Abstract
Background
Allergies to cashew are increasing in prevalence, with clinical symptoms ranging from oral pruritus to fatal anaphylactic reaction. Yet, cashew-specific T-cell epitopes and T-cell cross-reactivity amongst cashew and other tree nut allergens in humans remain uncharacterized.
Objectives
In this study, we characterized cashew specific T-cell responses in cashew allergic subjects and examined cross-reactivity of these cashew specific cells toward other tree nut allergens.
Methods
CD154 up-regulation assay was used to determine immunodominance hierarchy among cashew major allergens at the T cell level. The phenotype, magnitude and functionality of cashew-specific T-cells was determined by utilizing ex vivo staining with MHC class II tetramers. Dual tetramer staining and proliferation experiments were used to determine cross-reactivity to other tree nuts.
Results
CD4+ T-cell responses were directed towards cashew allergens Ana o 1 and Ana o 2. Multiple Ana o 1 and Ana o 2 T-cell epitopes were then identified. These epitopes elicited either TH2 or TH2/TH17 responses in allergic subjects, which were either cashew unique epitope or cross-reactive epitopes. For clones that recognized the cross-reactive epitope, T-cell clones responded robustly to cashew, hazelnut and/or pistachio but not to walnut.
Conclusions
Phylogenetically diverse tree nut allergens can activate cashew reactive T-cells and elicit a TH2 type response at an epitope specific level.
Clinical relevance
Lack of cross-reactivity between walnut and cashew suggest that cashew peptide immunotherapy approach may not be most effective for walnut.
Keywords: Food allergy, cashew, tree nuts, Ana o 1, Ana o 2, cross-reactivity, T-cells, MHC class II tetramers, epitopes
INTRODUCTION
Tree nut allergies, like peanut allergies, are increasing in prevalence. They now affect 1.1% of children younger than 18 years and 0.5% of adults in the United States [1]. After walnuts, cashew allergy is the next most commonly reported tree nut allergy (affecting 20% of tree nut allergic subjects) [2]. It has been shown that severe clinical reactions, including higher incidence of anaphylaxis, occur more frequently in cashew than peanut allergy [3–6]. The lack of a specific treatment for cashew allergy makes food avoidance the mainstay of therapy in cashew allergic subjects [7;8].
Three major cashew allergens have been reported; Ana o 1 (Anacardium occidentale) (7s vicilin-like protein), Ana o 2 (11s legumin-like protein) and Ana o 3 (2s albumin) [9–11]. All three are classified as seed storage proteins. Interestingly, this family of seed storage proteins is known to be allergenic in other tree nuts [12]. Furthermore, amino acid sequence alignments of vicilins, legumins and albumins indicate a high degree of amino acid identity between cashew allergens and allergens from other tree nuts including hazelnut, pistachio and walnut[13]. It has been estimated that at least 86% of subjects who are tree nut allergic are allergic to multiple tree nuts [14]. Cross-sensitization to pistachio, hazelnut and walnut are common within cashew allergic subjects [5]. Studies have shown high degree of cross-reactivity between pistachio and cashew at the sIgE (specific IgE) level, owing to their botanic relatedness [15–17]. This phenomenon has also been demonstrated between walnut, hazelnut and cashew allergens at moderate levels [15;18]. On the other hand, T-cell cross-reactivity between tree nuts in humans has not been well documented.
In this study, we used cashew as a model to study tree-nut cross-reactivity against hazelnut, pistachio and walnut at the T-cell level in humans. We initially investigated Ana o 1, Ana o2 and Ana o 3-specific T-cell responses using CD154 activation assay. Both Ana o 1 and Ana o 2 were identified as the predominant allergens that elicit CD4+ T-cell responses in allergic subjects. Several Ana o 1 and Ana o 2 derived epitopes were identified by using tetramer-guided epitope mapping (TGEM). Phenotypes for allergen-specific T-cells were analyzed by ex vivo tetramer staining. Our results showed that allergic subjects have a predominant TH2 (TH T-helper) phenotype, however, TH2/TH17 responses were also detected. T-cell clones (TCC) specific to these epitopes were generated to assess cross-reactivity by tetramer co-staining and proliferation experiments. We found that TCC specific to cashew allergen derived epitopes could readily proliferate with hazelnut and pistachio, but not with walnut allergen derived peptides.
METHODS
Subjects
Subjects were recruited from the Virginia Mason Medical Center Allergy Clinic and Benaroya Research Institute with informed consent and institutional review board approval (IRB title “Allergen and T-cell reagent resources for the study of allergic diseases”; approval number IRB7109). A total of 14 subjects, based on history of an acute reaction to cashew plus a positive ImmunoCAP score for cashew extract (>0.35 kU/L) (Phadia AB, Uppsala, Sweden), were recruited for this study. As an inclusion criteria, subjects with a low sIgE score to cashew need to have a large wheal size in the skin prick test (≥ 8 mm × 8 mm). Twelve non-atopic and 6 atopic subjects with no clinical symptoms to cashew, a negative ImmunoCAP score and HLA (Human histocompatibility leukocyte antigen)-matched were also recruited as controls for this study. The features of these subjects are shown in Table 1. DNA samples were HLA-typed using Dynal UnitrayTM SSP Kits (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions.
Table 1.
HLA and allergic status of recruited subjects
| ID | Age | Sex | HLA (DRB1*) | sIgE cashew (f202) kU/L | Skin Prick Test to cashew | sIgE hazelnut (f17) kU/L | sIgE pistachio (f203) kU/L | sIgE walnut (f256) kU/L | Symptoms after cashew ingestion | Asthma | Date of first reaction | Date of last reaction |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cashew allergics | ||||||||||||
| 1 | 28 | F | 07:01, 14:01 | >100 | Not tested | 58.9 | >100 | 38.6 | I, III, V, VII, VIII | yes | 2005 (walnut) | 2007 (pistachio/cashew) |
| 2 | 11 | F | 09:01, 15:01 | 31.4 | Not tested | 5.62 | 28.6 | 17.1 | I, III, IV, V | yes | 2013 (cashew) | No known ingestions since |
| 3 | 19 | F | 01:01, 13:01 | 1.73 | 12 × 12 mm | 0.67 | 2.2 | 0.58 | I, IV, V | yes | 1998 (cashew) | No known ingestions since |
| 4 | 24 | F | 01:01, 13:01 | 0.5 | 9 × 9 mm | <0.35 | 0.77 | <0.35 | I, II, III | yes | 2000 (cashew) | 2012 (walnut) |
| 5 | 8 | F | 15:01, 04:04 | >100 | Not tested | 26.9 | >100 | 6.61 | IV, V | yes | 2009 (cashew/pistahio) | No known ingestions since |
| 6 | 24 | F | 01:01, 15:01 | 32.6 | Not tested | 3.69 | 37 | 8.86 | I, II, III, IV, VI | no | 2013 (cashew) | No known ingestions since |
| 7 | 21 | M | 03:01, 15:01 | 19 | 10 × 10 mm | 61.2 | 25.4 | 28.2 | I, II, IV, V | yes | 2012 (unknown nut) | 2013 (pistachio/cashew) |
| 8 | 11 | M | 01:01, 10:01 | 9.78 | 15 × 15 mm | 1.03 | 13.9 | 6.79 | III, IV | no | 2012 (pecan) | 2014 (pecan/cashew) |
| 9 | 34 | F | 07:01, 15:01 | 7.66 | 15×10 mm | 21.3 | 6.58 | 77.7 | I, II, III, IV, V | no | 1984 (unknown nut) | 2014 (cashew) |
| 10 | 8 | F | 07:01, 11:01 | 2.95 | 8 × 8 mm | 7.38 | 7.4 | 3.93 | I, II, III, IV | no | 2007 (cashew) | 2010 (pistachio) |
| 11 | 11 | M | 01:01, 13:02 | 41.6 | Not tested | 0.72 | 4.94 | 15.8 | I, IV, V | yes | 2006 (cashew) | 2010 (cashew) |
| 12 | 36 | F | 01:01, 04:04 | 0.38 | 15×15 mm | 0.38 | 0.36 | 0.69 | II, III, IV | no | 1984 (walnut) | 2014 (cashew) |
| 13 | 10 | F | 01:01, 04:04 | >100 | 5 × 5 mm | 40.5 | >100 | 97.3 | III, IV, V | no | 2005 (walnut) | 2009 (walnut/cashew) |
| 14 | 15 | F | 11:01, 13:01 | 57.3 | Not tested | 87.02 | 65.9 | >100 | I, III | yes | 2001 (pecan/hazelnut/cashew) | No known ingestions since |
| Nonatopic subjects | ||||||||||||
| 18 | 29 | M | 01:01, 03:01 | 0 | Not tested | Not tested | Not tested | Not tested | Absent | no | No known reaction | No known reaction |
| 19 | 31 | F | 07:01, 07:01 | 0 | Not tested | Not tested | Not tested | Not tested | Absent | no | No known reaction | No known reaction |
| 20 | 41 | F | 04:01, 15:01 | 0 | Not tested | Not tested | Not tested | Not tested | Absent | no | No known reaction | No known reaction |
| 21 | 34 | M | 07:01, 13:01 | 0 | Not tested | Not tested | Not tested | Not tested | Absent | no | No known reaction | No known reaction |
| 22 | 32 | F | 15:01, 15:01 | 0 | Not tested | Not tested | Not tested | Not tested | Absent | no | No known reaction | No known reaction |
| 23 | 10 | F | 07:01, 11:01 | 0 | Not tested | Not tested | Not tested | Not tested | Absent | No | No known reaction | No known reaction |
| 24 | 30 | F | 01:01, 15:01 | 0 | Not tested | Not tested | Not tested | Not tested | Absent | yes | No known reaction | No known reaction |
| 25 | 31 | F | 04:04, 16: 01 | 0 | Not tested | Not tested | Not tested | Not tested | Absent | no | No known reaction | No known reaction |
| 26 | 23 | M | 01:01, 07:01 | 0 | Not tested | Not tested | Not tested | Not tested | Absent | yes | No known reaction | No known reaction |
| 27 | 22 | F | 07:01, 15:01 | 0 | Not tested | Not tested | Not tested | Not tested | Absent | yes | No known reaction | No known reaction |
| 28 | 23 | M | 07:01, 13:01 | 0 | Not tested | Not tested | Not tested | Not tested | Absent | yes | No known reaction | No known reaction |
| 29 | 31 | M | 01:01, 04:04 | 0 | Not tested | Not tested | Not tested | Not tested | Absent | yes | No known reaction | No known reaction |
| Atopic subjects without cashew and other tree nut allergy | ||||||||||||
| 30 * | 10 | M | 09:01, 15:01 | 0 | Not tested | Not tested | Not tested | Not tested | Absent | yes | No known reaction | No known reaction |
| 31 * | 6 | M | 10:01, 15:01 | 0 | Not tested | Not tested | Not tested | Not tested | Absent | no | No known reaction | No known reaction |
| 32 * | 63 | M | 01:01, 15:01 | 0 | Not tested | Not tested | Not tested | Not tested | Absent | no | No known reaction | No known reaction |
| 33 | 41 | F | 09:01, 15:01 | 0 | Not tested | Not tested | Not tested | Not tested | Absent | yes | No known reaction | No known reaction |
| 34 | 26 | F | 04:04, 15:01 | 0 | Not tested | Not tested | Not tested | Not tested | Absent | yes | No known reaction | No known reaction |
| 35 | 8 | F | 01:01, 15:01 | 0 | Not tested | Not tested | Not tested | Not tested | Absent | no | No known reaction | No known reaction |
I Itchy mouth, lips and / or pharynx
II Abdominal discomfort and / or diarrhea
III Nausea or vomiting
IV Acute or Severe skin itching, or hives, or angioedema
V Rhinitis and / or conjunctivitis and / or respiratory compromise
VI Dizziness (feeling loss of consciousness)
VII Syncope (loss of consciousness)
VIII Desaturation with respiratory compromise
Subjects also had history of peanut and positive IgE ImmunoCAP for peanut
TGEM
Peptide libraries were generated based on Ana o 1 and Ana o 2 sequences. The libraries consisted of overlapping peptides spanning the entire allergen, which were 20 amino acids in length with a 12 amino acid overlap synthetized by Mimotopes (Clayton, Australia). Peptide-loaded HLA-DR proteins were generated, as previously described [19;20]. The tetramer-guided epitope-mapping procedure was conducted as previously described [21].
Ex-vivo analysis of cashew-specific CD4+ T-cells
CD154+ detection assay was carried out as previously described [22]. Briefly, for detection of CD154+-reactive T-cells, 35 million PBMC (at 7 × 106 cells/mL) in culture medium (RPMI 1640 (Gibco) + 10% pooled human serum + 1% PenStrep) were stimulated with 5μg/mL of synthesized peptide pools (at a final concentration of 3 nM for Ana o 1 and Ana o 2 and 13 nM for Ana o 3), and 1 μg/ml anti-CD40 (Miltenyi Biotec, Auburn, CA) for 3 hours (for frequency and surface phenotype) at 37°C. Cells were also mock stimulated with DMSO (0.05% final concentration) as negative control. After stimulation, cells were stained with PE (phycoerythrin)-conjugated CD154 (Miltenyi Biotec, Auburn, CA) and labeled with anti-PE magnetic beads (Miltenyi Biotec, Auburn, CA) for 20 minutes at 4°C. A 1/100 fraction of cells was saved for analysis. The other fraction was passed through a Miltenyi magnetic column; magnetically enriched cells were next stained with a panel of antibodies of interest for 20 minutes at room temperature. After staining, cells were stained again with Via-probe+ (BD Biosciences, East Rutherford, NJ) for 10 minutes at 4°C before flow-cytometry. To set gates for each phenotypic marker, T-cells were gated within the naïve compartment, as these markers are not expressed on naive T-cells. Appropriate isotype antibody staining was also included to confirm positive staining of the marker used (Supplemental Figure 3A and 3B). Data acquisition was performed using a LSR II flow cytometer and data were analyzed utilizing FlowJo (Tree Star, Ashland, Ore). Frequency was calculated as previously described for tetramer analysis [23]. Ex vivo analysis with pMHC-II (Peptide/MHC class II) tetramers was carried out as previously described [23].
Basophil stimulation tests
Basophil activation was measured as previously described [24]. Briefly, heparinized whole blood from cashew allergic subjects was incubated with tree nut extract (2 μg/mL): Cashew (Anacardium occidentale, Ana o), Pistachio (Pistacia vera, Pis v), Hazelnut (Corylus americana, Cor a), Walnut (Juglans regia, Jug r) (Greer Laboratories, Lenoir, NC, USA) and mixture containing all tree nut extracts and simultaneously stained with anti-CD3 (eBioscience, San Diego, CA, USA), anti-CD203 (Beckman Coulter, Pasadena, CA, USA), anti-CRTH2 (Chemoattractant receptor-homologous molecule expressed on TH2 cells) (BD Biosciences, Franklin Lakes, NJ, USA) for 25 min at 37°C. Basophils were identified as CD3−CRTH2+ cells, and activation status was assessed following the detection of CD203c in the presence of the allergens tested [24]. Whole blood stimulated with buffer without allergen and a mixture of tree nut extract were used as negative and positive controls, respectively.
T-cell cloning procedure and proliferation analysis
T-cell clones were generated by staining T-cells with tetramer directly ex-vivo and sorting gated tetramer-positive CD4+ and CD45RA− cells using a FACSAria (at single-cell purity). Expansion was done in a 96-well plate in the presence of 1.0 × 105 irradiated PBMC and 2 μg/ml PHA (Remel, Lenexa, KS). T-cells were re-screened with tetramers loaded with antigenic epitopes to assess positivity for the corresponding specificity. T-cells were stimulated in parallel with the corresponding peptides (1 and 10 μg/ml) or tree nut extract (2 μg/ml), in the presence of autologous PBMC (peripheral blood mononuclear cells) as APC (Antigen presenting cells). After 48 hours, each well was pulsed for an additional 16 hours with 1 mCi [3H]thymidine (Amersham Biosciences, Piscataway, NJ), and uptake was measured with a scintillation counter to assess proliferation.
Phenotypical analysis of T cell clones
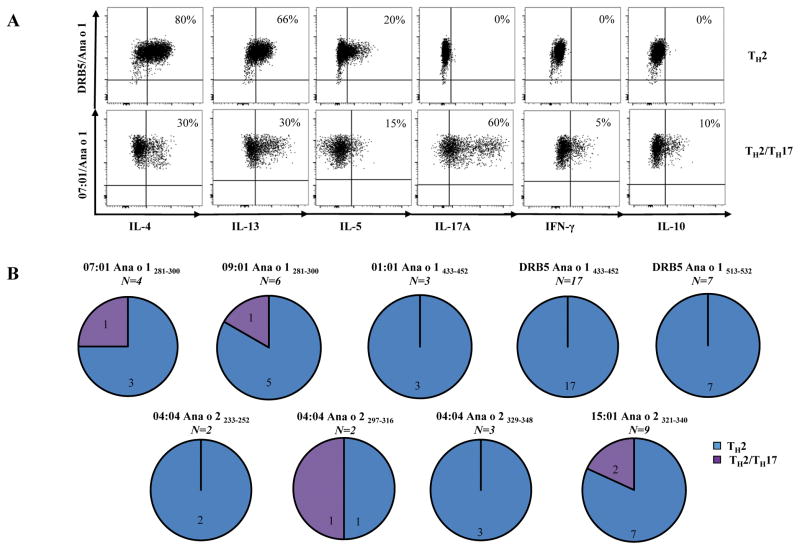
TCC ICS (intracellular cytokine staining) combined with tetramer staining was performed as previously described [25]. Cells were stained with a panel of antibodies directed against cytokines of interest, IL-4 (eBiosciences), IL-5 (Biolegend), IL-13 (Biolegend), IL-17A (Biolegend), IFN-γ (Biolegend) and IL-10 (BD Biosciences) for 20 minutes at room temperature. For phenotype analysis TCC were stained with a panel of antibodies directed against markers of interest, CCR4 (Biolegend), CRTH2 (BD Biosciences), CCR6 (Biolegend), CXCR3 (BD Biosciences) and CD27 (BD Biosciences). Two profiles (TH2 and TH2/TH17) of TCCs were arbitrarily defined as follows (Figure 6): TH2 profile was exemplified by CCR4+ with or without CRTH2 expression (data not shown) and production of IL-4 (≥10%), IL-5(≥10%), and IL-13 (≥10%); TH2/TH17 profile was characterized by co-expression CCR4 and CCR6 (data not shown) and co-production of both IL-4 and IL17A (≥10%) but no IFN-γ and IL-5 by individual cells within the clone.
Figure 6.
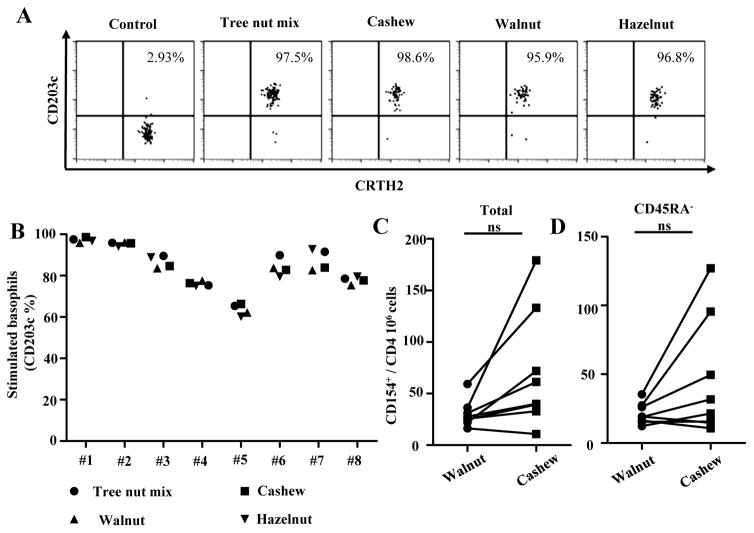
IgE and T-cells directed against other tree nut allergens. Cashew allergic subjects have IgE sensitivity to hazelnut, pistachio and walnut. Up-regulation of CD203c indicated activation of basophils. A, Representative results from a DRB1*15:01 cashew allergic subject. B, Summarized results for 8 cashew allergic subjects. Each data point represents the percentages of CD203c positive cells on stimulated basophils for each tree nut allergen. C, Frequencies of walnut-(filled circles) and cashew-reactive (filled squares) T-cells in subjects with cashew allergy (n=8). D, Frequencies of CD154+CD45RA− walnut-(filled circles) and cashew-reactive (filled squares) T-cells in subjects with cashew allergy (n=8; filled squares) and non-allergic subjects (n=8). Each data point represents the frequency of T-cells reactive to each allergen. A Student t test was used to compare expression of each marker in the statistical analysis. *P<0.05, **P<0.001, ***P<0.0001. NS. Not significant.
Cytokine detection with ELISA
For cytokine ELISA, IL-4 (clone 8D4-8) and IFN-γ (clone MD-1) capturing antibodies (BioLegend) were coated onto 96-well round bottom plates. Fifty μl of supernatants from co-cultures of T cell clones with autologous APC were collected after 48 h stimulation and added to each well. After overnight incubation, bound cytokines were detected by biotinylated, α-IL-4 (clone MP4-25D2) and α-IFN-γ (clone 4s.B3) antibodies (BioLegend) and quantified using a Victor2 D time resolved fluorometer (Perkin Elmer).
Statistical analysis
Statistical analysis was performed using the tests indicated in the figure legends utilizing Prism 5.0 software (Graphpad).
RESULTS
Ana o 1 and Ana o 2 as predominant cashew allergens recognized by CD4+ T-cells
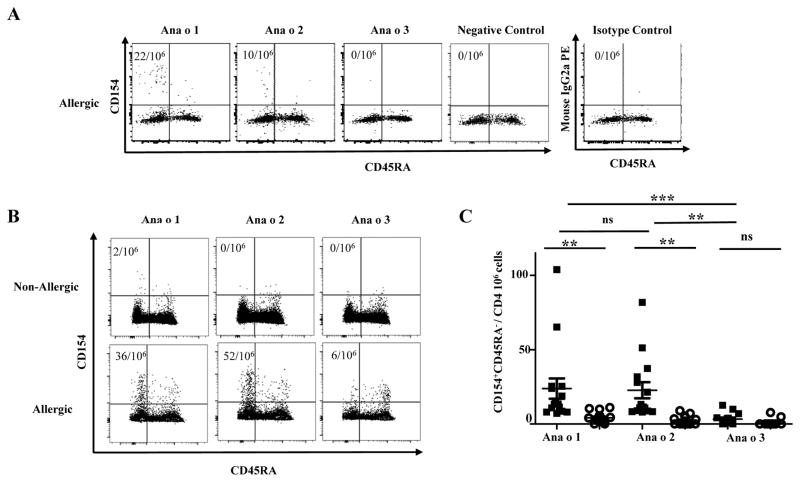
The CD154 up-regulation assay was used to examine Ana o 1, Ana o 2 and Ana o 3 reactive CD45RA−CD4+ memory T-cell responses in cashew non-allergic and allergic subjects. For non-allergic subjects, Ana o 1, Ana o 2 and Ana o 3 CD4+ T-cell responses were low (average frequencies of 6.87 ± 1.5, 3.88 ± 2.6 and <1 per million CD4+ T-cells, respectively). Conversely, in allergic subjects, strong Ana o 1, Ana o 2 and weak Ana o 3-specific responses were observed (average frequencies 23.81 ± 6.8, 22.70 ± 5.4 and 3.32 ± 1.1 per million CD4+ T-cells respectively) (Figure 1A, Figure 1B). The average frequency of Ana o 1 and Ana o 2 responses was 6 fold and 5 fold greater, respectively, than Ana o 3 T-cell immune responses in allergic subjects. No statistical differences were observed between Ana o 1 and Ana o 2 T-cell responses. Collectively, Ana o 1 and Ana o 2-reactive CD4+ T-cells played a predominant role in subjects with cashew allergy.
Figure 1.
Frequencies of cashew allergen reactive CD4+ T-cells with CD154 upregulation assay. A, Frequencies of Ana o 1-, Ana o 2- and Ana o 3-reactive T-cells within the memory compartment (CD45RA−) in a DRB1*01:01 subject with cashew allergy. Negative control was without peptide stimulation, and isotype control staining was with peptide stimulation. The frequencies of cashew allergen reactive T-cells per million CD4+ T-cells are as indicated. B, Frequencies of Ana o 1-, Ana o 2- and Ana o 3-reactive T-cells within the memory compartment (CD45RA−) in a DRB1*07:01 subject without allergy and a DRB1*07:01 subject with cashew allergy. The frequencies of cashew allergen reactive T-cells per million CD4+ T-cells are as indicated. C, Frequencies of CD154+CD45RA− Ana o 1-, Ana o 2- and Ana o 3-reactive T-cells in subjects with cashew allergy (n=14; filled squares) and non-allergic subjects (n=18; opened circles). Each data point represents the frequency of T-cells reactive to each allergen. An ANOVA test (with Dunnets correction) was used to compare all columns in the statistical analysis *P<0.05, **P<0.001, ***P<0.0001. NS. Not significant.
Both Ana o 1 and Ana o 2-specific CD4+ T-cells have a TH2 and TH2/TH17 phenotypes
The TGEM approach was utilized to identify CD4+ T-cell epitopes within cashew allergens Ana o 1 and Ana o 2 (Supplemental Figure 1). For Ana o 1, a total of 4 immunogenic epitopes restricted to DRB1*01:01, DRB1*07:01, DRB1*09:01 and DRB5*01:01 were identified and for Ana o 2, a total of 6 immunogenic epitopes restricted to DRB1*04:04, DRB1*07:01, DRB1*09:01, DRB1*15:01 and DRB4*01:01 were identified (Table 2).
Table 2.
Ana o 1 and Ana o 2 CD4+ T-cell epitopes
| HLA DRB1 Restriction | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ana o | Amino acid sequence | 01:01 | 04:04 | 07:01 | 09:01 | 11:01 | 13:01 | 15:01 | DRB4 | DRB5 |
| Ana o 1 281-300 | GPGGENPESFYRAFSWEILE | ● | ● | ● | ||||||
| Ana o 1 385-404 | MVVSYANITKGGMSVPFYNS | ● | ||||||||
| Ana o 1 433-452 | HPSYKKLRARIRKDTVFIVP | ● | ● | ● | ||||||
| Ana o 1 513-532 | VFGKQDEEFFFQGPEWRKEK | ● | ||||||||
| Ana o 2 233-252 | KVKDDELRVIRPSRSQSERG | ● | ● | |||||||
| Ana o 2 289-308 | PARADIYTPEVGRLTTLNSL | ● | ||||||||
| Ana o 2 297-316 | PEVGRLTTLNSLNLPILKWL | ● | ||||||||
| Ana o 2 321-340 | EKGVLYKNALVLPHWNLNSH | ● | ||||||||
| Ana o 2 329-348 | ALVLPHWNLNSHSIIYGCKG | ● | ● | |||||||
| Ana o 2 385-404 | QNFAVVKRAREERFEWISFK | ● | ● | |||||||
Frequency of Ana o 1 and Ana o 2-specific CD4+ T-cells was examined by direct ex vivo staining with Ana o 1- and Ana o 2-tetramers (Figure 2A and Figure 2B). For non-allergic subjects, CD4+ T-cells specific for Ana o 1- and Ana o 2 were barely detectable by tetramers (around 1 per million CD4+ T-cells). Conversely, in allergic subjects, the average frequency within the memory compartment (CD45RA−) was 16.42 ± 1.7 and 15.41 ± 2.8 per million CD4+ T-cells, respectively (Figure 2A and Figure 2B). These data confirmed the results from the CD154 assays that both Ana o 1 and Ana o 2 generate detectable T-cell responses ex vivo in cashew allergic subjects.
Figure 2.
Frequencies of cashew allergen reactive CD4+ T cells with tetramer assay. A, Frequencies of Ana o 1- and Ana o 2-specific T-cells within the memory compartment (CD45RA−) in a DRB1*15:01-DRB5*01:01 subject without allergy and a subject with cashew allergy. B, Frequencies of Tetramer+CD45RA− Ana o 1- and Ana o 2-specific T-cells in subjects with cashew allergy (n=12; filled squares) and non-allergic subjects (n=12; opened circles). Each data point represents the frequency of T-cells specific to each allergen. A Student t test was used in the statistical analysis. *P<0.05, **P<0.001, ***P<0.0001. NS. Not significant.
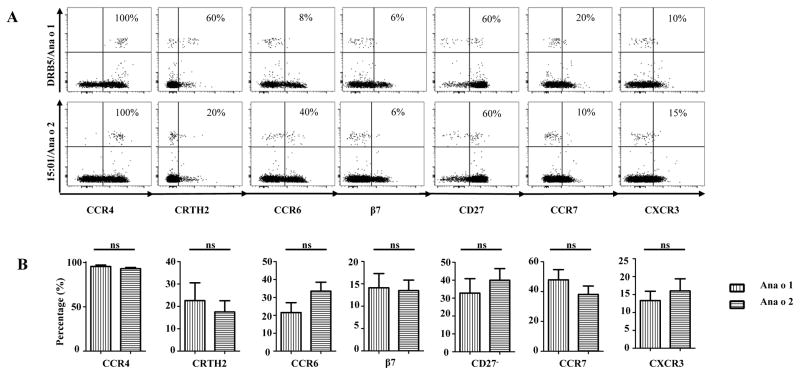
Chemokine receptor and differentiation marker expression of Ana o 1- and Ana o 2-specific T-cells were analyzed by ex vivo staining of PBMC in allergic subjects (Figure 3). A high percentage of Ana o 1- and Ana o 2-specific CD4+ T-cells expressed CCR4, while a low percentage expressed CXCR3. These data suggest a TH2 effector phenotype (Teff) for the Ana o 1- and Ana o 2-specific CD4+ T cells. The expression of CRTH2 and CD27 in these cells were heterogeneous. Although T-cells that lost CD27 expression were present, there were still higher percentages of T-cells that expressed CD27 for both Ana o 1 and Ana o 2-specific CD4+ T-cells. Additionally, the majority of these tetramer positive CD27+ T-cells also co-expressed CCR7, suggesting that a proportion of these T-cells have a central memory phenotype (TCM) (Supplemental Figure 2A). Variable expression of CCR6 with low β7 expression was detected, indicating the capacity of some cells homing to gut. In addition, all CCR6+ T-cells co-expressed CCR4, suggesting that a subset of T-cells are TH17 and/or TH2/TH17 (Supplemental Figure 2B). Conversely, for the non-allergic subjects with appreciable frequency, the main phenotype observed was characterized by CXCR3+CCR6+CD27+ (Supplemental Figure 3C). In conclusion, for allergic subjects, both Ana o and Ana o 2-specific cells expressed conventional TH2 markers. We did not observe a significant difference in the expression of different surface markers between Ana o 1 and Ana o 2.
Figure 3.
Phenotypes of Ana o 1- and Ana o 2-reactive T-cells. A, First row, profile for Ana o 1 in a DRB5*01:01 allergic subject. Second row, Ana o 2 profile in a DRB1*15:01 allergic subject. The percentages of surface markers expressed by Ana o 1- and Ana o 2-specific T-cells are as indicated. B, Ex vivo expression of CCR4, CRTH2, CCR6, β7,CD27−, CCR7 and CXCR3of Ana o 1- and Ana o 2- specific T-cells in subjects with cashew allergy (n=12) Each bar represents the percentage of tetramer positive T-cells with marker expression for each allergen. A Student t test was used to compare expression of each marker in the statistical analysis. NS. Not significant.
Cashew-specific CD4+ T-cells in allergic subjects are capable of cross-reacting with hazelnut and pistachio but not to walnut
It is likely that due to amino acid sequence identity among Ana o 1 with Cor a 11, Pis v 3 and Jug r 2 (44%, 78% and 36.6% respectively) and Ana o 2 with Cor a 9, Pis v 5 and Jug r 4 (60%, 80.5%, and 63.2% respectively) (http://fermi.utmb.edu/SDAP/), cross-reactivity between different tree nut allergens may be present at the T-cell level. For this purpose, peptides resembling homologous regions of cashew derived epitopes for other tree nut species were identified by selecting sequences with high amino acid sequence identity to Ana o 1 and Ana o 2 epitopes utilizing Blast alignments (Supplementary Table 1) [26]. Overall, the majority of Ana o 1 and Ana o 2 homologs from different tree nut species showed amino acid sequence identity to cashew derived epitopes, suggesting these are plausible T-cell epitopes.
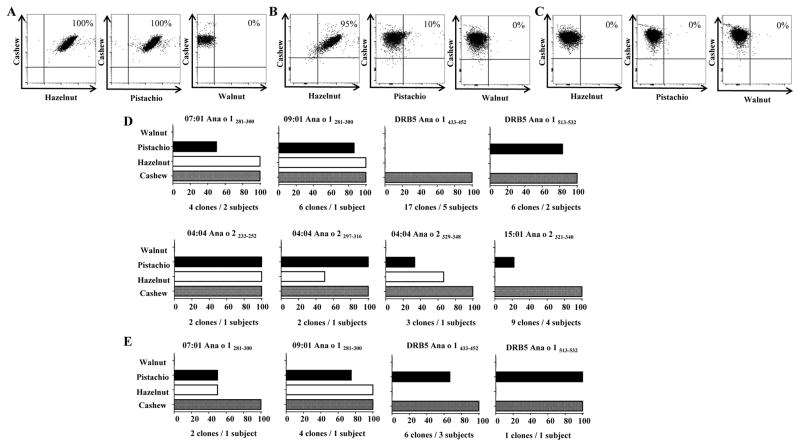
On account of these findings, Ana o 1- and Ana o 2-specific T-cells were single cell sorted directly ex-vivo for generation of TCC. TCC were obtained from 7 out of the 12 cashew epitopes identified. A total of 36 Ana o 1 and 16 Ana o 2 TCC were obtained and T-cell cross-reactivity was first evaluated by co-staining experiments using tetramers (Figure 4A, 4B, 4C and Supplementary Table 2). As several TCC with identical specificity were obtained from one subject (e.g. six 09:01 Ana o 1281-300-specific TCC) by ex vivo single cell sorting, it is still possible some of these TCC may in fact have been derived from the same identical clone in vivo and the some results described below should be interpreted with this caveat in mind. Three profiles were observed: (i) cross-reactivity with hazelnut and pistachio; (ii) cross-reactivity with hazelnut or pistachio; and (iii) no cross-reactivity with hazelnut or pistachio. Cross-staining with walnut was not observed. These profiles were later confirmed utilizing proliferation assay, where an SI>3 was defined as cut-off for positivity. Though clones that were co-stained by hazelnut tetramers and pistachio tetramers were also positive in proliferative assay, there were low affinity clones that did not co-stained but proliferated upon peptide specific stimulation. For example, 2 out of 9 TCC DRB1*15:01 restricted Ana o 2321-340 and 2 out of 4 DRB1*07:01 restricted Ana o 1281-300 did not co-stain with pistachio loaded tetramers, but these clones proliferated upon peptide specific activation. Low affinity CD4+ T-cells, below detection level with pMHC-II tetramers, with pathogenic properties have been previously observed for autoimmune disease[27]. Results from all these proliferation assays with peptides derived from different tree nut proteins were summarized in Figure 4D and Supplementary Table 2. At the epitope level, 7/8 of cashew epitopes screened could elicit cross-reactive response to pistachio, and 5/8 of cashew epitopes identified could elicit cross-reactive response to hazelnut. At the T cell level, 19.2% of the cashew reactive clones isolated were cross-reactive to both hazelnut and pistachio, 15.4 % of the clones were cross-reactive to pistachio and not to hazelnut, 11.5% of the clones were cross-reactive to hazelnut and not to pistachio, and 53.8% of the clones did not cross react to hazelnut or pistachio but did exclusively respond to cashew. To confirm our observations, TCC were stimulated with tree nut extracts and proliferation was analyzed. Similar profiles were observed for TCC tested (Figure 4E). Nonetheless, Ana o 1433-452 specific DRB5*01:01 restricted TCC proliferated to pistachio extract while it did not proliferate to pistachio peptide stimulation. We speculate that antigen processing of the pistachio extract leads to the generation of peptides of different lengths that are more potent stimuli compared to the 20mers used in the synthetic peptide stimulation. Notably, proliferation to walnut peptides and extract was not observed for all the TCC with different specificities.
Figure 4.
Cashew-specific CD4+ T-cells in allergic subjects are capable of cross-reacting with hazelnut and pistachio but not to walnut. T-cell clones isolated by DRB1*01:01, *07:01, *09:01, DRB5*01:01, *04:04 and *15:01 tetramers were assayed for specificity and functionality. The plots in A, B, and C, show representative results for different cross-reactivity profiles for the cashew-derived epitopes in Ana o 1 and Ana o 2, cells were co-stained with Allophycocyanin-labelled DRB1*/Ana o peptide loaded tetramers (y axis) and with PE-labelled DRB1*/homologous tree nut peptide-loaded tetramers (x axis). A, cross-reactivity to hazelnut and pistachio B, cross-reactivity to hazelnut or pistachio C, no cross-reactivity to hazelnut or pistachio. D, Summarized proliferation results for Ana o 1281-300, Ana o 1433-452, Ana o 1513-532, Ana o 2233-252, Ana o 2329-348 and Ana o 2321-340-specific T cell clones using APCs from their respective DRB1*specificities. Cells were stimulated with specific or irrelevant control peptide. E, Summarized proliferation results for Ana o 1281-300, Ana o 1433-452 and Ana o 1513-532-specific T cell clones using autologous APCs. Cells were stimulated with different tree nut extract. For D. and E. SI: stimulation index; cpm of specific peptide divided by cpm of irrelevant peptide. a SI>3 was defined as cut-off for positivity. Bars represent percentage of clones that had a SI>3. The number of clones and subjects from whom TCC were derived from are indicated under each graph.
The cytokine profiles of TCC was evaluated with ICS in addition to tetramer staining (Figure 5). In support of phenotyping experiments, both Ana o 1 and Ana o 2-specific CD4+ T-cells produced TH2 associated cytokines, including abundant IL-4, IL-5 and IL-13, while others only produced IL-4. TCCs that co-produced IL-4 and IL-17A with small amounts of IFN-γ were also observed. Cross-reactive epitopes also elicited almost identical cytokine profile when stimulated with peptides or tree nut extract (Supplemental Figure 4). TCC did not elicit cytokine responses to walnut peptides and extract. Collectively, these results suggest that cashew-specific T-cells can readily react to extract and T-cell epitopes with amino acid sequence identity from other tree-nut species.
Figure 5.
Cytokine profiles of Ana o 1 and Ana o 2-specific T-cell clones. A. ICS staining of a TH2 clone and a TH2/TH17 clone, and B, Phenotype of 53 T-cell clones derived from 9 allergic subjects. The numbers of T-cell clones for each profile and specificity are as indicated.
Cashew- and Walnut-reactive CD4+ T-cells are detected in cashew allergic subjects that have IgE sensitivity to other tree nuts
Majority of cashew allergic subjects had a positive Immunocap score for hazelnut, pistachio and walnut extract. Subjects were also evaluated for IgE reactivity to these tree nuts utilizing a basophil activation assay (Figure 6A and 6B). This confirmed that these cashew allergic subjects are sensitized to other tree nut species.
The observation that not all TCC co-stain and most lacked T-cell reactivity to both walnut peptides and extract suggested that walnut reactive T-cells represented a distinct population of T-cells, as most of these allergic subjects had a positive Immunocap score to walnut and positive basophil activation test. For this objective, we used CD154 assay and compared frequencies of Cashew- and Walnut-reactive T-cells in our cohort (Figure 6C and 6D). No differences in frequencies for walnut- and cashew-reactive T-cells were observed. These results suggested that these subjects are sensitized to both cashew and walnut and their respective allergen-reactive T-cells are distinct populations of T-cells.
DISCUSSION
Although allergic reactions to cashew nut are increasing in prevalence and adverse reactions may be more severe than peanut allergy [3–5], the role of CD4+ T-cell responses in cashew allergy has not yet been studied. In this study, both the class II tetramer and CD154 upregulation assays were used to examine cashew allergen-specific T-cells. The CD154 upregulation assay has the advantage that it enables the detection of global responses to an allergen in the absence of HLA-restriction information and epitope identification which is the prerequisite for class II tetramer-based experiments. On the other hand, tetramer assays are more specific and sensitive compared to the CD154 assay. In this study, both Ana o 1 and Ana o 2 were identified as the predominant cashew allergens that elicit CD4+ T-cell responses. T cell reactivity to Ana o 3 was essentially absent. As we did not assay for Ana o 3 sIgE, it remains a possibility that the selected subjects may not have been sensitized to Ana O 3. Similar to walnut allergen-specific T-cells [22], we observed a TH2 dominant population (including TCM and Teff), while a sub-population of T-cells that co-expressed CCR4 and CCR6 (TH2/TH17 and TH17-like) was also detected. This sub-population had been previously detected in both asthmatic and non-asthmatic subjects with peanut [28], walnut [22] and shrimp allergies [29]. ICS showed that the majority of TCC clones derived from cashew epitopes mainly produced IL-4, IL-13 and IL-5, while a minority of TCC co-produced IL-4 and IL-17A, implicating that these CCR4+CCR6− and CCR4+CCR6+ cells were bona fide TH2 and TH2/TH17 cells, respectively. Compared with peanut allergy, cashew nut allergy causes more gastro-intestinal symptoms [4]. Indeed these symptoms may be attributed to the presence of CCR6+β7+CCR7+ T-cells as they have the capacity to home to the GALT [30–33]. The association of CCR6 expression with food allergen specific T cells in both asthmatic and non-asthmatic allergic subjects may be unique as allergen specific T cells for aeroallergens in non-asthmatic subjects do not expressed CCR6 (Kwok WW, unpublished). The role of CCR6+ T-cells play a role in gastrointestinal allergic disease has also been documented in a murine model [34]. On the other hand, T-cell responses in non-allergic subjects were nearly absent in most subjects tested, for subjects with detectable frequency, the phenotype was CXCR3+CCR6+CD27+.
It has been estimated that the majority of cashew allergic subjects are poly-sensitized to different tree nuts [5]. Prior studies showed that vicilins and legumins from different tree nuts exhibit moderate cross-reactivity at the IgE level [15–18]. In contrast, cross-reactivity at the T-cell level in humans has not been thoroughly documented. In the present study, we detected basophil responses towards different tree nuts in subjects recruited on the basis of cashew allergy, confirming a poly-sensitization status. We also identified cashew derived epitopes and looked for cross-reactivity to epitopes with amino acid sequence identity from other species of tree nuts. We also found that the 46% of cashew-specific TCC from Ana o 1 and Ana o 2 reacted to peptides with amino acid identity from hazelnut and pistachio via proliferation assays. Importantly, these peptides with amino acid identity were able to elicit identical TH2 cytokine in cashew reactive T cells. Our observations inferred that phylogenetic relatedness among tree nuts reflects cross-reactivity. This is in agreement with the fact that cashew allergens Ana o 1, Ana o 2 and Ana o 3 and their corresponding allergens Pis v 1, Pis v 2 and Pis v 3 of the closely-related pistachio nut are almost identical in their aa sequences (78%, 80%,and 70%, respectively) and have a high degree of IgE-binding cross-reactivity[16;35]. Although Ana o 1 and the hazelnut allergen Cor a 11 are only 44% homologous, we were still able to detect cross-reactive epitopes between cashew and hazelnut. In contrast, though amino acid sequence identity is found in similar ranges (60%) between Ana o 2 and their corresponding allergens Jug r 4 in walnut, neither T-cell proliferation (peptide and/or extract) or class II tetramer co-staining were observed for all of the walnut homologous peptides. Additionally, all of the previously identified Jug r 2 epitopes [22] did not fall into the regions identified for Ana o 1 cashew derived epitopes. This was expected as Jug r 2 and Ana o 1 have low sequence homology 36%. Nonetheless, walnut T-cell responses were still detected in these subjects, suggesting that 1) walnut allergens contained species-specific T-cell epitopes and 2) walnut should be categorized within a tree nut allergens family that is distinct from cashew. Both walnut and pecan belong to the family of Juglandaceae [36] and their allergens are highly homologous. For example, Jug r 2 and Car i (Carya illinoinensis) 2 are 92% homologous; thus, we expect there will be extensive T cell cross reactivity between walnut and pecan. Similar profiles of IgE cross-reactivity have been published by Goetz et al [15]. They reported that cashew, pistachio and hazelnut form a group of tree nuts which were distinct from walnut and pecan [15].
From an immunotherapeutic standpoint, the presence of cross-reactive T-cell epitopes raises the possibility that single species peptide immunotherapy might be effective in modulating or depleting [38] these cross-reactive populations with both TH2 and TH2/TH17 phenotype. Indeed, linked epitope suppression has been previously shown where peptide immunotherapy with selected epitopes from a single allergen resulted in suppression of responses to other epitopes [39]. Additionally, Kulis et al [40] demonstrated that cashew immunotherapy prevented allergic responses to both cashew and pistachio in a cashew sensitized murine model as well as to cashew and walnut in a multisensitized murine model. Kulis et al also reported T cell cross-reactivity between walnut and cashew in a cashew sensitized mouse model [40]. We speculate that these cross-reactive epitopes between cashew and walnut are epitopes with low affinity to MHC-molecules. Low affinity cross-reactive epitopes may also be present in humans, but remain undetected in our tetramer approach. Indeed, the presence of walnut specific T-cell populations in walnut sensitized subjects of our cashew allergic cohort and their lack of cross-reactivity with cashew suggest that cashew peptide immunotherapy approach may not be most effective for walnut. Nonetheless, our results suggest that immunotherapy for cashew will be able to alleviate reactions toward pistachio and hazelnut.
Supplementary Material
Acknowledgments
NIH contract HHSN272200700046C
We thank Kavitha Gilroy and Sylvia Posso for help with subject recruitment.
List of nonstandard abbreviations used
- sIgE
specific IgE
- HLA
Human histocompatibility leukocyte antigen
- MHC
Major histocompatibility complex
- PBMC
Peripheral blood mononuclear cell
- Ana o
Anacardium occidentale
- Pis v
Pistacia vera
- Cor a
Corylus americana
- Car i
Carya illinoinensis
- Jug r
Juglans regia
- PE
Phycoerythrin
- pMHCII
Peptide/MHC class II
- TH
T helper
- CRTH2
Chemoattractant receptor-homologous molecule expressed on TH2 cells
- TGEM
Tetramer-guided epitope mapping
- ICS
Intracellular cytokine staining
- TCL
T-cell line
- TCC
T-cell clone
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Sicherer SH, Munoz-Furlong A, Godbold JH, Sampson HA. US prevalence of self-reported peanut, tree nut, and sesame allergy: 11-year follow-up. J Allergy Clin Immunol. 2010;125:1322–6. doi: 10.1016/j.jaci.2010.03.029. [DOI] [PubMed] [Google Scholar]
- 2.Sicherer SH, Furlong TJ, Munoz-Furlong A, Burks AW, Sampson HA. A voluntary registry for peanut and tree nut allergy: characteristics of the first 5149 registrants. J Allergy Clin Immunol. 2001;108:128–32. doi: 10.1067/mai.2001.115755. [DOI] [PubMed] [Google Scholar]
- 3.Davoren M, Peake J. Cashew nut allergy is associated with a high risk of anaphylaxis. Arch Dis Child. 2005;90:1084–5. doi: 10.1136/adc.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grigg A, Hanson C, Davis CM. Cashew Allergy Compared to Peanut allergy in a US Tertiary Care CEnter. Pedriatic Asthma, Allergy and Immunology. 2009;22:101–4. Ref Type: Journal (Full) [Google Scholar]
- 5.Rance F, Bidat E, Bourrier T, Sabouraud D. Cashew allergy: observations of 42 children without associated peanut allergy. Allergy. 2003;58:1311–4. doi: 10.1046/j.1398-9995.2003.00342.x. [DOI] [PubMed] [Google Scholar]
- 6.Clark AT, Anagnostou K, Ewan PW. Cashew nut causes more severe reactions than peanut: case-matched comparison in 141 children. Allergy. 2007;62:913–6. doi: 10.1111/j.1398-9995.2007.01447.x. [DOI] [PubMed] [Google Scholar]
- 7.Sicherer SH, Sampson HA. Food allergy: Epidemiology, pathogenesis, diagnosis, and treatment. J Allergy Clin Immunol. 2014;133:291–307. doi: 10.1016/j.jaci.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 8.van der Valk JP, Dubois AE, Gerth van WR, Wichers HJ, de Jong NW. Systematic review on cashew nut allergy. Allergy. 2014;69:692–8. doi: 10.1111/all.12401. [DOI] [PubMed] [Google Scholar]
- 9.Wang F, Robotham JM, Teuber SS, Tawde P, Sathe SK, Roux KH. Ana o 1, a cashew (Anacardium occidental) allergen of the vicilin seed storage protein family. J Allergy Clin Immunol. 2002;110:160–6. doi: 10.1067/mai.2002.125208. [DOI] [PubMed] [Google Scholar]
- 10.Robotham JM, Wang F, Seamon V, Teuber SS, Sathe SK, Sampson HA, Beyer K, Seavy M, Roux KH. Ana o 3, an important cashew nut (Anacardium occidentale L.) allergen of the 2S albumin family. J Allergy Clin Immunol. 2005;115:1284–90. doi: 10.1016/j.jaci.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 11.Robotham JM, Xia L, Willison LN, Teuber SS, Sathe SK, Roux KH. Characterization of a cashew allergen, 11S globulin (Ana o 2), conformational epitope. Mol Immunol. 2010;47:1830–8. doi: 10.1016/j.molimm.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 12.Roux KH, Teuber SS, Sathe SK. Tree nut allergens. Int Arch Allergy Immunol. 2003;131:234–44. doi: 10.1159/000072135. [DOI] [PubMed] [Google Scholar]
- 13.Radauer C, Breiteneder H. Evolutionary biology of plant food allergens. J Allergy Clin Immunol. 2007;120:518–25. doi: 10.1016/j.jaci.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 14.Clark AT, Ewan PW. The development and progression of allergy to multiple nuts at different ages. Pediatr Allergy Immunol. 2005;16:507–11. doi: 10.1111/j.1399-3038.2005.00310.x. [DOI] [PubMed] [Google Scholar]
- 15.Goetz DW, Whisman BA, Goetz AD. Cross-reactivity among edible nuts: double immunodiffusion, crossed immunoelectrophoresis, and human specific igE serologic surveys. Ann Allergy Asthma Immunol. 2005;95:45–52. doi: 10.1016/S1081-1206(10)61187-8. [DOI] [PubMed] [Google Scholar]
- 16.Noorbakhsh R, Mortazavi SA, Sankian M, Shahidi F, Tehrani M, Azad FJ, Behmanesh F, Varasteh A. Pistachio allergy-prevalence and in vitro cross-reactivity with other nuts. Allergol Int. 2011;60:425–32. doi: 10.2332/allergolint.10-OA-0222. [DOI] [PubMed] [Google Scholar]
- 17.Willison LN, Tawde P, Robotham JM, Penney RM, Teuber SS, Sathe SK, Roux KH. Pistachio vicilin, Pis v 3, is immunoglobulin E-reactive and cross-reacts with the homologous cashew allergen, Ana o 1. Clin Exp Allergy. 2008;38:1229–38. doi: 10.1111/j.1365-2222.2008.02998.x. [DOI] [PubMed] [Google Scholar]
- 18.Barre A, Sordet C, Culerrier R, Rance F, Didier A, Rouge P. Vicilin allergens of peanut and tree nuts (walnut, hazelnut and cashew nut) share structurally related IgE-binding epitopes. Mol Immunol. 2008;45:1231–40. doi: 10.1016/j.molimm.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 19.Novak EJ, Liu AW, Nepom GT, Kwok WW. MHC class II tetramers identify peptide-specific human CD4(+) T cells proliferating in response to influenza A antigen. J Clin Invest. 1999;104:R63–R67. doi: 10.1172/JCI8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwok WW, Liu AW, Novak EJ, Gebe JA, Ettinger RA, Nepom GT, Reymond SN, Koelle DM. HLA-DQ tetramers identify epitope-specific T cells in peripheral blood of herpes simplex virus type 2-infected individuals: direct detection of immunodominant antigen-responsive cells. J Immunol. 2000;164:4244–9. doi: 10.4049/jimmunol.164.8.4244. [DOI] [PubMed] [Google Scholar]
- 21.Novak EJ, Liu AW, Gebe JA, Falk BA, Nepom GT, Koelle DM, Kwok WW. Tetramer-guided epitope mapping: rapid identification and characterization of immunodominant CD4+ T cell epitopes from complex antigens. J Immunol. 2001;166:6665–70. doi: 10.4049/jimmunol.166.11.6665. [DOI] [PubMed] [Google Scholar]
- 22.Archila LD, Jeong D, Pascal M, Bartra J, Juan M, Robinson D, Farrington ML, Kwok WW. Jug r 2-reactive CD4 T cells have a dominant immune role in walnut allergy. J Allergy Clin Immunol. 2015 doi: 10.1016/j.jaci.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwok WW, Roti M, Delong JH, Tan V, Wambre E, James EA, Robinson D. Direct ex vivo analysis of allergen-specific CD4+ T cells. J Allergy Clin Immunol. 2010;125:1407–9. doi: 10.1016/j.jaci.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burtin D, Chabre H, Olagnier B, Didierlaurent A, Couret MN, Comeau D, Wambre E, Laparra H, Van OL, Montandon F, Batard T, Jonval V, Lorphelin A, Merle C, Berrouet C, Parry L, Gomord V, Van RR, Moingeon P. Production of native and modified recombinant Der p 1 molecules in tobacco plants. Clin Exp Allergy. 2009;39:760–70. doi: 10.1111/j.1365-2222.2009.03201.x. [DOI] [PubMed] [Google Scholar]
- 25.Archila LD, Delong JH, Wambre E, James EA, Robinson DM, Kwok WW. Grass-specific CD4(+) T-cells exhibit varying degrees of cross-reactivity, implications for allergen-specific immunotherapy. Clin Exp Allergy. 2014;44:986–98. doi: 10.1111/cea.12324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh H, Raghava GP. ProPred: prediction of HLA-DR binding sites. Bioinformatics. 2001;17:1236–7. doi: 10.1093/bioinformatics/17.12.1236. [DOI] [PubMed] [Google Scholar]
- 27.Sabatino JJ, Jr, Huang J, Zhu C, Evavold BD. High prevalence of low affinity peptide-MHC II tetramer-negative effectors during polyclonal CD4+ T cell responses. J Exp Med. 2011;208:81–90. doi: 10.1084/jem.20101574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delong JH, Simpson KH, Wambre E, James EA, Robinson D, Kwok WW. Ara h 1-reactive T cells in individuals with peanut allergy. J Allergy Clin Immunol. 2011;127:1211–8. doi: 10.1016/j.jaci.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renand A, Newbrough S, Wambre E, Delong JH, Robinson D, Kwok WW. Arginine kinase Pen m 2 as an important shrimp allergen recognized by T2 cells. J Allergy Clin Immunol. 2014;134:1456–9. doi: 10.1016/j.jaci.2014.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito T, Carson WF, Cavassani KA, Connett JM, Kunkel SL. CCR6 as a mediator of immunity in the lung and gut. Exp Cell Res. 2011;317:613–9. doi: 10.1016/j.yexcr.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunkel EJ, Campbell DJ, Butcher EC. Chemokines in lymphocyte trafficking and intestinal immunity. Microcirculation. 2003;10:313–23. doi: 10.1038/sj.mn.7800196. [DOI] [PubMed] [Google Scholar]
- 32.Campbell DJ, Butcher EC. Rapid acquisition of tissue-specific homing phenotypes by CD4(+) T cells activated in cutaneous or mucosal lymphoid tissues. J Exp Med. 2002;195:135–41. doi: 10.1084/jem.20011502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forster R, Schubel A, Breitfeld D, Kremmer E, Renner-Muller I, Wolf E, Lipp M. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 1999;99:23–33. doi: 10.1016/s0092-8674(00)80059-8. [DOI] [PubMed] [Google Scholar]
- 34.Blazquez AB, Knight AK, Getachew H, Bromberg JS, Lira SA, Mayer L, Berin MC. A functional role for CCR6 on proallergic T cells in the gastrointestinal tract. Gastroenterology. 2010;138:275–84. doi: 10.1053/j.gastro.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hasegawa M, Inomata N, Yamazaki H, Morita A, Kirino M, Ikezawa Z. Clinical features of four cases with cashew nut allergy and cross-reactivity between cashew nut and pistachio. Allergol Int. 2009;58:209–15. doi: 10.2332/allergolint.08-OA-0010. [DOI] [PubMed] [Google Scholar]
- 36.Gillespie DN, Nakajima S, Gleich GJ. Detection of allergy to nuts by the radioallergosorbent test. J Allergy Clin Immunol. 1976;57:302–9. doi: 10.1016/0091-6749(76)90086-5. [DOI] [PubMed] [Google Scholar]
- 37.Vidard L, Rock KL, Benacerraf B. Diversity in MHC class II ovalbumin T cell epitopes generated by distinct proteases. J Immunol. 1992;149:498–504. [PubMed] [Google Scholar]
- 38.Wambre E, Delong JH, James EA, LaFond RE, Robinson D, Kwok WW. Differentiation stage determines pathologic and protective allergen-specific CD4+ T-cell outcomes during specific immunotherapy. J Allergy Clin Immunol. 2012;129:544–51. 551. doi: 10.1016/j.jaci.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell JD, Buckland KF, McMillan SJ, Kearley J, Oldfield WL, Stern LJ, Gronlund H, van HM, Reynolds CJ, Boyton RJ, Cobbold SP, Kay AB, Altmann DM, Lloyd CM, Larche M. Peptide immunotherapy in allergic asthma generates IL-10-dependent immunological tolerance associated with linked epitope suppression. J Exp Med. 2009;206:1535–47. doi: 10.1084/jem.20082901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kulis M, Li Y, Lane H, Pons L, Burks W. Single-tree nut immunotherapy attenuates allergic reactions in mice with hypersensitivity to multiple tree nuts. J Allergy Clin Immunol. 2011;127:81–8. doi: 10.1016/j.jaci.2010.09.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.