firefly nuptial gifts: A multi-omics approach sheds light on postcopulatory sexual selection. Sci Rep. 2016;6:38556. doi: 10.1038/srep38556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takami T. In vitro culture of embryos in the silkworm, Bombyx Mori L. I. Culture in the silkworm egg extract, with special reference to some characteristics of the diapausing egg. J Exp Biol. 1958;35:286–296. [Google Scholar]
- 67.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 68.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 70.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 71.Bielawski JP, Yang Z. Maximum likelihood methods for detecting adaptive protein evolution. In: Nielsen R, editor. Statistical Methods in Molecular Evolution. Springer; Berlin: 2005. pp. 103–124. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.