Abstract
Dental caries can be described as a dysbiosis of the oral microbial community, in which acidogenic, aciduric, and acid-adapted bacterial species promote a pathogenic environment, leading to demineralization. Alkali generation by oral microbes, specifically via arginine catabolic pathways, is an essential factor in maintaining plaque pH homeostasis. There is evidence that the use of arginine in dentifrices helps protect against caries. The aim of the current study was to investigate the mechanistic and ecological effect of arginine treatment on the oral microbiome and its regulation of pH dynamics, using an in vitro multispecies oral biofilm model that was previously shown to be highly reflective of the in vivo oral microbiome. Pooled saliva from 6 healthy subjects was used to generate overnight biofilms, reflecting early stages of biofilm maturation. First, we investigated the uptake of arginine by the cells of the biofilm as well as the metabolites generated. We next explored the effect of arginine on pH dynamics by pretreating biofilms with 75 mM arginine, followed by the addition of sucrose (15 mM) after 0, 6, 20, or 48 h. pH was measured at each time point and biofilms were collected for 16S sequencing and targeted arginine quantification, and supernatants were prepared for metabolomic analysis. Treatment with only sucrose led to a sustained pH drop from 7 to 4.5, while biofilms treated with sucrose after 6, 20, or 48 h of preincubation with arginine exhibited a recovery to higher pH. Arginine was detected within the cells of the biofilms, indicating active uptake, and arginine catabolites citrulline, ornithine, and putrescine were detected in supernatants, indicating active metabolism. Sequencing analysis revealed a shift in the microbial community structure in arginine-treated biofilms as well as increased species diversity. Overall, we show that arginine improved pH homeostasis through a remodeling of the oral microbial community.
Keywords: dental caries, biofilms, dental plaque, microbiota, oral cavity, metabolomics
Introduction
Dental plaque is a multispecies microbial biofilm attached to the surface of the teeth. The dynamics of dental plaque pH were first described over 80 y ago (Stephan 1940), when Stephan observed that dental plaque universally undergoes a rapid drop in pH when exposed to a sugar source, followed by an eventual recovery to neutral. Dental caries is caused by acid demineralization of enamel; frequent exposure to sugars causes prolonged periods of acidification, leading to increased demineralization and risk of caries. Indeed, individuals with active caries display a lower plaque pH and a longer recovery time after exposure to a sugar source (Lingström et al. 2000; Gao et al. 2001). A healthy plaque microbial community exists in equilibrium with the host and allows for a quick recovery from the pH drop (McLean 2014; Edlund et al. 2015).
Although the microbiological basis of caries has been known for decades, modern technologies have led to an improved understanding of the complex etiology. Newer models better account for the underlying microbial ecological factors (Marsh 1994; Kleinberg 2002; Takahashi and Nyvad 2008) and suggest a dysbiosis in which acidogenic and aciduric bacterial species become abundant due to frequent acidification, creating a pathogenic environment. In contrast, the alkali-generating ability of plaque is fundamental for maintaining healthy conditions and pH homeostasis (Kleinberg 2002; Takahashi and Nyvad 2011).
Many current therapies for caries, such as fluoride treatment, primarily focus on protecting and repairing the tooth surface without altering the plaque microbiota (Cummins 2013). With the knowledge that the relative proportion of acid- and base-producing bacteria is an important determinant of caries risk, a potential therapeutic objective is to promote a shift in the microbial community to favor base-producing bacteria. The catabolism of arginine by oral bacteria is an important contributor to plaque alkalinity (Casiano-Colón and Marquis 1988; Burne and Marquis 2000); because of this, arginine has been investigated as a potential caries therapeutic. Studies have shown that high levels of free arginine in saliva are associated with caries resistance (Van Wuyckhuyse et al. 1995). Results from clinical studies have shown that arginine-containing dentifrices are effective at reversing caries and providing caries protection (Acevedo et al. 2008; Kraivaphan et al. 2013; Yin et al. 2013).
We have recently developed a specialized culture medium that captures 60% to 80% of the diversity present in the original saliva inoculum, including many previously uncultivated species (Tian et al. 2010; Edlund et al. 2013). This allows us to establish an in vitro multispecies oral community that is highly reflective of the in vivo oral microbiome, as well as maintaining the signature pH cycling of dental plaque (Edlund et al. 2013, 2015). In the current study, we used this model system to investigate the ability of arginine treatment to protect against the pH drop caused by sucrose exposure, with the goal of exploring the mechanistic and ecological basis for arginine regulation of plaque pH.
Materials and Methods
Growth of Biofilms and Treatment with Arginine and Sucrose
Saliva-derived, early-stage biofilms were established using a previously published protocol (Edlund et al. 2013). Briefly, saliva samples were collected and pooled from 7 healthy volunteers (approved under UCLA IRB 13-001075) and used to inoculate SHI medium (Tian et al. 2010) containing 15 mM sucrose. Then, 1 mL was added to each well of a 24-well plate that had been precoated with sterilized, cell-free saliva. Plates were incubated at 37°C for 16 h in microaerobic conditions (2% O2, 5% CO2, balanced with nitrogen) to allow biofilm formation, then were washed and starved (incubated without a carbon source) in chemically-defined medium (CDM), adjusted to pH 7, (McLean et al. 2012) for 2 h. Then, 75 mM L-arginine (adjusted to pH 7), 15 mM sucrose, or both, was added to the CDM. For arginine-treated biofilms, 15 mM sucrose was subsequently added after 6, 20, 48, or 54 h. Time 0 biofilms were collected prior to the addition of any arginine or sucrose. Negative controls were incubated in CDM only. pH was measured by pipetting 20 μL of each sample onto pH indicator strips. All experimental conditions were performed in duplicate. Full protocols for SHI medium and CDM preparation can be found at http://depts.washington.edu/jsmlab/downloads/protocols/.
Quantification of Arginine and Identification of Metabolites
Gas chromatography time-of-flight (GC TOF) mass spectrometry analysis of metabolites and targeted liquid chromatography mass spectrometry quantification of arginine were performed at the National Institutes of Health (NIH) West Coast Metabolomics Center, UC Davis Metabolomics Core, Davis, California. Detailed protocols are supplied in the Appendix.
Arginine Uptake and Utilization Assays
Biofilms were grown, washed, and starved as described above and then incubated with 75 mM arginine for 30 s, 2 m, 30 m, 1 h, and 3 h. At each time point, biofilms were thoroughly washed to remove the arginine, collected, and pelleted; pellets were then sent for targeted liquid chromatography–mass spectrometry (LC-MS) for arginine. A short-term utilization assay was performed in a similar manner. After washing, biofilms were incubated with 75 mM arginine for 2 h, followed by removal of the arginine and collection of the biofilm supernatants at varying time points. Supernatants were sent for GC TOF as described above to detect and quantify secreted metabolites.
Microbial Community Analysis
Detailed DNA extraction and sequencing protocol is supplied in the Appendix. Briefly, DNA was extracted from pelleted biofilms and sent to the Forsyth Institute (Cambridge, MA) for library prep and Illumina sequencing of the amplified V3 to V4 16S region.
Sequencing data were analyzed using QIIME (Quantitative Insights into Microbial Ecology) version 1.9.1 (Caporaso et al. 2010). Sequences were clustered into operational taxonomic units (OTUs) using UCLUST (Edgar 2010), then aligned and taxonomy assigned with the Human Oral Microbiome Database (HOMD) (Chen et al. 2010) as reference. For alpha (within-sample) diversity, OTU tables were rarefied to 35,000 reads and Chao1 values were calculated. For beta (between-sample) diversity, UniFrac distances (Lozupone and Knight 2005) were calculated, followed by principal coordinates analysis (PCoA).
Statistical Analysis
For the sequencing data, differences in the relative abundance of taxa were compared with the Kruskal-Wallis test controlling the false discovery rate to correct for multiple comparisons (Hochberg and Benjamini 1990), and a corrected P value of ≤0.05 was considered significant.
Results
Arginine Uptake and Utilization Assay
Saliva-derived, mixed-community biofilms were generated using a specialized growth medium (SHI medium) previously shown to preserve up to 80% of the species diversity present in the original inoculum (Edlund et al. 2013). Using saliva as the inoculum and culturing on a surface coated with saliva pellicle allows the biofilm to develop in succession, much like how plaque forms on the tooth surface, initiated by attachment of salivary early colonizers. In addition, saliva cultured in SHI medium as a multispecies biofilm has been previously shown to respond to a sugar source with a rapid pH drop, in a manner similar to plaque (Edlund et al. 2013, 2015).
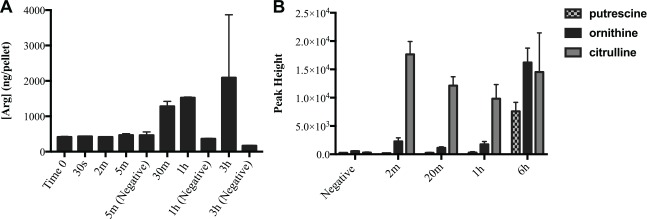
Using this system to generate early stage biofilms, we first assayed the uptake of arginine into the bacterial cells of the biofilm and the retention over time, while the biofilms were incubated in CDM. The amount of arginine inside the cells rose above baseline after 30 min of arginine exposure (Fig. 1A). We also performed an arginine utilization assay in which we quantified the metabolites present over time after incubation with arginine and subsequent removal. Results show an increase in the 3 major products of arginine catabolism (ornithine, putrescine, and citrulline) at the first time point (Fig. 1B). By 6 h, the amount of ornithine present in the arginine-treated biofilms was approximately 30 times higher than detected in negative controls, which were not incubated in arginine. Similar levels were detected for putrescine and citrulline, indicating that the bacteria within the biofilm were actively metabolizing the arginine.
Figure 1.
Arginine uptake and utilization assays. (A) Arginine uptake. Biofilms were incubated in 75 mM arginine for treatment time indicated above before washing and collection of cell pellets for liquid chromatography–mass spectrometry targeted arginine assay. Negative controls were not incubated in arginine and were collected at the indicated time point. Error bars = SD. (B) Arginine utilization. Biofilms were incubated in 75 mM arginine for 2 h, then washed and collected at the indicated time point after arginine removal. Supernatants were analyzed with gas chromatography–mass spectrometry, and the levels of the main arginine metabolites are shown. Negative controls were collected at 6 h. Error bars = SD.
Pretreatment with Arginine Prevents pH Drop Induced by Sucrose
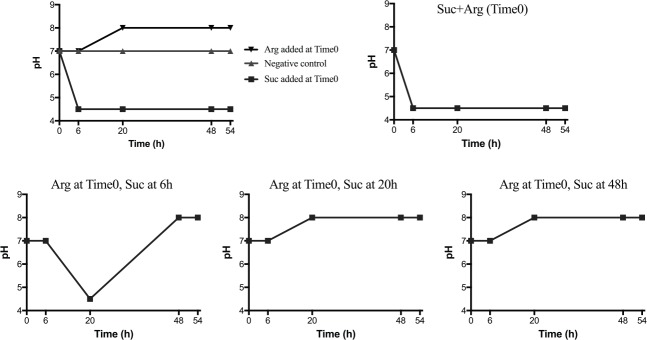
Using the biofilm model as described above, we tested whether treatment with 75 mM arginine could inhibit a pH drop induced by sucrose. Arginine (75 mM) was added to the biofilms 0, 6, 20, or 48 h prior to the addition of sucrose (15 mM). The pH change over time of each of the differentially treated biofilms is shown in Figure 2.
Figure 2.
Pretreatment with arginine prior to sucrose exposure protects against pH drop. Pooled saliva was inoculated into SHI media overnight in biofilm-inducing conditions (see Materials and Methods). Biofilms were then washed and incubated in chemically defined media (CDM) adjusted to pH 7, plus arginine and/or sucrose as indicated. pH was measured by pipetting 20 μL onto color-changing pH strips. Adding sucrose 6 h after adding arginine resulted in an initial drop, followed by an increase in pH. Adding sucrose at 20 h or 48 h prevented the drop completely. Arg = L-arginine (75 mM); Suc = sucrose (15 mM).
The pH of untreated biofilms (negative control) remained stable at the starting pH of 7 throughout the experiment when maintained in CDM. Biofilms that were exposed to sucrose at Time 0 experienced a rapid and sustained drop to pH 4.5, while treatment with arginine alone increased the starting pH to a final pH of 8. In biofilms that were pretreated with arginine before the addition of sucrose, the length of time before the sucrose exposure determined the extent of the pH response. Biofilms that were treated with both sucrose and arginine at the same time (Time 0) displayed a drastic drop in pH; the response was identical to the sucrose-only biofilm. The addition of sucrose after 6 h of preincubation with arginine resulted in an initial pH drop, followed by an increase to a final pH of 8. The addition of sucrose after 20h arginine pretreatment did not result in a sustained drop in pH.
Arginine Is Actively Metabolized during Treatment
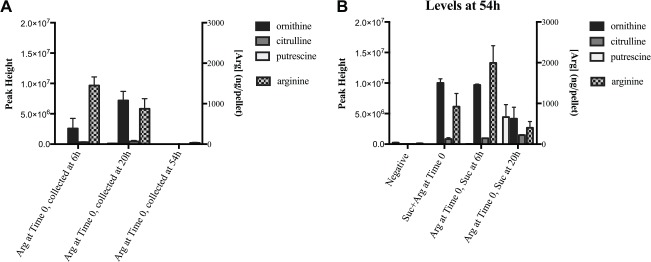
We investigated the extent to which the added arginine was taken up and metabolized by the differentially treated biofilms by quantifying the amount present in the cells as well as quantifying metabolites in the supernatant (Fig. 3). Targeted LC-MS of the biofilm pellets showed that arginine was internalized by the cells. Three major products of arginine catabolism—ornithine, putrescine, and citrulline—were detected in the supernatants, indicating that arginine was metabolized during the experiment. Interestingly, in biofilms treated with only arginine at Time 0 and not exposed to sucrose (Fig. 3A), the amount of arginine inside the cells at the end of the experiment (54 h) was almost 30-fold less than detected in the biofilms collected at 20 h, as well as much less (12- to 30-fold) than in biofilms subsequently treated with sucrose (Fig. 3B) collected at the same time point; the metabolites in the supernatant followed this same pattern. There is evidence that arginine uptake and metabolic pathways are regulated by pH (Burne and Marquis 2000). The arginine-only biofilms rose to a sustained pH of 8 (Fig. 2); therefore, arginine uptake may eventually become inhibited to avoid over-alkalinization. In the sucrose-treated biofilms (Fig. 3B), the pH was lower or fluctuated throughout the experiment, which may have activated specific arginine pathways to protect against the acidity, and a proportion of intermediates (ornithine, putrescine, and citrulline) was secreted into the supernatant. Arginine uptake is mediated by the arginine-ornithine antiporter found in many oral species (Liu and Burne 2009); thus, ornithine is expected to be secreted by the cells.
Figure 3.
Arginine and metabolite quantification. Biofilms were collected at each time point and centrifuged. Pellets were analyzed with targeted liquid chromatography–mass spectrometry for arginine. Supernatants were analyzed with gas chromatography–mass spectrometry to detect and quantify metabolites. Left y-axes represent metabolite levels in supernatants; right y-axes represent the concentration of arginine in pellets. (A) Levels of select arginine-related metabolites and concentration of arginine in biofilms treated with 75 mM arginine at Time 0 and collected at 6 h, 20 h, and 54 h. (B) Levels of select arginine-related metabolites and concentration of arginine in differentially treated biofilms collected at 54 h. Arg = L-arginine (75 mM); Suc = sucrose (15 mM). Experiments were performed in duplicate. Error bars = SD.
Microbial Community Analysis
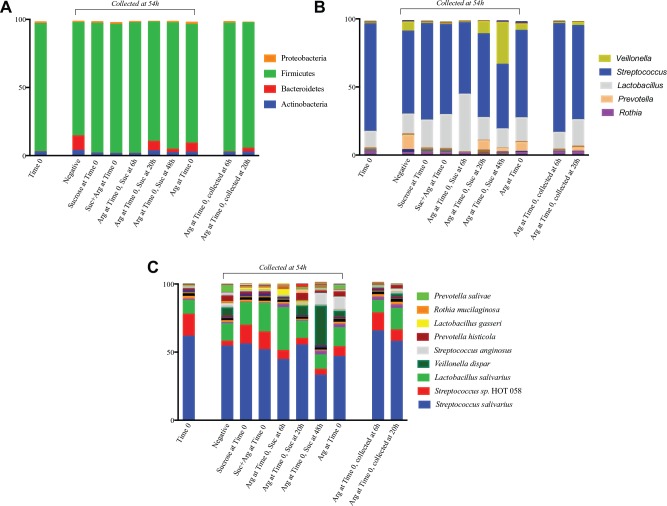
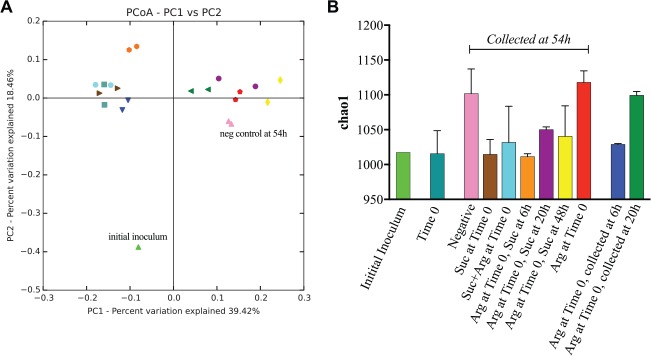
Sequencing generated a total of 2,179,843 sequences after quality filtering, with an average of 103,802 sequences per sample (range, 71,213–131,735) with a median sequence length of 429. Analysis revealed a total of 8 phyla, 54 genera, and 130 species-level OTUs across all biofilms. At the end point of the experiment (54 h), results show a shift in the community structure with increasing time of pretreatment with arginine before the addition of sucrose (Fig. 4). The microbial community results are confirmed by the PCoA plot of UniFrac distances (Fig. 5A). In general, biofilms in which protection from the sucrose-induced pH drop occurred clustered separately from the biofilms in which pH protection did not occur. The stacked bar graphs in Figure 4 show that at each taxonomic level, the biofilms in which sucrose and arginine were both added at Time 0 resemble the community in which only sucrose was added and also appear closer together on the PCoA plot. Conversely, the community that was pretreated with arginine 20 h prior to the addition of sucrose more closely resembled the community in which only arginine was added. The community treated with arginine 48 h before the addition of sucrose appears different from the rest of the biofilms due in part to the large increase in abundance of Veillonella dispar. Notably, treatment replicates clustered closely together on the PCoA, indicating that pretreatment with arginine and subsequent sucrose exposure had reproducible effects on the microbial community.
Figure 4.
Relative abundance of (A) phylum-level, (B) genus-level, and (C) species-level taxa detected in each biofilm. Experiments were performed in duplicate, and relative abundances were averaged for each taxon detected. Arg = L-arginine (75 mM); Suc = sucrose (15 mM).
Figure 5.
Diversity analyses. (A) Beta diversity was calculated by unweighted UniFrac, and the distances were plotted with principal coordinate analysis (PCoA). The colors of each sample correspond to the colors on the bar graph in (B), and samples of the same color represent replicate experiments. Biofilms in which protection from the sucrose-induced pH drop occurred clustered separately from the biofilms in which pH protection did not occur; this correlates with the amount of time the biofilms were incubated in arginine prior to sucrose exposure. (B) The alpha diversity measure Chao1 was calculated at a defined sequence depth of 35,000 for all samples. Values for duplicate samples were averaged, and the error bars represent the standard deviation. “Initial inoculum” refers to the pooled saliva used to inoculate SHI medium to generate the initial biofilms, and “Time 0” refers to the overnight biofilms prior to the addition of arginine or sucrose.
Alpha (within-sample) diversity was measured using Chao1, which is an estimate of species richness. Results show that biofilms treated with arginine only at Time 0 maintain the species richness of the negative control, while the addition of sucrose at any time point results in a less diverse community (Fig. 5B). Notably, the Time 0 community is as diverse in terms of species richness as the initial saliva inoculum. As the biofilm communities matured over the course of the 54-h experiment, the species richness increased, most likely due to initially present but undetectable organisms increasing in abundance to a detectable level over time.
At the phylum level, the relative abundance of Bacteroidetes was 24× higher in the biofilms pretreated with arginine that were protected from the sucrose-induced pH drop (“protected” biofilms, 20 and 48 h pretreatment) compared to those that were not (“unprotected,” 0 and 6 h pretreatment) (4.8% vs. 0.2%, P < 0.05); this phylum includes Prevotella, Alloprevotella, Tannerella, Porphyromonas, and Capnocytophaga; in total, these genera account for 27 species. In addition, the abundance of phylum Actinobacteria was significantly higher in the protected biofilms compared to the unprotected (2.7% vs. 1.4%, P > 0.05); this phylum includes Rothia, Bifidobacterium, Actinomyces, and Corynebacterium. At the genus level, Prevotella was 60× higher in the protected biofilms (4.7% vs. 0.08%); however, this did not reach statistical significance after correcting for multiple comparisons. At the species level, V. dispar and Streptococcus anginosus were dramatically increased in the protected biofilms (18% vs. 0.2% and 4.5% vs. 0.08%, respectively).
Discussion
An effective caries prevention needs to be able to protect against the pH drop induced by the influx of carbohydrates that occurs during meals by allowing for a quick recovery to shorten the amount of time enamel is exposed to demineralizing conditions (Nascimento and Burne 2014). Clinical studies with arginine-containing dentifrices have shown caries protection as well as an increase in plaque pH (Acevedo et al. 2008; Kraivaphan et al. 2013; Srisilapanan et al. 2013). In the current study, we tested the effects of arginine treatment on an in vitro oral microbial community using a biofilm model (Edlund et al. 2013; Edlund et al. 2015). With a specialized medium (SHI medium) developed in our laboratory (Tian et al. 2010), we are able to culture and maintain a diverse multispecies oral community and can mimic plaque pH dynamics, allowing for highly relevant investigations into the oral microbiome (Edlund et al. 2013). The goal of this study was to investigate the potential of arginine as a caries therapeutic and its effects on the ecology of the oral microbiota.
The results of the initial uptake and utilization assays (Fig. 1) show that arginine is taken up by the biofilms and metabolized, properties essential for modification of the microbiome. Pretreatment with arginine prior to sucrose exposure was able to protect the biofilms from the drastic pH drop that occurred with no arginine treatment (Fig. 2). We found that this protection correlates with changes that occur in the microbial community in the arginine-treated biofilms (Figs. 4 and 5).
Previous in vitro studies have shown that arginine treatment has positive effects on the community structure, in some cases increasing the species diversity (Kolderman et al. 2015) or stabilizing the community and preventing potential detrimental species such as Candida albicans from proliferating (Koopman et al. 2015). In addition, investigations of clinical samples have shown that microbial communities isolated from carious lesions are less diverse than those from healthy sites (Jiang et al. 2014; Kianoush et al. 2014; Xiao et al. 2016). In the current study, species richness increased over time in the untreated biofilms as well as in the arginine-treated biofilms; these samples also clustered together in the PCoA plot (Fig. 5). Conversely, in biofilms also treated with sucrose, the species diversity at the end of the experiment is much less. In our model system, it seems that arginine allows for the natural evolution of the community as would occur without any external perturbation, while the introduction of sucrose into the environment halts this progression and results in a less diverse community.
Beta-diversity analysis revealed that the biofilm communities in which protection from the sucrose-induced pH drop was observed were more alike than the biofilms that were not protected, indicating that the protection is due to differences in the microbial community between the biofilms. Specifically, the phylum Bacteroidetes, including the genus Prevotella, was increased in the biofilms incubated with arginine for 6, 20, or 48 h prior to the addition of sucrose compared to biofilms only treated with sucrose or treated with sucrose and arginine at the same time (Fig. 4). Increased abundance of Prevotella due to arginine treatment was also observed in a previous in vitro study (Kolderman et al. 2015), and earlier studies found that Prevotella intermedia has neutralizing activity at low pH in the presence of amino acids (Takahashi et al. 1997; Takahashi 2003). At the species level, V. dispar and S. anginosus were increased in the arginine-treated, pH-protected biofilms. V. dispar, usually metabolically supported by Streptococcus species, has been previously associated with arginine treatment in vitro (Kolderman et al. 2015; Koopman et al. 2015) and was also found to be an important contributor to pH recovery and neutralization by upregulation of activity and gene expression of arginine catabolic pathways (Edlund et al. 2015). S. anginosus was associated with neutral pH in another in vitro study (Kianoush et al. 2014), suggesting its increased abundance may be a result of the increased pH of the community.
According to the extended ecological plaque hypothesis (Takahashi and Nyvad 2008), the cariogenic disease process is driven by changing environmental conditions that lead to a shift in the microbial population structure. The plaque community adapts to changing pH; acidic conditions shift the community in the favor of acid producers and acid-tolerant species, while a higher pH favors a community with less cariogenic potential. As a carious lesion develops, the community evolves in a continuum that correlates with the changing pH (Takahashi and Nyvad 2011; Kianoush et al. 2014). The catabolism of arginine by oral bacteria is a major contributor to plaque alkalinity mainly due to the generation of ammonia and other alkaline products via the arginine deiminase system (ADS) and the agmatine deiminase system (AgDS) (Burne and Marquis 2000; Liu et al. 2012). Subjects in a clinical study with an arginine-containing dentifrice showed an increase in ADS activity within plaque as well as a shift in the microbial community (Nascimento et al. 2014); also, Nascimento et al. (2009) had previously demonstrated that caries-free individuals have higher levels of ADS activity compared to individuals with active caries, indicating that this pathway plays an important role in caries protection. In our experimental model, arginine treatment led to an increase in ornithine and citrulline, primary products of the ADS, as well as an eventual increase in putrescine, a product of the AgDS (Fig. 3). In the initial arginine utilization assay, our results show that these products were still detectable in the supernatants more than 6 h after the removal of arginine from the culture medium (Fig. 1), suggesting that the pH-raising effects can last long after the arginine treatment itself is removed. The changing pH gradually modifies the community, and eventually, the acidogenic organisms may no longer be present in large enough numbers to maintain the low pH when sucrose is added. In our model system, protection from a sustained pH drop occurred after 20 h of pretreatment with arginine before sucrose exposure. These results indicate that arginine treatment creates sustainable modifications of the microbial community via alkaline-generating metabolism.
We acknowledge some limitations to the model system used here, including the predominance of early colonizers such as Streptococcus salivarius. Because these are early stage (16 h) biofilms, this system reflects the conditions that occur after brushing in the evening, in which biofilm forms overnight. Therefore, addressing the impact of arginine on the early stages of biofilm growth provides valuable, practical information. Using this model, we are able to maintain a complex bacterial community that is highly diverse, reproducible, and well characterized (Edlund et al. 2013). Furthermore, the biofilms respond to a sugar source with a characteristic drop in pH, making the model useful and relevant to in vivo conditions. Overall, the oral biofilm system used here provides an excellent model to test the pH response of oral bacterial communities to various substances and potential therapeutics. Our results show that arginine is taken up by the cells within the biofilm, is readily metabolized, and improves pH homeostasis by altering the microbial ecology, demonstrating its potential as an effective caries therapeutic.
Author Contributions
M. Agnello, contributed to design, data acquisition, analysis, and interpretation, drafted the manuscript; L. Cen, N.C. Tran, contributed to conception, design, and data acquisition, critically revised the manuscript; W. Shi, contributed to conception, design, and data interpretation, critically revised the manuscript; J.S. McLean, contributed to conception, design, data analysis, and interpretation, critically revised the manuscript; X. He, contributed to conception, design, data acquisition, analysis, and interpretation, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Footnotes
A supplemental appendix to this article is available online.
This study was partially funded by Colgate-Palmolive, as well as in part by grants from the National Institutes of Health (NIH-1-R01-DE020102, NIH-1-R01-DE023810, R01-GM095373, and NIH-1-R01-DE026186-01) and NIDCR T90 (DE022734) training award to M.A.
W.S. is the founding scientist for C3 Jian, Inc., which has licensed an anti–S. mutans technology from UC Regents that could be indirectly related to this research project. The authors declare no other potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Acevedo AM, Montero M, Rojas-Sanchez F, Machado C, Rivera LE, Wolff M, Kleinberg I. 2008. Clinical evaluation of the ability of CaviStat in a mint confection to inhibit the development of dental caries in children. J Clin Dent. 19(1):1–8. [PubMed] [Google Scholar]
- Burne RA, Marquis RE. 2000. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol Lett. 193(1):1–6. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 7(5):335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casiano-Colón A, Marquis RE. 1988. Role of the arginine deiminase system in protecting oral bacteria and an enzymatic basis for acid tolerance. Appl Environ Microbiol. 54(6):1318–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. 2010. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford). 2010:baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins D. 2013. Dental caries: a disease which remains a public health concern in the 21st century–the exploration of a breakthrough technology for caries prevention. J Clin Dent. 24 Spec No. A:A1–A14. [PubMed] [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 26(19):2460–2461. [DOI] [PubMed] [Google Scholar]
- Edlund A, Yang Y, Hall AP, Guo L, Lux R, He X, Nelson KE, Nealson KH, Yooseph S, Shi W, et al. 2013. An in vitro biofilm model system maintaining a highly reproducible species and metabolic diversity approaching that of the human oral microbiome. Microbiome. 1(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund A, Yang Y, Yooseph S, Hall AP, Nguyen DD, Dorrestein PC, Nelson KE, He X, Lux R, Shi W, et al. 2015. Meta-omics uncover temporal regulation of pathways across oral microbiome genera during in vitro sugar metabolism. ISME J. 9(12):2605–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XJ, Fan Y, Kent RL, Jr, Van Houte J, Margolis HC. 2001. Association of caries activity with the composition of dental plaque fluid. J Dent Res. 80(9):1834–1839. [DOI] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y. 1990. More powerful procedures for multiple significance testing. Stat Med. 9(7):811–818. [DOI] [PubMed] [Google Scholar]
- Jiang W, Ling Z, Lin X, Chen Y, Zhang J, Yu J, Xiang C, Chen H. 2014. Pyrosequencing analysis of oral microbiota shifting in various caries states in childhood. Microb Ecol. 67(4):962–969. [DOI] [PubMed] [Google Scholar]
- Kianoush N, Adler CJ, Nguyen KA, Browne GV, Simonian M, Hunter N. 2014. Bacterial profile of dentine caries and the impact of pH on bacterial population diversity. PLoS One. 9(3):e92940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinberg I. 2002. A mixed-bacteria ecological approach to understanding the role of the oral bacteria in dental caries causation: an alternative to Streptococcus mutans and the specific-plaque hypothesis. Crit Rev Oral Biol Med. 13(2):108–125. [DOI] [PubMed] [Google Scholar]
- Kolderman E, Bettampadi D, Samarian D, Dowd SE, Foxman B, Jakubovics NS, Rickard AH. 2015. L-arginine destabilizes oral multi-species biofilm communities developed in human saliva. PLoS One. 10(5):e0121835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman JE, Röling WF, Buijs MJ, Sissons CH, ten Cate JM, Keijser BJ, Crielaard W, Zaura E. 2015. Stability and resilience of oral microcosms toward acidification and Candida outgrowth by arginine supplementation. Microb Ecol. 69(2):422–433. [DOI] [PubMed] [Google Scholar]
- Kraivaphan P, Amornchat C, Triratana T, Mateo LR, Ellwood R, Cummins D, DeVizio W, Zhang YP. 2013. Two-year caries clinical study of the efficacy of novel dentifrices containing 1.5% arginine, an insoluble calcium compound and 1,450 ppm fluoride. Caries Res. 47(6):582–590. [DOI] [PubMed] [Google Scholar]
- Lingström P, van Ruyven FO, van Houte J, Kent R. 2000. The pH of dental plaque in its relation to early enamel caries and dental plaque flora in humans. J Dent Res. 79(2):770–777. [DOI] [PubMed] [Google Scholar]
- Liu Y, Burne RA. 2009. Multiple two-component systems modulate alkali generation in Streptococcus gordonii in response to environmental stresses. J Bacteriol. 191(23):7353–7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YL, Nascimento M, Burne RA. 2012. Progress toward understanding the contribution of alkali generation in dental biofilms to inhibition of dental caries. Int J Oral Sci. 4(3):135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 71(12):8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh PD. 1994. Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res. 8(2):263–271. [DOI] [PubMed] [Google Scholar]
- McLean JS. 2014. Advancements toward a systems level understanding of the human oral microbiome. Front Cell Infect Microbiol. 4:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean JS, Fansler SJ, Majors PD, McAteer K, Allen LZ, Shirtliff ME, Lux R, Shi W. 2012. Identifying low pH active and lactate-utilizing taxa within oral microbiome communities from healthy children using stable isotope probing techniques. PLoS One. 7(3):e32219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento MM, Browngardt C, Xiaohui X, Klepac-Ceraj V, Paster BJ, Burne RA. 2014. The effect of arginine on oral biofilm communities. Mol Oral Microbiol. 29(1):45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento MM, Burne RA. 2014. Caries prevention by arginine metabolism in oral biofilms: translating science into clinical success. Current Oral Health Reports. 1(1):79–85. [Google Scholar]
- Nascimento MM, Gordan VV, Garvan CW, Browngardt CM, Burne RA. 2009. Correlations of oral bacterial arginine and urea catabolism with caries experience. Oral Microbiol Immunol. 24(2):89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisilapanan P, Korwanich N, Yin W, Chuensuwonkul C, Mateo LR, Zhang YP, Cummins D, Ellwood RP. 2013. Comparison of the efficacy of a dentifrice containing 1.5% arginine and 1450 ppm fluoride to a dentifrice containing 1450 ppm fluoride alone in the management of early coronal caries as assessed using quantitative light-induced fluorescence. J Dent. 41(Suppl 2):S29–S34. [DOI] [PubMed] [Google Scholar]
- Stephan RM. 1940. Changes in hydrogen-ion concentration on tooth surfaces and in carious lesions. J Am Dent Assoc. 27(5):718–723. [Google Scholar]
- Takahashi N. 2003. Acid-neutralizing activity during amino acid fermentation by Porphyromonas gingivalis, Prevotella intermedia and Fusobacterium nucleatum. Oral Microbiol Immunol. 18(2):109–113. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Nyvad B. 2008. Caries ecology revisited: microbial dynamics and the caries process. Caries Res. 42(6):409–418. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Nyvad B. 2011. The role of bacteria in the caries process: ecological perspectives. J Dent Res. 90(3):294–303. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Saito K, Schachtele CF, Yamada T. 1997. Acid tolerance and acid-neutralizing activity of Porphyromonas gingivalis, Prevotella intermedia and Fusobacterium nucleatum. Oral Microbiol Immunol. 12(6):323–328. [DOI] [PubMed] [Google Scholar]
- Tian Y, He X, Torralba M, Yooseph S, Nelson KE, Lux R, McLean JS, Yu G, Shi W. 2010. Using DGGE profiling to develop a novel culture medium suitable for oral microbial communities. Mol Oral Microbiol. 25(5):357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wuyckhuyse BC, Perinpanayagam HE, Bevacqua D, Raubertas RF, Billings RJ, Bowen WH, Tabak LA. 1995. Association of free arginine and lysine concentrations in human parotid saliva with caries experience. J Dent Res. 74(2):686–690. [DOI] [PubMed] [Google Scholar]
- Xiao C, Ran S, Huang Z, Liang J. 2016. Bacterial diversity and community structure of supragingival plaques in adults with dental health or caries revealed by 16S pyrosequencing. Front Microbiol. 7:1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W, Hu DY, Fan X, Feng Y, Zhang YP, Cummins D, Mateo LR, Pretty IA, Ellwood RP. 2013. A clinical investigation using quantitative light-induced fluorescence (QLF) of the anticaries efficacy of a dentifrice containing 1.5% arginine and 1450 ppm fluoride as sodium monofluorophosphate. J Clin Dent. 24 Spec No. A:A15–A22. [PubMed] [Google Scholar]