Abstract
Clinical data warehouses often contain analogous data from disparate sources, resulting in heterogeneous formats and semantics. We have developed an approach that attempts to represent such phenotypic data in its most atomic form to facilitate aggregation. We illustrate this approach with human blood antigen typing (ABO-Rh) data drawn from the National Institutes of Health’s Biomedical Translational Research Information System (BTRIS). In applying the method to actual patient data, we discovered a 2% incidence of changed blood types. We believe our approach can be applied to any institution’s data to obtain comparable patient phenotypes. The actual discrepant blood type data will form the basis for a future study of the reasons for blood typing variation.
Keywords: Clinical Data Repositories, Phenotype Detections, Blood Typing
Introduction
Clinical data warehouses are becoming a common tool for providing access to electronic health record data for various forms of reuse.[1] However, the reuse of historical data can be problematic, especially when data are pooled from multiple sources for which minimal documentation and metadata exist.[2] A challenge for clinical research informatics is to provide users with ways to reconcile such heterogeneous data.[2] For example, if one source codes gender as “male or female” and another source codes gender as “male, female, or other”, summarizing gender data across sources becomes problematic.
While normalzation procedures are standard for genetic sequence data,[3] they have been less well defined for phenotypic data, typically involving ad hoc mappings between patient characteristics.[4] The purpose of this paper is to present a method for normalization of heterogeneous data by reducing them to specific, well-characterized phenotypic traits. We take as our example common ABO and Rh blood typing such as is typically tested in blood bank laboratories.
Background
The Biomedical Translational Research Information System
The Clinical Center of the US National Institutes of Health (NIH) is a hospital in Bethesda, Maryland that has served as a site for clinical research since 1953. Over the years, data from clinical studies at NIH have been captured in a variety of clinical trials data management systems, as well as two electronic health records – one in operation from 1976 to 2004 and one in operation since 2004. These data, on over 500,000 human subjects from over 50 source systems are collected into a single database that forms the core of the NIH’s Biomedical Translational Research Information System (BTRIS).[5] All terms from source systems are assigned unique concept identifiers in BTRIS’s Research Entities Dictionary (RED), which is a unified ontology that includes hierarchical classifications of similar terms (such as tests that measure the same substance or medications that contain the same ingredients). BTRIS users select specific terms or classes of terms from the RED to use in retrieving identified data on their own clinical studies, as well as de-identified data across all clinical studies.[5]
Case Study: ABO and Rh Blood Type Antigens
Human red blood cells express a wide variety of antigens. Three in particular (A, B and Rh) are commonly identified in clinical laboratories for purposes such as cross-matching blood for transfusion. Blood donors and blood recipients are characterized as having A, B, AB or O blood and as being Rh positive or negative. Except in rare instances, an individual’s blood type does not change over his or her lifetime.
Today, most blood banks report these antigens together as the result of a single test; e.g., A+, B−, AB+, O−, etc. However, in the past, these results were reported across two tests: the ABO test to report types A, B, AB and O, and the Rh test with the possible results “positive” and “negative”. Even earlier, the ABO test results were reported as two separate tests for the A and B antigens. A patient could have a positive results for both of these tests (type AB), one of these tests (type A or B), or neither of these tests (implying type O).
This heterogeneity has been well-documented as a challenge for dealing with pooling data from, or sharing them between, multiple health care sites.([6], also: Huff SM, personal communication; inspiring [7]) As a repository of data from multiple sites over 40 years, BTRIS often presents users with this type of challenge. We chose it as a case study for the normalization of heterogeneous data, since proper normalization should result in patients having consistent blood types over time, in effect serving as their own gold standards.
Methods
Approach to Phenotype Normalization
Our approach to phenotypic characterization involves reducing complex findings to their most atomic forms and them assembling them in canonical ways into more complex phenotypic patterns. In the case of blood typing, we consider that each test provides evidence for the presence or absence of at least one red blood cell antigen. This can range from a single antigen (A, B, or Rh) to all of them (for example, type “AB+” indicates the presence of all three and type “O−” indicates the absence of all three).
Preliminary Analysis of Result Types
The first step in our normalization process was to identify the relevant tests panels, individual tests and actual results reported by those tests. We used the BTRIS Limited Data Set function[8] to retrieve de-identified data from BTRIS, using appropriate terms selected from the RED.
The second step was to review each unique panel-test-result triple to determine the antigenic evidence it provides. Each result was tagged with the letters A, B and R, with presence indicated by an upper-case letter and absence indicated by a lower case letter. Thus, the a B-Antigen test with the result “Positive” was labeled as “B” and a Type and Crossmatch test with the result “O+” was labeled as “abR”. A MUMPS data structure (PC-MUMPS, DataTree Inc., Waltham, MA), was created for each result and its assigned antigens.
Analysis of Patient Data for Phenotype Consistency
In the third step, the relevant data of individual patients were characterized based on the assigned antigens. Pooled evidence for the presence or absence of each antigen was stored for each patient in a second MUMPS global, such that a patient would typically have three letters (one each of “A” or “a”, “B” or “b”, and “R” or “r”). In the fourth step, we reviewed the results for each patient to determine situations where an individual did not have exactly one of each letter.
All data were obtained with oversight of the NIH Office of Human Subjects Research Protection (Agreement Number BTRIS_2014_835_CIMINO_J_CC). Only those data that did not require permission for reuse from the original investigators, as per NIH policy, were retrieved. The patients’ birthdates and test dates were included in the data but all other potentially identifying information was removed, as per NIH policy for limited-use data sets.
Results
Preliminary Analysis of Result Types
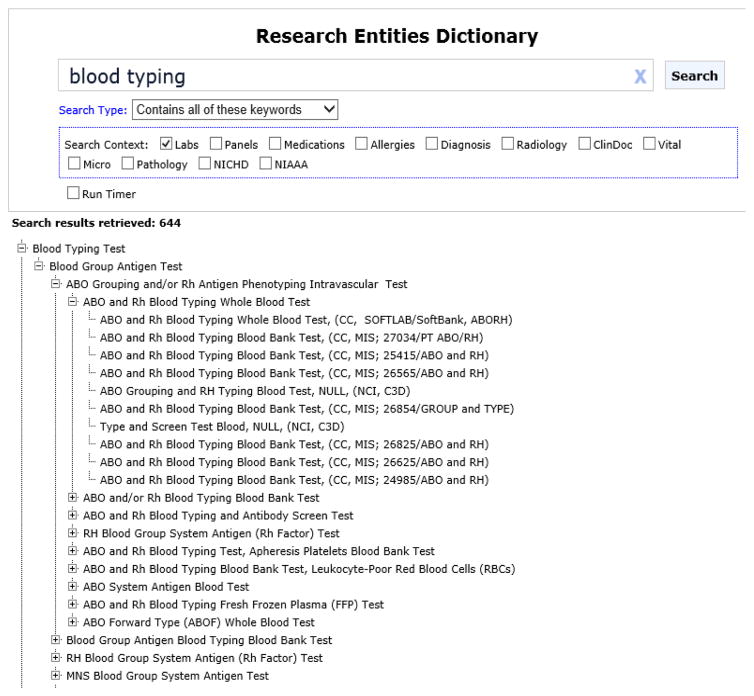
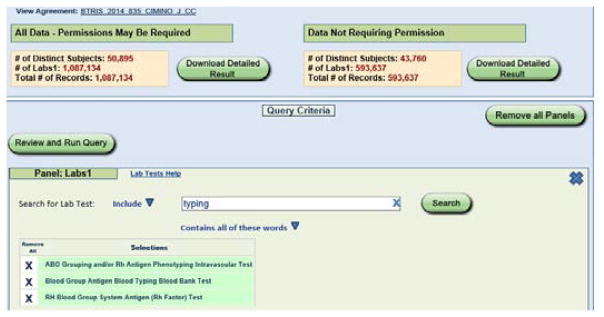
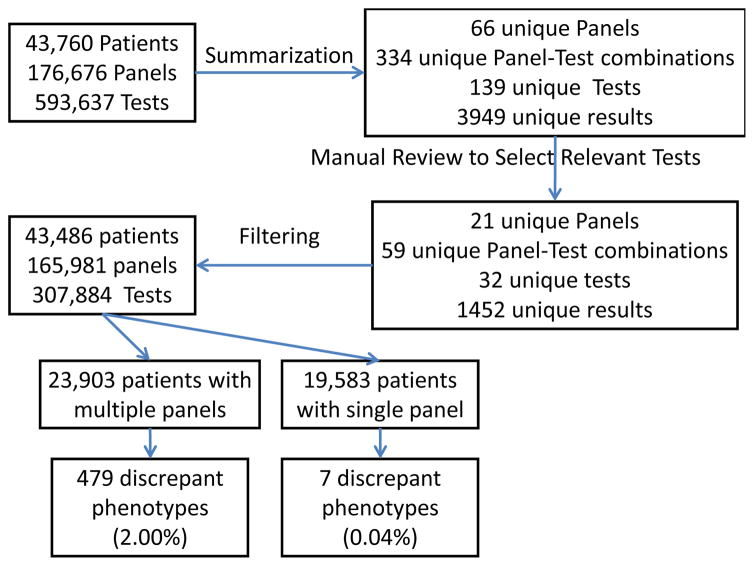
A search of the RED for tests with names containing the phrase “blood group” identified 644 terms, including three appropriate term classes as shown in Figure 1: ABO Grouping and/or Rh Antigen Phenotyping Intravascular Test, Blood Group Antigen Blood Typing Blood Bank Test, and RH Blood Group System Antigen (Rh Factor) Test. When BTRIS was queried with these three terms, 593,637 test results were found on 43,485 patients (see Figure 2). The data included results from 139 tests in 66 test panels, with 334 unique panel-test pairs that reported 3946 unique results (Figure 3).
Figure 1.
Blood type terms in the Research Entities Dictionary. A search for the phrase “blood typing”, restricted to the laboratory domain identified one class (“Blood Typing Test”) that subsumes 643 additional terms. The hierarchy is partially expanded.
Figure 2.
Query screen of BTRIS Limited Data Set application, showing three search terms at bottom and query summaries at top. Only data from the right (“Data Not Requiring Permissions”) were downloaded.
Figure 3.
Summary of analysis of initial data set.
Review of all panels and tests identified 21 panels and 59 tests within those panels that provide information on ABO and Rh blood typing. The data set included 1452 unique test results for these tests (many of which were misspellings, as in [6]). Each was reviewed manually to assign antigenic evidence. Table 1 shows a sample of panels, tests, results and assigned antigenic evidence.
Table 1.
Panel/test/result combinations with interpretations
| Panel | Tests | Result | Interpretation |
|---|---|---|---|
| ABO GRP-RH TYPE | ABO GRP-RH TYPE | O POSITIVE | abR |
| ABO GRP-RH TYPE | ABO GRP-RH TYPE | 0 POS | abR |
| ABO GRP-RH TYPE | ABO GRP-RH TYPE | A POSITIVE | AbR |
| ABO GRP-RH TYPE | ABO GRP-RH TYPE | A NEG | Abr |
| ABO GRP-RH TYPE | ABO GRP-RH TYPE | A NEGATIVE | Abr |
| ABO GRP-RH TYPE | ABO GRP-RH TYPE | AB NEG | ABr |
| ABO GRP-RH TYPE | ABO GRP-RH TYPE | B POS | aBR |
| ABO Group and Rh Type [ABORH] | ABO Group and Rh Type [ABORH] - A | 0 | A |
| ABO Group and Rh Type [ABORH] | ABO Group and Rh Type [ABORH] - A | 1+ | A |
| ABO Group and Rh Type [ABORH] | ABO Group and Rh Type [ABORH] - A | 2+ | A |
| ABO Group and Rh Type [ABORH] | ABO Group and Rh Type [ABORH] - A | 3+ | A |
| ABO Group and Rh Type [ABORH] | ABO Group and Rh Type [ABORH] - A | 4+ | A |
| ABO Group and Rh Type [ABORH] | ABO Group and Rh Type [ABORH] - A | M4 | A |
| ABO Group and Rh Type [ABORH] | ABO Group and Rh Type [ABORH] - B | 0 | B |
| ABO Group and Rh Type [ABORH] | ABO Group and Rh Type [ABORH] - B | 4+ | B |
| ABO Group and Rh Type [ABORH] | ABO Group and Rh Type [ABORH] - Rh | NEG | R |
| ABO Group and Rh Type [ABORH] | ABO Group and Rh Type [ABORH] - Rh | POS | R |
Panels are collections of one or more tests, each of which is associated with a result. We interpret the results to correspond to specific phenotypic characteristics: A=Antigen present; B=B antigen present, AB=A and B antigens present; a=A antigen absent; b=B antigen absent; ab=Neither A nor B antigens present (“Type O”); R=Rh antigen present (“Rh Positive”), r=Rh antigen absent (“Rh Negative”),
Assignment of Phenotype Based on Antigenic Evidence
After removal of irrelevant panels and tests, the patient data included 165,981 panel events with 307,884 test results for 43,485 patients. Pooling of all antigenic evidence for each patient identified 32 different phenotypes, of which 8 were complete (ABO and Rh designations), 6 were incomplete (ABO or R designation only) and 18 were discrepant (multiple ABO and/or Rh designations). Table 2 shows the antigenic evidence, and counts for each phenotypic designation.
Table 2.
Antigen patterns found in test results, classified as Complete (corresponding to an actual real phenotype found in nature), Incomplete (lacking sufficient information to determine the phenotype) or Aberrant (not corresponding to an actual phenotype)
| Antigenic Evidence | Phenotype | # Patients |
|---|---|---|
| Complete | ||
| abR | O+ | 17132 |
| AbR | A+ | 13925 |
| aBR | B+ | 4710 |
| abr | O− | 2538 |
| Abr | A− | 2316 |
| ABR | AB+ | 1441 |
| aBr | B− | 645 |
| ABr | AB− | 214 |
| Incomplete | ||
| r | − | 10 |
| R | + | 8 |
| ab | O | 7 |
| Ab | A | 5 |
| AB | AB | 1 |
| aB | B | 1 |
| bR | + | 1 |
| Aberrant | ||
| AabR | (aberrant) | 132 |
| abRr | (aberrant) | 89 |
| AbRr | (aberrant) | 67 |
| aBbR | (aberrant) | 51 |
| AaBbR | (aberrant) | 50 |
| AabRr | (aberrant) | 28 |
| ABbR | (aberrant) | 24 |
| aBRr | (aberrant) | 19 |
| AaBR | (aberrant) | 17 |
| Aabr | (aberrant) | 13 |
| aBbr | (aberrant) | 11 |
| ABRr | (aberrant) | 7 |
| AaBbRr | (aberrant) | 6 |
| aBbRr | (aberrant) | 6 |
| ABbr | (aberrant) | 6 |
| AaBbr | (aberrant) | 3 |
| ABbRr | (aberrant) | 2 |
Review of Aberrant Phenotypes
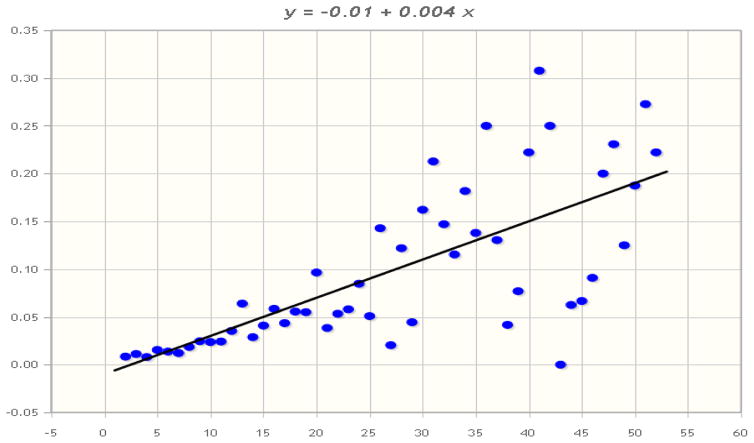
In all, 531 of the 43,485 patients (1.22%) had aberrant phenotypes, based on discrepant laboratory results. Some examples of their test results are shown in Table 3. Given that random, patient-independent laboratory errors could account for some of these discrepancies, and that the frequency of errors would be proportional to the number of tests run, we examined the frequency aberrant phenotypes versus number of test panels performed (Figure 4). The Pearson coefficient for this association is 0.7127 (P<.00001); however, the slope of the regression line is only 0.04.
Table 3.
Sample results showing discrepant blood type results for the same patient (dates are approximate for privacy reasons). Adjacent rows are results from tests in the same test panel; shaded rows separate panels for the same patient; white rows separate data from different patients. Note that last two subjects show discrepancies within a single panel, resulting in aberrant blood type interpretations.
| Subject | Date | Panel | Test | Result | Antigens | Interp. |
|---|---|---|---|---|---|---|
| 59 | 1/31/1989 2:34 | TYPE & SCREEN | ABO & RH | O POSITIVE | abR | abR (O+) |
| 59 | 1/31/1989 8:52 | TYPE & SCREEN | ABO & RH | A POSITIVE | AbR | AbR (O−) |
| 724 | 1/24/1989 9:55 | TYPE & SCREEN | ABO & RH | O NEG | abr | abr (O−) |
| 724 | 2/13/1989 2:22 | TYPE & SCREEN | ABO & RH | O POS | abR | abR (O+) |
| 986 | 1/2/1999 1:25 | Type and Antibody Screen | ABO Group and Rh Type - Rh | POS | R | ABR (AB+) |
| 986 | 1/2/1999 1:25 | Type and Antibody Screen | ABO Group and Rh Type - A | 4+ | A | |
| 986 | 1/2/1999 1:25 | Type and Antibody Screen | ABO Group and Rh Type - B | 4+ | B | |
| 986 | 1/18/2000 1:24 | Type and Antibody Screen | ABO Group and Rh Type - Rh | POS | R | AbR (A+) |
| 986 | 1/18/2000 1:24 | Type and Antibody Screen | ABO Group and Rh Type - A | 4+ | A | |
| 986 | 1/18/2000 1:24 | Type and Antibody Screen | ABO Group and Rh Type - B | 0 | b | |
| 1090 | 1/2/2002 12:39 | Type and Antibody Screen | ABO Group and Rh Type - ABO | A | Ab | AbR (A+) |
| 1090 | 1/2/2002 12:39 | Type and Antibody Screen | ABO Group and Rh Type - Rh | POS | R | |
| 1090 | 1/2/2002 12:39 | Type and Antibody Screen | ABO Group and Rh Type - A | 4+ | A | |
| 1090 | 1/2/2002 12:39 | Type and Antibody Screen | ABO Group and Rh Type - B | 0 | b | |
| 1090 | 1/28/2003 1:29 | Type and Antibody Screen | ABO Group and Rh Type - ABO | B | aB | aBR (B+) |
| 1090 | 1/28/2003 1:29 | Type and Antibody Screen | ABO Group and Rh Type - Rh | POS | R | |
| 1090 | 1/28/2003 1:29 | Type and Antibody Screen | ABO Group and Rh Type - A | 0 | a | |
| 1090 | 1/28/2003 1:29 | Type and Antibody Screen | ABO Group and Rh Type - B | 4+ | B | |
| 2185 | 1/11/1991 6:43 | TYPE & SCREEN | ABO & RH | O NEG | abr | Rabr (O+/O−) |
| 2185 | 1/11/1991 6:43 | TYPE & SCREEN | ABO & RH | O POS | abR | |
| 3986 | 1/18/2000 1:24 | Type and Antibody Screen | ABO Group and Rh Type - ABO | AB | AB | ABRb (AB+/A+) |
| 3986 | 1/18/2000 1:24 | Type and Antibody Screen | ABO Group and Rh Type - Rh | POS | R | |
| 3986 | 1/18/2000 1:24 | Type and Antibody Screen | ABO Group and Rh Type - A | 4+ | A | |
| 3986 | 1/18/2000 1:24 | Type and Antibody Screen | ABO Group and Rh Type - B | 0 | B |
A=Antigen present; B=B antigen present, AB=A and B antigens present; a=A antigen absent; b=B antigen absent; ab=Neither A nor B antigens present (“Type O”); R=Rh antigen present (“Rh Positive”), r=Rh antigen absent (“Rh Negative”)
Figure 4.
Pearson’s correlation between number of tests performed and incidence of at least one discrepant result between the tests. (http://www.mathportal.org/calculators/statistics-calculator/correlation-and-regression-calculator.php)
Initially, we did not consider patients with a single test panel, since we were looking for discrepancies between panels. Doing so would result in discrepancies in 479 of 23,903 patients (2.00%). However, we were surprised to find that seven of 19,583 patients (0.004%) with a single test panel had discrepancies, so although rare, this situation was not impossible. This led us to examine intra-panel discrepancies. We found that 109 patients (including the seven mentioned above) had 588 intra-panel discrepancies (see Table 3).
Discussion
Informatics Implications
As previously noted, heterogeneity of data syntax and semantics in clinical repositories complicates analysis of data aggregated from multiple sources over long time spans. The approach described here recasts clinical findings in terms of atomic phenotypic characteristics that are institution- and laboratory-independent. The case here concerns the presence or absence of specific human red blood cell antigens, but we believe this approach can be applied to many other data types, such as microorganism antigens and antibodies and clinical findings that are sometimes reported as collections of post-coordinated terms and sometimes as a single, precoordinated term. This approach has potential for use with other repositories or networks of repositories with heterogeneous data.
Biomedical Implications
We were surprised at the incidence of intra-patient blood type variation among patients with multiple sets of test results (2%). Human blood antigens do not normally change during a patient’s lifetime, except in unusual circumstances such as transplantation,[9] leukemia[10, 11] and, very rarely, viral infection[12]. It is possible – in fact probable – that some of this variation was due to laboratory error. The correlation (r=.72) between numbers of tests performed and likelihood is consistent with an independent influence such as systematic error; however the slope of the correlation (0.04) indicates that this would only account for about 4% of the variance. The data collected in this study are insufficient for determining what is likely to be a multifactorial cause for the variations. Discovering them will require a future study in which the entire patient records are accessed.
Conclusions
The reduction of heterogeneous laboratory results to their atomic phenotypic characteristics was an effective approach for normalizing patient test results from a longitudinal repository with 40 years of data from disparate sources systems. The normalization makes some patient characteristics explicit (e.g., by inferring Type O from the absence of A and B) as well as highlighting data that are discrepant. The approach seems extensible to other domains and repositories. The incidental finding of intra-patient blood type variation reminds us that the re-use of data constantly holds challenges and surprises, and that every question we seek to resolve seems to produce more new questions than answers.
Acknowledgments
This work was supported by intramural research funds from the NIH Clinical Center and the National Library of Medicine.
References
- 1.Prather JC, Lobach DF, Goodwin LK, Hales JW, Hage ML, Hammond WE. Medical data mining: knowledge discovery in a clinical data warehouse. Proc AMIA Annual Fall Symp. 1997:101–5. [PMC free article] [PubMed] [Google Scholar]
- 2.Hersh WR, Weiner MG, Embi PJ, Logan JR, Payne PR, Bernstam EV, Lehmann HP, Hripcsak G, Hartzog TH, Cimino JJ, Saltz JH. Caveats for the use of operational electronic health record data in comparative effectiveness research. Med Care. 2013 Aug;51(8 Suppl 3):S30–7. doi: 10.1097/MLR.0b013e31829b1dbd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xuan J, Wang Y, Hoffman E, Clarke R. Cross phenotype normalization of microarray data. Front Biosci (Elite Ed) 2010 Jan 1;2:171–86. doi: 10.2741/e80. [DOI] [PubMed] [Google Scholar]
- 4.Li D, Simon G, Chute CG, Pathak J. Using association rule mining for phenotype extraction from electronic health records. AMIA Jt Summits Transl Sci Proc. 2013 Mar 18;2013:142–6. [PMC free article] [PubMed] [Google Scholar]
- 5.Cimino JJ, Ayres EJ, Remennik L, Rath S, Freedman R, Beri A, Chen Y, Huser V. The National Institutes of Health’s Biomedical Translational Research Information System (BTRIS): Design, Contents, Functionality and Experience to Date. Journal of Biomedical Informatics. 2014 Dec;52:11–27. doi: 10.1016/j.jbi.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abhyankar S, Demner-Fushman D. A simple method to extract key maternal data from neonatal clinical notes. Proc AMIA Annual Fall Symp. 2013 Nov 16;2013:2–9. [PMC free article] [PubMed] [Google Scholar]
- 7.Baorto DM, Cimino JJ, Parvin CA, Kahn MG. Combining laboratory data sets from multiple institutions using the logical observation identifier names and codes (LOINC) Inter Journal Med Inform. 1998;51(1):29–37. doi: 10.1016/s1386-5056(98)00089-6. [DOI] [PubMed] [Google Scholar]
- 8.Cimino JJ, Ayres EJ, Beri A, Freedman R, Oberholtzer E, Rath S. Developing a self-service query interface for reusing de-identified electronic health record data. Stud Health Technol Inform. 2013;192:632–6. [PMC free article] [PubMed] [Google Scholar]
- 9.Bronson WR, McGinniss MH, Morse EE. Hematopoietic graft detected by a change in ABO group. Blood. 1964 Feb;23:239–49. [PubMed] [Google Scholar]
- 10.van der Hart, van der Veer, van Loghem J. Change of blood group B in a case of leukaemia. Vox Sang. 1962 Jul-Aug;7:449–53. doi: 10.1111/j.1423-0410.1962.tb03276.x. [DOI] [PubMed] [Google Scholar]
- 11.Hocking DR. Blood group change in acute myeloid leukaemia. Med J Aust. 1971 Oct 30;2(18):902–3. doi: 10.5694/j.1326-5377.1971.tb92614.x. [DOI] [PubMed] [Google Scholar]
- 12.Sherman LA, Silberstein LE, Berkman EM. Altered blood group expression in a patient with congenital rubella infection. Transfusion. 1984 May-Jun;24(3):267–9. doi: 10.1046/j.1537-2995.1984.24384225037.x. [DOI] [PubMed] [Google Scholar]