Summary
Molecular alterations involving PI3K/AKT/mTOR pathway (including mutation, copy number, protein, or RNA) were examined across 11219 human cancers representing 32 major types. Within specific mutated genes, frequency, mutation hotspot residues, in silico predictions, and functional assays were all informative in distinguishing the subset of genetic variants more likely to have functional relevance. Multiple oncogenic pathways including PI3K/AKT/mTOR converged on similar sets of downstream transcriptional targets. In addition to mutation, structural variations and partial copy losses involving PTEN and STK11 showed evidence for having functional relevance. A substantial fraction of cancers showed high mTOR pathway activity without an associated canonical genetic or genomic alteration, including cancers harboring IDH1 or VHL mutations, suggesting multiple mechanisms for pathway activation.
Introduction
The PI3K/AKT/mTOR signaling pathway is one of the main growth regulatory pathways in both normal cells and cancer (Hennessy et al., 2005; Mayer and Arteaga, 2016). This growth pathway begins with class IA phosphatidylinositol-3-kinases (PI3Ks), which are heterodimers consisting of p110 catalytic and p85 regulatory subunits. Growth factor receptor tyrosine kinases (RTKs) activate PI3K through phosphorylation of adaptor proteins such as IRS1/IRS2 (Engelman et al., 2006). These adaptor proteins bind the amino-terminal domain of the PI3K p85 regulatory subunits through YXXM motifs, to reverse its inhibition of the p110 catalytic subunit, and leads to movement of the p85–p110 heterodimer to the cell membrane where p110 can phosphorylate phosphatidylinositol-4,5-bisphosphate (PIP2) to generate phosphatidylinositol-3,4,5-trisphosphate (PIP3). RAS family members can also activate PI3K (Mayer and Arteaga, 2016). The primary negative regulator of PI3K activation is the phosphatase PTEN, which dephosphorylates PIP3 at the 3′ position (Keniry and Parsons, 2008), with a secondary negative regulator being INPP4B (inositol polyphosphate-4-phosphatase type II B). PIP3 recruits several pleckstrin homology (PH) domain–containing proteins to the membrane, including AKT and PDK1. AKT is phosphorylated at Thr308 by PDK1 and at Ser473 by mTOR complex 2 (mTORC2), which increases its kinase activity. AKT directly and indirectly phosphorylates many downstream proteins, including the GSKs, p27KIP1, FoxO transcription factors, MDM2, and BAD, to enhance cell survival and growth (Manning and Cantley, 2007). Furthermore, AKT phosphorylates TSC2 at multiple sites, to inhibit the GTPase-activating protein (GAP) function of the TSC protein complex (consisting of TSC1, TSC2, and TBC1D7) toward Rheb, a RAS family member (Dibble and Manning, 2013; Laplante and Sabatini, 2012). Rheb-GTP binds to mTOR complex 1 (mTORC1) to activate its kinase activity toward the S6Ks, 4E-BP1, and other substrates, leading to enhancement of multiple anabolic biosynthetic pathways that enable production of the building blocks (e.g. nucleotides) and macromolecules (e.g. ribosomes) required for cell size increase and mitosis (Dibble and Manning, 2013).
Multiple genetic events have been described that lead to activation of the PI3K/AKT/mTOR pathway in cancer (Thorpe et al., 2015). Activating mutations in PIK3CA, which encodes the PI3K p110a catalytic subunit, are common in many cancer types (Samuels et al., 2004; Thorpe et al., 2015). There are highly focal hotspots of mutation in PIK3CA, E542 and E545 in the helical domain and H1047 and G1049 in the kinase domain, which activate the kinase through different mechanisms. Other PI3K p110 isoforms are rarely mutated in cancer overall, but PIK3CA and PIK3CB, as well as the class II PI3K PIK3C2B are all amplified in one or more cancer types (Thorpe et al., 2015). PIK3R1 and less commonly PIK3R2, which encode the p85α and p85β regulatory subunits of PI3K, are commonly mutated, resulting in reduced ability to inhibit PI3K p110a (Cheung et al., 2011; Thorpe et al., 2015). PTEN is subject to both genomic deletion and small point mutations that inactivate its function, and is one of the most commonly mutated cancer genes overall (Keniry and Parsons, 2008). AKT1 is occasionally activated by mutation at a single site, E17K (Carpten et al., 2007). Inactivating mutations in both TSC1 and TSC2 have been identified in cancer at low frequency (Hornigold et al., 1999), as well as activating mutations in MTOR (Grabiner et al., 2014). RHEB mutations are rare but focal at Y35, suggesting a driver effect.
With the recent conclusion of the data generation phase of The Cancer Genome Atlas (TCGA), there is opportunity for systematic analyses of the entire TCGA pan-cancer cohort, including analyses focusing on specific oncogenic pathways. The aim of our study was to take a comprehensively examine the entire PI3K/AKT/mTOR pathway and its components in the over 10,000 human cancers and 32 cancer types profiled by TCGA, these data involving multiple molecular profiling platforms, including proteomics.
Results
Proteomic analysis of PI3K/AKT/mTOR pathway
Our study involved 11219 human cancer cases representing 32 different major types, for which TCGA generated data on one or more of the following molecular characterization platforms (Table S1): whole exome sequencing (WES, n=10224 cases), whole genome sequencing data (WGS, n=1363), somatic DNA copy by SNP array (n=10845), RNA-seq (n=10224), and Reverse Phase Protein Array (RPPA). We used RPPA proteomic platform to analyze 7663 patient samples from 31 cancer types (with no data available for AML patients). The RPPA dataset comprised 225 high-quality antibodies that target 166 total proteins and 56 phosphorylated proteins. In this study, we carried out data normalization and batch correction to allow for direct comparisons between different cancer types. In general, mRNA levels were significantly correlated with protein levels, but strong correlations between mRNA and phospho-protein levels involving PI3K/AKT/mTOR pathway members were not observed (Figure S1A). In this study, we regarded mTOR signaling as a separate pathway from PI3K/AKT, where the former integrates information from the PI3K/AKT, Ras/MAPK, and LKB1/AMPK pathways (Laplante and Sabatini, 2012). Following previous studies (Akbani et al., 2014), we developed pathway signatures for both PI3K/AKT and mTOR components, based on member proteins selected by literature review, as a means of assessing the overall level of pathway activity given the variations of individual members.
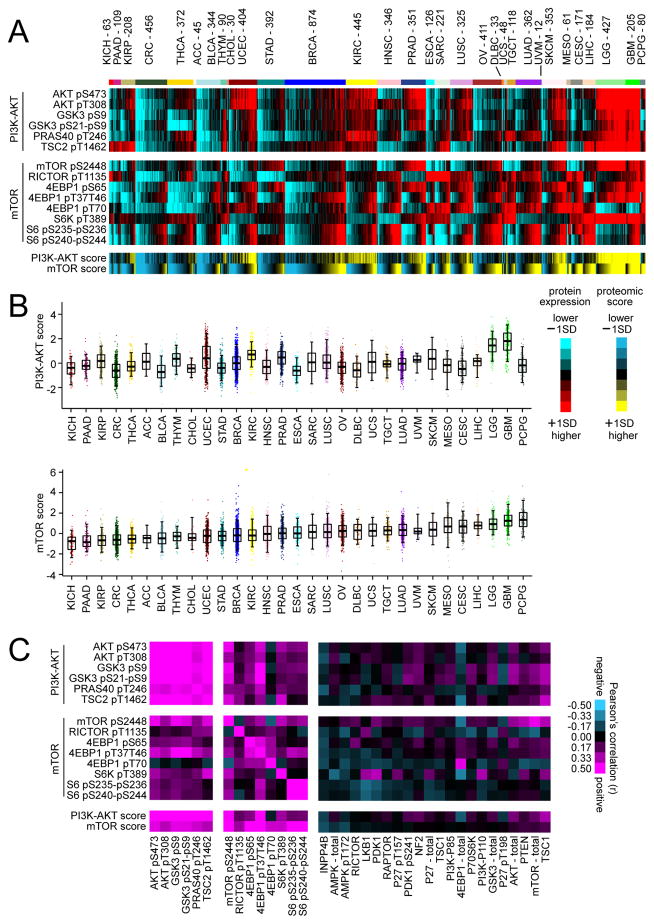
For each tumor, RPPA signatures for PI3K/AKT and mTOR were summarized into activity scores (Figure 1A and Table S2). On average, mTOR scores differed by tumor lineage, with, for example KICH (kidney chromophobe) tumors showing the lowest levels of mTOR activity, and with PCPG (pheochromocytoma and paraganglioma) showing the highest levels (followed by GBM and LGG, or glioblastoma and lower grade glioma, respectively); at the same time, within each tumor type a wide range of activity levels were evident (Figure 1B). Across tumor profiles, PI3K/AKT and mTOR activity scores were highly significantly correlated (Pearson’s r=0.50, p~0), though many cancer cases showed high mTOR activity but low PI3K/AKT activity or vice versa (Figures 1A and 1B), indicative of a certain degree of decoupling between the two pathway branches. Individual members of the PI3K/AKT signature were strongly correlated in protein expression with each other across cancers (Figure 1C). mTOR pathway-related members were also highly inter-correlated (Figure 1C), with distinct clusters involving 4EBP1- and S6-related features, respectively, and with phospho-RICTOR negatively correlated with phospho-mTOR (r=−0.14, p<1E-30). Other protein features strongly correlated with PI3K/AKT/mTOR signaling included members of the MAP Kinase pathway (Figures S1B and S1C). When considering a number of additional RPPA features for proteins understood to act peripherally on PI3K/AKT or mTOR signaling, these tended to show weaker correlations with PI3K/AKT and mTOR features (Figure 1C). INPP4B and AMPK were negatively correlated with mTOR activity as expected, while within subsets of tumors other proteins would presumably have pathway-related roles that may not be reflected in more global analyses.
Figure 1. Proteomic signatures of PI3K/AKT and mTOR across human cancers.
(A) Heat map of RPPA features considered core to either PI3K/AKT or mTOR pathways across 7663 cancers. Red, higher expression (values normalized to standard deviations from the median across all cancers); blue, lower expression. PI3K/AKT and mTOR features were each summarized into pathway activity scores for each tumor profile (yellow, higher inferred activity; blue, lower activity; bright yellow/blue denotes change of 1 standard deviation, or SD, from the median). Cancer types (denoted by TCGA project name) are ordered by low to high average mTOR pathway score. (B) Box plots of PI3K/AKT (top) and mTOR (bottom) pathway activities scores, as inferred using RPPA data. Box plots represent 5%, 25%, 50%, 75%, and 95%. (C) Pearson’s correlations between RPPA features across all cancers, involving features core to PI3K/AKT or mTOR pathways, as well as involving features representing proteins that may act peripherally upon either pathway. See also Figure S1 and Tables S1 and S2.
Somatic DNA alterations involving PI3K/AKT/mTOR pathway
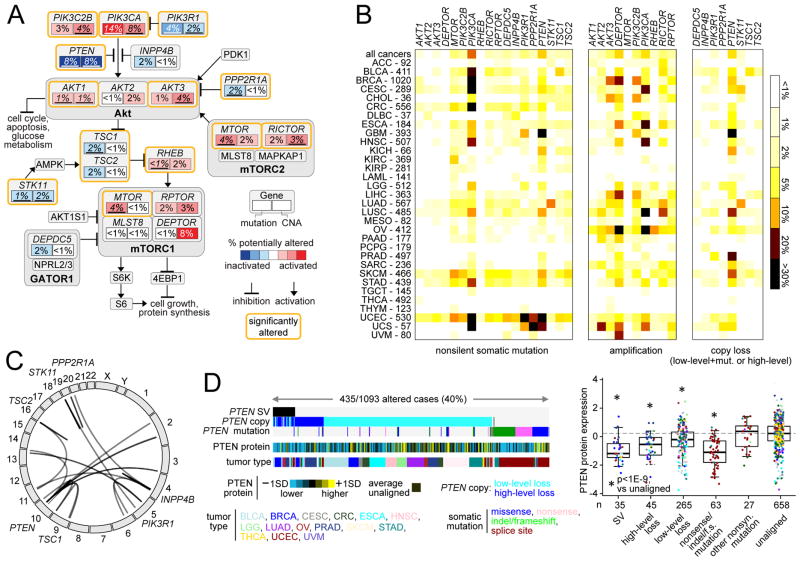
We examined gene mutations (using WES, n=10224 cases) and somatic DNA copy alterations (by SNP 6.0 arrays, n=10845 cases), focusing on genes in the canonical PI3K/AKT/mTOR pathway (Figure 2A and Table S3). Frequencies of somatic alteration for key genes in the pathway were tabulated across all cancers as well as within each cancer type according to TCGA project (Figure 2B and Table S4). A number of genes in the pathway were found significantly mutated or copy altered in pan-cancer analyses (Chang et al., 2016; Kandoth et al., 2013; Lawrence et al., 2014; Zack et al., 2013), including PIK3CA (14% mutated across all cancers; 6% amplified), PTEN (9% mutated; 7% deletion or two-hit loss), PIK3R1 (4% mutated), PPP2R1A (2% mutated), AKT1 (1% mutated), AKT1 (3% amplified), TSC1 (2% mutated), STK11 (2% mutated; 1% deletion or two-hit loss), RICTOR (3% amplified), and MTOR (4% amplified). With the notable exception of AKT3, copy number alterations of PI3K/AKT/mTOR pathway member genes were highly correlated with their mRNA expression (Figure S2A). When overlaid with mutation frequency data from human tumors, the model of PI3K/AKT/mTOR pathway (Figure 2A) can indicate which pathway members or interactions may be most relevant in the context of cancer. However, even genes with a low frequency of DNA alterations (e.g. AKTS1, MAPKAP1, MLST8, PDK1) may be critical in individual cancer cases or in specific cancer types or subtypes not included here in which they may be more commonly altered.
Figure 2. Somatic mutations and DNA copy and structural alterations involving components of the PI3K/AKT/mTOR pathway across human cancers.
(A) Diagram of somatic mutation and copy number alteration (CNAs) frequencies involving components of the PI3K/AKT/mTOR pathway. Key genes (with significant or sizable frequencies of alteration) are indicated by rectangles, with the percentages of somatic mutations and CNAs shown in the left and right portions of each rectangle, respectively. Significantly altered genes (from (Chang et al., 2016; Kandoth et al., 2013; Lawrence et al., 2014; Zack et al., 2013); percentages representing significant alterations are underlined) are bounded by orange lines. Red, potentially activating genetic alterations; blue, potentially inactivating genetic alterations. Copy loss represents either “high-level” deletion (approximating homozygous deletion) or mutation in combination with “low-level” deletion (partial loss). (B) By cancer type, percentages of somatic mutation or copy alteration for each indicated gene. Amplification denotes “high-level” copy gain. Numbers of cases denote representation on WES data platform. (C) Genomic rearrangements (represented in circos plot) involving PTEN, INPP4B, STK11, TSC1, TSC2, PIK3R1, or PPP2R1A, based on analysis of 1363 cases with WGS data. (D) Left: Alterations involving PTEN (somatic mutation, copy alteration, structural variation or SV) found in the set of 1093 cancers cases having both WGS and RPPA data available (protein values normalized to standard deviations, or SDs, from the median). Right: Box plot of PTEN protein expression by alteration class. Box plots represent 5%, 25%, 50%, 75%, and 95%. P values by t-test on log-transformed values. See also Figure S2 and Tables S3 and S4.
Genomic rearrangements represent another class of somatic alterations impacting gene function. Out of 1363 cases with WGS data available (1218 by low-pass sequencing), 63 cases (~5%) harbored a rearrangement within pathway suppressor genes PTEN (39 cases), INPP4B (14), STK11 (5), TSC1 (2), TSC2 (2), PIK3R1 (2), or PPP2R1A (2)(Figure 2C). By structural variation (SV), copy loss (partial or total), or mutation, PTEN was found altered in 40% of cancers with both RPPA and WGS data, with PTEN protein expression most impacted in tumors with SV, homozygous loss, or nonsense/indel/frameshift mutations (Figure 2D). In addition to PTEN, SVs within STK11 and TSC1 were also associated with decreased expression (Figure S2B). Furthermore, high-level and low-level copy number loss for several pathway genes were strongly correlated with reduced mRNA levels (Figure S2B), and 20 cases harbored candidate gene fusions involving PIK3CA, AKT1, AKT2, AKT3, or MTOR (Figure S2C and Table S3).
Recurrently mutated residues in key genes associated with protein activation
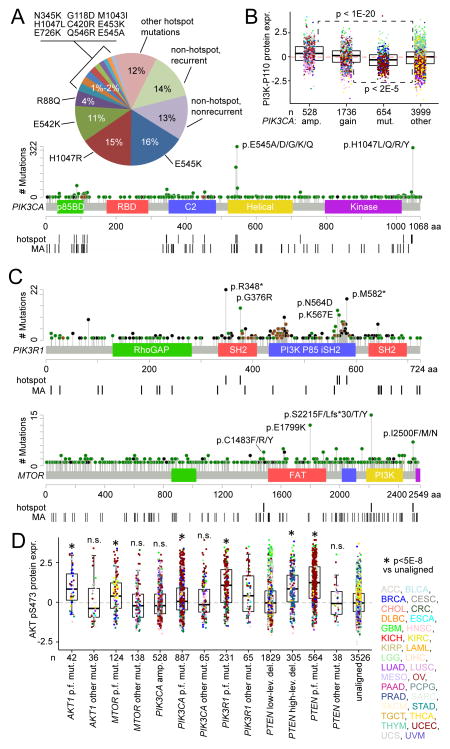
A large proportion of mutations identified in driver genes that activate PI3K/AKT/mTOR are of low occurrence, highlighting the need to functionally annotate the long tail of infrequent mutations present in heterogeneous cancers (Dogruluk et al., 2015). For example, PIK3CA is the gene most commonly activated by mutation in the cancer genome, with mutations being most frequent at positions E542, E545, and H1047 (Figure 3A); on the other hand, 13% of PIK3CA mutations observed occurred in a single case and showed no significant pattern of occurrence. Somatic copy alteration represents another potential mechanism for altering gene function, where for example, amplification of PIK3CA impacts p110α protein expression (Figure 3B). Previous pan-cancer sequence analyses (Chang et al., 2016) have identified recurrent mutational hotspots, where such hotspots would presumably have greater impact on protein function. In the case of PIK3CA, 73% of somatic, nonsilent mutation variants identified in TCGA pan-cancer cohort involved a hotspot residue as identified by Chang et al., while 13% of PIK3R1 mutations and 7% of MTOR mutations involved a hotspot residue (Figure 3C). In addition, algorithms such as Mutation Assessor (Reva et al., 2011) have predicted the likely functional impact of somatic mutation, e.g. based on evolutionary conservation of the affected amino acid in protein homologs.
Figure 3. Distributions of mutations in key PI3K/AKT/mTOR pathway genes and association with protein activation.
(A) PIK3CA nonsilent, somatic variant frequencies and distribution across domain-annotated p110α protein structure. “Recurrent” denotes mutation event observed in 2 or more tumor cases. “Hotspot” denotes recurrently mutated residues as identified by pan-cancer sequence analyses (Chang et al., 2016). “MA” denotes “medium” or “strong” functional prediction by Mutation Assessor algorithm (Reva et al., 2011). (B) Box plot of p110α expression by PIK3CA alteration class (gene amplification, gain of 1–2 copies, mutation, or none of the above, i.e. “unaligned”). P values by t-test on log-transformed values. (C) Distributions of nonsilent and somatic variants within PIK3R1 (top) and MTOR (bottom) across their respective domain-annotated protein structures. (D) Box plot of AKT pS473 phospho-protein expression by mutation (“mut.”) or copy alteration class, with the “unaligned” cases having none of the listed alteration types. “p.f.” denotes “predicted functional” mutations (by hotspot or by Mutation Assessor analysis or by literature review or by nonsense/frameshift/indel involving PTEN or PIK3R1); “amp.” denotes high-level gene amplification; “low-lev.” and “high-lev.”, low-level and high-level copy deletions, respectively. P values by t-test on log-transformed values. n.s., not significant (p>0.05). Box plots represent 5%, 25%, 50%, 75%, and 95%. Points in box plots are colored according to tumor type as defined by TCGA project as indicated in (D). See also Figure S3.
As the above genes as well as PTEN presumably act upon AKT (Figure 2A), phospho-protein expression of AKT was examined in relation to tumor groups as defined by somatic alteration of a key gene (Figure 3D). For each gene considered, mutations were separated on the basis of whether or not a prediction of mutation functionality could be made (by residue hotspot or by Mutation Assessor or by manual literature review or by nonsense/frameshift/indel involving PTEN or PIK3R1). For each of the genes considered (AKT1, MTOR, PIK3CA, PIK3R1, PTEN), tumors harboring mutations that were predicted to have functional effects had elevated phospho-AKT levels on average, as compared to tumors that did not harbor an alteration; in addition, tumors with mutations not predicted to be functional showed either a lesser effect or no significant effect on phospho-AKT. PTEN copy losses were also associated with AKT activation, while, interestingly, PIK3CA amplifications and copy alterations involving other specific genes (Figure S3) were not.
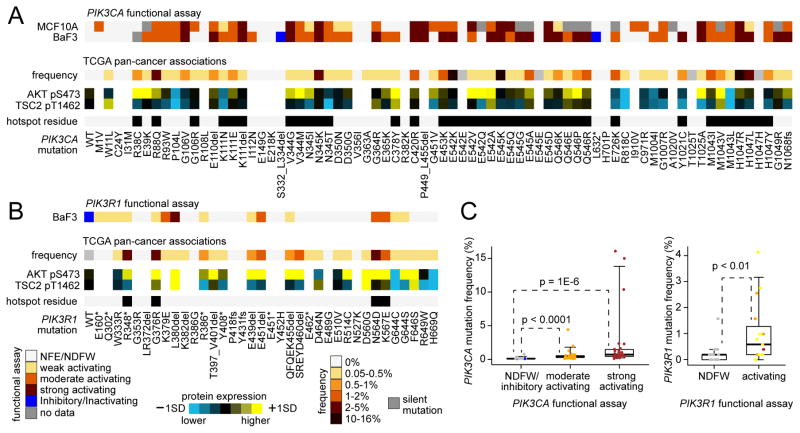
In addition to analysis of significantly mutated residues and of phospho-protein expression, functional studies using cell lines represents another way to annotate mutations in terms of their oncogenic potential. Using MCF10A and Ba/F3 cells, 69 different nonsilent PIK3CA mutation variants were functionally assessed in vitro for their activating potential (Figures 4A and S4 and Table S5). Most variants tested showed some level of functionality (from weak to strong) in at least one of the two cell lines, while 14 variants showed no functional effects and two showed inhibitory or inactivating effects. The degree of growth activation varied considerably, with the three highly recurrent PIK3CA site (E542, E545, H1047) mutants showing some of highest degree of activity in this assay. In another experiment, 35 different nonsilent PIK3R1 mutation variants were functionally interrogated in Ba/F3 cells (Figure 4B). When the results of the functional studies were aligned with data from TCGA, a significant trend was observed for both PIK3CA and PIK3R1, whereby variants that were associated with functionality in vitro had a higher frequency of occurrence in human tumors (Figure 4C), suggesting that natural selection favored tumor development for those variants with greater functional effects. Most variants showing some functionality also had higher phospho-AKT on average, as compared to tumors with the corresponding wild-type gene (Figures 4A and 4B), though variants associated with higher phospho-AKT were not necessarily associated with higher phospho-TSC2 (downstream in the pathway from AKT).
Figure 4. Functional assessment of specific PIK3CA and PIK3R1 variants by cell line viability assays.
(A, B) Ba/F3 or MCF-10A cells were transfected with wild-type (WT) or indicated mutant cDNA of PIK3CA (A) or PIK3R1 (B) then cultured for 4 weeks and harvested for viability assay. The extent of functionality conferred by the variant is indicated by colorgram. NFE/NDFW, no functional effect/no difference from wild-type. For the mutant variants assessed, corresponding human cancer data from TCGA are shown, including frequency of the variant (relative to other variants found for the same gene) and average protein expression for AKT pS473 and TSC2 pT1462. Hotspot residue, from ref (Chang et al., 2016). (C) For PIK3CA (left) and PIK3R1 (right), box plots of variant frequency in TCGA human tumors (relative to other variants found for the same gene) by functional assays results. P values by Mann-Whitney U-test. Box plots represent 5%, 25%, 50%, 75%, and 95%. See also Figure S4 and Table S5.
Transcriptomic analysis of PI3K/AKT/mTOR pathway
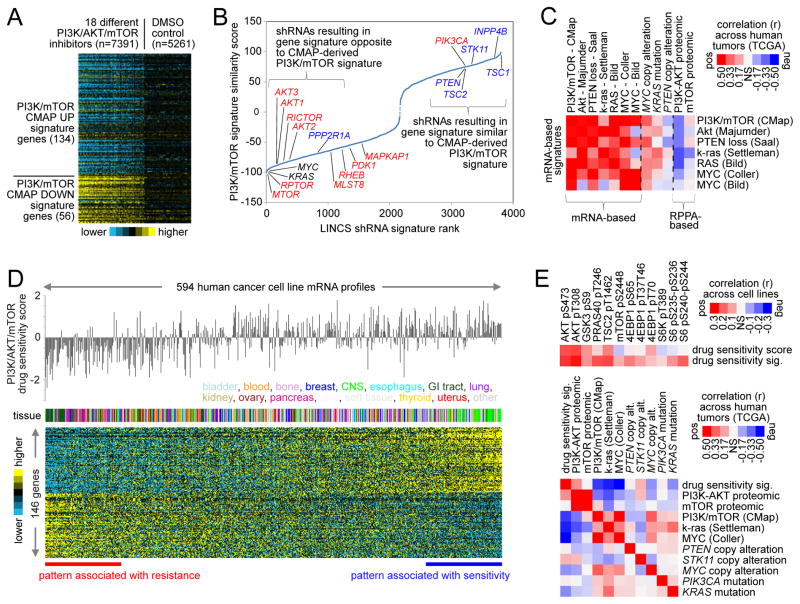
Signaling pathways that influence cell growth transduce signals to the nucleus, leading to activation or deactivation of the transcription of specific genes (Hanahan and Weinberg, 2000). Previously, we had defined a PI3K/AKT/mTOR transcriptional (mRNA) signature, based on the set of genes either induced or repressed by PI3K or mTOR inhibitors (Creighton et al., 2010). We applied this signature to the Library of Integrated Network-based Cellular Signatures (LINCS) database (Duan et al., 2014) of perturbational expression profiles across multiple cell and perturbation types. In the LINCS L1000 expression dataset (consisting of ~1000 genes), the PI3K/AKT/mTOR mRNA signature was inversely associated with the transcriptional responses of cell lines to PI3K/AKT/mTOR inhibitors (Figure 5A). We evaluated the signature against the LINCS expression profiles of cells treated with shRNAs for ~6K different genes; knockdown of pathway effectors (e.g. MTOR, RPTOR) resulted in gene signature patterns inversely correlated to those of our PI3K/AKT/mTOR signature, while knockdown of pathway suppressors (e.g. PTEN, INPP4B) resulted in signature patterns positively correlated with those of our signature (Figure 5B and Table S6). Notably, knockdown of MYC and KRAS also suppressed the PI3K/AKT/mTOR signature; furthermore, when scoring TCGA pan-cancer mRNA profiles for pre-defined signatures of PI3K/AKT/mTOR, MYC, and k-ras, cancers scoring high for PI3K/AKT/mTOR also tended to score high for MYC and k-ras (Figures 5C, S5A, and S5B), suggesting that multiple oncogenic signaling pathways may converge on similar sets of transcriptional targets. The above mRNA signatures would represent more than cell proliferation processes, given how the signatures were originally derived (Creighton et al., 2010), the lack of cell cycle regulators in the top LINCS shRNA results (Figure 5B), and the signature association with key alterations in human tumors (Figure 5C).
Figure 5. Survey of two distinct PI3K/AKT/mTOR-associated gene transcription signatures across human cancers.
(A) A previously defined gene transcription signature of PI3K/AKT/mTOR (Creighton et al., 2010)(originally derived using the Connectivity Map, or CMAP, dataset) was re-examined in the LINCS database of perturbational expression profiles, with PI3K/AKT/mTOR inhibitor treatment group compared with control group. (B) The PI3K/AKT/mTOR “CMAP” signature was evaluated against the LINCS expression profiles of cells treated with shRNAs for ~6K different genes. In the plot shown, shRNAs are ranked according to the overall similarities in their induced expression patterns with those of the PI3K/AKT/mTOR signature; for example, for shRNAs represented on the left of x-axis, knockdown of the gene results in a pattern inverse of that of the PI3K/AKT/mTOR signature. Red, canonical promoter of PI3K/AKT/mTOR pathway; blue, canonical suppressor. (C) TCGA pan-cancer mRNA profiles (n=10224 cases) were each scored for various transcriptional signatures associated with PI3K/AKT/mTOR, MYC, or k-ras pathways (defined previously using experimental models). Pearson’s correlations between indicated transcriptional and proteomic signature scores across the pan-cancer profiles are indicated, along with correlations of the signatures with specific genomic alterations. (D) A gene expression signature of sensitivity to PI3K/AKT/mTOR inhibition in cancer cell lines, consisting of 146 genes (p<0.01 by t-test and p<0.01 in regression model incorporating tumor type as a confounder), was derived using the Garnett et al. dataset (Garnett et al., 2012). (E) Top: For cell lines with both RPPA and mRNA data (n=231), Pearson’s correlations between key PI3K/AKT/mTOR proteins and PI3K/AKT/mTOR inhibition sensitivity (as defined by either drug treatment or gene signature from(D). Bottom: TCGA pan-cancer mRNA profiles were each scored for the drug sensitivity signature from (D); Pearson’s correlations across the pan-cancer profiles, involving transcriptional and proteomic signature scores and selected genomic features, are indicated. See also Figure S5 and Table S6.
As a means of identifying a transcriptional signature associated with PI3K/AKT/mTOR pathway, we examined datasets from Garnett et al. (Garnett et al., 2012), which included both gene expression and drug sensitivity data for 131 drugs on a set of 594 human cancer cell lines. In order to derive gene expression correlates of sensitivity to pathway inhibition, IC50 values for 11 different inhibitors to PI3K/AKT/MTOR were normalized and averaged to obtain a single drug sensitivity score across cell lines. After correcting for expression differences specific to tumor type, 146 genes were significantly associated (p<0.01, generalized linear model) with pathway inhibitor sensitivity (Figure 5D and Table S6). Across cell lines, this inhibitor sensitivity signature correlated significantly with PI3K/AKT phosphoprotein levels (Figure 5E), but showed little overlap with the above CMAP signature (from Figure 5A). Furthermore, when scoring TCGA pan-cancer mRNA profiles for the above signatures, tumors that scored high for the inhibitor signature tended to score low for the CMAP signature and vice versa, and PI3K/AKT proteomic score (but not mTOR score) was again highly correlated with the inhibitor signature score (Figures 5E, S5C, and S5D).
Molecular correlates of patient survival involving PI3K/AKT/mTOR pathway components
Molecular correlates of cancer patient survival can offer insights into the pathways and processes underlying more aggressive disease (The_Cancer_Genome_Atlas_Research_Network, 2013). For specific cancer types (e.g. breast and lung adenocarcinoma), the PI3K/AKT/mTOR pathway has been associated with aggressive disease (The_Cancer_Genome_Atlas_Network, 2012; The_Cancer_Genome_Atlas_Research_Network, 2013). In this present study, we sought to define survival correlates in pan-cancer analyses, leveraging the large numbers of patients available (these numbers helping to balance the relatively short patient follow-up times that characterize a number of individual TCGA projects). As some cancer types are inherently more aggressive than others (Hoadley et al., 2014), we carried two separate tests for each molecular feature examined: an “uncorrected” test across all cancers regardless of type and a “corrected” test incorporating cancer type (by TCGA project) as a covariate. Features more strongly associated with an aggressive cancer type but having a survival association that was not independent of cancer type (PIK3CA mutation, for example (Hoadley et al., 2014)) may show significance for the uncorrected but not the corrected survival test.
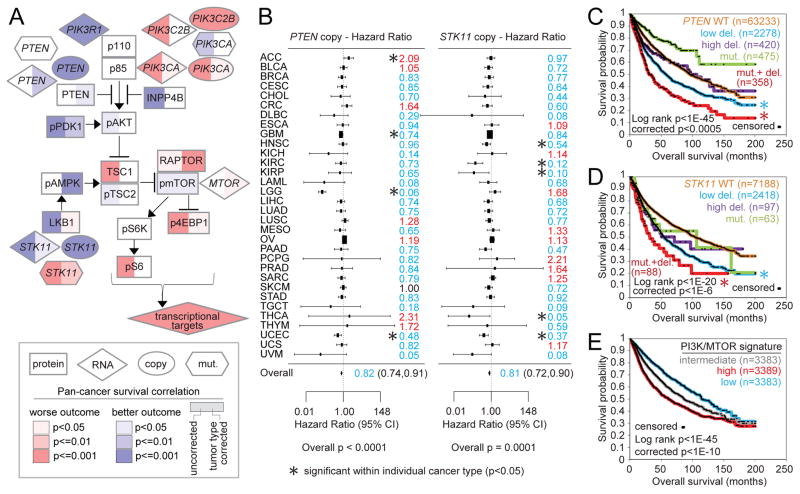
Numerous protein expression or genomic alteration features involving PI3K/AKT/mTOR pathway members were significantly associated with patient outcome in pan-cancer analyses (Figure 6A), a number of these features remaining significant after correcting for cancer type. Features significantly associated with worse patient outcome, independent of cancer type, included STK11 mutation, STK11 copy loss, PTEN copy loss, PIK3CA amplification, and higher phospho-4EBP1 expression. Focusing on PTEN and STK11 copy alterations, these features were found significant within several individual cancer types, with the aggregated patterns across cancer types denoting pan-cancer significance (Figure 6B). Interestingly, for both PTEN and STK11, low level deletion (approximating partial copy loss) but not high level deletion (approximating total loss) was associated with significantly worse outcome compared to wild-type (Figures 6C and 6D); loss of one copy combined with somatic mutation of the other copy was associated with the poorest outcome. For both PTEN and STK11, neither high-level deletion nor mutation without copy loss could be associated with worse outcome, where in this instance, survival differences by tumor type were a likely confounder (e.g. 65% of the PTEN mutation with no copy alteration group were UCEC—or uterine corpus endometrial carcinoma—cases). As a group, gene transcription targets of the PI3K/AKT/mTOR pathway (based on the signature described in Figure 5A) were also associated with worse patient outcome (Figure 6E).
Figure 6. Pan-cancer molecular correlates of patient survival involving PI3K/AKT/mTOR pathway components.
(A) Pathway diagram representing molecular features at the levels of mRNA (using n=10152 cancer cases in total with both mRNA and survival data), protein (n=7532), copy number (n=10685), and somatic mutation (n=10054). Red, significant correlation with worse patient outcome; blue, significant correlation with better outcome. “Tumor type corrected” survival p values denote significant correlation in model incorporating both the molecular feature and cancer type. P values < 0.05 correspond to an estimated False Discovery Rate (Storey and Tibshirani, 2003) of < 10%. (B) Forest plots of hazard ratios by tumor type (with 95% confidence intervals) for patient death for PTEN copy alteration (left) and for STK11 copy alteration (right). Hazard ratios based on log (tumor/normal) copy values; hazard ratio less than 1 (blue) denotes trend of copy loss with worse outcome. P value for overall survival correlation by meta-analysis fixed effects model. Asterisks denote cancer types that were individually significant (p<0.05). (C, D) Kaplan-Meier plot of overall survival of patients stratified by PTEN (C) or STK11 (D) alteration. Low del., low-level deletion (partial loss, no detected mutation); high del., high-level deletion (approximating total loss); mut., somatic nonsilent mutation (no copy loss); mut.+ del., copy loss combined with mutation. “Corrected” p values by stratified log-rank test incorporate cancer type as a confounder. Asterisks denote groups significantly different from wild-type (WT) group by stratified log-rank test. (E) Kaplan-Meier plot of overall survival of patients stratified by PI3K/AKT/mTOR transcriptional signature (“CMAP” signature). “Corrected” p values by stratified log-rank test incorporate cancer type as a confounder.
Genetic/genomic alteration classes in relation to PI3K/AKT/mTOR pathway activation
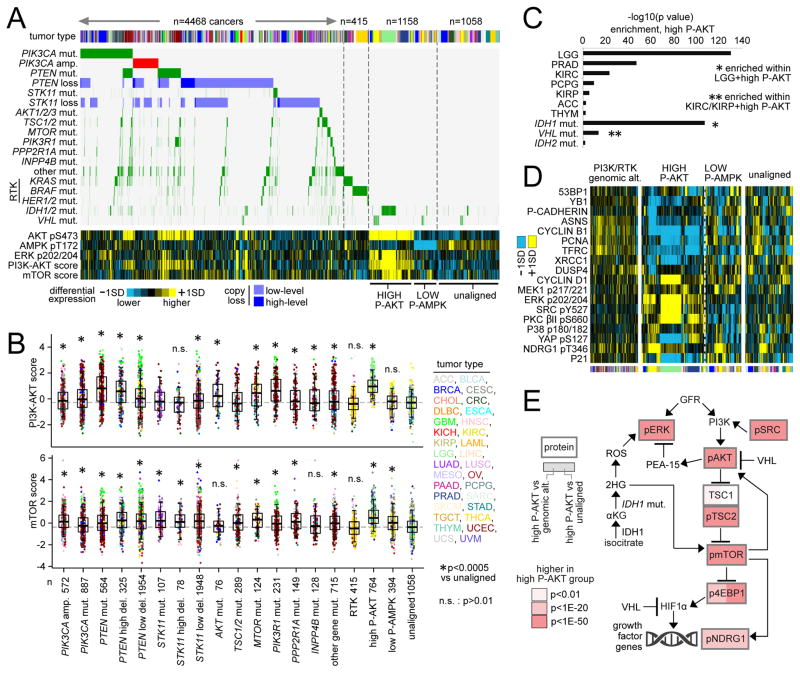
We then sought to examine the effects on pathway activation of some key genomic events in the tumors in which they occurred (including mutations represented in Figure 2A and copy alterations involving PIK3CA, PTEN, and STK11). Of the 7099 tumor cases examined (with both mutation and protein data), 4468 (63%) harbored at least one nonsilent somatic mutation or copy alteration involving PI3K/AKT/mTOR pathway (Figures 7A and S6A). Another set of 764 tumors showed high levels of phospho-AKT (>0.5 standard deviations, or SD, of pS473 from the median across samples) but without any of the genetic or genomic alterations associated with the above 4468 tumors, and another set of 394 tumors showed low levels of phospho-AMPK (<-0.5SD) without an associated genetic or genomic alteration. In comparison to a set of tumors that did not show pathway alteration at the DNA or protein level (an “unaligned” set, n=1058), mutation or copy alteration of individual PI3K/AKT/mTOR pathway members in general could be associated with higher PI3K/AKT or mTOR signaling as measured by protein arrays (Figures 7B and S6B). Notably, STK11 alteration or low phospho-AMPK was strongly associated with high mTOR signaling but not with high PI3K/AKT signaling, consistent with the LKB1/AMPK pathway acting on mTOR independently of PI3K/AKT (Figure 2A). Mutations associated with Receptor Tyrosine Kinase (RTK) signaling were not strongly associated with PI3K/AKT/mTOR activation (Figures 7A and 7B), indicative of decoupling between PI3K/AKT/mTOR and RTK. Low-level as well as high-level copy losses of PTEN and STK11 could be associated with greater mTOR signaling.
Figure 7. Tumor classes as defined by PI3K/AKT/mTOR-related alterations.
(A) Tumor cases were separated into distinct groups on the basis of genetic or genomic alteration and of protein expression: 1) cases with nonsilent somatic mutation or copy alteration involving selected PI3K/AKT/mTOR pathway members as shown (left side, n=4468 cases), 2) additional cases with nonsilent mutation involving selected Receptor Tyrosine Kinase (RTK)-associated genes (n=415 cases), 3) cases with high phospho-AKT (“HIGH P-AKT”) but with none of the above somatic alterations (n=764 cases), 4) cases with LOW phospho-AMPK (“LOW P-AMPK”) but with none of the above somatic alterations (n=394 cases), 5) cases not aligned with any of the above (“unaligned,” n=1058 cases). AKT/MTOR/PIK3CA/PIK3R1/PTEN mutations represent “predicted functional” mutations from Figure 3D. “Other mut.” track involves nonsilent mutations for other genes represented in Figure 2A (Methods and Figure S6). Protein values and proteomic scores normalized to standard deviations, or SDs, from the median. (B) Box plots of PI3K/AKT (top) and mTOR (bottom) pathway activity scores by alteration class. P values by t-test on log-transformed values. n.s., not significant (p>0.01). Box plots represent 5%, 25%, 50%, 75%, and 95%. (C) Enriched tumor types and mutations within the HIGH P-AKT group. P values by one-sided Fisher’s exact test. IDH1 and VHL mutation events were significant (p<1E-10 and p<0.01, respectively) when limiting the analysis to LGG and to KIRC/KIRP (renal) cases, respectively. (D) Top differentially expressed proteins in HIGH P-AKT group compared to unaligned and PI3K-altered groups (see Methods), not including core PI3K/AKT/mTOR members. (E) Diagram of interactions involving PI3K/AKT/MTOR pathway represented by selected features from (C) and (D) (Carbonneau et al., 2016; Dodd et al., 2015; Guo et al., 2016; Weiler et al., 2014), with differential protein expression patterns represented, comparing tumors in HIGH P-AKT group with tumors harboring PI3K/RTK genomic alteration or with unaligned tumors. P values by t-test on log-transformed data. See also Figure S6.
PI3K/AKT/mTOR pathway activity, when measured at the protein level, was explained by known mutations or copy alteration in most but not all of the cases examined, suggesting additional, unexplained or underappreciated mechanisms of pathway activation. Focusing on the “High P-AKT” tumor group (n=764), with high phospho-AKT but lacking a DNA alteration classically associated with PI3K/AKT activation, these tumors were highly enriched for specific cancer types including LGG, PRAD, KIRC, and PCPG (Figure 7C), as well as for IDH1 mutations (associated primarily with LGG, i.e. gliomas) and VHL mutations (associated with renal cancers). A set of microRNAs could also help distinguish the “High P-AKT” group (Figure S6C). Proteins that were highly expressed specifically within the High P-AKT group (Figure 7D) included phospho-ERK, phospho-SRC, and phospho-NDRG1. These mutations and proteins would suggest a model (Figure 7E), whereby mutant IDH1 may lead to high phospho-ERK (Chaturvedi et al., 2013), and SRC can activate PI3K (Chen et al., 2015; Su et al., 2016) and where activated mTOR signaling may activate transcription targets of hypoxia via HIF-1alpha (particularly in the absence of VHL), including NDRG1 and growth factors that may lead to a further increase ERK and PI3K signaling (Clark, 2009). Notably, VHL was recently found to directly suppress AKT activity (Guo et al., 2016), and generation of 2-hydroxyglutarate (2HG) by mutated IDH1/2 was also recently found to lead to the activation of mTOR (Carbonneau et al., 2016); our data here would highlight the importance of both of the above relationships in the setting of human cancer.
Discussion
TCGA pan-cancer datasets have enabled us to examine human tumor correlations in the context of PI3K/AKT/mTOR, to an extent not previously possible. Our current model of the PI3K/AKT/mTOR pathway has developed over the course of numerous independent molecular biology studies, spanning decades of research. In large part, our understanding of the pathway members and interactions involved has been derived from experimental systems, including cell lines. While cell lines may uncover cause-and-effect relationships in vitro, the relevance of such relationships in the setting of human diseases such as cancer may not always be clear from these data alone. On the other hand, molecular data from human tumors provide correlative (though not necessarily causal) relationships that would have relevance to disease in the human setting. Most of the correlations observed in our study fit well with our understanding of PI3K/AKT/mTOR signaling, in particular the genetic or genomic alteration of specific genes having an impact on phospho-protein expression of key downstream intermediates. Genes or alteration classes that were previously underappreciated would also be found relevant in our study, including partial loss of PTEN or STK11 (associated with both worse survival and increased mTOR signaling). Where gene mutation often inactivates one allele, loss of one allele by copy alteration, which is common across multiple cancer types for both PTEN and STK11, would presumably have the same impact on loss of gene function. IDH1 and VHL mutations would also be implicated here with PI3K/AKT/mTOR, where such alterations were associated with particularly high AKT/mTOR signaling, and which genes might be put forth for consideration as part of the “canon” of what would be recognized to constitute the core standard model of PI3K/AKT/mTOR pathway.
The multiplatform molecular datasets offered by TCGA allow for a more comprehensive view of PI3K/AKT/mTOR pathway. Pathway alterations in cancer may be manifested at different levels of molecular complexity, from DNA to protein to transcriptional consequences. Integration with RPPA proteomic data allows us to assess the impact on pathway activation of mutations or copy alterations observed at the DNA level. As observed in this study, multiple oncogenic pathways in addition to PI3K/AKT/mTOR may regulate similar sets of transcriptional targets, where transcriptional patterns would represent a degree of separation from the pathway as manifested at the protein level. Phospho-protein levels may only be assessed by protein data and not mRNA data, which also represents an advantage of RPPA as compared to other proteomic approaches (Creighton and Huang, 2015). Clear overall trends may be observed when integrating proteomic data with data from other platforms, though statistical trends (e.g. visualized as boxplots) would apply to groups of patients and not always to the individual patient, which has implications regarding personalized therapy. Various sources of biological noise, in addition to technical noise, may be present within human tumors, which give rise to variation in molecular signals. Widespread molecular aberrations involving numerous genes and pathways within a given tumor, clonal heterogeneity, microenvironmental influences, variable sample purity, and tissue-specific effects can all add noise to our ability to match protein signals with specific DNA alterations. The RPPA methodology may have limitations as well (e.g. antibody robustness, unknown history of sample material used to measure potentially labile phosphorylations, linearity of signal readout, etc.), and instances where proteomic signals would seem disconnected from other molecular profile features of a particular tumor may be difficult to interpret. The power of large sample numbers and the opportunities for data integration offered by TCGA pan-cancer cohort can aid greatly in detecting robust patterns relevant to our understanding aspects of pathway deregulation.
Results of this study include a comprehensive and annotated catalog of PI3K/AKT/mTOR-associated variants across over 10,000 tumors, which may serve as an additional resource for assessing variants in the clinical setting. One of the challenges of applying personalized and precision medicine approaches to cancer therapy is the large number of gene alterations that may be found within a given patient’s tumor. Stratifying patients by mutation status, e.g. PIK3CA mutation, has been shown to increase response rates in clinical trials testing inhibitors to PI3K/AKT/mTOR pathway, though non-responders are still common (Ilagan and Manning, 2016). Not all genetic variants impacting a given gene would necessarily have a similar impact on its function, including a large fraction of observed PIK3CA variants. Oncogenic variants that are found to occur frequently or are associated with a significant pattern would seem likely to be functionally relevant. Other measures of predicting variant functionality include in silico structural predictions, in vitro functional assays, domain-specific expertise, and protein expression, all of which were explored to varying extents in the present study. In practice, multiple measures may be needed, as no single measure may capture all of the variants likely to be functional. In addition, the RPPA proteomic platform would have potential for clinical applications to personalized therapy (Creighton and Huang, 2015), and transcriptional signatures associated with inhibitor sensitivity in cell lines may be defined (Singh et al., 2009). The focused, comprehensive analysis on the PI3K/AKT/mTOR pathway here will serve as a valuable resource for understanding its deregulation in cancers and how to maximize its clinical utility.
Experimental Procedures
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Chad J. Creighton (creighto@bcm.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Subjects
Cancer molecular profiling data were generated through informed consent as part of previously published studies and analyzed in accordance with each original study’s data use guidelines and restrictions.
Cell lines
Assay medium for survival assay were Advanced RPMI 1640 medium (Life Technologies) with 5% FBS (Life Technologies) and 1x GlutaMAX (Life Technologies) for Ba/F3 cells and MEBM Basal medium (Lonza) with 100 ng/ml Cholera toxin (Lonza) and 52 ng/ml Bovine Pituitary Extract (BPE) (Lonza) for MCF10A cells.
METHOD DETAILS
TCGA patient cohort
The results here are based upon data generated by TCGA Research Network (http://cancergenome.nih.gov/). Molecular data from 11219 human cancers were aggregated from public repositories (Table S1). Tumors spanned 32 different TCGA projects, each project representing a specific cancer type, listed as follows: LAML, Acute Myeloid Leukemia; ACC, Adrenocortical carcinoma; BLCA, Bladder Urothelial Carcinoma; LGG, Brain Lower Grade Glioma; BRCA, Breast invasive carcinoma; CESC, Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, Cholangiocarcinoma; CRC, Colorectal adenocarcinoma (combining COAD and READ projects); ESCA, Esophageal carcinoma; GBM, Glioblastoma multiforme; HNSC, Head and Neck squamous cell carcinoma; KICH, Kidney Chromophobe; KIRC, Kidney renal clear cell carcinoma; KIRP, Kidney renal papillary cell carcinoma; LIHC, Liver hepatocellular carcinoma; LUAD, Lung adenocarcinoma; LUSC, Lung squamous cell carcinoma; DLBC, Lymphoid Neoplasm Diffuse Large B-cell Lymphoma; MESO, Mesothelioma; OV, Ovarian serous cystadenocarcinoma; PAAD, Pancreatic adenocarcinoma; PCPG, Pheochromocytoma and Paraganglioma; PRAD, Prostate adenocarcinoma; SARC, Sarcoma; SKCM, Skin Cutaneous Melanoma; STAD, Stomach adenocarcinoma; TGCT, Testicular Germ Cell Tumors; THYM, Thymoma; THCA, Thyroid carcinoma; UCS, Uterine Carcinosarcoma; UCEC, Uterine Corpus Endometrial Carcinoma.
Datasets
Proteomic data were generated by RPPA across 7663 patient tumors obtained from TCGA. RPPA methodology and quality control procedures have been described previously (Akbani et al., 2014; Li et al., 2017). In total, 225 high-quality antibodies targeting total (n=166), cleaved (n=2), acetylated (n=1) and phosphoproteins (n=56) were used. The entire set of RPPA Pan-Cancer samples was run in several different batches, resulting in potential batch effects on merging the sets; replicates-based normalization (RBN) (Akbani et al., 2014), was therefore applied, using replicate samples run across multiple batches to adjust the data for batch effects. Data (“Level 4”) are available from The Cancer Proteome Atlas (http://tcpaportal.org/tcpa/).
RNA-seq and miRNA-seq data were obtained from The Broad Institute Firehose pipeline (http://gdac.broadinstitute.org/). All RNA-seq samples were aligned using the by UNC RNA-seq V2 pipeline (The_Cancer_Genome_Atlas_Research_Network, 2013). For miRNA-seq data, only sample profiles from the Hiseq platform were used (representing n=8690 cases).
DNA from each tumor or germline-derived sample was hybridized to Affymetrix SNP 6.0 arrays as previously described (The_Cancer_Genome_Atlas_Research_Network, 2013)(n=10845 tumor profiles in all). GISTIC 2.0 was applied to the transformed copy number data, with a noise threshold used to determine copy gain or loss. Low-level gene gain, high-level gene amplification, low-level copy loss, or high-level copy loss were inferred using the “thresholded” calls as made by Broad Firehose pipeline (using +1, +2, −1, or −2, respectively). High-level amplifications denotes amplifications above the threshold and larger than the arm level amplifications observed for the given sample. Low-level copy deletions represent deletion above the threshold (approximating heterozygous deletions in the absence of whole genome doubling); high-level copy deletions denote copy losses above the threshold and greater than the minimum arm-level deletion observed for the sample (approximating homozygous deletions in the absence of whole genome doubling). Log (tumor/normal) copy values were used to evaluate correlations with survival in Figure 6A.
Somatic mutation calls were obtained from the publicly-available “MC3” TCGA MAF file (covering n=10224 patients, https://www.synapse.org/#!Synapse:syn7214402). This MC3 set is a re-calling of uniform files from all TCGA projects, with variant calling using a standardized set of mutation callers. The BAM files used underwent a standardized local re-alignment to hg19 (Genome Reference Consortium GRCh37), six calling algorithms were applied, and a number of automated filters were applied. Variants called by two or more algorithms were used in the study. Whole genome sequence analysis was carried out for 1363 cases (with paired normal samples, high pass coverage for BRCA and OV cases, low pass for BLCA, CESC, CRC, ESCA, HNSC, LGG, LUAD, PRAD, SKCM, STAD, THCA, UCEC, and UVM). Genomic rearrangements were detected in all tumor and normal genomes by Meerkat (Yang et al., 2013). Five discordant read pairs support are required for each event. Each event was detected in tumor genome was filtered by all normal genomes to ensure it represented a somatic event.
Gene and protein signatures
Pan-cancer RPPA profiles were scored for a PI3K/AKT pathway signature, defined as the sum of normalized phosphoprotein levels of AKT (both S473 and T308 RPPA features), GSK3 (S9 and S21/S9 features), PRAS40, and phospho-TSC2. RPPA profiles were also scored for an mTOR pathway signature, defined as the sum of phosphoprotein levels of mTOR, 4EBP1 (S65, T37/T46, and T70 RPPA features), P70S6K, and S6 (S235/S236 and S240/S244 features).
Gene transcriptional signatures of PI3K/AKT/mTOR pathway were defined as described previously (Creighton et al., 2010): “Saal” PTEN loss signature, genes correlated with Pten protein levels in breast cancer; “CMap” PI3K/AKT/mTOR signature, genes modulated in vitro by inhibitors to PI3K or mTOR, according to CMap dataset (p<0.01, comparing PI3K/mTOR-inhibited cells with the rest of the Cmap profiles); “Majumder” Akt signature, genes modulated in a mouse model of inducible AKT (p<0.01). MYC signatures (Coller and Bild) and the Bild Ras signature were from ref (Creighton, 2008), and the Settleman k-ras sensitivity signature were from ref (Singh et al., 2009). For a given gene transcription signature, we extracted the expression values from the TCGA gene expression array dataset. For each gene, we normalized expression values to standard deviations from the median across tumors. For signatures with “up” versus “down” genes, we computed our previously described “t-score” (Creighton et al., 2010) to score each tumor profile for relative manifestation of the signature.
For deriving a PI3K/AKT/mTOR drug sensitivity signature in cell lines (Figure 5D), we utilized the dataset from Garnett et al. (Garnett et al., 2012). For the 11 inhibitors to PI3K/AKT/mTOR represented in Garnett (including Rapamycin:MTOR, JW-7-52-1:MTOR, A-443654:AKT1/2/3, CHIR-99021:GSK3B, AZD6482:PI3Kb (P3C2B), AKT inhibitor VIII:AKT1/2, Temsirolimus:MTOR, MK-2206:AKT1/2, NVP-BEZ235:PI3K (class 1) and mTORC1/2, GDC0941:PI3K (class 1), and AZD8055:mTORC1/2), we normalized IC50 values to standard deviations from median, then average to get single drug sensitivity score. Each gene was correlated in expression with the drug sensitivity score, first selecting for genes significant with p<0.01 by t-test on log-transformed data (1099 significant genes), then further selecting for genes remaining significant after correcting for tissue type differences using a regression model that incorporates tumor type as a confounder (146 genes with corrected p<0.01).
In silico mutation evaluation
In assessing whether mutations may be more or less likely to have a functional effect on the resulting protein, a number of factors were considered. Somatic substitution hotspots (470 in total involving 275 genes), based on a previous pan-cancer analysis of 11119 human tumors (Chang et al., 2016), were incorporated into the present study where noted. Mutation Assessor calls predicting the functional impact (medium to high) of somatic mutation (Reva et al., 2011) were obtained from cBioPortal (Cerami et al., 2012). Manual review of variants involving AKT1/2/3, MTOR, PIK3CA, PTEN, RHEB, TSC1/2 was also carried out by domain experts in the analysis group. Mutations that were predicted as potentially functional by any of the above—as well as mutations in tumor suppressor genes (e.g. PTEN, PIK3R1) classified as nonsense, frameshift, or indel—were evaluated separately with respect to comparing with AKT pS473 phospho-protein expression (Figure 3D).
Cell line viability assays
The effects of mutations on the function of PIK3CA and PIK3R1 were assessed in Ba/F3 and MCF10A by survival assay as previously described (Dogruluk et al., 2015) with lentiviral vector pHAGE used in the cloning. In Ba/F3, the PIK3CA mutations were assigned as “Strong activating (SA)” if the mutations have an activity higher than M1043I (known moderate driver); as “Moderate activating (MA)” if the mutations have a similar or lower activity than M1043I; as “No difference from WT (NDFW)” if the mutations have a similar activity with WT; or as “Inactivating (INA)” if the mutations have an activity similar to negative controls (GFP/mCherry/Luciferase). The PIK3R1 mutations were assigned as “SA” if the mutations have a relative level of activation higher than that of PIK3CA M1043I comparing to negative controls; as “MA” if the mutations have a relative level of activation between PIK3CA M1043I and WT; as “Weak activating (WA)” if the mutations have a relative level of activation between PIK3CA WT and negative controls; or as “NDFW” if the mutations have a similar activity with WT. In MCF10A, the PIK3CA mutations were assigned as “SA” and “NDFW” by the same mean as in Ba/F3 model. The mutations were assigned as “MA” and “WA” if the mutations have an activity above and lower than 50% of that of M1043I, respectively.
Tumor classes by gene alteration
Genetic/genomic alteration classes in relation to PI3K/AKT/mTOR pathway alteration were defined (Figure 7A), in order to relate these to PI3K/AKT and mTOR activation, as defined by protein signature score. For AKT/MTOR/PIK3CA/PIK3R1/PTEN mutations, “predicted functional” mutations from Figure 3D were used. An “other gene mutation” class of Figures 7A and 7B involved nonsilent mutations for other genes represented in Figure 2A (AKTS1, DEPDC5, DEPTOR, MAPKAP1, MLST8, NPRL2, NPRL3, PDK1, PRR5, RHEB, RICTOR, RPTOR, PIK3C2B). The RTK group represented cases with hotspot mutations in KRAS, BRAF, EGFR, or ERBB2, that were not also included in the other PI3K/AKT/mTOR-related groups. The set of genes previously found significantly mutated in pan-cancer analysis (Lawrence et al., 2014), were searched for enrichment of mutation events within the High P-AKT group (Figure 7C). When defining proteins that were highly expressed specifically within the High P-AKT group (Figure 7D), RPPA features were selected that were over- or under-expressed in the High P-AKT compared to unaligned cases (p<0.05, t-test on log-transformed data) for at least four of the seven cancer types, and differentially expressed in High P-AKT compared to unaligned and to PI3K/AKT/mTOR or RTK-altered cases across all cancer cases (p<0.01 for each).
QUANTIFICATION AND STATISTICAL ANALYSIS
All p values were two-sided unless otherwise specified. Statistical significance was defined at the 0.05 threshold. All available TCGA data in the public domain at the time of this study was utilized, and no patients were deliberately excluded. Differential expression between comparison groups was assessed using t-test on log-transformed values. For visualization using heat maps and box plots, mRNA and protein expression values were z-normalized to standard deviations from the median across all tumor sample profiles.
Individual gene and protein features were evaluated for correlation with patient survival by univariate Cox analysis; in addition, a stratified Cox model was used to evaluate survival association when correcting for tumor type. For PTEN and STK11 copy alteration features (Log [tumor/normal] ratios), Cox regression analysis within each individual cancer type was carried out; then, in order to aggregate the results across cancer types, we used “metafor” R package to conduct meta-analyses, with a random-effects model used to estimate the overall effectiveness of the molecular feature. For Kapan-Meier plots, a stratified Log-rank test evaluated differences between tumor groups after correction for tumor type. Patient survival data from TCGA were current as of March 31, 2016.
Supplementary Material
Significance.
Our current model of PI3K/AKT/mTOR pathway has largely been derived from experimental systems. The Cancer Genome Atlas (TCGA) pan-cancer cohort represents an opportunity to explore these pathway relationships in the setting of human cancer. Cause-and-effect relationships embodied by the pathway model can manifest as correlations in human disease. Integration of genomic with proteomic data may benefit personalized and precision medicine approaches in helping to assess variants for potential clinical relevance. Manifestation of pathways at the transcription level is distinct from manifestation at the phospho-protein level, highlighting the importance of proteomic approaches. Over time, previously unrealized or underappreciated members or connections may be incorporated into the standard pathway model, where TCGA data may aid in the process of discovery or confirmation.
Highlights.
Multiplatform-based survey of PI3K/AKT/mTOR across over 10,000 human cancers
Distinct classes of somatic alteration associated with greater pathway activation
Functional interrogation of specific mutations in PIK3CA and PIK3R1
Support for inclusion of IDH1 and VHL mutations within the canonical pathway model
Acknowledgments
This work was supported in part by Cancer Prevention and Research Institute of Texas (CPRIT) grant RP120713 C2 (C. Creighton), and by National Institutes of Health (NIH) grants 5P50CA098258 (G. Mills), 5U01CA168394 (G. Mills), U24CA143883 (G. Mills), 2P50CA101942-11A1 (D. Kwiatkowski), 1R21CA191687 (D. Kwiatkowski), 5P01CA120964 (D. Kwiatkowski), CA175486 (H. Liang), CA209851 (H. Liang and G. Mills), P30CA125123 (C. Creighton), and Cancer Center Support Grant (CCSG) P30CA016672.
Abbreviations
- PI3K
Phosphoinositide 3-kinase
- mTOR
mechanistic target of rapamycin
- TCGA
The Cancer Genome Atlas
- RNA-seq
RNA sequencing
- RPPA
reverse-phase protein arrays
Footnotes
Author Contributions
Conceptualization: C.J.C., D.J.K., G.B.M.; Methodology: C.J.C., D.J.K., G.B.M., Y.Z., M.K., K.L.S., J.N.W., P.J.P., R.K., H.L., R.A., Y.H.T., P.K-S.N.; Investigation: Y.Z., P.K-S.N., M.K., F.C., Y.L., Y.H.T., G. de V., C.S.S., T.K.C., M.I., C.V.W., T.F.W., E.P.H. A.K.G., G.B.M., C.J.C.; Formal Analysis: Y.Z., F.C., Y.L., M.K., P.K-S.N., C.J.C., D.J.K.; Data Curation: C.J.C., G.B.M., H.L., R.A., K.J.J., A.H., A.P., C.A.B., E.L., H.S.M., J.T., J.Z., L.Y., S.S., S.L., X.R., X.S., H.S., J.S., L.J.L., R.X., L.C., A.P., P.J.P., R.K., P.K-S.N.; Visualization; C.J.C., F.C.; Writing – Original Draft: C.J.C., D.J.K.; Writing – Review & Editing: G.B.M., C.J.C., D.J.K., P.K-S.N., G. de V., C.S.S., T.K.C., M.I., C.V.W., J.N.W., E.P.H., A.K.G., H.L.; Supervision: C.J.C., D.J.K., G.B.M.;
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akbani R, Ng P, Werner H, Shahmoradgoli M, Zhang F, Ju Z, Liu W, Yang J, Yoshihara K, Li J, et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat Commun. 2014 doi: 10.1038/ncomms4887. E-pub May 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonneau M, Gagne L, Lalonde M, Germain M, Motorina A, Guiot M, Secco B, Vincent E, Tumber A, Hulea L, et al. The oncometabolite 2-hydroxyglutarate activates the mTOR signalling pathway. Nat Commun. 2016;7:12700. doi: 10.1038/ncomms12700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage S, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross B, Sumer S, Aksoy B, Jacobsen A, Byrne C, Heuer M, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Asthana S, Gao S, Lee B, Chapman J, Kandoth C, Gao J, Socci N, Solit D, Olshen A, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34:155–163. doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi A, Araujo Cruz M, Jyotsana N, Sharma A, Yun H, Gorlich K, Wichmann M, Schwarzer A, Preller M, Thol F, et al. Mutant IDH1 promotes leukemogenesis in vivo and can be specifically targeted in human AML. Blood. 2013;122:2877–2887. doi: 10.1182/blood-2013-03-491571. [DOI] [PubMed] [Google Scholar]
- Chen B, Xu X, Luo J, Wang H, Zhou S. Rapamycin Enhances the Anti-Cancer Effect of Dasatinib by Suppressing Src/PI3K/mTOR Pathway in NSCLC Cells. PloS one. 2015;10:e0129663. doi: 10.1371/journal.pone.0129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung LW, Hennessy BT, Li J, Yu S, Myers AP, Djordjevic B, Lu Y, Stemke-Hale K, Dyer MD, Zhang F, et al. High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer Discov. 2011;1:170–185. doi: 10.1158/2159-8290.CD-11-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark P. The role of VHL in clear-cell renal cell carcinoma and its relation to targeted therapy. Kidney Int. 2009;76:939–945. doi: 10.1038/ki.2009.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton C. Multiple oncogenic pathway signatures show coordinate expression patterns in human prostate tumors. PloS one. 2008;3:e1816. doi: 10.1371/journal.pone.0001816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton C, Fu X, Hennessy B, Casa A, Zhang Y, Gonzalez-Angulo A, Lluch A, Gray J, Brown P, Hilsenbeck S, et al. Proteomic and transcriptomic profiling reveals a link between the PI3K pathway and lower estrogen-receptor (ER) levels and activity in ER+ breast cancer. Breast Cancer Res. 2010;12:R40. doi: 10.1186/bcr2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton C, Huang S. Reverse phase protein arrays in signaling pathways: a data integration perspective. Drug Des Devel Ther. 2015;9:3519–3527. doi: 10.2147/DDDT.S38375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nature cell biology. 2013;15:555–564. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd K, Yang J, Shen M, Sampson J, Tee A. mTORC1 drives HIF-1α and VEGF-A signalling via multiple mechanisms involving 4E-BP1, S6K1 and STAT3. Oncogene. 2015;34:2239–2250. doi: 10.1038/onc.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogruluk T, Tsang Y, Espitia M, Chen F, Chen T, Chong Z, Appadurai V, Dogruluk A, Eterovic A, Bonnen P, et al. Identification of Variant-Specific Functions of PIK3CA by Rapid Phenotyping of Rare Mutations. Cancer Res. 2015;75:5341–5354. doi: 10.1158/0008-5472.CAN-15-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q, Flynn C, Niepel M, Hafner M, Muhlich J, Fernandez N, Rouillard A, Tan C, Chen E, Golub T, et al. LINCS Canvas Browser: interactive web app to query, browse and interrogate LINCS L1000 gene expression signatures. Nucleic Acids Res. 2014;42:W449–460. doi: 10.1093/nar/gku476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Garnett M, Edelman E, Heidorn S, Greenman C, Dastur A, Lau K, Greninger P, Thompson I, Luo X, Soares J, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabiner BC, Nardi V, Birsoy K, Possemato R, Shen K, Sinha S, Jordan A, Beck AH, Sabatini DM. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov. 2014;4:554–563. doi: 10.1158/2159-8290.CD-13-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Chakraborty A, Liu P, Gan W, Zheng X, Inuzuka H, Wang B, Zhang J, Zhang L, Yuan M, et al. pVHL suppresses kinase activity of Akt in a proline-hydroxylation-dependent manner. Science. 2016;353:929–932. doi: 10.1126/science.aad5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg R. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hennessy B, Smith D, Ram P, Lu Y, Mills G. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- Hoadley K, Yau C, Wolf D, Cherniack A, Tamborero D, Ng S, Leiserson M, Niu B, McLellan M, Uzunangelov V, et al. Multiplatform Analysis of 12 Cancer Types Reveals Molecular Classification within and across Tissues of Origin. Cell. 2014;158:929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornigold N, Devlin J, Davies AM, Aveyard JS, Habuchi T, Knowles MA. Mutation of the 9q34 gene TSC1 in sporadic bladder cancer. Oncogene. 1999;18:2657–2661. doi: 10.1038/sj.onc.1202854. [DOI] [PubMed] [Google Scholar]
- Ilagan E, Manning B. Emerging Role of mTOR in the Response to Cancer Therapeutics. Trends in Cancer. 2016;2:241–251. doi: 10.1016/j.trecan.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, McLellan M, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael J, Wyczalkowski M, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keniry M, Parsons R. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene. 2008;27:5477–5485. doi: 10.1038/onc.2008.248. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Stojanov P, Mermel C, Robinson J, Garraway L, Golub T, Meyerson M, Gabriel S, Lander E, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhao W, Akbani R, Liu W, Ju Z, Ling S, Vellano C, Roebuck P, Yu Q, Eterovic A, et al. Characterization of Human Cancer Cell Lines by Reverse-phase Protein Arrays. Cancer Cell. 2017;31:225–239. doi: 10.1016/j.ccell.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer IA, Arteaga CL. The PI3K/AKT Pathway as a Target for Cancer Treatment. Annu Rev Med. 2016;67:11–28. doi: 10.1146/annurev-med-062913-051343. [DOI] [PubMed] [Google Scholar]
- Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, Settleman J. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Chen Y, Wang C, Ding X, Rwibasira G, Kong Y. Human cathelicidin LL-37 inhibits platelet aggregation and thrombosis via Src/PI3K/Akt signaling. Biochem Biophys Res Commun. 2016;473:283–289. doi: 10.1016/j.bbrc.2016.03.095. [DOI] [PubMed] [Google Scholar]
- The_Cancer_Genome_Atlas_Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The_Cancer_Genome_Atlas_Research_Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015;15:7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler M, Blaes J, Pusch S, Sahm F, Czabanka M, Luger S, Bunse L, Solecki G, Eichwald V, Jugold M, et al. mTOR target NDRG1 confers MGMT-dependent resistance to alkylating chemotherapy. Proc Natl Acad Sci U S A. 2014;111:409–414. doi: 10.1073/pnas.1314469111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Luquette L, Gehlenborg N, Xi R, Haseley P, Hsieh C, Zhang C, Ren X, Protopopov A, Chin L, et al. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell. 2013;153:919–929. doi: 10.1016/j.cell.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack T, Schumacher S, Carter S, Cherniack A, Saksena G, Tabak B, Lawrence M, Zhsng C, Wala J, Mermel C, et al. Pan-cancer patterns of somatic copy number alteration. Nature genetics. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.