Summary
We studied the Drosophila circadian neural circuit using whole brain imaging in vivo. Five major groups of pacemaker neurons display synchronized molecular clocks, yet each exhibits a distinct phase of daily Ca2+ activation. Light and neuropeptide PDF from morning cells (s-LNv) together delay the phase of the evening (LNd) group by ~12 h; PDF alone delays the phase of the DN3 group, by ~17 h. Neuropeptide sNPF, released from s-LNv and LNd pacemakers, produces latenight Ca2+ activation in the DN1 group. The circuit also features negative feedback by PDF to truncate the s-LNv Ca2+ wave and terminate PDF release. Both PDF and sNPF suppress basal Ca2+ levels in target pacemakers with long durations by cell autonomous actions. Thus, light and neuropeptides act dynamically at distinct hubs of the circuit to produce multiple suppressive events that create the proper tempo and sequence of circadian pacemaker neuronal activities.
Keywords: Circadian physiology, Drosophila, calcium, neuropeptide, modulation
INTRODUCTION
Circadian rhythms in behavior, helping animals to adapt to environmental day-night cycles, are generated by small groups of pacemaker neurons in the brain (Herzog, 2007), many of which display self-sustained oscillations in both gene expression and neural activity (Welsh et al., 1995). These pacemakers are coupled together into a highly interactive network, from which unique network properties emerge, including the synchronization of molecular oscillations, the coordination of responses to external inputs, and the generation of multiple temporal outputs, such as circadian rhythms in locomotor activity, feeding, and sleep (Welsh et al., 2010; Hastings et al., 2014). The network mechanisms promoting the synchronization of molecular oscillations have been well characterized (Lin et al., 2004; Aton and Herzog, 2005; Li et al., 2014). Recently, several studies have focused on the coordination of pacemakers’ molecular rhythms with their neuronal properties and the coordination with the network properties of inputs and outputs (e.g., Coomans et al., 2015; Flourakis et al., 2015), yet these critical aspects of circadian neuronal oscillators remain incompletely understood. Using the Drosophila circadian network as a model system, here we study how the neural network processes external light inputs and how it generates sequential outputs during the course of the 24-h day.
The Drosophila circadian network contains ~150 pacemaker neurons organized into seven groups (Nitabach and Taghert, 2008). Previously we reported (Liang et al., 2016) that five major groups from this network became sequentially active at specific times of day (Figure 1A): for several groups, these times can be associated with their unique functions in behavioral outputs. The small lateral neuron ventral (s-LNv) group, previously associated with the morning peak of locomotor activity (Renn et al., 1999), is active around dawn (Liang et al., 2016) and is operationally termed a Morning (M) cell. Likewise, the large lateral neuron ventral (l-LNv) group, that serves an arousal function and promotes the waking state (Parisky et al., 2008; Shang et al., 2008, 2013) is active just following the s- LNv in the morning. Activity by the Evening (E)-cell group (including the lateral neuron dorsal (LNd) and the 5th s-LNv) was previously associated with the evening peak of locomotor activity (Grima et al., 2004; Stoleru et al., 2004): its neural activity anticipates dusk (Liang et al., 2016). Two additional groups, the dorsal neuron 3 (DN3) and the dorsal neuron 1 (DN1) display distinct activity bouts during the nighttime. Thus the network generates sequential neural outputs to control different behavioral rhythms throughout the day and night.
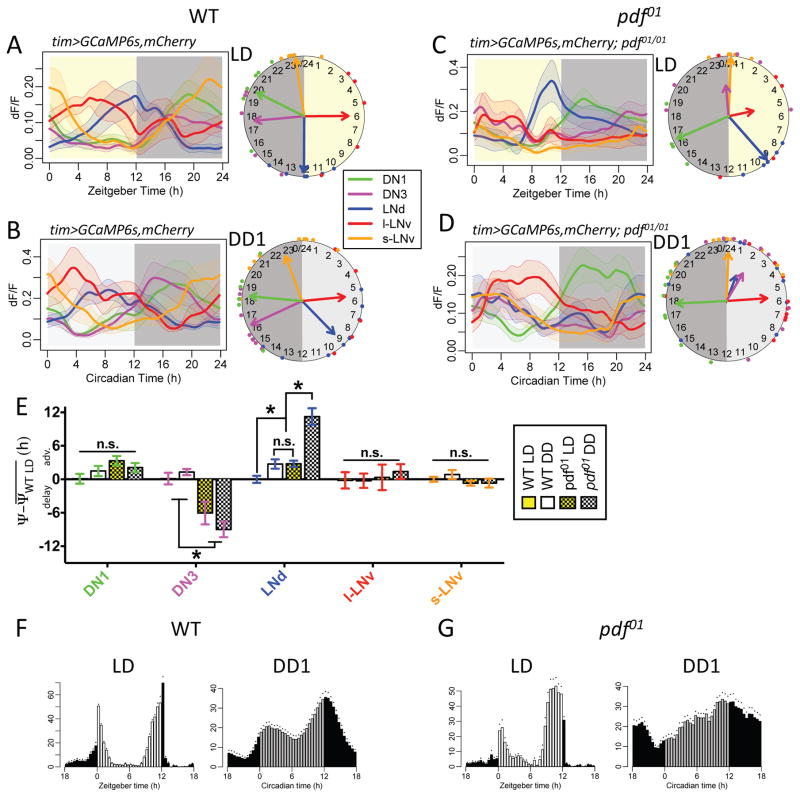
Figure 1. PDF and cyclic light-dark conditions phase-delay the Ca2+ rhythm of E-pacemaker LNd.
(A) Daily Ca2+ activity patterns of five major Drosophila circadian pacemaker groups in wild type (WT) flies under 12h light: 12h dark (LD) cycle (n = 6 flies). Left, average Ca2+ transients. Right, Ca2+ phase distribution: each colored dot represents calculated peak phase of one group in one fly; colored arrows are mean vectors for different groups; the arrow magnitude describes the Ca2+ phase coherence among different flies in a specific pacemaker group. Yellow aspect indicates 12 h period of light stimulation.
(B) Daily Ca2+ activity patterns in WT flies under constant darkness (DD) conditions (n = 12 flies). Darker gray aspect indicates subjective night.
(C) Daily Ca2+ activity patterns in pdf01 mutants under LD cycle (n = 5 flies).
(D) Daily Ca2+ activity patterns in pdf01 mutants under DD (n = 11 flies).
(E) Quantification of Ca2+ phase shifts described in panels (A–D). The mean phase of each group in WT controls under LD is set to zero (*p<0.01: Watson-Williams test). Error bars denote SEM.
(F) Average locomotor activity of WT flies (n = 16 flies) in LD cycles (averaged across 6 days) and in the first day under DD (DD1). Dots indicate SEM.
(G) Average locomotor activity of pdf01 mutants (n = 15 flies) in LD and DD1.
We previously reported that without neuropeptide pigment dispersing factor (PDF) receptor signaling, the daily neural activity patterns of the E-cell and DN3 groups became co-phasic with the M group around dawn (Liang et al., 2016). This observation suggested that such neuropeptide-mediated pacemaker interactions stagger neural activity patterns across the different groups in the circadian network. However several fundamental questions remain. By what mechanism does PDF signaling alter neural activity phases by several hours, and how does it promote different, group-specific phases? Also, the activity patterns of certain pacemaker groups (l-LNv and DN1) are strikingly indifferent to PDF signaling: do other neuropeptides released within the network contribute to setting their specific phasic activities? Interactions between pacemakers are not only crucial for generating multiple temporal outputs, but also for responding to external inputs. Light information can cell-autonomously adjust Drosophila molecular clocks through the internal photoreceptor Cryptochrome (CRY) (Ceriani et al., 1999; Emery et al., 2000) and non-cell-autonomously via pacemaker interactions (Cusumano et al., 2009; Guo et al., 2014). Yet, how such light information from different inputs eventually translates to multiple neural network outputs remains largely undefined.
To begin addressing these fundamental problems, here we investigate how the 24-h neural activity pattern of the entire Drosophila circadian network in vivo is regulated by light inputs and neuropeptides. We report three principal findings. First, PDF provides forward and feedback information within the network and that light cycles act in concert with PDF to provide the proper phase for E cell Ca2+ activity. Second, short light pulses rapidly phase-shift the 24-h neural activity patterns in all pacemaker groups, via both CRY and visual system pathways. Third, short Neuropeptide F (sNPF) is required for the nighttime phasing of the DN1 group. PDF and sNPF share phase-setting mechanisms by both suppressing Ca2+ activity in different pacemaker groups. Together, these results indicate that the Drosophila circadian pacemakers interact primarily by delay-based mechanisms, through light and neuropeptide-mediated inhibition. We propose this is a generalized process by which a neural network can variably produce sequentially-timed outputs.
RESULTS
Light inputs and PDF modulation converge to set Evening Cell Ca2+ phase
Previously we reported on pacemaker Ca2+ activities under constant conditions (DD: lacking external timing cues) (Liang et al., 2016). To consider the possible influence of light, we performed in vivo 24h Ca2+ imaging in a 12h light: 12h dark (LD) cycle. Wild type (WT) patterns of Ca2+ activity in the majority of pacemaker groups in LD were not altered, except in the E-cell group, LNd: its Ca2+ activity peak in LD was delayed by 3.8±0.6h (Figure 1AB), to the end the 12h photophase (ZT12). We next asked whether PDF is required for the delaying effect of light on LNd by comparing the pacemaker Ca2+ rhythms of pdf01 mutants under DD with those under LD cycles (Figure 1CD). Under DD and compared to WT controls, Ca2+ rhythms were phase-shifted in both LNd and DN3 of pdf01 mutants (Figure 1D). Both pacemaker groups became less consistent among animals and roughly co-active with the s-LNv around presumptive dawn, phenocopying Ca2+ patterns in pdfr mutants (Liang et al., 2016). However, under LD, pdf01 mutant LNd were selectively delayed compared to DD by 7.8±0.5h (Figure 1C–E). The results indicate that PDF/PDFR signaling is not required for the delaying effect of light on LNd Ca2+ activity.
The LNd group is operationally termed an Evening (E) cell group: in WT animals and as measured in DD, its Ca2+ activity peak is phase-locked to the evening behavioral peak (Liang et al., 2016). With the PDF-independent delay effect of light, under an LD cycle, the pdf01 mutant LNd peaked 2.7±0.5h earlier than WT, consistent with the 1–2h advanced evening behavioral phase of pdf/pdfr mutants (Renn et al., 1999; Hyun et al., 2005)(Figure 1FG). It is also worth noting that, for both WT and pdf mutants, LNd rhythms became more consistent across flies under cycling LD conditions than under DD (F-test for variances within genotypes, p<0.05). In contrast to the LNd group, the DN3 pacemaker group’s Ca2+ rhythms were not different between LD vs DD in either genotype (Figure 1A–D). Therefore, PDF>PDFR signaling alone sets the proper phase of DN3 Ca2+ rhythms, whereas PDF>PDFR signaling and light inputs together set the proper phase of LNd Ca2+ rhythms.
PDF receptor signaling alters Ca2+ rhythms in E cells and in M cells
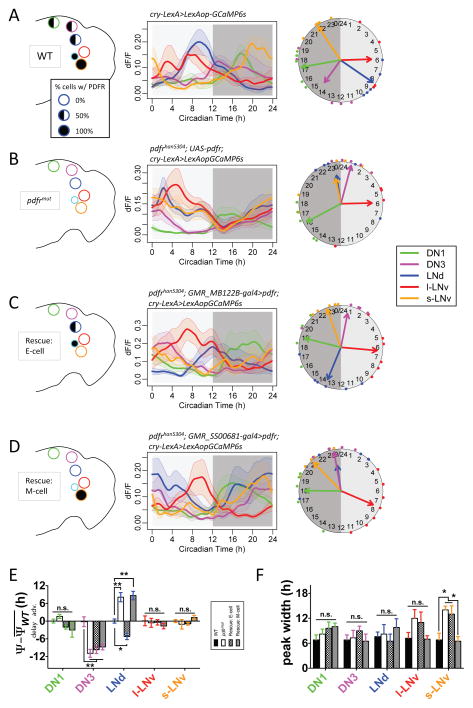
To define the mechanism by which neuropeptide PDF signaling sets Ca2+ phases, we first asked whether PDFR signaling acts cell autonomously in different pacemaker groups. We used the Gal4:UAS system to selectively express PDFR in alternate pacemaker subsets in the hypomorphic pdfr mutant han5304 (Hyun et al., 2005). In conjunction, we used cry-lexA>lexAop-GCaMP6s to independently express the calcium sensor throughout the pacemaker network. Using this lexA reporter system in both WT and the han5304 (Pdfr) mutant background, we recapitulated results we obtained previously with tim>GCaMP6s for WT and pdf01, respectively (Figure 2A and B). We first restricted PDFR rescue to a subset of E cells (the 5th sLNv and the three LNd, all four of which are normally CRY+/PDFR+) (Im and Taghert, 2010; Yao and Shafer, 2014)(Figure 2C and S1A). In this case, the LNd Ca2+ peaked 14.1±1.0h later than in the pdfr mutant (Figure 2B), and 5.2±1.0h later than in a control genotype (Figure 2B). These mosaic flies also displayed a later evening behavioral phase than in the pdfr mutant (Figure S1; cf. Lear et al., 2009). In this mosaic design, DN3 still lacked PDFR and did not display any rescue of the Ca2+ rhythm phase. In contrast, when we restricted PDF receptors to M cells only (s-LNv), the distribution of Ca2+ phases among the different pacemaker groups was the same as in the pdfr mutant (Figure 2D and S1B). However, the width of the Ca2+ wave in the s-LNv was longer in the pdfrmut, and also longer in the ‘Rescue: E cell’, than in the WT (Figure 2F). When PDFR expression was restored to the s-LNv (‘Rescue: M-cell’), the width of the s-LNv wave was shortened and no longer different from the WT value (Figure 2F). This result suggested that PDFR functions in s-LNv to help terminate the period of Ca2+ activation in s- LNv in cell-autonomous fashion. Collectively, these PDFR different rescues displayed distinct actions on Ca2+ activities in E versus in M cells: in the forward direction (i.e., to E cells), it delays the Ca2+ phase; in the feedback direction, (i.e., to M cells), it helps terminate the Ca2+ peak.
Figure 2. PDFR signaling regulates Ca2+ rhythms in M and E pacemakers by cell autonomous mechanisms.
(A–D) Daily Ca2+ activity patterns of five major pacemaker groups under DD (A) in WT flies (n = 5 flies), (B) in pdfr mutants (n = 6 flies), (C) in an E cell rescue design - wherein E pacemakers (three out of six LNd and the 5th s-LNv) express PDFR in the pdfr mutant background (n = 6 flies), and (D) in an M cell rescue – wherein flies with M pacemakers (four PDF-positive sLNv) express PDFR in the pdfr mutant background (n = 8 flies). Left, schematics of PDFR expression patterns in major pacemaker groups. Filled circles indicate all cells in the group express PDFR. Half circles indicate approximately half of cells in the group express PDFR. Middle, average Ca2+ transients. Right, Ca2+ phase distributions.
(E) Quantification of Ca2+ phase shifts described in panels (A–D). The mean phase of each group in WT controls under DD is set to zero (*p<0.05, **p<0.01, and n.s. - not significant: Watson-Williams test). Error bars denote SEM.
(F) Quantification of peak widths (the full width at half maximum) for Ca2+ transients in all groups and conditions (*p<0.05: Two-way ANOVA, followed by Bonferroni post hoc test).
Pharmacological tests of PDF-mediated suppression of Ca2+ in vivo
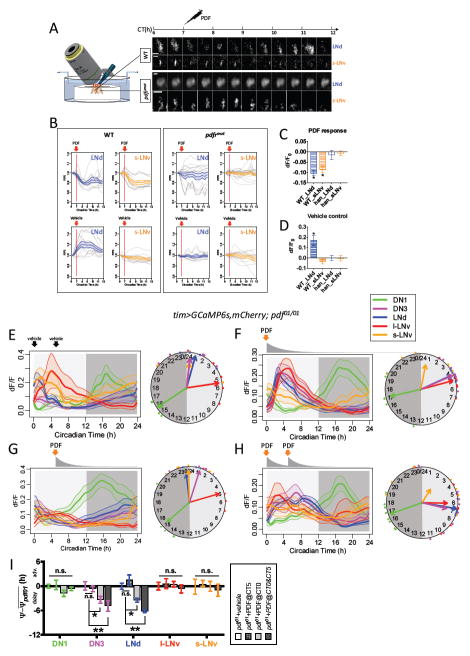
PDFR signaling in M vs. in E pacemakers may reflect a common mechanism - suppression of Ca2+ levels. To test this hypothesis, we applied synthetic PDF while imaging in vivo. We compared pacemaker responses between WT and pdfr (han5304) mutant genotypes by imaging them concurrently, as yoked pairs, to reduce measurement variance derived from non-genetic factors (Figure 3A). We restricted the GCaMP6s sensor expression to s-LNv and three out of six LNd with pdfr(B)-GAL4 (Im and Taghert, 2010). WT s-LNv and LNd both responded to PDF by 8.7±1.5% and 10.4±1.0% peak reductions in Ca2+ signals respectively (Figure 3B): these reductions developed over many hours. The net reduction of Ca2+ levels in WT LNd was ~25.5% compared to the vehicle control, as we performed the experiments during the normal Ca2+ peak of LNd (CT6-12) (Figure 3CD). In pdfr mutants, s- LNv and LNd showed no significant responses to the same applications of PDF. Together these results demonstrate PDF>PDFR signaling potently suppresses basal Ca2+ levels in both E cells (LNd) and M cells (s-LNv).
Figure 3. Synthetic PDF application suppresses and/or delays Ca2+ activity in vivo.
(A) Left, schematic illustrating yoked pairs of WT and pdfr mutant flies for pharmacological tests. Right, representative images of LNd and s-LNv pacemakers in such Drosophila pairs responding differentially to 10−05M synthetic PDF. Axis above denotes the circadian time of recordings and the CT7 time point of synthetic PDF application.
(B) Averaged Ca2+ transients of M pacemakers (s-LNv, orange) and E pacemakers (LNd, blue) responding to (top) PDF or (bottom) vehicle from (left) WT or (right) pdfr mutant flies. Gray traces represent individual cells in trials (PDF: n = 5 pairs; vehicle: n= 4 pairs).
(C–D) Ca2+ signal changes by (C) PDF treatment or (D) vehicle treatment measured at the point that WT pacemakers displayed a maximal reduction in response to PDF (*p<0.01: single-sample t-test). The average increase in Ca2+ signals in WT LNd with vehicle treatment represents their normal daily Ca2+ peak that occurs during these recording periods (cf. Figure 1C).
(E) Ca2+ activity responses in pdf01 mutants to application of vehicle saline at the time indicated by vertical arrows under DD (n = 6 flies).
(F) Ca2+ activity responses in pdf01 mutants to single synthetic PDF application under DD (n = 5 flies). PDF was present in the initial saline bath as indicated by the vertical orange arrow at a peak concentration of 10−5 M. This initial time point represents the peak time of Ca2+ in s-LNv (CT0 – orange arrow). PDF was slowly washed out by perfusion (gray curve below the orange arrow denotes PDF concentration).
(G) Ca2+ activity responses in pdf01 mutants to single synthetic PDF application at CT5 under DD (n = 6 flies).
(H) Ca2+ activity responses to two serial applications of synthetic PDF in pdf01 mutants under DD (n = 6 flies). The first dose was at CT0 (as in panel F), and the second at CT5 (~ at the l-LNv Ca2+ peak time) both denoted by vertical orange arrows; constant perfusion followed each application.
(I) Quantification of Ca2+ phase shifts produced by synthetic PDF; the mean phase of each group in pdf01 mutants under DD is set to zero (*p<0.05: Watson-Williams test).
We next asked whether suppression of Ca2+ levels by PDF>PDFR signaling is sufficient to delay the daily Ca2+ phases in LNd and DN3 by applying synthetic PDF, while imaging pdf mutants in vivo. PDF applied at the Ca2+ peak time (CT0) of the s-LNv delayed Ca2+ phases in both LNd by 3.4±0.5h and DN3 by 3.3±0.8h (Figure 3EF). Two serial applications of synthetic PDF at CT0 and at CT5 (at the Ca2+ peak time of the l-LNv PDF-releasing group) further delayed Ca2+ phases by a total of 6.2±0.3h (LNd) and by 4.9±1.3h (DN3) (Figure 3E–I). In contrast, a single application of PDF at CT5, after the peaks in LNd and DN3, did not induce delays in Ca2+ activity peaks in either group (Figure 3G). These results demonstrate that PDF can in principle affect Ca2+ phases in LNd and DN3 in vivo by delaying the onset of their incipient, clock-driven Ca2+activations.
Ectopic PDFR confers sensitivity to the phase-delaying effects of PDF
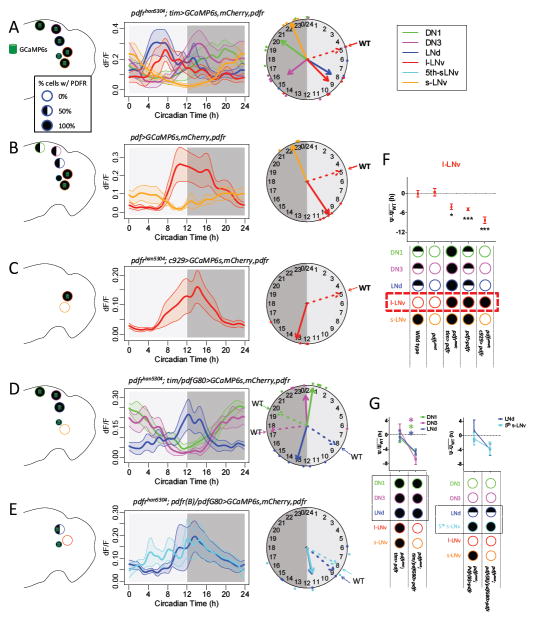
We also tested the cell autonomous effects of PDFR signaling on pacemaker Ca2+ activity with gain of function experiments. The l-LNv pacemaker group is not responsive to PDF, but becomes responsive with ectopic receptor expression (Shafer et al., 2008). Consistent with its lack of pharmacological sensitivity, the normal l-LNv Ca2+ phase is not altered in either the pdfr (Liang et al., 2016) or the pdf mutants (Figure 1B). When we overexpressed PDFR in all pacemaker neurons in the pdfr mutant background (thereby including the l-LNv), the phases of Ca2+ activity patterns that were aberrant in the mutant backgrounds (those of the LNd and DN3) were fully restored to their normal values. In addition, this experiment produced a 4.2±1.3h delay in the phase of l-LNv (Figure 4A). To test whether this PDFR-dependent delay in l-LNv Ca2+ was cell autonomous, we restricted ectopic PDFR expression within the pacemaker network to only the l-LNv in an otherwise WT background (pdf>pdfr, Figure 4B) and separately in a pdfr mutant background (han; c929>pdfr, Figure 4C). In both cases, ectopic PDFR expression in l-LNv delayed its Ca2+ phase: by 4.9±0.4h (compared to WT) and by 8.7±1.0h (compared to the pdfr mutant) (Figure 4F). The increased delay of the Ca2+ phase in the l-LNv in the han; c929>pdfr compared to that in the pdf>pdfr genotype is likely due to the lack of PDFR in the s-LNv. As described above, PDFR in the s-LNv mediates negative feedback to help terminate activity in that cell, hence a longer period of s-LNv activity produces greater delay in the l-LNv phase. In sum, gain-of-function genetic experiments support the hypothesis that PDFR signaling normally delays Ca2+ phases in diverse pacemakers by a cell-autonomous mechanism.
Figure 4. PDFR signaling delays Ca2+ activation in diverse pacemakers.
(A–E) Daily Ca2+ activity patterns of five major pacemaker groups under DD (A) in flies with all pacemakers over-expressing PDFR in pdfr mutants (n = 6 flies); (B) in flies with PDF-positive neurons (s-LNv and l-LNv) over-expressing PDFR in an otherwise WT background (n = 5 flies); (C) in flies with l-LNv over-expressing PDFR in pdfr mutants (n = 6 flies); (D) in flies with l-LNv alone over-expressing PDFR (by c929-gal4) in pdfr mutants (n = 4 flies); (D) in flies with all pacemakers except PDF-positive neurons over-expressing PDFR in pdfr mutants (n = 6 flies); and (E) in flies with E pacemakers (three out of six LNd and the 5th s- LNv) expressing PDFR in pdfr mutants (n = 5 flies). Dashed arrows on the clock face indicate the mean phases of those groups in WT flies (cf. Figure 1A).
(F) Quantification of l-LNv Ca2+ phase shifts described in panels (A–C). The mean phase of each group in WT controls under DD is set to zero (cf. Figure 1C; *p<0.05, ***p<0.001: Watson-Williams test).
(G) Quantification of Ca2+ phase shifts in PDF-negative, PDFR-positive pacemakers described in panels (D–E). The mean phase of each group in WT controls under DD is set to zero (cf. Figure 1C; the colors of the asterisks correspond to the cognate pacemaker groups; *p<0.001: Watson-Williams test).
PDFR signaling in M cells provides negative feedback
Based on the sensitivity of the Ca2+ wave duration in the s-LNv to cell autonomous pdfr signaling (Figure 2D&F), we speculated that a shorter period of Ca2+ activation would diminish neuropeptide PDF release and hence, diminish PDF signaling. To further examine this possibility, we used pdf- Gal80 to restrict tim>pdfr overexpression to only non-M pacemakers. Consistent with the predictions of the hypothesis (lack of M cell PDFR extends the duration of PDF cell signaling), we observed significantly delayed Ca2+ phases in DN1, DN3, and LNd, by an average of 4.5±0.6h (Figure 4D), as well as delayed evening behavioral phases (Figure S2). Independently, we observed similar E cell Ca2+ phase delays (3.8±1.1h) when PDFR expression was restricted to only a subset of E cells (PDFR-positive LNd and the 5th s-LNv) (Figure 4E&G). Collectively, these results support the hypothesis that PDFR signaling delays Ca2+ activations in non-M cells and, by negative feedback, terminates Ca2+ activation in M cells.
Light pulses rapidly phase-shift the Ca2+ rhythms
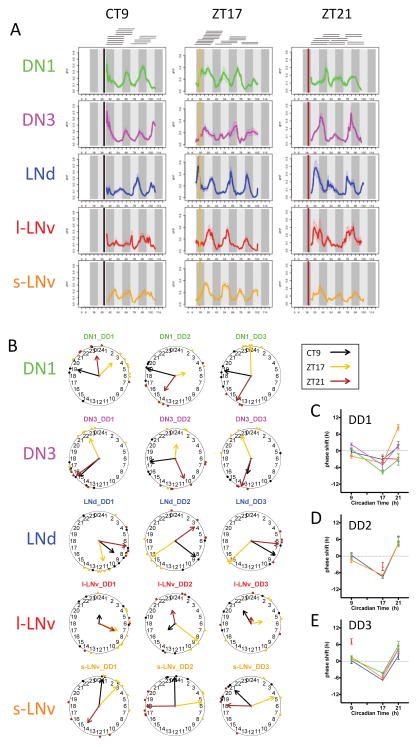
We further probed pacemaker interactions with the environment, we delivered light pulse stimuli at the phase-delaying zone (ZT17) or the phase-advancing zone (ZT21), and also in DD1 at the dead zone (CT9) (Figure S3). Before, or at different times after the light pulses, we performed 24h Ca2+ imaging sessions and tiled them together to unify the records across a three-day post-stimulus period (Figure 5AB & Figure S3B). A dead zone light pulse (CT9) did not phase-shift pacemaker Ca2+ rhythms (Figure 5B–E). In contrast, phase-delaying light pulses delayed Ca2+ rhythms in all pacemakers within the first circadian cycle (Figure 5C). During the second day, LNd and s-LNv further delayed their Ca2+ phases, while DN1 and DN3 became less coherent (Figure 5B&D). By the third day, the pacemaker network re-established a normal phase pattern with an overall 5.5±0.6h delay, compared to the pattern in flies that received no light pulse (Figure 5E). With phase-advancing light pulses, only the s-LNv responded during the first day post-stimulus, with a 8.5±1.0h phase advance (Figure 5C). During the second day, the s-LNv Ca2+ phase advance became smaller (4.6±0.8h) and all other pacemaker groups now displayed in Ca2+ phases advances that were roughly similar (5.3±0.4h) (Figure 5D); this pattern was maintained into the third day (Figure 5E). Together, these results show that the circadian pacemaker network responds rapidly to short light pulses, especially for phase delays.
Figure 5. Light pulses phase-shift circadian pacemaker Ca2+ rhythms in vivo.
(A) Averaged Ca2+ transients in the five circadian pacemaker groups in the three days following 15 min light pulses delivered either in the dead zone (CT9), or in the phase-delay zone (ZT17), or in the phase-advance zone (ZT21). Bars indicate the time of light pulses. Horizontal lines on top indicate separate 24 h imaging sessions for individual flies that were tiled to synthesize three-day patterns (CT9: n = 14 flies; ZT17: n = 18 flies; ZT21: n = 14 flies).
(B) Ca2+ phase distributions of five circadian pacemaker groups in three circadian cycles immediately following the three different light pulse stimuli: CT9, ZT17, and ZT21.
(C–E) Ca2+ phase response curves (PRC) plotted over the course of the three circadian cycles: (C) day one, (D) day two, and (E) day three. The Ca2+ phase shifts compared to Ca2+ phases in unstimulated flies (from Figure 1A) after three different light pulse stimuli. Phase-shifts that lacked coherence (p>0.05: Rayleigh test) were excluded.
Light inputs via CRY and PDF
We further investigated the mechanism for light inputs to regulate Ca2+ activity patterns of circadian pacemakers. The light-input pathways include the intracellular photoreceptor CRY and the visual systems (perhaps relayed by PDF signals). To examine these two pathways, we repeated the tests in cry null mutants and in pdf mutants independently, during the first post-stimulus cycle.
We previously found that in DD, the Ca2+ activity patterns of circadian pacemakers in cry nulls were not different from those of WT flies (Liang et al., 2016). However, phase-delaying light pulses in cry mutants failed to induce any Ca2+ phase shifts during the first circadian cycle (Figure S4A). In contrast phase-advancing light stimuli shifted cry mutant s- LNv, albeit by only half as much as WT controls (Figure S4B). In contrast, pacemaker groups in pdf null mutant flies responded rapidly to both phase-delaying and phase-advancing light pulses in the first circadian cycle. Of note, the phase-delaying light pulse caused a much larger Ca2+ phase shift in pdf mutant s-LNv (12.4±1.0h) than controls (4.4±0.6h) (Figure S4CD). Consistent with the poor rhythmicity in behavior under DD (Table S1), pdf null mutant flies had generally poorer coherence in Ca2+ phases among mutant flies than did control flies. These results indicate that the normal pattern of light induced Ca2+ phase shifts is partially modulated by PDF signaling.
sNPF modulation sets DN1 Ca2+ phase by suppressing daytime DN1 Ca2+ levels
PDF signaling is not required for the proper Ca2+ phases of the l-LNv or the DN1 (Liang et al., 2016 and see Figure 1). We therefore screened for the potential involvement of other neuropeptides (and cognate their receptors) that are known to be expressed by subsets of pacemaker neurons, including short neuropeptide F (sNPF - Johard et al., 2009), diuretic hormone 31 (DH31 - Kunst et al., 2014), ion transport peptide (ITP - Johard et al., 2009), and neuropeptide F (NPF - Hermann et al., 2012). Manipulation of DH31 signaling suggested possible changes: a strongly hypomorphic mutation of DH31 and the knockdown of one of the DH31 receptors (CG17415 – Johnson et al., 2005) displayed a trend toward lower coherence of l-LNv Ca2+ activity among different flies (p=0.16 (DH31) and p=0.38 (CG17415): Rayleigh test; Figure S5). However, neither the amplitudes of daily Ca2+ fluctuations (p>0.5: t-test) nor the mean Ca2+ phases (p>0.5: Watson-Williams test) were significantly altered compared to WT controls. Likewise, the expression of RNAi constructs of itp, npf, npfr did not cause major alterations in the daily Ca2+ activity patterns in the network or of locomotor activity rhythms (Figure S5; Table S1).
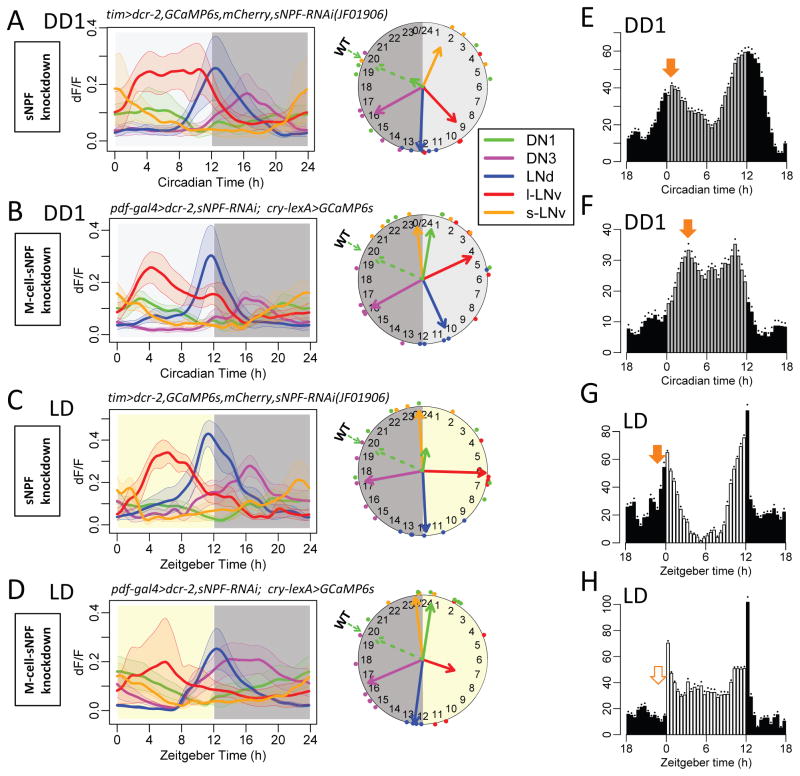
In contrast, we observed complete arrhythmicity in Ca2+ activity of the DN1 group following RNAi-knock down of the neuropeptide sNPF within the pacemaker network (tim>sNPF RNAi - Figure 6A). The amplitudes of Ca2+ fluctuations in DN1 were reduced (t-test, p<0.05) and coherence among different flies was lost (Rayleigh test, p=0.81). However, this manipulation did not alter the pacing of molecular clocks in DN1 cells (Figure S6A). RNAi knockdown of the sNPF receptor, using tim-Gal4, produced a very similar outcome (Figure S6D). These congruent effects suggested DN1 Ca2+ rhythms normally require neuropeptide sNPF signaling. Since sNPF was knocked down by an RNAi that targeted its 3′UTR sequence, we tested genetic specificity with a rescue experiment that overexpressed the sNPF coding sequence in the sNPF knockdown background (Figure S6E). In these flies, the phase and coherence of the Ca2+ activity in DN1 were fully rescued.
Figure 6. sNPF is required for DN1 Ca2+ rhythms in vivo.
(A) Daily Ca2+ activity patterns in tim > sNPF RNAi flies (sNPF knockdown) under DD conditions (n = 5 flies). Dashed arrows on the clock face indicate the mean phases of DN1 in WT flies (cf. Figure 1C). DN1 Ca2+ activity displayed poor phase coherence among flies (p>0.1: Rayleigh test).
(B) Daily Ca2+ activity patterns in flies with sNPF knockdown in M pacemakers, pdf > sNPF RNAi under DD conditions (n = 5 flies). DN1 Ca2+ activity was rhythmic but phase-shifted compared to WT controls (p<0.01: Watson-Williams test).
(C–D) Daily Ca2+ activity patterns under LD cycles in (C) tim > sNPF RNAi flies (n = 7 flies) and (C) pdf > sNPF RNAi flies (n = 6 flies).
(E–F) Average locomotor activity in the first day under DD of (E) tim > sNPF RNAi flies (n = 15 flies) and (F) pdf > sNPF RNAi flies (n = 32 flies). Dots indicate SEM. See Figure 1F for a comparison to locomotor profiles in a control (WT) genotype.
(G–H) Average locomotor activity in LD cycles (averaged across 6 days) of (G) tim > sNPF RNAi flies and (H) pdf > sNPF RNAi flies. See Figure 1F for a comparison to locomotor profiles in a control (WT) genotype.
Within the tim-GAL4 pattern of pacemakers, sNPF is normally expressed in both M (four s-LNv) and E (two of six LNd) cells (Johard et al., 2009). We therefore asked whether sNPF released by M versus E groups had different functions. We selectively knocked down sNPF in M cells and found surprisingly, that Ca2+ activity in DN1 was rhythmic and coherent, but now with a peak at dawn, co-phasic with M cells (Figure 6B). We observed this same effect under cyclic LD conditions (Figure 6CD). Together these results suggest that E-cell derived sNPF is sufficient for rhythmic Ca2+ in DN1, but is not sufficient for its proper Ca2+ phase.
When sNPF or sNPFR was ‘knocked-down’ in all pacemakers, we observed a slight increase of nighttime locomotor activity under LD (Figure 6EG, compared to WT in Figure 1F; Figure S6B; Table S1). However, when sNPF knockdown was restricted to M-cells (Pdf-cells), behavioral activity patterns were substantially altered (Figure 6FH, compared to WT in Figure 1F; Figure S6C). These flies lost morning anticipation under LD and displayed a phase-delayed morning peak within the first DD cycle. Therefore, M-cell specific knockdown of sNPF produces a strong behavioral phenotype correlated with the phase shift it produced in the DN1 Ca2+ activity.
When the sNPF knockdown was restricted to E cells, the DN1 Ca2+ rhythm was not different from control, and locomotor behavior likewise displayed no alterations (Figure S6F), suggesting that M-cell sNPF is sufficient to support rhythmic DN1 activity when E-cell sNPF is missing. Impairing sNPF or PDF signals from s-LNv caused the Ca2+ activity of DN1 in the case of lost sNPF, or LNd and DN3 in the case of lost PDF, to become co-phasic with s- LNv around dawn. When PDF neurons (s-LNv and l-LNv) were ablated by expressing the cell death genes (hid and rpr) (Renn et al., 1999), LNd and DN3 were phase-shifted to a morning peak, while DN1 still displayed a normal activity pattern, peaking at nighttime and remaining anti-phasic with LNd (Figure S7). We speculate that when LNd became active around dawn in this genotype, they could replace the role of s-LNv to set DN1 activity at the nighttime. Together these four sets of results suggest that sNPF from both M cells and E cells produces the normal Ca2+ activity pattern and behavioral output of DN1.
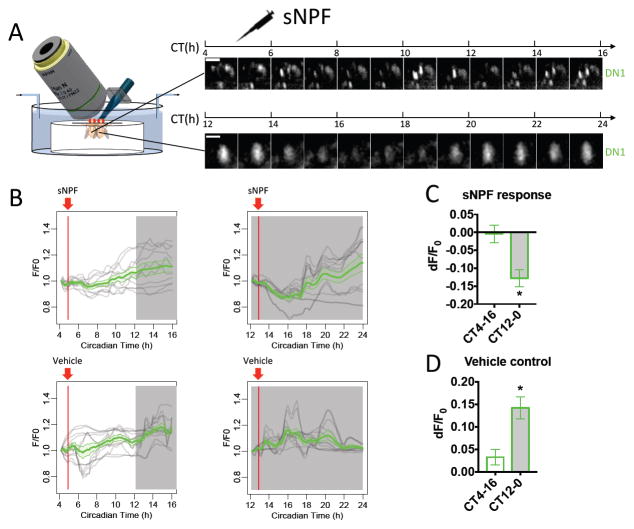
Pharmacological Tests of sNPF-Mediated Suppression in vivo
We hypothesized that the normal circadian function of sNPF, by analogy to that of PDF (Figure 3), is to suppress DN1 Ca2+ activity during the daytime, sequentially by M cells and then by E cells. To test this hypothesis, we pharmacologically applied sNPF onto WT brains during 12 hr episodes of Ca2+ imaging (Figure 7A). We again employed a design of ‘yoked’ fly pairs, now simultaneously testing flies that previously entrained to LD schedules with an 8h phase difference. Therefore, one fly of each pair began the recording session at CT4 and the other at CT12. CT4 pacemakers did not exhibit Ca2+ signal changes in response to single applications of synthetic sNPF (we propose this is because the intrinsic Ca2+ activity in DN1 was low due to intrinsic sNPF signaling at this time). Conversely, pacemakers staged to begin testing at CT12 exhibited a 12.8±2.3% reduction in Ca2+ signals in response to synthetic sNPF exposure, and they subsequently recovered to a higher level (Figure 7B–D). The net reduction of Ca2+ signal in DN1 pacemakers from CT12 was ~27.0%, compared to the vehicle control. These results demonstrated that sNPF can suppress the Ca2+ activity of DN1 in vivo.
Figure 7. sNPF suppresses DN1 Ca2+ activity.
(A–D) As in Figure 3A–D, yoked pairs of WT flies entrained to different circadian time (PDF: n = 3 pairs; vehicle: n= 3 pairs). DN1 Ca2+ signals from flies in subjective night were reduced by 10−05M synthetic sNPF application. The nighttime DN1 Ca2+ signals increased with vehicle treatment, reflecting their normal peak phase (cf. Figure 1C).
DISCUSSION
Each day, neuronal activity patterns in the Drosophila pacemaker network proceed sequentially in a wave from lateral to dorsal aspects of the brain: the relative phases are such that each pacemaker group occupies a distinct temporal niche (Liang et al., 2016). To understand how this 24-h neural activity pattern is organized and regulated, we tested many of the known interactions between pacemakers. We found that light and the neuropeptides PDF and sNPF are involved. PDF released from M cells affects the sequence in two ways: it terminates Ca2+ activity in M cells, and it delays Ca2+ activations in E cells and DN3 cells away from a morning phase. The proper phase of the E cells is in fact the product of delay from both PDF and light signals. sNPF, released from M and E cells, is required for Ca2+ rhythms in another pacemaker group, the DN1. We also found that light inputs arriving through both the internal photoreceptor CRYPTOCHROME and via the visual systems rapidly phase-shift Ca2+ waves. Therefore, neuropeptides and light coordinately produce sequentially-timed outputs from a network of otherwise synchronous molecular pacemakers.
Circadian pacemakers are coupled by neuropeptide-meditated inhibitions
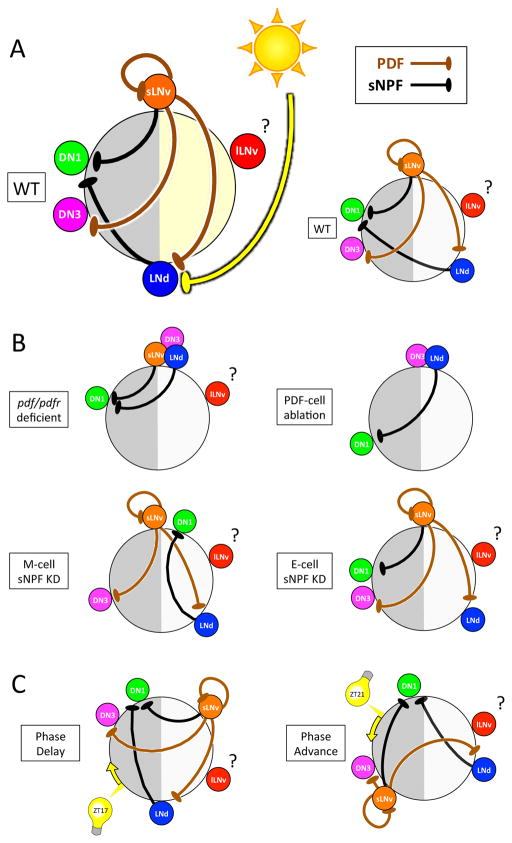
How can synchronous molecular clocks generate a pattern of sequential activities? We identified two neuropeptides, PDF and sNPF, that mediate much of the critical interactions between pacemaker groups to set the diverse Ca2+ activity phases (Figure 8AB). First, we suggest that neuropeptide PDF delays the Ca2+ activity peaks of the E cells (LNd/5th s-LNv) and DN3 groups, away from a morning phase they would otherwise share with the s-LNv. This model is based on four lines of evidence: (i) in both pdf− and pdfr− mutants, Ca2+ activation in E cells and in DN3 were co-active at prospective dawn (Figure 1D & 2B); (ii) restoring pdfr expression selectively to the LNd E cell-group autonomously delayed (rescued) their Ca2+ activation to an evening phase, but produced no rescue on the DN3 phase (Figure 2C); (iii) ectopic PDFR by l-LNv delayed its Ca2+ activation (Figure 4A–C); (iv) acute administration of synthetic PDF delayed Ca2+ activation in E cells (LNd) and DN3; this delay increased with serial PDF administration (Figure 3E–I). Therefore, PDF staggers Ca2+ activity phases between PDF-releasing neurons and PDF-receiving neurons by a cell autonomous, delay mechanism. However, our model cannot explain by what mechanism the same PDF signal causes different phase-delaying effects in Ca2+ activity in the E cells (7.4±1.4h) versus in the DN3 (16.4±1.5h).
Figure 8. Model of PDF-, sNPF- and light-mediated interactions that in concert set sequential Ca2+ activity phases of the different pacemaker groups.
(A) The position of each pacemaker group on the circle indicates its Ca2+ peak phase. Both PDF and sNPF signals suppress the receivers (LNd and DN3 for PDF; DN1 for sNPF) from being active when senders (s-LNv for PDF; s-LNv and LNd for sNPF) are active. Light cycles act together with PDF to delay LNd Ca2+ phases (Figure 1A&C). (B) Loss of neuropeptide-mediated interactions caused alterations in network Ca2+ activity patterns: pdf/pdfr deficient (Figure 1D and Figure 2B), M-cell sNPF knockdown (Figure 7B), E-cell sNPF knockdown (Figure S6F), and PDF cell ablation (Figure S7). (C) Ca2+ phase shifts occurring within 24 h following 15 min light pulses suggest that light also regulates PDF and sNPF signals (Figure 5).
We also found that neuropeptide sNPF determines the rhythmicity and phase of DN1 Ca2+ activity. Selectively impairing sNPF signals from M-cells (s-LNv) altered DN1 Ca2+ phase (Figure 6B), while impairing sNPF signals from both groups severely diminished the amplitude of the DN1 Ca2+ rhythms (Figure 6A). Together with the evidence that synthetic sNPF administration reduced DN1 Ca2+ level (Figure 7), we suggest the following model. Without sNPF inhibition, DN1 Ca2+ levels are constitutively high throughout the day with no evidence of a rhythmic change. A daily rhythm is imposed by sequential sNPF signals mediating daytime inhibition: strong inhibition from M cells in the morning is followed by more temperate inhibition from E cells in the afternoon. Thus, DN1 display an ‘activity peak’ during the nighttime, representing a period of dis-inhibition from sNPF released by the lateral pacemaker groups. These proposed sNPF effects on DN1 are further interesting in that both sNPF and the DN1 pacemakers have been implicated in the regulation of sleep (Shang et al., 2013; Guo et al., 2016). Consistent with previous observations (Hermann-Luibl, 2014), we found that all-pacemaker downregulation of sNPF increased night time locomotor activity (Figure 6G and S6B), which we interpret as the loss of night time sleep promotion by the mid-night activity peak of the DN1s. In addition, we found M-pacemaker specific downregulation of sNPF impaired morning anticipatory activity (Figure 6H and S6C). We interpret this to indicate a suppression/delay of the morning locomotor activity peak by DN1 activity coincident with s-LNv activity. DN1s inhibit s-LNv neurons via glutamatergic transmission (Collins et al., 2012). Previous findings also suggest that DN1s integrate multiple circadian outputs and they regulate both morning and evening behavior (Zhang et al., 2010a, Zhang et al., 2010b; Cavanaugh et al., 2014). Our results suggest that both M cells and E cells modulate DN1s, which represents a convergence of sNPF signals, and thus provides a mechanistic basis to help explain the integrating function of DN1.
The Ca2+ activity of the LNd appears highly correlated with the evening behavior. Yet, anomolously, in DD1 the LNd Ca2+ phase of the pdf01 mutant is advanced by ~ 8hr (Liang et al., 2016), while the phase of the evening behavior is only advanced by ~1.5 hr (Renn et al., 1999). How can this difference be reconciled with LNd control of the phase of the evening behavior? We propose the explanation involves a parallel delaying effect of light on the LNd pacemaker group, as revealed by measurements in pdf01 mutants in an LD cycle (Figure 1): the net advance in the LNd Ca2+ wave in that condition is only ~ 2 hr. Thus the phase of Ca2+ activity in the LNd remains highly correlated in both WT and pdf mutant conditions with the phase of the evening behavior. Likewise, the phase of the Ca2+ activity in the DN1 remains highly correlated in both WT and sNPF knockdowns with the presence, or the delay/absence of the morning peak of behavior. Together these data independently support the hypothesis that Ca2+ dynamics in pacemaker neurons are inextricably linked to specific behavioral outputs.
Furthermore, we found that PDF and sNPF may modulate daily activity patterns broadly among different pacemaker groups by suppressing Ca2+ activity (Figure 3A–D & Figure 7). In the literature, the effects of PDF on neuronal activity in insects vary (Seluzicki et al., 2014; Vecsey et al., 2014). With functional expression of PDFR in hEK-293 cells, PDF caused rapid elevations of both cAMP and Ca2+ (Mertens et al., 2005). In contrast, our present studies mainly focused on the in vivo effects of PDF that develop over many hours. Previous studies also suggested long term cell inhibition by PDF signaling (Choi et al., 2012). The documented effects of sNPF are likewise varied (Root et al., 2011; Shang et al., 2013). Future studies that detail the actual signaling pathways leading from PDF and sNPF receptors to suppress Ca2+ activities will be helpful.
We also found a suppressing effect of PDF on the M cell s-LNvs, mediated by the PDF receptor. We propose that negative feedback by PDF shortens the period during which s- LNv release PDF, and in so doing shortens the time window for PDF action across the pacemaker network. This model is based on two lines of evidence: selectively impairing the PDFR autoreceptor in s-LNv (i) prolonged s-LNv Ca2+ activity further into the morning (Figure 2F) and (ii) delayed the Ca2+ activation of LNd, DN3, and DN1, consistent with a longer duration of PDF-mediated delay (Figure 4G). Negative feedback by autoreceptors is a phenomenon common to many fast neurotransmitters. Choi et al. (2012) reported that chronic activation of PDFR autoreceptors (by a tethered-PDF design) increased morning behavioral activity. Thus, they argued that the PDFR autoreceptor normally regulates PDF secretion in a positive feedback mode. In that study, chronic PDFR autoreceptor activation also advanced the evening behavioral phase and shortened the free-running period under constant darkness. This phenotype is similar to those of both pdf and pdfr mutants (Renn et al., 1999; Hyun et al., 2005), two situations in which PDF signaling is diminished. By that analogy, we reason that PDFR autoreceptors may depress PDF secretion.
Our working model of phase control in the Drosophila circadian network suggests that morning (dawn) is the cardinal phase for periodic 24-h elevations of Ca2+. Yet all pacemaker groups except the s-LNv normally display a non-morning phase: we have shown that they are forced to do so by a series of delaying signals, mediated by light and neuropeptides. This model builds on a strong foundation of work highlighting the importance of cell interactions to generate circadian rhythmicity as an emergent property from the Drosophila neuronal network (e.g., Renn et al., 1999; Peng et al., 2003; Grima et al., 2004; Lin et al., 2004; Stoleru et al., 2004; Dissel et al., 2014; Yao and Shafer, 2014). We have so far identified signals that delay all pacemaker groups except the l-LNv to display group-specific, non-morning phases of activity (Figure 8A). The l-LNv Ca2+ phase was moderately altered by loss-of-function genetic manipulations for neuropeptide DH31 and the DH31-R1. However, the strength of the effects we observed were modest, suggesting other factor(s) regulating l-LNv Ca2+ activity remain to be identified.
Based on our observations, we generated a quantitative model of coupled oscillators. By optimizing parameters for PDF and sNPF inhibition strength to each pacemaker group, this model fits the sequential pattern of the WT pacemakers’ Ca2+ activities. Importantly, it predicts pacemakers’ Ca2+ activity patterns for nearly all genetic manipulations performed in this study (Figure S8). With only a single exception, the model supports the contention that PDF and sNPF neuropeptide-mediated inhibitions are sufficient to explain much of the details of the Ca2+ activity patterns in this pacemaker network. The exception is the failure to accurately predict the phenotype of PDFR overexpression in a pdfr mutant background that is shown in Figure 4D. Here DN3 and DN1 were both phase-shifted to a prospective dawn phase. Thus, DN1 Ca2+ phases were unaltered by pdf/pdfr loss-of-function mutants, but, anomalously, they were altered by gain-of-function PDFR signaling. In fact, seven of the 15 DN1s normally express endogenous PDFR (Im et al., 2011). Therefore, a possible explanation is that the altered Ca2+ activity pattern we observed in this gain-of-function state represents the anomalous responses of the eight DN1 that are normally PDFR-negative. This single inconsistency between the theoretical model and the experimental observation also suggested that the interactions between DN1 and s-LNv might be reciprocal, as indicated by previous work (Collins et al., 2012; Guo et al., 2016). We also noted that M-cell-specific sNPF knockdown in the pdfr mutant background restored their morning anticipation behavior (Figure S6G), which was impaired in either pdfr mutant background or the M-cell-specific sNPF knockdown in a pdfr+ background (Figure S1C & Figure 6H). This result reveals an unexpected interaction between sNPF and PDF signals and more complex interactions between pacemakers than have been so far described.
In the mammalian suprachiasmatic nucleus (SCN), circadian pacemaker neurons also show spatiotemporal patterns in Ca2+ activity (e.g. Ikeda et al., 2003; Enoki et al., 2012). The activity patterns in SCN were disrupted by manipulating neuropeptidergic G-coupled signaling (Aton et al., 2006), suggesting that such patterns might also be regulated by neuropeptide-mediated interactions. SCN pacemaker Ca2+ rhythms require ryanodine-sensitive Ca2+ stores (Ikeda et al., 2003), but a complete understanding of how neuropeptidergic G protein-coupled signaling regulates these internal Ca2+ stores is not yet at hand.
PDF and light signaling pathways converge to set pacemakers’ activity phases
We found that a 12:12 LD cycle delayed Ca2+ activation in the E-cell LNd (Figure 1 and Figure 8A). The functional significance of the LNd Ca2+ phase under the regulation of light is that LNd encodes the length of daytime (Liang et al., 2016). These results suggest that the PDF and light signaling pathways converge to set the phase of E-cell LNd Ca2+ activity. Previous studies have indicated a close interaction between PDF and light in the Drosophila circadian system. First, PDF receptor and the circadian photoreceptor CRY display coordinate expression by a subset of pacemakers (Im et al., 2011; Yao and Shafer, 2014). Second, cry interacts genetically with both pdf and pdfr, causing severe disruptions of molecular PER oscillations in E cells and behavioral rhythms (Cusumano et al., 2009; Zhang et al., 2009; Im et al., 2011). Third, PDF neurons use PDF to relay light inputs from visual systems to the rest of pacemaker network (Guo et al., 2014), suggesting that PDF signals may constitute critical parts of the network’s light signaling pathways.
The balance between PDF and CRY signals may be crucial for light input processing. Indeed, when one signal is reduced, the other may become stronger (Guo et al., 2014). Responses to light pulses in molecular clocks (Li et al., 2014) and Ca2+ rhythms (Figure S4CD) were larger in pdf01 than in WT. Therefore, the coordinate actions of PDF and light signals affect all levels of the pacemaker network, from molecules to neurons to behavior. VIP signaling in the mammalian SCN appears orthologous to Drosophila PDF signaling in many respects (Vosko et al., 2007). Pharmacologically, VIP induced phase delays and ‘phase tumbling’ of molecular clocks among the SCN cells (An et al., 2013). Furthermore, VIP signaling is needed to encode seasonal information, as VIP knock out mice show greatly diminished adaptation to either long or short days (Lucassen et al., 2012). Finally, Hughes et al. (2015) recently showed that constant light improves rhythmicity in mice deficient for the VIP receptor. These observations suggest further parallelism between PDF and VIP signaling in coordination with light inputs.
Light inputs change pacemaker interactions
Short light pulses regulate the molecular clock (Roberts et al., 2015); here we found they also regulate the 24-h neural activity patterns of circadian pacemakers within the first day post-stimulus. Both these changes preceded phase changes in rhythmic behavior, since the light-induced behavioral phase shift does not occur until in the second and third circadian cycle (Pittendrigh et al., 1958) (Figure S3A. Furthermore, in the first day post-stimulus, phase-delaying pulses rapidly delayed Ca2+ phases in all pacemaker groups, while a phase-advancing light pulse only advanced that of the s-LNv. In the following two days, other pacemaker groups were gradually advanced, possibly driven by s-LNv. Recently, Eck et al. (2016) reported that directly activating all pacemakers could induce both behavioral phase advances and delays, while activating s-LNv alone could only induce phase advances, but not phase delays. Those outcomes appear consistent with our observations: phase-delaying effects may need all pacemaker groups responding in coordination, while phase-advancing effects only involve s-LNv responses; the latter then recruits other groups to subsequently advance.
These data lead us to re-consider the relationship between Ca2+ dynamics and the molecular clock. We previously found that all Ca2+ dynamics are lost in the chronic absence of the PER-based clock (Liang et al., 2016). Therefore, the mechanism underlying differential light responsiveness in Ca2+ activities and behavioral effects (Figure 5) could be differential responsiveness in the molecular clocks (Lin et al., 2004; Yoshii et al., 2008). A phase-advancing light pulse instantaneously de-synchronizes molecular clocks among pacemaker groups ex vivo; they re-synchronize over the subsequent 2–3 cycles (Roberts et al. 2015). While more experiments are needed, the data at present lead us to suggest that phase shifts in molecular clocks cannot fully explain the phase shifts we observe in Ca2+ activity patterns. The generation of Ca2+ rhythms within a specific group may likewise be independent of that cell’s PER clock: the knockdown of sNPF (Figure 6) diminished the DN1 calcium activation without affecting the rhythm of the PER cycle in that pacemaker group. Thus Ca2+ rhythms could be imposed by other pacemaker groups in a manner that is independent of the target pacemaker’s PER-clock.
In summary, our study provides a new framework to help understand how 24-h neural activity patterns are generated in the Drosophila circadian pacemaker network. Among different groups of pacemakers, sequential oscillations in their neural activities are established by neuropeptide-mediated inhibition and delay (including by PDF and sNPF), and as well by light-mediated delay. These coupling mechanisms help ensure that dawn and dusk light pulses produce the appropriate advancing and delaying phase shifts for pacemaker entrainment (Meijer and Schwartz, 2003) and the proper sequencing of multiple, sequentially-timed neuronal outputs.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact, Paul H. Taghert (taghertp@wustl.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fly Rearing and Stocks
Flies were reared at room temperature on standard yeast-supplemented cornmeal/agar food. Males were entrained under 12 h light: 12 h dark (LD) cycles at 25°C for at least 3 days. The tim>GCaMPS6s, mCherry; cry01 flies were entrained for more than 6 days. The cry-LexA::GAD line was a gift from Dr. F Rouyer (CNRS Gyf, Paris): it is strongly expressed in CRY-expressing neurons and shows weaker expression in other clock neurons in addition to some non-clock cells, similarly to the cry-Gal439 pattern described by Klarsfeld et al. (2004). cry-LexA was recombined with LexAop-GCaMP6s into the same third chromosome. RNAi was tested along with UAS-dcr2 to increase the efficiency of RNAi-mediated knockdown. A stable line: UAS-dcr2; tim(UAS)-gal4; UAS-GCaMP6s, UAS-mCherry. NLS was created for RNAi screening of neuropeptides and their receptors. This line was then crossed with individual RNAi lines. In male progeny from these crosses, we measured locomotor behavior and calcium rhythms. Table S1 provides the complete genotype for flies used in this study. The age/developmental stage of experimental models was 3–10 days after eclosion.
METHOD DETAILS
Light stimulation
Light stimulation before or during in vivo calcium imaging was delivered by a white Rebel LED (Luxeon) that was powered by a LED driver (350 mA, externally dimmable, BuckPuck DC driver). The output of the driver was controlled by an Arduino UNO board (Smart Projects, Italy). The Arduino was timed by a real-time clock module (DS3231). The light intensity was measured by light meter (W/RS-232, VWR Scientific) and adjusted to ~3000 lux.
For the light stimulation before calcium imaging, the LED was programmed to deliver the first regular 12 h light: 12 h dark (LD) cycles followed with constant darkness (DD). During the dark period, 15 min light pulse was delivered 5 h (CT17), 9 h (CT21), or 21 h (CT9) after light off. The flies that received light stimulation remained in darkness for different periods of time (varying from 3 to 62 hr) and then underwent surgery. We previously described the surgical procedure (Liang et al., 2016): briefly, the pacemaker neurons on one side of the brain were exposed by removing a portion of the dorso-anterior head capsule on one side of the head. The entire surgery was typically 20 min in duration, and was performed under dim red light to avoid additional light stimulation. Immediately after surgery, in vivo 24-h calcium imaging was performed. Different 24h calcium imaging sessions were systematically tiled to create synthetic, uninterrupted ~72 h observation periods. For the light stimulation during calcium imaging, we performed surgery on flies that previously entrained to a 12:12 LD cycle and began imaging at the beginning of the dark period. During the imaging, the LED delivered a 15 min light pulse at CT17 (5 h after light-off). For the LD cycle stimulation during calcium imaging, the 24-h calcium imaging sessions began systematically at different Zeitgeber times (ZT). The LED delivered the 12 h light: 12 h dark cycles according to the original schedule by which the flies were entrained. During the light period, the LED was shut off for 10 seconds every 10 min, allowing the microscope to acquire complete volume brain scans.
In vivo calcium imaging
Imaging was performed on a custom Objective Coupled Planar Illumination (OCPI) microscope (Holekamp et al., 2008), as described in (Liang et al., 2016). Briefly, OCPI illuminated samples with a ~5μm thick light sheet set to the focal plane of the objective. The light sheet was generated from a 488nm laser that went through an optical fiber, a light collimator and light sheet–forming cylindrical lens. The objective was a water-immersion 0.5NA 20× (Olympus). Images were captured on an iXon DV885-KCS-VP cooled EM-CCD (Andor). The microscope was operated by a custom software (Imagine). To characterize the circadian properties of neural activity, GCaMP6s signals from circadian neurons were imaged every 10 min for 24 hr. Each stack of images was acquired by scanning the light sheet across the fly brain through the cranial window. Stacks contained 20 to 30 separate images with step size of 10 microns. The total scanning time for each brain stack was less than 3s. During the 24-h imaging, fresh haemolymph-like saline (HL3), containing 70 mM NaCl, 5 mM KCl, 1.5 mM CaCl2, 20 mM MgCl2, 10 mM NaHCO3, 5 mM trehalose, 115 mM sucrose, and 5 mM HEPES (pH 7.1), was perfused continuously (0.1–0.2 mL/min). For experiments that included light stimulation and pharmacological treatments, two brains were imaged simultaneously.
Pharmacology
For pharmacological tests of PDF during 24 h Ca2+ imaging in vivo (Fig 7A–B), imaging was started at CT0. ~10 minutes prior to the start of recording, PDF was pre-mixed in the bath at 10−5 M (the first dose at CT0). In the double PDF application experiments (Fig 7B), a second dose of PDF was added dropwise to the bath at CT5 (1mL of 10−4 M PDF solution added into the 10 ml bath over a ~10 s period). For pharmacological tests of PDF during 6 h Ca2+ imaging in vivo (Fig 7D–E), Ca2+ imaging started at CT6. PDF (1mL of 10−4 M PDF) was then added dropwise over a ~10 s period to the 10 ml bath at CT7.
For pharmacological tests of sNPF during 12 h Ca2+ imaging in vivo, flies were separately entrained to LD schedules with an 8h phase difference for at least 3 days before experiments. Therefore, for one fly of the pair, the Ca2+ imaging was started at CT4, while for the other it started at CT12. sNPF-2 (1mL of 10−4 M) was then added dropwise to the bath over a ~10 s period one hour after Ca2+ imaging was started (CT5 for one fly and CT13 for the other).
In all drug experiments, fresh HL3 saline was perfused continuously at a rate comparable to that of other experiments. Using bromophenol blue measurements, we estimate the perfusion system reduced the concentration of free PDF in the solution bathing the brain by 10-fold over 7 hr. PDF was purchased from Neo-MPS (San Diego, CA, USA) at a purity of 86%. sNPF-2 was kindly provided by Dr. P. Evans (Cambridge, U.K.).
Locomotor activity
The locomotor activity of 4–6 day old male flies was recorded using Trikinetics Activity Monitors. Flies were entrained to 12:12 h LD cycles for 6 days and then released into constant darkness (DD) for 9 days.
Mathematical Modeling
The quantitative model of oscillators coupled with PDF- and sNPF-mediated suppressions was generated in R 3.3.2. The Ca2+ rhythms of five circadian pacemaker groups were simplified as five identical simple harmonic oscillators. The Ca2+ level (x) for each group was set to oscillate with a 24 h period:
For pacemaker groups those release the designated neuropeptides (PDF and/or sNPF), the peptides were released when the Ca2+ level reached a threshold (θrelease). Levels of secreted neuropeptide (P) decayed at a constant rate (kdecay) and followed the equation:
N was a binary parameter representing whether the pacemaker group expresses a specific neuropeptide. The suppressing effect of neuropeptides on the Ca2+ level of a pacemaker group followed this equation once the neuropeptide level achieved a threshold (θeffect):
R was a binary parameter representing whether a pacemaker group expresses the specific PDF and/or the sNPF neuropeptide receptor. keffect was a constant for the strength of neuropeptide effects.
The model was fit to experimental data of Ca2+ phases in five pacemaker groups in WT controls shown in Figure 2B with parameter values: θrelease.PDF=0.17, kdecay.PDF=0.55, θeffect.PDF.s-LNv= 0.37, θeffect.PDF.l-LNv= 0.50, θeffect.PDF.LNd= 0.36, θeffect.PDF.DN3= 0.46, θeffect.PDF.DN1= 1.69, keffect.PDF.s-LNv=-0.79, keffect.PDF.LNd=-0.73, keffect.PDF.l-LNv=-2.00, keffect.PDF.DN3=-1.41, keffect.PDF.DN1=-1.18, θrelease.sNPF.s-LNv=0.37, θrelease.sNPF.LNd=0.91, kdecay.PDF=0.10, θeffect.sNPF.DN1=0.09, and keffect.sNPF.DN1=-1.03.
Based on these parameters and initial values of x for each groups (xs-LNv=1; xl-LNv=0.09, xLNd=1, xDN3=1, and xDN1=0), we added or removed neuropeptide interactions by manipulating the N and R values for each group to predict the Ca2+ phases in genetic manipulations done in this study. The differential equations were solved using lsoda solver.
Replication
For all in vivo calcium imaging experiments, at least 5 biological replications were performed for each genotype and/or condition. For all behavioral experiments, at least 8 biological replications were performed for each genotype and/or condition.
Strategy for randomization and/or stratification
24 hour recording sessions were randomly begun at different Zeitgeber times (ZT) except for pharmacological experiments, which began at the same ZT according to the experimental design. In light pulse stimulation experiments (Figure 5 and Figure S3), 24 hour recording sub-sessions were begun after the flies received light stimulation and had remained in darkness for random periods of time (varying from 3 to 62 hr).
Blinding at any stage of the study
The information about genotypes, light or pharmacological treatments, and the circadian time for each recording was blinded at the stage of image processes, ROIs selection, and time traces measurements.
Sample-size estimation and statistical method of computation
All the sample size information as well as specific statistical methods are listed in corresponding figure legends.
Inclusion and exclusion criteria of any data or subjects
The behavioral data of flies that had died during the analysis window (Day 1–6 in LD and Day 1–9 in DD) was excluded. In calcium imaging data, any group of pacemakers that didn’t display detectable signal at any point in the 24 hour recording period was not included.
QUANTIFICATION AND STATISTICAL ANALYSIS
Imaging data analysis
Calcium imaging data was analyzed as described in (Liang et al., 2016). Images were processed in MATLAB R2015b (MathWorks, Natick, MA, USA) using custom software described previously (Holekamp et al., 2008) and ImageJ-based Fiji. Regions of interest (ROIs) were manually selected over individual cells or groups of cells. Average intensities of ROIs were measured through the time course and divided by average of the whole image to subtract background noise. For each time trace, the calcium transient was then calculated as dF/F=(F-Fmin)/Fmean. The phase relationship between traces was estimated by cross-correlation analysis. Phases of traces were plotted on a 24-hr-clock circular plot reflecting both peak time and phase relationship of traces. This phase relationship was then tested statistically by the Rayleigh test of uniformity and the Watson-Williams test for homogeneity of means (Levine et al., 2002). For the light pulse stimulation experiments, as the Ca2+ dynamics were expected to vary among three circadian cycles, additional analysis was applied as described immediately following.
In light pulse stimulation experiments, traces were aligned by the time after the light pulse. 24-h traces from different animals, which were distributed and overlaid within three day period after light pulses, were then merged to simulate a continuous three day record. In other experiments, traces were aligned by Zeitgeber Time. Traces of the same cell type were then averaged across different animals.
Peak phases in three cycles after light pulses were estimated separately by cross-over analysis, because the circadian cycles might be phase-shifted and mismatched between cell types. For each cell type, averaged traces were first filtered by 8 hr, and then by 24 hr. The crossover points of two different filtered traces were identified as rising and falling phase markers for each cycle. In each cycle, the local maximum between the rising and falling phase markers was then identified as the peak phase. After identifying the peak phase of the averaged trace, traces from different animals falling in that cycle were then pooled together to estimate the phase coherence among these animals. In other experiments (no light pulses), all traces were considered within the same cycle. The phase relationship between traces from the same cycle was estimated by cross-correlation analysis. Phases of traces were plotted on a 24-hr-clock circular plot reflecting both peak time and phase relationship of traces. This phase relationship was then tested statistically by the Rayleigh test of uniformity and the Watson-Williams test for homogeneity of means (Levine et al., 2002). Trace analyses and statistics were performed using R 3.3.3 and Prism 7 (GraphPad, San Diego CA).
Behavioral experiment analysis
Locomotor activity from DD Days 1–9 was then normalized for aχ2 periodogram with a 95% confidence cutoff and SNR analysis, to estimate circadian rhythmicity (Levine et al., 2002). Arrhythmic flies were defined by a power value less than 10 and width lower than 1, or a period less than 18, or more than 30 h. Data were analyzed in R 3.3.3 and MATLAB R2015 (MathWorks, Natick, MA, USA).
Supplementary Material
Acknowledgments
We thank Weihua Li for technical assistance; the Holy and Taghert laboratories for advice. Erik Herzog and Orie Shafer offered comments on the manuscript; Heather Dionne, Gerry Rubin, Aljoscha Nern, and Francois Rouyer kindly shared fly stocks and unpublished information prior to publication. Erik Johnson, Orie Shafer, Peter Evans, Michael Rosbash and the DGRC kindly provided reagents, and the Bloomington Stock Center and Janelia Research Center provided fly stocks. The work was supported by the Washington University McDonnell Center for Cellular and Molecular Neurobiology, and by NIH grants R01 NS068409 and R01 DP1 DA035081 (T.E.H.), R01 MH067122 (P.H.T.) and R24 NS086741 (T.E.H. & P.H.T.).
Footnotes
Author Contributions
X.L., T.E.H., and P.H.T. conceived the experiments; X.L. performed and analyzed all experiments; X.L., T.E.H., and P.H.T. wrote the manuscript. T.E.H. has a patent on OCPI microscopy.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An S, Harang R, Meeker K, Granados-Fuentes D, Tsai Ca, Mazuski C, Kim J, Doyle FJ, Petzold LR, Herzog ED. A neuropeptide speeds circadian entrainment by reducing intercellular synchrony. Proc Natl Acad Sci U S A. 2013;110:E4355–61. doi: 10.1073/pnas.1307088110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aton SJ, Herzog ED. Come together, right…now: synchronization of rhythms in a mammalian circadian clock. Neuron. 2005;48:531–534. doi: 10.1016/j.neuron.2005.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aton SJ, Huettner JE, Straume M, Herzog ED. GABA and Gi/o differentially control circadian rhythms and synchrony in clock neurons. Proc Natl Acad Sci U S A. 2006;103:19188–19193. doi: 10.1073/pnas.0607466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau J, Young MW. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. doi: 10.1016/s0092-8674(00)81554-8. [DOI] [PubMed] [Google Scholar]
- Cavanaugh DJJ, Geratowski JDD, Wooltorton JRARA, Spaethling JMM, Hector CEE, Zheng X, Johnson ECC, Eberwine JHH, Sehgal A. Identification of a Circadian Output Circuit for Rest:Activity Rhythms in Drosophila. Cell. 2014;157:689–701. doi: 10.1016/j.cell.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriani MF, Darlington TK, Staknis D, Mas P, Petti AA, Weitz CJ, Kay SA. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science. 1999;285:553–556. doi: 10.1126/science.285.5427.553. [DOI] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr Ra, Orger MB, Jayaraman V, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C, Cao G, Tanenhaus AK, McCarthy Ev, Jung M, Schleyer W, Shang Y, Rosbash M, Yin JCP, Nitabach MN. Autoreceptor Control of Peptide/Neurotransmitter Corelease from PDF Neurons Determines Allocation of Circadian Activity in Drosophila. Cell Rep. 2012;2:332–344. doi: 10.1016/j.celrep.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins B, Kane Ea, Reeves DC, Akabas MH, Blau J. Balance of activity between LN(v)s and glutamatergic dorsal clock neurons promotes robust circadian rhythms in Drosophila. Neuron. 2012;74:706–718. doi: 10.1016/j.neuron.2012.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coomans CP, Ramkisoensing A, Meijer JH. The suprachiasmatic nuclei as a seasonal clock. Front Neuroendocrinol. 2015;37:29–42. doi: 10.1016/j.yfrne.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Cusumano P, Klarsfeld A, Chelot E, Picot M, Richier B, Rouyer F. PDF-modulated visual inputs and cryptochrome define diurnal behavior in Drosophila. Nat Neurosci. 2009;12:1431–1437. doi: 10.1038/nn.2429. [DOI] [PubMed] [Google Scholar]
- Dissel S, Hansen CN, Ozkaya O, Hemsley M, Kyriacou CP, Rosato E. The Logic of Circadian Organization in Drosophila. Curr Biol. 2014:2257–2266. doi: 10.1016/j.cub.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezelova E, Dolezel D, Hall JC. Rhythm defects caused by newly engineered null mutations in Drosophila’s cryptochrome gene. Genetics. 2007;177:329–345. doi: 10.1534/genetics.107.076513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P, Stanewsky R, Helfrich-Forster C, Emery-Le M, Hall JC, Rosbash M. Drosophila CRY is a deep brain circadian photoreceptor. Neuron. 2000;26:493–504. doi: 10.1016/s0896-6273(00)81181-2. [DOI] [PubMed] [Google Scholar]
- Enoki R, Kuroda S, Ono D, Hasan MT, Ueda T, Honma S, Honma K, Sens H. Topological specificity and hierarchical network of the circadian calcium rhythm in the suprachiasmatic nucleus. Proc Natl Acad Sci U S A. 2012;109:21498–21503. doi: 10.1073/pnas.1214415110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Reale V, Chatwin H, Kennedy K, Venard R, Ericsson C, Yu K, Evans PD, Hall LM. Functional characterization of a neuropeptide F-like receptor from Drosophila melanogaster. Eur J Neurosci. 2003;18:227–238. doi: 10.1046/j.1460-9568.2003.02719.x. [DOI] [PubMed] [Google Scholar]
- Flourakis M, Kula-Eversole E, Hutchison ALL, Han THH, Aranda K, Moose DLL, White KPP, Dinner ARR, Lear BCC, Ren D, et al. A Conserved Bicycle Model for Circadian Clock Control of Membrane Excitability. Cell. 2015;162:836–848. doi: 10.1016/j.cell.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B, Chelot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- Guo F, Cerullo I, Chen X, Rosbash M. PDF neuron firing phase-shifts key circadian activity neurons in Drosophila. Elife. 2014:e02780. doi: 10.7554/eLife.02780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Yu J, Jung HJ, Abruzzi KC, Luo W, Griffith LC, Rosbash M. Circadian neuron feedback controls the Drosophila sleep-activity profile. Nature. 2016;536:292–297. doi: 10.1038/nature19097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings MH, Brancaccio M, Maywood ES. Circadian pacemaking in cells and circuits of the suprachiasmatic nucleus. J Neuroendocrinol. 2014;26:2–10. doi: 10.1111/jne.12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann C, Yoshii T, Dusik V, Helfrich-Forster C. Neuropeptide F immunoreactive clock neurons modify evening locomotor activity and free-running period in Drosophila melanogaster. J Comp Neurol. 2012;520:970–987. doi: 10.1002/cne.22742. [DOI] [PubMed] [Google Scholar]
- Hermann-Luibl C. The Role of the Neuropeptides NPF, sNPF, ITP and PDF in the Circadian Clock of Drosophila melanogaster. Universitat Wurzburg; 2014. [Google Scholar]
- Herzog ED. Neurons and networks in daily rhythms. Nat Rev Neurosci. 2007;8:790–802. doi: 10.1038/nrn2215. [DOI] [PubMed] [Google Scholar]
- Holekamp TF, Turaga D, Holy TE. Fast three-dimensional fluorescence imaging of activity in neural populations by objective-coupled planar illumination microscopy. Neuron. 2008;57:661–672. doi: 10.1016/j.neuron.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Hughes ATL, Croft CL, Samuels RE, Myung J, Takumi T, Piggins HD. Constant light enhances synchrony among circadian clock cells and promotes behavioral rhythms in VPAC2-signaling deficient mice. Sci Rep. 2015;5:14044. doi: 10.1038/srep14044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun S, Lee Y, Hong ST, Bang S, Paik D, Kang J, Shin J, Lee J, Jeon K, Hwang S, et al. Drosophila GPCR Han is a receptor for the circadian clock neuropeptide PDF. Neuron. 2005;48:267–278. doi: 10.1016/j.neuron.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Sugiyama T, Wallace CS, Gompf HS, Yoshioka T, Miyawaki A, Allen CN. Circadian dynamics of cytosolic and nuclear Ca2+ in single suprachiasmatic nucleus neurons. Neuron. 2003;38:253–263. doi: 10.1016/s0896-6273(03)00164-8. [DOI] [PubMed] [Google Scholar]
- Im SH, Taghert PH. PDF Receptor Expression Reveals Direct Interactions between Circadian Oscillators in Drosophila. J Comp Neurol. 2010;518:1925–1945. doi: 10.1002/cne.22311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SH, Li W, Taghert PH. Pdfr and cry signaling converge in a subset of clock neurons to modulate the amplitude and phase of circadian behavior in Drosophila. PLoS One. 2011;6 doi: 10.1371/journal.pone.0018974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johard Ha D, Yoishii T, Dircksen H, Cusumano P, Rouyer F, Helfrich-Forster C, Nassel DR. Peptidergic clock neurons in Drosophila: Ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. J Comp Neurol. 2009;516:59–73. doi: 10.1002/cne.22099. [DOI] [PubMed] [Google Scholar]
- Johnson EC, Shafer OT, Trigg JS, Park J, Schooley DA, Dow JA, Taghert PH. A novel diuretic hormone receptor in Drosophila: evidence for conservation of CGRP signaling. J Exp Biol. 2005;208 doi: 10.1242/jeb.01529. [DOI] [PubMed] [Google Scholar]
- Klarsfeld A, Malpel S, Michard-Vanhee C, Picot M, Chelot E, Rouyer F. Novel Features of Cryptochrome-Mediated Photoreception in the Brain Circadian Clock of Drosophila. J Neurosci. 2004;24 doi: 10.1523/JNEUROSCI.3661-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst M, Hughes ME, Raccuglia D, Felix M, Li M, Barnett G, Duah J. Calcitonin Gene-Related Peptide Neurons Mediate Sleep-Specific Circadian Output in Drosophila. Curr Biol. 2014;24:2652–2664. doi: 10.1016/j.cub.2014.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear BC, Zhang L, Allada R. The neuropeptide PDF acts directly on evening pacemaker neurons to regulate multiple features of circadian behavior. PLoS Biol. 2009;7:e1000154. doi: 10.1371/journal.pbio.1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, You KH, Choo JK, Han YM, Yu K. Drosophila short neuropeptide F regulates food intake and body size. J Biol Chem. 2004;279:50781–50789. doi: 10.1074/jbc.M407842200. [DOI] [PubMed] [Google Scholar]
- Levine J, Funes P, Dowse H, Hall J. Signal analysis of behavioral and molecular cycles. BMC Neurosci. 2002;25:1–25. doi: 10.1186/1471-2202-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Guo F, Shen J, Rosbash M. PDF and cAMP enhance PER stability in Drosophila clock neurons. Proc Natl Acad Sci U S A. 2014;111:E1284–90. doi: 10.1073/pnas.1402562111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Holy TE, Taghert PH. Synchronous Drosophila circadian pacemakers display nonsynchronous Ca2+ rhythms in vivo. Science (80-) 2016;351:976–981. doi: 10.1126/science.aad3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Stormo GD, Taghert PH. The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J Neurosci. 2004;24:7951–7957. doi: 10.1523/JNEUROSCI.2370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucassen EA, van Diepen HC, Houben T, Michel S, Colwell CS, Meijer JH. Role of vasoactive intestinal peptide in seasonal encoding by the suprachiasmatic nucleus clock. Eur J Neurosci. 2012;35:1466–1474. doi: 10.1111/j.1460-9568.2012.08054.x. [DOI] [PubMed] [Google Scholar]
- Meijer JH, Schwartz WJ. In search of the pathways for light-induced pacemaker resetting in the suprachiasmatic nucleus. J Biol Rhythms. 2003;18:235–249. doi: 10.1177/0748730403018003006. [DOI] [PubMed] [Google Scholar]
- Mertens I, Vandingenen A, Johnson EC, Shafer OT, Li W, Trigg JS, De Loof A, Schoofs L, Taghert PH. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron. 2005;48:213–219. doi: 10.1016/j.neuron.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Taghert PH. Organization of the Drosophila circadian control circuit. Curr Biol. 2008;18:R84–93. doi: 10.1016/j.cub.2007.11.061. [DOI] [PubMed] [Google Scholar]
- Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJL, Kang K, Kang K, Liu X, Garrity Pa, et al. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron. 2008;60:672–682. doi: 10.1016/j.neuron.2008.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Stoleru D, Levine JD, Hall JC, Rosbash M. Drosophila free-running rhythms require intercellular communication. PLoS Biol. 2003;1:E13. doi: 10.1371/journal.pbio.0000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh C, Bruce V, Kaus P. on the Significance of Transients in Daily Rhythms. Proc Natl Acad Sci U S A. 1958;44:965–973. doi: 10.1073/pnas.44.9.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Roberts L, Leise TL, Noguchi T, Galschiodt AM, Houl JH, Welsh DK, Holmes TC. Light Evokes Rapid Circadian Network Oscillator Desynchrony Followed by Gradual Phase Retuning of Synchrony. Curr Biol. 2015;25:858–867. doi: 10.1016/j.cub.2015.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root CM, Ko KI, Jafari A, Wang JW. Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell. 2011;145:133–144. doi: 10.1016/j.cell.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluzicki A, Flourakis M, Kula-Eversole E, Zhang L, Kilman V, Allada R. Dual PDF signaling pathways reset clocks via TIMELESS and acutely excite target neurons to control circadian behavior. PLoS Biol. 2014;12:e1001810. doi: 10.1371/journal.pbio.1001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO, Lohse MJ, Taghert PH. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron. 2008;58:223–237. doi: 10.1016/j.neuron.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Griffith LC, Rosbash M. Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc Natl Acad Sci U S A. 2008;105:19587–19594. doi: 10.1073/pnas.0809577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Donelson NC, Vecsey CG, Guo F, Rosbash M, Griffith LC. Short neuropeptide F is a sleep-promoting inhibitory modulator. Neuron. 2013;80:171–183. doi: 10.1016/j.neuron.2013.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431 doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- Vecsey CG, Pirez N, Griffith LC. The Drosophila neuropeptides PDF and sNPF have opposing electrophysiological and molecular effects on central neurons. J Neurophysiol. 2014;111:1033–1045. doi: 10.1152/jn.00712.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosko AM, Schroeder A, Loh DH, Colwell CS. Vasoactive intestinal peptide and the mammalian circadian system. Gen Comp Endocrinol. 2007;152:165–175. doi: 10.1016/j.ygcen.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Yasar H, Funk NW, Giese M, Baz ES, Stengl M. Signaling of Pigment-Dispersing Factor (PDF) in the Madeira Cockroach Rhyparobia maderae. PLoS One. 2014;9:e108757. doi: 10.1371/journal.pone.0108757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh DK, Logothetis DE, Meister M, Reppert SM. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Takahashi JS, Kay Sa. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Shafer OT. The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science. 2014;343:1516–1520. doi: 10.1126/science.1251285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T, Todo T, Wulbeck C, Stanewsky R, Helfrich-Forster C. Cryptochrome is present in the compound eyes and a subset of Drosophila’s clock neurons. J Comp Neurol. 2008;508:952–966. doi: 10.1002/cne.21702. [DOI] [PubMed] [Google Scholar]
- Zhang L, Lear BC, Seluzicki A, Allada R. The CRYPTOCHROME photoreceptor gates PDF neuropeptide signaling to set circadian network hierarchy in Drosophila. Curr Biol. 2009;19:2050–2055. doi: 10.1016/j.cub.2009.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chung BY, Lear BC, Kilman VL, Liu Y, Mahesh G, Meissner R-A, Hardin PE, Allada R. DN1(p) circadian neurons coordinate acute light and PDF inputs to produce robust daily behavior in Drosophila. Curr Biol. 2010a;20:591–599. doi: 10.1016/j.cub.2010.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu Y, Bilodeau-Wentworth D, Hardin PE, Emery P. Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behavior. Curr Biol. 2010b;20:600–605. doi: 10.1016/j.cub.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.