Abstract
Here, we explore the potential of single-cell genomic analysis in blood for early detection of cancer; we consider a method that screens the presence of recurrent patterns of copy number (CN) alterations using sparse single-cell sequencing. We argue for feasibility, based on in silico analysis of existing single-cell data and cancer CN profiles. Sampling procedures from existing diploid single cells can render data for a cell with any given profile. Sampling from multiple published tumor profiles can interrogate cancer clonality via an algorithm that tests the multiplicity of close pairwise similarities among single-cell cancer genomes. The majority of common solid cancers would be detectable in this manner. As any early detection method must be verifiable and actionable, we describe how further analysis of suspect cells can aid in determining risk and anatomic origin. Future affordability rests on currently available procedures for tumor cell enrichment and inexpensive methods for single-cell analysis.
Keywords: cancer, early detection, circulating tumor cells, single-cell analysis
Advantages of Single-Cell Analysis for Early Detection of Cancer
Cancer kills by spreading to distant sites. At the time of the first clinical presentation, metastasis has often already occurred. Were it otherwise, most cancers would be curable by surgery. It follows that there may be a window of time when detection of cancer and its timely extirpation might result in a cure. Some -- perhaps most -- tumors spread to distant sites via blood. The evidence of this is that in patients with metastatic disease, cancer cells are found in blood [1] and bone marrow [2]. Even for non-metastatic malignancies, cancer cells may be present in the bone marrow [3]. Therefore, a periodic screen for cancer cells in the blood might detect disease prior to symptomatic clinical presentation, and at a stage before malignant cells have successfully colonized elsewhere. To be useful, such a test must have high sensitivity and specificity.
We argue that such screening should be based on a nearly universal signal in the genomic DNA of cancer cells: almost all cancers stem from a clonal population of cells each bearing a shared profile of DNA copy number variation (CNV). This profile is acquired somatically and through clonal expansion, and is distinct from the germ-line CNV profile of the patient. Several considerations distinguish screens based on enriched cell populations from a screens based on cell-free (cf) DNA in the circulation, and there are three potential advantages of analyzing cells over free DNA. First, both methods are plagued by a high background from the normal genome. But cancer cells can be enrichedfrom the blood[4–8], thus dramatically enhancing the signal of the cancer over the normal fraction. Second, once identified, suspect cancer cells can be exhaustively analyzed, either singly or in pools. For example, subsequent deeper sequencing of single or pooled cancer cell DNA can confirm clonality, enable risk assessment, identify coding mutations and help predict anatomic origin for subsequent image-based screening. The third advantage of cell-based screens is that they can start with a predictable, and an almost universal, cancer signature: CNV. Although cfDNA methods can also be based on CNV, the technical obstacles are enormous and the sensitivity is dubious [9]. To date, most cfDNAs in the blood of cancer patients have been tested through deep-sequencing of coding regions from known cancer-related genes [10]. This approach however is problematic; numerous target genes remain unknown and many mutations of functional consequence might not reside within coding domains. As a consequence, such a method would have low sensitivity. Moreover, an observed somatic coding variant in a target gene will typically be uninterpretable. Indeed, many sequence variants might arise by somatic mutation in tissues and be represented in the blood, yet not be harbingers of disease [11, 12]. Finally, it takes more than one gene mutation to generate a malignant cell [13], but observing many pathogenic mutations in the circulation does not allow us to infer if these mutations co-occur within the same cell. As a result, a gene-focused method would likely have low specificity. A method with low specificity would be a huge burden to the patient and medical system alike.
Powerful methods can now be applied to enrich cancer cells in the blood based on selecting for the cancer component, or by filtering out blood elements based on surface markers and size [5, 14]. Methods for analysis of single-cell DNA [15, 16], RNA [17, 18] and protein composition [19] are also increasing in power and decreasing in cost. From DNA analysis, we can infer if any atypical cells derive from a cancerous clonal population, and assess the malignant potential [20–22]. The tissue of origin might be inferred from RNA expression profiles [23] or from DNA methylation patterns [24, 25]. Once we identify the tissue of origin, more conventional imaging tools could be used to verify the localization of malignant cells.
Thus, practical questions emerge: (i) what algorithms allow the confident detection of sparse cancer cells in a much larger population of cells? And (ii) are there enough neoplastic cells in the circulation to be detected? In this opinion article, we illustrate and discuss a prospective protocol to test this (Key Figure, Figure 1). Indeed, most prevalent cancer types display extensive copy number (CN) alterations (Figure 2A and Table S1 in Supplemental Information) [26]. These alterations comprise CN profiles which must be shared by cancer cells from a given patient. From single-cell sequence analysis, and using CN data from 3,852 cases of solid tumors described in The Cancer Genome Atlas (TCGA) [26], we have concluded that characteristic tumor profiles might be detectable in patients carrying a low load of tumor cells in the blood.
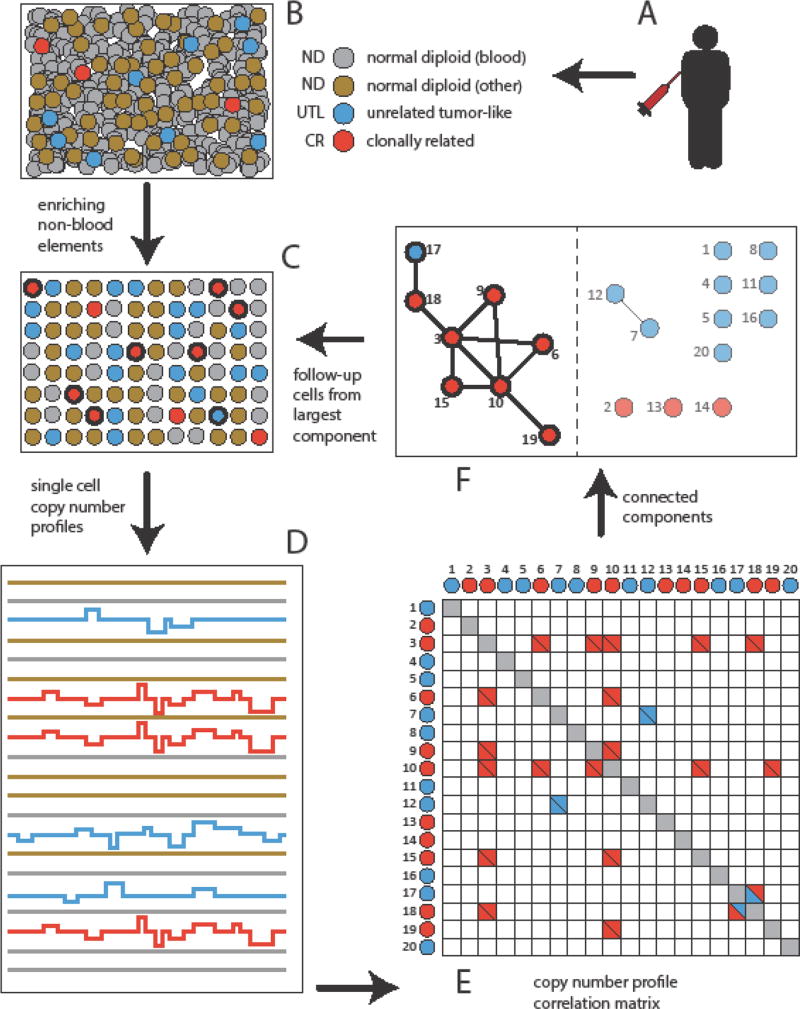
Figure 1. Schematic of Single-Cell Detection Procedure in Blood.
(A–B) Blood is drawn from a subject. (C) Blood elements are removed and cells of epithelial origin can be enriched. (D) Single-cell barcoded DNA libraries are prepared and sequenced, leading to single-cell copy number profiles. (E). Pairwise correlation analysis is performed. (F) A ‘graph’ constructed where ‘vertices’ represent single-cell profiles or ‘bin vectors’, and two vectors are connected by an ‘edge’ if they are highly correlated. The largest ‘connected component’ is identified (F), and resides on the left-hand side of the dotted vertical line. This identifies the clonally correlated cells, indicated in bold. In the illustrated example, the largest connected component of CN profiles is shown, and predominantly stems from the clonally related (CR) tumor cells (3, 6, 9, 10, 15, and 19 in red). Not all tumor cells might be identified this way (2, 13, and 14). In this example, one unrelated tumor like cell (17, UTL) is fortuitously linked to the largest connected component. Two UTLs (7 and 12) also form a small connected component. From the barcodes taken from the largest component, one can return to select cells in (C) (in bold) for deeper analysis; this might confirm cell clonal relations, potentially assess prognosis and therapeutic options, and determine the anatomic origin of the primary tumor.
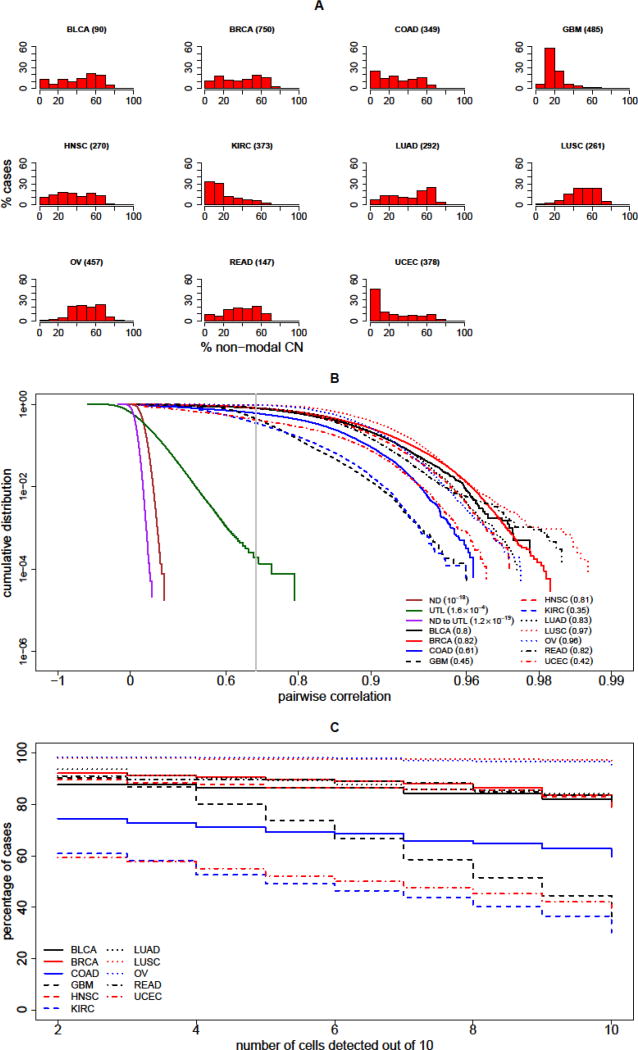
Figure 2. Detection of Clonal Cells in Circulation.
(A) Ubiquity of copy number (CN) variations across multiple cancer types; each histogram shows the percentage of patient cases (vertical axis) with a given percentage of the genome with an abnormal non-modal CN value (horizontal axis). The cancer types represented are (in alphabetical order) bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian cystadenocarcinoma (OV), rectum adernocarcinoma (READ) and uterine corpus endometrial carcinoma (UCEC). The number of cases for each type is listed parenthetically, and is based on data published by The Cancer Genome Atlas (TCGA) [26]. Thus, for example, 60% of glioblastoma multiforme (~500 cases) have CN alterations in more than 10–20% of the genome. (B) The graph presents the cumulative probability distribution (CDF) for correlation of pairs of single-cell CN profiles. The CDF is plotted for pairs of normal diploid cells (ND, solid brown); for pairs of clonally unrelated tumor-like cells (UTL, solid green); and for mixed pairs of an ND and a UTL cell (ND to UTL, solid magenta). For comparison, the CDF for pairs of profiles from two cells simulated from the same cancer are plotted, and the analysis of each of the 11 tumor types considered is shown, as described in the inset legend. The gray vertical line indicates the discriminatory threshold of 0.7 correlation. The values of the CDF at this threshold are indicated in the inset for each curve in parentheses. These values are taken from the simulations of the 11 tumor types and for UTL cells, and from estimates based on extreme-value theory for ND cells and for the mixed ND – UTL pairs. (C) To assess sensitivity of detection, for each tumor type, the cumulative percentage of patients are plotted against the number of clonal cells detected (out of 10 assumed to be present in the specimen). For example, for BRCA (solid red line) a sensitivity of close to 90% is achieved if the number of clonal cells detected is at least 6. For KIRC (dashed blue line), the sensitivity is approximately 50% for that threshold.
Performance of Single-Cell Based Early Detection: A Virtual Clinical Assessment
To conceptualize a single-cell based method of early detection of cancer, we envision the following procedure: Following a standard draw of 10 mL of blood, hematopoietic cell components could be depleted using surface antigens [5], or tumor-like epithelial cells directly selected for epithelial cell adhesion molecule (EpCAM) expression [14], or both (Key Figure, Figure 1). As we indicate below, very low sequence coverage (ultra-sparse whole genome sequencing) [15] on single cells enables sufficient DNA CN profiling to accomplish the goal of detecting a small sub-population of clonally-related tumor cells.
If the only challenge were to detect low numbers of a clonally-related (CR) subpopulation in an overwhelming population of normal diploid (ND) cells, the algorithmic task would be straightforward. However, troublesome subpopulations of unrelated tumor-like (UTL) cells can also be expected to be present. These, in all likelihood, result from chromosome degradation in dying normal cells, each with an abnormal and unique CN profile, ubiquitously observed in single-cell analyses. Conceptualizing a numerical method, we have assumed that some number of such UTL cells would be present. They are troublesome because they will not appear as normal, and thus, might be confused with tumor cells. However, we have also assumed that UTL cells might share CNVs with the tumor subpopulation only by chance. We argue that in the absence of an overwhelming number of UTL cells, it is possible to effectively assess the presence of a small population with a shared CN profile.
A detection procedure would be considered successful if it is both sensitive (low false negative detection rate), and highly specific (very low false positive detection rate). Success would critically depend on many factors: First, sensitivity would depend on the degree of CNV in the CR cells, their count, and their proportion in the population. Second, specificity would depend on the number and proportion of UTL cells. Lastly, success would depend on operational factors such as the accuracy of determining CN profiles of individual cells; and the computational protocol used to detect similar CN profiles in the presence of noise (the UTL profiles).
Modeling Single-Cell Sequence Data from Cells In Blood
To properly model a sample of cells that are subjected to an early detection procedure, we must specify for each category of cells in the sample, the expected CN properties and the expected cell count. For tumor cells, published integer CN profiles for 3,852 tumors, and representing eleven common solid tumor types, can be used in this analysis [26, 27]. We can assume that a sample obtained from a patient suffering from one of these tumors can contain CR cells, all sharing a CN profile with the tumor. We can further assume the CR cell count to be 10 cells per 10 mL, well within the range of published circulating tumor cell counts [6, 8, 28]. To model CR cells, one tumor profile is selected for each clone. The 3,852 published profiles are also used to model UTL cells: random chromosomes from genomes of archived tumor profiles are sampled. Modeling UTL cells as random tumor cells adds burden to the task, but it is a conservative procedure to follow. In the absence of experimental data on the number of UTL cells with substantial CNV, we can allow this number to vary in a broad range of 10 and 200 UTL cells per specimen. With these assumptions, one might prefer to err on the side of caution, as large numbers of cancer-like cells in a specimen can pose a challenge to achieve specificity of early detection. Finally, ND cells can be assumed to have strictly diploid genomes, and the specimen might be expected to contain 103 – 105 such cells. We anticipate that ND cells do not affect the specificity of early detection, even if present in large numbers.
Next, sparse single-cell sequence reads can be simulated. The methodology is outlined here (Key Figure, Figure 1) and described in full detail in the Supplemental Information. To model a read set with a specified coverage and CN profile, an empirical read set can be selected from an in-house (not yet published) collection of 1306 single-cell genomes of ND cells, with an average of 2×106 reads per genome. First, the needed number of 1,306 read sets are selected, and no read set is chosen twice. By not selecting the same read set twice, spurious correlations can be avoided, albeit limiting the simulation of sequence data to no more than that number of cells. The read set is then resampled with replacement to achieve a sparser read set that would correspond to a cell with the desired CN profile and coverage. The mean sampling rate per read can be set to reach an average overall coverage of 1.25×105 reads. This process might at first seem overly complicated, but captures nevertheless, the stochastic noise that is inherent of actual single-cell read data.
Clonal Detection Procedure
To derive CN profiles from simulated read data, a tested and published pipeline for sparse reads from cancer and diploid cells can be used [15, 29]: the genome is partitioned into contiguous bins, and the set of sequence reads is converted into read counts per bin. The output of the pipeline is thus a bin vector comprising counts whose values represent CN estimates in those bins. From two cells, each with bin vectors, a Pearson correlation can be calculated (Key Figure, Figure 1 D,E). Two CR cells would then exhibit high Pearson correlation, and UTL cells would have close to zero correlation. The distributions of pairwise correlation coefficients within the three populations (ND, UTL and CR) are shown in Figure 2B.
A simple heuristic is then introduced to detect the presence of a clonal population with a shared CN profile (Key Figure, Figure 1F). Individual cells in the specimen are considered vertices in a graph, and a pair of vertices is connected by an edge if the pairwise Pearson correlation of the bin vectors exceeds a given empirical test threshold. In this analysis, a correlation threshold of 0.7 is chosen because correlations of this magnitude are frequently observed among CR cells but never for ND cells (Figure 2B). Pairwise correlations of this magnitude might occur among the UTL cells, but at a frequency of approximately 1 per 104 pairs (Figure 2B). The largest component of the graph consists of the largest set of vertices and their connections such that a path of vertices and edges connects any two vertices (Key Figure, Figure 1F). The vertices of the largest component are selected to represent a clone of cells with shared CN profiles; and, the number of vertices in the connected component can provide an estimate for the number of clonal cells in the specimen. Tumor cells can be considered as ‘detected’ if the number of estimated clonal cells in the specimen exceeds a set value ‘S’, as described below (e.g. 8 detected cells are shown in Key Figure, Figure 1F).
Sensitivity and Specificity of Clonal Detection
With this procedure, the sensitivity and specificity of clone detection can be assessed. We consider these separately. For sensitivity we can ask how often a large connected component can be detected for 10 CR cells taken from each of the 3,852 tumors, each with published integer CN profiles [26]. Then, for specificity, we can ask how often ND or UTL cells -- present in given numbers -- result in a large connected component and therefore, represent a false positive signal.
For sensitivity, we can assume that 10 CR cells from the tumor are present in the specimen. Our summary analysis is depicted in Figure 2C, where for each of the eleven tumor types, the fraction of cases (on the Y-axis) with a detected clonal population of size S (on the X-axis) is depicted. Thus, the graph shows that this detection method would be highly sensitive in the majority of tumor types considered (Figure 2C). For example, a sensitivity of approximately 90% could be achieved in breast cancer if the size S of the largest connected component is set to be at least 6 (Figure 2B). For ovarian cancer, sensitivity could approach 100%, and this is relevant as this disease is most often diagnosed at a late stage, portending an adverse outcome.
To evaluate the specificity of detection, we can measure the frequency of false detection within a population containing only ND and UTL subpopulations. The criterion for a false positive would be defined by the presence of a connected component of highly correlated cells within the specimen. This false positive rate would therefore crucially depend on the frequency of high correlations in cell pairs within and between subpopulations. Pearson correlations of CN profiles of ND cells -- either among themselves or with CN profiles of UTL cells -- are highly unlikely to exceed the threshold value of 0.7. None of the empirical correlations of this kind are found to exceed this threshold. Furthermore, using methods of extreme-value theory [30–32], the probability of exceeding this threshold with at least one member of the pair being ND can be expressed in terms of the generalized Pareto distribution and estimated to be below 10−18. Consequently, even with approximately 5×109 pairwise correlations in a specimen of 105 cells, the ND cell subpopulation might present no challenge to specificity of detection, and could be safely dropped from the determination of the false positive rate.
By contrast, correlations among CN profiles of UTL cells might exceed the threshold of 0.7 with low, but non-negligible empirical probability (e.g. 1.6×10−4, Figure 2B). If present in sufficient numbers, these cells might occasionally form sizable connected components as defined above, leading to false discovery of a clonal population. The corresponding false positive rate can be determined empirically by simulating multiple random sets of 10, 20, 50, 100 or 200 of UTL cells and computing the size of the largest connected component for each set respectively (Table 1). In Table 1, the corresponding empirical false positive rate is listed for each of the five set sizes and for a range of threshold values ‘S’. For example, in a specimen with 100 UTL cells, a false discovery of a connected component comprising 6 or more cells would rarely occur, (approximately 1/3000 cases). Indeed, UTLs might occasionally contribute to the largest connected component (Key Figure, Figure 1 E, F), but could be eliminated upon follow-up (Key Figure, Figure 1 C–F).
Table 1.
Estimate of False Positive Ratea
| S | 10 UTL cells | 20 UTL cells | 50 UTL cells | 100 UTL cells | 200 UTL cells |
|---|---|---|---|---|---|
| 2 | 0.0086 | 0.028 | 0.15 | 0.43 | 0.84 |
| 3 | 3.00×10−4 | 0.0018 | 0.028 | 0.13 | 0.51 |
| 4 | 1.00×10−4 | 1.00×10−4 | 0.0036 | 0.044 | 0.3 |
| 5 | 0 | 1.00×10−4 | 4.00×10−4 | 0.012 | 0.16 |
| 6 | 0 | 0 | 2.00×10−4 | 0.0031 | 0.08 |
| 7 | 0 | 0 | 0 | 6.00×10−4 | 0.034 |
| 8 | 0 | 0 | 0 | 3.00×10−4 | 0.013 |
| 9 | 0 | 0 | 0 | 0 | 0.0049 |
| 10 | 0 | 0 | 0 | 0 | 0.0012 |
| 12 | 0 | 0 | 0 | 0 | 1.00×10−4 |
Assuming 10, 20, 50, 100 or 200 UTL cells are present in the specimen (columns), the frequency of observing a connected component of at least S = 2, 3, …, 10, or 12 cells is estimated (rows; cf Methods in the Supplemental Information).
Follow-up Analysis from Positive Findings
As discussed above, false positive rates can be low, but are not ‘zero’; consequently, any level of false positives is unwanted. Moreover, a true positive must be actionable. Therefore, detection of a clonal pattern in blood would require us to do more. Certainly, a repeat test, starting with larger volumes of blood would be warranted. Once the positive cells are identified, they can be further analyzed, either singly or in pools, provided one uses methods that enable the reexamination of single-cell nucleic acids (Key Figure, Figure 1 C–F). This is presently out of reach in our repertoire of molecular tools, but may be entirely possible using either addressable arrays of single-cell nucleic acids or, addressable nucleic acid libraries.
Deeper DNA sequencing of candidate cell nucleic acid can enable the virtual elimination of false positives, as UTLs with correlated profiles (low coverage and resolution) might rarely exhibit coincident individual CN events (higher coverage and resolution), whereas clonal tumor cells would. Moreover, details of CN profiles (overall ploidy, number and location of CN events, etc.) combined with knowledge of tissue origin, are likely to yield (ultimately) a good assessment of the malignant risk and source of the neoplasm [20–22]. Using pooled nucleic acid from tumor cells that are confirmed, we might reach sufficient sequence depth to observe therapeutically actionable mutation patterns. Finally, strong clues to the tissue of origin can in turn be discovered from methylation patterns, chromatin structure and gene expression data [24, 25]. Comprehensive databases with this type of information are currently being compiled and will be most useful [33, 34]. Indeed, defining an anatomical origin of neoplasia will be critical for diagnostics, where once the location of probable primary sites is identified, subsequent scanning/imaging at high resolution could validate any preliminary blood-based findings.
Concluding Remarks
We have outlined a sparse genomic sequencing method for detecting cancer from single-cells in blood (Key Figure, Figure 1). It comprises components that include: (i) enriching atypical cells from blood (Key Figure, Figure 1 A–C); (ii) separating individual cells (Key Figure, Figure 1C); (iii) performing inexpensive copy-number profiling (Key Figure, Figure 1D); and (iv) detecting clonal CN profiles computationally (Key Figure, Figure 1D–F). The technical components of (i)–(iii) are presently feasible, and we posit that based on simulation analysis under certain conditions, part (iv) could detect clonal cancer cells with low false positive rates. No false positive is without harm, and a true positive will require actionable information, which might be attained by tracing-back and performing deep-sequencing analysis of candidate single-cell nucleic acids, either singly, or in pools (Key Figure, Figure 1C–F). In this scenario, one might confirm risk of disease, lower false positive finding to negligible levels, discover critical mutations of possibly therapeutic significance, determine anatomic origin of the primary site of clonality. This information would then be actionable.
From another angle, we have set the sequence coverage for genomic analysis in a way that the procedure should be affordable. At approximately 125,000 reads per single-cell (given the least expensive high throughput sequence platform presently available), we anticipate that the cost basis for the sequencing alone might be constrained to approximately $1 USD per cell. Detecting ten tumor cells in the presence of a thousand diploid cells might cost approximately $1000 USD for the sequencing component. At this sequence depth, false positive rates may be acceptable, if no more than 100 non-clonal cells with abnormal genomes are present per enriched blood specimen. A more expensive test, with higher coverage, would reduce false positive rates. The follow-up analysis after a positive detection would undoubtedly be more expensive.
Of note, other conditions might arise to confound detection, such as the presence of benign populations in the blood carrying CN alterations (see Outstanding Questions and Box 1). We cannot presently answer this and many other critical questions, such as how often, in what numbers, and at what stage do tumors release malignant cells into the blood. One central question is whether this detection method in blood, reveals malignant neoplasms sufficiently early, so that appropriate interventions can take place, and the incidence of metastasis be reduced for any cancer type. The procedure might best benefit certain patients with increased risk, for example the elderly, those with genetic predispositions, or those with suspected lesions but for whom invasive surgical biopsy is not the best option. Of relevance, these ideas are not restricted to the detection of cancer signature in blood, but might also be of value in detecting low levels of non-specific cancer signatures in any biological specimen.
Outstanding Questions.
What is the specific abundance of malignant clonal cells in circulation, before the emergence of macroscopic disease, by tumor type?
Are non-clonal aneuploid cells in circulation rare enough to keep false detection rates low?
How frequently are non-malignant clonal cell populations bearing substantial copy number variation found in blood?
What categories of patients are most likely to benefit from, and should be a priority target for single-cell based early detection procedures?
Box 1. Clinician’s Corner.
Metastasis is by far the most lethal aspect of cancer. In all likelihood, cancer mortality would be significantly reduced if most tumors were detected early enough to preempt metastatic spread.
Early detection of tumors cannot be accomplished consistently by methods in present clinical practice, as none of these methods are simultaneously sensitive, specific and applicable across multiple tumor types. For example, MRI or CAT scans are able to detect a tumor at many anatomic locations, but not before the tumor is macroscopically large. Similarly, blood-based tests for antigens such as PSA (prostate specific antigen) focus on a single tumor type and lack specificity.
There is mounting evidence that tumor cells enter circulation and are disseminated from the primary site to distant organs early in disease. It is therefore likely that such cells are present in circulation even before the disease is detectable by imaging or by physical examination.
Here, we argue that for multiple tumor types, a combination of molecular techniques can be used to detect circulating tumor cells (CTC), even if the latter are as rare as one per milliliter of blood.
The procedure we envision involves three critical steps: i) candidate CTC are separated from blood cells using surface proteins that discriminate the two populations; ii) the genome of each candidate CTC is examined individually for aberrations in DNA CN; iii) CTC are identified from cell candidates based on a shared pattern of aberrations.
The entire detection procedure might be accomplished with existing molecular technologies, at a cost of approximately $1,000 per test.
Once CTC are identified, a more stringent molecular analysis can be used, and may include a combination of deep-sequencing and analysis of methylation or gene expression patterns. This information might be used to confirm malignancy risk and identify putative tissue of origin, thus guiding possible diagnosis and treatment.
Supplementary Material
Trends.
Metastasis is the most lethal aspect of cancer. To preempt metastatic spread tumors must be detected early.
There is ample experimental evidence that large scale DNA copy number (CN) variation is ubiquitous in multiple tumor types.
Clonal expansion is a hallmark of cancer; clonal expansion of cells with massively altered CN profiles is not observed outside of cancers and should be targeted for early detection.
Single-cell genomic analysis may be the best method to ascertain that multiple genomic alterations occur within the same cell.
Massively parallel CN profiling of individual cells by sparse sequencing is accurate and affordable.
Powerful microfluidic technologies exist to isolate small numbers (in the thousands) of candidate circulating tumor cells from blood samples.
Similarities among single-cell CN profiles may be exploited to identify clonal subpopulations.
Transcriptional, epigenetic or immunohistochemical examination of analytes pooled from suspicious malignant clonal cells found in blood, may offer an opportunity to determine the tissue of origin.
Acknowledgments
The authors are grateful to David Donoho for informative discussions in the early phases of this work. This work was supported by the National Cancer Institute (A.K. and M.W., award U01CA188590), the National Institutes of Health (M.W., award 5R01CA181595-03), by the Simons Foundation (A.K., D.L. and M.W.) and by the Breast Cancer Research Foundation (M.W.).
Glossary
- Bins
here, sequence intervals into which a genome is partitioned
- Cell-free (cf) DNA
DNA shed into the bloodstream as a result of cell disintegration
- Clonally related (CR) subpopulation
cells originating from a tumor clone
- Copy number (CN) profile
CN of a genomic DNA sequence as a function of genomic position
- Counts per bin
sequence read counts in each of the bins into which the genome is partitioned
- DNA methylation patterns
tissue-specific degree of DNA methylation as a function of genomic position
- DNA copy number variation
large-scale copy number losses and gains.
- Edge
a pairwise relationship among two vertices in a graph; if vertices are graphically represented by points, edges are represented by intervals connecting pairs of points
- Extreme-value theory
a branch of statistics dealing with extremely rare observations
- False negative rate
the fraction of patients, with at least one clonal tumor cell per milliliter of blood, missed by the test; equals one minus the sensitivity
- False positive rate
the fraction of tumor-free patients among those in whom the test points to the presence of a tumor clone in blood; equals one minus the specificity
- Graph
here, a set of objects called vertices and a (0, 1)-valued function on pairs of vertices, called adjacency; a pair of vertices with the adjacency of one are said to be connected by an edge
- Integer copy-number (CN) profile
an approximation of an observed CN profile by an integer function of the genomic position
- Largest (connected) component
the largest subset of vertices in a graph such that there is a path of edges between any two vertices in the subset
- Modal
here, a value of DNA CN observed in a greater portion of the genome
- Normal diploid (ND) cell
a cell with DNA CN of two throughout the genome, with a possible exception of the X and Y chromosomes
- RNA expression profiles
numerical data consisting of RNA expression levels for each gene in a given biological entity
- Sensitivity
the ratio of the number of patients testing positive for circulating tumor cells to the number of patients with circulating tumor cells being tested
- Sequence coverage
the total length of sequence reads divided by the length of the genome being sequenced
- Sequence read
a portion of an individual DNA molecule in which the nucleotide sequence is determined by a sequencing apparatus
- Shared copy number profile
CN profile common to several cells
- Specificity
the ratio of the number of true positives to the number of all positive test results
- Stochastic copy-number noise
variation in the CN profile due to random effects inherent in DNA preparation and sequencing procedures
- Unrelated tumor-like (UTL) cells
cells with significantly altered CN but not originating from a tumor clone
- Vertex
an object in a graph; vertices in a graph are customarily illustrated as points
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ignatiadis M, M Lee, Jeffrey SS. Circulating Tumor Cells and Circulating Tumor DNA: Challenges and Opportunities on the Path to Clinical Utility. Clinical Cancer Research. 2015;21(21):4786–4800. doi: 10.1158/1078-0432.CCR-14-1190. [DOI] [PubMed] [Google Scholar]
- 2.Shiozawa Y, et al. Bone marrow as a metastatic niche for disseminated tumor cells from solid tumors. Bonekey Rep. 2015;4:689. doi: 10.1038/bonekey.2015.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demeulemeester J, et al. Tracing the origin of disseminated tumor cells in breast cancer using single-cell sequencing. Genome Biol. 2016;17(1):250. doi: 10.1186/s13059-016-1109-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pantel K. Blood-Based Analysis of Circulating Cell-Free DNA and Tumor Cells for Early Cancer Detection. PLoS Med. 2016;13(12):e1002205. doi: 10.1371/journal.pmed.1002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarioglu AF, et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat Methods. 2015;12(7):685–91. doi: 10.1038/nmeth.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Theil G, et al. The Use of a New CellCollector to Isolate Circulating Tumor Cells from the Blood of Patients with Different Stages of Prostate Cancer and Clinical Outcomes - A Proof-of-Concept Study. PLoS One. 2016;11(8):e0158354. doi: 10.1371/journal.pone.0158354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Toom EE, et al. Technical challenges in the isolation and analysis of circulating tumor cells. Oncotarget. 2016;7(38):62754–62766. doi: 10.18632/oncotarget.11191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao L, et al. Enhanced and Differential Capture of Circulating Tumor Cells from Lung Cancer Patients by Microfluidic Assays Using Aptamer Cocktail. Small. 2016;12(8):1072–81. doi: 10.1002/smll.201503188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heitzer E, et al. Establishment of tumor-specific copy number alterations from plasma DNA of patients with cancer. Int J Cancer. 2013;133(2):346–56. doi: 10.1002/ijc.28030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bettegowda C, et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Science Translational Medicine. 2014;6(224) doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martincorena I, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348(6237):880–6. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Putten LJM, et al. Molecular profiles of benign and (pre) malignant endometrial lesions. Carcinogenesis. 2017;38(3):329–335. doi: 10.1093/carcin/bgx008. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Swennenhuis JF, et al. Improving the CellSearch(R) system. Expert Rev Mol Diagn. 2016;16(12):1291–1305. doi: 10.1080/14737159.2016.1255144. [DOI] [PubMed] [Google Scholar]
- 15.Baslan T, et al. Optimizing sparse sequencing of single cells for highly multiplex copy number profiling. Genome Research. 2015;25(5):714–724. doi: 10.1101/gr.188060.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gawad C, W Koh, Quake SR. Single-cell genome sequencing: current state of the science. Nature Reviews Genetics. 2016;17(3):175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 17.Kolodziejczyk AA, et al. The Technology and Biology of Single-Cell RNA Sequencing. Molecular Cell. 2015;58(4):610–620. doi: 10.1016/j.molcel.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 18.Trombetta JJ, et al. Preparation of Single-Cell RNA-Seq Libraries for Next Generation Sequencing. Curr Protoc Mol Biol. 2014;107:4 22 1–17. doi: 10.1002/0471142727.mb0422s107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spitzer MH, Nolan GP. Mass Cytometry: Single Cells, Many Features. Cell. 2016;165(4):780–91. doi: 10.1016/j.cell.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blegen H, et al. DNA amplifications and aneuploidy, high proliferative activity and impaired cell cycle control characterize breast carcinomas with poor prognosis. Anal Cell Pathol. 2003;25(3):103–14. doi: 10.1155/2003/491362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hicks J, et al. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res. 2006;16(12):1465–79. doi: 10.1101/gr.5460106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hieronymus H, et al. Copy number alteration burden predicts prostate cancer relapse. Proc Natl Acad Sci U S A. 2014;111(30):11139–44. doi: 10.1073/pnas.1411446111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stancel GA, et al. Identification of tissue of origin in body fluid specimens using a gene expression microarray assay. Cancer Cytopathol. 2012;120(1):62–70. doi: 10.1002/cncy.20167. [DOI] [PubMed] [Google Scholar]
- 24.Moran S, et al. Epigenetic profiling to classify cancer of unknown primary: a multicentre, retrospective analysis. Lancet Oncol. 2016;17(10):1386–1395. doi: 10.1016/S1470-2045(16)30297-2. [DOI] [PubMed] [Google Scholar]
- 25.Sun K, et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci U S A. 2015;112(40):E5503–12. doi: 10.1073/pnas.1508736112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45(10):1134–40. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30(5):413–21. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao W, et al. Analysis of circulating tumor cells from lung cancer patients with multiple biomarkers using high-performance size-based microfluidic chip. Oncotarget. 2016 doi: 10.18632/oncotarget.14203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kendall J, Krasnitz A. Computational methods for DNA copy-number analysis of tumors. Methods Mol Biol. 2014;1176:243–59. doi: 10.1007/978-1-4939-0992-6_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coles S. Springer series in statistics. London; New York: Springer. xiv; 2001. An introduction to statistical modeling of extreme values; p. 208. [Google Scholar]
- 31.Gilleland E, Katz RW. extRemes 2.0: An Extreme Value Analysis Package in R. Journal of Statistical Software. 2016;72(8):1–39. [Google Scholar]
- 32.Gilleland E, Ribatet M, Stephenson AG. A software review for extreme value analysis. Extremes. 2013;16(1):103–119. [Google Scholar]
- 33.Bujold D, et al. The International Human Epigenome Consortium Data Portal. Cell Syst. 2016;3(5):496–499 e2. doi: 10.1016/j.cels.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 34.Roadmap Epigenomics C, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.