Abstract
SIRT6 is a member of sirtuin family of deacetylases involved in diverse processes including genome stability, metabolic homeostasis and anti-inflammation. However, its function in the adipose tissue is not well understood. To examine the metabolic function of Sirt6 in the adipose tissue, we generated two mouse models that are deficient in Sirt6 using the Cre-lox approach. Two commonly used Cre lines that are driven by either the mouse Fabp4 or Adipoq gene promoter were chosen for this study. The Sirt6-knockout mice generated by the Fabp4-Cre line (Sirt6f/f:Fabp4-Cre) had a significant increase in both body weight and fat mass and exhibited glucose intolerance and insulin resistance as compared with the control wild-type mice. At the molecular levels, the Sirt6f/f:Fabp4-Cre-knockout mice had increased expression of inflammatory genes including F4/80, TNFα, IL-6, and MCP-1 in both white and brown adipose tissues. Moreover, the knockout mice showed decreased expression of the adiponectin gene in the white adipose tissue and UCP1 in the brown adipose tissue, respectively. In contrast, the Sirt6 knockout mice generated by the Adipoq-Cre line (Sirt6f/f:Adipoq-Cre) only had modest insulin resistance. In conclusion, our data suggest that the function of SIRT6 in the Fabp4-Cre-expressing cells in addition to mature adipocytes plays a critical role in body weight maintenance and metabolic homeostasis.
Keywords: Sirt6, Adipocyte, Fabp4-Cre, Adipoq-Cre, Conditional gene knockout
Introduction
Obesity has become a global epidemic, contributing to increased risk for type 2 diabetes, metabolic syndrome, cardiovascular disease and cancer (Bastien, et al. 2014). In obesity, excess expansion of adipose tissue is accompanied by low-grade, chronic inflammation characterized by an infiltration of inflammatory cells and an increase in production of proinflammatory cytokines (Tseng, et al. 2010). Adipose tissue is the main “storage site” of excess energy and plays as a key regulator of energy metabolism by secreting a diverse range of adipokines, such as leptin and adiponectin (Galic, et al. 2010). Therefore, adipose tissue is considered as highly important for systemic metabolic homeostasis.
The sirtuin deacetylase/deacylase family has seven members in mammals (Sirt1-7) (Dong 2012). SIRT6 is a chromatin-associated deacetylase that specifically deacetylates histone H3 at lysine 9 (H3K9), lysine 18 (H3K18) and lysine 56 (H3K56) residues (Michishita, et al. 2008; Michishita, et al. 2009; Tasselli, et al. 2016; Yang, et al. 2009). SIRT6 modulates multiple critical cellular processes, such as DNA damage repair, tumor suppression, inflammation and metabolism (Kugel and Mostoslavsky 2014). In recent years, SIRT6 has been implicated in a variety of metabolic processes including glycolysis (Zhong, et al. 2010), gluconeogenesis (Dominy, et al. 2012; Xiong, et al. 2013), hepatic lipid (Kim, et al. 2010) and cholesterol metabolism (Elhanati, et al. 2013; Tao, et al. 2013a, b), neuroendocrine regulation (Schwer, et al. 2010), circadian regulation of metabolism (Masri, et al. 2014), as well as pancreatic β-cell function (Xiong, et al. 2016a; Xiong, et al. 2016b). However, the role of SIRT6 in adipose tissue is not well understood.
Conditional gene knockout with the use of cell-type specific Cre lines is a powerful technique to ablate a specific gene in a target cell type, such as the adipocyte (Wang 2009). As a matter of fact, analysis of adipose function in vivo has been benefited from the development of several Cre mouse lines that are driven by different gene promoters (Lee, et al. 2013). For example, Fabp4-Cre mouse line developed by He et al. (He, et al. 2003) has been widely used for the intended adipose gene function studies; however, the Cre activity has been also detected in other cell types and tissues, such as macrophages, endothelial, intestine, muscle, pancreatic islets, and even central nervous system (Heffner, et al. 2012; Lee et al. 2013; Martens, et al. 2010). Another adipocyte-specific Cre line has been generated using the mouse adiponectin (Adipoq) gene promoter/enhancer (Eguchi, et al. 2011). The recombination from Adipoq-Cre mice has been shown very specific for mature adipocytes. (Eguchi et al. 2011). In order to assess the SIRT6 function in adipose tissue, we crossed Sirt6-floxed mice with either Fabp4-Cre or Adipoq-Cre to generate two different mouse models, Sirt6f/f:Fabp4-Cre (Sirt6-FKO) and Sirt6f/f:Adipoq-Cre (Sirt6-AKO), respectively.
Materials and Methods
Animal Studies
Sirt6 floxed mice (NIH black Swiss:129/Sv:FVB), Fabp4-Cre and Adipoq-Cre mice (C57BL/6J) were purchased from the Jackson Laboratory (Bar Harbor, ME). Sirt6-FKO and Sirt6-AKO mice were generated by crossing floxed mice with Fabp4-Cre or Adipoq-Cre mice, respectively. Genotyping was carried out as previously described (Xiong et al. 2016b). Blood glucose levels were measured using a glucose meter (Contour from Bayer). High fat diet (60% calories from fat) was purchased from Harlan Laboratories (Madison, WI). Glucose tolerance test (GTT) and insulin tolerance tests (ITT) were performed in mice fasted for 16h or 4h before injection of glucose (i.p. 2g/kg for chow diet mice and 1.5g/kg for HFD mice) or insulin (i.p. 0.5U/kg for chow diet mice and 0.75U/kg for HFD mice), respectively. For insulin stimulated AKT phosphorylation analysis in WAT, mice were starved overnight and injected intravenously with recombinant human insulin (1U/kg, Eli Lilly, Indianapolis, IN). WAT was rapidly excised after 3 min of stimulation and homogenized for preparation of tissue lysates. All animal procedures were performed in accordance with the Guide for Care and Use of Laboratory Animals of the National Institutes of Health and were approved by the Institutional Animal Use and Care Committee of Indiana University School of Medicine.
Real-time RT PCR
Total RNAs were isolated from tissues using Trizol reagent (Invitrogen) by following the manufacturer’s instructions and converted into cDNA using a cDNA synthesis kit (Applied Biosystems). Real-time PCR analysis was performed using SYBR Green Master Mix (Promega) in Eppendorf Realplex PCR system.
Western blot analysis
Protein extracts from tissues were made in tissue lysis buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM EDTA, 10 mM sodium pyrophosphate, 100 mM sodium fluoride, and freshly added 1 mM PMSF and an additional protease cocktail tablet from Roche at one tablet/10ml final buffer volume). Protein extracts were resolved on an SDS-PAGE gel and transferred to nitrocellulose membrane (Santa Cruz Biotechnology). The membrane was incubated with the following antibodies: SIRT6 (Sigma-Aldrich), Actinin (Santa Cruz Biotechnology), AKT, p-AKT(S473), and AdipoQ (Cell Signaling Technology). Detection of proteins was carried out by incubations with the HRP-conjugated secondary antibodies, followed by the ECL detection reagents (Thermo Fisher Scientific).
Histological analysis
Mouse adipose tissues were dissected and immediately fixed in 4% paraformaldehyde. Tissues were then routinely processed for paraffin embedding, and 5μm sections were cut and mounted on glass slides. Sections were stained with hematoxylin-eosin (H&E) according to the standard protocol.
Statistical Analysis
Results are expressed as means ± standard error of the mean (SEM). Group comparisons were performed using analysis of variance (ANOVA) and Student’s t-test. A probability value of p<0.05 was considered statistically significant.
Results
Generation of Fabp4-Cre-mediated Sirt6 conditional knockout mice
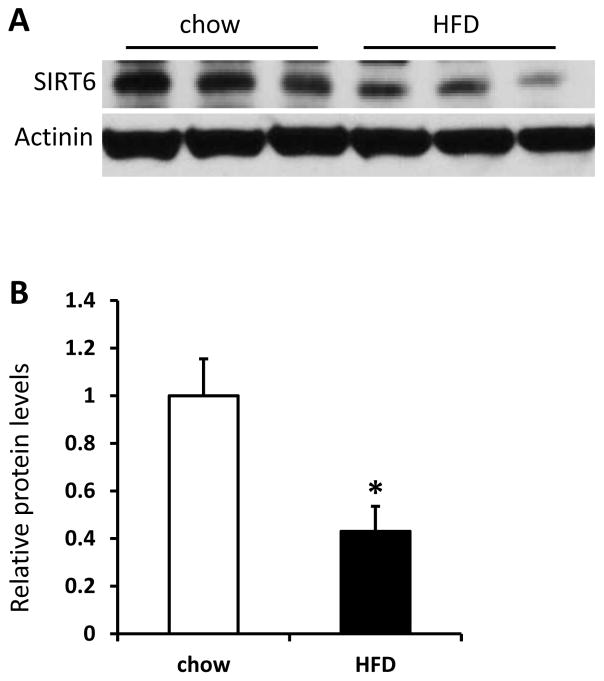
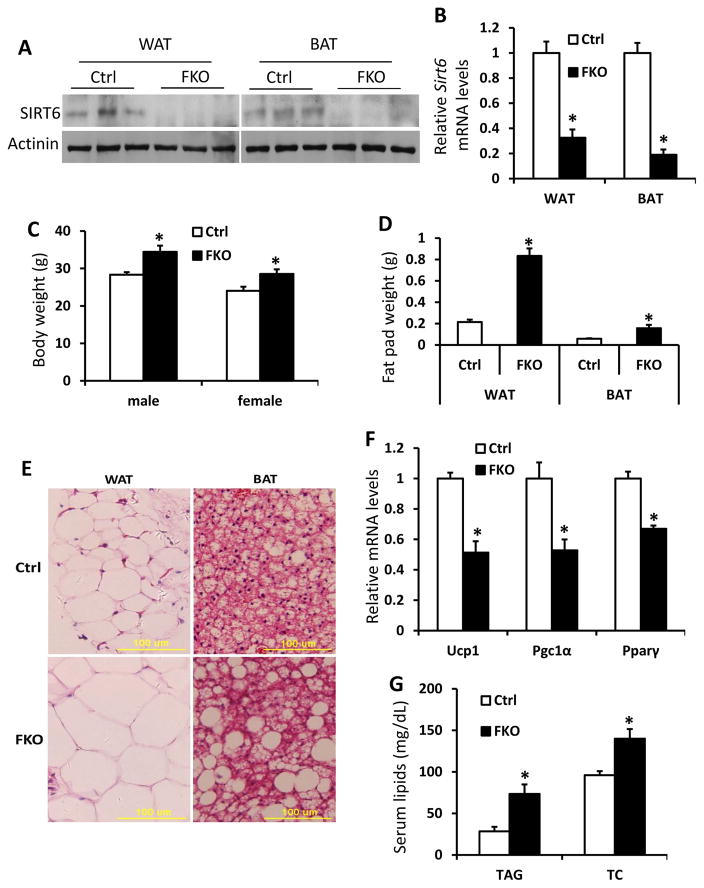
As we previously reported, Sirt6 is expressed in white adipose tissue (WAT) with an abundance similar to that in the liver of mouse (Xiong et al. 2016b). SIRT6 has been shown to play important roles in a number of organs/tissues, including liver, heart and islets; however, the function of SIRT6 in the adipose tissue is not well understood (Kugel and Mostoslavsky 2014). To examine whether SIRT6 protein is affected by diets, we analyzed SIRT6 protein in WAT of mice fed with 60% HFD for 5 months, which is commonly used to induce diet-induced obesity and diabetes. As shown in Figure 1A and 1B, SIRT6 protein was decreased in epididymal fat pads of HFD-fed C57BL/6J mice compared with regular chow-fed controls. We next sought to determine the effect of Sirt6 deletion in adipose tissue in vivo. In order to delete the Sirt6 gene in both pre-adipocytes and mature adipocytes, we crossed Sirt6 floxed mice with a widely used Fabp4-Cre line (He et al. 2003) to generate Sirt6fl/fl:Fabp4-Cre mice (Sirt6-FKO). As expected, Sirt6 was efficiently deleted in both epididymal WAT and interscapular BAT (Figure 2A and 2B).
Figure 1. Sirt6 is decreased in white adipose tissues (WAT) of high fat diet (HFD) fed mice.
A and B: Western blot (A) and densitometric analysis (B) of Sirt6 in epididymal fat pads of male mice fed with 60% HFD for 5 months (n=6 per group). Data are presented as mean ± SEM. *, p<0.05.
Figure 2. Fabp4-Cre mediated Sirt6 deletion in mice leads to obesity.
A and B: Western blot (A) and real-time PCR (B) analysis of Sirt6 protein and mRNA in WAT and BAT of 3-month-old control and Sirt6:Fabp4-Cre (FKO) mice (n=3 per genotype). C and D: Body weight (C) and epididymal fat pad weight (D) of 3-month-old male control and FKO mice (n=6–8 per genotype). E: H&E staining of WAT and BAT from 6-month-old control and FKO mice. F: Relative mRNA levels of thermogenic genes in BAT of 3-month-old control and FKO mice (n=3 per genotype). G: Plasma triglyceride and total cholesterol levels of 3-month-old male control and FKO mice (n=6–8 per genotype). Data are presented as mean ± SEM. *, p<0.05.
Increased fat mass and body weight gains in Sirt6-FKO mice
On normal chow, the body weight of Sirt6-FKO mice was increased by ~22% in males and ~19% in females at 3-month of age (Figure 2C). As expected, both epididymal white fat and interscapular brown fat depots were larger in Sirt6-FKO mice and the size of adipocytes was bigger in FKO mice than that in control wildtype mice (Figure 2D and 2E). To characterize the quality of brown adipose tissue, we analyzed expression of thermogenic genes, such as Ucp1, PGC1α and PPARγ, in the intersacpular BAT, and the data showed that all three genes were significantly downregulated in the FKO mice (Figure 2F). In addition, blood triglyceride and total cholesterol levels were also significantly elevated in Sirt6-FKO mice (Figure 2F).
Impaired glucose tolerance and decreased insulin sensitivity in Sirt6-FKO mice
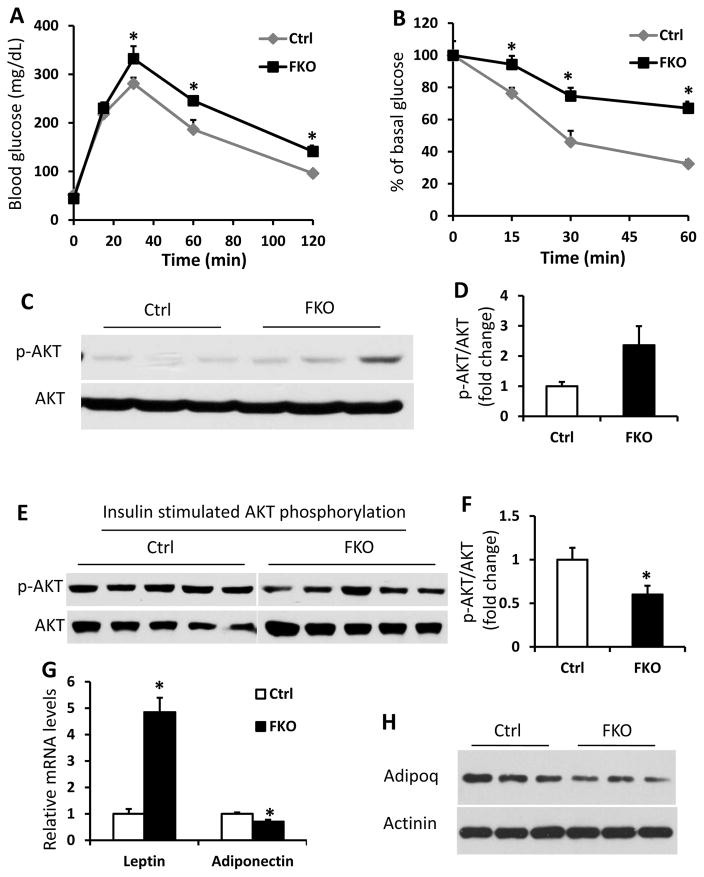
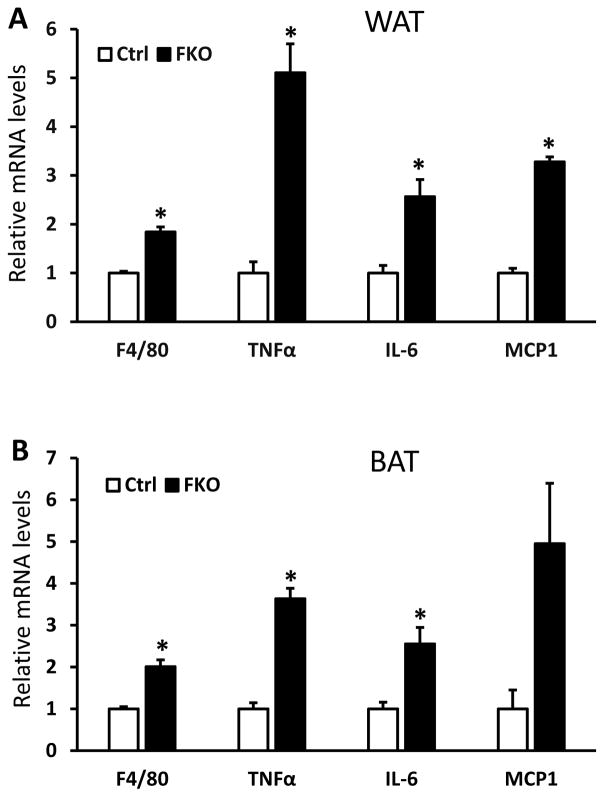
Obesity often leads to glucose intolerance and insulin resistance in human and mice (Tseng et al. 2010). To investigate the metabolic effects of Fabp4-Cre mediated deletion of Sirt6 in mice, we performed glucose tolerance tests (GTT) to assess glucose homeostasis in the Sirt6-FKO mice. As shown in Figure 3A, Sirt6-FKO mice exhibited significant glucose intolerance. Insulin sensitivity, as measured by insulin tolerance test, was also significantly decreased in Sirt6-FKO mice (Figure 3B). To further examine insulin resistance, we measured levels of AKT phosphorylation in WAT without or with insulin stimulation. Consistent with the impaired insulin sensitivity phenotype, phosphorylation of AKT was significantly decreased in WAT of Sirt6-FKO mice after insulin stimulation although the basal levels of phosphorylated AKT in WAT were a little higher in Sirt6-FKO mice (Figure 3C–3F). As hypertrophic adipocytes secrete more leptin and less adiponectin, the leptin/adiponectin ratio (LAR) has been proposed as a potentially useful measure of insulin resistance (Finucane, et al. 2009). We then further examined the expression levels of leptin and adiponectin in WAT of Sirt6-FKO mice. Our data showed that leptin mRNA levels were markedly elevated while adiponectin mRNA levels were significantly decreased in the WAT of Sirt6-FKO mice (Figure 3G). Leptin concentrations are generally proportional to body fat mass, however, adiponectin concentrations are not. We analyzed adiponectin protein levels in WAT of control and Sirt6-FKO mice and found that the FKO mice had lower adiponectin compared to the control mice (Figure 3H). Since obesity is associated with a state of chronic, low-grade inflammation, we also examined the inflammation status of adipose tissues in Sirt6-FKO and control mice. The mRNA levels of major inflammatory markers were analyzed in adipose tissues of chow fed control and Sirt6-FKO mice. As shown in Figure 4A and 4B, mRNA levels of F4/80 (Adgre1), TNFα, IL-6 and MCP-1 (Ccl2) in both WAT and BAT were significantly elevated, suggesting an increased macrophage infiltration in the adipose tissues of Sirt6-FKO mice.
Figure 3. Sirt6-FKO mice exhibit glucose intolerance and insulin resistance.
A and B: Glucose tolerance test (GTT, 2g/kg) (A) and insulin tolerance test (ITT, 0.5U/kg) (B) in control and FKO mice at 3–4 months of age (n=7–9 per genotype). C and D: Western blot for the phosphorylated and total AKT proteins in WAT (C) and quantification by densitometry of phosphorylated AKT (p-AKT473) normalized to total AKT (D) for control and FKO mice at 3 months of age (n=5 per genotype). E and F: Mice were injected with 0.5U/kg insulin for 3 minutes. Western blot (E) and quantification by densitometry of phosphorylated AKT (p-AKT473) normalized to total AKT (F) in WAT of control and FKO mice at 3 months of age (n=5 per genotype). G: Relative mRNA levels of leptin and adiponectin in WAT samples of 2-month-old male control and FKO mice (n=4 per genotype). H: Western blot analysis of adiponectin (Adipoq) protein level in 3-month-old control and FKO mice (n=3 per genotype). Data are presented as mean ± SEM. *, p<0.05.
Figure 4. Sirt6-FKO mice have increased adipose tissue inflammation.
A and B: Relative mRNA levels of inflammation related genes in WAT samples (A) and BAT samples (B) of 6-month-old male control and FKO mice (n=4–5 per genotype). Data are presented as mean ± SEM. *, p<0.05.
Deletion of the Sirt6 gene in mature adipocytes using the Adipoq-Cre mouse line
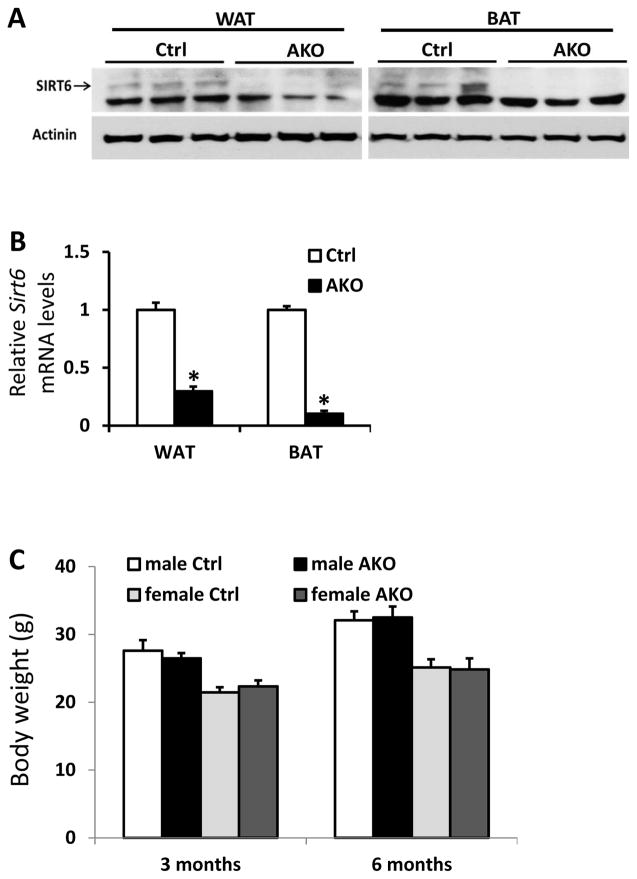
Fabp4-Cre mice have been reported to exhibit quite broad expression of Cre recombinase in both adipose and non-adipose tissues such as brain, muscle, macrophages, and intestine (Heffner et al. 2012; Lee et al. 2013; Martens et al. 2010). To further examine whether the obesity and insulin resistance phenotype in the Sirt6-FKO mice was due to loss of SIRT6 in adipocytes or not, we deleted the Sirt6 gene in mature adipocytes using an Adipoq-Cre mouse line (Eguchi et al. 2011). The generated Sirt6fl/fl:Adipoq-cre mice was named as Sirt6-AKO. As expected, the AKO mice displayed significantly reduced expression of Sirt6 in WAT and BAT (Figure 5A and 5B). Interestingly, there was no difference in body weight between Sirt6-AKO and control mice at either 3 months or 6 months of age on a chow diet (Figure 5C).
Figure 5. Deletion of Sirt6 in mature adipocytes using Adipoq-Cre.
A and B: Western blot (A) and real-time PCR (B) analysis of Sirt6 protein and mRNA in WAT and BAT of 2-month-old control and Sirt6:Adipoq-Cre (AKO) mice (n=3 per genotype). C: Body weight of control and AKO male mice with different ages (n=6–8 per genotype). Data are presented as mean ± SEM. *, p<0.05.
Sirt6-AKO mice do not exhibit any significant metabolic phenotype even under HFD feeding
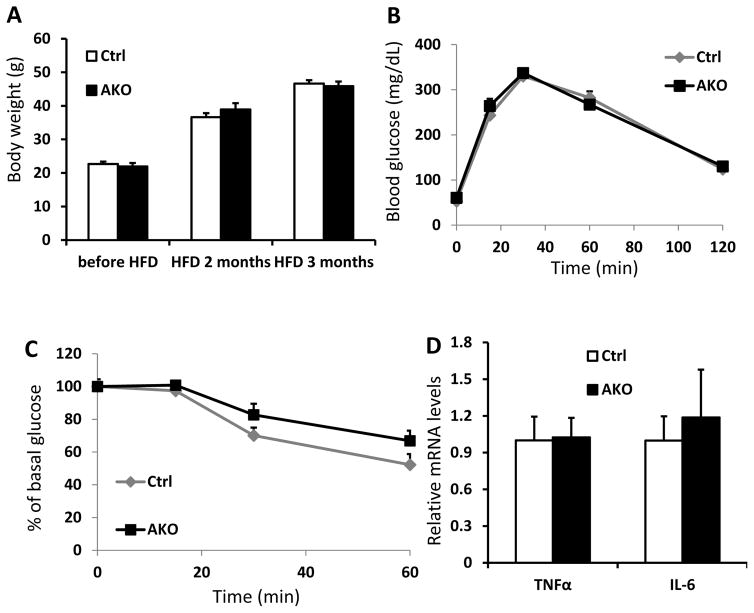
Compared to their littermate controls, Sirt6-AKO mice on chow diet exhibited normal body weight even at 6 months of age. To assess whether the Sirt6-AKO mice are sensitive to diet-induced obesity, control wildtype and Sirt6-AKO mice were fed with HFD (60% of calorie from fat) for up to 3 months. Surprisingly, HFD fed Sirt6-AKO mice had similar body weight and glucose tolerance compared to the control mice although the AKO mice showed moderate insulin resistance (Figure 6A–6C). Additionally, expression of common inflammatory genes, such as TNFα and IL-6, in WAT did not show any significant difference between the control and Sirt6-AKO mice (Figure 6D).
Figure 6. Challenge of Sirt6-AKO mice with a high fat diet.
Control and AKO male mice were fed with HFD (60% calorie from fat) for 3 months. A: Body weight of control and AKO mice before and after the HFD feeding (n=7–8 per genotype). B and C: Glucose tolerance test (GTT, 1.5g/kg) (B) and insulin tolerance test (ITT, 0.75U/kg) (C) in control and AKO mice fed with HFD for 3 months (n=7–8 per genotype). D: Relative mRNA levels of TNFα and IL-6 in WAT of HFD-fed control and AKO mice (n=4–5 per genotype). Data are presented as mean ± SEM. *, p<0.05.
Discussion
The increasing prevalence of obesity has been paralleled by an epidemic level of type 2 diabetes in the world, and these growing health problems underscore the need to elucidate the molecular basis for the obesity pathogenesis and to develop novel treatments for the obesity related metabolic disorders (Bastien et al. 2014). Adipose tissue is not only a primary depot for lipid storage but also an endocrine organ secreting adipokines for the control of energy homeostasis (Galic et al. 2010). In recent years, much effort has been made to understand the adipocyte-specific functions of genes of interest. To do so, adipocyte-expressing Cre transgenic mice can be used. Among those Cre strains, the Fabp4-Cre transgenic mouse created by the Ronald Evans group has been widely used in the field of adipocyte biology (He et al. 2003). Although FABP4 was originally identified as an adipocyte-specific protein, later studies have shown that FABP4 is also expressed in other cell types, including macrophages, the lymphatic system, and embryo (Lee et al. 2013). Thus, the Fabp4-Cre is believed to express in multiple cell types other than adipocytes. For instance, strong Cre activity has been also noticed in the central nervous system (Martens et al. 2010). Recently, another adipocyte-specific Cre line – Adipoq-Cre – has been developed with the Cre coding sequence driven by the mouse adiponectin gene promoter/enhancer (Eguchi et al. 2011). The recombination by the Adipoq-Cre is highly specific for mature adipocytes and absolutely no Cre activity has been observed in other tissues (Lee et al. 2013). Moreover, the Fabp4-Cre is expressed early on in adipocyte progenitor cells whereas the Adipoq-Cre is expressed only after adipocyte differentiation (Lee et al. 2013).
To understand the Sirt6 function in adipocytes, we have generated two mouse models with the Sirt6 gene deletion mediated by Fabp4-Cre (Sirt6-FKO) or Adipoq-Cre (Sirt6-AKO), respectively. Interestingly, Sirt6-FKO mice exhibit an obesity phenotype, manifested as overweight, insulin resistance, glucose intolerance, adipose inflammation. By contrast, no significant metabolic phenotype has been observed in Sirt6-AKO mice even challenged with HFD. As we obtained a similar Sirt6 knockout efficiency in both mouse models, the dramatic differences in the phenotype might be attributable to cell types other than adipocytes, for example, immune cells and neurons. Distinct phenotypes for some other gene knockouts using Fabp4-Cre and Adipoq-Cre mouse lines have been also reported (Lee, et al. 2016; Mullican, et al. 2013; Qiang, et al. 2016). Neural Sirt6 deletion promotes adult-onset obesity in mice likely through reduced growth hormone (GH) and hypothalamic neuropeptide levels (Schwer et al. 2010). It is noteworthy that Cre activity has been reported in neurons throughout the central nervous system in the Fabp4-Cre mice (Martens et al. 2010). Although the obese phenotype is not quite similar between neural Sirt6 deleted mice and Sirt6-FKO mice, Fabp4-Cre mediated neural deletion of Sirt6 may contribute, at least in part, to the metabolic dysregulation in the Sirt6-FKO mice.
It should be noted that during the revision process of our manuscript, Kuang and coworkers reported a fat-specific Sirt6-knockout study by using the same Adipoq-Cre line as ours (Kuang et al. 2017). They have shown that Sirt6 deletion in adipocytes exhibits normal glucose levels and body weight under chow diet, which is similar to the phenotype we have observed in the AKO mice. Interestingly, they have observed that fat-specific deletion of Sirt6 sensitizes mice to HFD-induced obesity and insulin resistance by inhibiting lipolysis (Kuang et al. 2017). Intriguingly, we have also treated our AKO mice with 60% HFD for 3 months, but we have not noticed any significant metabolic phenotypes (blood glucose levels, body weight, glucose tolerance, insulin resistance, and so forth). The discrepancy of the AKO mouse phenotype might be attributed to differences in animal genetic background, diet, facility environment, and others.
In conclusion, our data suggest that Sirt6 plays a critical role in the Fabp4+ cells for the regulation of adiposity, energy homeostasis, inflammation, insulin sensitivity, and glucose metabolism. Therefore, Sirt6 can be a useful therapeutic target for the treatment of obesity and diabetes.
Acknowledgments
Funding
This work was supported by grants from National Natural and Science Foundation of China (81670526, to XX), National Institute of Diabetes and Digestive and Kidney Diseases (R01DK091592, to XCD), and the Showalter Scholar Award (to XCD) from the Ralph W. and Grace M. Showalter Research Trust.
The Authors would like to thank Dr. Rongya Tao and Dr. Gaihong Wang for expert technical assistance.
Footnotes
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Author contributions
XX designed and carried out the study, interpreted data, analyzed data, and wrote the manuscript. CZ, YZ, RF, and QX contributed to data collection. XCD conceived the hypothesis, designed the experiments, analyzed and interpreted the data, and wrote the manuscript. All authors approved the manuscript.
References
- Bastien M, Poirier P, Lemieux I, Despres JP. Overview of epidemiology and contribution of obesity to cardiovascular disease. Prog Cardiovasc Dis. 2014;56:369–381. doi: 10.1016/j.pcad.2013.10.016. [DOI] [PubMed] [Google Scholar]
- Dominy JE, Jr, Lee Y, Jedrychowski MP, Chim H, Jurczak MJ, Camporez JP, Ruan HB, Feldman J, Pierce K, Mostoslavsky R, et al. The deacetylase Sirt6 activates the acetyltransferase GCN5 and suppresses hepatic gluconeogenesis. Mol Cell. 2012;48:900–913. doi: 10.1016/j.molcel.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong XC. Sirtuin biology and relevance to diabetes treatment. Diabetes Manag (Lond) 2012;2:243–257. doi: 10.2217/dmt.12.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi J, Wang X, Yu S, Kershaw EE, Chiu PC, Dushay J, Estall JL, Klein U, Maratos-Flier E, Rosen ED. Transcriptional control of adipose lipid handling by IRF4. Cell Metab. 2011;13:249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhanati S, Kanfi Y, Varvak A, Roichman A, Carmel-Gross I, Barth S, Gibor G, Cohen HY. Multiple regulatory layers of SREBP1/2 by SIRT6. Cell Rep. 2013;4:905–912. doi: 10.1016/j.celrep.2013.08.006. [DOI] [PubMed] [Google Scholar]
- Finucane FM, Luan J, Wareham NJ, Sharp SJ, O’Rahilly S, Balkau B, Flyvbjerg A, Walker M, Hojlund K, Nolan JJ, et al. Correlation of the leptin:adiponectin ratio with measures of insulin resistance in non-diabetic individuals. Diabetologia. 2009;52:2345–2349. doi: 10.1007/s00125-009-1508-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galic S, Oakhill JS, Steinberg GR. Adipose tissue as an endocrine organ. Mol Cell Endocrinol. 2010;316:129–139. doi: 10.1016/j.mce.2009.08.018. [DOI] [PubMed] [Google Scholar]
- He W, Barak Y, Hevener A, Olson P, Liao D, Le J, Nelson M, Ong E, Olefsky JM, Evans RM. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci U S A. 2003;100:15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffner CS, Herbert Pratt C, Babiuk RP, Sharma Y, Rockwood SF, Donahue LR, Eppig JT, Murray SA. Supporting conditional mouse mutagenesis with a comprehensive cre characterization resource. Nat Commun. 2012;3:1218. doi: 10.1038/ncomms2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Xiao C, Wang RH, Lahusen T, Xu X, Vassilopoulos A, Vazquez-Ortiz G, Jeong WI, Park O, Ki SH, et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 2010;12:224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang J, Zhang Y, Liu Q, Shen J, Pu S, Cheng S, Chen L, Li H, Wu T, Li R, et al. 2017 Fat-specific Sirt6 ablation sensitizes mice to high-fat diet-induced obesity and insulin resistance by inhibiting lipolysis. Diabetes. 2017 Mar 1; doi: 10.2337/db16-1225. pii:db161225. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Kugel S, Mostoslavsky R. Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem Sci. 2014;39:72–81. doi: 10.1016/j.tibs.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Russell SJ, Ussar S, Boucher J, Vernochet C, Mori MA, Smyth G, Rourk M, Cederquist C, Rosen ED, et al. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes. 2013;62:864–874. doi: 10.2337/db12-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PL, Tang Y, Li H, Guertin DA. Raptor/mTORC1 loss in adipocytes causes progressive lipodystrophy and fatty liver disease. Mol Metab. 2016;5:422–432. doi: 10.1016/j.molmet.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens K, Bottelbergs A, Baes M. Ectopic recombination in the central and peripheral nervous system by aP2/FABP4-Cre mice: implications for metabolism research. FEBS Lett. 2010;584:1054–1058. doi: 10.1016/j.febslet.2010.01.061. [DOI] [PubMed] [Google Scholar]
- Masri S, Rigor P, Cervantes M, Ceglia N, Sebastian C, Xiao C, Roqueta-Rivera M, Deng C, Osborne TF, Mostoslavsky R, et al. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell. 2014;158:659–672. doi: 10.1016/j.cell.2014.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, Chua KF. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009;8:2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullican SE, Tomaru T, Gaddis CA, Peed LC, Sundaram A, Lazar MA. A novel adipose-specific gene deletion model demonstrates potential pitfalls of existing methods. Mol Endocrinol. 2013;27:127–134. doi: 10.1210/me.2012-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang G, Whang Kong H, Xu S, Pham HA, Parlee SD, Burr AA, Gil V, Pang J, Hughes A, Gu X, et al. Lipodystrophy and severe metabolic dysfunction in mice with adipose tissue-specific insulin receptor ablation. Mol Metab. 2016;5:480–490. doi: 10.1016/j.molmet.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Schumacher B, Lombard DB, Xiao C, Kurtev MV, Gao J, Schneider JI, Chai H, Bronson RT, Tsai LH, et al. Neural sirtuin 6 (Sirt6) ablation attenuates somatic growth and causes obesity. Proc Natl Acad Sci U S A. 2010;107:21790–21794. doi: 10.1073/pnas.1016306107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Xiong X, DePinho RA, Deng CX, Dong XC. FoxO3 transcription factor and Sirt6 deacetylase regulate low density lipoprotein (LDL)-cholesterol homeostasis via control of the proprotein convertase subtilisin/kexin type 9 (Pcsk9) gene expression. J Biol Chem. 2013a;288:29252–29259. doi: 10.1074/jbc.M113.481473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Xiong X, DePinho RA, Deng CX, Dong XC. Hepatic SREBP-2 and cholesterol biosynthesis are regulated by FoxO3 and Sirt6. J Lipid Res. 2013b;54:2745–2753. doi: 10.1194/jlr.M039339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasselli L, Xi Y, Zheng W, Tennen RI, Odrowaz Z, Simeoni F, Li W, Chua KF. SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat Struct Mol Biol. 2016;23:434–440. doi: 10.1038/nsmb.3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YH, Cypess AM, Kahn CR. Cellular bioenergetics as a target for obesity therapy. Nat Rev Drug Discov. 2010;9:465–482. doi: 10.1038/nrd3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Cre transgenic mouse lines. Methods Mol Biol. 2009;561:265–273. doi: 10.1007/978-1-60327-019-9_17. [DOI] [PubMed] [Google Scholar]
- Xiong X, Sun X, Wang Q, Qian X, Zhang Y, Pan X, Dong XC. SIRT6 protects against palmitate-induced pancreatic beta-cell dysfunction and apoptosis. J Endocrinol. 2016a;231:159–165. doi: 10.1530/JOE-16-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Tao R, DePinho RA, Dong XC. Deletion of hepatic FoxO1/3/4 genes in mice significantly impacts on glucose metabolism through downregulation of gluconeogenesis and upregulation of glycolysis. PLoS One. 2013;8:e74340. doi: 10.1371/journal.pone.0074340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Wang G, Tao R, Wu P, Kono T, Li K, Ding WX, Tong X, Tersey SA, Harris RA, et al. Sirtuin 6 regulates glucose-stimulated insulin secretion in mouse pancreatic beta cells. Diabetologia. 2016b;59:151–160. doi: 10.1007/s00125-015-3778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Zwaans BM, Eckersdorff M, Lombard DB. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle. 2009;8:2662–2663. doi: 10.4161/cc.8.16.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]