SUMMARY
G protein-coupled receptor (GPCR)-mediated signal transduction is central to human physiology and disease intervention, yet the molecular mechanisms responsible for ligand-dependent signaling responses remain poorly understood. In Class A GPCRs, receptor activation and G protein coupling entail outward movements of transmembrane segment 6 (TM6). Using single-molecule Fluorescence Resonance Energy Transfer (smFRET) imaging, we examine TM6 motions in the β2 adrenergic receptor (β2AR) upon exposure to orthosteric ligands with different efficacies, in the absence and presence of the Gs heterotrimer. We show that partial and full agonists affect TM6 motions in a manner that differentially regulates the rate at which GDP-bound β2AR-Gs complexes are formed and the efficiency of nucleotide exchange leading to Gs activation. These data also reveal transient nucleotide-bound β2AR-Gs species distinct from known structures and single-molecule perspectives on the allosteric link between ligand and nucleotide binding pockets that shed new light on the G protein activation mechanism.
GPCRs regulate cellular responses to neurotransmitters and hormones and act as ligand-regulated guanosine nucleotide exchange factors (GEFs) for heterotrimeric G proteins1. Ligand efficacy has historically referred to a molecule’s capacity to elicit a specific physiological response downstream of receptor activation2,3. Although an important parameter in drug development, the molecular basis of efficacy with respect to a ligand’s impact on GPCR structure, dynamics and G protein coupling remains poorly understood.
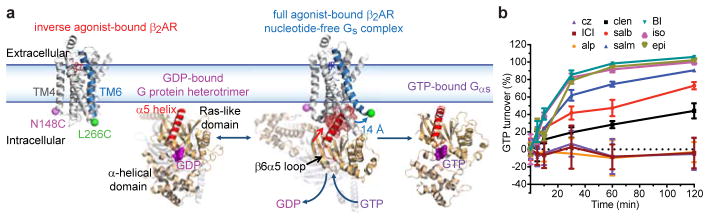
The β2 adrenergic receptor (β2AR), a paradigmatic, Class A GPCR couples preferentially to the heterotrimeric Gs protein, comprised of Gαs, Gβ and Gγ (Fig. 1a)4. Investigations of the β2AR activation mechanism have been enabled by synthetic ligands with efficacy profiles ranging from inverse agonists that suppress basal activity, and neutral antagonists that prevent agonist-induced activation, to partial and full agonists that differentially promote receptor-mediated Gs activation5. Recent crystallographic structures of distinct Class A GPCRs in both inactive and active states6–9 revealed that the largest conformational change associated with their activation is an outward movement of the cytoplasmic end of transmembrane helix 6 (TM6)6. In the nucleotide-free β2AR-Gs complex, TM6 is stabilized in an outward configuration by insertion of the C-terminal α5 helix of Gαs into a pocket formed by the cytoplasmic ends of TM3, TM5 and TM6 and intracellular loop 2 (ICL2).
Figure 1.
High-resolution perspective of β2AR-Gs activation. a, Labeling sites N148C (magenta) in TM4 and L266C (green) in TM6 (blue) are shown on the inactive β2AR structure (PDB:2RH1). Agonist activation leads to outward TM6 displacement (~14Å) and Gs coupling (shown GDP-bound Gi; PDB:1GP2). Within the GDP-free complex the α5 helix (red) of Gαs (wheat) engages the cytoplasmic face (red mesh) of β2AR (PDB:3SN6). GTP binding causes Gαs (PDB:1AZT) and Gβγ subunit (not shown) to separate. b, Ligand efficacy profiles determined using a GTP turnover assay (Methods; error bars=s.e.m., 3 replicates).
Ensemble techniques, including fluorescence10, electron paramagnetic resonance (EPR)11 and nuclear magnetic resonance (NMR) spectroscopy11,12 reveal that even the most potent agonists fail to fully stabilize β2AR in its activated conformation in the absence of G protein or stabilizing nanobodies. The molecular basis of ligand efficacy may therefore be defined by changes in receptor dynamics and conformation that impact the probability of G-protein coupling, productive nucleotide exchange and subsequent dissociation. Hence, we used total internal reflection fluorescence (TIRF) smFRET imaging to track TM6 movements in β2AR bound to ligands with distinct efficacy profiles to determine the impacts on receptor structure, dynamics, and G protein coupling.
Site-specific labeling of β2AR
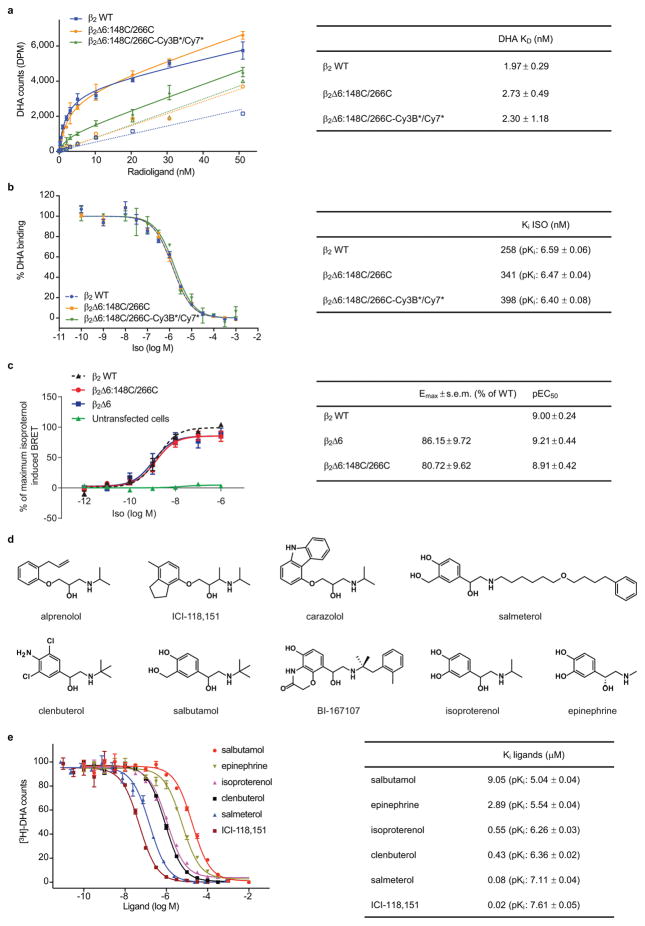
We site-specifically attached donor and acceptor fluorescent probes at the cytoplasmic ends of TM6 (L266C6.28) and TM4 (N148C4.40), respectively, within a full-length, minimal cysteine β2AR mutant (Fig. 1a; Methods). This construct (β2Δ6-148C/266C) exhibits wild-type ligand binding and Gs coupling (Extended Data Fig. 1a–c).
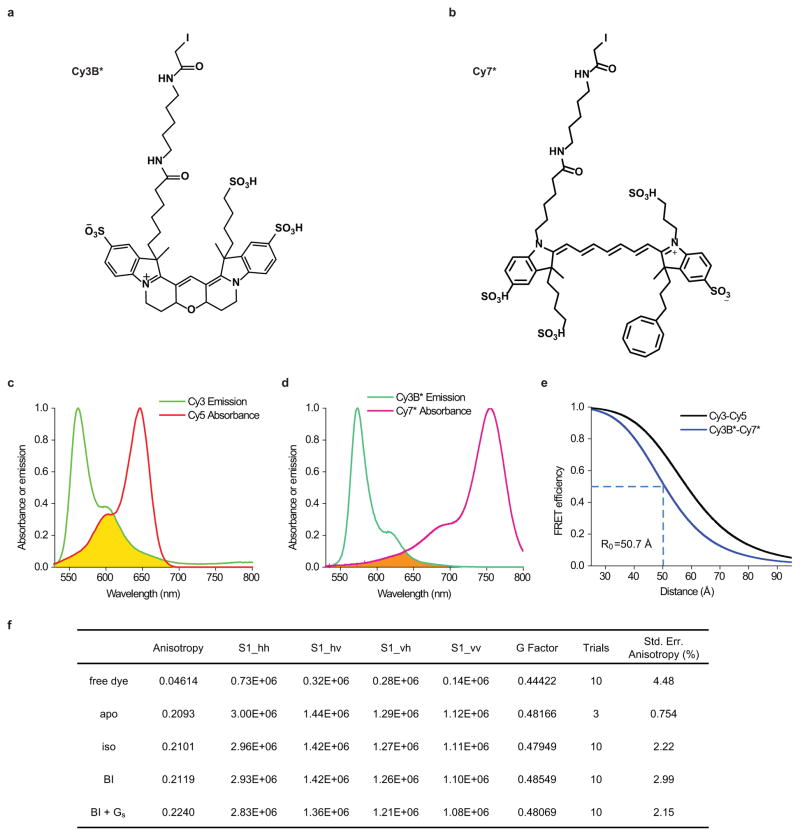
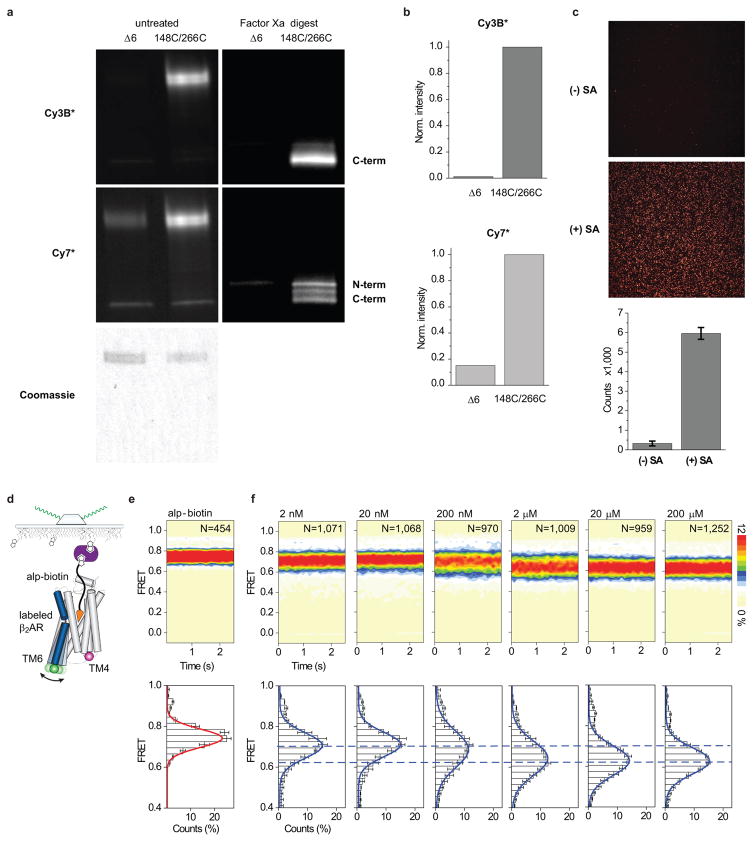
Given β2AR’s relatively small size (~34Å lateral dimension) and TM6’s anticipated displacement (~14Å) upon activation (Fig. 1a)6, β2Δ6-148C/266C was labeled with an optimized Cy3B and Cy7 fluorophore pair (Cy3B* and Cy7*, Extended Data Figs. 2a,b and 3a,b; Methods), which exhibit high quantum yields and a small R0 (~50.7Å) (Extended Data Fig. 2a–e) and should be relatively insensitive to chemical environment13. Labeled receptors showed wild-type activities with respect to both antagonist and agonist binding (Extended Data Fig. 1a,b).
Ligand-induced TM6 displacement
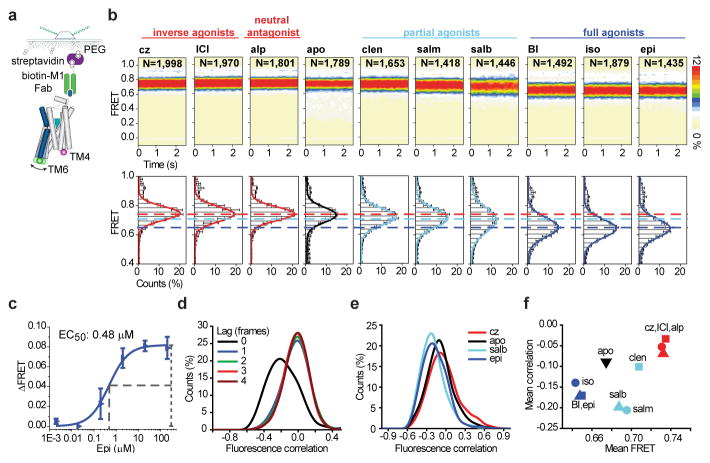
The impacts of saturating concentrations of nine ligands with distinct efficacies (Fig. 1b, Extended Data Fig. 1d,e) were examined by imaging Cy3B*/Cy7*-labeled β2Δ6-148C/266C immobilized with an M1 Fab fragment (Fig. 2a,b; Extended Data Figs. 3c and 4a,b). In the presence of the neutral antagonist alprenolol (alp), the inverse agonists carazolol (cz), and ICI-118,151 (ICI), β2AR exhibited indistinguishable population FRET efficiencies (FRET) centered at 0.74±0.01 (~0.74) and full-widths at half maximum height (FWHM) of 0.1±0.01 (~0.1; mean of biological replicates ±s.d.) (Fig. 2b). Inspection of individual trajectories from alp-, cz- and ICI-bound receptors revealed relatively stable fluorescence and FRET (Extended Data Fig. 4a). Similar results were obtained when β2AR was immobilized via biotinylated alp (Extended Data Fig. 3d,e). These data predict an average inter-dye distance of approximately 42Å (Extended Data Fig. 2d–f), in good agreement with molecular dynamics (MD) simulations (Extended Data Fig. 5a,b).
Figure 2.
The extent of TM6 motions correlate with efficacy. a, Schematic of ligand binding experiments (Methods). b, Population FRET contour plots (top; scale bar=relative population) and cumulative population histograms (bottom; error bars=s.d., 3 replicates with N total molecules). Dashed lines indicate the distinct mean FRET values observed. c, Apparent EC50 for epinephrine (Methods; error bars=s.d., 3 replicates). d, Pearson’s correlation histograms (Methods) describing anticorrelated donor and acceptor fluorescence fluctuations in epinephrine with increasing donor lag time. e, Comparison of zero-lag Pearson’s correlation histograms for representative ligands. f, Scatter plot of mean FRET efficiencies vs. mean correlation coefficients observed for each ligand.
The unliganded (apo) β2AR exhibited a mean FRET value and FWHM similar to that of alp, cz, and ICI (Fig. 2b). In the presence of the partial agonists clenbuterol (clen), salmeterol (salm) or salbutamol (salb), β2AR exhibited modestly lower, and more broadly distributed FRET values (~0.70–0.72; FWHM:~0.15) (Fig. 2b). Reductions in FRET were more pronounced in the presence of the full agonists epinephrine (epi), isoproterenol (iso) and BI-167107 (BI), where the mean FRET value shifted to ~0.64 (FWHM:~0.15) (Fig. 2b). In agreement with radioligand binding studies (Extended Data Fig. 1e), smFRET experiments showed that epi exhibited a half-maximum effective concentration (EC50) of 0.48±0.09 μM (mean of biological replicates ±s.e.m.) (Fig. 2c; Extended Data Fig. 3f). Hence, we infer that these epi-induced FRET changes (ΔFRET apo/epi:~0.1), corresponding to an increase in average interdye distance of ~4Å (Extended Data Fig. 2e), reflect those of a fully functional receptor.
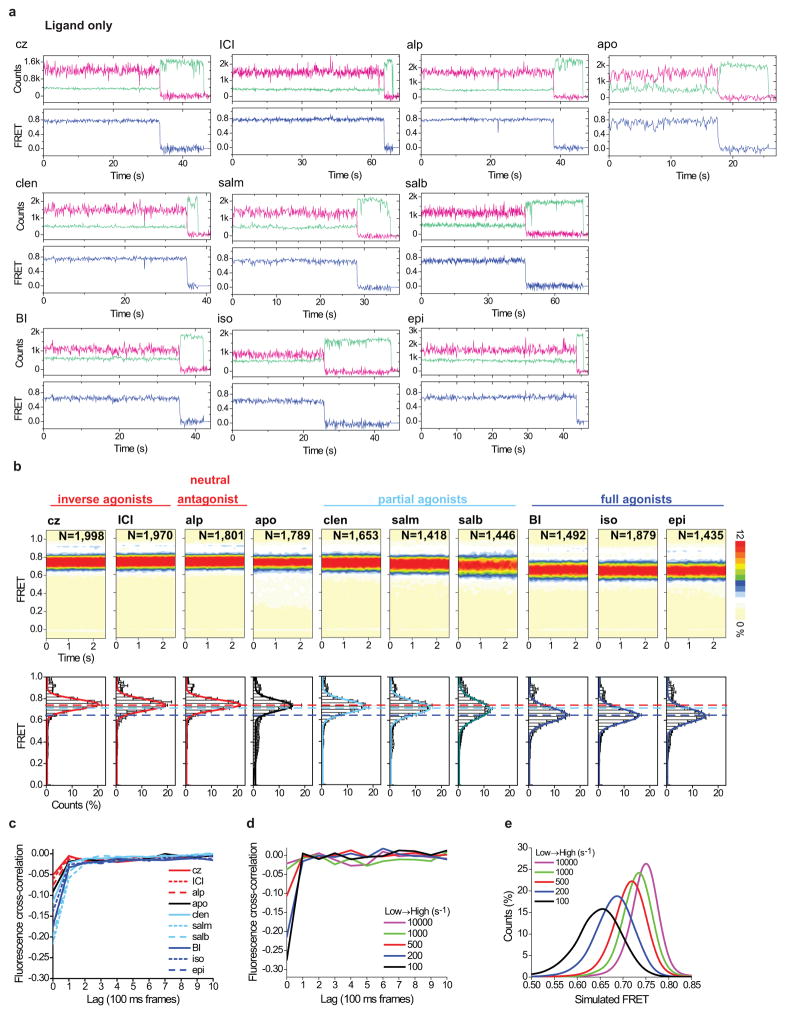
While the ~14Å outward TM6 movement observed in fully activated β2AR (Fig. 1a; Extended Data Fig. 5) is anticipated to yield a FRET change of ~0.3 (Extended Data Fig. 2e), inspection of individual FRET trajectories revealed only rare fluctuations (ca. 1 min−1) of this amplitude (Extended Data Fig. 4a). However, correlation analyses14,15 revealed clear signatures of anticorrelated fluorescence within the ensemble of individual molecules (Fig. 2d–f; Extended Data Fig. 4c–e; Methods), indicating fast (>10 s−1), reversible TM6 movements. By this measure, more rapid TM6 dynamics (more negative mean correlation) were observed in agonist-bound samples than antagonist-bound samples (Fig 2e,f). Simulations revealed that the observed fluorescence correlation and FRET distributions could be recapitulated by TM6 deflections to lower FRET states at rates of 100–500 s−1 (Extended Data Fig. 4d,e). Previous reports of slower TM6 movements (ca.~0.5–5 s−1)11,16 may reflect differences in experimental conditions or the nature of their probes, which detect changes in environment, not distance. Hence, we conclude that the distinct FRET values observed for each agonist reflect differences in the underlying rates and/or amplitudes of TM6 motions into, and out of, active-like conformations, which are time averaged at the present imaging resolution.
The rate of β2AR-Gs complex formation
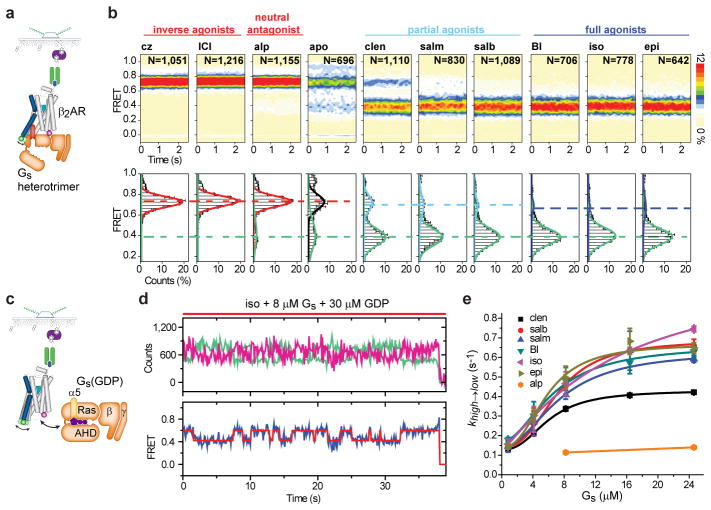
To ascertain directly the extent to which the ligand-induced changes observed in smFRET correlate with the coupling efficiency of β2AR to G protein, labeled β2AR (1 nM) was incubated with Gs (8 μM) and apyrase (0.2 nM). Individual β2AR molecules were subsequently imaged via M1 immobilization to determine the extent of β2AR-Gs complex formation (Fig. 3a).
Figure 3.
Ligand efficacy affects β2AR-Gs coupling efficiency. a, Schematic of Gs-coupling experiments (Methods). b, Population FRET contour plots (top; scale bar=relative population) and cumulative population histograms (bottom; error bars=s.d., 2 replicates with N total molecules) observed for apyrase-treated (nucleotide free) β2AR-Gs complexes. Dashed lines show correspondence between high- and low-FRET values. c, Schematic of Gs titration experiments (Methods). d, Example fluorescence (donor [light green]; acceptor [magenta]), and FRET (blue) time traces overlaid with predicted state sequence (red) in the presence of iso, GDP and Gs. e, Transition rates from high- to low-FRET states (khigh→low; error bars=s.d., 2 replicates) with increasing Gs(GDP) concentrations.
In the presence of cz and ICI, β2AR exhibited FRET behaviors indistinguishable from those observed in the absence of the Gs heterotrimer (Fig. 2b,3b). By contrast, agonist-activated β2AR complexes exhibited a distinct low-FRET (~0.4) state with similar FWHM values (~0.14) (Fig. 3b). This low-FRET value reflects an inter-dye distance of approximately 55Å (Extended Data Fig. 2e), in close agreement with molecular dynamics simulations of the β2AR-Gs complex (Extended Data Fig. 5a–c). Analogous low-FRET states were observed when agonist-activated receptor was immobilized via a biotinylated Gs heterotrimer (Extended Data Fig. 6a–d). The proportion of receptors exhibiting low FRET (Fig. 3b), as well as the fraction of time individual receptors occupied low-FRET states (Extended Data Fig. 7a), correlated with ligand efficacy (Fig. 1b). Consistent with the known basal activity of β2AR17, low levels (5–20%) of Gs coupling were evidenced in the absence of ligand and in the presence of alp (Fig. 3b; Extended Data Fig. 7a).
To examine the role of ligand efficacy on the rates of β2AR-Gs complex formation, M1-immobilized β2AR (Fig. 3c) was imaged in the presence of 30 μM GDP at increasing Gs concentrations, where reversible transitions between high- and low-FRET states could be quantified using a two-state hidden Markov Model (HMM) (Fig. 3d; Methods). Consistent with a bimolecular reaction entailing at least one ligand-dependent, rate-determining process that precedes complete Gs coupling, we found that rates of low-FRET state formation increased with Gs concentration and then plateaued (Fig. 3e; Extended Data Table 1).
Based on the initial slope of the Gs-dependent increase in the rate of low-FRET state formation, we estimate the apparent Gs on rate to β2AR at ~0.03 and ~0.05 μM−1s−1 for clen and epi, respectively (Fig. 3e; Extended Data Table 1). These rates are orders of magnitude slower than expected for the binding of large entities18. They are, however, similar to bioluminescence resonance energy transfer measurements of β2AR-mediated Gs activation in living cells (ca. 2–3 s−1)19, where Gs lipidation localizes the heterotrimer to cellular membranes. They are also consistent with the observation that rate-limiting conformational changes within the rhodopsin-Gt complex precede GTP loading, although Gt activation occurs at a faster rate20. Hence, either initial interactions between Gs and β2AR preceding low-FRET state formation are highly transient (≪100ms) or do not immediately precipitate a FRET change. We therefore hypothesized that Gs coupling may be rate-limited by one or more ligand-dependent conformational processes that occur within a high-FRET β2AR-Gs complex.
Ligand impacts on β2AR-Gs stability
To examine the stability of apyrase-treated β2AR-Gs complexes in the presence of partial and full agonists, we measured the dissociation rate of β2AR from immobilized Gs heterotrimers (Extended Data Fig. 6a). Under low illumination intensity, where photobleaching was negligible, immobilized β2AR-Gs complexes exhibited lifetimes of 5–10 minutes (Extended Data Table 2). Notably, these lifetimes decreased ~20–100 fold in the presence of physiological concentrations of GDP (30 μM) or GTP (100 μM)21 (Extended Data Table 2), to become similar to those observed in cell-based studies of β2AR-Gs19 and β1AR-Gs complexes22. Hence, β2AR-Gs complexes generally persist for multiple seconds in the presence of nucleotide and dissociate approximately two-fold faster in the presence of GTP compared to GDP. These observations are consistent with specific interactions between the receptor and nucleotide-bound G protein and suggest nucleotide-specific dissociation pathways.
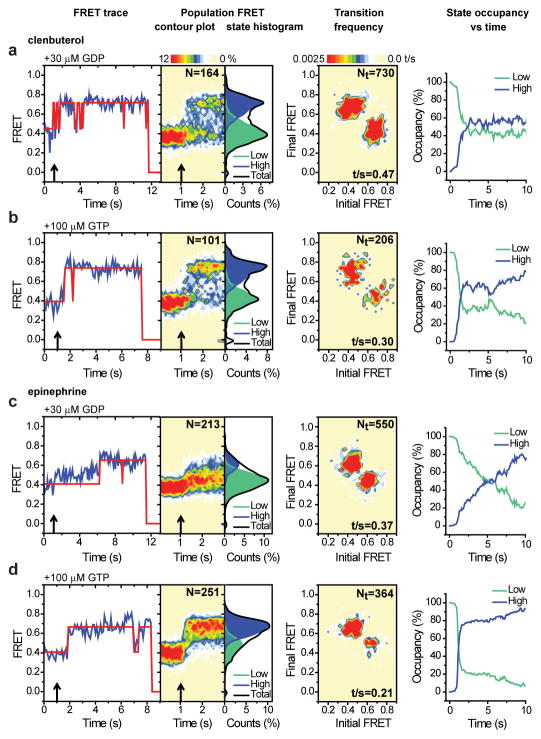
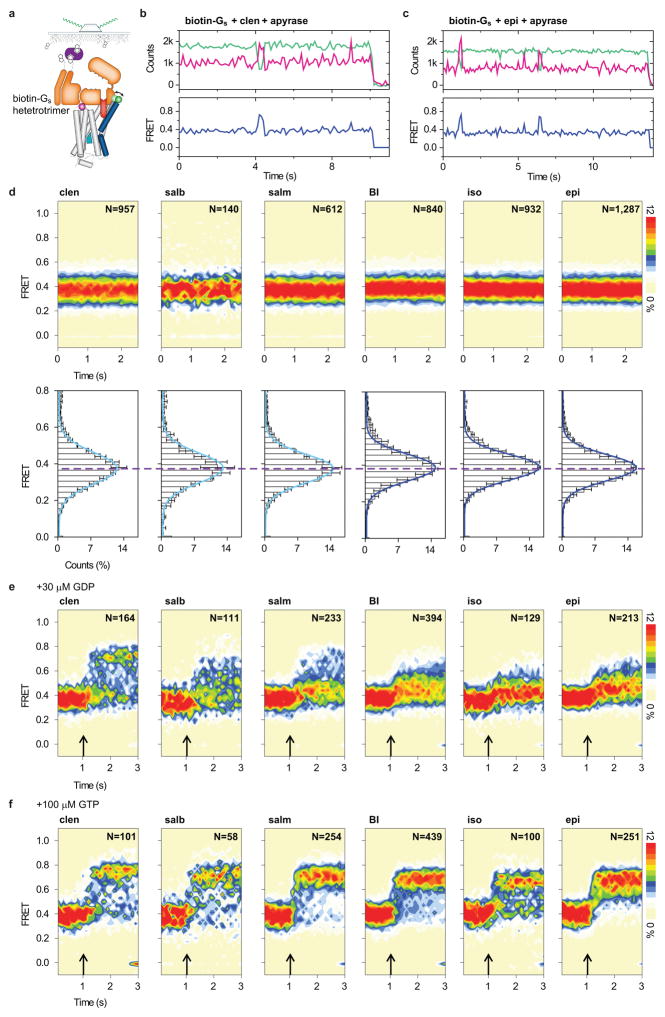
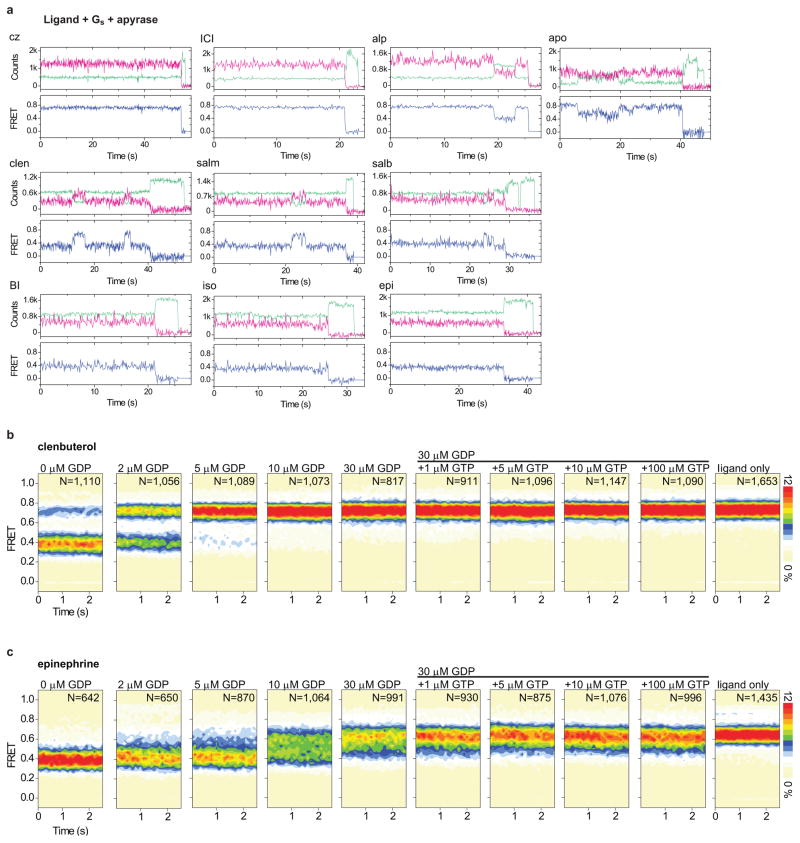
To track changes in FRET efficiency within the nucleotide-depleted β2AR-Gs complex upon GDP or GTP nucleotide binding, we performed analogous experiments at high spatial and temporal resolution (Fig. 4; Extended Data Fig. 6a,e,f). In contrast to the relatively slow rates of complex dissociation, the rapid introduction (ca. 100 ms mixing time) of either GDP (30 μM) or GTP (100 μM) gave rise to an immediate increase, or broadening, of the low-FRET state (ΔFRET ~0.05–0.1) (Fig. 4; Extended Data Fig. 6e,f). These data indicate that nucleotides bind the nucleotide-free β2AR-Gs complex at, or near, the diffusion limit, and that binding results in structural and/or dynamic changes in TM6 that initiate its return towards the helix bundle. Strikingly, these rapid changes were followed by formation of a relatively long-lived, predominantly high-FRET configuration from which reversible transitions to lower-FRET states occurred with frequencies and durations that were efficacy dependent (Fig. 4; Extended Data Fig. 6e,f).
Figure 4.
Pre-steady state measurements of nucleotide binding to clen- and epi-bound β2AR-Gs complexes. a–d (panels from left to right), Sample FRET traces (blue) with state idealizations (red line) following nucleotide addition (arrow). Population FRET contour plots (scale bar=relative population; N=total molecules) before and after nucleotide addition (arrow). Population histograms showing the relative occupancies of low- (green) and high- (blue) FRET states post-nucleotide addition. Transition density plots displaying the mean FRET values before (x axis) and after (y axis) each transition (scale bar=transitions per second t/s; Nt=total transitions). Time-dependent changes in the relative occupancies of low- (green) and high- (blue) FRET states.
The addition of GTP triggered transitions to high-FRET that were more rapid and complete than observed for GDP, and both the rate and efficiency of high-FRET state formation were greater for full agonists than for partial agonists (Fig. 4b,d; Extended Data Fig. 6f). These findings provide direct evidence for nucleotide-specific dissociation pathways, and suggest that β2AR-Gs(GNP) complexes can access multiple conformations distinct from the nucleotide-free state observed crystallographically6. The observed persistence of high-FRET, GDP- and GTP-loaded configurations implies the existence of relatively long-lived, β2AR-Gs(GDP) complexes during initial binding that precede low-FRET state formation, as well as long-lived β2AR-Gs(GTP) complexes after G protein activation. While the absolute rates we observe may be influenced by interactions between the lipid modifications on Gs and the detergent micelle surrounding β2AR, pre- and post-coupled GEF-G protein complexes have been evidenced in distinct systems using independent methods23–26 and may have important consequences for GPCR signaling.
Ligand impact on G protein activation
The effect of ligand efficacy on the allosteric link between β2AR and Gs, and its role in nucleotide exchange was further examined in steady-state experiments using M1-immobilized receptor in the presence of activating ligands, nucleotide, and 8μM Gs (Fig. 5a; Extended Data Fig. 7b,c). Under such conditions, Gs dissociation events are expected to be slow (ca. ~0.1–0.2 s−1; Extended Data Table 2), while dynamic processes within the β2AR-Gs complex should occur rapidly (Fig. 4). As noted above, β2AR-Gs complexes can exhibit three distinct FRET states: (1) a low-FRET, nucleotide-free state (0.4); (2) a high-FRET, agonist- (0.64) or partial agonist- (0.72) bound state while the receptor remains associated with Gs; and (3) an intermediate-FRET state (~0.5) reflecting a GDP-bound β2AR-Gs complex with a distinct mode of interaction between TM6 and the α5-helix that is relatively short lived (Fig. 4c). Because the two lower-FRET states can be seen clearly only in pre-steady-state experiments, we analyzed our steady state data as a two-state system where states 1 and 3 are collapsed into a single, broadly defined (~0.4–0.5) low-FRET state.
Figure 5.
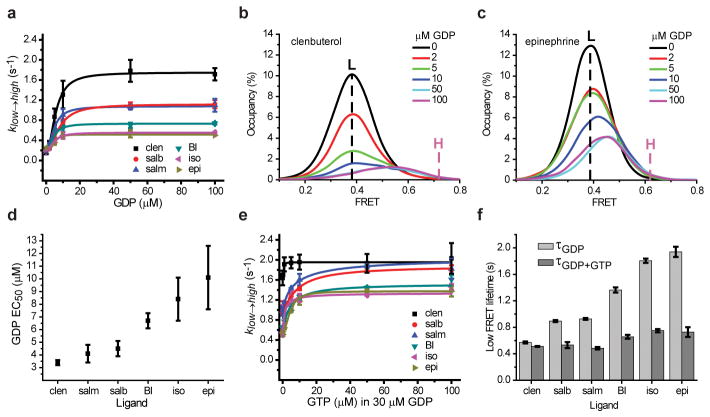
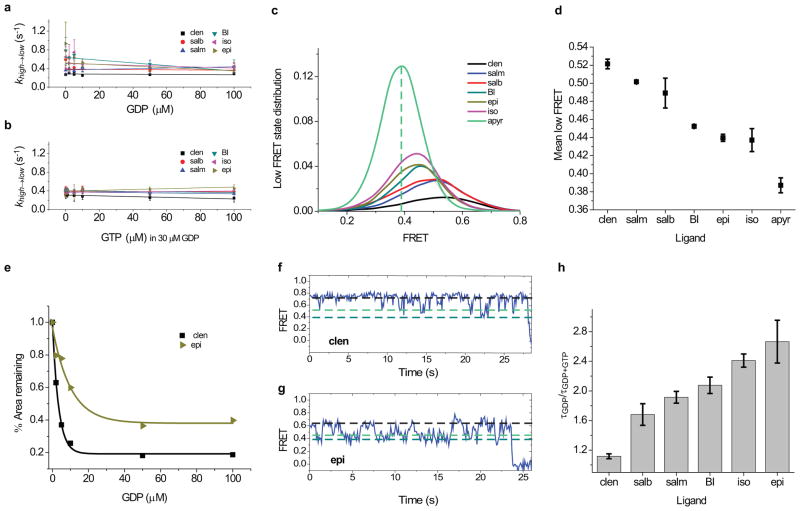
Nucleotide exchange efficiency is ligand dependent. a, Transition rates from low- to high-FRET states (klow→high) with agonists, Gs and increasing GDP concentrations. b–c, Low-FRET state distributions with increasing GDP in Gs and clen (b) or epi (c). Vertical dashed lines indicate the mean low-FRET value observed without GDP (black; L) and the mean high-FRET value at 100 μM GDP (purple; H). d, Apparent EC50 for GDP binding to the β2AR-Gs complex with different agonists. e, Transition rates from low- to high-FRET states (klow→high) with agonists, Gs, GDP, and increasing GTP concentrations. f, Histograms of low-FRET state lifetimes (τ) for each agonist with Gs and saturating GDP (light gray) or saturating GTP and GDP (dark gray). Error bars represents s.d. (two replicates) except (d), which represents s.e.m.
To learn about the rate-limiting features of β2AR-Gs complex formation, we examined the rates of low- and high-FRET state formation (khigh→low; klow→high, respectively) over a range of GDP concentrations. As expected for binding of a GDP-bound Gs heterotrimer, khigh→low was largely independent of GDP concentration for all agonists (Extended Data Fig. 8a). By contrast, klow→high increased with GDP and plateaued at concentrations above ~20 μM (Fig. 5a). Consistent with TM6 dynamics occurring within the β2AR-Gs complex, the maximum rates exiting low FRET were ~5–15-fold more rapid than the apparent Gs dissociation rate (Extended Data Table 2). The rank order of the low- to high-FRET state transition was: clen> salb>salm>BI>iso>epi. Given that GDP binding to the nucleotide-free β2AR-Gs complex is rapid (Fig. 4a,c; Extended Data Fig. 6e), we conclude that the transition out of low-FRET states into high FRET is rate-limited by one or more ligand-dependent processes within the β2AR-Gs (GDP) complex.
As low FRET includes both nucleotide-free and GDP-bound complexes (Fig. 4a,c), we speculated that the slower rates of return to high FRET observed for full agonists (Fig. 5a) may reflect higher proportions of the relatively stable nucleotide-free state. We therefore undertook an evaluation of differences in the proportion of nucleotide-free β2AR-Gs complexes in the presence of distinct agonists using the experimentally observed mean value of the low-FRET state as a function of GDP concentration (Fig. 5b,c). This analysis revealed that the low-FRET state values observed at saturating GDP concentration (100 μM) were considerably lower for full agonists compared to partial agonists, more closely approximating the FRET value (~0.4) observed for the nucleotide-free β2AR-Gs complex (Extended Data Fig. 8c,d). Consistent with GDP binding promoting return to high FRET, increasing GDP concentrations increased the mean values of the low-FRET states, while decreasing their time-averaged occupancies (Fig. 5b,c). The concentration dependence of these effects revealed that full agonists exhibited EC50 values that were approximately 2–3-fold higher than for partial agonists (Fig. 5d; Extended Data Fig. 8e). These data suggest that β2AR-Gs complexes exhibit higher affinity for GDP when bound to partial agonists compared to full agonists. They also support the notion that low-FRET states represent a mixture of nucleotide-free and GDP-bound β2AR-Gs configurations, where complexes activated by full agonists spend more time on average in the relatively stable, nucleotide-free state. These findings may help explain why epi promotes a greater extent of [3H]GDP release from β2AR compared to salb27, despite both agonists promoting low FRET states at similar rates (Fig. 3e). As the rates of GDP binding to nucleotide-free β2AR-Gs complexes are rapid, and appear indistinguishable at the present time resolution (Fig. 4a,c), we conclude that more efficacious agonists increase the probability of GDP release and thus the likelihood that nucleotide-free states are achieved.
To test this model directly, we performed analogous GTP titrations in the presence of a fixed, saturating GDP concentration (30 μM) (Extended Data Fig. 7b,c). As anticipated, the transition rate from low-to-high FRET (klow→high) was in all cases specifically increased at even the lowest GTP concentrations tested (100nM) (Fig. 5e; Extended Data Fig. 8b). While the absolute values of klow→high were greater for partial agonists in the presence of GTP (Fig. 5e), the fold increase in rate, and hence the magnitude reduction in low-FRET state lifetime, correlated with ligand efficacy in the order: epi>iso>BI>salm>salb>clen (Fig. 5e,f; Extended Data Fig. 8h). Taken together with the rapid rates of GDP and GTP binding to nucleotide-free β2AR-Gs complexes (Fig. 4), we conclude that the relatively short-lived, active-like, low-FRET β2AR conformations evidenced in the presence of partial agonists (Fig. 5a,e,f) predominantly reflect failed attempts at nucleotide release, and that full agonists more efficiently promote relatively long-lived, nucleotide-free configurations (Extended Data Fig. 8f,g). Hence, ligands with greater efficacy preferentially promote GDP release, and under competitive conditions, rapid and efficient GTP loading to the subpopulation of nucleotide-free complexes. The more rapid return of β2AR-Gs(GTP) complexes to high-FRET states following GTP loading argues that the terminal phosphate of GTP lowers the barrier for the rate-limiting conformational transition that enables the return of TM6 to its position adjacent to the helix bundle. This distinction may reflect GTP-specific impacts on Gs heterotrimer stability.
DISCUSSION
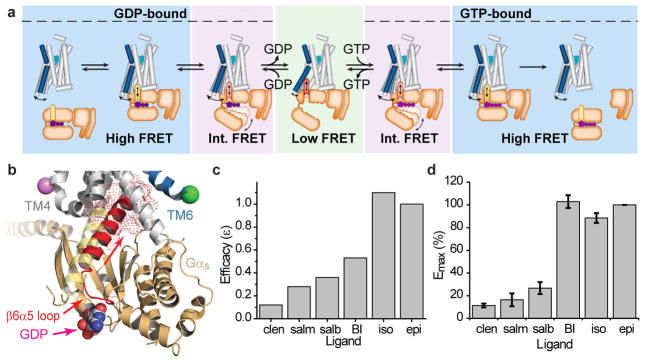
We have examined ligand-activated Gs binding, nucleotide exchange, and Gs release, from the perspective of time-dependent changes in β2AR conformation. The results illuminate the established concept of ligand efficacy in terms of a specific kinetic framework for the activation pathway (Fig. 6a). Quantifying the ligand-dependence of both the rate and the efficiency of Gs coupling in the presence of physiological GDP concentrations revealed that the process is achieved by rate-limiting conformational processes intrinsic to the β2AR-Gs complex (Fig. 3e). Although the nature of the interactions preceding excursions to low-FRET, active-like conformations are not presently known, the rates evidenced at saturating Gs concentration (Fig. 3e) suggest ligand-specific impacts on the probability that Gαs productively engages the intracellular face of β2AR after the complex has formed (Fig. 6a). The molecular events underpinning these early activation steps minimally include the remodeling of the β6α5 loop proximal to the Gαs switch regions, and translation and rotation of the α5 helix of Gα away from the GDP binding pocket towards the intracellular vestibule in β2AR created by the outward movement of TM628–35 (Fig. 6b).
Figure 6.
Proposed kinetic framework underlying β2AR-Gs coupling and nucleotide exchange. a, Schematic of β2AR TM6 conformational states within the G protein activation cycle (Note: the low-FRET β2AR-Gs(GTP) complex was not experimentally observed but is inferred). b, Gαs α5 helix engaging the intracellular face of β2AR disrupts helix-proximal β6α5 loop interactions with GDP. c, Ranking of agonist molecular efficacies (ε) relative to epi in terms of the effective rate of generating Gs(GTP) from Gs(GDP) (Methods). d, Ligand efficacy-based cAMP measurements in living cells (Methods; error bars=s.e.m, 3 replicates).
The apparent differences in initial engagement and the rate-limiting conformational changes leading to active-like, lower-FRET states (Fig. 3e; Extended Data Table 1), provide an estimate of the relative ligand-dependent efficiencies (η1) in these hidden early steps (Methods). A second determinant of ligand efficacy (η2) – which likely arises from differences in how efficiently the α5 helix C-terminus of Gαs productively engages, and forms stabilizing interactions, with the intracellular β2AR surface34 – can be estimated from the propensity of full agonists to more effectively promote GDP release (Methods). Using the normalized parameters (η1, η2), the efficacies of activating ligands relative to epi (ε) can be ranked in terms of their effective rates of generating Gs(GTP) from Gs(GDP) (Fig. 6c; Methods). In doing so, we estimate that iso is ~8-times more efficacious than clen. While these predictions suggest greater efficacy differences than inferred from in vitro GTP turnover assays (Fig. 1b), they are in close agreement with cyclic AMP production in living cells (Fig. 6d). Ligand-specific disparities in our calculated values versus the measured ligand efficacy values may reflect agonist-specific differences in GTP binding rates and/or affinities, which could not be accurately quantified.
In addition to providing quantitative insights into ligand efficacy, the present smFRET studies shed light on β2AR-Gs conformations that are structurally distinct from the nucleotide-free complex (Fig. 1a). While GDP- and GTP-bound complexes may be too unstable for crystallographic study, in-depth characterizations of these states are expected to provide important insights into both G protein coupling rates and specificities. Quantitative single-molecule imaging investigations will be critical in such efforts, as well as for delineating distinct ligand-dependent GPCR signaling pathways.
Extended Data
Extended Data Figure 1.
Ligand binding properties of β2Δ6-148C/266C. a, [3H]-dihydroalprenolol (DHA) saturation binding on purified β2AR, comparing wild-type (WT; blue squares) versus unlabeled (orange circles) and labeled (green triangles) β2Δ6-148C/266C. Affinities (Kd) are shown in the table. Non-specific binding controls are shown as corresponding open symbols. Error bars represent the standard error of the mean (s.e.m.) from triplicate measurements. b, [3H]-DHA–iso competition binding on purified β2AR, comparing wild-type (WT; blue circles) versus unlabeled (orange squares) and labeled (green triangles) β2Δ6-148C/266C. Affinities (Ki) are reported in the table. Error bars represent the s.e.m. from triplicate measurements. c, Dose-response curves of the BRET-based cAMP biosensor CAMYEL with wild-type (WT) β2AR (black triangles) and the mutants β2Δ6 (blue squares) and β2Δ6-148C/266C (red circles). Data from untransfected cells are shown in green triangles. Data from five independent experiments were normalized to the maximal isoproterenol response by WT β2AR in each experiment and globally fit to the entire data set, with the error bars representing the s.e.m., as reported in the table. d, Skeletal structures of ligands used in the current study. e, [3H]-DHA competition binding on unlabeled β2Δ6-148C/266C for ligands used in the current study, except for cz and BI, for which the ultra-high affinities reported (32 pM and 84 pM, respectively) would not allow us to determine them accurately in our assay. Instead, we used concentrations of 1 μM for both in all our measurements. The calculated Kis are shown on the table.
Extended Data Figure 2.
Fluorophore structures and properties. a–b, Skeletal structures of the modified Cy3B* donor (a) and Cy7* acceptor (b) fluorophores. c–d, Normalized donor fluorophore emission (Cy3: green; Cy3B*: dark green) and acceptor fluorophore absorbance (Cy5: red; Cy7*: dark magenta) spectra for Cy3/Cy5 (c) and Cy3B*/Cy7* (d) FRET pairs. The spectral overlap integral (shaded region) was calculated and used to determine the Förster distance (R0) values for each pair. e, Inter-dye FRET efficiencies of the Cy3/Cy5 (black) and Cy3B*/Cy7* (blue) donor and acceptor fluorophore pairs as a function of inter-dye distances calculated based on R0 values. f, Bulk anisotropy measurements on Cy3B*-labeled β2Δ6-148C/266C.
Extended Data Figure 3.
smFRET experimental controls. a, Site-specific labeling. SDS-PAGE gels under green (540 nm) or near IR (740 nm) epi-illumination for fluorescence visualization of Cy3B* or Cy7* labeling of β2Δ6 and β2Δ6-148C/266C. Coomassie-stained gel image is shown as a gel-loading control. Digestion with Factor Xa protease and deglycosylation with PNGase F leads to separation of the 148C and 266C labeling sites on generated N-terminal and C-terminal fragments, respectively. For gel source data, see Supplementary Figure 1. b, Quantification of Cy3B*/Cy7*-labeling specificity of full-length β2Δ6-148C/266C. Data is normalized to β2Δ6-148C/266C labeling. c, Specificity of streptavidin-mediated receptor immobilization. Frame capture from immobilization movies showing labeled β2Δ6-148C/266C on streptavidin-free (-SA) or streptavidin-coated (+SA) surfaces. Bar graph shows the number of immobilized, labeled β2AR in these conditions. Error bars represent the standard deviation from two replicates. d, Schematic of labeled β2AR immobilization via biotinylated alp (alp-biotin). e, FRET population contour plot and histogram for alp-biotin-immobilized receptor shows correspondence with the FRET population distribution of biotin-M1-Fab-immobilized, alp-bound, labeled β2AR (Fig. 2b). Histogram error bars represent the standard deviation from four replicates with N total molecules analyzed. f, FRET population contour plots (top row) and histograms (bottom row) for epi titration on biotin-M1-Fab-immobilized, labeled β2Δ6-148C/266C (Fig. 2c). Dashed lines (blue) highlight the mean FRET values for the lowest (2 nM; top dashed line) and highest (200 μM; bottom dashed line) epi concentrations tested. Histogram error bars represent the standard deviation from three replicates with N total molecules analyzed. The scale bar on the right indicates relative populations for the contour plots.
Extended Data Figure 4.
a, Sample fluorescence (green for Cy3B*; magenta for Cy7*) and FRET (blue) time traces for biotin-M1-Fab-immobilized, labeled β2Δ6-148C/266C imaged in the absence and presence of saturating ligands. b, Same as Fig. 2b but with the full FRET efficiency range (0–1) shown for the population histograms (bottom row). c, Plots showing the mean cross correlation values of donor and acceptor fluorescence as a function of lag time for the ensemble of individual fluorescence traces obtained from experiments shown in Figure 2b. d, Plots showing the mean cross correlation values of donor and acceptor fluorescence as a function of lag time for an ensemble of simulated fluorescence trajectories rapidly fluctuating between high (0.75) and intermediate (0.55) FRET values with varying low- to high-FRET transition rate constants (colored lines), where the high- to low-FRET transition rate is held constant at 100 s−1 (Methods). e, FRET distributions of the simulated data, as described in panel d.
Extended Data Figure 5.
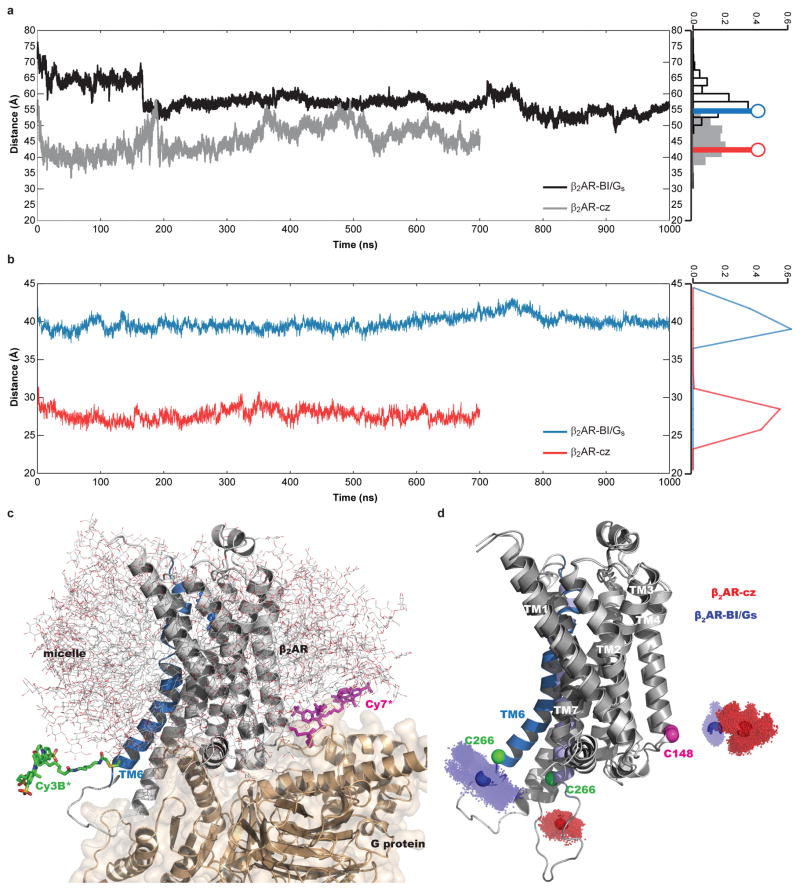
All-atom molecular dynamics simulations of the Cy3B*/Cy7*-labeled β2AR in a detergent micelle. a, Time evolution of the distance between the dyes. Time dependence of the distances between the midpoints of the dyes along the simulation trajectories is shown for the β2AR-cz (gray) and β2AR-BI/Gs (black) systems. The distributions are displayed as histograms on the right; gray bars: β2AR-cz; clear bars: β2AR-BI/Gs. The estimated inter-dye distances derived from the experimental mean FRET values (Figs. 2b,3b; Extended Data Fig. 2e) are indicated by solid lines topped with circles (β2AR-cz: red; β2AR-BI/Gs: blue). b, Time evolution of the distance between Calpha carbons at the labeling site. Calpha-Calpha distances for β2AR-cz: red and β2AR-BI/Gs: blue. c, The simulated dye-tethered β2AR-BI/Gs system embedded in a DDM micelle (gray sticks). β2AR is rendered in gray, with TM6 and the agonist (BI) highlighted in blue. The Gs protein is rendered in wheat color surrounded by its molecular surface to indicate the excluded volume for dye movements. The Cy3B* and Cy7* dyes are colored green and magenta, respectively. Water molecules, ions, and detergent molecules distant from the β2AR structure are omitted. d, Positions explored by the midpoints of the dyes during the simulations are shown as clusters of dots in the context of the β2AR-cz (transparent red dots) and β2AR-BI/Gs (transparent blue dots) complexes; the center of mass of each collection of dots is indicated by a solid sphere. Calpha carbons for labeled positions 1484.40 and 2666.28, are shown as magenta and green spheres, respectively.
Extended Data Figure 6.
smFRET imaging of biotin-Gs-immobilized, labeled β2AR. a, Schematic of labeled β2AR immobilization via biotinylated Gs heterotrimer. b–c, Representative fluorescence (green for Cy3B*; magenta for Cy7*) and FRET (blue) time traces for biotin-Gs-immobilized, labeled β2Δ6-148C/266C imaged in the presence of clen (b) or epi (c) in nucleotide-free conditions (apyrase-treated). d, FRET population contour plots (top row) and histograms (bottom row) for biotin-Gs-immobilized β2Δ6-148C/266C imaged in the presence of partial and full agonists in nucleotide-free conditions. The dashed line indicates the invariant mean FRET value (~0.38) for all agonists tested. Histogram error bars represent the standard deviation from three replicates with N total molecules analyzed. e–f, FRET population contour plots of biotin-Gs-immobilized β2Δ6-148C/266C in the presence of agonists exhibiting FRET transitions upon rapid addition (arrow) of 30 μM GDP (e) or 100 μM GTP (f). Scale bar on the right indicates the relative population for the contour plots.
Extended Data Figure 7.
a, Sample fluorescence and FRET time traces for biotin-M1-Fab-immobilized, labeled β2Δ6-148C/266C imaged in the absence and presence of saturating ligands plus 8 μM Gs in nucleotide-free (after apyrase treatment) conditions. b–c, FRET population contour plots for biotin-M1-Fab-immobilized, labeled β2Δ6-148C/266C imaged in the presence of the agonists clenbuterol (b) or epinephrine (c), 8 μM Gs and increasing concentrations of GDP (Fig. 5a) or GTP in the presence of saturating GDP (30 μM) (Fig. 5e) with N indicating the total number of molecules analyzed from two replicates. Nucleotide-free (0 μM GDP; from Fig. 3b) and ligand only (from Fig. 2b) conditions are included as references. Scale bar on the right indicates the relative population for the contour plots.
Extended Data Figure 8.
a–b, Transition rates from high- to low-FRET states (khigh→low) of labeled β2AR with different agonists, saturating Gs (8 μM) and increasing GDP concentrations (2–100 μM)(a) or increasing GTP concentrations (0–100 μM) in 30 μM GDP (b). c, Low-FRET state distributions for labeled β2AR with different agonists in 100 μM GDP and saturating Gs showing their overlap with the distribution in the presence of epi and saturating Gs in nucleotide-free conditions (apyr) (Fig. 3b). The dashed line shows the mean FRET value for apyr. d, Mean low FRET values from c. e, The percent change in the area of the low-FRET state population distributions for clen (black squares) and epi (dark yellow triangles)(as shown in Fig. 5b and c, respectively) was plotted with increasing GDP concentration (0–100 μM) and fitted to a single exponential decay function to derive the GDP EC50. f–g, Sample FRET traces (blue line) of labeled β2AR in the presence of clen (f) or epi (g) plus 100 μM GDP and saturating Gs. Dashed lines indicate each ligand’s corresponding mean high FRET value (black), mean low FRET value (light green) and mean FRET value in nucleotide-free conditions (dark green). h, The ratio of the low-FRET state lifetime of β2AR in the presence of saturating Gs and GDP (τGDP) over the low-FRET state lifetime in saturating GTP plus 30 μM GDP (τGDP+GTP)(Fig. 5f) is shown for different agonists. All error bars in the figure represent the standard deviation from two replicates.
Extended Data Table 1.
Apparent on rate and transition rates from high- to low-FRET states calculated from the data presented in Fig. 3e.
| apparent kon (μM−1s−1) | khigh→low (s−1) | |
|---|---|---|
| alp | ≪0.02 | - |
| clen | 0.03 | 0.4 |
| salm | 0.04 | 0.6 |
| salb | 0.04 | 0.7 |
| BI | 0.04 | 0.7 |
| epi | 0.05 | 0.7 |
| iso | 0.04 | 1.1 |
Extended Data Table 2.
Lifetimes of biotin-Gs-immobilized β2AR in the nucleotide-free state and upon addition of GDP or GTP and the lifetimes of biotin-M1-Fab-immobilized β2AR in the absence of Gs.
| Ligand | Lifetime | |||
|---|---|---|---|---|
| M1-immobilized (min) | Biotin-Gs-immobilized | |||
| GNP-free (min) | +GDP (s) | +GTP (s) | ||
| clen | 14.4 ± 2.1 | 8.3 ± 1.3 | 9.8 ± 1.6 | 6.8 ± 0.5 |
| salb | 18.1 ± 8.4 | 5.0 ± 0.4 | 8.2 ± 0.9 | 6.4 ± 0.1 |
| salm | 10.8 ± 4.1 | 3.5 ± 1.6 | 9.0 ± 0.4 | 6.6 ± 0.2 |
| BI | 11.0 ± 5.5 | 8.0 ± 3.2 | 9.4 ± 0.6 | 6.2 ± 0.1 |
| iso | 12.7 ± 3.1 | 6.0 ± 5.2 | 9.8 ± 1.3 | 5.8 ± 0.1 |
| epi | 13.2 ± 3.4 | 11.2 ± 5.1 | 11.8 ± 1.2 | 6.4 ± 0.1 |
Supplementary Material
Acknowledgments
We thank Dr. Mark Howarth for the kind gift of trans-divalent streptavidin, and Chaya Stern in the lab of Prof. John Chodera at MSKCC for constructing the CHARMM-consistent parameters for the dyes used in the MD simulations. Computational resources are gratefully acknowledged: an XSEDE allocation at the Texas Advanced Computing Center at the University of Texas at Austin (Stampede supercomputer, project TG MCB120008), support from resources at the Oak Ridge Leadership Computing Facility (ALCC allocation BIP109) at the Oak Ridge National Laboratory that is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC05-00OR22725; and the resources of the David A. Cofrin Center for Biomedical Information in the HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine at Weill Cornell Medicine. This work was supported in part by National Institutes of Health Grants GM098859 (S.C.B), R21DA0354585 (J.A.J., S.C.B. and G.G.G.), K05DA022413 and R01 MH54137 (J.A.J.), R01GM083118 and R01NS028471 (B.K.K.), and U54GM087519 (H.W. and J.M.P-A.), the German Academic Exchange Service (DAAD) (D.H.), the American Heart Association Postdoctoral fellowship (15POST22700020) (M.M.), and the Novo Nordisk Foundation Center for Basic Metabolic Research (M.H.).
Footnotes
Data availability. The data that support the findings of this study are available from the corresponding author upon reasonable request.
Author Contributions. G.G.G., M.M., D.H., B.K.K. and S.C.B. designed single-molecule experiments. G.G.G. labeled receptor and performed all single-molecule experiments. G.G.G. analyzed single-molecule data, with support from D.S.T. M.J. and D.S.T. developed the imaging and analysis platform. M.M. expressed, purified and characterized receptor constructs. D.H. expressed, purified and biotinylated Gs, and performed GTP turnover assays. H.Z. and Z.Z. synthesized the fluorophores. J.M.P-A. performed MD simulations under supervision of H.W. M.H. and S.M. performed cell-based G protein-coupling assays under supervision of J.A.J.. G.G.G., M.M., D.H., J.A.J., H.W., B.K.K., and S.C.B. interpreted all the data and wrote the manuscript. B.K.K. and S.C.B. provided overall project supervision.
The authors declare competing financial interest: details accompany the full-text HTML version of the paper (nature.com).
References
- 1.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 2.Vilardaga JP, et al. GPCR and G proteins: drug efficacy and activation in live cells. Mol Endocrinol. 2009;23:590–599. doi: 10.1210/me.2008-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kenakin T. New concepts in pharmacological efficacy at 7TM receptors: IUPHAR review 2. Br J Pharmacol. 2013;168:554–575. doi: 10.1111/j.1476-5381.2012.02223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manglik A, Kobilka B. The role of protein dynamics in GPCR function: insights from the beta2AR and rhodopsin. Curr Opin Cell Biol. 2014;27:136–143. doi: 10.1016/j.ceb.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker JG. The selectivity of beta-adrenoceptor agonists at human beta1-, beta2- and beta3-adrenoceptors. Br J Pharmacol. 2010;160:1048–1061. doi: 10.1111/j.1476-5381.2010.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kruse AC, et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 2013;504:101–106. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang W, et al. Structural insights into micro-opioid receptor activation. Nature. 2015;524:315–321. doi: 10.1038/nature14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpenter B, Nehme R, Warne T, Leslie AG, Tate CG. Structure of the adenosine A2A receptor bound to an engineered G protein. Nature. 2016;536:104–107. doi: 10.1038/nature18966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao XJ, et al. The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proc Natl Acad Sci U S A. 2009;106:9501–9506. doi: 10.1073/pnas.0811437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manglik A, et al. Structural Insights into the Dynamic Process of beta2-Adrenergic Receptor Signaling. Cell. 2015;161:1101–1111. doi: 10.1016/j.cell.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nygaard R, et al. The dynamic process of beta(2)-adrenergic receptor activation. Cell. 2013;152:532–542. doi: 10.1016/j.cell.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper M, et al. Cy3B: improving the performance of cyanine dyes. J Fluoresc. 2004;14:145–150. doi: 10.1023/b:jofl.0000016286.62641.59. [DOI] [PubMed] [Google Scholar]
- 14.Vafabakhsh R, Levitz J, Isacoff EY. Conformational dynamics of a class C G-protein-coupled receptor. Nature. 2015;524:497–501. doi: 10.1038/nature14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim HD, et al. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc Natl Acad Sci U S A. 2002;99:4284–4289. doi: 10.1073/pnas.032077799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamichhane R, et al. Single-molecule view of basal activity and activation mechanisms of the G protein-coupled receptor beta2AR. Proc Natl Acad Sci U S A. 2015;112:14254–14259. doi: 10.1073/pnas.1519626112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bond RA, et al. Physiological effects of inverse agonists in transgenic mice with myocardial overexpression of the beta 2-adrenoceptor. Nature. 1995;374:272–276. doi: 10.1038/374272a0. [DOI] [PubMed] [Google Scholar]
- 18.Ferguson A, et al. Functional Dynamics within the Human Ribosome Regulate the Rate of Active Protein Synthesis. Mol Cell. 2015;60:475–486. doi: 10.1016/j.molcel.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gales C, et al. Real-time monitoring of receptor and G-protein interactions in living cells. Nat Methods. 2005;2:177–184. doi: 10.1038/nmeth743. [DOI] [PubMed] [Google Scholar]
- 20.Ernst OP, Gramse V, Kolbe M, Hofmann KP, Heck M. Monomeric G protein-coupled receptor rhodopsin in solution activates its G protein transducin at the diffusion limit. Proc Natl Acad Sci U S A. 2007;104:10859–10864. doi: 10.1073/pnas.0701967104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 22.Hein P, et al. Gs activation is time-limiting in initiating receptor-mediated signaling. J Biol Chem. 2006;281:33345–33351. doi: 10.1074/jbc.M606713200. [DOI] [PubMed] [Google Scholar]
- 23.Gales C, et al. Probing the activation-promoted structural rearrangements in preassembled receptor-G protein complexes. Nat Struct Mol Biol. 2006;13:778–786. doi: 10.1038/nsmb1134. [DOI] [PubMed] [Google Scholar]
- 24.Qin K, Dong C, Wu G, Lambert NA. Inactive-state preassembly of G(q)-coupled receptors and G(q) heterotrimers. Nat Chem Biol. 2011;7:740–747. doi: 10.1038/nchembio.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westfield GH, et al. Structural flexibility of the G alpha s alpha-helical domain in the beta2-adrenoceptor Gs complex. Proc Natl Acad Sci U S A. 2011;108:16086–16091. doi: 10.1073/pnas.1113645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Damian M, et al. Ghrelin receptor conformational dynamics regulate the transition from a preassembled to an active receptor:Gq complex. Proc Natl Acad Sci U S A. 2015;112:1601–1606. doi: 10.1073/pnas.1414618112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murayama T, Ui M. [3H]GDP release from rat and hamster adipocyte membranes independently linked to receptors involved in activation or inhibition of adenylate cyclase. Differential susceptibility to two bacterial toxins. J Biol Chem. 1984;259:761–769. [PubMed] [Google Scholar]
- 28.Ceruso MA, Periole X, Weinstein H. Molecular dynamics simulations of transducin: interdomain and front to back communication in activation and nucleotide exchange. J Mol Biol. 2004;338:469–481. doi: 10.1016/j.jmb.2004.02.064. [DOI] [PubMed] [Google Scholar]
- 29.Herrmann R, et al. Sequence of interactions in receptor-G protein coupling. J Biol Chem. 2004;279:24283–24290. doi: 10.1074/jbc.M311166200. [DOI] [PubMed] [Google Scholar]
- 30.Herrmann R, et al. Rhodopsin-transducin coupling: role of the Galpha C-terminus in nucleotide exchange catalysis. Vision Res. 2006;46:4582–4593. doi: 10.1016/j.visres.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 31.Oldham WM, Van Eps N, Preininger AM, Hubbell WL, Hamm HE. Mechanism of the receptor-catalyzed activation of heterotrimeric G proteins. Nat Struct Mol Biol. 2006;13:772–777. doi: 10.1038/nsmb1129. [DOI] [PubMed] [Google Scholar]
- 32.Kapoor N, Menon ST, Chauhan R, Sachdev P, Sakmar TP. Structural evidence for a sequential release mechanism for activation of heterotrimeric G proteins. J Mol Biol. 2009;393:882–897. doi: 10.1016/j.jmb.2009.08.043. [DOI] [PubMed] [Google Scholar]
- 33.Kaya AI, et al. A conserved phenylalanine as a relay between the alpha5 helix and the GDP binding region of heterotrimeric Gi protein alpha subunit. J Biol Chem. 2014;289:24475–24487. doi: 10.1074/jbc.M114.572875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dror RO, et al. SIGNAL TRANSDUCTION. Structural basis for nucleotide exchange in heterotrimeric G proteins. Science. 2015;348:1361–1365. doi: 10.1126/science.aaa5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flock T, et al. Universal allosteric mechanism for Galpha activation by GPCRs. Nature. 2015;524:173–179. doi: 10.1038/nature14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.