Scientists have endeavored to understand sigma receptors for over 40 y. Although most agree that they are important, there is little agreement on anything else. In their behavioral classification of opioid receptors in 1976, Martin et al. (1) proposed three groups of compounds illustrating three distinct opioid receptor classes (mu, kappa, and sigma) based upon morphine, ketocyclazocine, and SKF10,047, respectively, and noted that the opioid antagonist naltrexone antagonized them all. Since then, the sigma receptor story has undergone many twists and turns. Although the SKF10,047 stereoisomer used in the initial description is not stated, subsequent investigators used (+)SKF10,047 to define sigma receptors, identifying sites that clearly were not opioid. Extensive binding studies associated (+)SKF10,047 with many putative receptors, including phencyclidine (2), but these ligands proved quite promiscuous, labeling a multitude of sites. As more ligands became available, sigma receptors were dissociated into two categories: sigma1 and sigma2 (3). Functional studies strongly suggested that both classes were important. Our understanding of sigma1 receptors took a major leap forward with the cloning of the protein in 1996 (4) and its subsequent crystallization in 2016 (5). However, these structural insights have not answered many fundamental questions regarding how these proteins work. In PNAS, Alon et al. (6) present compelling information for the cloning of the sigma2 receptor, completing the molecular characterization of this class of receptor and opening the door to more studies exploring mechanisms of action.
Hundreds of publications have addressed the functions of small molecules with affinity for the sigma2 receptor, which has been implicated in cancer and neurodegenerative diseases (7–9), with a significant focus on the former. Through pharmacological studies, sigma2 has been implicated in tumor biology (10) and has been proposed as a potential drug target in cancer therapy (11, 12), and sigma2 radiotracers have been developed for tumor imaging (6). However, these associations were functionally based, with little molecular foundation. Identifying the gene for the sigma2 receptor brings us much closer to unraveling fundamental questions about the pharmacology of this target. The association of sigma receptors with lipids and steroids goes back many years, and, in 2011, it was proposed that the sigma2 receptor corresponded to a part of the progesterone receptor membrane component 1 (PGRMC1) complex (13). Although initial studies were encouraging, subsequent work questioned this identification (14–16). Most compelling was the fact that overexpression or knockdown of PGRMC1 failed to affect prototypic sigma2 ligand binding.
The paper by Alon et al. (6) now resolves the question. Using classical affinity purification approaches, they isolated the sigma2-binding site and identified it as the endoplasmic reticulum (ER)-resident membrane protein TMEM97, also known as MAC30. TMEM97 shows similar characteristics to the classical sigma2 receptor-binding site, with the appropriate affinity and selectivity for a range of prototypic sigma2 compounds. Mutagenesis studies then established the importance of two aspartate residues in the binding pocket. Although a significant step forward, identifying TMEM97 still leaves many questions. It has been associated with cholesterol homeostasis (17) and has been implicated in Niemann–Pick disease (18). Like sigma2, TMEM97 is highly expressed in a range of cancers and has been associated with poor prognosis and even metastasis (19–23). However, much remains unknown about the functional role(s) of this protein and how the protein actually produces its actions. Major efforts in developing sigma2 therapeutics have focused upon cancer, and, hopefully, these efforts will continue to shed light on the underlying mechanisms of TMEM97/sigma2 actions.
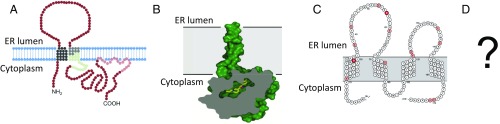
Structurally, the sequence of TMEM97/sigma2 predicts an integral membrane protein with an ER retention signal and four transmembrane domains with the N and C termini extending into the cytoplasm. The two aspartate residues important in binding are predicted to reside near the ER luminal surface of TMEM97/sigma2. Initial predictions of the sigma1 receptor structure from its sequence suggested two transmembrane domains with intracellular tails. However, the crystal structure of the protein was quite different, showing a single transmembrane domain with a short extracellular N terminus and with most of the C terminus extending into the cytoplasm (5). It will be quite interesting to see if the crystal structure of TMEM97 corresponds to its prediction (Fig. 1). This next step is important in our endeavor to understand sigma2 receptor function. Many have questioned the term “receptor” when referring to sigma. There is clearly a binding pocket with established structure–activity relationships. However, there is no endogenous ligand for either sigma receptor and no indication of a transduction system, with neither structure corresponding to any established receptor class. Presumably, as with sigma1, sigma2 ligand binding may lead to conformational changes that influence other, associated protein systems. The sigma1 receptor shows an allosteric-like effect on the functions of proteins as diverse as the androgen receptor (24) and G protein-coupled receptors (25). It also interacts with a wide range of other classes of signaling proteins, receptors, and channels. Will sigma2 receptors also have widespread activity? Are they primarily structural or modulatory? These questions are important, but remain to be answered.
Fig. 1.
Comparison of predicted topology and crystal structure of the sigma1 and sigma2 receptors. (A) Topology model of the sigma1 receptor. Reproduced from Laurini et al. (26). (B) Space-fill model of the sigma1 crystal structure with the bound ligand in yellow. Reproduced from Schmidt et al. (5). (C) Topology model of TMEM97/sigma2 receptor. Reproduced from Alon et al. (6). (D) Question regarding crystal structure of TMEM97/sigma2 receptor.
Sigma receptors have had a long and nebulous history. Initially defined functionally, understanding them was limited by the selectivity of the ligands, which were often “dirty.” However, extensive evidence suggested that they had the potential of yielding novel, important therapeutic agents in a range of disease areas, including cancer. The current cloning of the sigma2 receptor is a major step forward in uncovering the molecular mechanisms responsible for sigma2 ligand activity and brings us closer to understanding the true physiological role of these important proteins.
Acknowledgments
This work was supported, in part, by an American Cancer Society institutional research grant through the Sidney Kimmel Cancer Center at Thomas Jefferson University, a Drexel University Clinical and Translational Research Institute grant, a Drexel University professional enrichment grant, and a Sidney Kimmel Cancer Center Consortium Pilot Study Award to F.J.K.; by grants from the National Institute on Drug Abuse (DA06241 and DA07242), the Mayday Foundation, and The Peter F. McManus Charitable Trust (to G.W.P.); and by a grant from the National Cancer Institute (CA08748) to Memorial Sloan–Kettering Cancer Center.
Footnotes
Conflict of interest statement: G.W.P. declares no conflict of interest. F.J.K. is cofounder of Context Therapeutics.
See companion article on page 7160.
References
- 1.Martin WR, Eades CG, Thompson JA, Huppler RE, Gilbert PE. The effects of morphine- and nalorphine- like drugs in the nondependent and morphine-dependent chronic spinal dog. J Pharmacol Exp Ther. 1976;197:517–532. [PubMed] [Google Scholar]
- 2.Kushner L, Zukin SR, Zukin RS. Characterization of opioid, sigma, and phencyclidine receptors in the neuroblastoma-brain hybrid cell line NCB-20. Mol Pharmacol. 1988;34:689–694. [PubMed] [Google Scholar]
- 3.Hellewell SB, Bowen WD. A sigma-like binding site in rat pheochromocytoma (PC12) cells: Decreased affinity for (+)-benzomorphans and lower molecular weight suggest a different sigma receptor form from that of guinea pig brain. Brain Res. 1990;527:244–253. doi: 10.1016/0006-8993(90)91143-5. [DOI] [PubMed] [Google Scholar]
- 4.Hanner M, et al. Purification, molecular cloning, and expression of the mammalian sigma1-binding site. Proc Natl Acad Sci USA. 1996;93:8072–8077. doi: 10.1073/pnas.93.15.8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt HR, et al. Crystal structure of the human sigma receptor. Nature. 2016;532:527–530. doi: 10.1038/nature17391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alon A, et al. Identification of the gene that codes for the σ2 receptor. Proc Natl Acad Sci USA. 2017;114:7160–7165. doi: 10.1073/pnas.1705154114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng C, McDonald ES, Mach RH. Molecular probes for imaging the sigma-2 receptor: In vitro and in vivo imaging studies. Handb Exp Pharmacol. February 8, 2017 doi: 10.1007/164_2016_96. [DOI] [PubMed] [Google Scholar]
- 8.Guo L, Zhen X. Sigma-2 receptor ligands: Neurobiological effects. Curr Med Chem. 2015;22:989–1003. doi: 10.2174/0929867322666150114163607. [DOI] [PubMed] [Google Scholar]
- 9.Narayanan S, Bhat R, Mesangeau C, Poupaert JH, McCurdy CR. Early development of sigma-receptor ligands. Future Med Chem. 2011;3:79–94. doi: 10.4155/fmc.10.279. [DOI] [PubMed] [Google Scholar]
- 10.Wheeler KT, et al. Sigma-2 receptors as a biomarker of proliferation in solid tumours. Br J Cancer. 2000;82:1223–1232. doi: 10.1054/bjoc.1999.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crawford KW, Bowen WD. Sigma-2 receptor agonists activate a novel apoptotic pathway and potentiate antineoplastic drugs in breast tumor cell lines. Cancer Res. 2002;62:313–322. [PubMed] [Google Scholar]
- 12.Crawford KW, Coop A, Bowen WD. sigma(2) Receptors regulate changes in sphingolipid levels in breast tumor cells. Eur J Pharmacol. 2002;443:207–209. doi: 10.1016/s0014-2999(02)01581-9. [DOI] [PubMed] [Google Scholar]
- 13.Xu J, et al. Identification of the PGRMC1 protein complex as the putative sigma-2 receptor binding site. Nat Commun. 2011;2:380. doi: 10.1038/ncomms1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abate C, Niso M, Infantino V, Menga A, Berardi F. Elements in support of the ‘non-identity’ of the PGRMC1 protein with the σ2 receptor. Eur J Pharmacol. 2015;758:16–23. doi: 10.1016/j.ejphar.2015.03.067. [DOI] [PubMed] [Google Scholar]
- 15.Chu UB, et al. The sigma-2 receptor and progesterone receptor membrane component 1 are different binding sites derived from independent genes. EBioMedicine. 2015;2:1806–1813. doi: 10.1016/j.ebiom.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pati ML, et al. Sigma-2 receptor and progesterone receptor membrane component 1 (PGRMC1) are two different proteins: Proofs by fluorescent labeling and binding of sigma-2 receptor ligands to PGRMC1. Pharmacol Res. 2017;117:67–74. doi: 10.1016/j.phrs.2016.12.023. [DOI] [PubMed] [Google Scholar]
- 17.Bartz F, et al. Identification of cholesterol-regulating genes by targeted RNAi screening. Cell Metab. 2009;10:63–75. doi: 10.1016/j.cmet.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Ebrahimi-Fakhari D, et al. Reduction of TMEM97 increases NPC1 protein levels and restores cholesterol trafficking in Niemann-pick type C1 disease cells. Hum Mol Genet. 2016;25:3588–3599. doi: 10.1093/hmg/ddw204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding H, et al. Prognostic value of MAC30 expression in human pure squamous cell carcinomas of the lung. Asian Pac J Cancer Prev. 2016;17:2705–2710. [PubMed] [Google Scholar]
- 20.Zhao ZR, et al. Significance of mRNA and protein expression of MAC30 in progression of colorectal cancer. Chemotherapy. 2011;57:394–401. doi: 10.1159/000331716. [DOI] [PubMed] [Google Scholar]
- 21.Yan BY, et al. Overexpression of MAC30 in the cytoplasm of oral squamous cell carcinoma predicts nodal metastasis and poor differentiation. Chemotherapy. 2010;56:424–428. doi: 10.1159/000317582. [DOI] [PubMed] [Google Scholar]
- 22.Wilcox CB, et al. Coordinate up-regulation of TMEM97 and cholesterol biosynthesis genes in normal ovarian surface epithelial cells treated with progesterone: Implications for pathogenesis of ovarian cancer. BMC Cancer. 2007;7:223. doi: 10.1186/1471-2407-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moparthi SB, et al. Expression of MAC30 protein is related to survival and biological variables in primary and metastatic colorectal cancers. Int J Oncol. 2007;30:91–95. [PubMed] [Google Scholar]
- 24.Thomas JD, et al. Sigma1 targeting to suppress aberrant androgen receptor signaling in prostate cancer. Cancer Res. 2017;77:2439–2452. doi: 10.1158/0008-5472.CAN-16-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim FJ, et al. Sigma 1 receptor modulation of G-protein-coupled receptor signaling: Potentiation of opioid transduction independent from receptor binding. Mol Pharmacol. 2010;77:695–703. doi: 10.1124/mol.109.057083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laurini E, Marson D, Fermeglia M. Pricl S, 3-D homology model of the sigma1 receptor. Handb Exp Pharmacol. doi: 10.1007/164_2017_35. in press. [DOI] [PubMed] [Google Scholar]