Abstract
Data that defines IGHV (immunoglobulin heavy chain variable) germline gene inference using sequences of IgM-encoding transcriptomes obtained by Illumina MiSeq sequencing technology are described. Such inference is used to establish personalized germline gene sets for in-depth antibody repertoire studies and to detect new antibody germline genes from widely available immunoglobulin-encoding transcriptome data sets. Specifically, the data has been used to validate (Parallel antibody germline gene and haplotype analyses support the validity of immunoglobulin germline gene inference and discovery (DOI: 10.1016/j.molimm.2017.03.012) (Kirik et al., 2017) [1]) the inference process. This was accomplished based on analysis of the inferred germline genes’ association to the donors’ different haplotypes as defined by their different, expressed IGHJ alleles and/or IGHD genes/alleles. The data is important for development of validated germline gene databases containing entries inferred from immunoglobulin-encoding transcriptome sequencing data sets, and for generation of valid, personalized antibody germline gene repertoires.
Keywords: Antibody, Gene inference, Germline repertoire, Immunoglobulin germline gene, Transcriptome, Validation
Specifications Table
| Subject area | Biology, Medicine |
| More specific subject area | Immunobiology |
| Type of data | Sequence reads, tables, figures |
| How data was acquired | Next generation sequencing using Illumina MiSeq technology; analysis using immunoglobulin repertoire inference software |
| Data format | Raw data, analyzed data |
| Experimental factors | Data processing was performed using pRESTO, Change-O, TIgGER, IgDiscover, GIgGle |
| Experimental features | Immunoglobulin M heavy chain variable domain-encoding genes were amplified by RT-PCR, sequenced by next generation sequencing technology, and analyzed by bioinformatics approaches. |
| Data accessibility | FASTQ raw sequence data files are available from the European Nucleotide Archive, study accession number: PRJEB18926. Data is within this article. Code available athttps://github.com/ukirik/giggle |
Value of the data
-
•
The data is valuable for development of computational inference approaches that feature improved confidence in the outcomes of the inference process.
-
•
The data is valuable for development of validated immunoglobulin germline gene databases.
-
•
The data is valuable for validation of computational inference of personalized antibody germline gene repertoires.
-
•
The data is valuable for the analytical process preceding studies of evolution of immune responses.
1. Data
The data of this article summarize the identity and accession numbers of sequencing data files (Table 1), the sizes of the sequence sets during the different stages of data processing (Table 2), and the outcome of validation of new inferred genes/alleles (Table 3), identified by use of IgDiscover and TIgGER. The frequencies of readily inferable [2] IGHD (Immunoglobulin heavy D-gene) genes used by the two haplotypes of five subjects are summarized (Table 4). Furthermore the data illustrate the effect of using a germline gene database that extends beyond codon 105 on gene inference (Fig. 1), and summarizes the outcome of TIgGER-based germline gene inference of six transcriptoms (Fig. 2). The data also illustrates how low sequencing quality scores are associated with some, but certainly not all, inferred germline gene alleles (Fig. 3), and summarizes IGHJ (Immunoglobulin heavy J-gene) alleles used by transcriptomes of six subjects (Fig. 4). The link between inferred IGHV (Immunoglobulin heavy V-gene) germline genes/alleles and different alleles of IGHJ6 in bone marrow (BM)- and peripheral blood (PB)-derived transcriptomes of two heterozygous subjects is shown (Fig. 5). The data summarizes linkage of different IGHD genes to two different haplotypes defined by alleles of IGHJ6 or defined by heterozygous IGHV genes (Fig. 6). The linkage of IGHV1-8, IGHV3-9, IGHV5-10-1, and IGHV3-64D germline genes to different haplotypes in subjects with two different IGHD gene-defined haplotypes (Fig. 7) is shown. Association of IGHV germline genes/alleles with particular IGHD genes in five subjects with different IGHD-defined haplotypes is shown (Fig. 8), as is the extent of association of alleles of IGHV4-59 to particular IGHD genes (Fig. 9). Finally, data describing assessment of alleles of IGHD genes detected in IgM-encoding transcriptomes of six subjects (Fig. 10), and of IGHV germline genes associated to the different alleles of IGHD genes in two subjects (Fig. 11) is shown.
Table 1.
Summary of identity of sequenced samples of the study (European Nucleotide Archive (ENA) accession number PRJEB18926).a
| Subject | Sample originb | Replicate | Sequencing sample ID | Isotypes | ENA sample accession number | ENA experiment accession number |
|---|---|---|---|---|---|---|
| 1 | BM | 1 | P1882_1001 | IgA, IgE, IgG, IgM | ERS1531209 | ERX1875309 |
| 1 | BM | 2 | P1882_1002 | IgA, IgE, IgG, IgM | ERS1531210 | ERX1875310 |
| 1 | PB | 1 | P1882_1007 | IgA, IgE, IgG, IgM | ERS1531215 | ERX1875315 |
| 1 | PB | 2 | P1882_1008 | IgA, IgE, IgG, IgM | ERS1531216 | ERX1875316 |
| 2 | BM | 1 | P1882_1003 | IgA, IgE, IgG, IgM | ERS1531211 | ERX1875311 |
| 2 | BM | 2 | P1882_1004 | IgA, IgE, IgG, IgM | ERS1531212 | ERX1875312 |
| 2 | PB | 1 | P1882_1009 | IgA, IgE, IgG, IgM | ERS1531217 | ERX1875317 |
| 2 | PB | 2 | P1882_1010 | IgA, IgE, IgG, IgM | ERS1531218 | ERX1875318 |
| 3 | BM | 1 | P1882_1005 | IgA, IgE, IgG, IgM | ERS1531213 | ERX1875313 |
| 3 | BM | 2 | P1882_1006 | IgA, IgE, IgG, IgM | ERS1531214 | ERX1875314 |
| 3 | PB | 1 | P1882_1011 | IgA, IgE, IgG, IgM | ERS1531219 | ERX1875319 |
| 3 | PB | 2 | P1882_1012 | IgA, IgE, IgG, IgM | ERS1531220 | ERX1875320 |
| 4 | BM | 1 | P1882_1013 | IgA, IgE, IgG, IgM | ERS1531221 | ERX1875321 |
| 4 | BM | 2 | P1882_1014 | IgA, IgE, IgG, IgM | ERS1531222 | ERX1875322 |
| 4 | PB | 1 | P1882_1019 | IgA, IgE, IgG, IgM | ERS1531227 | ERX1875327 |
| 4 | PB | 2 | P1882_1020 | IgA, IgE, IgG, IgM | ERS1531228 | ERX1875328 |
| 5 | BM | 1 | P1882_1015 | IgA, IgE, IgG, IgM | ERS1531223 | ERX1875323 |
| 5 | BM | 2 | P1882_1016 | IgA, IgE, IgG, IgM | ERS1531224 | ERX1875324 |
| 5 | PB | 1 | P1882_1021 | IgA, IgE, IgG, IgM | ERS1531229 | ERX1875329 |
| 5 | PB | 2 | P1882_1022 | IgA, IgE, IgG, IgM | ERS1531230 | ERX1875330 |
| 6 | BM | 1 | P1882_1017 | IgA, IgE, IgG, IgM | ERS1531225 | ERX1875325 |
| 6 | BM | 2 | P1882_1018 | IgA, IgE, IgG, IgM | ERS1531226 | ERX1875326 |
| 6 | PB | 1 | P1882_1023 | IgA, IgG, IgMc | ERS1531231 | ERX1875331 |
| 6 | PB | 2 | P1882_1024 | IgA, IgG, IgMc | ERS1531232 | ERX1875332 |
Read numbers representing each sample/isotype are available in Supplementary Table EIV of Ref. [3].
BM: bone marrow; PB: peripheral blood.
No PCR product was derived using IgE-specific 3′-primers.
Table 2.
Number of IgM-encoding sequences at different stages of the analysis process.
| Donor | Tissuea | # of reads after filteringb | # of sequences after PRESTO pipelinec | # of unique sequencesd | # of unique sequences with V_errors=0d | # of unique sequences with V_errors=0 & D_coverage>35%d |
|---|---|---|---|---|---|---|
| 1 | BM | 258,988 | 261,967 | 86,135 | 47,233 | 43,006 |
| PB | nd | 1,068,050 | 370,114 | 233,786 | 212,414 | |
| 2 | BM | 194,555 | 197,949 | 90,181 | 58,685 | 52,815 |
| PB | nd | 548,228 | 241,853 | 152,456 | 136,060 | |
| 3 | BM | 278,426 | 281,711 | 70,515 | 28,827 | 26,400 |
| PB | nd | 1,285,522 | 394,304 | 172,864 | 157,687 | |
| 4 | BM | 339,935 | 345,021 | 91,511 | 45,510 | 40,850 |
| PB | nd | 456,175 | 201,889 | 124,741 | 111,341 | |
| 5 | BM | 318,207 | 324,269 | 106,047 | 63,924 | 57,998 |
| PB | nd | 511,142 | 96,357 | 48,325 | 43,553 | |
| 6 | BM | 406,893 | 412,689 | 152,125 | 85,956 | 77,603 |
| PB | nd | 693,033 | 208,311 | 122,685 | 109,770 |
nd – not done.
BM: bone marrow; PB peripheral blood
Number of sequences used for initiation of the workflow towards TIgGER-based analysis.
Number of sequences used for initiation of the workflow towards IgDiscover-based analysis.
Number of unique sequences in the final filtered output obtained using IgDiscover as inference method
Table 3.
Summary of sequence variants of germline genes not present in the IMGT germline gene database but inferred from BM transcript data using IgDiscover or TIgGER.
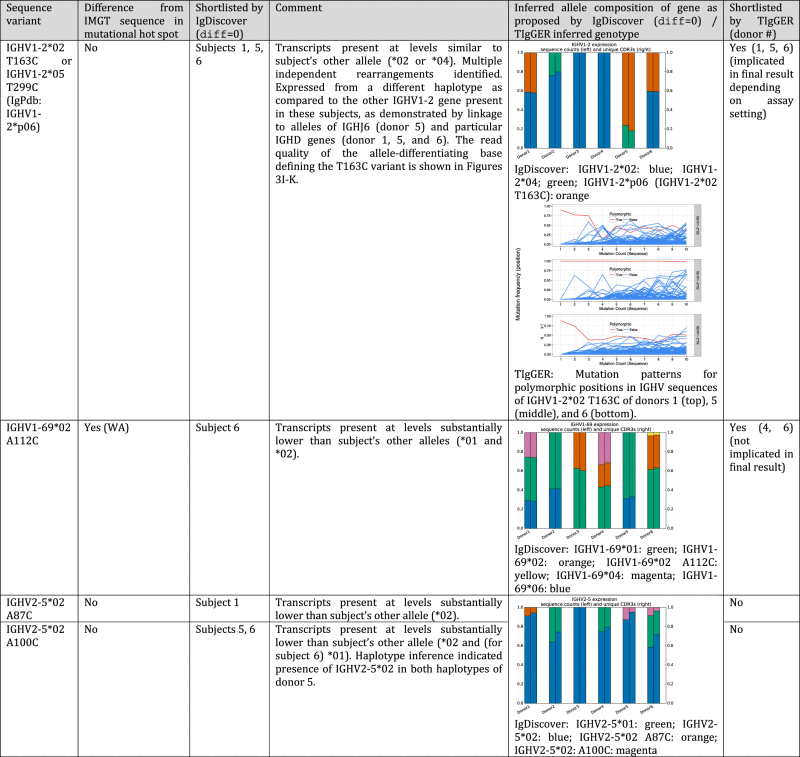 |
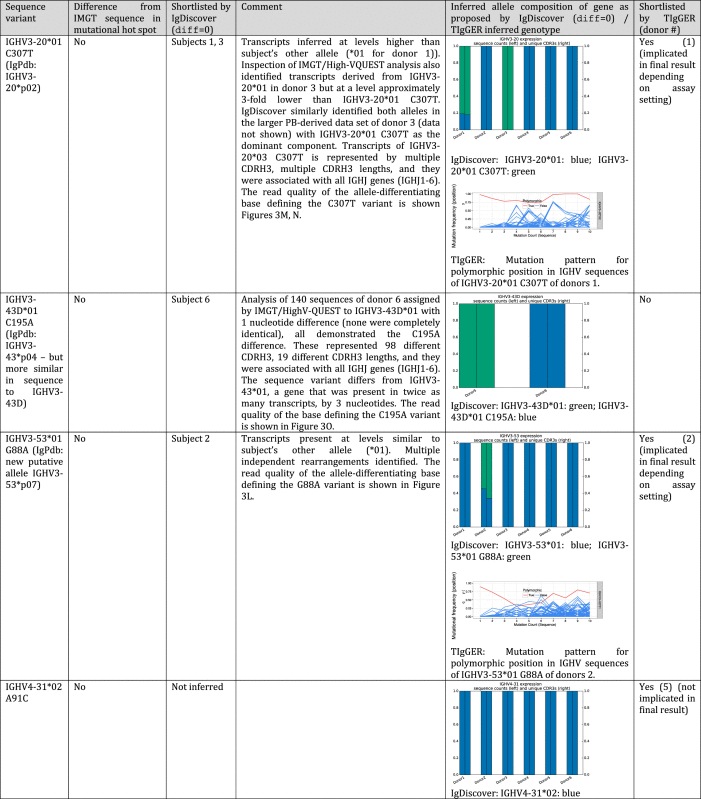 |
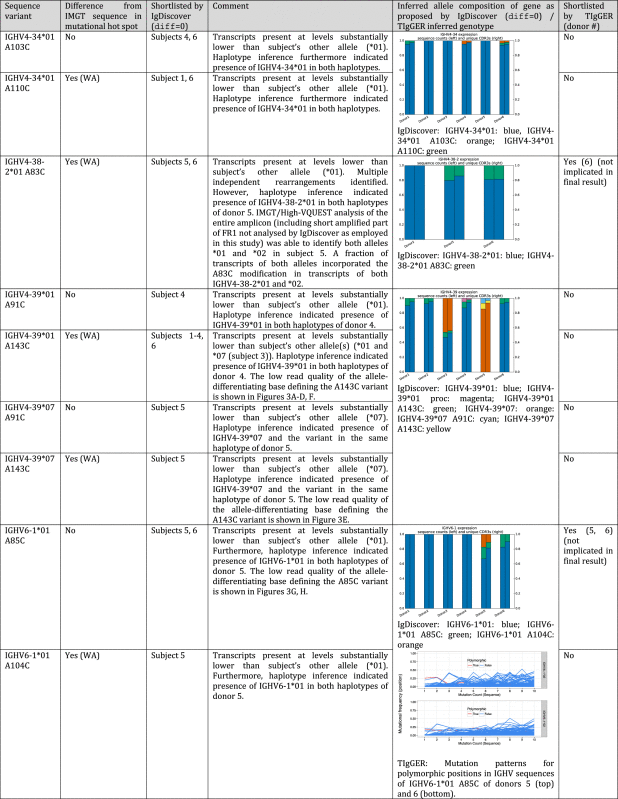 |
Table 4.
Estimated frequency* of use of readily identified IGHD germline genes [2] in haplotypes of five lymphocyte donors, and the ratio of estimated frequency† of these genes in the two haplotypes.
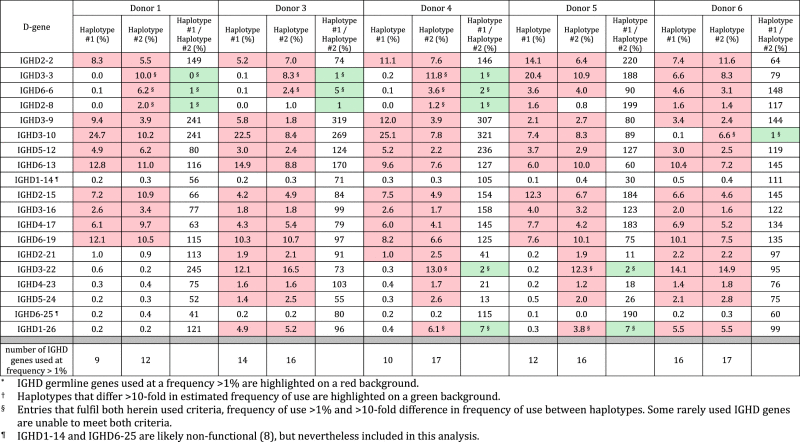 |
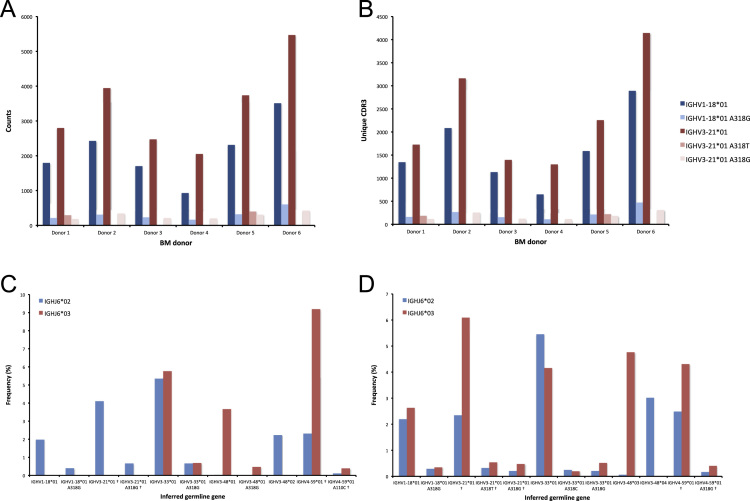
Fig. 1.
Germline gene variants of IGHV1-18 and IGHV3-21 inferred by IgDiscover when a starting germline database extending beyond codon 105 was used to initiate the process. The number of sequence counts (A) and unique CDRH3 (B) are shown. Examples (IGHV1-18, IGHV3-21, IGHV3-33, IGHV3-48, and IGHV4-59) of germline genes with new inferred variants, mostly in codon 106, and their similar association to the two different alleles of IGHJ6 of donor 4 (C) and donor 5 (D) are shown. Segregation of different established alleles of IGHV3-48 to the two alleles of IGHJ6 is also shown for comparison. † defines that the name of only one of a set of different alleles of the gene that cannot be differentiated by the analysis approach is shown.
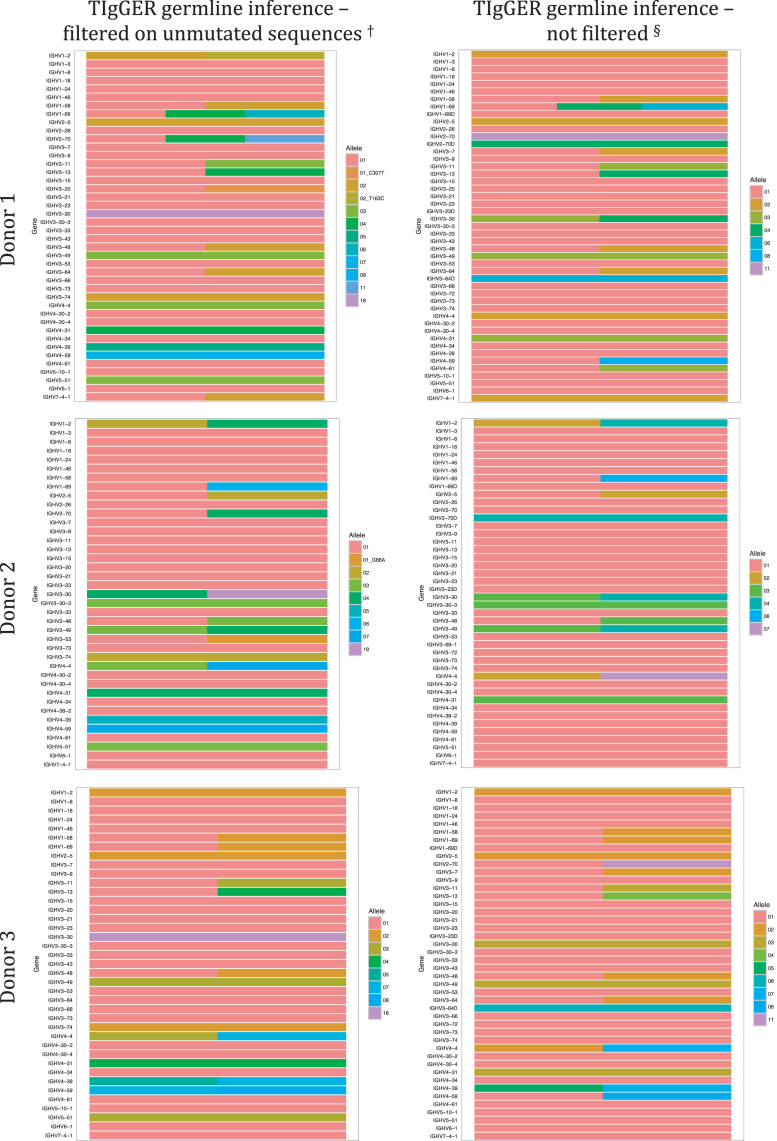
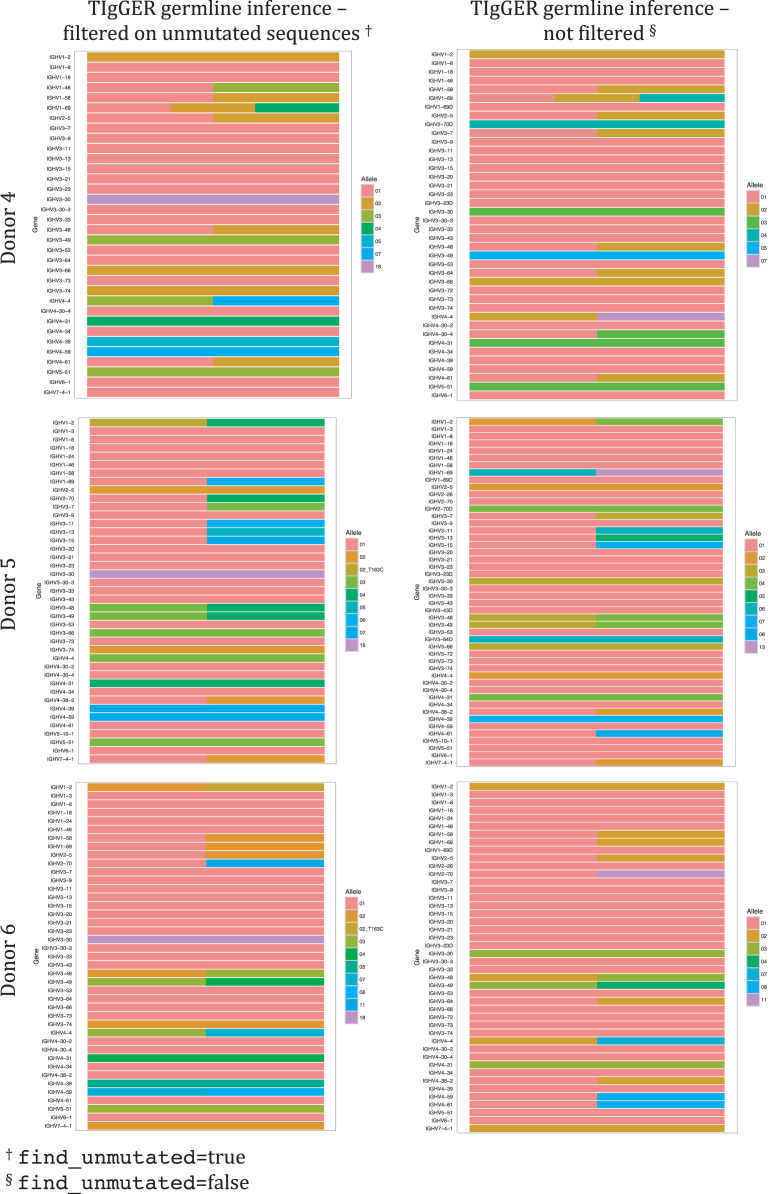
Fig. 2.
Genotype inferred by TIgGER using IgM-encoding transcripts of BM. Note difference in the calling of IGHV1-2. Heterozygous state of IGHV1-2 (*02/*p06) is inferred in subjects 1 and 6 only when argument find_unmutated=true while it is inferred in subject 2 (*02/*04) independently of the setting of find_unmutated. Heterozygous state of IGHV3-7 (*01/*02) is inferred in subjects 1, 3, and 4 only when argument find_unmutated=false while it is inferred in subject 5 (*01/*03) independently of the setting of find_unmutated. Heterozygous state of IGHV3-20 (*01/*01 C307T) is inferred in subject 1 only when argument find_unmutated=true and the allele variant is not at all inferred in donor 3. Heterozygous state of IGHV3-64 is inferred in donors 1, 3, 4, and 6 when argument find_unmutated=false and in donor 1 when argument find_unmutated=true. ,
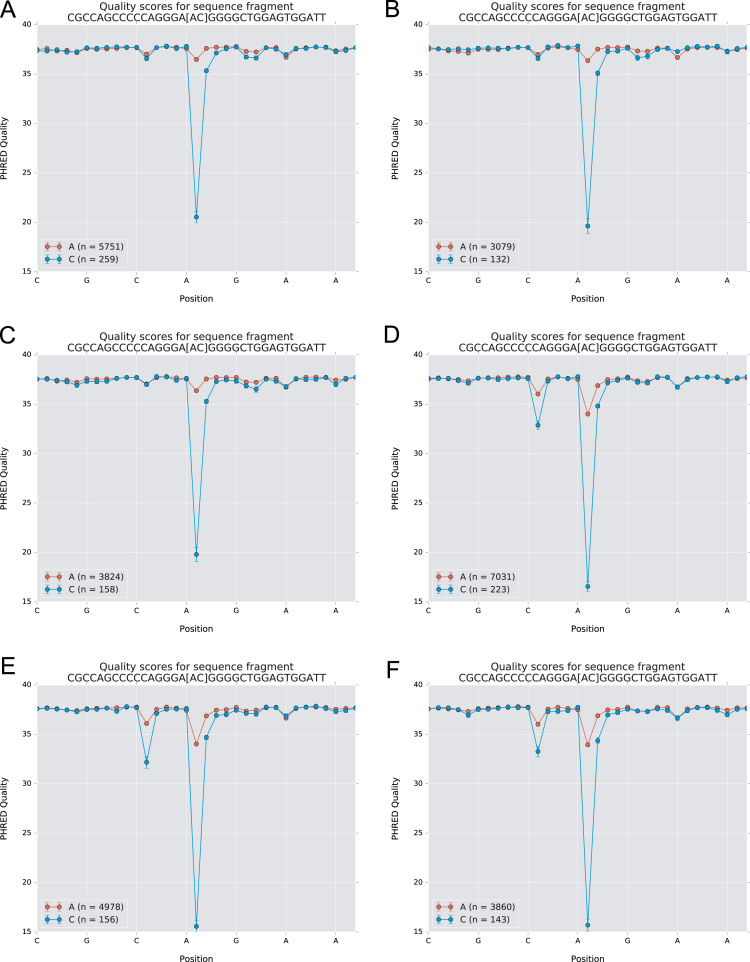
Fig. 3.
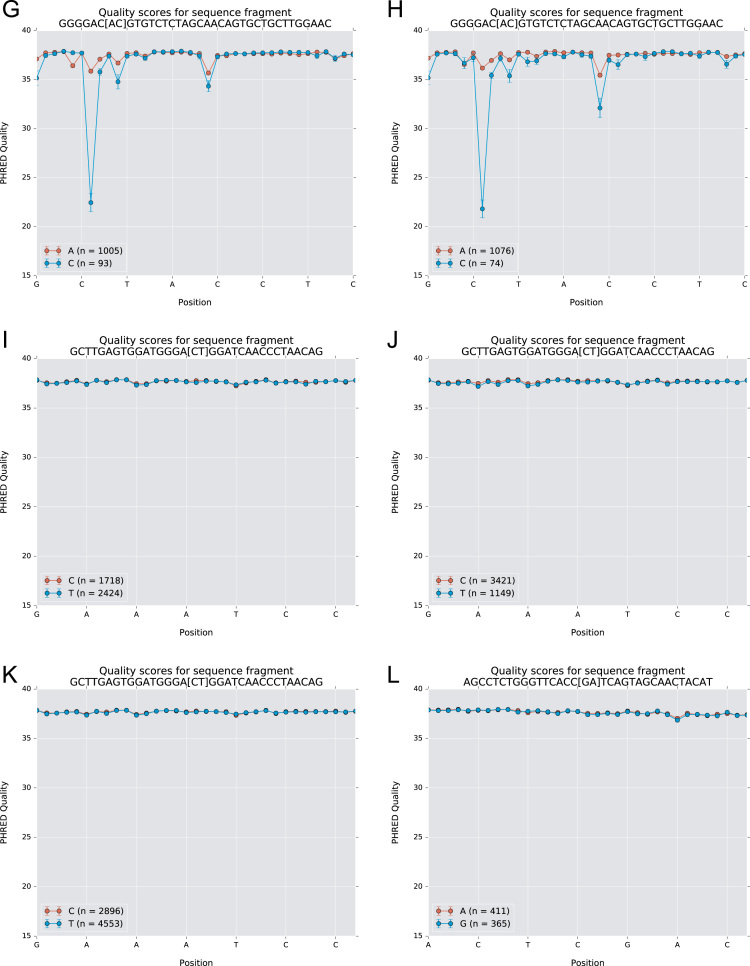
Quality score of sequencing reads representing germline genes inferred by IgDiscover. Sequence reads representing sequence variant A143C of IGHV4-39 show lower read quality (donors 1 (A), 2 (B), 3, (C), 4 (D), 5 (E), and 6 (F)) of the nucleotide representing the allele-differentiating base as opposed to reads defining the corresponding unmutated alleles (IGHV4-39*01 and *07). Similarly, inferred allele IGHV6-1 A85C shows low read quality of the allele-differentiating base (donor 5 (G), donor 6 (H)). Sequence reads representing parts of the sequences of alleles of IGHV1-2*02 and IGHV1-2*04 (represented by nucleotide T163) and IGHV1-2*p06 (C163) of donors 1 (I), 5 (J), and 6 (K) show highly similar read quality. Sequences representing IGHV3-53*01 and IGHV3-53*01 G88A of donor 2 (L), IGHV3-20*01 and IGHV3-20*01 C307T of donor 1 (M) and donor 3 (N), and IGHV3-43D*01 C195A of donor 6 (O) show high quality of the allele-differentiating base calls. The analysed sequence is shown above each graph and the allele-differentiating base is highlighted within square brackets.
Fig. 4.
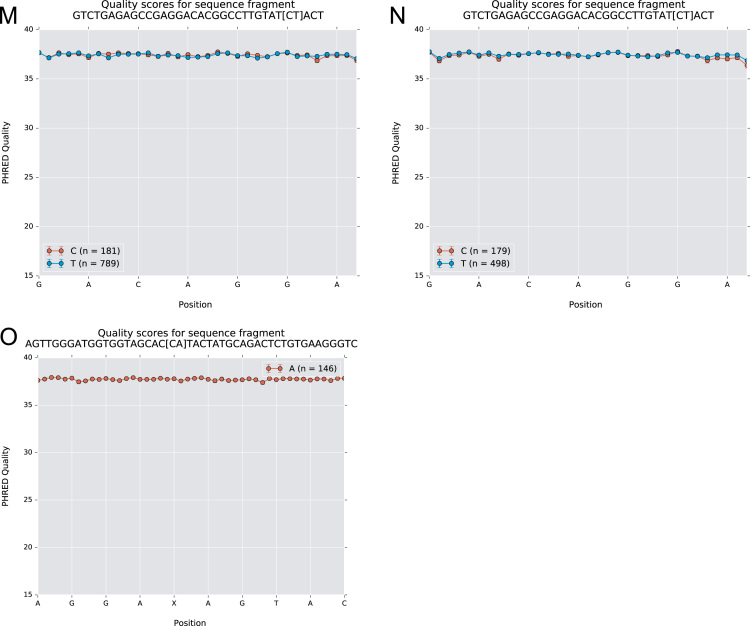
Perceived frequency of IGHJ gene usage in transcripts derived from donors 1–6, as analysed by IgDiscover.
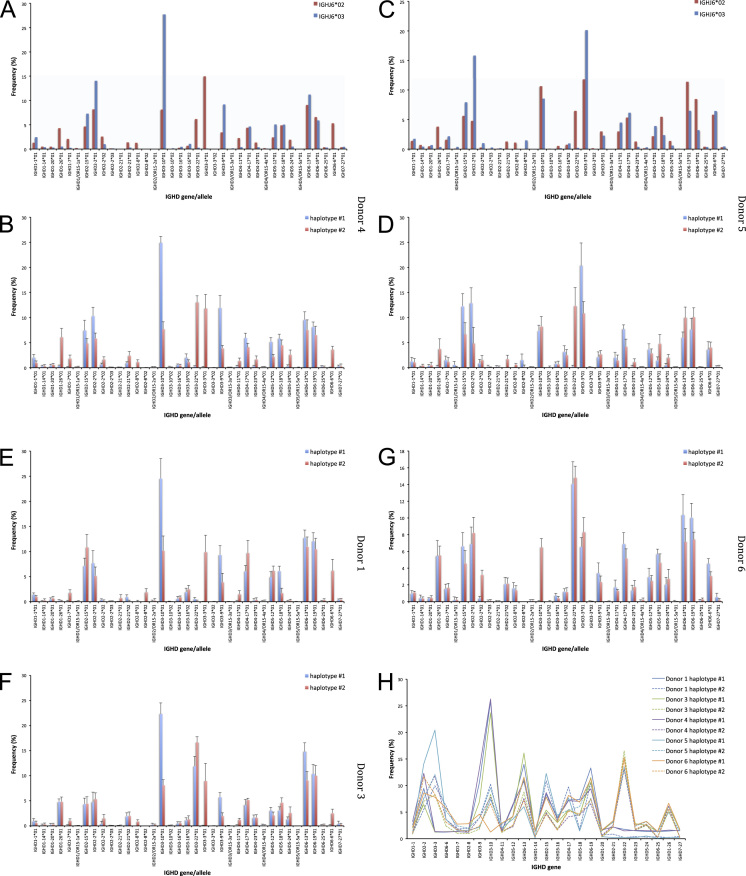
Fig. 5.
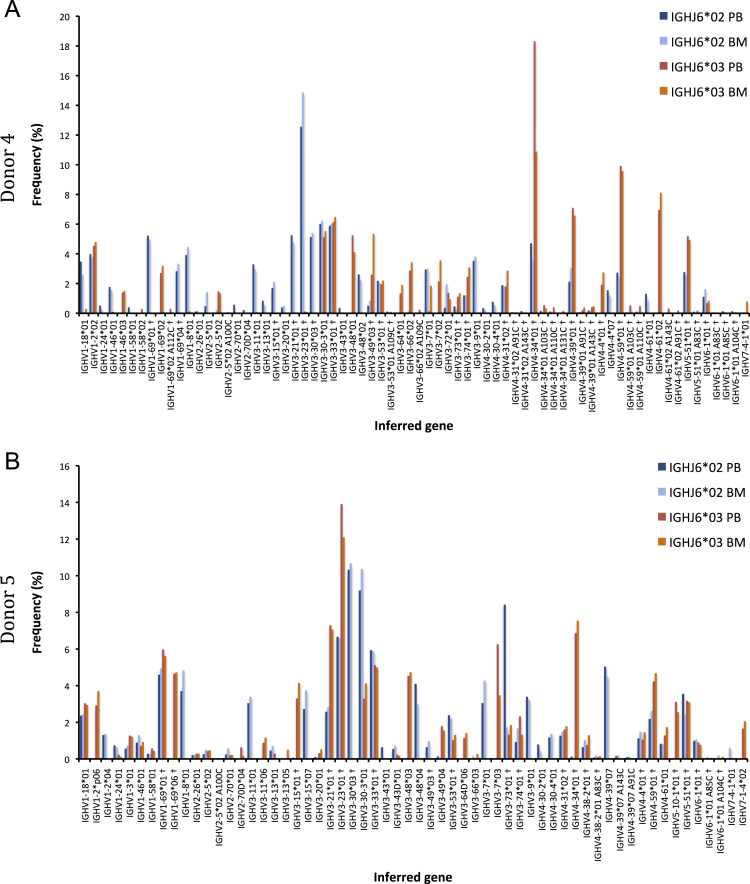
Summary of linkage of inferred germline genes/alleles of donors 4 (A) and 5 (B) to IGHJ6*02 and *03, as indicators of the donors’ two haplotypes, after analysis of transcripts found in bone marrow (BM) (also shown in Fig. 2 in Ref. [1]) and peripheral blood (PB). † defines that the name of only one of a set of different alleles of the gene that cannot be differentiated by the analysis approach is shown.
Fig. 6.
Association of IGHD gene expression IGHJ expression in unique IgM-encoding transcripts (at V_errors=0 and D_coverage>35 as defined by IgDiscover) derived from PB of donor 4 (A) and donor 5 (C). Association of IGHD gene expression (average±SD) to that of IGHV genes inferred as being present as two different alleles in transcripts derived from PB donors 1 (E), 3 (F), 4 (B), 5 (D), and 6 (G). A summary of IGHD gene usage (irrespective of allele call) based on association to expression of IGHV genes is shown (H).
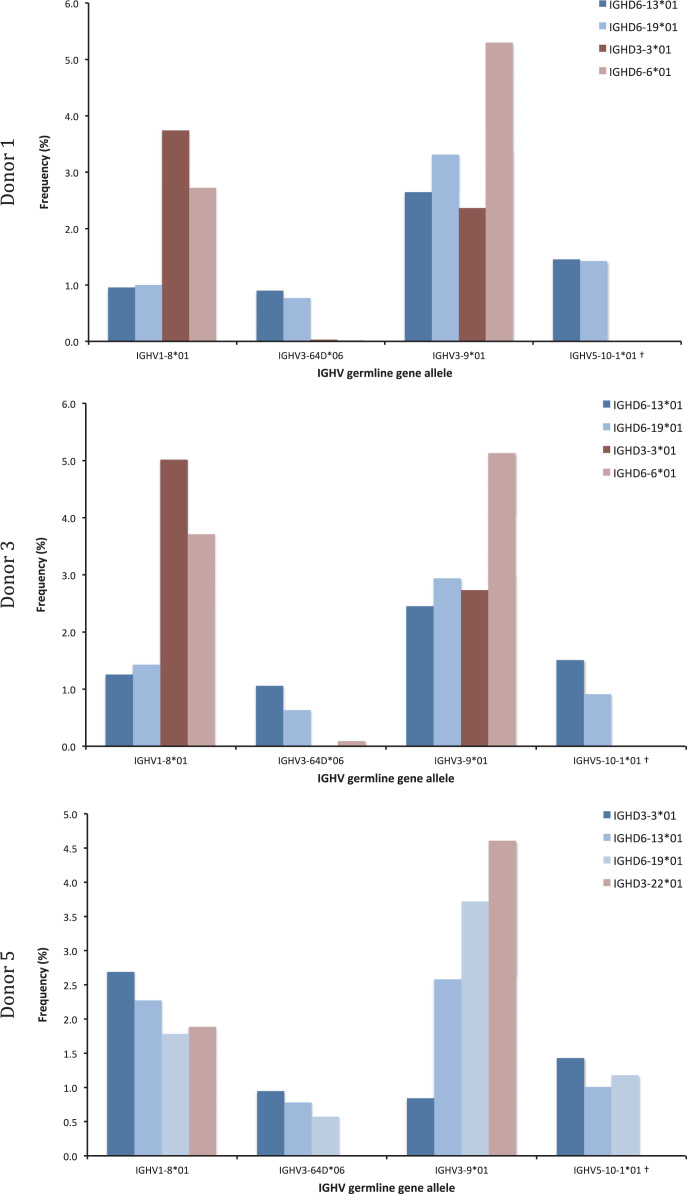
Fig. 7.
Linkage of IGHV1-8*01, IGHV3-64D*06, IGHV3-9*01, and IGHV5-10-1*01 to different IGHD genes in transcripts of donor 1, 3, and 5. While germline genes IGHV1-8*01 and IGHV3-9*01 were linked to the haplotype also carrying IGHD genes not present on both haplotypes, IGHV3-64D*06 and IGHV5-10-1*01 were not.
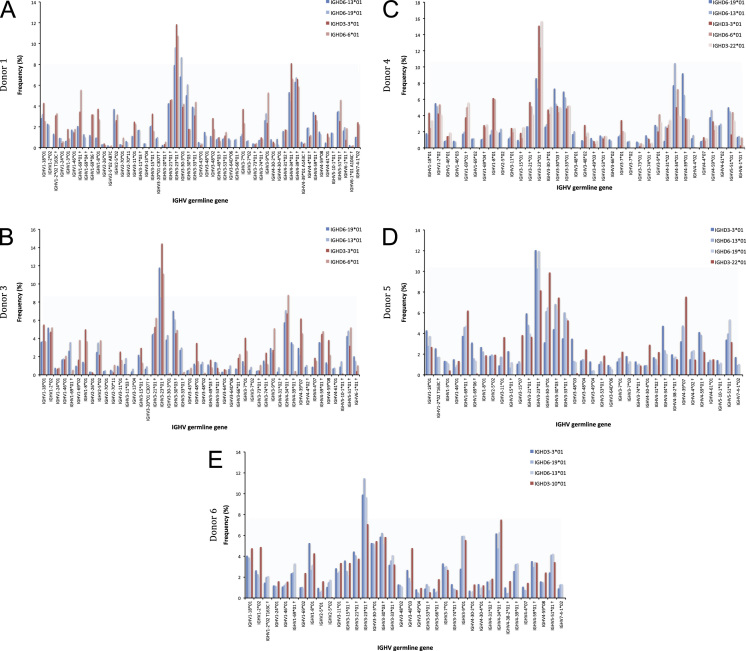
Fig. 8.
Association of IGHV genes/alleles of donors 1 (A), and 3–6 (B-E) with different IGHD genes as indicators of association with different haplotypes represented by IGHD. Analysis was performed on sequences found in cells of PB using the final filtered output of IgDiscover (diff=0). Only IGHV genes/alleles represented by at least 50 sequences with V_errors=0 and D_coverage>35 in the IGHD gene set shown in dark blue are shown. The frequencies of IGHV sequences associated to IGHD genes found in both haplotypes are shown in blue while the corresponding frequencies of IGHV sequences associated to IGHD genes expressed from only one of the inferred haplotypes are shown in red. † defines that the name of only one of a set of different alleles of the gene that cannot be differentiated by the analysis approach is shown.
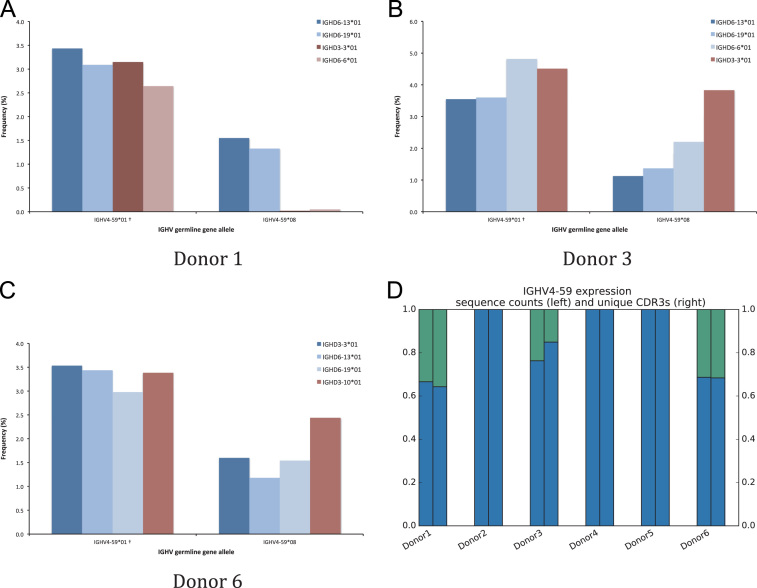
Fig. 9.
Differential association of inferred alleles of IGHV4-59 with different haplotypes of IGHD of donors 1 (A), 3 (B), and 6 (C). The frequencies of sequences associated to IGHD genes apparently expressed from both haplotypes are shown in blue while the frequencies of sequences associated to IGHD genes apparently expressed from only one of the haplotypes are shown in red. The fraction of reads represented by IGHV4-59*01 (blue) and *08 (green) in all three subjects is shown (fraction of sequences to the left and fraction of unique CDR3 to the right) (D). † defines that the name of only one of a set of different alleles of the gene that cannot be differentiated by the analysis approach is shown.
Fig. 10.
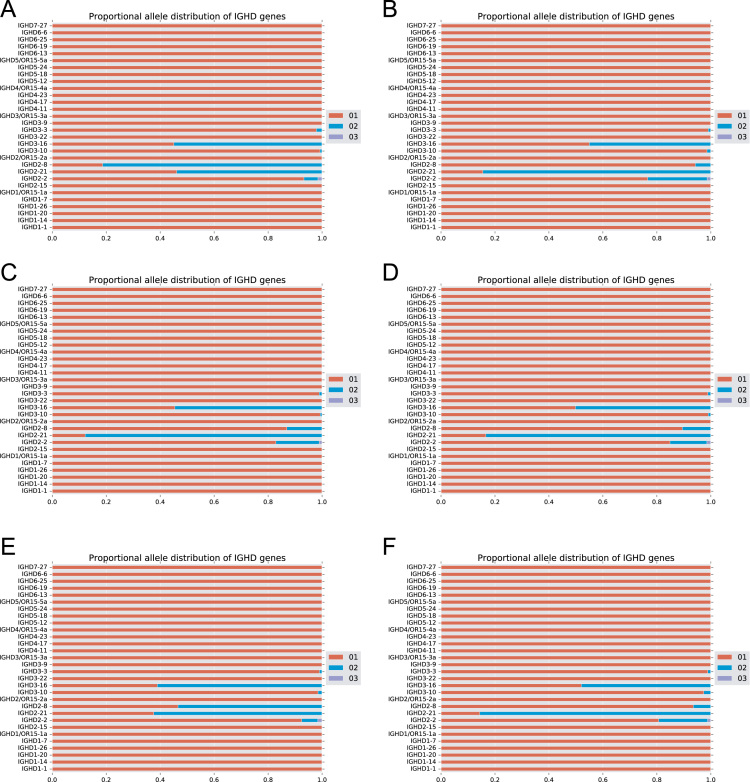
Apparent utilization of alleles of IGHD genes in IgM-encoding transcripts of BM of donors 1 (A), 2 (B), 3 (C), 4 (D), 5 (E), and 6 (F), as annotated by IgDiscover.
Fig. 11.
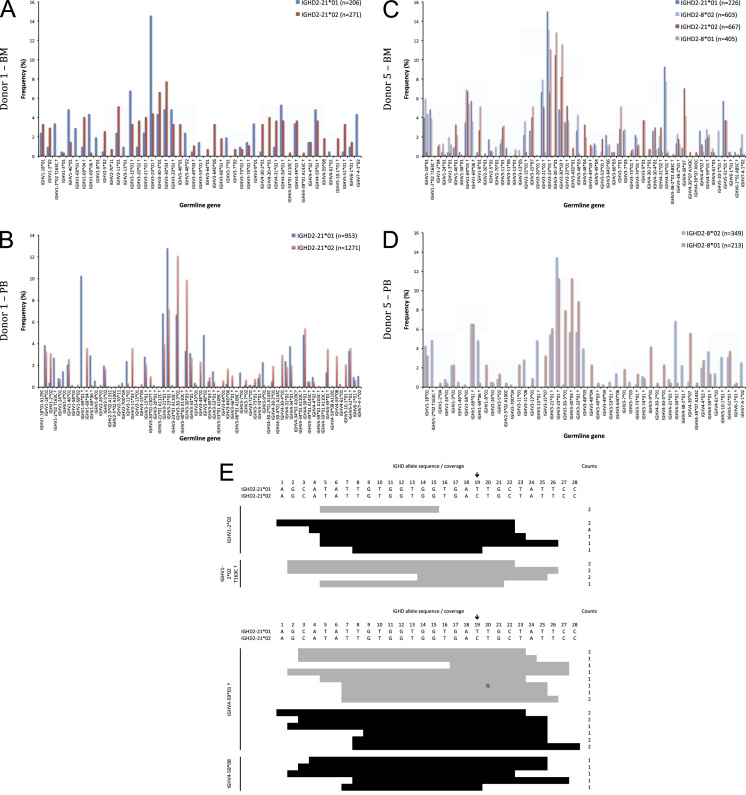
Immunoglobulin IGHV gene haplotype analysis based on heterozygous presence of IGHD alleles of donor 1 (A, B) and donor 5 (C, D). Transcripts found in BM (A, C) and PB (B, D) were analysed. The analysis of transcripts derived from PB employing IGHD2-21 was not included due to the low number of such sequences. Detailed sequence analysis (E) may be used to define whether or not IGHD allele assignments are appropriate. The rare association of reads of IGHV1-2*02 to IGHD2-21*01 (grey) instead of the expected IGHD2-21*02 (black) in some BM-derived transcripts of donor 1 (see A) does not cover the base within the IGHD that defines the individual alleles. IGHD2-21 allele calls for both alleles of IGHV4-59*01 include the allele-differentiating base, and rearrangements involving IGHV4-59*08 include the base identifying IGHD2-21*02. The arrow indicates the only base that differentiate IGHD2-21*01 and *02. Mutated bases within the sequences derived from IGHD genes are spelled out. † defines that the name of only one of a set of different alleles of the gene that cannot be differentiated by the analysis approach is shown.
2. Experimental design, materials and methods
IgM heavy chain variable domain-encoding gene repertoires were isolated by RT-PCR from transcriptomes of PB and BM collected out of season of most seasonal allergens from six allergic subjects [3]. Ethical approval and informed consent had been obtained from all donors. Sequencing was performed using the 2 × 300 bp MiSeq technology (Illumina, Inc., San Diego, CA, USA) at the National Genomics Infrastructure (SciLifeLab, Stockholm, Sweden) [3]. Details of sequence output and availability are outlined in Table 1. Data was pre-processed using pRESTO [4] and Change-O [5] as summarized in Fig. 1 in Ref. [1]. Germline gene inference was performed using TIgGER [6] and IgDiscover [7]. Additional bioinformatics analysis was performed as outlined elsewhere [1] including analysis performed using GIgGle (release 0.2) that is available under Apache License at https://github.com/ukirik/giggle. Immunoglobulin gene names and sequence numbering complies with the nomenclature defined by the International ImMunoGeneTics information system® (IMGT) (http://www.imgt.org) [8], [9].
Acknowledgements
The collection and analysis of the data set was supported by Lund University (ALF), the Swedish Research Council (Grant number 2016-01720), the Crafoord Foundation. Science for Life Laboratory, the Knut and Alice Wallenberg Foundation, the National Genomics Infrastructure funded by the Swedish Research Council, and Uppsala Multidisciplinary Center for Advanced Computational Science assisted with NGS and access to the UPPMAX computational infrastructure.
Footnotes
Transparency data associated with this article can be found in the online version at doi:10.1016/j.dib.2017.06.031.
Transparency document. Supplementary material
Supplementary material
.
References
- 1.Kirik U., Greiff L., Levander F., Ohlin M. Parallel antibody germline gene and haplotype analyses support the validity of immunoglobulin germline gene inference and discovery. Mol. Immunol. 2017;87:12–22. doi: 10.1016/j.molimm.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Kidd M.J., Jackson K.J., Boyd S.D., Collins A.M. DJ pairing during VDJ recombination shows positional biases that vary among individuals with differing IGHD locus immunogenotypes. J. Immunol. 2016;196:1158–1164. doi: 10.4049/jimmunol.1501401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levin M., Levander F., Palmason R., Greiff L., Ohlin M. Antibody-encoding repertoires of bone marrow and peripheral blood-a focus on IgE. J. Allergy Clin. Immunol. 2017;139:1026–1030. doi: 10.1016/j.jaci.2016.06.040. [DOI] [PubMed] [Google Scholar]
- 4.Vander Heiden J.A., Yaari G., Uduman M., Stern J.N., O׳Connor K.C., Hafler D.A., Vigneault F., Kleinstein S.H. pRESTO: a toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics. 2014;30:1930–1932. doi: 10.1093/bioinformatics/btu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta N.T., Vander Heiden J.A., Uduman M., Gadala-Maria D., Yaari G., Kleinstein S.H. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 2015;31:3356–3358. doi: 10.1093/bioinformatics/btv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gadala-Maria D., Yaari G., Uduman M., Kleinstein S.H. Automated analysis of high-throughput B-cell sequencing data reveals a high frequency of novel immunoglobulin V gene segment alleles. Proc. Natl. Acad. Sci. USA. 2015;112:E862–E870. doi: 10.1073/pnas.1417683112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corcoran M.M., Phad G.E., Nestor V.B., Stahl-Hennig C., Sumida N., Persson M.A., Martin M., Karlsson Hedestam G.B. Production of individualized V gene databases reveals high levels of immunoglobulin genetic diversity. Nat. Commun. 2016;7:13642. doi: 10.1038/ncomms13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lefranc M.P. IMGT unique numbering for the variable (V), constant (C), and groove (G) domains of IG, TR, MH, IgSF, and MhSF. Cold Spring Harb. Protoc. 2011;2011:633–642. doi: 10.1101/pdb.ip85. [DOI] [PubMed] [Google Scholar]
- 9.Lefranc M.P., CLASSIFICATION Axiom From I.M.G.T.-O.N.T.O.L.O.G.Y. to IMGT standardized gene and allele nomenclature: for immunoglobulins (IG) and T cell receptors (TR) Cold Spring Harb. Protoc. 2011;2011:627–632. doi: 10.1101/pdb.ip84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material